Abstract
Genome-wide association studies (GWAS) of asthma have yielded exciting results and identified novel risk alleles and loci. But, like other common complex diseases, asthma-associated alleles have small effect sizes and account for little of the prevalence of asthma. In this review, I discuss the limitations of GWAS approaches and the major challenges facing geneticists in the post-GWAS era and propose alternative strategies to address these challenges. In particular, I propose that focusing on genetic variations that influences gene expression and using cell models of gene–environment interactions in cell types that are relevant to asthma will allow us to more completely characterize the genetic architecture of asthma.
Keywords: asthma, genetic variation, gene-environment interaction, epigenomics, quantitative trait loci
Asthma is a heterogeneous disease that results from complex interactions between both genetic and nongenetic factors. In addition, the exact timing of specific exposures during critical developmental windows influences individual risk trajectories that ultimately lead to the development of asthma (reviewed in Reference 1). Given its complex etiology and the importance of early-life exposures, therefore, it is not surprising that estimates of asthma heritability from twin studies range from 35 to 70% (2, 3) and are inversely correlated with age of disease onset (4). Overall, these heritability estimates indicate that, on average, genetic variation among individuals accounts for one-half of the risk of asthma and that genetics plays a greater role in childhood-onset asthma. Despite this significant role of genetics, the importance of environmental exposures in asthma risk is evidenced by the dramatic rise in asthma prevalence in developed countries over the past 50 years (5–7) and the striking differences in asthma prevalence between farming and nonfarming children in central Europe (8–12). Thus, it is likely that the effects of genetic variation on asthma risk will vary among individuals raised in different environments and among those with specific exposures during key developmental windows.
In this review, I summarize results from genome-wide association studies (GWAS) and the limitations of that approach to gene discovery, discuss the challenges facing us in the post-GWAS era and the importance of studying gene–environment interactions, and suggest additional strategies for more comprehensively characterizing the genetic architecture of asthma.
GWAS of Asthma
Given the clinical heterogeneity and important role of environmental exposures in the development of asthma, it is actually remarkable that GWAS in very large samples of children and adults have identified asthma-associated single nucleotide polymorphisms (SNPs) at genome-wide levels of significance (P < 10−8), nearly all of which have been replicated across studies (13–18). The results of these GWAS have highlighted the importance of genetic variants in or near genes that were already implicated in asthma or allergic diseases (e.g., TSLP, IL33, SMAD3, and HLA region genes), and also led to the discovery of novel loci, such as the 17q12–21 locus including the ORMDL3 and GSDML genes and the CDHR3 gene, which was shown subsequently to be the elusive receptor for rhinovirus (RV)-C (18, 19). Overall, GWAS have yielded exciting results and have revealed unexpected genes with variants that contribute to asthma risk. Yet the contribution of individual variants to risk is small, with odds ratios (ORs) of around 1.2 for even the most significant loci (for examples, see References 13, 14, and 20), and the combined risk across all associated variants accounts for little of the asthma prevalence (13).
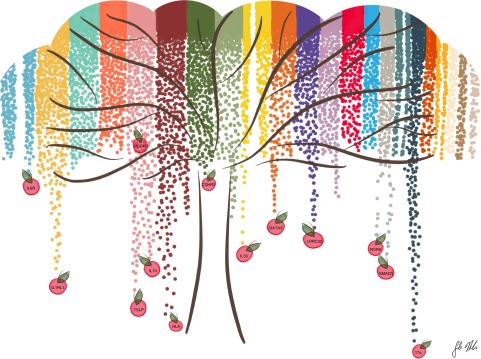
The observation that the variants identified by GWAS account for a small fraction of the heritability for most complex diseases, including asthma, was initially referred to as “missing heritability” (21, 22) and then as “hidden heritability” (23). This “missingness” was attributed largely to limitations of GWAS. In particular, the genotyping platforms used for GWAS interrogate common SNPs, but not rare variants or copy number variants. In addition, the statistical models used in GWAS are simplistic and do not take into account polygenic models or interactions, such as gene–gene (i.e., epistasis) and gene–environment interactions. Furthermore, controlling the false-discovery rate in GWAS that test for associations with several million SNPs requires exceedingly small P values (<10−8) to declare significance, revealing associations that have been referred to as the “low-hanging fruit” (Figure 1). However, the thousands of SNPs with P values lower than 10−4, for example, also include many true associations. Sorting through this “mid-hanging fruit” to differentiate the true from the false positives remains challenging (24). Although more recently the very concept of missing or hidden heritability has been disputed, instead attributing the appearance of “missingness” to underestimating the contribution of common variants to disease risk, overestimating heritability, and misunderstanding of the term itself (25–28), nearly all investigators agree that there are more asthma risk variants to be discovered and that more comprehensive surveys of variation and more complex models are required to fully characterize the genetic risk profiles for complex phenotypes, such as asthma.
