Abstract
Hsp70s use ATP hydrolysis to disrupt protein:protein associations or move macromolecules. One example is Hsc70-mediated disassembly of clathrin coats that form on vesicles during endocytosis. We exploit the exceptional features of these coats to test three models—Brownian ratchet, power-stroke and entropic pulling—proposed to explain how Hsp70s transform their substrates. Our data rule out the ratchet and power-stroke models, and instead support a collision pressure mechanism whereby collisions between clathrin coat walls and Hsc70s drive coats apart. Collision pressure is the complement to the pulling force described in the entropic pulling model. We also find that self-association can augment collision pressure to allow disassembly of clathrin lattices predicted to resist disassembly. These results illuminate how Hsp70s generate the forces that transform their substrates.
Introduction
Hsp70s inhibit formation of non-functional protein complexes and aggregates by sequestering interactive (primarily hydrophobic) segments of both native and misfolded protein substrates1,2. The conformational changes that allow Hsp70s to alternately bind and release their substrates are understood3–7, but don't explain how Hsp70s disassociate aggregates or complexes8–11, or pull proteins into organelles12,13. The latter require that Hsp70s move their substrates (i.e., through translocation pores or away from other proteins). The power-stroke model proposes that during ATP hydrolysis Hsp70 undergoes a conformational change that pulls on its substrate12. The Brownian ratchet model proposes that Hsp70 asymmetrically traps spontaneous fluctuations that move the substrate in a particular direction14,15. Finally, the entropic pulling model16,17 notes that Hsp70 recruitment to its substrate results in an Hsp70 bound to a floppy polypeptide abutting a structural wall (e.g., an aggregate or translocation pore). Movement of Hsp70 away from the wall, dragging the polypeptide with it, is thermodynamically favored because it increases Hsp70 freedom of motion/entropy.
Clathrin uncoating, in which constitutively expressed Hsc70 is recruited to coated vesicles by the J co-chaperone auxilin, and then drives coat disassembly to triskelia (trimers of clathrin heavy- and light-chain dimers)2 provides an exceptional system to study how Hsp70s alter their substrates. Clathrin cage assembly and disassembly in vitro18 can be precisely monitored in both single molecule19,20 and ensemble experiments21. Cage stability can be modulated by pH22. Structures of cages and other reaction components are known3,23–26, and a single Hsc70 binding site at the clathrin heavy chain (CHC) C-terminus mediates disassembly27.
We set out to test models for Hsp70 force generation by exploiting these features. Our results argue against the power-stroke or Brownian ratchet, and support entropic pulling. They also show that Hsp70 oligomerization, a ubiquitous phenomenon with no known active biological function28–30, can augment the force that Hsp70s generate when acting on their substrates.
Results
The 3 models in the context of uncoating
Clathrin cage disassembly is driven by Hsc70 binding to a sequence (`QLMLT') in the flexible CHC C-terminal tail 10 residues downstream of each of the 3 helices at the center of the triskelion under each cage vertex (fig. 1A, B)23,27. Based on structural studies it was proposed that Hsc70 doesn't induce, but sterically locks in, fluctuations that loosen the clathrin lattice until they accumulate to a point that disassembly ensues23. This Brownian/steric wedge model can be tested by moving the Hsc70 binding site to relieve steric clashes between the Hsc70 and cage walls. If disassembly persists even as steric clashes are relieved, it would argue against this mechanism.
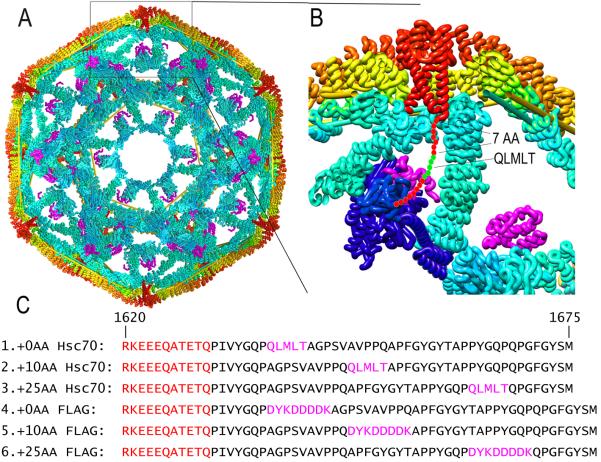
Figure 1. Structural context of approaches to test Hsc70 disassembly mechanisms.
A: Cut-away view of clathrin cage (interior surface in cyan; exterior in yellow/orange) with auxilin (magenta; pdb 1XI523). C-termini of CHCs form a helical tripod (red) under each vertex. B: Expanded view of boxed region from A. Hsc70 (pdb 4B9Q3; blue) modeled into the clathrin:auxilin cage based on an Hsp70 NBD:auxilin J domain structure26 positions its protein binding domain (PBD) to bind the terminal tail (red circles with Hsc70 binding QLMLT sequence in green). C: Sequences of the termini of the CHCs used (#1 is WT), with helical segments in red, and Hsc70 binding and FLAG sites in magenta.
The entropic pulling model emphasizes thermodynamics—the entropy change—in the reaction16,17, but the complementary molecular kinetic description would be collision pressure: it's collisions and repulsion between Hsp70s and closely apposed structural walls that impart momentum, away from the walls31,32, to the Hsp70s. Necessarily, such collisions generate equal and opposite momentum on the walls. Thus, Hsc70s bound to flexible tethers under each vertex would impart pressure against cage walls to drive the triskelia apart. In contrast to the steric wedge model, the collision pressure mechanism predicts that moving the Hsc70 binding site away from the walls will slow, but not stop, disassembly, and predicts that moving this site by 10 and 25 amino acids will slow disassembly by ~10- and ~60-fold, respectively (Supplementary Note 1)17.
The collision pressure and Brownian/steric wedge models predict that any protein similar in size to Hsc70 and binding at the same location should drive disassembly, while the power-stroke model requires an Hsc70-specific conformational change. Therefore, unlike the steric wedge and collision pressure models, the power-stroke mechanism predicts that if the Hsc70 binding site is replaced with an antibody binding site, the antibody should not drive disassembly.
To test the mutually exclusive predictions of these models we assembled cages from CHCs in which the Hsc70 binding site was at its WT position (`+0AA'), or 10 or 25 AA C-terminal to this position (`+10AA' and +25AA'; fig. 1C). We also prepared cages with this site replaced with an antibody binding site (a FLAG tag) at either the WT position, or 10 or 25 AA C-terminal to this position (`+0AA FLAG', `+10AA FLAG' and `+25AA FLAG'; fig. 1C).
Moving the Hsc70 binding site slows disassembly
Disassembly was measured by light-scattering in a stopped-flow fluorometer (supplemental figure 1) with cages reacted with varying concentrations of Hsc70. With +0AA cages, reaction profiles (fig. 2A) were like those seen by Rothnie et al. who used a similar approach21. Reaction of cages with 2 μM Hsc70 resulted in an initial ~15% increase in scattering, followed by a drop to ~20% of the initial level. The large drop is due to disassembly21,24. The nature of the initial smaller increase is unknown but was assigned by Rothnie et al., to binding of the 1st Hsc70 to one of the 3 CHC termini in their sequential mechanism, in which 3 Hsc70s must bind all 3 sites in a triskelion before that triskelion is released from the cage. A subsequent single molecule study cast doubt on this mechanism as it showed that disassembly began when only one (or even fewer20) Hsc70(s) were bound for every 2 triskelia, and that Hsc70s continued to bind and accelerate disassembly even after it had begun19.
