Abstract
TNF-related apoptosis-inducing ligand (TRAIL) is a promising antitumor agent for its remarkable ability to selectively induce apoptosis in cancer cells, without affecting the viability of healthy bystander cells. The TRAIL tumor suppressor pathway is deregulated in many human malignancies including lung cancer. In human non-small cell lung cancer (NSCLC) cells, sensitization to TRAIL therapy can be restored by increasing the expression levels of the tumor suppressor microRNA-212 (miR-212) leading to inhibition of the anti-apoptotic protein PED/PEA-15 implicated in treatment resistance. In this study, we exploited a previously described RNA aptamer inhibitor of the tyrosine kinase receptor Axl (GL21.T) expressed on lung cancer cells, as a means to deliver miR-212 into human NSCLC cells expressing Axl. We demonstrate efficient delivery of miR-212 following conjugation of the miR to GL21.T (GL21.T-miR212 chimera). We show that the chimera downregulates PED and restores TRAIL-mediate cytotoxicity in cancer cells. Importantly, treatment of Axl+ lung cancer cells with the chimera resulted in (i) an increase in caspase activation and (ii) a reduction of cell viability in combination with TRAIL therapy. In conclusion, we demonstrate that the GL21.T-miR212 chimera can be employed as an adjuvant to TRAIL therapy for the treatment of lung cancer.
Keywords: aptamer, microRNA, non-small cell lung cancer, TNF-related apoptosis-inducing ligand
Introduction
Members of the tumor necrosis factor (TNF) superfamily of cytokines bind to cognate receptors, called death receptors, on the surface of cells. Since their first discovery, more than 20 human TNF ligands and more than 30 corresponding receptors have been identified.1 Members of this superfamily have a wide tissue distribution and regulate broad physiological processes such as immune responses, hematopoiesis, morphogenesis, and cell death, thus playing a key role in homeostasis, up to their role in tumorigenesis.2 Key members of this family include TNF, CD95L (FasL), and TNF-related apoptosis-inducing ligand (TRAIL).
The clinical application of TNF ligands as cytotoxic agents for cancer is limited due to their toxicity. For example, TNF induces systemic toxicity.3 In vivo use of CD95L is also limited by its lethal hepatotoxicity resulting from massive hepatocyte apoptosis.4,5 TRAIL, instead, has been developed as a promising antitumor agent because it induces apoptosis in several tumor-derived cell types, but not in normal cells.6,7 However, tumors often develop resistance to TRAIL monotherapy. Resistance to drug treatment is mainly due to deregulation of apoptosis-related proteins such as PED, a death effector domain (DED) family member of 15 KDa having a variety of effects on cell growth and metabolism.8 PED has a broad anti-apoptotic function, being able to inhibit both the intrinsic and the extrinsic apoptotic pathways. In the extrinsic pathway, its interaction with Fas-associated protein with death domain (FADD) and pro-caspase-8 acts as competitive inhibitor of these pro-apoptotic molecules during the assembly of the death-inducing signaling complex (DISC).9,10,11,12,13 PED has been shown to be overexpressed in TRAIL-resistant human non-small cell lung cancer (NSCLC) cells.14 An important mechanism of protein expression regulation involves microRNAs (miRNAs).15,16 Toward this end, we found that miR-212 negatively modulates PED expression and sensitizes NSCLC cells to TRAIL-induced apoptosis. In fact, miR-212 levels in resistant cell lines of NSCLC were downregulated and inversely correlated with PED levels.17 Consistently, transfection of a miR-212 mimic resulted in sensitization of resistant cancer cells to TRAIL-induced apoptosis. This occurred, at least in part, through PED downregulation.17
A major obstacle to the translation of RNAi drugs (e.g., miRNA mimics) into the clinic is the absence of an effective targeted delivery system. In addition to their ability to inhibit the function of their targets, in the past decade much attention has been focused on aptamers as delivery vehicles for targeted therapy.18,19,20 Aptamers are highly structured single-stranded RNA molecules that bind to their cognate molecular targets (including transmembrane receptors) with high affinity and selectivity.21,22 Aptamers have been successfully adapted for the targeted delivery of active molecules both in vitro and in vivo, including anticancer drugs, toxins, radionuclides, siRNAs, and, more recently, miRNAs.23, 24, 25 Aptamer-siRNA or aptamer-miRNA chimeras are characterized by low immunogenicity, easy chemical synthesis and modification, and superior target selectivity.23,26,27,28
In previous studies, an internalizing RNA aptamer (GL21.T)29 has been identified, through a cell-SELEX (systematic evolution of ligands by exponential enrichment) methodology.30 GL21.T aptamer is able to bind and inhibit the signaling of Axl receptor, belonging to the TAM family of tyrosine kinase receptors. Axl family members are activated by growth-arrest-specific gene 6 (GAS6), a member of the vitamin K–dependent protein family, that resembles blood coagulation factors rather than typical growth factors.31 Axl overexpression has been reported in many human cancers and is associated with invasiveness and/or metastasis in lung,32 prostate,33 breast,34 gastric,35 and pancreatic cancers,36 renal cell carcinoma,37 as well as glioblastoma.38 Importantly, we have recently described the combinatorial potential of a chimera composed of GL21.T aptamer and a miRNA combining the clinical benefits of both moieties.24 Here, we demonstrate selective delivery of miR-212 to Axl+ lung cancer cells with GL21.T resulting in restoration of TRAIL-mediated sensitivity in NSCLC cells. Treatment of Axl+ cells with the GL21.T-miR212 chimera resulted in caspase activation and in a concomitant reduction of cancer cell viability. In conclusion, we describe a novel aptamer-miRNA chimera as a means to sensitize lung cancers to TRAIL therapy.
Results
Chimera design
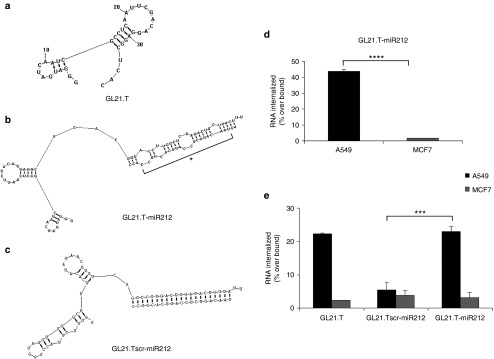
To conjugate GL21.T aptamer and miR-212, a molecular chimera (termed GL21.T-miR212) was designed using the RNA structure 5.3 program. GL21.T is a 34-mer truncated version of the original GL21 aptamer, corresponding to the functional portion of the aptamer able to bind to and to antagonize Axl receptor.29 GL21.T was used as a delivery carrier of human miR-212. For this purpose, the GL21.T sequence was elongated at its 3′ end, by a covalent bond, with the sequence of the passenger strand of miR-212, and annealed to the guide strand. Even if full complementary miRNA sequences have been shown to be sufficient for targeted gene silencing,27,28 several recent reports on the use of molecular aptamer-siRNA chimeras have shown that silencing efficacy and specificity can be improved by introducing internal partial complementarities and increased length extension with respect to the mature sequence in order to obtain a more effective Dicer substrate.26,39,40 Therefore, in order to encourage correct strand selection and thereby encourage target specificity, passenger and guide strands presented an imperfect pairing, consisting in a portion of stem-loop structure making the double strand similar to the pre-miR. A scrambled chimera, GL21.Tscr-miR212, was also designed, with the GL21.T sequence substituted by an unrelated sequence of the same length elongated with the miR-212 mimic passenger strand and annealed to the full complementary miR-212 guide strand. In both types of chimera, the antisense strand presented two overhanging bases (UU) at 3′ end necessary for Dicer processing (Figure 1). Since, based on its predicted structure, the folding of GL21.T appears to be preserved also in the context of the chimera, we experimentally assessed the selective binding and the internalization potential of GL21.T-miR212 on Axl-expressing cells. Binding and internalization assays were performed using A549 (Axl+) cells, while MCF7 cells were used as negative control since they do not express Axl. As shown in Figure 1, GL21.T-miR212 was able to bind to and internalize into A549 respect to the scrambled chimera used as control, but not in MCF7 cells, as assessed by two different methods (Figure 1d,e). Noteworthy, a similar percentage of internalization was obtained comparing GL21.T-miR212 and GL21.T alone (Figure 1e). These results indicate that, as previously reported for the GL21.T aptamer,29 in the GL21.T-miR212 conjugate, the binding specificity of the GL21.T aptamer moiety is preserved and the conjugate is internalized into target cells in a receptor-dependent manner.
