Abstract
Oncogenic PIK3CA (p110α), the catalytic subunit of class IA PI3K, plays a major role in PI3K-related cancer progression. The mechanisms underlying the dynamic regulation of PIK3CA protein levels remain unknown. Here we demonstrated that PIK3CA is regulated by polyubiquitination. We identified NEDD4L as the E3 ligase that catalyzes PIK3CA polyubiquitination, leading to its proteasome-dependent degradation. NEDD4L ubiquitinates both the free and regulatory subunit-bound PIK3CA but does not ubiquitinate the regulatory subunit of PI3K. Overexpression of NEDD4L accelerates the turnover rate of PIK3CA, whereas suppression of NEDD4L results in not only the accumulation of PIK3CA but also a paradoxical decrease of AKT activation. Thus, we propose that NEDD4L negatively regulates PIK3CA protein levels via ubiquitination and is required for the maintenance of PI3K-AKT signaling pathway.
Keywords: Akt PKB, E3 ubiquitin ligase, phosphatidylinositide 3-kinase (PI 3-kinase), proteasome, protein degradation, ubiquitylation (ubiquitination), NEDD4L, PIK3CA
Introduction
Class IA phosphoinositide 3-kinases (PI3Ks) are lipid kinases that integrate signals from growth factors and hormones to regulate multiple cellular process including growth, proliferation, migration, and survival (1). In response to activation of various growth factor receptors, PI3K is recruited to the cell membrane to catalyze the production of phosphoinositide-3,4,5-trisphosphate, a second messenger required for the activation of a series of downstream kinases (1, 2). Class IA PI3K is composed of a catalytic subunit (p110) and a regulatory subunit (p85), which are demonstrated to form an obligatory heterodimer (3). PIK3CA (p110α), a major isoform of the catalytic subunits, is widely expressed in various tissues (4). Somatic mutations in PIK3CA occur frequently in cancers of the breast, colon, endometrium, and prostate as well as glioblastomas (5–7). Gain of PIK3CA copy number leads to increased PIK3CA protein levels and is also associated with human cancers (4, 8, 9). These observations indicate that dysregulation of PI3K plays an important role in cancer emergence and development, underscoring the significance of understanding the regulation mechanisms of PI3K signaling pathway.
PI3K function can be modulated via its gene expression or its association with Ras, receptor-tyrosine kinases and other adaptor proteins such as insulin receptor substrate1/2 (IRS1/2). It can also be regulated through post-translational modifications. For instance, previous reports demonstrate that the p85 regulatory subunit can be polyubiquitinated and negatively regulated by the E3 ubiquitin ligase Cbl-b in T cells without affecting its protein level (10, 11). In addition, recent work shows that dephosphorylation of the p110-free p85β regulatory subunit leads to its degradation through F-box protein FBXL2 (12). Importantly, the catalytic subunit PIK3CA may also be subject to a similar regulation: that is, a dynamic cycle of proteasome-dependent degradation and resynthesis of PIK3CA was observed in response to the stimulation of epidermal growth factor (13).
To further understand the regulation of PIK3CA, we developed an in vitro ubiquitination assay for PIK3CA. Using this assay and biochemical fractionation, we identified NEDD4L (neural precursor cell expressed, developmentally down-regulated 4-like),3 the E3 ubiquitin ligase responsible for PIK3CA polyubiquitination. NEDD4L polyubiquitinates PIK3CA to promote its proteasomal degradation at both its regulatory subunit free and bound states. Knockdown of NEDD4L increases the protein level of PIK3CA but, paradoxically, impairs the activation of AKT. We propose that NEDD4L is a PIK3CA E3 ubiquitin ligase that controls the stability of PIK3CA and is required for the maintenance of PI3K-AKT signaling pathway.
Results
Ubiquitination of PIK3CA in Vivo
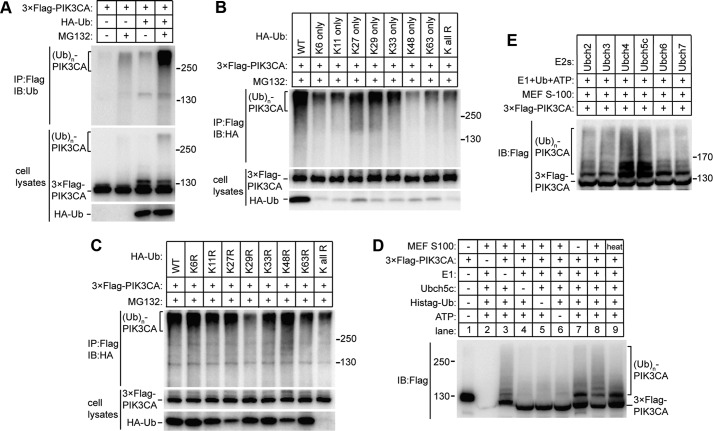
To examine whether PIK3CA is regulated by ubiquitination in vivo, we transfected human embryonic kidney (HEK)-293T cells with FLAG-tagged PIK3CA alone or in combination with HA-tagged ubiquitin followed by treatment with DMSO or proteasome inhibitor MG132. PIK3CA was then immunoprecipitated and detected with ubiquitin antibody. Proteasome inhibition resulted in an accumulation of ubiquitinated PIK3CA, which were robustly increased by co-expression with an exogenous ubiquitin (Fig. 1A, upper panel). To verify that PIK3CA is indeed directly polyubiquitinated, we immunoblotted PIK3CA in whole cell lysates using an antibody against the FLAG epitope (Fig. 1A, middle panel). The results suggest that PIK3CA is itself polyubiquitinated in vivo; PIK3CA formed high molecular weight shifts in the presence of MG132, and expression of an exogenous ubiquitin resulted in an increase of the polyubiquitinated PIK3CA (Fig. 1A, middle and lower panels).
FIGURE 1.
Ubiquitination of PIK3CA in vivo and in vitro. A, visualization of the ubiquitinated PIK3CA in vivo. The indicated plasmids were transfected into HEK293T cells. Twenty-four hours later cells were treated with DMSO or MG132 for 6 h. The ubiquitination were detected either by immunoprecipitation (IP) of FLAG-tagged PIK3CA followed by Western blot analysis with anti-ubiquitin (Ub) antibody (upper panel) or directly immunoblotting (IB) the cell lysates with anti-FLAG antibody (middle panel). The expression of HA-tagged ubiquitin was detected by anti-HA antibody (lower panel). B, determination of the linkage type of ubiquitin chain. HEK293T cells were co-transfected with PIK3CA and ubiquitin mutants that have only one lysine residues. Twenty-four hours later cells were treated with MG132 for 6 h. FLAG-tagged PIK3CA was immunoprecipitated, and ubiquitination was detected with anti-HA antibody. C, determination of the linkage type of ubiquitin chain. Western samples were prepared as in B except the afore-used ubiquitins were substituted with single lysine to arginine mutants. D, in vitro ubiquitination of PIK3CA. Recombinant FLAG-tagged PIK3CAs were incubated alone or with the combination of various components as indicated in a total volume of 15 μl. The assay was performed at 37 °C for 2 h, then boiled and applied to immunoblot using anti-FLAG antibody. E, in vitro ubiquitination test for E2 preference. Recombinant FLAG-tagged PIK3CA, E1, ubiquitin, MEF S-100, and ATP were mixed with equal moles of recombinant E2s. The assay was performed, and samples were processed as in D.
Proteasome-dependent protein degradation is classically mediated by covalent modification of the Lys-48-linked polyubiquitin chain (14). Ubiquitin chains linked through other alternative lysine residues are generally thought to function in signaling transduction even though they may also serve as a proteasome-targeting signal (15). To determine the chain type involved in PIK3CA polyubiquitination and degradation, we co-expressed PIK3CA with a series of ubiquitin mutants possessing only one lysine residue (Fig. 1B). By immunopurification of PIK3CA and detection of polyubiquitin, we found that PIK3CA was mainly modified by Lys-27-, Lys-29-, and Lys-33-linked chain, among which Lys-29 linkage was the most favored (Fig. 1B). To confirm this, we did similar experiments by substituting the afore-used ubiquitins with single lysine to arginine mutants (Fig. 1C). Consistent with the above results, K29R mutation resulted in the greatest reduction in the mount of the polyubiquitinated PIK3CA (Fig. 1C). Taken together, these data suggest Lys-29-linked chain is the major form for PIK3CA polyubiquitination and degradation.
