Abstract
Anopheles melas is a member of the recently diverged An. gambiae species complex, a model for speciation studies, and is a locally important malaria vector along the West-African coast where it breeds in brackish water. A recent population genetic study of An. melas revealed species-level genetic differentiation between three population clusters. An. melas West extends from The Gambia to the village of Tiko, Cameroon. The other mainland cluster, An. melas South, extends from the southern Cameroonian village of Ipono to Angola. Bioko Island, Equatorial Guinea An. melas populations are genetically isolated from mainland populations. To examine how genetic differentiation between these An. melas forms is distributed across their genomes, we conducted a genome-wide analysis of genetic differentiation and selection using whole genome sequencing data of pooled individuals (Pool-seq) from a representative population of each cluster. The An. melas forms exhibit high levels of genetic differentiation throughout their genomes, including the presence of numerous fixed differences between clusters. Although the level of divergence between the clusters is on a par with that of other species within the An. gambiae complex, patterns of genome-wide divergence and diversity do not provide evidence for the presence of pre- and/or postmating isolating mechanisms in the form of speciation islands. These results are consistent with an allopatric divergence process with little or no introgression.
Keywords: Anopheles melas, Anopheles gambiae, malaria, population genomics, Pool-seq
The Anopheles gambiae complex of African malaria mosquitoesis a model system for the study of speciation (Fontaine et al. 2015; Mallet et al. 2015; Neafsey et al. 2015; Nosil 2012). This is partly due to its importance to human health, but also because varying levels of reproductive isolation and introgression are found between its member species (Besansky et al. 1994; Davidson 1962; Fontaine et al. 2015; Lanzaro and Lee 2013; Marsden et al. 2011; Powell et al. 1999; Slotman et al. 2004, 2005a,b; Weetman et al. 2014), chromosomal and molecular forms occur within species (Coluzzi et al. 2002; della Torre et al. 2001; Favia et al. 2001; Gentile et al. 2001; White et al. 2011), and contrasting patterns of intraspecific population structure have been observed between species (Deitz et al. 2012; Donnelly and Townson 2000; Lehman et al. 2003; Loaiza et al. 2012). The recent evolutionary analyses of 16 Anopheles genomes highlighted the role of adaptive introgression in the divergence of the An. gambiae complex (Clarkson et al. 2014; Fontaine et al. 2015; Norris et al. 2015), and how biological factors involved in their capacity to vector human malaria parasites have influenced the evolution of these species (Neafsey et al. 2015).
Eight species have now been formerly described within the An. gambiae complex, including two recent additions: An. coluzzii, formerly An. gambiae M molecular form, and An. amharicus, formerly An. quadriannulatus B (Coetzee et al. 2013). The elevation of the An. gambiae M form to species rank was based on ecological divergence, assortative mating (della Torre et al. 2001; Simard et al. 2009; Tripet et al. 2005; Aboagye-Antwi et al. 2015), and genetic divergence that appears to be limited to several small regions of the genome (Turner et al. 2005; White et al. 2010). The description of An. coluzzii therefore broke with the tradition of describing new species in the complex based on the presence of hybrid sterility (Davidson 1962; Hunt et al. 1998), as hybrids between An. gambiae and An. coluzzii are fully fertile (Diabaté et al. 2007). Thus, the description of An. coluzzii is aligned more with a genotypic cluster species concept (Mallet 1995) rather than a biological species concept (Mayr 1970).
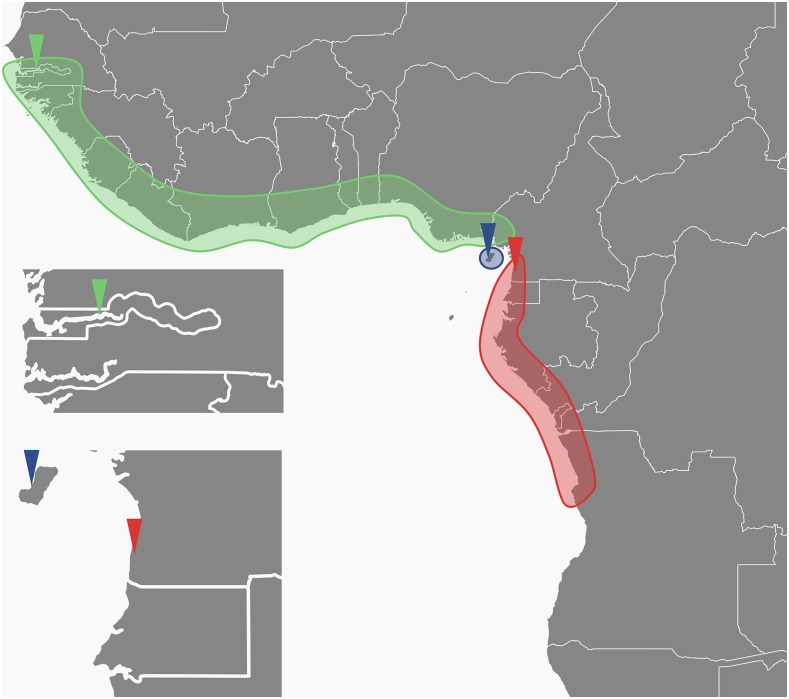
A recent study on the population structure of An. melas throughout its range uncovered species-level genetic divergence between three population clusters (Deitz et al. 2012). An. melas is distributed along the west coast of Africa as its larval ecology is tied to brackish water, mangrove forests, and salt marshes. Nonetheless, it is an important vector of human malaria where it is found (Bryan et al. 1987; Caputo et al. 2008), with the average number of malaria infective An. melas bites/person/year sometimes reaching 130 (Overgaard et al. 2012). Coluzzi et al. (2002) found that some chromosomal inversions were nonrandomly distributed between An. melas populations, suggesting the presence of some reproductive barriers. Deitz et al. (2012) showed that An. melas is in fact divided into three genetic clusters that appear to be mostly isolated from each other. Two of these clusters are distributed on the African mainland: An. melas West ranges from The Gambia to Northwest Cameroon, and An. melas South ranges from Southeast Cameroon to Angola. A third cluster, An. melas Bioko, is limited to Bioko Island, Equatorial Guinea, located approximately 40 km off the Cameroonian coast (Figure 1).
Figure 1.
This map of West Africa illustrates the distributions of An. melas genetic clusters. Ranges of An. melas West (green), South (red), and Bioko (blue) are shown as shaded regions. Triangles show the sample locations of An. melas populations used to represent each An. melas genetic cluster. The top inset shows the collection location of Ballingho, The Gambia (green triangle, An. melas West), and the bottom inset shows the collection locations of Arena Blanca, Bioko Island, Equatorial Guinea (blue triangle, An. melas Bioko) and Ipono, Cameroon (red triangle, An. melas South).
No mtDNA haplotypes are shared between An. melas clusters, and microsatellite data indicates almost complete genetic isolation, with the exception of limited introgression into An. melas West from the South and Bioko, which was identified through a Bayesian analysis of population structure. Additionally, the level of genetic divergence (FST) between An. melas West and South equaled or exceeded levels previously observed between An. gambiae and An. arabiensis (Slotman et al. 2005a; Fontaine et al. 2015). Interestingly, An. melas West and South populations are only separated by approximately 190 km of unsampled terrain along the Cameroonian coast. The high level of isolation of the An. melas Bioko Island population is also remarkable given the short distance to the mainland, and the very low level of genetic differentiation between Bioko Island and mainland populations of both An. gambiae and An. coluzzii (Moreno et al. 2007; Deitz et al. 2012).
An analysis of the demographic history of An. melas populations using approximate Bayesian computation analysis indicated that a larger ancestral An. melas population split into two mainland clusters through a vicariance event sometime during the last several hundred thousand years. Similarly, An. melas Bioko was once connected to An. melas West populations, but became isolated around 90,000 years before the present day, presumably due to rising sea levels (Deitz et al. 2012).