Figure 1.
Overview of results of genome-wide association studies (GWAS) for asthma. Figure is shown as a stylized inverted Manhattan plot, which was created on the basis of published data. Colors represent different autosomal chromosomes (1–22, left to right). Many asthma-associated loci have been discovered by GWAS. These associations are generally robust across ethnic and racial groups and represent the “low-hanging fruit” (shown as apples). However, separating signal from noise among the variants with small P values that do not reach the genome-wide criteria for significance (i.e., the “mid-hanging fruit” among the branches, not shown) remains challenging. These loci likely include many exposure-specific associations and may be validated using cell culture models of gene–environment interactions. Figure drawn by S. Mozaffari.
Lessons Learned from GWAS
Although GWAS and metaanalyses of GWAS of asthma and allergic disease phenotypes will continue, the large number of published studies of common diseases can inform future research agendas for genetic studies. For example, there is overwhelming evidence from more than 1,000 GWAS of common diseases that (1) more than 90% of associated variants reside outside the protein coding regions of genes (29); (2) the locations of associated variants are enriched for DNase I hypersensitivity sites, which represent genomic regions of open chromatin that are poised for transcription (30); and (3) associated variants are more likely to be associated with gene expression levels (31–36). Overall, these data indicate that a significant portion of the genetic risk for common diseases is caused by variants that perturb gene expression. This conclusion is further supported by our recent genome-wide study of rare (<1%) and low-frequency (1–4%) putatively functional exonic variants in nearly 5,000 asthma cases and 5,000 control subjects (37). We designed this study to test the hypothesis that rare and low-frequency variants that alter the protein coding sequence of genes accounted for some of the missing heritability in asthma. Using an exome genotyping platform that included more than 60% of the variants present in whole genome sequences of a subset of the asthma cases, only modest associations with variants in just a few genes were discovered (ORs, 1.10–1.25, similar to those for common variants), nearly all of which were exclusive to specific ethnic or racial groups. This study suggested that protein-coding variants in these frequency ranges do not contribute to a significant portion of the heritability. Overall, the combined available data indicate that studies focusing on noncoding variants that are associated with the regulation gene expression or with epigenetic markers that regulate gene expression will yield important insights on the genetic architecture of asthma.
Gene–Environment Interactions (or Genotype-specific Responses to Environmental Exposures)
Given the important role of environmental risk factors in the development of asthma, and the prominence of regulatory variation among GWAS SNPs, I suggest that a significant portion of the genetic risk for asthma results from genotype-specific responses to environmental exposures throughout life, particular during early life and potentially beginning in utero. Thus, an individual’s risk of developing asthma may be mediated, in fact, by his or her genotype-specific responses to the many exposures that ultimately influence asthma risk. The following examples illustrate this concept with respect to two important early-life microbial exposures that influence asthma risk trajectories.