Figure 1.
Chimeras structure prediction and binding and internalization analysis. Secondary structure prediction of chimeras using RNA structure 5.3 program. (a) GL21.T aptamer; (b) GL21.T-miR212; (c) GL21.Tscr-miR212. MiR mature sequence is indicated with an asterisk. (d) Internalization assay for the 5′-[32P]-labeled GL21.Tscr-miR212 and GL21.T-miR212 chimeras performed on A549 (Axl+) and MCF7 (Axl−) cells. The percentage of the RNA internalized over bound was obtained subtracting the counts relative to the scrambled chimera GL21.Tscr-miR212 used as negative control. Each bar shows the mean ± SD values from three wells. (e) Internalization analysis of GL21.T, GL21.Tscr-miR212, and GL21.T-miR212 was monitored using quantitative RT-PCR (qRT-PCR) and normalizing to an internal RNA reference control for the PCR. The percentage of internalization has been expressed as the amount of internalized RNA relative to total bound RNA. Statistics were calculated using Student's t-test, ****p < 0.0001; ***p < 0.001. Each bar shows the mean ± SEM values from three wells.
Dose–response effects and dicer processing of GL21.T-miR212 chimera
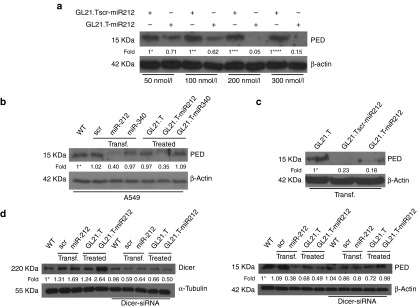
In order to characterize the effects of the chimera treatment on the miR-212 target, PED protein, A549 cells were treated with increasing amounts of GL21.T-miR212 and of control, GL21.Tscr-miR212, for 48 hours (Figure 2a). By western blot, we observed that PED levels were reduced in a dose–response manner by a concentration of 200 nM.
Figure 2.
MiR-212 effect, aptamer-mediated specific delivery, and Dicer processing of GL21.T-miR212 chimera. (a) A549 cells were treated with different final concentrations (50, 100, 200, and 300 nM) of chimera and scrambled chimera for 48 hours. (b) A549 cells were incubated with GL21.T-miR340, GL21.T, and GL21.T-miR212 or alternatively were transfected with pre-miR-212 and pre-miR-340. (c) A549 cells were transfected with the aptamer alone, GL21.Tscr-miR212 and GL21.T-miR212. (d) A549 cells were transfected with control scrambled or pre-miR-212 or treated with the aptamer alone or GL21.T-miR212 in presence or absence of Dicer-siRNA. After 48 hours, the efficiency of si-Dicer transfection (left panel) was controlled by immunoblotting using anti-Dicer and anti-α-tubulin antibodies. The effect on the downregulation of target protein (right panel) was analyzed by immunoblotting with anti-PED and anti-β actin antibodies. Values below the blots indicate signal levels relative to (a) scrambled chimera-treated cells, arbitrarily set to 1 (with a different number of asterisks for each dose), or (b,d) to untreated cells (indicated as “WT”), and (c) to GL21.T-treated cells arbitrarily set to 1 (with asterisk). Intensity of bands was calculated using ImageJ (v1.46r). For a, b, and c, cell lysates were immunoblotted with anti-PED and anti-β actin antibodies.
To test the specificity of the GL21.T-miR212 chimera and simultaneously evaluate the broad applicability of our delivery platform, we conjugated the Axl aptamer to a different miR, miR-340. We have recently described that miR-340 has an onco-suppressive role in NSCLC by targeting PUM1, PUM2, and SKP2. The downregulation of these three genes was inversely correlated to p27 expression.41 Treatment of A549 cells with GL21.T-miR340 resulted in an increase in miR-340 expression levels suggesting the proper internalization of the chimera. The effect on SKP2 downregulation, as well as on the increase of p27 levels, confirmed the effectiveness of the conjugate (Supplementary Figure S1). This effect was similar to that observed with transfection of A549 cells with miR-340 (used as positive control). In contrast, as anticipated, the aptamer alone and the aptamer conjugated to miR-340 (GL21.T-miR340) did not reduce PED protein levels under the same experimental conditions. By western blot, results showed that GL21.T-miR340 was not able to modify PED levels, thus indicating that PED downregulation was merely dependent on miR-212 moiety (Figure 2b).
To demonstrate that GL21.Tscr-miR212 was not functional due to the aptamer portion and not to inactivation of the miR sequence, A549 cells were transfected with the aptamer alone, GL21.Tscr-miR212 and GL21.T-miR212. As shown, following transfection, the scrambled chimera was as effective as the GL21.T-miR212 at downregulating PED protein levels (Figure 2c).
In order to investigate the mechanism by which the chimera was functional, A549 cells were transfected with a Dicer-specific siRNA and, then, treated with GL21.T-miR212. The co-transfection of pre-miR-212 was used as positive control. The efficiency of si-Dicer transfection and the effect on the downregulation of target protein were determined by immunoblotting (Figure 2d). As shown, in the presence of a Dicer-specific siRNA, GL21.T-miR212 was not able to reduce PED protein level, suggesting that Dicer was necessary for chimera processing.
Cell-type specificity of chimera treatment
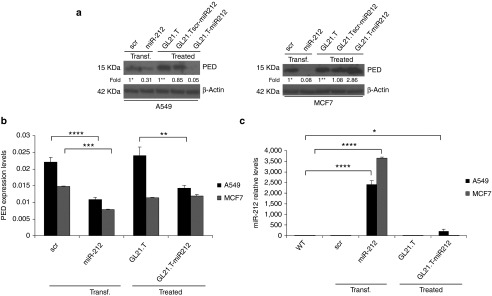
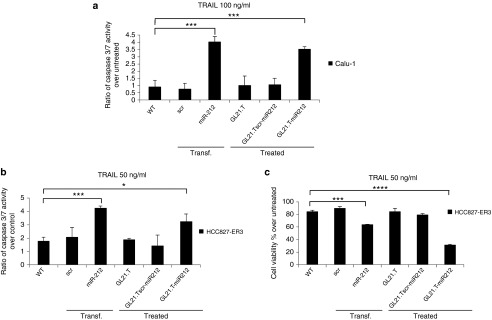
To test whether PED downregulation was cell-type specific, A549 (Axl+) and MCF7 (Axl-) cells were treated with GL21.T-miR212 and GL21.Tscr-miR212. Transfection of pre-miR-212 and treatment with GL21.T aptamer were used as positive and negative controls, respectively. In A549 cells, GL21.T-miR212 downregulated PED both at mRNA level (measured using qRT-PCR) and protein level (assessed by immunoblotting with specific antibodies). As expected, no effect of the chimeras on PED expression was observed in MCF7 (Axl-negative) cells (Figure 3a,b). To confirm that the effects on PED protein levels were mediated by miR-212 upregulation, the same samples were evaluated by qRT-PCR to analyze miR-212 expression (Figure 3c). GL21.T delivered miR-212 inside the target cells, resulting in miR-212 upregulation. Furthermore, despite the fact that the intracellular levels of miR-212 were lower in cells treated (no transfection reagent used) with GL21.T-miR212 compared with cells transfected with the pre-miR, the effects on PED downregulation were comparable. Results indicate that the efficiency of the conjugate to deliver functional miR-212 and thus modulate the expression of miRNA target genes is similar to that observed with transfection. We also validated the effect of GL21.T-miR212 on PED downregulation in other NSCLC Axl+ cell lines, Calu-1 and HCC827-ER3 (Figure 4).
