Abstract
The sterol regulatory element-binding protein (SREBP) transcription factors have become attractive targets for pharmacological inhibition in the treatment of metabolic diseases and cancer. SREBPs are critical for the production and metabolism of lipids and cholesterol, which are essential for cellular homeostasis and cell proliferation. Fatostatin was recently discovered as a specific inhibitor of SREBP cleavage-activating protein (SCAP), which is required for SREBP activation. Fatostatin possesses antitumor properties including the inhibition of cancer cell proliferation, invasion, and migration, and it arrests cancer cells in G2/M phase. Although Fatostatin has been viewed as an antitumor agent due to its inhibition of SREBP and its effect on lipid metabolism, we show that Fatostatin's anticancer properties can also be attributed to its inhibition of cell division. We analyzed the effect of SREBP activity inhibitors including Fatostatin, PF-429242, and Betulin on the cell cycle and determined that only Fatostatin possessed antimitotic properties. Fatostatin inhibited tubulin polymerization, arrested cells in mitosis, activated the spindle assembly checkpoint, and triggered mitotic catastrophe and reduced cell viability. Thus Fatostatin's ability to inhibit SREBP activity and cell division could prove beneficial in treating aggressive types of cancers such as glioblastomas that have elevated lipid metabolism and fast proliferation rates and often develop resistance to current anticancer therapies.
Keywords: cancer, cell death, cell division, lipid metabolism, mitosis, Betulin, Fatostatin, PF-429242, SREBP
Introduction
Cancer cell proliferation relies on the ability of cancer cells to reprogram their metabolic pathways to meet the biosynthetic and bioenergetic demands required for rapid cell divisions (1–3). Consequently, the lipid metabolic pathway, which is critical for lipid homeostasis, is often dysregulated in cancer (4, 5). Key regulators of the lipid metabolic pathway include SREBP1a,2 SREBP1c, and SREBP2, which function as transcription factors to control the expression of genes involved in fatty acid and cholesterol synthesis (6). SREBP is synthesized as an inactive precursor that is inserted into the endoplasmic reticulum membrane, and when the cellular lipid levels decline, SREBP is transported to the Golgi membrane through a SREBP cleavage-activating protein (SCAP)-dependent process (6, 7). The N-terminal transcription factor domain of SREBP is then proteolyzed by two site-specific proteases (site 1 protease (S1P) and site 2 protease (S2P) and released from the Golgi membrane (8, 9). This mature form of SREBP is transported to the nucleus, where it binds to sterol regulatory elements (SREs) upstream of lipid genes to transactivate their expression (6, 10, 11).
Due to the importance of SREBP in lipid homeostasis and cancer proliferation, several approaches have been taken to inhibit the SREBP maturation pathway (6, 12). A screen for modulators of adipogenesis identified Fatostatin, which inhibited SREBP in cell culture and in vivo by binding to SCAP and inhibiting the ability of SCAP to transport SREBP to the Golgi (13, 14). Similarly, Betulin was shown to inhibit SREBP in cell culture and in vivo by binding to SCAP and stimulating the interaction between SCAP and insulin-induced gene (Insig), which inhibited the ability of SCAP to transport SREBP to the Golgi (15). Additionally, a screen for site 1 protease inhibitors identified PF-429242, which also inhibited SREBP in cell culture and in vivo (16, 17).
Several studies showed that Fatostatin has anticancer properties in cell culture and in vivo mouse models of prostate and brain cancers (18–20). Additionally, Fatostatin arrested cancer cells in G2/M (18), indicating that inhibition of SREBP activity leads to a G2/M arrest and/or that Fatostatin was inhibiting a second target that was critical for G2/M progression. In this study, we analyzed the mechanism of Fatostatin's anticancer activity and determined that Fatostatin not only targets SCAP but also the mitotic microtubule spindle that is critical for cell division.