In some, but not all, studies, environmental exposures to bacterial products, such as LPS and its bioactive moiety endotoxin, have an inverse dose–response relationship to the prevalence of asthma and allergic diseases (reviewed in Reference 38). LPS signals through a complex receptor that includes CD14, and a polymorphism in the promoter of the CD14 gene (rs2569190; C to T at position 159) is associated with serum levels of soluble CD14 (39), suggesting that this SNP regulates expression of the CD14 gene. In studies of four different populations, the C allele at this SNP was protective against asthma and allergic disease among children with high exposure to endotoxin (such as those living on traditional farms or with pets in their homes), whereas the C allele conferred a risk of asthma or allergic disease among children living in homes with low endotoxin exposure (40–46). Moreover, the age at the time of exposure may be critical because in one study, the effects of this genotype-by-environment interaction were observed only when exposure occurred in early life (47). These data support the idea that neither genotype nor exposure alone is sufficient to influence the risk of asthma or allergic disease but that an interaction between the two is required to provide protection or risk and, importantly, that genotype-specific effects can vary in different environments (38).
Viral-associated respiratory wheezing illness in the first few years of life is often the earliest clinical manifestation of asthma, and wheezing with respiratory syncytial virus (RSV), RV, and overall number of viral infections are significant predictors of asthma diagnosis later in childhood (48–52). But not all children wheeze during respiratory viral infections, and among those with wheezing illness, only some develop asthma, suggesting that the host genotype plays a role in both. To explore this possibility, we studied children in the Childhood Origins of ASThma birth cohort from Madison, Wisconsin (53), in whom wheezing illness with both RSV and RV in the first 3 years of life was a significant predictor of asthma by age 6 years (51). Genetic variation at the 17q12–21 asthma locus was shown previously to be associated specifically with childhood-onset asthma (13, 54, 55), and with larger genotype-specific effect sizes among children with early-life exposure to environmental tobacco smoke (54, 56, 57), respiratory infections (58), or exacerbations (18, 59). We asked if genotype at this important asthma locus was associated specifically with RV- or RSV-wheezing illness during the first 3 years of life in the Childhood Origins of ASThma children. Indeed, genotype at SNP rs7216389 was a strong predictor of the number of childhood wheezing illnesses during an RV infection but, surprisingly, not with wheezing illness during an RSV infection (60). The number of T alleles at this SNP was associated with both the occurrence of RV wheezing illness (P = 0.01) and the number of RV wheezing illnesses (P < 0.001), but not with either occurrence (P = 0.22) or number (P = 0.54) of RSV wheezing illnesses, during the first 3 years of life. Strikingly, the association between the T allele and asthma, which has been associated robustly with childhood-onset asthma (discussed above), was observed only in the children who wheezed with RV infection in the first 3 years of life (P = 0.006); there was no association between genotype at this SNP and asthma among children who did not wheeze with an RV infection (P = 0.70), yielding an interaction P value of 0.004. This interaction was replicated in a second cohort of children from Denmark (60). Overall, the OR for developing asthma was significantly higher in children with both the high-risk genotype and RV wheezing illness in the first 3 years of life than in children with either the high-risk genotype or RV wheezing illness by itself (Table 1). Thus, stratifying by an “exposure” (RV wheezing illness) that influences asthma risk significantly impacts the 17q12–21 genotype-specific ORs and illustrates the importance of considering the joint effects of genetic and environmental risk factors. Presumably, other asthma risk alleles will also have larger effects among subsets of those with asthma with common causes.
Table 1.
ORs for developing asthma by age 6 years in the Childhood Origins of ASThma cohort, caused by genotype at the 17q SNP rs7216389 and wheezing illness with RV infection in the first 3 years of life
| Risk Factor | OR for Asthma (95% CI) |
|---|---|
| Genotype TT | 2.3 (1.0–5.2) |
| RV-wheezing illness | 5.2 (2.8–9.9) |
| RV-wheezing illness × genotype TT | 26.1 (5.1–133.0) |
Definition of abbreviations: CI = confidence interval; OR = odds ratio; RV = rhinovirus; SNP = single nucleotide polymorphism.
Interaction P = 0.004. Data from Reference 60.
Future Directions
The complexity of asthma and allergic disease phenotypes makes studies of cause and pathogenesis challenging, but as a disease model it affords unparalleled opportunities to study gene–environment interactions (1). The sheer number of known environmental risk factors and the importance of the timing of these exposures during development make asthma an exceptional model for understanding genotype-specific responses to key exposures, as illustrated in the examples above for CD14 genotype-endotoxin and 17q12–21 genotype-RV wheezing illness interactions. Although large-scale GWAS-type studies of gene–environment interactions may be impractical, or even impossible, cell models of gene–environment interactions provide a flexible approach to studying global transcriptional and epigenetic responses (e.g., References 61–63) as well as genotype-specific transcriptional and epigenetic responses (e.g., References 64–69) to key exposures in relevant cell types.
