Summary
Background
Chronic injury in kidney transplants remains a major cause of allograft loss. The aim of this study was to identify a gene set capable of predicting renal allografts at risk of progressive injury due to fibrosis.
Methods
This Genomics of Chronic Allograft Rejection (GoCAR) study is a prospective, multicentre study. We prospectively collected biopsies from renal allograft recipients (n=204) with stable renal function 3 months after transplantation. We used microarray analysis to investigate gene expression in 159 of these tissue samples. We aimed to identify genes that correlated with the Chronic Allograft Damage Index (CADI) score at 12 months, but not fibrosis at the time of the biopsy. We applied a penalised regression model in combination with permutation-based approach to derive an optimal gene set to predict allograft fibrosis. The GoCAR study is registered with ClinicalTrials.gov, number NCT00611702.
Findings
We identified a set of 13 genes that was independently predictive for the development of fibrosis at 1 year (ie, CADI-12 ≥2). The gene set had high predictive capacity (area under the curve [AUC] 0·967), which was superior to that of baseline clinical variables (AUC 0·706) and clinical and pathological variables (AUC 0·806). Furthermore routine pathological variables were unable to identify which histologically normal allografts would progress to fibrosis (AUC 0·754), whereas the predictive gene set accurately discriminated between transplants at high and low risk of progression (AUC 0·916). The 13 genes also accurately predicted early allograft loss (AUC 0·842 at 2 years and 0·844 at 3 years). We validated the predictive value of this gene set in an independent cohort from the GoCAR study (n=45, AUC 0·866) and two independent, publically available expression datasets (n=282, AUC 0·831 and n=24, AUC 0·972).
Interpretation
Our results suggest that this set of 13 genes could be used to identify kidney transplant recipients at risk of allograft loss before the development of irreversible damage, thus allowing therapy to be modified to prevent progression to fibrosis.
Funding
National Institutes of Health.
Introduction
Kidney transplantation is the most common type of solid organ transplant surgery in the USA, with more than 16 900 transplants performed in 2013.1 Despite reduced incidence of acute rejection over the past two decades, improvements in long-term allograft survival have not been realised.2–3
Chronic allograft damage, or interstitial fibrosis and tubular atrophy of unknown cause, is the major cause of allograft loss in the first year after transplantation.4 Clinical and histological events associated with interstitial fibrosis and tubular atrophy are poorly predictive of allograft loss,5 making it difficult to identify allografts that could benefit from early interventions to prevent progression of fibrosis. Allograft biopsies in response to renal dysfunction remain the current approach for the diagnosis of chronic injury, by which stage irreversible fibrosis has developed. Substantial evidence suggests that pathological changes in the renal graft precede functional changes.6 Studies using surveillance or protocol biopsies—ie, biopsies performed at predefined timepoints for surveillance—have shown that about 50% of allografts with stable renal function show evidence of interstitial fibrosis and tubular atrophy by 1 year.7 The development of a predictive assay to identify allografts at risk of chronic damage early after transplantation (ie, within the first 3 months after transplantation) will be essential for the design of targeted therapeutic interventions. We hypothesised that the molecular changes noted in protocol biopsies early after transplantation would reflect the processes that lead to fibrosis and would precede any pathological evidence of fibrosis. By use of gene expression profiling of protocol biopsies obtained at 3 months after transplantation, we aimed to develop a predictive gene set that is able to identify allografts at risk of progressive injury, thereby enabling the identification of recipients at risk of allograft loss at a time when therapeutic intervention could still prevent the development of interstitial fibrosis and tubular atrophy.
Methods
Study design and patients
This part of the Genomics of Chronic Allograft Rejection (GoCAR) study is a prospective, multicentre study done at five clinical sites and central pathological and immunological cores in the USA and Australia to examine the use of differential gene expression to predict the development of chronic allograft injury. Exclusion criteria included a positive T lymphocyte or B lymphocyte complement-dependent cytotoxicity cross match, desensitisation for donor-specific antibodies, paediatric transplant recipients (age <18 years), and inability to give consent. Protocol renal allograft biopsies were obtained at 0, 3, 12, and 24 months after transplantation at all five clinical sites. Patients included in the study were prospectively enrolled from May 12, 2007, to July 30, 2011. The study was approved by the institutional review boards of the participating institutions (appendix p 2). Written informed consent was obtained from all enrolled patients. Protocol biopsies at 3 months after transplantation were collected from 204 patients. Microarray analysis was done on the first 159 biopsies (discovery set), and the remaining 45 biopsies were used for validation.
Histopathological classification
Two biopsy cores were taken at each protocol biopsy, with one used for histology and the other used for mRNA analysis. Biopsies were processed and evaluated centrally blinded to local reports if these were available and scored separately by two of three renal pathologists who alternated in serving as the first and second lines of scoring (RC, RNS, and IAR) in accordance with the Chronic Allograft Damage Index (CADI)6 and Revised Banff 2009 Classification,8 which are measures of allograft damage. If diagnoses were discordant between pathologists, a third pathologist (RC, RNS, or IAR) provided a consensus diagnosis. Whole slide images prepared from formalin-fixed paraffin-embedded sections were scanned, analysed by the pathologists, and entered into a customised database that calculated the Banff categories and CADI scores6 (appendix p 3). We calculated CADI scores for biopsies obtained at 3 months (CADI-3) and 12 months (CADI-12) after transplantation. High CADI-12 scores, which indicate chronic allograft damage, were defined as scores of 2 or more, which we based on the previous association of such scores with adverse allograft outcomes.5,6,9
Microarray, data analysis, and cross-validation
Details of the microarray analysis are described in the appendix (pp 3–9). Briefly, we extracted total RNA from fresh frozen biopsies and did the microarray analysis with the Affymetrix human exon 1.0 ST array (Affymetrix, Santa Clara, CA, USA). We identified correlations between gene expression in 3-month biopsies and CADI-3 or CADI-12 scores by use of Spearman’s correlation analysis and the genes associated with CADI-3 or CADI-12 were subsequently subjected to enrichment analysis based on Gene Ontology Biological Process (BP) term and immune cell type.
To identify a minimum gene set to predict future kidney fibrosis, as represented by a high CADI-12 score, we identified genes that were specifically correlated with CADI-12 scores, then did a 100 time random shuffling analysis with adjustment for confounding clinical parameters. We identified the gene set with the best area under the curve (AUC) for the prediction of high and low CADI-12 scores. We then applied this optimum gene set to predict which patients have CADI-3 of 3 or less and increase of CADI-12 scores by at least 2 points (CADI progression) and early allograft loss (within 2 years or 3 years). We also investigated the prediction of CADI-12 with a threshold of 3 or 4, the contribution of inflammation to prediction of high CADI-12 by the gene set, and lastly, we assessed the prediction of the kidney function by calculating the AUC for estimated glomerular filtration rate (eGFR) at 12 and 24 months with the gene set.
We independently validated the gene set with quantitative PCR (qPCR) data for the remaining biopsies from the GoCAR study (n=45) for prediction of kidney fibrosis at 12 months and in two external datasets. These datasets consisted of 282 samples from a study by Einecke and colleagues investigating graft loss prediction (GSE21374)10 and 24 samples from a study by Naesens and colleagues investigating progression prediction based on CADI at 24 months (GSE25902).11 Microarray data are available from the Gene Expression Omnibus (GSE57387).
