Abstract
Rapid advances in the discovery of long noncoding RNAs (lncRNAs) have identified lineage- and cancer-specific biomarkers that may be relevant in the clinical management of prostate cancer (PCa). Here we assembled and analyzed a large RNA-seq dataset, from 585 patient samples, including benign prostate tissue and both localized and metastatic PCa to discover and validate differentially expressed genes associated with disease aggressiveness. We performed Sample Set Enrichment Analysis (SSEA) and identified genes associated with low versus high Gleason score in the RNA-seq database. Comparing Gleason 6 versus 9+ PCa samples, we identified 99 differentially expressed genes with variable association to Gleason grade as well as robust expression in prostate cancer. The top-ranked novel lncRNA PCAT14, exhibits both cancer and lineage specificity. On multivariate analysis, low PCAT14 expression independently predicts for BPFS (P = .00126), PSS (P = .0385), and MFS (P = .000609), with trends for OS as well (P = .056). An RNA in-situ hybridization (ISH) assay for PCAT14 distinguished benign vs malignant cases, as well as high vs low Gleason disease. PCAT14 is transcriptionally regulated by AR, and endogenous PCAT14 overexpression suppresses cell invasion. Thus, Using RNA-sequencing data we identify PCAT14, a novel prostate cancer and lineage-specific lncRNA. PCAT14 is highly expressed in low grade disease and loss of PCAT14 predicts for disease aggressiveness and recurrence.
Introduction
Early detection of prostate cancer, largely facilitated by the advent of PSA screening, has also been attributed to over-diagnosis and overtreatment of this disease [1], [2], [3]. While coupling PSA screening with other biomarkers such as the long non-coding RNA (lncRNA) transcript PCA3 or gene fusions events (such as TMPRSS2-ERG) have increased specificity of cancer diagnosis, these biomarkers have limited utility in stratifying patients in terms of prognosis [4], [5]. While stratifying patients into risk groups based on clinicopathologic features is currently used to guide treatment decisions [6], it is clear that current stratification approaches need to be further refined to allow better personalization of therapy. Thus, identifying molecular biomarkers to distinguish indolent versus aggressive disease would address an unmet need in the clinical management of prostate cancer.
Advances in next-generation sequencing technologies have enabled thorough characterization of cancer transcriptomes, especially in unraveling the realm of non-coding RNAs (ncRNAs) [7], [8]. In particular, lncRNAs, a class of ncRNAs, have gained increasing attention as biomarkers due to their tissue- and cancer-specific expression profile [9]. In this study, we assembled and analyzed a large RNA-seq compendium compiled from recent publications from consortiums such as The Cancer Genome Atlas (TCGA), the Prostate Cancer Foundation/Stand Up to Cancer international team, and others to identify differentially expressed genes (both protein coding and non-coding genes), that are associated with indolent versus aggressive disease [10], [11]. Our results identify PCAT14, a prostate cancer- and lineage-specific lncRNA, as a top differentially expressed gene in this context. We characterize PCAT14 preclinically and demonstrate that it correlates inversely in expression with disease aggressiveness and adds to conventional clinicopathologic risk factors in predicting prognosis in prostate cancer patients. Finally, we develop a novel in-situ hybiridation (ISH)-based approach for detecting PCAT14 in clinical samples.
Material and Methods
RNA-Seq Data Set
Prostate RNA-seq cohort (n = 585) containing 52 benign prostate tissues, 501 primary prostate cancers, and 132 metastatic prostate cancers was used in this study. For nomination of Gleason associated genes, we compared low Gleason tumors (Gleason 6, n = 45) to high Gleason tumors (Gleason 9+, n = 140).
RNA-seq Data Processing
TCGA prostate Fastq files were obtained from the CGhub. Reads were aligned using STAR version 2.4.2 [12] and read abundance was calculated using FeatureCounts version 1.4.6 [13].
RNA-Seq Differential Expression Testing
Differential expression testing was performed using the Sample Set Enrichment Analysis (SSEA) tool described previously [7]. Briefly, following count data normalization, SSEA performs the weighted KS-test procedure described in GSEA [14]. The resulting enrichment score (ES) statistic describes the enrichment of the sample set among all samples being tested. To test for significance, SSEA enrichment tests are performed following random shuffling of the sample labels. These shuffled enrichment tests are used to derive a set of null enrichment scores (1000 null enrichment scores computed). The nominal p value reported is the relative rank of the observed enrichment score within the null enrichment scores. Multiple hypothesis testing is performed by comparing the enrichment score of the test to the null normalized enrichment score (NES) distributions for all transcripts in a sample set. This null NES distribution is used to compute FDR q values in the same manner used by GSEA [14]. SSEA percentile score determined by ranking the genes in each analysis by their NES score.
Tissue Expression Heatmap Generation
The “gplots” R-package was used to generate heatmaps using the heatmap.2 function. Expression was normalized as log2 of the fold-change over the median of the normal samples for each transcript. Unsupervised hierarchical clustering was performed with the hclust function, using Pearson correlation as the clustering distance, using the “ward” agglomeration method.
Identification of Genes Differentially Expressed in Prostate Cancer of Varying Gleason Score
Differentially expressed Gleason associated genes were identified as any gene with an SSEA FDR< 0.01 when comparing Gleason 6 primary tumors to Gleason 9+ primary tumors. Filtering for expression levels in tissues was done by enforcing that each gene had >5FPKM expression in the top 5% of prostate tumor samples. Filtering for overexpression in cancers versus normal was done by enforcing an SSEA FDR of <0.0001 in an analysis comparing the TCGA prostate cancer vs normal tissues. Tissue specificity percentile was determined as the SSEA percentile for each gene in an SSEA analysis comparing the TCGA prostate samples to all other TCGA tumors in our multi-tissue compendium [7].
Clinical Analysis
To assess the prognostic value of PCAT14, microarray data was obtained from the Johns Hopkins University (JHU) (N = 355). Patients were treated with prostatectomy and subsequently received no adjuvant or salvage treatment until metastasis. Microarray processing and normalization was performed as described previously [15]. PCAT14 expression was calculated by taking the mean expression of probe sets mapping to exons. High/low PCAT14 was determined by splitting on the median expression level. Kaplan–Meier curves are shown and statistical inference was performed using the Log-rank test. Multivariate analysis was performed using Cox regression. Age was treated as a continuous variable. PSA was grouped into low (<10 ng/ml), intermediate (10–20 ng/ml), and high (>20 ng/ml). Surgical margin status (SMS), seminal vesicle invasion (SVI), extracapsular extension (ECE), and lymph node invasion (LNI) were treated as binary variables. Gleason score was grouped into low (≤7) or high (8–10). Association of PCAT14 and clinicopathologic variables was evaluated using a t-test for continuous variables, and a chi-squared test for categorical variables. Statistical significance was set as a two-sided p-value <0.05. All analyses were performed in R 3.1.2.