Ubiquitination of PIK3CA in Vitro
The above findings suggest there should be at least an E3 ubiquitin ligase in cells that catalyze the conjugation of ubiquitin to PIK3CA. To test this hypothesis, we first developed an in vitro ubiquitination assay. As shown in Fig. 1D, after incubation of purified recombinant human ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), ubiquitin and FLAG-tagged PIK3CA together with mouse embryonic fibroblast (MEF) cell extracts (S-100) and ATP, PIK3CA showed at least two clear upshifted ladders and a decreased level of native protein (Fig. 1D, lane 8). Dropping off one of the any components in the reactions resulted in only one upshifted ladder or even none at all. Heating of the cell extract decreased the ubiquitination activity to basal level (Fig. 1D, lane 9), suggesting the PIK3CA E3 ligase is likely to be a temperature-sensitive protein. Moreover, the E2 preference was tested using the same assay except equal moles of different E2 enzymes (including Ubch2, -3, -4, -5C, -6, -7) were used in each reaction (Fig. 1E). We observed that Ubch4 and Ubch5C conferred the strongest ubiquitination activity for PIK3CA.
Mapping the Ubiquitination Sites on PIK3CA
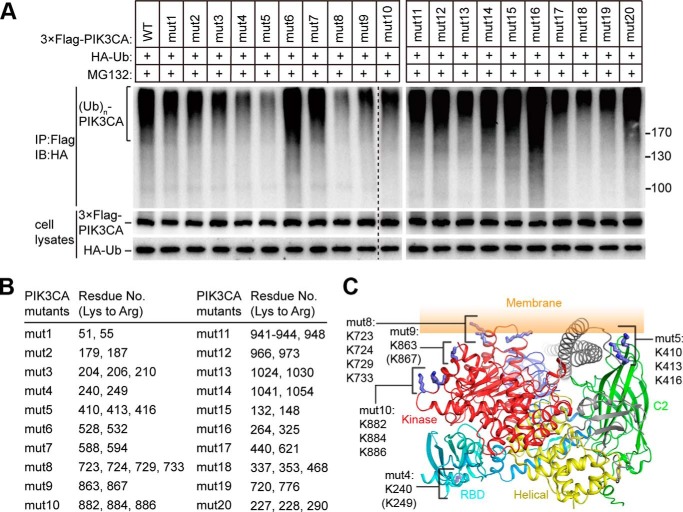
We used the mutagenesis strategy to map the ubiquitination sites on PIK3CA. Taking into account the fact that there are total of 72 lysine residues on PIK3CA, we first analyzed the structures of PIK3CA-PIK3R1 heterodimeric complex (16, 17) and selected 50 lysine residues that are exposed to the solvent as our mutational targets. We generated 20 PIK3CA mutants, each covering 2–5 lysine to arginine mutations (Fig. 2, A and B). These mutants were co-transfected with ubiquitin in HEK293T cells, treated with MG132, and then immunoprecipitated for detection of ubiquitination (Fig. 2A). We found five PIK3CA mutants (mut4, 5, 8, 9, 10) showed a dramatic decrease in ubiquitination (Fig. 2A). Based on the membrane binding model of PI3K (17), with the exception of mut4 that locates in the Ras binding domain, all of the ubiquitination-deficient lysine mutations are located in the membrane-contact or membrane-adjacent region (Fig. 2C). Lysine residues in mut5 are on the top of the C2 domain, which mediates the interaction of PI3K with membrane, whereas those in mut8 locate in the N-lobe of kinase domain, which also interacts with membrane. Lysine residues in mut9 and mut10 locate in the C-lobe of kinase domain and, if they do not directly interact, are very close to the membrane. These results indicate lysine residues that either interact with or are close to the cell membrane in PIK3CA are hot spots for ubiquitination.
FIGURE 2.
Mapping the ubiquitination sites on PIK3CA. A, mapping the possible ubiquitination residues on PIK3CA. A series of PIK3CA lysine to arginine mutants was co-transfected with HA-tagged ubiquitin. Twenty-four hours after transfection cells were treated with MG132 for 6 h and collected. PIK3CA was immunoprecipitated (IP) from the cleared cell lysates using anti-FLAG beads. Ubiquitination (Ub) was detected by immunoblotting (IB) the precipitated PIK3CA with anti-HA antibody. B, a list of the mutated residues on each PIK3CA mutants. C, structural presentation of the ubiquitination hot spots on PIK3CA. The complex structure of PIK3CA-PIK3R1 (Protein Data Bank code 4JPS) is presented as ribbon with possible ubiquitinated lysine residues shown as sticks. Residues in parenthesis are not seen in the structure. The Ras binding domain is colored cyan, the C2 domain is colored green, the helical domain is colored yellow, and the kinase domain is colored red. PIK3R1 is colored gray. The figure was generated using PyMOL software.
Purification of the E3 Ubiquitin Ligase for PIK3CA
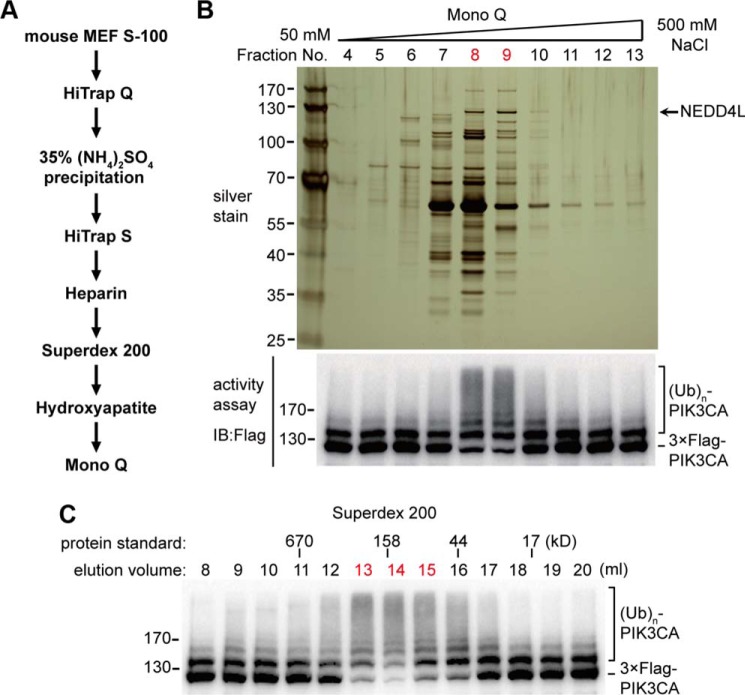
To identify the E3 ligase for PIK3CA ubiquitination, we fractionated the MEF cell extract S-100 by an ion exchange (Q) column and analyzed the E3 ligase activity by using the aforementioned in vitro ubiquitination assay. The activity was eluted out as a wide peak with a salt concentration between 200 and 350 mm NaCl. The active fractions were collected and subjected to a serious of sequential purification steps (Fig. 3A). At the final purification step, PIK3CA E3 ligase activity from the Mono Q column mainly existed in two fractions, which efficiently promoted the polyubiquitination of PIK3CA (Fig. 3B). The fractions were applied to silver staining, and inspection of the gel revealed one band, indicated by an arrow in Fig. 3B, correlated perfectly with the activity. The observed molecular weight of this band was in agreement with the corresponding elution volumes on gel filtration (Fig. 3C). Mass spectrum analysis revealed this protein to be NEDD4L (neural precursor cell expressed, developmentally down-regulated 4-like), an E3 ubiquitin ligase known to associate with the plasma membrane and previously shown to ubiquitinate multiple membrane proteins (18).
FIGURE 3.