In the present study, we used a whole-genome, pooled-population sequencing (Pool-seq) approach (Schlötterer et al. 2014) to examine genome-wide patterns of diversity within, and divergence between, a representative population sample of An. melas West, South, and Bioko. Such an analysis may reveal whether the geographically isolated forms of An. melas harbor any genetically highly diverged regions of the genomes, similar to those that have been tied to premating isolation between An. gambiae s.s. and An. coluzzii (Aboagye-Antwi et al. 2015). The genome-wide single nucleotide polymorphism (SNP) data show that An. melas population clusters have high levels of genome-wide genetic differentiation, as evidenced by numerous high-FST and fixed SNPs in each population comparison. Genetic differentiation is particularly high on the X chromosome, which also carries the largest number of fixed differences. Additionally, we identified candidate regions under positive selection within each An. melas population cluster. A lack of narrow, highly differentiated genomic regions is consistent with allopatric divergence with little or no introgression.
Materials and Methods
Population genomic analysis
Pool-seq was performed on DNA of Anopheles melas females collected from Ballingho, The Gambia (N = 20), Ipono, Cameroon (N = 23), and Arena Blanca, Bioko Island, Equatorial Guinea (N = 20). These populations fall within An. melas West, South, and Bioko Island genetic clusters, respectively (Figure 1) (Deitz et al. 2012). Populations were chosen based upon the high quality of DNA available to create pooled libraries for sequencing, and the lack of gene flow observed between them and neighboring An. melas clusters (Deitz et al. 2012). Mosquito collection and DNA extraction methods are as described in Deitz et al. (2012). We pooled equal amounts of DNA from each individual, and sequencing libraries were constructed from 1.0 µg of pooled DNA. Covaris shearing (Fisher et al. 2011) was used to produce approximately 200 bp inserts for each library. Libraries were bar-coded, combined, and paired-end sequenced on a single lane of the Illumina HiSequation 2000 DNA sequencing platform.
Sequencing reads were trimmed to a minimum Phred quality score of 20 and a minimum length of 50 base pairs using Trimmomatic version 0.35 (Bolger et al. 2014), and then mapped to the An. gambiae PEST P4.3 genome assembly (Holt et al. 2002) using Stampy (Lunther and Goodson 2011) with a substitution rate = 0.02. Stampy is designed to map DNA sequencing reads to a divergent reference genome and has been previously used for this purpose in the An. gambiae species complex (Smith et al. 2015). Sequencing reads were mapped to the An. gambiae genome rather than the An. melas genome (Neafsey et al. 2015) because the former is assembled into chromosomes and at the present time the An. melas genome is comprised of 20,229 scaffolds (Giraldo-Calderón et al. 2015; Neafsey et al. 2015). No coordinate lift-over file is available to convert the coordinates of the An. melas scaffolds to those of the An. gambiae P4.3 chromosomes. As such, we aligned our data to the An. gambiae genome because it allowed us to interpret population genetic statistics in the context of chromosomal location. SAM alignment files were sorted, converted to BAM format, filtered to a minimum mapping quality value (MAPQ) of 20, and converted to pileup files using SAMtools version 0.1.19 (Li et al. 2009).
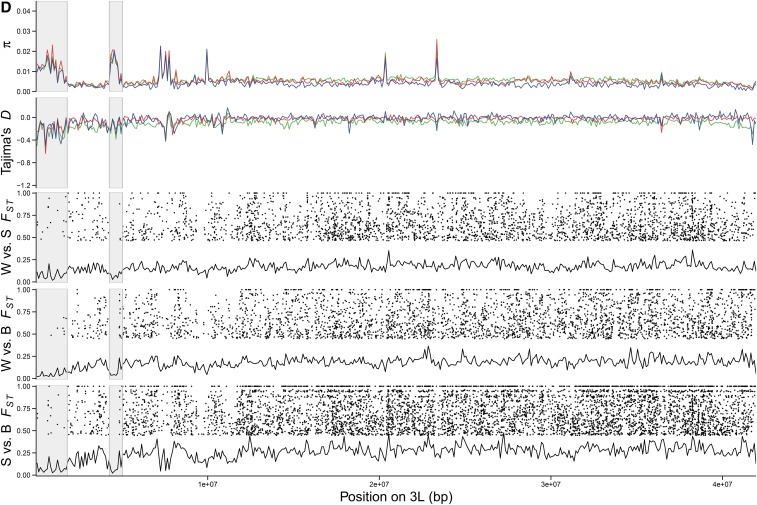
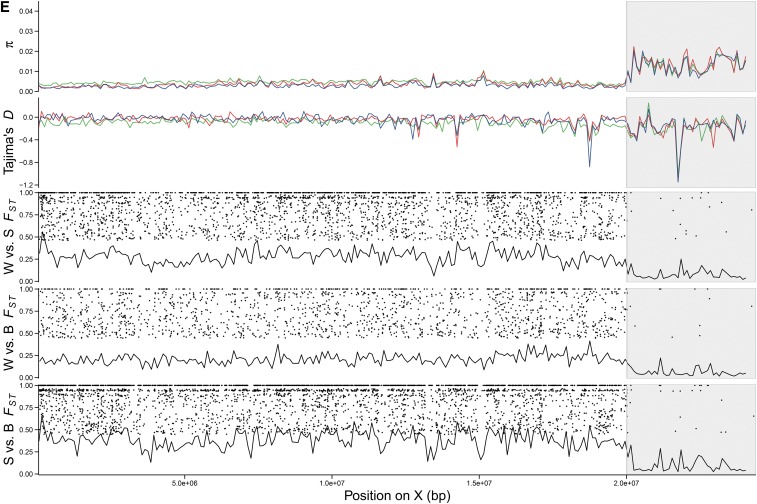
Pileup files were used to calculate nucleotide diversity (π, Nei and Li 1979) and Tajima’s D (Tajima 1989) using the PoPoolation package (Kofler et al. 2011a). Both statistics were calculated using 100 kb, nonoverlapping sliding-windows using a minimum sequence coverage of four reads and maximum coverage of 40. We required a minimum of two reads for each allele at a polymorphic site to retain the site for further analysis. The highly repetitive nature of heterochromatic genomic regions leads to inaccurate read mapping, which biases population genetic statistics. Heterochromatic regions of the An. gambiae reference genome (Sharakhova et al. 2010) were removed for the calculation of π, Tajima’s D, and FST summary statistics. Vertical gray bars in Figure 3 and Figure 4 highlight heterochromatic regions.
Figure 3.