Results
Fatostatin Induces Spindle Damage and Mitotic Arrest
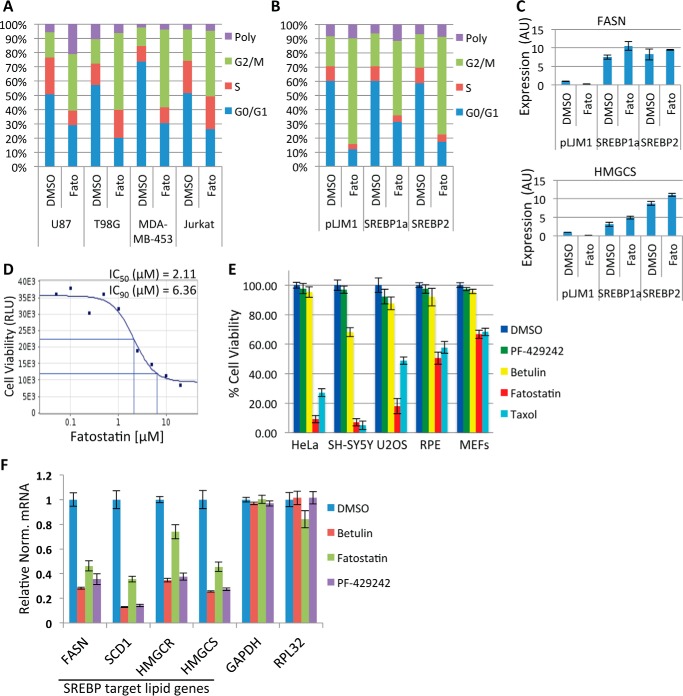
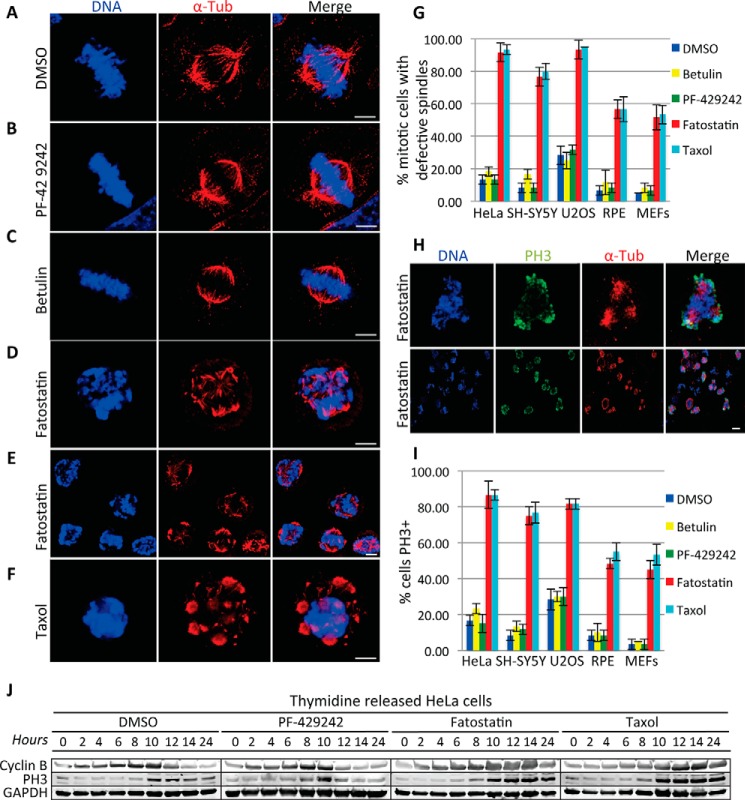
To explore the mechanism of Fatostatin's anticancer property, we first verified that Fatostatin was able to induce a G2/M arrest as described previously (18). U87, T98G, MDA-MB-453, and Jurkat T-cells were treated with DMSO or Fatostatin (5 μm) for 24 or 48 h and stained with propidium iodide, and the percentage of cells in G2/M was quantified by FACS. Indeed, Fatostatin arrested all cell lines in G2/M (Fig. 1A). However, cell lines that overexpressed active forms of hSREBP1 or hSREBP2 (devoid of the C-terminal regulatory domains and constitutively transactivated SREBP target genes independent of SCAP) were still sensitive to Fatostatin and arrested in G2/M (Fig. 1, B and C). This indicated that the Fatostatin-induced G2/M arrest was independent of SCAP inhibition and SREBP maturation. Next, we verified Fatostatin's anticancer properties. HeLa cells were treated with increasing concentrations of Fatostatin for 48 h, and cell viability was measured using the CellTiter-Glo cell viability assay. Fatostatin increased cell death in a dose-dependent manner (Fatostatin IC50 = 2.11 μm, IC90 = 6.36 μm) (Fig. 1D). Herein all experiments were carried out with 2.11 μm Fatostatin, 10 μm Betulin, 10 μm PF-429242, 100 nm Taxol, or 330 nm Nocodazole, unless otherwise noted. Next, we asked whether Fatostatin was specifically targeting cancer cells or all types of cells. Cancer (HeLa, SH-SY5Y, and U2OS) and normal (RPE and MEFs) cells were treated with Fatostatin, and the percent viability was measured after 60 h. Interestingly, Fatostatin reduced the viability of cancer cells (∼10–20% cell viability) more than normal cells (∼50–62% cell viability) (Fig. 1E). Next, we verified that Fatostatin was able to inhibit SREBP activity by analyzing the gene expression levels of SREBP target lipid genes (FASN, SCD1, HMGCR, and HMGCS) and non-SREBP genes (GAPDH and RPL32) in HeLa cells that had been treated with DMSO, Fatostatin, Betulin, or PF-429242 for 24 h. Indeed, Fatostatin was able to inhibit the expression of SREBP target genes similar to Betulin and PF-429242 (Fig. 1F). Next, we probed the nature of the Fatostatin-induced G2/M arrest. Cancer (HeLa, SH-SY5Y, and U2OS) and normal (RPE and MEFs) cells were treated with DMSO, Fatostatin, Betulin, PF-429242, or Taxol for 20 h. Cells were stained for DNA and α-tubulin and imaged by immunofluorescence (IF) microscopy. DMSO-, Betulin-, and PF-429242-treated cells displayed a normal interphase cell morphology (data not shown) and a normal mitosis with a bipolar spindle and chromosomes properly aligned at the metaphase plate (Fig. 2, A–C), However, Fatostatin-treated cells arrested in mitosis with aberrant spindles (fragmented and multipolar) (Fig. 2, D and E). This arrest was different from the mitotic arrest induced by Taxol, which arrests cells in prometaphase with stabilized microtubule asters (Fig. 2F). Quantification of these data showed that Fatostatin-treated mitotic cells had defective spindles, similar to Taxol, which was more apparent in cancer cells (∼80–90% defective spindles) than in normal cells (∼50–58% defective spindles) (Fig. 2G). In contrast, PF-429242 and Betulin had no effect on the percentage of mitotic cells with defective spindles in cancer or normal cell lines (Fig. 2G). These results indicated that unlike other SREBP activity inhibitors, only Fatostatin exhibited antimitotic properties.