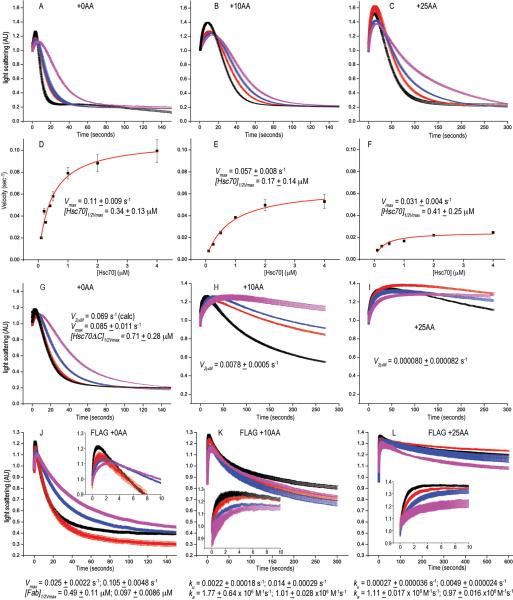
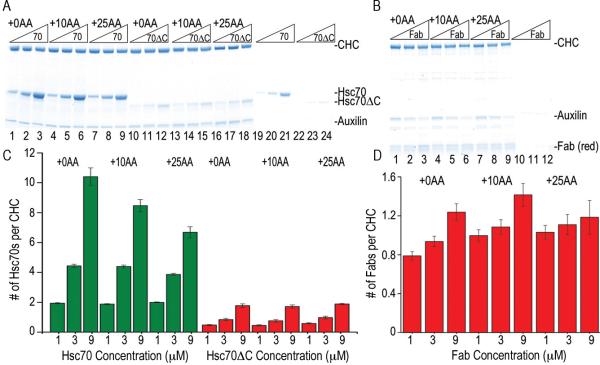
Figure 2. Moving the Hsc70 binding site slows disassembly and reveals a reaction intermediate of large scattering amplitude, and replacing it with a FLAG-tag allows disassembly by anti-FLAG Fabs.
A: Scattering (normalized to starting value of 1) vs. time for reactions with WT (+0AA) cages reacted with 0.25 (magenta), 0.5 (blue), 1.0 (red) or 2.0 (black) μM Hsc70. B: As in A, but with Hsc70 binding site moved 10 AA downstream (+10AA). C: As in A, but with site moved 25 AA (+25AA). D: Hyperbolic fits of WT cage disassembly rates vs. [Hsc70]. E: As in D, but for +10AA cages. F: As in D, but for +25AA cages. G: As in A, but using Hsc70ΔC. H: As in G, but with +10AA cages. I: As in G, but with +25AA cages. J: As in A, but using anti-FLAG Fab and cages with Hsc70 binding site replaced with a FLAG tag. K: As in J, but with tag shifted 10AA. L: As in J, but with tag moved 25 AA. In J–L, insets show initial 10 sec. of reactions to resolve Fab binding phase. For all plots, trace thickness, error bars and fitted values in figs 2, 3, 5 and 6 reflect +/− sem. The number of replicates for each experimental condition in these figures is specified in Supplemental Table 2.
We tried fitting our data using Rothnie et al., sequential scheme but concluded that neither the amplitude of the initial increase in scattering, nor the rates of this initial increase or subsequent decrease could be accurately determined as these parameters are coupled (supplemental figure 2). We therefore used the simplest possible scheme: C+H→CH→T, where C, H, and T correspond to cages, Hsc70 and triskelia respectively (see Supplementary Note 2) to fit values for the rate (ka) of Hsc70 association, the amplitude of scattering from the intermediate (CH) and the cage disassembly rate (kd). However, because these parameters are coupled, we didn't attach meaning to these values but derived a parameter which was independent of the amplitude of the intermediate and incorporated both association and disassembly rates: kov=1/(1/([Hsc70]*ka)+1/kd)). This corresponds to the overall rate of disassembly (k3+=0.105 μM−1s−1 of Bocking et al.,19. However, Bocking et al., concluded that this rate increased linearly with [Hsc70] (brackets denote concentration). This is inconsistent with their data, which show large divergence from linearity at [Hsc70]>1 μM (ref.19), and, as also indicated by our data (fig. 2D–F) and that of Rothnie et al21, which show hyperbolic kinetics. We therefore fit overall rates at different [Hsc70] to a hyperbolic equation (fig. 2D). With +0AA cages we obtained a Vmax of 0.11 s−1 with an [Hsc70]1/2Vmax of 0.34 μM, and a rate of 0.065 s−1 at 0.5 μM Hsc70, similar to the Bocking et al. estimate at 0.5 μM Hsc70 (where divergence from their linear model is small) of 0.053 s−1.
With +10AA and +25AA cages disassembly was, respectively, 2- and 4-fold slower than with +0AA cages (fig. 2). Disassembly of cages missing Hsc70 binding site was ~20x slower (supplemental figure 1), indicating that disassembly of +10AA or +25AA cages is predominately due to Hsc70 binding to the displaced site. The initial scattering increase becomes larger as the Hsc70 binding site is moved, reaching ~70% of the starting value in reactions with +25AA cages (fig. 2C).
That disassembly slows only 4-fold even when the Hsc70 binding site is moved 25AA argues strongly against the Brownian/steric wedge model, but is also inconsistent with the collision pressure model which predicts a ~60-fold decrease when the binding site is moved this much. We wondered if this is due to Hsc70 self-association28, since the collision pressure model assumes identical numbers of Hsc70s binding to cages with different tether lengths. We tested this with Hsc70ΔC, which is functional in disassembly but bears a C-terminal deletion that reduces self-association33. With Hsc70ΔC and +0AA cages, the maximal disassembly rate was ~2-fold slower and the 1/2Vmax concentration was ~2-fold greater than with Hsc70 (fig. 2G). However, with +10AA and +25AA cages, disassembly with Hsc70ΔC was much slower than with Hsc70 (figs. 2H, 2I), and we couldn't determine Vmax as rates didn't plateau over the tested concentration range, so we instead compare rates at 2 μM Hsc70ΔC and obtained values of 0.051 s−1, 0.0078 s−1 and 0.00080s−1 for the +0AA, +10AA and +25AA cages respectively (fig. 2G–I; supplementary table 1). Thus, with Hsc70ΔC rate decreases are in acceptable agreement with the collision pressure model. With Hsc70ΔC the initial scattering increase was also smaller than with Hsc70, being maximally ~40% (with +25AA cages; fig. 2I) but reaching ~70% with Hsc70 (fig. 2C).
Anti-FLAG Fabs disassemble FLAG-tag cages
To test the power-stroke model we evaluated the ability of anti-FLAG Fab to disassemble FLAG-tag cages. Fab (50 kD), rather than IgG, was used because Fab is similar in MW to Hsc70 (70 kD) or Hsc70ΔC (60 kD), and because divalent binding may cross-link CHCs and inhibit disassembly (supplemental figure 3). When the tag was at the same position as the WT Hsc70 binding site, Fab disassembled cages at rates similar to Hsc70 or Hsc70ΔC (fig. 2J). However, when the tag was moved 10 or 25 AA downstream (fig. 2K, L) rates slowed ~8- or ~40-fold, respectively, similar to what was seen with Hsc70ΔC and in agreement with the collision pressure model. The amplitude of the initial increase in scattering with Fab is also similar to that seen with Hsc70ΔC. However, the rate of this increase is faster, being complete within 1-2 seconds with Fab but extending over 5–25 seconds with Hsc70ΔC. This is likely due to Hsc70 first having to bind ATP and then auxilin, followed by transfer to clathrin in a step limited by an ATP hydrolysis rate of ~0.1 s−1,34. The rate of the initial increase with Fab indicates a binding rate of 1-2×106M−1s−1, consistent with known antibody binding rates35.
Hsc70 vastly increases scattering by low pH stabilized cages
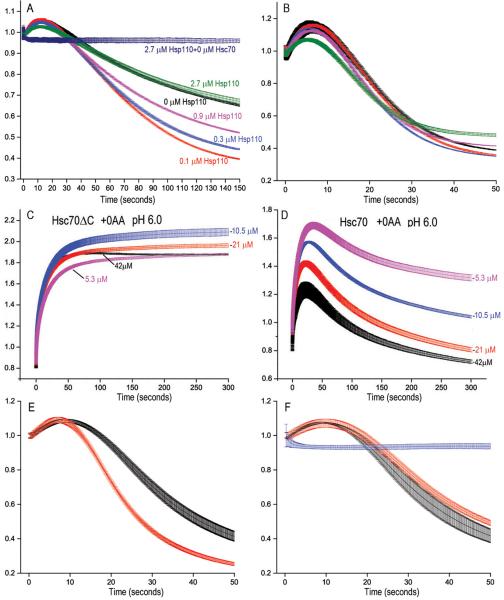
Figs. 2A–C show that, as the Hsc70 binding site is shifted, disassembly slows and the initial scattering increase becomes larger. We expect this occurs because when disassembly is fast, the potential amplitude of this increase is obscured since, before reaching its maximum, scattering drops as disassembly ensues. However, kinetic coupling (supplemental figure 2) precludes estimating its maximum amplitude by fitting rate data. To measure this we carried out reactions at pH 6.0, where cages resist disassembly23. With Hsc70 we observed an ATP- and auxilin-dependent increase in scattering of ~200% of the starting value (fig. 3A–C). With Fab (fig. 3D–F) or Hsc70ΔC (fig. 3G, H) the maximal increases was only ~50% and ~70%, respectively.
