Abstract
Allopolyploidization is a biological process that has played a major role in plant speciation and evolution. Genomic changes are common consequences of polyploidization, but their dynamics over time are still poorly understood. Coffea arabica, a recently formed allotetraploid, was chosen to study genetic changes that accompany allopolyploid formation. Both RNA-seq and DNA-seq data were generated from two genetically distant C. arabica accessions. Genomic structural variation was investigated using C. canephora, one of its diploid progenitors, as reference genome. The fate of 9047 duplicate homeologous genes was inferred and compared between the accessions. The pattern of SNP density along the reference genome was consistent with the allopolyploid structure. Large genomic duplications or deletions were not detected. Two homeologous copies were retained and expressed in 96% of the genes analyzed. Nevertheless, duplicated genes were found to be affected by various genomic changes leading to homeolog loss or silencing. Genetic and epigenetic changes were evidenced that could have played a major role in the stabilization of the unique ancestral allotetraploid and its subsequent diversification. While the early evolution of C. arabica mainly involved homeologous crossover exchanges, the later stage appears to have relied on more gradual evolution involving gene conversion and homeolog silencing.
Keywords: polyploidy, evolution, gene conversion, homoeologous recombination, genome dominance
Polyploidy (the complete doubling of a genome) has long been recognized as an important mechanism in plant speciation and genome evolution (Soltis et al. 2014). It is now well established that polyploidy occurred frequently during angiosperm evolution, and that all flowering plant species have undergone one or more rounds of genome duplication in their history (Jiao et al. 2011). In particular, allopolyploidization [arising from interspecific hybridization accompanied by whole-genome duplication (WGD)] is considered to have contributed to the adaptation to broader and novel environmental niches, and is thought to have played a fundamental role in the evolutionary history of speciation (Comai 2005; Soltis et al. 2014). In addition, many major agricultural crop plants, including wheat (Triticum aestivum), cotton (Gossypium hirsutum), rapeseed (Brassica napus), sugarcane (Saccharum officinarum), and coffee (Coffea arabica) are allopolyploids.
The establishment of a new allopolyploid species is not a trivial feat. The merging of two or more divergent genomes, and the presence of these parental genomes in duplicate in a single nucleus, can set the stage for dynamic changes to the genome, transcriptome, and phenotype of the new polyploid species (Doyle et al. 2008). However, the newly formed allopolyploid faces several immediate challenges. It must secure exclusive intragenomic pairing at meiosis that will lead to full fertility and disomic inheritance. Meiosis can therefore have a dual impact on the evolution of many newly formed polyploids by (1) enabling sexual propagation, and (2) generating, through meiotic errors, large-scale chromosomal variation upon which genetic drift and/or selection can act (Leitch and Leitch 2008). In addition, the newly formed allopolyploid must orchestrate intergenomic interactions and regulate gene expression to adapt to its environment (Jackson and Chen 2010). Hence, successful allopolyploidizations are those that trigger an array of genomic changes that confer evolutionary advantages (Arrigo and Barker 2012).
In the last decade, molecular data from resynthesized and natural allopolyploids has shown that genetic and epigenetic changes are common consequences of polyploidization across a wide range of species (Madlung and Wendel 2013). Nevertheless, little is known about the mechanisms that lead to these changes, and even less about their directed or random nature. In addition, genomic responses to WGD appear to be extremely diverse, and both genomic rearrangements and gene expression changes vary to different degrees depending on the polyploid species concerned, thus preventing simple generalizations. For example, rapid genomic changes have been reported in many new allopolyploids (Doyle et al. 2008), but not in allopolyploid cotton (Liu et al. 2001). Some polyploid genomes appear to undergo rapid homeolog loss (Gaeta et al. 2007; Soltis et al. 2010; Buggs et al. 2012), whereas, in other polyploids, changes in gene expression appear to dominate (Lee and Chen 2001; Wang et al. 2006; Flagel and Wendel 2010; Wang et al. 2016). To understand allopolyploid genome evolution in a broad context, genomic data from many more allopolyploids are required.
Among allopolyploid plants, Coffea arabica is an interesting case (Lashermes et al. 1999, 2000). C. arabica is a recent (< 100,000 yr ago) allotetraploid (CaEa genome) formed by hybridization between two diploid species: C. canephora (C genome) and C. eugenioides (E genome). The two parental species are closely related, and the two subgenomes have low sequence divergence (i.e., 1.3% average difference for genes, Cenci et al. 2012). In spite of the close relationship between the two constitutive subgenomes, C. arabica displays diploid-like meiotic behavior with bivalent formation. In addition, the most recent molecular analyses (Lashermes et al. 2014) showed that genomic rearrangements involving homeologous exchanges occurred in C. arabica, and could be a major source of genetic diversity. Evidence for a large number of homeologous exchange events (HEEs) shared by all accessions of C. arabica strongly supports the hypothesis of a single allopolyploidization event. The highlands of south west Ethiopia are considered as the primary center of genetic diversity of C. arabica (Sylvain 1955). The presence of wild populations has been also reported in forest on the Boma Plateau and Mount Imantong of Sudan (Thomas 1942), and in Mount Marsabit of Kenya (Anthony et al. 1987). Furthermore, although C. arabica accessions exhibit very low genetic diversity (as estimated through molecular markers), they display marked phenotypic/adaptive variation, and there is strong evidence of hybrid vigor when particular accessions are crossed (Van der Vossen et al. 2015).
In the present study, C. arabica was used to investigate genomic changes in allopolyploid using high-throughput sequencing approaches. Both RNA-seq and DNA-seq data were generated from two genetically distant accessions, and analyzed using C. canephora as a reference genome (Denoeud et al. 2014). On the one hand, genomic structural variation was investigated based on mapping patterns of DNA sequence reads, and on the detection of copy number alterations (CNA). On the other hand, genes affected by homeolog loss or silencing were inferred by comparing the number of single nucleotide polymorphisms (SNPs) detected in C. arabica and between its two diploid progenitor species at both DNA and RNA level. To validate these results, Sanger direct sequencing of cDNA and DNA amplicons from various C. arabica accessions was performed. In particular, the distribution of two homeologous crossover exchange events among accessions from the C. arabica primary center of diversity was investigated. Genomic rearrangements involving homeologous DNA exchanges, as well as gene conversion and homeolog silencing were evidenced that could have played a major role in the evolution and diversification of C. arabica.
Materials and Methods
Plant material, library construction, and sequencing
The plant material came from two C. arabica accessions, including the commercial cultivar Caturra (an inbred line), one wild accession (AR41) from the ORSTOM (Guillaumet and Hallé 1978) collection mission in Ethiopia, and one accession from each of the two modern-day diploid progenitors, C. canephora (acc. DH200-94) and C. eugenioides (acc. DA58). Young leaf tissues were collected at the same time from individuals plants grown in a greenhouse in Montpellier (France), and were immediately flash frozen in liquid nitrogen and stored at –80° until DNA and RNA extractions were performed as previously reported (Combes et al. 2013; Lashermes et al. 2014). For genomic DNA sequencing of the two C. arabica accessions, nonindexed paired-ends (PE) libraries were constructed using the TruSeq DNA sample preparation kit (Illumina, San Diego, CA), which includes a fragmentation step by sonication, and, after end-repair and adapter ligation, selection of 470 ± 60 bp DNA fragments by band excision after gel electrophoresis. PE sequencing was carried out at 2 × 100 bp using the Illumina HiSequation 2000, according to the manufacturer’s instructions at the MGX platform (Montpellier Genomix, Montpellier, France). DNA data were collected from two lanes (one lane per library, and per accession) in the same sequencing run. For RNA sequencing of C. arabica and C. eugenioides accessions, the method and data were as previously reported in Lashermes et al. (2014). Briefly, total RNA was isolated from 1 g of material from each plant, and mRNA libraries were constructed using the Illumina “RNA-seq sample prep” kit (Illumina). Single-reads (∼72 nt) were generated with the Illumina HiSequation 2000. For the accession Caturra (C. arabica), four independent libraries were constructed from four different plants. The entire sequence dataset has been deposited with the European Nucleotide Archive under the study accession numbers PRJEB5543 and PRJEB9368 for RNA-seq and DNA-seq, respectively.
