Abstract
The contrast between the stochasticity of biochemical networks and the regularity of cellular behavior suggests that biological networks generate robust behavior from noisy constituents. Identifying the mechanisms that confer this ability on biological networks is essential to understanding cells. Here we show that queueing for a limited shared resource in broad classes of enzymatic networks in certain conditions leads to a critical state characterized by strong and long-ranged correlations between molecular species. An enzymatic network reaches this critical state when the input flux of its substrate is balanced by the maximum processing capacity of the network. We then consider enzymatic networks with adaptation, when the limiting resource (enzyme or cofactor) is produced in proportion to the demand for it. We show that the critical state becomes an attractor for these networks, which points toward the onset of self-organized criticality. We suggest that the adaptive queueing motif that leads to significant correlations between multiple species may be widespread in biological systems.
Introduction
Transcription, translation, and signaling are stochastic processes often dominated by small-number effects. Yet overall, cellular behaviors proceed with remarkable predictability and regularity. How are such robust and reliable systems built from noisy elements? Previous work has suggested that some networks actively suppress noise and others harness it (1, 2, 3), with particular attention paid to noise in the concentrations of protein species. Certain regulatory networks achieve high sensitivity by exploiting mechanisms such as substrate competition and molecular titration in which only relative levels of molecular species matter (4, 5). Then, correlations between different proteins are important and the noise in the level of a single species is less relevant. Here we show that competition for limited shared resources in a broad class of enzymatic networks can lead to such strong correlations and furthermore, that network adaptation can make such highly correlated states robust to changes in parameters. The state of an enzymatic network characterized by strong and long-ranged correlations can naturally be interpreted as a critical state, and the adaptation leading toward this regime can likewise be interpreted as a mechanism for self-organized criticality. In a recent work, Ray et al. (6) demonstrated that expression of a single enzyme may have a profound effect on the physiology of the whole cell by driving the metabolic network across a threshold above which cells undergo growth arrest due to the toxicity of overabundant metabolite. They showed that cells may optimize biomass production by balancing the cell growth and toxicity caused by the metabolite overproduction that occurs in the vicinity of the critical state of the metabolic network.
Critical phenomena associated with phase transitions have received much attention as possible explanations for the complexity observed in nature, because critical systems exhibit large fluctuations, slow dynamics, and strong correlations. In particular, self-organized critical systems—those that naturally tend to their critical states—have been suggested to explain phenomena as diverse as earthquakes (7) and evolution (8). Recent work has indicated a possibility of near-criticality in single-enzyme systems (9) and suggested that multicellular organisms harness criticality in development (10).
Traditionally, enzymatic networks have been modeled deterministically using the Michaelis-Menten formalism (11). More recently, the statistical properties of enzymatic pathways have begun to attract significant attention (12, 13, 14, 15, 16). Levine and Hwa (12) theoretically studied stochastic fluctuations in different classes of metabolic pathways and found that steady-state fluctuations of intermediaries are effectively uncorrelated. This result, however, is linked to the important assumption that different enzymatic steps are catalyzed by different enzymes. While many enzymes are highly substrate-specific, many also target multiple substrates. For example, RNA transcripts must compete for translation by a limited number of ribosomes (17, 18). Bacterial sigma factors are coupled by their competition for RNA polymerases (19). In yeast, ultrasensitivity of Wee1 inactivation is believed to be generated by competition between Wee1 and other Cdk1 substrates for phosphorylation by Cdk1 (4). In mice, two F-box protein paralogs FBXL3 and FBXL21 (as part of an SCF complex) compete for binding of CRY proteins that act as circadian clock inhibitors (20). Degradation of many different proteins within the same cell is often enabled by a small group of enzymes such as the ClpXP protease in bacteria or the 26S proteasome in eukaryotes.
Previous work has shown that proteins degraded by a common protease exhibit strong correlations near the balance point where the total synthesis rate of the proteins matches the processing capacity of the protease (13, 14). This coupling mechanism has been recently used to tightly synchronize two independent genetic oscillators (21). It has been shown (22, 23) that posttranslation regulation via microRNA also leads to strong correlations among competing endogenous RNAs.
Here we consider a broad class of enzymatic networks in which different protein species either are interconverted by a common enzyme or share a common cofactor. These enzymatic networks with limited shared resources model a variety of phenomena in complex biological systems, including enzyme promiscuity in metabolic networks (24) and multisite phosphorylation of a protein (25). We use the mathematical theory of multiclass queues (26, 27, 28) to describe the statistical properties of protein fluctuations in steady state and demonstrate that in certain parameter regimes these networks exhibit critical behavior characterized by strong and long-ranged correlations between molecular species.