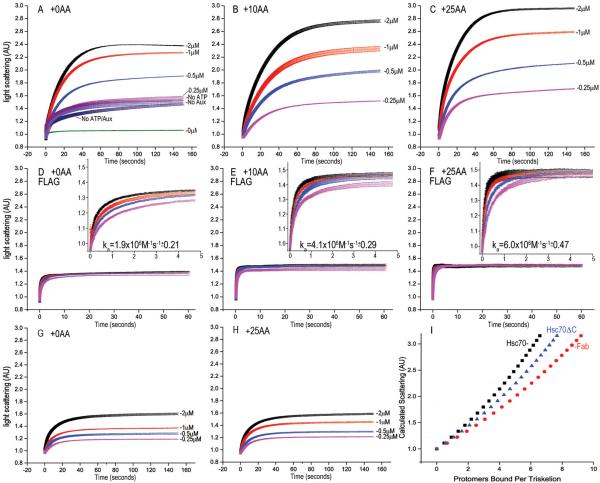
Figure 3. Hsc70 binding to cages under conditions that block disassembly leads to a massive increase in light scattering.
A: Hsc70 reacted with cages at pH 6.0; other conditions as in fig. 2A; AU=Arbitrary Units. Also plotted are reactions without Hsc70 (green), with 2 μM Hsc70 but no ATP (purple), without auxilin (indigo) or absent ATP and auxilin (dark blue). B: As in A, but with +10 AA cages. C: As in A, but with +25AA cages. D: As in A, but with FLAG cages and Fab (inset shows first 5 sec of reaction). E: As in D, but with tag moved 10 AA. F: As in D, but with tag moved 25 AA. G: As in A but using Hsc70ΔC. H: As in G but with +25AA cages. I. Calculated scattering of 70 nm diameter cages bound by the indicated number of protomers per triskelion of either Hsc70 (black squares), Hsc70ΔC (blue triangles), or Fab (red circles).
Light scattering by a protein is a function of its dimensions and mass. To determine if the initial increases in scattering were due to increases in cage mass, we adapted a hollow sphere model developed to analyze scattering from vesicles36. For Hsc70ΔC and Fab, this model accurately predicts the amplitude of the scattering increases to be ~70% and ~55%, respectively, assuming that each of these proteins binds 1:1 to their binding sites in a cage (fig. 3I). However it predicts a scattering increase of only ~80% for binding of Hsc70 at the same stoichiometry, suggesting either that Hsc70 induces a larger increase in cage dimensions than does Hsc70ΔC or Fab, that a larger number of Hsc70s bind, or that Hsc70 causes cages to aggregate. We ruled out the latter as we saw no change in either the rate or amplitude of the Hsc70 driven scattering increase in experiments in which the cage concentration was varied (supplemental figure 4).
Scattering increases are not due to increases in cage size
If only one Hsc70 binds the single Hsc70 site in each CHC, cage diameters would have to increase ~30% to explain the 3-fold increase in scattering seen when reacted at pH 6 with Hsc70 (fig. 3A–C). However, Xing et al.23 observed that Hsc70ΔC binding decreased cage dimensions by 0.8% along one axis, and increased them by 1.4% along the other two. Since Hsc70 might drive larger changes in size than Hsc70ΔC, we carried out cryoEM reconstructions of pH 6.0 cages with or without Hsc70 or Hsc70ΔC23. Our reconstructions and difference maps were similar to those published previously23 (fig. 4A, B), with the density due to Hsc70ΔC attributable to envelopes of similar volume under each cage vertex. With the isosurface representation contoured at 2.5σ -a common value used for highly symmetric structures- this envelope could accommodate only a single ~25 kD PBD (fig. 4D–F), suggesting that much of the Hsc70ΔC density is averaged out due to dynamic or static disorder. Cages with Hsc70 were similar in size to cages alone or with Hsc70ΔC (fig. 4A–C), indicating that size changes can't explain the large scattering increases seen when Hsc70 binds. Difference maps between cages −/+ Hsc70 revealed an element positioned similarly to the density ascribed to Hsc70ΔC in the Hsc70ΔC cages (fig. 4F). However, while Hsc70 is only ~16% larger than Hsc70ΔC, this density was >2-fold larger with Hsc70 vs. Hsc70ΔC, suggesting either reduced disorder or binding of a larger number of molecules with Hsc70 vs. Hsc70ΔC.
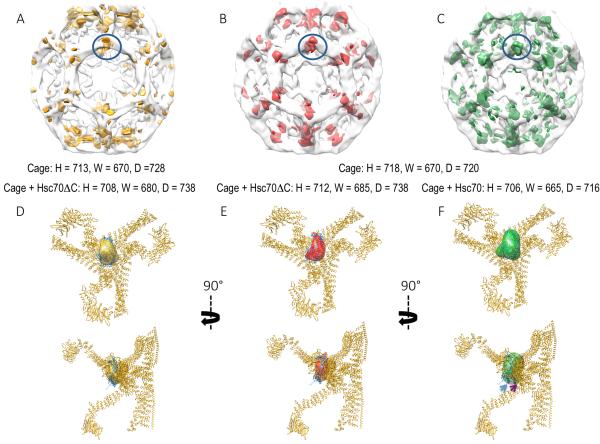
Figure 4. CryoEM reconstructions of cages with or without Hsc70ΔC or Hsc70.
A: Partially transparent cage image reconstructed previously by Xing et al.23, with difference density attributable to Hsc70ΔC [(Cages+Hsc70ΔC)-(Cages)] in gold. B. Partially transparent cage image from our reconstructions with difference density attributable to Hsc70ΔC in red. C. Partially transparent cage image from our reconstructions with difference density attributable to Hsc70 in green. In each image the bulk of the difference density is centered under each cage vertex and highlighted by being circled in blue. Cage dimensions +/− Hsc70ΔC or Hsc70 (+/− 40 Å) are specified under each image. D–F: Two orthogonal views of the ribbon model of the asymmetric unit of the clathrin cage (pdb 1XI423) with the difference density attributable to Hsc70ΔC from previous reconstructions (D; gold), our reconstruction with Hsc70ΔC (E; red), or our reconstructions with Hsc70 (F; green) shown. View is from inside the cage centered on a vertex. The Hsc70ΔC density is similarly sized and positioned in both our reconstruction and the previous one, and can accommodate one 25 kD Hsc70 PBD (docked in the volume; pdb 1DKX48). Hsc70 density is similarly positioned but ~2× as large and able to accommodate 2×25 kD PBDs (indicated by blue and magenta arrows).
Multiple Hsc70s bind per CHC to low pH stabilized cages
Since neither cage expansion, aggregation, nor binding of one Hsc70 to each CHC in a cage explained the Hsc70 induced scattering increases at pH 6, we determined if multiple Hsc70s might be binding each CHC by reacting Hsc70, Hsc70ΔC or Fab with cages at pH 6 and then pelleting the cages and their associated proteins (figs. 5A, B). In agreement with the observed scattering and our model predictions (fig. 3I), ~1 Fab per CHC was bound to cages at all concentrations tested (fig. 5D), while ~0.5, ~0.8, and ~2 Hsc70ΔCs were bound at 1, 3, and 9 μM, respectively (fig. 5C). However, Hsc70 was bound in molar excess of CHC, with ~10 Hsc70s per CHC at 9 μM Hsc70 (fig. 5C). Binding of such a large number of Hsc70s was dependent on auxilin, ATP and, critically, the presence of an Hsc70 binding site in the CHC (supplemental figure 5). We conclude that the large scattering increases seen when Hsc70 is reacted with pH 6.0 cages reflect multiple Hsc70s binding per CHC, which is likely due to Hsc70 self-association since it is not seen with Hsc70ΔC.
