Abstract
Cells engage numerous signaling pathways in response to oxidative stress that together repair macromolecular damage or direct the cell toward apoptosis. As a result of DNA damage, mitochondrial DNA or nuclear DNA has been shown to enter the cytoplasm where it binds to “DNA sensors,” which in turn initiate signaling cascades. Here we report data that support a novel signaling pathway in response to oxidative stress mediated by specific guanine-rich sequences that can fold into G-quadruplex DNA (G4DNA). In response to oxidative stress, we demonstrate that sequences capable of forming G4DNA appear at increasing levels in the cytoplasm and participate in assembly of stress granules. Identified proteins that bind to endogenous G4DNA in the cytoplasm are known to modulate mRNA translation and participate in stress granule formation. Consistent with these findings, stress granule formation is known to regulate mRNA translation during oxidative stress. We propose a signaling pathway whereby cells can rapidly respond to DNA damage caused by oxidative stress. Guanine-rich sequences that are excised from damaged genomic DNA are proposed to enter the cytoplasm where they can regulate translation through stress granule formation. This newly proposed role for G4DNA provides an additional molecular explanation for why such sequences are prevalent in the human genome.
Keywords: DNA repair, G-quadruplex, oxidative stress, signaling, stress granule, stress response, telomere
Introduction
Cells generally respond to DNA in the cytosol as a warning signal. Numerous cytosolic proteins are known to play a role in sensing and responding to DNA (1). The STING adaptor is a major component of the innate immune response toward cytosolic DNA, whether the source is external, such as bacterial or viral infection, or degradation of endogenous nuclear or mitochondrial DNA (2). Oligonucleotides composed of telomeric sequences (TTAGGG) have been introduced into cells, resulting in the suppression of a variety of inflammatory responses including blocking of Toll-like receptor 9 (3). These sequences contain appropriately spaced guanine residues such that folding forms structures referred to as G-quadruplex DNA or G4DNA.2
G4DNA is made of four guanines that form Hoogsteen hydrogen bonds in a planar ring referred to as a G-quartet (Fig. 1A). Multiple stacks of these G-quartets associate through pi bond interactions to form highly stable structures (4) (Fig. 1B). Further stabilization is supported through the coordination of monovalent ions in the central channel. Formation of quadruplex structures has been suggested to mediate the biological response to telomeric oligonucleotides introduced into cells by transfection (5). Cells that undergo apoptosis have been suggested as a natural source of telomeric DNA presented to neighboring cells to reduce potential autoimmune responses (6). Thus far, evidence has not been presented for the appearance of endogenous G4DNA in the cytoplasm.
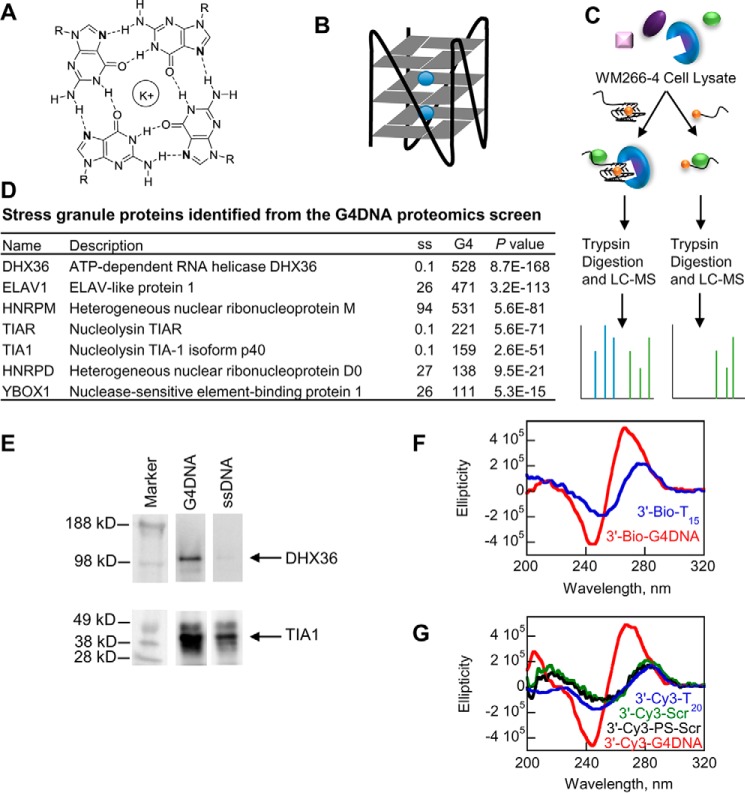
FIGURE 1.
Proteomics screen for G4DNA-binding proteins. A, structure of a single G-quadruplex tetrad. B, stacks of tetrads, usually three, form a G-quadruplex, which is stabilized by monovalent cations (blue) that bind in the central channel. C, strategy for proteomics screen for binders to G4DNA. To determine the enrichment of proteins with the G4DNA, a separate experiment was conducted with single-stranded DNA as the bait. Cell lysates were incubated with G4DNA or ssDNA followed by SDS-PAGE and then LC-MS. Relative quantities of proteins were determined by spectral counting. D, proteins previously found in stress granules that were identified in the G4DNA proteomics screen (24) (full list is in supplemental Table 1). Data are the result of a single experiment in WM266-4 cells. E, Western blotting analysis of cell lysates incubated with G4DNA or ssDNA. The G4DNA and ssDNA affinity purifications contained identical quantities of the cell lysate. 30% of the cell lysate volume was affinity-purified on ssDNA or an identical concentration of G4DNA-coated beads as described under “Experimental Procedures.” 20% of the elution volume was loaded in each lane for SDS-PAGE. F and G, circular dichroism indicates that the G4DNA oligonucleotides used in the proteomics screen and Western blotting analysis (F) and for immunofluorescence (G) form parallel quadruplexes, whereas the ssDNAs do not form quadruplexes.
Recent reports provide convincing evidence strongly supporting the formation of G4DNA in the nuclear and mitochondrial genomes of human cells. Antibodies to G4DNA have been used to visualize G4DNA in cells (7–10). Small molecules designed to interact specifically with G4DNA have been found to alter gene expression in a G4DNA-selective manner (11, 12). The Pif1 helicase localizes to G4DNA sequence motifs and binds and unwinds G4DNA structures (13, 14). G4DNA is proposed to play specific roles in DNA metabolism including replication (14), gene expression (15), and mitochondrial DNA metabolism (16).
In relation to human disease, the involvement of G-quadruplexes has been observed in neurological disorders. Expansion of the DNA repeat sequence GGGGCC in the C9orf72 gene is associated with amyotrophic lateral sclerosis and frontotemporal dementia (17, 18). Transcription of the repeat sequences results in RNA that can fold into a stable parallel G-quadruplex, leading to disruption of transcription and/or translation. The fragile X mental retardation protein binds specifically to G-quadruplex RNA through a conserved RGG motif (19). Studies on fragile X syndrome indicate that repeat expansion of the CGG triplet can result in dysregulation of translation in neuronal cells through the binding of transcripts by fragile X mental retardation protein (20).
The in vivo existence and impact of G4DNA on normal and pathological processes is now accepted, but questions regarding the specific functions and mechanisms of action remain to be addressed. We report a new role for DNA sequences that have the capacity to fold into G-quadruplex structures. We used hydrogen peroxide (H2O2) to damage the DNA. Under oxidative stress conditions, we found that endogenous G4DNA sequences appear in the cytoplasm. G4DNA from this endogenous source can participate in the assembly of stress granules, which are known to alter mRNA translation. Stress granules are cytoplasmic aggregates of mRNA and proteins that regulate mRNA translation and decay (21, 22). They have been proposed to function as sites of mRNA triage (23), which govern the composition and function of ribonucleoprotein complexes in order to determine whether individual mRNAs are stored, degraded, or translation is reinitiated. There are clear connections between stress granule function and the pathogenesis of cancer (24). The pathway proposed in this work provides a new mechanism by which cells can respond to the effects of DNA damage during oxidative stress.
Results
Identification of G4DNA-binding Proteins
We performed quantitative proteomics analysis using G4DNA as bait to identify G4DNA-binding proteins in WM266-4 melanoma cells (Fig. 1C). G4DNA is characterized by the sequence motif G3N1–xG3N1–xG3N1–xG3. However, G4DNA can vary in terms of the direction of each strand, the number of different strands, and the length of the DNA loops that occur between runs of guanines. A biotinylated DNA sequence based on a region of the c-MYC promoter that readily forms a stable, parallel quadruplex (18) bound to streptavidin-coupled Dynabeads (25) was incubated with human cell lysate to isolate G4DNA-bound proteins. To account for nonspecific interactions, a single-stranded DNA (ssDNA) was incubated with lysate in a separate experiment. Isolated proteins were identified by quantitative mass spectrometry to determine those proteins enriched upon binding to G4DNA (Fig. 1D and supplemental Table 1 and Fig. S1) (26).
The most highly enriched protein in the G4DNA sample was the DHX36/RHAU/G4R1 RNA helicase, previously identified as the major source of G4DNA binding and unwinding activity in the cell (27–29). DHX36 is a multifunctional enzyme involved in transcription (30) and translation (31) and has been suggested to serve as a sensor for DNA in the cytoplasm (32). Surprisingly, other enriched proteins are ELAV1/HUR, YB-1/YBOX1, and TIA1, as well as other proteins known to regulate translation and assemble into stress granules (21, 24). Stress granules are cytosolic ribonucleoprotein aggregates of stalled translation complexes that form in response to various stresses. DHX36 has also been shown to be associated with stress granules (33). The formation of stress granules was recently shown to result from the introduction of exogenous G4DNA into cells (34). The proteomic results here support the hypothesis that endogenous G4DNA binds tightly to proteins that play important roles in translation, such as DHX36 and YB-1, thereby leading to the assembly of stress granules.
The proteomic results do not serve as definitive proof that specific proteins do or do not bind to G4DNA. For example, nucleolin, a protein known to interact with G4DNA through its RGG motif (35), was not identified as a G4DNA-interacting protein. However, nucleolin also binds tightly to ssDNA (36). Consistent with these results, we identified peptides from nucleolin in both the G4DNA and ssDNA samples. Although nucleolin was not classified as a G4DNA-interacting protein due to its presence at similar levels in both the G4DNA and ssDNA samples, this does not exclude the possibility that it interacts with G4DNA; it only indicates that it did not interact with the G4DNA bait significantly more than with the ssDNA bait. The most highly enriched protein in the ssDNA sample was acetyl-CoA carboxylase 1; its Escherichia coli homolog is a known ssDNA- and dsDNA-binding protein (37). The identification of DHX36 as the most highly enriched protein in the G4DNA sample and nucleolin in both the G4DNA and ssDNA samples provides evidence that our pulldown and data analysis generally identified G4DNA-interacting proteins. The results of the proteomics screen were also confirmed by incubating additional cell lysates with G4DNA or ssDNA and probing for DHX36 or TIA1 by Western blotting (Fig. 1E). In both cases, the proteins were enriched in the G4DNA sample, consistent with the proteomics experiment.
The proteomics screen allowed us to develop the hypothesis that some cytoplasmic proteins involved in stress granule formation can bind to G4DNA in the cell. We surmised that G4DNA might enter the cytoplasm during episodes of DNA damage. We then developed a series of experiments to test this hypothesis working with the knowledge that DNA damage has been shown previously to increase the concentration of DNA in the cytoplasm.
Exogenous G4DNA Co-localizes with Stress Granule Markers
Initially, we wished to repeat previous work indicating that exogenous G4DNA can induce stress granule formation in U2OS cells (34), so we conducted similar experiments by transfecting G4DNA into melanoma cells. Fluorescently labeled G4DNA was introduced into cells and then visualized by fluorescence microscopy along with selected proteins identified from the proteomics screen. G4DNA was found predominantly in the nucleus, with some punctate staining in the cytoplasm (Fig. 2A). TIA1, a stress granule marker (38), was found to co-localize with G4DNA-induced stress granules in the cytoplasm (Fig. 2A). In contrast, the introduction of fluorescently labeled ssDNA did not lead to stress granule formation, as seen by the lack of TIA1 foci in the cytoplasm (Fig. 2, B–D). Three different Cy3-ssDNAs were used for comparison: Cy3-T20 (Fig. 2B), as the G4DNA bait in the pulldown contained a single-stranded thymidine region; Cy3-Scr (Fig. 2C), a scrambled version of the G4DNA sequence, to account for any proteins that bind preferentially to G-rich DNA; and Cy3-PS-Scr (Fig. 2D). Because ssDNA is more sensitive than G4DNA to degradation by cytoplasmic nucleases such as TREX1, a scrambled sequence with phosphorothioate (PS) at the ends of the oligonucleotide to prevent degradation was also used for comparison. G4DNA is refractory to the nuclease activity of TREX1 (39). Although Cy3-ssDNA was visible in the cytosol, the lack of cytoplasmic TIA1 foci indicated that stress granules were not present (Fig. 2, B–D). Transfection efficiency was 100% for all of the oligonucleotides (G4DNA, T20, ScrDNA, and PS-ScrDNA) based on counting the fractions of cells identified by DAPI-stained nuclei that exhibited red fluorescence for at least 100 cells transfected with 500 nm Cy3-DNA.
