Abstract
Although the cellular prion protein (PrPC) is concentrated at synapses, the factors that target PrPC to synapses are not understood. Here we demonstrate that exogenous PrPC was rapidly targeted to synapses in recipient neurons derived from Prnp knock-out(0/0) mice. The targeting of PrPC to synapses was dependent upon both neuronal cholesterol concentrations and the lipid and glycan composition of its glycosylphosphatidylinositol (GPI) anchor. Thus, the removal of either an acyl chain or sialic acid from the GPI anchor reduced the targeting of PrPC to synapses. Isolated GPIs (derived from PrPC) were also targeted to synapses, as was IgG conjugated to these GPIs. The removal of sialic acid from GPIs prevented the targeting of either the isolated GPIs or the IgG-GPI conjugate to synapses. Competition studies showed that pretreatment with sialylated GPIs prevented the targeting of PrPC to synapses. These results are consistent with the hypothesis that the sialylated GPI anchor attached to PrPC acts as a synapse homing signal.
Keywords: cholesterol, glycosylphosphatidylinositol (GPI anchor), prion, sialic acid, synapse
Introduction
The cellular prion protein (PrPC)2 gained prominence when it was identified as the normal isoform of the disease-associated protein (PrPSc) that accumulates in the brains of humans and animals with transmissible spongiform encephalopathies (1). That observation increased interest in the role of PrPC in neurons. Reports that PrPC is concentrated at synapses (2, 3) and that transgenic mice in which the gene for PrP had been knocked-out (Prnp(0/0)) showed synaptic and memory deficits (4, 5) suggest that it plays a role in neurotransmission. The targeting of proteins to specific cellular locations may be critical for their function. For example, the proteins involved in synaptic vesicle recycling need to be delivered to the synapse in neurons. Little is known about the molecular mechanism(s) by which PrPC accumulates within synapses. PrPC is linked to cell membranes by a glycosylphosphatidylinositol (GPI) anchor (6), which affects the cellular distribution and function of PrPC (7–9). PrPC is found within cholesterol-rich, membrane microdomains that are commonly called lipid rafts (10, 11). Because only a small proportion of proteins that are found within lipid rafts are subsequently transported to synapses, the factors affecting the targeting of PrPC to synapses were studied.
The GPI anchor that links PrPC to the cell membrane targets it to lipid rafts (7). There are many different lipid rafts that demonstrate heterogeneity in their protein, glycolipid, and lipid composition (12), and different GPI-anchored proteins are targeted to different lipid rafts. For example, Thy-1 and PrPC occupy separate domains on the neuronal surface (13). Although all GPI anchors contain a conserved core, variations on this core structure are common (14), and the GPI attached to PrPC differs from that of Thy-1 (15, 16). It has been hypothesized that the localization of some GPI-anchored proteins to specific lipid rafts and hence specific cell membranes is due to the composition of the GPI anchor. Thus, the composition of GPI anchors directs antigens to different rafts in the absence of interactive external domains (17, 18), and the chemical composition of the GPI anchor alters the intracellular trafficking of proteins (19). Many GPI-anchored molecules are rapidly incorporated into living cells (20, 21). In this study, PrPC was transferred to recipient cortical neurons derived from Prnp knock-out(0/0) mice. We demonstrate that the targeting of PrPC to synapses was dependent upon cholesterol concentrations in the recipient neurons. Critically, we also demonstrate that the composition of the GPI anchor attached to PrPC is a key factor that affects the targeting of PrPC to synapses. More specifically, we show that the presence of sialic acid on the GPI anchor is necessary for the targeting of PrPC to synapses.
Results
PrPC Is Targeted to Synapses
To study the factors that control the targeting of PrPC to synapses, PrPC (10 nm) was introduced into neurons derived from Prnp(0/0) mice (22). PrPC preparations run in gels and stained with Coomassie Brilliant Blue did not show any contaminants (Fig. 1A). After 2 h, recipient neurons were collected, and organelles were separated upon a Percoll density gradient. The synaptic protein synaptophysin, used to identify synaptosomes, was concentrated in fractions 42–46 (Fig. 1B). These fractions also contained high concentrations of the synaptic proteins synapsin-1 and synaptobrevin-1 and were consequently pooled as synaptosomes. Following introduction to neurons, PrPC showed a time-dependent (Fig. 1C) and concentration-dependent (Fig. 1D) accumulation within synaptosomes. Not all GPI-anchored proteins were targeted to synapses because GPI-anchored CD14 added to neurons did not accumulate within synapses.
FIGURE 1.
PrPC is targeted to the synapses. A, Coomassie Brilliant Blue-stained gel showing protein ladder (lane 1) and neuronal PrPC (lane 2). B, amounts of synaptophysin in fractions derived from Prnp(0/0) neurons separated on a Percoll density gradient. Values are means ± S.D. (error bars) from an experiment measured in quadruplicate. C, concentrations of PrPC (□) or CD14 (■) in synaptosomes derived from Prnp(0/0) neurons incubated with 10 nm proteins for time periods as shown. Values are means ± S.D. from duplicate experiments performed five times (n = 10). D, concentrations of PrPC (□) or CD14 (■) in synaptosomes from Prnp(0/0) neurons incubated with proteins as shown for 2 h. Values are means ± S.D. from duplicate experiments performed five times (n = 10).
Targeting of PrPC to Synapses Is Sensitive to Cholesterol Depletion
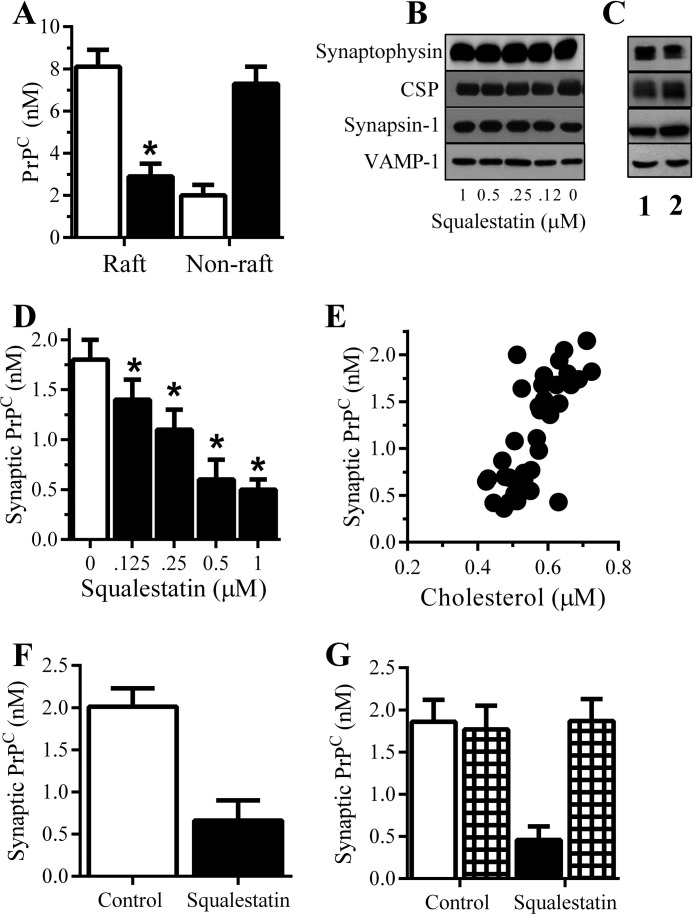
PrPC is found in lipid rafts that contain high concentrations of cholesterol (11). Because cholesterol depletion affects the expression of PrPC (7, 23, 24), the effects of cholesterol depletion on the targeting of PrPC to synapses were examined. Prnp(0/0) neurons were treated with squalestatin, a selective squalene synthase inhibitor that reduced neuronal cholesterol concentrations (25). Although treatment with 1 μm squalestatin did not affect the uptake of exogenous PrPC into neurons (9.3 nm PrPC ± 0.6 compared with 9.2 nm ± 0.5, n = 9, p = 0.68), it significantly reduced the concentrations of PrPC found in lipid rafts (detergent-resistant membranes (DRMs)) (Fig. 2A). Although cholesterol is required for the formation and maintenance of synapses (26, 27), the mild cholesterol depletion caused by 1 μm squalestatin did not significantly alter the concentrations of synaptophysin, cysteine string protein (CSP), synapsin-1, or VAMP-1 (vesicle-associated membrane protein 1) in neurons (Fig. 2B) or isolated synaptosomes (Fig. 2C), indicating that it did not damage synapses. When Prnp(0/0) neurons were treated with squalestatin for 24 h and incubated with 10 nm PrPC, squalestatin caused a dose-dependent reduction in the concentrations of PrPC found within synaptosomes (Fig. 2D). There was a significant correlation between cholesterol concentrations in squalestatin-treated neurons and the concentrations of PrPC found in synapses (Pearson's coefficient = 0.73, p < 0.01) (Fig. 2E). Treatment of Prnp(+/+) wild-type neurons with 500 nm squalestatin reduced the concentrations of PrPC in synaptosomes (Fig. 2). The addition of 5 μm squalene reversed the effects of squalestatin upon neuronal cholesterol concentrations (23) and PrPC trafficking to synapses (Fig. 2G).