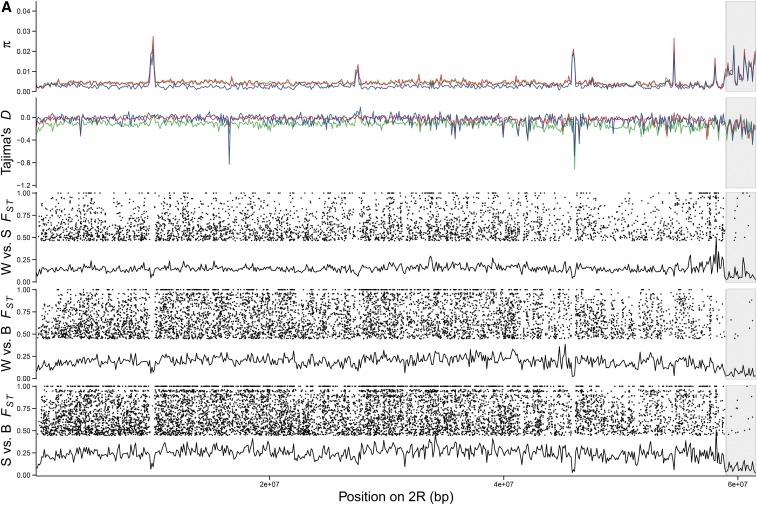
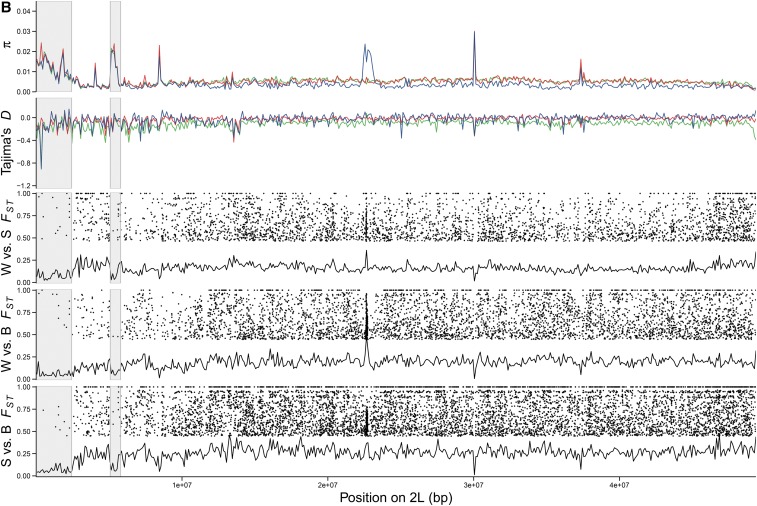
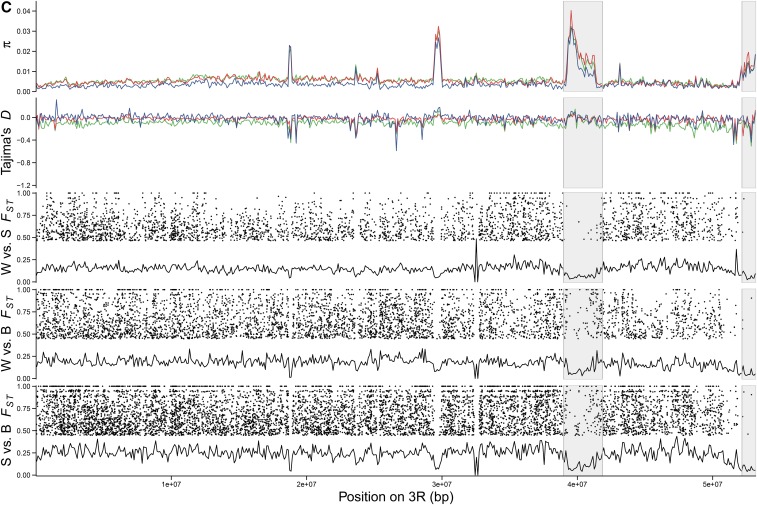
Line plots illustrate genome-wide nucleotide diversity (π) and Tajima’s D estimates for each chromosome arm and population based upon nonoverlapping, 100 kb sliding windows. (A–E) Green lines represent An. melas West, red lines represent An. melas South, and blue lines represent An. melas Bioko. FST plots are presented for each pairwise population comparison: An. melas West vs. South (W vs. S), West vs. Bioko (W vs. B), and South vs. Bioko (S vs. B). The solid line indicates FST calculated for nonoverlapping, 100 kb sliding windows, and dots indicate significant FST SNPs. Vertical gray bars indicate regions of heterochromatin in the An. gambiae genome that were not included in the calculation of summary statistics.
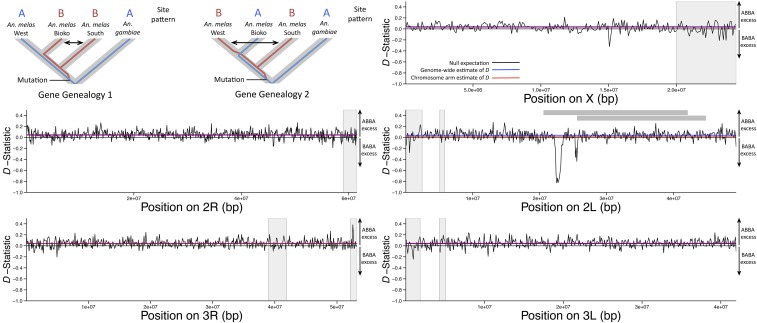
Figure 4.
Line plots illustrate genome-wide values of Patterson’s D-statistic for each chromosome arm for the An. melas population tree {[(West,Bioko)South]An. gambiae}. Positive values indicate an excess of ABBA patterns and negative values indicate a biased proportion of BABA patterns. Horizontal black lines indicate the null expectation, no ABBA or BABA excess (D = 0). Horizontal blue lines indicate the genome-wide estimate of Patterson’s D, and horizontal red lines indicate the average Patterson’s D for each chromosome arm. Vertical gray bars indicate regions of heterochromatin in the An. gambiae genome that were not included in the calculation of summary statistics. Horizontal gray bars in the chromosome arm 2L panel indicate the locations of the 2La/+ (top) and 2La2/+ (bottom) inversions. The top left panel demonstrates the ABBA vs. BABA patterns in the context of the An. melas tree, where an ABBA pattern indicates introgression between An. melas Bioko and South, and a BABA pattern indicates introgression between An. melas West and South (arrows).
Multiple pileup files were created with SAMtools version 0.1.19 (Li et al. 2009) and transformed into synchronized pileup files using PoPoolation2 (Kofler et al. 2011b). This program was then used to calculate pair-wise FST values for each SNP, and for 100 kb, nonoverlapping sliding-windows using a minimum sequencing depth of 30 × and a maximum equal to the top 2% of the sequencing depth distribution of each pool. Reads exceeding the top 2% sequencing depth threshold were excluded from our analysis to reduce the effect of sequencing and mapping bias.
We chose 30 × coverage to measure SNP and window-based FST because it allows us to have enough coverage in both populations in a comparison to provide a genome-wide distribution of informative loci for population genomic analysis, and have enough the power to detect significant differentiation. In our initial FST null distribution simulations, we found that coverage below this value incorporates a high level of variation in the allele frequency and FST estimates at a single locus. Thus, a high coverage threshold allows us to be confident that differences in read coverage between populations in a comparison is not biasing our FST calculation. We used a lower threshold for π and Tajima’s D (above) because these values are averaged over a 100 kb window and inaccuracy in estimates for individual loci should cancel out within each window and not introduce bias.
If significant SNPs fell within the bottom 5% of the Tajima’s D distribution in both populations in a pair-wise comparison (e.g., An. melas West and South), the SNP was subjected to gene ontology analyses. These analyses excluded SNPs and low Tajima’s D regions that fell inside regions of heterochromatin in the An. gambiae reference genome. SNPs were compared to the An. gambiae AgamP4.4 gene set (Holt et al. 2002; Sharakhova et al. 2007) to determine if they fell within a known gene exon. The molecular function, biological process, and protein class of these genes was determined using the Panther Classification System (Thomas et al. 2003; Mi et al. 2010).
To identify regions of introgression between An. melas forms, we calculated Patterson’s D-statistic, i.e., the ABBA/BABA test (Green et al. 2010; Durand et al. 2011), using the program ANGSD (Korneliussen et al. 2014). We used 100 kb windows to analyze patterns of introgression between An. melas populations throughout the genome. The ABBA/BABA test compares biased proportions of ABBA vs. BABA patterns across a four species lineage to identify regions of introgression between populations P3 and P1 or P3 and P2, given the following topology: {[(P1, P2)P3]O}, where O signifies the outgroup. Positive values of Patterson’s D-statistic indicate biased proportions of ABBA patterns, indicating introgression between P3 and P2, whereas negative Patterson’s D-statistic values indicate a biased proportion of BABA patterns, and introgression between species P3 and P1. It is important to note that this test cannot determine the direction of introgression (i.e., from P3 to P1, or P1 to P3).
Patterson’s D-statistic was calculated using An. gambiae as an outgroup and using the following tree topology: {[(West, Bioko) South] An. gambiae}. This tree topology is strongly supported by an approximate Bayesian computation analysis of the demographic history of An. melas populations based upon microsatellite data (posterior probability = 0.97) (Deitz et al. 2012). This tree topology allowed us to test which scenario is more likely, introgression between An. melas South and Bioko (P3 and P2) or between An. melas South and West (P3 and P1). ABBA/BABA sites were included in this analysis if sequence reads had a minimum map quality score of 30, and the SNP had a minimum base quality score of 30. The ANGSD implementation of the ABBA/BABA test uses one allele sampled from each population. While this could result in a loss of power when implemented using Pool-seq data, it will not bias the number of ABBA vs. BABA sites (R. Nielsen, personal communication). A delete-m jackknife approach (Busing et al. 1999) was used to determine the standard error of the mean Patterson’s D-statistic on each chromosome arm, and the entire genome. We calculated a Z-score to test if ABBA or BABA counts on each chromosome arm differed significantly from the null hypothesis of Patterson’s D-statistic = 0 (no excess of ABBA or BABA sites), indicating introgression between two of the populations.