Such cell-based models will directly address the post-GWAS challenges discussed above. First, they will yield functional insights into many known asthma loci by establishing their role as regulatory SNPs in specific tissues or cells and in response to key exposures, as well as the genes or epigenetic marks that they influence. This would significantly extend previous studies of expression quantitative trait loci (QTLs) (70, 71) to include response expression QTLs and additional types of QTLs, such as methylation QTLs (68), DNase I hypersensitivity QTLs (69), and so on. Second, because the genotype effects for many regulatory variants may be context or environment specific, these variants may fail to reach genome-wide thresholds of significance in typical GWAS. Characterizing the variants that regulate the transcriptional or epigenetic response to asthma-relevant exposures in cell models will provide a framework for prioritizing the “mid-hanging fruit” identified by GWAS. In addition, identifying regulatory variants in cells and tissues that are relevant to asthma and in response to exposures that are associated with asthma will ultimately yield a set of genome-wide variants for ‘functional GWAS’ and novel gene discovery. The smaller set of variants (compared with the millions of SNPs currently used in GWAS) combined with their known functional effects increases the power to discover disease-associated variants, possibly in smaller samples than those currently required for GWAS (72). Ultimately, integrating these multiomic data sets from cell models of gene–environment interactions will yield a more complete picture of the genetic, epigenetic, and transcriptional architecture of asthma.
Finally, discoveries of genetic variation associated with asthma either through GWAS or through gene–environment interactions will ultimately provide insights into the genes that contribute to the pathogenesis and identify potential therapeutic targets. Thus, linking associated variation to genes and pathways, and the biological follow-up of their role in asthma, are critical next steps that will facilitate the translation of genetic study results to clinical care.
Acknowledgments
Acknowledgment
The author thanks S. Mozaffari, J. Nicodemus-Johnson, and M. Stein for helpful discussions and comments on drafts of this manuscript.
Footnotes
Supported by National Institutes of Health grants R01 HL085197, U19 AI095230, P01 HL070831, and P01 AI106683.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Ober C, Vercelli D. Gene-environment interactions in human disease: nuisance or opportunity? Trends Genet. 2011;27:107–115. doi: 10.1016/j.tig.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duffy DL, Martin NG, Battistutta D, Hopper JL, Mathews JD. Genetics of asthma and hay fever in Australian twins. Am Rev Respir Dis. 1990;142:1351–1358. doi: 10.1164/ajrccm/142.6_Pt_1.1351. [DOI] [PubMed] [Google Scholar]
- 3.Nieminen MM, Kaprio J, Koskenvuo M. A population-based study of bronchial asthma in adult twin pairs. Chest. 1991;100:70–75. doi: 10.1378/chest.100.1.70. [DOI] [PubMed] [Google Scholar]
- 4.Thomsen SF, Duffy DL, Kyvik KO, Backer V. Genetic influence on the age at onset of asthma: a twin study. J Allergy Clin Immunol. 2010;126:626–630. doi: 10.1016/j.jaci.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 6.Beasley R. The burden of asthma with specific reference to the United States. J Allergy Clin Immunol. 2002;109:S482–S489. doi: 10.1067/mai.2002.122716. [DOI] [PubMed] [Google Scholar]
- 7.Burr ML, Butland BK, King S, Vaughan-Williams E. Changes in asthma prevalence: two surveys 15 years apart. Arch Dis Child. 1989;64:1452–1456. doi: 10.1136/adc.64.10.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun-Fahrländer C, Gassner M, Grize L, Neu U, Sennhauser FH, Varonier HS, Vuille JC, Wüthrich B Swiss Study on Childhood Allergy and Respiratory Symptoms with Respect to Air Pollution. Prevalence of hay fever and allergic sensitization in farmer’s children and their peers living in the same rural community. SCARPOL team. Clin Exp Allergy. 1999;29:28–34. doi: 10.1046/j.1365-2222.1999.00479.x. [DOI] [PubMed] [Google Scholar]