Figure 5. Scattering increases due to Hsc70 binding reflect multiple Hsc70s binding per CHC, not cage expansion.
A. SDS PAGE of pellets of the indicated cages incubated with 1, 3, or 9 μM Hsc70 (lanes 1–9) or Hsc70ΔC (lanes 10–18; [Hsc70] and [Hsc70ΔC] increase from left to right as indicated). Lanes 19–24 show experiments without cages. B: As in A, but using FLAG cages and Fab (lanes 10–12 are no cage control). C: Hsc70/CHC or Hsc70ΔC/CHC ratios plotted vs. [Hsc70] or [Hsc70ΔC], as indicated. D: As in C, but using FLAG cages and Fab.
Hsc70 self-association augments its disassembly force
The collision pressure mechanism predicts that reducing the number of cage-bound Hsc70s will reduce the net force they generate against cage walls and slow disassembly. To test this, we used Hsp110, a nucleotide exchange factor (NEF) for Hsp7037. Hsp110 accelerates disassembly at low Hsc70 concentrations by releasing Hsc70 from released triskelia so that it can cycle back to cages38. With 20 nM Hsc70, low Hsp110 levels enhanced disassembly, but higher levels resulted in less enhancement or modest inhibition, together with a reduction in the initial scattering increase (fig. 6A). With 250 nM Hsc70, increasing Hsp110 progressively reduced the initial scattering increase and slowed disassembly (fig. 6B). Thus, when Hsc70 is limiting, Hsp110 accelerates disassembly by releasing Hsc70 from triskelia, but at higher Hsc70 concentrations, Hsp110 inhibits disassembly by unloading Hsc70s from cages.
Figure 6. Accumulation of Hsc70 association with cages drives their disassembly and Hsc70 self-association augments its cage disassembly force.
A: Reactions carried out as in panel 2A, but with only 20 nM Hsc70 and either 0 (black trace), 0.1 μM (red), 0.3 μM (blue), 0.9 μM (magenta) or 2.7 μM (green) Hsp110. The dark blue trace shows a reaction with no Hsc70 and 2.7 μM Hsp110. B: As in panel A but with 250 nM Hsc70. C: Reactions with +0AA cages at pH 6 carried out as in panel 4A, but with the indicated concentrations of Hsc70ΔC. D: As in C, but using Hsc70. E: Cage disassembly by 125 nM His-tagged Hsc70 in the presence (red) or absence (black) of 125 nM anti-His Fab. F: Red and black traces as in panel E but using untagged Hsc70; blue trace shows reaction of cages with anti-His Fab and no Hsc70. Experiments in panels E and F used recombinant GST-auxilin (with GST cleaved post-purification) and clathrin purified from bovine brain clathrin coated vesicles rather than recombinant his-tagged auxilin and CHC to eliminate complications due to Fab binding to auxilin or CHC.
If reducing the number of Hsc70s bound to cages reduces the forces driving disassembly, then self-association, by increasing the number of Hsc70s, should increase these forces. This could explain why Hsc70 is more effective than Hsc70ΔC in disassembling +10AA and +25AA cages, while both disassemble +0AA cages (fig. 2), and why we observe gradual drops in scattering with +0AA cages in pH 6 reactions with Hsc70 (fig. 3A), but not Hsc70ΔC (fig. 3G), suggesting that only the former slowly disassembles these cages. To test this we used higher Hsc70ΔC and Hsc70 concentrations to increase self-association. When we reacted pH 6 cages with 5 to 42 μM Hsc70ΔC, scattering doubled (fig. 6C), consistent with the scattering model's predictions (fig. 3I) and data that, at these concentrations, ~2 Hsc70ΔCs bind per CHC (fig 5C). With Hsc70, scattering increased but then dropped (fig. 6D), revealing disassembly. EM of pH 6 cages reacted with excess Hsc70 or Hsc70ΔC confirmed not only that cages were disassembled by the former, but not the latter, but also that Hsc70 formed oligomers in such reactions while Hsc70ΔC did not (supplemental figure 6). Hsc70, but not Hsc70ΔC, therefore disassembles pH 6 cages, probably because of the greater self-association activity of the former. That reduced disassembly by Hsc70ΔC vs. Hsc70 was not due to weaker binding was shown by observations that Hsc70ΔC competed effectively with Hsc70 for binding to cages (supplemental figure 7). The collision pressure mechanism also predicts that increasing Hsc70 bulk can increase its disassembly activity. To test this we compared disassembly by His-tagged Hsc70 +/− anti-His Fab. Addition of Fab 1:1 with His-tagged Hsc70 accelerated disassembly (fig. 6E) but had no effect on disassembly by untagged Hsc70 (fig. 6F).
Hsc70 makes cages rigid but prone to catastrophic deformation
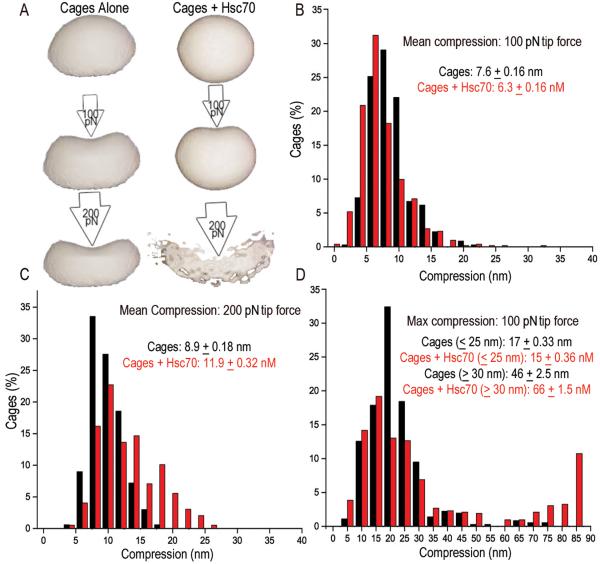
The collision pressure model's predictions for Hsc70's effects on cage compressibility may be understood by analogy to balloons inflated to high vs. low pressure. Just as gas molecule collisions with a balloon's interior generate outwardly directed forces, bound Hsc70s may generate such forces on the interior walls of the cage. Cages with Hsc70s should therefore be less compressible than Hsc70-free cages when probed with a low force, just as high pressure balloons are less compressible than low pressure balloons (fig. 7A). However, a high pressure balloon is more prone to burst--to undergo catastrophic deformation--when probed with a strong force, while a low pressure balloon can deform more rather than burst (fig. 7A). To test this we used AFM to measure sizes, and mean and maximum deformations of cages, cages with auxilin, and cages with auxilin and Hsc70ΔC or Hsc70 at pH 6.0. In agreement with our cryoEM, the sizes of all of these were similar (supplemental figure 8; supplemental table 3). With a 100 pN tip force, Hsc70 reduced mean compressibility from 7.6±0.16 to 6.3±0.16 nm (fig. 7B), but with a 200 pN force, Hsc70 increased compressibility from 8.9±0.18 to 11.9±0.32 nm (fig. 7C). While the distribution of deformations was approximately Gaussian without Hsc70, addition of Hsc70 led to a non-Gaussian distribution and appearance of a population of large (>15 nm) deformations (fig. 7C), indicating that catastrophic deformation events were more frequent with Hsc70. To test this we measured the maximum deformation seen for each cage at a 100 pN force. Without Hsc70, 8% of cages exhibited max deformations of >30 nm with a mean of 46+2.5nm, but with Hsc70, 30% of the cages exhibited deformations >30 nm with a mean of 66+1.5nm (fig. 7D). With the smaller (<30 nm) deformations, Hsc70 had the opposite effect: mean max deformations for this group were 17+0.33 nm and 15+0.36 nm −/+ Hsc70, respectively. The effect of Hsc70 was therefore consistent with collision pressure predictions: Hsc70 made cages less compressible when probed at low force but markedly increased the frequency of catastrophic deformations.
Fig. 7. Hsc70 binding makes cages less compressible but more prone to catastrophic deformations.