Figure 3.
Cell-type specificity of chimera treatment. (a) A549 and MCF7 cells were treated with 300 nM of GL21.T-miR212 for 48 hours. GL21.Tscr-miR212 and GL21.T aptamer were used as negative controls, whereas transfection with 100 nM of pre-miR-212 was used as positive control. Control scrambled was used to assure transfection efficiency. Cell lysates were immunoblotted with anti-PED and anti-β actin antibodies for PED protein levels while (b) PED expression levels were analyzed by qRT-PCR. (c) The same samples were subjected to qRT-PCR for miR-212 expression levels analysis. Bands' intensity has been calculated as in Figure 2. In b and c each bar shows the mean ± SD values from three wells. Statistics were calculated using Student's t-test, ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05.
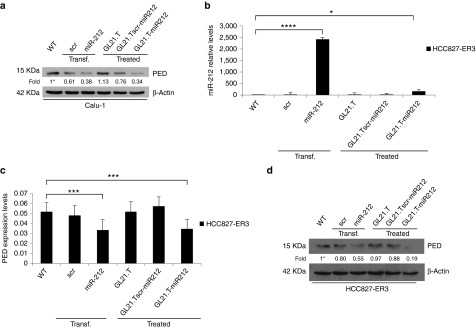
Figure 4.
Effects of GL21.T-miR212 on additional Axl+ non-small cell lung cancer (NSCLC) cell lines. (a,d) Calu-1 and HCC827-ER3 cells were treated with 300 nM of GL21.T-miR212, GL21.Tscr-miR212, and GL21.T aptamer or, alternatively, transfected with 100 nM of pre-miR-212. After 72 hours for Calu-1 or 48 hours for HCC827-ER3, cells were collected and cell lysates were immunoblotted with anti-PED and anti-β actin antibodies for PED protein levels. (b) The same samples of HCC827-ER3 were subjected to qRT-PCR for miR-212 and (c) for PED expression levels analysis. In a,d values below the blots indicate signal levels relative to untreated cells (indicated as “WT”), arbitrarily set to 1 (with asterisk). Bands' intensity has been calculated as in Figure 2. In b and c each bar shows the mean ± SD values from three wells. Statistics were calculated using Student's t-test, ****p < 0.0001; ***p < 0.001; *p < 0.05.
Receptor-dependent internalization of GL21.T-miR212 chimera
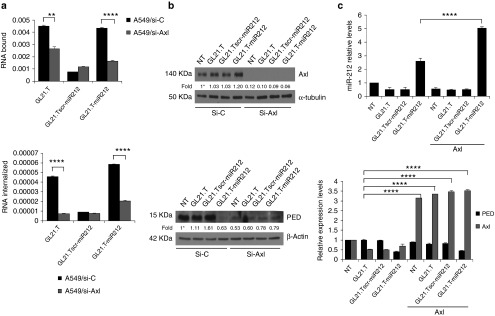
To confirm receptor-dependent internalization of GL21.T-miR212 chimera, we silenced Axl levels in A549 cells with RNAi. Following 48 hours of si-Axl transfection, we tested the binding and internalization potential of GL21.T-miR212. As expected, we observed a statistically significant decrease in bound/internalized GL21.T aptamer and chimera in A549 (siAxl)-treated cells. In contrast, no differences in binding/internalization were observed for the scrambled chimera (Figure 5a). The efficiency of si-Axl transfection and the effect on the downregulation of target protein were determined by immunoblotting (Figure 5b). Alternatively, Axl levels were transiently upregulated transfecting Axl cDNA. Following Axl overexpression, the treatment with GL21.T-miR212 increased miR-212 levels by twofold compared with parental A549. Simultaneously, the conjugate decreased PED levels to the same extent in parental and transfected A549 cells (Figure 5c). Thus, we conclude that the functional delivery of miR-212 is dependent on the amount of Axl on the cell surface and that the additional miR-212 delivered is not necessary to increase the effect on PED downregulation. We next assessed whether internalization of the conjugate was Axl mediated. MCF7 cells were transiently transfected with Axl cDNA and levels of miR-212 evaluated following treatment with the conjugate. As predicted, miR-212 levels were higher in cells treated with GL21.T-miR212, compared with the treatment with GL21.Tscr-miR212 or the aptamer alone, thus indicating that internalization of GL21.T-miR212 chimera is receptor dependent (Supplementary Figure S2).
Figure 5.
Receptor-dependent internalization of GL21.T-miR212 chimera. (a) A549 cells were transfected with si-Axl or siRNA control for 24 hours and, then, treated with GL21.T-miR212, the scrambled chimera or with the aptamer alone to perform the binding (upper panel) and internalization (lower panel) assays. Each bar shows the mean ± SEM values from three wells. (b) The efficiency of si-Axl transfection and the effect on the downregulation of target protein were evaluated after 48 hours of treatment by immunoblotting with anti-Axl and anti-tubulin, in the upper panel, and with anti-PED and anti-β actin antibodies, in the lower panel. Values below the blots indicate signal levels relative to untreated cells (indicated as “WT”), arbitrarily set to 1 (with asterisk). Bands' intensity has been calculated as in Figure 2. (c) A549 (Axl+) cells, following 24-hour transfection with Axl TruClone (Axl), were treated with 300 nM of GL21.T-miR212, GL21.Tscr-miR212, or GL21.T for additional 48 hours. miR-212 (upper panel), PED and Axl (lower panel) levels were quantified by RT-qPCR. Each bar shows the mean ± SD values from three wells. Statistics were calculated using Student's t-test, ****p < 0.0001; **p < 0.01.
