Summary
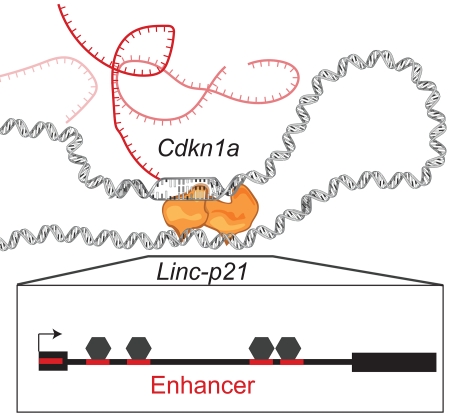
The Linc-p21 locus, encoding a long non-coding RNA, plays an important role in p53 signalling, cell cycle regulation, and tumour suppression. However, despite extensive study, confusion exists regarding its mechanism of action: is activity driven by the transcript acting in trans, in cis, or by an underlying functional enhancer? Here, using a knockout mouse model and a massively parallel enhancer assay, we delineate the functional elements at this locus. We observe that even in tissues with no detectable Linc-p21 transcript, deletion of the locus significantly affects local gene expression, including of the cell cycle regulator Cdkn1a. To characterize this RNA-independent regulatory effect, we systematically interrogated the underlying DNA sequence for enhancer activity at nucleotide resolution, and confirmed the existence of multiple enhancer elements. Together, these data suggest that, in vivo, the cis-regulatory effects mediated by Linc-p21, in the presence or absence of transcription, are due to DNA enhancer elements.
eTOC blurb
Using a knockout mouse model and massively parallel reporter assay, Groff et al. show that the lincp21 gene, which was previously thought to produce a cis-acting lincRNA, actually contains multiple enhancer elements that are directly responsible for regulating transcription of nearby genes, including Cdkn1a.
Introduction
It has long been known that transcription occurs at many more sites in the genome than encode proteins. Among the main constituents of the resulting non-coding transcriptome are long non-coding (lnc)RNAs, which are more than 200 nucleotides in length and exhibit tissue-specific expression (ENCODE Project Consortium et al., 2007, Mercer et al 2009, Cabili et al., 2011, St Laurent et al., 2015, Quinn and Chang, 2016). Although originally dismissed as transcriptional noise, it is now clear that several lncRNAs have important biological functions (Wang and Chang, 2011, Guttman and Rinn, 2012). However, the rapid creation of entire catalogs of lncRNAs – made possible by RNA sequencing – has meant that our knowledge of where lncRNA genes are located far exceeds our understanding of their functions. Indeed, it is even unclear as to whether the functional element at these loci is the RNA transcript itself or the underlying DNA sequence, which could have enhancer activity. For example, while the Lockd locus fulfils all of the requirements of a lncRNA, the phenotype associated with deletion of this locus is actually due to loss of the underlying DNA element and not the RNA transcript (Paralkar et al., 2016).
Linc-p21 is one of the most-studied lncRNAs due to its role in p53 signalling and relevance to human disease (Huarte et al., 2010, Dimitrova et al., 2014, Yoon et al., 2012, Tang et al., 2015). Indeed, since its discovery in 2010, dozens of studies have examined Linc-p21 in human and/or mouse cell-based assays, and have collectively identified roles in a range of biological processes including cell cycle control, reprogramming, apoptosis, and energy metabolism (Dimitrova et al., 2014, Bao et al., 2015, Huarte et al., 2010, Yang et al., 2014). However, despite being the subject of extensive study, this locus has not yet been examined in tissues or in vivo. Moreover, confusion exists regarding the mechanism by which the Linc-p21 locus functions in any context. For example, at different times this locus has been thought to produce a trans-acting lncRNA (Huarte et al., 2010), a cis-acting lncRNA (Dimitrova et al., 2014), or an enhancer-derived RNA (Allen et al., 2014). Here, we aimed to resolve this confusion by characterizing the functional elements at the Linc-p21 locus in vivo. Using a Linc-p21 knockout mouse model (Sauvageau et al., 2013, Goff et al., 2015), we demonstrate that deletion of Linc-p21 results in the cis-dysregulation of several genes, including Cdkn1a. Interestingly, this dysregulation was observed across multiple tissues, even those in which Linc-p21 RNA was not expressed, and thus cannot be due to an RNA-dependent mechanism. To better understand how a DNA-dependent effect might be mediated, we comprehensively surveyed the entire Linc-p21 locus for enhancer activity using a massively parallel reporter assay, and identified multiple enhancer elements including a conserved p53-binding site. Collectively, we show that the Linc-p21 locus harbours DNA enhancer elements that are directly responsible for the cis-regulation of multiple genes in vivo.