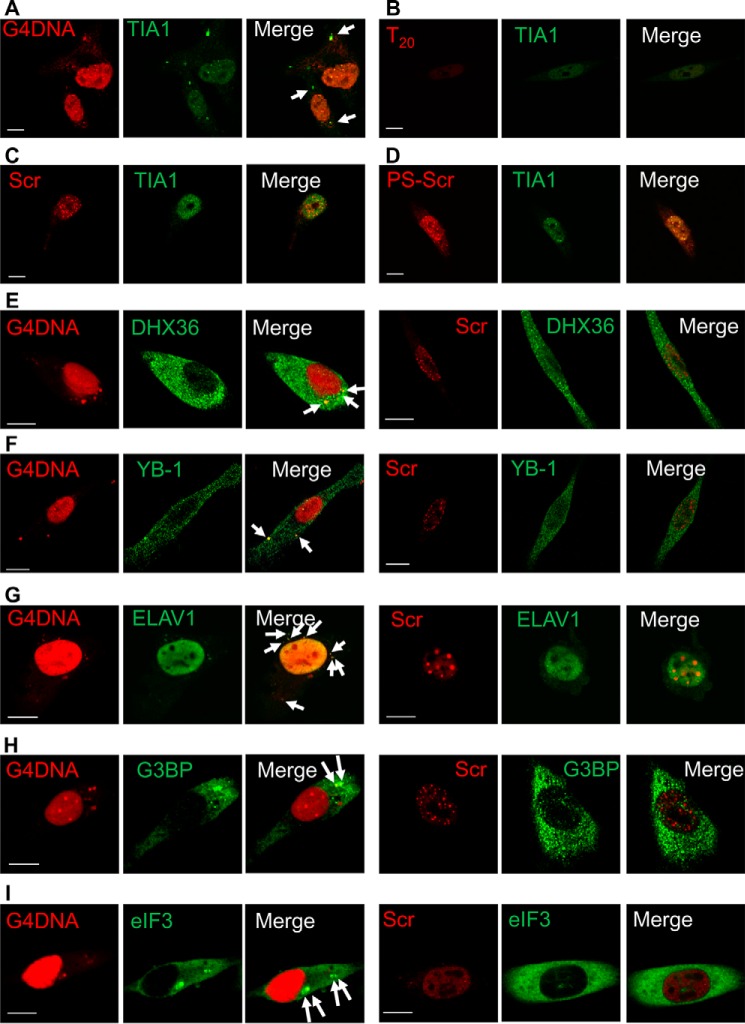
FIGURE 2.
G4DNA introduced into cells co-localizes with stress granule markers in WM266-4 cells. A, transfected Cy3-G4DNA induces stress granule formation and co-localizes with TIA1 (90% of TIA1 foci contain G4DNA, indicated by arrows). Three different Cy3-ssDNA were used for comparison: Cy3-T20 (B), because the G4DNA bait in the pulldown contained a single-stranded thymidine region; Cy3-Scr (C), a scrambled version of the G4DNA sequence, to account for any proteins that bind preferentially to G-rich DNA; and Cy3-PS-Scr (D), which contains phosphororthioate linkages at each end of the oligonucleotide to prevent degradation by cellular nucleases. Transfected Cy3-ssDNA does not cause stress granule formation. Transfected G4DNA co-localizes with additional stress granule proteins. DXH36 (E), YB-1 (F), ELAV1 (G), G3BP (H), and eIF3 (I) co-localize with transfected Cy3-G4DNA in the cytosol (indicated by arrows) but not Cy3-Scr DNA. Quantitation of foci containing G4DNA showed that 7% of the DHX36 foci, 75% of the YB-1 foci, 96% of the ELAV1 foci, 17% of the G3BP foci, and 83% of the eIF3 foci contained G4DNA. Bar equals 10 μm.
Three additional proteins identified in the G4DNA proteomics screen that have been shown to associate with stress granules (33, 34, 40) were examined for co-localization with G4DNA-induced stress granules. DHX36 (Fig. 2E), YB-1 (Fig. 2F), and ELAV1/HuR (Fig. 2G) were also found in punctate regions of the cytoplasm; these proteins co-localize with DNA after transfection with G4DNA but not after transfection with ssDNA. In addition to DHX36, which is known to bind quadruplexes (27, 41), YB-1 was recently reported to bind to G4RNA and G4DNA (34). Two stress granule markers that were not identified by the proteomics screen, due to low mass spectral coverage of the proteins, were G3BP and eIF3. However, both G3BP (Fig. 2H) and eIF3 (Fig. 2I) were also found in stress granules induced by G4DNA. The fraction of stress granule protein foci that contain G4DNA foci was determined to be 90, 7, 75, 96, 17, and 83% of the TIA1, DHX36, YB-1, ELAV1, G3BP, and eIF3 foci, respectively. The fraction of DHX36 and G3BP foci containing G4DNA was significantly lower than the other stress granule proteins. Although DHX36 is known to associate with stress granules (33), it also has other roles in the cytoplasm such as sensing double-stranded RNA (42). The cytoplasmic localization of G3BP, combined with the inherent punctate staining observed with this antibody, likely results in reduced co-localization of G4DNA with G3BP foci. Quantitation of cells with punctate foci containing both DNA and TIA1 indicated an increase in cells exhibiting stress granules with increasing G4DNA concentration (Fig. 3). Overall, the fluorescence microscopy results support the proteomics screen and a previous report (34) indicating that G4DNA introduced into cells can participate in stress granule formation.
FIGURE 3.
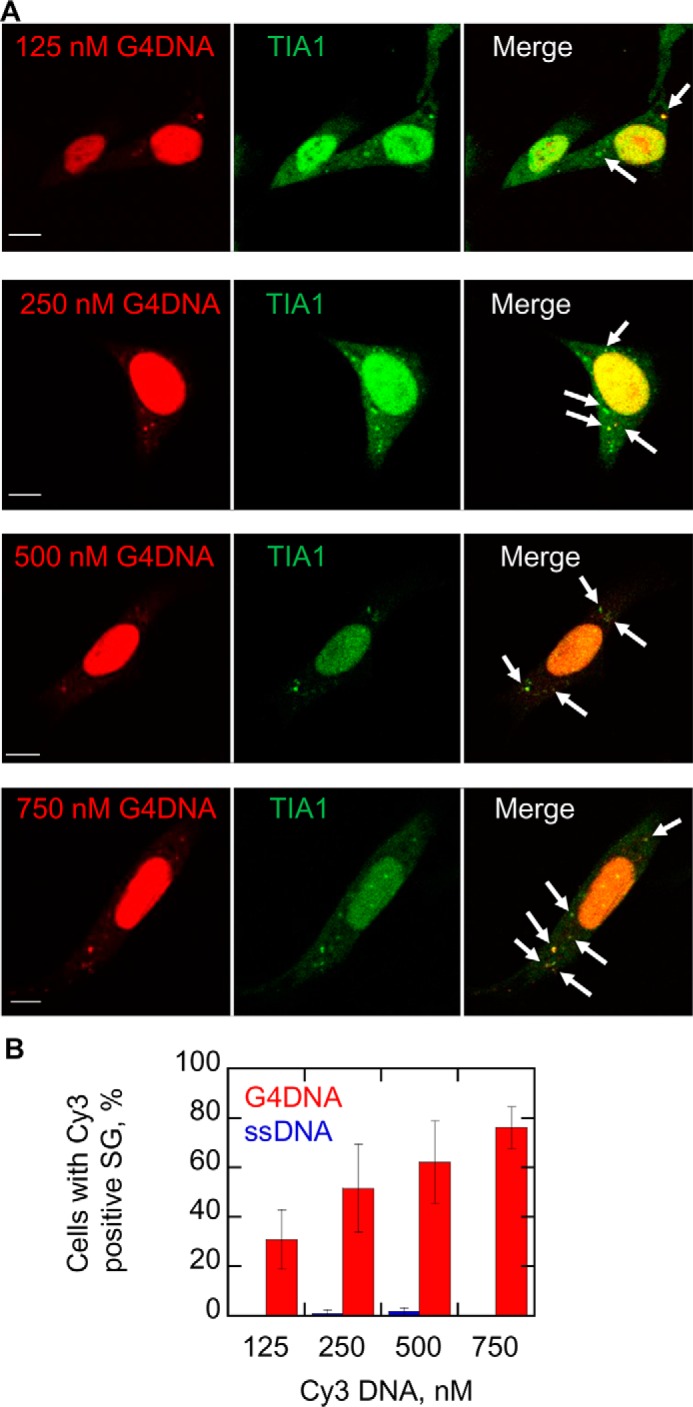
Increasing quantities of G4DNA introduced into cells increase the quantity of stress granules formed. A, G4DNA (125, 250, 500, or 750 nm) was transfected into WM266-4 cells. Increasing quantities of transfected Cy3-G4DNA (red) induced more stress granule formation as determined by TIA1 foci (green). The merged images indicate co-localization as marked by arrows. The fraction of TIA1 foci that contain G4DNA was 81, 80, 81, and 84% for cells transfected with 125, 250, 500, and 750 nm G4DNA, respectively. B, the plot indicates that the fraction of cells that contain cytoplasmic TIA1 foci co-localized with DNA increases as the concentration of transfected G4DNA increases (red bars). For comparison, the fraction of cells containing TIA1 foci co-localized with transfected ssDNA is shown (blue bars). Experiments were performed three separate times in WM266-4 cells. Bar equals 10 μm.
Endogenous DNA Sequences Capable of Forming G4DNA Structures Accumulate in the Cytoplasm during Oxidative Stress
We next examined whether G4DNA from endogenous cellular sources could participate in stress granule formation. Stress granules are reported to form under conditions of oxidative stress (24, 43, 44). We reasoned that oxidative stress could damage DNA, leading to the excision of nuclear or mitochondrial DNA followed by transfer of the excised sequences into the cytoplasm.
DNA sequences containing runs of guanine are hot spots for oxidative damage (45–47), which could serve as an endogenous source of G4DNA after partial degradation or excision repair. We used H2O2 because it is known to cause oxidative damage to DNA (48–50). We applied a G-quadruplex structure-specific antibody, BG4 (7, 10, 51), to determine whether levels of endogenous G4DNA would increase in the cytoplasm with oxidative stress. Cells were treated with H2O2 and harvested, and the cytosol was isolated. DNA from RNase-treated cytosol was then passed through a nylon membrane using a dot blot apparatus so that all DNA in the cytosol would bind to the membrane. The membrane was probed with the G4DNA antibody, and the bound antibody was visualized by chemiluminescence. Cells treated with H2O2 for 2 h exhibited a 9-fold increase in G4DNA compared with untreated cells (Fig. 4A). Therefore, we concluded that quadruplex DNA structures increase dramatically in the cytoplasm under conditions of oxidative stress.
FIGURE 4.
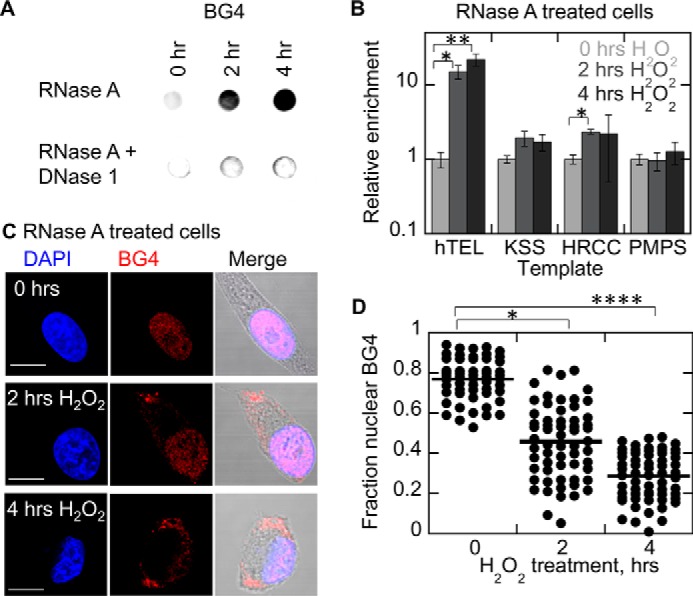
Treatment of cells with hydrogen peroxide leads to accumulation of G4DNA in the cytosol. A, dot blot of cytosolic fractions with BG4 indicates G4DNA in the cytosol increases 8.7 ± 1.8-fold after 2 h with H2O2 and 16.9 ± 3.5-fold after 4 h with H2O2 (mean ± S.D. of biological triplicates) in WM266-4 cells. B, the relative copy numbers of four quadruplex-forming sequences in the cytosol were determined by qPCR. Results shown are relative to untreated cells for each template. qPCR indicates a dramatic increase of telomeric DNA in the cytoplasm upon H2O2 treatment (mean ± S.D. of biological triplicates). C, BG4 indicates that G4DNA is present in the nucleus in untreated cells but appears in the cytoplasm in the presence of oxidative stress. Cell morphology changes upon treatment with 0.5 mm H2O2, consistent with previous reports (113). D, quantitation of BG4 fluorescence localization in C. The p values were calculated using a two-tailed paired t test. Plot shows the mean of 60 cells/condition from three or more independent biological experiments/condition in WM266-4 cells. Bar equals 10 μm.
