Abstract
Langerhans cells (LC) represent a specialized subset of evolutionarily conserved dendritic cells (DC) that populate stratified epithelial tissues, which are essential for the induction of skin and mucosal immunity and tolerance, including allergy. TGF-β1 has been confirmed to be a predominant factor involved in LC development. Despite great advances in the understanding of LC ontogeny and diverse replenishment patterns, the underlying molecular mechanisms remain elusive. This review focuses on the recent discoveries in TGF-β1-mediated LC development and maintenance, with special attention to the involved transcription factors and related regulators.
Keywords: Langerhans cells, development, TGF-β1, transcription factors
INTRODUCTION
Dendritic cells (DC) are arguably the most potent antigen-presenting cells, capable of initiating adaptive immune responses. Located within epidermis and mucosae, Langerhans cells (LC) represent a unique subset of evolutionarily conserved DCs, which play an essential role in cutaneous and mucosal immunity and tolerance, including allergy (1, 2). Unlike bone marrow (BM)-derived conventional DCs (cDC), adult mouse LCs mainly stem from embryonic fetal liver monocytes with a minor contribution from yolk sac (YS)-derived macrophages (3, 4). Likewise, the heterogeneous human LC progenitors appear at 7 weeks estimated gestational age when hematopoiesis is still inactive in the BM (5, 6). After the initial establishment of skin LC pool, the LC replenishment patterns are differentially referred as “steady state” and “inflamed state”. Under the steady state, epidermal LCs self-renew at an extremely low speed by scattered in situ proliferative precursors without any influx of circulating precursors (7); when the local inflammation induces a loss of epidermal LCs, inflamed-state LCs including short-term” LCs, which develop from circulating Gr-1hi monocytes, and BM-derived “long-term” LCs would transiently or stably reconstitute the LC compartment, respectively (8).
Transforming growth factor-β1 (TGF-β1) is a crucial factor for LC development and maintenance. The presence of TGF-β1 is a prerequisite for in vitro LC differentiation from various sources. For examples, human CD34+ hematopoietic progenitor cells (HPC) expanded and developed into LCs in a stringent TGF-β1-dependent manner under serum-free conditions (9); TGF-β1 was indispensable for human CD14+ blood monocyte-derived DCs, which were induced by granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin-4 (IL-4), to further differentiate into LCs (10); and human dermal CD14+ cells maximally acquired LC features when cultured with TGF-β1 alone (11). Moreover, in vivo studies demonstrated that both TGF-β1 and TGF-β receptor II (TGFβRII) null mice exhibited a profound LC loss (12, 13). In DC-specific TGFβRI knockout (KO) mice (CD11c-Cre.TGFβRIfl/fl) mice, LCs gradually disappeared within the first week after birth with a spontaneous mature and migratory phenotype, suggesting that TGF-β1 is also crucial for maintaining the epidermal pool of immature LCs (14). Unlike macrophage-colony stimulating factor receptor (M-CSFR), which affects both steady-state and inflamed-state LC replenishment, the deletion of TGF-β1 exerted no impairment on the BM’s potential to generate LCs in vivo (15). Lack of bone morphogenetic protein 7 (BMP7), another TGF-β/BMP superfamily member, also substantially diminished LC numbers in mice (16).
Despite recent great advances in the understanding of LC development and homeostasis mediated by TGF-β1, the underpinning mechanisms remain elusive. Canonically, TGF-β1 activates a Smad signaling pathway. However, we have found that Smad3 (17), Smad2 and Smad4 (unpublished data) are not required for LC differentiation, indicating that TGF-β1-mediated non-Smad pathways are involved in the development of LCs. This review will mainly focus on the transcription factors and related regulators engaged in TGF-β1-related LC development and maintenance.
Transcription regulation in LC development and maintenance
ID2
The inhibitors of DNA-binding (ID) proteins bind E proteins, which constitute a family of class I basic helix loop helix transcription factors, to prevent target gene regulation. As a major member, ID2 expression is induced by TGF-β1 in the human cord blood CD34+ HPCs undergoing LC commitment (18). ID2 null mice essentially lack epidermal LCs and Langerin+CD103+ dermal DCs, with a reduced splenic CD8α+ DC subset (19, 20). Although ID2 is indispensable for steady-state LCs, its role during inflammation remains variant. Recently, Seré et al (8) discovered that “short-term” inflammatory LCs were ID2-independent, whereas “long-term” counterparts stringently depended on ID2. In contrast, Chopin et al (21) claimed that the repopulation of injured epidermis with BM-derived LCs was generally ID2-independent. This divergence may result from the different experimental setup. Seré et al (8) used conventional ID2 null mice and their recipients were immunodeficient mice, whereas Chopin et al (21) applied DC-specific ID2 KO (CD11c-Cre.ID2fl/fl) mice and their hosts were wild-type mice.