Results
Deletion of Linc-p21 results in quantifiable effects on whole organ gene expression
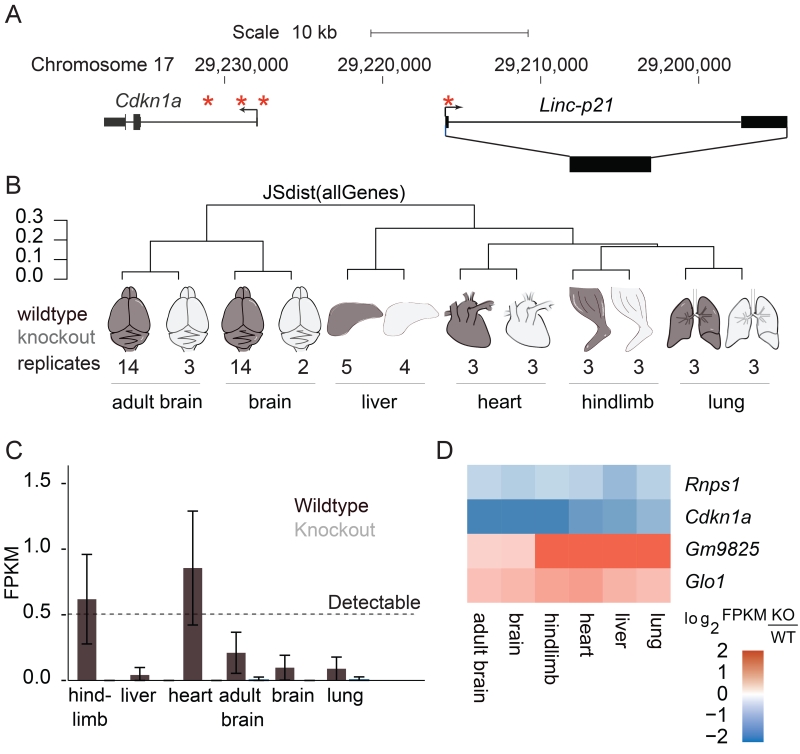
The mouse Linc-p21 gene resides in a 21.6 kb locus on chromosome 17, approximately 15 kb upstream of the cell cycle regulator Cdkn1a. We previously targeted this locus for deletion using homologous recombination in which the entire gene body and a portion of the promoter, including the known p53-binding motif, were replaced with a lacZ reporter that maintains transcription at the locus (Figure 1A, Sauvageau et al., 2013). Using this reporter mouse, we initially investigated the spatial and temporal expression of Linc-p21 in organ development in order to identify relevant tissues for downstream experiments (Figure S1). Importantly, because deletion of the p53 motif meant that endogenous Linc-p21 expression may not have been faithfully reported by the model, we validated the observed expression pattern in wildtype mice, confirming that Linc-p21 expression was detectable in the tissues where lacZ was present in the reporter mouse.
Figure 1. Linc-p21 in vivo deletion overview.
(A) Linc-p21/Cdkn1a locus on mouse chromosome 17. Asterisks indicate known p53 binding sites. (B) Dendrogram showing the type and number of samples sequenced and the Jenson-Shanon distance, a measure of total transcriptome similarity, between their expression profiles. Wildtype shown in black, knockout shown in grey. (C) Average Linc-p21 expression profile in each tissue. Error bars represent 95% confidence interval. (D) Heatmap depicting expression of genes significantly differentially expressed in every tissue. Log2 fold-change was calculated using the average FPKMs for all replicates (KO/WT). See also Figure S1.
Overall, we detected differential expression of the Linc-p21 locus in different tissues, with expression being highest in striated muscle – an observation that may relate to the role that Cdkn1a is known to play in muscle development (Halevy et al., 1995, Guo et al., 1995). Interestingly, we also found that transcription at this locus was strongly decreased in adulthood (Figure S1).
Based on these results, we selected a range of embryonic tissues for high throughput RNA sequencing analyses (Figure 1B, File S1), including some in which Linc-p21 was expressed in wildtype mice (e.g. hindlimb and heart) and others in which there was no detectable expression (e.g. liver and lung) (Figure 1C, S1). We also included adult and embryonic brain samples from our previous study of whole brain sequencing (Goff et al., 2015). For each tissue, we conducted a comprehensive analysis of whole-genome transcription between wildtype and Linc-p21 knockout (File S2). We then performed pathway analysis using the list of genes that were differentially expressed in one or more tissues and observed enrichment for genes involved in cell cycle and muscle-related processes, consistent with the observed expression pattern of Linc-p21 (Figure S2A,B).
Identification of DNA regulatory elements within the Linc-p21 locus in vivo
To better characterize the transcriptional perturbations that occurred in the absence of Linc-p21, we next identified the genes that were significantly differentially expressed in all of the tissues examined. Strikingly, 3 of the 4 genes that met these criteria (Glo1, Rnps1, and Cdkn1a) are located on chromosome 17 – the same chromosome as Linc-p21 (Figure 1D). Cdkn1a is a well-known regulator of the cell cycle, and its dysregulation is thus in keeping with the observation that the genes involved in cell cycle were consistently upregulated in the knockout (Figure S2D).