Purification of PIK3CA E3 ubiquitin ligase. A, diagram of the purification scheme for PIK3CA E3 ligase. B, the final step of purification. The indicated Mono Q fractions (20 μl per fraction) were resolved by a 4–12% Bis-Tris gel and silver-stained (upper panel). The PIK3CA E3 activity was assayed by incubation of FLAG-tagged PIK3CA with 1.5 μl Mono Q fractions combined with other ubiquitination components for 2 h at 37 °C. The samples were boiled and applied to a Western blot (IB) using anti-FLAG antibody. The active fraction numbers are colored red. C, the activity assay of the gel filtration step of purification. 3 μl of the eluted fractions from a Superdex 200 column were used in the in vitro ubiquitination assay.
NEDD4L Ubiquitinates PIK3CA in Vitro
To confirm that NEDD4L is involved in ubiquitination of PIK3CA, we expressed and purified recombinant NEDD4L protein from insect cells (Fig. 4A, upper and lower panel). The addition of recombinant NEDD4L greatly enhanced in vitro ubiquitination of PIK3CA (Fig. 4B, lanes 7 and 8). At low concentrations, NEDD4L protein levels were positively correlated with the amount of ubiquitin molecules attached to PIK3CA (Fig. 4C); at high concentrations, NEDD4L promoted polyubiquitination of PIK3CA, which showed a dramatically delayed shift on gel and even partially stacked at the bottom of the loading wells (Fig. 4C). Heat treatment (50 °C for 10 min) of recombinant NEDD4L protein largely abrogated its E3 ligase activity (Fig. 4C).
FIGURE 4.
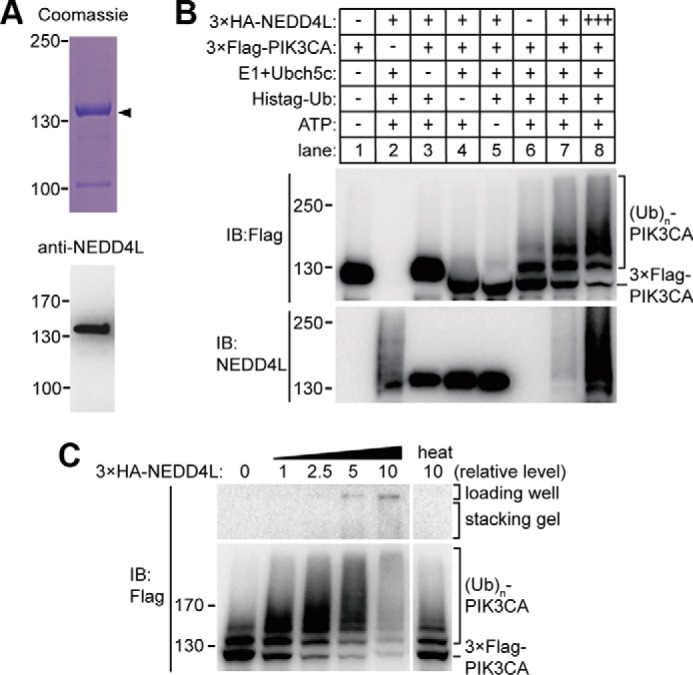
Recombinant NEDD4L protein ubiquitinated PIK3CA in vitro. A, Coomassie Blue staining of the purified recombinant NEDD4L from baculovirus expression system (upper panel) and Western blot (IB) verification using anti-NEDD4L antibody (lower panel). B, in vitro ubiquitination (Ub) of PIK3CA with recombinant NEDD4L as the E3 ligase. Recombinant FLAG-tagged PIK3CA were incubated with the combination of various components as indicated. NEDD4L protein in lane 2, 7, and 8 were observed as smeared bands because of the self-ubiquitination. C, different concentrations of recombinant NEDD4L were used in the in vitro ubiquitination assay with FLAG-tagged PIK3CA as substrates.
NEDD4L Ubiquitinates PIK3CA and Regulates Its Stability in Vivo
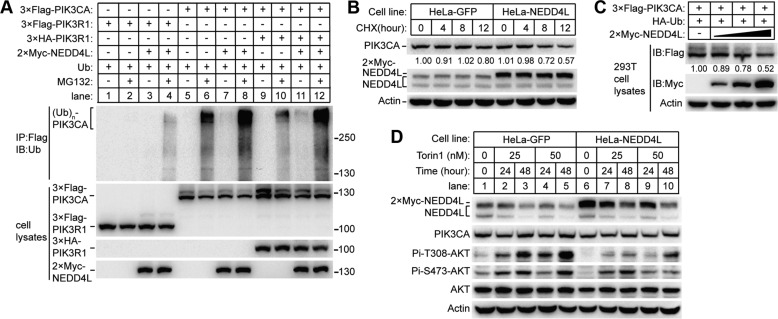
Because PIK3CA forms obligate heterodimers with p85 regulatory subunits (3), we asked whether NEDD4L can also ubiquitinate p85 subunits and whether heterodimerization affects the ubiquitination of PIK3CA. To address this question, we co-transfected plasmids encoding PIK3R1 (one of the p85 subunits) and PIK3CA, either alone or in combination with NEDD4L in HEK293T cells. We pulled down the target proteins and analyzed their ubiquitination level. As shown in Fig. 5A, NEDD4L co-transfection with the addition of MG132 greatly promoted the accumulation of polyubiquitinated PIK3CA (Fig. 5A, lane 8) compared with NEDD4L absent control (Fig. 5A, lane 6), whereas we did not observe an ubiquitination signal on immunoprecipitated PIK3R1 in the presence of MG132 (Fig. 5A, lane 2). Co-transfection of PIK3R1 with NEDD4L in the presence of MG132 caused a detection of a rather low level of ubiquitination on immunoprecipitated PIK3R1 (Fig. 5A, lane 4), which cannot be distinguished with the endogenous ubiquitinated PIK3CA that co-pulled-down with FLAG-tagged PIK3R1. In addition, the PIK3CA E3 ligase activity was not affected by the co-transfection of PIK3R1 (Fig. 5A, compare lane 8 with lane 12), suggesting NEDD4L can ubiquitinate heterodimeric PI3K. In summary, these results suggest that NEDD4L mainly targets PIK3CA but not PIK3R1 for ubiquitination. Dimerization of PIK3CA with PIK3R1 does not show obvious impact on the efficiency of NEDD4L-mediated ubiquitination.
FIGURE 5.
NEDD4L targeted PIK3CA for polyubiquitination and proteasome-dependent degradation in vivo. A, NEDD4L ubiquitinates PIK3CA and PIK3CA-PIK3R1 heterodimer but not individual PIK3R1. HEK293T cells were co-transfected with the indicated plasmids. Twenty-four hours later cells were treated with DMSO or MG132 for 6 h and then lysed. The FLAG-tagged proteins were immunoprecipitated (IP), and ubiquitination (IB) was detected by anti-ubiquitin (Ub) antibody. B, NEDD4L accelerates the turnover rate of PIK3CA in HeLa cells. HeLa cells stably expressing GFP or Myc-tagged NEDD4L were chased with 25 μg/ml cycloheximide and collected at the indicated times. Cleared cell lysates were normalized and immunoblotted by anti-PIK3CA and anti-NEDD4L antibody as indicated. C, NEDD4L promotes the degradation of PIK3CA. Different amounts of Myc-tagged NEDD4L were co-transfected with FLAG-tagged PIK3CA and HA-tagged ubiquitin in HEK293T cells. Cleared cell lysates were normalized and immunoblotted. The protein levels of PIK3CA and NEDD4L were detected by anti-FLAG and anti-Myc antibody, respectively. D, overexpression of NEDD4L down-regulates the PI3K-AKT pathway. HeLa cells stably overexpressing GFP or NEDD4L were not treated (lane 1 and 6) or were treated with Torin1 for the indicated times and concentrations. Normalized lysates were applied to Western blots against the antibodies as indicated.