FIGURE 2.
Cholesterol depletion reduced the targeting of PrPC to synapses. A, concentrations of PrPC in raft and non-raft membranes from Prnp(0/0) neurons pretreated with a vehicle control (□) or 1 μm squalestatin (■) and incubated with 10 nm PrPC for 2 h. Values are means ± S.D. (error bars) from triplicate experiments performed four times (n = 12). *, raft PrPC significantly less than in control neurons. B, immunoblots showing the amounts of synaptophysin, CSP, synapsin-1, and VAMP-1 in neurons incubated with squalestatin as shown. C, immunoblotting showing the amounts of synaptic proteins in synaptosomes treated with control medium (lane 1) or 1 μm squalestatin (lane 2). D, concentrations of PrPC in synaptosomes from Prnp(0/0) neurons pretreated with a vehicle control (□) or squalestatin as shown (■) and incubated with 10 nm PrPC for 2 h. Values are means ± S.D. from triplicate experiments performed four times (n = 12). *, synaptic PrPC significantly less than in control neurons. E, there was a significant correlation between cholesterol concentrations in Prnp(0/0) neurons treated with squalestatin (range from 1 μm to 125 nm) and the concentrations of PrPC in synaptosomes following incubation with 10 nm PrPC for 2 h (Pearson's coefficient = 0.73, p < 0.01). F, concentrations of PrPC in synaptosomes from Prnp(+/+) neurons treated with a vehicle control (□) or 1 μm squalestatin (■) for 2 h. Values are means ± S.D. from triplicate experiments performed three times (n = 9). G, concentrations of PrPC in synaptosomes from Prnp(0/0) neurons pretreated with a vehicle control or 1 μm squalestatin as shown (■) mixed with 5 μm squalene (checkered bars) and incubated with 10 nm PrPC for 2 h. Values are means ± S.D. from triplicate experiments performed three times (n = 9).
Monoacylated PrPC Is Not Targeted to Synapses
PrPC was digested with PLA2, an enzyme that targets acyl chains contained within GPIs to form monoacylated PrPC (Fig. 3B) and isolated using an immunoaffinity column and reverse phase chromatography as described (8). Although similar amounts of monoacylated PrPC and PrPC bound to recipient Prnp(0/0) neurons (9.4 nm PrPC ± 0.7 compared with 9.2 nm ± 0.5, n = 12, p = 0.3), they were differently distributed between membranes. Whereas the majority of PrPC was found within lipid rafts (DRMs), most of the monoacylated PrPC was found within detergent-soluble membranes (DSMs) (Fig. 3C). Monoacylated PrPC added to Prnp(0/0) neurons was not targeted to synaptosomes (Fig. 3D).
FIGURE 3.
Monoacylated PrPC is not targeted to synapses. Schematics show the putative structure of the GPIs attached to PrPC (A) and monoacylated PrPC (B). Glycan residues shown include mannose (Man), sialic acid (SA), galactose (Gal), N-acetyl galactosamine (GalNAc), inositol (Inos), and glucosamine (GlcN). C, concentrations of PrPC (□) and monoacylated PrPC (■) in raft and non-raft membranes derived from Prnp(0/0) neurons incubated with 10 nm PrPC/monoacylated PrPC for 2 h. Values are means ± S.D. (error bars) from triplicate experiments performed three times (n = 9). D, the concentrations of PrPC in synaptosomes derived from Prnp(0/0) neurons incubated with PrPC (□) or monoacylated PrPC (■) as shown for 2 h. Values are means ± S.D. from duplicate experiments performed five times (n = 10).
Glia-derived PrPC Did Not Target Synapses
Because PrPC is also expressed by glial cells, we sought to determine whether glia-derived and neuron-derived PrPC had the same properties. Prnp(0/0) neurons were incubated with 10 nm glia-derived PrPC or 10 nm neuron-derived PrPC for 2 h. There were no significant differences in the concentrations of glial and neuronal PrPC incorporated into recipient neurons; nor were there any significant differences in the concentrations found within DRMs (Table 1). However, glial PrPC did not accumulate within synaptosomes (Fig. 4A). By comparing neuron and glia-derived PrPC we sought to determine the factors that affect synaptic targeting. Because the glycosylation of PrPC affects the trafficking of PrPC (28, 29), the possibility that cell-specific glycosylation affected the targeting of PrPC to synapses was examined. PNGase removed the N-linked glycans from PrPC (30). When Prnp(0/0) neurons were pulsed with 10 nm neuronal PrPC or 10 nm PNGase-digested neuronal PrPC for 2 h, we found that the removal of N-linked glycans did not alter the concentrations of PrPC that bound to recipient neurons or the concentrations of PrPC found within synaptosomes (Table 1 and Fig. 4B). PNGase-digested glial PrPC was not found within synaptosomes, indicating that N-linked glycans did not stop glial PrPC from targeting synapses (Table 1). To eliminate the possible effects of N-linked glycans on the trafficking of PrPC, all subsequent experiments were carried out on PNGase-digested PrPC preparations.
TABLE 1.
N-linked glycans do not affect the targeting of PrPC to synapses
The concentrations of PrPC in cell extracts, DRMs, DSMs, and synaptosomes isolated from Prnp(0/0) neurons were incubated with 10 nm neuron-derived PrPC, 10 nm PNGase-digested neuron-derived PrPC, 10 nm glia-derived PrPC, or 10 nm PNGase-digested glia-derived PrPC for 2 h. Values are means ± S.D. from triplicate experiments performed four times (n = 12).
| Concentration PrPC |
||||
|---|---|---|---|---|
| Total | DRM | DSM | Synapse | |
| nm | ||||
| Neuronal PrPC | 9.2 ± 0.9 | 8.5 ± 0.8 | 1.0 ± 0.4 | 1.8 ± 0.32 |
| PNGase-digested neuronal PrPC | 8.9 ± 0.6 | 8.2 ± 0.8 | 0.9 ± 0.3 | 1.9 ± 0.35 |
| Glial PrPC | 9.2 ± 1.0 | 8.6 ± 0.8 | 1.1 ± 0.3 | 0.29 ± 0.24a |
| PNGase-digested glial PrPC | 9.0 ± 0.5 | 8.0 ± 0.9 | 1.1 ± 0.4 | 0.24 ± 0.18a |
a Concentration of PrPC in synaptosomes significantly less than in neurons incubated with neuron-derived PrPC.