FIGURE 1.
Fatostatin inhibits SREBP maturation and arrests cancer cells in G2/M. A, U87, T98G, MDA-MB-453, and Jurkat T-cells treated with DMSO or 5 μm Fatostatin (Fato) for 24 h (U87 cells were treated for 48 h). Cells were stained with propidium iodide, and their cell cycles were analyzed by FACS. B, same as in A, except that U87 cells were infected with lentivirus expressing truncated hSREBP1a (aa 1–490), hSREBP2 (aa 1–484), or an empty vector (pLJM1). Poly, polyploid. C, a fraction of cells in B were collected and analyzed for SREBP target lipid gene expression (FASN and HMGCS1) by RT-qPCR. AU, arbitrary units. D, HeLa cells were treated with increasing concentrations of Fatostatin for 48 h. Cell viability was measured, and IC50s and IC90s were generated. E, cancer (HeLa, SH-SY5Y, and U2OS) and normal (RPE and MEFs) cells were treated with DMSO, 2.11 μm Fatostatin, 10 μm Betulin, or 10 μm PF-429242 for 60 h, and cell viability was measured as in D. F, HeLa cells were treated as in E for 24 h, and gene expression levels of SREBP target lipid genes and non-SREBP genes were quantified by RT-qPCR. Data are presented as relative normalized (Relative Norm.) mRNA levels to the 36B4 control gene. Data represent average ± S.D. deviation of three independent experiments.
FIGURE 2.
Fatostatin induces spindle damage and mitotic arrest. A–F, immunofluorescence microscopy of HeLa cells treated with DMSO (A), 10 μm PF-429242 (B), 10 μm Betulin (C), 2.11 μm Fatostatin (D and E), or 100 nm Taxol (F) for 20 h. Cells were stained for DNA and tubulin (Tub). Scale bar = 5 μm. G, quantification of the percentage of DMSO-, PF-429242-, Betulin-, Fatostatin-, or Taxol-treated mitotic cells with defective spindles in cancer (HeLa, SH-SY5Y, and U2OS) and normal (RPE and MEFs) cells. Data represent average ± S.D. of three independent experiments, 100 cells counted for each. H, HeLa cells treated with 2.11 μm Fatostatin for 20 h and stained for DNA, tubulin, and phospho-Ser-10 histone H3. The top row shows a zoomed in view of a cell in the bottom row. Scale bar = 5 μm. I, quantification of the percentage of DMSO-, PF-429242-, Betulin-, Fatostatin-, or Taxol-treated cells that stained positive for PH3 in cancer (HeLa, SH-SY5Y, and U2OS) and normal (RPE and MEFs) cells. Data represent average ± S.D. deviation of three independent experiments, 100 cells counted for each. J, HeLa cells were arrested in G1/S and released into DMSO, 10 μm PF-429242, 2.11 μm Fatostatin, or 100 nm Taxol. Cell extracts were prepared at the indicated time points after release and immunoblotted for cyclin B (degraded during mitosis), PH3 (mitotic marker), and GAPDH (loading control).
To verify Fatostatin's antimitotic activity, cancer (HeLa, SH-SY5Y, and U2OS) and normal (RPE and MEFs) cells were treated with DMSO, Fatostatin, Betulin, PF-429242, or Taxol for 20 h. Cells were fixed and stained for DNA, α-tubulin, and PH3 (phosphorylated Ser-10 on histone H3, a biochemical marker of mitotic cells) and imaged by IF microscopy. Fatostatin treatment led to an increase in PH3-positive cells, similar to Taxol, which was more apparent in cancer cells (∼75–85% PH3+ cells) than in normal cells (∼45–48% PH3+ cells) (Fig. 2, H and I). In contrast, PF-429242 and Betulin had no effect on the percentage of PH3-positive cells (Fig. 2I). These results indicated that unlike other SREBP activity inhibitors, Fatostatin was arresting cells in mitosis. To further confirm this, HeLa cells were synchronized in G1/S and released into DMSO, Fatostatin, PF-429242, or Taxol. Protein extracts were prepared every 2 h for 24 h and immunoblotted for markers of mitosis. Consistently, Fatostatin arrested cells in mitosis with high PH3 and cyclin B levels, similar to Taxol, as compared with DMSO and PF-429242, which had no effect (Fig. 2J).