ISH Analysis
PCAT14 ISH was performed on thin (approximately 4 μm thick) TMA sections (Advanced Cell Diagnostics, Inc., Hayward, CA), as described previously [16], [17]; in parallel, PCAT14 ISH was performed on previously identified positive and negative control index formalin-fixed paraffin embedded (FFPE) tissue sections. All slides were examined for PCAT14 ISH signals in morphologically intact cells and scored manually by a study pathologist (Rohit Mehra). Specific PCAT14 ISH signal was identified as brown, punctate dots, and expression level was scored as follows: 0 = no staining or less than 1 dot per 10 cells, 1 = 1 to 3 dots per cell, 2 = 4 to 9 dots per cell (few or no dot clusters), 3 = 10 to 14 dots per cell (less than 10% in dot clusters), and 4 = greater than 15 dots per cell (more than 10% in dot clusters). For each evaluable tissue core, a cumulative ISH product score was calculated as the sum of the individual products of the expression level (0 to 4) and percentage of cells (0 to 100) (i.e., [A% × 0] + [B% × 1] + [C% × 2] + [D% × 3] + [E% × 4]; total range = 0 to 400). For each tissue sample, the ISH product score was averaged across evaluable TMA tissue cores. All quantitative data were shown as mean ± S.D. To obtain significance in the difference between two groups was performed by two-sided t test using Graph Pad Prism 6.02 software.
Cell Lines, Tissues and Reagents
All prostate cell lines used in this study were purchased from the American Type Culture Collection (ATCC), cultured according to their recommendations and were periodically checked for mycoplasma contamination and genotyped to confirm identity. For androgen treatment experiments, VCaP cells were pre-cultured in androgen-free charcoal-stripped medium for 48 hours and treated with 10 nM dihydrotestosterone (DHT) or 10 μM MDV3100 or vehicle (ethanol) for indicated time points before cells were harvested for RNA isolation. For drug treatment experiments, LNCaP cells were treated with the 5–20 μM DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-aza) (catalog: A3656-5MG, Sigma), or DMSO for 5 days. RNA was isolated 24 h after drug treatment and expression was analyzed by qRT-PCR.
Prostate specimens were acquired from the patients who underwent radical prostatectomy and from the Rapid Autopsy Program at the tissue core of University of Michigan as part of the University of Michigan Prostate Cancer Specialized Program Of Research Excellence (S.P.O.R.E.). Informed consents were obtained from each patient.
RNA Isolation and qPCR Analysis
Total RNA was extracted using Trizol reagent and an RNeasy Micro Kit (Qiagen) with DNase I digestion according to the manufacturer's protocols. RT-PCR was performed from total RNA using Superscript III (Invitrogen) with random primers (Invitrogen). Quantitative PCR (qPCR) was performed using Fast SYBR Green Master Mix (Applied Biosystems) on a 7900HT Fast Real-Time PCR system (Applied Biosystems). All oligonucleotide primers were purchased from Integrated DNA Technologies (Coralville, IA) are sequence of each primer is listed in Supplementary Table 4. Primer specificity was determined by sequence verifying the PCR products using the University of Michigan Sequencing Core facility.
Rapid Amplification of cDNA Ends (RACE)
5′ and 3′ RACE was performed using the GeneRacer RLM-RACE kit (Invitrogen) according to the manufacturer's protocols. RACE PCR products obtained using Platinum Taq high-fidelity polymerase (Invitrogen), were resolved on a 1.5% agarose gel. Individual bands were gel purified using a Gel Extraction kit (Qiagen), and cloned into PCR4 TOPO vector, and sequenced using M13 primers.
Knock Down Studies
MDA-PCa-2b and VCaP cells were seeded in biocoated 6-well plates at 60% confluency, incubated overnight, and transfected with 50 nM siRNAs targeting different exons of PCAT14 or non-targeting siRNAs, using RNAi MAX reagent (Invitrogen) per manufacturer's instructions. RNA was harvested 48 h after transfection. Functional experiments were performed at indicated time points. Sequence of all the siRNA used in shown in Supplementary Table 4.
Nuclear-Cytoplasmic Subcellular Fractionation
Nuclear-cytoplasmic fraction of MDA-PCa-2b and VCaP cells was performed using an NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo Scientific) following manufacturer's instructions, followed by RNA isolation and qPCR analysis.
CRISPR Based Overexpression of PCAT14
Stable cell lines overexpressing PCAT14 endogenously were made using previously published protocol [18]. Briefly, guide RNAs targeting promoter region of PCAT14 (Supplementary Table 4) were designed using online tool at http://crispr.mit.edu/ and cloned into sgRNA-MS2 vector using lenti sgRNA(MS2) zeo backbone. Lentiviral particles expressing PCAT14 sgRNA-MS2 were generated by the University of Michigan vector core. To generate LNCaP or PC3 cell over expressing PCAT14, first cells were seeded into 100 mm dish and transduced with Lenti dCAS-VP64 (blasticidin) and Lenti-MS2-p65-HSF1 (hygromycin) vectors. After 2 days, cells were selected with 4 μg/ml Blasticidin and 200 μg/ml Hygromycin. Cells stably expressing dCAS-VP64 and MS2-p65-HSF1 cells were then seeded in 6-well plates and infected with PCAT14 sgRNA-MS2 lentivirus. 24 hours later, cells were selected with triple antibiotics: 4 μg/ml Blasticidin, 200 μg/ml Hygromycin and 800 μg/ml Zeomycin for 1 week. Expression of PCAT14 in these cells was verified by qPCR.
In Vitro FluoroBlok Tumor Invasion Assay
The In vitro FluoroBlok Tumor Invasion Assay (BD) was performed as previously described [19]. Briefly, after rehydration of the BD FluoroBlok membrane, 500 ul of serum-free RPMI medium resuspended prostate cancer cells (PC3, 50,000 cells per well, or LNCaP, 100,000 cells per well) were seeded into the apical chambers. 750 ul RPMI medium containing 10% FBS were added to the basal chamber as chemoattractant. Then plates were incubated at 37 °C, 5% CO2 for 24 hours. Following incubation, medium from the apical chambers were removed, and the inserts were transferred to a 24-well plate containing 500ul/well of 4ug/mL Calcein AM (Invitrogen) in Hanks buffered saline. Plates were incubated for 1 hour at 37 °C, 5% CO2, then pictures of invaded cells were taken by using inverted fluorescence microscope (Olympus), and quantified by ImageJ software [20].
Oncomine Concepts Analysis of the PCAT14 Signature
Gene that positively correlated (R2 > 0.35, n = 591) with PCAT14 in TCGA RNA-seq data were selected and uploaded into Oncomine database [21] as custom concepts (Supplementary Table 2). All the prostate cancer concepts with odds ratio> 2.0 and p-value <1 × 10−4 were selected. For simplicity, top 4 concepts (based on odds ratios) were selected for representation. We exported these results as the nodes and edges of a concept association network and visualized the network using Cytoscape version 3.3.0. Node positions were computed using the Edge-weighted force directed layout in Cytoscape using the odds ratio as the edge weight. Node positions were subtly altered manually to enable better visualization of node labels.
Statistics
All quantitative data were shown as mean ± S.D. To obtain significance in the difference between two groups was performed by two-sided t test or ANOVA using Graph Pad Prism 6.02 software.