FIGURE 4.
Glia-derived PrPC is not targeted to synapses. A, concentrations of PrPC in synaptosomes from Prnp(0/0) neurons incubated for 2 h with neuronal PrPC (□) or glial PrPC (hatched bars) as shown. Values are means ± S.D. (error bars) from triplicate experiments performed three times (n = 9). B, Coomassie Brilliant Blue-stained gel showing protein ladder (lane 1), deglycosylated glial PrPC (lane 2), and deglycosylated neuronal PrPC (lane 3). C, concentrations of PrPC in synaptosomes from Prnp(0/0) neurons incubated for 2 h with neuronal PrPC (□) or endoglycosidase F-digested neuronal PrPC (checkered bars) as shown. Values are means ± S.D. from duplicate experiments performed five times (n = 10). D, dot blots showing the binding of mAb 5AB3-11 (phosphatidylinositol), concanavalin A (mannose), or S. nigra lectin (sialic acid) to GPIs isolated from neuronal PrPC (lane 1) or glial PrPC (lane 2).
Because the composition of the GPI anchor is dependent upon the cell type (31, 32), the nature of GPIs attached to glial and neuronal PrPC was determined. Isolated GPIs from neuronal and glial PrPC were blotted onto nitrocellulose membranes. Detection with the phosphatidylinositol-reactive mAb 5AB3-11 and biotinylated concanavalin A (which binds to mannose, a core component of all GPIs) showed that similar amounts of GPIs were loaded onto membranes. However, Sambucus nigra lectin (which reacts with terminal sialic acid bound either α-2,6 or α-2,3 to galactose) bound to GPIs isolated from neuronal PrPC but not glial PrPC (Fig. 4C), indicating that only the GPI attached to neuronal PrPC contained a terminal sialic acid.
Desialylated PrPC Does Not Target Synapses
The composition of GPI anchors affects the targeting of proteins to specific membranes (20, 33). This sialic acid on GPIs derived from neuronal PrPC is susceptible to neuraminidase digestion (34), resulting in desialylated PrPC (Fig. 5A). Digestion of neuronal PrPC with neuraminidase reduced the binding of S. nigra lectin without affecting the binding of concanavalin A or the phosphatidylinositol-reactive mAb 5AB3-11. Neuraminidase removal of terminal sialic acid would be expected to reveal a galactose residue (Fig. 5A). The lectin RC1 (which reacts with terminal galactose) binds to GPIs derived from desialylated PrPC but not to GPIs from neuronal PrPC, consistent with the putative structure of PrPC GPIs as proposed by Stahl et al. (15) (Fig. 5B). Similar amounts of PrPC and desialylated PrPC bound to Prnp(0/0) neurons and were found within DRMs (Table 1). Although both neuronal and desialylated PrPC were targeted to rafts, desialylated PrPC did not accumulate in synapses (Fig. 5C).
FIGURE 5.
Desialylated PrPC is not targeted to synapses. A, putative structure of the GPI attached to neuronal PrPC after digestion with neuraminidase. B, dot blots showing the binding of mAb 5AB3-11 (phosphatidylinositol), concanavalin A (mannose), S. nigra lectin (sialic acid), or RC1 (galactose) to GPIs isolated from PrPC (lane 1) and desialylated PrPC (lane 2). C, concentrations of PrPC in synaptosomes from Prnp(0/0) neurons incubated for 2 h with PrPC (□) or desialylated PrPC (striped bars) as shown. Values are means ± S.D. (error bars) from triplicate experiments performed three times (n = 9). D, GPIs derived from neuronal PrPC isolated from synaptosomes (lane 1) or from the cell body (lane 2) separated by HPTLC. E, dot blots showing the binding of mAb 5AB3-11 (phosphatidylinositol), concanavalin A (mannose), or S. nigra lectin (terminal sialic acid) to GPIs attached to PrPC isolated from synaptosomes (lane 1) or from the neuronal cell body (lane 2).
To determine whether the GPI attached to PrPC found at synapses differed from that found in the cell body, synaptosomes from Prnp(+/+) wild-type neurons were isolated. PrPC was purified, and GPIs were isolated. There were no obvious differences between GPIs isolated from synaptic PrPC or from cell body PrPC when analyzed by high performance thin layer chromatography (HPTLC) (Fig. 5D) or by dot blots (Fig. 5E). The S. nigra lectin (which binds terminal sialic acid) bound to GPIs isolated from PrPC derived from the cell body.
Isolated GPIs Are Targeted to Synapses
GPIs from neuronal PrPC and desialylated PrPC were isolated on C18 columns (Fig. 6A) and HPTLC (Fig. 6B) and labeled with FITC. Prnp(0/0) neurons were incubated with 10 nm FITC or 10 nm FITC-labeled GPIs for 2 h and washed, and florescence was measured. Neurons incubated with 10 nm FITC contained only 0.6 nm ± 0.2 FITC compared with neurons incubated with FITC-GPI (8.2 nm ± 0.88) or FITC-desialylated GPI (7.7 nm ± 0.98) (Table 2). Similar concentrations of GPIs and desialylated GPIs were found in lipid rafts (DRMs) (Table 2). GPIs derived from neuronal PrPC accumulated in synaptosomes in a dose-dependent manner (Fig. 6C). FITC-GPIs accumulated in synaptosomes at higher concentrations than FITC alone or desialylated GPIs (Fig. 6D).
FIGURE 6.
Desialylated GPIs do not target synapses. A, GPIs derived from neuronal PrPC (○) and desialylated GPIs (●) eluted from C18 columns. Values are means of duplicates. B, GPIs derived from neuronal PrPC (lane 1) and desialylated GPIs (lane 2) separated by HPTLC. C, amounts of FITC in synaptosomes from neurons incubated for 2 h with FITC-labeled GPIs as shown. Values are means ± S.D. (error bars) from triplicate experiments performed twice (n = 6). D, amounts of FITC in synaptosomes from neurons incubated for 2 h with 10 nm FITC alone (□) or with 10 nm FITC-labeled GPIs (■) or 10 nm FITC-desialylated GPIs (striped bar). Values are means ± S.D. from triplicate experiments performed twice (n = 6). E, concentrations of PrPC in synaptosomes isolated from Prnp(0/0) neurons pretreated with control medium (□) or GPIs as shown (■) and incubated with 10 nm PrPC. Values are means ± S.D. from triplicate experiments performed two times (n = 6). *, significantly less synaptic PrPC than in control synaptosomes. F, concentrations of PrPC in synaptosomes isolated from Prnp(0/0) neurons pretreated with control medium (□), 10 nm GPIs (■), or 10 nm desialylated GPIs (striped bar) and incubated with 10 nm PrPC. Values are means ± S.D. from triplicate experiments performed two times (n = 6). *, significantly less synaptic PrPC than in control synaptosomes.
TABLE 2.
