Summary
The ability for the obligate anaerobe, Clostridium difficile, to form a metabolically dormant spore is critical for the survival of this organism outside of the host. This spore form is resistant to a myriad of environmental stresses, including heat, desiccation and exposure to disinfectants and antimicrobials. These intrinsic properties of spores allow C. difficile to survive long-term in an oxygenated environment, to be easily transmitted from host-to-host and to persist within the host following antibiotic treatment. Because of the importance of the spore form to the C. difficile lifecycle and treatment and prevention of C. difficile infection (CDI), the isolation and purification of spores are necessary to study the mechanisms of sporulation and germination, investigate spore properties and resistances, and for use in animal models of CDI. This chapter provides basic protocols, in vitro growth conditions and additional considerations for purifying C. difficile spores for a variety of downstream applications.
Keywords: Clostridium difficile, sporulation, endospore, anaerobe, anaerobic chamber, antibiotic-associated diarrhea
1. Introduction
Clostridium difficile is a significant gastrointestinal pathogen of humans and other animals and is the primary agent of antibiotic-associated diarrhea. C. difficile infection (CDI) is responsible for billions of dollars in healthcare costs and causes greater than 14,000 deaths annually in the United States alone [1, 2]. C. difficile was recently named an urgent public health threat by the United States Centers for Disease Control and Prevention [1]. This anaerobic organism survives outside of its host by forming a metabolically inactive spore. Spore formation is a complex process in which a vegetative cell forms a dormant structure that protects C. difficile from exposure to oxygen, heat, disinfectants and desiccation and provides inherent resistance to antibiotics and other antimicrobials. Once ingested by the host, the spore germinates in the presence of bile salts [3], producing the vegetative form of the bacterium. The vegetative cell reproduces and secretes toxins, which cause disease symptoms. As the vegetative cells transit through the gastrointestinal tract, unknown host and intracellular signals spur sporulation. Once shed from the host in feces, C. difficile spores can survive in the environment for long periods of time and are an enormous obstacle in preventing CDI in short-term and long-term healthcare facilities.
Because C. difficile spores are the infectious form of the organism, they are used to study infection in animal models of CDI [4-6] and to perform molecular biological experiments to discover sporulation and germination characteristics, resistances and susceptibilities [7-9]. The emergence of the epidemic 027 ribotype strain [10] has underscored the need for further studies on spore properties and the development of antimicrobials that directly target the C. difficile spore.
Here, we describe how to elucidate the best conditions to optimize the production of C. difficile spores in vitro. We provide protocols that allow for the isolation of C. difficile spores and for further spore preparation purification, necessary for some downstream applications, of spore preparations. These protocols rely on the intrinsic heat, cold and chemical resistances of C. difficile spores to kill vegetative cells and separate these and other debris from the spores. Finally, conditions and additional considerations for the use and long-term storage of C. difficile spores are discussed. Propagation and purification of C. difficile spores is relatively simple as long as a strict anaerobic environment is provided during C. difficile growth and spore formation.
2. Materials
For reliable growth of C. difficile, is it critical to pre-reduce all liquid and solid media in the anaerobic chamber for at least two hours before use. Handle all materials with appropriate aseptic technique to prevent contamination from other prevalent spore forming species (e.g. Clostridium perfringens).
2.1. Culture Media
Brain Heart Infusion Supplement (BHIS) Medium: Dissolve 37 g brain heart infusion extract (BD Difco) and 5 g yeast extract (BD Difco) in 800 ml dH2O. Bring to 1 L with dH2O. Autoclave for 30 min and store at room temperature. Add 15 g agar per liter before autoclaving for 1.5% (w/v) agar plates.
BHIS medium supplemented with 0.1% taurocholate: Prepare medium as described above and autoclave. To prevent the growth of contaminating Mycoplasma species on the surface of plates containing taurocholate (which is a cholesterol derivative [11]), add 10 ml of 10% taurocholate to 1 L BHIS when the autoclaved medium reaches ~60-65°C. Incubate medium with 0.1% taurocholate for 20 min as the medium cools, before pouring plates.