Analysis of DNA-seq data
Sequences were first cleaned to remove adapter sequences and quality filtered (phred score > 28). After trimming, reads of < 50 bp were discarded. PE sequences were then mapped onto the total C. canephora (acc. DH200-94) reference genome (http://coffee-genome.org, Denoeud et al. 2014) using the BWA-MEM algorithm (Li and Durbin 2009; http://bio-bwa.sourceforge.net/) with the PE mode and default parameters. The resulting BAM files of unambiguously aligned sequences were then analyzed for SNP discovery with the GATK toolkit (http://www.broadinstitute.org/gatk/), using the Unified-Genotyper module with default parameters. For the detection of copy number changes, the BAM files were analyzed with FREEC (control-Free Copy number caller, Boeva et al. 2011) using a nonoverlapping 50-kb sliding window. The main steps are (1) normalization of the copy number profiles using GC content, (2) segmentation of normalized profiles, and (3) assignment of copy number changes to losses and gains.
SNP density was estimated along the 11 homeologous chromosome groups, and regions exhibiting homeologous SNP deficit (HSD) were identified using in-house Perl-scripts (available upon request). Nonoverlapping 10-kb sliding windows were used to estimate the SNP density along the C. canephora reference genome. To minimize the rate of false-positive SNPs, a minimum depth coverage of 10 was required for a position to be considered, while positions exhibiting depth coverage greater than twice the overall sample mean were discarded. Since the genome sequence of C. eugenioides is not yet available, expected local SNP density (in the absence of genetic changes in C. arabica) along the homeologous chromosome groups could not be determined. So, the overall mean SNP density was used as reference value, and the hypothesis of HSD was retained when at least five consecutive 10-kb windows (presenting a minimum of 8000 positions that satisfied the depth criteria) along the C. canephora reference genome were identified as exhibiting a highly significant SNP deficit using a test of proportions with R software (prop.test; Newcombe 1998). To control for multiple testing, the resulting P values were corrected using the Benjamini and Hochberg (1995) procedure. A cumulative window size of 50 kb was found to produce a good compromise between informativeness and accuracy.
RNA-seq data processing
The 72-nt reads of each library were mapped to a C. canephora coding DNA sequence reference (25574 CDS) as transcriptome reference using BWA MEM with the default parameters (http://coffee-genome.org; Denoeud et al. 2014). The aligned sequences of each library were then analyzed to find SNPs with the GATK toolkit using the Unified-Genotyper module with default parameters to obtain a list of SNPs and allelic data, and the Depth-Of-Coverage module to obtain information on depth coverage. Regarding the accession Caturra, analyses were performed using either a pool of reads from the four libraries or each individual library. To avoid artifacts due to reads from pseudogenes or repeat sequences, only CDS identified as single copy were used for subsequent analyses.
Comparison of SNPs determined by RNA-seq and DNA-seq
SNPs were quantified and compared using SNiPlay (http://www.southgreen.fr/)—a dedicated web-based tool (Dereeper et al. 2011). In particular, SNiPlay authorizes the user to set minimum depth coverage for a sequence position to be taken into consideration. To allow combined analysis of SNP-gene data generated by both RNA-seq and DNA-seq, the list of SNPs, allelic data, and depth coverage corresponding to the 25574 CDS reference were extracted from the whole genome DNA-seq data using the in-house Perl-script (available upon request) and the GFF file of the C. canephora reference genome (http://coffee-genome.org). For each gene, the SNPs detected in each of accessions analyzed by either DNA-seq or RNA-seq were determined and common sequence positions were compared. A minimum depth coverage of 12 was required for the DNA-seq in C. arabica to ensure good coverage of the two subgenomes, while the RNA-seq read cutoff was set at four and 16 for C. eugenioides and C. arabica, respectively, to account for the possibility of SNP allele low frequency due to homeolog expression bias in the allopolyploid (Yoo et al. 2014; Combes et al. 2013). In addition, SNP cut-off parameters were applied throughout the analyses in order to take into account the occurrence of sequencing (or SNP detection) errors, and the fact that the two diploid parental accessions used in this experiment are not the true diploid progenitors of the allotetraploid C. arabica.
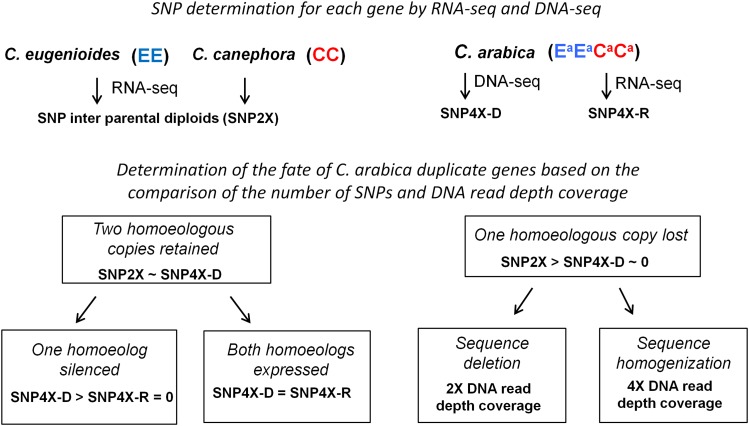
The fate of homeologous genes in C. arabica was investigated as shown in Figure 1. For each gene, SNP number and DNA read depth coverage comparisons were carried out. A minimum of three SNPs between the two diploid species, as well as in the allopolyploid, as determined by DNA-seq were selected as the cutoff to set a lower bound on the resolution of SNP number change for a given gene. First, the number of SNPs determined by DNA-seq in the allopolyploid (SNP4X-D) was compared to the number of SNPs between the two diploid species (SNP2X). For a given gene, when SNP2X and SNP4X-D were comparable, two homeologous copies were considered to have been retained. In contrast, genes displaying at least a fourfold decrease between SNP2X and SNP4X-D, and no more than one SNP classified as homeologous (SNP4X-D ≤ 1) were considered as exhibiting homeolog loss in the C. arabica accession concerned (when SNP4X-D was null, a minimum value of three was required for SNP2X). Second, for each gene presenting two homeologous copies, the number of SNPs determined in the allopolyploid by RNA-seq (SNP4X-R) and DNA-seq (SNP4X-D) were compared. Genes in the allopolyploid displaying at least three SNPs at the DNA level (SNP4X-D ≥ 3) and no SNPs at the RNA level (SNP4X-R = 0; presence of one SNP was tolerated if different than those detected by DNA-seq) were considered as exhibiting homeolog silencing in the C. arabica accession concerned. Others genes displaying similar SNPs at the DNA and RNA levels were considered as expressing both homeologs. Finally, for each gene presenting one homeolog loss, the average read coverage was compared to the expected values (99% confidence interval) for a gene with two copies and four copies, respectively, as reported previously (Lashermes et al. 2014). According to the gene read depth coverage categorization, the homeolog loss was inferred to result from either sequence deletion (i.e., two copies) or sequence homogenization (i.e., four copies).
Figure 1.
Flow chart of methods used to analyze the fate of duplicate genes in allotetraploid C. arabica.
Analysis of genes exhibiting homeolog loss and homeolog silencing
In each gene involved in homeolog loss and homeolog silencing, the numbers of SNPs shared by C. arabica and either C. canephora or C. eugenioides were compared to identify the subgenome in which the event occurred. Homeolog loss/silencing was attributed to the subgenome deriving from the diploid progenitor species with the smallest number of SNPs shared with C. arabica. A difference between the two SNP categories of at least three SNPs was required to determine the subgenome in which the homeolog loss or silencing occurred.
Computational mapping and Plant GO-slim annotation were performed using Blast2GO software v2.6.4 (http://www.blast2go.org; Conesa and Gotz 2008) as described in Combes et al. (2013). Functional enrichments in groups of interesting genes were investigated using Fisher’s exact test, applying a false discovery rate (FDR), and a correction for multiple testing.