Fatostatin Destabilizes the Mitotic Microtubule Spindle
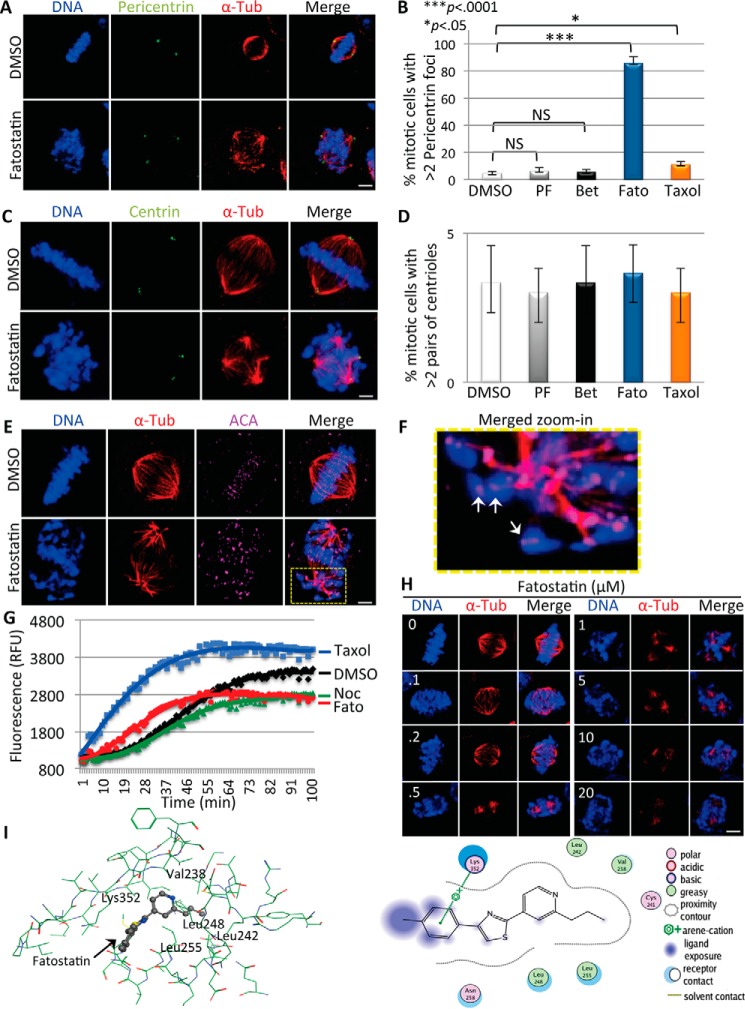
Defective multipolar spindles can arise through multiple mechanisms, including centrosome over-duplication and pericentriolar material (PCM) fragmentation (23, 24). To probe the nature of the Fatostatin-induced defective spindles, we analyzed the number of centrosomes (centriole pairs) and the status of the PCM in DMSO-, Fatostatin-, Betulin-, PF-429242-, or Taxol-treated HeLa cells. Cells were fixed and stained for DNA, α-tubulin, and centrin (centriole marker) or for pericentrin (PCM marker) and imaged by IF microscopy. Fatostatin-treated mitotic cells had more than two pericentrin foci, whereas only a few Taxol-, DMSO-, Betulin-, and PF-429242-treated cells had more than two pericentrin foci (percentage of more than two pericentrin foci for Fatostatin = 85 ± 4.8, p < 0.0001; Taxol = 11 ± 2.2, p < 0.0001; Betulin = 5.7 ± 1.7, NS; PF-429242 = 6.3 ± 2.5, NS; as compared with DMSO = 4.7.0 ± 1.2) (Fig. 3, A and B), whereas all cells had a similar number of centriole pairs (Fig. 3, C and D). These results indicated that the increased PCM fragmentation in Fatostatin-treated cells could be generating multipolar spindles. However, cells treated with microtubule destabilizers such as Vinblastine appear to generate multipolar spindles through spindle destabilization (25). Consistent with this, Fatostatin-treated cells also displayed defects in kinetochore-microtubule attachment, where lagging chromosomes had unattached kinetochores (Fig. 3, E and F). Therefore we asked whether Fatostatin was targeting the microtubule spindle. First, we performed in vitro tubulin polymerization reactions in the presence of DMSO, Taxol, Nocodazole, and Fatostatin. Although Taxol promoted tubulin polymerization, Nocodazole and Fatostatin inhibited tubulin polymerization when compared with DMSO (Fig. 3G). Additionally, treatment of HeLa cells with increasing concentrations of Fatostatin led to a concentration-dependent decrease in polymerized microtubules (Fig. 3H). These results indicated that Fatostatin was directly targeting tubulin, leading to tubulin depolymerization, spindle fragmentation, and the generation of multipolar spindles with PCM centers. To probe the mechanism by which Fatostatin was inhibiting tubulin polymerization, we asked whether Fatostatin was able to dock favorably into the Colchicine- or Vinblastine-binding sites on β-tubulin, both tubulin-destabilizing sites. Fatostatin docked favorably into the Colchicine-binding site, with a docking score 80% that of Colchicine, and showed several interactions including an arene-cation interaction between methyl-benzene and Lys-352 and lipophilic interactions between the propylpyridine scaffold and Leu-242, Val-238, Leu-248, and Leu-255 (Fig. 3, I and J). Therefore Fatostatin was likely binding to the Colchicine-binding site of β-tubulin to inhibit tubulin polymerization.
FIGURE 3.