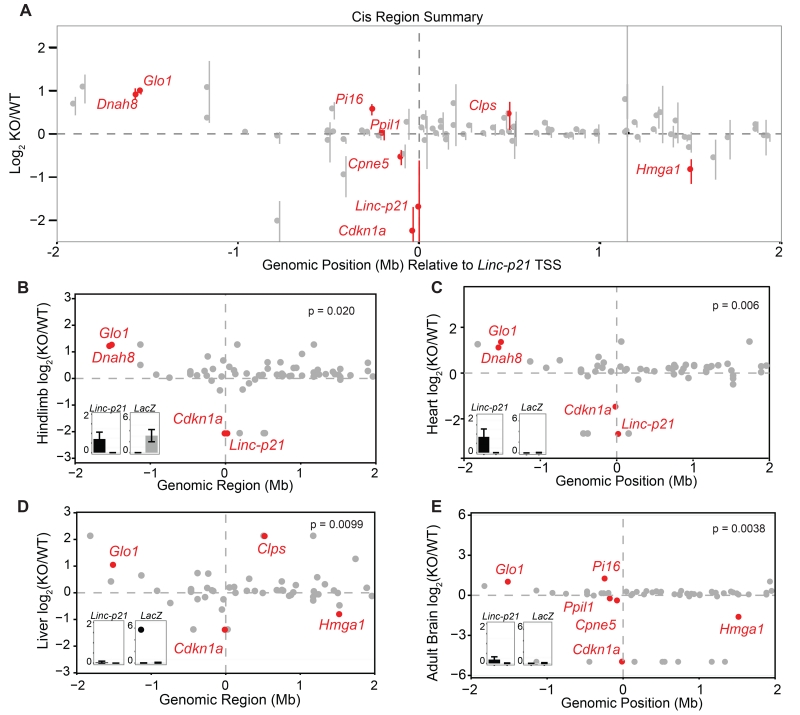
To further investigate this regulatory effect upon local gene expression, we examined whether other nearby genes within a 4Mb region centred on Linc-p21 were similarly dysregulated in different tissues. Of the 84 genes within this window, we observed that 8 were significantly dysregulated in one or more of the tissues examined (4 upregulated and 4 downregulated) and that a substantial proportion of the remainder (34 of 76 genes) showed non-significant expression effects of a similar direction in all tissues in response to Linc-p21 deletion (Figure 2A). Notably, in each of the tissues examined, the number of significantly dysregulated genes in proximity to the Linc-p21 locus was higher than would have been expected by chance (based on permutation testing using 10,000 randomly selected size-matched regions in each tissue; Figure 2B-E and File S3). To provide a genome-wide context, we identified those genes whose expression changed in the same direction across all tissues (irrespective of statistical significance) and observed that a highly significant fraction of these were located on chromosome 17 (91 of 698, p<2.1×10−19; Figure S2C,E). Such a chromosomal bias for gene expression effects is consistent with multiple cis-regulatory effects arising from this locus.
Figure 2. Linc-p21 transcript is not required for local gene regulation.
(A) Summary of local transcriptional changes upon Linc-p21 deletion (+/− 2Mb of Linc-p21). Dots represent average log2 fold change across all 6 tissues and error bars represent standard error. Red indicates significant differential expression in at least one tissue. (B-E) Expression of genes in local region (+/− 2Mb of Linc-p21) for E14.5 hindlimbs (B), heart (C), liver (D), and adult brain (E). In each plot, the Y-axis represents the log2 FPKM fold change and genes marked in red were significantly differentially expressed. The p value represents the probability that this number of genes would be differentially expressed within a region of this size. Insets show Linc-p21 and lacZ expression (FPKM) in wildtype (black) and knockout (gray). See also Figure S2.
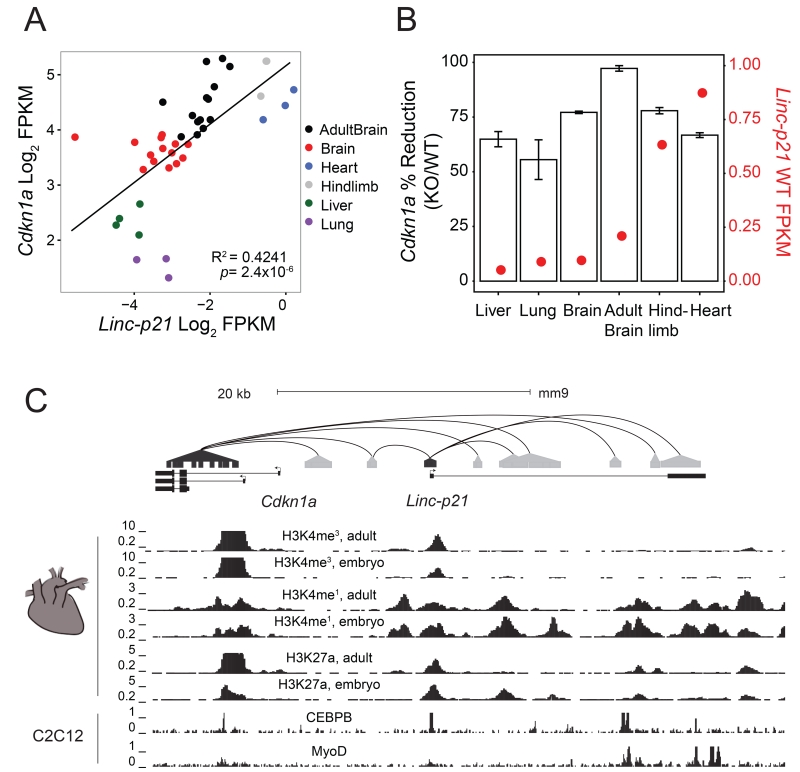
To delineate the relative roles of the DNA element and the RNA transcript in mediating these cis-regulatory effects, we next examined the expression of Linc-p21 and Cdkn1a in wildtype tissues and detected a positive correlation between their RNA abundances (Figure 3A, R2=0.42, p=2.4×10−6). For each tissue, we then compared the reduction in Cdkn1a expression that was observed in the knockout mouse with the expression of Linc-p21 in that tissue in the wildtype. We hypothesized that if Linc-p21 RNA was responsible for activating Cdkn1a, then the reduction in Cdkn1a expression should be proportional to the endogenous abundance of Linc-p21 RNA. However, this relationship was not observed. In fact, in all of the tissues examined, the magnitude of the change in expression of Cdkn1a was wholly unrelated to the wildtype expression level of Linc-p21 in that tissue (Figure 3B, p=0.96). To consider whether this phenomenon was limited to Cdkn1a or more widespread, we performed pathway analysis between individual wildtype and knockout tissues. This demonstrated similar enrichment for cell cycle processes and muscle-related processes across all tissues, even those in which Linc-p21 was not endogenously expressed (Figure S2D, File S3). Together this suggested that the regulatory effects mediated by this locus were not due to either transcription of Linc-p21 or the mature RNA transcript, and implied that another, RNA-independent regulatory mechanism must be present.