A: Collision pressure model analogizes cages and cages+Hsc70 to balloons inflated to low and high pressure, respectively. Internal pressure generated by Hsc70s makes cages less deformable, but more prone to catastrophic deformation (bursting) especially as probing force is increased. B: Percent of cages +/− Hsc70 exhibiting indicated mean compressions with 100 pN force (mean compression for each cage is determined from 9–12 measurements obtained from probing each cage on a 3×3 or 4×4 grid, depending on cage size). C: As in B, but using a 200 pN tip force. D: Percent of cages exhibiting indicated maximum compressions during probing. Average max compressions for cage populations with max compressions <30 nm or >30nM are given (statistics in supplemental table 3).
Discussion
Support for a power-stroke rested, in part, on the conclusion that a Brownian ratchet can't generate the force to unfold the proteins that Hsp70s pull through translocation pores12. This conclusion was undermined when it was shown that J proteins deposit Hsp70s on translocating substrates but then disengage, leaving the Hsp70 without a platform on which to push while pulling15,39. Our observations that Fab binding at the same position as Hsc70 causes disassembly, or that Fab bound to Hsc70 can enhance its disassembly activity, further argues against a power-stroke and for a mechanism in which the mass/volume of a protein at this position causes disassembly. This supports either the collision pressure or Brownian/steric wedge mechanisms. However, the latter is undermined by the observation that disassembly persists even if Hsc70 is displaced by 25 amino acids, which should relieve any direct steric clashes. By elimination, this leaves collision pressure as the sole viable mechanism. But there's also positive evidence for this mechanism: it predicts that interposition of 10 or 25 residues between Hsp70 and a wall should, respectively, slow disassembly ~10- and ~60-fold (Supplemental note 1), in agreement with decreases seen with Hsc70ΔC or Fab. Our observation that Hsc70 makes cages more rigid but more likely to experience catastrophic deformations also supports this model. In addition, while our gel analyses and scattering data indicate that ~2 Hsc70ΔCs and multiple Hsc70s (per CHC) bind cages under conditions like those used for cryoEM, the volume under the vertex attributable to the chaperones in the reconstructions accommodates only one or two PBDs for, respectively, Hsc70ΔC or Hsc70. It's unlikely this reflects most of the Hsc70s binding elsewhere on the cages, as binding of multiple Hsc70s requires an Hsc70 binding site in the CHC C-termini (supplemental figure 5). Density for most of the Hsc70s is therefore likely being averaged out due to static or dynamic disorder, consistent with their being mobile and able to exert collision pressure on cage walls.
The maximum calculated Hsp70 entropic pulling/pushing force of ~20 pN16,17 may be compared to experimentally measured osmotic pressures of concentrated protein solutions, as osmotic pressure is the macroscopic product of microscopic collisions between proteins and membranes31,32: a 450 mg/ml solution of 67 kD albumin generates 0.5 pN/nm2 of osmotic pressure at pH 7.440. If Hsc70 generates similar pressures, then an Hsc70 bound to the CHC tail under a cage vertex could generate a pushing force of ~20 pN (Supplemental note 1). By comparison, the kinesin power-stroke generates 5 pN41 with a step size of 8 nm42, while the force generated by an Hsp70 drops precipitously as it moves 1-2 nanometers away from a wall16,17. The work done by these systems is therefore similar, but the force mechanism is appropriately adapted to the function of each.
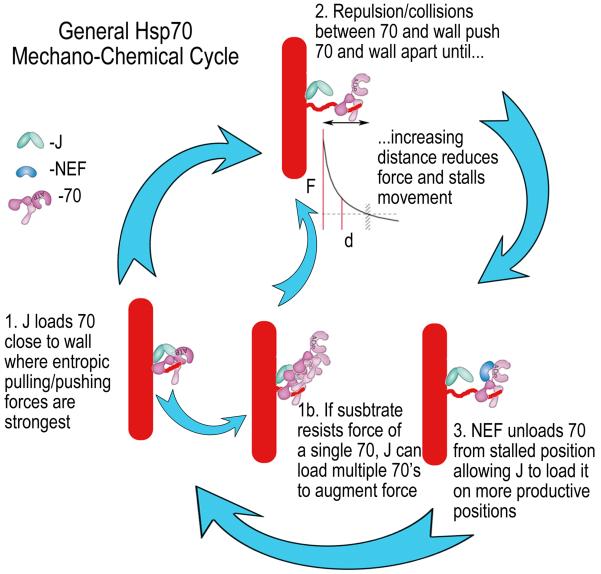
While anti-FLAG Fabs disassemble FLAG-tag cages, they differ from Hsc70 since they can't actively cycle from released triskelia to cages. Auxilin loads Hsc70 on cages in preference to triskelia both because cages present a higher density and/or better positioning of binding sites for the multiple clathrin binding motifs on the auxilin molecule43, and because cage architecture places auxilin closer to Hsc70 binding sites23,44. Hsp110, the NEF in this system, stimulates disassembly when Hsc70 is limiting (fig. 6A) by unloading Hsc70 from triskelia38, but when Hsc70 isn't limiting, high concentrations of Hsp110 inhibit disassembly by also unloading Hsc70 from cages (fig. 6B). It is similarly seen that Hsp70-mediated protein disaggregation reactions are usually optimally stimulated by low Hsp110 concentrations9,45,46, an observation that may be explained by invoking the role of geometry in the entropic pulling/pushing model. In this model, the role of the J co-chaperone isn't simply to load the Hsp70 onto substrate, but to load it onto substrate at an optimal position: close to a wall where entropic pulling/pushing forces are greatest (fig. 8, step 1). Collisions/repulsion between the wall and Hsp70 push the two apart, but as predicted by the entropic pulling/pushing model and validated by our observations of the effects of shifting the Hsc70 binding sites in the clathrin cage, these pushing forces diminish with distance (fig. 8, step 2). The reaction may stall if this distal Hsp70 sterically blocks loading of another Hsp70 at the more forcefully productive position, near the wall. The role of the NEF in the cycle is to unload the Hsp70 from the non-productive position (step 3), but high NEF concentrations can be inhibitory if they begin to also unload Hsp70s from productive positions. In addition, if the substrate is recalcitrant and does not yield to the force of single Hsp70s (e.g., clathrin cages at pH 6 vs. pH 6.8) then the Hsp70 will remain close to the J co-chaperone, allowing the latter to exploit the self-association properties of the chaperone to load additional Hsp70s onto the first to augment the forces (step 1b). While this model doesn't argue that Hsp110 cannot contribute to disaggregation via other mechanisms47, it can account for the observed Hsp110 concentration dependence and can explain how the established functions of J co-chaperones and NEFs in, respectively, loading and unloading of substrates in the Hsp70 chemical cycle, are harnessed in a mechanical cycle to generate the forces by which Hsp70s move and transform their substrates.
Figure 8. General model for Hsp70 mechano-chemical cycle.
Step 1: J co-chaperone loads Hsp70 on substrate, close to a structural wall where entropic pulling/pushing forces are strongest. Step 2: Collisions/repulsion between Hsp70 and the wall push Hsp70 and the associated substrate away from the wall, but with increasing distance the repulsive interactions, frequency of collisions and force diminish so movement stalls (the F vs. d exponential decay curve is adapted from17). Step 3: A NEF unloads Hsp70 from the stalled position, allowing J to again load an Hsp70 close to the wall so the cycle can continue. Step 1b: If the substrate does not yield to the force exerted by a single Hsp70 then the Hsp70 persists near the J co-chaperone, allowing the latter to load additional Hsp70s onto the first so as to augment the force that is generated.
Methods
DNA constructs
DNA encoding rat clathrin heavy chain 1 (CHC) was PCR amplified from DNA obtained from the Kirchhausen lab19 with primer pair 1/2, and cloned into pET28a+ (Novagen). The +10AA and +25AA CHC DNAs were constructed by deleting nucleotides encoding the QLMLT sequence using the QuikChange mutagenesis kit (Agilent Technologies) by amplification with primer pair 3/4, then adding back the QLMLT coding sequence 10 or 25 aa downstream using, respectively, primer pairs 5/6 and 7/8. CHC FLAG, +10AA FLAG and +25AA FLAG DNAs were constructed by replacing the sequence encoding QLMLT in CHC, CHC+10AA and CHC+25AA, respectively, with a sequence encoding DYKDDDDK (FLAG tag) using primer pairs 9/10, 11/12, and 13/14 (primer sequences in Supplemental data set 1). DNA encoding clathrin light chain A1 (CLCA1) was amplified from pPPA1-2 rat LCA1 with primer pair 15/16 and cloned into pBAT4 (EMBL).