The source of the G4DNA in the cytoplasm was investigated by selecting a few candidate sequences and performing quantitative PCR. The relative copy numbers of four G4DNA targets in the cytosol were examined: telomeric DNA and three sequences from mitochondrial DNA (mtDNA). These mtDNA sequences (KSS, HRCC, and PMPS) (52) and telomeric DNA (hTEL) (4) have been shown to form stable quadruplexes in vitro. The results from quantitative PCR (qPCR) showed a dramatic increase in telomeric DNA in the cytoplasm of treated cells (Fig. 4B), which is consistent with previous results showing that telomeres are shortened during treatment of cells with H2O2 (53). Small increases in the mtDNA sequences were also observed. Because the PCR reports only on the level of each DNA sequence observed in the treated cells relative to the untreated cells, but not the absolute quantity of each DNA sequence present in the cytosol, these data do not suggest that most of the DNA is telomeric in origin, only that telomeric DNA sequences significantly increase in the cytosol in response to oxidative stress.
The appearance in the cytoplasm of endogenous G4DNA was next examined by fluorescence microscopy using BG4. This antibody revealed punctate fluorescence staining primarily in the nucleus after treatment with RNase (Fig. 4C), which is consistent with published results (7, 10). Interestingly, upon treatment of cells with H2O2, there is an increase in punctate G4DNA fluorescent foci appearing in the cytoplasm and a reduction of foci in the nucleus (Fig. 4C). The fluorescence data are quantified in Fig. 4D. Control experiments confirmed that the observed quadruplex signal in response to oxidative stress is indeed DNA. The inclusion of RNase treatment prior to BG4 antibody in all experiments excludes observation of G4RNA, and the addition of DNase eliminates the observed G4DNA foci (Fig. 5). Additional control experiments lacking primary antibodies show the specificity of the antibodies (Fig. 5E).
FIGURE 5.
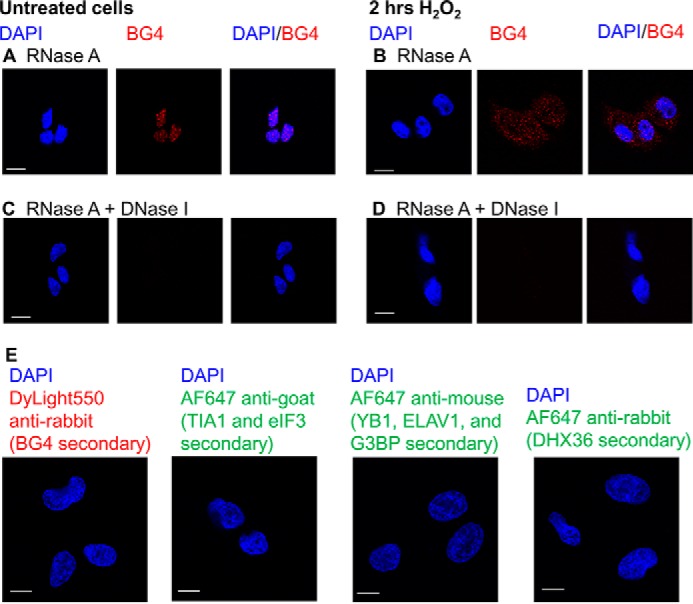
Control experiments to confirm the quadruplex is DNA. In WM266-4 cells treated with RNase A, only G4DNA in the nucleus is detected by BG4 in cells without H2O2 treatment (A). In WM266-4 cells treated with H2O2 for 2 h, G4DNA is visible throughout the cell in RNase A-treated cells (B). When cells are treated with both RNase A and DNase I, little G4DNA is detected in either untreated cells (C) or cells treated with H2O2 for 2 h (D). E, control experiments lacking primary antibodies. WM266-4 cells were treated with 0.5 mm H2O2 for 2 h and then fixed and stained with secondary antibodies without primary antibodies. Bar equals 10 μm.
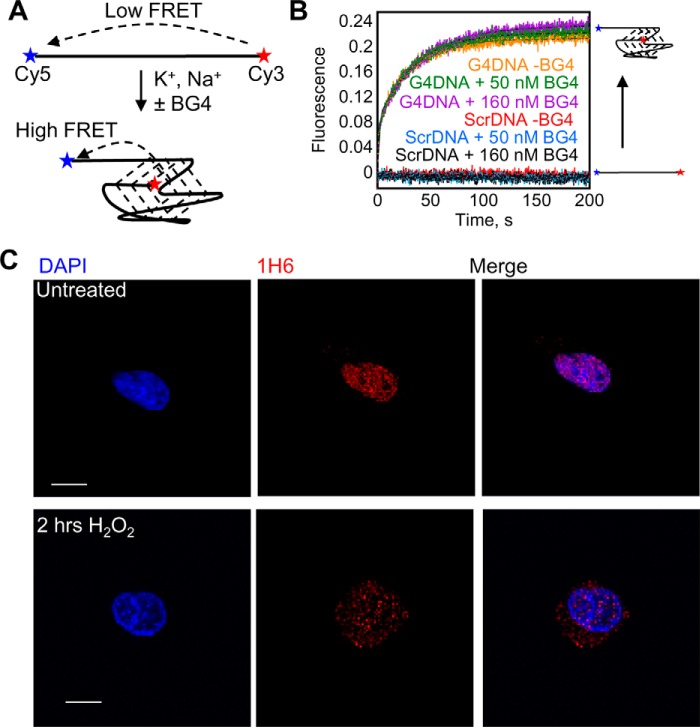
Because there is a potential for antibodies that bind to a specific DNA conformation to induce folding of the recognized conformation, the effect of BG4 on the folding of G4DNA was investigated. A quadruplex-forming oligonucleotide with Cy5 and Cy3 at the termini was rapidly mixed with salt in a stopped flow apparatus, and the increase in FRET due to G4DNA folding was monitored (Fig. 6, A and B). Folding in the absence of BG4 with an equivalent concentration of BG4 and DNA, and with an excess of BG4 (ten times the BG4 concentration used in the immunofluorescence experiments), all occurred at the same rate. When a Cy5 and Cy3-labeled scrambled DNA was mixed with salt, no increase in FRET was observed in the presence or absence of BG4. The lack of effect of BG4 on G4DNA folding indicates that BG4 does not cause quadruplex folding from unstructured DNA. Additionally, to confirm that the species detected in the cytosol of H2O2-treated cells was indeed a quadruplex, the localization of G4DNA in untreated and H2O2-treated cells was visualized using a different quadruplex-specific antibody, 1H6 (Fig. 6C) (9). Similar to what was observed with BG4, 1H6 detects G4DNA in the nucleus of untreated cells, but G4DNA appears in the cytosol after H2O2 treatment.
FIGURE 6.
G4DNA folding. A, folding of Cy3-Cy5-G4DNA can be monitored by the increase in FRET upon the addition of salt. B, folding of 50 nm Cy3-Cy5-G4DNA upon the addition of salt was measured in the absence of BG4 (orange), in the presence of 50 nm BG4 (green), and in the presence of 160 nm BG4 (purple). Data were fit to a sum of 2 exponentials, and the rate constants were 0.083 and 0.032 s−1, 0.067 and 0.025 s−1, and 0.084 and 0.029 s−1 for 0, 50, and 160 nm BG4, respectively. Folding of a Cy3-Cy5-ScrDNA was not observed in the absence of BG4 (red) or in the presence of 50 nm (blue) or 160 nm BG4 (black). C, staining with another quadruplex-specific antibody, 1H6, indicates that G4DNA is present in the nucleus in untreated cells but appears in the cytoplasm in the presence of oxidative stress.
These results suggest that DNA sequences capable of forming G4DNA structures, as indicated by the BG4 and 1H6 antibodies, accumulate in the cytoplasm. The specific source of the DNA is not clear from these data; however the results are consistent with the data from the dot blot assay and the qPCR (Fig. 4). When the medium containing H2O2 was removed and replaced with fresh medium, G4DNA reappeared in the nucleus within 6 h (Fig. 7, A and B), suggesting that as the cell progresses through the cell cycle, G-quadruplexes reform. Some BG4 staining was still evident in the cytosol even 12 h after removing the H2O2. These results indicate that after folding, G4DNA has a relatively long lifetime in the cell, which is consistent with G4DNA serving as a signaling molecule. The appearance of G4DNA in the cytoplasm is not a consequence of apoptotic DNA fragmentation, as H2O2 treatment under these conditions resulted in no detectable level of DNA fragmentation (Fig. 7C). Staurosporine, a common apoptosis inducer (54), was used as a positive control.
FIGURE 7.
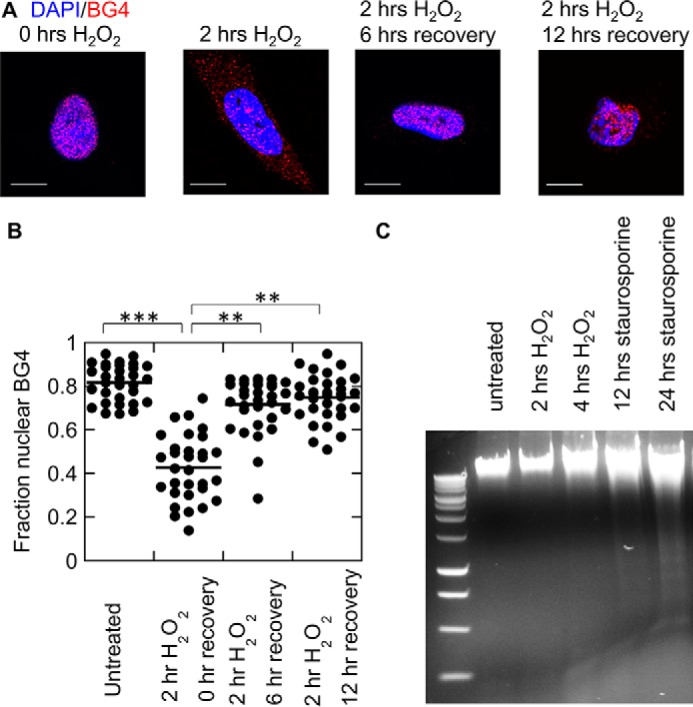
Removal of hydrogen peroxide from the medium leads to reappearance of G4DNA in the nucleus. A, after 2 h of treatment with 0.5 mm H2O2, the medium was removed and replaced with fresh medium for 6–12 h before fixing the cells. B, quantification of BG4 fluorescence localization shown in A. The p values were calculated using a two-tailed paired t test. Plot shows the mean of 30 cells/condition from three independent biological experiments/condition in WM266-4 cells. Bar equals 10 μm. C, DNA from untreated and 0.5 mm H2O2-treated cells runs as a large species on an agarose gel, whereas DNA from cells treated with 1 μm staurosporine for 24 h shows some fragmentation, indicative of apoptosis.
The increase in BG4 fluorescence in the cytoplasm upon oxidative stress was also observed in HeLa cells (Fig. 8A) and primary melanocytes (Fig. 8B) treated with H2O2. To generate oxidative stress through a different mechanism, melanoma cells were treated with menadione (Fig. 8C). Menadione generates oxidative stress through an unstable semiquinone metabolite that redox cycles in the presence of oxygen to produce reactive oxygen species (55). Treatment with menadione produced cytoplasmic BG4 foci, consistent with what was observed with H2O2 treatment.
FIGURE 8.
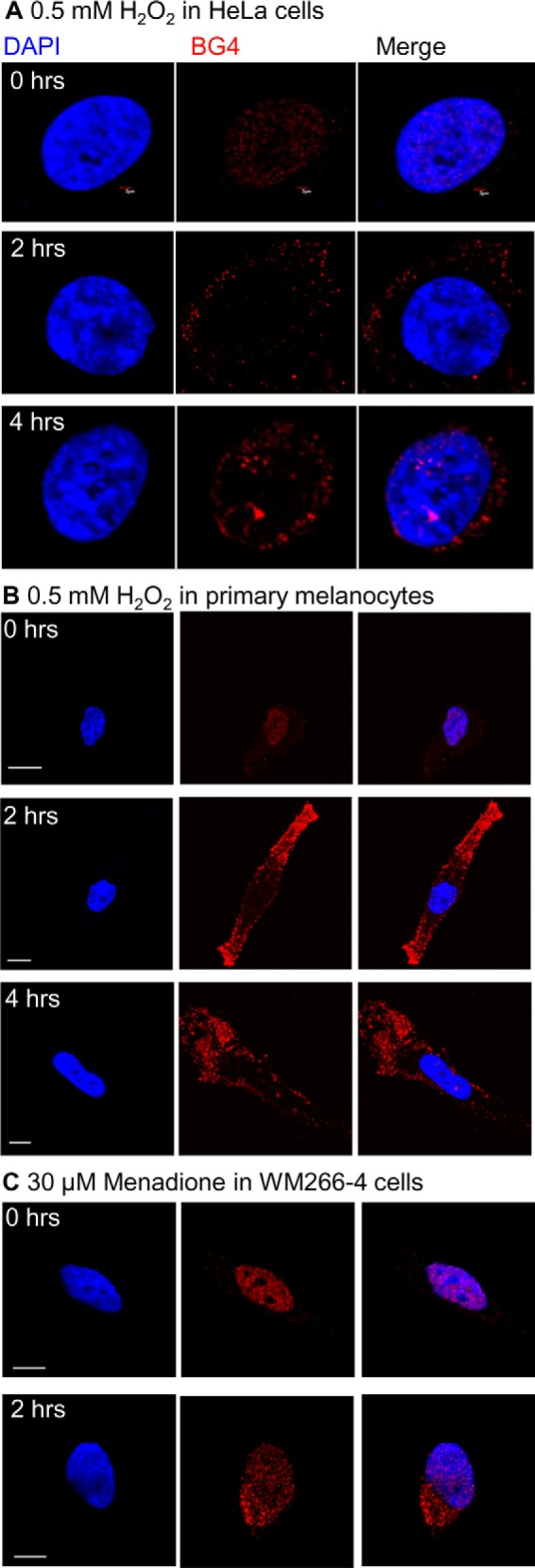
G4DNA appears in the cytosol of cells during oxidative stress. A, treatment of HeLa cells with 0.5 mm hydrogen peroxide leads to accumulation of G4DNA in the cytosol, similar to WM266-4 cells. Experiments were performed twice. Bar equals 2 μm. B, treatment of primary melanocytes with hydrogen peroxide leads to G4DNA accumulation in the cytosol. Bar equals 10 μm. C, treatment of WM266-4 cells with 30 μm menadione for 2 h results in the appearance of G4DNA in the cytosol. Bar equals 10 μm.