Validation of genes exhibiting homeolog loss and silencing
Experimental validation of bioinformatically inferred genes exhibiting either homeolog lost or silencing was performed using traditional Sanger sequencing, as previously reported in Combes et al. (2012). For homeolog loss analysis, three genes from the two main regions (i.e., B and C, see Table 2) showing contiguous genes exhibiting homeolog losses in both accessions analyzed were selected (Supplemental Material, Table S4). Primer pairs were designed to amplify DNA fragments containing species-specific SNPs that differentiate the two diploid progenitor species, C. canephora and C. eugenioides (Combes et al. 2015). SNP detection assays were performed based on Sanger direct sequencing of DNA amplicons from 96 accessions originating from the main regions of C. arabica primary center of diversity (Labouisse et al. 2008) (Table S5). For silencing validation, a subset of five genes was selected, and primer pairs were designed to amplify single-exon fragments containing several homeologous SNP (Table S4). The expression of homeologs was analyzed using a SNP ratio quantification method based on dideoxy-terminated sequences of cDNA and DNA amplicons from C. arabica (acc. Caturra, AR41 and AR59) as described in Combes et al. (2012). PCR reactions were performed twice in a volume of 25 µl, with either 1 µl of the diluted (one-tenth) cDNA generated by the first-strand synthesis or 25 ng of genomic DNA, 0.4 µM of each primer, 2.5 mM MgCl2, and 1 U of Taq DNA Polymerase. Cycling was done in a GeneAmp PCR 9700 thermocycler for 2 min at 94° followed by five cycles of 10 sec at 94°, 30 sec at 60° to 55° (–1°/cycle), and by 30 cycles of 10 sec at 94°, 30 sec at 55°, 30 sec at 72°, and a final 8-min extension at 72°. Sequencing was performed by Beckman Coulter Genomics (Takeley UK). Sequence chromatograms of exon portions amplified on cDNA and DNA were analyzed and compared using BioEdit (Hall 1999).
Table 2. Characterization of four regions in C. arabica (acc. Caturra) carrying contiguous genes exhibiting homeolog losses, using the 11 chromosomes of C. canephora as genomic reference sequence.
| Region Code | Homeologous Chromosome Groups | Number of Genes Exhibiting Homeolog Loss | Gene Identifier (Start/End) | Size (kb) |
|---|---|---|---|---|
| A | 2 | 4 | Cc02g15410/Cc02g15440 | 15.4 |
| B | 2 | 7 | Cc02g39930/Cc02g39990 | 68.0 |
| C | 7 | 142 | Cc07g00010/Cc07g01770 | 1197.9 |
| D | 10 | 3 | Cc10g00010/Cc10g00030 | 27.8 |
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The entire sequence dataset has been deposited at European Nucleotide Archive under the study accession numbers PRJEB5543 and PRJEB9368 for RNA-seq and DNA-seq, respectively.
Results
Genomic structural variation
Genome sequencing of the two genetically distant accessions of C. arabica yielded 286 million 100-bp PE reads comprising 57.3 Gb of raw data (Table S1). After cleaning, 267 million (i.e., 93%) of the reads remained, and were mapped to the high-quality draft C. canephora genome sequence. The number of reads mapped onto the genome ranged from 106 million in accession AR41 to 159 million in accession Caturra. The average sequence depth of coverage of the reference canephora genome varied from 36× to 51× depending on the C. arabica accession.
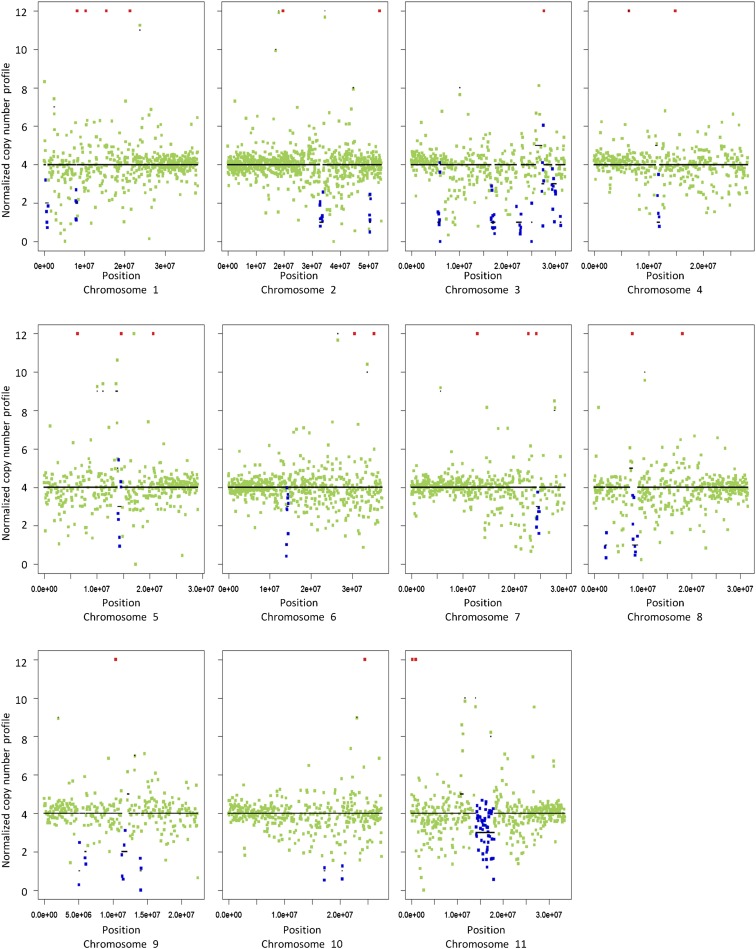
CNA based on the depth of coverage was assessed using FREEC (Boeva et al. 2011). Copy number profiles were established for the two C. arabica accessions using a nonoverlapping 50-kb sliding window, and normalization of GC content. The outcome for the accession Caturra is shown in Figure 2 as an example. Although few CNAs were predicted for each accession, the overall profiles appeared to be consistent with a 4× copy number along the 11 C. canephora chromosomes used as reference. Gross copy number changes due to large genomic duplications or deletions were not detected. These results are coherent with the expected allotetraploid structure of C. arabica.
Figure 2.
GC-content normalized DNA copy number profile, and FREEC-predicted copy number alteration (Red: gains; blue: losses) for the C. arabica genome (acc. Caturra) using a nonoverlapping 50-kb sliding window and the 11 chromosomes of C. canephora as genomic reference sequence. Automatically predicted copy numbers are shown in black (line).