GL21.T-miR212 regulates TRAIL-induced cell death
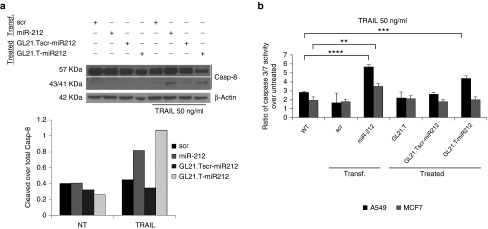
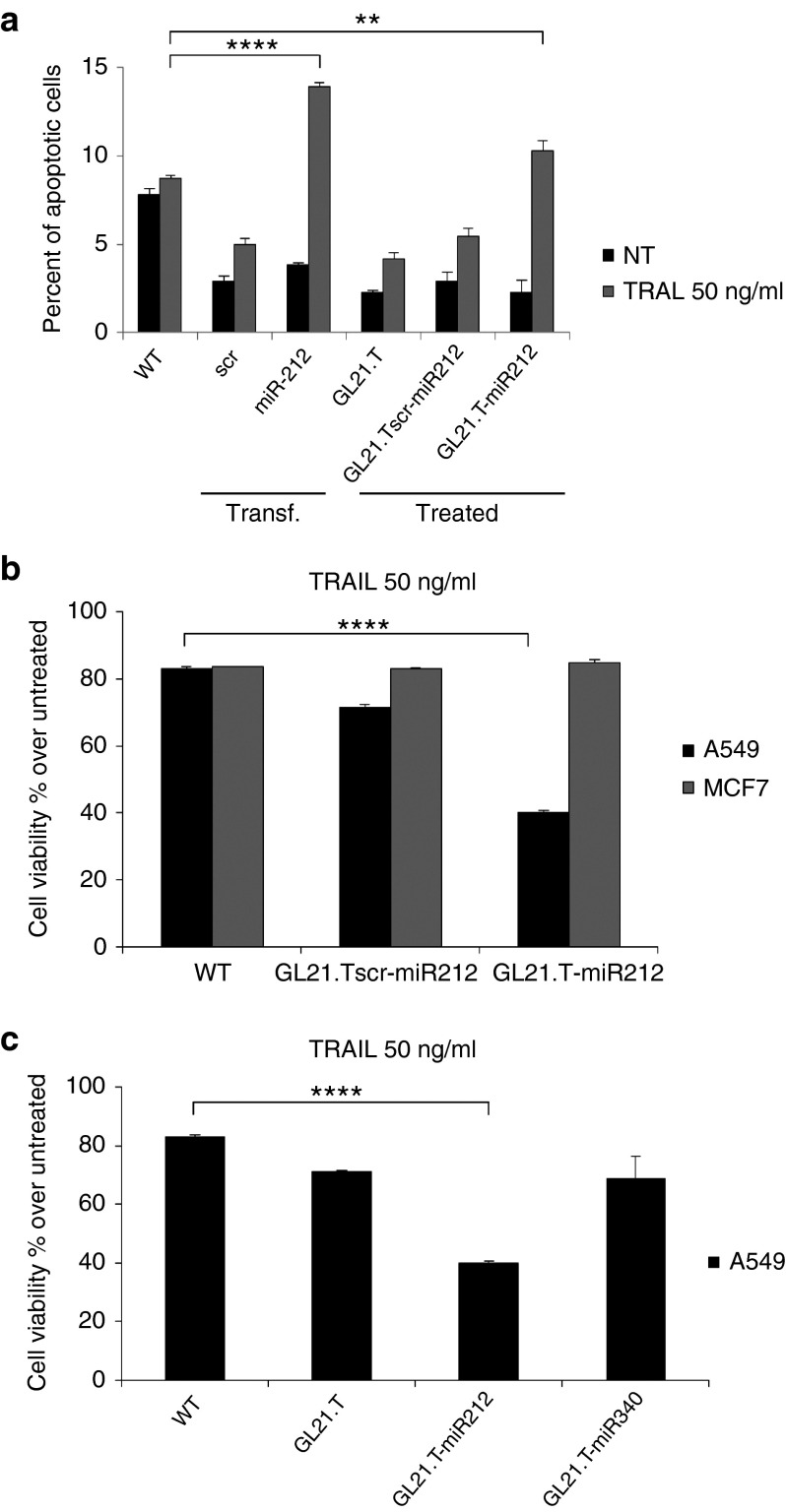
We have previously shown that the TRAIL-resistant phenotype in NSCLC is related to aberrant elevated levels of PED. Furthermore, we showed that ectopic expression of miR-212 (achieved with a miR mimic) downregulates PED and re-establishes sensitivity to TRAIL.14,17 To investigate whether treatment with the chimera induced sensitivity to TRAIL, we treated A549 cells with GL21.T-miR212. GL21.Tscr-miR212 and transfected pre-miR-212 were included as negative and positive controls, respectively. Caspase-8 activation was evaluated following treatment with TRAIL for 3 hours by western blot (Figure 6a). As shown, cleavage of caspase-8 was evident in cells treated with GL21.T-miR212 or, alternatively, transfected with pre-miR-212, but not with the GL21.Tscr-miR212, and accompanied by the activation of caspase 3/7, assessed by Caspase-Glo® 3/7 Assay (Figure 6b). Thus, sensitization to TRAIL upon treatment with the chimera was selective for Axl-expressing cells as demonstrated by the activation of caspase 3/7. To further confirm that the chimera sensitizes cancer cells to TRAIL-induced apoptosis, we evaluated the percentage of apoptotic cells after TRAIL treatment (Figure 7a). Transfected or treated cells were labeled with Annexin V-FITC and propidium iodide and analyzed using flow cytometry. GL21.T-miR212 increased the percentage of apoptotic (Annexin V—positive, PI—negative) cells following TRAIL treatment, as miR-212 was used as positive control. In addition, we measured cell viability using an MTT assay that showed the same results (Figure 7b). GL21.T-miR340 treatment did not produce any gain in TRAIL sensitivity (Figure 7c). In summary, the GL21.T-miR212 chimera was able to increase the activation of caspase3/7 and, consequently, TRAIL-induced cell death in A549 cells, but not in MCF7 cells. TRAIL sensitization mediated by GL21.T-miR212 treatment was also confirmed on additional NSCLC cell lines, Calu-1 and HCC827-ER3, which display a TRAIL-resistant phenotype (Figure 8).
Figure 6.
Caspases activation induced by GL21.T-miR212. (a) A549 cells were transfected with 100 nM of pre-miR-212 or alternatively treated with 300 nM of GL21.T-miR212 for 48 hours. Scrambled miR and scrambled chimera were used as negative controls. Cells were, then, treated for 3 hours with TNF-related apoptosis-inducing ligand (TRAIL) 50 ng/ml, and cell lysates were immunoblotted with anti-caspase-8 antibody (upper panel). Band intensity is represented in the diagram of the lower panel as a ratio of cleaved over total caspase-8, both quantization normalized over β-actin. (b) A549 and MCF7 cells were transfected with pre-miR-212 or treated with the unconjugated aptamer, the scrambled chimera and GL21.T-miR212 for 48 hours and then incubated with 50 ng/ml of TRAIL for 6 hours. The activation of caspase 3/7 was measured by Caspase-Glo® 3/7 Assay. Each bar shows the mean ± SD values from three wells. Statistics were calculated using Student's t-test, ****p < 0.0001; ***p < 0.001; **p < 0.01.
Figure 7.
TNF-related apoptosis-inducing ligand (TRAIL) sensitization induced by GL21.T-miR212. (a) A549 cells were transfected with pre-miR-212 and control scrambled or treated with the chimera, the scrambled chimera and the unconjugated aptamer for 48 hours. Cells were, then, incubated with TRAIL for 24 hours, and the percentage of apoptotic cells was evaluated by flow cytometry. (b) A549 and MCF7 cells were treated with 300 nM of GL21.T-miR212 and GL21.Tscr-miR for 48 hours and were exposed to TRAIL for 24 hours at 50 ng/ml as final concentration. (c) A549 cells were treated with the unconjugated aptamer, GL21.T-miR212 or GL21.T-miR340 for 48 hours and were exposed to TRAIL for 24 hours at 50 ng/ml as final concentration. For b and c cell viability was evaluated with MTT assay. In a, b, and c each bar shows the mean ± SD values from three wells. Statistics were calculated using Student's t-test, ****p < 0.0001; **p < 0.01.
Figure 8.
TNF-related apoptosis-inducing ligand (TRAIL) sensitization induced by GL21.T-miR212 in additional Axl+ non-small cell lung cancer (NSCLC) cell lines. Calu-1 and HCC827-ER3 cells were transfected with pre-miR-212 or treated with the unconjugated aptamer, the scrambled chimera and GL21.T-miR212. (a,b) After 72 hours for Calu-1 or 48 hours for HCC827-ER3 of treatment or transfection, cells were incubated with TRAIL (100 ng/ml in Calu-1 and 50 ng/ml in HCC827-ER3) for 6 hours. The activation of caspase 3/7 was measured by Caspase-Glo® 3/7 Assay. (c) The same samples of HCC827-ER3 were exposed to TRAIL for 24 hours, and cell viability was evaluated with MTT assay. Each bar shows the mean ± SD values from three wells. Statistics were calculated using Student's t-test, ****p < 0.0001; ***p < 0.001; *p < 0.05.