For a system with fixed limited resources, the critical state emerges only when the system is tuned to be near the balance point. However, allowing enzyme or cofactor levels to adapt to the size of the protein queue makes critical behavior robust to changes in system parameters. This type of adaptivity has been found in a number of similar enzymatic contexts, such as bacterial chemotaxis (29), signal transduction in the retina (30), calcium homeostasis (31), yeast osmoregulation (32), and temperature compensation in circadian clocks (33). Furthermore, in several recent works, adaptivity of general enzymatic networks was considered analytically and numerically in deterministic approximations (30, 34). We find that augmenting our enzymatic networks with adaptive feedback regulation gives rise to critical behavior in a broad region of parameter space, eliminating the need to tune a system to its balance point and indicating that these systems may exhibit self-organized criticality. Together, our results suggest that adaptive queueing may be a general principle that plays a pivotal role in conferring robustness on native biological circuits and a designer’s tool for constructing synthetic networks.
Materials and Methods
Shared resource-limited enzymatic networks
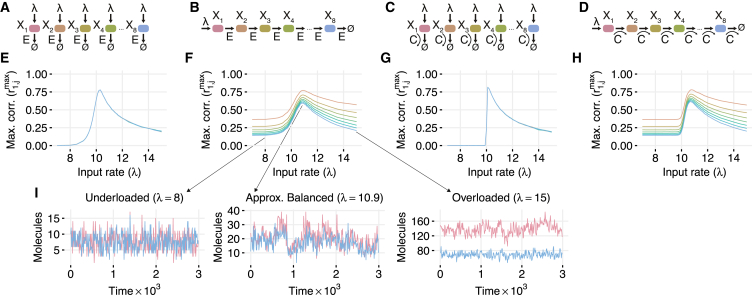
We study four classes of enzymatic networks in which reactions are rate-limited by a common resource (Fig. 1, A–D). Specifically, we consider competition for a fixed number of enzyme molecules that perform all enzymatic reactions (Fig. 1, A and B), or competition for a consumable cofactor produced at a fixed rate that is required for all enzymatic reactions (Fig. 1, C and D). For the parallel network with a shared enzyme (Fig. 1 A), we assume that L copies of the enzyme E catalyze degradation of n proteins . However, the same model can be applied to conversion of substrates into their corresponding products . The biochemical reactions in this system are:
| (1) |
Figure 1.
Generality of correlation resonance in biochemical networks. For each different type of network (A–D), levels of all species are highly correlated at the critical point where the input rate λ is balanced by removal of molecules from the system. (A) Parallel network of proteins degraded by a common enzyme. (B) Serial network of proteins interconverted by a common enzyme. (C) Parallel network of molecules processed by different abundant enzymes that all use a common cofactor. (D) Serial network of molecules processed by different abundant enzymes that all use a common cofactor. (E–H) Maximum correlations between and (denoted ) for simulations of each network. For the two serial networks, these maximal correlations occur at λ- and j-dependent time delays . Line color corresponds to the diagrams in (A–D). (I) Sample trajectories from the underloaded (left), approximately balanced (center), and overloaded (right) regimes of the serial network shown in (B). For networks with shared enzymes, the number of enzymes was fixed at . For the networks with a shared cofactor, the cofactor was synthesized at rate and diluted at rate γ. Other parameters were γ = 0.01, μ = 1, η+ = 1000, η− = 0, and . To see this figure in color, go online.
Here ; λ is the synthesis rate of all proteins; are binding/unbinding rates of proteins to the enzyme; μ is the rate at which an enzyme processes a protein; and γ is the dilution rate of all proteins (bound and unbound).
For the serial network with a shared enzyme (Fig. 1 B), we assume that the only input to the system is synthesis of protein at rate λ, a common pool of L copies of the enzyme E converts into , and the last stage of enzymatic processing degrades the protein :
| (2) |
Note that a similar system of enzymatic reactions has been studied in a recent work (35) where the authors found a critical slowdown of the cascade response to external perturbations when the system approached maximum capacity.
The last two networks have shared cofactors instead of shared enzymes. For them, we assume that enzymes are not rate-limiting and exclude them from consideration. Instead, we assume that the enzymatic reactions consume a shared cofactor C that is produced at rate and is degraded at rate . We consider both a parallel and a serial network. For the parallel network (Fig. 1 C), the biochemical reactions are:
| (3) |
where . For the serial network (Fig. 1 D), the reactions are:
| (4) |
We performed numerical simulations of these four systems of biochemical reactions using the direct Gillespie algorithm (36) and computed cross correlations among different substrates in the statistically stationary regime:
| (5) |
where is the covariance between the total number of protein (both free and enzyme-bound) at time t, , and the total number of protein at time t + τ, Qj(t + τ), and is the standard deviation of . For simplicity, we consider the case of irreversible binding where η− = 0. Unless noted otherwise, parameters for all simulations described here and below are n = 8, γ = γC = 0.01, μ = 1, η+ = 1000, η− = 0, and L = 80.