To confirm the hypothesis that polyubiquitination of PIK3CA by NEDD4L leads to protein degradation, we performed a cycloheximide (CHX) chase assay (Fig. 5B). HeLa cells stably overexpressing GFP or NEDD4L were treated with cycloheximide and collected at interval time points. The endogenous PIK3CA protein levels in the normalized cell lysates were visualized by Western blot. We observed that NEDD4L overexpression caused a higher turnover rate of PIK3CA in contrast to the GFP control (Fig. 5B). Consistent with this observation, the decrement of PIK3CA protein levels were positively correlated with the amount of transfected NEDD4L plasmid in HEK293T cells (Fig. 5C).
PI3K recruitment to the cell membrane in response to activation of receptor-tyrosine kinases leads to the phosphorylation of downstream kinase AKT (1, 2). Because NEDD4L targets PIK3CA for proteasomal degradation, we tested whether overexpression NEDD4L down-regulates the PI3K-AKT pathway. We examined the phosphorylation levels of AKT in HeLa cells overexpressing GFP or NEDD4L. As expected, NEDD4L overexpression resulted in decreased level of AKT phosphorylation (Fig. 5D, compare lane 6 with lane 1). Because the phosphorylation signal of AKT in HeLa cells was relatively low, we treated the cells with Torin1, an inhibitor of mammalian target of rapamycin (mTOR) that at low concentrations up-regulates PI3K-AKT signaling by relieving the feedback inhibition of this pathway from mTOR complex 1 (19, 20). In accordance with the above results, treatment of Torin1 caused a dramatic increase in AKT phosphorylation in HeLa cells overexpressing GFP (Fig. 5D, lane 2–5), whereas the increase of AKT phosphorylation in NEDD4L-overexpressing cells was very limited (Fig. 5D, lanes 6–10).
NEDD4L Knockdown Caused an Increase in PIK3CA Protein Level but Impaired PI3K-AKT Signaling Pathway
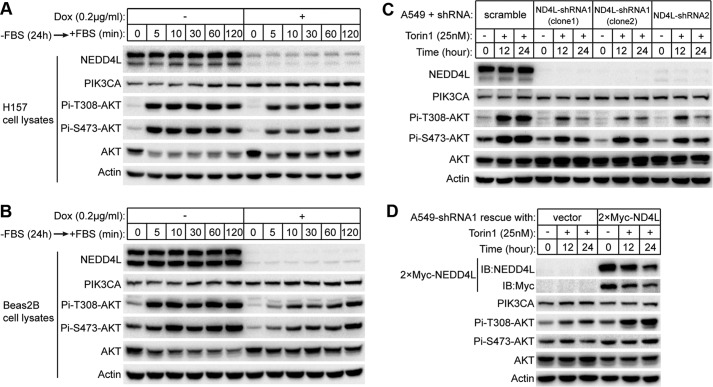
As overexpression of NEDD4L down-regulates PI3K-AKT signaling, we expected suppression of NEDD4L expression might have opposite effects. To test this, we stably expressed short hairpin RNA (shRNA), which targets NEDD4L mRNA in a doxycycline (Dox)-inducible manner in both H157 and Beas2B cells. The two cell lines were treated with Dox for 5 days and deprived of fetal bovine serum (FBS) for 24 h followed by treatment with FBS for different duration times (Fig. 6, A and B). In both cells, NEDD4L knockdown resulted in an overall increased levels of PIK3CA whose differences were especially large at time points 0–10 min. We noticed there was a down-regulation of PIK3CA protein level in the absence of FBS, which was less apparent in NEDD4L-knockdown cells, suggesting a dysregulation of the PIK3CA dynamics. Unexpectedly, although PIK3CA protein levels were elevated in NEDD4L-knockdown cells, phosphorylation of AKT at both Thr-308 and Ser-473 in response to FBS stimulation was impaired (Fig. 6, A and B). In Dox absent controls, phosphorylation of AKT reached a peak within 5 min after FBS treatment, whereas in NEDD4L-knockdown cells, the overall AKT phosphorylation level was much lower, and the time point for peak phosphorylation was delayed for 30 min and 120 min in H157 and Beas2B cells, respectively. These results suggest NEDD4L is involved in the regulation of PIK3CA dynamics, which is required for efficient signaling transduction.
FIGURE 6.
NEDD4L is required for the maintenance of PI3K-AKT pathway. A, inducible knockdown of NEDD4L in H157 cells impairs AKT phosphorylation. H157 cells containing pLKO.1-Tet-on-NEDD4L-shRNA1 were treated with or without Dox for 5 days and then plated in serum-free DMEM and cultured for 24 h followed by the addition of 10% FBS and collection of the cells at the indicated time points. Cleared cell lysates were normalized and applied to Western blot for the level of indicated proteins and phosphorylation states. B, inducible knockdown of NEDD4L in Beas2B cells impairs AKT phosphorylation. Beas2B cells stably transfected with pLKO.1-Tet-on-NEDD4L-shRNA1 were treated, and samples were processed as in A. C, stable knockdown of NEDD4L in A549 cells led to a decrease in AKT phosphorylation. A549 cells stably expressing NEDD4L-shRNA1, NEDD4L-shRNA2, or scramble shRNA were treated with Torin1 for the indicated times. Normalized cell lysates were applied to Western blot against the antibodies as indicated. ND4L, NEDD4L. D, decrease of AKT phosphorylation caused by NEDD4L knockdown is rescued by expression of shRNA-resistant NEDD4L. A549 cells stably expressing NEDD4L-shRNA1 were transfected with pcDNA3.1 vector or pcDNA3.1–2×Myc-NEDD4L with synonymous mutations in shRNA1 targeting region. Cells were treated, and samples were processed as in C. IB, immunoblot.
To further confirm the functional role of NEDD4L in PI3K-AKT signaling pathway, we engineered A549 cells to stably express two different sets of shRNAs against NEDD4L. Because the phosphorylation signal of AKT in A549 cells was relatively low in our preliminary examination (as previously reported; Ref. 21), we treated the cells with Torin1 to up-regulate PI3K-AKT signaling and then compared the phosphorylation levels of AKT between control and NEDD4L-knockdown cells. Similar to the previously described effects of NEDD4L knockdown (Fig. 6, A and B), NEDD4L suppression in A549 cells resulted in a very limited increase in the AKT phosphorylation level in the presence of Torin1 (Fig. 6C). To exclude the possibility that these observations were off-target effects of shRNA, we rescued the knockdown cells with a codon-modified NEDD4L (Fig. 6D). Stable expression of an exogenous NEDD4L, which is resistant to the shRNA, greatly increased the phosphorylation level of AKT, underscoring the importance of NEDD4L in maintenance of the robustness of PI3K-AKT signaling.
NEDD4L Promotes Ubiquitination of Endogenous PIK3CA in Vivo
To examine the ubiquitination states of PIK3CA in vivo and to verify the importance of NEDD4L in PIK3CA ubiquitination, we treated Beas2B-inducible knockdown cells as illustrated in Fig. 7A. The endogenous PIK3CA protein was then immunoprecipitated from cell lysates and detected with ubiquitin antibody. We observed that in NEDD4L-expressing cells, PIK3CA was polyubiquitinated under both serum-absent and insulin-like growth factor (IGF)-present conditions (Fig. 7B, lane 1 and 2). In NEDD4L-deficient cells, PIK3CA ubiquitination showed a dramatic decrease in the absence of serum compared with control cells, suggesting a major role of NEDD4L in catalyzing PIK3CA ubiquitination under serum deprivation (Fig. 7B, lane 3). This result is consistent with the previous observation that in the absence of serum PIK3CA protein was down-regulated in control but not NEDD4L-deficient cells (Fig. 6, A and B). In contrast, NEDD4L knockdown did not affect the ubiquitination level of PIK3CA under IGF stimulation (Fig. 7B, lane 4), suggesting that in addition to NEDD4L, there is another E3 ligase in cells that polyubiquitinates PIK3CA when PI3K-AKT pathway is activated.
FIGURE 7.