RUNX3
The RUNX3 belongs to the runt domain transcription factors which are determinants of lineage-specific gene expressions in the major developmental processes. In RUNX3 null mice, the epidermal LCs were absent whereas splenic cDCs displayed accelerated maturation along with increased efficacy in T cell priming (22). Furthermore, the mutant Runx3 protein, which lacked the C-terminal VWRPY motif required for Runx3 interaction with the corepressor Groucho/transducin-like Enhancer-of-split (TLE), had normally developed LCs but failed to prevent their spontaneous maturation, implying that the ability of Runx3 to tether Groucho/TLE was required for preserving the immature status of epidermal LCs other than their development (23). In addition, the LC defect in RUNX3 null mice was irrelevant to ID2 expression (22). Recently, Chopin et al (21) uncovered that TGF-β1-inducible transcription factor PU.1 directly upregulated RUNX3 during in vitro BM culture, and ectopic expression of RUNX3 could rescue LC differentiation in the absence of PU.1 and promote LC generation from PU.1-sufficient progenitors. In general, RUNX3 is crucial in mediating LC development as well as restraining LC maturation, presumably under the control of a TGF-β1-PU.1-RUNX3 transcription axle.
IRF8
The interferon regulatory family (IRF) plays a major role in myeloid cell commitment. IRF8, a key member of the IRF family, is indispensable for driving DC commitment and blocking alternative myeloid lineage potential (24). IRF8 was lowly expressed in epidermal LCs but its expression was upregulated upon migration to draining lymph nodes (LN) (21). Schiavoni et al (25) first reported that deletion of IRF8 reduced LC ratio, suppressed cutaneous DC migration and impaired contact hypersensitivity (CHS). However, IRF8 was recently found to be redundant for LC homeostasis and motility in both IRF8 null and DC-specific IRF8 KO (CD11c-Cre. IRF8fl/fl) mice (21). Despite the unsolved discrepancy, IRF8 is very likely to be involved in LC development, given that IRF8 could activate TGF-β1 signaling (26) and PU.1 directly regulates its expression through chromatin remodeling (27).
IRF2
IRF2 is another member of the IRF family. IRF2 null mice exhibited a selective cell-autonomous deficiency in the CD4+ DC subset, including epidermal CD4+ LCs and splenic CD4+CD11b+ DCs (28). These DC abnormalities diminished in the mice that lacking both IRF-2 and the interferon (IFN)-α/β receptor, suggesting that IRF-2 acted through negatively regulating IFN-α/β signals during DC development (28). Moreover, IRF2 might cooperate with IRF8 or with both IRF8 and PU.1 to co-regulate gene expressions in LC development (29).
β-catenin
β-catenin is a transcriptional regulator of the Wnt signaling pathway. Typically, β-catenin forms tight connections with the intracellular tail of cadherins such as E-cadherin, which profoundly affects LC migration and maturation (30, 31). Among all the skin DC subsets, β-catenin is uniquely expressed by epidermal LCs (32). TGF-β1 upregulated β-catenin expression in the human CD34+ HPC-derived LCs, and ectopic β-catenin expression in the HPCs further heightened LC commitment. Vitamin D, another epidermal signal, enhanced TGF-β1-mediated β-catenin induction, while a truncated vitamin D receptor (VDR) would diminish the positive effects of ectopic β-catenin on LC differentiation, demonstrating that β-catenin is a positive regulator of LC differentiation in response to TGF-β1 through a functional interaction with VDR (32). Moreover, an early research has proposed that ID2 might be a potential downstream target of β-catenin during in vitro HPC culture (31). Nevertheless, it is still unclear if E-cadherin is involved in β-catenin or TGF-β1/β-catenin interaction mediated LC development.
