SUMMARY
Co-option of RAG1 and RAG2 for antigen receptor gene assembly by V(D)J recombination was a crucial event in the evolution of jawed vertebrate adaptive immunity. RAG1/2 are proposed to have arisen from a transposable element, but definitive evidence for this is lacking. Here we report the discovery of ProtoRAG, a DNA transposon family from lancelets, the most basal extant chordates. A typical ProtoRAG is flanked by 5 bp target site duplications and a pair of terminal inverted repeats (TIRs) resembling V(D)J recombination signal sequences. Between the TIRs reside tail-to-tail oriented, intron-containing RAG1-like and RAG2-like genes. We demonstrate that ProtoRAG was recently active in the lancelet germline and that the lancelet RAG1/2-like proteins can mediate TIR-dependent transposon excision, host DNA recombination, transposition, and low efficiency TIR rejoining using reaction mechanisms similar to those used by vertebrate RAGs. We propose that ProtoRAG represents a molecular “living fossil” of the long-sought RAG transposon.
INTRODUCTION
The immunoglobulin and T cell receptor genes of jawed vertebrates are assembled from variable (V), diversity (D), and joining (J) gene segments during B and T lymphocyte development (Davis et al., 1984; Tonegawa, 1983). This assembly process, known as V(D)J recombination, is initiated by the RAG1 and RAG2 proteins (referred to collectively as RAG), which form a complex that excises the DNA between the V, D, and J gene segments (Gellert, 2002). DNA binding and cleavage by RAG are guided by recombination signal sequences (RSSs) that flank each gene segment (Figure 1A) and that are composed of conserved heptamer and nonamer sequences separated by a poorly conserved spacer sequence of either 12 or 23 bp (termed the 12RSS and 23RSS, respectively) (Ramsden et al., 1994). RAG-mediated DNA cleavage occurs preferentially in a complex containing one 12RSS and one 23RSS, a restriction known as the 12/23 rule (Lewis, 1994), and involves a nick-hairpin mechanism characteristic of several cut-and-paste DNA transposases (Fugmann, 2010). After cleavage, the hairpin-tipped coding segments are processed and joined imprecisely to form a coding joint (CJ) (Figure 1B) while the cleaved RSSs are joined precisely to form a signal joint (SJ) (Figure 1C). End processing and joining are carried out by the non-homologous end joining (NHEJ) DNA repair pathway (Figure 1B–C) (Lieber et al., 2004).
Figure 1. Schematic of V(D)J recombination and transposition.
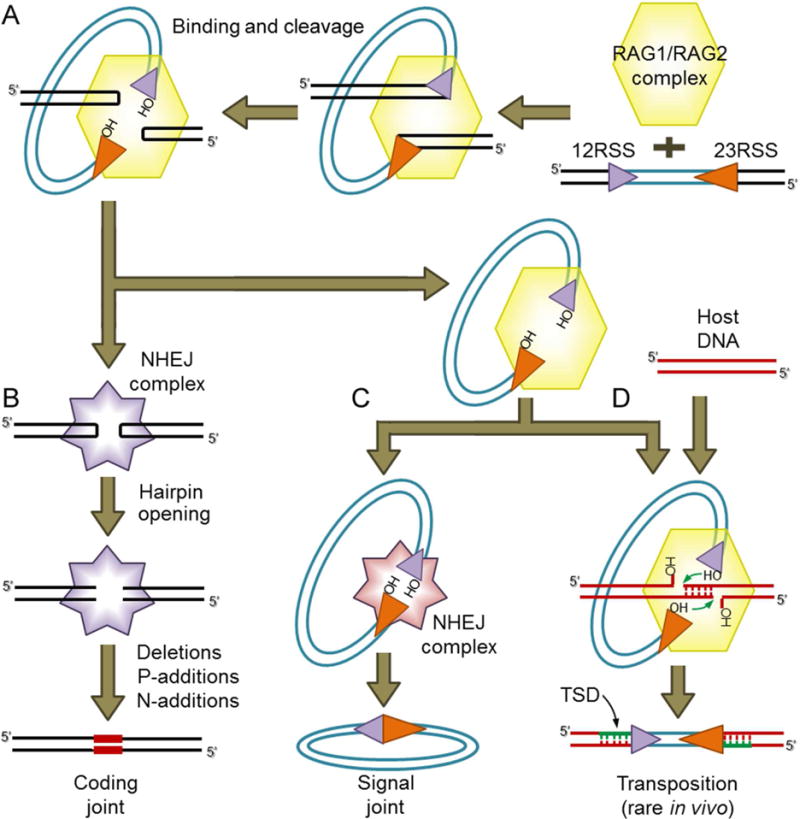
(A) V(D)J recombination is initiated when the RAG complex binds a 12RSS/23RSS pair and cleaves the DNA, generating hairpin sealed coding ends and blunt RSS ends with a 3′ hydroxyl (OH) group.
(B) The coding ends are nicked open by NHEJ DNA repair factors and then processed and joined, resulting in imprecise coding joints that can contain added nucleotides (red bars).
(C) The cleaved RSS ends are thought to be bound initially by RAG, and subsequently are ligated together precisely by NHEJ repair factors to form a signal joint.
(D) An alternative fate for the cleaved RSS ends bound to RAG is staggered attack by the 3′OH groups on a target DNA duplex (host DNA) resulting in insertion of the cleaved RSS fragment into the target and the creation of a flanking target site duplication (TSD). For RAG, this transposition outcome is rare in vivo but efficient in vitro.
Cut and paste transposition and V(D)J recombination are similar in their early steps but have different outcomes. Like RAG, the transposase cleaves adjacent to terminal inverted repeats (TIR) structures, but thereafter, instead of allowing TIR end joining, the transposase inserts the transposon into target DNA (Figure 1D). The sites of insertion on the two strands of the target are typically staggered, and after repair of the insertion junctions, this yields target site duplications (TSDs) immediately adjacent to the TIRs whose length is determined by the amount of the stagger and is distinctive for different TE superfamilies (Craig, 2002; Hencken et al., 2012).
The emergence of RAG is considered a milestone event in the genesis of the adaptive immune system of jawed vertebrates (Flajnik and Kasahara, 2010). Host domestication of a DNA transposon has been the prevailing hypothesis for the origin of “split” antigen receptor genes (that is, divided into V, D, and J segments) since the late 1970s, when Tonegawa and colleagues noticed that the inverted pairing of the 12RSS and 23RSS was reminiscent of the TIRs flanking a DNA transposon (Sakano et al., 1979). The discovery of RAG1 and RAG2 revealed that the two genes lie immediately adjacent to one another in the jawed vertebrate genome (Oettinger et al., 1990; Schatz et al., 1989). This and biochemical insights into RAG function led to the hypothesis that a “RAG transposon”, composed of adjacent RAG1 and RAG2 genes flanked by RSS-like TIRs, was the source of jawed vertebrate RAG genes and was responsible for creating the initial split antigen receptor gene (McBlane et al., 1995; Thompson, 1995). This hypothesis received support from the finding that RAG is able to mediate efficient transposition of RSS-flanked DNA in vitro, yielding insertions flanked by characteristic 5 bp TSDs (Figure 1D) (Agrawal et al., 1998; Hiom et al., 1998). RAG-mediated transposition is very inefficient in living cells (Chatterji et al., 2006; Reddy et al., 2006), perhaps reflecting the ability of RAG to channel the cleaved DNA ends into the NHEJ repair pathway (Lee et al., 2004).
