Summary
The clustering of neurons sharing similar functional properties and connectivity is a common organizational feature of vertebrate nervous systems. Within motor networks, spinal motor neurons (MNs) segregate into longitudinally arrayed subtypes, establishing a central somatotopic map of peripheral target innervation. MN organization and connectivity relies on Hox transcription factors expressed along the rostrocaudal axis; however, the developmental mechanisms governing the orderly arrangement of MNs are largely unknown. We show that Pbx genes, which encode Hox cofactors, are essential for the segregation and clustering of neurons within motor columns. In the absence of Pbx1 and Pbx3 function, Hox-dependent programs are lost and the remaining MN subtypes are unclustered and disordered. Identification of Pbx gene targets revealed an unexpected and apparently Hox-independent role in defining molecular features of dorsally-projecting medial motor column (MMC) neurons. These results indicate Pbx genes act in parallel genetic pathways to orchestrate neuronal subtype differentiation, connectivity, and organization.
Introduction
In many regions of the central nervous system groups of neurons targeting common peripheral targets are centrally organized within topographic maps. The ordered spatial relationship between neuronal position and target specificity is a prominent anatomical feature of primary sensory and motor systems, including the retinotectal map of the visual system and the somatotopic representation of the body surface within the cortex (Kania, 2014; Levine et al., 2012). While topographical maps appear to be critical in establishing appropriate connectivity and functionality within neural circuits, the underlying genetic mechanisms governing their formation are poorly understood.
Within the vertebrate spinal cord, the cell bodies of MNs innervating specific muscle targets are somatotopically organized within columnar, divisional, and pool subtypes (Lance-Jones and Landmesser, 1981; Landmesser, 1978a, b; Romanes, 1951). The topographical arrangement of spinal MNs appears to be a unique attribute of vertebrate motor systems, as MN subtypes of invertebrates lack somatotopic organization, although MN dendrites in Drosophila are highly structured (Baek and Mann, 2009; Landgraf et al., 2003; Thor and Thomas, 2002). While the purpose of MN clustering in vertebrates is not fully understood, it likely evolved to simplify the task of coordinating limb muscle activation sequences during locomotion (Fetcho, 1987), acting in part by enabling and constraining MN access to specific premotor circuits (Goetz et al., 2015; Hinckley et al., 2015; Surmeli et al., 2011).
An early step in establishing MN topographical organization involves the separation of dorsally and ventrally projecting subtypes along the mediolateral axis of the spinal cord. Neurons within the medial motor column (MMC) project dorsally to innervate axial muscles and occupy a ventromedial position. All other MN subtypes typically reside more laterally and initially pursue ventral trajectories. These highly diverse “non-MMC” populations are generated at specific segmental levels of the spinal cord as a consequence of Hox gene activity along the rostrocaudal axis (Philippidou and Dasen, 2013). At forelimb and hindlimb levels a network of Hox genes specifies the identity of the lateral motor columns (LMCs) as well as its resident ~50 MN pools targeting individual limb muscles. At thoracic levels the Hoxc9 gene determines the identity of preganglionic motor column (PGC) neurons, and contributes to the positioning of the hypaxial motor column (HMC) (Jung et al., 2010). In contrast MMC neurons are generated at all segmental levels and appear to differentiate in a Hox-independent manner (Dasen et al., 2008; Dasen et al., 2003; Sharma et al., 1998).
The cell fate determinants that facilitate the clustering of MNs into columns are largely unknown. Mutation of the transcription factor Pea3 leads to a disorganization of neurons within a subset of LMC pools (Livet et al., 2002). Downstream targets of Pea3 include type II cadherins, which appear to be critical for the clustering of neurons within motor pools. MN pools express specific cadherin profiles, and manipulating cadherin expression alters MN settling position (Price et al., 2002). Genetic removal of α- and γ-catenin, which mediate signal transduction through cadherins, leads to a disorganization of neurons within the LMC (Demireva et al., 2011). Nevertheless, the separation of MMC and non-MMC populations persists in the absence of Pea3 and catenins, suggesting an earlier program governs MN columnar organization.
Hox transcription factors are essential during MN subtype diversification and are plausible candidates for governing the coalescence and somatotopic organization of MMC and non-MMC populations. Disruption of Hox gene function however typically leads to a transformation of ventrally-projecting MNs, while preserving their separation from the MMC. For example, in mice mutant for the Hoxc9 gene thoracic-level specific motor columns are converted to an LMC fate, but the distribution and position of MMC neurons is unchanged (Jung et al., 2010). Similarly, the relative position of MMC and non-MMC neurons is retained after depletion of the Foxp1 gene, which encodes an accessory factor required for Hox-dependent programs of LMC and PGC differentiation (Dasen et al., 2008; Rousso et al., 2008). Certain Hox activities in MNs are unaffected by Foxp1 mutation, including the initial induction of the Foxp1 gene and the cross-repressive interactions necessary to establish Hox expression boundaries (Dasen et al., 2008; Jung et al., 2014). The retention of early Hox function in Foxp1 mutants raises the possibility that Hox-dependent pathways contribute to the segregation and clustering of spinal MNs.
We reasoned that insight into the contribution of subtype determinants during MN columnar topographic organization might emerge through manipulations that disrupt Hox activities, but otherwise preserve basic features of MN class identity. Hox proteins are known to rely on interactions with both cell type-restricted and broadly expressed cofactors (Mann et al., 2009; Moens and Selleri, 2006). In most cellular contexts, the three amino acid loop extension (TALE) class of homeodomain proteins plays prominent roles in shaping Hox protein specificity. Pbx proteins, vertebrate homologs of the Drosophila TALE protein extradenticle, are essential for Hox proteins to select gene targets with high affinity. Analysis of Pbx gene function has been constrained due to the existence of multiple gene homologs and the early lethality of mice lacking individual Pbx genes (Moens and Selleri, 2006). Nevertheless, studies in zebrafish and mice have demonstrated essential roles for Pbx genes in rostrocaudal patterning and cell type specification in the hindbrain (Cooper et al., 2003; Vitobello et al., 2011; Waskiewicz et al., 2002). Interpretation of these results is however confounded by a non-cell autonomous role of Pbx genes in controlling expression of morphogens during rhombomere development.
Here we investigated the role of Pbx genes in neuronal differentiation and topographical organization by selectively removing their activities from spinal MNs. We found that Pbx genes are essential for the specification and connectivity of Hox-dependent MN columnar, divisional, and pool subtypes. Unexpectedly, the remaining dorsally and ventrally projecting neurons were intermixed in Pbx mutants, indicating Pbx genes are also responsible for governing the basic program of MN clustering and columnar segregation. Identification of Pbx gene targets in MNs revealed an unanticipated Hox-independent role in defining molecular features of dorsally-projecting MMC neurons. These findings indicate that Pbx genes operate in parallel MN subtype-specific pathways to govern the formation of spinal motor columns.
Results
Pbx Genes Are Dispensable in Early MN Differentiation and Rostrocaudal Patterning
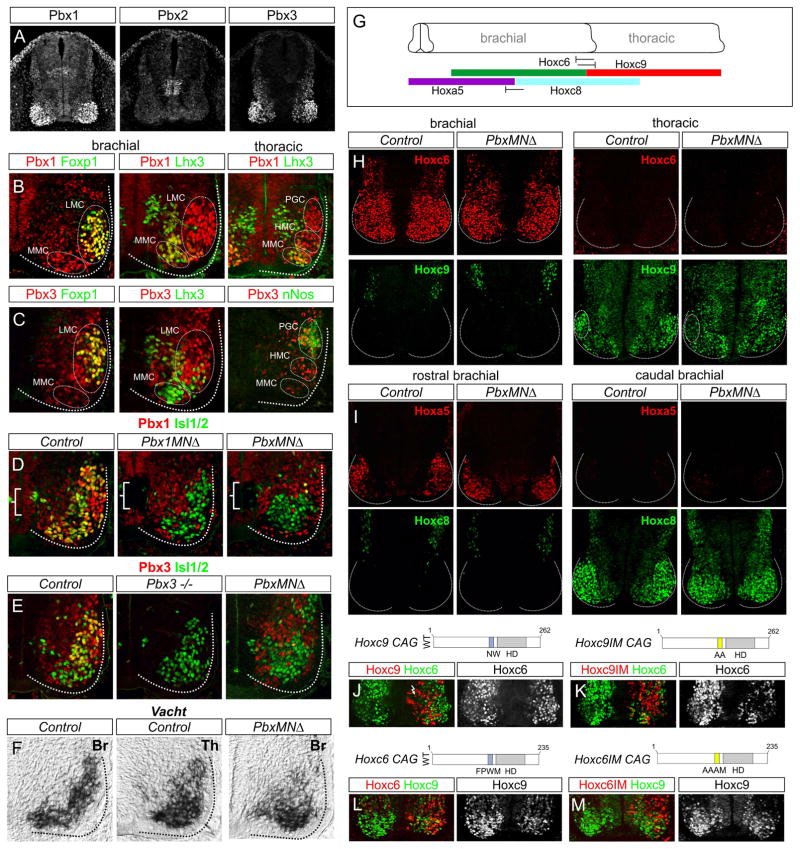
To assess the involvement of Pbx genes in MN subtype diversification we first examined the profile of Pbx protein expression within the spinal cord. Three of the four mammalian Pbx proteins; Pbx1, Pbx2, and Pbx3, were expressed by spinal neurons at embryonic day (e) 11.5 (Figure 1A). Pbx1 was ubiquitously expressed with elevated expression in progenitors and postmitotic MNs (Figure 1A,B, S1A). Pbx2 was detected at low levels throughout the spinal cord, while Pbx3 was expressed in postmitotic neurons, with prominent expression in MNs (Figure 1A,C). Pbx3 was also restricted within Hox-dependent MN populations, including thoracic PGC neurons and subsets of limb-level LMC neurons, but was excluded from axially projecting MMC neurons (Figure 1C, S1B–D). Thus, in mice two of the four mammalian Pbx proteins, Pbx1 and Pbx3 are highly expressed by spinal MNs.