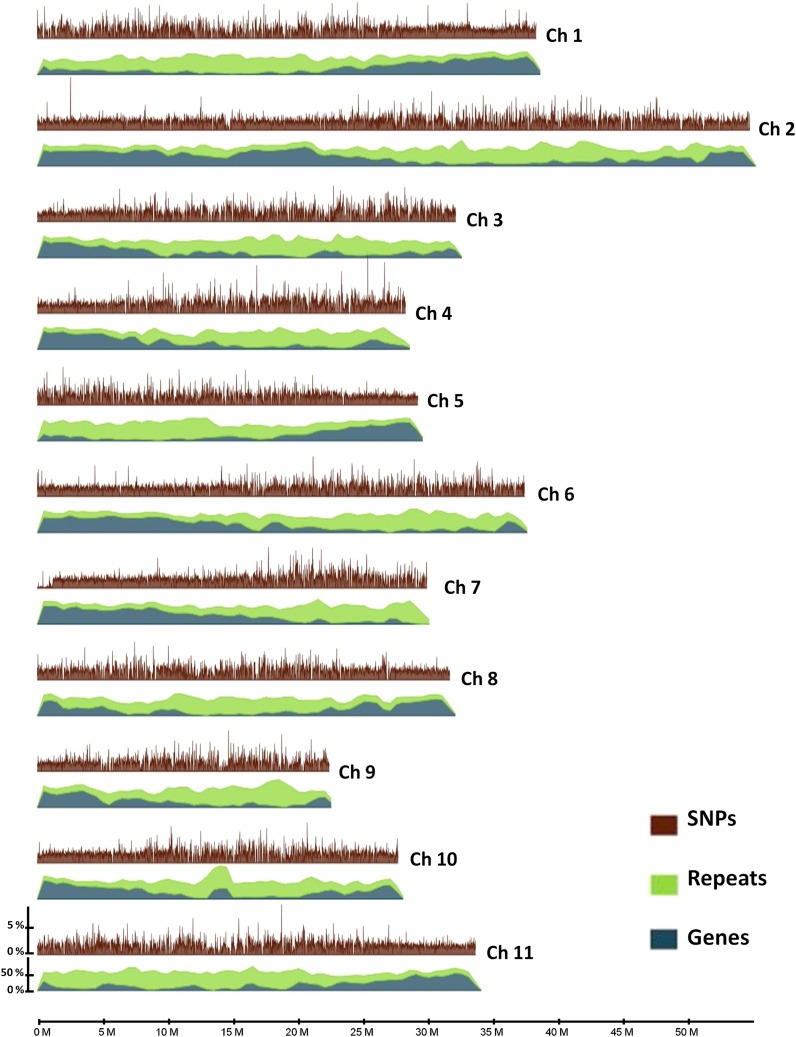
SNPs were detected in each of the two accessions of C. arabica. In allopolyploids such as C. arabica, sequence variations between subgenomes (homeologous SNPs) coexist with allelic variations within subgenomes (homologous SNPs). However, given the high level of homozygosity in C. arabica (Lashermes et al. 2014), it was assumed that most of the identified SNPs are homeologous SNPs. Nonoverlapping 10-kb sliding windows and coverage criteria required to consider a position to minimize the rate of false SNPs were applied to estimate the density of SNPs along the 11 homeologous chromosome groups. Whatever the accession considered, the overall SNP density appeared to be relatively constant along the different reference chromosomes, with a slight increase in the genomic regions enriched in transposable elements (Figure 3). In the two accessions and the regions analyzed, the structural heterozygosity observed was in line with what is expected in an allotetraploid. Nevertheless, regions exhibiting homeologous SNP deficit (HSD) were identified (Figure 4), e.g., 39 regions exhibiting HSD were revealed in C. arabica acc. Caturra (Table S2). Of these regions, 37 (i.e., 95%) were shared by the two accessions analyzed. In addition, except the region (1170 kb in size) identified on chromosome 7, the regions were rather small, ranging from 50 kb to 160 kb with an average of 81 kb. The overall size of these regions was 4230 kb, representing 1.5% of the genome windows analyzed that satisfied the depth criteria.
Figure 3.
SNP density along the 11 homeologous chromosome groups in C. arabica (acc. Caturra). The 11 chromosomes of C. canephora (acc. DH200-94) were used as reference genome. Nonoverlapping 10-kb sliding windows, and coverage criteria required to consider a position to minimize the rate of false SNPs, were applied to estimate the density of SNPs in C. arabica. The relative proportion (percentage nucleotides) in C. canephora (1-Mb sliding window) of transposable elements (green) and genes (blue) are shown at the bottom.
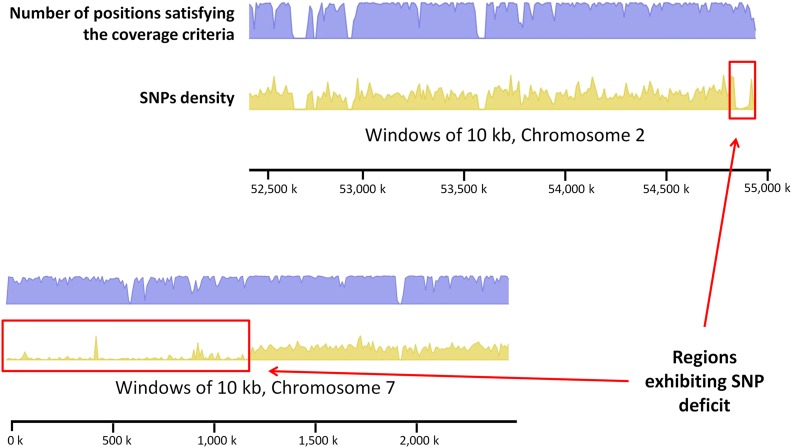
Figure 4.
Examples of regions exhibiting homeologous SNP deficit in C. arabica (acc. Caturra) on homeologous chromosome groups 2 and 7. Nonoverlapping 10-kb sliding windows were used to estimate SNP density in C. arabica (Yellow track). To minimize the rate of false-positive SNPs, a minimum depth coverage of 10 was required for a position to be taken into consideration, and positions with a depth coverage more than twice the overall sample mean were discarded (Blue track).
Evidence of homeolog loss
The fate of homeologous genes in C. arabica was inferred as described in Figure 1. The number of SNPs per gene detected by DNA-seq in individual accessions of C. arabica were compared to those detected between accessions of its two diploid progenitor species in common sequence positions. A set of 9047 genes that satisfied depth and quality requirements was analyzed in the two arabica accessions. While two homeologous copies appeared to be retained in 98% of the genes, 179 genes (2.0%) exhibiting homeolog loss were identified (Table 1). Although the number of homeolog losses varied from 148 (Caturra) to 174 (AR41) among the two C. arabica accessions, a large proportion (80%) of genes exhibiting homeolog loss was shared by the two accessions.
Table 1. Determination of the subgenome origin of homeolog loss (A) and silencing events (B) detected in two accessions of C. arabica.
| (A) C. arabica Accession | Number of Homeolog Loss Events | Subgenome Origin of Homeolog Losses | ||
|---|---|---|---|---|
| Ca | Ea | Not Determined | ||
| AR41 | 174 | 120 | 39 | 15 |
| Caturra | 148 | 110 | 27 | 11 |
| Loss shared by both accessions | 143 | 108 | 25 | 10 |
| Loss not shared | 36 | 14 | 17 | 5 |
| (B) C. arabica Accession | Number of Silenced Genes | Subgenome Origin Of Silenced Genes | ||
|---|---|---|---|---|
| Ca | Ea | Not Determined | ||
| AR41 | 120 | 61 | 37 | 22 |
| Caturra | 116 | 60 | 39 | 17 |
| Silencing shared by both accessions | 80 | 47 | 24 | 9 |
| Silencing not shared | 76 | 30 | 33 | 13 |
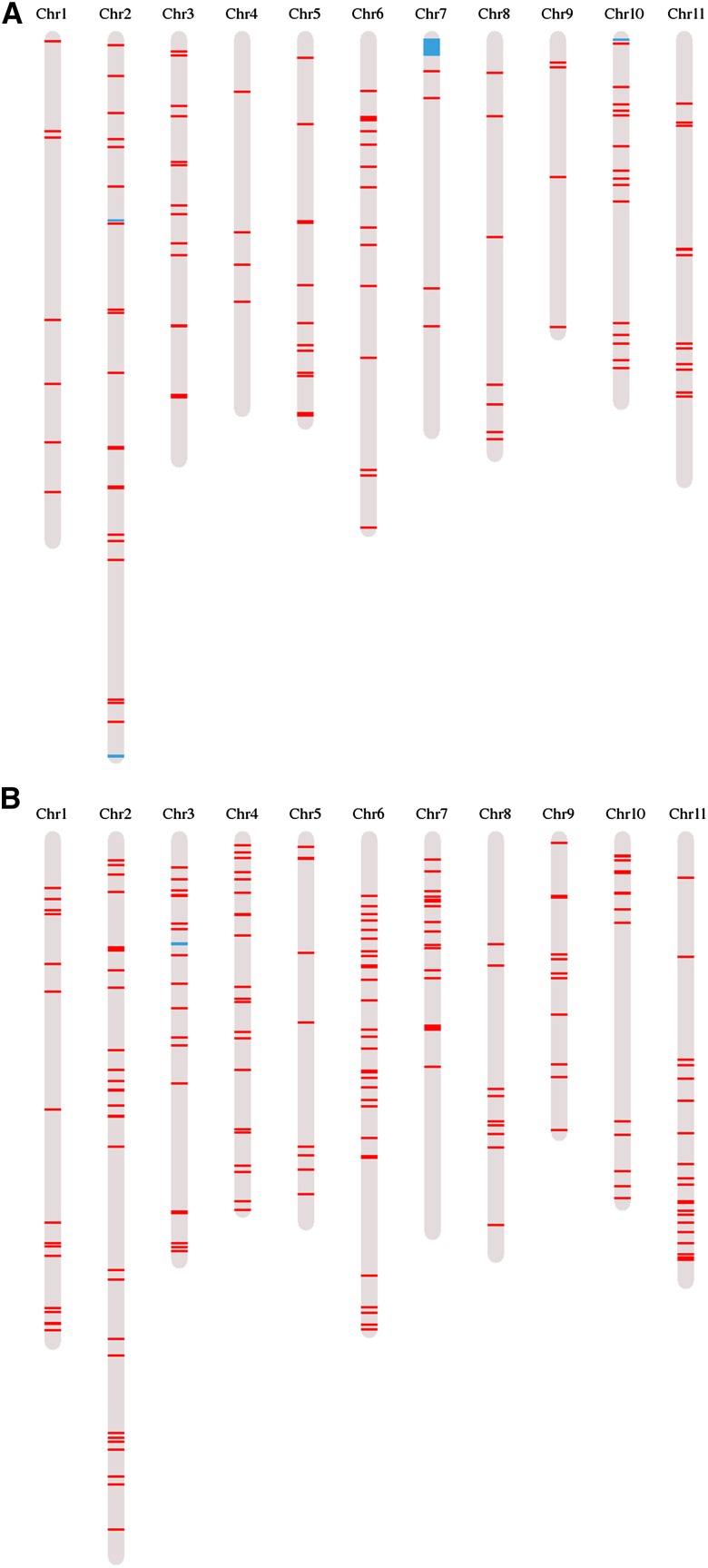
The distribution across the reference genome of loci exhibiting homeolog loss was next investigated (Figure 5). While homeolog loss events were observed across the 11 reference chromosomes, four regions carrying contiguous genes exhibiting homeolog losses were detected (Table 2). The number of genes per region in the accession Caturra ranged from three to 142, and the corresponding genome fragment size ranged from 15 kb to 1198 kb. The four regions were shared by the two accessions, and corresponded to genomic regions previously identified as exhibiting HSD. In contrast, all the single genes exhibiting homeolog losses that did not belong to the four identified clusters corresponded to genome regions (10 kb windows) exhibiting standard SNP density.