Each RAG protein is composed of a “core” region essential for DNA cleavage activity and non-core regions that appear to function primarily in a regulatory capacity (Little et al., 2015). The evolutionary origins of the core and non-core portions of RAG have been the subject of substantial interest. The RAG1 core has been suggested to derive from the widely dispersed Transib transposon family while its N-terminal non-core region has been proposed to derive from a different “N-RAG-TP” transposon (Kapitonov and Jurka, 2005; Panchin and Moroz, 2008). The RAG2 protein is composed of an N-terminal core region containing six Kelch-like repeats that adopt a six-bladed β-propeller structure and a C-terminal region containing a plant homeodomain (PHD) (Callebaut and Mornon, 1998; Kim et al., 2015). RAG2 lacks similarity to any known transposon protein, but Kelch-like repeats and PHDs are present in many eukaryotic proteins, leading to the hypothesis that RAG2 arose through exon shuffling that brought two domains together (Fugmann, 2010). Other theories, distinct from the RAG transposon hypothesis, have also been proposed for the origins of RAG, for example suggesting links between RAG1 and herpes virus recombinases or a retroviral nuclease (Dreyfus, 2009; Zhang et al., 2014). A closely-linked gene pair encoding RAG1/RAG2-like proteins has been identified in an invertebrate, the sea urchin (an echinoderm), but this gene pair lacks signatures of transposons such as TIRs and TSDs (Fugmann et al., 2006). Other echinoderms also appear to harbor adjacent RAG1-RAG2 gene pairs (Kapitonov and Koonin, 2015), but their relationship to transposons is uncertain. Therefore, there is currently no definitive evidence that the RAG transposon exists or is active in the animal kingdom.
Lancelet (amphioxus) represents the most basal extant chordate (cephalochordates) that diverged from the other two chordate lineages (urochordates and vertebrates) half a billion years ago (Delsuc et al., 2006). Comparative genomic analysis reveals a huge diversity of ancient transposable elements (TEs) in lancelet genomes (Huang et al., 2014). Here we demonstrate that this includes an TE superfamily, ProtoRAG, which meets the structural criteria for the long-sought RAG transposon. We demonstrate that ProtoRAG encodes RAG1-like and RAG2-like proteins that constitute an active endonuclease and transposase in vitro and in living cells with striking mechanistic similarities to vertebrate RAG. Our findings strongly implicate ProtoRAG as an evolutionary relative of RAG and provide powerful evidence in favor of the RAG transposon hypothesis for the origins of jawed vertebrate adaptive immunity.
RESULTS
Discovery of the RAG transposon
Since they preserve a huge ancient TE diversity, the genomes of lancelet Branchiostoma belcheri and B. floridae were promising places to find the RAG transposon (Huang et al., 2014). Initial homology searches identified RAG1/2-like gene fragments in the lancelet genomes, but failed to detect the signature features of transposons (TSDs and TIRs) near the fragments. Previously, we had reconstructed two haploid assemblies (named the reference and the alternative) for the diploid genome of B. belcheri. We surmised that by comparing the two haploid assemblies, we might discover allele-specific copies of an unknown but active TE family. Using this method, we identified ~24,000 polymorphic TE insertions (Huang et al., 2014), one of which was a strong candidate to be a RAG transposon. This insertion, located on scaffold5 of the alternative assembly, encoded partial RAG1/2-like proteins and appeared to be flanked by TSDs and TIRs. To recover intact copies, we used the RAG1/2-like sequences as probes to screen lancelet (B. belcheri) BAC libraries, yielding fourteen positive clones which were completely sequenced. The most complete TE copy was identified on BAC plasmid clone 73 and contained intact TSD-TIR structures and coding regions for both RAG1-like and RAG2-like proteins. The sequence of this clone was used to scrutinize the available lancelet genomes and assembled BAC sequences, yielding a set of 53 TE copies (Table S1) that define a superfamily of cut-and-paste DNA transposons that we designate as “ProtoRAG”.
TSD and TIR structures of ProtoRAG
ProtoRAG elements are flanked by 5 bp TSDs (Figure 2A), a length which distinguishes them from all other cut-and-paste DNA transposons except Transib and vertebrate RAG (Agrawal et al., 1998; Hencken et al., 2012; Hiom et al., 1998). The target sites of ProtoRAG insertion exhibit a bias toward GC base pairs (62.5% versus 41% average genomic GC content), as is also observed for Transib and vertebrate RAG (Kapitonov and Jurka, 2005; Tsai et al., 2003). The 5′-TIR and 3′-TIR of ProtoRAG have weak sequence similarity with one another and between lancelet species, with the terminal 47–64 bp exhibiting the highest conservation and containing several regions of high identity (Figure 2B). The terminal seven bp (5′-CACTATG-3′) are identical in all ProtoRAG TIRs and resemble the consensus RSS heptamer (5′-CACAGTG-3′) and the terminal seven bp of Transib TIRs (5′-CACWRTG-3′). A second highly conserved block of nine bp exists at the 3′ end of the conserved region (Figure 2B). We refer to this as the TIR “nonamer”, but we note that its sequence does not resemble the consensus RSS nonamer. The B. belcheri 5′-TIR (defined as that lying upstream of the RAG1-like gene) and 3′-TIR typically have 27 bp and 31 bp, respectively, separating the heptamer from the nonamer. Unlike RSS spacers, these regions contain multiple well conserved nucleotides (Figure 2B).
Figure 2. Genomic sequence features of ProtoRAG.
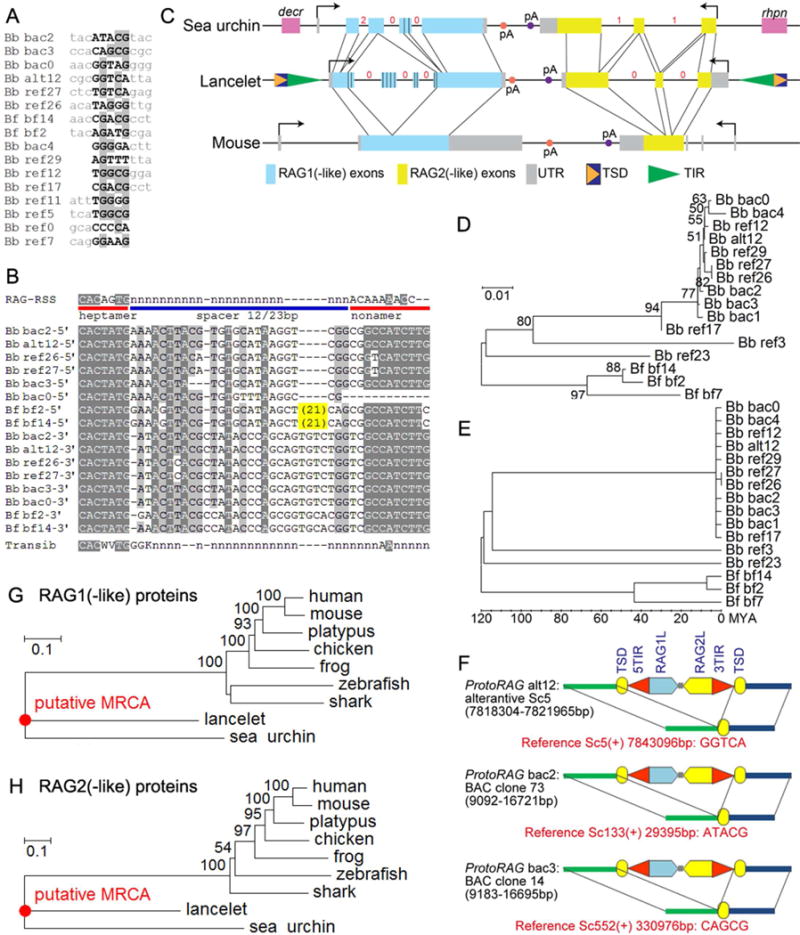
(A) Alignment of ProtoRAG TSDs and flanking sequences from the lancelet genome.
(B) Alignment of ProtoRAG TIR sequences with the consensus RSS and Transib TIR. IUPAC codes used in the alignment: N=A, C, G or T; K=G or T; W=A, T; V=A, C or G. Lower case n indicates an undetermined nucleotide. Shading indicates sequence conservation, with darker gray indicating a higher degree of conservation. Bb: B. belcheri; Bf: B. floridae. ProtoRAG copy identification numbers correspond to those listed in Table S1.