- 9.Ege MJ, Frei R, Bieli C, Schram-Bijkerk D, Waser M, Benz MR, Weiss G, Nyberg F, van Hage M, Pershagen G, et al. PARSIFAL Study team. Not all farming environments protect against the development of asthma and wheeze in children. J Allergy Clin Immunol. 2007;119:1140–1147. doi: 10.1016/j.jaci.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 10.Riedler J, Braun-Fahrländer C, Eder W, Schreuer M, Waser M, Maisch S, Carr D, Schierl R, Nowak D, von Mutius E ALEX Study Team. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. 2001;358:1129–1133. doi: 10.1016/S0140-6736(01)06252-3. [DOI] [PubMed] [Google Scholar]
- 11.Riedler J, Eder W, Oberfeld G, Schreuer M. Austrian children living on a farm have less hay fever, asthma and allergic sensitization. Clin Exp Allergy. 2000;30:194–200. doi: 10.1046/j.1365-2222.2000.00799.x. [DOI] [PubMed] [Google Scholar]
- 12.Von Ehrenstein OS, Von Mutius E, Illi S, Baumann L, Böhm O, von Kries R. Reduced risk of hay fever and asthma among children of farmers. Clin Exp Allergy. 2000;30:187–193. doi: 10.1046/j.1365-2222.2000.00801.x. [DOI] [PubMed] [Google Scholar]
- 13.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO GABRIEL Consortium. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, Himes BE, Levin AM, Mathias RA, Hancock DB, et al. Mexico City Childhood Asthma Study (MCAAS); Children’s Health Study (CHS) and HARBORS study; Genetics of Asthma in Latino Americans (GALA) Study, Study of Genes-Environment and Admixture in Latino Americans (GALA2) and Study of African Americans, Asthma, Genes & Environments (SAGE); Childhood Asthma Research and Education (CARE) Network; Childhood Asthma Management Program (CAMP); Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-Ethnicity (SAPPHIRE); Genetic Research on Asthma in African Diaspora (GRAAD) Study. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramasamy A, Kuokkanen M, Vedantam S, Gajdos ZK, Couto Alves A, Lyon HN, Ferreira MA, Strachan DP, Zhao JH, Abramson MJ, et al. Australian Asthma Genetics Consortium Collaborators. Genome-wide association studies of asthma in population-based cohorts confirm known and suggested loci and identify an additional association near HLA. PLoS One. 2012;7:e44008. doi: 10.1371/journal.pone.0044008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan YI, Shrine NR, Soler Artigas M, Wain LV, Blakey JD, Moffatt MF, Bush A, Chung KF, Cookson WO, Strachan DP, et al. Australian Asthma Genetics Consortium. Genome-wide association study to identify genetic determinants of severe asthma. Thorax. 2012;67:762–768. doi: 10.1136/thoraxjnl-2011-201262. [DOI] [PubMed] [Google Scholar]
- 17.Ferreira MA, Matheson MC, Tang CS, Granell R, Ang W, Hui J, Kiefer AK, Duffy DL, Baltic S, Danoy P, et al. Australian Asthma Genetics Consortium Collaborators. Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J Allergy Clin Immunol. 2014;133:1564–1571. doi: 10.1016/j.jaci.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bønnelykke K, Sleiman P, Nielsen K, Kreiner-Møller E, Mercader JM, Belgrave D, den Dekker HT, Husby A, Sevelsted A, Faura-Tellez G, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–55. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 19.Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, Palmenberg AC, Gern JE. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci USA. 2015;112:5485–5490. doi: 10.1073/pnas.1421178112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bønnelykke K, Matheson MC, Pers TH, Granell R, Strachan DP, Alves AC, Linneberg A, Curtin JA, Warrington NM, Standl M, et al. Australian Asthma Genetics Consortium (AAGC); EArly Genetics and Lifecourse Epidemiology (EAGLE) Consortium. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat Genet. 2013;45:902–906. doi: 10.1038/ng.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson G. Hints of hidden heritability in GWAS. Nat Genet. 2010;42:558–560. doi: 10.1038/ng0710-558. [DOI] [PubMed] [Google Scholar]