Endogenous G4DNA Co-localizes with Cytoplasmic Stress Granule Proteins
To determine whether endogenous G4DNA can bind to stress granule proteins in the cytoplasm, as was already shown for exogenous G4DNA (Figs. 2 and 3) (34), co-localization between the BG4 antibody and the stress granule protein TIA1 was examined. We chose TIA1 because it is found primarily in the nucleus prior to treatment with H2O2, and it was identified as a G4DNA-interacting protein (Fig. 1). Untreated cells exhibited no stress granule formation (Fig. 9A). Increasing amounts of G4DNA are observed in the cytoplasm after treatment with H2O2, and cytoplasmic stress granules, indicated by punctate staining with TIA1 antibodies, clearly co-localize with a subpopulation of the BG4 antibody after treatment (Fig. 9A, merged panels). The co-localization of a subset of the endogenous G4DNA and stress granule proteins was confirmed with G3BP, a stress granule marker, which was not identified as a G4DNA-binding protein in the proteomics experiment (Fig. 9B). Because stress granules contain both proteins and translationally stalled mRNAs, the presence of RNA in the granules was confirmed (Fig. 9C). Co-localization of BG4 with TIA1 was also observed in HeLa cells (Fig 10A) and in primary melanocytes (Fig. 10B) treated with H2O2. In addition, melanoma cells that were treated with menadione also exhibited foci containing BG4 and TIA1 antibodies (Fig. 10C).
FIGURE 9.
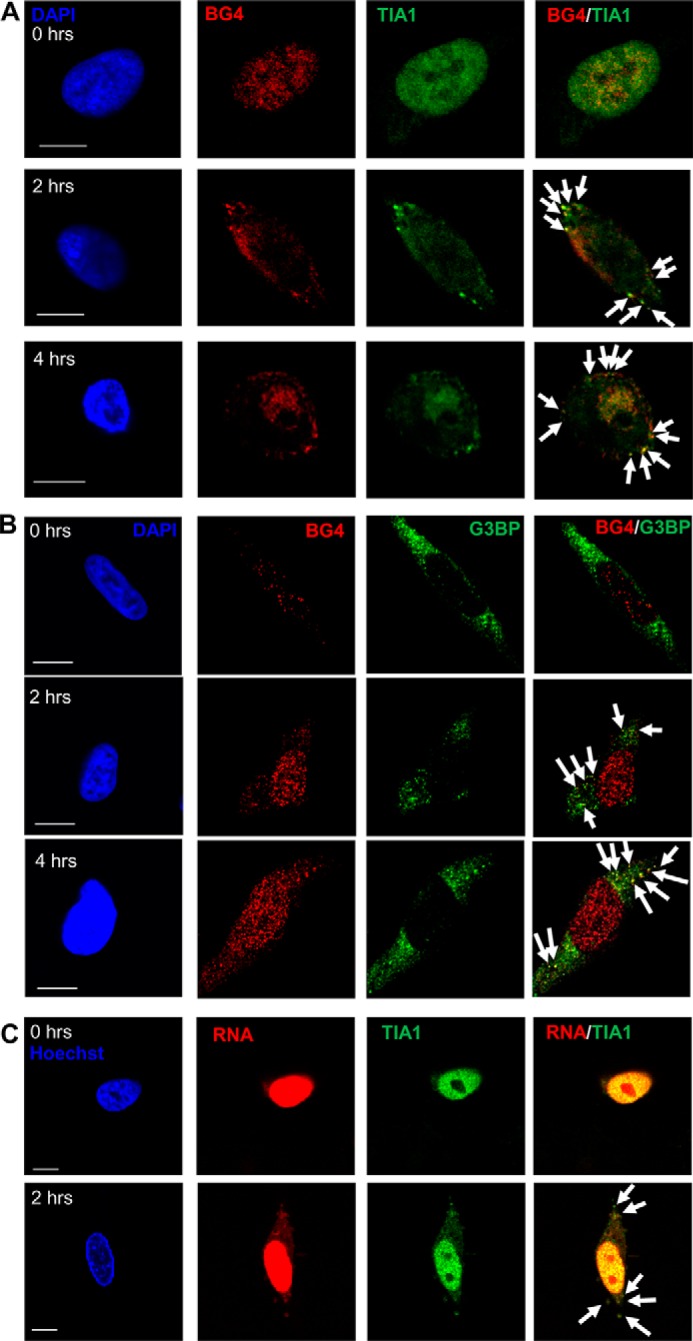
Endogenous G4DNA co-localizes with stress granule markers with H2O2 treatment. The G4DNA-binding antibody, BG4, co-localizes with TIA1 (A) and G3BP (B) in stress granules after treatment with H2O2 (indicated by arrows). For TIA1, 79% of the TIA1 foci co-localize with BG4 after 2 h of H2O2 treatment, and 81% co-localize after 4 h of H2O2 treatment. For G3BP, 76% of the G3BP foci co-localize with BG4 after 2 or 4 h of H2O2 treatment. C, cytosolic TIA1 positive foci that form in response to H2O2 treatment also contain RNA (96% of TIA1 foci contain RNA, indicated by arrows). Images are shown for treatment with 0.5 mm H2O2 in WM266-4 cells. Bar equals 10 μm.
FIGURE 10.
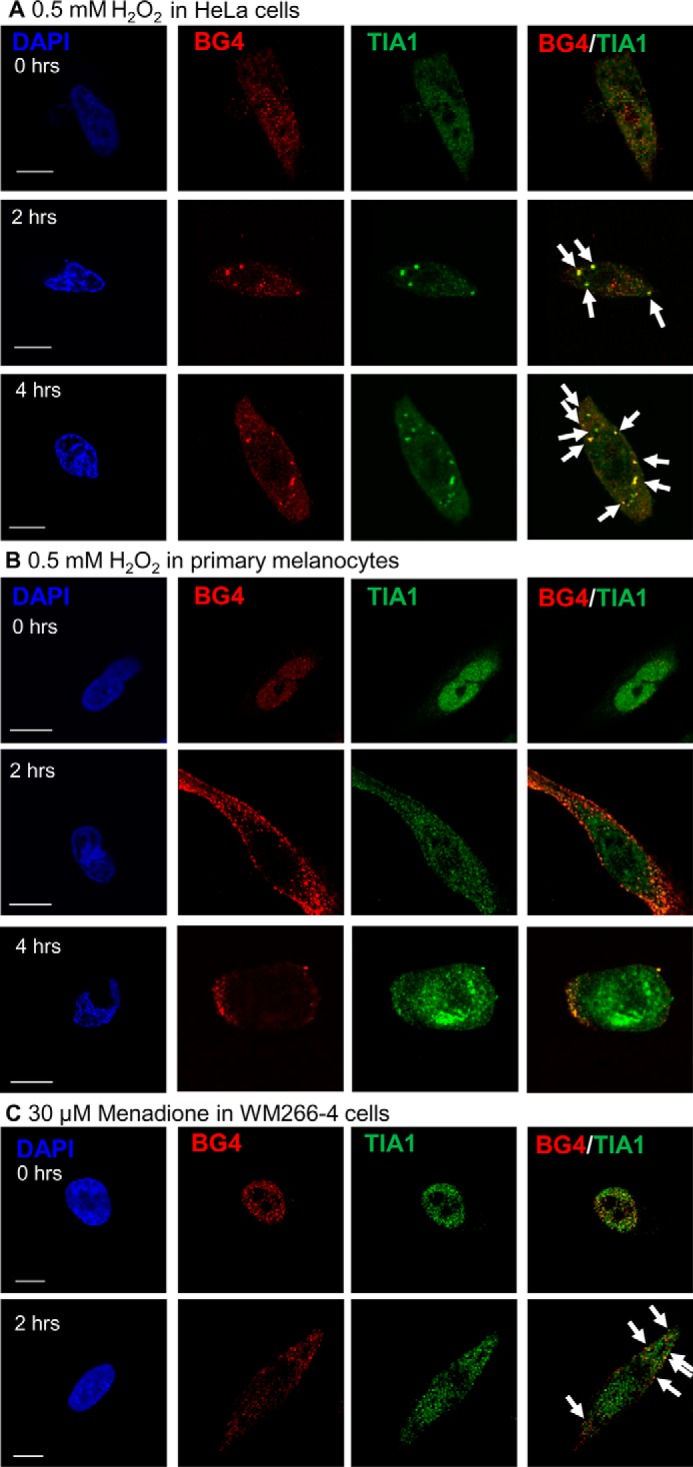
Endogenous G4DNA co-localizes with TIA1 in different cell types with oxidative stress. The G4DNA-binding antibody, BG4, co-localizes with TIA1 (indicated by arrows) in HeLa cells (A) and primary melanocytes (B) treated with 0.5 mm H2O2. In HeLa cells, 68% of the TIA1 foci co-localize with BG4 after 2 h of H2O2 treatment, and 71% co-localize after 4 h of H2O2 treatment. In primary melanocytes, 68% of the TIA1 foci co-localize with BG4 after 2 h of H2O2 treatment, and 72% co-localize after 4 h of H2O2 treatment. C, treatment of WM266-4 cells with 30 μm menadione for 2 h results in co-localization of BG4 with TIA1 in the cytosol. Treatment with menadione for 2 h results in 77% of the TIA1 foci co-localized with BG4. Bar equals 10 μm.
Discussion
The data presented here support the conclusion that DNA sequences that can fold into G4DNA structures accumulate in the cytoplasm as a result of oxidative stress. G4DNA binds to proteins such as DHX36 and YB-1 and participates in the formation of stress granules (Fig. 11). The source of the DNA is not known, but DNA sequences from both nuclear and mitochondrial genomes are implicated. Multiple results support the conclusion that the observed molecule, which co-localizes with stress granule proteins after oxidative stress, is indeed G4DNA. 1) The quadruplex-specific antibodies BG4 and 1H6 bind to the molecule (Figs. 4 and 6). 2) The molecule is not sensitive to RNase (Figs. 4–10). 3) The molecule is sensitive to excess DNase (Fig. 5). 4) The reduction in nuclear foci with H2O2 treatment (Fig. 4, C and D) followed by the re-emergence in the nucleus after removal of H2O2 (Fig. 7) is consistent with DNA excision followed by DNA synthesis and chromatin reorganization. Finally, 5) qPCR indicates an increase in specific G4DNA sequences in the cytoplasm (Fig. 4B). G4DNA structures from damaged G-rich sequences provide a direct mechanism for the cell to sense oxidative stress-induced DNA damage (56). The formation of stress granules is known to modulate RNA translation during oxidative stress (24, 57). Although the translation of some mRNAs is stalled by stress granule formation, other transcripts are actively excluded from stress granules (23), making the effects of stress granule formation on individual protein levels difficult to predict.
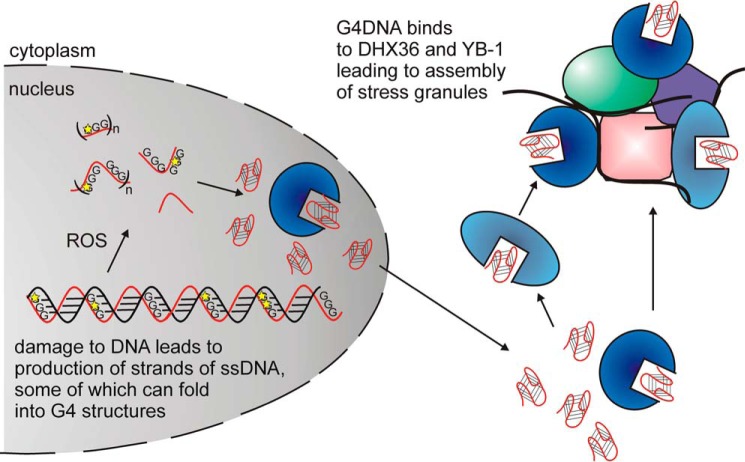
FIGURE 11.