(C) Genomic organization of the lancelet ProtoRAG copy on BAC plasmid clone 73, sea urchin RAG1/2-like gene locus, and mouse RAG locus. Corresponding coding regions are indicated by thin lines. The phases of introns in coding regions are shown by red numbers. Repetitive regions in lancelet RAG1-like and sea urchin RAG1-like are marked by vertical bars. Terminal exons of flanking genes (decr, dienoyl-CoA reductase; rhpn, rhophilin) for the sea urchin RAG1/2-like locus are shown as purple boxes.
(D) Neighbor-Joining trees of lancelet ProtoRAG copies assembled using Mega v5.2 (see Supplementary Methods).
(E) Molecular dating analysis of lancelet ProtoRAG copies. This linearized tree with clock calibration was calculated using Mega v5.2. The root, or the divergence between B. belcheri and B. floridae, was set to 120 million years ago.
(F) Three unfixed (polymorphic) ProtoRAG transposition events identified in B. belcheri genomes. Red text provides the coordinates of target sites and the sequence of TSDs on the reference genome. See Table S1 and Data S1 for further details.
(G, H) Neighbor-joining trees of RAG1 (G) and RAG2 (H) protein homologs. MCRA, most common recent ancestor.
See also Data S1 and Table S1.
Genomic organization and expression of ProtoRAG
The most complete copy of ProtoRAG (7639 bp long) from BAC plasmid clone 73 contains TSDs and TIRs flanking a pair of genes lying in tail-to-tail orientation, the same orientation as RAG1 and RAG2 in the vertebrate and the sea urchin RAG-like loci (Figure 2C). The complete sequences of ProtoRAG from BAC plasmid clones 73 and 14 are displayed in Data S1. Unlike typical transposase genes, both the RAG1-like and RAG2-like genes of B. belcheri (bbRAG1L and bbRAG2L hereafter) are interrupted by multiple introns, and their intron sites and phases are different from those in vertebrates and sea urchin (Figure 2C). Notably, mammalian RAG1/2 has no introns in the coding regions. It is not known whether the ancestral RAG1/2 lacked introns, or contained introns that were lost in mammals. RT-PCR and sequencing suggested that bbRAG1L/2L each can generate several alternatively-spliced transcript isoforms (data not shown). The presence of two genes containing introns makes lancelet ProtoRAG an unusual transposon.
Phylogenetic analysis of the 3′-terminal 700 bps of ProtoRAG indicates that most copies in B. belcheri are nearly identical (Figure 2D). Molecular dating analysis suggests that 11 of the 13 ProtoRAG copies arose during the last 2.7 million years (Figure 2E). Moreover, three polymorphic transposition-type insertions of ProtoRAG were identified in the lancelet genome sequences (Figure 2F). These lines of evidence suggest recent in vivo transposition activity of ProtoRAG in lancelet.
Quantitative RT-PCR assays revealed weak expression of both bbRAG1L and bbRAG2L in different lancelet tissues and developmental stages (Figure S1A–B). When transfected into human 293T cells, the plasmid of BAC clone 73 generated both bbRAG1L and bbRAG2L mRNA (Figure S1C), indicating that this copy of ProtoRAG retains functional transcriptional regulatory elements. When expressed in human HeLa cells, GFP-bbRAG1L accumulated predominantly in the nucleus (Figure S1D) while GFP-bbRAG2L was detected in both the cytoplasm and nucleus (Figure S1D). Nuclear-cytoplasmic fractionation analysis showed no increase of GFP-bbRAG2L in the nucleus in the presence of bbRAG1L as compared to its absence (data not shown). Weak expression and the inefficient nuclear localization of bbRAG2L might suggest low in vivo activity of ProtoRAG.
Features of the bbRAG1/2L proteins
Phylogenetic analysis shows that both bbRAG1L and 2L have the shortest branch from the most recent common ancestor (MRCA), suggesting either that RAG1L/2L evolved more slowly in lancelets or that vertebrate RAG underwent particularly rapid evolution during their host domestication process (Figure 2G–H). The 1136 aa-long bbRAG1L protein shares 29% and 36% sequence identity with vertebrate RAG1 and sea urchin RAG1L, respectively, whereas sea urchin RAG1L and Transib transposase have only 26% and 16–18% identity, respectively, with vertebrate RAG1 (Figure 3A and S2). Blocks of identity between bbRAG1L and vertebrate RAG1 are found along much of their length, suggesting conservation of multiple functional elements (Figure 3A and S2). Vertebrate RAG1 uses four acidic residues to coordinate critical active site divalent cations (Ru et al., 2015), and all four are conserved in bbRAG1L (Figure 3A, red highlight). In addition, many cysteine and histidine residues that coordinate zinc ions and play a critical role in proper folding of RAG1 (Kim et al., 2015; Yin et al., 2009) are conserved between bbRAG1L and vertebrate RAG1 (Figure 3A, * and # symbols). However, bbRAG1L has little similarity to vertebrate RAG1 in the region corresponding to the nonamer binding domain, consistent with the fact that ProtoRAG TIRs have no clear similarity to the RSS nonamer (Figure 2B). The N-terminal portion of bbRAG1L contains a repetitive region not found in vertebrate RAG1, consisting of variants of a 12-aa sequence (PPTADVRATTSQ). Sea urchin RAG1L also contains repeats (TAPLPPTA) in its N-terminal region, although they are inserted into a different position in the protein as compared to bbRAG1L (Figure 3A). The function of these repetitive regions is unknown.
Figure 3. The features of the proteins encoded by ProtoRAG.
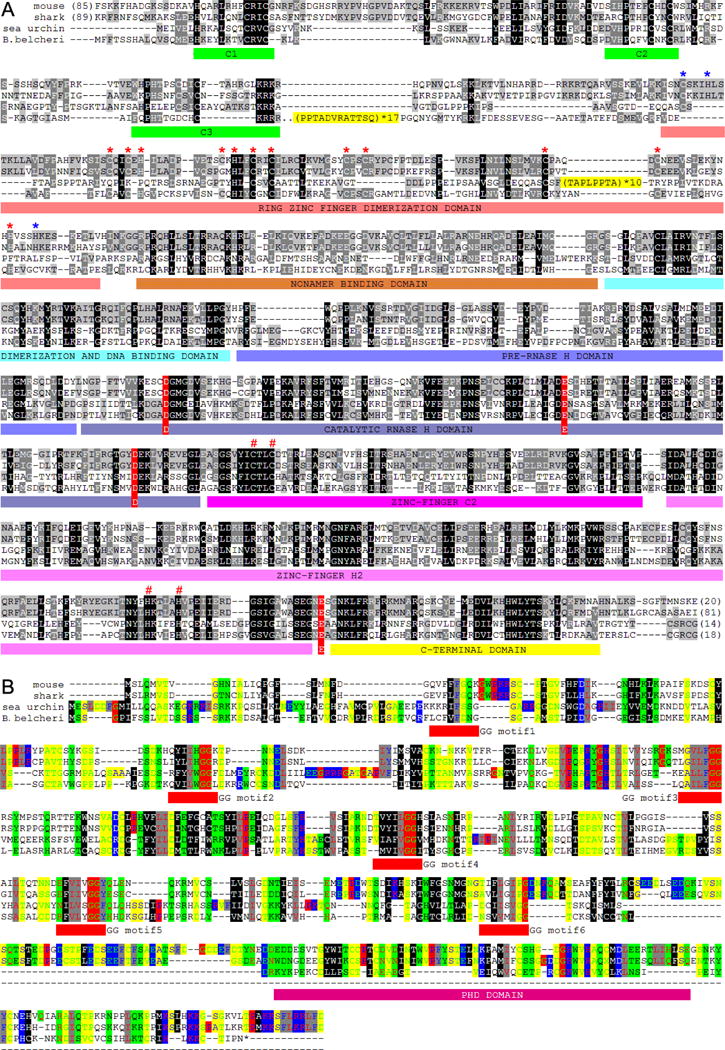
(A) Protein alignment of lancelet RAG1L with mouse RAG1, shark RAG1 and sea urchin RAG1L. Repeat motifs are highlighted in yellow. Three regions of conserved cysteine and histidine residues that might bind zinc are underlined with green bars. The N-terminal zinc binding dimerization domain is underlined with dark-red bars. The subdomains of the RAG1 core region are indicated with colored bars and labeled according to (Kim et al., 2015). The four conserved zinc ligand residues that contribute to proper folding of the RAG1 catalytic region are labeled with a red “#” (C727, C730, H937, H942 on mouse RAG1). The conserved acidic catalytic residues are highlighted with red shading (D600, E662, D708 and E962 on mouse RAG1). In the N-terminal RING-zinc finger dimerization domain (ZDD), fifteen zinc-coordinating residues (C266, H270, C290, C293, H295, C305, H307, C310, C313, C325, C328, C355, C360, H372 and H376 on mouse RAG1) that are conserved in vertebrate RAG1s are labeled with asterisks, with red asterisks indicating residues conserved in both vertebrate RAG1s and bbRAG1L, and blue asterisks indicating residues conserved in vertebrate RAG1s but not in lancelet RAG1L. GenBank accessions for mouse RAG1, shark RAG1, lancelet RAG2L and sea urchin RAG1L are NP_033045, XP_007886047, KJ748699 and NP_001028179, respectively.