Discussion
NSCLC represents about 80% of all lung cancers and is mostly diagnosed at an advanced stage (either locally advanced or metastatic disease). Because of resistance to therapeutic drugs, standard treatment of this tumor has only a 20% to 30% positive clinical response. Over the last years, the discovery of the pivotal of epidermal growth factor receptor (EGFR) in tumorigenesis has opened the way to a new class of targeted therapeutic agents: the EGFR tyrosine kinase inhibitors (EGFR TKIs). Since their introduction in therapy, in advanced NSCLC patients harboring EGFR mutations, the use of EGFR TKIs in first-line treatment has provided an unusually large progression-free survival benefit with a negligible toxicity when compared with cytotoxic chemotherapy. Nevertheless, resistance invariably occurs.42 In this setting, TRAIL emerged as a novel therapeutic agent. TRAIL (ApoL/TNF-related apoptosis-inducing ligand) is a relatively new member of the tumor necrosis factor (TNF) ligand family, which induces apoptosis in a variety of cancers.
Initial promising studies demonstrated its remarkable specificity in inducing apoptosis in tumor cell lines, but not in normal cells both in vitro and in vivo.7 This unique property makes TRAIL an attractive candidate for targeted cancer therapy.43,44 However, resistance to TRAIL-induced apoptosis poses a challenge for effective anticancer strategies. To overcome this problem, drug cocktails in combination with TRAIL therapy have been proposed in order to induce synergism or sensitize resistant cancer cells. Toward this end, a number of combinatorial treatments with chemotherapeutic agents are in phase 1/2 of clinical studies.45,46,47 More recently, aptamer-siRNA/miRNA chimeras have been proposed as novel adjuvants to standard chemotherapy.48,49 Unlike nontargeted drugs, the advantage of these new class of biodrugs is that they are specifically delivered into target cells where they release their therapeutic cargo, thus limiting toxicity to normal cells.
In this study, we designed a chimera composed of a RNA aptamer to Axl (GL21.T) and miR-212 as a means to deliver functional miR-212 into TRAIL-resistant Axl+ A549 cells, but not into Axl- MCF7 cells. Indeed, GL21.T-miR212 selectively sensitizes the A549 cells to TRAIL-induced apoptosis, proving to be a unique tool to synergize with TRAIL in mediating cell death.
To increase specificity and facilitate large-scale chemical synthesis,24,26,50 we conjugated a truncated version of the GL21 aptamer (GL21.T) to the tumor suppressor miR-212 duplex sequence. We demonstrated that in the context of the chimera, the active sequence (sequence required for binding to Axl) of GL21 is preserved, thus providing high binding affinity and the subsequent selective internalization of the conjugate into Axl+ cells. The miRNA moiety is a 25/27mer duplex having two overhanging bases (UU) at the 3′ end of the passenger strand, thus adopting the conformation described as Dicer substrate for duplex siRNAs.51 By using a similar approach with miRNAs, we have recently shown that nonperfect duplex miRNAs are correctly processed by Dicer, increasing the gene target specificity of the miRNA moiety.24 Indeed, the optimal loading of the guide strand into RNA-induced silencing complex (RISC) is thought to reduce off-target effects that result from inappropriate incorporation of both miRNA strands into the silencing complex.52
A major limitation to the use of RNA-based drugs in vivo is the rapid degradation (within few minutes) of natural RNAs in serum or blood. As previously described, in order to protect the GL21.T aptamer from degradation, it was generated as a 2′-F-Py containing RNA.29 Therefore, in order to increase the stability of the entire GL21.T-miR212 molecule, we substituted the pyrimidines with 2′-F-Py at all positions. This modification is well characterized in humans and is reported to be well tolerated with little toxicity.53 RNA aptamers with this modification have already been approved for their use in humans (Macugen), with many more quickly moving through the clinical pipeline.54 Although we cannot completely rule out potential intracellular toxicity of 2′-F-Py-modified RNAs leading to nonspecific immunostimulation, experiments in vivo demonstrated that problematic toxicity in humans is not expected.24,26
The silencing moiety of the chimera is constituted by the miR-212, a tumor suppressor miRNA that acts by negatively modulating PED expression, an onco-protein with a broad anti-apoptotic action. Indeed, the presence of elevated cellular levels of PED has been shown to contribute to resistance to TRAIL-induced cell death in several human tumors, including breast and lung cancer.17,55 The DED domain of PED acts as a competitive inhibitor for pro-apoptotic molecules during the assembly of a functional DISC and inhibiting the activation of caspase-8, which take place following treatment with different apoptotic cytokines (CD95/FasL, TNF-α, and TRAIL). These data demonstrate that GL21.T-miR212 is a functional molecule that upon internalization, downregulates PED in a dose-dependent manner, reaching a plateau at around 200 nM. In turn, target cells become sensitive to TRAIL and upon treatment undergo apoptosis following caspase-8 and caspase-3 activation.
Based on the suppressive action of miR-212 on PED expression as a means to sensitize cancer cells to TRAIL, here we demonstrated that GL21.T-miR212 chimera can sensitize target cells in a high selective manner. Specifically, exogenous miR-212 delivered by GL21.T aptamer led to TRAIL sensitization via activation of the apoptotic cascade selectively in A549, NSCLC Axl+ cells. In conclusion, the approach presented in this work indicates an innovative tool for a combined therapy that makes use of an aptamer-based molecular chimera to selectively sensitize TRAIL-resistant target tumor cells.
Materials and methods
Cell lines and transfection. A549 and HCC827-ER3 cells were grown in RPMI 1640 while MCF7 and Calu-1 cells were grown in Dulbecco's modified Eagle's medium. A549, MCF7, and Calu cells were from American Type Culture Collection, while HCC827-ER3 were kindly provided by Dr. Balazs Halmos (Columbia University Medical Center, New York, NY). Their media were supplemented with 10% heat-inactivated fetal bovine serum, 2 mM of glutamine, and 100 U/ml of penicillin/streptomycin. For miRNAs transient transfection, cells were transfected with 100 nM (final concentration) of miRNA stem-loop precursor hsa-miR212, hsa-miR340, or negative control 1 (Ambion, Foster City, CA) using Oligofectamine (Invitrogen, Carlsbad, CA). Also si-control and si-Axl (Santa Cruz Biotechnology, Santa Cruz, CA) were transfected using Oligofectamine (Invitrogen), according to the manufacturer's protocol. For aptamer and chimeras transient transfection, cells were transfected with 100 nM (final concentration) of RNAs, using Lipofectamine 2000 (Invitrogen). Also Axl TruClone (Origene, Rockville, MD) and si-Dicer (Cell Signaling Technology, Beverly, MA) were transfected with Lipofectamine 2000, according to the manufacturer's protocol.
Aptamer-miRNA chimeras. The following sequences were used for the chimera production: GL21.T-miR212 passenger strand: 5′GGGAUGAUCAAUCGCCUCAAUUCGACAGGAGGCUCACGGUACCUUGGCUCUAGACUGCUUACUUU. miR-212 guide strand: 5′ AGUAACAGUCUCCAGUCACGGCCACC. GL21.Tscr-miR212 passenger strand: 5′GGGUUCGUACCGGGUAGGUUGGCUUGCACAUAGAACGUGUCAGGCCGUGACUGGAGACUGUUAUU. miR-212 (1g) guide strand: 5′ UAACAGUCUCCAGUCACGGCC. GL21.T: 5′ GGGAUGAUCAAUCGCCUCAAUUCGACAGGAGGCUCAC. GL21.T-miR340 passenger strand: 5′GGGAUGAUCAAUCGCCUCAAUUCGACAGGAGGCUCACAAUCAGUCUCAUUGCUUUAUAAUU. miR-340 guide strand: 5′ UUAUAAAGCAAUGAGACUGAUU.