70:30 Sporulation Medium [12; Note 1]: Dissolve 63 g Bacto peptone (BD Difco), 3.5 g proteose peptone (BD Difco), 0.7 g ammonium sulfate ((NH4)2SO4), 1.06 g Tris base, 11.1 g brain heart infusion extract (BD Difco) and 1.5 g yeast extract (BD Difco) in 800 ml dH2O. Bring to 1 L with dH2O. Autoclave for 30 min; once medium is cool to touch, add 3 ml 10% (w/v) cysteine (dissolve 0.4 g cysteine in 4 ml dH2O and filter sterilize with a 0.45 μm filter; see Note 2). Add 15 g agar per liter before autoclaving for 1.5% (w/v) agar plates; use 35 ml per plate for consistent sporulation results. Important: 70:30 sporulation medium provides the best conditions for efficient C. difficile sporulation if used fresh (within a week; see Note 3).
2.2. Media Supplements
Taurocholate: To induce germination of C. difficile spores, 0.1% (w/v) taurocholate must be supplemented in BHIS medium [13, 14]. Prepare a stock solution of 10% (w/v) taurocholate (1 g taurocholate dissolved in 10 ml dH2O) and filter sterilize with a 0.22 μm filter. Store at room temperature.
Fructose: To prevent high rates of sporulation of C. difficile in overnight cultures (unpublished data, Edwards and McBride), 0.2% (w/v) fructose may be added to BHIS medium. Prepare a stock solution of 20% (w/v) D-fructose (2 g fructose dissolved in 10 ml dH2O) and filter sterilize with a 0.45 μm filter. Store at room temperature.
Antibiotic supplements: The use of antibiotics can influence spore formation in C. difficile [15]; however, antibiotic supplements are necessary for maintaining plasmids. Using the lowest concentration of antibiotic necessary for stable plasmid maintenance is strongly recommended (e.g. use a final concentration of 2 μg/mL thiamphenicol [15] instead of the commonly used 10-15 μg/mL thiamphenicol [16, 17]).
2.3. Solutions for C. difficile spore stocks
10X Phosphate Buffered Saline (PBS) Stock Solution: Dissolve 25.6 g Na2HPO4 · 7H2O, 80 g NaCl, 2 g KCl and 2 g KH2PO4 in 800 ml dH2O. Bring to 1 L. Autoclave for 30 min and store at room temperature.
1X PBS: Dilute 10X PBS Stock Solution 1:10 (100 ml 10X PBS into 900 ml dH2O). Sterilize by autoclaving for 30 min or using a 0.45 μm vacuum filter. Store at room temperature.
1X PBS + 1% (w/v) BSA: Dissolve 10 g bovine serum albumin (BSA) into 1 L 1X PBS. Sterilize only by using a 0.45 μm vacuum filter as BSA denatures and precipitates in high heat and pressure. Store at room temperature.
2.4. Additional reagents
95% ethanol
0.7% (w/v) agarose: Dissolve 0.7 g agarose in 100 ml of dH2O and heat by stir plate or microwave to fully solubilize agarose. The agarose solution can be stored as a solid at room temperature and be reheated for subsequent use or as a liquid at 60°C for long-term storage.
50% (w/v) sucrose: Dissolve 50 g in 100 ml dH2O. Filter sterilize.
3. Methods
One of the more vital considerations to remember when working with C. difficile spores is that their hydrophobic and anionic properties allow them to adhere to many surfaces, including pipet tips and polypropylene. When working with spore stocks, it is important to pipet up and down several times, especially when making dilutions, to ensure as few spores as possible are left behind. The use of polypropylene materials, such as conical tubes, is sufficient when working with high concentrations of spore stocks (greater than 107 colony forming units (CFU)/ml) for a short period of time; however, for long-term storage of high concentration spore stocks and when diluting spores to a concentration of 106 CFU/ml or less, the use of glass [15] or Teflon-coated tubes [18] is highly recommended to prevent loss and obtain reproducible spore counts. Finally, the use of 1% BSA in final spore preparations decreases clumping of spores, prevents loss and facilitates accurate enumeration [15].
While it is not necessary to use phase contrast microscopy to track sporulation, as described in Section 3.1, the use of this method is a reliable and efficient way to determine the progress of sporulation without removing the plates from the anaerobic chamber and interrupting an experiment. This technique allows for the accurate enumeration of sporulation efficiency in various conditions and at different time points before embarking on the more time-consuming spore isolation and purification protocol.
3.1 Growth and sporulation efficiency of C. difficile on various media
This protocol describes the general methods needed for determining the best sporulation conditions in preparation for isolating and purifying spore stocks. Methods for general cultivation of C. difficile and anaerobic chamber maintenance can be found elsewhere [19, 20].