(B) Protein alignment of lancelet RAG2L with mouse RAG2, shark RAG2 and sea urchin RAG2L. Color shading shows the conservation of physiochemical properties. The N-terminal amino acid sequence can be grouped into Kelch-like repeats similar to Callebaut and Mornon (Callebaut and Mornon, 1998) and Fugmann et al (Fugmann et al., 2006). The central conserved GG motifs of the six Kelch-like repeats are underlined in red. The plant homeodomain (PHD) is also underlined below the alignment. GenBank accessions for mouse RAG2, shark RAG2, lancelet RAG2L and sea urchin RAG2L are NP_033046, XP_007885835, KJ748699 and NP_001028184, respectively.
See also Figure S2.
The 366 aa-long bbRAG2L shares weak sequence identity with vertebrate RAG2 (20%) and sea urchin RAG2L (24%). The N-terminal six-bladed β-propeller domain (six Kelch-like repeats), which is conserved in both vertebrate RAG2 and sea urchin RAG2L, can be discerned in bbRAG2L, though the first and last repeats in bbRAG2L are barely conserved (Figure 3B). Strikingly, bbRAG2L lacks the entire RAG2 C-terminal region, including the PHD that is present in both the sea urchin and vertebrate RAG2 proteins (Figure 3B). The vertebrate RAG2 PHD binds trimethylated histone 3 lysine 4 (H3K4me3) (Liu et al., 2007; Matthews et al., 2007), and thereby helps localize the RAG complex to active chromatin (Ji et al., 2010; Teng et al., 2015), while the extreme C-terminal region of RAG2 is important for nuclear localization (Corneo et al., 2002). In addition, the C-terminal portion of mouse RAG2 has been shown to suppress RAG transposase activity (Elkin et al., 2003; Tsai and Schatz, 2003). Thus, the absence of this region might be advantageous for the mobility and survival of ProtoRAG.
bbRAG1L and bbRAG2L form a TIR-dependent endonuclease ex vivo
A fluorescent reporter assay was designed to investigate whether bbRAG1L/2L constitute an active endonuclease capable of TIR-dependent transposon excision in 293T cells. Artificial transposons, consisting of an inverted pair of ProtoRAG 5′-TIR and 3′-TIR sequences flanking a transcription stop sequence, were inserted into a reporter plasmid between a promoter and a GFP gene (Figure 4A). Transposon excision allows GFP expression if the remaining plasmid DNA ends can be rejoined appropriately. By analogy with the CJs formed during V(D)J recombination (Figure 1B), we refer to the resealed plasmid DNA sites left behind after transposon excision as host DNA joints (HDJs). Full length bbRAG1L and bbRAG2L were expressed from cDNA expression vectors. Evidence of transposon excision was obtained both by flow cytometry for GFP expression (Figure 4B) and by PCR (Figure 4C). In both cases, a signal above background was detected only when the reporter plasmid (pTIRG8) was co-transfected with both bbRAG1L and bbRAG2L, suggesting that co-expression of the two proteins is required for transposon excision.
Figure 4. TIR-dependent transposon excision mediated by bbRAG1L/2L ex vivo.
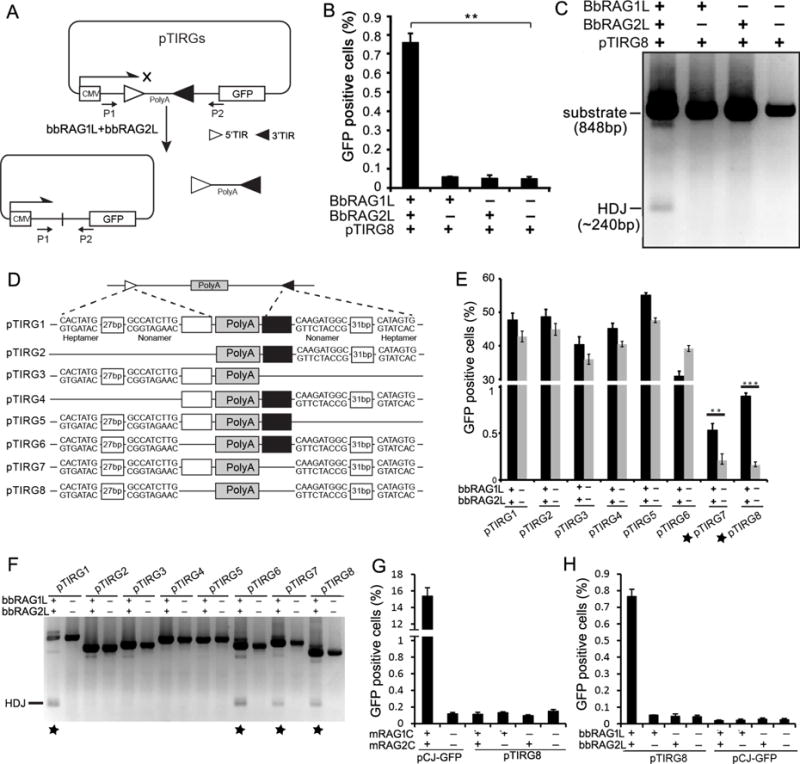
(A) Diagram of the cell-based fluorescent reporter and PCR assay for DNA excision and recombination. Filled and unfilled triangles, 5′- and 3′-TIR sequences of ProtoRAG, respectively; P1/P2, PCR primers; CMV: cytomegalovirus promoter; PolyA: polyadenylation signal sequence.
(B) Quantification of GFP positive cells by flow cytometry after transfection of 293T cells with pTIRG8 (containing the minimal ProtoRAG TIRs) with bbRAG1L and bbRAG2L expression vectors, as indicated.
(C) PCR detection of HDJs from transfections of pTIRG8 as in (B).
(D) Diagram of truncated TIR-containing substrates. Unfilled and filled boxes indicate the remainder of the 5′- and 3′-TIRs, respectively.
(E) Quantification of GFP positive cells by flow cytometry after transfection of 293T cells with truncated TIR-containing substrates.
(F) PCR detection of HDJs for truncated TIR-containing substrates.
(G) Quantitation of GFP positive cells by flow cytometry after transfection of 293T cells with mouse RAG1 core and RAG2 core expression vectors with pTIRG8 (containing the minimal ProtoRAG TIRs) or pCJ-GFP (containing RSSs), as indicated.
(H) Quantitation of GFP positive cells by flow cytometry after transfection of 293T cells with bbRAG1L and bbRAG2L expression vectors with pTIRG8 or pCJ-GFP (which contains a 12/23 RSS pair instead of the TIRs of pTIRG8), as indicated.