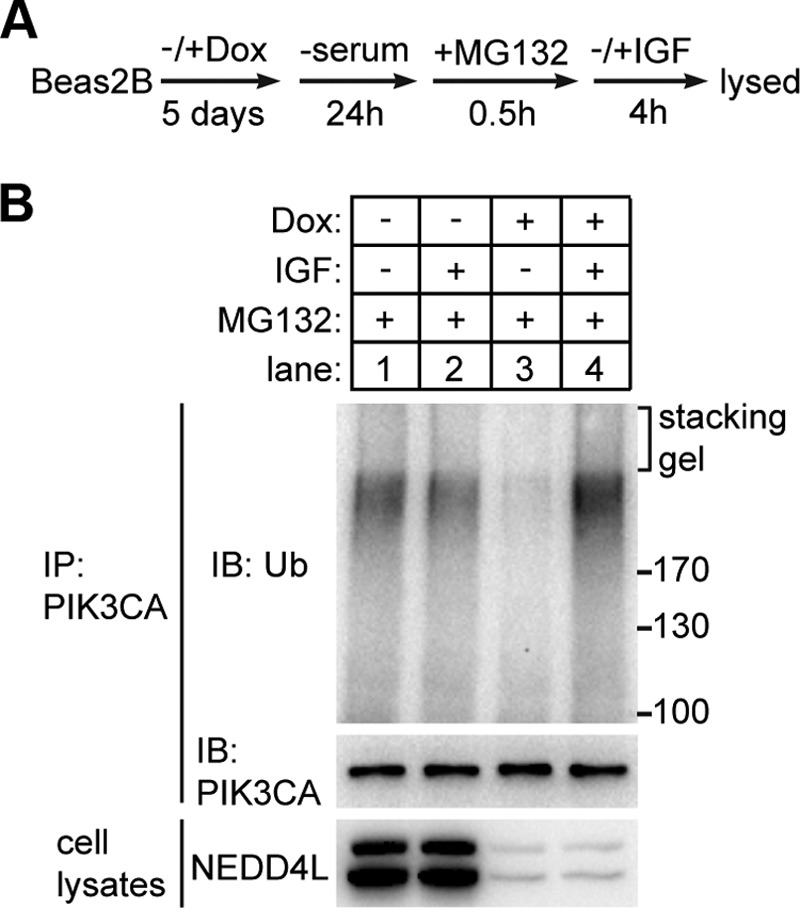
NEDD4L promoted ubiquitination of PIK3CA in vivo under serum deprivation. A, treatment of Beas2B-inducible knockdown cells before immunoprecipitation of endogenous PIK3CA protein. The cells were cultured with or without Dox for 5 days and then changed to serum free DMEM medium for 24 h. MG132 (20 μm) was added in the medium 0.5 h before the addition of 100 ng/ml IGF. Cells were collected and lysed 4 h after the addition of IGF. B, ubiquitination (Ub) states of endogenous PIK3CA under different conditions. Endogenous PIK3CAs were immunoprecipitated (IP) from each sample using PIK3CA antibody and protein A/G beads. The washed beads were boiled, and the supernatant was immunoblotted (IB) by ubiquitin antibody.
NEDD4L Knockdown Decreases Cell Size, Lactate Production, and Colony Formation Ability
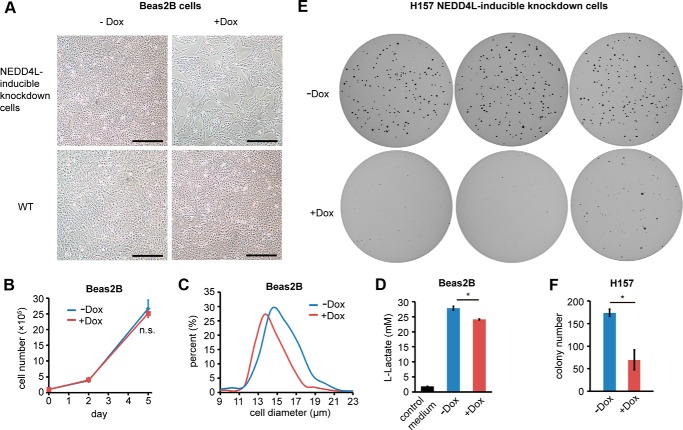
To examine whether the effects of NEDD4L loss on PI3K-AKT pathway are physiologically relevant, we measured the cell growth, a major output of PI3K-AKT pathway, in control and NEDD4L-knockdown Beas2B cells. Equal numbers of cells were seeded in the dish and grew for 5 days in the presence or absence of Dox. We observed that cells with reduced NEDD4L expression had an obvious decrease in cell confluency, and this phenomenon was not caused by the addition of Dox as the confluency of wild type Beas2B cells was not affected (Fig. 8A). We further found that the decrease in cell confluency was due to a dramatic decrease in cell size of NEDD4L-deficient Beas2B cells but not a decrease in cell number (Fig. 8, B and C). Because PI3K promotes glycolysis and thus promotes lactate production (22), we compared the lactate concentration in the medium of control and NEDD4L-knockdown Beas2B cells under equal culture conditions. As expected, NEDD4L-knockdown resulted in relatively low lactate production (Fig. 8D), suggesting a decrease in PI3K-AKT signaling.
FIGURE 8.
NEDD4L knockdown decreased cell size, lactate production, and colony formation ability. A, knockdown of NEDD4L in Beas2B cells decreased the cell confluency. Equal cells (1 × 105/dish) were seeded in a 6-cm dish and cultured with or without Dox for 5 days. Then the cell pictures were taken. The scale bar indicates 0.5 mm. B, cell number of the culture described in A was counted at the indicated time points. n.s., no significance. C, NEDD4L knockdown decreased cell size. Cell sizes of the culture described in A were measured at the 5 day. D, NEDD4L knockdown decreases lactate production. Beas2B cells were induced with or without Dox for 5 days. Then 2 × 106 cells were seeded in a 6-cm dish and cultured for 2 days. The lactate concentration in the medium was measured. The data are shown as the mean ± S.D. *, p value < 0.05 (unpaired Student's t test). E, NEDD4L knockdown in H157 cells resulted in a dramatic decrease in colony formation ability in soft agar. F, statistics of the colony number in soft agar plates. The data are shown as the mean ± S.D. *, p value < 0.05 (unpaired Student's t test).
To further verify the negative effect of NEDD4L knockdown on PI3K-AKT pathway, we did a soft agar colony formation assay. Equal numbers of H157-inducible knockdown cells were seeded in soft agar and cultured in the presence or absence of Dox. We found the colony number was dramatically decreased when NEDD4L was suppressed (Fig. 8, E and F), suggesting an important role of NEDD4L in maintaining PI3K-AKT signaling.
Discussion
NEDD4L is a HECT family E3 ubiquitin ligase containing four WW domains for substrate interaction (23). We did not, however, observe an interaction between NEDD4L and PIK3CA by co-immunoprecipitation analysis. Nonetheless, our results support the notion that NEDD4L ubiquitinates PIK3CA and targets it for proteasome degradation in vivo. First, co-transfection of NEDD4L increased the amount of the polyubiquitinated PIK3CA in the presence of proteasome inhibitor (Fig. 5A). Second, overexpression of NEDD4L accelerated the turnover rate of PIK3CA, whereas knockdown of it led to the increased level of endogenous PIK3CA (Figs. 5B and 6). Third, NEDD4L knockdown greatly impaired the ubiquitination of endogenous PIK3CA under serum deprivation conditions (Fig. 7B). Moreover, NEDD4L contains a C2 domain that is involved in plasma membrane association (24, 25). A recent structural study suggests NEDD4L likely exhibits maximal activity when localized to membrane (26). By interaction with membrane phospholipid phosphatidylinositol 4,5-bisphosphate (PIP2), NEDD4L undergoes a conformational change, thus relieved from a self-inhibiting state (26). These results provide a rationale for NEDD4L to catalyze the ubiquitination of membrane or membrane-associated proteins such as PIK3CA.