All RNAs were custom synthesized by TriLink Biotechnologies (San Diego, CA) as 2′-fluoropyrimidine RNAs. UU in bold are 3′-overhang. The control conjugate is composed of an unrelated aptamer sequence linked to the fully complementary miR-212 duplex. In the context of the control conjugate, it was necessary to use a fully complementary miR-212 to stabilize the functional miR duplex and prevent unwanted intramolecular interactions with the scrambled aptamer sequence.
To prepare GL21.T-miR212, GL21.Tscr-miR212, and GL21.T-miR340, 5 µM of aptamer-passenger RNA strand was denatured at 98 °C for 20 minutes, combined with 5 µM of the appropriate guide strand at 55 °C for 10 minutes in binding buffer 10× (200 mM N-2-Hydroxyethylpiperazine-N'-2-Ethanesulfonic Acid, pH 7.4, 1.5 M NaCl, 20 mM CaCl2) and then warmed up to 37 °C for 20 minutes.
Cell binding and internalization assays. Aptamer binding and internalization have been assessed by two different methods, by radioactivity labeling or by quantitative reverse transcription-PCR (qRT-PCR)
Radioactivity labeling. A549 and MCF7 cells were plated in 24 multiwell plates in triplicate. RNAs were 5′-[32P]-labeled and incubated at 200 nM as final concentration on cells at 37 °C for 15 minutes. After several washings, the amount of 32P-labeled RNA recovered in SDS 1% was determined by scintillation counting. In contrast, to check the endocytosis rate, after the incubation with radiolabeled chimeras, the cells were subjected to a stringent high-salt wash, with High Salt phosphate-buffered saline (PBS; 0.5 M NaCl), to remove any unbound RNAs or RNAs bound to the cell surface. Following 5-minute treatment at 4 °C, the amount of 32P-labeled RNA internalized was recovered in Sodium Dodecyl Sulfate 1% and determined by scintillation counting. In both assays, results were normalized for cell number. The background values obtained with the scrambled chimera were subtracted from the values obtained with GL21.T-miR212. Finally, the resulting recovered RNAs were plotted as percent of RNA internalized over RNA bound.
qRT-PCR method. Target (A549) and nontarget (MCF7) cells were incubated with 100 nM of aptamer or chimeras for 15 minutes at 37 °C with 5% CO2. Cells were washed with ice-cold PBS or incubated with High Salt PBS (0.5 M NaCl) at 4 °C for 5 minutes, and RNA was recovered using TRIzol reagent (Invitrogen). Samples were normalized to an internal RNA reference control. Specifically, 0.5 pmol per sample CL4 aptamer56 was added to each sample along with TRIzol as a reference control. Recovered RNAs were quantitated using Reverse Transcriptase M-MuLV (Roche Life Science, Basel, Switzerland) with SYBR Green (BioRad) with a Biorad iCycler. All reactions were done in a 25-ml volume in triplicate with specific primers (GL21.T 5′: TAATACGACTCACTATAGGGATGATC; 3′: GTGAGCCTCCTGTcGAAT; GL21.Tscr 5′: TTCGTACCGGGTAGGTT; 3′: TGACACGTTCTATGTGCA) and CL4 reference control (CL4 5′: TAATACGACTCACTATAGGGGCCTTA; 3′: GCCTCCTGTCGAATCG).
For each cell line, the percentage of internalization has been expressed as the amount of internalized RNA relative to total bound RNA without normalizing for background. The same protocol was used for the experiment on A549 cells upon transfection with si-control and si-AXL.
Protein isolation and immunoblotting. Cells were treated with chimeras for 48 hours, or alternatively Calu-1 for 72 hours, and then were washed twice in ice-cold PBS and lysed in Lysis buffer (50 mM N-2-Hydroxyethylpiperazine-N'-2-Ethanesulfonic Acid pH 7.5 containing 150 mM NaCl, 1% GLYCEROL, 1% Triton 100×, 1.5 mM MgCl2, 5 mM ethylene glycol tetraacetic acid, 1 mM Na3VO4 and 1X protease inhibitor cocktail). Protein concentration was determined by the Bradford assay (Bio-Rad, Hercules, CA) using bovine serum albumin as the standard, and equal amounts of protein were analyzed by Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (15% acrylamide). Gels were electroblotted onto nitrocellulose membrane (Merck Millipore, Billerica, MA). For immunoblot experiments, membranes were blocked for 1 hour with 5% non-fat dry milk in tris-buffered saline containing 0.1% Tween-20 and incubated at 4 °C overnight with primary antibody. Detection was performed by peroxidase-conjugated secondary antibodies using the enhanced chemiluminescence system (GE Healthcare Life Sciences, Pittsburgh, PA). Primary antibodies used were anti-PED,57 anti-Caspase-8, anti-p27, and anti-Dicer from Cell Signaling Technology, anti-α tubulin from Santa Cruz Biotechnology, anti-β actin from Sigma-Aldrich (St. Louis, MO), and anti-Axl from R&D Systems (Minneapolis, MN).
RNA extraction and Real-time PCR. Cells were treated with 300 nM of chimeras for 48 hours, and then total RNAs (miRNA and mRNA) were extracted using TRIzol (Invitrogen) according to the manufacturer's protocol. Reverse transcription of total miRNA was performed starting from equal amounts of total RNA/sample (1 µg) using miScript reverse Transcription Kit (Qiagen, Hilden, Germany). Quantitative analysis of miRNAs and RNU6B (as an internal reference) was performed by real-time PCR using specific primers (Qiagen) and miScript SYBR Green PCR Kit (Qiagen). The reaction for detection of miRNAs was performed as follows: 95 °C for 15 minutes, 40 cycles of 94 °C for 15 seconds, 55 °C for 30 seconds, and 70 °C for 30 seconds. All reactions were run in triplicate. For reverse transcription of mRNA, we used SuperScript® III Reverse Transcriptase (Life Technologies). Quantitative analysis of PED, AXL, and actin (as an internal reference) was performed by real-time PCR using specific primers and iQTM SYBR Green Supermix (BioRad). The threshold cycle (CT) is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. For quantization has been used the 2(-ΔCT) method, where ΔCt is the difference between the amplification fluorescent thresholds of the miRNA of interest and the miRNA of U6 used as an internal reference. Instead, fold changes were calculated with 2(-ΔΔCT) method as previously described.58 Experiments were carried out in triplicate for each data point, and data analysis was performed by using software (Bio-Rad).
Cell death quantification. A549, MCF7, and HCC827-ER3 cells were treated with GL21.T-miR212 and GL21.Tscr-miR212 300 nM for 3 hours. Then, cells were plated in 96 multiwell plates in triplicate for 48 hours and incubated with TRAIL (Vinci-Biochem, Firenze, Italy) at a final concentration of 50 ng/ml for 24 hours. Cell viability was assessed with CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega, Madison, WI). Metabolically active cells were detected by adding 20 µl of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) to each well, and plates were analyzed in a Multilabel Counter (BioTek, Winooski, VT).
Caspase 3/7 assay. The assay was performed with the use of Caspase-Glo ® 3/7 Assay (Promega) according to the manufacturer's protocol. Briefly, A549, MCF7, and HCC827-ER3 cells were before transfected with pre-miR-212 or treated with GL21.T, GL21.Tscr-miR212, GL21.T-miR212 for 48 hours, or alternatively Calu-1 for 72 hours, and, then, incubated for 6 hours with TRAIL. An equal volume of Caspase-Glo 3/7 reagent was added to each well for 30 minutes in the dark, and luminescence was measured by luminometer (Turner BioSystems-Promega). The ratio of caspase 3/7 activity over control was calculated normalizing treated samples over untreated ones.