Streak strain(s) from glycerol stock(s) onto BHIS medium agar plates and incubate overnight at 37°C.
Inoculate 10 ml BHIS supplemented with 0.1% taurocholate and 0.2% fructose from a single colony using a sterile inoculating loop (see Note 4). Incubate overnight at 37°C.
In the morning, while the overnight culture is still in active growth (OD600 < 1.0; again, see Note 4), backdilute the cells to an OD600 of 0.5 with BHIS. Spread 250 μl of culture onto a fresh, pre-reduced 70:30 agar plate and incubate anaerobically at 37°C for a minimum of 24 h.
After a minimum of 24 h, take a small sample from the plate with a sterile inoculating loop and resuspend in 500 μl BHIS in a microcentrifuge tube. Remove sample(s) from the anaerobic chamber (see Note 5).
Centrifuge the cells at 13000 rpm for 30 sec at room temperature and decant the supernatant. Resuspend the cells in 10 – 50 μl BHIS.
Prepare a microscope slide by applying a small amount (< 0.5 ml) of 0.7% agarose in the center of slide (see Note 6). Allow to solidify for ~2 min.
Apply 5 μl of resuspended culture to the agarose pad and carefully place a coverslip over the sample.
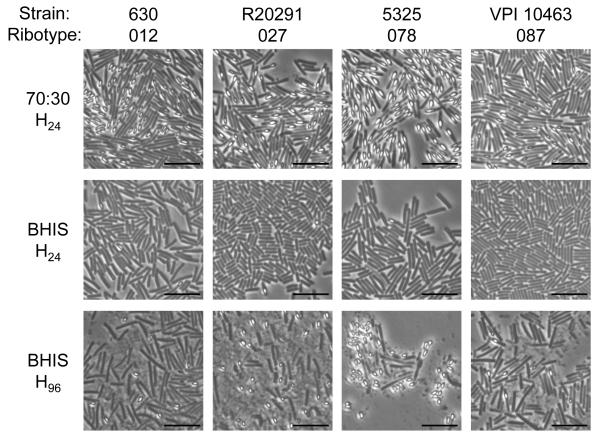
Observe cells by phase contrast microscopy (see Note 7). Mature spores will appear phase bright while vegetative cells and prespores will appear phase dark and phase gray (partially refractile), respectively (Figure 1). Ensure that the field of view contains only a single layer of cells, as overlapping vegetative cells may falsely appear phase bright.
- To determine the sporulation efficiency, enumerate the number of phase bright spores and phase dark vegetative cells and use the equation detailed below:
Count at least 1000 cells to obtain an accurate representation of the population. This protocol can be performed and repeated on different types of medium and at multiple time points to determine the best conditions for sporulation for the strain or mutant of interest (Figure 1; see Note 8).
Figure 1.
Phase contrast micrographs of C. difficile strains 630 [32, 33], R20291 [34], 5325 (ATCC BAA-1875) and VPI 10463 (ATCC 43255) grown on 70:30 sporulation medium agar plates or BHIS agar plates for the indicated amount of time (h). All strains were obtained from the ATCC except R20291, which was a gift from Linc Sonenshein. Images were corrected for exposure differences, which was applied uniformly across each image.
3.2 Isolation of C. dfficile spores from solid medium
This method relies on the intrinsic alcohol and heat resistance of C. difficile spores to kill vegetative cells present in the spore stock. Importantly, this protocol does not remove lysed or killed vegetative cells or debris; however, this is not necessary for many downstream applications, including most animal studies [15]
Once the optimal sporulation conditions are determined for the C. difficile strain(s) of interest (Section 3.1), inoculate 3-10 fresh agar plates with an actively growing overnight culture, as outlined in Section 3.1, and incubate for the appropriate amount of time.
At the appropriate time point(s), verify spore formation via phase contrast microscopy as detailed above.
Remove plates from the anaerobic chamber. Scrape off the entire bacterial lawn using a sterile inoculating loop and resuspend in 10 ml 1X PBS. Vortex well to thoroughly resuspend the cells.
Centrifuge the cells at 3000 rpm for 10 min at room temperature.
Decant the supernatant and thoroughly resuspend the cell pellet in 10 ml 1X PBS.
Add 10 ml of 95% ethanol to the resuspended cell pellet, vortex well and incubate at room temperature for 1 h.