Protein expression and purification
BL21(DE3) cells (Agilent) with plasmid encoding His tagged CHC were grown to OD600 of 1.0 at 30° C in 2x YT, transferred to 12°C, induced with IPTG to 1mM and incubated for 24 hours. Cells from 1L culture were pelleted and re-suspended in 50 mL 0.5M Tris pH8.0, 10mM β-ME (lysis buffer) with one Roche protease inhibitor cocktail tablet added, and lysed by sonication and addition of 2.5mL 20% Triton X-100, and centrifuged at 125,000g for 30 min. Supernatants were loaded onto 20-mL of Ni-NTA agarose (Qiagen), washed with 200 mL lysis buffer, and eluted with 100 mL of 0.4M Tris pH8.0, 10mM β-ME, 200mM imidazole. Protein was precipitated with equal volume of saturated (NH4)2SO4 and fractionated on a Superpose 6 (GE Healthcare) column in 0.5M Tris pH7.0, 1mM EDTA and 3mM DTT (storage buffer). Pure proteins were concentrated by NH4SO4 precipitation, dialyzed into storage buffer, flash frozen in liquid N2 and stored at −80°C. CLCA1 was co-expressed with His-tagged CHC as expression of CLCA1 alone led to its degradation, and purified by Ni-NTA agarose as described above. Co-purified proteins were incubated at 95°C for 5min to precipitate CHC, which was removed by centrifugation. Pure CHC and CLCA1 were mixed 1:1 for subsequent experiments. Hsc70 and auxilin (547-910) were prepared as described38. Anti-FLAG Fab was obtained by digesting anti-FLAG M2 (Sigma; cat # F-1804; lot#SLBG5673V) using mouse IgG1 Fab preparation kit (Thermo Sci.). Validation of this monoclonal antibody is provided in49. For the experiments utilizing the anti-his Fab, clathrin was purified from bovine brain clathrin coated vesicles24, auxilin (547-910) was prepared by thrombin cleavage of GST-auxilin (547-910)25, and bovine Hsc70 with an N-terminal 10-his tag was utilized so that Hsc70 would be the only his-tagged protein in the reaction. Anti-His Fab was obtained by digesting anti-His Mab (Miltenyi Biotec; cat # 130-095-212; monoclonal antibody GG11-8F3.5.1) using mouse IgG1 Fab preparation kit (Thermo Sci.). Validation of this antibody is provided on the manufacturer's website: http://www.miltenyibiotec.com/~/media/Images/Products/Import/0006500/IM0006556.ashx.
Clathrin polymerization
CHC and CLCA1 were mixed 1:1 at 4.5uM in storage buffer and dialyzed against 100mM MES pH6.2, 1.5mM MgAc2, 2mM DTT (polymerization buffer) for 7hrs, followed by an O/N dialysis against 20mM imidazole pH6.8, 10mM (NH4)2SO4, 25mM KCl, 2mM MgAc2, 2mM DTT or 20 mM MES pH 6.0, 10mM (NH4)2SO4, 25mM KCl, 2mM MgCl2, 2 mM DTT (DTT was omitted from all experiments with Fab). After dialysis, the sample was centrifuged at 9,000g for 10min to remove aggregated material. EM of cages revealed that cages prepared from recombinant clathrin were indistinguishable from those prepared with clathrin purified from bovine brain clathrin coated vesicles (data available on request).
Stoichiometry of protein binding to cages
Cages polymerized as described above were dialyzed O/N against 20mM MES pH6.0, 2mM MgCl2, 25mM KPO4, 10mM (NH4)2SO4, diluted to 0.15 μM CHC, and mixed with 0.23 μM His-Aux547-910m, 0.5mM ATP and the indicated concentrations of Hsc70, Hsc70ΔC or anti-FLAG Fab. Reactions (200 μl) were centrifuged at 400,000g at 4°C for 10min to pellet cages and associated proteins. Pellets were suspended in 75ul of 1X SDS sample buffer and resolved by denaturing PAGE. Proteins were imaged using a BIO RAD Criterion Stain Free Imager and quantified using BIO RAD Image Lab 2.0 software by comparison to lanes in which defined amounts of each protein were loaded. Experiments in which cages were omitted were run side-by-side to determine background pelleting of each protein in the absence of cages. Uncropped images of gels used in assembling fig. 5 are presented in Supplemental data set 2.
Stopped-flow experiments
Light scattering experiments were carried out in an Applied Photosystems stopped-flow fluorometer with excitation/emission wavelengths of 395 nm. Cages corresponding to 0.3 μM CHC (unless otherwise indicated in figure legends) with 1mM ATP+0.45 μM auxilin in 20mM imidazole pH6.8, 10mM (NH4)2SO4, 25mM KCl, 2mM MgAc2 or 20 mM MES pH 6.0, 10mM (NH4)2SO4, 25mM KCl, 2mM MgCl2 were reacted with an equal volume of 0.04 μM to 8 μM Hsc70 in the same buffer. Background scattering determined from reactions without cages was subtracted from measured scattering values which were normalized by dividing by the starting scattering value so that the initial scattering in all reactions equaled 1.0. Measures of scattering by varying concentrations of cages or triskelia showed that scattering was linear with cage or triskelia concentration over the ranges used in these experiments (supplemental figure 1). As indicated in figure legends, auxilin and/or ATP were omitted from some reactions to control for the effects of these molecules. Removal of ATP (or substitution with ADP or ATPγS), auxilin, the Hsc70 NBD, or Hsc70 ATP binding or hydrolysis activity severely slowed or abrogated disassembly (supplemental figure 1).
Atomic force and electron microscopy
QNM peak force AFM experiments were carried out on a multimode 8-nanoscope V instrument (Bruker, CA) under pH 6.0 buffer using an MSCT cantilever with its calibrated spring constant designed between 0.01 to 0.03 N/m and peak force setpoints of 100–200 pN; 5 μl drops of fresh cages +/− auxilin, ATP and Hsc70 or Hsc70ΔC were deposited on freshly peeled mica and imaged under the same buffer following routine optimization for biological AFM. Data were analyzed with instrument software (Nanoscope ver8.15, Bruker, CA) and exported as ascii files for further analysis with Excel (Microsoft, Richmond, WA), and displayed with ImageJ (ver 1.4x, NIH, Bethesda, MD). Samples as described for AFM were absorbed for 20 seconds to a Formvar-carbon grid, followed by a rinse and 20 second incubation with 1% uranyl acetate. Grids were wicked, air dried, and examined at 25,000× magnification in a JEOL JEM 1200EX-II.
Cryo-electron microscopy reconstructions
CHC and CLCA1 were mixed 1:1 at 4.5uM in storage buffer, the clathrin assembly domain of AP180 was added at 4.5 μM (GST-C58)50, followed by dialysis against 20 mM MES pH 6.0, 10mM (NH4)SO4, 25mM KCl, 2mM MgCl2, 2 mM DTT. After dialysis, the sample was centrifuged at 9,000g for 10min to remove aggregates. Cages were then incubated with 1 mM ATP, auxilin (1.3:1 molar ratio to CHC) and either Hsc70ΔC or Hsc70 (at a 10:1 molar ratio to CHC) and aliquots of each sample were applied to glow-discharged, holey carbon grids (Quantifoil R 1.2/R 1.3 300 mesh grids) containing an additional continuous thin layer of carbon, blotted, and frozen in liquid ethane. Images were acquired under minimal dose conditions with a FEI Tecnai G2 FEG200 electron microscope at 200 kV with a Gatan side-entry cryo-holder at a nominal magnification of 41000× and underfocus values from 2 to 8 um using a 16 megapixel FEI Eagle CCD camera with a step size of 15 um, thus the pixel size of the acquired images was 3.65 Å (representative images of the cages are in supplementary data set 3). The contrast transfer function of each image was estimated using CTFFIND351. Single particles were manually selected and extracted using XMIPP352. Particles were classified using a free-pattern maximum-likelihood method (Relion 2D-Classification)53. To evaluate the structural homogeneity of the different data sets, Relion 3D classifications were performed54 and the D6 particles were selected and used for the 3D reconstruction using projection matching from XMIPP55. A total of 3468, 5609 and 4891 particles, were used for the 3D reconstruction of cages, cages+Hsc70ΔC and cages+Hsc70, respectively. The resolution obtained for the 3D reconstruction of the cages, cages+Hsc70ΔC and cages+Hsc70 was 28.5 Å, 28.5 Å and 28.7 Å, respectively56. For the subtraction maps, the volumes used were first normalized and filtered to the same resolution (30 Å), then scaled to approximately the same dimensions and finally subtracted using the “vop subtract” option in the Chimera package57 (Pettersen, et al, 2004). The same procedure was used to subtract the clathrin D6 coat with Hsc70 and Auxilin (EMD-5118;23) from the clathrin D6 coat (EMD-5119; 44), as shown in Fig. 4A and D. The program Chimera was also used for the visualization of the volumes and docking of the atomic structures.