Figure 3. In vivo evidence of enhancer activity in the Linc-p21 locus.
(A) Correlation of Cdkn1a expression with Linc-p21 transcript expression. X-axis represents Linc-p21 expression and Y-axis represents Cdkn1a expression in each wildtype replicate. Color indicates the tissue of origin, and the linear regression line is shown. (B) Average change in Cdkn1a expression in knockout tissues (1-KO/WT, bars, left y-axis) plotted against average wildtype Linc-p21 expression levels (red dots, right y-axis); error bars represent standard error. (C) Publically available chromatin interaction data between Linc-p21 and Cdkn1a from HiCap (plotted as black lines between capture probes and gray distal regions). Publically available histone mark and transcription factor binding data at this locus. All panels are from embryonic or adult heart tissue, except CEBPB and MyoD binding which are from C2C12 cells. See also Figure S3.
The Linc-p21 locus contains multiple enhancer elements
Based on these results, we investigated whether the Linc-p21 DNA sequence might contain functional enhancer elements that could explain the observed cis-regulatory effects. We first examined histone modifications and transcription factor binding sites across the locus using publically available datasets derived from murine heart tissue. We found that the Linc-p21 gene body and promoter have multiple features typically associated with enhancer activity, including monomethylation of histone 3 at lysine 4 (H3K4me) and acetylation of histone 3 at lysine 27 (H3K27ac, Figure 3C, data from Rosenbloom et al., 2013). Moreover, chromatin contact data from a genome-wide promoter capture method, HiCap, indicated that the Linc-p21 locus and Cdkn1a promoter physically interact through intra-chromosomal looping (upper panel, Figure 3C, Sahlén et al., 2015). To assess if Linc-p21 interacts with other nearby loci, we analysed all of the interactions within a 4Mb region using available data from a complementary method, Hi-C (Dixon et al., 2012). We found that genes which physically interact with the Linc-p21 locus were significantly enriched for those which were dysregulated following Linc-p21 deletion, compared to the background interaction rate (Figure S3A). Similar interactions were also identified in human capture Hi-C data (Mifsud et al., 2015) between LINC-p21 and the promoters of genes whose orthologs were significantly dysregulated in the knockout mouse (including CDKN1A, PPIL1 and CPNE5; Figure S3B). These findings are consistent with the Linc-p21 locus containing regulatory DNA elements that mediate conserved intra-chromosomal cis-regulatory contacts.
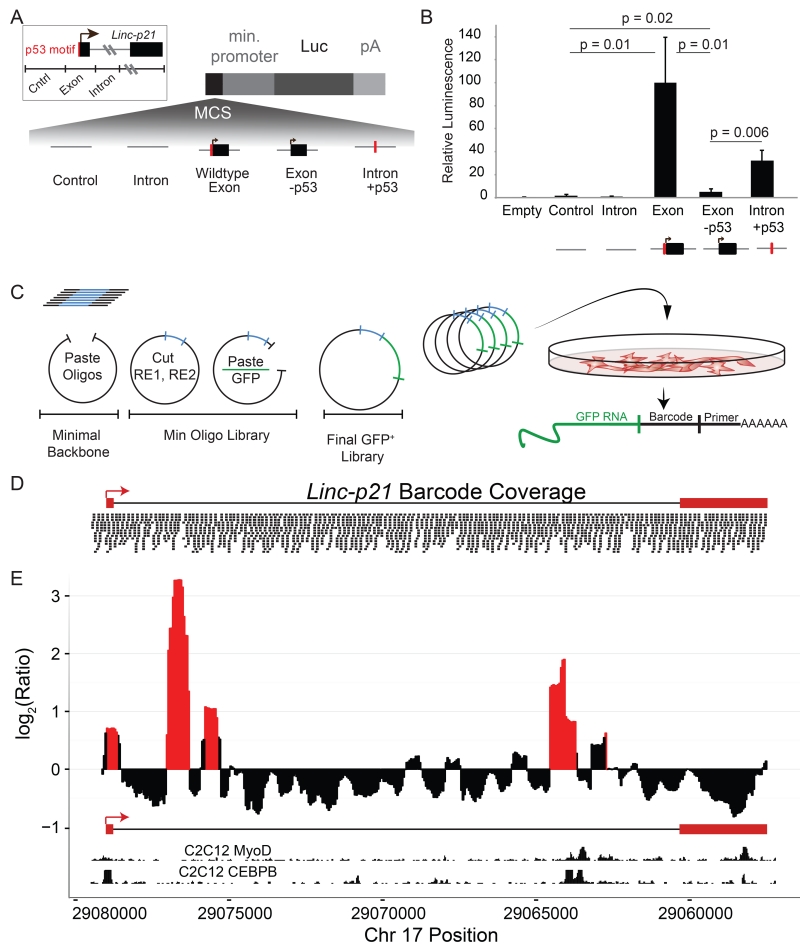
To test for functional DNA regulatory elements, we assessed whether the following 1kb regions could promote transcription of luciferase from a reporter vector in C2C12 cells, a mouse myoblast cell line: (1) a region surrounding the promoter and first exon, including the conserved p53 motif, (2) a region from intron 1, and (3) a negative control from an upstream intergenic region that has neither enhancer histone marks nor evidence of transcription (Figure 4A). We observed strong activation of luciferase activity from the Linc-p21 promoter but not the intronic region (Figure 4B). Much of this signal was shown to be due to the p53-binding site, as a strong decrease in luciferase activity was observed if the 16-bp p53 motif was disrupted by site-directed mutagenesis. Moreover, if this motif was inserted into intron 1 (the region that previously lacked enhancer activity), a 29-fold increase in luciferase activity was observed. Notably, however, this increased activity level was still much lower than that of the native promoter (Figure 4B). Together these results suggest that the p53 motif in the Linc-p21 promoter is a major factor in driving enhancer activity. However, because ectopic insertion of this motif into an intronic site did not fully recapitulate the enhancer activity of the promoter, and because presence of enhancer-related histone marks are present throughout the gene body, we hypothesized that other as-yet-unidentified DNA regulatory elements might be present. To investigate this possibility, we used a massively parallel reporter assay (MPRA; Melnikov et al., 2012) to systematically and comprehensively interrogate the Linc-p21 locus – at nucleotide resolution – and establish whether additional enhancer elements were present. To do this, we synthesized a library of 2,225 individually-tagged 140-bp oligos that redundantly tiled the entire Linc-p21 locus (including the gene body and promoter; Figure 4C). This oligo library was cloned into a GFP+ reporter vector (Melnikov et al., 2012, Melnikov et al., 2014), and the coverage in the final pooled library was checked by high throughput sequencing (Figure 4D). The plasmid library was then transfected into C2C12 cells and after 24 hours RNA was extracted and indexed libraries were constructed and sequenced.