Clinical data and statistical analysis
In accordance with the protocol, we collected clinical and laboratory data about the kidney transplant donors and recipients (appendix p 2) at 0, 3, 12, and 24 months post-transplantation, and we summarised these data with descriptive statistics. We used GraphPad Prism 5.03 to plot survival curves for the duration of the study, with allograft loss as the outcome. Graft losses were entered by the study sites into the electronic research database (eRAP). To establish predictive clinical factors for high CADI-12 scores (≥2) and CADI progression or non-progression outcomes, we used multiple logistic regression models that included donor age, recipient race and sex, donor vital status, expanded-criteria donor (ECD) status, cold ischaemia time (h), induction therapy, presence of human leucocyte antigen (HLA) antibodies, eGFR at 3 months post-transplantation, acute cellular rejection at or before 3 months, delayed graft function, HLA mismatch, and CADI-3 score. We used SAS version 9.2 to build logistic models with these predictors to assess factors predictive for the development of high CADI-12 scores. The GoCAR study is registered with ClinicalTrials.gov, number NCT00611702.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. All the authors had full access to the data and made the decision to submit the manuscript for publication.
Results
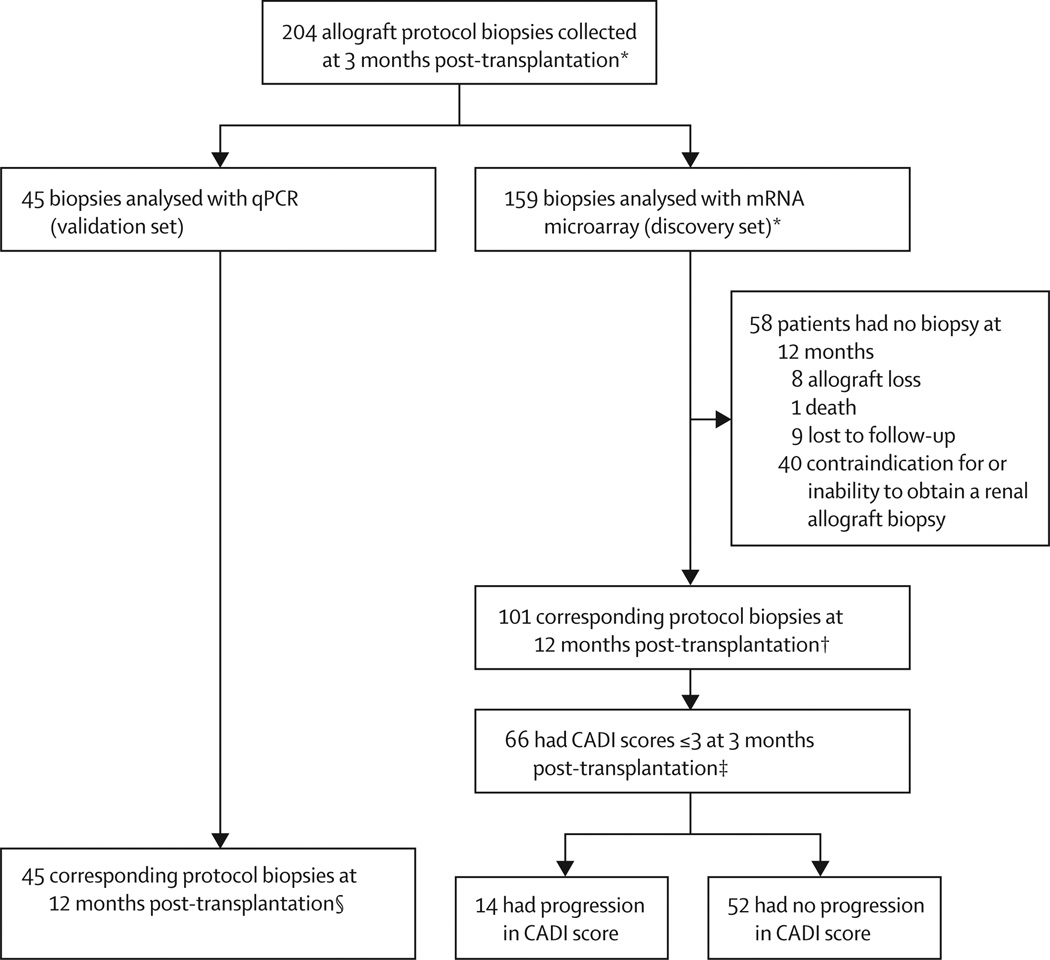
We included 204 patients from the GoCAR cohort who had a protocol allograft biopsy at 3 months post-transplantation in the current study (figure 1). Microarray analysis was done on the first 159 samples collected (discovery set), whereas qPCR experiments were done on the remaining 45 biopsies as a validation set. All 45 patients included in the validation set and 101 patients from the discovery set had a corresponding protocol biopsy at 12 months. Reasons for the absence of a 12 month biopsy included allograft loss (n=8), death (n=1), loss to follow-up (n=9), and contraindication or inability to obtain biopsy (n=40). Demographics and clinical variables of the 159 patients included in the microarray analysis were similar to those of the 101 patients with biopsies at 12 months (table 1) and the 45 patients in the validation set (appendix p 30). 57 (57%) biopsies collected at 12 months had a CADI-12 score of 0–1, 30 (30%) had a score of 2–4, and 14 (14%) had scores more than 4. CADI-12 scores of 2 or more were associated with reduced 3-year allograft survival compared with scores less than 2 (log rank p=0·0187; appendix p 16).
Figure 1. Patient eligibility.
*Expression microarray was done on RNA extracted from the first 159 patients based on date of enrolment. †The 101 corresponding protocol biopsies at 12 months post-transplantion were used to identify the optimal gene set based on CADI score at 12 months. ‡These patients were included in analysis of progression versus non-progression of CADI scores, where progression was defined as an increase in CADI score of at least 2 points. §This cohort was used for independent qPCR validation of the 13 gene set. CADI=Chronic Allograft Damage Index. qPCR=quantitative PCR.
Table 1.