Centrifuge the cells at 3000 rpm for 10 min at room temperature.
Decant the supernatant and resuspend in 20 ml 1X PBS. Wash twice more in 20 ml 1X PBS to ensure complete removal of residual ethanol.
Resuspend the cell pellet in 10 ml 1X PBS + 1% BSA after final wash.
Heat spore stock to 70°C for 20 min (see Note 9). Allow spore stock to cool to room temperature. Time permitting, immediately enumerate the spore stock (see Section 3.4).
3.3 Purification of C. difficile spores
Some downstream applications, including spore germination and outgrowth studies [7, 9], require pure spore stocks in which the spores are physically separated from vegetative cells, lysed mother cells and other debris. This protocol describes a slightly different purification method than described in Section 3.2, and utilizes a density gradient centrifugation step to obtain free C. difficile spores [7, 9, 21].
Propagate spores as described in Section 3.1 to achieve the highest sporulation efficiency possible. At the appropriate time point(s), verify spore formation via phase contrast microscopy as detailed above.
Remove plates from the chamber and resuspend cells from one-half of a plate in 1 ml sterile, ice cold dH2O. Repeat for the remaining half.
Centrifuge at 13000 rpm for 2 min at 4°C. Carefully remove supernatant and resuspend in 1 ml sterile, ice cold dH2O. Wash with sterile, ice cold dH2O two more times.
Incubate the spore preparation at −20°C for 48 h to facilitate the lysis of mother cells and subsequent release of mature endospores.
After incubation, centrifuge at 13000 rpm for 2 min at 4°C, decant the supernatant and resuspend the spore preparation with 1 ml sterile, ice cold dH2O. Repeat for a total of five washes in sterile, ice-cold dH2O as described in Step 3.
After the final wash, combine both cell pellets in 3 ml sterile, ice cold dH2O.
Apply the entire 3 ml spore preparation slowly to the top of a 10 ml 50% sucrose gradient in a 15 ml polypropylene conical tube.
Centrifuge in a swinging bucket rotor at 4000 × g for 20 min at 4°C. The vegetative cells and debris will collect at the interface and throughout the gradient, while the spores move through the gradient and form a pellet at the bottom of the centrifuge.
Remove the cell debris at the interface, then remove the rest of the solution, leaving the spore pellet.
Resuspend the spore pellet in 1 ml of sterile dH2O. Centrifuge at 13000 rpm for 2 min at room temperature, decant the supernatant and resuspend the spore preparation in 1 ml sterile dH2O.
Repeat step 10 for a total of five washes in sterile dH2O and three washes in 1X PBS + 1% BSA.
Resuspend the final spore preparation in 1 ml 1X PBS + 1% BSA and quantify as described in Section 3.4.
3.4 Enumeration of initial spore stock and serial dilutions
To determine the number of viable spores per ml for the spore preparation, a range of serial dilutions of the primary, highly concentrated spore stock are plated and enumerated. The range of serial dilutions required to achieve accurate enumeration of a spore stock depends on the sporulation efficiency of the strain, the type and number of agar plates used for spore propagation, and the length of time the plates are incubated before beginning spore isolation and purification. Remember: when making serial dilutions of spores, vortex each dilution well and pipet up and down multiple times to reduce the retention of spores on tube walls and in pipet tips.
Pre-reduce the same number of BHIS supplemented with 0.1% taurocholate agar plates as the number of serial dilutions performed.
Vortex the spore stock vigorously for 15 s.
Prepare a series of 10-fold dilutions in 1X PBS + 1% BSA. Dilute 100 μl of the spore stock into 900 μl 1X PBS + 1% BSA (which results in the 10−1 dilution). Dilute 100 μl of the 10−1 dilution into 900 μl 1X PBS + 1% BSA (which results in the 10−2 dilution). Continue until the appropriate dilutions have been made (see Note 10 for an example).
Heat the dilutions to 55°C for 20 min. Important: If you are performing these dilutions immediately after Step 10 in Section 3.2, the dilutions do not have to be reheated. However, a small proportion of spores become “inactivated” over time, and reheating the spore stocks immediately before plating assists in in vitro germination and provides more accurate representation of spores.
Cycle the reheated, serial dilutions into the anaerobic chamber. Apply 100 μl of each serial dilution onto individual pre-reduced BHIS supplemented with 0.1% taurocholate agar plates and evenly spread the dilution onto the surface of the plate using a sterile inoculating loop.