Figure 4. MPRA of the entire Linc-p21 locus reveals enhancer activity.
(A) Experimental design for luciferase reporter assay using an intergenic control and regions from the Linc-p21 promoter and exon 1 (“exon”), intron 1 (“intron”), exon with mutated p53 binding sequence (“exon -p53”) and intron with p53 binding sequence (“intron+p53”). Each region was cloned into a luciferase reporter construct. P53 motif indicated in red. (B) Relative luminescence for each construct, normalized against the signal from exon fragment and averaged across triplicate samples. P values were calculated using unpaired one-tailed t-tests. Error bars represent standard error of the mean. (C) Massively Parallel Reporter Assay (MPRA) experimental design: oligos were synthesized, subcloned into a minimal backbone, opened by enzymatic digestion and re-ligated with a GFP cDNA insert. Pooled constructs were transfected into C2C12 cells in triplicate and libraries were made from GFP+ RNA. (D) Coverage of final pooled GFP+ vector library across the Linc-p21 locus and promoter (assessed by high throughput sequencing). (E) MPRA signal across the Linc-p21 locus. Y-axis represents the log2 ratio of normalized RNA to control signal per base (averaged in 500bp sliding windows every 50bp). Significance (p<0.01) is calculated by comparing this signal to 1000 random shuffles of the input data. Significant peaks are shown in red. Inlayed tracks are CEBPB and MyoD ChIP-seq signals from UCSC genome browser. See also Figure S4.
In each sample, the number of tags (indicative of a transcriptional event) was quantified, normalized for sequencing depth, and used to calculate the signal for each base-pair of the region (Melnikov et al., 2012, Kheradpour et al., 2013, Figure S4). Using this approach, we confirmed the enhancer activity of the p53-binding site across multiple independent oligos. Strikingly, we also observed 4 other regions of enhancer activity within the Linc-p21 locus, two of which displayed stronger enhancer activity than that of the known p53-motif (Figure 4E). Using available data from the same cell line, we observed that the first of these regions overlapped with an experimentally confirmed CCAAT/Enhancer Binding Protein (CEBPB) binding site and the second was located proximal to a MyoD ChIP peak (Figure 4E, Rosenbloom, et al., 2013) – a finding that connects the known role of MyoD in muscle development with our finding that the Linc-p21 locus is most highly expressed in muscle (Figures 1C, S1). Collectively, these data indicate that the Linc-p21 locus is a complex genomic environment containing several functional DNA elements that interact with, and regulate the transcription of, multiple local genes including Cdkn1a.
Discussion
Since its discovery in 2010, Linc-p21 has been the subject of intense study due to its reported roles in important biological processes (Huarte et al., 2010, Dimitrova et al., 2014, Yang et al., 2014, Hall et al., 2015, Bao et al., 2015, Yoon et al., 2012, Wu et al., 2014, Wang et al., 2014, Tran et al., 2015, Tang et al., 2015). However, despite extensive investigation, the nature of transcription at the Linc-p21 locus and the mechanism by which the gene functions have not been tested outside of cell-based assays. Here we present, to our knowledge, the first study of the Linc-p21 locus in vivo, and by implementing a whole gene deletion and reporter knock-in, are able to disentangle the relative contributions of DNA and RNA to the observed cis-regulatory effects (Bassett et al., 2014, Goff and Rinn, 2015). Moreover, by combining this approach with MPRA, we demonstrate that, in vivo, the Linc-p21 locus is a complex DNA enhancer element that regulates the expression of multiple genes in a range of tissues in cis.