Figure 5.
Distribution of loci exhibiting either homeolog loss (A) or homeolog silencing (B) identified in C. arabica (acc. Caturra) across C. canephora reference chromosomes. Single events of either homeolog loss or homeolog silencing are in red, and regions carrying contiguous genes exhibiting either homeolog loss or homeolog silencing are in blue.
The subgenome origin of the detected homeolog loss events was further investigated (Table 1). Homeolog loss was attributed to the subgenome deriving from the diploid progenitor species with the least SNPs shared with C. arabica. Whatever the accession, homeolog losses were attributed to the two subgenomes, but with a marked preference (at least 75%) for subgenome Ca. Contrasted behavior was observed between the groups of genes exhibiting homeolog loss shared by the two accessions, or restricted to one accession. While the homeolog losses shared by accessions were attributed mainly to subgenome Ca (i.e., 81%), the subgenome origin of homeolog loss events specific to one accession appeared to be balanced between Ca and Ea.
The mechanisms behind the homeolog loss events were further investigated. To distinguish between sequence deletion and sequence homogenization, the gene copy numbers in C. arabica were estimated using the DNA-seq read depth of coverage (Figure S1). Indeed, homeolog sequence deletion in an allotetraploid is expected to be associated with a decrease in gene copy number from 4× to 2×. The distribution of genes exhibiting homeolog loss according to their average read depth was very similar to the distribution of the overall genes, supporting the hypothesis that most of the observed homeolog losses are not associated with mere gene loss. Nevertheless, genes that exhibited an average read depth corresponding to the expected coverage for gene in two copies were overrepresented in the group of genes exhibiting homeolog loss (Chi-squared test, P-value < 0.0001) suggesting that a few homeolog loss events could result from sequence deletion. For instance in C. arabica acc. Caturra, of the 148 genes identified as exhibiting homeolog loss, 12 (8.1%) displayed an average read depth corresponding to the expected coverage for gene in two copies.
Homeolog loss was validated using direct Sanger sequencing of amplicons for three genes (Table S4) from the two main regions showing contiguous genes exhibiting homeolog losses in the two examined arabica accessions (i.e., regions B and C, see Table 2). The frequency of these homeolog loss events among the C. arabica germplasm was further investigated. These events appeared shared by all of the 96 analyzed accessions that represent the main coffee growing regions in the C. arabica primary center of diversity.
Evidence of homeolog silencing
The occurrence of homeolog silencing in C. arabica accessions was inferred by comparing the number of SNPs determined by DNA-seq (SNP4X-D) and RNA-seq (SNP4X-R) in common sequence positions in each individual gene (Figure 1). It was assumed that, in the absence of homeolog silencing in C. arabica, the number of SNPs determined by DNA-seq and RNA-seq would be equivalent, whereas the failure to detect homeologous SNPs by RNA-seq would reveal homeolog silencing. For most of the 9047 genes analyzed in the two accessions, the number of SNPs determined by DNA-seq and RNA-seq was comparable, and, overall, 95% of the SNPs identified with the two methods were similar. Taking into account the two C. arabica accessions, 156 genes (1.7%) exhibiting homeolog silencing were identified (Table 1). The number of genes exhibiting homeolog silencing varied from 116 (Caturra) to 120 (AR41) between the two accessions of C. arabica, with 80 genes (51.3%) shared by the two accessions, and 49% of the silenced genes specific to one of the accessions. Homeolog silencing were examined among the four individual plants of the accession Caturra. After filtering for the depth coverage, 66 of 120 genes exhibiting homeolog silencing in Caturra were investigated. For all of them, homeolog silencing was observed in all individual plants.
The distribution across the reference genome of loci exhibiting homeolog silencing was next investigated. The distribution observed for accession Caturra is shown in Figure 5 as an example. Homeolog silencing events were observed across the 11 reference chromosomes, and two small regions (92 kb and 59 kb respectively on chromosome 2 and unanchored scaffold) carrying a cluster of genes exhibiting homeolog silencing were detected in two accessions.
The subgenome origin of the homeolog silencing events was further investigated (Table 1). Homeolog silencing was attributed to the subgenome deriving from the diploid progenitor species displaying the smallest number of SNPs shared with C. arabica. Whatever the accession, homeolog silencing was attributed to the two subgenomes with a marked preference for subgenome Ca (from 60.6% to 62.2% depending on the accession). This preference (i.e., 66.2% of Ca silenced homeologs) was observed particularly among the group of genes exhibiting homeolog silencing shared by the two accessions.
Homeolog silencing was validated using direct Sanger sequencing of cDNA and DNA amplicons from C. arabica (acc. Caturra, AR41 and AR59). Primer pairs were designed to amplify single-exon fragments of five genes for which homeolog silencing was bioinformatically inferred (Table S2). For all the genes assayed, expression of only one homeolog from either the subgenomes Ca or Ea was detected as expected from the previous analyses.
Gene ontology enrichment analysis
Gene ontology (GO) enrichment was analyzed in groups of genes showing either homeolog loss or homeolog silencing using the full set of analyzed genes as reference group. No significant variation was observed in the group of genes exhibiting homeolog loss, whereas statistically highly significant impoverishments in very general GO terms were observed in the group of genes exhibiting homeolog silencing (Table S3). In particular, specific functions related to macromolecular biosynthesis and organic cyclic compound metabolism were under-represented in the group of genes exhibiting homeolog silencing shared by the two accessions (Table 3).
