Abstract
Polysialic acid (polySia) is a unique carbohydrate polymer expressed on the surface of NCAM (neuronal cell adhesion molecule) in a number of cancers where it modulates cell-cell and cell-matrix adhesion, migration, invasion and metastasis and is strongly associated with poor clinical prognosis. We have carried out the first investigation into the effect of polySia expression on the behaviour of cancer cells in hypoxia, a key source of chemoresistance in tumours. The role of polysialylation and associated tumour cell migration and cell adhesion were studied in hypoxia, along with effects on cell survival and the potential role of HIF-1. Our findings provide the first evidence that polySia expression sustains migratory capacity and is associated with tumour cell survival in hypoxia. Initial mechanistic studies indicate a potential role for HIF-1 in sustaining polySia-mediated migratory capacity, but not cell survival. These data add to the growing body of evidence pointing to a crucial role for the polysialyltransferases (polySTs) in neuroendocrine tumour progression and provide the first evidence to suggest that polySia is associated with an aggressive phenotype in tumour hypoxia. These results have significant potential implications for polyST inhibition as an anti-metastatic therapeutic strategy and for targeting hypoxic cancer cells.
Polysialic acid (polySia) is an α-2,8-glycosidically linked polymer of sialic acid, and a developmentally regulated post-translational modification of NCAM (neuronal cell adhesion molecule)1. Cancers of neuroendocrine-origin exhibit selective high level expression of polySia-NCAM as part of the tumour glycocalyx, a term used to describe the myriad of functionally-important carbohydrates that are to be found on the surface of cancer cells2. Tumours where polySia expression has been identified notably include neuroblastoma3,4, lung cancer5,6 and many others1,7,8,9,10,11. Crucially, whilst high levels are expressed during embryonic development, peripheral adult organs do not express polySia-NCAM. This means that the polysialyltransferase (polyST) enzymes (ST8SiaII and ST8SiaIV) responsible for polySia biosynthesis12 have received considerable interest as novel anti-metastatic drug targets, particularly ST8SiaII, which is thought to be the prominent enzyme in tumours1.
PolySia-NCAM expression strongly correlates with the migration and invasion of tumour cells13 and with aggressive, metastatic disease and poor clinical prognosis in the clinic1. Its detailed roles in tumour growth and dissemination continue to emerge, but involve disruption of homo- and heterophilic NCAM interactions, and in modulation of key intracellular signalling pathways, notably FGFR-1, ERK1/2, FAK and c-MET/ALK1,14,15. Furthermore, it has long been proposed that polySia-NCAM expression may protect the tumour cell from immunosurveillance mechanisms, in a manner analogous to bacteria expressing polySia16 and that it is closely associated with tumour chemoresistance17.
The tumour microenvironment is intimately connected with the evolution of cancers and the limited success of cancer treatments. Hypoxia, a condition of low oxygen tension occurring in poorly vascularised areas of tumours, has profound effects on cancer cell growth18,19, metastasis20,21, susceptibility to apoptosis22,23 and resistance to radiotherapy and chemotherapy24,25. Within solid tumours, oxygen delivery to neoplastic and stromal cells in different regions of the tumour varies considerably due to the chaotic nature of the tumour vasculature and the diffusion limit of oxygen of just a few hundred micrometres. Oxygen gradients exist across the tumour with decreasing levels of oxygen as distance from a blood vessel increases. Whilst different levels of hypoxia are thus likely to exist in different parts of the tumour, in general, hypoxic cancer cells are associated with a more aggressive, invasive phenotype26,27,28. The altered glycosylation of cancer cells appears to play a key role in this; promoting loss of cell-cell adhesion and cell migration29,30. However, how glycosylation changes under hypoxia and what effect, if any, this has on the behaviour of cancer cells, such as their growth, survival and invasive potential remain largely unexplored. Given the key role played by polySia in neuroendocrine tumour progression, we hypothesised that polySia may play a crucial role in tumour cell behaviour under hypoxic conditions.
Materials and Methods
Cell lines
Human neuroblastoma SH-SY5Y (ATCC® CRL2266™) and DLD-1 colorectal adenocarcinoma (ATCC® CCL221™) cell lines were obtained from the American Type Culture Collection (ATCC). Human neuroblastoma SH-SY5Y cells were maintained in MEM medium and nutrient mixture F-12 Ham (1:1), supplemented with 10% foetal bovine serum, 1% sodium pyruvate and 1% glutamine. DLD-1 colorectal adenocarcinoma cell lines were maintained in RPMI media supplemented with 10% foetal bovine serum, 1% sodium pyruvate and 1% glutamine. C6-STX and C6-WT cells were obtained from the Fukuda group, Sanford-Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA (for full details, see Suzuki et al.)31. Briefly, C6-STX glioma cells were prepared by transfecting wild-type C6 cells (C6-WT) with the pcDNA3-STX plasmid inserted with cDNA encoded full length human ST8SiaII (also known as STX)31. Both cell lines were maintained in minimum essential medium (MEM Alpha Eagle) with ultraglutamine I, deoxyribonucleoside and ribonucleosides supplemented with 10% foetal bovine serum. All cell lines were maintained in a humid atmosphere of 5% CO2 and 95% air at 37 °C.
For studies carried out under conditions of low oxygen tension (“hypoxic conditions”), all media used was incubated for at least 48 hours under hypoxic conditions (0.1% oxygen) before being added to the cells.
MTT cell viability assay
C6 glioma cells (5 × 103 cells/well) were seeded in 96-well plates. After overnight attachment, the cells were treated with different concentrations of cobalt chloride (Sigma–Aldrich) to induce pseudohypoxia. Following 24 h treatment, cells were washed and fresh media was added. Cells were incubated for another 72 hours and MTT (0.5 mg/ml) was then added followed by incubation for a further 4 hours. The MTT metabolic product (formazan) was dissolved in DMSO (200 μL), and its absorbance was measured at 560 nm. Cell survival was calculated in relation to control (vehicle-treated) cells.
In vitro cell migration assay
Effects on tumour cell migration were analysed using a simple 2D scratch assay13,32. Cells were seeded into six-well plates at different concentrations (1 × 106 cells for SH-SY5Y, 0.5 × 106 cells for C6 and 0.8 × 106 cells for DLD-1), and plates were then incubated overnight at 37 °C in a 5% CO2 humidified atmosphere. Once a confluent monolayer had formed, a 200 μl pipette tip was used to create a scratch wound. The monolayer was then washed with growth medium (1 ml) to remove floating cells and replaced with fresh medium (2 ml) containing only 2% FBS to limit cell proliferation. For experiments under hypoxia, the scratch was performed in a hypoxia station (Whitley H35 hypoxystation; Don Whitley, UK) at 0.1% O2 and hypoxia-equilibrated media was used. Images of the scratch at the start of the experiment were acquired using a Lumascope 500 microscope (Etaluma, USA) and reference points were marked to obtain the same field during the image acquisition after the incubation.