GPIs are targeted to rafts
Shown are the concentrations of GPIs in Prnp(0/0) neurons incubated with 10 nm FITC (as a control) or 10 nm FITC conjugated to GPI or desialylated GPI for 2 h when cell extracts, DRMs, and DSMs were isolated. Values are means ± S.D. from triplicate experiments performed 4 times (n = 12).
| FITC (Fluorescence) |
|||
|---|---|---|---|
| Total | DRM | DSM | |
| nm | |||
| FITC-control | 0.6 ± 0.2 | 0.2 ± 0.2 | 0.4 ± 0.2 |
| FITC-GPI | 8.2 ± 0.88 | 7.6 ± 0.64a | 0.53 ± 0.21 |
| FITC-desialylated GPI | 7.7 ± 0.98 | 7.2 ± 0.71a | 0.51 ± 0.25 |
a Concentration of GPIs in DRMs (rafts) significantly less than in neurons incubated with control GPIs.
To determine whether GPIs inhibited the targeting of neuronal PrPC to synaptosomes, Prnp(0/0) neurons were pretreated with GPIs derived from PrPC (10 to 1.25 nm) and incubated with 10 nm neuronal PrPC for 2 h. These GPIs reduced the concentrations of PrPC found within synaptosomes in a dose-dependent manner (Fig. 6E). This effect was structure-dependent because desialylated GPIs did not affect the concentrations of PrPC found within synaptosomes (Fig. 6F).
GPIs Target IgG to Synapses
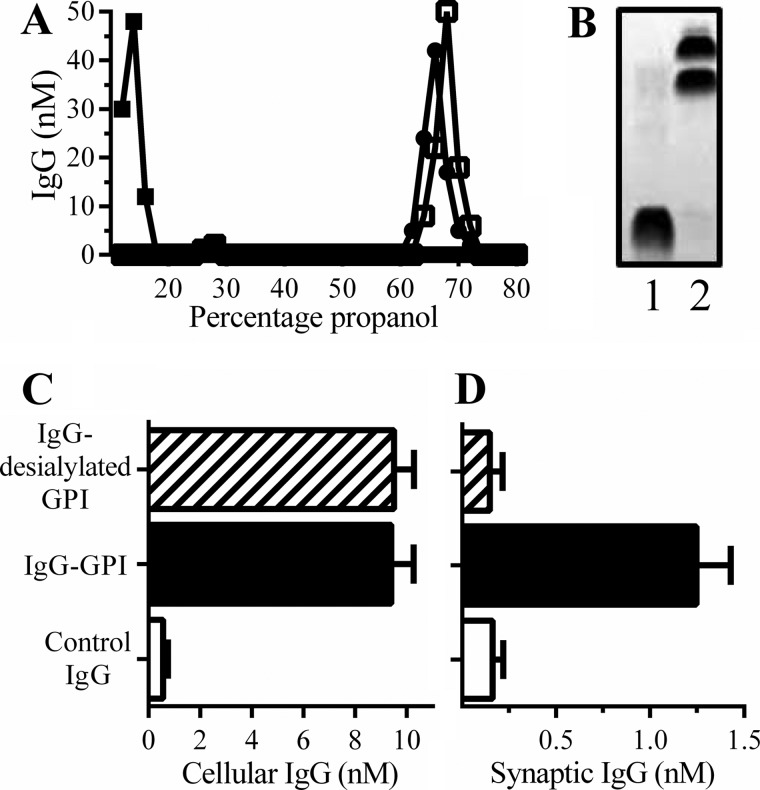
To test the hypothesis that specific GPIs could target other proteins to synapses, rabbit IgG was conjugated to GPIs (derived from neuronal PrPC) or desialylated GPIs. The GPI-modified rabbit IgG was isolated by reverse phase chromatography on C18 columns. IgG conjugated to GPIs or desialylated GPIs eluted from C18 columns in concentrations between 60 and 70%, whereas control IgG did not bind and eluted in the void volume (Fig. 7A). IgG conjugated to GPIs was differentiated from IgG by HPTLC (Fig. 7B). Subsequently, neurons were incubated with 10 nm IgG, 10 nm IgG-GPI, or 10 nm IgG-desialylated GPI for 2 h. Only low concentrations of IgG (0.3 nm) bound to neurons. Similar concentrations of IgG-GPI and IgG-desialylated GPI were found in neurons, indicating that the composition of the GPI did not affect the binding to neurons (Fig. 7C). Both IgG-GPI and IgG-desialylated GPI were targeted to lipid rafts (DRMs) (Table 3). Both were released from neurons by digestion with phosphatidylinositol-specific phospholipase C (PI-PLC), indicating that they were expressed at the surface of neurons (Table 3). Critically, IgG-GPI, but not IgG-desialylated GPI, was found in synaptosomes (Fig. 7D).
FIGURE 7.
Sialylated GPIs targeted IgG to synapses. A, concentrations of IgG (■), IgG-GPI (●), or IgG-desialylated GPI (□) in fractions eluted off of C18 columns under a gradient of propanol and water. B, IgG (lane 1) and IgG-GPI (lane 2) separated by HPTLC. C, concentrations of IgG in neurons incubated with 10 nm IgG (□), 10 nm IgG-GPI (■), or 10 nm IgG-desialylated GPI (striped bar) for 2 h. Values are means ± S.D. (error bars) from triplicate experiments performed three times (n = 9). D, concentrations of IgG in synaptosomes derived from neurons incubated with 10 nm IgG (□), 10 nm IgG-GPI (■), or 10 nm IgG-desialylated GPI (striped bar) for 2 h. Values are means ± S.D. from triplicate experiments performed three times (n = 9).
TABLE 3.
Sialylated GPIs target IgG to rafts
Shown are the concentrations of rabbit IgG in neurons incubated with 10 nm rabbit IgG (as a control), 10 nm rabbit IgG-GPI, or 10 nm IgG-desialylated GPI for 2 h when cell extracts, DRMs, and DSMs were collected. Also shown are the concentrations of IgG in neurons following digestion with PI-PLC. Values are means ± S.D. from triplicate experiments performed four times (n = 12).
| Rabbit IgG |
||||
|---|---|---|---|---|
| Total | DRM | DSM | PI-PLC | |
| nm | ||||
| Control IgG | 0.3 ± 0.1 | 0.1 ± 0.1 | 0.1 ± 0.1 | 0.1 ± 0.1 |
| IgG-GPI | 9.1 ± 0.4 | 8.4 ± 0.5 | 0.8 ± 0.2 | 0.2 ± 0.2 |
| IgG-desialylated GPI | 9.2 ± 0.5 | 8.5 ± 0.7 | 0.8 ± 0.2 | 0.1 ± 0.1 |
Discussion
Our study examined the factors that affect the distribution of PrPC in neurons. We report that PrPC added to recipient Prnp(0/0) neurons rapidly accumulated at synapses. The targeting of PrPC to synapses was dependent upon two key factors: the cholesterol concentration of cell membranes and the composition of the GPI attached to PrPC. More specifically, PrPC was not targeted to synapses following the removal of either sialic acid or an acyl chain from the GPI attached to PrPC.
The targeting of proteins to specific membrane domains is of key importance in neurons. PrPC is concentrated in synapses (2, 3), and in this study, neuron-derived PrPC was targeted to synapses after its inclusion in specific, cholesterol-sensitive, lipid rafts. The mild cholesterol depletion caused by squalestatin did not affect the number of synapses. However, this cholesterol depletion reduced the targeting of neuronal PrPC to lipid rafts and the trafficking of PrPC to synapses (there was a significant correlation between the concentrations of cholesterol in squalestatin-treated neurons and the concentrations of PrPC found within synaptosomes), results consistent with the hypothesis that the inclusion of PrPC into lipid rafts precedes delivery of PrPC to synapses. Squalestatin also reduced the concentrations of PrPC found in synaptosomes from wild type neurons. The effect of squalestatin on cholesterol concentrations and the targeting of PrPC to rafts and synaptosomes were reversed by the addition of squalene.