Table 3. Gene ontology enrichment analysis of genes exhibiting homeolog silencing shared by the two accessions of C. arabica analyzed.
| GO-ID | Term | Category | FDR | P-Value | Test | Ref | notAnnotTest | notAnnotRef | Over/Under |
|---|---|---|---|---|---|---|---|---|---|
| GO:0043229 | Intracellular organelle | C | 4.151711E-5 | 3.610184E-7 | 13 | 3977 | 53 | 3898 | Under |
| GO:0043226 | Organelle | C | 4.151711E-5 | 3.610184E-7 | 13 | 3977 | 53 | 3898 | Under |
| GO:0043227 | Membrane-bounded organelle | C | 7.544177E-5 | 1.312031E-6 | 13 | 3881 | 53 | 3994 | Under |
| GO:0043231 | Intracellular membrane-bounded organelle | C | 7.544177E-5 | 1.312031E-6 | 13 | 3881 | 53 | 3994 | Under |
| GO:0005623 | Cell | C | 1.361508E-4 | 3.219604E-6 | 24 | 5116 | 42 | 2759 | Under |
| GO:0044464 | Cell part | C | 1.361508E-4 | 3.561238E-6 | 24 | 5098 | 42 | 2777 | Under |
| GO:0044424 | Intracellular part | C | 1.361508E-4 | 4.143721E-6 | 19 | 4510 | 47 | 3365 | Under |
| GO:0034641 | Cellular nitrogen compound metabolic process | P | 1.882623E-4 | 6.944386E-6 | 2 | 1852 | 64 | 6023 | Under |
| GO:0005622 | Intracellular | C | 1.882623E-4 | 7.366786E-6 | 21 | 4679 | 45 | 3196 | Under |
| GO:0006725 | Cellular aromatic compound metabolic process | P | 5.411463E-4 | 3.058653E-5 | 2 | 1720 | 64 | 6155 | Under |
| GO:1901360 | Organic cyclic compound metabolic process | P | 5.411463E-4 | 3.058653E-5 | 2 | 1720 | 64 | 6155 | Under |
| GO:0046483 | Heterocycle metabolic process | P | 5.411463E-4 | 3.058653E-5 | 2 | 1720 | 64 | 6155 | Under |
| GO:0006139 | Nucleobase-containing compound metabolic p. | P | 5.411463E-4 | 3.058653E-5 | 2 | 1720 | 64 | 6155 | Under |
| GO:0005634 | Nucleus | C | 8.456478E-3 | 5.147422E-4 | 2 | 1413 | 64 | 6462 | Under |
| GO:0044444 | Cytoplasmic part | C | 8.769347E-3 | 6.260661E-4 | 14 | 3281 | 52 | 4594 | Under |
| GO:0010467 | Gene expression | P | 8.769347E-3 | 6.331017E-4 | 0 | 912 | 66 | 6963 | Under |
| GO:0003676 | Nucleic acid binding | F | 8.769347E-3 | 6.481691E-4 | 1 | 1146 | 65 | 6729 | Under |
| GO:0034645 | Cellular macromolecule biosynthetic process | P | 1.039839E-2 | 9.925819E-4 | 0 | 860 | 66 | 7015 | Under |
| GO:0044249 | Cellular biosynthetic process | P | 1.039839E-2 | 9.925819E-4 | 0 | 860 | 66 | 7015 | Under |
| GO:0009059 | Macromolecule biosynthetic process | P | 1.039839E-2 | 9.925819E-4 | 0 | 860 | 66 | 7015 | Under |
| GO:0044271 | Cellular nitrogen compound biosynthetic proc. | P | 1.039839E-2 | 9.925819E-4 | 0 | 860 | 66 | 7015 | Under |
| GO:1901576 | Organic substance biosynthetic process | P | 1.039839E-2 | 9.946287E-4 | 0 | 861 | 66 | 7014 | Under |
| GO:0006807 | Nitrogen compound metabolic process | P | 1.267849E-2 | 1.267849E-3 | 7 | 2175 | 59 | 5700 | Under |
| GO:0005737 | Cytoplasm | C | 2.685331E-2 | 2.802085E-3 | 19 | 3719 | 47 | 4156 | Under |
| GO:0044237 | Cellular metabolic process | P | 2.979136E-2 | 3.238192E-3 | 14 | 3065 | 52 | 4810 | Under |
Analysis was performed using the full set of 9047 analyzed genes as reference group and Fisher’s exact test with a false discovery rate (FDR) correction for multiple testing.
Discussion
Plant allopolyploidy appeared to be associated with an array of rapid genomic changes in genetic/epigenetic, transcriptomic, and proteomic layers that may affect the fitness of the newly formed allopolyploid and increase its competitiveness, leading to its successful establishment in nature (Arrigo and Barker 2012). In the present study, several approaches based on high-throughput sequencing technologies were used to investigate these genetic changes in C. arabica—a model allopolyploid perennial plant. The use of a high-quality draft genome sequence of C. canephora (Denoeud et al. 2014) as genome and transcriptome references offered new opportunities for analyses compared with previous works (Lashermes et al. 2014). However, inherent limitations remained such as the impossibility to study putative genomic regions of C. arabica that do not have a counterpart in the C. canephora reference genome, and the fact that interpretation of the subgenome (i.e., parental) of the identified homeologous SNPs was limited to the part of the genome for which reference sequences of both diploid parents were available. Nevertheless, whole genome analyses and comparison of two geographically and genetically distant accessions of C. arabica provided compelling evidence for the efficiency of sequencing based methods for the investigation of genetic changes in allopolyploid, and of the mechanisms that lead to these changes, their timing, and their directed or random nature.
Fixed heterozygosity and allopolyploid structure
Although challenging because of the low divergence between the two diploid constitutive subgenomes, the allotetraploid genome organization of C. arabica was first revealed by genomic in situ hybridization (Lashermes et al. 1999). The present data confirm a state of fixed heterozygosity related to the presence of two complete sets of homeologous chromosomes in C. arabica. Whatever the accession considered, the observed pattern of SNP density along the 11 homeologous chromosome groups in C. arabica is consistent with its assumed allopolyploid structure. Furthermore, large genomic duplications or deletions were not detected, confirming low chromosomal divergence between genomes E and C of the two diploid progenitor species (Pinto-Maglio 2006), and suggesting that, following the original hybridization event, overall genome organization is stable in C. arabica.
Evidence for homeologous exchanges
Homeologous exchanges appear to have played an important role in the evolution of the C. arabica genome. Indeed, regions exhibiting homeologous SNP deficit (HSD) and 4× copy number were identified across the genome. These regions, which ranged from 50 kb to 1170 kb in size, suggest major genetic changes. Although certainly underestimated, since the method used here cannot resolve small regions (a minimum of 50 kb was required), these regions represented nearly 1.5% of the portion of C. arabica genome analyzed. Furthermore, among the 9047 duplicate homeologous genes whose fate was successfully inferred in the two accessions of C. arabica analyzed, 2.0% were found to be affected by genomic changes leading to homeolog loss without sequence deletion in most cases sequence. Two mechanisms have been suggested to lead to such exchanges of homeologous chromatids: via crossovers and the subsequent segregation of one parental and one recombinant chromatid, or via noncrossover exchange, also called gene conversion (Gaeta and Pires 2010). Due to their large size, homeologous crossover exchanges are the most likely mechanism at the origin of the C. arabica regions exhibiting HSD. In contrast, both mechanisms are hypothesized to be involved in the occurrence of genes exhibiting homeolog losses. While the regions carrying contiguous genes exhibiting homeolog losses correspond to regions exhibiting HSD, most single homeolog loss events likely result from gene conversion. The high frequency of homeologous contact across the 11 homeologous chromosome groups is a surprising result given the apparently complete bivalent formation at meiosis. However, meiotic abnormalities have been repeatedly observed in C. arabica (Grassias and Kammacher 1975; Owuor 1985). Although exceptional, weakness in the control of the chromosome pairing in C. arabica could therefore enable homeologous exchanges. A possibility is that most of these homeologous exchanges, having occurred early, could be the result of possible multivalent (or less strict bivalent) pairing at meiosis at inception. Similar high frequencies of noncrossover homeologous exchanges have been reported in other genome-wide analyzed allotetraploids such as Gossypium hirsutum and Brassica napus (Salmon et al. 2010; Guo et al. 2014; Chalhoub et al. 2014), suggesting an important mechanism by which allopolyploid genomes adapt to the duplicate state.