The plates were incubated in either normoxic (i.e. normal laboratory conditions) or hypoxic conditions for 16, 24 and 41 hours for C6, SH-SY5Y and DLD-1 cells respectively. After incubation, plates were placed under the Lumascope 500 microscope, reference points were matched, the photographed region acquired at the first image was aligned and a second image was acquired.
The images acquired were analysed quantitatively by using Image J software. For each image, distances between one side of scratch and the other were measured at certain intervals (μm). By comparing the images from time 0 to the last time point the % wound closure was calculated.
Chromatographic cell-based analysis of polySia expression
C6 cells were seeded at the density of 2 × 105 cells/well in a six-well plate and incubated at 37 °C in either hypoxic or normoxic conditions. After 48 hours, cells were collected by trypsinisation followed by centrifugation for 5 minutes at 1000 rpm. Using routine mild acidic hydrolysis for polySia solubilisation33,34, cell pellets were treated with 100 μL of 0.5% Igepal CA-630 (in distilled water) followed by incubation at 37 °C for 0.5–1 h and vortex mixing for 5 minutes. The insoluble material was removed by centrifugation and the supernatant was used for analysis. For Endo-N treatment for polySia solubilisation, cells were treated with Endo-N (3 μg/ml) for 24 hours prior collecting the media. One well was used as negative control where no Endo-N was added.
For determination of polySia content, samples were hydrolysed in 0.1 M trifluoroacetic acid (TFA) for 2 h at 80 °C. TFA was removed under vacuum and the resulting sialic acid residues were incubated with the DMB reagent for 24 h at 4 °C as described previously13.
Mass spectrometric analysis of DMB-sialic acid
DMB-sialic acid was subjected to analysis by electrospray UPLC-MS using an Acquity UPLC Waters system and a Quattro Premier XE (Waters corporation, USA) mass spectrometer and an Acquity UPLC BEH C18 column (2.1 × 100 mm). The mobile phases used were: ammonium formate (5 mM; mobile phase A) and methanol (mobile phase B), with a flow rate of 0.3 ml/min and isocratic method of 20% mobile phase B. The peaks and masses were integrated using Masslynx software. The source temperature was set to 120 °C. MRM transitions acquired at unit resolution in both the Q1 and Q3 quadrupoles to maximise specificity. Cone voltage (CV) was adjusted to 25 V while collision energy (CE) was adjusted to 20 V using positive ionisation mode. The transitions detected are summarised in Table 1.
Table 1. Transition data for mass spectrometric analysis of DMB-sialic acid.
| Transition | Molecular weight (MW) | Charge Z | Parent ion (m/z) | Daughter ion (m/z) | CV (V) | CE (eV) | Dwell time (sec) |
|---|---|---|---|---|---|---|---|
| 1 | 425.4 | +1 | 426 | 229.23 | 25 | 20 | 0.2 |
| 2 | 425.4 | +1 | 426 | 313.02 | 25 | 20 | 0.2 |
Cell-cell adhesion assay
C6 cells were incubated under hypoxic or normoxic conditions for 48 hours at 37 °C. Matrigel© (200 μl, 40 μg/ml) (VWR, USA) solution was added to each well of a 24-well plate and incubated at 37 °C. After two hours, excess Matrigel was removed and the wells were washed twice with cold PBS. Blocking solution (DMEM media with %10 FBS, 500 μl) was added to each well. The plates were then incubated at 37 °C. After 30 minutes the wells were washed once with cold PBS and fresh culture medium (200 μl) was added and the plates were incubated at 37 °C.
2 × 104 cells in 100 μl medium were added to the pre-coated 24-well plate. Culture medium (300 μl) was added to one well to be used as a blank to calculate the background staining. The plates were incubated at 37 °C for 30 minutes. Non-adherent cells were removed by inverting the plates on a plastic reservoir. Wells were washed two times with 300 μl ice-cold PBS containing 1 mM CaCl2 and 1 mM MgCl2. Cells were fixed by adding ice-cold 100% methanol (300 μl). After 10 minutes, wells were washed with PBS three times and of 0.5% crystal violet solution, prepared in 20% ethanol (100 μl), was added. Plates were placed on a shaker and incubated with crystal violet solution for 10 minutes at room temperature. Excess crystal violet solution was then removed. Crystal violet was then recovered by adding 200 μl of 100% methanol to each well. 100 μl of the extracted crystal violet was transferred to 96-well plate. Absorbance of the samples was measured spectrophotometrically at a wavelength of 540 nm. The background absorbance of multi-well plates was also measured and subtracted from the 540 nm measurements.
Absorbance readings acquired for each sample were analysed quantitatively. Background absorbance of the multi-well plates was subtracted and the % cell adhesion relative to C6-WT cells was calculated (% adhesion = mean absorbance of the C6-WT cells /mean absorbance of C6-STX cells in the same condition × 100). Three independent experiments were carried out, each comprising 4 technical replicates.
Trypan blue exclusion assay for cell viability
C6 cells were seeded in T75 flasks with seeding density of 2.5 × 104 cells/flask, with one flask per treatment incubated in normoxic conditions and the other in hypoxic (0.1% O2) conditions for 96 hours. 0.4% solution of trypan blue was prepared in PBS solution (pH 7.2) and 0.1 mL of trypan blue stock solution was added to 1 mL of trypsinised cells. Each haemocytometer chamber was loaded with 10 μl of the solution and examined immediately under a microscope at low magnification. The number of trypan blue-stained cells and the number of total cells was counted. The % trypan blue-stained cells was calculated as follows: % dead cells = (Average number of trypan blue-stained cells count ÷ average total cell count) × 100.% cell viability was also calculated as follows: % Viable cells = [1.00 – (Number of blue cells ÷ Number of total cells)] × 100.
Flow cytometry (Annexin V apoptosis detection assay)
C6 cells were seeded (8 × 105 cells) in T75 flasks under normoxic or hypoxic (0.1% O2) conditions at 37 °C. After incubation for 96 h, media was collected and cells were trypsinised. Trypsinised cells, along with the collected media, were then centrifuged at 1000 g for 5 minutes. After centrifugation, media was discarded and cell pellets were re-suspended in HEPES buffer (1 ml) as provided with the Annexin V_FLUOS staining kit (Roche) in accordance with the manufacturer’s instructions. Cells were further centrifuged (1500 g, 5 minutes) and pellets were re-suspended in 100 μl HEPES buffer containing Propidium iodide and Annexin V as previously described35. After 15 minutes, 0.5 ml of Annexin-binding buffer was added and cell samples were analysed by flow cytometry.
Western blot analysis
For CoCl2 treatment, C6-STX and C6-WT cells were seeded at a density of 2 × 105 cells/well in a six-well plate and incubated for 24 hours at 37 °C in a 5% CO2 humidified atmosphere to allow cells to adhere. After 24 hours, medium was removed and fresh medium containing different concentrations of CoCl2 was added. Two wells were used as a control where no CoCl2 was added. After 16 hours cells were collected.
Samples were resolved on 12% polyacrylamide gels and blotted onto PVDF membranes (Amersham). Non-specific antibody binding was blocked via incubation with skimmed milk (0.05 g/ml) and the blot was probed with rat anti-HIF-1α antibody (R&D systems, UK) (1:100 dilution) or rat anti-LDH-A antibody (Abcam, UK) (1:500 dilution) overnight at 4 °C. Antibody reactivity was detected by horseradish peroxidase (HRP)-conjugated antibody and chemiluminescence using ECL-Plus (Amersham).