- 24.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era--concepts and misconceptions. Nat Rev Genet. 2008;9:255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 26.Clarke AJ, Cooper DN. GWAS: heritability missing in action? Eur J Hum Genet. 2010;18:859–861. doi: 10.1038/ejhg.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golan D, Lander ES, Rosset S. Measuring missing heritability: inferring the contribution of common variants. Proc Natl Acad Sci USA. 2014;111:E5272–E5281. doi: 10.1073/pnas.1419064111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: genetic interactions create phantom heritability. Proc Natl Acad Sci USA. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gusev A, Lee SH, Trynka G, Finucane H, Vilhjálmsson BJ, Xu H, Zang C, Ripke S, Bulik-Sullivan B, Stahl E, et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium; SWE-SCZ Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; SWE-SCZ Consortium. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torres JM, Gamazon ER, Parra EJ, Below JE, Valladares-Salgado A, Wacher N, Cruz M, Hanis CL, Cox NJ. Cross-tissue and tissue-specific eQTLs: partitioning the heritability of a complex trait. Am J Hum Genet. 2014;95:521–534. doi: 10.1016/j.ajhg.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RK, Hua Y, Gueroussov S, Najafabadi HS, Hughes TR, et al. RNA splicing: the human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Igartua C, Myers RA, Mathias RA, Pino-Yanes M, Eng C, Graves PE, Levin AM, Del-Rio-Navarro BE, Jackson DJ, Livne OE, et al. Ethnic-specific associations of rare and low-frequency DNA sequence variants with asthma. Nat Commun. 2015;6:5965. doi: 10.1038/ncomms6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson A, Martinez FD. The role of lipopolysaccharide in the development of atopy in humans. Clin Exp Allergy. 2010;40:209–223. doi: 10.1111/j.1365-2222.2009.03391.x. [DOI] [PubMed] [Google Scholar]
- 39.Baldini M, Lohman IC, Halonen M, Erickson RP, Holt PG, Martinez FDA. A polymorphism* in the 5′ flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol. 1999;20:976–983. doi: 10.1165/ajrcmb.20.5.3494. [DOI] [PubMed] [Google Scholar]
- 40.Simpson A, John SL, Jury F, Niven R, Woodcock A, Ollier WE, Custovic A. Endotoxin exposure, CD14, and allergic disease: an interaction between genes and the environment. Am J Respir Crit Care Med. 2006;174:386–392. doi: 10.1164/rccm.200509-1380OC. [DOI] [PubMed] [Google Scholar]
- 41.Smit LA, Bongers SI, Ruven HJ, Rijkers GT, Wouters IM, Heederik D, Omland Ø, Sigsgaard T. Atopy and new-onset asthma in young Danish farmers and CD14, TLR2, and TLR4 genetic polymorphisms: a nested case-control study. Clin Exp Allergy. 2007;37:1602–1608. doi: 10.1111/j.1365-2222.2007.02831.x. [DOI] [PubMed] [Google Scholar]
- 42.Smit LA, Siroux V, Bouzigon E, Oryszczyn MP, Lathrop M, Demenais F, Kauffmann F Epidemiological Study on the Genetics and Environment of Asthma, Bronchial Hyperresponsiveness, and Atopy (EGEA) Cooperative Group. CD14 and toll-like receptor gene polymorphisms, country living, and asthma in adults. Am J Respir Crit Care Med. 2009;179:363–368. doi: 10.1164/rccm.200810-1533OC. [DOI] [PubMed] [Google Scholar]
- 43.Bottema RW, Reijmerink NE, Kerkhof M, Koppelman GH, Stelma FF, Gerritsen J, Thijs C, Brunekreef B, van Schayck CP, Postma DS. Interleukin 13, CD14, pet and tobacco smoke influence atopy in three Dutch cohorts: the allergenic study. Eur Respir J. 2008;32:593–602. doi: 10.1183/09031936.00162407. [DOI] [PubMed] [Google Scholar]