PU.1
PU.1, belonging to the ETS-domain family of transcription factors, is generally upregulated among the myeloid and lymphoid lineages with a repressive role in granulopoiesis. An early study reported that TGF-β1 upregulated PU.1 expression in BM-derived DCs (BMDC), and ectopic expression of PU.1 enhanced TGF-β1-mediated LC differentiation without altering LC maturation (18). However, neither PU.1 nor Id2 alone, nor both together, could substitute for TGF-β1 in the in vitro BM culture, suggesting that these transcription factors only favor LC development within the proper microenvironment (18). Additionally, DC-specific PU.1 KO (CD11c-Cre.PU.1fl/fl) mice present a severe defect in LC development (21). PU.1 probably controls LC differentiation by directly regulating the aforementioned RUNX3, and the physical binding of PU.1 with the latter depends on the presence of TGF-β1, suggesting that TGF-β1 is a prerequisite for PU.1-mediated LC development (21). However, unlike TGF-β1, PU.1 deficiency not only affects steady-state LCs, but also disturbs LC repopulation under inflammatory conditions, indicating that PU.1 is not an exclusive mediator of TGF-β1 signaling during LC differentiation and homeostasis (21). Indeed, PU.1 directs M-CSFR expression under various conditions (33, 34). Thus, it is possible that PU.1 might mediate its role in inflammatory-state LC repopulation through the regulation of M-CSFR signaling.
STAT5
The signal transducer and activator of transcription 5 (STAT5) plays an important role in hematopoiesis, especially the development of multiple myeloid lineages. Dominant-negative STAT5 expression in the human CD34+ HPCs produced a loss of pre-interstitial DCs (35). However, TGF-β1-induced inhibition of STAT5 activity and subsequent upregulation of PU.1 are required for the initiation of LC commitment, although the terminal differentiation of already committed pre-LCs demands a higher level of STAT5 (35). Hence, STAT5 appears to dynamically regulate LC development.
AHR
The aryl hydrocarbon receptor (AhR) belongs to the basic Helix-Loop-Helix/Per-Arnt-Sim family of transcriptional regulators, which is known to mediate the toxic effects of pollutants as well as modulate cell differentiation. Using human CD34+ HPCs, all analyzed AhR ligands (including β-naphtoflavone, tetrachlorodibenzo-p-dioxin, and VAF347) demonstrated similar efficacy in hindering monocyte and LC generation without affecting other DC subsets (36). Furthermore, AhR activation reduced PU.1 expression, and ectopic PU.1 expression could overcome the impairment of AhR on LC and monocyte development (36). However, epidermal LC frequency was not affected in AhR null mice (37). Instead, AhR-deficient LCs displayed compromised maturation and enhanced phagocytic capacity.
C/EBP
The CCAAT/Enhancer Binding Protein (C/EBP) comprises a family of transcription factors with a basic region-leucine zipper (bZIP) structure. C/EBPα, C/EBPβ, C/EBPδ and C/EBPε share a transcriptional activation domain and collaborate with PU.1 to regulate a variety of myeloid-specific genes. Dominant-negative C/EBP switched myeloid cell fate from granulocytes/macrophages to LCs, while wild-type C/EBP would completely block TNFα-dependent LC development (38). Furthermore, co-expressed C/EBP would abrogate PU.1-induced LC differentiation from human CD34+ HPCs (38). Therefore, counter-regulation between C/EBP and PU.1 might be engaged in LC development.
Other regulators involved in TGF-β1-mediated LC differentiation
p14
The p14 adaptor molecule belongs to the lysosomal adaptor and mitogen-activated protein kinases (MAPK) and mTOR activator/regulator (LAMTOR) complex, thereby contributing to the signaling pathways of “extracellular signaling-regulated kinase” (ERK) and mTOR cascade. DC-specific p14 deficiency caused an almost complete loss of LCs, and the few remaining LCs exhibited spontaneous maturation (39). Furthermore, the LC-specific p14 KO mice (Langerin-Cre.p14fl/fl) totally lacked LCs and displayed reduced CHS to the contact sensitizer 2,4,6-trinitrochlorobenzene (TNCB) (40). Intriguingly, they observed a transient recruitment of MHCII+ cells via hair follicles after the application of TNCB, and believed that these MHCII+ cells were “short-term” LCs (39). Thus, loss of p14 mainly impairs normal LC development and maturation status other than LC repopulation during inflamed state, which resemble the phenotype of TGF-β1 deficiency. Indeed, p14 deficiency in DCs/LCs interfered with TGF-β1 pathway, by reducing TGF-βRII expression on BMDCs and LCs, as well as impeding the surface binding of TGF-β1 on BMDCs (40). Over all, p14 controls LC differentiation through direct influence on TGF-β1 signaling.