We also obtained analytical results for these systems for the underloaded regime in the zero dilution limit.
Modeling of adaptive enzymatic networks
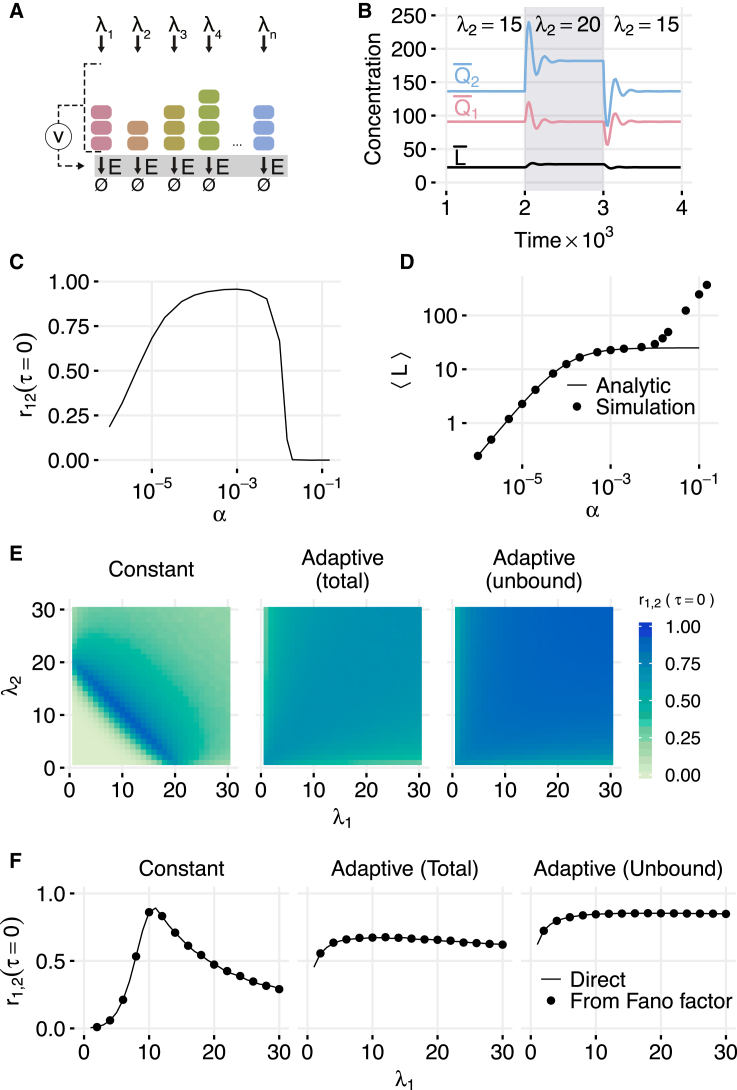
To investigate the role of adaptation in the dynamics of enzymatic networks with shared resources, we generalize our previous models. Instead of keeping the number of enzymes fixed, we allow molecules of the enzyme to be produced and diluted, with the production rate of the enzymes dependent on the numbers of proteins in the system. We first consider parallel degradation of multiple species (see Fig. 4 A). We assume that n proteins are synthesized at rates and degraded by the shared enzyme E that is synthesized at a rate ν that depends on the amount of the proteins present in the system (see below). All species (including E) are diluted at the rate γ. The reactions for the model are:
| (6) |
where the enzyme synthesis rate ν is allowed to take various forms as a function of . Here we consider only the simple case where . Similar systems of enzymatic reactions with feedback have been explored in the literature (see, for example, Furusawa and Keneko (34) and He et al. (37)), but only with a single class of substrate for each enzyme.
Figure 4.
Adaptive queueing in a parallel enzymatic network. (A) Diagram of the system. (B) Perfect adaptation in the deterministic model of the adaptive queueing network with two proteins (n = 2) with λ1 = 10, λ2 = 15, Km = 0.1, μ = 1, γ = 0.01, and α = 0.001. When λ2 is transiently changed from 15 to 20 (shaded region), all species initially respond. The other species 1 then settles to its original steady-state value. (C) Dependence of correlation on α in stochastic simulations of a two-species system with λ1 = 10, λ2 = 15, μ = 1, γ = 0.01, η+ = 1000, and η− = 0. Adaptation is effective for α across roughly three orders of magnitude. (D) Dependence of (mean enzymes including those bound to proteins) on α in stochastic simulations compared with the analytical expression given in Eq. 11. Mean enzyme level is constant for α in the range that gives effective adaptation. (E) Same-time correlations in stochastic simulations for different combinations of λ1 and λ2 with α = 0.01 and other parameters as in (C). In the constant enzyme case (left), correlations are only high in the vicinity of the balance line λ1+ λ2 = 20. In the systems adapting to either total species (left) or unbound species (right), correlations are high for nearly all combinations of rates. Same-time correlations are used because the absence of spatial structure in the parallel network removes the delays seen in serial networks. (F) Same-time correlations between and in stochastic simulations of nonadaptive and adaptive networks. Here λ2 = 10 with λ1 varying. For a constant number of enzymes, the correlation peaks near the balance point (left); however, when enzyme levels adapt to total number of proteins (center) or the number of unbound proteins (right), correlations are high for nearly all inputs. (Lines) Results of full stochastic simulations of the expressions in Eq. 6. (Points) Correlations calculated using Eq. S23 with the Fano factor computed from numerical simulation of the 2D Markov process given in the expressions in Eq. 21. To see this figure in color, go online.