- 44.Leynaert B, Guilloud-Bataille M, Soussan D, Benessiano J, Guénégou A, Pin I, Neukirch F. Association between farm exposure and atopy, according to the CD14 C-159T polymorphism. J Allergy Clin Immunol. 2006;118:658–665. doi: 10.1016/j.jaci.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 45.Gern JE, Reardon CL, Hoffjan S, Nicolae D, Li Z, Roberg KA, Neaville WA, Carlson-Dakes K, Adler K, Hamilton R, et al. Effects of dog ownership and genotype on immune development and atopy in infancy. J Allergy Clin Immunol. 2004;113:307–314. doi: 10.1016/j.jaci.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 46.Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, Braun-Fahrländer C, Nowak D, Martinez FD Allergy And Endotoxin Alex Study Team. Opposite effects of CD 14/-260 on serum IgE levels in children raised in different environments. J Allergy Clin Immunol. 2005;116:601–607. doi: 10.1016/j.jaci.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Lau MY, Dharmage SC, Burgess JA, Lowe AJ, Lodge CJ, Campbell B, Matheson MC. CD14 polymorphisms, microbial exposure and allergic diseases: a systematic review of gene-environment interactions. Allergy. 2014;69:1440–1453. doi: 10.1111/all.12454. [DOI] [PubMed] [Google Scholar]
- 48.Sigurs N, Bjarnason R, Sigurbergsson F, Kjellman B. Respiratory syncytial virus bronchiolitis in infancy is an important risk factor for asthma and allergy at age 7. Am J Respir Crit Care Med. 2000;161:1501–1507. doi: 10.1164/ajrccm.161.5.9906076. [DOI] [PubMed] [Google Scholar]
- 49.Stein RT, Sherrill D, Morgan WJ, Holberg CJ, Halonen M, Taussig LM, Wright AL, Martinez FD. Respiratory syncytial virus in early life and risk of wheeze and allergy by age 13 years. Lancet. 1999;354:541–545. doi: 10.1016/S0140-6736(98)10321-5. [DOI] [PubMed] [Google Scholar]
- 50.Kotaniemi-Syrjänen A, Vainionpää R, Reijonen TM, Waris M, Korhonen K, Korppi M. Rhinovirus-induced wheezing in infancy: the first sign of childhood asthma? J Allergy Clin Immunol. 2003;111:66–71. doi: 10.1067/mai.2003.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, Printz MC, Lee WM, Shult PA, Reisdorf E, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178:667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonnelykke K, Vissing NH, Sevelsted A, Johnston SL, Bisgaard H. Association between respiratory infections in early life and later asthma is independent of virus type. J Allergy Clin Immunol. 2015;136:81–86.e84. doi: 10.1016/j.jaci.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lemanske RF., Jr The childhood origins of asthma (COAST) study. Pediatr Allergy Immunol. 2002;13:38–43. doi: 10.1034/j.1399-3038.13.s.15.8.x. [DOI] [PubMed] [Google Scholar]
- 54.Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, Chateigner N, Gormand F, Just J, Le Moual N, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–1994. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 55.Halapi E, Gudbjartsson DF, Jonsdottir GM, Bjornsdottir US, Thorleifsson G, Helgadottir H, Williams C, Koppelman GH, Heinzmann A, Boezen HM, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010;18:902–908. doi: 10.1038/ejhg.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flory JH, Sleiman PM, Christie JD, Annaiah K, Bradfield J, Kim CE, Glessner J, Imielinski M, Li H, Frackelton EC, et al. 17q12-21 variants interact with smoke exposure as a risk factor for pediatric asthma but are equally associated with early-onset versus late-onset asthma in North Americans of European ancestry. J Allergy Clin Immunol. 2009;124:605–607. doi: 10.1016/j.jaci.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 57.van der Valk RJ, Duijts L, Kerkhof M, Willemsen SP, Hofman A, Moll HA, Smit HA, Brunekreef B, Postma DS, Jaddoe VW, et al. Interaction of a 17q12 variant with both fetal and infant smoke exposure in the development of childhood asthma-like symptoms. Allergy. 2012;67:767–774. doi: 10.1111/j.1398-9995.2012.02819.x. [DOI] [PubMed] [Google Scholar]