Flow cytometry. Apoptosis was analyzed via Annexin V-FITC Apoptosis Detection kit I (BD Biosciences, San Diego, CA). A549 cells were transfected with pre-miR-212 or treated with GL21.T, GL21.Tscr-miR212, GL21.T-miR212 for 48 hours and, then, incubated for 24 hours with TRAIL. The cells were washed in PBS, resuspended in binding buffer 10×, and labeled with Annexin V-FITC and propidium iodide according to the manufacturer's protocol. After incubation at room temperature for 15 minutes in the dark, cells were analyzed with a BD AccuriTM C6 Flow cytometry (BD Biosciences). To calculate the percent of apoptotic cells, the gate was placed on annexin V-positive, PI-negative cells, and thus double positive cells were excluded from the analysis.
SUPPLEMENTARY MATERIAL Figure S1. GL21.T-miR340 characterization. Figure S2. Internalization of the GL21.T-miR212 conjugate in MCF7 exogenously expressing Axl.
Acknowledgments
This work was partially supported by funds from Associazione Italiana Ricerca sul Cancro, AIRC (grant n.ro 10620) to G.C. and AIRC (grant n.ro 13345) to V.d.F.; MERIT (RBNE08E8CZ_002) to G.C., POR Campania FSE 2007–2013, Project CREME to G.C., Fondazione Berlucchi to G.C. This work was partially supported by grants to P.H.G. from the National Institutes of Health (R01CA138503 and R21DE019953), Mary Kay Foundation (9033-12 and 001-09), Elsa U Pardee Foundation (E2766) and the Roy J Carver Charitable Trust (RJCCT 01-224). M.I. was supported by the ‘Federazione Italiana Ricerca sul Cancro' (FIRC) Post-Doctoral Research Fellowship. G.R. was supported by a MERIT project Fellowship. The authors declare no conflict of interest.
Supplementary Material
References
- Aggarwal, BB, Gupta, SC and Kim, JH (2012). Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood 119: 651–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal, IS (2009). Overview of TNF superfamily: a chest full of potential therapeutic targets. Adv Exp Med Biol 647: 1–7. [DOI] [PubMed] [Google Scholar]
- Wielockx, B, Lannoy, K, Shapiro, SD, Itoh, T, Itohara, S, Vandekerckhove, J et al. (2001). Inhibition of matrix metalloproteinases blocks lethal hepatitis and apoptosis induced by tumor necrosis factor and allows safe antitumor therapy. Nat Med 7: 1202–1208. [DOI] [PubMed] [Google Scholar]
- Ni, R, Tomita, Y, Matsuda, K, Ichihara, A, Ishimura, K, Ogasawara, J et al. (1994). Fas-mediated apoptosis in primary cultured mouse hepatocytes. Exp Cell Res 215: 332–337. [DOI] [PubMed] [Google Scholar]
- Galle, PR, Hofmann, WJ, Walczak, H, Schaller, H, Otto, G, Stremmel, W et al. (1995). Involvement of the CD95 (APO-1/Fas) receptor and ligand in liver damage. J Exp Med 182: 1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi, A, Pai, RC, Fong, S, Leung, S, Lawrence, DA, Marsters, SA et al. (1999). Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest 104: 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak, H, Miller, RE, Ariail, K, Gliniak, B, Griffith, TS, Kubin, M et al. (1999). Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med 5: 157–163. [DOI] [PubMed] [Google Scholar]
- Fiory, F, Formisano, P, Perruolo, G and Beguinot, F (2009). Frontiers: PED/PEA-15, a multifunctional protein controlling cell survival and glucose metabolism. Am J Physiol Endocrinol Metab 297: E592–E601. [DOI] [PubMed] [Google Scholar]
- Zanca, C, Cozzolino, F, Quintavalle, C, Di Costanzo, S, Ricci-Vitiani, L, Santoriello, M et al. (2010). PED interacts with Rac1 and regulates cell migration/invasion processes in human non-small cell lung cancer cells. J Cell Physiol 225: 63–72. [DOI] [PubMed] [Google Scholar]
- Quintavalle, C, Di Costanzo, S, Zanca, C, Tasset, I, Fraldi, A, Incoronato, M et al. (2014). Phosphorylation-regulated degradation of the tumor-suppressor form of PED by chaperone-mediated autophagy in lung cancer cells. J Cell Physiol 229: 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo, M, Romano, G, Quintavalle, C, Romano, MF, Chiurazzi, F, Zanca, C et al. (2007). Selective inhibition of PED protein expression sensitizes B-cell chronic lymphocytic leukaemia cells to TRAIL-induced apoptosis. Int J Cancer 120: 1215–1222. [DOI] [PubMed] [Google Scholar]
- Garofalo, M, Quintavalle, C, Di Leva, G, Zanca, C, Romano, G, Taccioli, C et al. (2008). MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene 27: 3845–3855. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani, L, Pedini, F, Mollinari, C, Condorelli, G, Bonci, D, Bez, A et al. (2004). Absence of caspase 8 and high expression of PED protect primitive neural cells from cell death. J Exp Med 200: 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanca, C, Garofalo, M, Quintavalle, C, Romano, G, Acunzo, M, Ragno, P et al. (2008). PED is overexpressed and mediates TRAIL resistance in human non-small cell lung cancer. J Cell Mol Med 12(6A): 2416–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo, M, Leva, GD and Croce, CM (2014). MicroRNAs as anti-cancer therapy. Curr Pharm Des 20: 5328–5335. [DOI] [PubMed] [Google Scholar]
- Garofalo, M, Condorelli, GL, Croce, CM and Condorelli, G (2010). MicroRNAs as regulators of death receptors signaling. Cell Death Differ 17: 200–208. [DOI] [PubMed] [Google Scholar]
- Incoronato, M, Garofalo, M, Urso, L, Romano, G, Quintavalle, C, Zanca, C et al. (2010). miR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res 70: 3638–3646. [DOI] [PubMed] [Google Scholar]
- Yan, AC and Levy, M (2009). Aptamers and aptamer targeted delivery. RNA Biol 6: 316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farokhzad, OC, Karp, JM and Langer, R (2006). Nanoparticle-aptamer bioconjugates for cancer targeting. Expert Opin Drug Deliv 3: 311–324. [DOI] [PubMed] [Google Scholar]
- Zhou, J and Rossi, JJ (2014). Cell-type-specific, aptamer-functionalized agents for targeted disease therapy. Mol Ther Nucleic Acids 3: e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J and Li, G (2011). Aptamers against cell surface receptors: selection, modification and application. Curr Med Chem 18: 4107–4116. [DOI] [PubMed] [Google Scholar]
- Catuogno, S, Esposito, CL, de Franciscis, V, (2016). Developing Aptamers by Cell-Based SELEX. Methods Mol Bioll 1380: 33–46. [DOI] [PubMed] [Google Scholar]
- Zhou, J and Rossi, JJ (2010). Aptamer-targeted cell-specific RNA interference. Silence 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, CL, Cerchia, L, Catuogno, S, De Vita, G, Dassie, JP, Santamaria, G et al. (2014). Multifunctional aptamer-miRNA conjugates for targeted cancer therapy. Mol Ther 22: 1151–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, F, Zhang, Y, Zhu, X, Shan, N and Chen, Y (2012). Anticancer role of MUC1 aptamer-miR-29b chimera in epithelial ovarian carcinoma cells through regulation of PTEN methylation. Target Oncol 7: 217–225. [DOI] [PubMed] [Google Scholar]