RT-PCR analysis
RNA purification and quantification
C6 were incubated in normoxic or hypoxic conditions for 48 h, after which RNA was extracted using the RNeasy Mini Kit (Qiagen, UK) according to the manufacturer’s instructions. The concentration and quality of the eluted RNA were determined by measuring absorbance at 260 nm and 280 nm using a NanodropTM2000 (Thermo Fisher Scientific) spectrophotometer and determining the 260/280 ratio.
2000 ng of total RNA was reverse transcribed into single-stranded cDNA using the High Capacity cDNA Reverse Transcription Kit (Life technologies) according to manufacturer’s handbook. Once the reaction finished, samples were stored at −20 °C.
Real-time quantitative PCR
qPCR was carried out using the TaqMan Universal PCR Master Mix (Applied Biosystems). The PCR reaction consisted of 12.5 μl of 2X master mix, 1.25 μl of 20X TaqMan Gene Expression Assay Mix, and 11.25 μl cDNA diluted in dH2O. The cDNA samples were diluted 1:5 and 2 μl of diluted cDNA was used per reaction. A 96-well MicroAmp Optical plate (Applied Biosystems) was used and all reactions were performed in triplicate. The validated Taqman primers (Applied Biosystems) utilised are listed in Table 2. qPCR reactions were performed using a AB7500 Real time PCR system (Applied Biosystems). The fold-change in gene expression was determined using the 2-ddCT method36.
Table 2. Validated Taqman primers utilised in PCR experiments.
| Target gene (C6 cell lines) | TaqMan gene expression assay |
|---|---|
| GLUT1 | Rn01402419_g1 |
| HK2 | Rn00562457_m1 |
| LDH-A | Rn00820751_g1 |
Statistics
A minimum of three independent biological experiments were performed for all studies. Statistical analyses were performed using Excel software. Differences between two groups were evaluated with Student’s t test (Two tailed).
Results and Discussion
Evaluation of polySia expression in C6-STX cells under hypoxic conditions
Cells were initially evaluated to assess the effect of hypoxia on polySia expression. The isogenic C6 glioma cells served as an ideal model in this regard. First reported by the Fukuda group31, C6-STX cells have been engineered to express ST8SiaII and thus express polysialylated NCAM (polySia-NCAM)13. The merits of these cells as a model for polySia-expressing cancer cells are well established13,31,37. C6-STX cells migrate and invade significantly faster than C6-WT (wild type) cells in vitro13,31 and in vivo31. Our own experiments have additionally confirmed that levels of polySia expression in C6-STX cells are similar to those observed in naturally expressing cell lines (e.g. SH-SY5Y, data not shown). C6-STX and C6-WT cells are otherwise genetically identical (expressing NCAM, the acceptor for polySia), only differing in the expression of ST8SiaII and associated polySia31. We were thus able to reasonably attribute any effects observed in the subsequent experiments to the presence of polySia.
C6-STX cells were incubated at 37 °C in either 0.1% oxygen, 5% carbon dioxide and 95% nitrogen (hypoxia) or 5% carbon dioxide and 95% air (normoxia). After 48 h, cells were harvested and polySia expression was quantified using a cell-based chromatographic assay (see Methods).
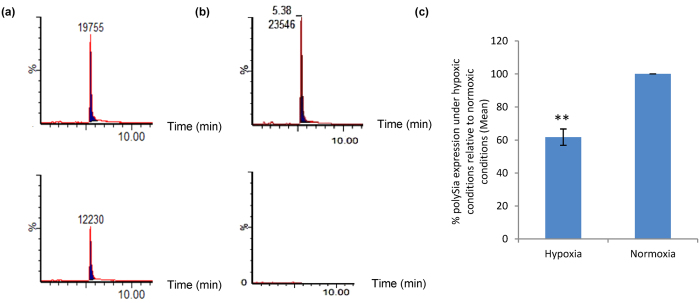
Interestingly, a comparison of the polySia expression of C6-STX cells under hypoxia and normoxia revealed a marked difference. C6-STX cells exhibited a significant reduction in polySia expression of polySia under hypoxia conditions (61.8 ± 4.8% of that measured under normoxia, P = 0.01, Fig. 1). The use of C6-WT cells as a negative control confirmed a lack of polySia expression under both conditions (data not shown) and validated the assay given the isogenic nature of the cells. Cell-based quantification of cell-surface polySia was carried out using both endoneuraminidase-N (Endo-N, an enzyme that selectively cleaves polySia) and mild acidic hydrolysis for the removal of polySia from the cell surface, two techniques routinely available to us in our laboratory. Similar results were obtained from both experiments, giving confidence in the result obtained (SD ± 10%).
Figure 1. Effect of hypoxia (48 hours, 0.1% O2) on polySia expression in C6-STX cells.
(a) Mild acidic hydrolysis combined with UPLC/MS/MS analysis used to detect polySia in C6-STX cells under normoxia (upper trace) and hypoxia (lower trace, after 48 hours). Y-axis represents % abundance of polySia, x-axis represents time. (b) Elution standard for standard DMB-labelled sialic acid (upper trace) and negative control sample where no Endo-N was added as described in the material and methods section (lower trace). (c) Quantification of the mean polySia levels as determined by HPLC. (**P ≤ 0.01).
These data show that steady-state levels of exogenously expressed polySia are reduced in C6-STX cells under hypoxic conditions. These findings are exciting and represent the first such evidence that hypoxia affects polySia expression in tumour cells. That said, these results are consistent with previous studies that have more broadly suggested that acute hypoxia produces a degree of glycocalyx degradation38.
Effect of hypoxia on polySia-mediated tumour cell migration
PolySia has been shown to play a key role in tumour cell migration. Generally speaking, there are many reports demonstrating that hypoxia modulates cancer cell migration39,40. Given our determination that polySia levels were reduced under hypoxic conditions, implications for tumour cell migration were investigated.
The effect of polySia expression on tumour cell migration under hypoxic conditions was once again analysed using the C6 glioma cell line pair: C6-STX (polySia+/NCAM+) and C6-WT (polySia−/NCAM+). We, along with our collaborators before us, have previously demonstrated that C6-STX cells migrate significantly faster than C6-WT cells in normoxia13,31. We additionally evaluated naturally polySia-expressing neuroblastoma cell line SH-SY5Y (polySia+/NCAM+), and utilised DLD-1 colorectal cancer cells as a negative control (polySia−/NCAM−). Migration was evaluated using a simple scratch assay32, routinely used in our laboratory. For the two polySia-expressing cell lines (C6-STX and SH-SY5Y), no difference in cell migration was observed between the hypoxic and normoxic conditions. This is intriguing given our finding that polySia expression is reduced in C6-STX cells under hypoxia. Interestingly, the control cells (C6-WT and DLD-1), which do not express polySia, showed a significant and marked reduction in cell migration under hypoxia (Fig. 2a,b).