Generation of an FST null distribution and false discovery rate
Previous studies using Pool-seq identified divergent genomic regions by visually inspecting sliding-window FST graphs for high peaks (e.g., Karlsen et al. 2013), or considered SNPs to be significant if they were four standard deviations above the mean value of the Z-transformed FST distribution (e.g., Montague et al. 2014). Others considered SNPs to be significantly differentiated between populations if their pair-wise FST values fell in the top 0.5% of the FST distribution, and had a Bonferroni-corrected p-value lower than 0.05 when subjected to a Fisher’s exact test (Kofler et al. 2011b; Fabian et al. 2012). While conservative approaches such as a Bonferroni correction reduce type I error, they may exclude a large number of biologically significant SNPs from downstream analyses (Darum 2006). Additionally, relying on the Fisher’s exact test implemented in PoPoolation2 for detecting significant differences in allele frequencies does not take into account pool size, which can influence allele frequency estimates. Thus, it only works well for studies in which pool size is considerably larger than sequencing coverage and can be ignored. In cases of small pool size, it will lead to a potentially large number of false positive results.
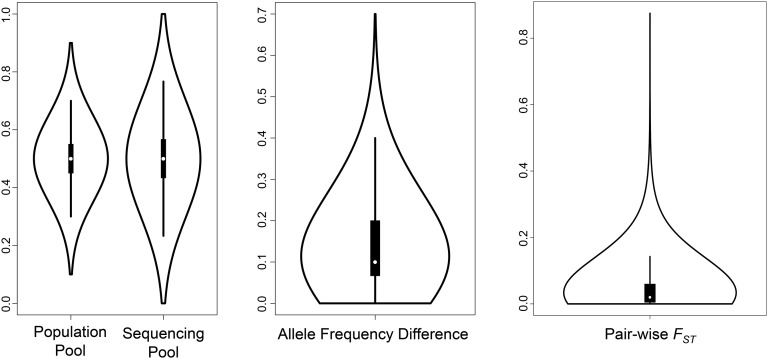
Therefore, we created a FST null distribution by simulating FST values observed between two samples drawn from a single population, given our pool size and sequence coverage. This null distribution allows us to determine which SNPs are significantly differentiated in our data. We created this null distribution by performing simulations in R (https://www.r-project.org). First, we drew 40 alleles (N = 20) from a population of 1000 individuals with a single SNP at an allele frequency of 0.5. We used an initial allele frequency of 0.5 because this value results in the largest variance of the estimated allele frequency. This step was repeated 10 million times to create our “population pool” allele distribution (Figure 2). This step simulates the pooling of individuals. We then drew 30 alleles (the minimum sequencing coverage (30 ×) used for SNP-wise and window-based FST estimation) from our population pool allele distribution. This step was repeated 10 million times to create the “sequencing pool” allele distribution (Figure 2). This step simulates the random generation of sequencing reads from the Pool-seq DNA library. The simulation of these two sampling steps combined provides the distribution of possible allele frequency estimates.
Figure 2.
Summary violin plots of the FST null distribution and false discovery rate simulation. The left plots show the allele frequency distribution of population and sequencing pools. The middle plot represents the difference between two randomly sampled allele frequencies drawn from the sequencing pool. The right plot shows the distribution of FST values calculated from the distribution of allele frequency differences.
To obtain the FST null distribution, we drew two allele frequency values from this allele frequency distribution 10 million times and calculated the allele frequency difference between them (Figure 2). We calculated the FST value for each of these pairs using FST = (HT − HS) / (HT), where HT is the total population heterozygosity and HS is the subpopulation heterozygosity. This process was also repeated 10 million times to create the “pair-wise FST” distribution. This FST null distribution was used to find the FST value for which the false discovery rate (FDR) ≤ 0.05. For each pair-wise population comparison, this was done by finding the threshold FST-value for which: (p-value × Total SNP number) / (significant SNP number) = 0.05. Here, the “p-value” is the proportion of FST values above the threshold FST value in the null distribution, “total SNP number” is the number of SNPs in the population data set, and “significant SNP number” is the number of SNPs in the population data set with an FST value above the threshold. In other words, the numerator is the expected number of false positives, and the denominator is the number of significantly differentiated SNPs in the data set.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Accession numbers for raw sequence reads are provided in Table S1.
Results
Sequence read quality control
The sequencing effort resulted in 78,025,712 paired-end reads for An. melas West (Ballingho, The Gambia), 52,594,743 for An. melas South (Ipono, Cameroon), and 56,776,632 for An. melas Bioko (Arena Blanca, Bioko Island, Equatorial Guinea) (Supplemental Material, Table S1). Paired-end reads were mapped to the genome only if both forward and reverse reads survived quality and length trimming (Phred ≥ 20, length ≥ 50 bp). Mapped reads with MAPQ values greater than 20, and that mapped to chromosomes X, 2, or 3, were retained for further analysis (West = 52.31%, South = 26.16%, and Bioko = 38.38% of original, raw reads). These reads had a mean length of 98.7–99.1 bp for each population (Table S1). However, the mean, genome-wide read coverage per base pair varied between populations (West = 34.44, South = 17.27, and Bioko = 25.41). This factor limited the number of SNPs that met our criteria of 30 × coverage for analysis of FST between population pools.
Nucleotide diversity and evolution
While we used lower thresholds (minimum coverage of 4 ×) for the calculation of nucleotide diversity and Tajima’s D, our results show that the mean reads/bp far exceed these values on all chromosome arms in all populations (Table S1). For example, the lowest observed mean reads/bp (15.63) was on chromosome arm 3L of An. melas South. The 4 × threshold was used to maximize the number of variable sites within a 100 kb window included in the calculation of nucleotide diversity and Tajima’s D. On chromosome arm 3L of An. melas South, on average 36.34% of a 100 kb window exceeded the minimum coverage threshold.
Genome-wide nucleotide diversity across 100 kb windows was very similar in An. melas West from Ballingho, The Gambia (mean π = 0.0052, SEM = 4.78 × 10−5), and An. melas South from Ipono, Cameroon (mean π = 0.0048, SEM = 5.31 × 10−5), but perhaps not unexpectedly, was somewhat lower in An. melas Bioko from Arena Blanca, Bioko Island (mean π = 0.0034, SEM = 5.12 × 10−5, Table 1). This pattern was consistent across all chromosomes (An. melas West π > An. melas South π > An. melas Bioko π) (Figure 3 and Table 1). In each population, mean chromosomal nucleotide diversity was higher on the third chromosome, and lowest on 2R or X (Figure 3 and Table 1). Interestingly, the patterns of nucleotide diversity are remarkably concordant between An. melas populations when viewed across their genomes, with the exception of a peak of high nucleotide diversity on chromosome 2L in An. melas Bioko (Figure 3).
Table 1. Estimates of mean nucleotide diversity (π) and Tajima’s D for each chromosome arm and An. melas population, measured in 100 kb, nonoverlapping sliding windows.
| Population | X | 2R | 2L | 3R | 3L | Genome-Wide | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| π | Tajima’s D | π | Tajima’s D | π | Tajima’s D | π | Tajima’s D | π | Tajima’s D | π | Tajima’s D | |
| West | 0.0046 | −0.100 | 0.0045 | −0.126 | 0.0053 | −0.108 | 0.0058 | −0.107 | 0.0058 | −0.093 | 0.0052 | −0.1092 |
| (0.00008) | (0.0050) | (0.00009) | (0.0029) | (0.00009) | (0.0032) | (0.00013) | (0.0028) | (0.00011) | (0.0028) | (4.78 × 10−5) | (0.0014) | |
| South | 0.0035 | −0.035 | 0.0045 | −0.030 | 0.0050 | −0.025 | 0.0053 | −0.032 | 0.0051 | −0.026 | 0.0048 | −0.0291 |
| (0.00010) | (0.0054) | (0.00010) | (0.0024) | (0.00010) | (0.0026) | (0.00014) | (0.0025) | (0.00012) | (0.0025) | (5.31 × 10−5) | (0.0012) | |
| Boiko | 0.0029 | −0.042 | 0.0029 | −0.038 | 0.0035 | −0.024 | 0.0037 | −0.022 | 0.0039 | −0.021 | 0.0034 | −0.0287 |
| (0.00008) | (0.0070) | (0.00009) | (0.0037) | (0.00013) | (0.0037) | (0.00012) | (0.0039) | (0.00012) | (0.0038) | (5.12 × 10−5) | (0.0018) | |
Values in parentheses indicate the standard error of the mean for each statistic. Regions of heterochromatin in the An. gambiae genome were removed from summary statistics.