Furthermore, the two main homeologous crossover exchanges detected in the two sequenced accessions of C. arabica were also evidenced in all analyzed accessions (i.e., 96 accessions) originating from the main regions of C. arabica primary center of diversity. This observation strongly support the hypothesis of a single origin for C. arabica (Lashermes et al. 2014). All C. arabica populations would derive from a unique allopolyploidization event associated with large and specific homeologous crossover exchanges.
Homeolog silencing suggests epigenetic changes
Gene silencing is a common response to polyploidy and has been described in many allopolyploids, including Arabidopsis (Wang et al. 2006), cotton (Chaudhary et al. 2009), Tragopogon (Buggs et al. 2011), and wheat (Bottley et al. 2006). Silencing can occur as early as the first generation following polyploidy, and some duplicated genes may be silenced in some organs of the plant but continue to be expressed in other organs (Adams and Wendel 2005). By combining RNA-seq and DNA-seq data, homeolog silencing was inferred for 1.7% of genes in the two accessions of C. arabica analyzed in the present study. Since gene expression was estimated only in leaves, partition of the expression of duplicate genes to specialized tissue-specific expression activity was not investigated. Silencing mechanisms almost certainly vary with the gene. In the absence of sufficient time for point mutations to accumulate in promoter regions or other cis-regulatory elements, most gene silencing is believed to be induced epigenetically (Adams and Wendel 2005). Intergenomic interactions between progenitor genomes in allopolyploids are predicted to induce epigenetic changes, including histone modifications and DNA methylation (Song and Chen 2015). In the last few years, gene silencing via epigenetic changes has been documented in several plants including Tragopogon (Sehrish et al. 2014), Arabidopsis (Chen et al. 2008), and Brassica rapa (Cheng et al. 2016).
Two temporally distinct phases of evolution
The implication of homeolog loss and silencing events in the establishment and diversification of C. arabica is questionable (Madlung and Wendel 2013). Comparison of the two analyzed accessions revealed the occurrence of shared genomic changes, whereas other events appeared specific to one accession. While 80% of genes exhibiting homeolog loss were shared by the two accessions, only half of genes exhibiting homeolog silencing seemed shared by the two accessions. In particular, the four detected genomic regions exhibiting HSD and contiguous homeolog losses were shared by the two accessions. Two temporally distinct phases of evolution can therefore be hypothesized. A first phase accompanying the allopolyploidization process, and involving mainly homeologous crossover exchanges, could have been followed by a more gradual phase of duplicate gene evolution involving gene conversion and homeolog silencing.
Furthermore, patterns of gene loss and retention could be explained by the gene-balance hypothesis (Freeling 2009; Feldman et al. 2012). Under this hypothesis, the evolution of genes is linked to their function within networks (Edger and Pires 2009), so genes coding for products that are closely connected would be dosage-sensitive genes, and are thought to be retained or eliminated together to preserve stoichiometry. This evolution mechanism was recently proposed for homeologs in T. miscellus, a young natural allopolyploid species (Buggs et al. 2012). This process is probably marginal in the pattern of gene loss observed in C. arabica given the lack of significant GO term enrichment in the subset of genes showing homeolog loss. However, an equivalent trend might play a role in the gene silencing pattern of C. arabica. Indeed, genes related to macromolecular biosynthesis and organic cyclic compound metabolism tended to be preserved from gene silencing, suggesting gene dosage requirements (Veitia et al. 2013).
Asymmetric evolution
Studies on duplicated genomes or genomic regions in ancient polyploids showed that they have often experienced unequal gene losses (or genome fractionation), with one genome or genomic region retaining more genes (dominant) than the other (more fractionated). More interestingly, genes located on the dominant genome or genomic region tend to have higher expression levels (Schnable et al. 2011; Cheng et al. 2012; Garsmeur et al. 2014; Li et al. 2014). Such genome dominance phenomena have been reported in a few plants including Arabidopsis thaliana (Wang et al. 2006), Brassica (Cheng et al. 2012), maize (Schnable et al. 2011), and wheat (Li et al. 2014; Wang et al. 2016). In C. arabica, homeolog loss and silencing were attributed to both subgenomes, with a marked preference for subgenome Ca, suggesting dominance of the Ea genome. However, neither of the two subgenomes appeared to be preferentially expressed in C. arabica (Combes et al. 2013), or in interspecific hybrids between C. canephora and C. eugenioides (Combes et al. 2015). The absence of relationships between the observed preferential changes in the Ca genome and overall homeologous expression could be linked to the recent origin and the intertwined homeolog regulations occurring in C. arabica (Combes et al. 2013).
Conclusions
The present results point to overall structural genome stability in C. arabica following the original hybridization event. However, homeologous DNA exchanges and homeolog silencing were evidenced that could have played a major role in the stabilization and survival of the ancestral allotetraploid, and in its subsequent diversification. While the early phase of evolution involved mainly homeologous crossover exchanges, the later phase appears to have relied on a more gradual phase of duplicate gene evolution involving gene conversion and homeolog silencing. As suggested in C. arabica, the evolutionary success of a newly formed polyploidy may require a delicate balance between genetic and epigenetic changes.
Supplementary Material
Acknowledgments
We thank Dr. V. Boeva for advice on the use of the FreeC tool to identify copy number alteration. Also, we acknowledge M. S. Seyoum (Rothamsted International African Fellow-2006) from the Awada Coffee Research Centre (Ethiopia) for providing leaf samples. This work was partially supported by a grant from the Direction de la Valorisation au Sud de l’Institut de Recherche pour le Développement (IRD, France).
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.030858/-/DC1
Communicating editor: A. H. Paterson
Literature Cited
- Adams K. L., Wendel J. F., 2005. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Anthony F., Berthaud J., Guillaumet J. L., Lourd M., 1987. Collecting wild Coffea species in Kenya and Tanzania. Plant Genet. Resour. Newsl. 69: 23–29. [Google Scholar]
- Arrigo N., Barker M. S., 2012. Rarely successful polyploids and their legacy in plant genomes. Curr. Opin. Plant Biol. 15: 140–146. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Boeva V., Zinovyev A., Bleakley K., Vert J. P., Janoueix-Lerosey I., et al. , 2011. Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics 27: 268–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottley A., Xia G. M., Koebner R. M. D., 2006. Homoeologous gene silencing in hexaploid wheat. Plant J. 47: 897–906. [DOI] [PubMed] [Google Scholar]
- Buggs R. J. A., Zhang L., Miles N., Tate J. A., Gao L., et al. , 2011. Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr. Biol. 21: 551–556. [DOI] [PubMed] [Google Scholar]
- Buggs R. J. A., Chamala S., Wu W., Tate J. A., Schnable P. S., et al. , 2012. Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Curr. Biol. 22: 248–252. [DOI] [PubMed] [Google Scholar]
- Cenci A., Combes M. C., Lashermes P., 2012. Genome evolution in diploid and tetraploid Coffea species as revealed by comparative analysis of orthologous genome segments. Plant Mol. Biol. 78: 135–145. [DOI] [PubMed] [Google Scholar]
- Chalhoub B., Denoeud F., Liu S., Parkin I. A., Tang H., et al. , 2014. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345: 950–953. [DOI] [PubMed] [Google Scholar]