Figure 1. Pbx Genes are Not Required for MN Generation or Establishing Hox Boundaries.
(A) Pbx protein expression in mouse spinal cord at e11.5. Pbx1 is expressed in progenitors and postmitotic MNs. Pbx2 is expressed at low levels in progenitors and postmitotic neurons, and a population of interneuron progenitors. Pbx3 is expressed by postmitotic spinal neurons. (B) Pbx1 colocalizes with Foxp1+ LMC neurons at brachial levels, Lhx3+ MMC neurons, and thoracic HMC and PGC neurons. (C) Pbx3 is restricted to rostral brachial Foxp1+ LMC neurons and excluded from Lhx3+ MMC neurons. At thoracic levels Pbx3 is expressed in HMC and nNos+ PGC neurons. (D) Pbx1 expression is lost in progenitors (brackets) and postmitotic MNs in Pbx1MNΔ and PbxMNΔ mice. (E) Pbx3 expression is lost in Pbx3−/− and PbxMNΔ mice. Pbx3 staining in PbxMNΔ section is from the Pbx3 conditional allele. (F) Vacht mRNA expression at e12.5 in control brachial (Br) and thoracic (Th) MNs and in Br MNs of PbxMNΔ mice. Loss of Br MNs in Pbx mutants does not appear to be due to increased apoptosis (Figure S1H). (G) Summary of Hox expression boundaries in MNs at brachial and thoracic levels. (H) Hoxc6 and Hoxc9 boundaries are maintained in PbxMNΔ mutants, but Hoxc9 levels are reduced in PGC neurons. (I) Hoxa5 and Hoxc8 boundaries are maintained in PbxMNΔ mutants. (J, K) Misexpression of Hoxc9 or a Hoxc9-Pbx interaction mutant (Hoxc9IM) at brachial levels represses Hoxc6. (L, M) Misexpression of Hoxc6 or a Hoxc6IM at thoracic levels represses Hoxc9. See also Figure S1.
To determine the role of Pbx genes in MN differentiation we devised genetic strategies to inactivate their function. To circumvent the early lethality of Pbx1 null mutant mice (Selleri et al., 2001), we bred a floxed Pbx1 allele to Olig2::Cre mice to generate a MN-specific knockout line (referred to henceforth as Pbx1MNΔ mice) (Koss et al., 2012). Pbx3 mutants are viable during embryogenesis (Rhee et al., 2004a), and we therefore generated Pbx1 and Pbx3 double mutants by introducing a Pbx3 null line into the Pbx1MNΔ background. The phenotypes of Pbx1MNΔ; Pbx3−/− mice were not enhanced by introduction of a Pbx2−/− allele (data not shown) (Selleri et al., 2004), and we therefore refer to Pbx1/Pbx3 double mutants as PbxMNΔ mice. In Pbx1MNΔ, Pbx3−/−, and PbxMNΔ lines the respective Pbx proteins were effectively depleted from MNs (Figure 1D,E), and of these alleles, only a rare Pbx1MNΔ mutant survived until adulthood. We also generated embryos in which both Pbx1 and Pbx3 were selectively eliminated from MNs by combining a floxed Pbx3 allele (Rottkamp et al., 2008) with Pbx1MNΔ and Pbx3 heterozygote alleles (Figure 1E). The phenotypes of Pbx1 flox/flox; Pbx3-/flox; Olig2::Cre+ embryos were indistinguishable from that of Pbx1MNΔ;Pbx3−/− embryos at e12.5 (data not shown).
We assessed how loss of individual and multiple Pbx genes affects core features of MN identity and early rostrocaudal patterning. In Pbx1MNΔ, Pbx3, and PbxMNΔ embryos markers for MN neurotransmitter synthesis including choline acetyltransferase (Chat) and vesicular choline acetyltransferase transporter (Vacht) were present at e12.5, indicating Pbx genes are not required for generating MNs as a class (Figure 1F, S1E and data not shown). In addition the MN progenitor determinants Olig2 and Nkx6.1, and the early postmitotic markers Isl1/2, Hb9 and Lhx3, were grossly unaltered in Pbx mutants (Figure 1D,E, S1F). At thoracic levels the total number of MNs was similar between control and PbxMNΔ mutants, although there was ~30% reduction in MNs at limb levels, similar to the loss observed in Foxp1 mutants (Figure 4I) (Dasen et al., 2008; Rousso et al., 2008). In Pbx1MNΔ mutants we also observed an expansion of the Pbx3 expression domain at caudal brachial levels (Figure S1G), although no changes in Pbx2 and Pbx4 expression were observed in any of the Pbx mutant alleles we analyzed (data not shown).
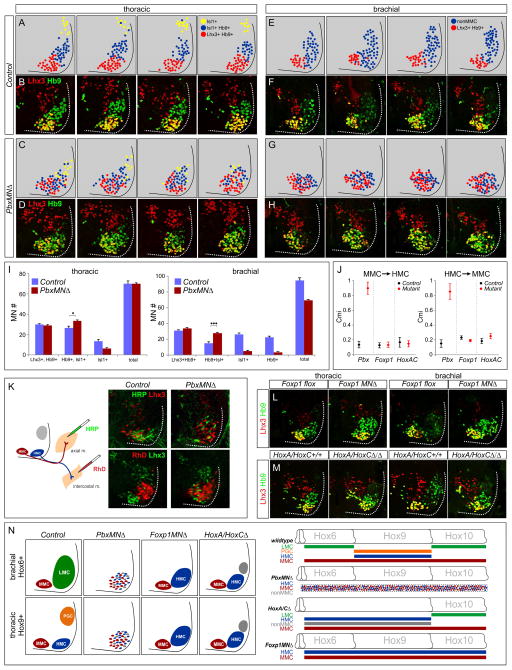
Figure 4. Motor Neuron Columnar Disorganization in PbxMNΔ Mice.
(A–D) Position of molecularly defined thoracic MN subtypes in control and PbxMNΔ mice at e12.5. Schematics represent serial thoracic sections of control and Pbx mutants indicating the position of MN subtypes defined by Lhx3, Hb9, and Isl1 expression. MMC neurons are Lhx3+, Hb9+; HMC neurons are Hb9+, Isl1+, Lhx3−; and PGC neurons are Isl1+. (E–H) Position of MN subtypes at brachial levels. Non-MMC neurons are defined as MNs that are Lhx3−, and express Hb9 and/or Isl1. Brachial sections shown are from segments C6–C8. (I) Quantification of MN subtypes in control and Pbx mutants at brachial and thoracic levels. Molecular codes for columnar subtypes are indicated. MN counts show average of indicated subtype on one side of spinal cord +/− SEM. *p<0.05, ***p<0.001. (J) Quantification of columnar mixing indices (Cmi) for the indicated mutant at thoracic levels. Data are shown as mean Cmi +/ SEM, averaged from n>4 sections, from n>3 animals of indicated genotype. (K) Retrograde labeling of HMC and MMC neurons in control and PbxMNΔ mice at e12.5. MMC neurons were labeled by injection of horseradish peroxidase (HRP) into axial muscles, HMC neurons by injection of intercostal nerves with rhodamine dextran (RhD). (L–M) Comparison of MN organization at thoracic levels in control, PbxMNΔ, Foxp1 and HoxA/C mutants. In Foxp1 and HoxA/C mutants MMC neurons are organized and segregated from non-MMC neurons. (N) Summary of defects in MN specification and positioning in Pbx, Foxp1, and Hox cluster mutants. See also Figure S4
In the hindbrain Pbx function has been implicated in positive autoregulatory interactions that maintain expression of Hox genes in specific rhombomeres (Tumpel et al., 2009). At spinal levels a major determinant of Hox gene expression is cross-repressive interactions between Hox proteins and Hox genes (Philippidou and Dasen, 2013). We therefore investigated a possible function of Pbx genes in controlling the pattern of Hox expression within the spinal cord. In PbxMNΔ mutants the rostrocaudal boundaries between Hoxc6/Hoxc9 (brachial/thoracic boundary) and Hoxa5/Hoxc8 (rostral brachial/caudal brachial boundary) were preserved (Figure 1G–I, S1I–K). In thoracic segments Hoxc9 protein levels were reduced in MNs of PbxMNΔ mice at e12.5, while the overall patterns of Hoxa5, Hoxc6, and Hoxc8 were similar to controls (Figure 1H,I,S1I–K). These observations indicate that Pbx proteins contribute to sustaining the levels of certain Hox proteins, but are dispensable for cross-repressive interactions in spinal MNs.
To further assess whether Hox cross-repressive interactions can occur independent of Pbx function, we determined the effects of misexpressing Hox mutant derivatives in which the canonical Pbx interaction motifs have been mutated. We generated mutations within motifs N-terminal to the homeodomain which are necessary for high affinity interactions with Pbx proteins on DNA. Misexpression of a Hoxc9 derivative with a mutated Pbx interaction motif (Hoxc9IM NW→AA) in chick neural tube was able to repress Hoxc6, Hoxa5 and LMC specification at brachial levels (Figure 1J,K, S1L). However, unlike wildtype Hoxc9, this mutation failed to induce PGC neurons at brachial levels (Figure S1L). As previously reported Hoxc6IM repressed Hoxc9 at thoracic levels (Figure 1L,M). Hoxc6IM can induce an LMC fate at thoracic levels (Lacombe et al., 2013), likely as a consequence of Hoxc9 attenuation. Although we cannot rule out the possibility that Hox proteins interact with Pbx proteins through additional domains (Merabet and Mann, 2016), these results indicate that Pbx genes are not required for establishing the overall rostrocaudal pattern of Hox expression in MNs.