Demographic and clinical characteristics of comparison groups
| All patients vs patients with biopsy at 12 months |
High vs low CADI-12 scores | Progression of CADI scores vs wno progression |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Patients included in microarray analysis (n=159) |
Patients with biopsy at 12 months (n=101) |
p value |
Patients with high CADI-12 scores (n=44) |
Patients with low CADI-12 scores (n=57) |
p value | Progression by 12 months (n=14) |
No progression by 12 months (n=52) |
p value |
|
| Donor age (years) | 41·13 (16·80) | 40·73 (16·80) | 0·87 | 45·45 (18·05) | 37·07 (14·91) | 0·0121 | 46·50 (18·85) | 37·69 (17·51) | 0·1 |
| Donor sex female | 77 (48%) | 46 (46%) | 0·7 | 23 (52%) | 28 (49%) | 0·84 | 6 (43%) | 28 (54) | 0·76 |
| Donor race | 0·64 | 1 | 0·72 | ||||||
| Caucasian | 121 (76%) | 80 (79%) | 35 (80%) | 46 (81%) | 11 (79%) | 43 (83%) | |||
| African American | 17 (11%) | 8 (8%) | 4 (9%) | 3 (5%) | 3 (21%) | 2 (4%) | |||
| Hispanic | 12 (8%) | 5 (5%) | 1 (2%) | 4 (7%) | 0 | 3 (6%) | |||
| Other/unreported | 9 (6%) | 8 (8%) | 4 (9%) | 4 (7%) | 0 | 4 (8%) | |||
| Donor vital status | 0·79 | 0·31 | 0·23 | ||||||
| Deceased | 94 (59%) | 58 (57%) | 28 (63%) | 30 (53%) | 10 (71%) | 28 (54%) | |||
| Alive | 65 (41%) | 43 (43%) | 16 (7%) | 27 (43%) | 4 (29%) | 4 (8%) | |||
| Recipient age (years) | 48·84 (13·27) | 46·90 (12·38) | 0·24 | 49·86 (11·23) | 44·43 (12·80) | 0·0281 | 49·92 (8·69) | 46·90 (12·29) | 0·39 |
| Recipient sex female | 45 (28%) | 32 (32%) | 0·57 | 17 (39%) | 15 (26%) | 0·18 | 6 (43%) | 17 (33%) | 0·53 |
| Recipient race | 0·33 | 0·45 | 0·48 | ||||||
| White | 92 (58%) | 66 (65%) | 26 (59%) | 40 (70%) | 8 (57%) | 17 (73%) | |||
| African American | 37 (23%) | 15 (15%) | 7 (16%) | 8 (14%) | 3 (21%) | 6 (12%) | |||
| Hispanic | 14 (9%) | 7 (7%) | 4 (9%) | 3 (5%) | 0 (0%) | 2 (4%) | |||
| Other/unreported | 16 (10%) | 13 (13%) | 7 (16%) | 6 (11%) | 3 (21%) | 6 (12%) | |||
| Recipient end stage renal disease diagnosis |
0·52 | 0·84 | 0·9 | ||||||
| Diabetic nephropathy | 57 (36%) | 33 (33%) | 13 (30%) | 20 (35%) | 3 (21%) | 17 (33%) | |||
| Hypertension | 24 (15%) | 17 (17%) | 7 (16%) | 10 (18%) | 3 (21%) | 7 (13%) | |||
| Glomerulonephritis | 28 (18%) | 22 (22%) | 9 (20%) | 13 (23%) | 3 (21%) | 12 (23%) | |||
| Polycystic kidney | 14 (9%) | 13 (13%) | 6 (14%) | 7 (12%) | 2 (14%) | 7 (13%) | |||
| Other | 36 (23%) | 16 (16%) | 9 (20%) | 7 (12%) | 3 (21%) | 9 (17%) | |||
| Previous renal transplant | 4 (3%) | 2 (2%) | 1 | 1 (2%) | 1 (2%) | 1 | 1 (7%) | 1 (2%) | 0·38 |
| eGFR at 3 months | 59·18 (18·25) | 59·48 (18·11) | 0·9 | 53·23 (16·87) | 64·36 (17·68) | 0·0029 | 47·68 (15·59) | 61·69 (17·33) | 0·008 |
| CADI score at 3 months | |||||||||
| Mean (SD) | 1·78 (1·67) | 1·74 (1·65) | 0·89 | 2·45 (1·80) | 1·20 (1·30) | 0·0005 | 1·36 (1·28) | 1·07 (1·06) | 0·45 |
| Median (IQR) | 1 (0–3) | 1·5 (0–3) | 2 (1–3) | 1 (0–2) | 1·5 (0–3) | 1 (1–3) | |||
| Cold ischaemia time (h)* | 14·64 (6·78) | 13·81 (6·74) | 0·46 | 13·99 (7·26) | 12·64 (5·39) | 0·45 | 13·62 (6·72) | 13·78 (7·37) | 0·95 |
| Delayed graft function* | 21 (13%) | 9 (9%) | 0·32 | 7 (25%) | 2 (7%) | 0·12 | 2 (20%) | 4 (14%) | 0·64 |
| Anti-HLA antibodies† | 36 (23%) | 26 (27%) | 0·53 | 12 (29%) | 14 (25%) | 0·75 | 6 (17%) | 9 (17%) | 0·33 |
| Donor-specific antibody positive |
7 (4%) | 4 (4%) | 3 (7%) | 1 (2%) | 2 (14%) | 4 (8%) | |||
| Class I | 5 (3%) | 3 (3%) | 2 (5%) | 1 (2%) | 2 (14%) | 4 (8%) | |||
| Class II | 3 (2%) | 1 (1%) | 0 (0%) | 1 (2%) | 1 (7%) | 1 (2%) | |||
| Non-donor-specific antibody positive |
29 (20%) | 22 (23%) | 9 (21%) | 13 (24%) | 4 (29%) | 5 (10%) | |||
| Class I | 29 (20%) | 22 (23%) | 9 (21%) | 13 (24%) | 4 (22%) | 5 (10%) | |||
| Class II | 16 (11%) | 9 (9%) | 3 (7%) | 6 (11%) | 1 (7%) | 3 (6%) | |||
| Induction therapy | 0·67 | 0·5 | 0·75 | ||||||
| Thymoglobulin | 49 (31%) | 27 (27%) | 13 (30%) | 14 (25%) | 4 (29%) | 13 (25%) | |||
| Anti-CD25 therapy | 52 (33%) | 39 (39%) | 19 (43%) | 20 (35%) | 6 (43%) | 23 (44%) | |||
| Anti-CD52 therapy | 9 (6%) | 4 (4%) | 2 (5%) | 2 (4%) | 0 | 1 (2%) | |||
| None | 49 (31%) | 31 (31%) | 10 (23%) | 21 (37%) | 4 (29%) | 15 (29%) | |||
| 12-month maintenance immunosuppression |
072 | 0·84 | 0·52 | ||||||
| Mycophenolate mofetil, calcineurin inhibitors, steroids |
139 (87%) | 90 (89%) | 39 (89%) | 51 (89%) | 13 (93%) | 50 (96%) | |||
| Mycophenolate mofetil, calcineurin inhibitors |
12 (8%) | 8 (8%) | 2 (5%) | 6 (11%) | 1 (7%) | 1 (2%) | |||
| Others | 8 (5%) | 3 (3%) | 3 (7%) | 0 | 0 | 1 (2%) | |||
| Acute rejection within 1 year |
22 (14%) | 22 (22%) | NA | 17 (39%) | 5 (9%) | 0·0005 | 5 (36%) | 6 (12%) | 0·36 |
Data are mean (%) or mean (SD) unless noted otherwise. p values are calculated with the Mann-Whitney test (for non-parametric comparisons) or unpaired t test for continuous variables and χ2 or Fisher’s exact test for categorical data. CADI-12 scores of 2 or more were deemed to be high and scores of less than 2 were deemed to be low. Progression was shown by a CADI-3 score of 3 or less and an increase of CADI score of at least 2 points. Delayed graft function was defined as need for dialysis within the first 7 days after transplantation. CADI-12=chronic allograft damage index at 12 months. eGFR=estimated glomerular filtration rate. HLA=human leucocyte antigen.
Deceased donors only.
Only 148 of 159 patients included in microarray analysis and 97 of 101 patients with biopsies at 12 months had baseline anti-HLA antibodies reported in the database: 42 patients with high CADI and 55 with low CADI had anti HLA antibodies reported; all 14 progressors and 52 non-progressors had anti-HLA antibodies reported.