Model for G4DNA signaling. DNA repair and excision of oxidatively damaged DNA can release short segments of DNA, some of which fold into quadruplex structures that are resistant to nuclease activity and can diffuse or be transported out of the nucleus. Quadruplex folding may occur before excision, after excision in the nucleus, or in the cytosol after transport out of the nucleus. Cytoplasmic G4DNA binds to proteins that assemble into stress granules such as DHX36 and YB-1 (33, 34), thereby regulating translation. Excised G4DNA in the nucleus is available to bind to and potentially regulate any other G4DNA-binding protein, thereby impacting additional biological pathways.
It is not clear from the data whether oxidation of G4DNA sequences themselves results in the appearance of G4DNA in the cytoplasm or whether DNA damage in general up-regulates DNA repair, resulting in increased excision. It is known that DNA sequences containing runs of guanine residues are more reactive with hydroxyl radicals than guanine residues scattered throughout the genome (45, 46, 58). Guanine oxidation does reduce the thermal stability of G-quadruplexes; however, G4DNA structures can still form in the presence of 8-oxo-dG (59, 60). Naturally occurring G4DNA-forming sequences can have additional guanines that are not involved in quadruplex formation. In telomeres, the formation of more stable G4DNA structures may occur via the expulsion of the damage-containing telomeric repeat from the quadruplex (60, 61). Promoter G4 sequences often contain five tracks of guanines, leading to swapping of the spare guanine track for the track containing the damaged guanine (47). Additionally, it is known that DNA double strand breaks occur more frequently at G4 sequences (62). During repair of dsDNA breaks, resection generates ssDNA regions in which quadruplexes can form when the appropriate sequences are present. Thus, dsDNA breaks could also lead to the formation of G4DNA, and DNA damage in general may lead to an increase in G4DNA in the cytoplasm.
Sequences that can form G4DNA are found predominantly in mitochondrial DNA, telomeres, and promoters of proto-oncogenes. Hence, the appearance of G4DNA sequences in these regions provides an opportunity for an early alarm system for the DNA repair machinery to focus on these damaged regions. As shown in Fig. 4, the G4DNA that is observed in the nucleus is reduced within 4 h of treatment with H2O2. A concomitant increase in the G4DNA signal appears in the cytoplasm. We suggest that DNA excision repair of G-rich genomic DNA produces short segments of DNA that fold into stable quadruplex structures in response to episodes of oxidative stress and possibly other forms of DNA damage (Fig. 11). Although a clear reduction in nuclear G4DNA after H2O2 treatment was observed (Fig. 4, C and D), the origin of the G4DNA in the cytoplasm is unknown. It is possible that these G4DNA sequences are excised from nuclear or mitochondrial genomes or that they are synthesized by telomerase in the cytoplasm. Telomerase is known to be present in the cytosol under some conditions, including oxidative stress, which could result in synthesis of the G4DNA sequences in the cytosol (63–66). Although the source of the G4DNA (nuclear genome, mitochondrial genome, or synthesized in the cytoplasm) is not known at this time, it appears that these short strands of G4DNA serve as a signaling agent to inform the cellular machinery of DNA damage.
Numerous lines of evidence indicate that cells have evolved to respond to G-quadruplex structures. Several laboratories have reported that many cell types, usually cancer cells, spontaneously take up exogenous G4DNA (67–70). G4DNA taken up by cells causes various biological responses including apoptosis in tumor cells (69, 71). The c-MYC quadruplex was recently shown to be readily taken up by cells, where it destabilizes proteins that normally bind G4DNA in the cell (72). Indeed, the G4DNA oligonucleotide AS1411 has been developed as a possible treatment for cancer (67, 73). G4 structures made up of DNA and RNA have also been proposed as important regulators of transcription (74–76). These facts suggest a fundamental biological response to G4DNA structures, even when excised from genomes. Our work is the first to show that G4DNA from an endogenous source can be observed within stress granules, thereby participating in a known response to cellular stress.
Deletions of mtDNA and the resulting loss of mitochondrial function are thought to be a major cause of cancer, aging, and genetic disorders (77). Human mtDNA contains 270 potential G4-forming sequences (52), many of which correlate with DNA breakpoints associated with diseases (52, 78, 79). It is of note that hypersensitive DNA cleavage sites have been described adjacent to G4DNA sequences in mitochondria (52, 78, 79) and proto-oncogenes (80, 81). Recent results from the Balasubramanian laboratory support the conclusion that a very large number of DNA sequences throughout the genome can fold into quadruplex structures in addition to telomeric sequences (82). Hence, nuclear and mitochondrial human DNA has great potential for producing G4DNA during repair or excision of damaged DNA.
The signaling role proposed here for G4DNA may explain in part why nature has preserved these sequences at such high propensity in the mitochondrial and nuclear genomes. The selection of G4DNA sequences for mitochondria, telomeres, and promoters of proto-oncogenes provides an additional survival advantage if these sequences and the resulting quadruplex structures play multiple roles, first in regulating DNA metabolism and then in alerting the cell to DNA damage of critical regions of the genome. Telomeres are reportedly favored targets of persistent DNA damage (83, 84), which may be related in part to the high reactivity and chemical breakdown products observed for specific guanine residues in telomeric sequences (45). Oxidative stress can cause telomere shortening (53, 85). Under conditions of mild oxidative stress, single strand breaks in telomeres accumulate (86), and the rate of telomere shortening can be altered by changes in the amount of oxidative stress (87).
As the name implies, stress granules are known to form in response to many different cellular stress events (88) through more than one mechanism. One route involves phosphorylation of eIF2α (reviewed in Ref. 21). Assembly occurs through aggregation of proteins such as TIA1 or G3BP, which can interact with mRNA leading to translationally stalled aggregates. The DDX3 RNA helicase can associate with stress granules and suppress translation through the inhibition of eIF4E (89). Additionally, molecules that inhibit eIF4A can promote stress granule formation independently of eIF2α (reviewed in Ref. 90). The observation that G4DNA can participate in the assembly of stress granules suggests another mechanism for their formation. The fact that TIA1 and G3BP are found co-localized with cytoplasmic G4DNA supports this conclusion. We anticipate finding that G4DNA acts through RNA-binding proteins to promote stress granule formation.
A recent report found that oxidative stress from H2O2 reduces stress granule formation due to TIA1 oxidation (91). However, others have reported that stress granules can form in the presence of H2O2 in an eIF2α-independent manner (44, 92). It is possible that the concentration and exposure time for H2O2 can modulate the appearance of stress granules. Our work here used similar H2O2 concentrations and exposure times to that detailed in Ref. 44, lower H2O2 concentrations than found in Ref. 92, and longer exposure times and higher concentrations than in Ref. 91.
Stress granule formation reduces the development of reactive oxygen species, thereby inhibiting apoptosis and favoring repair (93). The YB-1 protein has been proposed to play a key role in translation regulation by interacting with quadruplexes. tRNA-derived, stress-induced RNA (tiRNA)-derived quadruplexes, bind to YB-1, leading to YB-1 inhibition of cap-dependent translation initiation, thereby altering gene expression in response to cellular stress (34). Based on data shown here, it appears that endogenous G4DNA sequences (Fig. 11) can link oxidative stress and DNA damage directly to translation regulation through stress granule formation, perhaps in part through YB-1 binding.
The DExD/H RNA helicase family has many members that play roles in cellular stress response (90). DHX36, a member of this family, is capable of binding and unwinding RNA, which combined with its ability to be recruited to stress granules (33) likely plays a role in regulating translation. It is an essential gene (94) and is the primary enzyme responsible for unwinding G4DNA in cells (27). Another cytosolic RNA helicase, DDX41 (95), binds to DNA and activates STING, which ultimately leads to the activation of NF-kB and interferon regulatory factor 3 (IRF3). G4DNA may serve as a signal through binding to multiple proteins that process RNA.
The proposed G4DNA signaling response to DNA oxidation reported here could be especially valuable to the cell by directly reporting the level of DNA damage to proteins that regulate translation (21, 22, 24). The fact that many cells can readily take up G4DNA indicates that this mechanism may also allow the cell to respond to its external environment as well as internal stress (67, 72). Although our results suggest important roles for G4DNA in the regulation of cytoplasmic cellular processes, other studies have suggested roles for G4DNA in modulating transcription through the binding of helicases such as DHX36 (30, 96, 97), the RecQ family helicases (98, 99), and the XPB and XPD helicases (75). It is possible that any G4DNA-binding protein may be regulated by freely diffusing G4DNA, leading to various cellular responses based on the large number of proteins that are reported to have G4DNA binding capacity (4, 100–102). G4DNA has been proposed as a drug target for treatment of cancer and other diseases (74). Our work here expands the rationale for targeting G4DNA and provides further support for G4DNA mimics as potential drugs (67, 68).
Experimental Procedures
Oligonucleotides and Antibodies
All oligonucleotides were purchased from Integrated DNA Technologies and purified (103). The sequences used were: Cy3-G4DNA (5′-T15 GAG GGT GGG TAG GGT GGG TAA-Cy3–3′), a well characterized G4DNA-forming sequence based on a region of the c-MYC promoter (104); Cy3-T20 (5′-T20-Cy3–3′); Cy3-Scr (5′-TGT TGT TGT TGT TGT TGT TGT TGA GTG TAG TTG AAG-Cy3–3′), a scrambled version of the G4DNA sequence; Cy3-PS-Scr (5′-T*G*T TGT TGT TGT TGT TGT TGT TGA GTG TAG TTG A*A*G -Cy3–3′), where the asterisk indicates a phosphorothioate linkage; 3′-Bio-G4DNA (5′-T15 GAG GGT GGG TAG GGT GGG TAA-BioTEG-3′); 3′-Bio-ssDNA (5′-T15-BioTEG-3′); Cy3-Cy5-G4DNA (5′-Cy5-T15 GAG GGT GGG TAG GGT GGG TAA-Cy3–3′); and Cy3-Cy5-Scr (5′-Cy5-TGT TGT TGT TGT TGT TGT TGT TGA GTG TAG TTG AAG-Cy3–3′). Intramolecular quadruplexes were formed as described (105), and structures were confirmed by circular dichroism of 10 μm DNA in 10 mm Hepes-Li+, pH 7.5, 1 mm EDTA-Li+, and 100 mm KCl in a JASCO 715 spectropolarimeter (Fig. 1, F and G). The PCR primers sequences used were: hTEL forward, 5′-GGT TTT TGA GGG TGA GGG T-3′; hTEL reverse, 5′-TCC CGA TAT CCC TAT CCC TA-3′ (modified from Ref. 106); KSS forward, 5′-GAC AAT TAT ACC CTA GCC AAC C-3′; KSS reverse, 5′-CTT GAT GTG GGG AGG GGT G-3; HRCC forward, 5′-CTA TGA ACC CCC CTC CCC-3′; HRCC reverse, 5′-GTT GAC CAG GGG GTTGGG TA-3′; PMPS forward, 5′-GCT TCG ACC CTA TAT CCC CC-3′; and PMPS reverse, 5′-GGA GAA AGG GAC GCG GGG-3′.
pSANG10–3F-BG4 plasmid encoding the BG4 single chain antibody (7, 10, 51) was a gift from Shankar Balasubramanian (Addgene plasmid 55756). Monoclonal antibody 1H6 was kindly provided by Peter Lansdorp (9). BG4 was used at 16 nm and was detected using rabbit anti-FLAG (1:800 dilution, 2368P, Cell Signaling) and DyLight 550-conjugated goat anti-rabbit (1:100 dilution, 84541, Thermo Fisher Scientific) antibodies or HRP-conjugated goat anti-rabbit antibody (1:3000 dilution, NEF812, PerkinElmer Life Sciences). 1H6 was used at 1 μg/ml and was detected using Cy3-conjugated donkey anti-mouse antibodies (1:50 dilution, 715-165-151, Jackson ImmunoResearch). DHX36 was detected using rabbit anti-DHX36 (1:100 dilution for immunofluorescence, 1:5000 dilution for Western blotting, ab70269, Abcam) and Alexa Fluor 647-conjugated donkey anti-rabbit antibodies (1:100 dilution, 711-605-152, Jackson ImmunoResearch) or HRP-conjugated goat anti-rabbit antibody (1:3000 dilution, NEF812, PerkinElmer Life Sciences). TIA1 was detected using goat anti-TIA1 (1:100 dilution for immunofluorescence, 1:1000 dilution for Western blotting, sc-1751, Santa Cruz) and Alexa Fluor 647-conjugated donkey anti-goat antibodies (1:100 dilution, 705-605-147, Jackson ImmunoResearch) or HRP-conjugated donkey anti-goat antibody (1:5000 dilution, sc-2020, Santa Cruz Biotechnology). YB-1 was detected using mouse anti-YB-1 (1:50 dilution, sc-101198, Santa Cruz Biotechnology) and Alexa Fluor 647-conjugated donkey anti-mouse antibodies (1:50 dilution, 715–605-150, Jackson ImmunoResearch). ELAV1 was detected using mouse anti-HuR (1:50 dilution, sc-5261, Santa Cruz Biotechnology) and Alexa Fluor 647-conjugated donkey anti-mouse antibodies (1:50 dilution, 715-605-150, Jackson ImmunoResearch). G3BP was detected using mouse anti-G3BP (1:200 dilution, 611126, BD Transduction Laboratories) and Alexa Fluor 647-conjugated donkey anti-mouse antibodies (1:50 dilution, 715-605-150, Jackson ImmunoResearch). eIF3 was detected using goat anti-eIF3η (1:50 dilution sc-16377, Santa Cruz Biotechnology) and Alexa Fluor 647-conjugated donkey anti-goat antibodies (1:100 dilution, 705-605-147, Jackson ImmunoResearch).