Fatostatin inhibits tubulin polymerization and perturbs the microtubule spindle. A and C, immunofluorescence microscopy of HeLa cells treated with DMSO, 10 μm PF-429242, 10 μm Betulin, 2.11 μm Fatostatin, or 100 nm Taxol for 20 h and stained for DNA, tubulin, and pericentrin (A) or centrin (C). Scale bar = 5 μm. B and D, quantification of the percentage of DMSO-, PF-429242 (PF)-, Betulin (Bet)-, Fatostatin (Fato)-, or Taxol-treated mitotic cells that have more than two pericentrin foci (B) or more than two pairs of centrioles (D). Data represent average ± S.D. of three independent experiments, 100 cells counted for each. NS, not significant. E, HeLa cells were treated with DMSO or 2.11 μm Fatostatin and stained for DNA, tubulin, and kinetochores (anti-centromere antibody (ACA)). Scale bar = 5 μm. F, zoom-in view of a Fatostatin-treated cell in E. See the yellow box, showing unattached kinetochores (white arrows). G, summary of tubulin polymerization reactions carried out with 3 μm Nocodazole (Noc), 3 μm Fatostatin, 3 μm Taxol, or DMSO. Polymerization was monitored for 100 min, and relative fluorescence units (RFU) were quantified over time. H, HeLa cells were treated with increasing concentrations of Fatostatin (0–20 μm) and stained for DNA and tubulin. Scale bar = 5 μm. I and J, docking of Fatostatin into the β-tubulin Colchicine-binding site (PDB ID: 1Z2B) in stick figure representation (I) and highlighting the Fatostatin molecular interactions including an arene-cation interaction between methyl-benzene and Lys-352 and lipophilic interactions between the propylpyridine scaffold and Leu-242, Val-238, Leu-248, and Leu-255 (J).
Fatostatin Activates the Spindle Assembly Checkpoint
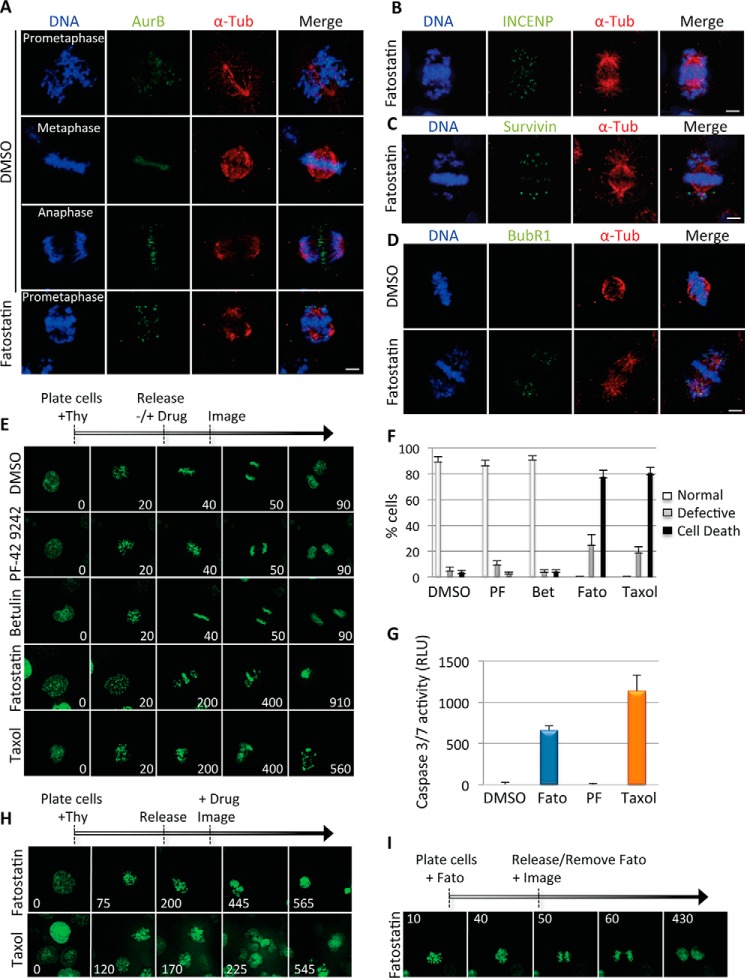
To further understand the nature of the Fatostatin-induced mitotic arrest, we analyzed whether Fatostatin activated the spindle assembly checkpoint to arrest cells in mitosis. HeLa cells were treated with DMSO or Fatostatin for 20 h and then stained for DNA, α-tubulin, and AurB/INCENP/survivin (components of the chromosome passenger complex) or BubR1 (component of the mitotic checkpoint complex) and imaged by IF microscopy. All Fatostatin-treated mitotic cells had AurB/INCENP/survivin and BubR1 staining at the kinetochores, as compared with only a few metaphase DMSO-treated cells (Fig. 4, A–D). These results indicated that Fatostatin activated the spindle assembly checkpoint to arrest cells in mitosis.
FIGURE 4.