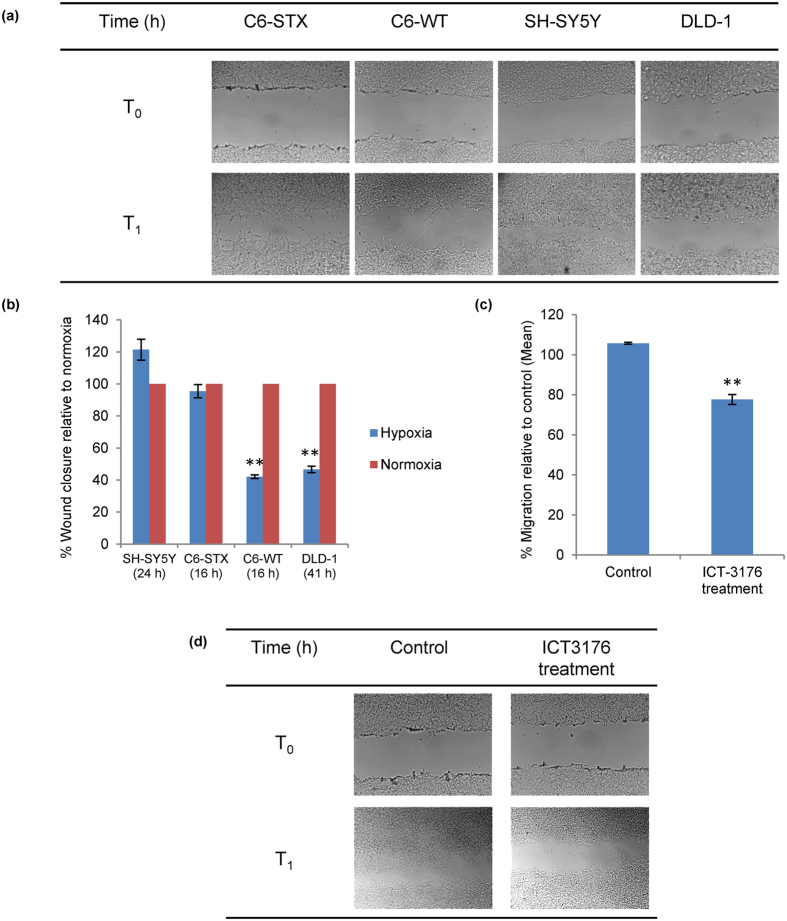
Figure 2. Effect of polySia on the migration of tumour cells under hypoxic conditions.
(a) Representative images of the migration of SH-SY5Y, C6-STX cells (expressing polySia), and control cell lines C6-WT and DLD-1 using time-lapse microscopy under hypoxia, showing the wound size immediately and 16, 24 and 41 hours after scratching; (b) Graph representing the quantification of wound closure calculated by measuring the reduction of wound area over time, using Image J software. Wound closure under hypoxia is calculated as a percentage of wound closure under normoxia for each cell line; (c) Effect of ICT-3176 treatment on C6-STX cell migration under hypoxia; (d) Representative images of the migration of C6-STX cells after treatment with ICT-3176 compared to control untreated cells under hypoxia. T1 = 16 h (C6-STX, C6-WT); 24 h (SH-SY5Y); 41 h (DLD-1). (**P ≤ 0.01).
Given these results, we hypothesised that a polyST inhibitor would reduce polySia expression further and thus may reduce migration of polySia-expressing cells under hypoxia. We have previously demonstrated that polyST inhibitor ICT-3176 reduces tumour cell migration in polySia-expressing cells (including C6-STX and SH-SY5Y cells) with high selectivity41. C6-STX cells were thus treated with ICT-3176 (250 μM) under hypoxic conditions. Cell migration was compared with non-treated cells. Treatment of the cells with ICT-3176 resulted in significant inhibition of C6-STX cell migration (22.4 ± 2.5%, Fig. 2c,d) in hypoxia. As expected, ICT-3176 treatment showed no effect on C6-WT cells.
Taken together, these results demonstrate that the polySia-expressing tumour cells evaluated under hypoxia are able to maintain the migratory capacity observed in normoxia, and this is despite the significant reduction in polySia expression levels noted earlier. Furthermore, polyST inhibition results in reduced migration in hypoxia, as it does in normoxia.
Effect of hypoxia on polySia-mediated cancer cell-matrix adhesion
It was previously reported that polySia modulates cancer cell adhesion through modulation of NCAM-NCAM interactions and NCAM interaction with other molecules in addition to exerting a repulsive force created by the negative charge on the polySia chains. Furthermore, evidence has been reported suggesting that polySia and hypoxia modulate the c-MET signalling pathway, which has a significant role on cancer cell adhesion42.
In order to investigate the effect of hypoxia on polySia-mediated tumour cell-matrix adhesion, an assay was performed with C6-STX and C6-WT cells after 48 hour growth in either normoxic or hypoxic conditions. C6-STX cells were less able to adhere to Matrigel© than C6-WT cells, irrespective of oxygen conditions. Adhesion capacity in polySia-expressing C6-STX cells was approximately 15% less than that observed in control C6-WT cells. There was no difference, however, in cell adhesiveness between normoxic and hypoxic conditions for either C6-STX or C6-WT cells (P = 0.9 for C6-STX cell adhesion in hypoxia compared to normoxia; P = 0.3 for C6-WT adhesion in hypoxia compared to normoxia, see Supplementary data S1).
These results suggest that unlike migration, hypoxia does not appear to have any significant effect on polySia-mediated cell-matrix adhesion, at least to Matrigel in this case. The lack of a difference between adhesion observed in hypoxia and normoxia is perhaps surprising given the lower quantity of polySia expressed by C6-STX cells under hypoxic conditions. It may be that polySia modulates cell adhesion by interacting with different signalling pathways under hypoxic conditions, such as c-MET which is over expressed in hypoxic conditions43,44. This is interesting and certainly worthy of further exploration.