Pbx Genes are Essential for the Differentiation of Hox-Dependent MN Subtypes
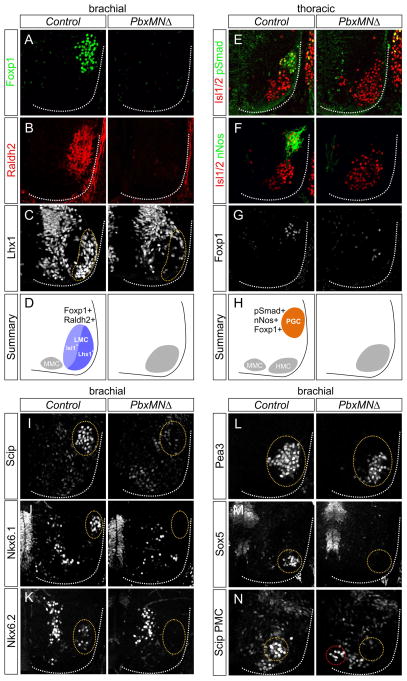
We next examined the function of Pbx genes in the diversification of segmentally-restricted MN subtypes. At limb levels LMC neurons are defined by expression of the transcription factor Foxp1 and the retinoic acid (RA) synthetic enzyme Raldh2 (Dasen et al., 2008). In Pbx3 mutants Foxp1 and Raldh2 expression was unaltered at brachial levels (Figure S2A). In Pbx1MNΔ embryos we observed a marked reduction in the number of brachial Foxp1+, Raldh2+ neurons (Figure S2D,E), although the penetrance of this phenotype was apparently offset by the upregulation of Pbx3 within this region (Figure S1G). Analysis of mice lacking both Pbx1 and Pbx3 revealed a dramatic reduction in Foxp1 and Raldh2 expression (Figure 2A,B). Raldh2-dependent RA synthesis in LMC neurons provides a feed-back signal that promotes the proliferation of MN progenitors (Sockanathan and Jessell, 1998), and the loss of this signal likely accounts for the decrease in brachial MNs in Pbx mutants. These results indicate Pbx genes are essential to establish the identity and normal number of LMC neurons.
Figure 2. Pbx Genes are Essential for MN Columnar, Divisional, and Pool Specification.
(A,B) Expression of Foxp1 and Raldh2 is reduced in PbxMNΔ mutants at e12.5. (C) Expression of Lhx1 within the lateral division of the LMC is depleted at brachial levels in PbxMNΔ mice. (D) Summary of LMC neuron organization at brachial levels in control and PbxMNΔ mice. (E–G) At thoracic levels there is a loss of nNos, Foxp1, and pSmad expression in PbxMNΔ mutants. (H) Summary of MN organization at thoracic levels in control and PbxMNΔ mice. (I) Loss of Scip expression from median and ulnar MN pools in PbxMNΔ mice (J,K) Loss of Nkx6.1 and Nkx6.2 expression from rostral brachial pools in PbxMNΔ mutants. (L) Expression of Pea3 is detected in PbxMNΔ mice, but mispositioned ventromedially. (M) Loss of the non-LMC Sox5+ pool in PbxMNΔ mice. (N) Expression of Scip in phrenic MNs is reduced and mislocalized (red circle) in PbxMNΔ mice. In panels I–N circled areas discriminate MNs that express indicated factors from other spinal populations. In PbxMNΔ mice circled areas represent position where these pools would be present normally. See also Figure S2.
Hox genes are critical for the further differentiation of LMC neurons into divisional and pool subtypes which determine how motor axons select muscle targets in the limb (Dasen et al., 2005). Expression of Lhx1 in the lateral division of the LMC defines MNs that project to the dorsal limb compartment (Kania, 2014), and this population of Lhx1+, Foxp1+ neurons was depleted in PbxMNΔ mutants (Figure 2C,D). Within the LMC, neurons targeting individual limb muscles segregate into MN pools, some of which can be defined by expression of specific transcription factors. In PbxMNΔ mice, we observed a loss of MN pools marked by Scip, Nkx6.1 and Nkx6.2 expression (Figure 2I–K). Expression of the pool marker Pea3 was preserved in PbxMNΔ mutants (Figure 2L), consistent with studies suggesting Pbx-independent regulation of its expression (Catela et al., 2016; Lacombe et al., 2013), and reliance on peripheral signaling to control its induction (Figure S2F–H) (Haase et al., 2002). Pbx genes are therefore essential for the appearance of multiple molecular features of LMC subtypes.
At thoracic levels Hox genes are necessary for the differentiation of PGC neurons that innervate the sympathetic chain ganglia. PGC neurons express neuronal nitric oxide synthase (nNos), phospho (p) Smad1/5/8, Isl1, low levels of Foxp1, and settle dorsolaterally (Dasen et al., 2008). In PbxMNΔ mutants nNos and pSmad expression is lost from MNs and Isl1+ MNs fail to migrate to a dorsolateral position (Figure 2E–H). Interestingly, the differentiation of PGC neurons also relied on the net level of Pbx expression in MNs as Pbx1MNΔ; Pbx3+/− mutants (which retain a single copy of Pbx3) also display dramatically reduced numbers of PGC neurons (Figure S2B,C). The dose-dependent phenotypes of Pbx mutant alleles likely reflect differences in the level and pattern of Pbx1 and Pbx3 within specific MN subtypes (Figure S1A–D).
In addition to these well-characterized Hox-dependent MN populations, other segmentally-restricted subtypes were affected in Pbx mutants. At rostral cervical levels two non-LMC (Foxp1−) populations of MNs can be defined by the expression of the transcription factors Sox5 and Scip (Philippidou et al., 2012). Sox5 is expressed by a laterally-positioned Lhx3+, Hb9+, Isl1+ MN pool, and this population is lost in Pbx MNΔ mice (Figure 2M). Motor neurons within the phrenic motor column (PMC) rely on the activities of Hox5 genes and coexpress Scip and Isl1. In PbxMNΔ embryos this population is reduced, disorganized, and shifted to a more medial position (Figure 2N). Interestingly both Scip+ PMC and Sox5+, Lhx3+ MNs are retained in Foxp1 mutants (Dasen et al., 2008; Rousso et al., 2008), indicating that loss of Pbx genes affects most segmentally-restricted MNs subtypes.
Peripheral Innervation Defects in Pbx Mutants
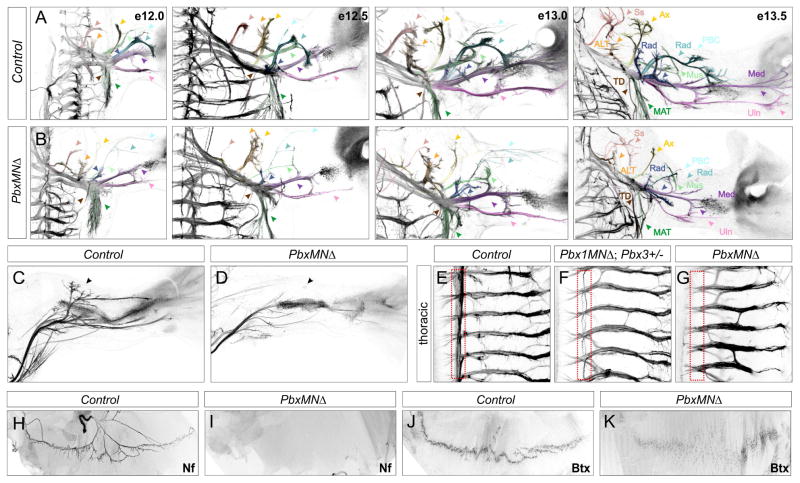
Because molecular signatures of Hox-dependent MN subtypes are lost in PbxMNΔ mice, we next determined the impact of Pbx mutation on the trajectory and target selectivity of motor axons. We bred PbxMNΔ mutant mice to Hb9::GFP mice, in which all motor axons are GFP labeled, and analyzed the overall pattern of peripheral innervation. Projections into the limb were detectable in PbxMNΔ; Hb9::GFP mutants at e12.0, but at subsequent time points the distal nerve branches were thinner, and specific target regions lacked innervation (Figure 3A,B). Nerve branches to muscles in the proximal forelimb were missing or stunted in PbxMNΔ mutants, including a severe reduction in the density of projections along the radial nerve (Figure 3A,B). PbxMNΔ; Hb9::GFP mice also displayed pronounced innervation defects within the hindlimb. The most striking defect was observed along the tibialis anteroir nerve which was completely absent at in PbxMNΔ mice at e13.5 (Figure 3C,D)
Figure 3. Motor Axon Projections in PbxMNΔ Mice.
(A,B) Dorsal view of forelimb innervation in control and PbxMNΔ mice between e12.0 and e13.5. PbxMNΔ mice have thinner axons and display defects in axonal branching and nerve trajectories. Color coding: Suprascapular Nerve (N.) (Ss, red), Anterior Lateral Thoracic N. (ALT, orange), Axillary N. (Ax, Yellow), Musculocutaneous N. (Mus, neon green), Radial N. (Rad, blue), Posterior Brachial Cutaneous N. (PBC, aqua), Radial/Musculospiral N. (Rad, dark blue), Median N. (Med, dark purple) Ulnar N. (Uln, light purple), Thoracodorsal nerve to Lattismus Dorsi (LD) (TD, brown) and Medial Anterior Thoracic N. to the Cutaneous Maximus (CM) (MAT, dark green). (C,D) In PbxMNΔ; Hb9::GFP mice the dorsal tibialis anterior nerve fails to form in the hindlimb. (E–G) Motor axon projections at thoracic levels showing loss of sympathetic chain ganglia innervation (outlined in red). Mice retaining one allele of Pbx3 also display projection defects. Projections along the intercostal nerves are retained in Pbx mutants, although some aberrant branching is observed. (H,I) Diaphragm innervation defects in PbxMNΔ mutants at e16.5. Motor axons are labeled using Neurofilament (Nf) staining. In PbxMNΔ mice phrenic axons fail to innervate the diaphragm. (J,K) Staining with α-bungarotoxin (Bgt) showing acetylcholine receptor (AChR) clustering in control animals and absence of concentrated clusters in PbxMNΔ mice. See also Figure S3.