mTOR1
The mammalian target of rapamycin (mTOR) is a conserved serine/threonine kinase constituted in two different multiprotein complexes termed mTORC1 and mTORC2, with Raptor and Rictor being their essential components, respectively (41). Kellersch et al (42) recently reported that mTORC1 but not mTORC2 is required for epidermal LC homeostasis. Approximately a10-fold decrease of epidermal LCs with enhanced migration, faster turnover rate and increased apoptosis was observed in DC-specific Raptor KO (CD11c-Cre.Raptorfl/fl) mice (42). The LCs lacking Raptor displayed decreased expression of Langerin, E-cadherin, β-catenin and CCR7 but normal levels of MHC-II, which did not support a spontaneous mature phenotype. A previous study reported that the translational regulation of TGF-β1 production in DCs required mTOR activity (43). Thus, mTORC1 is very likely to be involved in TGF-β1-mediated LC differentiation.
SIRPα
The signal regulatory protein α (SIRPα) is an immunoglobulin superfamily protein that is predominantly expressed by DCs. The SIRPα mutant mice exhibited a markedly decrease of epidermal LC frequency together with its phagocytic and migratory dysfunction (44). Furthermore, the mRNA expression of TGFβRII in LCs of SIRPα mutant mice was markedly decreased compared with that of WT mice. Thus, SIRPα probably regulates LC development and homeostasis through the regulation of TGFβRII mRNA expression.
Axl
Axl belongs to the TAM (Tyro3, Axl, and Mer) receptor tyrosine kinase family, which plays an essential role in clearing apoptotic cells and suppressing DC inflammatory responses in innate immunity (45). Axl is highly expressed in monocyte-derived LCs both in vivo and in vitro, and Axl-positive human CD34+ HPCs showed an enrichment of LC differentiation potential (46). Besides, Axl expression is induced by TGF-β1 during LC differentiation. TAM triple-deficient mice, instead of Axl, Mer or Tysro3 single-deficient mice, had a substantial reduction in epidermal LC frequency, suggesting that there might be redundancy within the TAM system. Overall, Axl serves as a downstream effector of TGF-β1 during LC development.
MicroRNAs
MicroRNAs (miRNA), a relatively new class of small non-coding RNAs, play pivotal roles in the regulation of gene expression at a post transcriptional level. MiRNAs bind to the 3′ untranslated region or coding sequence of target mRNAs, triggering translational repression or mRNA cleavage (47, 48). Accumulated studies indicated that miRNAs are involved in immune cell development and function (47, 49, 50). DC-specific deletion of Dicer, the enzyme processing mature miRNAs, caused a 12-fold reduction of epidermal LCs with increased apoptosis and compromised maturation (51). Notably, Dicer-deficient LCs expressed considerably decreased levels of TGF-βRII, indicating that miRNAs might influence LC biology via TGF-β1 signaling (51). Indeed, aged epidermal LCs with reduced frequency and altered function expressed a distinct set of miRNAs that putatively target LC-related TGF-β1 signaling (52). Nonetheless, the individual miRNA(s) in charge of LC homeostasis is yet undetermined. Although high miR-146a expression in the human CD34+ HPC-derived LCs was induced by PU.1 in response to TGF-β1, loss of miR-146a did not affect LC differentiation (53). Using specific miRNA KO mice, we have identified that miR-150, miR-223 and miR-17-92 cluster were not required for LC development and maintenance regardless of their substantial expressions in LCs (50, 54, 55). However, miR-150 and miR-223 regulated LC-mediated CD8+ T cell activation and cytokine production (50, 54).
Conclusions and perspective
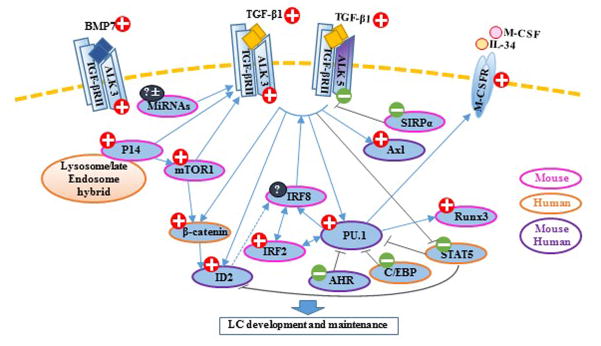
With the advance in the mice with gene mutations and a tracing system in vivo, some facets of LC development and maintenance have been revealed in spite of its complexity. Involved transcription factors of TGF-β1-mediated LC development and maintenance are summarized in the Table 1, and the plausible regulatory network of TGF-β1-mediated LC differentiation is depicted in Figure 1. As shown in the figure, PU.1 appears to be the core transcription factor downstream of LC-related TGF-β1 signaling pathways, and multiple aforementioned transcriptional regulators might exert their own functions though the interactions with PU.1.