Tajima’s D was calculated to identify genomic regions that may be evolving under positive selection in each population. Mean Tajima’s D was negative for all populations, indicating a deviation from neutral evolution (D = 0) (Figure 3 and Table 1). Various low Tajima’s D regions are shared between all three populations, although some low Tajima’s D windows are unique to a single population (Figure 3). While broad patterns of Tajima’s D for each population are similar across their genomes, the genome-wide mean Tajima’s D of An. melas West is over three times lower than that of An. melas South and Bioko (Figure 3 and Table 1).
FST null distribution
To determine significance thresholds for genetic differentiation (FST) between the three An. melas populations, the null distribution of allele frequency differences was determined based on our pooling and sequencing coverage using simulations. Next, two values were randomly drawn from this distribution to calculate an FST value. Each step of the simulation was repeated 10 million times to create each distribution. The first step in this simulation created a population pool with a mean allele frequency of 0.5 and a range of 0.1–0.9 (Figure 2 and Table 2). The second step created a sequencing pool distribution with a mean allele frequency of 0.5 and a range of 0.0–1.0. The final pair-wise FST null distribution ranges from 0.0 to 0.875 and has a mean of 0.046 (Figure 2 and Table 2). For each An. melas pair-wise population comparison, the FST value corresponding to FDR = 0.05 was determined and set as the significance threshold for the SNP-wise FST analyses. These significance thresholds between the populations are FST = 0.463 for West-South, FST = 0.446 for West-Bioko, and FST = 0.402 for South-Bioko. While these values are high due to relatively small pool sizes and low sequencing coverage, this conservative approach reduces the number of false positive results.
Table 2. Summary statistics of the FST null distribution and false discovery rate simulation.
| Summary Statistic | Population Pool Distribution | Sequencing Pool Distribution | Allele Frequency Difference Distribution | Pairwise FST Distribution |
|---|---|---|---|---|
| Minimum | 0.100 | 0.000 | 0.000 | 0.000 |
| Q1 | 0.450 | 0.433 | 0.067 | 0.005 |
| Median | 0.500 | 0.500 | 0.100 | 0.020 |
| Mean | 0.500 | 0.500 | 0.135 | 0.046 |
| Q3 | 0.550 | 0.567 | 0.200 | 0.060 |
| Maximum | 0.900 | 1.000 | 0.700 | 0.875 |
Genetic differentiation and introgression
Significant genetic differentiation between the three An. melas population clusters extends across the entire genome (Table 3 and Table S2), and includes fixed SNPs on all chromosome arms (Figure 3 and Table 3). Even though the Ipono, Cameroon and Arena Blanca, Bioko Island populations, which represent An. melas South and Bioko, respectively, are geographically close compared to the Ballingho, The Gambia (An. melas West), they are the most differentiated (Q1 = 0.018, median FST = 0.033, mean FST = 0.114, Q3 = 0.091), followed by the West and Bioko (Q1 = 0.016, median FST =0.028, mean FST = 0.076, Q3 = 0.055), and West and South (Q1 = 0.021, median FST = 0.034, mean FST = 0.075, Q3 = 0.062) (Table S2). An. melas South and Bioko also have the highest number of significantly differentiated (39,730, 8.56% of total) and fixed SNPs (5387, 1.16% of total) between them (total SNPs = 463,910), followed by West and Bioko [significant = 21,427 (3.81% of total), fixed = 1724 (0.31% of total), total SNPs = 562,493], and West and South [significant = 17,117 (2.76% of total), fixed = 1602 (0.26% of total), total SNPs = 621,184] (Table 3). It should be noted that the number of SNPs in each population comparison is influenced by differences in mapping coverage between the populations (Table 3 and Table S1). However, divergence between An. melas South and the other populations was largest, whereas this population has the lowest number of mapped reads.
Table 3. Number of significant and fixed SNPs per chromosome in each pair-wise An. melas population comparison.
| X | 2R | 2L | 3R | 3L | Genome-Wide | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Comparison | Fixed | Sig. | Fixed | Sig. | Fixed | Sig. | Fixed | Sig. | Fixed | Sig. | Fixed | Sig. |
| West - South | 879 | 3028 | 185 | 3853 | 202 | 3624 | 116 | 3340 | 220 | 3272 | 1602 | 17,117 |
| West - Bioko | 319 | 1810 | 439 | 6373 | 403 | 5061 | 299 | 4671 | 264 | 3512 | 1724 | 21,427 |
| South - Bioko | 1725 | 4324 | 981 | 10,396 | 1110 | 9197 | 692 | 8825 | 879 | 6988 | 5387 | 39,730 |
Regions of heterochromatin in the An. gambiae genome were removed from summary statistics. Sig., significant.
The X chromosome has a disproportionately large number of fixed and significant SNPs (Figure 3 and Table 3) in both West and South and South and Bioko population comparisons. This pattern of elevated FST extends across the entire X chromosome (Figure 3). This could potentially be the result of increased genetic drift acting on polymorphisms due the lower effective population size of the X chromosome. Interestingly, however, this X chromosome effect is not obvious between An. melas West and Bioko, the two most recently diverged groups.
We performed a gene ontology analysis on genes within windows that show evidence of nonneutral evolution (low Tajima’s D). First we identified 100 kb sliding windows with the lowest 5% Tajima’s D values for each population (genome-wide, excluding heterochromatic regions) (D < −0.200, −0.096, and −0.148 for An. melas West, South, and Bioko, respectively). Next, we identified genes inside these windows that harbored SNPs with significant FST values in each pair-wise comparison. The West-South comparison yielded 95 significant SNPs located inside the exons of 64 genes. The molecular functions of these genes are associated with binding, catalytic activity, nucleic acid binding transcription factor activity, and receptor activity, among others (Table S3). The West-Bioko comparison yielded 79 significant SNPs located inside exons of 62 genes and the South-Bioko comparison yielded 188 significant SNPs located inside exons of 127 genes (Table S3). The molecular functions associated with these genes are similar to those found in the West-South example. The most commonly found molecular functions (across all comparisons) include binding, catalytic activity, and nucleic acid binding transcription factor activity, and some genes are common among population comparisons (Table S3).
Common biological processes in all population comparisons include biological regulation, cellular processes, localization, and metabolic processes (Table S4). The South-Bioko comparison had 161 biological process gene ontology hits associated with the 127 genes in this analysis. The most frequent hits to protein classes across all comparisons were found in the hydrolase category, followed by proteases, nucleic acid binding proteins, proteases, and transcription factors (Table S5).
Our analysis of introgression between An. melas populations was based on the topology {[(West, Bioko) South] An. gambiae} (Deitz et al. 2012), and screened for introgression between An. melas South and Bioko or South and West. This test found a genome-wide, positive deviation of the D-statistic (mean D-statistic = 0.040, Z-score = 21.80, Table S6), indicating an excess of ABBA sites and ancient or weak introgression between An. melas South and Bioko. An exception to this pattern was found on chromosome 2L (∼22.25–23.45 Mb), where D-statistic windows with a strong, negative deviation from zero (as low as −0.83) suggest recent An. melas South and West introgression (Figure 4). Interestingly, this introgression block overlaps precisely with a region of high nucleotide diversity in An. melas Bioko (Figure 3), and falls between the proximal breakpoint of the 2La chromosomal inversion (which is fixed for the standard arrangement in An. melas) and the proximal breakpoint of the 2La2 chromosomal inversion (which is polymorphic within An. melas) (Coluzzi et al. 2002: Sharakhov et al. 2006; White et al. 2007). The 2La2 inversion is specific to An. melas and is polymorphic within it (Coluzzi et al. 2002). An. melas collected from Guinea Bissau and Cotonou, Benin (inside the range of the An. melas West cluster, Figure 1) share the standard arrangement (2L+a2), while An. melas collected from Democratic Republic of the Congo (likely belonging to the An. melas South genetic cluster) are polymorphic for the standard and inverted arrangements (2La2 and 2L+a2) (Coluzzi et al. 2002).