Fatostatin induces spindle assembly checkpoint activation and mitotic catastrophe. A–D, immunofluorescence microscopy of HeLa cells treated with DMSO or Fatostatin for 20 h and stained for DNA, tubulin, and AurB (A), INCENP (B), survivin (C), or BubR1 (D). Scale bar = 5 μm. E, HCT116-GFP-H2B cells were arrested in G1/S and released into media with DMSO, 10 μm PF-429242, 10 μm Betulin, 2.11 μm Fatostatin, or 100 nm Taxol. Cells were imaged every 10 min using time-lapse microscopy, and snapshots of representative cell divisions are shown. Time is in minutes. Thy, thymidine. F, quantification of the percentage of cells undergoing normal or defective (multipolar) cell divisions or cell death (during mitosis or after a failed mitosis) that were treated with the drugs indicated in E. PF, PF-429242; Fato, Fatostatin; Bet, Betulin. Data represent the average ± S.D. of three independent experiments, 40 cells counted for each. G, HeLa cells were treated with DMSO, 2.11 μm Fatostatin, 10 μm PF-429242, or 100 nm Taxol for 24 h, and caspase 3/7 activity was monitored. Data represent the average caspase 3/7 activity in RLU ± S.D. of three independent experiments. H, same as in E, except that 2.11 μm Fatostatin or 100 nm Taxol was added 6 h after G1/S release, just prior to mitotic entry, and cells were imaged every 5 min. I, same as in E, except that cells were arrested in mitosis with 2.11 μm Fatostatin for 16 h and released into fresh medium before imaging. See also supplemental Movies S1–S8.
Fatostatin Induces Mitotic Catastrophe
Next, we sought to understand the cellular consequences of arresting cells in mitosis with Fatostatin. HCT116-GFP-H2B cells were synchronized in G1/S and released into DMSO, Fatostatin, Betulin, PF-429242, or Taxol, and cells were imaged by time-lapse microscopy 6 h after release (Fig. 4E and supplemental Movies S1–S5). Fatostatin-treated cells arrested in mitosis, failed to divide, and underwent mitotic catastrophe (cell death during mitosis or failed cell division followed by cell death), similar to Taxol-treated cells, whereas DMSO-, Betulin-, and PF-429242-treated cells were able to divide normally (Fig. 4F). To determine whether the Fatostatin-induced mitotic catastrophe was caspase-dependent, we analyzed the activity of caspase 3/7 in cells treated with DMSO, Fatostatin, PF-429242, or Taxol for 24 h. Fatostatin induced caspase 3/7 activation similar to Taxol, whereas DMSO and PF-429242 had no effect (Fig. 4G). Next, we sought to determine whether Fatostatin's effect on mitosis could occur in an acute temporal manner. HCT116-GFP-H2B cells were released from G1/S arrest, and 6 h after release, cells were treated with Fatostatin or Taxol and imaged just before mitotic entry. Both Fatostatin and Taxol arrested cells in mitosis and induced mitotic catastrophe as shown previously (Fig. 4H and supplemental Movies S6 and S7). This indicated that Fatostatin's induced mitotic arrest was independent of its inhibition of SREBP maturation, consistent with our previous data showing that cells overexpressing active forms of hSREBP1 or hSREBP2 were still sensitive to Fatostatin and arrested in G2/M (Fig. 1, B and C). Interestingly washing out the Fatostatin from Fatostatin-arrested cells allowed cells to continue through mitosis and divide (Fig. 4I and supplemental Movie S8), indicating that it was a reversible inhibitor. Together, these results indicate that independent of its inhibition of SREBP maturation and expression of lipid metabolism target genes, Fatostatin arrests cells in mitosis, which leads to caspase 3/7 activation, mitotic catastrophe, and reduced cell viability.
Discussion
In vivo Fatostatin is very well tolerated; it inhibits lipid metabolism through its inhibition of SREBP maturation, and has great antitumor activity (13, 18–20). Here we explored the mechanism of Fatostatin's antitumor activity and determined that in addition to inhibiting lipid metabolism, Fatostatin also inhibits tubulin polymerization, which perturbs mitotic spindle assembly and leads to mitotic catastrophe. Therefore we have uncovered a new targeting mechanism for Fatostatin that explains its effective antitumor activity. Additionally, Fatostatin's ability to inhibit lipid metabolism and cell division could be particularly useful for targeting aggressive types of cancers that reprogram their metabolic pathways and undergo rapid cell divisions such as glioblastomas, where Fatostatin has already been shown to be effective at reducing tumor burden in vivo (20). These findings may motivate future studies aimed at targeting more than one critical pathway that cancer cells rely on to proliferate and the development of Fatostatin for treating cancer.
Experimental Procedures
Cell Culture
HeLa, SH-SY5Y, and RPE cells were grown in DMEM/F12, and U2OS and HCT116 cells were grown in McCoy's 5A (Gibco: 11320-033 and 16600-082). U87 and T98G cells were grown in Iscove's modified Dulbecco's medium, MDA-MB-453 and MEFs were grown in DMEM, and Jurkat T-cells were grown in RPMI 1640 (Fisher: 12440053, 11965092, and 11875093) with 10% FBS and penicillin-streptomycin (Fisher: 15140122) in 5% CO2 at 37 °C.
Fixed and Live Cell Imaging
Fixed and live cell imaging was carried out as described previously (21, 26). For quantifying spindle and mitotic defects, 100 cells from three independent experiments were counted, and the data are presented as the average ± S.D. For time-lapse microscopy, HCT116-GFP-H2B cells were arrested in G1/S with 2 mm thymidine (Sigma: T9250) for 18 h, washed three times with PBS and twice with complete medium, and then either 1) released into fresh medium with or without the indicated drugs or 2) released into the cell cycle after which drugs were added 6 h after release. For Fatostatin reversibility tests, cells were treated with Fatostatin for 16 h, washed, released, and imaged. Maximum intensity projection images were converted to AVI movies. Each frame represents a 5- or 10-min interval as indicated.