BG4 Purification
BL21(DE3) E. coli containing the pSANG10-3F-BG4 plasmid were grown, and BG4 expression was induced at 25 °C overnight with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were pelleted at 4500 × g for 1 h at 4 °C. The cell pellet was suspended in TS buffer (50 mm Tris-Cl, pH 8.0, 20% sucrose) with 1 mm PMSF and then placed on ice for 15 min followed by microfluidization. Lysate was centrifuged at 8000 × g for 30 min at 4 °C followed by centrifugation at 110,000 × g for 1.5 h at 4 °C. Imidazole was added to the supernatant to 25 mm followed by incubation with Ni2+-Sepharose resin overnight at 4 °C. The slurry was loaded into a column and washed with TS buffer containing 25 mm imidazole, and the bound proteins were eluted with elution buffer (PBS, pH 8.0, with 250 mm imidazole). Purified BG4 was dialyzed to PBS, pH 8.0, 20% glycerol and stored in aliquots at −80 °C.
Cell Culture
WM-266-4 cells (melanoma, ATCC CRL-1676) or HeLa cells (cervical adenocarcinoma, ATCC CCL2) were cultured in DMEM, 10% fetal bovine serum, 1× MEM non-essential amino acids (Thermo Fisher Scientific), and 100 units/ml penicillin/streptomycin at 37 °C with 5% CO2. Adult primary melanocytes (ATCC PCS-200-013) were cultured in dermal cell basal medium (ATCC) supplemented with an adult melanocyte growth kit (ATCC) and 100 units/ml penicillin/streptomycin at 37 °C with 5% CO2. Cells treated with H2O2 were cultured in serum-free medium for 30 min before the addition of 0.5 mm H2O2. Cells treated with menadione were treated with 30 μm menadione in serum-free DMEM for 2 h. For comparison, experiments with 30 μm menadione in DMEM containing 10% serum produced similar results. Some cells were transfected for 6 h with Cy3-G4DNA or Cy3-ssDNA using 0.8 μl of Lipofectamine 2000 in 200 μl of serum free medium.
Conjugation of DNA to Dynabeads
M-280 streptavidin-coupled Dynabeads (Life Technologies) were conjugated with 3′-biotinylated oligonucleotides (3′-Bio-G4DNA or 3′-Bio-ssDNA). The beads were washed, resuspended in 5 mm Tris-Cl, pH 7.5, 0.5 mm EDTA, and 150 mm KCl followed by the addition of DNA (at a ratio of 1 mg of beads/200 pmol of DNA), and incubated at 25 °C for 20 min with gentle rotation. The beads were washed three times with binding and washing buffer followed by a final wash in protein-binding buffer (300 mm KCl, 20 mm Tris, and 0.1% Tween, pH 7.5).
Affinity Purification of Proteins for Mass Spectrometry
WM266-4 cells were grown to ∼80% confluence in 75-cm2 flasks and harvested by centrifugation at 225 × g for 5 min. The cell pellets were resuspended in lysis buffer (300 mm KCl, 20 mm Tris, and 0.1% Tween, pH 7.5, in the proportion of 1 ml/200 mg of cells) and sonicated at 4 °C using a Bioraptor UCD-200 sonicator at 200 watts for 15 min. Mammalian protease inhibitor mixture was added to the cell lysates at a 1:166 dilution (Sigma-Aldrich). Cell lysates were incubated with DNA-conjugated Dynabeads (5 ml/mg) at 4 °C with gentle rotation for 4 h. The beads were collected with magnets and washed four times with protein-binding buffer. Proteins were eluted by boiling in Laemmli sample buffer for 5 min and resolved on 4–15% Mini-Protean TGX precast gels (Bio-Rad).
LC-MS Analysis
Protein gel bands were excised, then digested with trypsin (107). Tryptic peptides were separated using a nanoAcquity UPLC system (Waters) coupled to a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) (107, 108). Proteins were identified by a database search using PEAKS Studio v7 (Bioinformatics Solutions). A Data Refine routine was performed with the following parameters: no merge, corrected precursor, charge options: no correction, filter quality > 0.65. De novo and Peaks database searches were done with 10 ppm parent mass error tolerance and 0.85 Da fragment mass error tolerance. Methionine oxidation, asparagine deamidation, glutamine deamidation, and pyroglutamine formation were set as variable modifications. Cysteine carbamidomethylation was set as a fixed modification. Glutamine was set to equal to lysine. Leucine was set to equal to isoleucine. The searches were done against the reviewed Swiss-Prot human proteome database (20,187 total accessions), which was downloaded from the UniProt Website. Peaks PTM and Spider searches were performed, with the maximum number of post-translational modifications per peptide set to three. Semi-tryptic peptides were included. The peptide-to-spectrum match false identification rate was set at 1%, which corresponded approximately to a −10LogP score of 15. Proteins identified by only one unique peptide were included in the identification results, but the protein hit −10LogP threshold was set to 30. Proteins that had no unique peptides were reported as a group. Proteins that were subsets without unique peptides were not reported. G4DNA-interacting proteins were determined using spectral counting (26, 109, 110) by comparing spectral counts (SPCs) in the G4DNA-bound sample with the ssDNA-bound sample. For quantification by spectral counting, each accession was scored for total SPCs, unique SPC (uniquely matching to an accession), and adjusted SPC. The latter category assigns shared peptides to accessions in proportion to their relative abundance using unique SPCs for each accession as a basis. Proteins with adjusted p values less than 0.05 were judged significant (p values were derived from a G-test of independence and adjusted for multiple hypotheses testing using the Bonferroni correction). Zero spectral count values, which indicated the absence of a protein in a given condition, were replaced with 0.1 SPCs to allow log operation to be performed and G-statistics to be derived. Significant proteins, which were enriched at least four times in the G4DNA-bound sample compared with the ssDNA-bound proteins and were represented by at least 14 adjusted SPCs, were considered to be G4DNA-interacting proteins. The particular cutoff of 14 for adjusted SPCs was chosen based on the results of confidence testing by G-test; namely we found that given the absence of protein in one condition, 14 or more SPCs are required in the other condition to claim significant over accumulation with 95% confidence (after Bonferroni correction for multiple hypothesis testing). The Perl script for the calculation of adjusted SPCs, as well as the Excel template for the calculation of G-statistics and p values, is available from the authors by request.
Western Blotting Analysis
Proteins were separated by 10% SDS-PAGE and transferred to PVDF. Membranes were blocked with 5% nonfat dry milk (DHX36) or 5% chicken ovalbumin (TIA1) in TBS-T (20 mm Tris-Cl pH 7.5, 150 mm NaCl, 0.1% Tween 20). Membranes were probed with antibody dilutions in 1% nonfat dry milk or ovalbumin in TBS-T. Proteins were detected using horseradish peroxidase-conjugated secondary antibodies and Amersham Biosciences ECL Plus (GE Healthcare). Blots were imaged using a LAS4000 imager (GE Healthcare).
Immunofluorescence
WM266-4 cells, unless otherwise stated, were grown on chamber slides and fixed with 3.7% formaldehyde for 15 min at room temperature followed by 10 min with ice cold methanol at −20 °C. Slides were blocked with 2 mg/ml chicken ovalbumin, and 0.02% sodium azide in PBS overnight at 4 °C. Cells were further permeabilized with a buffer containing 0.2% Triton X-100, 2 mg/ml chicken ovalbumin, and 0.02% sodium azide in PBS. Before staining with BG4, cells were treated with 0.1 mg/ml RNase A for 30 min as described (7–10) so that only DNA quadruplexes would be detected. After staining with antibodies, coverslips were mounted with 2.5 mg/ml DABCO (1,4-diazobicyclo[2.2.2]octane) and 0.1 μg/ml DAPI in 25% PBS, 75% glycerol.
RNA Labeling
RNA was fluorescently labeled with Alexa Fluor 488 using the Click-iT RNA imaging kit (Invitrogen) according to the manufacturer's instructions. Briefly, WM266-4 cells were grown on an 8-chamber slide and incubated for 24 h in complete medium containing 1 mm 5-ethynyl uridine to allow incorporation of the nucleoside analog into cellular RNA pools. The cells were pulsed for 10 min with serum-free medium containing 2 mm 5-ethynyl uridine followed by the addition of an equal volume of serum-free medium with or without 1 mm H2O2 resulting in final concentrations of 1 mm 5-ethynyl uridine and 0.5 mm H2O2. Cultures were incubated at 37 °C for an additional 2 h, washed with PBS, and fixed for 15 min in 3.7% formaldehyde. For the labeling of modified RNA, the cells were washed with PBS and incubated with 1× Click-iT reaction buffer containing 4 mm CuSO4 and Alexa Fluor 488 azide for 30 min at room temperature. Cells were rinsed once with Click-iT reaction rinse buffer and twice with PBS followed by staining for TIA1 as described above. Nuclear DNA was stained with Hoechst 33342 for 15 min, and slides were mounted with a 75% glycerol/PBS solution containing 2.5 mg/ml DABCO.
Fluorescence Microscopy
Images were captured using an Olympus IX81 confocal microscope at room temperature with a 60× PLAPON60XO objective (1.42 numerical aperture) using FV10-ASW 3.1a software. DAPI was excited at 405 nm, and fluorescence was measured after a 430–470-nm band-pass filter. Alexa Fluor 488 was excited at 488 nm, and emission was measured after a 505–605-nm band-pass filter. Cy3 and DyLight550 were excited at 543 nm, and emission was measured after a 560–620-nm band-pass filter. Alexa Fluor 647 was excited at 635 nm and emission was measured after a 655–755 nm band-pass filter. Images were acquired with an Olympus FV1000 camera. Fluorescence was quantitated using Equation 1 with Fiji software (111), where Fn is the fraction nuclear BG4, Dn is the integrated density of BG4 in the nucleus, An is the area of the nucleus, Db is the mean density of the background, Dc is the integrated density of BG4 in the whole cell, and Ac is the area of the cell.
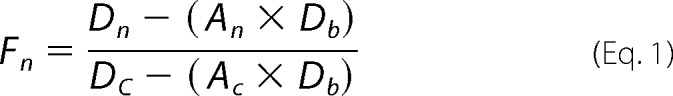 |
The p values were calculated using a two-tailed paired t test.
Isolation of Cytosol
Approximately 107 cells were harvested after treatment with 0.5 mm H2O2 and lysed using a Dounce homogenizer in a buffer containing 50 mm Hepes, pH 7.5, 20 mm NaCl, 1% Nonidet P-40, and 0.1 mg/ml RNase A with or without 0.05 mg/ml DNase I. Samples were centrifuged at 700 × g for 10 min at 4 °C to remove the nuclei. After determining the protein concentration of the lysates using a Bradford assay, samples were normalized for protein concentration and volume. The mitochondria were depleted by centrifuging them at 10,000 × g for 30 min at 4 °C. Proteins were removed by the addition of 0.5 mg/ml proteinase K with 0.5 mg/ml glycogen in 20 mm Tris, pH 7.5, 1 ml CaCl2 for 30 min at 50 °C. The enzyme was inactivated at 65 °C for 10 min.
Dot Blot Assay
G4DNA in cytosolic samples was refolded by the addition of 100 mm KCl and incubation at 60 °C for 30 min followed by slow cooling to room temperature. Samples were applied to an Ambion BrightStar-Plus positively charged nylon membrane pre-wet in 25 mm Hepes, pH 7.5, 0.1 mm EDTA, and 50 mm KCl using a dot blot apparatus. The membrane was blocked with 5% milk in TBS-T for 1 h before probing sequentially with BG4 in 1% milk for 1 h at room temperature, rabbit anti-FLAG antibody in 1% milk for 1 h at room temperature, and HRP-conjugated goat anti-rabbit antibody for 1 h at room temperature. Samples were detected using ECL Plus (GE Healthcare), imaged with an ImageQuant LAS4000 (GE Healthcare) and quantitated using ImageQuant software.
qPCR
DNA was isolated from cytosolic fractions by phenol-chloroform extraction and ethanol precipitation. qPCR was performed using SsoAdvanced Universal SYBR Green supermix (Bio-Rad) with the primers listed above. DNA was amplified with 40 cycles of 10 s at 90 °C and 30 s at 50 °C in a MiniOpticon real-time PCR detection system (Bio-Rad). The p values were calculated using a two-tailed paired t test.
DNA Fragmentation
Approximately 2 × 106 cells were harvested after treatment with 0.5 mm H2O2 or 1 μm staurosporine. Both the medium and the adherent cells were collected to ensure that both live and dead cells were retained. Cell pellets were resuspended in PBS. Proteinase K and RNase A were added to 6.7 mg/ml each, and samples were incubated at room temperature for 2 min followed by the addition of an equal volume of lysis buffer AL (Qiagen). Samples were incubated at 56 °C for 15 min, and 1 μg of DNA was loaded into each lane of a 1.5% agarose gel containing 0.5 μg/ml ethidium bromide.