Incubate plates at 37°C for at least 24 hours to allow spore germination and sufficient growth of a colony.
Remove plates from chamber. For most accurate enumeration, use a plate that has between 30-300 colonies with few to no overlapping or clustered colonies (see Note 11).
- Calculate the number of spores per ml of spore stock using the following equation:
Finally, one last concern with the high concentration spore stocks is the presence of contaminants. Optionally, a small amount of the spore stock (50-100 μl) may be plated onto i) a BHIS agar plate and incubated aerobically and ii) a pre-reduced BHIS agar plate and incubated anaerobically to ensure no growth of non-C. difficile bacteria occurs. It is important to note here that a small number of C. difficile spores will germinate in the absence of taurocholate, so it is possible to see some C. difficile colonies on the BHIS agar plate incubated anaerobically. This will vary based on the strain of C. difficile used.
3.5 Preparation of final working spore stocks
To create a working spore stock, the original spore preparation needs to be diluted to the desired concentration. The working spore stock concentration will vary depending upon the downstream application (e.g. germination studies, spore outgrowth inhibition concentration (OIC) assays and animal studies all use a wide range of spore concentrations [3, 7, 9, 15, 22-24]).
- Calculate the dilution required to achieve the needed working spore stock concentration using the following equation (solve for X):
Of note: it will likely be necessary to perform a few dilutions before making the final dilution of the final working spore stock. Perform all dilutions using 1X PBS + 1% BSA as the diluent. Store the final working spore stock in glass or Teflon-coated tubes, as mentioned above, at room temperature. The shelf-life of spores varies by strain and stock, and should be retested if more than two weeks old. Remember to always reheat the spore stock to 55°C for 20 min before replating.
4. Notes
70:30 sporulation medium is a mixture of 70% SMC medium [14, 25] and 30% BHIS medium and was specifically developed to enhance and reliably determine the sporulation frequency of C. difficile [12].
A stock solution of 10% cysteine must be prepared fresh as cysteine will precipitate when stored at room temperature or cooler.
At H24 (24 h after plates are inoculated with an actively growing C. difficile culture), sporulation efficiency is ~30% for most strains tested when grown on 70:30 sporulation medium agar plates (Figure 1; unpublished data, Edwards and McBride), which is similar to previously observed sporulation frequencies on 70:30 sporulation plates for similar strains [12]. For unknown reasons, the sporulation efficiency significantly decreases after 70:30 sporulation medium is a week old. To obtain consistent results, we suggest using plates that are no more than three days old.
We achieve the most reliable and reproducible results in a variety of molecular biological assays by starting with C. difficile cultures that are in exponential phase. Oftentimes, overnight cultures of C. difficile will have already reached stationary phase after 12-16 h of growth. To combat this, we immediately dilute our initial, already-inoculated overnight culture 1:500 into another test tube with 10 ml BHIS supplemented with 0.1% taurocholate and 0.2% fructose. This results in two overnight cultures: the original culture and a 1:500 dilution of the original culture.
Because the protocol from this point is performed outside of the chamber, the use of pre-reduced BHIS medium is not important in this step. However, if the final time point has been reached or the plates will no longer be needed, the plates can be removed from the chamber, and this step may be performed in aerobic conditions.
The use of a thin agarose pad (0.7-1.0% agarose) immobilizes the bacteria for microscopy. We caution against the use of higher percentages of agarose as this decreases the amount of visible light that can been seen through the sample.
Phase contrast microscopy allows the observation of the later stages of sporulation (Stage IV+) in endospore-forming bacteria [26, 27] and is a frequently used technique to observe and quantitate sporulation frequencies in C. difficile [12, 15, 28-31]. Although this technique requires special equipment, it is easy to perform, does not require special dyes, sample fixation or preparation and is relatively inexpensive beyond the initial cost of the equipment. For the most accurate results, 100X magnification with oil immersion is needed.
Some strains do not always grow or sporulate well on 70:30 sporulation medium. Spores for these strains can instead be propagated on BHIS or TY agar plates. To increase the production and recovery of spores on these richer media, incubate plates for 4-5 days before harvesting.
To ensure that the 10 ml volume of spores is thoroughly heated to 70°C, we have found it easier to divide the spores into 10 – 1 ml aliquots in microcentrifuge tubes and heat the spore preparation using a benchtop heat block. Once this step is completed, the aliquots are then recombined into a fresh conical tube.