Discussion
Population genomic analysis of An. melas West, South, and Bioko Island identified significant, genome-wide genetic differentiation, including the presence of numerous fixed SNPs throughout the genome in all An. melas population comparisons. Previous work based on microsatellites and mtDNA markers indicated levels of differentiation between An. melas forms that are on a par with, or exceed, those observed between An. gambiae and An. arabiensis (Deitz et al. 2012). Species pairs in the An. gambiae complex with comparable genetic differentiation are separated by strong pre- and postmating isolation (Marchand 1983; Okereke 1980; Slotman et al. 2004; Weetman et al. 2014). Recently, the M and S molecular forms of An. gambiae were raised to species level (Coetzee et al. 2013) based on well-documented ecological and some behavioral differences. These species have diverged considerably less than the three An. melas genetic clusters throughout most of their genomes but have several regions of high differentiation. This is not the case for the three An. melas forms where, with the exception of a chromosome-wide X effect, genetic differentiation is distributed mostly evenly across the genome. This is consistent with a process of allopatric divergence with little gene flow/introgression. No evidence for “speciation islands”, genomic regions with high levels of divergence that are maintained in the face of extensive hybridization gene flow (Turner et al. 2005), was found in this study.
We used a simulation approach to construct an FST null distribution and FDR that incorporates both pool-size and sequencing coverage. To our knowledge, this is the first time that this approach has been applied to a Pool-seq study. This allowed us to determine the FST significance threshold for each pair-wise population comparison. In doing so, we assumed a starting allele frequency of 0.5, which results in the largest variance in the subsequent sampling steps of the simulation. In addition, we used a sequencing coverage of 30 × for our simulations, which was the minimum sequencing coverage we required for FST calculations in our empirical analysis. Therefore, our approach is conservative. A downside of our approach is that it does not provide q-values for individual SNPs, though our method could be adapted to do so in the future.
Intrapopulation nucleotide diversity in An. melas revealed remarkably similar patterns of variation across the genomes of each population (Figure 3 and Table 1). This shared pattern may be attributed to shared ancestry and genome organization (e.g., chromosomal inversions). Additionally, selective constraints on many genes may be similar between these populations, as the ecology may be largely shared between forms. A single peak in nucleotide diversity on chromosome 2L of An. melas Bioko is the exception. Interestingly, the results of the ABBA/BABA test suggested that this exact region introgressed between An. melas South and West (Figure 4). This highly surprising overlap suggests to us an alternative explanation: recent introgression of this region from An. gambiae (or more likely, the closely related An. coluzzii, see below), the outgroup species in the ABBA/BABA test, into An. melas Bioko. This would also create a pattern of BABA excess (suggesting introgression between An. melas South and West) and could explain the remarkably high nucleotide diversity in Bioko Island in this particular region. Both An. coluzzi and An. melas are present on Bioko Island (Overgaard et al. 2012), female hybrids between the two species are fertile (Davidson 1962), and extensive introgression between various species in the complex was recently documented (Fontaine et al. 2015). An. gambiae s.s. (i.e., An. gambiae S form) was eliminated from Bioko Island through a malaria control campaign, and only An. coluzzii (i.e., An. gambiae Forest-M form) remains (Overgaard et al. 2012).
Genome-wide Patterson’s D-statistic values from the ABBA/BABA test also suggests a slight bias toward a low level of ancestral introgression between An. melas South and Bioko (vs. between West and South). This finding is perhaps not surprising considering the geographical proximity of the An. melas South and Bioko populations used in this study (Ipono, Cameroon and Arena Blanca, Bioko Island, Equatorial Guinea, respectively) (Figure 1) in comparison to An. melas from Ballingho, The Gambia, which was our representative population of An. melas West.
Measures of nucleotide diversity in An. melas populations are less than half of the mean chromosomal nucleotide diversity values observed in An. gambiae (S form) populations collected from the north and south of Cameroon (0.008–0.15, Cheng et al. 2012). This may reflect a lower Ne due to the patchy distribution of An. melas populations compared to An. gambiae (Athrey et al. 2012; Deitz et al. 2012). Genome-wide nucleotide diversity is the lowest in An. melas Bioko, which likely reflects a smaller effective population size (Ne) compared to the other An. melas populations. Previous findings also found that the Bioko Island population harbors lower levels of rarefied allelic richness at microsatellite loci, far fewer mitochondrial DNA haplotypes, and a much lower Ne compared to mainland populations (Deitz et al. 2012). An alternative explanation of lower diversity due to founder effects is not supported by a previous Approximate Bayesian Computation analysis of the demographic history of these populations, which indicated that all three An. melas forms separated through vicariance events (Deitz et al. 2012).
Mean chromosomal Tajima’s D and nucleotide diversity were lowest on the X chromosome for An. melas South and Bioko (Table 1), and nucleotide diversity of the An. melas X chromosome was the second lowest of any chromosome arm. This may be due to positive selection on (partially) recessive alleles acting more strongly on the X chromosome. These findings are in agreement with an effects model (SnIPRE) analysis of natural selection between An. melas West, South, and Bioko Island populations, which found an increased selection effect of the An. melas X chromosome (Struchiner et al., unpublished results). Low diversity on the X chromosome of An. melas populations is consistent with findings in An. gambiae s.s. (Cohuet et al. 2008; Holt et al. 2002; Wilding et al. 2009) and An. arabiensis (Marsden et al. 2014). Introgression between member species of the An. gambiae complex is well documented (Fontaine et al. 2015), but is limited between the X chromosome of An. gambiae s.s. and other members of the complex due to the Xag inversion, which covers ∼60% of the An. gambiae s.s. X chromosome. The Xag inversion suppresses recombination between the An. gambiae and An. arabiensis X chromosomes, and plays a large role in their postzygotic reproductive isolation (Slotman et al. 2004, 2005b), preventing introgression. This suppressed introgression of the X chromosome between An. gambiae and An. arabiensis may have contributed to reduced nucleotide diversity on the X in these species (Marsden et al. 2014). Reduced introgression of the X chromosome may also contribute to its lower nucleotide diversity in An. melas, although its lower effective population size resulting in higher levels of genetic drift is probably a more important factor.
Mean Tajima’s D was over three times lower in An. melas West as compared to the South and Bioko. As this is a genome-wide effect, it likely is the result of demographic factors, such as a recent population bottleneck in the An. melas West population analyzed. Windows of low Tajima’s D are found throughout the genomes of the An. melas populations, which may indicate that these regions harbor genes under positive selection. Notably, very similar patterns of genome-wide Tajima’s D are found in each An. melas population cluster. This suggests that while geographic isolation of An. melas clusters has greatly reduced gene flow between them, their resulting genetic differentiation is likely not a result of diverging selection pressures, which is expected to result in diverging Tajima’s D patterns. The similar patterns of genome-wide Tajima’s D likely also mean that genetic drift has not yet greatly impacted ancestral signatures of selection in these genomes.
Our gene ontology analysis explored the molecular and biological functions, and protein classes associated with genes found in low Tajima’s D regions that also harbored significant or fixed SNPs. These included molecular functions associated with binding, catalytic, and nucleic acid binding transcription factor activity, biological functions including metabolic and cellular processes, localization and biological regulation, and protein classes such as enzyme modulators, nucleic acid binding, transcription factors, and transferases, among others (Table S4, Table S5, and Table S6). Future analyses of the functions of these genes might be able to reveal a link to their biological significance in An. melas.