We hypothesized that the GPI also targeted PrPC to synapses. This hypothesis was supported by the observation that monoacylated PrPC was neither targeted to rafts nor found in synapses. It is thought that saturated acyl chains attached to GPIs allow tight molecular packing and increase the solubilization of cholesterol (35, 36). Consequently, the loss of a saturated acyl chain disrupts the membrane surrounding monoacylated PrPC (8). Not all GPI-anchored proteins found within rafts are found within synapses, and neither the GPI-anchored CD14 nor glial PrPC was found within synaptosomes. The observation that glial PrPC was not found within synapses indicated that the essential synaptic targeting signal in PrPC lay in a post-translational modification. Because digestion with PNGase did not affect the trafficking of PrPC to synapses, we concluded that N-linked glycosylation of neuronal and glial PrPC was not a significant factor in targeting PrPC to synapses.
The composition of the GPI anchor is dependent upon the cell type (31, 32), and the structure of the GPI attached to PrPC can vary (15). Whereas the GPI attached to neuronal PrPC contains sialic acid, GPIs derived from glial PrPC did not, suggesting that the presence of sialic acid on the GPI attached to neuronal PrPC was a key factor that targeted it to synapses. This hypothesis was supported by observations that the removal of sialic acid from neuronal PrPC reduced its migration to synapses. Notably, desialylated neuronal PrPC and glial PrPC had similar properties; both targeted lipid rafts and were predominantly expressed at the surface of recipient neurons. The composition of GPI targets proteins to different lipid rafts (17, 33, 37) that traffic via different pathways (9, 19), indicating how the composition of the GPI can affect the trafficking of PrPC. Thus, whereas PrPC with a sialylated GPI enters a pathway that ends at the synapse, desialylated PrPC targeted a different raft and trafficked to a different cellular location.
GPI-anchored proteins are surrounded by lipid shells that are dependent on multiple protein-lipid, glycan-lipid, and lipid-lipid interactions (38). The concept that the molecular structure of the GPI may regulate the composition of the underlying membrane was supported by observations that Thy-1 and PrPC have different GPI anchors (15, 16) and occupy separate domains on the neuronal surface (13), which have different lipid compositions (40). Similarly, desialylated PrPC is associated with membrane rafts containing greater concentrations of gangliosides and cholesterol than native PrPC (41). It is noteworthy that gangliosides help sequester cholesterol and stabilize rafts (42–44) and affect the distribution and trafficking of GPI-anchored proteins (45, 46), including PrPC (47). The increased association of desialylated PrPC with gangliosides helps explain why desialylated PrPC showed greater resistance to cholesterol depletion than did neuronal PrPC.
These results suggested that the synapse-targeting information is contained within the GPI alone, which is consistent with reports that the glycan composition of GPIs direct antigens to specific membrane microdomains in the absence of interactive external domains (18, 33). The targeting of isolated GPIs (derived from PrPC) also required the presence of sialic acid. The role of the GPI as a major “synapse-targeting” signal was demonstrated when isolated GPIs were conjugated to rabbit IgG. IgG was not readily taken up by neurons and was not found within synapses. In contrast, IgG conjugated to GPIs derived from neuronal PrPC inserted into recipient neurons and was targeted to synapses. Although IgG conjugated to desialylated GPIs also inserted into neurons, it was not found within synapses. Such results suggest a novel mechanism of cell engineering whereby proteins could be targeted to synapses by the attachment of specific GPIs.
The maximum concentration of PrPC measured in synapses of recipient neurons, regardless of how much PrPC was added, was ∼2 nm. Similar concentrations of PrPC were found in synaptosomes from Prnp wild-type neurons, suggesting that the process by which PrPC traffics to synapses is limited. In a similar manner, the concentrations of isolated GPIs found at synapses were also limited. The observation that pretreatment with isolated GPIs reduced the targeting of neuronal PrPC to synapses supported the hypothesis that PrPC and isolated GPIs compete for the same synapse-targeting pathways. This hypothesis is supported by the observation that the capacity of GPIs to block the targeting of neuronal PrPC to synapses was also dependent upon the presence of sialic acid.
In conclusion, we demonstrated the role of GPIs as targeting signals in neurons. The targeting of PrPC to synapses was dependent upon the structure of the GPI anchor requiring the presence of two acyl chains and sialic acid. The composition of the GPI anchor alone contained sufficient information to target synapses in the absence of the external protein domain.
Experimental Procedures
Primary Neuronal Cultures
Cortical neurons were prepared from the brains of mouse embryos (day 15.5) from both Prnp wild-type(+/+) and Prnp knock-out(0/0) mice. Neurons were plated at 1 × 106 cells/well in 6-well plates (precoated with poly-l-lysine) in Ham's F-12 containing 5% fetal calf serum for 2 h. Cultures were then shaken (600 rpm for 5 min), and non-adherent cells were removed by three washes in PBS. Neurons were grown in neurobasal medium containing B27 components and nerve growth factor (5 ng/ml) (Sigma) for 10 days. Immunohistochemistry showed that the cells were greater than 95% neurofilament-positive. For cell-targeting studies, neurons were incubated with PrPC preparations for different time periods. In some experiments, neurons were pretreated with test compounds (drugs, PrP preparations, and isolated GPIs) and incubated with test preparations.
Cell Extracts
Neurons were homogenized in an extraction buffer containing 150 mm NaCl, 10 mm Tris-HCl, pH 7.4, 10 mm EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 0.2% SDS, and mixed protease inhibitors (4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, aprotinin, leupeptin, bestatin, pepstatin A, and E-46) (Sigma) and a phosphatase inhibitor mixture including PP1, PP2A, microcystin LR, cantharidin, and p-bromotetramisole (Sigma) at 106 cells/ml. Nuclei and cell debris were removed by centrifugation (500 × g for 5 min).
Isolation of Synaptosomes
Synaptosomes were prepared on a discontinuous Percoll gradient as described (48). Briefly, neurons were homogenized at 4 °C in 1 ml of SED solution (0.32 m sucrose, 5 mm Tris-HCl, pH 7.2, 1 mm EDTA, and 0.25 mm dithiothreitol) and centrifuged at 1000 × g at 4 °C. The supernatant was transferred to a four-step gradient of 3, 7, 15, and 23% Percoll in SED solution and centrifuged at 16,000 × g for 30 min at 4 °C. The synaptosomes were collected from the interface between the 15 and 23% Percoll and washed (16,000 × g for 10 min at 4 °C) and suspended in neurobasal medium containing B27 components. All synaptosomes were used on the same day of preparation; freshly prepared synaptosomes were incubated with peptides for 1 h. After the test period, synaptosomes were homogenized in either extraction buffer (as above) or in the DRM extraction buffer (as below). All synaptosome preparations contained equal amounts of synaptophysin.
Western Blotting
Samples were mixed with Laemmli buffer containing β-mercaptoethanol and heated to 95 °C for 5 min, and proteins were separated by electrophoresis on 15% polyacrylamide gels. Proteins were transferred onto a Hybond-P polyvinylidene fluoride membrane by semidry blotting. Membranes were blocked using 10% milk powder; synapsin-1 was detected with goat polyclonal antibody (Santa Cruz Biotechnology), synaptophysin with MAB368 (Abcam), CSP with rabbit polyclonal anti-CSP (sc-33154, Santa Cruz Biotechnology), VAMP-1 with mAb 4H302 (Abcam), caveolin with rabbit polyclonal antibodies to caveolin (Upstate), and PrPC with mAb 4F2 (J. Grassi). These were visualized using a combination of biotinylated anti-mouse/goat/rat/rabbit IgG (Sigma), extravidin-peroxidase, and enhanced chemiluminescence.