Results
Identification of Genes Associated With Gleason Grade in Prostate Cancer
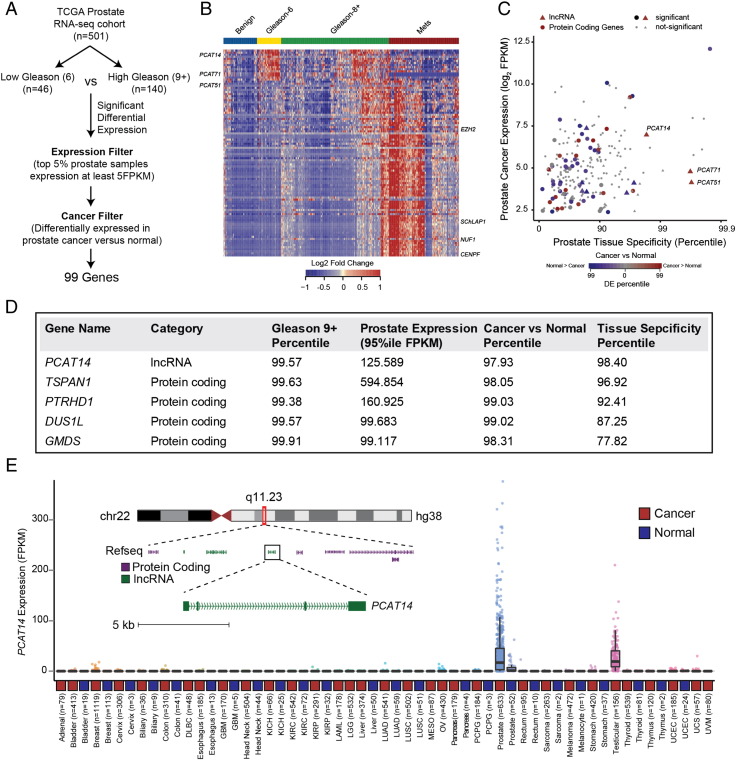
Comprehensive molecular characterization of common cancer types has become feasible with the recent availability of large next generation sequencing datasets on tumor tissues. To identify genes (both coding and non-coding) associated with aggressive prostate cancer, we assembled a large prostate RNA-seq cohort (n = 585) containing 52 benign prostate tissues, 501 primary prostate cancers, and 132 metastatic prostate cancers. We performed differential expression testing utilizing a non-parametric tool we developed for RNA-seq data called Sample Set Enrichment Analysis [7]. In order to nominate the most intriguing biomarkers associated with aggressive disease, we compared low Gleason tumors (Gleason 6, n = 45) to high Gleason tumors (Gleason 9+, n = 140) and applied filters for substantial expression in prostate tumor tissue (>5PKM in the top 5% of prostate samples), and significant differential expression in prostate cancers versus normal (SSEA, FDR <0.0001) leaving a total of 99 candidates genes (Figure 1A, Supplementary Table 1). Interestingly, clustering analysis revealed signature expression patterns, specifically associated with low, high Gleason and metastatic status and included both novel and previously characterized genes (Figure 1B). CENPF and EZH2, protein coding genes with a known association with high grade prostate cancer were rediscovered through this analysis [22], [23]. Similarly, we rediscovered SChLAP1 a long non-coding RNA (lncRNA) associated with aggressive prostate cancers [15], [17] in our analysis (Figure 1B). With the goal of identifying potential biomarkers that distinguish indolent prostate cancers, we focused on genes enriched in low grade disease that are expressed highly in prostate tissue and that also show prostate cancer and tissue specificity (Figure 1C). Interestingly, a lncRNA, PCAT14 appeared to be one of the top low-Gleason-associated genes with robust prostate tissue expression, substantial prostate tissue specificity, and significant overexpression in prostate cancers versus normal (Figure 1D). In fact, among all genes (coding and non-coding), PCAT14 ranked among the top 5 in terms of expression level, Gleason 6 versus 9+ association, and cancer versus normal association (Figure 1D). Additionally, among the top 5 candidate genes, PCAT14 was the only gene to exhibit striking prostate tissue specificity, a particularly relevant metric for a potential biomarker (Figure 1E). The remaining 4 genes exhibited variable prostate tissue specificity (Supplementary Figure 1). PCAT14 is a poly-exonic gene found within a gene desert on chromosome 22, with a striking prostate cancer and lineage specific expression pattern across the >10,000 TCGA cancer and normal tissue samples (Figure 1E). For these reasons, we elected to pursue PCAT14 as a promising biomarker that can identify low grade prostate cancer.
Figure 1.
Identification of lncRNA PCAT14 as a prostate cancer biomarker. A. Schematic representation of the workflow utilized to identify highly-expressed, prostate cancer specific genes associated with low-Gleason disease. B. Heatmap depiction of the lncRNA and protein coding genes differentially expressed (n = 99) between Gleason 6 versus 9+ analysis in TCGA prostate RNA-seq data. Relative expression of these genes in benign and metastatic prostate cancer tissues [11] are also displayed alongside for comparison. Expression is depicted as log2 of the fold-change over the median of the Gleason 6 samples for each gene. Patients grouped by cancer progression/Gleason score. Rows represent genes and columns represent samples. C. Scatterplot showing the expression level, prostate tissue specificity, and prostate cancer association of protein coding (solid circle) and lncRNA (solid triangles) genes identified in 1 A. Expression is represented by the FPKM value for the 95th percentile prostate cancer sample. Cancer versus normal and prostate tissue specificity are represented by the percentile score for each gene in an SSEA analysis. D. The top five, Gleason 6 associated genes listed in the order of prostate tissue specificity. E. Expression of PCAT14 across all cancer and normal tissue type represented in the TCGA. Inset shows genome browser view of PCAT14 genomic location.
Genomic Organization and Regulation of PCAT14
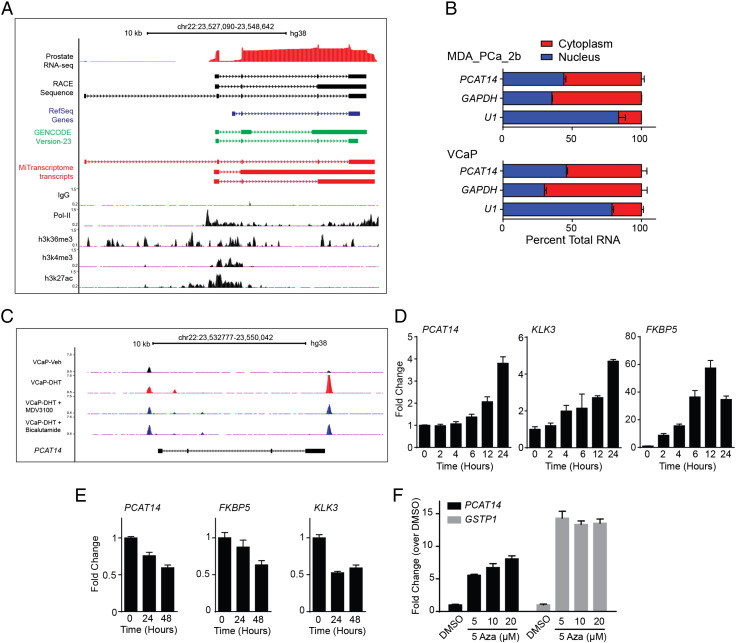
We collected multiple lines of evidence from both experimental data and available annotations to consolidate the genomic organization of PCAT14. Based on assembled reads from RNA-seq data assembled in the MiTranscriptome [7], we predicted the structure of the PCAT14 transcript variants (Supplementary Figure 1A). Additionally, as an independent approach to define the exon structure of PCAT14, we performed rapid amplification of cDNA ends (RACE) in two prostate cancer cell lines VCaP and MDA-PCa-2b that express PCAT14 at high levels (Supplementary Figure 1B and C). Our analyses show that the PCAT14 gene is located on chr22-q11.2 and contains 4 exons. Among the four transcript isoforms, the 2.3 kb variant-1 demonstrates the highest expression (Supplementary Figure 1D). Next, using published ChIP-seq data in VCaP cells [24], we show that PCAT14 has all the histone marks (H3K4me3, H3K36me3, H3K27ac) associated with actively transcribed genes (Figure 2A). We further performed subcellular fractionation followed by qPCR to show that PCAT14 is distributed equally between nuclear and cytoplasmic compartments (Figure 2B).