The transcriptome obtained from the discovery set was analysed to obtain an optimal gene set predictive of CADI-12 score. Initially, we applied Spearman correlation analysis to identify significantly associated genes, pathways, and corresponding functions associated with high CADI-3 and CADI-12 scores. We noted that the transcripts specifically associated with CADI-3 were related to alloimmunity, including T-cell activation, whereas genes involved in programmed cell death or apoptosis and cell adhesion were only associated with CADI-12. Furthermore, immune cell gene enrichment analysis revealed that dendritic cell genes are specifically associated with CADI-3, however stromal cell (mostly fibroblast cell) genes are the most over-represented genes significantly associated with CADI-12 according to Fisher’s exact test, in addition to genes from macrophage, dendritic, and CD4-positive T cells (appendix pp 11, 17–23). To identify an optimal predictive gene set, we filtered genes significantly associated with CADI-3 or CADI-12 scores via a two-group re-sampling approach to derive a smaller set of 149 genes that correlated specifically with CADI-12 scores, but not CADI-3 scores (appendix p 5). After excluding genes with low expression (log2 of the intensity <5) and adjustment for clinical parameters, we further reduced the gene set to 84 genes (appendix pp 31–33). Through iterative application of penalised logistic regression fitting on expression data for these 84 genes (appendix p 6), we identified an optimal set of 13 genes from 3-month biopsies that was able to differentiate high CADI-12 scores from low CADI-12 scores with an AUC of 0·967 (table 2, appendix p 24). To avoid overfitting of the prediction model on the training set from which the gene set was derived, the 101 patient cohort was randomly divided into thirds and assigned into training and test sets (threefold cross-validation), and the performance of the selected gene set in these random subsets was evaluated. This process was repeated 100 times and these test sets were calculated to have an average sensitivity of 81% and specificity of 79%, with an average cross-validated AUC of 0·889 (95% CI 0·886–0·897; appendix pp 6–7, 24). To assess whether the gene set that we identified was an optimal gene set for the prediction of high or low CADI-12 scores, we compared the original prediction AUC with prediction AUCs from the gene sets that were identified from high and low CADI-12 score groups, with random reassignment of CADI scores to the patients; the original AUC was higher than any AUC from 2000 permutations (p=0·0015; appendix pp 7, 24). Finally, we did a complete leave-one-out cross-validation to validate our approach, which involved gene reselection and model building based on the original training set with one patient being left out and validation on the left-out patient. Most of the new gene sets identified from this cross-validation overlapped with the original set of 13 genes, and the prediction AUC of the probabilities of all possible testing sets was 0·774, confirming that the originally selected gene set was the optimal set for the prediction of CADI-12 scores (appendix pp 7, 24).
Table 2.
Genes included in the prediction set by array probeID
| Symbol | Gene description | Cytoband | mRNA accession | CADI-12 correlation |
p value | |
|---|---|---|---|---|---|---|
| 3954887 | CHCHD10 | Coiled-coil-helix-coiled-coil-helix domain containing 10 | 22q11.23 | NM_213720 | 0·404 | 2·85 × 10−5 |
| 4019160 | KLHL13 | Kelch-like family member 13 (Drosophila) | Xq23-q24 | NM_001168302 | 0·369 | 1·49 × 10−4 |
| 3326826 | FJX1 | Four jointed box 1 (Drosophila) | 11p13 | NM_014344 | 0·367 | 1·60 × 10−4 |
| 3020343 | MET | Met proto-oncogene (hepatocyte growth factor receptor) | 7q31 | NM_001127500 | 0·352 | 3·01 × 10−4 |
| 2864449 | SERINC5 | Serine incorporator 5 | 5q14.1 | NM_001174072 | 0·318 | 0·0012 |
| 2567583 | RNF149 | Ring finger protein 149 | 2q11.2 | NM_173647 | 0·280 | 0·0046 |
| 2879105 | SPRY4 | Sprouty homolog 4 (Drosophila) | 5q31.3 | NM_030964 | 0·270 | 0·0062 |
| 3776504 | TGIF1 | TGFB-induced factor homeobox 1 | 18p11.3 | NM_170695 | 0·244 | 0·0140 |
| 2898441 | KAAG1 | Kidney associated antigen 1 | 6p22.1 | NM_181337 | 0·240 | 0·0154 |
| 3361971 | ST5 | Suppression of tumorigenicity 5 | 11p15 | NM_005418 | 0·232 | 0·0197 |
| 2459352 | WNT9A | Wingless-type MMTV integration site family member 9A | 1q42 | NM_003395 | 0·212 | 0·0332 |
| 3021696 | ASB15 | Ankyrin repeat and SOCS box-containing 15 | 7q31.31 | NM_080928 | −0·263 | 0·0079 |
| 3193339 | RXRA | Retinoid X receptor alpha | 9q34.3 | NM_002957 | −0·300 | 0·0023 |
CADI-12=chronic allograft damage index at 12 months.
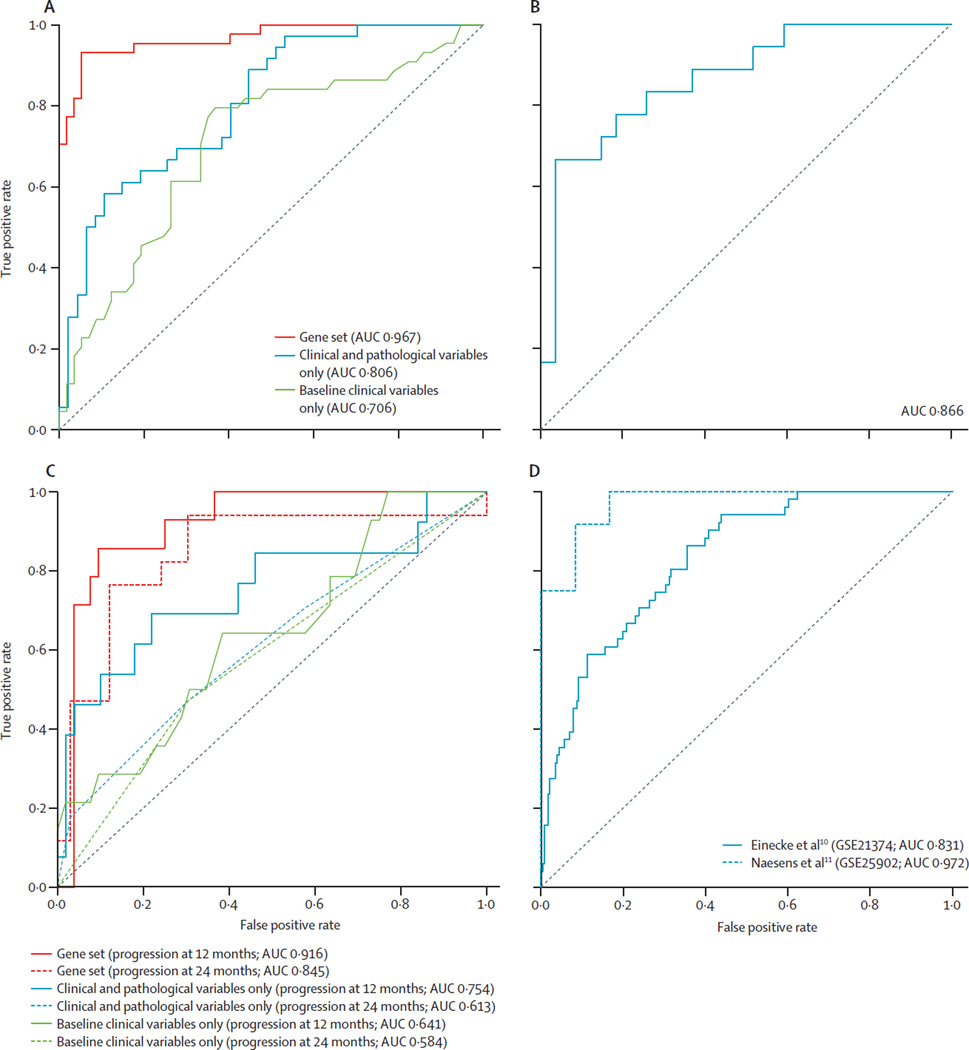
The gene set also performed well when higher cutoffs to define a high CADI-12 score were used, with an AUC of 0·934 for a CADI-12 cutoff of 3 points or more and an AUC of 0·928 for a CADI-12 cutoff of 4 points or more (appendix p 25), which supported the robustness of our gene set. When validated with the qPCR data from the independent GoCAR cohort (n=45), which had similar demographics to the training set (appendix p 30), the gene set accurately differentiated between patients with high and low CADI-12 scores (18 patients classified as having high CADI-12, 27 patients classified as having low CADI-12, AUC 0·866; figure 2B).