See also Figure S3 and S4.
To determine the minimal TIR sequences needed for transposon excision, a series of truncated TIR pairs (Figure 4D) were tested using the GFP and PCR assays. As suggested by the TIR sequence alignment (Figure 2B), the first 43 bp of a 5′-TIR paired with the first 47 bp of a 3′-TIR (plasmid pTIRG8) were sufficient for transposon excision (Figures 4E–F). The TIR heptamer and nonamer sequences were each essential for excision by bbRAG1L/2L (Figure S3A–B), as is the case for the RSS (Hesse et al., 1989). The minimal 5′-TIR/3′-TIR pair (pTIRG8) was used in most subsequent ex vivo functional analyses because it yielded the best signal/noise ratio in the reporter assay. (Substrates containing longer TIRs exhibited very high background in the GFP assay (Figure 4E), perhaps because of promoter activity associated with long TIRs (data not shown).
The excision efficiency of mouse RAG1/2 on a 12RSS/23RSS substrate was nearly 20-fold higher than that of bbRAG1L/2L on a ProtoRAG 5′-TIR/3′-TIR substrate (Figure 4G–H). It is not known which step(s) of the reaction are less efficient with bbRAG1L/2L. Notably, mouse RAG could cleave the RSS substrate but not the ProtoRAG TIR substrate (Figure 4G), while the opposite was observed for bbRAG1L/2L (Figure 4H). These ex vivo observations, reinforced by in vitro data described below, suggest that the two RAG systems are no longer functionally compatible with one other after a divergence of over half a billion years.
The essential mouse RAG1 core region (aa 384–1008 of 1040 aa) corresponds to aa 468–1110 of bbRAG1L. This portion of bbRAG1L was inactive, but regained activity to a level above that of full-length bbRAG1L upon addition of the terminal 36 aa (bbRAG1C; Figure S3C–D). Therefore, we tentatively define the bbRAG1L core region as aa 468–1136. In addition, reporter assays showed that single, double, or triple mutations of putative acidic active site residues in bbRAG1L (D701, D811 and E1063) eliminated transposon excision activity without dramatically affecting protein expression (Figure S3E–F). This observation, together with in vitro experiments described below, argues that bbRAG1L and mouse RAG1 use similar catalytic residues for cleavage activity.
BbRAG1L and bbRAG2L form a TIR-dependent endonuclease in vitro
We co-purified full-length bbRAG1L and bbRAG2L, each fused at its N-terminus to maltose binding protein (MBP), from mammalian cells (Figure 5A) and performed cleavage reactions using substrates containing either a 5′-TIR/3′-TIR pair or a 12RSS/23RSS pair (Figure 5B). Reactions contained Mg2+ and human HMGB1 (a DNA binding protein). Co-expressed bbRAG1L/bbRAG2L exhibited robust cleavage activity on the TIR substrate, with cleavage products detectable as early as 2 minutes (Figure 5C, lanes 1–5), while co-expressed RAG1/2 cleaved the RSS substrate with similar kinetics (lanes 6–10). Cleavage products had the sizes expected for double strand breaks at the borders of the TIRs or RSSs (see diagrams in Figure 5C), with both enzymes generating substantial amounts of the double cleavage product (double asterisks). Some differences were noted in the pattern of cleavage products generated by bbRAG1L/2L as compared to RAG1/2. BbRAG1L/2L generated a product corresponding to single cleavage at the 3′-TIR (asterisk), which was visible at 2 min and accumulated to high levels over the 30 min time course. In contrast, RAG1/2 generated predominantly the double cleavage product, with single cleavage products visible as minor species only at later time points, consistent with RAG’s well known propensity to perform coordinated cleavage at a 12/23 RSS pair (Eastman et al., 1996).
Figure 5. Biochemical analysis of DNA cleavage by bbRAG1L/2L in vitro.
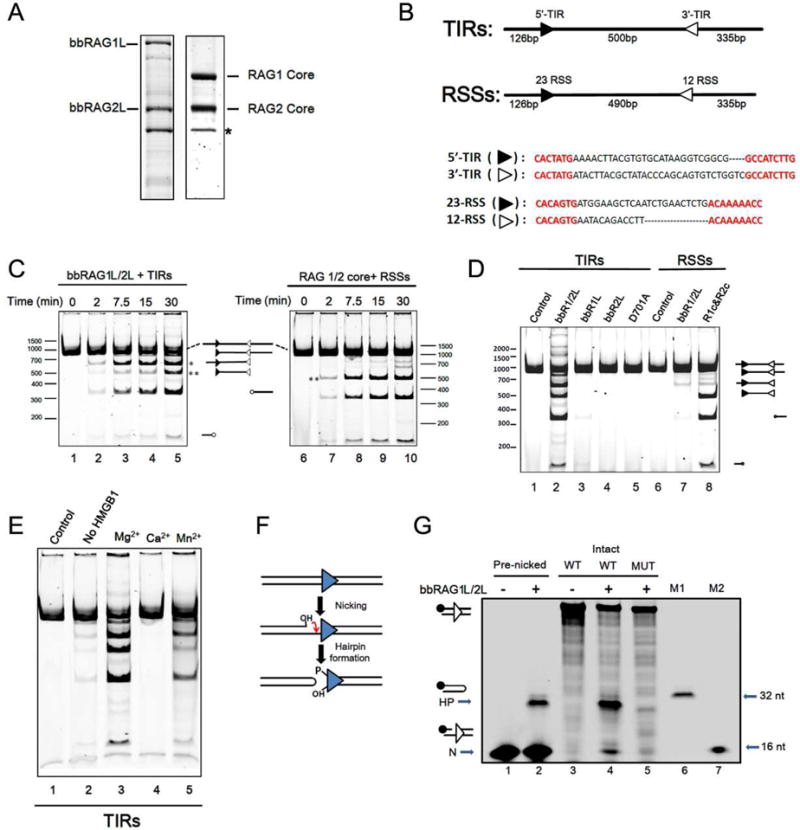
(A) Co-expressed, single-step purified bbRAG1L/2L and RAG1/2 proteins. *: background protein that elutes from amylose columns.
(B) DNA cleavage substrates, with expected sizes of cleavage products indicated.
(C) Time course of cleavage by bbRAG1L/2L (left) and RAG (right) as assessed by native polyacrylamide gel electrophoresis. *: prominent product corresponding to single cleavage at the 3′-TIR; **: central fragment resulting from double cleavage. All reactions contain HMGB1 and Mg2+ unless otherwise indicated.
(D) Cleavage of TIR substrate (lanes 1–5) or RSS substrate (lanes 6–8) by the indicated proteins. D701A, bbRAG1L containing a D701A mutation combined with bbRAG2L. Lane 8, RAG1 core and RAG2 core proteins.
(E) Cleavage by bbRAG1L/2L with different divalent cations (lanes 3–5) and in Mg2+ but in the absence of HMGB1 (lane 2).
(F) Diagram of nick-hairpin mechanism of DNA cleavage.
(G) Nicking and hairpinnning by bbRAG1L/2L as assessed by denaturing polyacrylamide gel electrophoresis. 3′-TIR DNA substrates, with 16 bp flanking the TIR on each side, were fluorescently 5′ end labeled on the top strand (filled circle) and were either intact (lanes 3–5) or pre-nicked immediately adjacent to the TIR (lanes 1–2). MUT: 3′-TIR substrate with a scrambled heptamer. M1 and M2: 16 nt and 32 nt markers. N: nick; HP: hairpin product. The hairpin product runs slightly faster than the 32 nt marker, likely due to a propensity to partially reanneal.
In vitro cleavage by bbRAG1L/2L required the presence of both bbRAG proteins (Figure 5D, lanes 2–4) and was eliminated by mutation of one of the predicted active site residues (D701) (lane 5). As expected from the ex vivo results (Figure 4H), bbRAG1L/2L exhibited very weak activity on the RSS substrate, with no detectable double cleavage product generated (lane 7). Much like RAG1/2, cleavage activity of bbRAG1L/2L was strongly dependent on HMGB1 (Figure 5E, lane 2) and was supported by Mg2+ and Mn2+, but not Ca2+, divalent cations (lanes 3–5).