Figure 2.
Subcellular localization and regulation of PCAT14. A. Genome browser view of PCAT14 locus. ChIP-seq tracks for H3K4me3, H3K27ac, H3K36me3 and Pol-II generated in prostate cancer VCaP cells are shown. Prostate RNA-seq reads, transcript schematic based on RACE results and Refseq, GENCODE, MiTranscriptome assembly annotations are also provided. Solid blocks indicate exons while thin lines intron and arrows indicate the genomic orientation. B. Bar plots represent the subcellular localization of PCAT14 in prostate cell lines. PCAT14 transcript was equally found in both cytoplasmic (red) and nuclear (blue) compartments in both MDA-PCa-2b and VCaP cell lines. GAPDH and U1 RNA were used as controls. C. Genome browser view of the PCAT14 genomic locus for AR ChIP-seq data tracks obtained from VCaP cells treated with either vehicle (black) or dihydrotestosterone (DHT) alone (Red) or combinations (dark blue) including DHT + MDV3100 and DHT + Bicalutamide. Significant AR binding observed in each data track are represented as peaks. D-E. Histograms represent the expression of PCAT14, TMPRSS2 and KLK3 in VCaP cells after treatment with 10 nM DHT or with MDV3100 for indicated time points. F. Bar plots represent re-expression of PCAT14 and GSTP1 in LNCaP cells after treatment with 5-Aza deoxycytidine (5-Aza) for 5 days at indicated concentrations.
Androgen receptor plays a major role To identify any potential regulation of PCAT14 gene by androgen, we assessed the presence of AR peaks in PCAT14 genomic region using AR-ChIP-seq data generated in VCaP cells [24] and saw significant AR peaks in PCAT14 loci. Some of these peaks were also enhanced upon treatment with DHT and were suppressed upon treatment with AR antagonist MDV3100 or bicalutamide (Figure 2C). To corroborate this finding, we assessed the expression of PCAT14 mRNA in VCaP cells upon AR stimulation. Similar to the canonical AR targets such as KLK3 and FKBP5, PCAT14 expression was also significantly elevated (four fold in 24 hours) upon DHT stimulation (Figure 2D) and suppressed by MDV3100 treatment (Figure 2E). In another line of investigation, we queried if epigenetic regulation might play a role in the prostate cancer and lineage specific expression of PCAT14 observed in tissue samples (Figure 1E). Using a prostate cancer cell line (LNCaP) model we show significant elevation of PCAT14 expression when treated with 5-azacytidine (5-Aza), a DNA demethylation agent, suggesting a potential role for promoter methylation in regulation of PCAT14 (Figure 2F). However, our attempt to capture this event in TCGA tissue samples where Infinium 450 K DNA methylation array data is available was inconclusive, due to the lack of probes in PCAT14 promoter region. Taken together we show PCAT14 is an AR target gene that may also be subjected to epigenetic regulation in prostate cancer.
Clinical Association of PCAT14
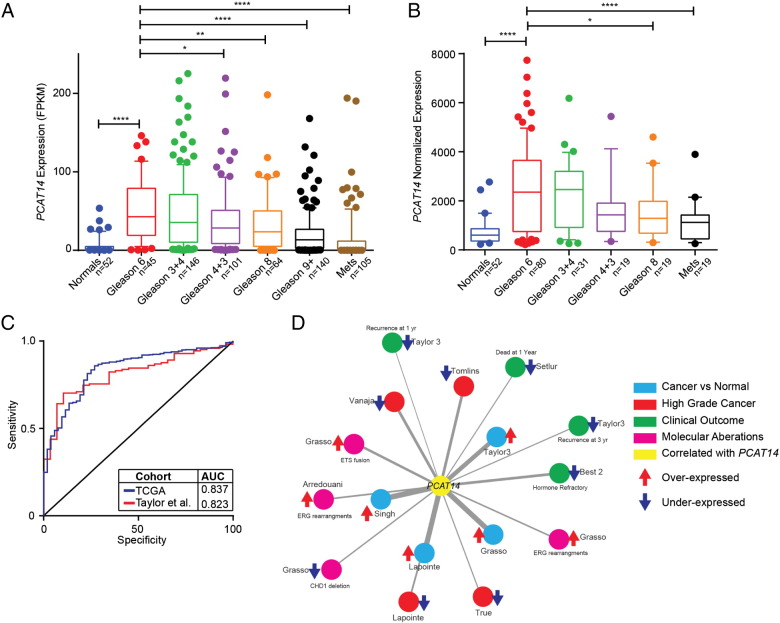
Having observed an inverse correlation of PCAT14 with Gleason Score (GS) in our RNA-seq cohort, we next assessed the association of PCAT14 expression with clinical outcomes in prostate cancer. For this analysis we first divided samples into 7 groups (benign, GS-6, GS-7 (3 + 4), GS7 (4 + 3), GS-8, GS-9 and Mets) and examined the expression of PCAT14 using two different datasets (TCGA and Taylor et al.). We identified a significant decrease in PCAT14 expression as Gleason grade increased in both cohorts (Figure 3A and B). Importantly, in the large TCGA dataset, expression was significantly different between GS6 and all other groups except GS7 (3 + 4). We next assessed the diagnostic ability of PCAT14 to identify prostate cancers versus normal. In both the TCGA and Taylor prostate cancer cohorts, PCAT14 expression was able to significantly distinguish cancer from normal with an AUC of 0.837 and 0.823 respectively (Figure 3C) supporting its utility as a diagnostic biomarker.
Figure 3.
PCAT14 is marker of low grade tumros. A-B. Expression of PCAT14 in samples distinguished by Gleason grade in TCGA (A), Taylor (B) cohorts. (* = P < .05, ** = P < .01, **** = P < .0001; compared to Gleason 6). C. ROC analysis of PCAT14 expression in the TCGA and Taylor cohorts. D. Network representation of genes positively correlated with PCAT14 in localized prostate cancers using Oncomine concepts analysis and visualized with the Force-Directed Layout algorithm in the Cytoscape tool [29]. Node names are assigned according to the author of the primary study [25], [30], [31], [32], [33], [34], [35], [36], [37], [38]. Nodes are colored according to the concept categories indicated in the figure legend. Thickness of the edges implies higher odds ratio.
Using an alternate approach to further characterize the clinical associations of PCAT14, we performed a “guilt-by-association” analysis, assessing the clinical significance of the protein-coding genes most correlated with PCAT14 (Supplementary Table 2) in the TCGA prostate cancer cohort, leveraging cancer microarray data from the Oncomine resource [21]. As expected, genes positively correlated with PCAT14 were upregulated in cancer vs normal analysis and were downregulated in clinically advanced prostate cancer (Figure 3D). Interestingly, we found a striking association of PCAT14 correlated genes with concepts related to better prognosis (Figure 3D), and these genes were under-expressed in recurrent and hormone refractory prostate cancer suggesting that PCAT14 may be a marker of better clinical outcomes in prostate cancer. In contrast, genes that positively correlated with SChLAP1, a lncRNA known to be associated with clinically aggressive prostate cancer, were found to be overexpressed in advanced prostate cancer as well as in cancer with poor outcomes [15], [17].