Dysregulation of NEDD4L occurs in the progression of various cancers. Decreased levels of NEDD4L has been seen in gastric cancer (27), malignant glioma (28), non-small cell lung cancer (29), colorectal cancer (30), and prostate cancer (31). However, there are also reports showing an increased expression of NEDD4L in prostate cancer (32). Expression of NEDD4L was also seen in a portion of melanoma samples but not detected in non-tumorous melanocytes or benign nevi tissues (33). These results indicate a potentially complex role of NEDD4L in the development of cancer. NEDD4L knockdown impaired AKT phosphorylation (Fig. 6), decreased cell size and colony formation ability (Fig. 8), and significantly reduced the growth of melanoma cells (33), suggesting a positive role of NEDD4L in maintaining the strength of PI3K-AKT signaling. However, partial down-regulation of NEDD4L seen in many cancers may lead to an increase in PIK3CA protein level that probably promotes cell growth, suggesting a tumor suppressing role of NEDD4L through promoting PIK3CA degradation. The oncogenic state induced by NEDD4L down-regulation may reach a compromise between increasing the PIK3CA stability and maintaining a proper turnover rate of PIK3CA. Thus, small molecule inhibitors against NEDD4L, which presumably will disturb the normal turnover of PIK3CA, may open up a new avenue for anti-cancer drug development.
Experimental Procedures
Reagents
The following antibodies were used: anti-PIK3CA (#4249), anti-NEDD4L (#4249), anti-AKT (#9272), anti-phospho-Thr-308-AKT (#2965), anti-phospho-Ser-473-AKT (#9271), and anti-ubiquitin (#3933) are from Cell Signaling Technology; anti-β actin (sc-47778), anti-HA (sc-7392), anti-c-Myc HRP conjugated (sc-40), and anti-ubiquitin HRP conjugated (sc-8017) are from Santa Cruz Biotechnology; anti-FLAG M2 HRP-conjugated (A8592) is from Sigma. Immunoprecipitation of the FLAG-tagged proteins was performed using anti-FLAG M2 affinity gel (Sigma). Puromycin (P7255), hygromycin B (H7772), doxycycline (D9891), phosphatase inhibitor mixture 3 (P0044) was from Sigma. MG132 (Ser-2619) was from Selleck. Cycloheximide (#2112) was from Cell Signaling Technology. Protease inhibitor mixture tablets (#04693116001) were from Roche Applied Science.
Transfection and Immunoprecipitation
For transient expression of proteins in HEK293T cells, the cDNAs for human PIK3CA were cloned into pCI vector (Promega), and human ubiquitin, PIK3R1, and NEDD4L were cloned into pcDNA3.1 vector (Invitrogen) with the indicated tags at the N terminus. HEK293T cells were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% FBS and antibiotics (100 units/ml penicillin, 100 μg/ml streptomycin) at 37 °C in a 5% CO2 incubator. The day before transfection HEK293T cells were seeded in 6-cm dishes at a density of 1.2 × 106 per dish, cultured for 24 h, and then transfected using Lipofectamine 2000 reagent with the plasmids as indicated. Twenty-four hours after transfection cells were treated with 20 μm MG132 (dissolved in DMSO) or DMSO for 6 h, washed once with PBS (phosphate-buffered saline), and collected by centrifugation. Cells were lysed on ice for 30 min in 200 μl of buffer A (20 mm HEPES, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 0.02% CHAPS, pH 7.5) supplemented with 0.3% CHAPS, 150 mm NaCl, protease inhibitor mixture, and phosphatase inhibitor mixture 3 and centrifuged at 20,000 × g for 10 min at 4 °C. To check protein expression, 30 μl of the cleared cell lysates were boiled with loading buffer and applied to Western blots. The remaining lysates were transferred to a new tube and diluted to 800 μl for immunoprecipitations. The anti-FLAG M2 beads (25 μl of the slurry beads per sample) were washed with lysis buffer 3 times, added to the cell lysates, and then incubated with rotation for 2 h at 4 °C. After washing with lysis buffer 3 times, 100 μl of a 1× protein loading buffer was added to the beads and boiled for 5 min at 95 °C. The supernatant of the boiled samples were resolved by SDS-PAGE and analyzed by Western blot.
Expression and Purification of Recombinant Proteins
The cDNAs encoding human E1, six E2 (Ubch2, -3, -4, -5c, -6, and -7) and ubiquitin were inserted into pET-28a (Novagen) vector with an N-terminal His tag. The first 18 amino acids of Ubch6, which contains a polyserine sequence, were deleted to enhance the protein expression. The proteins were overexpressed in Escherichia coli strain Rosetta at 18 °C for 18 h by induction with 0.5 mm β-d-thiogalactopyranoside (IPTG). The harvested cells were resuspended in buffer B (300 mm NaCl, 20 mm Tris-HCl, pH 7.5) supplemented with protease inhibitor mixture and lysed by sonication. After centrifugation at 75,000 × g for 2 h, the supernatant was loaded on a Ni2+ affinity column, washed, and eluted by 20 and 80% buffer C (300 mm NaCl, 500 mm imidazole, 20 mm Tris-HCl, pH 7.5), respectively. The human E1 was further purified by a Q column; E1 protein eluted from Ni2+ column was desalted into buffer A, loaded on a 1-ml HiTrap Q HP column, and eluted by a linear gradient of 25 times of column volumes (CV) from 0 to 600 mm NaCl in buffer A.
Recombinant human PIK3CA and NEDD4L proteins were overexpressed using the Bac-to-Bac baculovirus expression system. Full-length of the respective cDNAs were inserted into a modified pFastBac-HTB vector (Invitrogen) containing an N-terminal tandem His tag and 3×FLAG tag (for PIK3CA) or a tandem His tag and 3×HA tag (for NEDD4L). Recombinant progeny 1 (P1) baculovirus was produced by transfecting one well of Sf9 cells (grown in a 6-well plate, 60% confluence) with the recombinant bacmid DNA derived from 1.5 ml of DH10Bac E. coli cell culture and harvested after 4 days. The P2 virus was generated by infecting 10 ml of the suspended SF9 cells (2 × 106/ml) with 0.5 ml P1 virus. The supernatant from P2 culture was harvested 3 days post-infection, and 1 ml was used to infect 30 ml of suspended SF9 cells (2 × 106/ml) followed by a 2-day culture to generate P3 virus. For protein expression, 1 liter of High5 cells (2 × 106/ml) was infected with 10 ml of P3 virus, cultured in suspension for 2 days, and then collected by centrifugation. The recombinant proteins was purified by a Ni2+ column and a Q column as described above except that NEDD4L protein was further purified using a Superdex 200 10/300 GL column equilibrated with 200 mm NaCl in buffer A. All the purified recombinant proteins were desalted into buffer A containing 5% glycerol, aliquoted, and stored at −80 °C.
Preparation of MEF S-100
The eternalized MEF cells were cultured as a monolayer in DMEM supplemented with 10% FBS and antibiotics at 37 °C in a 5% CO2 incubator. Cells at ∼80% confluence were scraped from the dishes, washed once with PBS, and harvested by centrifugation at 1000 × g for 5 min. The cell pellets were resuspended in buffer A (0.7 ml/10-cm dish) containing 0.3% CHAPS and protease inhibitor mixture and lysed on ice for 30 min. The cell lysates were centrifuged at 20,000 × g for 10 min at 4 °C using an Eppendorf 5417R centrifuge, and the supernatant was further centrifuged at 100,000 × g for 1 h at 4 °C using a Beckman Optima MAX-XP centrifuge. The supernatant from the second centrifugation (S-100) was stored at −80 °C and used as the starting material for the purification of PIK3CA E3 ligase.
In Vitro Ubiquitination Assay
The assay was performed at 37 °C for 2 h in a 15-μl reaction volume containing final concentrations of 0.4 μm E1, 5 μm E2, 70 μm ubiquitin, 5 mm ATP, pH 7.5, 2 nm FLAG-tagged PIK3CA, and an appropriate amount of MEF S-100 or chromatographic fraction. Buffer A supplemented with 10 mm MgCl2 and 50 mm KCl was used as the reaction buffer. The assay was stopped by boiling with protein loading buffer followed by immunoblotting with anti-FLAG antibody.