Data fitting
We followed Rothnie et al., in using Dynafit—which fits reaction schemes as sets of simultaneous differential equations—to fit our light scattering data. In Dynafit scripts a “?” after a parameter value indicates the value is to be varied during fitting. The sequential aspect of the scheme used by Rothnie et al.,21 to globally fit their data is expressed in by having 3 steps for Hsc70*ATP binding to cages and 3 steps for Hsp70*ATP to Hsp70*ADP transformation, corresponding to sequential loading of an Hsc70 onto each CHC in a triskelion. It also contains 5 global adjustable parameters (ka and kd, corresponding to, respectively, the bimolecular association rate for Hsc70 binding to cages and the monomolecular rate of cage disassembly; kr, corresponding to the rate of ATP hydrolysis; and scattering amplitudes for the starting (CA) and intermediate (CAT) species); and 3 locally adjustable parameters: an offset (the scattering value of the end state), and the concentrations of the clathrin cages (CA) and Hsc70 (T) for each data set with different Hsc70 concentrations (i.e., a total of 21 adjustable parameters for the example shown with data for 6 different Hsc70 concentrations: one cage and one Hsc70 concentration is fixed to anchor the fitting). We could obtain good fits to our data with this script (Rothnie et al., fit; Supplementary Note 2), but this required cage and Hsc70 concentrations to vary as much as 2× from their experimentally input values, which seemed unreasonable. When concentrations were fixed as experimental inputs, the quality of the fits was poor, especially at lower Hsc70 concentrations. We therefore used a script (Simplefit, Supplementary Note 2) corresponding to the simplest scheme that could still capture our reaction profiles to fit our averaged data individually at each Hsc70 concentration (all reactant concentrations fixed as experimental inputs). It contains only 3 adjustable parameters (ka and kd as defined above, and the amplitude of the scattering of the intermediate (CAT) species), and was used simply to allow extraction of the global rate (1/(1/ka*[Hsc70]+1/kd)) of the reaction at different Hsc70 concentrations as determination of this global rate was robust, but the 3 adjustable parameters were too tightly coupled (supplemental figure 2) to allow confident determinations of their values. Global disassembly rates at different Hsc70, Hsc70ΔC or Fab concentrations were then fit using a Hill equation with the cooperativity parameter set to 1.0 to extract the maximal (Vmax) disassembly rate and the Hsc70/Hsc70ΔC/Fab concentration at which rates were half-maximal, as described in Results. In those cases where velocities did not plateau at the concentrations tested (e.g., with Hsc70ΔC and +10AA or +25AA cages in fig. 2, panel H and I, we compare velocities at the highest (2 μM) Hsc70ΔC concentrations used). With Fab and +0AA FLAG cages, disassembly appeared incomplete so fitting was also done by fixing final scattering to correspond to complete disassembly (20% of starting intensity), resulting in the smaller Vmax and [Fab]1/2Vmax values reported in fig. 2J. With Fab and +10AA FLAG or +25AA FLAG cages, the initial binding and subsequent disassembly phases were well separated, allowing the separate bimolecular association (ka) and single-exponential decay (disassembly) rates (ku) to be individually fit as presented in figs. 2K and 2L (again values were fit by either allowing the final scattering intensity to vary, or fixing it at 20% of the initial value, resulting in two values for each parameter).
Statistics
The minimal number of experimental repetitions was set so that the +/− 1 SD ranges for experimentally determined values (that were concluded to differ) were non-overlapping, though in almost all cases the # of repetitions (as specified in individual figure legends or supplementary table 2) exceeded this by several-fold. A small number (<5%) of data from experimental replicates were excluded when the experimental outcome (e.g., value of a rate determination) differed by >2.5 SD from the mean of experiments carried out under ostensibly identical conditions and when a reason for the variation (e.g., bubbles in stopped-flow apparatus; aging of labile reagents) was clear.
Supplementary Material
Acknowledgements
We thank L. Wang of UTHSCSA for technical assistance; C. Brantner and D. Sackett of NICHD, the NHLBI EM Core Facility for help with EM at an early stage of the work; and D. Sackett and R. Nossal at NICHD, D. Luque from ISCIII, and J. Stachowiak from UT-Austin for helpful discussions.
Accession codes the cryo-EM data are EMD-3442, EMD-4036 and EMD-4035 for, respectively, clathrin cages, cages+Hsc70ΔC and cages+Hsc70.
Funding: Supported by NS029051 (to EML) and GM118933 (to EML and RS); the NIH Intramural Research Program (NIBIB); and by grants BFU2013-44202 from the Spanish Ministry of Economy and Innovations and S2013/MIT-2807 from the Madrid Regional Government to JMV. HSL supported in part by a scholarship from the Taiwan National Science Council (NSC103-2917-I-564-072).
Footnotes
Author Contributions: R.S designed and performed experiments, analyzed data and wrote the paper. H.S.L., J.C., S.J., A.J.J. and E.M.L. designed and performed experiments, and analyzed data. J.M.V. designed experiments and analyzed data.
Competing interests: The authors declare no competing interests.