To determine whether bbRAG1L/2L cleave DNA by a nick/hairpin mechanism similar to that used by RAG (Figure 5F), cleavage reactions were performed using an end-labeled 3′-TIR DNA substrate, with the cleavage products analyzed by denaturing gel electrophoresis. BbRAG1L/2L were found to generate cleavage products of the size expected for nicking and hairpin formation immediately adjacent to the TIR (Figure 5G, lane 4), with cleavage eliminated by scrambling of the 3′-TIR heptamer (lane 5). Furthermore, bbRAG1L/2L could generate hairpin product from a pre-nicked substrate (Figure 5G, lane 2), demonstrating that the nicked species is an intermediate in hairpin formation, as is the case for RAG (McBlane et al., 1995).
Ex vivo host DNA rejoining after transposon excision
HDJ-containing PCR products (e.g., Figure 4C) were cloned and sequenced. The 48 junctions obtained were consistent with cleavage occurring at the junction between TIRs and the flanking host DNA and argued that cleavage did not occur within the TIRs. Most HDJs (>90%) contained small deletions, and a few contained short insertions, almost all of which appeared to be palindromic- (P-) nucleotide additions (Figure S4). P-nucleotides are well-known feature of V(D)J recombination that are thought to arise when the hairpin sealed DNA ends generated by RAG (Figure 1A–B) are opened asymmetrically (Lewis, 1994).
Because of potential biases associated with PCR, we used an established bacterial colony assay (Hesse et al., 1987) to recover HDJ-containing plasmids from 293T cells after transposon excision (Figure 6A). Sequencing of the plasmids revealed small deletions and P-nucleotides in a large fraction of the HDJ junctions (Figure S5A). The presence of P-nucleotides suggests that bbRAG1L/2L generates hairpins on the host DNA ends during cleavage ex vivo, consistent with the in vitro data (Figure 5G). CJs generated by mouse RAG on an RSS substrate in a similar ex vivo assay also contained small deletions and P-nucleotide additions (Figure S5B). Hence, HDJ formation after bbRAG1L/2L-mediated transposon excision recapitulates key features of CJ formation during V(D)J recombination. More careful analyses will be needed to determine if there are subtle differences in the fine structure of HDJs generated by bbRAG1L/2L and CJs generated by RAG.
Figure 6. Ex vivo analysis of the host DNA rejoining and transposon self-resealing after transposon excision by bbRAG1L/2L.
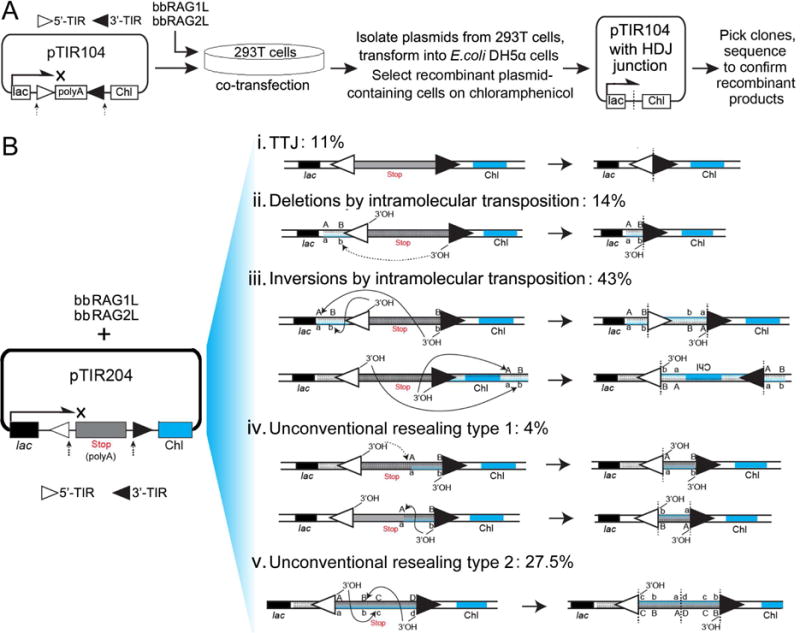
(A) The bacterial colony assay and the plasmid (pTIR104) designed to detect bbRAG1L/bbRAG2L mediated transposon excision and HDJ rejoining. The results are shown in Figure S5.
(B) The bacterial colony assay designed to detect bbRAG1L/bbRAG2L mediated complex transposon self-resealing events after transposon excision. The resulted recombinants are categorized by the junctions identified by DNA sequencing. The left panel shows the plasmid (pTIR204) designed for the detection, in which the TIRs are retained with the backbone of the plasmid after cleavage.
See also Figure S5.
Ex vivo transposon resealing by intracellular transposition and TIR-TIR joining
The results above address the fate of the host DNA ends generated by bbRAG1L/2L cleavage but not that of the cleaved TIR ends, which we anticipated would be handled in a manner distinct from the cleaved RSSs generated by RAG. To address this issue, the TIRs in pTIRG8 and pTIR104 were reoriented so that they would be retained on the plasmid backbone after cleavage, yielding new reporter substrates pTIRG13 (Figure S6A) and pTIR204 (Figure 6B). When pTIRG13 was transfected into 293T cells, a clear bbRAG1L/2L-dependent increase in GFP expression was observed (Figure S6B) but the PCR assay for the resealed junctions yielded a broad and indistinct band (Figure S6C). These results suggested that bbRAG1L/2L cut the substrate but that the products were not simply precise TIR-TIR joints (TTJs).
Use of plasmid pTIR204 and the bacterial colony assay (Figure 6B) allowed recovery of plasmids containing a range of different outcomes of transposon resealing. Sequence analysis of 127 recombinant plasmids recovered from 293T cells revealed that only 11% carried perfect end-to-end TTJs (Figure 6B, i). Another 57% appeared to be the products of intramolecular transposition, which occurred to generate deletions (14%; Figure 6B, ii) or inversions (43%; Figure 6B, iii) depending on the DNA strand that was attacked by the TIR ends. The inversion products contained TSDs (predominantly 5 bp in length) adjacent to the TIRs and hence were unambiguously the result of transposition. Because only one TIR is recovered in deletion products, a similar strong conclusion cannot be drawn about their mechanism of generation. The remaining plasmids (31.5%) arose from two types of unconventional resealing events. Type 1 plasmids (4%) contained a deletion of inter-TIR sequences (Figure 6B, iv), while type 2 plasmids (27.5%) appeared to result from transposition into the excised inter-TIR sequences (Figure 6B, v). When a similar experiment was performed using mouse RAG and a substrate containing RSSs, most (30/35) of the recovered recombinant plasmids contained precise SJs. Together, these data demonstrate that bbRAG1L/2L is capable of performing intramolecular transposition ex vivo and generates precise TTJs at a relatively low frequency.
We note that this assay likely substantially overestimates the frequencies of TTJ and unconventional resealing events because most intramolecular transposition events would be expected to involve attack of the TIRs on the long plasmid backbone (4.7 kb; thick line in the substrate diagram in Figure 6B), and such events are not recovered because they separate the lac promoter from the chloramphenicol resistance gene (deletional transposition) or invert the resistance gene relative to the promoter (inversional transposition).
In vitro and ex vivo intermolecular transposition by ProtoRAG
To determine whether bbRAG1L/2L could mediate intermolecular transposition in vitro, a linear artificial ProtoRAG transposon donor containing a tetracycline resistance gene, a target plasmid harboring a kanamycin resistance gene, and purified bbRAG1L/2L proteins were incubated together (Figure 7A) followed by transformation of bacteria. Analysis of 35 plasmids from KanR/TetR colonies revealed that all contained the intact artificial transposon, flanked by TSDs (80% 5 bp in length), inserted into many different sites in the target plasmid (Figure 7B). Parallel reactions using mouse RAG proteins and an artificial transposon flanked by a 12/23RSS pair revealed a similar distribution of target sites and TSDs (Figure S6D–E). The efficiency of transposition mediated by full length bbRAG1L/2L was about half that mediated by mouse core RAG1/2 (Figure 7C). These results demonstrate that bbRAG1L/2L mediates bone fide transposition in vitro at frequencies roughly comparable to those of RAG1/2.