Isolation of DRMs
These membranes were isolated by their insolubility in non-ionic detergents as described (49). Briefly, samples were homogenized in an ice-cold buffer containing 1% Triton X-100, 10 mm Tris-HCl, pH 7.2, 150 mm NaCl, 10 mm EDTA, and mixed protease and phosphatase inhibitors, and nuclei and large fragments were removed by centrifugation (300 × g for 5 min at 4 °C). The postnuclear supernatant was incubated on ice (4 °C) for 1 h with intermittent shaking and centrifuged (16,000 × g for 30 min at 4 °C). The supernatant was reserved as the DSM, whereas the pellet was homogenized in an extraction buffer (as above) and centrifuged (10 min at 16,000 × g), and the soluble material was reserved as the DRM fraction.
Synaptophysin ELISA
The amounts of synaptophysin in neurons were measured by ELISA (50). Maxisorb immunoplates were coated with 0.5 μg/ml mouse anti-synaptophysin mAb (MAB368, Chemicon, Darmstadt, Germany) and blocked with 5% milk powder. Samples were added for 1 h, and bound synaptophysin was detected with rabbit polyclonal anti-synaptophysin (Abcam) followed by biotinylated anti-rabbit IgG, extravidin-alkaline phosphatase, and 1 mg/ml 4-nitrophenol phosphate (Sigma). Absorbance was measured on a microplate reader at 405 nm. Samples were expressed as “units of synaptophysin,” where 100 units was defined as the amount of synaptophysin in 106 control neurons.
Cholesterol
The concentrations of cholesterol in samples were measured using the Amplex Red cholesterol assay kit (Invitrogen) according to the manufacturer's instructions. Briefly, cholesterol was oxidized by cholesterol oxidase to yield hydrogen peroxide and ketones. The hydrogen peroxide reacted with 10-acetyl-3,7-dihydroxyphenoxazine (Amplex Red reagent) to produce highly fluorescent resorufin, which was measured by excitation at 550 nm and emission detection at 590 nm.
Isolation of PrPC
PrPC molecules were isolated from neurons or from N9 glial cells that had been homogenized in an extraction buffer (as above), and cell debris and nuclei were removed by centrifugation. The postnuclear supernatant was incubated with an affinity column loaded with mAb ICSM35 (M. Tayebi), and PrPC was eluted using glycine-HCl at pH 2.7, neutralized with 1 m Tris, and desalted. These PrPC preparations were further purified using size exclusion chromatography (Superdex 200 PC column). Some PrPC preparations were digested with 2 units/ml endoglycosidase F (Elizabethkingia meningoseptica), 0.2 units/ml PI-PLC (Bacillus cereus), 10 units/ml bee venom PLA2, or 0.2 units/ml neuraminidase (Clostridium perfringens) (all from Sigma) for 2 h at 37 °C. Samples were centrifuged through a 50-kDa filter to remove enzymes, and samples were loaded onto C18 columns (Waters). Proteins were purified using reverse phase chromatography with a gradient of propanol in water. Fractions were tested by ELISA; PrP-containing fractions were pooled, desalted, and stored at −80 °C. For bioassays, samples were thawed on the day of use and solubilized in culture medium by sonication.
PrP ELISA
The concentrations of PrP in samples were determined by ELISA as described (8). Briefly, Maxisorb immunoplates were coated with mAb ICSM18 and blocked with 5% milk powder. Samples were applied and detected with biotinylated mAb ICSM35, followed by extravidin-alkaline phosphatase and 1 mg/ml 4-nitrophenyl phosphate. Absorbance was measured on a microplate reader at 405 nm, and the amount of PrP in samples was calculated by reference to a standard curve of recombinant murine PrP (Fisher).
Isolation of CD14
CD14 was isolated from N9 glial cells that had been homogenized in an extraction buffer (as above). Insoluble debris and nuclei were removed by centrifugation. CD14 in the postnuclear supernatant was isolated using an affinity column loaded with rat anti-mouse CD14 mAb (clone RmC5-3) (BD Biosciences). CD14 was eluted with glycine-HCl at pH 2.7, neutralized with 1 m Tris, and loaded onto C18 columns. Proteins were eluted under a gradient of acetonitrile in water and 0.1% TFA. Fractions were tested by ELISA, and CD14-containing fractions were pooled, lyophilized, and stored at −80 °C. For bioassays, samples were thawed on the day of use and solubilized in culture medium by sonication.
CD14 ELISA
Concentrations of CD14 were measured by ELISA as described (39). Briefly, Maxisorb immunoplates were coated with 0.5 μg/ml rat IgG1 anti-mouse CD14 mAb (clone RmC5-3). Samples were applied, and bound CD14 was detected using a goat polyclonal IgG anti-mouse CD14 (R&D Systems), followed by anti-goat IgG conjugated to alkaline phosphatase (Sigma) and 1 mg/ml 4-nitrophenyl phosphate. Absorbance was measured on a microplate reader at 405 nm and compared with a titration of recombinant mouse CD14 (Enzo Lifesciences).
Isolation and Analysis of GPI Anchors
PrPC preparations were digested with 100 μg/ml proteinase K for 24 h at 37 °C, resulting in GPI anchors attached to the terminal amino acid. The released GPIs were extracted with water-saturated butan-1-ol and washed with water a further three times before being loaded onto C18 columns. GPIs were eluted using reverse phase chromatography under a gradient of acetonitrile and water. GPIs were detected by ELISA (see below), and positive fractions were pooled and lyophilized. Stock solutions were dissolved in ethanol at 2 μm. For some experiments, isolated GPIs were conjugated to FITC using the free amino group on the remaining amino acid and the homobifunctional cross-linking agent dimethyl pimelimidate (Fisher) using the manufacturer's instructions. Stock solutions were diluted in tissue culture medium for bioassays. GPIs were detected by immunoblotting as described with a mAb to phosphatidylinositol (34). The concentrations of FITC-labeled GPIs in samples were calculated by measuring fluorescence, excitation at 490 nm, and emission at 525 nm and compared with a dose response of FITC. Extracted GPIs were applied to silica gel 60 HPTLC plates (Whatman) and developed using a mixture of chloroform/methanol/water (4:4:1, v/v/v). Plates were soaked in 0.1% polyisobutyl methacrylate in hexane, dried, and blocked with 5% milk powder and probed with 1 μg/ml of mAb 5AB3-11, followed by biotinylated anti-mouse IgM and extravidin-horseradish peroxidase (Sigma), and visualized using chemiluminescence.
GPI ELISA
Maxisorb immunoplates were coated with 0.5 μg/ml concanavalin A (which binds mannose) and blocked with 5% milk powder in PBS-Tween. Samples were added, and any bound GPI was detected by the addition of the phosphatidylinositol-reactive mAb 5AB3-11, followed by a biotinylated anti-mouse IgM (Sigma), extravidin-alkaline phosphatase, and 1 mg/ml 4-nitrophenyl phosphate. Absorbance was measured at 405 nm.