G4DNA Folding
All concentrations listed are final after mixing. Cy3-Cy5-G4DNA or Cy3-Cy5-Scr in 50 mm Tris-OAc, pH 7.2, was mixed with 135 mm KOAc, 4 mm KCl, 12 mm NaHCO3, and 0.8 mm Mg(OAc)2 with or without BG4 in an SX.18MV stopped flow reaction analyzer (Applied Photophysics). The samples were excited at 550 nm, and FRET was measured after a 665-nm cut-on filter (Newport Corp., catalogue No. 51330). Data were fit to a sum of 2 exponentials to obtain rate constants for folding.
Author Contributions
K. D. R., R. L. E., G. B., and A. K. B. designed and supervised the research. B. L .Z. and J. G. performed the proteomics experiment, and B. L. Z., M. J., and S. G. M. analyzed the data for the proteomics experiments. A. K. B., L. M., J. C. M., M. R. B., M. R. R., and S. C. conducted the immunofluorescence experiments. A. K. B. performed the circular dichroism, DNA fragmentation, G4DNA folding, dot blot, and qPCR experiments. W. C. G. purified BG4. A. K. B., B. L. Z., L. M., J. G., J. C. M., A. M. M., G. B., R. L. E., and K. D. R. interpreted the data. K. D. R., A. K. B., B. L. Z., and J. C. M. prepared the manuscript. All authors edited the manuscript.
Supplementary Material
Acknowledgments
The University of Arkansas for Medical Sciences (UAMS) Proteomics Core is supported by the Arkansas IDeA Network for Biomedical Research Excellence (National Institutes of Health Grant P20GM103429), the University of Arkansas Center for Protein Structure and Function (National Institutes of Health Grant P30GM103450), the UAMS Center for Microbial Pathogenesis and Host Inflammatory Responses (National Institutes of Health Grant P20GM103625), and the UAMS Translational Research Institute (National Institutes of Health Grant UL1TR000039). We thank Shankar Balasubramanian, University of Cambridge, for the gift of the pSANG10-3F-BG4 plasmid encoding the BG4 single chain antibody. Monoclonal antibody 1H6 was kindly provided by Peter Lansdorp, European Research Institute for the Biology of Aging, University of Groningen. We thank Timothy Chambers for helpful discussions and Andrea Edwards for technical assistance.
This work was supported by National Institutes of Health Grants R01 GM098922 and R01 GM117439 (to K. D. R.) and by the Arkansas Biosciences Institute, major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (112) partner repository with the data set identifier PXD003501. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table 1 and Fig. S1.
- G4DNA
- G-quadruplex DNA
- Scr
- scrambled
- SPC
- spectral count
- PS
- phosphorothioate
- qPCR
- quantitative PCR
- SPC
- spectral count
- ssDNA
- single-stranded DNA
- mtDNA
- mitochondrial DNA.
References
- 1. Lemos H., Huang L., Chandler P. R., Mohamed E., Souza G. R., Li L., Pacholczyk G., Barber G. N., Hayakawa Y., Munn D. H., and Mellor A. L. (2014) Activation of the STING adaptor attenuates experimental autoimmune encephalitis. J. Immunol. 192, 5571–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. West A. P., Khoury-Hanold W., Staron M., Tal M. C., Pineda C. M., Lang S. M., Bestwick M., Duguay B. A., Raimundo N., MacDuff D. A., Kaech S. M., Smiley J. R., Means R. E., Iwasaki A., and Shadel G. S. (2015) Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaminski J. J., Schattgen S. A., Tzeng T. C., Bode C., Klinman D. M., and Fitzgerald K. A. (2013) Synthetic oligodeoxynucleotides containing suppressive TTAGGG motifs inhibit AIM2 inflammasome activation. J. Immunol. 191, 3876–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Murat P., and Balasubramanian S. (2014) Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 25, 22–29 [DOI] [PubMed] [Google Scholar]
- 5. Gursel I., Gursel M., Yamada H., Ishii K. J., Takeshita F., and Klinman D. M. (2003) Repetitive elements in mammalian telomeres suppress bacterial DNA-induced immune activation. J. Immunol. 171, 1393–1400 [DOI] [PubMed] [Google Scholar]
- 6. Klinman D. M., Tross D., Klaschik S., Shirota H., and Sato T. (2009) Therapeutic applications and mechanisms underlying the activity of immunosuppressive oligonucleotides. Ann. N.Y. Acad. Sci. 1175, 80–88 [DOI] [PubMed] [Google Scholar]
- 7. Biffi G., Tannahill D., McCafferty J., and Balasubramanian S. (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 5, 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lam E. Y., Beraldi D., Tannahill D., and Balasubramanian S. (2013) G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 4, 1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Henderson A., Wu Y., Huang Y. C., Chavez E. A., Platt J., Johnson F. B., Brosh R. M. Jr., Sen D., and Lansdorp P. M. (2014) Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 42, 860–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Biffi G., Tannahill D., Miller J., Howat W. J., and Balasubramanian S. (2014) Elevated levels of G-quadruplex formation in human stomach and liver cancer tissues. PLoS One 9, e102711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hershman S. G., Chen Q., Lee J. Y., Kozak M. L., Yue P., Wang L. S., and Johnson F. B. (2008) Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 36, 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rodriguez R., Miller K. M., Forment J. V., Bradshaw C. R., Nikan M., Britton S., Oelschlaegel T., Xhemalce B., Balasubramanian S., and Jackson S. P. (2012) Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 8, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paeschke K., Capra J. A., and Zakian V. A. (2011) DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145, 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paeschke K., Bochman M. L., Garcia P. D., Cejka P., Friedman K. L., Kowalczykowski S. C., and Zakian V. A. (2013) Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 497, 458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fernando H., Sewitz S., Darot J., Tavaré S., Huppert J. L., and Balasubramanian S. (2009) Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 37, 6716–6722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wanrooij P. H., Uhler J. P., Shi Y., Westerlund F., Falkenberg M., and Gustafsson C. M. (2012) A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 40, 10334–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fratta P., Mizielinska S., Nicoll A. J., Zloh M., Fisher E. M., Parkinson G., and Isaacs A. M. (2012) C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haeusler A. R., Donnelly C. J., Periz G., Simko E. A., Shaw P. G., Kim M. S., Maragakis N. J., Troncoso J. C., Pandey A., Sattler R., Rothstein J. D., and Wang J. (2014) C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vasilyev N., Polonskaia A., Darnell J. C., Darnell R. B., Patel D. J., and Serganov A. (2015) Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP. Proc. Natl. Acad. Sci. U.S.A. 112, E5391–E5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simone R., Fratta P., Neidle S., Parkinson G. N., and Isaacs A. M. (2015) G-quadruplexes: emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 589, 1653–1668 [DOI] [PubMed] [Google Scholar]
- 21. Kedersha N., Ivanov P., and Anderson P. (2013) Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waris S., Wilce M. C., and Wilce J. A. (2014) RNA recognition and stress granule formation by TIA proteins. Int. J. Mol. Sci. 15, 23377–23388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Anderson P., and Kedersha N. (2008) Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150 [DOI] [PubMed] [Google Scholar]
- 24. Anderson P., Kedersha N., and Ivanov P. (2015) Stress granules, P-bodies, and cancer. Biochim. Biophys. Acta 1849, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao J., Zybailov B. L., Byrd A. K., Griffin W. C., Chib S., Mackintosh S. G., Tackett A. J., and Raney K. D. (2015) Yeast transcription co-activator Sub1 and its human homolog PC4 preferentially bind to G-quadruplex DNA. Chem. Commun. (Camb.) 51, 7242–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zybailov B., Coleman M. K., Florens L., and Washburn M. P. (2005) Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal. Chem. 77, 6218–6224 [DOI] [PubMed] [Google Scholar]
- 27. Vaughn J. P., Creacy S. D., Routh E. D., Joyner-Butt C., Jenkins G. S., Pauli S., Nagamine Y., and Akman S. A. (2005) The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 280, 38117–38120 [DOI] [PubMed] [Google Scholar]
- 28. Giri B., Smaldino P. J., Thys R. G., Creacy S. D., Routh E. D., Hantgan R. R., Lattmann S., Nagamine Y., Akman S. A., and Vaughn J. P. (2011) G4 resolvase 1 tightly binds and unwinds unimolecular G4-DNA. Nucleic Acids Res. 39, 7161–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meier M., Patel T. R., Booy E. P., Marushchak O., Okun N., Deo S., Howard R., McEleney K., Harding S. E., Stetefeld J., and McKenna S. A. (2013) Binding of G-quadruplexes to the N-terminal recognition domain of the RNA helicase associated with AU-rich element (RHAU). J. Biol. Chem. 288, 35014–35027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iwamoto F., Stadler M., Chalupníková K., Oakeley E., and Nagamine Y. (2008) Transcription-dependent nucleolar cap localization and possible nuclear function of DExH RNA helicase RHAU. Exp. Cell Res. 314, 1378–1391 [DOI] [PubMed] [Google Scholar]
- 31. Booy E. P., Howard R., Marushchak O., Ariyo E. O., Meier M., Novakowski S. K., Deo S. R., Dzananovic E., Stetefeld J., and McKenna S. A. (2014) The RNA helicase RHAU (DHX36) suppresses expression of the transcription factor PITX1. Nucleic Acids Res. 42, 3346–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim T., Pazhoor S., Bao M., Zhang Z., Hanabuchi S., Facchinetti V., Bover L., Plumas J., Chaperot L., Qin J., and Liu Y. J. (2010) Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 107, 15181–15186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chalupníková K., Lattmann S., Selak N., Iwamoto F., Fujiki Y., and Nagamine Y. (2008) Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J. Biol. Chem. 283, 35186–35198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ivanov P., O'Day E., Emara M. M., Wagner G., Lieberman J., and Anderson P. (2014) G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. U.S.A. 111, 18201–18206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hanakahi L. A., Sun H., and Maizels N. (1999) High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 274, 15908–15912 [DOI] [PubMed] [Google Scholar]
- 36. Dickinson L. A., and Kohwi-Shigematsu T. (1995) Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol. Cell. Biol. 15, 456–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Benson B. K., Meades G. Jr., Grove A., and Waldrop G. L. (2008) DNA inhibits catalysis by the carboxyltransferase subunit of acetyl-CoA carboxylase: implications for active site communication. Protein Sci. 17, 34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kedersha N., and Anderson P. (2007) Mammalian stress granules and processing bodies. Methods Enzymol. 431, 61–81 [DOI] [PubMed] [Google Scholar]
- 39. Leung C. H., Chan D. S., Man B. Y., Wang C. J., Lam W., Cheng Y. C., Fong W. F., Hsiao W. L., and Ma D. L. (2011) Simple and convenient G-quadruplex-based turn-on fluorescence assay for 3′ 224 5′ exonuclease activity. Anal. Chem. 83, 463–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brennan C. M., and Steitz J. A. (2001) HuR and mRNA stability. Cell. Mol. Life Sci. 58, 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Creacy S. D., Routh E. D., Iwamoto F., Nagamine Y., Akman S. A., and Vaughn J. P. (2008) G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 283, 34626–34634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fullam A., and Schröder M. (2013) DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication. Biochim. Biophys. Acta 1829, 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A., Chen J. J., Anderson P., and Kaufman R. J. (2005) Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280, 16925–16933 [DOI] [PubMed] [Google Scholar]
- 44. Emara M. M., Fujimura K., Sciaranghella D., Ivanova V., Ivanov P., and Anderson P. (2012) Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 423, 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fleming A. M., and Burrows C. J. (2013) G-quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA. Chem. Res. Toxicol. 26, 593–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ming X., Matter B., Song M., Veliath E., Shanley R., Jones R., and Tretyakova N. (2014) Mapping structurally defined guanine oxidation products along DNA duplexes: influence of local sequence context and endogenous cytosine methylation. J Am. Chem. Soc. 136, 4223–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fleming A. M., Zhou J., Wallace S. S., and Burrows C. J. (2015) A role for the fifth G-track in G-quadruplex-forming oncogene promoter sequences during oxidative stress: do these “spare tires” have an evolved function? ACS Cent. Sci. 1, 226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Phillips L. L. (1956) Effect of free radicals on chromosomes of barley. Science 124, 889–890 [DOI] [PubMed] [Google Scholar]
- 49. Marnett L. J. (2000) Oxyradicals and DNA damage. Carcinogenesis 21, 361–370 [DOI] [PubMed] [Google Scholar]
- 50. Hussain S. P., Hofseth L. J., and Harris C. C. (2003) Radical causes of cancer. Nat. Rev. Cancer 3, 276–285 [DOI] [PubMed] [Google Scholar]