In the limit of large numbers of all molecules and fast binding-unbinding reactions (the Michaelis-Menten approximation), the system can be described by the mass-action equations for the deterministic variables and that denote ensemble averages of total proteins and enzyme L, respectively:
| (7) |
| (8) |
where . The stable stationary solution of Eqs. 7 and 8 in implicit form is:
| (9) |
| (10) |
where . In the strong binding limit , these expressions simplify to explicit formulae:
| (11) |
| (12) |
Interestingly, despite the coupling of all species by enzymatic degradation, the steady-state value of depends only on its own synthesis rate and not on any other . This is a manifestation of the perfect adaptation caused by the integral feedback via the regulated enzyme synthesis (37, 38). Similar perfect adaptation is known to play a key role in making bacterial chemotaxis robust against changes in overall concentration of chemoattractants (29). However, an abrupt change in synthesis rate of one of the proteins transiently affects the rate (per molecule) at which proteins are removed from the system, and therefore the abundances of all proteins, as the amount of enzyme evolves to a new balance point (see Fig. 4 B).
Substituting from Eq. 11 into , we obtain that, in the stationary state, . Thus, for sufficiently large , the stationary regime is close to balance; however, for smaller α there are not enough copies of the enzyme due to dilution, and the system becomes overloaded. On the other hand, for sufficiently large α, the Michaelis-Menten approximation used in Eq. 7 no longer holds. Indeed, in the strong binding limit the rate of enzymatic degradation is limited by μmin {Q, L}. Therefore, from Eqs. 11 and 12 for and , there are more copies of the enzyme than arriving proteins, and the system becomes strongly underloaded in the steady state. Therefore, enzymatic adaptation should be effective within the range .
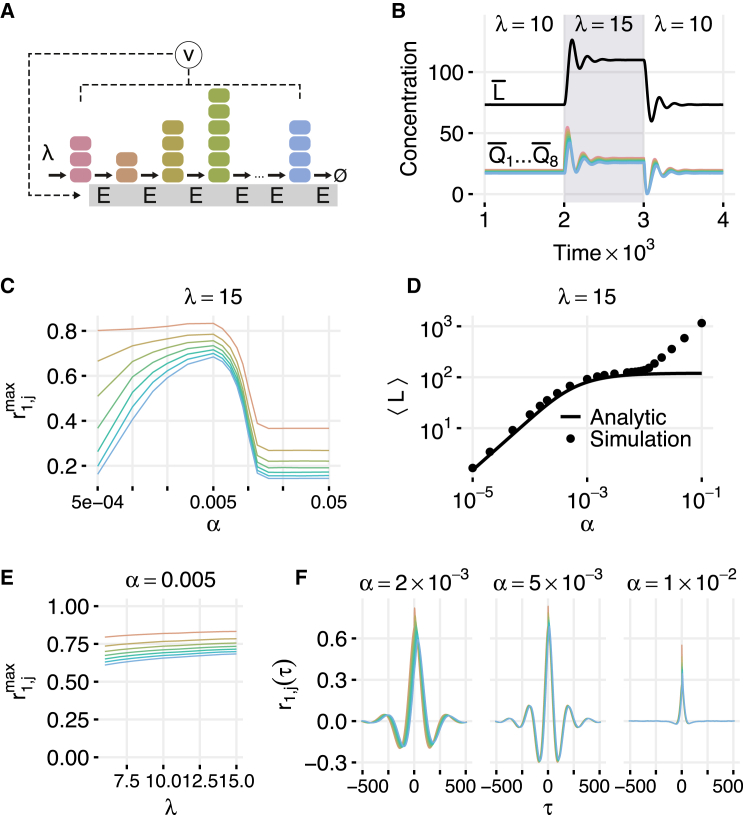
We next consider enzymatic adaptation for the serial enzymatic network introduced earlier in Fig. 1 B. In addition to the reactions in Eq. 2, we assume that enzymes are synthesized with a rate that is proportional to the total number of proteins in the system Q and are diluted at rate , with for (see Fig. 5 A). The reactions in this system are
| (13) |
Figure 5.