Purification of PIK3CA E3 Ligase from MEF S-100
The purification was performed at 4 °C, and all chromatography steps were carried out using an AKTA Purifier 10 FPLC system (GE Healthcare). MEF S-100 (total 500 ml) was loaded on four tandem connected 5-ml HiTrap Q columns (250 ml S-100 for one run) equilibrated with buffer A. The columns were equilibrated with 5 CVs of buffer A after loading and then eluted with a linear gradient of 15 CVs from 0 to 600 mm NaCl in Buffer A. Fractions of 0.5 ml were dialyzed against buffer A, and 10 μl were used for activity assay using the in vitro ubiquitination system. The active fractions were pooled, and ammonium sulfate was added to reach 15% saturation followed by centrifugation at 8000 × g for 10 min. The supernatant was transferred to a new tube, and the activity was precipitated by the addition of ammonium sulfate to 35% saturation. After rotation at 4 °C for 30 min and centrifugation at 8000 × g for 10 min, the protein pellet was resolubilized in buffer A and loaded on three tandem-connected 1-ml HiTrap SP columns that were washed with 5 CVs of buffer A after loading and eluted with a linear gradient of 10 CVs from 0 to 600 mm NaCl in buffer A. Fractions of 0.5 ml were dialyzed, and 10 μl of each were assayed for activity. The active fractions were pooled, dialyzed against buffer A, and loaded on two tandem-connected 1-ml HiTrap Heparin columns, washed, and eluted by the same procedures as S column. All fractions were dialyzed, and 3 μl of each were assayed for activity. The active fractions were pooled, concentrated to 0.5 ml, subjected to a Superdex 200 10/300 GL column, and eluted with 200 mm NaCl in buffer A. The eluted fractions were dialyzed, and 3 μl were assayed for activity. The active fractions were pooled and loaded on a 1-ml hydroxyapatite column (Bio-Rad), washed with 5 CV of buffer A, and eluted with a linear gradient of 10 CVs from 0 to 500 mm phosphate in buffer A. All fractions were dialyzed, and 3 μl of each fraction were assayed for activity. The active fractions were pooled, loaded on a Mono Q 5/50 GL column, and eluted with a linear gradient of 15 CVs from 0 to 600 mm NaCl in buffer A. The fractions were dialyzed against buffer A, and eventually 1.5 μl of each dialyzed fraction were assayed for the PIK3CA E3 ubiquitin ligase activity.
Overexpression of NEDD4L or GFP in HeLa Cells
For lentiviral-based stable overexpression, NEDD4L or GFP cDNA was cloned into a modified pLVX (Clontech) vector. Lentivirus was produced by co-transfection of packaging plasmid pCMV-dR8.2 (900 ng), envelope plasmid pCMV-VSV-G (100 ng), and pLVX-NEDD4L (1 μg) into HEK293T cells in a 6-cm dish at ∼50% confluence. Eighteen hours after transfection the medium was changed to high serum medium (DMEM containing 30% FBS and antibiotics) and culturing was continued for another 24 h followed by harvesting the virus by passing the medium through a 0.22-μm filter. HeLa cells cultured in 6-cm dishes were infected by adding 1 ml of lentiviral medium and incubated for 24 h. Then the medium was changed to fresh growth medium with 2 μg/ml puromycin and cultured for 5 days to select stably transduced cells. The puromycin-resistant HeLa cells were trypsinized, seeded in a 96-well plate with ∼1 cell per well, and cultured for 10 days in growth medium with 2 μg/ml puromycin. Individual clones were picked out to test the expression of exogenous NEDD4L by Western blot. To examine the turnover rate of PIK3CA, HeLa cells were treated with 25 μg/ml cycloheximide and collected at the indicated time points. Cells were lysed for 15 min in buffer A containing 0.3% CHAPS, 150 mm NaCl, protease inhibitor mixture, and phosphatase inhibitor mixture 3. After centrifugation at 20,000 × g for 10 min at 4 °C, the protein concentrations of cleared cell lysates were measured by BCA (bicinchoninic acid) assay (Pierce). The normalized lysates were applied to Western blot analysis.
NEDD4L Knockdown and Rescue
The shRNA sequences for NEDD4L knockdown were obtained from the collection of The RNAi Consortium (TRC). NEDD4L shRNA1 (TRCN0000000905) was used for inducible knockdown in H157 and Beas2B cells. Both shRNA1 and shRNA2 (TRCN0000000906) were used for stable knockdown in A549 cell. The scramble shRNA sequence used was 5′-CCGGGTGGACTCTTGAAAGTACTATCTCGAGATAGTACTTTCAAGAGTCCACTTTTT-3′.
For inducible knockdown of NEDD4L, shRNA1 was inserted into a pLKO.1-tet-on vector. Lentiviral based transfection and puromycin selection were described as above. Individual clones picked from 96-well plate were tested for the efficiency of Dox induced NEDD4L knockdown. To examine the AKT phosphorylation in response to FBS stimulation, a positive clone was induced with 0.2 μg/ml Dox for 5 days. Then cells were trypsinized, washed with PBS for once, and plated in a 6-cm dish (2 × 105 cells/dish) in serum-free DMEM. Twenty-four hours later 10% FBS was added to the medium. At the indicated time points, the medium was aspirated, and the cells were washed with 1 ml of PBS for 10 s followed by the addition of 100 μl of lysis buffer. Cells were rapidly scraped off the dishes, lysed, and centrifuged. The normalized supernatant was applied to Western blot analysis.
The methods for stable knockdown of NEDD4L in A549 cells were generally the same as described above, except the NEDD4L shRNA1, shRNA2, and scramble shRNAs were inserted into pLKO.1 vector. To compare the AKT phosphorylation level under the treatment of Torin1, positive and control cells were plated in 6-well plate (5 × 104 cells/well), cultured overnight, and treated with 25 nm Torin1 for the indicated times.
For rescue assay, pcDNA3.1 vector or pcDNA3.1-2×Myc-NEDD4L with synonymous mutations in the shRNA1 targeting region were transfected into A549 cells stably expressing NEDD4L shRNA1. Twenty-four hours after transfection cells were selected with 250 μg/ml hygromycin for 4–5 days and seeded in 96-well plate. Individual clones were picked out to test the expression of NEDD4L. The positive clone was used for testing AKT phosphorylation in the presence of Torin1.
Cell Growth, Lactate Production, and Soft Agar Colony Formation Assay
WT or NEDD4L-inducible knockdown Beas2B cells were seeded in 6-cm dishes at a density of 1 × 105 cells/dish and cultured with or without 0.5 μg/ml Dox. Cell pictures were taken after 5 days of culture. Cell number was counted at the indicated times. Cell size was measured using a Cellometer Auto T4 machine after 5 days of culture. To measure the lactate production, 2 × 106 Beas2B cells that had been induced with or without Dox were seeded in 6-cm dishes and cultured for 2 days. The medium was collected, and lactate concentration was measured using a lactate assay kit (Abcam, #ab65331) according to the manufacturer's protocol.
To perform colony formation assay, H157-inducible knockdown cells were cultured with or without Dox for 5 days, and then 7.5 × 103 cells were seeded in the upper layer of soft agar medium (0.7% agarose in DMEM plus 10% FBS) with or without 1 μg/ml Dox in a 6-cm dish. The plates were incubated for 5 weeks. The colonies were stained using crystal violet solution, and digital pictures were taken. The colony numbers were counted using Clono-Counter software (34).
Author Contributions
M. F. designed and supervised the study. Z. W. designed and performed most of the experiments, interpreted the experimental results, and prepared the figures. T. D. and T. L. were involved in the molecular and cell biology experiments. S. C. and L. L. conducted the mass spectrometry analysis. S. H. provided all the cDNAs. Z. W. and M. F. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Yanru Guo (the Fang laboratory at Peking University) for technical assistance, Dr. Zhirong Shen (National Institute of Biological Science Beijing (NIBS)), and Dr. Xuejun Jiang (Program of Cell Biology, Memorial Sloan-Kettering Cancer Center) for fruitful discussions. Dr. Yeguang Chen (College of Life Sciences, Tsinghua University) provided the plasmids encoding various ubiquitin mutants. We are grateful to Dr. Deepak Nijhawan (Department of Biochemistry, UT Southwestern Medical Center at Dallas) for critical reading of the manuscript. We are in debt to Dr. Yi Rao (Division of Physiology, College of Life Sciences and Joint Center of Life Sciences at Peking University) for enthusiastic and continuous encouragement.