At thoracic levels MNs within the PGC pursue a ventromedial trajectory towards sympathetic chain ganglia (scg), and subsequently send collateral projections that extend along the rostrocaudal axis. In wholemount staining of control Hb9::GFP mice, these projections are visible as a medial GFP+ band that extends parallel to the spinal cord (Figures 3E, S3C,D). In PbxMNΔ mutants projections towards and between sympathetic chain ganglia were dramatically reduced (Figures 3G, S3A,B,G,H). Similarly, in Pbx1 conditional mutants retaining one copy of Pbx3 there was a pronounced decrease in sympathetic chain ganglia innervation (Figures 3F, S3E,F). In contrast motor nerves projecting to axial and hypaxial muscles (which derive from MMC and thoracic HMC neurons, respectively) were preserved in PbxMNΔ mutants (Figure 3G).
One of the most severely affected nerve branches in Pbx mutants derives from phrenic MNs that extend to the diaphragm muscle. At e12.5 the phrenic nerve was visible in PbxMNΔ; Hb9::GFP mutants, but was dramatically thinner and shorter than in control littermates (Figure S3I,J). By e16.5 there was a severe loss of synapses at the diaphragm, with the majority of muscle fibers lacking innervation and postsynaptic acetyl choline receptor clusters (Figure 3H–K). The severe defects in diaphragm innervation likely account for the perinatal lethality of PbxMNΔ mutants.
Motor Columns Are Disorganized in Pbx Mutants
What are the fates of the remaining MN populations after deletion of Pbx genes? In mice mutant for the Hox accessory factor Foxp1, LMC and PGC neurons acquire the identity of thoracic HMC neurons, while axially projecting MMC neurons are unaffected (Dasen et al., 2008). In Pbx mutants the number of Lhx3+, Hb9+ MMC neurons was not significantly altered at brachial and thoracic levels (Figure 4A–I). The remaining populations consisted predominantly of MNs with an HMC-like molecular profile (Hb9+; Isl1/2+) as well a smaller group that expressed Isl1/2 alone (Figure 4I, S4A–D). These results indicate that in the absence of Pbx function, the remaining MNs display molecular features of MMC and HMC subtypes.
Analysis of the distribution of MNs in Pbx mutants revealed a striking disorganization in their settling position. In PbxMNΔ mice Lhx3+, Hb9+ (MMC-like); Hb9+, Isl1/2+ (HMC-like) and Isl1/2+ MNs were intermixed at e12.5 (Figure 4A–H). This phenotype was observed at all rostrocaudal levels of the spinal cord, and was particularly prominent at thoracic levels. Comparison of serial sections in Pbx mutants revealed no positional preference of MMC and non-MMC neurons indicating they are stochastically positioned within the ventral spinal cord (Figure 4A–H). In contrast, the organization and specification of ventral interneurons was not affected in PbxMNΔ mice (Figure S4G). To quantify the degree of MN intermixing in Pbx mutants, we calculated a columnar mixing index (Cmi) for thoracic MMC and HMC neurons in control and mutant mice at e12.5 (Demireva et al., 2011). This allowed us to determine the extent to which MMC neurons invade the confines of the HMC, and HMC invasion into the MMC. In control thoracic sections Cmi values for MMC→HMC and HMC→MMC averaged 0.14 and 0.15 respectively. In Pbx mutants this value increased to 0.90 for MMC→HMC and 0.85 for HMC→MMC (Figure 4J).
Because the transcription factors used to discriminate columnar identities (Lhx3, Hb9, and Isl1/2) are expressed by the precursors to all MN subtypes, we considered the possibility that the observed intermixing is not due to migratory or clustering defects, but rather a failure of MNs to fully differentiate in PbxMNΔ mice. If, however the remaining columnar subtypes are properly differentiated, they would be predicted to target muscles appropriate for their molecular identity. Because loss of an LMC identity leads to a random targeting of limb muscles by MNs (Dasen et al., 2008; Rousso et al., 2008), we assessed the targeting of MMC and HMC neurons at thoracic levels, where these two populations are present normally. We injected tracers into the intercostal nerves (targets of HMC neurons) or axial muscles (targets of MMC neurons) in control and PbxMNΔ mutants at e12.5 and monitored the transcriptional profile of retrogradely labeled MNs. After tracer injection into the intercostal nerves of PbxMNΔ mutants, labeled MNs exhibited an HMC profile (Isl1/2+, Hb9+) and lacked Lhx3 (Figure 4K). After injection into axial muscles, labeled MNs exhibited an MMC profile (Lhx3+, Hb9+, Isl1/2low) (Figure 4K). In agreement with the analysis of MMC and HMC molecular profiles, retrogradely labeled neurons lacked any clear columnar organization. These results indicate that despite their altered position, the remaining thoracic MMC and HMC neurons in PbxMNΔ mice differentiate and select appropriate muscle targets. In contrast, retrograde labeling from the forelimb ulnar nerve labeled HMC-like profile (Isl1+, Lhx3−) that were dispersed within the spinal cord (Figure S4H), similar to the targeting defects observed in Foxp1 mutants (Dasen et al., 2008).
Several lines of evidence indicate that the intermixing of HMC and MMC neurons reflect a unique function of Pbx genes during MN differentiation. Analysis of Foxp1 mutants revealed a loss of Hox-dependent subtypes at limb and thoracic levels, with the remaining MNs consisting predominantly of HMC and MMC neurons (Dasen et al., 2008; Rousso et al., 2008). Despite the similarity in the loss of MN identities in Foxp1 and Pbx mutants, in the absence of Foxp1 the remaining HMC and MMC neurons were clustered and well segregated (Figures 4L, S4E). In mice lacking the HoxA and HoxC gene clusters, Hox-dependent MN subtypes are similarly lost at brachial and thoracic levels (Jung et al., 2014). The HoxA and HoxC clusters encode the majority of Hox proteins expressed at these levels, and therefore approximate a Hox-less ground state. In combined HoxA and HoxC mutants, the clustering and segregation of the remaining subtypes (MMC, HMC, and Isl1/2+ MNs) was unaffected (Figures 4M, S4F) (Jung et al., 2014). Calculation of columnar mixing indices in Foxp1 and HoxA/C mutants revealed no increase in thoracic MMC/HMC intermixing relative to Cmi values of control littermates (Figure 4J). These observations suggest a unique, and possibly Hox-independent, function for Pbx proteins in the organization of columnar subtypes projecting to epaxial and hypaxial muscles (Figure 4N).
Identification of Genes Selectively Depleted in Pbx Mutants
The intermixing of the remaining MMC and non-MMC neurons in PbxMNΔ mice prompted us to consider whether Pbx genes might selectively regulate target effectors present in these populations. To explore this possibility, we analyzed a panel of genes demonstrated to be downstream of MN fate determinants. We reasoned genes which facilitate the segregation and clustering of MMC and non-MMC populations would be specifically lost in Pbx mutants, but maintained under conditions where columnar segregation is preserved, such as in Foxp1 mutants. We screened over two dozen genes, including members of the cadherin and ephrin/Eph genes families, which have been implicated in neuronal migration and clustering within the hindbrain and spinal cord (Kania, 2014). This analysis identified a number of motor pool-restricted genes that are diminished in PbxMNΔ mice, although the majority of these genes were also downregulated in Foxp1 mutants (Figure S5A–E and data not shown).
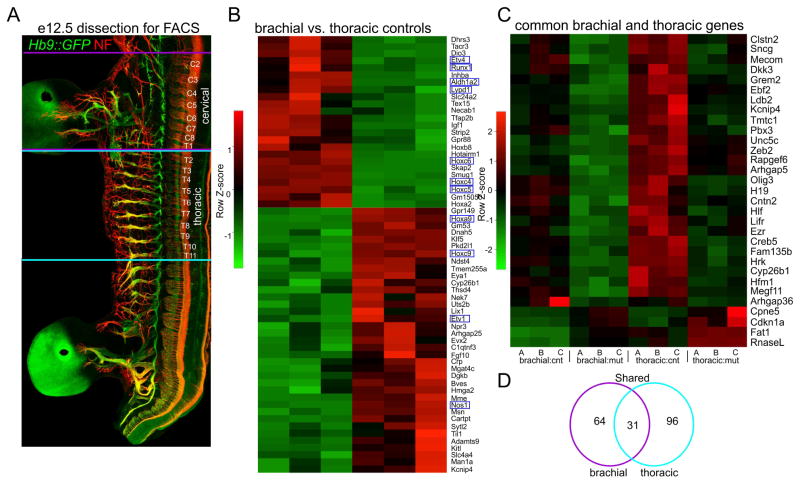
These results encouraged us to initiate an unbiased screen to identify genes selectively lost in PbxMNΔ mice. We compared gene expression profiles in MNs isolated from control and PbxMNΔ embryos at e12.5. We purified MNs from PbxMNΔ; Hb9::GFP and control Hb9::GFP embryos by fluorescence-activated cell sorting (FACS) (Figure 5A). Due to the distinct molecular profiles of MNs generated at brachial and thoracic levels, we independently profiled both populations. We extracted RNA from MNs purified from 9 PbxMNΔ; Hb9::GFP and 9 Hb9::GFP embryos at brachial and thoracic levels, pooled 3 RNA samples of each genotype, and prepared 12 bar coded libraries. We then performed expression profiling by RNA-seq. The samples were mixed into two pools and run on two 50-nucleotide paired-end read rapid run flow cell lanes with the Illumina HiSeq 2500 sequencer.
Figure 5. Identification of Pbx Gene Targets in Motor Neurons.