Lectin Analysis of GPI Anchors
The presence of specific glycans in GPI anchors was determined using biotinylated lectins and dot blotting as described (34). Isolated GPIs were blotted onto nitrocellulose membranes, which were blocked (10% milk powder). Samples were incubated with biotinylated S. nigra (which detects terminal sialic acid residues bound α-2,6 or α-2,3 to galactose) or biotinylated concanavalin A to detect mannose (Vector Laboratories). Bound biotinylated lectins were visualized using extravidin peroxidase and enhanced chemiluminescence. The presence of phosphatidylinositol in GPIs was determined using mAb (5AB3-11), which was visualized using a horseradish peroxidase-conjugated anti-murine IgG and chemiluminescence.
Conjugation of GPIs to Rabbit IgG
Rabbit IgG (Sigma) was conjugated to GPIs by a two-step process. First, GPIs were conjugated to protein A (Innova Biosciences). The protein A complex was then cross-linked to rabbit IgG, and both steps used the homobifunctional cross-linking agent dimethyl pimelimidate (Pierce) according to the manufacturer's instructions. IgG-GPI conjugates were purified using reverse phase chromatography on C18 columns. The concentrations of rabbit IgG in samples were measured by ELISA. Maxisorb immunoplates were coated with mAb anti-rabbit IgG, clone L27A9 (New England Biolabs) and blocked with 5% milk powder. Samples were added, and rabbit IgG was detected with biotinylated goat anti-rabbit IgG (Sigma) followed by extravidin-alkaline phosphatase and 1 mg/ml 4-nitrophenyl phosphate. Absorbance was measured on a microplate reader at 405 nm, and the amount of rabbit IgG in samples was calculated by reference to a standard curve of rabbit IgG (Sigma). IgG and IgG-GPI conjugates were separated by HPTLC on silica gel 60 plates and developed using a mixture of chloroform/methanol/water (4:4:1, v/v/v).
Statistical Methods
Comparison of treatment effects was carried out using Student's paired t tests and one-way and two-way analysis of variance with Bonferroni's post hoc tests (IBM SPSS Statistics version 20). Error values are S.D., and significance was determined where p was <0.01. Correlations between data sets were analyzed using Pearson's bivariate coefficient (IBM SPSS Statistics version 20).
Author Contributions
C. B., W. N., and H. M.-O. were responsible for carrying out the experiments and data analysis. C. B. and A. W. were responsible for planning experiments and writing the manuscript.
This work was supported by the European Commission FP6 “Neuroprion,” Network of Excellence. The authors declare that they have no conflicts of interest with the contents of this article.
- PrPC
- cellular prion protein
- GPI
- glycosylphosphatidylinositol
- DRM
- detergent-resistant membrane
- DSM
- detergent-sensitive membrane
- HPTLC
- high performance thin layer chromatography
- PI-PLC
- phosphatidylinositol-specific phospholipase C
- CSP
- cysteine string protein.
References
- 1. Prusiner S. B. (1998) Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Herms J., Tings T., Gall S., Madlung A., Giese A., Siebert H., Schürmann P., Windl O., Brose N., and Kretzschmar H. (1999) Evidence of presynaptic location and function of the prion protein. J. Neurosci. 19, 8866–8875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown D. R. (2001) Prion and prejudice: normal protein and the synapse. Trends Neurosci. 24, 85–90 [DOI] [PubMed] [Google Scholar]
- 4. Lledo P. M., Tremblay P., DeArmond S. J., Prusiner S. B., and Nicoll R. A. (1996) Mice deficient for prion protein exhibit normal neuronal excitability and synaptic transmission in the hippocampus. Proc. Natl. Acad. Sci. U.S.A. 93, 2403–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maglio L. E., Martins V. R., Izquierdo I., and Ramirez O. A. (2006) Role of cellular prion protein on LTP expression in aged mice. Brain Res. 1097, 11–18 [DOI] [PubMed] [Google Scholar]
- 6. Stahl N., Borchelt D. R., Hsiao K., and Prusiner S. B. (1987) Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 51, 229–240 [DOI] [PubMed] [Google Scholar]
- 7. Taraboulos A., Scott M., Semenov A., Avrahami D., Laszlo L., Prusiner S. B., and Avraham D. (1995) Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J. Cell Biol. 129, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bate C., and Williams A. (2011) Monoacylated cellular prion protein modifies cell membranes, inhibits cell signaling and reduces prion formation. J. Biol. Chem. 286, 8752–8758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Puig B., Altmeppen H. C., Thurm D., Geissen M., Conrad C., Braulke T., and Glatzel M. (2011) N-Glycans and glycosylphosphatidylinositol-anchor act on polarized sorting of mouse PrP(C) in Madin-Darby canine kidney cells. PLoS One 6, e24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vey M., Pilkuhn S., Wille H., Nixon R., DeArmond S. J., Smart E. J., Anderson R. G., Taraboulos A., and Prusiner S. B. (1996) Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc. Natl. Acad. Sci. U.S.A. 93, 14945–14949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Taylor D. R., and Hooper N. M. (2006) The prion protein and lipid rafts. Mol. Membr. Biol. 23, 89–99 [DOI] [PubMed] [Google Scholar]
- 12. Pike L. J. (2004) Lipid rafts: heterogeneity on the high seas. Biochem. J. 378, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Madore N., Smith K. L., Graham C. H., Jen A., Brady K., Hall S., and Morris R. (1999) Functionally different GPI proteins are organized in different domains on the neuronal surface. EMBO J. 18, 6917–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ikezawa H. (2002) Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol. Pharm. Bull. 25, 409–417 [DOI] [PubMed] [Google Scholar]
- 15. Stahl N., Baldwin M. A., Hecker R., Pan K. M., Burlingame A. L., and Prusiner S. B. (1992) Glycosylinositol phospholipid anchors of the scrapie and cellular prion proteins contain sialic acid. Biochemistry 31, 5043–5053 [DOI] [PubMed] [Google Scholar]
- 16. Homans S. W., Ferguson M. A. J., Dwek R. A., Rademacher T. W., Anand R., and Williams A. F. (1988) Complete structure of the glycosyl phosphatidylinositol membrane anchor of rat brain Thy-1 glycoprotein. Nature 333, 269–272 [DOI] [PubMed] [Google Scholar]
- 17. Premkumar D. R., Fukuoka Y., Sevlever D., Brunschwig E., Rosenberry T. L., Tykocinski M. L., and Medof M. E. (2001) Properties of exogenously added GPI-anchored proteins following their incorporation into cells. J. Cell Biochem. 82, 234–245 [DOI] [PubMed] [Google Scholar]
- 18. Zacharias D. A., Violin J. D., Newton A. C., and Tsien R. Y. (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 [DOI] [PubMed] [Google Scholar]
- 19. Sunyach C., Jen A., Deng J., Fitzgerald K. T., Frobert Y., Grassi J., McCaffrey M. W., and Morris R. (2003) The mechanism of internalization of glycosylphosphatidylinositol-anchored prion protein. EMBO J. 22, 3591–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Legler D. F., Doucey M. A., Schneider P., Chapatte L., Bender F. C., and Bron C. (2005) Differential insertion of GPI-anchored GFPs into lipid rafts of live cells. FASEB J. 19, 73–75 [DOI] [PubMed] [Google Scholar]