Several lines of evidence come together to support this conclusion. First, we observed that Linc-p21 deletion consistently led to changes in the expression of local genes irrespective of whether the Linc-p21 locus itself was transcribed – thereby excluding an RNA-dependent regulatory mechanism. Second, MPRA data revealed that multiple enhancer elements are present within the Linc-p21 locus, including a known p53 motif which was confirmed to have strong enhancer activity. Third, the Linc-p21 locus physically interacts in 3D space with the promoters of local genes that are dysregulated following Linc-p21 deletion, including Cdkn1a – an observation that is consistent with studies that have shown that p53-bound DNA elements can interact with local genes via DNA looping (Link et al., 2013, Melo et al., 2013, Younger et al., 2015). Accordingly, we believe that Linc-p21 represents an example of a primed p53 enhancer, in which the structural contact and even the enhancer activity is established independent of p53, but is further activated upon p53 binding. In keeping with this conclusion, global run-on sequencing analysis has previously identified transcription at the Linc-p21 locus in p53-null cells, suggesting that the enhancer is functional even without p53 binding (Allen et al., 2014).
It is important to consider how these data fit with other studies in which Linc-p21 has been proposed to be a trans- or cis-acting lincRNA. While several of the initial studies of Linc-p21 reported trans-regulatory effects, many of these are now thought to be mediated indirectly via genes that Linc-p21 regulates in cis- (Dimitrova et al., 2014). Indeed, Dimitrova and colleagues demonstrated that Cdkn1a expression was reduced by ~50% following deletion of the Linc-p21 promoter and concluded that Linc-p21 is a cis-regulatory lncRNA (Dimitrova et al., 2014). However, this deletion included the p53-binding site that we have shown has important enhancer activity. These data would therefore also be consistent with a functional DNA element, and further highlight both the specific role of this element in the control of Cdkn1a expression and the general importance of genetic strategies that can definitively delineate the relative contributions of DNA and RNA to any observed function.
The Linc-p21 locus exemplifies the difficulties in elucidating the relative functions of DNA, RNA, and the act of transcription at non-coding loci. While there are clear examples of lncRNAs that function through their RNA transcript (Brown et al. 1992, Rinn et al., 2007), there are also examples where the RNA is dispensable and either transcription itself or elements within the DNA sequence are responsible for observed functions (Paralkar et al., 2016). These potential mechanisms cannot be resolved through deletion of the entire locus (Basset et al. 2014, Goff and Rinn, 2015) and thus other methods are needed to dissect their relative contributions.
To date, most of these methods have focused upon disrupting the RNA transcript by reducing transcription or truncating the transcript using terminator sequences (Paralkar et al., 2016, Basset et al. 2014). Here we show that MPRA can be used to detect functional DNA elements within non-coding loci, and thus provides an important complementary technique. Indeed, using this approach, we discover several previously uncharacterized regions within the Linc-p21 locus that have enhancer activity and overlap histone and protein ChIP peaks – consistent with DNA enhancer elements. Moreover, these regions of enhancer activity lie within the vicinity of structural chromosome contact points that would bring them to the promoter of Cdkn1a and other nearby genes. Interestingly, many direct targets of p53 are known to associate with primed enhancers that are further activated by p53 binding (Melo et al., 2013, Allen et al., 2014, Younger et al., 2015). It could therefore be hypothesized that these novel enhancer regions might be pioneer-factor binding sites that could organize local chromatin contacts and bring the Linc-p21 p53 motif into the vicinity of the nearby promoters, where it can simultaneously activate transcription in both regions.
The Linc-p21 promoter region (but not the transcript) is well conserved between human and mouse, both in terms of sequence homology and physical interactions with orthologous genes (Figure S3B). Intriguingly, this suggests that enhancer contacts between this locus and nearby genes may be evolutionarily conserved, which might also explain why genetic variation in this region has been associated with human disease, including colorectal cancer and cardiac defects (Sotoodehnia et al., 2010, Dunlop et al., 2012, Ritchie et al., 2013, Hong et al., 2014). Collectively, these data provide a starting point to better understand the exact nature of the regulatory interactions at this locus, which could ultimately provide important insights into the pathogenesis of several human diseases.
In summary, our data demonstrate that the cis-regulatory effects mediated by Linc-p21 are due to a functional DNA element rather than the RNA transcript. As such, we would support the reclassification of the Linc-p21 RNA as an eRNA, and note that although this term usually implies some cis-regulatory function, in this example neither the transcript nor transcription are required for local gene regulation by the locus. It is important to note, however, that the RNA transcript may still have other functions that are not related to cis-regulation, and which have not been identified in this study. For example, we did not seek to explore the mechanism underlying any potential trans-regulatory effects, and we also cannot exclude the possibility that the RNA has a function at an earlier developmental stage which is subsequently lost as tissues differentiate. Moreover, we suspect that many lncRNA loci probably contain both functional DNA elements and RNA transcripts, and that these possibilities should not be considered mutually exclusive. Indeed, just as coding gene loci can contain functional intragenic enhancers (Li et al., 2012, Zhang et al., 2013) and still produce translated mRNA transcripts, we expect that a similarly complex situation will exist at many non-coding loci. Unravelling the respective functions of DNA and RNA at these loci is likely to be an on-going challenge for the field, and one that may ultimately lead to a revision of our classification of non-coding transcripts, along with the catalogs of transcripts themselves.
Experimental Procedures
Mice
Mice were housed under pathogen-free conditions in Harvard University’s Biological Research Infrastructure. All procedures were approved by the Harvard University Committee on the Use of Animals in Research and Teaching and performed in accordance with the National Institutes of Health guidelines.
RNA isolation and RNA-Seq library preparation and sequencing
Global gene expression was assessed by RNA sequencing (RNAseq) of different organs and tissues from at least 3 Linc-p21 knockout and 3 wildtype embryos. Hindlimbs, liver, lungs, and heart were harvested from E14.5 embryos and immediately homogenized in TRIzol (Life Technologies). Total RNA was extracted by chloroform extraction followed by spin-column purification (RNeasy mini kit, Qiagen). RNA-Seq libraries (TruSeq RNA Sample Preparation Kit v2; Illumina) were prepared as previously described using 500ng of total RNA and a 10-cycle PCR enrichment to minimize PCR artifacts (Sauvageau et al., 2013, Goff et al., 2015). Knockout and wildtype samples from different litters were processed within each library preparation. The indexed libraries were sequenced in pools of six (Illumina HiSeq 2000, 101-bp paired-end reads).