Table 1.
Transcription factors in LC development and maintenance
| Transcription Factor | Transcription Factor Family | Human (H)/Mouse (M) | Steady-state LCs | Inflamed-state LCs | Phenotype | Ref |
|---|---|---|---|---|---|---|
| ID2 | Inhibitor of DNA binding family protein containing HLH domains | H, M | ✓ | Controversial results | ID2 null mice lack LCs and Langerin+ DCs; its role in LC repopulation during inflammatory-state remains variant. | (8, 19–21) |
| RUNX3 | RUNT domain family of transcription factors | M | ✓ | Unknown | In RUNX3 null mice, epidermal LCs are absent; ectopic expression of RUNX3 could rescue LC differentiation in the absence of PU.1 during in vitro BM culture. | (21, 22) |
| IRF8 (ICSBP) | Interferon-regulatory factor; interferon consensus sequence-binding protein | M | Controversial results | Unknown | IRF8 null mice had decreased LC ratio, suppressed cutaneous DC migration, and impaired CHS response. Controversial results. | (21, 25) |
| IRF2 | Interferon-regulatory factor; interferon consensus sequence-binding protein | M | ✓ | Unknown | IRF2 null mice exhibited a selective cell autonomous deficiency in epidermal CD4+ LCs and splenic CD4+CD11b+ DCs. | (28) |
| PU.1 (SFPI1, SPI1) | ETS-domain transcription factor; binds to PU box sequences | H,M | ✓ | ✓ | PU.1 deficiency affects both steady-state LCs and BM-mediated LC repopulation in skin under inflammatory conditions. | (21) |
| STAT5 | Signal transducer and activator of transcription | H | ✓ | Unknown | TGF-β1-induced inhibition of STAT5 activity is required for initial LC commitment, while terminal differentiation of already committed pre-LCs calls for a higher level of STAT5. | (35) |
| C/EBP | CCAAT/Enhancer Binding Protein (C/EBP) | H | ✓ | Unknown | Dominant-negative C/EBP switched myeloid cell fate from granulocytes/macrophages to LCs. Co-expressed C/EBP would abrogate PU.1-induced LC differentiation from human CD34+ HPCs. | (38) |
LC, Langerhans cell; DC, dendritic cell; CHS, contact hypersensitivity; BM, bone marrow; TGF-β1, transforming growth factor-β1; HPC, hematopoietic progenitor cells.
Figure 1. TGF-β1-mediated LC differentiation.
A number of transcription factors and regulators form a network in TGF-β1-regulated LC development and maintenance, in which transcription factor PU.1 is a core factor. Red plus indicates a promoting role in LC differentiation, and green minus indicates an inhibitive role. Solid blue arrows indicate positive stimulatory functions that have been proven or suggested by previous reports. Dotted arrows indicate hypothesized relationships. Dark-grey lines indicate negative regulation. Peachblow rims indicate that the results derive from mouse experiments. Orange rims indicate that the results derive from human experiments. Purple rims indicate that the results derived from both mouse and human experiments.
Several questions related to LC development and function need further investigation. First, what are the downstream targets of TGF-β1-induced transcription factors involved in LC development and homeostasis? Second, what are the specific extrinsic and intrinsic triggers of various LC replenishment patterns? Third, given that LCs are capable of mediating immune defense or tolerance in a context-dependent manner, is it possible that “steady-state” and “inflamed-state” LCs, which are regulated by distinct transcription factors, differ in their functions and have a preference for inflammation or tolerance? The precise answers of these questions might bring promising solutions to the different ends of skin and mucosal disease spectrum, including infection, allergy, as well as cancer and autoimmune disease.
Acknowledgments
The authors thank Xiaofan Mi and Matthew Weiland for their critical reading. This study is supported by in part by a grant from National Institutes of Health Grant R21AR059976 and RO1AR069681 (QSM), and the Henry Ford Health System Research Grant for the Immunology Program T71016 (QSM) and T71017 (LZ).
Footnotes
DISCLOSURE
The authors declared no conflict of interest.