Figure 7. In vitro and ex vivo intermolecular transposition mediated by bbRAG1L/2L.
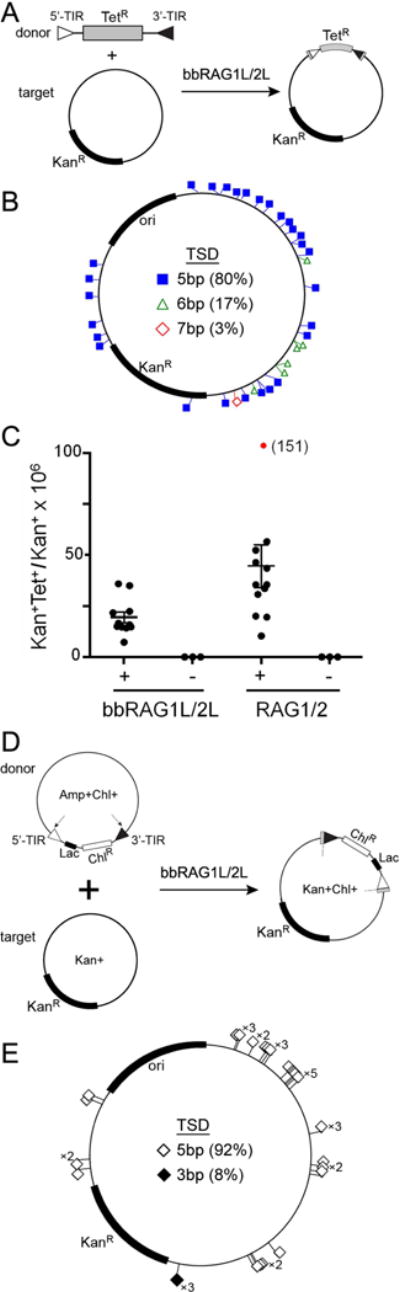
(A) Schematic diagram of the assay used to detect in vitro transposition mediated by purified bbRAG1L and bbRAG2L proteins.
(B) The distribution of in vitro transposition target sites in the recipient plasmid.
(C) Quantitation of in vitro transposition efficiency of bbRAG1L/2L and mouse RAG1/2. Each dot represents the results of an independent reaction with the horizontal bar indicating the mean (+/− SEM). For RAG1/2, one data point (red) was outside of the range of the y-axis and its value is indicated in parentheses. Means for bbRAG1L/2L and RAG1/2 were 19.4 and 44.6, respectively.
(D) Schematic diagram of the assay used to detect in vivo transposition mediated by bbRAG1L and bbRAG2L in 293T cells.
(E) The distribution of in vivo transposition target sites in the recipient plasmid.
See also Figure S6.
To confirm that ProtoRAG is also capable of intermolecular transposition ex vivo in human cells, we transfected 293T cells with a ProtoRAG transposon donor plasmid, a target plasmid, and bbRAG1L/2L expression vectors (Figure 7D). After transformation of cell lysates into bacteria, dual antibiotic resistant (KanR/ChlR) bacterial colonies were readily recovered. Analysis of 39 plasmids confirmed insertion of the artificial ProtoRAG transposon into numerous target sites in the recipient plasmid, generating TSDs that were predominantly (92%) 5 bp in length (Figure 7E) and enriched in GC bp (Figure S6F). Notably, when this same assay was performed with mouse RAG and an RSS-flanked transposon, transposition events were exceedingly rare (Chatterji et al., 2006). These results indicate that despite the vast species difference, lancelet ProtoRAG is capable of bone fide transposition in human cells.
DISCUSSION
Here we provide extensive evidence that ProtoRAG, a cut-and-paste DNA transposon from lancelets, is an evolutionarily relative of the RAG transposon, which our findings suggest dates back as far as basal chordates, some 550 million years ago. We propose that this element was transmitted vertically through chordate and vertebrate evolution, remaining an active in lancelets while being co-opted in jawed vertebrates for the assembly and diversification of antigen receptor loci by V(D)J recombination. A recent report proposed that RAG1/2-like gene loci from echinoderms might belong to a putative transposon superfamily (Kapitonov and Koonin, 2015), although without definitive evidence. We have thus far found no RAG-like sequences in tunicates or jawless vertebrates. This, combined with the absence of the RAG2 C-terminal region in bbRAG2L and the appearance of introns in novel locations in bbRAG1L and bbRAG2L, indicate that pieces remain missing from the puzzle of the evolution and domestication of the RAG transposon.
bbRAG2L is the only known RAG2-like protein that lacks a PHD finger, raising the question of whether RAG2 in the ancestral RAG transposon contained a PHD or not. If the original RAG transposon did not contain a PHD, then echinoderm and vertebrate RAG2 must each have acquired a PHD independently. Alternatively, if the ancestral RAG2 contained a PHD, then it was lost from RAG2L in the lancelet lineage. The C-terminal portion of mouse RAG2 that encodes the PHD has been shown to suppress RAG transposase activity (Elkin et al., 2003; Tsai and Schatz, 2003) and we find that TIRs cleaved by bbRAG1L/2L preferentially undergo transposition rather than TTJ formation. Thus, the absence of the C-terminal region in bbRAG2L might have been advantageous for the mobility and survival of ProtoRAG.
DNA cleavage by bbRAG1L/2L and RAG1/2 exhibit striking mechanistic similarities. In both systems, as well as in Transib transposase (Hencken et al., 2012), DNA cleavage occurs by a nick-hairpin mechanism adjacent to the sequence 5′-CAC-3′, the only perfectly conserved portion of RSSs. Furthermore, RAG and bbRAG1L/2L have similar divalent metal ion requirements, rely on a parallel group of acidic catalytic residues, generate 5 bp TSDs, and prefer CG-rich transposition target sites. Furthermore, both are stimulated by HMGB1, which is thought to facilitate RAG DNA binding and cleavage by stabilizing bends in the RSS (Schatz and Swanson, 2011). Together, the extensive mechanistic parallels between lancelet bbRAG1L/2L and vertebrate RAG1/2 strongly suggest a common origin for the two pairs of proteins.
ProtoRAG fails to function with RSSs while mouse RAG cannot act on ProtoRAG TIRs, indicating that two RAG machineries have become incompatible after evolving independently for over half a billion years. One likely explanation for this incompatibility is the dissimilarities that exist between the RSS nonamer and the ProtoRAG TIR and between the RAG1 nonamer binding domain and the corresponding region in bbRAG1L. Notably, ProtoRAG can produce correctly spliced mRNA and carry out bona fide transposition in human cells, suggesting that the lancelet transposon is able to adapt to the human cellular environment despite the vast evolutionary distance between humans and lancelets. Such flexibility might have been important for the RAG transposon, serving as a prerequisite for transposon domestication by vertebrates and for its co-option for V(D)J recombination, which now involves cooperation between RAG and DNA repair factors (Little et al., 2015). Moreover, although excised ProtoRAG elements exhibit a strong bias toward transposition, they also allow for a low frequency of precise end-to-end (TIR-TIR) joining, the molecular equivalent of SJ formation during V(D)J recombination. This suggests that the RAG transposon was permissive for TTJ/SJ formation long before its domestication by vertebrates. Future biochemical, structural, and functional analyses of ProtoRAG will likely provide additional insights into the origins of RAG and jawed vertebrate adaptive immunity.