- Dassie, JP, Liu, XY, Thomas, GS, Whitaker, RM, Thiel, KW, Stockdale, KR et al. (2009). Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol 27: 839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, JO 2nd, Andrechek, ER, Wang, Y, Viles, KD, Rempel, RE, Gilboa, E et al. (2006). Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol 24: 1005–1015. [DOI] [PubMed] [Google Scholar]
- Dai, F, Zhang, Y, Zhu, X, Shan, N and Chen, Y (2013). The anti-chemoresistant effect and mechanism of MUC1 aptamer-miR-29b chimera in ovarian cancer. Gynecol Oncol 131: 451–459. [DOI] [PubMed] [Google Scholar]
- Cerchia, L, Esposito, CL, Camorani, S, Rienzo, A, Stasio, L, Insabato, L et al. (2012). Targeting Axl with an high-affinity inhibitory aptamer. Mol Ther 20: 2291–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington, AD and Szostak, JW (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822. [DOI] [PubMed] [Google Scholar]
- Stitt, TN, Conn, G, Gore, M, Lai, C, Bruno, J, Radziejewski, C et al. (1995). The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell 80: 661–670. [DOI] [PubMed] [Google Scholar]
- Shieh, YS, Lai, CY, Kao, YR, Shiah, SG, Chu, YW, Lee, HS et al. (2005). Expression of axl in lung adenocarcinoma and correlation with tumor progression. Neoplasia 7: 1058–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainaghi, PP, Castello, L, Bergamasco, L, Galletti, M, Bellosta, P and Avanzi, GC (2005). Gas6 induces proliferation in prostate carcinoma cell lines expressing the Axl receptor. J Cell Physiol 204: 36–44. [DOI] [PubMed] [Google Scholar]
- Zhang, YX, Knyazev, PG, Cheburkin, YV, Sharma, K, Knyazev, YP, Orfi, L et al. (2008). AXL is a potential target for therapeutic intervention in breast cancer progression. Cancer Res 68: 1905–1915. [DOI] [PubMed] [Google Scholar]
- Wu, CW, Li, AF, Chi, CW, Lai, CH, Huang, CL, Lo, SS et al. (2002). Clinical significance of AXL kinase family in gastric cancer. Anticancer Res 22(2B): 1071–1078. [PubMed] [Google Scholar]
- Koorstra, JB, Karikari, CA, Feldmann, G, Bisht, S, Rojas, PL, Offerhaus, GJ et al. (2009). The Axl receptor tyrosine kinase confers an adverse prognostic influence in pancreatic cancer and represents a new therapeutic target. Cancer Biol Ther 8: 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, BI, Malkowicz, SB, Nguyen, TB, Libertino, JA and McGarvey, TW (2003). Expression of the proto-oncogene Axl in renal cell carcinoma. DNA Cell Biol 22: 533–540. [DOI] [PubMed] [Google Scholar]
- Hutterer, M, Knyazev, P, Abate, A, Reschke, M, Maier, H, Stefanova, N et al. (2008). Axl and growth arrest-specific gene 6 are frequently overexpressed in human gliomas and predict poor prognosis in patients with glioblastoma multiforme. Clin Cancer Res 14: 130–138. [DOI] [PubMed] [Google Scholar]
- Wu, X, Ding, B, Gao, J, Wang, H, Fan, W, Wang, X et al. (2011). Second-generation aptamer-conjugated PSMA-targeted delivery system for prostate cancer therapy. Int J Nanomedicine 6: 1747–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarzguioui, M, Lundberg, P, Cantin, E, Hagstrom, J, Behlke, MA and Rossi, JJ (2006). Rational design and in vitro and in vivo delivery of Dicer substrate siRNA. Nat Protoc 1: 508–517. [DOI] [PubMed] [Google Scholar]
- Fernandez, S, Risolino, M, Mandia, N, Talotta, F, Soini, Y, Incoronato, M et al. (2014). miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene 34: 3240–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgambato, A, Casaluce, F, Maione, P, Rossi, A, Rossi, E, Napolitano, A et al. (2012). The role of EGFR tyrosine kinase inhibitors in the first-line treatment of advanced non small cell lung cancer patients harboring EGFR mutation. Curr Med Chem 19: 3337–3352. [DOI] [PubMed] [Google Scholar]
- Stuckey, DW and Shah, K (2013). TRAIL on trial: preclinical advances in cancer therapy. Trends Mol Med 19: 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst, RS, Eckhardt, SG, Kurzrock, R, Ebbinghaus, S, O'Dwyer, PJ, Gordon, MS et al. (2010). Phase I dose-escalation study of recombinant human Apo2L/TRAIL, a dual proapoptotic receptor agonist, in patients with advanced cancer. J Clin Oncol 28: 2839–2846. [DOI] [PubMed] [Google Scholar]
- Falschlehner, C, Ganten, TM, Koschny, R, Schaefer, U and Walczak, H (2009). TRAIL and other TRAIL receptor agonists as novel cancer therapeutics. Adv Exp Med Biol 647: 195–206. [DOI] [PubMed] [Google Scholar]
- Soria, JC, Smit, E, Khayat, D, Besse, B, Yang, X, Hsu, CP et al. (2010). Phase 1b study of dulanermin (recombinant human Apo2L/TRAIL) in combination with paclitaxel, carboplatin, and bevacizumab in patients with advanced non-squamous non-small-cell lung cancer. J Clin Oncol 28: 1527–1533. [DOI] [PubMed] [Google Scholar]
- Hotte, SJ, Hirte, HW, Chen, EX, Siu, LL, Le, LH, Corey, A et al. (2008). A phase 1 study of mapatumumab (fully human monoclonal antibody to TRAIL-R1) in patients with advanced solid malignancies. Clin Cancer Res 14: 3450–3455. [DOI] [PubMed] [Google Scholar]
- Thiel, KW, Hernandez, LI, Dassie, JP, Thiel, WH, Liu, X, Stockdale, KR et al. (2012). Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res 40: 6319–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, N, Zhou, C, Zhao, J and Chen, Y (2012). Reversal of paclitaxel resistance in epithelial ovarian carcinoma cells by a MUC1 aptamer-let-7i chimera. Cancer Invest 30: 577–582. [DOI] [PubMed] [Google Scholar]
- Esposito, CL, Catuogno, S and de Franciscis, V (2014). Aptamer-mediated selective delivery of short RNA therapeutics in cancer cells. J RNAi Gene Silencing 10: 500–506. [PMC free article] [PubMed] [Google Scholar]
- Ma, JB, Ye, K and Patel, DJ (2004). Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429: 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz, CA, Holko, M, de Veer, MJ, Silverman, RH and Williams, BR (2003). Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 5: 834–839. [DOI] [PubMed] [Google Scholar]
- Behlke, MA (2008). Chemical modification of siRNAs for in vivo use. Oligonucleotides 18: 305–319. [DOI] [PubMed] [Google Scholar]
- Keefe, AD, Pai, S and Ellington, A (2010). Aptamers as therapeutics. Nat Rev Drug Discov 9: 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassi, G, Garofalo, M, Zerilli, M, Ricci-Vitiani, L, Zanca, C, Todaro, M et al. (2005). PED mediates AKT-dependent chemoresistance in human breast cancer cells. Cancer Res 65: 6668–6675. [DOI] [PubMed] [Google Scholar]
- Esposito, CL, Passaro, D, Longobardo, I, Condorelli, G, Marotta, P, Affuso, A et al. (2011). A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death. PLoS One 6: e24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli, G, Vigliotta, G, Iavarone, C, Caruso, M, Tocchetti, CG, Andreozzi, F et al. (1998). PED/PEA-15 gene controls glucose transport and is overexpressed in type 2 diabetes mellitus. EMBO J 17: 3858–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, KJ and Schmittgen, TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.