Since early studies of host preference, parasitemia rate, and ecology of An. melas (Gelfand 1955), and the original taxonomic, genetic, and descriptive studies of the An. gambiae complex (Davidson 1962; White 1974), An. melas has been considered a malaria vector of minor importance due to its limited distribution and broad host preference. However, early studies focused on populations representing An. melas West alone. Recent studies have shown that on Bioko Island, Equatorial Guinea, An. melas populations readily feed on humans both indoors and outdoors (Redd et al. 2011), and are responsible for up to 130 malaria infectious bites/person/year in the village of Arena Blanca (Overgaard et al. 2012). These studies highlight the important role that An. melas plays in malaria transmission. The results of this study, in combination with previous work (Deitz et al. 2012), indicate that An. melas is undergoing an allopatric divergence process. Therefore, what we know about the ecology and behavior of An. melas West populations, which have been the focus of the handful of studies on the species (Bryan 1983; Bryan et al. 1987; Bogh et al. 2007; Caputo et al. 2008), may not hold true for the other An. melas forms. Additionally, as a member of a species complex that serves as a model for the speciation process, a better understanding of the population genomics of An. melas populations enhances our view of how the evolution of the An. gambiae species complex is influenced by the diverse host preferences, ecologies, distributions, and demographic histories of its member species.
Supplementary Material
Acknowledgments
We thank Michael C. Fontaine (University of Groningen), Daniel E. Neafsey (Broad Institute of Massachusetts Institute of Technology and Harvard), and Nora J. Besansky (University of Notre Dame) for their helpful comments and feedback regarding this manuscript. Additionally, we thank the Anopheles Genome Consortium for data availability. We are grateful to Parfait H. Awono-Ambene, Christophe Antonio-Nkondjo, and Frederic Simard for assistance with collections in Ipono, Cameroon. Collections in Ballingho, The Gambia were supported by a research grant to M.A.S. by the Bioko Island Malaria Control Project (BIMCP). The BIMCP is funded by a consortium led by Marathon Oil Corporation (Houston, TX) and the Government of Equatorial Guinea. Collections on Bioko Island were conducted as part of the vector monitoring efforts under the BIMCP. We are grateful for the entomology staff of the BIMCP and the local volunteers for conducting these collections. Partial support of this work was provided by National Institutes of Health/National Institute of Allergy and Infectious Diseases grants R01AI085079 and R21 AI115175 to M.A.S. K.C.D. was partially supported by the J.H. Benedict, Sr. Memorial Graduate Student Scholarship and the Herb Dean ’40 Endowed Scholarship, through the Department of Entomology at Texas A&M University during the term of this project.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.031906/-/DC1
Communicating editor: H. Tachida
Literature Cited
- Aboagye-Antwi F., Alhafez N., Weedall G. D., Brothwood J., Kandola S., et al. , 2015. Experimental swap of Anopheles gambiae’s assortative mating preferences demonstrates key role of X–chromosome divergence island in incipient sympatric speciation. PLoS Genet. 11: e1005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athrey G., Hodges T. K., Reddy M. R., Overgaard H. J., Matias A., et al. , 2012. The effective population size of malaria mosquitoes: large impact of vector control. PLoS Genet. 8: e1003097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky N. J., Powell J. R., Caccone A., Hamm D. M., Scott J. A., et al. , 1994. Molecular phylogeny of the Anopheles gambiae complex suggests genetic introgression between principal malaria vectors. Proc. Natl. Acad. Sci. USA 91: 6885–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogh C., Lindsay S. W., Clarke S. E., Dean A., Jawara M., et al. , 2007. High spatial resolution mapping of malaria transmission risk in The Gambia, West Africa using TM satellite imagery. Am. J. Trop. Med. Hyg. 76: 875–881. [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan J. H., 1983. Anopheles gambiae and Anopheles melas at Brefet, The Gambia, and their role in malaria transmission. Ann. Trop. Med. Parasitol. 77: 1–2. [DOI] [PubMed] [Google Scholar]
- Bryan J. H., Petrarca V., Di Deco M. A., Coluzzi M., 1987. Adult behavior of members of the Anopheles gambiae complex in the Gambiae with special reference to An. melas and its chromosomal variants. Parassitologia 29: 221–249. [PubMed] [Google Scholar]
- Busing F. M. T. A., Meijer E., Van Der Leeden R., 1999. Delete-m jackknife for unequal m. Stat. Comput. 9: 3–8. [Google Scholar]
- Caputo B., Nwakanma D., Jawara M., Adiamoh M., Dia I., et al. , 2008. Anopheles gambiae complex along The Gambia river, with particular reference to the molecular forms of An. gambiae s.s. Malar. J. 7: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., White B. J., Kamdem C., Mockaitis K., Constantini C., et al. , 2012. Ecological genomics of Anopheles gambiae along a latitudinal cline: a population-resequencing approach. Genetics 190: 1417–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson C. S., Weetman D., Essandoh J., Yawson A. E., Maslen G., et al. , 2014. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat. Commun. 5: 4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee M., Hunt R. H., Wikerson R., della Torre A., Coulibaly M. B., et al. , 2013. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 3619: 246–274. [PubMed] [Google Scholar]
- Cohuet A., Krishnakumar S., Simard F., Morlais I., Koutsos A., et al. , 2008. SNP discovery and molecular evolution in Anopheles gambiae, with special emphasis on innate immune system. BMC Genomics 9: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluzzi M., Sabatini A., della Torre A., Di Deco M. A., Petrarca V., 2002. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298: 1415–1418. [DOI] [PubMed] [Google Scholar]
- Darum S. R., 2006. Beyond Bonferroni: less conservative analyses for conservation genetics. Conserv. Genet. 7: 783–787. [Google Scholar]
- Davidson G., 1962. Anopheles gambiae complex. Nature 196: 907. [Google Scholar]
- Deitz K. C., Athrey G., Reddy M. R., Overgaard H. J., Matias A., et al. , 2012. Genetic isolation within the malaria mosquito Anopheles melas. Mol. Ecol. 18: 4498–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- della Torre A., Fanello C., Akogbeto M., Dossou-yovo J., Favia G., et al. , 2001. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol. Biol. 10: 9–18. [DOI] [PubMed] [Google Scholar]
- Diabaté A., Dabire R. K., Millogo N., Lehmann T., 2007. Evaluating the effect of postmating isolation between molecular forms of Anopheles gambiae (Diptera: Culicidae). J. Med. Entomol. 44: 60–64. [DOI] [PubMed] [Google Scholar]
- Donelly M. J., Townson H., 2000. Evidence for extensive genetic differentiation among populations of the malaria vector Anopheles arabiensis in Eastern Africa. Insect Mol. Biol. 9: 357–367. [DOI] [PubMed] [Google Scholar]
- Durand E. Y., Patterson N., Reich D., Slatkin M., 2011. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28: 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian D. K., Kapun M., Nolte V., Kofler R., Schmidt P. S., et al. , 2012. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol. Ecol. 21: 4748–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favia G., Lanfrancotti A., Spanos L., Siden-Kiamos I., Louis C., 2001. Molecular characterization of ribosomal DNA polymorphisms discriminating among chromosomal forms of Anopheles gambiae s.s. Insect Mol. Biol. 10: 19–23. [DOI] [PubMed] [Google Scholar]
- Fisher S., Barry A., Abreu J., Minie J., Delorey T. M., et al. , 2011. A scaleable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 12: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, et al. 2015 Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347 Available at: http://science.sciencemag.org/content/347/6217/1258524.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand H. M., 1955. Anopheles gambiae giles and An. melas Theobald in a coastal area of Liberia, West Africa. Trans. R. Soc. Trop. Med. Hyg. 49: 508–527. [DOI] [PubMed] [Google Scholar]
- Gentile G., Slotman M., Ketmaier V., Powell J. R., Caccone A., 2001. Attempts to molecularly distinguish cryptic taxa in Anopheles gambiae s.s. Insect Mol. Biol. 10: 25–32. [DOI] [PubMed] [Google Scholar]
- Giraldo-Calderón GI, Emrich S. J., MacCallum R. M., GMaslen G, EDialynas, et al. , 2015. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43(Database issue): D707–D713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. E., Krause J., Briggs A. W., Maricic T., Stenzel U., et al. , 2010. A draft sequence of the neandertal genome. Science 328: 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt R., Subramanian G., Halpern A., Sutton G., Charlab R., et al. , 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149. [DOI] [PubMed] [Google Scholar]