(A) Wholemount staining of Hb9::GFP mouse at e12.5 showing dorsal root ganglia (DRG) and spinal segmental levels used for gene profiling. Neurofilament (Nf) staining highlights the segments isolated for FACS. Brachial MNs were isolated from cervical (C) level C2 to thoracic (T) level T1 and thoracic MNs from T2 to T11. (B) Heatmap showing comparison of gene expression differences between brachial and thoracic MNs in controls. Known differentially expressed genes are outlined in blue (C) Heatmap showing expression differences between control and PbxMNΔ mutants. Heatmap lists genes that are common to both brachial and thoracic samples and that are differentially expressed with a padj.<0.05 cutoff. Heat maps for each of the three pools are shown, and are labeled A, B, C (D) Venn diagram of differentially expressed genes shared between brachial and thoracic levels. See also Figure S5.
To evaluate the quality of this screen we examined the expression of genes known to be differentially expressed between brachial and thoracic levels. Comparison of expression profiles between purified control samples yielded known cervical/brachial- (Aldh1a2/Raldh2, Etv4/Pea3, Runx1, Lypd1/Lynx2, Hoxc6) and thoracic- level (nNos, Etv1/Er81, Hoxc9) restricted MN determinants (Figure 5B, Table S1). Read counts of HoxC cluster genes were similar between control and PbxMNΔ mice at both brachial and thoracic levels (Figure S6E), reinforcing the conclusion that the Hox patterns are grossly preserved in PbxMNΔ mice.
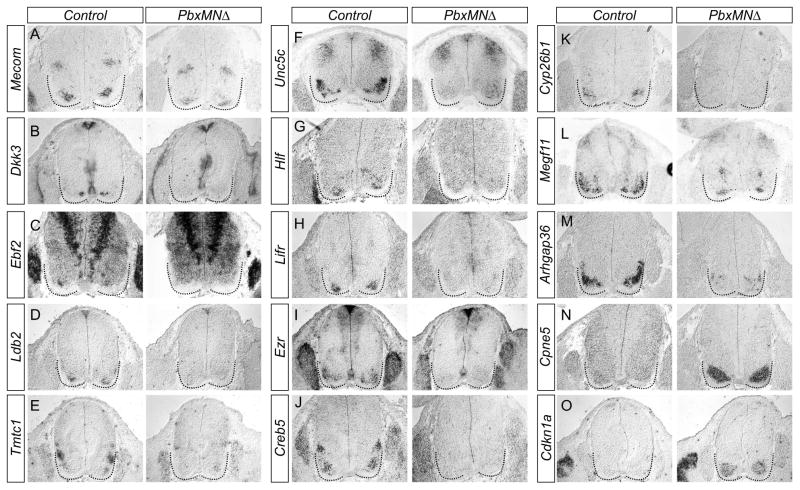
Because loss of Pbx genes affects MN organization at all rostrocaudal levels, we focused on genes whose profiles were altered at both brachial and thoracic levels. Comparison of gene profiles within brachial and thoracic MNs identified 31 transcripts (27 downregulated and 4 upregulated genes) that were common to both populations and were differentially expressed between control and Pbx mutant mice (Figure 5C,D, S6A–D, Table S2,S3). To validate these targets, we used in situ hybridization to compare expression patterns of candidates between control and PbxMNΔ mice at e12.5. Analysis of the 27 downregulated candidates identified 13 that were expressed by MNs in control animals (Figures 6A–M, data not shown). Each of these genes was undetectable or markedly downregulated in MNs of PbxMNΔ mice, confirming them as Pbx targets (Figure 6A–M). Novel genes that were upregulated in PbxMNΔ mice and validated by in situ hybridization included Cpne5 and Cdkn1a (Figure 6N,O). In control embryos Cpne5 was weakly expressed by MNs with elevated expression in MMC neurons (Figure 6N). In PbxMNΔ mice Cpne5 expression was markedly upregulated in all remaining MNs (Figure 6N). Additional genes from this list were either not detected by in situ hybridization or expressed by non-MN populations (data not shown).
Figure 6. Characterization of Pbx Target Gene Expression in Motor Neurons.
(A–O) Analysis of genes common to brachial and thoracic levels that are differentially expressed between control and PbxMNΔ mice. Panels show in situ hybridization of indicated genes in spinal cord sections of e12.5 mice. All sections are brachial except J and K, which are thoracic. Sections are derived from embryos which are Pbx1 flox/flox; Pbx3-/flox; Olig2::Cre+. (A) Mecom is downregulated in ventromedial neurons but preserved in dorsal interneurons. (B–K) Expression of Dkk3, Ebf2, Ldb2, Tmtc1, Unc5c, Hlf, Lifr, Ezr, Creb5, and Cyp26b1 is markedly decreased in MNs of Pbx mutants. (L) Megf11 is downregulated in LMC neurons but maintained in a subset of ventromedial MNs. (M) Arhgap36 expression is markedly reduced in PbxMNΔ mice. (N,O) Cpne5 and Cdkn1a are upregulated in Pbx mutants. See also Figure S6.
Among the confirmed downregulated genes in Pbx mutants were a number that were restricted to ventral spinal populations. Included in this group were the cell surface proteins Lifr and Megf11, the secreted protein Dkk3, the intracellular protein Ezr, and the transcription regulators Mecom, Ebf2, Ldb2, Hlf, and Creb5 (Figure 6A–D,G–I,J,L). Expression of Lbd2, Hlf, Lifr, and Creb5 was not detectable in the spinal cords of PbxMNΔ mice at e12.5, while expression of Mecom, Dkk3, Ezr, and Megf11 was selectively lost from subsets of MNs (Figure 6). Mecom was expressed in ventromedial MN populations as well as a dorsal interneuron population at all rostrocaudal levels. In PbxMNΔ mutants expression of Mecom was markedly diminished in MNs, while its pattern in dorsal interneurons was unaffected (Figure 6A). Expression of Mecom, Dkk3, Ebf2, Ldb2, Hlf, Lifr, and Cyp26b1 were also maintained in Foxp1 mutants, while Cpne5 and Cdkn1a were not upregulated (Figure S5F–T), suggesting regulation of these targets independent of the Hox/Foxp1-dependent programs acting in non-MMC neurons.
Mecom Defines a Pbx-Dependent Population of MNs Targeting Axial Muscle
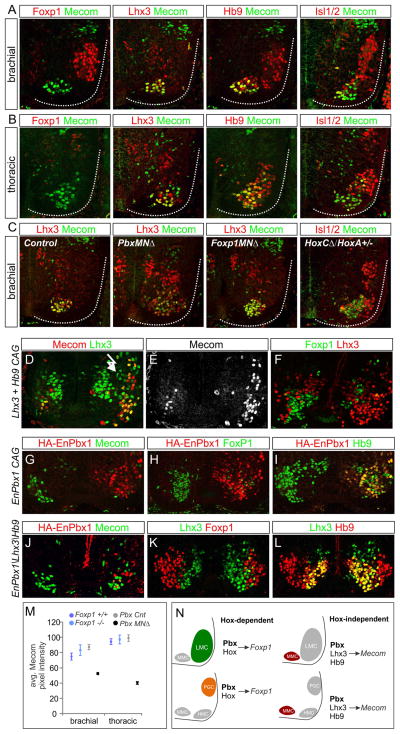
The identification of MN subtype-restricted genes that are selectively downregulated in Pbx mutants enabled us to further assess a potential Hox-independent function of Pbx proteins during MN specification. Due to its restricted expression in ventromedial MN populations we chose Mecom for further analysis. We characterized the pattern of Mecom expression relative to MN determinants between e9.5-e12.5. Mecom protein was first detected at e9.5 as the first brachial postmitotic MNs appear, and was subsequently maintained in a subset of medial MNs (Figures 7A,B, S7A). At e12.5 Mecom colocalized with markers for MMC identity at all rostrocaudal levels of the spinal cord, but was excluded from Foxp1+ LMC and PGC neurons (Figure 7A,B). In addition, Mecom was absent from rostral brachial Sox5+, Lhx3+ MNs, but was present in a small subset of thoracic Lhx3−, Foxp1− MNs (Figure 7B, S7B). The MMC-restricted pattern of Mecom was also conserved in MNs of chick embryos (Figure S7C). Mecom therefore defines a novel postmitotic marker labeling MMC neurons projecting to dorsal axial muscles.
Figure 7. Hox-Independent Regulation of Mecom by Pbx Proteins and Lhx3.
(A) Mecom protein expression at brachial levels at e12.5. Mecom is detected in Lhx3+, Hb9+ MMC neurons but excluded from Foxp1+ LMC neurons. (B) Mecom expression at thoracic levels. Mecom is detected in MMC neurons, a subset of Lhx3−, Hb9+ MNs, but is excluded from Foxp1+ PGC neurons. (C) Analysis of Mecom expression in Pbx, Foxp1, and HoxC mutants. Mecom is reduced in MMC neurons of PbxMNΔ mice, but is unaffected in Foxp1MNΔ and HoxC mutants. (D,E) Chick electroporations at thoracic levels showing Mecom expression is induced after Lhx3 and Hb9 misexpression. (F) Lhx3/Hb9 also represses Foxp1 expression. (G–I) Expression of EnPbx1 represses Mecom and Foxp1 in MNs. (J–L) Coexpression of EnPbx1, Lhx3, and Hb9 fails to generate ectopic Mecom+ MNs. (M) Quantification of Mecom levels in Pbx and Foxp1 mutants. Data are shown as average pixel intensities of Mecom immunofluorescence +/− SEM. (N) Summary MN columnar specification by Pbx proteins. Pbx proteins act in Hox-dependent pathways to induce expression of columnar determinants such as Foxp1. Pbx proteins also act in a Hox-independent manner to regulate expression of MMC-restricted genes including Mecom. See also Figure S7.