- 21. Li R., Liu T., Yoshihiro F., Tary-Lehmann M., Obrenovich M., Kuekrek H., Kang S.-C., Pan T., Wong B.-S., Medof M. E., and Sy M.-S. (2003) On the same cell type GPI-anchored normal cellular prion and DAF protein exhibit different biological properties. Biochem. Biophys. Res. Commun. 303, 446–451 [DOI] [PubMed] [Google Scholar]
- 22. Liu T., Li R., Pan T., Liu D., Petersen R. B., Wong B. S., Gambetti P., and Sy M. S. (2002) Intercellular transfer of the cellular prion protein. J. Biol. Chem. 277, 47671–47678 [DOI] [PubMed] [Google Scholar]
- 23. Bate C., Salmona M., Diomede L., and Williams A. (2004) Squalestatin cures prion-infected neurons and protects against prion neurotoxicity. J. Biol. Chem. 279, 14983–14990 [DOI] [PubMed] [Google Scholar]
- 24. Gilch S., Kehler C., and Schätzl H. M. (2006) The prion protein requires cholesterol for cell surface localization. Mol. Cell Neurosci. 31, 346–353 [DOI] [PubMed] [Google Scholar]
- 25. Crick D. C., Suders J., Kluthe C. M., Andres D. A., and Waechter C. J. (1995) Selective inhibition of cholesterol biosynthesis in brain cells by squalestatin 1. J. Neurochem 65, 1365–1373 [DOI] [PubMed] [Google Scholar]
- 26. Hering H., Lin C. C., and Sheng M. (2003) Lipid rafts in the maintenance of synapses, dendritic spines, and surface AMPA receptor stability. J. Neurosci. 23, 3262–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mauch D. H., Nägler K., Schumacher S., Göritz C., Müller E.-C., Otto A., and Pfrieger F. W. (2001) CNS synaptogenesis promoted by glia-derived cholesterol. Science 294, 1354–1357 [DOI] [PubMed] [Google Scholar]
- 28. Cancellotti E., Wiseman F., Tuzi N. L., Baybutt H., Monaghan P., Aitchison L., Simpson J., and Manson J. C. (2005) Altered glycosylated PrP proteins can have different neuronal trafficking in brain but do not acquire scrapie-like properties. J. Biol. Chem. 280, 42909–42918 [DOI] [PubMed] [Google Scholar]
- 29. Tuzi N. L., Cancellotti E., Baybutt H., Blackford L., Bradford B., Plinston C., Coghill A., Hart P., Piccardo P., Barron R. M., and Manson J. C. (2008) Host PrP glycosylation: a major factor determining the outcome of prion infection. PLoS Biol. 6, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haraguchi T., Fisher S., Olofsson S., Endo T., Groth D., Tarentino A., Borchelt D. R., Teplow D., Hood L., and Burlingame A. (1989) Asparagine-linked glycosylation of the scrapie and cellular prion proteins. Arch. Biochem. Biophys. 274, 1–13 [DOI] [PubMed] [Google Scholar]
- 31. Chen R., Walter E. I., Parker G., Lapurga J. P., Millan J. L., Ikehara Y., Udenfriend S., and Medof M. E. (1998) Mammalian glycophosphatidylinositol anchor transfer to proteins and posttransfer deacylation. Proc. Natl. Acad. Sci. U.S.A. 95, 9512–9517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McConville M. J., and Ferguson M. A. (1993) The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem. J. 294, 305–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nicholson T. B., and Stanners C. P. (2006) Specific inhibition of GPI-anchored protein function by homing and self-association of specific GPI anchors. J. Cell Biol. 175, 647–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bate C., and Williams A. (2012) Neurodegeneration induced by the clustering of sialylated glycosylphosphatidylinositols of prion proteins. J. Biol. Chem. 287, 7935–7944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maeda Y., Tashima Y., Houjou T., Fujita M., Yoko-o T., Jigami Y., Taguchi R., and Kinoshita T. (2007) Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Mol. Biol. Cell 18, 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schroeder R., London E., and Brown D. (1994) Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc. Natl. Acad. Sci. U.S.A. 91, 12130–12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Medof M. E., Nagarajan S., and Tykocinski M. L. (1996) Cell-surface engineering with GPI-anchored proteins. FASEB J. 10, 574–586 [DOI] [PubMed] [Google Scholar]
- 38. Anderson R. G. W., and Jacobson K. (2002) A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 296, 1821–1825 [DOI] [PubMed] [Google Scholar]
- 39. Ingham V., Williams A., and Bate C. (2014) Glimepiride reduces CD14 expression and cytokine secretion from macrophages. J. Neuroinflammation 11, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brügger B., Graham C., Leibrecht I., Mombelli E., Jen A., Wieland F., and Morris R. (2004) The membrane domains occupied by glycosylphosphatidylinositol-anchored prion protein and Thy-1 differ in lipid composition. J. Biol. Chem. 279, 7530–7536 [DOI] [PubMed] [Google Scholar]
- 41. Bate C., Nolan W., and Williams A. (2016) Sialic acid on the glycosylphosphatidylinositol anchor regulates PrP-mediated cell signaling and prion formation. J. Biol. Chem. 291, 160–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cantù L., Del Favero E., Sonnino S., and Prinetti A. (2011) Gangliosides and the multiscale modulation of membrane structure. Chem. Phys. Lipids 164, 796–810 [DOI] [PubMed] [Google Scholar]
- 43. Simons M., Friedrichson T., Schulz J. B., Pitto M., Masserini M., and Kurzchalia T. V. (1999) Exogenous administration of gangliosides displaces GPI-anchored proteins from lipid microdomains in living cells. Mol. Biol. Cell 10, 3187–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ohmi Y., Ohkawa Y., Yamauchi Y., Tajima O., Furukawa K., and Furukawa K. (2012) Essential roles of gangliosides in the formation and maintenance of membrane microdomains in brain tissues. Neurochem. Res. 37, 1185–1191 [DOI] [PubMed] [Google Scholar]
- 45. Crespo P. M., Zurita A. R., and Daniotti J. L. (2002) Effect of gangliosides on the distribution of a glycosylphosphatidylinositol-anchored protein in plasma membrane from Chinese hamster ovary-K1 cells. J. Biol. Chem. 277, 44731–44739 [DOI] [PubMed] [Google Scholar]
- 46. Fujinaga Y., Wolf A. A., Rodighiero C., Wheeler H., Tsai B., Allen L., Jobling M. G., Rapoport T., Holmes R. K., and Lencer W. I. (2003) Gangliosides that associate with lipid rafts mediate transport of cholera and related toxins from the plasma membrane to endoplasmic reticulum. Mol. Biol. Cell 14, 4783–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Galvan C., Camoletto P. G., Dotti C. G., Aguzzi A., and Ledesma M. D. (2005) Proper axonal distribution of PrPC depends on cholesterol-sphingomyelin-enriched membrane domains and is developmentally regulated in hippocampal neurons. Mol. Cell Neurosci. 30, 304–315 [DOI] [PubMed] [Google Scholar]
- 48. Dunkley P. R., Jarvie P. E., and Robinson P. J. (2008) A rapid Percoll gradient procedure for preparation of synaptosomes. Nat. Protoc. 3, 1718–1728 [DOI] [PubMed] [Google Scholar]
- 49. London E., and Brown D. A. (2000) Insolubility of lipids in Triton X-100: physical origin and relationship to sphingolipid/cholesterol membrane domains (rafts). Biochim. Biophys. Acta 1508, 182–195 [DOI] [PubMed] [Google Scholar]
- 50. Lipton A. M., Cullum C. M., Satumtira S., Sontag E., Hynan L. S., White C. L. 3rd, and Bigio E. H. (2001) Contribution of asymmetric synapse loss to lateralizing clinical deficits in frontotemporal dementias. Arch. Neurol. 58, 1233–1239 [DOI] [PubMed] [Google Scholar]