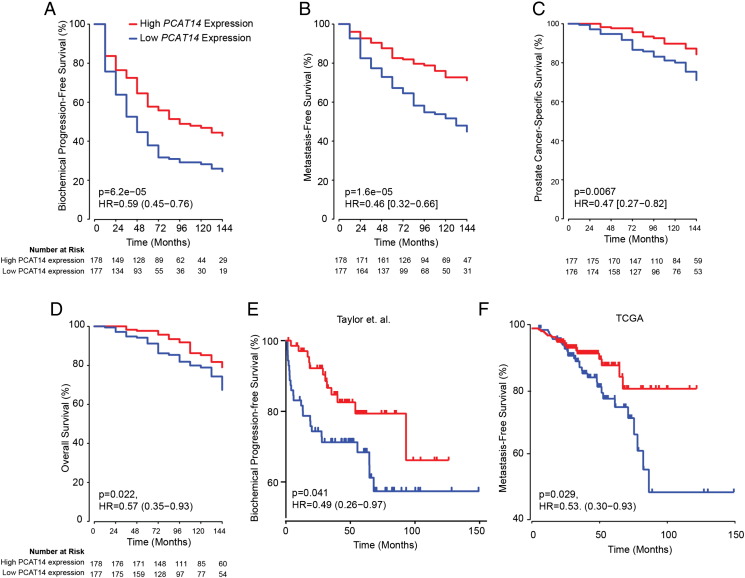
To further investigate the association of PCAT14 with favorable clinical outcomes in prostate cancer, we performed Cox regression analysis on a cohort of 355 patients (John Hopkins University (JHU) cohort) who did not receive treatment prior to metastasis (median follow-up 9 years). Univariate analysis showed that, patients with high PCAT14 expression were significantly associated with better BPFS (P = .000062; HR = 0.59 [0.45–0.76]), MFS (P = .00016; HR = 0.46 [0.32–0.66]), PSS (P = .0067; HR = 0.47[0.27–0.82]) and OS (P = .022; HR = 0.57 [0.35–0.93]) (Figure 4A-D). In a Cox multivariate analysis including clinicopathologic variables, PCAT14 stands out as a significant independent predictor of PSS (P = .0385; HR = 0.55 [0.31–0.97]), MFS (P = .000609; HR = 0.52[0.36–0.76]) and BRFS (P = .00126, HR = 0.64 [0.49–0.84]), with borderline significance for OS (Table 1, Supplementary Table 3). In addition, we also analyzed the association of PCAT14 expression with clinical outcome in two independent data sets of 140 (Taylor et al) and 377 (TCGA) patients using the statistical approaches mentioned above [25]. Similar to JHU cohort, high PCAT14 expression predicted for better BRFS (Figure 4E) and MFS (Figure 4F). We also show that high PCAT14 expression was predictor of better prognosis in lower Gleason grade samples (Supplementary Figure 3B).
Figure 4.
PCAT14 is a prognostic biomarker. A-D. Kaplan–Meier analyses of prostate cancer outcomes in the John Hopkins cohort. PCAT14 expression was measured using Affymetrix exon arrays, and subjects were stratified according to their PCAT14 expression level. Subject outcomes were analyzed for biochemical progression (D) and Metastasis free survival (E), Prostate cancer-specific survival (F) and overall survival (G). Subject outcomes were analyzed for Kaplan–Meier curves, P values determined using a log-rank test. E-F. Kaplan–Meier analyses of biochemical progression free survival in the Taylor (E) and Metastasis Free survival in the TCGA (F) cohorts of prostate cancer. Patients were divided into two groups based on the expression level of PCAT14. P values for Kaplan–Meier curves were determined using a log-rank test.
Table 1.
Multivariate Analysis in JHU Cohort
| Biochemical Recurrence Free Survival |
Metastasis Free Survival |
Prostate Cancer Free Survival |
Overall Survival |
|||||
|---|---|---|---|---|---|---|---|---|
| P-Value | HR [95% CI] | P-Value | HR [95% CI] | P-Value | HR [95% CI] | P-Value | HR [95% CI] | |
| PCAT14 High vs. Low | .00126 | 0.64 [0.49–0.84] | .000609 | 0.52 [0.36–0.76] | .0385 | 0.55 [0.31–0.97] | .0567 | 0.62 [0.38–1.01] |
| Age | .818 | 1 [0.98–1.02] | .65 | 0.99 [0.96–1.02] | .338 | 0.98 [0.93–1.02] | .151 | 0.97 [0.93–1.01] |
| PSA Int vs. Low | .241 | 0.83 [0.62–1.13] | .353 | 0.83 [0.55–1.24] | .385 | 0.75 [0.4–1.42] | .366 | 0.77 [0.44–1.35] |
| PSA High vs. Low | .916 | 0.98 [0.63–1.52] | .574 | 0.84 [0.47–1.52] | .463 | 0.73 [0.31–1.7] | .582 | 0.81 [0.39–1.7] |
| Gleason High vs. Low | 2.98E-05 | 1.83 [1.38–2.43] | 1.00E-08 | 3.08 [2.1–4.52] | .000224 | 3.1 [1.7–5.65] | .000988 | 2.38 [1.42–3.99] |
| Seminal vesicle invasion | .0042 | 1.52 [1.14–2.03] | .453 | 1.16 [0.79–1.69] | .774 | 0.92 [0.51–1.66] | .82 | 0.94 [0.56–1.59] |
| Surgical margin status | .000533 | 1.78 [1.28–2.47] | .000276 | 2.15 [1.42–3.25] | .0487 | 1.93 [1–3.7] | .0825 | 1.67 [0.94–2.99] |
| Extracapsular extension | .456 | 1.14 [0.81–1.58] | .459 | 1.21 [0.73–2.03] | .636 | 0.83 [0.39–1.77] | .816 | 0.93 [0.48–1.77] |
| Lymph node invasion | 8.98E-12 | 3.23 [2.31–4.52] | .000164 | 2.21 [1.46–3.35] | .0616 | 1.86 [0.97–3.57] | .254 | 1.42 [0.78–2.6] |
| HR: Hazard Ratio | ||||||||
PCAT14 Expression In-Situ
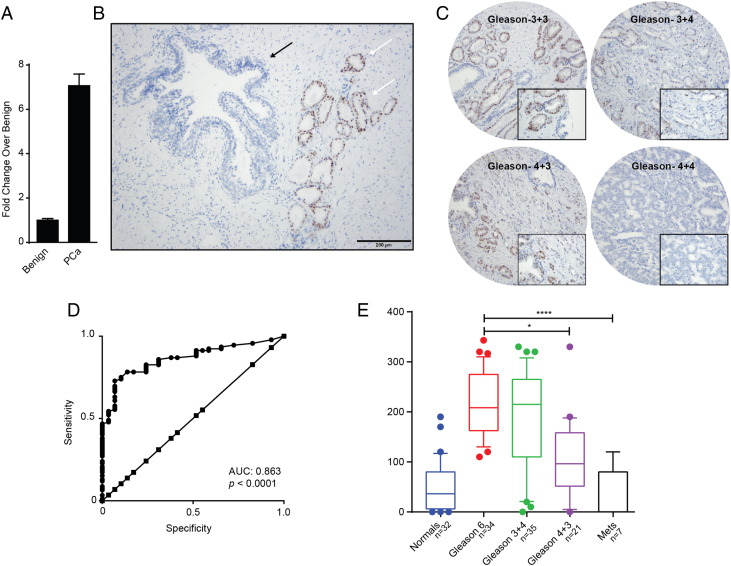