When initially enumerating a new spore stock, we typically plate 100 μl of 10−4, 10−5 and 10−6 dilutions of the original spore stock. With most strains, we easily obtain spore stocks of 1 × 108 CFU/ml from three to four 70:30 sporulation medium agar plates incubated for 48-72 h. For example, with a spore stock of this concentration, we obtain approximately 100 CFU on a BHIS supplemented with 0.1% taurocholate agar plate inoculated with 100 μl of a 10−5 dilution.
Before removing plates from the chamber, ensure that all of the colonies are large enough to be accurately counted. If not, allow colonies to grow longer in the anaerobic chamber. While counting, we find placing the plate on top of a light source and using a digital colony counter allows the accurate enumeration of smaller and clustered colonies and reduces user error.
5. References
- 1.CDC Control CfD, editor. Antibiotic Resistance Threats in the United States, 2013. http://wwwcdcgov/features/AntibioticResistanceThreats/2013.
- 2.Dubberke ER, Olsen MA. Burden of Clostridium difficile on the healthcare system. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2012;55(Suppl 2):S88–92. doi: 10.1093/cid/cis335. PMCID: 3388018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. Journal of bacteriology. 2008;190(7):2505–12. doi: 10.1128/JB.01765-07. PMCID: 2293200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, Kelly CP. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 2008;135(6):1984–92. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Goulding D, Thompson H, Emerson J, Fairweather NF, Dougan G, Douce GR. Distinctive profiles of infection and pathology in hamsters infected with Clostridium difficile strains 630 and B1. Infection and immunity. 2009;77(12):5478–85. doi: 10.1128/IAI.00551-09. PMCID: 2786451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douce G, Goulding D. Refinement of the hamster model of Clostridium difficile disease. Methods Mol Biol. 2010;646:215–27. doi: 10.1007/978-1-60327-365-7_14. [DOI] [PubMed] [Google Scholar]
- 7.Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. Journal of bacteriology. 2010;192(19):4983–90. doi: 10.1128/JB.00610-10. PMCID: 2944524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francis MB, Allen CA, Shrestha R, Sorg JA. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS pathogens. 2013;9(5):e1003356. doi: 10.1371/journal.ppat.1003356. PMCID: 3649964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu R, Suarez JM, Weisblum B, Gellman SH, McBride SM. Synthetic Polymers Active against Clostridium difficile Vegetative Cell Growth and Spore Outgrowth. Journal of the American Chemical Society. 2014 doi: 10.1021/ja506798e. PMCID: 4210120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuijper EJ, Coignard B, Tull P. Emergence of Clostridium difficile-associated disease in North America and Europe. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2006;12(Suppl 6):2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 11.Berg JM, Tymoczko JL, Stryer L, Stryer L. Biochemistry. 5th ed W.H. Freeman; New York: 2002. [Google Scholar]
- 12.Putnam EE, Nock AM, Lawley TD, Shen A. SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. Journal of bacteriology. 2013;195(6):1214–25. doi: 10.1128/JB.02181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.George WL, Sutter VL, Citron D, Finegold SM. Selective and differential medium for isolation of Clostridium difficile. Journal of clinical microbiology. 1979;9(2):214–9. doi: 10.1128/jcm.9.2.214-219.1979. PMCID: 272994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson KH, Kennedy MJ, Fekety FR. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. Journal of clinical microbiology. 1982;15(3):443–6. doi: 10.1128/jcm.15.3.443-446.1982. PMCID: 272115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards AN, Nawrocki KL, McBride SM. Conserved Oligopeptide Permeases Modulate Sporulation Initiation in Clostridium difficile. Infection and immunity. 2014;82(10):4276–91. doi: 10.1128/IAI.02323-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouillaut L, McBride SM, Sorg JA. Genetic manipulation of Clostridium difficile. Current protocols in microbiology. 2011 doi: 10.1002/9780471729259.mc09a02s20. Chapter 9:Unit 9A 2. PMCID: 3615975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuehne SA, Heap JT, Cooksley CM, Cartman ST, Minton NP. ClosTron-mediated engineering of Clostridium. Methods Mol Biol. 2011;765:389–407. doi: 10.1007/978-1-61779-197-0_23. [DOI] [PubMed] [Google Scholar]