RNA-Seq analysis
Reads from fastq samples were aligned to the mouse genome (mm10) using Tophat2 with non-standard options “--no-coverage-search --max-multihits 10 -p 8” (Kim et al. 2013). Each sample was quantified using Cuffquant with nonstandard options “-p 8 --no-update-check”, and differential analysis was performed for each wildtype-vs-knockout tissue comparison using Cuffdiff2 with nonstandard option “-p 8”. We also performed a Cuffdiff2 analysis in which all wildtype-vs-knockout samples were assessed together using nonstandard option “-p 8” (Trapnell et al. 2012). All analysis scripts are available as Files S2 and S4, all code is available on Github, and we frequently used Cummerbund for analysis and to generate figures (Goff et al 2013, https://github.com/rinnlab/lincp21).
Cloning and mutagenesis
The Linc-p21 locus was cloned using a BAC plasmid (RP24-248L4) obtained from Children’s Hospital Oakland Research Institute. Acc651 and Xho1 restriction sites were added to amplification primers to enable ligation into the multiple cloning site of the pGL4.23 vector (Promega) containing the luciferase gene. The p53 binding site was perturbed using inverse PCR and 5′ phosphorylated primers containing mutations amplified off of the Exon 1 clone in pGL4.23 similar to the Quick Change protocol (Agilent Technologies). The pGL4.73 vector was used in co-transfection as a transformation control expressing Renilla luciferase.
Cell culture and Transfection
The C2C12 cell line was obtained from ATCC (CRL-1772) and maintained according to the recommended guidelines. Transfections were performed using TransfeX reagent (ATCC ACS-4005) for luciferase assays and Lipofectamine 3000 (Thermo Fisher) for the MPRA experiment. All experiments were performed in triplicate.
Massively Parallel Reporter Assay (MPRA)
We designed 140bp oligos to redundantly tile the genomic region spanning the Linc-p21 locus and 500bp of its promoter, as previously described (Melnikov et al., 2012, Melnikov et al., 2014, Kheradpour et al., 2013). Our pool consisted of 90bp genomic regions starting every 50bp. Each genomic region was represented by 5 unique barcodes. Oligos were synthesized by the Broad Institute Technology Core and cloned into final GFP+ constructs as previously described (Melnikov et al., 2012, Melnikov et al., 2014, Kheradpour et al., 2013). 10ug of GFP+ pooled construct was used to transfect C2C12 cells in 6-well plates. After 24 hours, cells were harvested in Trizol and total RNA was extracted as described and treated with DNase. MPRA libraries were constructed from 1ug input as previously described RNA (Melnikov et al., 2014). Libraries from the pooled vector construct were used as a control. All libraries were purified by a triple SPRI bead cleanup (0.65×, 0.8×, 0.8×, Agencourt AMPure XP, Beckman Coulter), quantified by Qubit, and size-checked on a BioAnalyzer before deep sequencing (HiSeq 2500). Experiments were performed in triplicate.
MPRA Analysis
We counted tags originating from reads containing GFP sequence and with a perfect match to the barcodes we designed, and then normalized each sample to the total number of counts from that sample. We calculated the median ratio of RNA library signal to vector library signal for each base-pair and calculated rolling signal means across the locus with different windows and slides (File S5). For all analysis reported in this paper, we used a window of 500bp and a slide of 50bp. To generate p-values for significance of any given region, we permuted the signal ratio values across the entire locus 1000 times and repeated the sliding window analysis for each permutation, generating a randomized permutation p-value for each 500bp window.
Supplementary Material
Highlights.
Linc-p21 regulates transcription of multiple local genes in vivo, including Cdkn1a
Linc-p21 RNA is entirely dispenable for this cis-regulatory function
Massively parallel reporter assay identifies several enhancer elements at this locus
The Linc-p21 locus physically interacts with cis-regulated genes in human and mouse
Acknowledgements
We thank members of the Rinn lab for experimental help, scientific discussion, and critical feedback during the course of this project. In particular we thank Jordan Lewandowski for manuscript feedback, David Shechner for assistance with luciferase assays, and Meryem Gonzalez-Celeiro for assistance in the mouse facility. A.F.G was supported by the NSF graduate research fellowship and is currently supported by the HHMI Gilliam Fellowship. J.C.L. is supported by a Wellcome Trust Intermediate Clinical Fellowship (105920/Z/14/Z). J.L.R is the Alvin and Esta Star Associate Professor of Stem Cell and Regenerative Biology. This work was supported by the National Institutes of Health (NIH)/Institute of Mental Health grant R01MH102416-03 and the NIH/National Institute of General Medical Sciences grant P01GM099117.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
For extended experimental procedures, please see supplemental materials online.
Accession Numbers
All newly sequenced data has been deposited in GEO with accession number GSE73472. Data on embryonic and adult brains was processed from publically available data set GSE61716. Hi-C data was processed from data set GSE35156.
Author Contributions
A.F.G. and J.L.R. conceived the experiments. A.F.G., D.B.S.G., M.M.L.S., C.G., E.L., L.E., O.P., and L.V.S. performed the experiments. A.F.G., A.R.B., and J.C.L. analyzed the data. A.F.G., J.C.L., and J.L.R. wrote the manuscript with input from M.S., C.G., and A.R.B.. All authors approved the final version of the manuscript.