LncRNA detection in cancer tissue sections by RNA in-situ hybridization (RNA-ISH) technology has similar clinical utility as immunohistochemical evaluation of protein biomarkers [16], [26]. Hence we evaluated PCAT14 transcript levels in PCa FFPE tissues using specific probes to perform a RNA-ISH. We first probed a panel of FFPE sections derived from either murine prostate, kidney, lung (negative controls) or xenografts from MDA-PCa-2b cells, a cell line that expresses PCAT14 at high levels (positive control). As expected, high levels of specific signal was present in MDA-PCa-2b xenografts while no expression/staining was seen in the negative control murine tissues (Supplementary Figure 5A and B). Consistent with the cell fractionation data, expression of PCAT14 was seen in both nuclear and cytoplasmic compartments. Next we obtained frozen and matched formalin fixed paraffin embedded (FFPE) tissues sections derived from a patient radical prostatectomy specimen with Gleason score 3 + 3 = 6 disease. q-PCR analysis on cDNA from frozen tissues derived from this specimen shows a 7–8 fold increase in PCAT14 expression in cancer compared to the adjacent benign tissue (Figure 5A). RNA-ISH also demonstrated that PCAT14 is differentially expressed in PCa as we saw striking difference of transcript expression with high signals located in the prostatic adenocarcinoma glands and with no/minimum staining in the benign section (Figure 5B). To further expand these results, we performed RNA-ISH on a PCa tissue microarray (TMA, n = 129) (Figure 5C) and found that PCAT14 expression was able to distinguish tumor from normal (AUC 0.863) (Figure 5D) and was high in Gleason-6 with minimal expression noted in benign tissue or Gleason 8 disease (Figure 5E).
Figure 5.
PCAT14 RNA-ISH in prostate cancer tissues. A. Barplot to show the expression of PCAT14 in tumor tissue and adjacent benign by qRT-PCR. B. A representative PCAT14 RNA in-situ hybridization image. White arrows indicate Gleason score 6 disease and black arrows indicate benign glands. C. Representative PCAT14 In situ hybridization images of human prostate cancer samples of different Gleason grades. D. ROC analysis of PCAT14 expression in the prostate TMAs. E. Representation of mean PCAT14 ISH product score for benign prostatic glands (benign), Gleason score 6, Gleason score 3 + 4 = 7, Gleason score 4 + 3 = 7 and Gleason score 8+ clinically localized prostate cancer in a TMA cohort. (** = P < .01; compared to benign).
Functional Evaluation of PCAT14
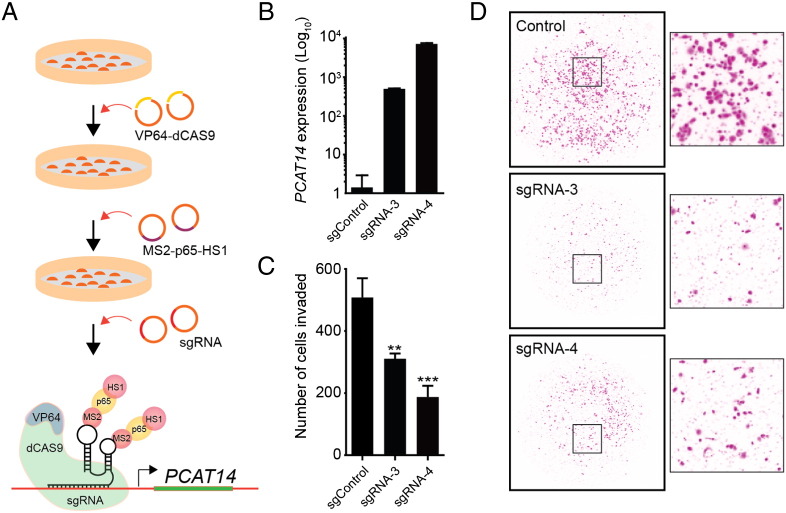
Since expression of PCAT14 was lower in high grade prostate cancer and its expression predicted better outcomes, we hypothesize that PCAT14 may have tumor suppressive effects. To test this hypothesis, we performed overexpression studies in PC3 and LNCaP cells, prostate cancer cell lines that do not express PCAT14 (Supplementary Figure 2B, C). To overexpress PCAT14, we used a CRISPR (clustered regularly interspaced short palindromic repeat)-Cas9 Synergistic Activation Mediator (SAM) complex [18]. This method allows endogenous overexpression of a gene by recruiting artificial transcriptional factors to the promoter using single-guide RNA (sgRNA-MS2) (See method section for details). We designed 6 sgRNAs targeting the PCAT14 promoter and tested their ability to induce PCAT14 expression using HEK293 cells stably expressing transcription factors. We found three sgRNAs that significantly increased PCAT14 expression in HEK293 cells (Supplementary Figure 5A). We next used these sgRNA to construct PC3 and LNCaP cells stable expressing PCAT14 (Figure 6A). Using two independent sgRNAs we were able to achieve 500 to 1000-fold endogenous overexpression of PCAT14 in PC3 cells (Figure 6B) and 20–100 fold overexpression in LNCaP cells (Supplementary Figure 5B). While we observed no significant effect of PCAT14 overexpression on proliferation of PC3 or LNCaP cells (Figure 6C and Supplementary Figure 5C), overexpression of PCAT14 lead to suppression of invasion capacity of both PC3 and LNCaP cells (Figure 6C, D; Supplementary Figure 5E, F), in line with its prior identified association with clinically indolent disease. We then looked at the effects of PCAT14 knockdown on cell expressing PCAT14 at high levels (VCaP and MDA-PCa-2B). In both MDA-PCa-2b and VCaP cells using 2 independent siRNA as well as 8 independent ASOs we were able to achieve more than 80% knockdown efficiency (Supplementary Figure 5G-J). However, we did not observe a consistent effect on cell proliferation as well as cell invasion (Supplementary Figure 5K-N and data not shown).
Figure 6.
Functional analysis is PCAT14.
A. Schematic representation of the workflow to endogenously overexpress PCAT14 in prostate cancer cells using CRISPR/SAM system. B. Bar plots represent fold increase in PCAT14 level in PC3 cells expressing dCas9–VP64 and MS2–p65–HSF1 with control or 2 independent PCAT14 sgRNAs. C. Bar plot represent quantification of invaded PC3 cells with or without PCAT14 expression. D. Representative images of invaded PC3 cells with or without PCAT14 expression.