EXPERIMENTAL PROCEDURES
Sequence identification and analysis
A whole genome alignment method was designed to identify recently-transposed ProtoRAG copies from lancelet draft genomes. BAC plasmids containing ProtoRAG, identified from B. belcheri BAC libraries, were sequenced and assembled, and the genomic and protein structures of ProtoRAG were determined by aligning the obtained sequences to each other and to vertebrate RAG sequences. Phylogenetic and molecular dating analyses were performed using MEGA5. DNA motifs were identified using custom scripts and BLAST. See Supplemental Experimental Procedures for details of the sequence analysis.
Expression and functional analyses
Transcriptional activities of ProtoRAG were analyzed using RT-PCR and quantitative RT-PCR. Subcellular localization of ProtoRAG proteins was examined using human 293T cells or HeLa cells transfected with appropriate expression constructs. The ex vivo fluorescent reporter assay used to investigate bbRAG1L/2L-mediated transposon excision and host DNA rejoining was based on a previously described assay for RAG function (Corneo et al., 2007). bbRAG1L/2L-mediated ex vivo transposition was examined using a bacterial colony assay as described previously (Chatterji et al., 2006; Zhang et al., 2014). This assay is less subject to bias and limitations imposed by the positioning of primers than is the PCR assay. RAG(-like) proteins for in vitro cleavage and transposition assays were expressed in and purified from mammalian Expi293F™ cells (Gibco). The in vitro cleavage assay might not capture certain aspects of protein function in vivo, but it is conducted under well-defined conditions and hence avoids various uncontrolled factors present in a cellular context. DNA products generated by RAG (-like) proteins after transient transfection of mammalian cells were recovered by PCR or bacterial transformation, and subjected to sequence analysis. For detailed descriptions of functional analyses, see Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Dr. Yiquan Wang and Dr. Guan Li for providing BAC libraries, and Marius Surleac and Dr. Andrei Petrescu for sequence and structural analyses of RAG2-like genes. This work was supported by Project 2013CB835305 (973), 91231206 (NNSF), 2011CB946101 (973), 31171193 (NNSF), 31470846 (NNSF) and 2014J2200017 (New Star of Pearl River), projects from the Guangdong Province Key Laboratory of Computational Science and the Guangdong Province Computational Science Innovative Research Team, and NIH R37AI32524 (D.G.S.). D.G.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
All genome assemblies and sequencing data have been deposited at NCBI under the genome project PRJNA214454. The ProtoRAG sequence of plasmid clone BAC73 has been deposited at DDBJ/ENA/GenBank under the accession KJ748699. The shotgun sequencing data of thirteen ProtoRAG-containing BAC plasmid clones have been deposited at DDBJ/ENA/GenBank under the accessions SRR1565422-SRR1565432, SRR1565434 and SRR1565436.
Supplemental Information
Supplemental information includes Supplemental Experimental Procedures, seven figures, and one table and can be found with this article online at (URL to be supplied).
Author contributions
A.X., S.H. and S.Y. conceived of the study and A.X., S.H., S.Y., X.T., Yuhang Z., and D.G.S. designed the experiments. A.X., S.Y. and S.H. coordinated the project. S.H. carried out phylogenomics and genome informatics. S.H., X.T., Yuhang Z., S.Y., and D.G.S. performed sequence analyses. X.T., Yuhang Z., S.Y., S.H., P.L., H.A.B., Ya Z., W.Y., Y.L.P., and H.E. conducted and analyzed functional experiments. S.H., S.Y., D.G.S. and A.X. wrote the manuscript. D.G.S., P.P., X.T., S.C. and X.L. reviewed and edited the manuscript.
References
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci. 1998;54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji M, Tsai CL, Schatz DG. Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Mol Cell Biol. 2006;26:1558–1568. doi: 10.1128/MCB.26.4.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneo B, Benmerah A, Villartay JP. A short peptide at the C terminus is responsible for the nuclear localization of RAG2. Eur J Immunol. 2002;32:2068–2073. doi: 10.1002/1521-4141(200207)32:7<2068::AID-IMMU2068>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- Craig NL. Mobile DNA: an Introduction. Mobile DNA II (American Society of Microbiology) 2002 [Google Scholar]
- Davis MM, Chien YH, Gascoigne NR, Hedrick SM. A murine T cell receptor gene complex: isolation, structure and rearrangement. Immunol Rev. 1984;81:235–258. doi: 10.1111/j.1600-065x.1984.tb01113.x. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Dreyfus DH. Paleo-immunology: evidence consistent with insertion of a primordial herpes virus-like element in the origins of acquired immunity. PLoS One. 2009;4:e5778. doi: 10.1371/journal.pone.0005778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman QM, Leu TM, Schatz DG. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- Elkin SK, Matthews AG, Oettinger MA. The C-terminal portion of RAG2 protects against transposition in vitro. EMBO J. 2003;22:1931–1938. doi: 10.1093/emboj/cdg184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD. The origins of the Rag genes–from transposition to V(D)J recombination. Semin Immunol. 2010;22:10–16. doi: 10.1016/j.smim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci U S A. 2006;103:3728–3733. doi: 10.1073/pnas.0509720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- Hencken CG, Li X, Craig NL. Functional characterization of an active Rag-like transposase. Nat Struct Mol Biol. 2012;19:834–836. doi: 10.1038/nsmb.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse JE, Lieber MR, Gellert M, Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 1987;49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- Hesse JE, Lieber MR, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- Huang S, Chen Z, Yan X, Yu T, Huang G, Yan Q, Pontarotti PA, Zhao H, Li J, Yang P, et al. Decelerated genome evolution in modern vertebrates revealed by analysis of multiple lancelet genomes. Nat Commun. 2014;5:5896. doi: 10.1038/ncomms6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Koonin EV. Evolution of the RAG1-RAG2 locus: both proteins came from the same transposon. Biol Direct. 2015;10:20. doi: 10.1186/s13062-015-0055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Lapkouski M, Yang W, Gellert M. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature. 2015;518:507–511. doi: 10.1038/nature14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- Lewis SM. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Little AJ, Matthews A, Oettinger M, Roth DB, Schatz DG. Chapter 2 – The Mechanism of V(D)J Recombination A2 – Reth, Frederick W AltTasuku HonjoAndreas RadbruchMichael In Molecular Biology of B Cells. Second. London: Academic Press; 2015. pp. 13–34. [Google Scholar]
- Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- Panchin Y, Moroz LL. Molluscan mobile elements similar to the vertebrate Recombination-Activating Genes. Biochem Biophys Res Commun. 2008;369:818–823. doi: 10.1016/j.bbrc.2008.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden DA, Baetz K, Wu GE. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 1994;22:1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy YV, Perkins EJ, Ramsden DA. Genomic instability due to V(D)J recombination-associated transposition. Genes Dev. 2006;20:1575–1582. doi: 10.1101/gad.1432706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru H, Chambers MG, Fu TM, Tong AB, Liao M, Wu H. Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell. 2015;163:1138–1152. doi: 10.1016/j.cell.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- Teng G, Maman Y, Resch W, Kim M, Yamane A, Qian J, Kieffer-Kwon KR, Mandal M, Ji Y, Meffre E, et al. RAG Represents a Widespread Threat to the Lymphocyte Genome. Cell. 2015;162:751–765. doi: 10.1016/j.cell.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CB. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Tsai CL, Chatterji M, Schatz DG. DNA mismatches and GC-rich motifs target transposition by the RAG1/RAG2 transposase. Nucleic Acids Res. 2003;31:6180–6190. doi: 10.1093/nar/gkg819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CL, Schatz DG. Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2. EMBO J. 2003;22:1922–1930. doi: 10.1093/emboj/cdg185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, Schatz DG. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol. 2009;16:499–508. doi: 10.1038/nsmb.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu K, Deng A, Fu X, Xu A, Liu X. An amphioxus RAG1-like DNA fragment encodes a functional central domain of vertebrate core RAG1. Proc Natl Acad Sci U S A. 2014;111:397–402. doi: 10.1073/pnas.1318843111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