Effect of polySia expression on cancer cell survival in hypoxia
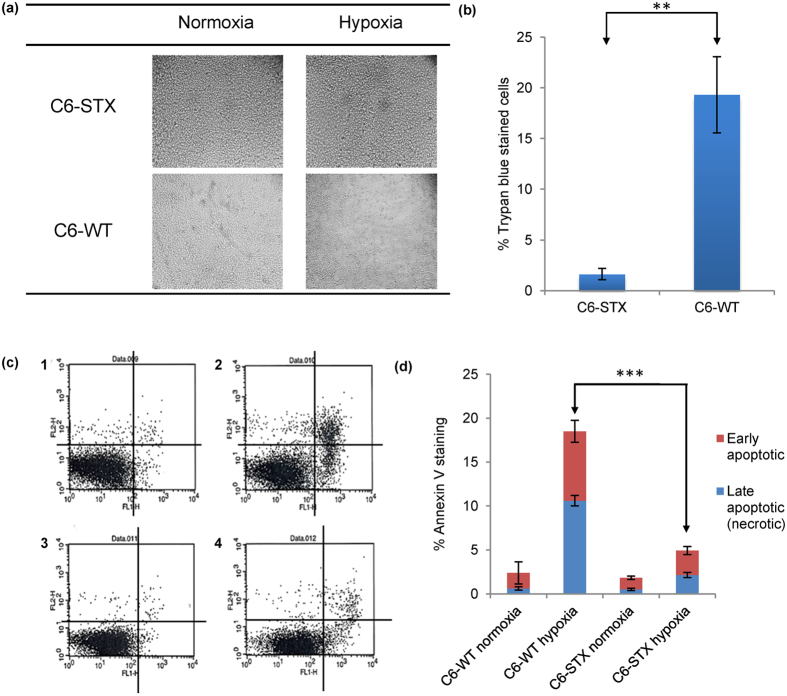
We next considered whether polySia plays any role in cell survival under hypoxic conditions. In order to determine this, C6-STX and C6-WT cells were subjected to hypoxia for 96 h while control cells were incubated for the same duration under normoxic conditions. After 96 h, cells were examined under the microscope and were subsequently collected. Viable and dead cell counts were determined using trypan blue staining. Interestingly, polySia-expressing cells were associated with significantly higher cancer cell viability under hypoxic conditions compared to controls (Fig. 3a). The trypan blue exclusion assay revealed that after 96 h under hypoxia, C6-WT control cells demonstrated significantly reduced viability, as evidenced by a greater number of trypan blue-stained cells (Fig. 3b). C6-WT cells demonstrated cell viability of 80% (i.e. 19.7 ± 3.8% trypan blue-stained cells, Fig. 3b) compared to C6-STX polySia-expressing cells with cell viability of 98% (i.e. 1.6 ± 0.6% trypan-blue stained cells, Fig. 3b).
Figure 3. Effect of polySia expression on the survival of cancer cells in hypoxia.
(a) Morphological change in C6-STX and C6-WT cells after incubation under hypoxic or normoxic conditions for 96 h; (b) Trypan blue exclusion assay, showing the effect of hypoxia on the viability of C6 cells. The graph represents the mean of three independent experiments; (c) Flow cytometry analysis of C6-STX and C6-WT cells apoptosis under hypoxic and normoxic conditions after 96 h. The x-axis shows levels of annexin V cellular staining whereas levels of propidium iodide staining are shown on the y-axis; lower left quadrant of dot plots 1–4: live cells (annexin V-negative, propidium iodide-negative); lower right quadrant: early apoptotic cells (annexin V-positive, propidium iodide-negative); upper right quadrant: late apoptotic or necrotic cells (annexin V-positive, propidium iodide-positive); upper left quadrant: mechanically damaged cells (annexin V-negative, propidium iodide-positive). C1 represents C6-WT in normoxia, C2 represents C6-WT cells in hypoxia, C3 represents C6-STX cells in normoxia, C4 represents C6-STX cells in hypoxia; (d) Percentage of annexin V staining of cell lines under each condition. (**P ≤ 0.01; ***P ≤ 0.001).
In order to further analyse this reduction in cell survival observed under hypoxia and potential mechanisms of increased cell death in the C6-WT cells, levels of apoptosis in hypoxia-cultured C6-STX and C6-WT cells were determined. As shown in Fig. 3c, the apoptotic fraction of C6-WT cells was significantly higher than that of C6-STX cells under hypoxic conditions (18.5% apoptotic cells in C6-WT compared to 4.9% apoptotic cells in C6-STX, i.e. 73.5% less apoptosis in C6-STX cells, Fig. 3d, Table 3), while no significant difference was observed in normoxia. These results strongly suggest that polySia expression plays a key role in enhancing C6 glioma cell survival in hypoxia.
Table 3. Summary percentages of annexin V-stained C6 cells under normoxia and hypoxia.
| Cell line and conditions | Apoptotic cells (%) |
Viable cells (%) | |
|---|---|---|---|
| Late apoptotic (necrotic) | Early apoptotic | ||
| C6-WT (Normoxia) | 0.64 ± 0.19 | 1.75 ± 1.24 | 96.22 ± 1.14 |
| C6-WT (Hypoxia) | 10.6 ± 0.60 | 7.89 ± 1.24 | 79.03 ± 1.50 |
| C6-STX (Normoxia) | 0.53 ± 0.13 | 1.33 ± 0.19 | 97.48 ± 0.37 |
| C6-STX (Hypoxia) | 2.16 ± 0.29 | 2.75 ± 0.46 | 94.45 ± 0.04 |
For comparison, we additionally performed this experiment using SH-SY5Y cells, which naturally express polySia. The levels of apoptosis in SH-SY5Y cells cultured in hypoxia and normoxia were similarly compared. No significant difference in the percentage of viable cells cultured in hypoxia or normoxia was identified, mirroring the results obtained using C6-STX cells (Supplementary data S2).
Investigating a potential role of hypoxia-inducible factor-1 (HIF-1) in the altered behaviour of polySia-expressing tumour cells under hypoxic conditions
A critical mediator of the hypoxic response is the transcription factor hypoxia-inducible factor 1 (HIF-1) that upregulates expression of proteins that promote angiogenesis, anaerobic metabolism, metastasis and many other survival pathways45. Glucose metabolism is upregulated by many cancer cells under hypoxia, generating lactate through the enzyme lactate dehydrogenase A (LDH-A)46, an established HIF-1a target gene47,48. HIF-1 activity in tumours depends on the availability of the HIF-1α subunit, the levels of which increase under hypoxic conditions and through the activation of oncogenes and/or inactivation of tumour suppressor genes49,50.
In order to investigate whether HIF-1α plays any role in the altered polySia-mediated cancer cell behaviour observed under hypoxic conditions, we utilised an established experimental tool to investigate HIF-1 dependency of hypoxia-induced phenomena, namely cobalt chloride (CoCl2). CoCl2 administration under normoxia is thus a simple means by which to mimic the HIF-1-dependent processes that normally occur under hypoxia. CoCl2 stabilises HIF-1α by blocking post-translational modification by prolyl hydroxylase51,52. Whereas CoCl2 lacks exquisite selectivity for HIF-1α, it is routinely used in this context as a tool prior to more complex experiments53,54. In the next few experiments, the effect of HIF-1α induction using CoCl2 on the key polySia-induced behavioural changes of cancer cells under hypoxia were examined.
Determination of CoCl2 cytotoxicity
In order to determine the most suitable CoCl2 concentrations for studying the effect of HIF1α induction on both migration and survival of C6 cell lines, an MTT viability assay was performed.