Simulations of the adaptive serial enzymatic network. (A) Schematic of the adaptive network. The production rate of the enzyme E is set by ν. Here we consider ν = αQ, where Q is the total number of proteins (free and enzyme-bound) in the system. (B) Results from a deterministic model of the adaptive queueing system with μ = 1, γ = 0.01, α = 0.005, and . When the input rate of is changed, levels of all proteins transiently oscillate before reaching a new steady-state level. (C) Maximum correlations between and as a function of the adaptation parameter α for λ = 15. For very low α, not enough enzyme is produced and for high α, enough enzyme is produced that the system is always underloaded. (D) Mean total enzyme level for different values of α. The range where adaptation occurs can be seen as the flat region of the curve. (E) Maximum correlations between and as a function of λ for α = 0.005. (F) Correlations as a function of τ for different values of α with λ = 15. Oscillations increase in strength and frequency for larger α but eventually disappear for high α. In (C–F), other simulation parameters are μ = 1, γ = 0.01, η+ = 1000, and η− = 0. To see this figure in color, go online.
The mass-action model of adaptive queueing in a serial network with the continuous variables for ensemble averages and is:
| (14) |
| (15) |
| (16) |
In the strong binding limit , the stationary solution of Eqs. 14–16 is given by
| (17) |
| (18) |
For large , the number of enzymes in the stationary state approaches the exact balance level . A typical transient regime for the eight-stage chain is shown in Fig. 5 B. As in the case of adaptive enzymatic degradation in the parallel network, the system adapts to be very near the balance point, but the negative feedback leads to transient ringing in the mean number of proteins and the number of enzymes in the system when parameters are changed.
Results
Criticality in nonadaptive resource-limited enzymatic networks
Fig. 1, E–H, depicts the maximum value of the cross correlations as a function of λ for the four types of enzymatic networks shown in Fig. 1, A–D. All four networks exhibited well-pronounced correlation resonance: when the rate of resource utilization approached the rate of resource availability, all species in the network became highly correlated. This correlation resonance phenomenon is not specific to these examples. We observed similar behavior in networks with reversible binding (Figs. S1 and S2 in the Supporting Material), even when binding constants η± are significantly varied (Fig. S3), as well as in more complicated networks with multiple shared cofactors (Fig. S4).
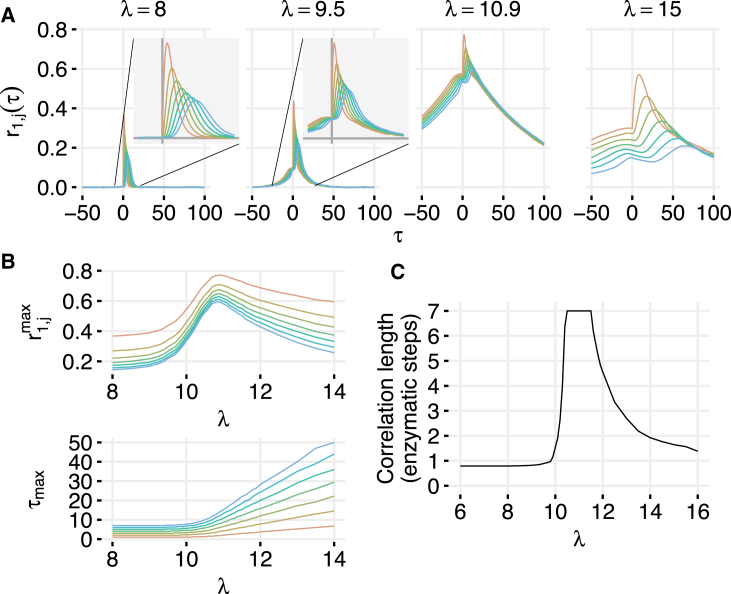
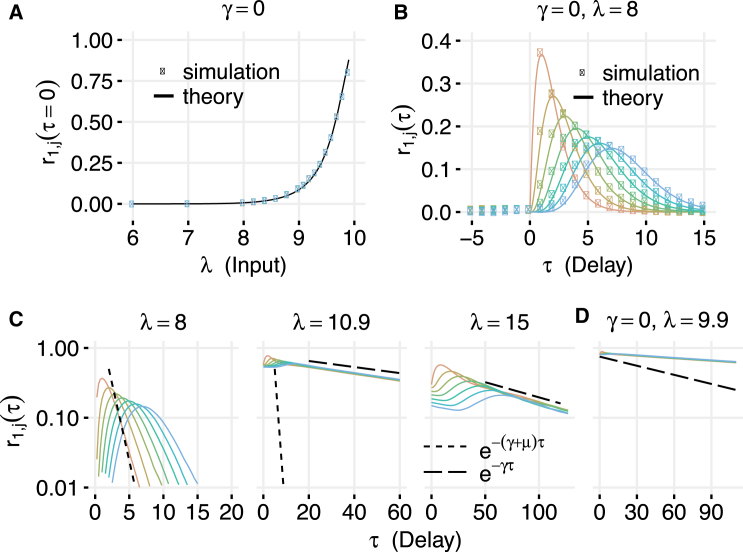
For a detailed analysis, we focus now on the serial shared enzyme network (Fig. 1, B, F, and I). The other three networks behave in a qualitatively similar manner. This system can, for example, represent sequential phosphorylation of a protein by a kinase. When , the network is strongly underloaded and the correlation between and is very close to zero for τ ≤ 0. However, the system can still exhibit time-delayed correlations that emerge due to the propagation of fluctuations in downstream via its processing by E and reaches a maximum at a small positive delay τmax (Fig. 2 A, left). Interestingly, while correlations at τ = 0 are independent of position in the network, time-shifted correlations decay along the chain (Fig. 2 A, center-left). Near the balance point, where the input rate λ is equal to the processing capacity , even the same-time correlations between all pairs of species are high. This is remarkable, given that can only be introduced at the expense of (Fig. 2 A, center-right).