- Hunt R. H., Coetzee M., Fettene M., 1998. The Anopheles gambiae complex: a new species from Ethiopia. Trans. R. Soc. Trop. Med. Hyg. 92: 231–235. [DOI] [PubMed] [Google Scholar]
- Karlsen B. O., Klingan K., Emblem A., Jorgensen T. E., Jueterbock A. J., et al. , 2013. Genomic divergence between migratory and stationary ecotypes of Atlantic cod. Mol. Ecol. 22: 5098–5111. [DOI] [PubMed] [Google Scholar]
- Kofler R., Orozco-terWengel P., De Maio N., Pandey R. V., Nolte V., et al. , 2011a PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 6: e15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R., Pandey R. V., Schlotterer C., 2011b PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27: 3435–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen T. S., Albrechtsen A., Nielsen R., 2014. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics 15: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzaro G. C., Lee Y., 2013. Speciation in Anopheles gambiae. The Distribution of Genetic Polymorphism and Patterns of Reproductive Isolation Among Natural Populations, Anopheles mosquitoes - New insights into malaria vectors, edited by Prof. Sylvie Manguin, ISBN: 978-953-51-1188-7, InTech, DOI: 10.5772/56232. [DOI] [Google Scholar]
- Lehman T., Licht M., Elissa N., Maega B. T., Chimumbwa J. M., et al. , 2003. Population structure of Anopheles gambiae in Africa. J. Hered. 94: 133–147. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loaiza J. R., Bermingham E., Sanjur O. I., Scott M. E., Bickersmith S. A., et al. , 2012. Review of genetic diversity in malaria vectors (Culicidae: Anophilinae). Infect. Genet. Evol. 12: 1–12. [DOI] [PubMed] [Google Scholar]
- Lunther G., Goodson M., 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21: 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J., 1995. A species definition for the modern synthesis. Trends Ecol. Evol. 10: 294–299. [DOI] [PubMed] [Google Scholar]
- Mallet J., Besansky N., Hahn M. W., 2015. How reticulated are species? BioEssays 38: 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand R. P., 1983. Field observations on swarming and mating in Anopheles gambiae mosquitoes in Tanzania. Neth. J. Zool. 34: 367–387. [Google Scholar]
- Marsden C. D., Lee Y., Nieman C. C., Sandford M. R., Dinis J., et al. , 2011. Asymmetric introgression between the M and S forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridization. Mol. Ecol. 20: 4983–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden C. D., Lee Y., Kreppel K., Weakley A., Cornel A., et al. , 2014. Di-versity, differentiation, and linkage disequilibrium: prospects for association mapping in the malaria vector Anopheles arabiensis. G3 (Bethesda) 4: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E., 1970. Populations, Species, and Evolution, Belknap Press of Harvard University Press, Cambridge. [Google Scholar]
- Mi H., Dong Q., Muruganujan A., Gaudet P., Lewis S., et al. , 2010. PANTHER version 7: improved phylogenetic trees, orthologs, and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38: D204–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague M. J., Li G., Gandolfi B., Khan R., Aken B. L., et al. , 2014. Com-parative analysis of the domestic cat genome reveals genetic signatures underlying feline biology and domestication. Proc. Natl. Acad. Sci. USA 111: 17230–17235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno M., Salgueiro P., Vicente J. L., Cano J., Berzosa P. J., et al. , 2007. Genetic population structure of Anopheles gambiae in Equatorial Guinea. Malar. J. 6: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey D. E., Waterhouse R. M., Abai M. R., Aganezov S. S., Alekseyev M. A., et al. , 2015. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347: 1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H., 1979. Mathematical model for studying genetic variation in terms of restriction enzyme endonucleases. Proc. Natl. Acad. Sci. USA 10: 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris L. C., Main B. J., Lee Y., Collier T. C., Fofana A., et al. , 2015. Ada-ptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proc. Natl. Acad. Sci. USA 112: 815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P., 2012. Ecological Speciation, Oxford University Press, Oxford. [Google Scholar]
- Okereke T. A., 1980. Hybridization studies on sibling species of the Anopheles gambiae Giles complex (Diptera, Culicidae) in the laboratory. Bull. Entomol. Res. 70: 391–398. [Google Scholar]
- Overgaard H. J., Reddy V. P., Abaga S., Matias A., Reddy M. R., et al. , 2012. Malaria transmission after five years of vector control on Bioko Island, Equatorial Guinea. Parasit. Vectors 5: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. R., Petrarca V., della Torre A., Caccone A., Coluzzi M., 1999. Population structure, speciation, and introgression in the Anopheles gambiae complex. Parassitologia 41: 101–113. [PubMed] [Google Scholar]
- Redd M. R., Overgaard H. J., Abaga S., Reddy V. P., Caccone A., et al. , 2011. Outdoor host seeking behavior of Anopheles gambiae mosquitoes following initiation of malaria vector control on Bioko Island, Equatorial Guinea. Malar. J. 10: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer C., Tobler R., Kofler R., Nolte V., 2014. Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nat. Rev. Genet. 15: 749–763. [DOI] [PubMed] [Google Scholar]
- Sharakhov I. V., White B. J., Sharakhova M. V., Kayondo J., Lobo N. F., et al. , 2006. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc. Natl. Acad. Sci. USA 103: 6258–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova M. V., Hammond M. P., Lobo N. F., Krzywinski J., Unger M. F., et al. , 2007. Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 8: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova M. V., George P., Brunsentsova I. V., Leman S. C., Bailey J. A., et al. , 2010. Genome mapping and characterization of the Anopheles gambiae heterochromatin. BMC Genomics 11: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard F., Ayala D., Kamdem G. C., Pombi M., Etouna J., et al. , 2009. Eco-logical niche partitioning between Anopheles gambiae molecular forms in Cameroon: the ecological side of speciation. BMC Ecol. 9: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman M. A., della Torre A., Powell J. R., 2004. The genetics of inviability and male sterility in hybrids between Anopheles gambiae and Anopheles arabiensis. Genetics 167: 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman M. A., della Torre A., Calzetta M., Powell J. R., 2005a Diff-erential introgression of chromosomal regions between Anopheles gambiae and An. arabiensis. Am. J. Trop. Med. Hyg. 73: 326–335. [PubMed] [Google Scholar]
- Slotman M. A., della Torre A., Powell J. R., 2005b Female sterility in hybrids between Anopheles gambiae and An. arabiensis and the causes of Haldane’s rule. Evolution 59: 1016–1026. [PubMed] [Google Scholar]
- Smith H. A., White B. J., Kundert P., Cheng C., Romero-Severson J., et al. , 2015. Genome-wide QTL mapping of saltwater tolerance in sibling species of Anopheles (malaria vector) mosquitoes. Heredity 115: 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. D., Campbell M. J., Kejariwal A., Mi H., Karlak B., et al. , 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13: 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet R., Thiemann T., Lanzaro G. C., 2005. Effect of seminal fluids in mating between M and S forms of Anopheles gambiae. J. Med. Entomol. 42: 596–603. [DOI] [PubMed] [Google Scholar]
- Turner T. L., Hahn M. W., Nuzhdin S. V., 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 9: e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D., Steen K., Rippon E. J., Mawejje H. D., Donelly M. J., et al. , 2014. Contemporary gene flow between wild An. gambiae s.s. and An. arabiensis. Parasit. Vectors 7: 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. J., Santolamazza F., Kamau L., Pombi M., Grushko O., et al. , 2007. Molecular karyotyping of the 2La inversion in Anopheles gambiae. Am. J. Trop. Med. Hyg. 76: 334–339. [PubMed] [Google Scholar]
- White B. J., Cheng C., Simard F., Simard F., Constantini C., et al. , 2010. Ge-netic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae. Mol. Ecol. 19: 925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. J., Collins F. H., Besansky N. J., 2011. Evolution of Anopheles gambiae in relation to humans and malaria. Annu. Rev. Ecol. Evol. Syst. 42: 111–132. [Google Scholar]
- White G. B., 1974. Anopheles gambiae complex and disease transmission in Africa. Trans. R. Soc. Trop. Med. Hyg. 68: 278–298. [DOI] [PubMed] [Google Scholar]
- Wilding C., Weetman D., Steen K., Donnelly M., 2009. High, clustered, nucleotide diversity in the genome of Anopheles gambiae revealed through pooled-template sequencing: implications for high-throughput genotyping protocols. BMC Genomics 10: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Accession numbers for raw sequence reads are provided in Table S1.