Figure 2. Prediction of high and low CADI scores and progression or non-progression at 12 months post-transplantation.
(A) Prediction of high or low CADI-12 scores with the 13-gene set and clinical and pathological variables. (B) Internal validation of the ability of the set of 13 genes to predict high or low CADI-12 was done with qPCR of biopsies collected at 3 months post-transplantation in an independent cohort of 45 patients within the GoCAR study. (C) Prediction of fibrosis progression versus non-progression at 12 and 24 months with the 13-gene set and clinical and pathological variables. (D) ROC curves for external validation of the 13-gene set in two publically available biopsy microarray datasets.10,11 CADI-12 scores of 2 or more were deemed high and scores of less than 2 were deemed low. Progression was shown with CADI-3 of 3 or less and an increase in CADI-12 score of at least 2 points. AUC=area under the curve. CADI-12=Chronic Allograft Damage Index at 12 months. ROC=receiver operating characteristic.
To examine the contribution of allograft inflammation (represented by the i score in CADI) in both the identification of the gene set and the prediction of high CADI-12 scores, we tested the gene set against the sum of Banff chronic interstitial fibrosis (Ci) and chronic tubular atrophy (Ct) scores obtained at 12 months. The gene set accurately predicted fibrosis based on Banff score, with an AUC of 0·922 for the sum of 12-month Ci and Ct scores (appendix p 26) and an AUC of 0·923 for interstitial fibrosis and tubular atrophy by diagnosis (interstitial fibrosis and tubular atrophy >1; appendix p 26). Subclinical rejection was present in 20 patients in the discovery set and was associated with high CADI-12 scores (p=0·0003) and high Banff scores (Ci plus Ct; p=0·002) at 12 months. However, exclusion of these patients with rejection or Banff interstitial inflammation (i) plus tubulitis (t) scores of more than two did not alter the ability of the gene set to predict high CADI-12 scores (AUC 0·975; appendix p 26). These data suggest that inflammation was not the predominant driver for the derivation of the gene set.
We also investigated whether the 13-gene set was predictive of kidney function at 12 or 24 months. We measured the creatinine concentrations of transplant recipients at each biopsy timepoint and we calculated eGFR based on the Modification of Diet in Renal Disease (MDRD) Study equation.12 As expected, eGFR was negatively correlated with CADI-12 (Pearson’s r correlation of –0·38, p=0·0001; appendix p 27). We analysed whether the set of 13 genes could predict high and low eGFR at 12 and 24 months in all 159 patients in the discovery cohort. Our gene set predicted the occurrence of low or high eGFR at 12 months with an AUC of 0·872 (appendix p 27), and predicted high or low eGFR at 24 months with an AUC of 0·928 at an eGFR cutoff of 30 mL per min (appendix p 27).
To study the clinical variables associated with high CADI-12 scores, we did multivariate analyses in the discovery set including clinical and pathological characteristics. The ability of our set of 13 genes to predict CADI-12 scores was superior to that of baseline clinical variables alone (donor age, recipient race and sex, donor vital status, ECD status, cold ischaemia time, induction therapy, presence of anti-HLA antibodies, delayed graft function, and HLA mismatch; AUC 0·706; figure 2A). Furthermore, when baseline and clinical variables (the variables listed for the previous analysis and eGFR at 3 months) were combined with pathological variables at 3 months (acute cellular rejection at or before 3 months and CADI-3 score), the gene set was still superior (AUC 0·806 for clinical and pathological variables vs AUC 0·967 for the gene set; figure 2A). In multivariate analysis with a logistic regression model incorporating clinical and pathological variables alone or combined with the gene set, the gene set remained significantly associated with high CADI-12 scores whereas the clinical parameters did not (appendix p 34).
Predicting the pathological course of allografts that have little or no fibrosis is clinically very difficult. We investigated whether the gene set could be used to categorise patients in the discovery set with minimal or no fibrosis into those who would or would not develop progressive fibrosis by 12 months. From the original 101 patients with protocol biopsies at 12 months, we identified 66 patients with CADI-3 scores of 3 or less, which indicated minimal or no fibrosis. We separated these patients into those with a change from CADI-3 to CADI-12 of less than 2 points (non-progression; n=52) versus those who had an increase of 2 points or more (progression; n=14). Most patients with progression at 12 months had further progression at 24 months, and most patients who had not had progression at 12 months did not have progression by 24 months (table 1; appendix p 28). Comparisons of biopsy pathology CADI subscores at 2 months and 12 months are shown in the appendix (p 35) for patients who had progression versus those without progression. Compared with patients without progression, those who had progression had no significant differences in CADI-3 subscores. CADI-3 scores did not predict which patients would have progression or not by 12 months. Clinical and pathological parameters were poor predictors of progression by 12 months or 24 months (baseline clinical variables: AUC at 12 months 0·641; AUC at 24 months 0·584; clinical plus pathological variables: AUC at 12 months 0·754; AUC at 24 months 0·613; figure 2C). The gene set accurately predicted which patients would have progression in CADI scores and those who would not at both 12 months (AUC 0·916) and 24 months (AUC 0·845; figure 2C). Multivariate analysis showed that the gene set was significantly associated with progression at 12 months, whereas most demographic, clinical, and pathological parameters were not, except for eGFR at 3 months (appendix p 36). The gene set, but not demographic, clinical, and pathological variables, was associated with progression at 24 months (appendix p 37). Furthermore, the gene set was also able to predict CADI progression by 12 months and 24 months when the cutoff for CADI-3 was lowered to 2 points or less (AUC at 12 months 1·00 and AUC at 24 months 0·834; appendix p 28). These findings are of clinical relevance because patients with progression in CADI scores had reduced allograft survival compared with those without progression at 36 months (p=0·0369; appendix p 29).
15 patients who had improvements in CADI scores between 3 and 12 months were excluded from our initial progression versus non-progression analysis. We retrospectively applied the progression versus non-progression model to this set of patients, and 13 of these 15 patients with apparent improvements were accurately predicted to not have progression. Conversely, two of these patients were predicted by our gene set to have progression, despite the improvements in CADI-12 score. On review, these two patients had high CADI scores at 3 months and 12 months (one patient had a CADI-3 score of 7 and a CADI-12 score of 4 and the other had a CADI-3 score of 7 and a CADI-12 score of 3), suggesting that these two patients, irrespective of the changes in their CADI scores, behaved biologically as progressors, as predicted by the gene set.