References
- 1.Igyarto BZ, Kaplan DH. Antigen presentation by Langerhans cells. Curr Opin Immunol. 2013;25:115–119. doi: 10.1016/j.coi.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romani N, Clausen BE, Stoitzner P. Langerhans cells and more: langerin-expressing dendritic cell subsets in the skin. Immunol Rev. 2010;234:120–141. doi: 10.1111/j.0105-2896.2009.00886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoeffel G, Wang Y, Greter M, et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med. 2012;209:1167–1181. doi: 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomez Perdiguero E, Klapproth K, Schulz C, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuster C, Mildner M, Mairhofer M, et al. Human embryonic epidermis contains a diverse Langerhans cell precursor pool. Development. 2014;141:807–815. doi: 10.1242/dev.102699. [DOI] [PubMed] [Google Scholar]
- 6.Tavian M, Peault B. Embryonic development of the human hematopoietic system. Int J Dev Biol. 2005;49:243–250. doi: 10.1387/ijdb.041957mt. [DOI] [PubMed] [Google Scholar]
- 7.Ghigo C, Mondor I, Jorquera A, et al. Multicolor fate mapping of Langerhans cell homeostasis. J Exp Med. 2013;210:1657–1664. doi: 10.1084/jem.20130403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sere K, Baek JH, Ober-Blobaum J, et al. Two distinct types of Langerhans cells populate the skin during steady state and inflammation. Immunity. 2012;37:905–916. doi: 10.1016/j.immuni.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 9.Strobl H, Bello-Fernandez C, Riedl E, et al. flt3 ligand in cooperation with transforming growth factor-beta1 potentiates in vitro development of Langerhans-type dendritic cells and allows single-cell dendritic cell cluster formation under serum-free conditions. Blood. 1997;90:1425–1434. [PubMed] [Google Scholar]
- 10.Geissmann F, Prost C, Monnet JP, Dy M, Brousse N, Hermine O. Transforming growth factor beta1, in the presence of granulocyte/macrophage colony-stimulating factor and interleukin 4, induces differentiation of human peripheral blood monocytes into dendritic Langerhans cells. J Exp Med. 1998;187:961–966. doi: 10.1084/jem.187.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larregina AT, Morelli AE, Spencer LA, et al. Dermal-resident CD14+ cells differentiate into Langerhans cells. Nat Immunol. 2001;2:1151–1158. doi: 10.1038/ni731. [DOI] [PubMed] [Google Scholar]
- 12.Borkowski TA, Letterio JJ, Farr AG, Udey MC. A role for endogenous transforming growth factor beta 1 in Langerhans cell biology: the skin of transforming growth factor beta 1 null mice is devoid of epidermal Langerhans cells. J Exp Med. 1996;184:2417–2422. doi: 10.1084/jem.184.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan DH, Li MO, Jenison MC, Shlomchik WD, Flavell RA, Shlomchik MJ. Autocrine/paracrine TGFbeta1 is required for the development of epidermal Langerhans cells. J Exp Med. 2007;204:2545–2552. doi: 10.1084/jem.20071401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kel JM, Girard-Madoux MJ, Reizis B, Clausen BE. TGF-beta is required to maintain the pool of immature Langerhans cells in the epidermis. J Immunol. 2010;185:3248–3255. doi: 10.4049/jimmunol.1000981. [DOI] [PubMed] [Google Scholar]
- 15.Borkowski TA, Letterio JJ, Mackall CL, et al. A role for TGFbeta1 in langerhans cell biology. Further characterization of the epidermal Langerhans cell defect in TGFbeta1 null mice. J Clin Invest. 1997;100:575–581. doi: 10.1172/JCI119567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasmin N, Bauer T, Modak M, et al. Identification of bone morphogenetic protein 7 (BMP7) as an instructive factor for human epidermal Langerhans cell differentiation. J Exp Med. 2013;210:2597–2610. doi: 10.1084/jem.20130275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu YP, Shi Y, Cui ZZ, et al. TGFbeta/Smad3 signal pathway is not required for epidermal Langerhans cell development. J Invest Dermatol. 2012;132:2106–2109. doi: 10.1038/jid.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinz LX, Platzer B, Reisner PM, et al. Differential involvement of PU. 1 and Id2 downstream of TGF-beta1 during Langerhans-cell commitment. Blood. 2006;107:1445–1453. doi: 10.1182/blood-2005-04-1721. [DOI] [PubMed] [Google Scholar]