This work was supported by the National Basic Program of China 973 Program (2013CB910104), National Natural Science Foundation of China (31271524), and the start-up foundation from the Joint Center for Life Sciences at Peking University. The authors declare that they have no conflicts of interest with the contents of this article.
- NEDD4L
- neural precursor cell expressed, developmentally down-regulated 4-like
- MEF
- mouse embryonic fibroblast
- Dox
- doxycycline
- CV
- column volume
- P1
- progeny 1
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Cantley L. C. (2002) The phosphoinositide 3-kinase pathway. Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 2. Downward J. (2004) PI 3-kinase, Akt and cell survival. Semin. Cell Dev. Biol. 15, 177–182 [DOI] [PubMed] [Google Scholar]
- 3. Geering B., Cutillas P. R., Nock G., Gharbi S. I., and Vanhaesebroeck B. (2007) Class IA phosphoinositide 3-kinases are obligate p85-p110 heterodimers. Proc. Natl. Acad. Sci. U.S.A. 104, 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kok K., Geering B., and Vanhaesebroeck B. (2009) Regulation of phosphoinositide 3-kinase expression in health and disease. Trends Biochem. Sci. 34, 115–127 [DOI] [PubMed] [Google Scholar]
- 5. Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S. M., Riggins G. J., Willson J. K., Markowitz S., Kinzler K. W., Vogelstein B., and Velculescu V. E. (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science 304, 554. [DOI] [PubMed] [Google Scholar]
- 6. Engelman J. A. (2009) Targeting PI3K signalling in cancer: opportunities, challenges, and limitations. Nat. Rev. Cancer 9, 550–562 [DOI] [PubMed] [Google Scholar]
- 7. Yuan T. L., and Cantley L. C. (2008) PI3K pathway alterations in cancer: variations on a theme. Oncogene 27, 5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Woenckhaus J., Steger K., Werner E., Fenic I., Gamerdinger U., Dreyer T., and Stahl U. (2002) Genomic gain of PIK3CA and increased expression of p110α are associated with progression of dysplasia into invasive squamous cell carcinoma. J. Pathol. 198, 335–342 [DOI] [PubMed] [Google Scholar]
- 9. Yamamoto H., Shigematsu H., Nomura M., Lockwood W. W., Sato M., Okumura N., Soh J., Suzuki M., Wistuba I. I., Fong K. M., Lee H., Toyooka S., Date H., Lam W. L., Minna J. D., and Gazdar A. F. (2008) PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 68, 6913–6921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fang D., Wang H. Y., Fang N., Altman Y., Elly C., and Liu Y. C. (2001) Cbl-b, a RING-type E3 ubiquitin ligase, targets phosphatidylinositol 3-kinase for ubiquitination in T cells. J. Biol. Chem. 276, 4872–4878 [DOI] [PubMed] [Google Scholar]
- 11. Fang D., and Liu Y. C. (2001) Proteolysis-independent regulation of PI3K by Cbl-b-mediated ubiquitination in T cells. Nat. Immunol. 2, 870–875 [DOI] [PubMed] [Google Scholar]
- 12. Kuchay S., Duan S., Schenkein E., Peschiaroli A., Saraf A., Florens L., Washburn M. P., and Pagano M. (2013) FBXL2- and PTPL1-mediated degradation of p110-free p85beta regulatory subunit controls the PI(3)K signalling cascade. Nat. Cell Biol. 15, 472–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuan T. L., Wulf G., Burga L., and Cantley L. C. (2011) Cell-to-cell variability in PI3K protein level regulates PI3K-AKT pathway activity in cell populations. Curr. Biol. 21, 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hershko A., and Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 15. Saeki Y., Kudo T., Sone T., Kikuchi Y., Yokosawa H., Toh-e A., and Tanaka K. (2009) Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 28, 359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang C. H., Mandelker D., Schmidt-Kittler O., Samuels Y., Velculescu V. E., Kinzler K. W., Vogelstein B., Gabelli S. B., and Amzel L. M. (2007) The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science 318, 1744–1748 [DOI] [PubMed] [Google Scholar]
- 17. Mandelker D., Gabelli S. B., Schmidt-Kittler O., Zhu J., Cheong I., Huang C. H., Kinzler K. W., Vogelstein B., and Amzel L. M. (2009) A frequent kinase domain mutation that changes the interaction between PI3Kα and the membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 16996–17001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goel P., Manning J. A., and Kumar S. (2015) NEDD4–2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene 557, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peterson T. R., Laplante M., Thoreen C. C., Sancak Y., Kang S. A., Kuehl W. M., Gray N. S., and Sabatini D. M. (2009) DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137, 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., and Gray N. S. (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brognard J., Clark A. S., Ni Y., and Dennis P. A. (2001) Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res. 61, 3986–3997 [PubMed] [Google Scholar]
- 22. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang B., and Kumar S. (2010) Nedd4 and Nedd4–2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ. 17, 68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nalefski E. A., and Falke J. J. (1996) The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 5, 2375–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Plant P. J., Yeger H., Staub O., Howard P., and Rotin D. (1997) The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca2+-dependent plasma membrane localization. J. Biol. Chem. 272, 32329–32336 [DOI] [PubMed] [Google Scholar]
- 26. Escobedo A., Gomes T., Aragón E., Martín-Malpartida P., Ruiz L., and Macias M. J. (2014) Structural basis of the activation and degradation mechanisms of the E3 ubiquitin ligase Nedd4L. Structure 22, 1446–1457 [DOI] [PubMed] [Google Scholar]
- 27. Gao C., Pang L., Ren C., and Ma T. (2012) Decreased expression of Nedd4L correlates with poor prognosis in gastric cancer patient. Med. Oncol. 29, 1733–1738 [DOI] [PubMed] [Google Scholar]
- 28. He S., Deng J., Li G., Wang B., Cao Y., and Tu Y. (2012) Down-regulation of Nedd4L is associated with the aggressive progression and worse prognosis of malignant glioma. Jpn. J. Clin. Oncol. 42, 196–201 [DOI] [PubMed] [Google Scholar]
- 29. Sakashita H., Inoue H., Akamine S., Ishida T., Inase N., Shirao K., Mori M., and Mimori K. (2013) Identification of the NEDD4L gene as a prognostic marker by integrated microarray analysis of copy number and gene expression profiling in non-small cell lung cancer. Ann. Surg. Oncol. 20, S590–S598 [DOI] [PubMed] [Google Scholar]
- 30. Tanksley J. P., Chen X., and Coffey R. J. (2013) NEDD4L is downregulated in colorectal cancer and inhibits canonical WNT signaling. PLoS ONE 8, e81514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu X. Y., Xu Y. M., Fu Q., Yu J. J., and Huang J. (2009) Nedd4L expression is downregulated in prostate cancer compared to benign prostatic hyperplasia. Eur. J. Surg. Oncol. 35, 527–531 [DOI] [PubMed] [Google Scholar]
- 32. Hellwinkel O. J., Asong L. E., Rogmann J. P., Sültmann H., Wagner C., Schlomm T., and Eichelberg C. (2011) Transcription alterations of members of the ubiquitin-proteasome network in prostate carcinoma. Prostate Cancer Prostatic Dis. 14, 38–45 [DOI] [PubMed] [Google Scholar]
- 33. Kito Y., Bai J., Goto N., Okubo H., Adachi Y., Nagayama T., and Takeuchi T. (2014) Pathobiological properties of the ubiquitin ligase Nedd4L in melanoma. Int. J. Exp. Pathol. 95, 24–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Niyazi M., Niyazi I., and Belka C. (2007) Counting colonies of clonogenic assays by using densitometric software. Radiat. Oncol. 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]