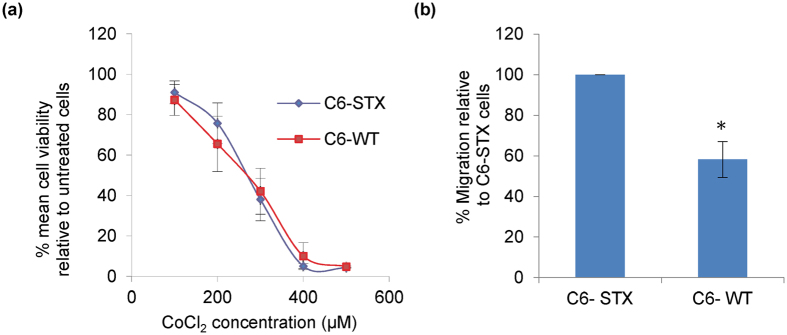
In this experiment, the viability of C6-STX and C6-WT cells was examined after incubation with different concentrations of CoCl2 for 24 hours to determine IC50 values. These were determined to be 258 ± 10 μM for C6-STX cells and 289 ± 12 μM and C6-WT cells (Fig. 4a). For the migration assay, a concentration of CoCl2 that did not cause a significant reduction in the viability of either cell line was used, i.e. 100 μM. For studying the ability of cells to survive under stress conditions, 200 μM of CoCl2 was utilised, a concentration causing approximately 20% loss in viability of cell population.
Figure 4. Effect of CoCl2 treatment on C6 cell lines.
(a) Determination of IC50 of CoCl2 in C6-STX and C6-WT cell lines; (b) Quantification of migration of C6-STX and C6-WT cells after treatment with CoCl2, calculated by measuring the reduction of wound area over time using Image J software. The percentage of C6-WT wound closure after CoCl2 treatment is calculated as a percentage of the C6-STX wound closure. (*P ≤ 0.05).
In the absence of being able to reliably detect changes in HIF-1α expression directly, we focused instead on induction of LDH-A expression to confirm effects of CoCl2, which are well established. Whilst important in itself, LDH-A induction by CoCl2 has additionally been routinely utilised by others as an indirect indicative marker of transcriptionally active and functional HIF-1a expression, given its strong correlation with HIF-1α expression55,56. Expression of LDH-A in both C6-STX and C6-WT CoCl2-treated cells was significantly higher than control untreated cells. On this basis, we could additionally assume induction of HIF-1α expression and pseudohypoxia (Supplementary data S4).
Effect of CoCl2 on polySia-mediated tumour cell migration
In order to investigate whether the effect of polySia expression on enhancing migration of cancer cells under hypoxic conditions is HIF-1α-related, the migration of C6-STX and C6-WT cells was examined following treatment with 100 μM CoCl2 (a non-toxic concentration, as determined above). Both C6-STX and C6-WT cells exhibited a significant reduction in tumour cell migration following CoCl2 treatment (Supplementary data S3). However, CoCl2 had a greater effect on C6-WT cells than C6-STX cells (35 ± 15% greater migration in C6-STX cells compared with C6-WT cells, P = 0.02, Fig. 4b). When considered in conjunction with the results obtained under hypoxia (Fig. 2a), this suggests a link between polySia-mediated migration of tumour cells under hypoxic conditions and LDH-A induction, with implications for HIF-1α.
Effect of CoCl2 on polySia-expressing cancer cell survival
Following our observation the polySia-expressing tumour cells exhibit enhanced cell survival under hypoxia, we evaluated the effect of CoCl2 on cell survival. Percentage cell survival of C6-STX and C6-WT cells was examined after incubation with CoCl2 (200 μM; a concentration selected to study the ability of cells to survive under stress conditions) using the trypan blue exclusion assay. In contrast to the results obtained under hypoxia, no significant difference in cell survival was observed between C6-STX and C6-WT cells (P = 0.26, Supplementary data S5). This suggests that the enhancement of cell survival induced by polySia expression under hypoxic conditions may not be correlated to HIF-1α upregulation.
Differential expression of selected HIF-1α induced genes in C6-STX versus C6-WT cells under hypoxia
Possible cross-talk between polySia and glucose transporters has been previously suggested: N-Acetyl-D-mannosamine (ManNAc) and N-acetyl-D-glucosamine (GlcNAc) are the essential precursors of N-acetylneuraminic acid (NeuAc), the precursor monomer of polySia. Studies on the specificity of the ManNAc transport system have revealed that addition to the uptake mixture of glucose, fructose, mannose, glucosamine, mannosamine and GlcNAc (5 mM), but not other sugars, caused a marked inhibition in ManNAc transport (70, 60, 70, 60, 65 and 85%, respectively)57,58. Although this effect was observed in E. coli, given the link between hypoxia and increased glucose metabolism in cancer cells, this raised the question of possible cross-talk between polySia and these processes. In initial studies, we selected three key glucose metabolism regulators, namely glucose transporter 1 (GLUT-1), hexokinase 2 (HK-2) and the aforementioned LDH-A55 for evaluation59,60,61. The change in expression of these genes was measured under hypoxia and normalised to expression under normoxia.
Gene analysis of the C6 isogenic cells revealed a significant increase in LDH-A gene expression in C6-STX cells under hypoxic conditions as compared to WT cells. This is a significant finding, suggesting a possible role for LDH-A in polySia-mediated changes in cancer cell behaviour in hypoxia (Fig. 5). LDH-A has previously been reported to promote cancer cell survival in multiple cancer cell lines35,46,62, and has also been shown to promote cancer cell migration under hypoxic conditions63.
Figure 5.
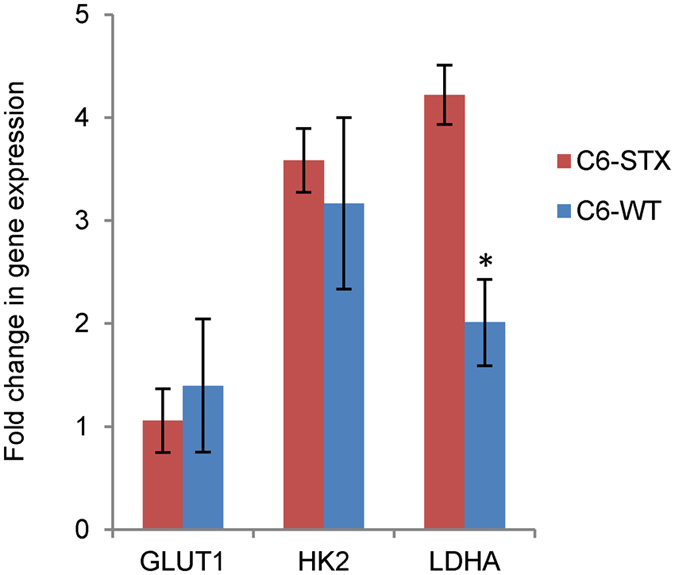
Fold-change in gene expression of selected hypoxia target genes under hypoxic conditions in C6-STX compared to C6-WT cells. (*P ≤ 0.05).
Conclusion
The role of polySia in neuroendocrine tumours in tumour cell growth, dissemination and metastasis has now been widely reported. Whilst polySia expression has previously been proposed to be associated with chemoresistance, the significance of polySia expression in a hypoxic environment has not been studied. We have shown that while hypoxia leads to a reduction in polySia expression, this is strongly associated with increased cell survival, and sustains a migratory capacity. Initial mechanistic studies indicate a potential role for HIF-1 in sustaining polySia-mediated migratory capacity, but not cell survival. Furthermore, our study additionally provides the first evidence of a role for LDH-A, a key regulator of glucose metabolism under hypoxia in cancer cells, in these processes.