- 18.Francis MB, Sorg JA. Virulence Studies of Clostridium difficile. Bio-protocol. 2013;3(24):e1002. [Google Scholar]
- 19.Edwards AN, Suarez JM, McBride SM. Culturing and Maintaining Clostridium difficile in an Anaerobic Environment. Journal of visualized experiments : JoVE. 2013;(79) doi: 10.3791/50787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorg JA, Dineen SS. Laboratory maintenance of Clostridium difficile. Current protocols in microbiology. 2009 doi: 10.1002/9780471729259.mc09a01s12. Chapter 9:Unit9A 1. [DOI] [PubMed] [Google Scholar]
- 21.Lawley TD, Croucher NJ, Yu L, Clare S, Sebaihia M, Goulding D, Pickard DJ, Parkhill J, Choudhary J, Dougan G. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. Journal of bacteriology. 2009;191(17):5377–86. doi: 10.1128/JB.00597-09. PMCID: 2725610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorg JA, Sonenshein AL. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. Journal of bacteriology. 2009;191(3):1115–7. doi: 10.1128/JB.01260-08. PMCID: 2632082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis MB, Allen CA, Sorg JA. Spore cortex hydrolysis precedes DPA release during Clostridium difficile spore germination. Journal of bacteriology. 2015 doi: 10.1128/JB.02575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paredes-Sabja D, Bond C, Carman RJ, Setlow P, Sarker MR. Germination of spores of Clostridium difficile strains, including isolates from a hospital outbreak of Clostridium difficile-associated disease (CDAD) Microbiology. 2008;154(Pt 8):2241–50. doi: 10.1099/mic.0.2008/016592-0. [DOI] [PubMed] [Google Scholar]
- 25.Permpoonpattana P, Tolls EH, Nadem R, Tan S, Brisson A, Cutting SM. Surface layers of Clostridium difficile endospores. Journal of bacteriology. 2011;193(23):6461–70. doi: 10.1128/JB.05182-11. PMCID: 3232898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashimoto T, Black SH, Gerhardt P. Development of fine structure, thermostability, and dipicolinate during sporogenesis in a bacillus. Canadian journal of microbiology. 1960;6:203–12. doi: 10.1139/m60-022. [DOI] [PubMed] [Google Scholar]
- 27.Hitchins AD, Kahn AJ, Slepecky RA. Interference contrast and phase contrast microscopy of sporulation and germination of Bacillus megaterium. Journal of bacteriology. 1968;96(5):1811–7. doi: 10.1128/jb.96.5.1811-1817.1968. PMCID: 315245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burns DA, Minton NP. Sporulation studies in Clostridium difficile. Journal of microbiological methods. 2011;87(2):133–8. doi: 10.1016/j.mimet.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Pereira FC, Saujet L, Tome AR, Serrano M, Monot M, Couture-Tosi E, Martin-Verstraete I, Dupuy B, Henriques AO. The Spore Differentiation Pathway in the Enteric Pathogen Clostridium difficile. PLoS genetics. 2013;9(10):e1003782. doi: 10.1371/journal.pgen.1003782. PMCID: 3789829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, Lawley TD, Shen A. Global Analysis of the Sporulation Pathway of Clostridium difficile. PLoS genetics. 2013;9(8):e1003660. doi: 10.1371/journal.pgen.1003660. PMCID: 3738446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saujet L, Pereira FC, Serrano M, Soutourina O, Monot M, Shelyakin PV, Gelfand MS, Dupuy B, Henriques AO, Martin-Verstraete I. Genome-Wide Analysis of Cell Type-Specific Gene Transcription during Spore Formation in Clostridium difficile. PLoS genetics. 2013;9(10):e1003756. doi: 10.1371/journal.pgen.1003756. PMCID: 3789822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nature genetics. 2006;38(7):779–86. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 33.Monot M, Boursaux-Eude C, Thibonnier M, Vallenet D, Moszer I, Medigue C, Martin-Verstraete I, Dupuy B. Reannotation of the genome sequence of Clostridium difficile strain 630. Journal of medical microbiology. 2011;60(Pt 8):1193–9. doi: 10.1099/jmm.0.030452-0. [DOI] [PubMed] [Google Scholar]
- 34.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome biology. 2009;10(9):R102. doi: 10.1186/gb-2009-10-9-r102. PMCID: 2768977. [DOI] [PMC free article] [PubMed] [Google Scholar]