Compound Potency
Cells were treated with increasing concentrations of Fatostatin (0–20 μm) for 48 h and subjected to the CellTiter-Glo assay (Promega: G7570) as described previously (22). Data are presented in relative light units (RLU). The CDD Vault software (Collaborative Drug Discovery, Inc.) was used for generating IC50 and IC90 values.
Caspase Assay
Cells were treated with drugs for 24 h, and the Caspase-Glo 3/7 assay (Promega: G8090) was used to measure caspase 3/7 activity. Plates were read with a Tecan M1000 plate reader, and the data were presented in RLU.
Tubulin Polymerization Reactions
Tubulin polymerization reactions were carried out as described previously (22) with DMSO or 3 μm Nocodazole, Fatostatin, or Taxol. Data are presented in relative fluorescence units.
Antibodies
The following antibodies were used: anti-centromere antibody (Cortex Biochem: CS1058); phospho-Ser-10-H3 (Millipore: 06570); α-tubulin (Serotec: mca77g); cyclin B (Santa Cruz Biotechnology: sc-245); survivin (Zymed Laboratories Inc.: 37-2000); INCENP and pericentrin (Abcam: ab12183 and ab4448); AurB (BD Biosciences: 611082); and BubR1 (a gift from Hongtao Yu). FITC and CY3 secondary antibodies were from Jackson ImmunoResearch Laboratories.
Compounds
DMSO, Taxol, Fatostatin, PF-429242, Betulin, Hoechst 33342, and Nocodazole were from Sigma (D4540, T7402, F8932, SML0667, B9757, B2261, and M1404).
FACS
Cells were treated with 5 μm Fatostatin for 24 h (U87 cells were treated for 48 h), stained with propidium iodide for 20 min, and analyzed on a LSRII flow cytometer (BD Biosciences) with the FlowJo software. For SREBP overexpression experiments, U87 cells were infected with lentivirus expressing truncated hSREBP1a (aa 1–490), hSREBP2 (aa 1–484), or empty vector (pLJM1) as described previously (20).
RT-qPCR
Cells were treated for 24 h with the indicated drugs and collected for gene expression analysis. The RNA preparation procedure and qPCR primers were described previously (20).
Molecular Modeling
Molecular modeling was performed using the MOE (molecular operating environment) software (Chemical Computing Group) as described previously (27). Fatostatin, Colchicine, Vinblastine, and β-tubulin structures were from the PubChem and Protein Data Bank (PDB) databases (PDB ID: 1Z2B), respectively.
Author Contributions
A. A. G., K. C., S. A. H., C. N., C. K., K. J. W., Y. C. L., S. J. B., and J. Z. T. performed the experiments, discussed the results, and wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. Y. Kidani for flow cytometry and P. Mischel, H. Christofk, and S. Smale for cell lines.
This work was supported by NCI, National Institutes of Health Grant P30CA016042, Jonsson Comprehensive Cancer Center (JCCC) seed and University of California Cancer Research Coordinating Committee (UC-CRCC) grants, and a Cottrell Scholar Award from the Research Corporation for Science Advancement (to J. Z. T.); National Institutes of Health Grants AI093768 and HL126556 (to S. J. B.); and a Whitcome fellowship (to K. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Movies S1–S8.
- SREBP
- sterol regulatory element-binding protein
- SCAP
- SREBP cleavage-activating protein
- RPE
- retinal pigment epithelium
- MEF
- mouse embryonic fibroblast
- IF
- immunofluorescence
- PCM
- pericentriolar material
- qPCR
- quantitative PCR
- aa
- amino acids
- RLU
- relative light units
- h
- human
- NS
- not significant.