References
- Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA, Sullivan KD, Galbraith MD, Luo X, Kraus WL, et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. Elife. 2014;3:e02200. doi: 10.7554/eLife.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Wu H, Zhu X, Guo X, Hutchins AP, Luo Z, Song H, Chen Y, Lai K, Yin M, et al. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Nature Publishing Group. 2015;25:80–92. doi: 10.1038/cr.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W, et al. Considerations when investigating lncRNA function in vivo. Elife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, et al. LincRNA-p21 Activates p21 In cis to Promote Polycomb Target Gene Expression and to Enforce the G1/S Checkpoint. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop MG, Dobbins SE, Farrington SM, Jones AM, Palles C, Whiffin N, Tenesa A, Spain S, Broderick P, Ooi L-Y, et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat. Genet. 2012;44:770–776. doi: 10.1038/ng.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res. 2015;25:1456–1465. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Groff AF, Sauvageau M, Trayes-Gibson Z, Sanchez-Gomez DB, Morse M, Martin RD, Elcavage LE, Liapis SC, Gonzalez-Celeiro M, et al. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2015;112:6855–6862. doi: 10.1073/pnas.1411263112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo K, Wang J, Andrés V, Smith RC, Walsh K. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol. Cell. Biol. 1995;15:3823–3829. doi: 10.1128/mcb.15.7.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, Lassar AB. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- Hall JR, Messenger ZJ, Tam HW, Phillips SL, Recio L, Smart RC. Long noncoding RNA lincRNA-p21 is the major mediator of UVB-induced and p53-dependent apoptosis in keratinocytes. 2015;6:e1700–e1709. doi: 10.1038/cddis.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K-W, Lim JE, Kim JW, Tabara Y, Ueshima H, Miki T, Matsuda F, Cho YS, Kim Y, Oh B. Identification of three novel genetic variations associated with electrocardiographic traits (QRS duration and PR interval) in East Asians. Hum. Mol. Genet. 2014;23:6659–6667. doi: 10.1093/hmg/ddu374. [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P, Ernst J, Melnikov A, Rogov P, Wang L, Zhang X, Alston J, Mikkelsen TS, Kellis M. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–811. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link N, Kurtz P, O’Neal M, Garcia-Hughes G, Abrams JM. A p53 enhancer region regulates target genes through chromatin conformations in cis and in trans. Genes & Development. 2013;27:2433–2438. doi: 10.1101/gad.225565.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG, Kinney JB, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nature Biotechnology. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Zhang X, Rogov P, Wang L, Mikkelsen TS. Massively parallel reporter assays in cultured mammalian cells. J Vis Exp. 2014:e51719–e51719. doi: 10.3791/51719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Vrielink JAFO, Elkon R, Melo SA, Léveillé N, Kalluri R, et al. eRNAs Are Required for p53-Dependent Enhancer Activity and Gene Transcription. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- Paralkar VR, Taborda CC, Huang P, Yao Y, Kossenkov AV, Prasad R, Luan J, Davies JOJ, Hughes JR, Hardison RC, et al. Unlinking an lncRNA from Its Associated cis Element. Mol. Cell. 2016;62:104–110. doi: 10.1016/j.molcel.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nature Publishing Group. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Denny JC, Zuvich RL, Crawford DC, Schildcrout JS, Bastarache L, Ramirez AH, Mosley JD, Pulley JM, Basford MA, et al. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation. 2013;127:1377–1385. doi: 10.1161/CIRCULATIONAHA.112.000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013 Jan;41(Database issue):D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlén P, Abdullayev I, Ramsköld D, Matskova L, Rilakovic N, Lötstedt B, Albert TJ, Lundeberg J, Sandberg R. Genome-wide mapping of promoter-anchored interactions with close to single-enhancer resolution. Genome Biol. 2015;16:1. doi: 10.1186/s13059-015-0727-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotoodehnia N, Isaacs A, de Bakker PIW, Dörr M, Newton-Cheh C, Nolte IM, van der Harst P, Müller M, Eijgelsheim M, Alonso A, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 2010;42:1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S-S, Zheng B-Y, Xiong X-D. LincRNA-p21: Implications in Human Diseases. Ijms. 2015;16:18732–18740. doi: 10.3390/ijms160818732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran UM, Rajarajacholan U, Soh J, Kim T-S, Thalappilly S, Sensen CW, Riabowol K. LincRNA-p21 acts as a mediator of ING1b-induced apoptosis. 2015;6:e1668–10. doi: 10.1038/cddis.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Li Z, Zhao Q, Zhu Y, Zhao C, Li X, Ma Z, Li X, Zhang Y. LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/β-catenin signaling pathway. Oncol Rep. 2014:1–7. doi: 10.3892/or.2014.3047. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Cai J, Han Y, Chen J, Huang Z-P, Chen C, Cai Y, Huang H, Yang Y, Liu Y, et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130:1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Zhang H, Mei Y, Wu M. Reciprocal Regulation of HIF-1α and LincRNA-p21 Modulates the Warburg Effect. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Yoon J-H, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 Suppresses Target mRNA Translation. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger ST, Kenzelmann-Broz D, Jung H, Attardi LD, Rinn JL. Integrative genomic analysis reveals widespread enhancer regulation by p53 in response to DNA damage. Nucleic Acids Research. 2015;43:4447–4462. doi: 10.1093/nar/gkv284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wong C-H, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.