Figure 2.
(A) Time-shifted correlations in different regimes for the serial network with a shared enzyme (see Fig. 1B). The broadening of the curves indicates that the correlation time increases as criticality is approached. (B) The value of the maximum correlation between and and the time delay at which that correlation is found . In the underloaded regime, the delay is short and correlations are small. Near the balance point, delay increases slightly while correlation increases dramatically. In the overloaded regime, the delay becomes very large and correlations decrease. (C) Correlation length in the serial biochemical network with a shared enzyme. Correlation length was calculated as interpolated distance in reaction steps at which (see Section S4 in the Supporting Material). Parameters for all simulations were γ = 0.01, μ = 1, η+ = 1000, η− = 0, and . To see this figure in color, go online.
Fig. 2 B shows the dependences of and on the input rate λ. As λ increases, correlations become larger, and become nonzero even for τ ≤ 0. In the strongly overloaded regime (ρ ≫ 1), the correlations decay much more slowly than in the underloaded case (Fig. 2 A, right). The time lag at which the cross-correlation coefficients are maximal also becomes much longer. The maximal correlation itself reaches a sharp peak near balance. As λ is increased substantially beyond the balance point, decreases, while grows linearly (Fig. 2 B).
High correlations and slowing of the dynamics are strong indicators of emergent criticality in this enzymatic network. The serial network also allows us to define a distance between two species: the number of enzymatic steps between them. We therefore computed the correlation length Lc for the serial networks that we define as the (linearly interpolated) number of stages at which the maximum cross-correlation coefficient with the first protein is >0.5 (see Section S4 in the Supporting Material). We find that the correlation length Lc reaches a sharp maximum near balance where all species are highly correlated (Fig. 2 C).
We also computed the susceptibility of the serial network as a function of the input rate λ. We defined the susceptibility in the usual way as the normalized rate of change of the output in response to a small stepwise change of input. The results presented in Section S5 in the Supporting Material show that susceptibility has a strong peak near the balance point, similar to the correlation peak. This gives further evidence that the enzymatic network near balance indeed can be characterized as critical.
We next sought analytical results describing the behavior of this network. For the underloaded regime (ρ < 1), it can be rigorously shown that in the absence of dilution (γ = 0) and in the limit of instant irreversible binding (η+ → ∞, η− = 0), the steady-state distribution for in the serial network is the same as that for a parallel network where each of the proteins is produced independently at the rate λ and they all share the same pool of L enzymes that process them (see Section S6 in the Supporting Material). Therefore, in the underloaded regime with no dilution, steady-state same-time correlations among any pair of different proteins in the serial network are the same. From our previous results (13), it follows that the same-time correlation between any two different proteins and is equal to:
| (19) |
where F is the Fano factor (the ratio of the variance to the mean) of the steady-state distribution of the total number of all proteins, . The moments of Q and its Fano factor can be found analytically in closed form (13). For L = 1, the expressions simplify and . For other values of L, one can obtain the leading term in the Taylor expansion of the correlation for small ρ (strongly underloaded regime), . It can also be shown that the correlation approaches 1 as ρ → 1. Fig. 3 A demonstrates excellent agreement between our analytical formulae and stochastic simulations.
Figure 3.
Comparison of analytical results to simulation for a serial shared-enzyme network. (A) Same-time correlations between species as a function of the input rate λ for the no-dilution network. (Points) Correlations from stochastic simulations; (lines) correlation predicted using Eq. 19. (B) Time-delayed correlations in the underloaded regime with no dilution. (Points) Correlations from stochastic simulations; (lines) correlations predicted from Eq. 20. (C) Temporal decay of correlations in different regimes with γ = 0.01. In the underloaded regime (λ = 8, left), correlations decay as . In the overloaded regime (λ = 15, right), they decay as . Near balance (center), correlation decay slows dramatically, but for nonzero γ cannot decay slower than . (D) In the absence of dilution, correlations near balance decay very slowly. Vertical scale shared with (C). Parameters not shown were μ = 1, η+ = 1000, η− = 0, and . To see this figure in color, go online.