- Chaudhary B., Flagel L., Stupar R. M., Udall J., Verma N., et al. , 2009. Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium). Genetics 182: 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Ha M., Lackey E., Wang J., Chen Z. J., 2008. RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics 178: 1845–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Wu J., Fang L., Sun S., Liu B., et al. , 2012. Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One 7: e36442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Sun C., Wu J., Schnable J., Woodhouse M. R., et al. , 2016. Epigenetic regulation of subgenome dominance following whole genome triplication in Brassica rapa. New Phytol. DOI: 10.1111/nph.13884. [DOI] [PubMed] [Google Scholar]
- Comai L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Combes M. C., Cenci A., Baraille H., Bertrand B., Lashermes P., 2012. Homeologous gene expression in response to growing temperature in a recent allopolyploid (Coffea arabica L.). J. Hered. 103: 36–46. [DOI] [PubMed] [Google Scholar]
- Combes M. C., Dereeper A., Severac D., Bertrand B., Lashermes P., 2013. Contribution of subgenomes to the transcriptome and their intertwined regulation in the allopolyploid Coffea arabica L. grown at contrasted temperatures. New Phytol. 200: 251–260. [DOI] [PubMed] [Google Scholar]
- Combes M. C., Hueber Y., Dereeper A., Rialle S., Herrera J. C., et al. , 2015. Regulatory divergence between parental alleles determines gene expression patterns in hybrids. Genome Biol. Evol. 7: 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Gotz S., 2008. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008: 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F., Carretero-Paulet L., Dereeper A., Droc G., Guyot R., et al. , 2014. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345: 1181–1184. [DOI] [PubMed] [Google Scholar]
- Dereeper A., Nicolas S., Lecunff L., Bacilieri R., Doligez A., et al. , 2011. SNiPlay: a web-based tool for detection, management and analysis of SNPs. Application to grapevine diversity projects. BMC Bioinformatics 12(1): 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Flagel L. E., Paterson A. H., Rapp R. A., Soltis D. E., et al. , 2008. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42: 443–461. [DOI] [PubMed] [Google Scholar]
- Edger P. P., Pires J. C., 2009. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 17: 699–717. [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A., Fahima T., Korol A., 2012. Genomic asymmetry in allopolyploid plants: wheat as a model. J. Exp. Bot. 63: 5045–5059. [DOI] [PubMed] [Google Scholar]
- Flagel L. E., Wendel J. F., 2010. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 186: 184–193. [DOI] [PubMed] [Google Scholar]
- Freeling M., 2009. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60: 433–453. [DOI] [PubMed] [Google Scholar]
- Gaeta R.T., Pires J.C., Iniguez-Luy F., Leon E., Osborn T.C., 2007. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta R. T., Pires J. C., 2010. Homoeologous recombination in allopolyploids: the polyploid ratchet. New Phytol. 186: 18–28. [DOI] [PubMed] [Google Scholar]
- Garsmeur O., Schnable J. C., Almeida A., Jourda C., D’Hont A., et al. , 2014. Two evolutionarily distinct classes of paleopolyploidy. Mol. Biol. Evol. 31: 448–454. [DOI] [PubMed] [Google Scholar]
- Grassias M., Kammacher P., 1975. Observations sur la conjugaison chromosomique de Coffea arabica L. Cafe, Cacao, The (Paris) 19: 177–190. [Google Scholar]
- Guillaumet J.L., Hallé F., 1978. Echantillonnage du matériel récolté en Ethiopie. Bull IFCC 14: 13–18. [Google Scholar]
- Guo H., Wang X., Gundlach H., Mayer K. F. X., Peterson D. G., et al. , 2014. Extensive and biased intergenomic nonreciprocal DNA exchanges shaped a nascent polyploid genome, Gossypium (Cotton). Genetics 197: 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium 41: 95–98. [Google Scholar]
- Jackson S., Chen Z. J., 2010. Genomic and expression plasticity of polyploidy. Curr. Opin. Plant Biol. 13: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y. N., Wickett N. J., Ayyampalayam S., Chanderbali A. S., Landherr L., et al. , 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Labouisse J. P., Bellachew B., Kotecha S., Bertrand B., 2008. Current status of coffee (Coffea arabica L.) genetic resources in Ethiopia: implications for conservation. Genet. Resour. Crop Evol. 55: 1079–1093. [Google Scholar]
- Lashermes P., Combes M. C., Robert J., Trouslot P., D’Hont A., et al. , 1999. Molecular characterisation and origin of the Coffea arabica L. genome. Mol. Genet. Genomics 261: 259–266. [DOI] [PubMed] [Google Scholar]
- Lashermes P., Paczek V., Trouslot P., Combes M. C., Couturon E., et al. , 2000. Single-locus inheritance in the allotetraploid Coffea arabica L. and interspecific hybrid C. arabica x C. canephora. J. Hered. 91: 81–85. [DOI] [PubMed] [Google Scholar]
- Lashermes P., Combes M. C., Hueber Y., Severac D., Dereeper A., 2014. Genome rearrangements derived from homoeologous recombination following allopolyploidy speciation in coffee. Plant J. 78: 674–685. [DOI] [PubMed] [Google Scholar]
- Lee H. S., Chen Z. J., 2001. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc. Natl. Acad. Sci. USA 98: 6753–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch A. R., Leitch I. J., 2008. Perspective—Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483. [DOI] [PubMed] [Google Scholar]
- Li A., Liu D., Wu J., Zhao X., Hao M., et al. , 2014. mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26: 1878–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Brubaker C. L., Mergeai G., Cronn R. C., Wendel J. F., 2001. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome 44: 321–330. [PubMed] [Google Scholar]
- Madlung A., Wendel J. F., 2013. Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet. Genome Res. 140: 270–285. [DOI] [PubMed] [Google Scholar]
- Newcombe R. G., 1998. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat. Med. 17: 857–872. [DOI] [PubMed] [Google Scholar]
- Owuor J. B. O., 1985. Interspecific hybridization between Coffea arabica L. and tetraploid C. canephora P. Ex Fr. II. Meiosis in F1 hybrids and back crosses to C. arabica. Euphytica 34: 355–360. [Google Scholar]
- Pinto-Maglio C. A. F., 2006. Cytogenetics of coffee. Braz. J. Plant Physiol. 18: 37–44. [Google Scholar]
- Salmon A., Flagel L., Ying B., Udall J. A., Wendel J. F., 2010. Homoeologous nonreciprocal recombination in polyploid cotton. New Phytol. 186: 123–134. [DOI] [PubMed] [Google Scholar]
- Schnable J. C., Springer N. M., Freeling M., 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehrish T., Symonds V. V., Soltis D. E., Soltis P. S., Tate J. A., 2014. Gene silencing via DNA methylation in naturally occurring Tragopogon miscellus (Asteraceae) allopolyploids. BMC Genomics 15: 701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D. E., Buggs R. J. A., Doyle J. J., Soltis P. S., 2010. What we still don’t know about polyploidy. Taxon 59: 1387–1403. [Google Scholar]
- Soltis D. E., Visger C. J., Soltis P. S., 2014. The polyploidy revolution then...and now: Stebbins revisited. Am. J. Bot. 101: 1057–1078. [DOI] [PubMed] [Google Scholar]
- Song Q., Chen Z. J., 2015. Epigenetic and developmental regulation in plant polyploids. Curr. Opin. Plant Biol. 24: 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvain P. G., 1955. Some observations on Coffea arabica L. in Ethiopia. Turrialba 5: 37–53. [Google Scholar]
- Thomas A. S., 1942. The wild arabica coffee on the Boma Plateau, Anglo-Egyptian Sudan. Empire Journal Experimental Agriculture 10: 207–212. [Google Scholar]
- Van der Vossen H. A. M., Bertrand B., Charrier A., 2015. Next generation variety development for sustainable production of arabica coffee (Coffea arabica L.): a review. Euphytica DOI: 10.1007/s10681-015-1398-z. [DOI] [Google Scholar]
- Veitia R. A., Bottani S., Birchler J. A., 2013. Gene dosage effects: nonlinearities, genetic interactions, and dosage compensation. Trends Genet. 29: 385–393. [DOI] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H. S., Wei N. E., Jiang H., et al. , 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang H., Li Y., Zhang Z., Li L., et al. , 2016. Transcriptome asymmetry in synthetic and natural allotetraploid wheats, revealed by RNA-sequencing. New Phytol. 209: 1264–1277. [DOI] [PubMed] [Google Scholar]
- Yoo M. J., Liu X., Pires J. C., Soltis P. S., Soltis D. E., 2014. Nonadditive gene expression in polyploids. Annu. Rev. Genet. 48: 485–517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The entire sequence dataset has been deposited at European Nucleotide Archive under the study accession numbers PRJEB5543 and PRJEB9368 for RNA-seq and DNA-seq, respectively.