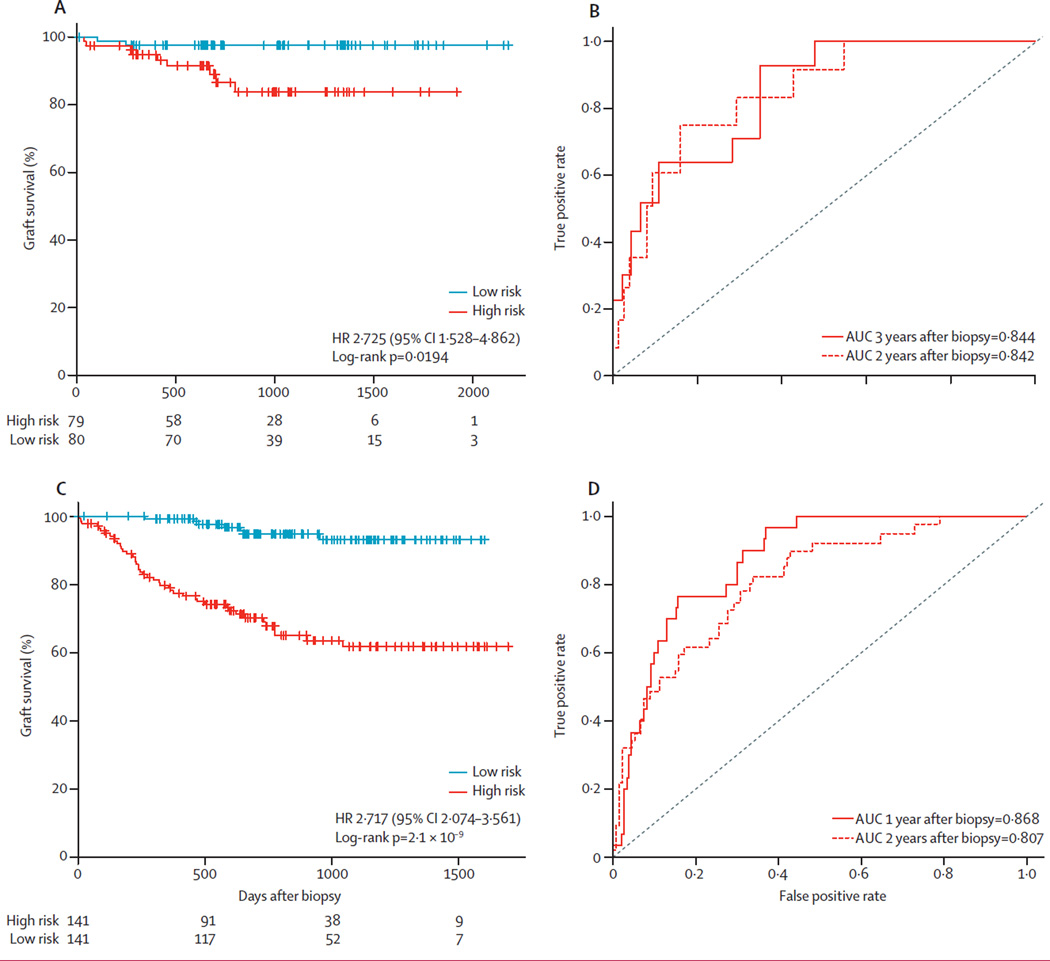
We next generated Cox models with our gene set and clinical variables to predict death-censored graft loss in the total discovery set of 159 patients (11 cases of allograft loss). We did principle component analysis (PCA) on the expression data of the 13 genes in the set, and two principle components (P4, P6) were significantly associated with allograft loss (p=0·0032 for P4 and p=0·0120 for P6; overall p=0·0287; appendix p 38). We used these two principle components to derive the gene set risk score (appendix p 8), and patients were stratified into two groups based on this gene set risk score: high risk and low risk. We found that a higher score was associated with significantly greater risk of allograft loss (hazard ratio for graft loss 2·725, 95% CI 1·528–4·862; log-rank p=0·0194; figure 3A). With prediction based on the gene set risk score, the AUCs for time-dependent allograft loss were 0·844 by 2 years and 0·842 by 3 years post-biopsy (figure 3B). Demographic and clinical variables were not significantly associated with allograft loss (overall p=0·4010; appendix p 39).
Figure 3. Survival analysis of time to allograft loss.
(A) Kaplan-Meier plot of time to allograft loss for patients stratified into high-risk and low-risk groups according to the gene set risk score, which was calculated by the linear combination of eigenvalues of significant principle components multiplied by their coefficiencies in a Cox proportional hazard model. Hazard ratio of graft loss was estimated from the coefficiency of the gene risk score in the Cox proportional model. (B) ROC curves for prediction of allograft loss within 2 years or 3 years after the 3-month biopsy. (C) Kaplan-Meier plot of time to allograft loss for patients from a publically available dataset (GSE21374)10 who were stratified into high-risk and low-risk groups according to the gene set risk score. (D) ROC curves for prediction of time to allograft loss by 1 year and 2 years post-biopsy through application of the gene set risk score to the publically available dataset.10 AUC=area under the curve. ROC=receiver operating characteristic. HR=hazard ratio.
To confirm the usefulness of the 13-gene set in other populations of patients, we interrogated two independent, publically available datasets in which the endpoints were graft loss in Einecke and colleagues’ study10 (GSE21374; 282 samples) and CADI score at 24 months in Naesens and colleagues’ study11 (GSE25902; 24 samples; appendix p 40). Our gene set accurately predicted the respective endpoints for each of the datasets (figure 2D), performing favourably when compared with the original studies (0·831 vs 0.83 [using a 30-gene set by Einecke and colleagues10]; 0·974 vs 0·82 to 0·926 [using various immune response genesets by Naesens and colleagues11], respectively. Furthermore, our gene set is substantially smaller and able to predict the fibrosis at an early time post-transplant. Survival analysis of Einecke and colleagues’ cohort10 stratified with our gene set into high-risk and low-risk groups showed significant differences between these risk groups with respect to graft survival (hazard ratio of graft loss 2·717, 95% CI 2·074–3·561 p=2·1 × 10−9; figure 3C); In this dataset, the AUCs were 0·865 for allograft loss within 1 year and 0·807 for graft loss within 2 years after biopsy (figure 3D).
Discussion
We have identified a set of 13 genes from biopsies of stably functioning renal allografts that predicts the development and progression of chronic allograft damage and subsequent allograft loss. Our results show that this molecular gene risk profile has superior predictive ability to the clinicopathological variables currently used in practice for the prediction of these outcomes.
Many renal allografts show early and rapid histological deterioration by 12 months after transplantation.4 Fibrosis at 12 months has been correlated with adverse long-term allograft outcomes in all but the highest risk recipients.13–15 The pathological findings from our study lend further support to evidence showing that an ongoing cycle of subclinical inflammation and injury leads to fibrosis, loss of function, and organ failure.16 Data from our discovery cohort and previous studies have shown that CADI-12 scores of 2 or more are associated with increased risk of allograft loss.6,17 Interventions started after chronic damage has already become established are unlikely to alter outcomes.4 The clinicopathological variables currently used to identify allografts at risk of histological deterioration perform poorly. For example, although 60% of our cohort had low CADI-3 scores (0–1), more than half of the patients with progression in CADI scores at 12 months belonged to this group, and were not identified as being at risk of deterioration by histology alone at 3 months. The identification of early markers to detect the initiation of pro-fibrotic pathways of inflammation and injury at the molecular level within the graft would offer the potential to interrupt the process.
As has been shown previously, the causes of chronic kidney allograft injury are diverse and cumulative.4,18 This diversity is reflected in the large number of genes that were associated with adverse allograft outcomes in the two publically available validation cohorts. Einecke and colleagues10 identified a tissue gene signature (886 genes related to tissue injury and the effects of TGF-β) that was predictive of allograft loss in clinically indicated allograft biopsies obtained between 1 and 31 years after transplantation. A 601-probe set signature identified from protocol biopsies collected at 6 months from low-risk paediatric patients receiving kidney transplants showed upregulation of genes for immune response in patients who had histological progression compared with those who did not.11 Our set of 13 genes was validated in these cohorts, showing high predictive value in the prediction of allograft outcomes, despite differences in demographics, timing of biopsies after transplantation, presence of pre-existing fibrosis, and endpoints (appendix p 40). Additionally, the previous study11 used limited gene sets based on previous data or predicted pathological pathways. By contrast, we took an all-inclusive, non-hypothesis-driven approach that was enabled by our sample size and based on the multifaceted nature of chronic injury. Since the CADI-12 score is a continuous variable, we derived the list of genes on the basis of indications of correlation, identifying genes with higher expression at 3 months that were correlated with higher CADI-12 scores, rather than comparing differential gene expression between two well-defined clinical cohorts, as in previous studies. This approach avoids the use of cutoffs predetermined for experimental purposes and hence more accurately represents the continuous nature of pathological processes seen in clinical settings, enhancing the usefulness of the gene set in clinical cohorts.