Discussion
In this study, we perform a large-scale RNA-sequencing-based analysis of biomarkers associated with indolent versus aggressive prostate cancer and identify the long noncoding RNA PCAT14 as a marker of low grade and indolent disease. We define the exon structure of PCAT14 and demonstrate that PCAT14 is an AR-regulated lncRNA. Using two independent data sets, we show that PCAT14 is highly upregulated in prostate cancer compared to benign tissue and is able to distinguish prostate cancer from normal tissue with high sensitivity and specificity, suggesting that PCAT14 can be an excellent diagnostic biomarker. Moreover, we demonstrate that expression of PCAT14 is prognostic of outcome and is associated with better biochemical progression-free survival, metastases-free survival, and prostate cancer-specific survival. Importantly, we find that PCAT14 expression is a prognostic biomarker which adds to standard clinicopathologic variables.
As such, PCAT14 represents a unique biomarker. Most diagnostic biomarkers, such as PCA3, can distinguish cancer from normal tissue, but are not prognostic [4]. Conversely, many prognostic biomarkers, such as Ki-67, hold little diagnostic value. It is unclear why PCAT14 increases significantly in expression during the initial formation of cancer, but then subsequently decreases in expression in disease aggressiveness; this observation requires follow up with further mechanistic studies but is also a feature that gives PCAT14 value as a biomarker across multiple clinical contexts. Of note, PCAT14 was also found to be expressed in testicular cancer samples along with prostate cancer, suggesting the role of PCAT14 in the testicular cancer pathogenesis. However, due to lack of normal testis samples in the TCGA database, it is unclear, at this point, whether PCAT14 is differentially regulated in testicular cancer compared to normal testis. Recently, the Genotype-Tissue Expression (GTEx) program has generated a large amount of high throughput sequencing data on normal tissue including testis [27]. This data would be useful to look at the role of PCAT14 in testicular carcinoma.
In an attempt to develop a clinical grade assay to detect expression of PCAT14, we developed a novel assay, using ISH probes, which can be applied to formalin fixed paraffin-embedded tissues. This ISH assay provides an opportunity to validate our findings in larger cohorts with associated clinical data in the future. Ultimately, an optimized approach for predicting indolent versus aggressive disease will include both clinicopathologic parameters integrated with molecular biomarkers. It is likely that this molecular assay will involve multiplexing multiple biomarkers, and may require combining both tissue-based and urine-based biomarkers. Potential intriguing subsequent studies include the assessment of PCAT14 and other candidate lncRNAs, in addition to PCA3, as urine biomarkers.
There are a several limitations to our study. While we demonstrate the potential value of PCAT14 expression as a biomarker, it is unclear how PCAT14 is modulating oncogenic phenotypes, from a mechanistic perspective. Additionally, while we demonstrate the relative specificity of PCAT14 for both prostate and testicular cancers, the molecular basis underlying this specificity remains to be elucidated. It is known that AR can regulate expression of genes in both prostatic and testicular tissues, but we do not know whether the relative cancer-specificity can be attributed to AR. Clearly, these are important areas for future study.
Overall, our study highlights the need to look at both conventional protein-coding genes and noncoding genes in the search for optimal biomarkers. To our knowledge, there are approximately 20,000 protein coding genes [28], which comprise 2% of the genome. Given our recent study demonstrating that there are close to 60,000 long noncoding RNAs (lncRNAs) [7], many of which are specific to certain cancers, it is clear that these lncRNAs present a relatively underexplored frontier for biomarker development, and that PCAT14 may represent an initial candidate to be further explored along this frontier.
Conclusion
By performing differential expression analysis between prostate cancer with low vs high Gleason scores, we identified lncRNA PCAT14 as a prostate cancer- and lineage- specific biomarker of indolent disease. We show that PCAT14 is an AR-regulated transcript and its overexpression suppresses invasion of prostate cancer cells. Moreover, in multiple independent datasets, PCAT14 expression associates with favorable outcomes in prostate cancer and adds prognostic value to standard clinicopathologic variables.
The following are the supplementary data related to this article.
Supplementary Figure 1. Expression pattern of other Gleason associated genes. Box plots represent the expression of TSPAN1, PTRHD1, DUSL1, and GMDS genes across the TCGA cancer/normal tissue RNA-seq compendia. All genes showed increased expression in prostate cancer compared to normal, but unlike PCAT14, they lacked prostate tissue specificity and differential expression is observed in several other cancer types. Blue: normal; red: Cancer.
Supplementary Figure 2. PCAT14 genomic organization, transcript variants, and expression in prostate cancer cell lines. (A) Schematic representation of genomic organization of various PCAT14 isoforms determined by RACE in MDA-PCa-2b and VCaP cells. Solid blocks indicate exons, while thin lines indicate intron and arrows indicate the genomic orientation. (B) VCaP and MDA-PCa-2b showed highest expression among the five androgen receptor–positive cell lines (VCaP, MDA-PCa-2b, 22RV1, LNCaP, and H660), whereas the 2 AR-negative cells lines (DU145 and PC3) lacked expression, as determined by RNA-seq (B) and qPCR. Inset shows the amplicon size on a 2% agarose gel. (C). D. The deduced intron/exon junction and mRNA sequence of PCAT14 by RACE and Sanger sequencing.
Supplementary Figure 3. Concept enrichment analysis. (A) Expression of PCAT14 in all the samples from the JHU cohort. (B) Kaplan-Meier analyses of MFS in the low-grade Gleason (Gleason 6 and 7) samples from the TCGA cohorts of prostate cancer. P values for Kaplan-Meier curves were determined using a log-rank test.
Supplementary Figure 4. (A–B) In situ hybridization of PCAT14 on FFPE sections from MDA-PCa-2b xenografts (A). Inset shows expression of PCAT14 in both nucleus and cytoplasm of MDA-PCa-2b cells. (B) Mouse prostate, kidney, and Lung.
Supplementary Figure 5. Functional analysis is PCAT14.
(A) Expression of PCAT14 in HEK293 cells expressing dCas9-VP64 and MS2-p65-HSF1 with control or six independent PCAT14 sgRNAs. (B) Expression of PCAT14 in LNCaP cells expressing dCas9-VP64 and MS2-p65-HSF1 with control or two independent sgRNA. (C–D) Effect of endogenous PCAT14 overexpression on proliferation of PC3 (C) and LNCaP cells (D). (E) Effect of PCAT14 overexpression on invasion capacity of LNCaP cells. (F) Representative images of invaded LNCaP cells with or without PCAT14 overexpression. (G–J) Expression of PCAT14 in MDA-PCa-2b (G) or VCaP (E) cells transfected with control siRNA or siRNA targeting PCAT14. Expression of PCAT14 in MDA-PCa-2b (H) or VCaP (J) cells transfected with control ASO or ASOs targeting PCAT14. (K–N) Effect of siRNA mediates knockdown of PCAT14 on proliferation of MDA-PCa-2b (K) or VCaP (M) cells. Effect of ASO mediates knockdown of PCAT14 on proliferation of MDA-PCa-2b (L) or VCaP (N) cells.
List of genes differentially expressed between Gleason 6 and Gleason 9+ prostate cancer.
Oncomine analysis: Genes correlated with PCAT14 (S4a) and concepts enriched (S4b).
Demographics of JHU cohort.
Sequences of primers, siRNA, and Guide RNA used in this study.
Acknowledgments
We thank Sethuramasundaram Pitchiaya, Xia Jiang, Fengyun Su and Ingrid Apel for technical assistance; K. Giles for critically looking over the manuscript and the submission of documents; the University of Michigan Viral Vector Core for generating the lentiviral constructs.