In the strongly underloaded regime (ρ ≪ 1), the enzymatic reactions are approximately first-order (the propensities depend only on the number of proteins and not on the number of enzymes, because free enzymes are nearly always available). Using this approximation, the time-delayed correlations between protein levels are given by
| (20) |
and are 0 for negative τ-values (see Section S7 in the Supporting Material). For large positive τ, correlations decay exponentially with rate μ. Fig. 3 B shows good agreement between this theoretical formula and direct numerical simulations of an eight-stage enzymatic chain. This calculation can be straightforwardly extended to the case of nonzero dilution (see Section S7 in the Supporting Material).
Adaptation in a parallel enzymatic network
The critical behavior of enzymatic networks near the balance point described above is striking, but requires precise tuning of system parameters. However, living organisms employ numerous adaptive strategies to optimize resource allocation in the face of uncertain and variable environmental conditions. Indeed, having too many enzymes to process arriving proteins would be wasteful, while an insufficient number of enzymes would create an excess of unprocessed molecular species. We wondered whether adaptation of enzyme levels would drive the system toward the balance point and lead to the critical state, eliminating the need for parameter tuning.
We performed stochastic simulations of the adaptive system associated with Eq. 6 with two types of proteins and with . Fig. 4, C and D, shows the correlations between different proteins, as well as mean enzyme levels, as functions of α. The range of α values that effectively leads to adaptation can be seen as the region where the enzyme level is flat in Fig. 4 D. In agreement with the above estimate, for γ = 0.01 and μ = 1 it spans the range from 10−4 to 10−2. Fig. 4 E shows the heat maps of the steady-state correlations for different values of λ1,λ2 with or without adaptation. While in the nonadaptive case the correlations are high only in the vicinity of the balance line λ1 + λ2 = μL, in the adaptive cases the correlations are strong through nearly the whole range of synthesis rates. Similarly strong correlation in a broad range of input rates is obtained for other forms of the adaptation function; for example, when the adaptation rate is proportional to the number of unbound proteins of only one kind, (see Section S9 in the Supporting Material).
If the adaptation rate ν depends only on the sum of all protein counts, and the binding of proteins to enzyme is very fast, an extension of the method used in Mather et al. (13) allows us to approximately express the multidimensional steady-state distribution for the protein counts in terms of that for a two-dimensional (2D) birth-death process that tracks the sum Q and the number L of the enzyme copies that are in the system. Under an instant binding assumption, this finding reduces the dimension of the natural Markovian state descriptor from 2n+1 to 2 and allows us to explore steady-state correlations using numerical methods for the 2D process (Q,L). Furthermore, it can be rigorously shown (see Section S8 in the Supporting Material) that the correlation coefficient can still be expressed as a function of the Fano factor of the one-dimensional distribution of the total number of proteins using Eq. 19 when all production rates are equal. For a system with different production rates for , Eq. S23 (Supporting Material) can be used.
Under the instant binding assumption, the stochastic dynamics of protein X (which denotes any type of protein free or bound to E) and enzyme E (also both free or bound to protein) can be described by the following set of biochemical reactions (where the quantities above the arrows now indicate propensities of the corresponding reactions):
| (21) |
where Q and L values are the total numbers of protein X and enzyme E at time t, respectively; indicates the smaller of Q and L; and and values are the number of unbound copies of protein and enzyme, respectively. The last reaction denotes simultaneous removal of one protein and one enzyme when a protein-enzyme complex is diluted.
This set of reactions describes the dynamics of a 2D nonlinear birth-death process for which the steady-state distribution cannot be found analytically. An approximate solution can be found assuming that the adaptation rate and dilution rate γ are small compared with protein synthesis and enzymatic degradation rates . In this case, the number of enzymes changes slowly compared with the number of proteins, and the marginal probability distribution for Q equilibrates toward the stationary distribution corresponding to a fixed value of L (see Mather et al. (13)). The slow dynamics of L can be approximated as a nonlinear birth-death process with the birth rate and the death rate γ. In the general case of nonsmall α, we can perform numerical simulations of the reduced 2D birth-death process (Q, L) to compute the Fano factor F and then use Eq. 19 to compute the correlations between and . Fig. 4 F compares this method with the results of direct simulations of the system described by Eq. 6. As expected, there is excellent agreement between the two.