Despite the focus of our approach on prediction, we identified functional implications of the differentially expressed transcripts, including pathogenic and protective mechanisms that merit further investigation. Genes involved in cell growth and tumour development or suppression were significantly over-represented, including MET (MET proto-oncogene), ST5 (suppression of tumorigenicity 5) and KAAG1. Also over-represented were genes that are involved in ubiquitination either as E3 ligases or as interacting proteins, including RNF149 (ring finger protein 149), ASB15 (ankyrin repeat and SOCS box containing 15), KLH13 (Kelch-like family member 13); genes involved in significant developmental or growth pathways such as in the NOTCH/Wnt pathway and the RAR pathway through SMAD, including the genes TGIF1 (TGFB-induced factor homeobox 1), SPRY4 (sprouty homolog 4), WNT9A (Wnt family member 9A), RXRA (retinoid X receptor alpha), and FJX1 (four jointed box 1); and genes involved in energy and membrane repair, such as the mitochondrial gene CHCHD10 (coiled-coil-helix-coiled-coil-helix domain containing 10) and SERINC5 (serine incorporator 5), which phosphorylates membrane proteins. The representation of genes involved in embryonic growth and malignancy, gene regulation through ubiquitination, developmental signalling pathways, and genes that regulate membrane formation and mitochondrial function suggests representation of active repair and regeneration pathways, as might be expected for markers of injury and fibrosis. For example, c-MET and TGIF proteins have been reported to mediate the anti-fibrotic effects of hepatocyte growth factor (HGF),19 whereas SPRY4 and ST5 proteins have been found to exert a range of regulatory effects on ERK/MAPK activation.20–22 The SPRY4 gene encodes a member of a family of cysteine-rich and proline-rich proteins, and might have a similar function in organ fibrosis to that of SPRY1.23 SPRY4 protein is an inhibitor of the receptor-transduced MAPK signalling pathway. Inhibition of the p38 MAPK24 and MAPK/ERK25 signalling pathways and treatment with recombinant HGF26 have shown benefit in experimental chronic allograft damage, showing that these pathways could be potential early therapeutic targets. By comparing gene expression data from stromal cells (mostly fibroblast cells) with that from other immune cell types (eg, macrophages, dendritic cells, monocytes, T cells, and B cells), we found that most of the 13 genes in our panel, including KLHL13, MET, SPRY4, SERINC5, FJX1, ST5, and RXRA are highly expressed in fibroblast cells (appendix p 22), suggesting that dysregulation of fibrotic genes at 3 months after transplantation are associated with the development of kidney fibrosis.
Contrary to the widely held belief that scar tissue is permanent, growing evidence suggests that it is in fact an actively remodelled tissue that, under certain circumstances, can regress.16 The development of a predictive indicator to identify those at risk early after transplantation allows for the possibility to slow, arrest, or even reverse the progression of tissue fibrogenesis. Furthermore, the identification of a gene set that is predictive of progressive fibrosis and declining renal function has the potential to guide immunosuppression therapy and stratify allografts for risk. At the very least, early identification of patients with progressive fibrosis would allow a review of their immunosuppression therapy and concomitant medications such as angiotensin-converting-enzyme inhibitors. Maximising use of anti-proliferative drugs such as mycophenolate mofetil where additional immunosuppresion is needed or switching to an mTOR inhibitor or belatacept where avoidance of calcineurin inhibitors is safe and appropriate are two possible options. Stratification of risk with the gene signature could also be used as an enrichment strategy to identify patients for inclusion in an inter ventional clinical trial.
Despite our validation of the predictive gene set, there are limitations to the study. The cohort size was limited by the stringent requirements for inclusion. Additionally, roughly 20% of participants who underwent a biopsy at 3 months did not have a biopsy at 12 months. Although this loss of patients might have biased the development of the gene set and the study outcomes, we showed that the demographics of the original cohort of 159 patients did not differ from those of the 101 patients who had a 12-month biopsy. The patients in the study all received calcineurin inhibitors and anti-proliferative agents such as mycophenolate mofetil or azathioprine. The validity of this gene set has not been established in the context of other immunosuppressive regimens. Finally, although our gene set obtained at 3 months is highly predictive of adverse allograft outcomes, histological progression from delayed causes of allograft injury such as late-onset antibody-mediated rejection and recurrent glomerulonephritis might not be captured by our gene set.
We used a novel, non-biased approach to identify a gene signature that is able to differentiate allografts at risk of early histological progression, and has further applications in the identification of kidneys at risk of long-term allograft injury and allograft failure. Although further studies are needed, the ability to identify patients at risk of allograft loss has important clinical and therapeutic applications in an area where progress has so far been limited.
Supplementary Material
Research in context.
Evidence before this study
We have previously systematically followed up and reviewed the scientific literature related to the use and application of transcriptional genomic information to chronic kidney allograft injury. This literature has substantially expanded within the duration of our GoCAR study (2007–13) and the preparation of this manuscript. No large prospective transcriptional allograft biopsy datasets such as we report here have been published between 2013 and the writing of this manuscript. So far, the application of gene expression analysis to allograft transcriptional data to predict subsequent adverse outcomes has had limited generalisability. Previous studies have involved either retrospective cohorts of clinically indicated biopsies obtained at widely varying timepoints, or very early protocol biopsies (ie, at 6 weeks) where gene expression profiles might have been confounded by early ischaemia and reperfusion injury signals or small sample sizes of highly selected cohorts such as paediatric patients receiving kidney allografts.
Added value of this study
In this study, we prospectively enrolled adult kidney transplant recipients from five clinical centres and collected protocol biopsies at 3 months to minimise early injury signals after transplantation. We then took a non-hypothesis-driven, inclusive approach to identify a gene signature that was significantly correlated with the development of subsequent histological and functional decline. We identified a set of 13 genes in protocol allograft biopsies collected at 3 months after transplantation, which was independently predictive of the development of histological injury at 1 year. The predictive capacity of the gene set was superior to that of clinical indicators or routine histological parameters. Furthermore, the gene set accurately identified allografts that would have histological progression by 1 year or 2 years, as well as early allograft loss. We validated these data in an independent GoCAR cohort and two independent, publically available expression datasets.
Implications of all the available evidence
Our results suggest that those kidney transplant recipients who are at risk of allograft loss can be identified before the development of irreversible damage, thus offering the potential to modify therapeutic approaches before the onset of fibrosis. Future clinical trials in renal graft recipients could use our data to stratify patients by risk before enrolment to target specific interventional strategies to high-risk or low-risk groups, thereby improving efficiency by reducing sample sizes and costs.
Acknowledgments
This work is a substudy of the GoCAR study sponsored by NIH 5U01AI070107-03. The cost of clinical and genomic experiments and the effort of all the co-authors involved in patient enrolment, data management and analysis, and manuscript preparation were paid through this grant. MCM acknowledges support from the American Heart Association Scientist Development award 15SDG25870018, and past support from a research fellowship from the American Society of Nephrology. IAR would like to acknowledge the ISN Fellowship Program for her participation in this project. PJO’C was a recipient of a Senior Practitioner Fellowship from the National Health and Medical Research Council of Australia.