We next investigated the regulation of Mecom by Pbx proteins and other MN subtype determinants. In PbxMNΔ mutants the level of Mecom protein expression was markedly reduced, with low levels detected in the remaining scattered Lhx3+ MMC cells (Figure 7C,M). In contrast the pattern of Mecom expression was unaffected in Foxp1 and HoxC cluster mutants, and its restriction to MMC neurons was retained (Figure 7C). These observations provide additional in vivo evidence that Pbx proteins function independently of the Hox/Foxp1 program to regulate expression of MMC-restricted genes such as Mecom.
If Mecom is regulated by Pbx proteins independent of Hox function, its expression could reflect an output of an earlier MMC-specific differentiation program. We therefore tested if determinants of MMC identity can induce expression of Mecom in non-MMC neurons. Misexpression of the Hb9 transcription factor has been shown to impose a MN fate on interneurons, while expression of Lhx3 in all MNs directs an MMC fate (Sharma et al., 2000; William et al., 2003). Because expression of Lhx3 in non-MNs produces predominantly V2a interneurons, while expression of Hb9 generates MNs with either MMC or HMC-like properties (Dasen et al., 2008; Thaler et al., 2002), we coexpressed both factors to produce ectopic MMC neurons. We found that misexpresion of Lhx3 and Hb9 induced ectopic Mecom+ MNs (Figure 7D,E, S7D). Lhx3 and Hb9 coexpression also extinguished expression of Foxp1, a known target of Hox proteins in MNs (Figure 7F).
To test whether the supernumerary Mecom+ MNs induced by Lhx3 and Hb9 requires Pbx function, we expressed these factors in conjunction with a dominant-negative Engrailed-repressor fusion with Pbx1 (EnPbx1). Expression of EnPbx1 alone repressed Mecom and Foxp1, but not Hb9, consistent with Pbx1 activity being required for the differentiation of MMC and LMC neurons (Figure 7G–I). In contrast, coexpression of Lhx3, Hb9, and EnPbx1 failed to generate Mecom+ MMC neurons (Figure 7J–L). Collectively, these results show that Pbx genes are essential for the normal expression of Mecom in MNs, and act in concert with Lhx3 to determine its MMC-restricted pattern (Figure 7N). Pbx proteins therefore appear to act in parallel Hox-dependent and independent programs to control the subtype differentiation and organization of MN subtypes.
Discussion
The clustering of motor neurons into longitudinally arrayed columnar groups is a defining feature of topographical maps within tetrapod motor systems, but the underlying genetic mechanisms governing their formation has remained elusive. We found that Pbx genes are essential for the formation and differentiation of spinal motor columns. Consistent with roles as Hox cofactors, Pbx genes are required for the specification of MN subtypes along the rostrocaudal axis and the establishment of appropriate patterns of peripheral innervation. Unexpectedly, our studies show that Pbx genes are also critical for the coalescence of MNs into columns, revealing a novel molecular program mediating the partitioning of dorsally projecting MMC neurons from all other MN subtypes. These studies could provide a foundation for resolving the role of MN position in locomotor circuit connectivity and exploring the origins of topographic organization within motor systems.
Pbx Genes as Cofactors for Hox-Dependent Steps in MN Differentiation
Hox genes are essential for the specification of neuronal classes along the rostrocaudal axis where they contribute to the diversification and connectivity of MN columnar, divisional, and pool subtypes. Pbx cofactors are well known to enhance the affinity and binding selectivity of Hox proteins to target sites, but their precise roles during neuronal subtype specification are poorly defined. In the hindbrain, mutation of Pbx genes disrupts expression of extrinsic signaling factors, leading to non-cell autonomous defects in neuronal specification and connectivity (Cooper et al., 2003; Vitobello et al., 2011). By eliminating Pbx genes selectively from MNs, we found that Pbx proteins are cell autonomously required for the differentiation of limb and thoracic-specific MN subtypes. In contrast to the role of the Hox accessory factor Foxp1, which is necessary for subtype differentiation of LMC and PGC neurons (Dasen et al., 2008; Rousso et al., 2008), loss of Pbx genes affects all ventrally-projecting MN subtypes. These results indicate that Pbx genes are essential for the differentiation of the majority of Hox-dependent subtypes.
While Pbx genes are necessary for the differentiation of MNs, not all Hox activities are lost in their absence. In spinal MNs Hox cross-repressive interactions define the position of columns and pools along the rostrocaudal axis, as exemplified by the phenotype of Hoxc9 mutants, where brachial-level Hox genes are derepressed and thoracic MNs are transformed to an LMC fate (Jung et al., 2010). We find that Pbx genes are dispensable for this repressive activity, as in their absence Hox boundaries are preserved. Moreover, expression of the majority of Hox genes is not affected by loss of Pbx genes, suggesting positive-autoregulatory interactions are not critical in most spinal MNs. Thus while the ability of Hox genes to promote neuronal diversity relies on Pbx activity, early Hox patterns appear to be established in a Pbx-independent manner.
Hox-Independent Roles of Pbx Genes in MN Columnar Organization
In tetrapods MNs projecting to functionally-related peripheral targets cluster into columnar and pool groups, establishing a central somatotopic map of peripheral innervation (Kania, 2014; Levine et al., 2012). The topographical organization of MNs can be revealed at a molecular level by expression of certain classes of transcription factors including Hox, Lim HD, and Foxp1 proteins (Dasen et al., 2008; Tsuchida et al., 1994). While Lim HD proteins define many MN subtypes, their specific role in establishing columnar organization is unclear. In mice lacking Lhx3/4 and Isl1/2 Lim HD proteins, MNs lose basic features of their identity or are transformed to an interneuron fate (Sharma et al., 1998; Thaler et al., 2004), confounding any potential role in MN clustering. In the absence of Hox genes or Foxp1, MNs still retain core features of their identity, and the remaining columnar subtypes are well clustered. In contrast, Pbx genes appear to have a relatively specific role in segregating MMC from non-MMC populations. In the absence of Pbx genes, MNs still retain general features of their identity but the remaining MMC and HMC populations are intermixed. These observations indicate that pathways acting within MMC and non-MMC populations ensure MN coalescence and appropriate settling position (Figure 8A).
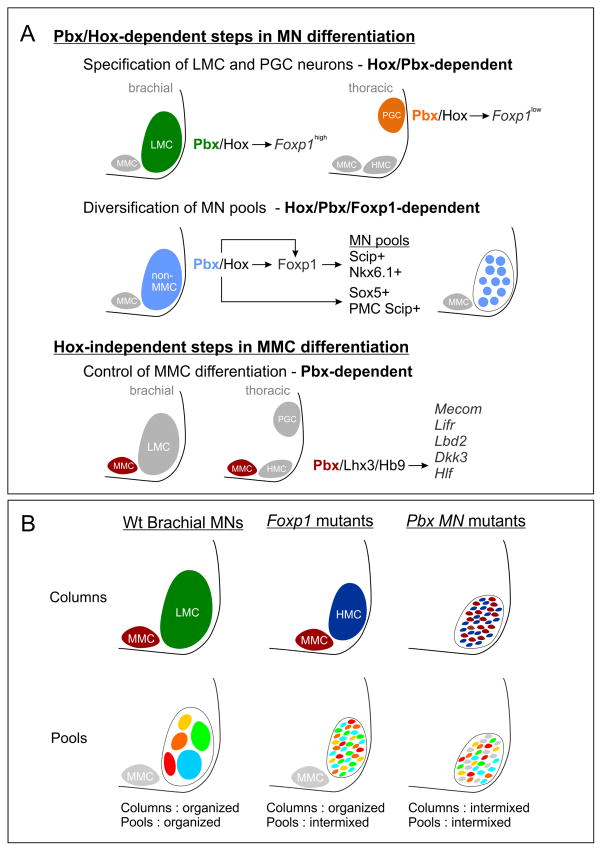
Figure 8. Pbx Genes Function in Parallel MN Differentiation Pathways.
(A) Pbx genes are essential for Hox-dependent and -independent steps in MN differentiation. Pbx genes are required for two critical Hox-dependent steps: the specification of LMC and PGC columnar subtypes, and the diversification of LMC motor pools. Pbx genes are also necessary for the maturation of MMC neurons and govern expression of a subset of MMC-restricted genes, including Mecom. (B) Summary of defects in MN organization in Foxp1 and Pbx mutants at brachial levels. MN organization in wildtype (Wt) mice at brachial levels is shown. In Foxp1 mutants LMC neurons revert to an HMC-like identity but are clustered and segregated from MMC neurons. However, the position of MN pools projecting to individual muscles is disordered. In Pbx mutants the remaining HMC and MMC neurons are intermixed, leading to severe defects in MN clustering.
In principle the segregation of MMC and non-MMC populations could be governed by specific molecular programs acting within these groups, or a consequence of the migratory paths of MNs as a function of their relative birth order. Our analysis of Pbx gene targets suggest the absence of columnar organization is due to the combined loss of molecular signatures of MMC and non-MMC neurons. We found that Pbx genes are essential to regulate a set of MMC-restricted genes, including the transcription factor Mecom. Mecom is selectively lost in Pbx mutants but retained in the absence of Foxp1, suggesting Mecom is regulated independently of Hox protein activity. Consistent with this idea misexpression of determinants of MMC fates such as Lhx3 can induce expression of Mecom at all rostrocaudal levels. Lhx3 is known to suppress Hox-dependent programs in MNs (Dasen et al., 2008), providing further evidence that Mecom induction does not rely on specific Hox proteins. In the cortex, a CNS region that lacks Hox gene expression, Pbx1 has been recently shown to bind within regions of the Mecom locus, suggesting direct regulation of Mecom by Pbx proteins (Golonzhka et al., 2015). These observations are in agreement with studies in Drosophila and mice, showing Pbx proteins have essential functions independent of their roles as Hox cofactors (Merabet and Mann, 2016). Our studies indicate that Pbx proteins can act within a single neuronal class to facilitate both Hox-dependent and Hox-independent programs of neuronal organization and connectivity.