Footnotes
Funding/Support: This work was supported in part by the Prostate Cancer Foundation (F.Y.F, A.M.C), the National Institutes of Health Prostate SPORE (P50CA186786 to A.M.C) and the Early Detection Research Network (U01CA111275 and U01CA113913 to A.M.C). A.M.C. is supported by the Alfred A. Taubman Institute, Howard Hughes Medical Institute and the American Cancer Society. R.M is supported by a Department of Defense postdoctoral award (W81XWH-13-1-0284). R.M. and M.C. are supported by a Prostate Cancer Foundation Young Investigator award. The sponsors played no role in the design and conduct of the study.
References
- 1.Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, Minasian LM, Ford LG, Lippman SM, Crawford ED. Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N Engl J Med. 2004;350:2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 2.Andriole GL, Crawford ED, Grubb RL, 3rd, Buys SS, Chia D, Church TR, Fouad MN, Gelmann EP, Kvale PA, Reding DJ. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 4.Leyten GH, Hessels D, Jannink SA, Smit FP, de Jong H, Cornel EB, de Reijke TM, Vergunst H, Kil P, Knipscheer BC. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur Urol. 2014;65:534–542. doi: 10.1016/j.eururo.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 5.Tomlins SA, Aubin SM, Siddiqui J, Lonigro RJ, Sefton-Miller L, Miick S, Williamsen S, Hodge P, Meinke J, Blase A. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohler JL, Armstrong AJ, Bahnson RR, D'Amico AV, Davis BJ, Eastham JA, Enke CA, Farrington TA, Higano CS, Horwitz EM. Prostate Cancer, Version 1.2016. J Natl Compr Canc Netw. 2016;14:19–30. doi: 10.6004/jnccn.2016.0004. [DOI] [PubMed] [Google Scholar]
- 7.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends Cancer. 2015;1:93–109. doi: 10.1016/j.trecan.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network. Electronic address scmo, Cancer Genome Atlas Research N The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM, Magi-Galluzzi C, Mehra R, Sahu A, Siddiqui J. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15:1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehra R, Shi Y, Udager AM, Prensner JR, Sahu A, Iyer MK, Siddiqui J, Cao X, Wei J, Jiang H. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia. 2014;16:1121–1127. doi: 10.1016/j.neo.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Partridge J, Flaherty P. 2009. An in vitro FluoroBlok tumor invasion assay. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A, Pienta KJ, Shen MM. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25:638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 24.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehra R, Udager AM, Ahearn TU, Cao X, Feng FY, Loda M, Petimar JS, Kantoff P, Mucci LA, Chinnaiyan AM. Overexpression of the Long Non-coding RNA SChLAP1 Independently Predicts Lethal Prostate Cancer. Eur Urol. 2015 doi: 10.1016/j.eururo.2015.12.003. pii: S0302-2838(15)01211-7. doi: 10.1016/j.eururo.2015.12.003. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium GT Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–3882. [PubMed] [Google Scholar]
- 33.Arredouani MS, Lu B, Bhasin M, Eljanne M, Yue W, Mosquera JM, Bubley GJ, Li V, Rubin MA, Libermann TA. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res. 2009;15:5794–5802. doi: 10.1158/1078-0432.CCR-09-0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Best CJ, Gillespie JW, Yi Y, Chandramouli GV, Perlmutter MA, Gathright Y, Erickson HS, Georgevich L, Tangrea MA, Duray PH. Molecular alterations in primary prostate cancer after androgen ablation therapy. Clin Cancer Res. 2005;11:6823–6834. doi: 10.1158/1078-0432.CCR-05-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 36.True L, Coleman I, Hawley S, Huang CY, Gifford D, Coleman R, Beer TM, Gelmann E, Datta M, Mostaghel E. A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proc Natl Acad Sci U S A. 2006;103:10991–10996. doi: 10.1073/pnas.0603678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Setlur SR, Mertz KD, Hoshida Y, Demichelis F, Lupien M, Perner S, Sboner A, Pawitan Y, Andren O, Johnson LA. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008;100:815–825. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Expression pattern of other Gleason associated genes. Box plots represent the expression of TSPAN1, PTRHD1, DUSL1, and GMDS genes across the TCGA cancer/normal tissue RNA-seq compendia. All genes showed increased expression in prostate cancer compared to normal, but unlike PCAT14, they lacked prostate tissue specificity and differential expression is observed in several other cancer types. Blue: normal; red: Cancer.
Supplementary Figure 2. PCAT14 genomic organization, transcript variants, and expression in prostate cancer cell lines. (A) Schematic representation of genomic organization of various PCAT14 isoforms determined by RACE in MDA-PCa-2b and VCaP cells. Solid blocks indicate exons, while thin lines indicate intron and arrows indicate the genomic orientation. (B) VCaP and MDA-PCa-2b showed highest expression among the five androgen receptor–positive cell lines (VCaP, MDA-PCa-2b, 22RV1, LNCaP, and H660), whereas the 2 AR-negative cells lines (DU145 and PC3) lacked expression, as determined by RNA-seq (B) and qPCR. Inset shows the amplicon size on a 2% agarose gel. (C). D. The deduced intron/exon junction and mRNA sequence of PCAT14 by RACE and Sanger sequencing.
Supplementary Figure 3. Concept enrichment analysis. (A) Expression of PCAT14 in all the samples from the JHU cohort. (B) Kaplan-Meier analyses of MFS in the low-grade Gleason (Gleason 6 and 7) samples from the TCGA cohorts of prostate cancer. P values for Kaplan-Meier curves were determined using a log-rank test.
Supplementary Figure 4. (A–B) In situ hybridization of PCAT14 on FFPE sections from MDA-PCa-2b xenografts (A). Inset shows expression of PCAT14 in both nucleus and cytoplasm of MDA-PCa-2b cells. (B) Mouse prostate, kidney, and Lung.
Supplementary Figure 5. Functional analysis is PCAT14.
(A) Expression of PCAT14 in HEK293 cells expressing dCas9-VP64 and MS2-p65-HSF1 with control or six independent PCAT14 sgRNAs. (B) Expression of PCAT14 in LNCaP cells expressing dCas9-VP64 and MS2-p65-HSF1 with control or two independent sgRNA. (C–D) Effect of endogenous PCAT14 overexpression on proliferation of PC3 (C) and LNCaP cells (D). (E) Effect of PCAT14 overexpression on invasion capacity of LNCaP cells. (F) Representative images of invaded LNCaP cells with or without PCAT14 overexpression. (G–J) Expression of PCAT14 in MDA-PCa-2b (G) or VCaP (E) cells transfected with control siRNA or siRNA targeting PCAT14. Expression of PCAT14 in MDA-PCa-2b (H) or VCaP (J) cells transfected with control ASO or ASOs targeting PCAT14. (K–N) Effect of siRNA mediates knockdown of PCAT14 on proliferation of MDA-PCa-2b (K) or VCaP (M) cells. Effect of ASO mediates knockdown of PCAT14 on proliferation of MDA-PCa-2b (L) or VCaP (N) cells.
List of genes differentially expressed between Gleason 6 and Gleason 9+ prostate cancer.
Oncomine analysis: Genes correlated with PCAT14 (S4a) and concepts enriched (S4b).
Demographics of JHU cohort.
Sequences of primers, siRNA, and Guide RNA used in this study.