- 51. Biffi G., Di Antonio M., Tannahill D., and Balasubramanian S. (2014) Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 6, 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bharti S. K., Sommers J. A., Zhou J., Kaplan D. L., Spelbrink J. N., Mergny J. L., and Brosh R. M. Jr. (2014) DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative Twinkle helicase. J. Biol. Chem. 289, 29975–29993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. von Zglinicki T. (2002) Oxidative stress shortens telomeres. Trends Biochem. Sci. 27, 339–344 [DOI] [PubMed] [Google Scholar]
- 54. Belmokhtar C. A., Hillion J., and Ségal-Bendirdjian E. (2001) Staurosporine induces apoptosis through both caspase-dependent and caspase-independent mechanisms. Oncogene 20, 3354–3362 [DOI] [PubMed] [Google Scholar]
- 55. Criddle D. N., Gillies S., Baumgartner-Wilson H. K., Jaffar M., Chinje E. C., Passmore S., Chvanov M., Barrow S., Gerasimenko O. V., Tepikin A. V., Sutton R., and Petersen O. H. (2006) Menadione-induced reactive oxygen species generation via redox cycling promotes apoptosis of murine pancreatic acinar cells. J. Biol. Chem. 281, 40485–40492 [DOI] [PubMed] [Google Scholar]
- 56. Kawane K., Motani K., and Nagata S. (2014) DNA degradation and its defects. Cold Spring Harb. Perspect. Biol. 6, a016394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lian X. J., and Gallouzi I. E. (2009) Oxidative stress increases the number of stress granules in senescent cells and triggers a rapid decrease in p21waf1/cip1 translation. J. Biol. Chem. 284, 8877–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rhee D. B., Ghosh A., Lu J., Bohr V. A., and Liu Y. (2011) Factors that influence telomeric oxidative base damage and repair by DNA glycosylase OGG1. DNA Repair (Amst.) 10, 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vorlícková M., Tomasko M., Sagi A. J., Bednarova K., and Sagi J. (2012) 8-Oxoguanine in a quadruplex of the human telomere DNA sequence. FEBS J. 279, 29–39 [DOI] [PubMed] [Google Scholar]
- 60. Zhou J., Fleming A. M., Averill A. M., Burrows C. J., and Wallace S. S. (2015) The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 43, 4039–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. An N., Fleming A. M., and Burrows C. J. (2016) Human telomere G-quadruplexes with five repeats accommodate 8-oxo-7,8-dihydroguanine by looping out the DNA damage. ACS Chem. Biol. 11, 500–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Capra J. A., Paeschke K., Singh M., and Zakian V. A. (2010) G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 6, e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Seimiya H., Sawada H., Muramatsu Y., Shimizu M., Ohko K., Yamane K., and Tsuruo T. (2000) Involvement of 14–3-3 proteins in nuclear localization of telomerase. EMBO J. 19, 2652–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Armbruster B. N., Banik S. S., Guo C., Smith A. C., and Counter C. M. (2001) N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol. 21, 7775–7786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Haendeler J., Hoffmann J., Brandes R. P., Zeiher A. M., and Dimmeler S. (2003) Hydrogen peroxide triggers nuclear export of telomerase reverse transcriptase via Src kinase family-dependent phosphorylation of tyrosine 707. Mol. Cell. Biol. 23, 4598–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Haendeler J., Hoffmann J., Diehl J. F., Vasa M., Spyridopoulos I., Zeiher A. M., and Dimmeler S. (2004) Antioxidants inhibit nuclear export of telomerase reverse transcriptase and delay replicative senescence of endothelial cells. Circ. Res. 94, 768–775 [DOI] [PubMed] [Google Scholar]
- 67. Bates P. J., Laber D. A., Miller D. M., Thomas S. D., and Trent J. O. (2009) Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 86, 151–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jing N., Zhu Q., Yuan P., Li Y., Mao L., and Tweardy D. J. (2006) Targeting signal transducer and activator of transcription 3 with G-quartet oligonucleotides: a potential novel therapy for head and neck cancer. Mol. Cancer Ther. 5, 279–286 [DOI] [PubMed] [Google Scholar]
- 69. Qi H., Lin C. P., Fu X., Wood L. M., Liu A. A., Tsai Y. C., Chen Y., Barbieri C. M., Pilch D. S., and Liu L. F. (2006) G-quadruplexes induce apoptosis in tumor cells. Cancer Res. 66, 11808–11816 [DOI] [PubMed] [Google Scholar]
- 70. Sedoris K. C., Thomas S. D., Clarkson C. R., Muench D., Islam A., Singh R., and Miller D. M. (2012) Genomic c-Myc quadruplex DNA selectively kills leukemia. Mol. Cancer Ther. 11, 66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Collie G. W., and Parkinson G. N. (2011) The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 40, 5867–5892 [DOI] [PubMed] [Google Scholar]
- 72. Islam M. A., Thomas S. D., Murty V. V., Sedoris K. J., and Miller D. M. (2014) c-Myc quadruplex-forming sequence Pu-27 induces extensive damage in both telomeric and nontelomeric regions of DNA. J. Biol. Chem. 289, 8521–8531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rosenberg J. E., Bambury R. M., Van Allen E. M., Drabkin H. A., Lara P. N. Jr., Harzstark A. L., Wagle N., Figlin R. A., Smith G. W., Garraway L. A., Choueiri T., Erlandsson F., and Laber D. A. (2014) A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest. New Drugs 32, 178–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Balasubramanian S., Hurley L. H., and Neidle S. (2011) Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 10, 261–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gray L. T., Vallur A. C., Eddy J., and Maizels N. (2014) G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol. 10, 313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xiao S., Zhang J. Y., Wu J., Wu R. Y., Xia Y., Zheng K. W., Hao Y. H., Zhou X., and Tan Z. (2014) Formation of DNA:RNA hybrid G-quadruplexes of two G-quartet layers in transcription: expansion of the prevalence and diversity of G-quadruplexes in genomes. Angew. Chem. Int. Ed. Engl. 53, 13110–13114 [DOI] [PubMed] [Google Scholar]
- 77. Tuppen H. A., Blakely E. L., Turnbull D. M., and Taylor R. W. (2010) Mitochondrial DNA mutations and human disease. Biochim. Biophys. Acta 1797, 113–128 [DOI] [PubMed] [Google Scholar]
- 78. Dong D. W., Pereira F., Barrett S. P., Kolesar J. E., Cao K., Damas J., Yatsunyk L. A., Johnson F. B., and Kaufman B. A. (2014) Association of G-quadruplex forming sequences with human mtDNA deletion breakpoints. BMC Genomics 15, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Oliveira P. H., da Silva C. L., and Cabral J. M. (2013) An appraisal of human mitochondrial DNA instability: new insights into the role of non-canonical DNA structures and sequence motifs. PLoS One 8, e59907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. González V., and Hurley L. H. (2010) The c-MYC NHE III(1): function and regulation. Annu. Rev. Pharmacol. Toxicol. 50, 111–129 [DOI] [PubMed] [Google Scholar]
- 81. Huppert J. L., and Balasubramanian S. (2007) G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 35, 406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chambers V. S., Marsico G., Boutell J. M., Di Antonio M., Smith G. P., and Balasubramanian S. (2015) High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 33, 877–881 [DOI] [PubMed] [Google Scholar]
- 83. Hewitt G., Jurk D., Marques F. D., Correia-Melo C., Hardy T., Gackowska A., Anderson R., Taschuk M., Mann J., and Passos J. F. (2012) Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat. Commun. 3, 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Oikawa S., and Kawanishi S. (1999) Site-specific DNA damage at GGG sequence by oxidative stress may accelerate telomere shortening. FEBS Lett. 453, 365–368 [DOI] [PubMed] [Google Scholar]
- 85. Wang Z., Rhee D. B., Lu J., Bohr C. T., Zhou F., Vallabhaneni H., de Souza-Pinto N. C., and Liu Y. (2010) Characterization of oxidative guanine damage and repair in mammalian telomeres. PLoS Genet. 6, e1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. von Zglinicki T., Saretzki G., Döcke W., and Lotze C. (1995) Mild hyperoxia shortens telomeres and inhibits proliferation of fibroblasts: a model for senescence? Exp. Cell Res. 220, 186–193 [DOI] [PubMed] [Google Scholar]
- 87. von Zglinicki T., Pilger R., and Sitte N. (2000) Accumulation of single-strand breaks is the major cause of telomere shortening in human fibroblasts. Free Radic. Biol. Med. 28, 64–74 [DOI] [PubMed] [Google Scholar]
- 88. Anderson P., and Kedersha N. (2009) Stress granules. Curr. Biol. 19, R397–R398 [DOI] [PubMed] [Google Scholar]
- 89. Shih J. W., Wang W. T., Tsai T. Y., Kuo C. Y., Li H. K., and Wu Lee Y. H. (2012) Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. J. 441, 119–129 [DOI] [PubMed] [Google Scholar]
- 90. Shih J. W., and Lee Y. H. (2014) Human DExD/H RNA helicases: emerging roles in stress survival regulation. Clin. Chim. Acta 436, 45–58 [DOI] [PubMed] [Google Scholar]
- 91. Arimoto-Matsuzaki K., Saito H., and Takekawa M. (2016) TIA1 oxidation inhibits stress granule assembly and sensitizes cells to stress-induced apoptosis. Nat. Commun. 7, 10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Thedieck K., Holzwarth B., Prentzell M. T., Boehlke C., Kläsener K., Ruf S., Sonntag A. G., Maerz L., Grellscheid S. N., Kremmer E., Nitschke R., Kuehn E. W., Jonker J. W., Groen A. K., Reth M., et al. (2013) Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 154, 859–874 [DOI] [PubMed] [Google Scholar]
- 93. Takahashi M., Higuchi M., Matsuki H., Yoshita M., Ohsawa T., Oie M., and Fujii M. (2013) Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol. Cell. Biol. 33, 815–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lai J. C., Ponti S., Pan D., Kohler H., Skoda R. C., Matthias P., and Nagamine Y. (2012) The DEAH-box helicase RHAU is an essential gene and critical for mouse hematopoiesis. Blood 119, 4291–4300 [DOI] [PubMed] [Google Scholar]
- 95. Zhang Z., Yuan B., Bao M., Lu N., Kim T., and Liu Y. J. (2011) The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Huang W., Smaldino P. J., Zhang Q., Miller L. D., Cao P., Stadelman K., Wan M., Giri B., Lei M., Nagamine Y., Vaughn J. P., Akman S. A., and Sui G. (2012) Yin Yang 1 contains G-quadruplex structures in its promoter and 5′-UTR and its expression is modulated by G4 resolvase 1. Nucleic Acids Res. 40, 1033–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kim H. N., Lee J. H., Bae S. C., Ryoo H. M., Kim H. H., Ha H., and Lee Z. H. (2011) Histone deacetylase inhibitor MS-275 stimulates bone formation in part by enhancing Dhx36-mediated TNAP transcription. J. Bone Miner. Res. 26, 2161–2173 [DOI] [PubMed] [Google Scholar]
- 98. Johnson J. E., Cao K., Ryvkin P., Wang L. S., and Johnson F. B. (2010) Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 38, 1114–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Nguyen G. H., Tang W., Robles A. I., Beyer R. P., Gray L. T., Welsh J. A., Schetter A. J., Kumamoto K., Wang X. W., Hickson I. D., Maizels N., Monnat R. J. Jr., and Harris C. C. (2014) Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc. Natl. Acad. Sci. U.S.A. 111, 9905–9910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Rhodes D., and Lipps H. J. (2015) G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 43, 8627–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Maizels N. (2015) G4-associated human diseases. EMBO Rep. 16, 910–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Vatovec S., Kovanda A., and Rogelj B. (2014) Unconventional features of C9ORF72 expanded repeat in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Neurobiol. Aging 35, 2421. [DOI] [PubMed] [Google Scholar]
- 103. Morris P. D., Tackett A. J., Babb K., Nanduri B., Chick C., Scott J., and Raney K. D. (2001) Evidence for a functional monomeric form of the bacteriophage T4 DdA helicase: Dda does not form stable oligomeric structures. J. Biol. Chem. 276, 19691–19698 [DOI] [PubMed] [Google Scholar]
- 104. Hatzakis E., Okamoto K., and Yang D. (2010) Thermodynamic stability and folding kinetics of the major G-quadruplex and its loop isomers formed in the nuclease hypersensitive element in the human c-Myc promoter: effect of loops and flanking segments on the stability of parallel-stranded intramolecular G-quadruplexes. Biochemistry 49, 9152–9160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Byrd A. K., and Raney K. D. (2015) A parallel quadruplex DNA is bound tightly but unfolded slowly by pif1 helicase. J. Biol. Chem. 290, 6482–6494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cawthon R. M. (2002) Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Byrum S. D., Raman A., Taverna S. D., and Tackett A. J. (2012) ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Rep. 2, 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Byrum S. D., Taverna S. D., and Tackett A. J. (2013) Purification of a specific native genomic locus for proteomic analysis. Nucleic Acids Res. 41, e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zybailov B., Mosley A. L., Sardiu M. E., Coleman M. K., Florens L., and Washburn M. P. (2006) Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 [DOI] [PubMed] [Google Scholar]
- 110. Zybailov B., Friso G., Kim J., Rudella A., Rodríguez V. R., Asakura Y., Sun Q., and van Wijk K. J. (2009) Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol. Cell. Proteomics 8, 1789–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Chen Q. M., Tu V. C., Catania J., Burton M., Toussaint O., and Dilley T. (2000) Involvement of Rb family proteins, focal adhesion proteins and protein synthesis in senescent morphogenesis induced by hydrogen peroxide. J. Cell Sci. 113, 4087–4097 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.