This study paves the way for more detailed studies in a broader array of neuroendocrine cancer cell lines, and invites further investigations using more complex in vitro models (e.g. 3D spheroids) and ultimately in vivo. Our ability to modulate polySia expression using small molecule polyST inhibitor ICT-3176, in conjunction with our striking finding that cells engineered to express polySia exhibit significantly enhanced survival in a hypoxic environment, raises exciting therapeutic implications for the targeting of hypoxic cells and for future anti-metastatic and combination therapies.
Additional Information
How to cite this article: Elkashef, S. M. et al. Polysialic acid sustains cancer cell survival and migratory capacity in a hypoxic environment. Sci. Rep. 6, 33026; doi: 10.1038/srep33026 (2016).
Supplementary Material
Acknowledgments
Prof Minoru Fukuda (Sanford Burnham Prebys Medical Discovery Institute, USA) is thanked for providing the C6 cells utilised in this study. Members of the ICT team are thanked for technical assistance. Dr. Lina Elsalem is thanked for useful discussions. This work was primarily supported by a PhD studentship for SME (RAF) and partly by Yorkshire Cancer Research (PML, KP, RAF), a Prostate Cancer UK studentship for MS (KP) and a Wellcome Trust grant (RAF).
Footnotes
Author Contributions S.M.E. designed the experiments in conjunction with R.A.F., performed all experiments except those carried out by the individuals listed, analysed data, prepared the figures and co-wrote/edited the manuscript. S.J.A. assisted with flow cytometry experiments, provided input into experimental design and edited the manuscript. M.S. performed the RT-PCR experiments. H.A.B. performed the western blots. G.R.M. synthesised ICT-3176. P.M.L. co-supervised the work and edited the manuscript. K.P. provided input into experimental design, co-supervised elements of the work and edited the manuscript. R.A.F. oversaw the project, contributed to data analysis/discussion and co-wrote and edited the manuscript.
References
- Falconer R. A., Errington R. J., Shnyder S. D., Smith P. J. & Patterson L. H. Polysialyltransferase: a new target in metastatic cancer. Curr. Cancer Drug Targets 12, 925–939 (2012). [DOI] [PubMed] [Google Scholar]
- Pinho S. S. & Reis C. A. Glycosylation in cancer: mechanisms and clinical implications. Nature Rev. Cancer 15, 540–555, (2015). [DOI] [PubMed] [Google Scholar]
- Hildebrandt H. et al. Polysialic acid on the neural cell adhesion molecule correlates with expression of polysialyltransferases and promotes neuroblastoma cell growth. Cancer Res. 58, 779–784 (1998). [PubMed] [Google Scholar]
- Valentiner U., Muhlenhoff M., Lehmann U., Hildebrandt H. & Schumacher U. Expression of the neural cell adhesion molecule and polysialic acid in human neuroblastoma cell lines. Int. J. Oncol. 39, 417–424 (2011). [DOI] [PubMed] [Google Scholar]
- Tanaka F. et al. Prognostic significance of polysialic acid expression in resected non-small cell lung cancer. Cancer Res. 61, 1666–1670 (2001). [PubMed] [Google Scholar]
- Tanaka F. et al. Expression of Polysialic Acid and STX, a Human Polysialyltransferase, Is Correlated with Tumor Progression in Non-Small Cell Lung Cancer.Cancer Res. 60, 3072–3080 (2000). [PubMed] [Google Scholar]
- Roth J. et al. Presence of the long chain form of polysialic acid of the neural cell adhesion molecule in Wilms’ tumor. Identification of a cell adhesion molecule as an oncodevelopmental antigen and implications for tumor histogenesis. Am. J. Pathol. 133, 227–240 (1988). [PMC free article] [PubMed] [Google Scholar]
- Figarella-Branger D., Durbec P. & Rougon G. Differential spectrum of expression of neural cell adhesion molecule isoforms and L1 adhesion molecules on human neuroectodermal tumors. Cancer Res. 50, 6364–6370 (1990). [PubMed] [Google Scholar]
- Lahr G. et al. Neural cell adhesion molecules in rat endocrine tissues and tumor cells: distribution and molecular analysis. Endocrinology 132, 1207–1217 (1993). [DOI] [PubMed] [Google Scholar]
- Komminoth P. et al. Polysialic Acid of the Neural Cell Adhesion Molecule in the Human Thyroid: A Marker for Medullary Thyroid Carcinoma and Primary C-Cell Hyperplasia: An Immunohistochemical Study on 79 Thyroid Lesions. Am. J. Surg. Pathol. 18, 399–411 (1994). [DOI] [PubMed] [Google Scholar]
- Trouillas J. et al. Polysialylated neural cell adhesion molecules expressed in human pituitary tumors and related to extrasellar invasion. J. Neurosurg. 98, 1084–1093 (2003). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. Enhanced expression of polysialic acid correlates with malignant phenotype in breast cancer cell lines and clinical tissue samples. Int. J. Mol. Medicine 37, 197–206 (2016). [DOI] [PubMed] [Google Scholar]
- Al-Saraireh Y. M. et al. Pharmacological inhibition of polysialyltransferase ST8SiaII modulates tumour cell migration. PLoS One 8, e73366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Dai G., Cheng Y. B., Qi X. & Geng M. Y. Polysialylation promotes neural cell adhesion molecule-mediated cell migration in a fibroblast growth factor receptor-dependent manner, but independent of adhesion capability. Glycobiology 21, 1010–1018 (2011). [DOI] [PubMed] [Google Scholar]
- Seidenfaden R., Krauter A., Schertzinger F., Gerardy-Schahn R. & Hildebrandt H. Polysialic acid directs tumor cell growth by controlling heterophilic neural cell adhesion molecule interactions. Mol. Cell. Biol. 23, 5908–5918 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiberger F. et al. Biochemical characterization of a Neisseria meningitidis polysialyltransferase reveals novel functional motifs in bacterial sialyltransferases. Mol. Microbiol. 65, 1258–1275 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaheta R. A. et al. Chemoresistance induces enhanced adhesion and transendothelial penetration of neuroblastoma cells by down-regulating NCAM surface expression. BMC Cancer 6, 294 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A. L. Hypoxia–a key regulatory factor in tumour growth. Nat. Rev. Cancer 2, 38–47 (2002). [DOI] [PubMed] [Google Scholar]
- Kunz M. & Ibrahim S. M. Molecular responses to hypoxia in tumor cells. Mol. Cancer 2, 23 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subarsky P. & Hill R. P. The hypoxic tumour microenvironment and metastatic progression. Clin. Exp. Metastasis 20, 237–250 (2003). [DOI] [PubMed] [Google Scholar]
- Gilkes D. M., Semenza G. L. & Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat. Rev. Cancer 14, 430–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greijer A. E. & van der Wall E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J. Clin. Pathol. 57, 1009–1014 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sermeus A. et al. Hypoxia-induced modulation of apoptosis and BCL-2 family proteins in different cancer cell types. PLoS One 7, e47519 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardin P. et al. A biology-driven approach identifies the hypoxia gene signature as a predictor of the outcome of neuroblastoma patients. Mol. Cancer 9, 185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. M. Tumor hypoxia in cancer therapy. Methods Enzymol. 435, 297–321 (2007). [DOI] [PubMed] [Google Scholar]