References
- 1.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–55. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 2.Sousa R, Lafer EM. The role of molecular chaperones in clathrin mediated vesicular trafficking. Front Mol Biosci. 2015;2:26. doi: 10.3389/fmolb.2015.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kityk R, Kopp J, Sinning I, Mayer MP. Structure and Dynamics of the ATP-Bound Open Conformation of Hsp70 Chaperones. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 4.Qi R, et al. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat Struct Mol Biol. 2013;20:900–7. doi: 10.1038/nsmb.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuravleva A, Clerico EM, Gierasch LM. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell. 2012;151:1296–307. doi: 10.1016/j.cell.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Misselwitz B, Staeck O, Rapoport TA. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 7.Zuiderweg ER, et al. Allostery in the Hsp70 chaperone proteins. Top Curr Chem. 2013;328:99–153. doi: 10.1007/128_2012_323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sousa RJ. Structural Mechanisms of Chaperone Mediated Protein Disaggregation. Frontiers in Molecular Biosciences. 2014;1 doi: 10.3389/fmolb.2014.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rampelt H, et al. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31:4221–35. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iosefson O, Sharon S, Goloubinoff P, Azem A. Reactivation of protein aggregates by mortalin and Tid1--the human mitochondrial Hsp70 chaperone system. Cell Stress Chaperones. 2012;17:57–66. doi: 10.1007/s12192-011-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkler J, Tyedmers J, Bukau B, Mogk A. Chaperone networks in protein disaggregation and prion propagation. J Struct Biol. 2012;179:152–60. doi: 10.1016/j.jsb.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Voisine C, et al. The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell. 1999;97:565–74. doi: 10.1016/s0092-8674(00)80768-0. [DOI] [PubMed] [Google Scholar]
- 13.Matlack KE, Misselwitz B, Plath K, Rapoport TA. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell. 1999;97:553–64. doi: 10.1016/s0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- 14.Misselwitz B, Staeck O, Matlack KE, Rapoport TA. Interaction of BiP with the J-domain of the Sec63p component of the endoplasmic reticulum protein translocation complex. J Biol Chem. 1999;274:20110–5. doi: 10.1074/jbc.274.29.20110. [DOI] [PubMed] [Google Scholar]
- 15.Sousa R, Lafer EM. Keep the traffic moving: mechanism of the Hsp70 motor. Traffic. 2006;7:1596–603. doi: 10.1111/j.1600-0854.2006.00497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De los Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P. Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6166–6171. doi: 10.1073/pnas.0510496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goloubinoff P, De Los Rios P. The mechanism of Hsp70 chaperones: (entropic) pulling the models together. Trends in Biochemical Sciences. 2007;32:372–380. doi: 10.1016/j.tibs.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Ungewickell E, et al. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378:632–5. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- 19.Bocking T, Aguet F, Harrison SC, Kirchhausen T. Single-molecule analysis of a molecular disassemblase reveals the mechanism of Hsc70-driven clathrin uncoating. Nat Struct Mol Biol. 2011;18:295–301. doi: 10.1038/nsmb.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bocking T, et al. Key interactions for clathrin coat stability. Structure. 2014;22:819–29. doi: 10.1016/j.str.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothnie A, Clarke AR, Kuzmic P, Cameron A, Smith CJ. A sequential mechanism for clathrin cage disassembly by 70-kDa heat-shock cognate protein (Hsc70) and auxilin. Proc Natl Acad Sci U S A. 2011;108:6927–32. doi: 10.1073/pnas.1018845108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Jaarsveld PP, Nandi PK, Lippoldt RE, Saroff H, Edelhoch H. Polymerization of clathrin protomers into basket structures. Biochemistry. 1981;20:4129–35. doi: 10.1021/bi00517a028. [DOI] [PubMed] [Google Scholar]
- 23.Xing Y, et al. Structure of clathrin coat with bound Hsc70 and auxilin: mechanism of Hsc70-facilitated disassembly. EMBO J. 2010;29:655–65. doi: 10.1038/emboj.2009.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang J, Prasad K, Lafer EM, Sousa R. Structural Basis of Interdomain Communication in the Hsc70 Chaperone. Molecular Cell. 2005;20:513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang J, et al. Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface. Biochemistry. 2003;42:5748–53. doi: 10.1021/bi034270g. [DOI] [PubMed] [Google Scholar]
- 26.Jiang J, et al. Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell. 2007;28:422–33. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rapoport I, Boll W, Yu A, Bocking T, Kirchhausen T. A motif in the clathrin heavy chain required for the hsc70/auxilin uncoating reaction. Mol Biol Cell. 2008;19:405–13. doi: 10.1091/mbc.E07-09-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benaroudj N, Batelier G, Triniolles F, Ladjimi MM. Self-association of the molecular chaperone HSC70. Biochemistry. 1995;34:15282–90. doi: 10.1021/bi00046a037. [DOI] [PubMed] [Google Scholar]
- 29.Aprile FA, et al. Hsp70 oligomerization is mediated by an interaction between the interdomain linker and the substrate-binding domain. PLoS One. 2013;8:e67961. doi: 10.1371/journal.pone.0067961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preissler S, et al. Physiological modulation of BiP activity by trans-protomer engagement of the interdomain linker. Elife. 2015;4:e08961. doi: 10.7554/eLife.08961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kramer EM, Myers DR. Osmosis is not driven by water dilution. Trends Plant Sci. 2013;18:195–7. doi: 10.1016/j.tplants.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Kramer EM, Myers DR. Five popular misconceptions about osmosis. American Journal of Physics. 2012;80:694–699. [Google Scholar]
- 33.Wilbanks SM, Chen L, Tsuruta H, Hodgson KO, McKay DB. Solution small-angle X-ray scattering study of the molecular chaperone Hsc70 and its subfragments. Biochemistry. 1995;34:12095–106. doi: 10.1021/bi00038a002. [DOI] [PubMed] [Google Scholar]
- 34.Ha JH, McKay DB. ATPase kinetics of recombinant bovine 70 kDa heat shock cognate protein and its amino-terminal ATPase domain. Biochemistry. 1994;33:14625–35. doi: 10.1021/bi00252a031. [DOI] [PubMed] [Google Scholar]
- 35.Northrup SH, Erickson HP. Kinetics of protein-protein association explained by Brownian dynamics computer simulation. Proc Natl Acad Sci U S A. 1992;89:3338–42. doi: 10.1073/pnas.89.8.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zanten JHV, Monbouquette HG. Characterization of Vesicles by Classical Light Scattering. Journal of Colloid and Interface Science. 1991;146:330–336. [Google Scholar]
- 37.Andreasson C, Fiaux J, Rampelt H, Mayer MP, Bukau B. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J Biol Chem. 2008 doi: 10.1074/jbc.M710063200. [DOI] [PubMed] [Google Scholar]
- 38.Schuermann JP, et al. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol Cell. 2008;31:232–43. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q, D'Silva P, Walter W, Marszalek J, Craig EA. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science. 2003;300:139–41. doi: 10.1126/science.1083379. [DOI] [PubMed] [Google Scholar]
- 40.Vilker VL, Colton CK, Smith KA. The Osmotic-Pressure of Concentrated Protein Solutions - Effect of Concentration and Ph in Saline Solutions of Bovine Serum-Albumin. Journal of Colloid and Interface Science. 1981;79:548–566. [Google Scholar]
- 41.Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–84. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 42.Coy DL, Wagenbach M, Howard J. Kinesin takes one 8-nm step for each ATP that it hydrolyzes. J Biol Chem. 1999;274:3667–71. doi: 10.1074/jbc.274.6.3667. [DOI] [PubMed] [Google Scholar]
- 43.Scheele U, Kalthoff C, Ungewickell E. Multiple interactions of auxilin 1 with clathrin and the AP-2 adaptor complex. J Biol Chem. 2001;276:36131–8. doi: 10.1074/jbc.M106511200. [DOI] [PubMed] [Google Scholar]
- 44.Fotin A, et al. Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating. Nature. 2004;432:649–53. doi: 10.1038/nature03078. [DOI] [PubMed] [Google Scholar]
- 45.Ben-Zvi A, De Los Rios P, Dietler G, Goloubinoff P. Active solubilization and refolding of stable protein aggregates by cooperative unfolding action of individual hsp70 chaperones. J Biol Chem. 2004;279:37298–303. doi: 10.1074/jbc.M405627200. [DOI] [PubMed] [Google Scholar]
- 46.Gao X, et al. Human Hsp70 Disaggregase Reverses Parkinson's-Linked alpha-Synuclein Amyloid Fibrils. Mol Cell. 2015;59:781–93. doi: 10.1016/j.molcel.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mattoo RU, Sharma SK, Priya S, Finka A, Goloubinoff P. Hsp110 is a bona fide chaperone using ATP to unfold stable misfolded polypeptides and reciprocally collaborate with Hsp70 to solubilize protein aggregates. J Biol Chem. 2013;288:21399–411. doi: 10.1074/jbc.M113.479253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu X, et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–14. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brizzard BL, Chubet RG, Vizard DL. Immunoaffinity purification of FLAG epitope-tagged bacterial alkaline phosphatase using a novel monoclonal antibody and peptide elution. Biotechniques. 1994;16:730–5. [PubMed] [Google Scholar]
- 50.Ye W, Lafer EM. Clathrin binding and assembly activities of expressed domains of the synapse-specific clathrin assembly protein AP-3. J Biol Chem. 1995;270:10933–9. doi: 10.1074/jbc.270.18.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 52.Abrishami V, et al. A pattern matching approach to the automatic selection of particles from low-contrast electron micrographs. Bioinformatics. 2013;29:2460–8. doi: 10.1093/bioinformatics/btt429. [DOI] [PubMed] [Google Scholar]
- 53.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–4. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de la Rosa-Trevin JM, et al. Xmipp 3.0: an improved software suite for image processing in electron microscopy. J Struct Biol. 2013;184:321–8. doi: 10.1016/j.jsb.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 56.Penczek PA. Three-dimensional spectral signal-to-noise ratio for a class of reconstruction algorithms. J Struct Biol. 2002;138:34–46. doi: 10.1016/s1047-8477(02)00033-3. [DOI] [PubMed] [Google Scholar]
- 57.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.