- 19.Hacker C, Kirsch RD, Ju XS, et al. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat Immunol. 2003;4:380–386. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- 20.Ginhoux F, Liu K, Helft J, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chopin M, Seillet C, Chevrier S, et al. Langerhans cells are generated by two distinct PU. 1-dependent transcriptional networks. J Exp Med. 2013;210:2967–2980. doi: 10.1084/jem.20130930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fainaru O, Woolf E, Lotem J, et al. Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004;23:969–979. doi: 10.1038/sj.emboj.7600085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yarmus M, Woolf E, Bernstein Y, et al. Groucho/transducin-like Enhancer-of-split (TLE)-dependent and -independent transcriptional regulation by Runx3. Proc Natl Acad Sci U S A. 2006;103:7384–7389. doi: 10.1073/pnas.0602470103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura T, Ozato K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J Interferon Cytokine Res. 2002;22:145–152. doi: 10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- 25.Schiavoni G, Mattei F, Borghi P, et al. ICSBP is critically involved in the normal development and trafficking of Langerhans cells and dermal dendritic cells. Blood. 2004;103:2221–2228. doi: 10.1182/blood-2003-09-3007. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida Y, Yoshimi R, Yoshii H, et al. The transcription factor IRF8 activates integrin-mediated TGF-beta signaling and promotes neuroinflammation. Immunity. 2014;40:187–198. doi: 10.1016/j.immuni.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schonheit J, Kuhl C, Gebhardt ML, et al. PU. 1 level-directed chromatin structure remodeling at the Irf8 gene drives dendritic cell commitment. Cell Rep. 2013;3:1617–1628. doi: 10.1016/j.celrep.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Ichikawa E, Hida S, Omatsu Y, et al. Defective development of splenic and epidermal CD4+ dendritic cells in mice deficient for IFN regulatory factor-2. Proc Natl Acad Sci U S A. 2004;101:3909–3914. doi: 10.1073/pnas.0400610101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W, Horvath E, Eklund EA. PU.1, interferon regulatory factor (IRF) 2, and the interferon consensus sequence-binding protein (ICSBP/IRF8) cooperate to activate NF1 transcription in differentiating myeloid cells. J Biol Chem. 2007;282:6629–6643. doi: 10.1074/jbc.M607760200. [DOI] [PubMed] [Google Scholar]
- 30.Van den Bossche J, Malissen B, Mantovani A, De Baetselier P, Van Ginderachter JA. Regulation and function of the E-cadherin/catenin complex in cells of the monocyte-macrophage lineage and DCs. Blood. 2012;119:1623–1633. doi: 10.1182/blood-2011-10-384289. [DOI] [PubMed] [Google Scholar]
- 31.Jiang A, Bloom O, Ono S, et al. Disruption of E-cadherin-mediated adhesion induces a functionally distinct pathway of dendritic cell maturation. Immunity. 2007;27:610–624. doi: 10.1016/j.immuni.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yasmin N, Konradi S, Eisenwort G, et al. beta-Catenin promotes the differentiation of epidermal Langerhans dendritic cells. J Invest Dermatol. 2013;133:1250–1259. doi: 10.1038/jid.2012.481. [DOI] [PubMed] [Google Scholar]
- 33.Zhang DE, Hetherington CJ, Chen HM, Tenen DG. The macrophage transcription factor PU. 1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol. 1994;14:373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aikawa Y, Katsumoto T, Zhang P, et al. PU.1-mediated upregulation of CSF1R is crucial for leukemia stem cell potential induced by MOZ-TIF2. Nat Med. 2010;16:580–585. doi: 10.1038/nm.2122. 581p following 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van de Laar L, van den Bosch A, Wierenga AT, Janssen HL, Coffer PJ, Woltman AM. Tight control of STAT5 activity determines human CD34-derived interstitial dendritic cell and langerhans cell development. J Immunol. 2011;186:7016–7024. doi: 10.4049/jimmunol.1003977. [DOI] [PubMed] [Google Scholar]
- 36.Platzer B, Richter S, Kneidinger D, Waltenberger D, Woisetschlager M, Strobl H. Aryl hydrocarbon receptor activation inhibits in vitro differentiation of human monocytes and Langerhans dendritic cells. J Immunol. 2009;183:66–74. doi: 10.4049/jimmunol.0802997. [DOI] [PubMed] [Google Scholar]