Footnotes
See Online for appendix
Contributors
BM conceived the study. BM and PJO’C designed the study. BM, WZ, MCM, BS, PJO’C, and IG drafted the manuscript. WZ, ZY, MCM, YG, BL, KLK, CWe, JO, EB, NN, and SIA analysed and interpreted the data. BS, LG, MS, AD, BJN, and JRC enrolled and followed up the patients. YL and CWo prepared the samples. RC, IAR, and RNS reported pathology results. CX managed clinical data. All authors were involved in revising the Article and approved the final version of the manuscript.
Declaration of interests
We declare no competing interests.
References
- 1.Matas AJ, Smith JM, Skeans MA, et al. OPTN/SRTR 2013 annual data report: kidney. Am J Transplant. 2015;15(suppl 2):1–34. doi: 10.1111/ajt.13195. [DOI] [PubMed] [Google Scholar]
- 2.Meier-Kriesche HU, Schold JD, Srinivas TR, Kaplan B. Lack of improvement in renal allograft survival despite a marked decrease in acute rejection rates over the most recent era. Am J Transplant. 2004;4:378–383. doi: 10.1111/j.1600-6143.2004.00332.x. [DOI] [PubMed] [Google Scholar]
- 3.Wolfe RA, Roys EC, Merion RM. Trends in organ donation and transplantation in the United States, 1999–2008. Am J Transplant. 2010;10:961–972. doi: 10.1111/j.1600-6143.2010.03021.x. [DOI] [PubMed] [Google Scholar]
- 4.Nankivell BJ, Borrows RJ, Fung CL, O’Connell PJ, Allen RD, Chapman JR. The natural history of chronic allograft nephropathy. N Engl J Med. 2003;349:2326–2333. doi: 10.1056/NEJMoa020009. [DOI] [PubMed] [Google Scholar]
- 5.Yilmaz S, McLaughlin K, Paavonen T, et al. Clinical predictors of renal allograft histopathology: a comparative study of single-lesion histology versus a composite, quantitative scoring system. Transplantation. 2007;83:671–676. doi: 10.1097/01.tp.0000262015.77625.90. [DOI] [PubMed] [Google Scholar]
- 6.Yilmaz S, Tomlanovich S, Mathew T, et al. Protocol core needle biopsy and histologic Chronic Allograft Damage Index (CADI) as surrogate end point for long-term graft survival in multicenter studies. J Am Soc Nephrol. 2003;14:773–779. doi: 10.1097/01.asn.0000054496.68498.13. [DOI] [PubMed] [Google Scholar]
- 7.Baboolal K, Jones GA, Janezic A, Griffiths DR, Jurewicz WA. Molecular and structural consequences of early renal allograft injury. Kidney Int. 2002;61:686–696. doi: 10.1046/j.1523-1755.2002.00149.x. [DOI] [PubMed] [Google Scholar]
- 8.Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 2008;8:753–760. doi: 10.1111/j.1600-6143.2008.02159.x. [DOI] [PubMed] [Google Scholar]
- 9.Isoniemi H, Taskinen E, Hayry P. Histological chronic allograft damage index accurately predicts chronic renal allograft rejection. Transplantation. 1994;58:1195–1198. [PubMed] [Google Scholar]
- 10.Einecke G, Reeve J, Sis B, et al. A molecular classifier for predicting future graft loss in late kidney transplant biopsies. J Clin Invest. 2010;120:1862–1872. doi: 10.1172/JCI41789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naesens M, Khatri P, Li L, et al. Progressive histological damage in renal allografts is associated with expression of innate and adaptive immunity genes. Kidney Int. 2011;80:1364–1376. doi: 10.1038/ki.2011.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 13.Cosio FG, Grande JP, Wadei H, Larson TS, Griffin MD, Stegall MD. Predicting subsequent decline in kidney allograft function from early surveillance biopsies. Am J Transplant. 2005;5:2464–2472. doi: 10.1111/j.1600-6143.2005.01050.x. [DOI] [PubMed] [Google Scholar]
- 14.Stegall MD, Park WD, Larson TS, et al. The histology of solitary renal allografts at 1 and 5 years after transplantation. Am J Transplant. 2011;11:698–707. doi: 10.1111/j.1600-6143.2010.03312.x. [DOI] [PubMed] [Google Scholar]
- 15.Mannon RB, Matas AJ, Grande J, et al. Inflammation in areas of tubular atrophy in kidney allograft biopsies: a potent predictor of allograft failure. Am J Transplant. 2010;10:2066–2073. doi: 10.1111/j.1600-6143.2010.03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rockey DC, Bell PD, Hill JA. Fibrosis—a common pathway to organ injury and failure. N Engl J Med. 2015;372:1138–1149. doi: 10.1056/NEJMra1300575. [DOI] [PubMed] [Google Scholar]
- 17.Hayry P, Paavonen T, Taskinen E, et al. Protocol core needle biopsy and histological chronic allograft damage index as surrogate endpoint for long-term graft survival. Transplant Proc. 2004;36:89–91. doi: 10.1016/j.transproceed.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 18.El-Zoghby ZM, Stegall MD, Lager DJ, et al. Identifying specific causes of kidney allograft loss. Am J Transplant. 2009;9:527–535. doi: 10.1111/j.1600-6143.2008.02519.x. [DOI] [PubMed] [Google Scholar]
- 19.Dai C, Liu Y. Hepatocyte growth factor antagonizes the profibrotic action of TGF-beta1 in mesangial cells by stabilizing Smad transcriptional corepressor TGIF. J Am Soc Nephrol. 2004;15:1402–1412. doi: 10.1097/01.asn.0000130568.53923.fd. [DOI] [PubMed] [Google Scholar]
- 20.Majidi M, Gutkind JS, Lichy JH. Deletion of the COOH terminus converts the ST5 p70 protein from an inhibitor of RAS signaling to an activator with transforming activity in NIH-3T3 cells. J Biol Chem. 2000;275:6560–6565. doi: 10.1074/jbc.275.9.6560. [DOI] [PubMed] [Google Scholar]
- 21.Sasaki A, Taketomi T, Kato R, et al. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat Cell Biol. 2003;5:427–432. doi: 10.1038/ncb978. [DOI] [PubMed] [Google Scholar]
- 22.Lee SH, Schloss DJ, Jarvis L, Krasnow MA, Swain JL. Inhibition of angiogenesis by a mouse sprouty protein. J Biol Chem. 2001;276:4128–4133. doi: 10.1074/jbc.M006922200. [DOI] [PubMed] [Google Scholar]
- 23.Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 24.Tan HB, Feng Y, Liu M, Wu YC. Protective effects of FR167653 on chronic allograft nephropathy by inhibiting p38 MAPK in rats. Transplant Proc. 2008;40:1685–1689. doi: 10.1016/j.transproceed.2008.01.073. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Jiang J, Guan Q, et al. Reduction of chronic allograft nephropathy by inhibition of extracellular signal-regulated kinase 1 and 2 signaling. Am J Physiol Renal Physiol. 2008;295:F672–F679. doi: 10.1152/ajprenal.90285.2008. [DOI] [PubMed] [Google Scholar]
- 26.Azuma H, Takahara S, Matsumoto K, et al. Hepatocyte growth factor prevents the development of chronic allograft nephropathy in rats. J Am Soc Nephrol. 2001;12:1280–1292. doi: 10.1681/ASN.V1261280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.