References
- 1. DeBerardinis R. J., Lum J. J., Hatzivassiliou G., and Thompson C. B. (2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20 [DOI] [PubMed] [Google Scholar]
- 2. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ward P. S., and Thompson C. B. (2012) Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21, 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Murai T. (2015) Cholesterol lowering: role in cancer prevention and treatment. Biol. Chem. 396, 1–11 [DOI] [PubMed] [Google Scholar]
- 5. Notarnicola M., Tutino V., and Caruso M. G. (2014) Tumor-induced alterations in lipid metabolism. Curr. Med. Chem. 21, 2729–2733 [DOI] [PubMed] [Google Scholar]
- 6. Soyal S. M., Nofziger C., Dossena S., Paulmichl M., and Patsch W. (2015) Targeting SREBPs for treatment of the metabolic syndrome. Trends Pharmacol. Sci. 36, 406–416 [DOI] [PubMed] [Google Scholar]
- 7. Nohturfft A., DeBose-Boyd R. A., Scheek S., Goldstein J. L., and Brown M. S. (1999) Sterols regulate cycling of SREBP cleavage-activating protein (SCAP) between endoplasmic reticulum and Golgi. Proc. Natl. Acad. Sci. U.S.A. 96, 11235–11240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown M. S., and Goldstein J. L. (1997) The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 [DOI] [PubMed] [Google Scholar]
- 9. Sakai J., Duncan E. A., Rawson R. B., Hua X., Brown M. S., and Goldstein J. L. (1996) Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell 85, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 10. Briggs M. R., Yokoyama C., Wang X., Brown M. S., and Goldstein J. L. (1993) Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. I. Identification of the protein and delineation of its target nucleotide sequence. J. Biol. Chem. 268, 14490–14496 [PubMed] [Google Scholar]
- 11. Hua X., Yokoyama C., Wu J., Briggs M. R., Brown M. S., Goldstein J. L., and Wang X. (1993) SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. U.S.A. 90, 11603–11607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo D., Bell E. H., Mischel P., and Chakravarti A. (2014) Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr. Pharm. Des. 20, 2619–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kamisuki S., Mao Q., Abu-Elheiga L., Gu Z., Kugimiya A., Kwon Y., Shinohara T., Kawazoe Y., Sato S., Asakura K., Choo H. Y., Sakai J., Wakil S. J., and Uesugi M. (2009) A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem. Biol. 16, 882–892 [DOI] [PubMed] [Google Scholar]
- 14. Choi Y., Kawazoe Y., Murakami K., Misawa H., and Uesugi M. (2003) Identification of bioactive molecules by adipogenesis profiling of organic compounds. J. Biol. Chem. 278, 7320–7324 [DOI] [PubMed] [Google Scholar]
- 15. Tang J. J., Li J. G., Qi W., Qiu W. W., Li P. S., Li B. L., and Song B. L. (2011) Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab. 13, 44–56 [DOI] [PubMed] [Google Scholar]
- 16. Hawkins J. L., Robbins M. D., Warren L. C., Xia D., Petras S. F., Valentine J. J., Varghese A. H., Wang I. K., Subashi T. A., Shelly L. D., Hay B. A., Landschulz K. T., Geoghegan K. F., and Harwood H. J. Jr. (2008) Pharmacologic inhibition of site 1 protease activity inhibits sterol regulatory element-binding protein processing and reduces lipogenic enzyme gene expression and lipid synthesis in cultured cells and experimental animals. J. Pharmacol. Exp. Ther. 326, 801–808 [DOI] [PubMed] [Google Scholar]
- 17. Hay B. A., Abrams B., Zumbrunn A. Y., Valentine J. J., Warren L. C., Petras S. F., Shelly L. D., Xia A., Varghese A. H., Hawkins J. L., Van Camp J. A., Robbins M. D., Landschulz K., and Harwood H. J. Jr. (2007) Aminopyrrolidineamide inhibitors of site-1 protease. Bioorg Med. Chem. Lett. 17, 4411–4414 [DOI] [PubMed] [Google Scholar]
- 18. Li X., Chen Y. T., Hu P., and Huang W. C. (2014) Fatostatin displays high antitumor activity in prostate cancer by blocking SREBP-regulated metabolic pathways and androgen receptor signaling. Mol. Cancer Ther. 13, 855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li X., Wu J. B., Chung L. W., and Huang W. C. (2015) Anti-cancer efficacy of SREBP inhibitor, alone or in combination with docetaxel, in prostate cancer harboring p53 mutations. Oncotarget 6, 41018–41032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Williams K. J., Argus J. P., Zhu Y., Wilks M. Q., Marbois B. N., York A. G., Kidani Y., Pourzia A. L., Akhavan D., Lisiero D. N., Komisopoulou E., Henkin A. H., Soto H., Chamberlain B. T., Vergnes L., et al. (2013) An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity. Cancer Res. 73, 2850–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Torres J. Z., Ban K. H., and Jackson P. K. (2010) A specific form of phospho protein phosphatase 2 regulates anaphase-promoting complex/cyclosome association with spindle poles. Mol. Biol. Cell 21, 897–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Senese S., Lo Y. C., Huang D., Zangle T. A., Gholkar A. A., Robert L., Homet B., Ribas A., Summers M. K., Teitell M. A., Damoiseaux R., and Torres J. Z. (2014) Chemical dissection of the cell cycle: probes for cell biology and anti-cancer drug development. Cell Death Dis. 5, e1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fukasawa K. (2007) Oncogenes and tumour suppressors take on centrosomes. Nat. Rev. Cancer 7, 911–924 [DOI] [PubMed] [Google Scholar]
- 24. Oshimori N., Ohsugi M., and Yamamoto T. (2006) The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat. Cell Biol. 8, 1095–1101 [DOI] [PubMed] [Google Scholar]
- 25. Singh A., Zapata M. C., Choi Y. S., and Yoon S. O. (2014) GSI promotes vincristine-induced apoptosis by enhancing multi-polar spindle formation. Cell Cycle 13, 157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gholkar A. A., Senese S., Lo Y. C., Vides E., Contreras E., Hodara E., Capri J., Whitelegge J. P., and Torres J. Z. (2016) The X-linked-intellectual-disability-associated ubiquitin ligase Mid2 interacts with Astrin and regulates Astrin levels to promote cell division. Cell Rep. 14, 180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lo Y. C., Senese S., Li C. M., Hu Q., Huang Y., Damoiseaux R., and Torres J. Z. (2015) Large-scale chemical similarity networks for target profiling of compounds identified in cell-based chemical screens. PLoS Comput. Biol. 11, e1004153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.