- 58.Smit LA, Bouzigon E, Pin I, Siroux V, Monier F, Aschard H, Bousquet J, Gormand F, Just J, Le Moual N, et al. EGEA Cooperative Group. 17q21 variants modify the association between early respiratory infections and asthma. Eur Respir J. 2010;36:57–64. doi: 10.1183/09031936.00154509. [DOI] [PubMed] [Google Scholar]
- 59.Bisgaard H, Bønnelykke K, Sleiman PM, Brasholt M, Chawes B, Kreiner-Møller E, Stage M, Kim C, Tavendale R, Baty F, et al. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am J Respir Crit Care Med. 2009;179:179–185. doi: 10.1164/rccm.200809-1436OC. [DOI] [PubMed] [Google Scholar]
- 60.Calışkan M, Bochkov YA, Kreiner-Møller E, Bønnelykke K, Stein MM, Du G, Bisgaard H, Jackson DJ, Gern JE, Lemanske RF, Jr, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Solberg OD, Ostrin EJ, Love MI, Peng JC, Bhakta NR, Hou L, Nguyen C, Solon M, Nguyen C, Barczak AJ, et al. Airway epithelial miRNA expression is altered in asthma. Am J Respir Crit Care Med. 2012;186:965–974. doi: 10.1164/rccm.201201-0027OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, Ellwanger A, Sidhu SS, Dao-Pick TP, Pantoja C, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA. 2007;104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nicodemus-Johnson J, Naughton KA, Herman C, Sudi J, Hogarth DK, Naureckas ET, Nicolae DL, Sperling AI, Solway J, White SR, Ober C.Genome-wide methylation study identifies an IL-13 induced epigenetic signature in asthmatic airwaysAm J Respir Crit Care MedIn press [DOI] [PMC free article] [PubMed]
- 64.Çalışkan M, Baker SW, Gilad Y, Ober C. Host genetic variation influences gene expression response to rhinovirus infection. PLoS Genet. 2015;11:e1005111. doi: 10.1371/journal.pgen.1005111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barreiro LB, Tailleux L, Pai AA, Gicquel B, Marioni JC, Gilad Y. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc Natl Acad Sci USA. 2012;109:1204–1209. doi: 10.1073/pnas.1115761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Romanoski CE, Lee S, Kim MJ, Ingram-Drake L, Plaisier CL, Yordanova R, Tilford C, Guan B, He A, Gargalovic PS, et al. Systems genetics analysis of gene-by-environment interactions in human cells. Am J Hum Genet. 2010;86:399–410. doi: 10.1016/j.ajhg.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye CJ, Feng T, Kwon HK, Raj T, Wilson MT, Asinovski N, McCabe C, Lee MH, Frohlich I, Paik HI, et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science. 2014;345:1254665. doi: 10.1126/science.1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banovich NE, Lan X, McVicker G, van de Geijn B, Degner JF, Blischak JD, Roux J, Pritchard JK, Gilad Y. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 2014;10:e1004663. doi: 10.1371/journal.pgen.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McVicker G, van de Geijn B, Degner JF, Cain CE, Banovich NE, Raj A, Lewellen N, Myrthil M, Gilad Y, Pritchard JK. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo W, Obeidat M, Di Narzo AF, Chen R, Sin DD, Paré PD, Hao K.Airway epithelial expression quantitative trait loci reveal genes underlying asthma and other airway diseases Am J Respir Cell Mol BiolIn press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hao K, Bossé Y, Nickle DC, Paré PD, Postma DS, Laviolette M, Sandford A, Hackett TL, Daley D, Hogg JC, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Das K, Li J, Wang Z, Tong C, Fu G, Li Y, Xu M, Ahn K, Mauger D, Li R, et al. A dynamic model for genome-wide association studies. Hum Genet. 2011;129:629–639. doi: 10.1007/s00439-011-0960-6. [DOI] [PMC free article] [PubMed] [Google Scholar]