- 37.Jux B, Kadow S, Esser C. Langerhans cell maturation and contact hypersensitivity are impaired in aryl hydrocarbon receptor-null mice. J Immunol. 2009;182:6709–6717. doi: 10.4049/jimmunol.0713344. [DOI] [PubMed] [Google Scholar]
- 38.Iwama A, Osawa M, Hirasawa R, et al. Reciprocal roles for CCAAT/enhancer binding protein (C/EBP) and PU.1 transcription factors in Langerhans cell commitment. J Exp Med. 2002;195:547–558. doi: 10.1084/jem.20011465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sparber F, Scheffler JM, Amberg N, et al. The late endosomal adaptor molecule p14 (LAMTOR2) represents a novel regulator of Langerhans cell homeostasis. Blood. 2014;123:217–227. doi: 10.1182/blood-2013-08-518555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sparber F, Tripp CH, Komenda K, et al. The Late Endosomal Adaptor Molecule p14 (LAMTOR2) Regulates TGFbeta1-Mediated Homeostasis of Langerhans Cells. J Invest Dermatol. 2015;135:119–129. doi: 10.1038/jid.2014.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kellersch B, Brocker T. Langerhans cell homeostasis in mice is dependent on mTORC1 but not mTORC2 function. Blood. 2013;121:298–307. doi: 10.1182/blood-2012-06-439786. [DOI] [PubMed] [Google Scholar]
- 43.Kushwah R, Wu J, Oliver JR, et al. Uptake of apoptotic DC converts immature DC into tolerogenic DC that induce differentiation of Foxp3+ Treg. Eur J Immunol. 2010;40:1022–1035. doi: 10.1002/eji.200939782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwamura H, Saito Y, Sato-Hashimoto M, et al. Essential roles of SIRPalpha in homeostatic regulation of skin dendritic cells. Immunol Lett. 2011;135:100–107. doi: 10.1016/j.imlet.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen KQ, Tsou WI, Kotenko S, Birge RB. TAM receptors in apoptotic cell clearance, autoimmunity, and cancer. Autoimmunity. 2013;46:294–297. doi: 10.3109/08916934.2013.794515. [DOI] [PubMed] [Google Scholar]
- 46.Bauer T, Zagorska A, Jurkin J, et al. Identification of Axl as a downstream effector of TGF-beta1 during Langerhans cell differentiation and epidermal homeostasis. J Exp Med. 2012;209:2033–2047. doi: 10.1084/jem.20120493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou L, Park JJ, Zheng Q, Dong Z, Mi Q. MicroRNAs are key regulators controlling iNKT and regulatory T-cell development and function. Cell Mol Immunol. 2011;8:380–387. doi: 10.1038/cmi.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiland M, Gao XH, Zhou L, Mi QS. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9:850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 49.Zhou L, Seo KH, He HZ, et al. Tie2cre-induced inactivation of the miRNA-processing enzyme Dicer disrupts invariant NKT cell development. Proc Natl Acad Sci U S A. 2009;106:10266–10271. doi: 10.1073/pnas.0811119106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng Q, Zhou L, Mi QS. MicroRNA miR-150 is involved in Valpha14 invariant NKT cell development and function. J Immunol. 2012;188:2118–2126. doi: 10.4049/jimmunol.1103342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuipers H, Schnorfeil FM, Fehling HJ, Bartels H, Brocker T. Dicer-dependent microRNAs control maturation, function, and maintenance of Langerhans cells in vivo. J Immunol. 2010;185:400–409. doi: 10.4049/jimmunol.0903912. [DOI] [PubMed] [Google Scholar]
- 52.Xu YP, Qi RQ, Chen W, et al. Aging affects epidermal Langerhans cell development and function and alters their miRNA gene expression profile. Aging (Albany NY) 2012;4:742–754. doi: 10.18632/aging.100501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jurkin J, Schichl YM, Koeffel R, et al. miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J Immunol. 2010;184:4955–4965. doi: 10.4049/jimmunol.0903021. [DOI] [PubMed] [Google Scholar]
- 54.Mi QS, Xu YP, Wang H, Qi RQ, Dong Z, Zhou L. Deletion of microRNA miR-223 increases Langerhans cell cross-presentation. Int J Biochem Cell Biol. 2013;45:395–400. doi: 10.1016/j.biocel.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou L, Qi RQ, Liu M, et al. microRNA miR-17-92 cluster is highly expressed in epidermal Langerhans cells but not required for its development. Genes Immun. 2014;15:57–61. doi: 10.1038/gene.2013.61. [DOI] [PubMed] [Google Scholar]