MN Clustering and Topographic Organization Within the Motor System
While multiple classes of transcription factors contribute to the formation of MN topographical maps, the developmental mechanisms through which columnar organization is achieved are not well understood. Reelin and its receptor Disabled play essential roles in the migration and final positioning of LMC and PGC subtypes, and are downstream targets of Foxp1 and Lhx1 (Palmesino et al., 2010). In mice lacking Reelin or Disabled neurons occupy inappropriate positions within the spinal cord but are otherwise well clustered (Kania, 2014). The transcription factor Pea3 is necessary for the organization of MNs targeting the cutaneous maximus (CM) muscle, and in Pea3 mutants CM neurons are interspersed with MN pools occupying the same segment. Targets of Pea3 include cadherin8, and type II cadherins have been implicated in the clustering of MN pools (Demireva et al., 2011; Livet et al., 2002). However, the early genetic pathways that ensure the clustering and segregation of MMC and non-MMC neurons are not known.
Our results indicate that Pbx genes operate in parallel pathways to govern MMC and non-MMC differentiation, and that these two programs coordinate the coalescence and organization of motor columns. In non-MMC neurons, including limb-innervating LMC populations, loss of Pbx genes prevents the differentiation of Hox-dependent divisional and motor pool subtypes. These phenotypes are highly reminiscent of mutation in the Hox accessory factor Foxp1, where LMC neurons are transformed to an HMC fate, and the position of LMC pools is scrambled, likely as a consequence of altered cadherin expression (Figure 8B). Nevertheless the segregation of MMC and HMC neurons persists in Foxp1 mutants, due to the preservation of organizational systems acting within MMC neurons.
In Pbx mutants there is a selective depletion in a subset of MMC-restricted genes, and loss of these factors likely contributes to their disorganization. Importantly, loss of the Pbx-dependent program does not affect the ability of the remaining MMC and HMC neurons to select appropriate muscle targets, suggesting a unique function of Pbx targets in governing MN columnar organization. It is unlikely that this MMC-specific program governs MN coalescence alone, but rather acts in concert with the Pbx/Hox-dependent network. Consistent with this idea, a preliminary analysis of existing Mecom mutants indicated a grossly normal segregation of MMC and non-MMC neurons (Hanley, unpublished observations), likely due to the preservation of Hox-dependent clustering programs. The disordering of MMC and non-MMC neurons therefore appears to be due to the loss of both Pbx-dependent programs, a condition that is achieved through removal of Pbx genes from all MN subtypes.
Columnar Organization and the Evolution of Motor Circuits
What is the purpose of organizing MNs into columns? The segregation of MNs into columnar groups appears to be a unique organizational feature of vertebrates, and is conserved in all tetrapod classes that have been examined including birds, reptiles and mammals (Jung and Dasen, 2015). A basic step in establishing MN topography involves the separation of dorsally-projecting MMC neurons from ventrally projecting subtypes. In contrast, MNs targeting dorsal and ventral axial muscle compartments in zebrafish are largely intermixed with each other (Ampatzis et al., 2013; Menelaou and McLean, 2012). Nevertheless, axial MNs of zebrafish appear to be functionally organized along the dorsoventral axis, where specific “pools” of MMC-like neurons are recruited at distinct locomotor speeds (Ampatzis et al., 2014; McLean et al., 2007). This organizational feature may have evolved to coordinate the activation of axial MNs that drive specific types of undulatory locomotor behaviors, such as slow swimming or predator escape responses. In contrast, in tetrapods MMC neurons are typically associated with postural stabilization, and locomotion is driven predominantly by LMC neurons. Although the origin of the Pbx-dependent MMC program in tetrapods is unclear, it may have appeared during the transition of vertebrates to terrestrial habitats, or was selectively lost in lineages adapted to undulatory forms of locomotion.
The organization of MNs into columnar groups could impact the assembly and function of motor networks by restricting the neuronal populations that a MN has access to. It has been demonstrated that LMC and MMC neurons engage distinct populations of spinal premotor interneurons (Goetz et al., 2015). LMC neurons receive a preponderance of inputs from ipsilaterally located inhibitory interneurons, while MMC neurons connect with premotor populations that are evenly distributed across both sides of the spinal cord. The medial location of MMC neurons could enable access to the contralateral side of the spinal cord, allowing the MMC to capture a greater proportion of inputs from commissural interneurons. Similarly, the inputs that MNs receive from proprioceptive sensory neurons appears to be shaped by the relative position of motor pools within the LMC (Surmeli et al., 2011). The Pbx-dependent pathways described here may have evolved as a means to separate MMC premotor circuits required for postural stabilization from the LMC-directed networks that govern locomotion.
Experimental Procedures
Mouse Genetics
Pbx1 flox (Koss et al., 2012), Pbx3−/− (Rhee et al., 2004b), and Pbx3 flox (Rottkamp et al., 2008), have been described previously. Pbx3 flox and Hb9::eGFP mice were obtained from Jackson Laboratories. Animal procedures were performed in accordance with the US National Institutes of Health Animal Protection Guidelines and approved by the Institutional Animal Care and Use Committee of the New York University School of Medicine.
Wholemount, Immunohistochemistry and In Situ Hybridization
Immunohistochemistry was performed on 16 μm cryostat sections as described (Dasen et al., 2005). Primary antibodies were generated as described (Dasen et al., 2008; Dasen et al., 2005; Tsuchida et al., 1994). Additional antibodies are described in Supplemental Experimental Procedures. Wholemount antibody staining was performed as described (Dasen et al., 2008) and GFP-labeled motor axons were visualized in projections of confocal Z-stacks (400–600 μm). Dissections and wholemounts of diaphragm muscles from E14–18.5 mice were stained as described (Philippidou et al., 2012). Unless indicated otherwise, immunohistological data shown in figures are representative of n>3 mutants analyzed and are taken from animals which are Pbx1MNΔ; Pbx3−/−. Images for control animals are from age matched littermates that are Cre−, and either Pbx3+/+, or Pbx3+/−. Further information on histological analyses are described in Supplemental Experimental Procedures.
In Ovo Chick Embryo Electroporation
Chick neural tube electroporations were performed at Hamburger and Hamilton (HH) st12–14 and analyzed at st27–28 as previously described (Dasen et al., 2003). Plasmid concentrations ranged from 100–500 ng/μl and pBKS was used as carrier DNA to achieve a final concentration of 1 μg/μl. Results for each experiment are representative of five or more embryos in which the electroporation efficiency in MNs was >50%. The Hoxc9IM-pCAGGS construct was generated by mutation in the conserved Pbx interaction domain (ANWI→AAAI), and Hoxc6IM-pCAGGS has been described previously (Lacombe et al., 2013).
RNAseq and Computational Analysis
Details on acquisition of RNAseq data are described in Supplemental Experimental Procedures. The alignment program, Bowtie (version 1.0.0) was used with reads mapped to the Ensemble NCBIM37/mm9 (iGenome version) with two mismatches allowed. The uniquely-mapped reads were subjected to subsequent necessary processing, including removal of PCR duplicates, before transcripts were counted with htseq-count. Counts files were imported into the R statistical programming environment and analyzed with the DESeq2 R/Bioconductor package (Love et al., 2014). Analyses were done on the NYULMC high performance computing cluster. Reproducible pipeline scripts are available: https://github.com/dasenlab/Pbx-Neuron-Paper.
Supplementary Material
Acknowledgments
We thank Myungin Baek, Gord Fishell, Holger Knaut, Hyung Don Ryoo, Brett Spurrier, and Hynek Wichterle. We thank NYULMC’s Office of Collaborative Science Cytometry core for FAC sorting, the Rodent Genetic Engineering Core for mouse rederivation, and the Genomics Technology Core (GTC) for RNA-seq library preparation, sequencing, and bioinformatics. The GTC is partially supported by the Cancer Center Support Grant, P30CA016087, at the Laura and Isaac Perlmutter Cancer Center. The Bioinformatics core is supported in part by grant UL1 TR00038 from the NIH National Center for Advancing Translational Sciences (NCATS). L.S. is supported by NIDCR R01 DE024745, J.S.D by NINDS R01 NS062822 and R01 NS097550.
Footnotes
ACCESSION NUMBERS
The GEO accession number for the RNAseq data reported in this paper is GSE84271.