- Padhani A. R., Krohn K. A., Lewis J. S. & Alber M. Imaging oxygenation of human tumours. Eur. Radiol. 17, 861–872 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel P., Kallinowski F. & Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 49, 6449–6465 (1989). [PubMed] [Google Scholar]
- Hockel M. & Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J. Natl. Cancer Inst. 93, 266–276 (2001). [DOI] [PubMed] [Google Scholar]
- Fujimoto I., Bruses J. L. & Rutishauser U. Regulation of cell adhesion by polysialic acid. Effects on cadherin, immunoglobulin cell adhesion molecule, and integrin function and independence from neural cell adhesion molecule binding or signaling activity. J. Biol. Chem. 276, 31745–31751 (2001). [DOI] [PubMed] [Google Scholar]
- Guan F., Wang X. & He F. Promotion of cell migration by neural cell adhesion molecule (NCAM) is enhanced by PSA in a polysialyltransferase-specific manner. PLoS One 10, e0124237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. et al. Polysialic acid facilitates tumor invasion by glioma cells. Glycobiology 15, 887–894 (2005). [DOI] [PubMed] [Google Scholar]
- Liang C. C., Park A. Y. & Guan J. L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2, 329–333 (2007). [DOI] [PubMed] [Google Scholar]
- Inoue S. & Inoue Y. Ultrasensitive analysis of sialic acids and oligo/polysialic acids by fluorometric high-performance liquid chromatography. Methods Enzymol. 362, 543–560 (2003). [DOI] [PubMed] [Google Scholar]
- Nishimura S. et al. Comparison of Analytical Methods to Detect Polysialic Acid. J. Glycomics Lipidomics 4, 113 (2014). [Google Scholar]
- Allison S. J. et al. Identification of LDH-A as a therapeutic target for cancer cell killing via (i) p53/NAD(H)-dependent and (ii) p53-independent pathways. Oncogenesis 3, e102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Grant C. A. et al. The Effect of Polysialic Acid Expression on Glioma Cell Nano-mechanics. Bionanoscience 6, 81–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P. I. et al. Effect of acute hypobaric hypoxia on the endothelial glycocalyx and digital reactive hyperemia in humans. Front. Physiol. 5, 459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J. V. et al. Hypoxia enhances migration and invasion in glioblastoma by promoting a mesenchymal shift mediated by the HIF1alpha-ZEB1 axis. Cancer Lett. 359, 107–116 (2015). [DOI] [PubMed] [Google Scholar]
- Nagelkerke A. et al. Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response. Breast Cancer Res. 15, R2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkashef S. M. et al. Polysialyltransferase ST8SiaII: A novel target for the treatment of neuroblastoma. Cancer Res. 75 (Suppl 15), 5431 (2015). [Google Scholar]
- Tate A. et al. Met-Independent Hepatocyte Growth Factor-mediated regulation of cell adhesion in human prostate cancer cells. BMC Cancer 6, 197 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ S. L. & Tsao M. S. An overview of the c-MET signaling pathway. Ther. Adv. Med. Oncol. 3, S7–S19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra J. R. & Tsao M. S. c-MET as a potential therapeutic target and biomarker in cancer. Ther. Adv. Med. Oncol. 3, S21–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh M. Y., Spivak-Kroizman T. R. & Powis G. HIF-1alpha and cancer therapy. Recent Results Cancer Res. 180, 15–34 (2010). [DOI] [PubMed] [Google Scholar]
- Le A. et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl. Acad. Sci. USA 107, 2037–2042 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G. L. et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271, 32529–32537 (1996). [DOI] [PubMed] [Google Scholar]
- Semenza G. L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest. 123, 3664–3671 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziello J. E., Jovin I. S. & Huang Y. Hypoxia-Inducible Factor (HIF)-1 regulatory pathway and its potential for therapeutic intervention in malignancy and ischemia. Yale J. Biol. Med. 80, 51–60 (2007). [PMC free article] [PubMed] [Google Scholar]
- Powis G. & Kirkpatrick L. Hypoxia inducible factor-1alpha as a cancer drug target. Mol. Cancer Ther. 3, 647–654 (2004). [PubMed] [Google Scholar]
- An W. G. et al. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature 392, 405–408 (1998). [DOI] [PubMed] [Google Scholar]
- Yeom C. J. et al. Visualization of hypoxia-inducible factor-1 transcriptional activation in C6 glioma using luciferase and sodium iodide symporter genes. J. Nucl. Med. 49, 1489–1497 (2008). [DOI] [PubMed] [Google Scholar]
- Ardyanto T. D., Osaki M., Tokuyasu N., Nagahama Y. & Ito H. CoCl2-induced HIF-1alpha expression correlates with proliferation and apoptosis in MKN-1 cells: a possible role for the PI3K/Akt pathway. Int. J. Oncol. 29, 549–555 (2006). [PubMed] [Google Scholar]
- Piret J. P., Mottet D., Raes M. & Michiels C. CoCl2, a chemical inducer of hypoxia-inducible factor-1, and hypoxia reduce apoptotic cell death in hepatoma cell line HepG2. Ann. N. Y. Acad. Sci. 973, 443–447 (2002). [DOI] [PubMed] [Google Scholar]
- Denko N. C. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nature Rev. Cancer 8, 705–713 (2008). [DOI] [PubMed] [Google Scholar]
- Hu C. J., Wang L. Y., Chodosh L. A., Keith B. & Simon M. C. Differential Roles of Hypoxia-Inducible Factor 1α (HIF-1α) and HIF-2α in Hypoxic Gene Regulation. Mol. Cell. Biol. 23, 9361–9374 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla-Nuin B., Reglero A., Ferrero M. A. & Rodriguez-Aparicio L. B. Uptake of N-acetyl-D-mannosamine: an essential intermediate in polysialic acid biosynthesis by Escherichia coli K92. FEBS Lett. 449, 183–186 (1999). [DOI] [PubMed] [Google Scholar]
- Revilla-Nuin B. et al. Transport of N-acetyl-D-mannosamine and N-acetyl-D-glucosamine in Escherichia coli K1: effect on capsular polysialic acid production. FEBS Lett. 511, 97–101 (2002). [DOI] [PubMed] [Google Scholar]
- Jiang S. et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 31, 1985–1998 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airley R. et al. Glucose transporter glut-1 expression correlates with tumor hypoxia and predicts metastasis-free survival in advanced carcinoma of the cervix. Clin Cancer Res 7, 928–934 (2001). [PubMed] [Google Scholar]
- Han T. et al. How does cancer cell metabolism affect tumor migration and invasion? Cell Adh. Migr. 7, 395–403 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin V. R., St-Pierre J. & Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9, 425–434 (2006). [DOI] [PubMed] [Google Scholar]
- Maftouh M. et al. Synergistic interaction of novel lactate dehydrogenase inhibitors with gemcitabine against pancreatic cancer cells in hypoxia. Br. J. Cancer 110, 172–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.