Author Contributions
O.H. and J.S.D conceived the project, designed the experiments, and wrote the paper; R.Z. and L.S. generated Pbx mutants, shared them prior to publication, and helped us recover lost lines after superstorm Sandy; L.J.C. and O.H. analyzed RNAseq data; O.H., H.J., J.L., P.P., and D.H.L. performed experiments. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ampatzis K, Song J, Ausborn J, El Manira A. Pattern of innervation and recruitment of different classes of motoneurons in adult zebrafish. J Neurosci. 2013;33:10875–10886. doi: 10.1523/JNEUROSCI.0896-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampatzis K, Song J, Ausborn J, El Manira A. Separate microcircuit modules of distinct v2a interneurons and motoneurons control the speed of locomotion. Neuron. 2014;83:934–943. doi: 10.1016/j.neuron.2014.07.018. [DOI] [PubMed] [Google Scholar]
- Baek M, Mann RS. Lineage and birth date specify motor neuron targeting and dendritic architecture in adult Drosophila. J Neurosci. 2009;29:6904–6916. doi: 10.1523/JNEUROSCI.1585-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catela C, Shin MM, Lee DH, Liu JP, Dasen JS. Hox Proteins Coordinate Motor Neuron Differentiation and Connectivity Programs through Ret/Gfralpha Genes. Cell Rep. 2016;14:1901–1915. doi: 10.1016/j.celrep.2016.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KL, Leisenring WM, Moens CB. Autonomous and nonautonomous functions for Hox/Pbx in branchiomotor neuron development. Dev Biol. 2003;253:200–213. doi: 10.1016/s0012-1606(02)00018-0. [DOI] [PubMed] [Google Scholar]
- Dasen JS, De Camilli A, Wang B, Tucker PW, Jessell TM. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell. 2008;134:304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Liu JP, Jessell TM. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425:926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Demireva EY, Shapiro LS, Jessell TM, Zampieri N. Motor neuron position and topographic order imposed by beta- and gamma-catenin activities. Cell. 2011;147:641–652. doi: 10.1016/j.cell.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetcho JR. A review of the organization and evolution of motoneurons innervating the axial musculature of vertebrates. Brain Res. 1987;434:243–280. doi: 10.1016/0165-0173(87)90001-4. [DOI] [PubMed] [Google Scholar]
- Goetz C, Pivetta C, Arber S. Distinct limb and trunk premotor circuits establish laterality in the spinal cord. Neuron. 2015;85:131–144. doi: 10.1016/j.neuron.2014.11.024. [DOI] [PubMed] [Google Scholar]
- Golonzhka O, Nord A, Tang PL, Lindtner S, Ypsilanti AR, Ferretti E, Visel A, Selleri L, Rubenstein JL. Pbx Regulates Patterning of the Cerebral Cortex in Progenitors and Postmitotic Neurons. Neuron. 2015;88:1192–1207. doi: 10.1016/j.neuron.2015.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase G, Dessaud E, Garces A, de Bovis B, Birling M, Filippi P, Schmalbruch H, Arber S, deLapeyriere O. GDNF acts through PEA3 to regulate cell body positioning and muscle innervation of specific motor neuron pools. Neuron. 2002;35:893–905. doi: 10.1016/s0896-6273(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Hinckley CA, Alaynick WA, Gallarda BW, Hayashi M, Hilde KL, Driscoll SP, Dekker JD, Tucker HO, Sharpee TO, Pfaff SL. Spinal Locomotor Circuits Develop Using Hierarchical Rules Based on Motorneuron Position and Identity. Neuron. 2015;87:1008–1021. doi: 10.1016/j.neuron.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Dasen JS. Evolution of patterning systems and circuit elements for locomotion. Dev Cell. 2015;32:408–422. doi: 10.1016/j.devcel.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Lacombe J, Mazzoni EO, Liem KF, Jr, Grinstein J, Mahony S, Mukhopadhyay D, Gifford DK, Young RA, Anderson KV, et al. Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron. 2010;67:781–796. doi: 10.1016/j.neuron.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Mazzoni EO, Soshnikova N, Hanley O, Venkatesh B, Duboule D, Dasen JS. Evolving Hox activity profiles govern diversity in locomotor systems. Dev Cell. 2014;29:171–187. doi: 10.1016/j.devcel.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A. Spinal motor neuron migration and the significance of topographic organization in the nervous system. Adv Exp Med Biol. 2014;800:133–148. doi: 10.1007/978-94-007-7687-6_8. [DOI] [PubMed] [Google Scholar]
- Koss M, Bolze A, Brendolan A, Saggese M, Capellini TD, Bojilova E, Boisson B, Prall OW, Elliott DA, Solloway M, et al. Congenital asplenia in mice and humans with mutations in a Pbx/Nkx2–5/p15 module. Dev Cell. 2012;22:913–926. doi: 10.1016/j.devcel.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe J, Hanley O, Jung H, Philippidou P, Surmeli G, Grinstein J, Dasen JS. Genetic and functional modularity of Hox activities in the specification of limb-innervating motor neurons. PLoS Genet. 2013;9:e1003184. doi: 10.1371/journal.pgen.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lance-Jones C, Landmesser L. Pathway selection by chick lumbosacral motoneurons during normal development. Proc R Soc Lond B Biol Sci. 1981;214:1–18. doi: 10.1098/rspb.1981.0079. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Jeffrey V, Fujioka M, Jaynes JB, Bate M. Embryonic origins of a motor system: motor dendrites form a myotopic map in Drosophila. PLoS Biol. 2003;1:E41. doi: 10.1371/journal.pbio.0000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmesser L. The development of motor projection patterns in the chick hind limb. J Physiol. 1978a;284:391–414. doi: 10.1113/jphysiol.1978.sp012546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmesser L. The distribution of motoneurones supplying chick hind limb muscles. J Physiol. 1978b;284:371–389. doi: 10.1113/jphysiol.1978.sp012545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Lewallen KA, Pfaff SL. Spatial organization of cortical and spinal neurons controlling motor behavior. Curr Opin Neurobiol. 2012;22:812–821. doi: 10.1016/j.conb.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Sigrist M, Stroebel S, De Paola V, Price SR, Henderson CE, Jessell TM, Arber S. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron. 2002;35:877–892. doi: 10.1016/s0896-6273(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Lelli KM, Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean DL, Fan J, Higashijima S, Hale ME, Fetcho JR. A topographic map of recruitment in spinal cord. Nature. 2007;446:71–75. doi: 10.1038/nature05588. [DOI] [PubMed] [Google Scholar]
- Menelaou E, McLean DL. A gradient in endogenous rhythmicity and oscillatory drive matches recruitment order in an axial motor pool. J Neurosci. 2012;32:10925–10939. doi: 10.1523/JNEUROSCI.1809-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabet S, Mann RS. To Be Specific or Not: The Critical Relationship Between Hox And TALE Proteins. Trends Genet. 2016;32:334–347. doi: 10.1016/j.tig.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Palmesino E, Rousso DL, Kao TJ, Klar A, Laufer E, Uemura O, Okamoto H, Novitch BG, Kania A. Foxp1 and lhx1 coordinate motor neuron migration with axon trajectory choice by gating Reelin signalling. PLoS Biol. 2010;8:e1000446. doi: 10.1371/journal.pbio.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P, Dasen JS. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P, Walsh CM, Aubin J, Jeannotte L, Dasen JS. Sustained Hox5 gene activity is required for respiratory motor neuron development. Nat Neurosci. 2012;15:1636–1644. doi: 10.1038/nn.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SR, De Marco Garcia NV, Ranscht B, Jessell TM. Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell. 2002;109:205–216. doi: 10.1016/s0092-8674(02)00695-5. [DOI] [PubMed] [Google Scholar]
- Rhee JW, Arata A, Selleri L, Jacobs Y, Arata S, Onimaru H, Cleary ML. Pbx3 deficiency results in central hypoventilation. Am J Pathol. 2004a;165:1343–1350. doi: 10.1016/S0002-9440(10)63392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee JW, Arata A, Selleri L, Jacobs Y, Arata S, Onimaru H, Cleary ML. Pbx3 Deficiency Results in Central Hypoventilation. The American Journal of Pathology. 2004b;165:1343–1350. doi: 10.1016/S0002-9440(10)63392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanes GJ. The motor cell columns of the lumbo-sacral spinal cord of the cat. J Comp Neurol. 1951;94:313–363. doi: 10.1002/cne.900940209. [DOI] [PubMed] [Google Scholar]
- Rottkamp CA, Lobur KJ, Wladyka CL, Lucky AK, O’Gorman S. Pbx3 is required for normal locomotion and dorsal horn development. Dev Biol. 2008;314:23–39. doi: 10.1016/j.ydbio.2007.10.046. [DOI] [PubMed] [Google Scholar]
- Rousso DL, Gaber ZB, Wellik D, Morrisey EE, Novitch BG. Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron. 2008;59:226–240. doi: 10.1016/j.neuron.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleri L, Depew MJ, Jacobs Y, Chanda SK, Tsang KY, Cheah KS, Rubenstein JL, O’Gorman S, Cleary ML. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- Selleri L, DiMartino J, van Deursen J, Brendolan A, Sanyal M, Boon E, Capellini T, Smith KS, Rhee J, Popperl H, et al. The TALE homeodomain protein Pbx2 is not essential for development and long-term survival. Mol Cell Biol. 2004;24:5324–5331. doi: 10.1128/MCB.24.12.5324-5331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Leonard AE, Lettieri K, Pfaff SL. Genetic and epigenetic mechanisms contribute to motor neuron pathfinding. Nature. 2000;406:515–519. doi: 10.1038/35020078. [DOI] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95:817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- Sockanathan S, Jessell TM. Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell. 1998;94:503–514. doi: 10.1016/s0092-8674(00)81591-3. [DOI] [PubMed] [Google Scholar]
- Surmeli G, Akay T, Ippolito GC, Tucker PW, Jessell TM. Patterns of spinal sensory-motor connectivity prescribed by a dorsoventral positional template. Cell. 2011;147:653–665. doi: 10.1016/j.cell.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JP, Koo SJ, Kania A, Lettieri K, Andrews S, Cox C, Jessell TM, Pfaff SL. A postmitotic role for Isl-class LIM homeodomain proteins in the assignment of visceral spinal motor neuron identity. Neuron. 2004;41:337–350. doi: 10.1016/s0896-6273(04)00011-x. [DOI] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- Thor S, Thomas JB. Motor neuron specification in worms, flies and mice: conserved and ‘lost’ mechanisms. Curr Opin Genet Dev. 2002;12:558–564. doi: 10.1016/s0959-437x(02)00340-4. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Tumpel S, Wiedemann LM, Krumlauf R. Hox genes and segmentation of the vertebrate hindbrain. Curr Top Dev Biol. 2009;88:103–137. doi: 10.1016/S0070-2153(09)88004-6. [DOI] [PubMed] [Google Scholar]
- Vitobello A, Ferretti E, Lampe X, Vilain N, Ducret S, Ori M, Spetz JF, Selleri L, Rijli FM. Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev Cell. 2011;20:469–482. doi: 10.1016/j.devcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Moens CB. Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev Cell. 2002;3:723–733. doi: 10.1016/s1534-5807(02)00319-2. [DOI] [PubMed] [Google Scholar]
- William CM, Tanabe Y, Jessell TM. Regulation of motor neuron subtype identity by repressor activity of Mnx class homeodomain proteins. Development. 2003;130:1523–1536. doi: 10.1242/dev.00358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.