Adaptation in serial enzymatic networks
To investigate the statistical properties of the adaptive serial network, we performed direct numerical simulations of the full stochastic model associated with Eq. 13. As with the parallel network, for a broad range of intermediate values of α, the cross correlations between and other are high (Fig. 5 C) and nearly identical independent of λ (Fig. 5 E). As before, the mean number of enzymes exhibits a plateau in the region of adaptation in excellent agreement with Eq. 18, but for larger α > γ deviates up when the Michaelis-Menten approximation loses its validity (Fig. 5 D). Interestingly, the temporal cross-correlation functions demonstrate nonmonotonous behavior that is due to stochastic ringing caused by the negative feedback loop. The frequency of ringing is, as expected, dependent on α (Fig. 5 F).
We obtained similar results for a network with enzyme synthesis rate proportional to the abundance of the first or the last protein in the series (see Sections S10 and S11 in the Supporting Material). These results show that adaptation and approach to criticality also takes place in these cases; however, in the second case the adaptation is less robust. In particular, it is possible for enzyme production to permanently cease if both E and are zero at the same time. However, if we add small basal synthesis of E, adaptation becomes robust again. We also considered the case when ν is proportional to the total abundance of unbound proteins (see Section S12 in the Supporting Material). The latter form of adaptation generated even higher correlations than those using total proteins, because the number of unbound proteins is more sensitive to queueing.
We also computed the network susceptibility to small input perturbations using the deterministic model Eqs. 14–16 (and see Section S5 in the Supporting Material). Again similarly to the cross-correlation analysis, the susceptibility becomes virtually independent of λ because the network adapts to the neighborhood of the balance point. However, generally the susceptibility of the adaptive network is lower than for the nonadaptive one at the balance because of the negative feedback, which is known to reduce the magnitude of the response to external perturbations.
Discussion
Much attention has been given to the question of how cells are able to function reliably when biological networks are seemingly so noisy. Here we used a queueing approach to study a broad class of networks in which protein species are processed by a common enzyme or utilize a common cofactor. Despite the noisiness of the networks’ dynamics due to intrinsic stochasticity of the underlying biochemical reactions, we found strong and long-range correlations among multiple protein species near the balance between the substrate input rates and the processing capacity of the network. Furthermore, the correlation time also increases dramatically as the system approaches the balance point. This phenomenology suggests that, at the balance point, enzymatic networks are poised near a critical state.
We also considered network adaptation where the enzyme (or cofactor) synthesis is regulated by the protein species processed by the enzyme. Our theoretical analysis shows that for sufficiently slow adaptation, the network automatically approaches the balance point with large and slowly decaying correlations. An adaptive queueing network therefore behaves much like a self-organized critical system where the critical point is an attractor (39). It is interesting to note that self-organized critical states characterized by a power-law distribution in the abundances of different reactants in random autocatalytic metabolic networks with active transport of nutrients were found in Furusawa and Kaneko (34) and Awazu and Kaneko (40), where this was shown to lead to optimal cell growth. The tendency of adaptive networks to spontaneously approach a critical state may also explain recent experimental observations of criticality in neuronal systems (41) and development (10). The ability of signal transduction cascades to respond to changes in the statistical properties of the input and maximize their information capacity also relies on enzymatic adaptation (30). Taken together with ours, these results point toward the ubiquity and wide-ranging implications of the tendency of enzymatic networks toward criticality.
Emergence of adaptivity of the kind considered here appears to be evolutionarily advantageous, because the balance between the input flux and the processing capacity provides optimal use of enzymatic resources and maximizes cell growth. It can be achieved if one of the species in the queue acts as a transcription factor or posttranscriptional regulator of the limiting enzyme or cofactor synthesis. This motif is in fact observed in natural systems. For example, the Saccharomyces cerevisiae transcription factor Rpn4 stimulates production of proteasome genes but is itself degraded by the proteasome (42). In a population context, when overproduction of certain species becomes toxic, the adaptation toward balance may lead to complex, possibly bimodal distributions of cellular phenotypes (6).
The balance point also appears to be optimal for synchronizing levels of the interacting species in a network. In the underloaded regime, changes in protein levels propagate across the network very rapidly, but with low fidelity (small correlations). In the overloaded regime correlations are high, but the delay in their propagation is large. At the balance point, correlations are maximized, while delay times are only slightly increased compared to the underloaded regime. Given the generality of our model and the ubiquity of molecular competition, we anticipate that understanding these more general cases might provide useful insights for whole cell models with many substrates connected by a core of common enzymes and cofactors.
Author Contributions
R.J.W., J.H., and L.S.T. conceived and designed research; P.J.S. and L.S.T. performed and analyzed numerical simulations; and all authors participated in theoretical analysis and contributed to writing the article.
Acknowledgments
This work was supported by the National Science Foundation and the National Institutes of Health under the Joint DMS/NIGMS Initiative to Support Research at the Interface of the Biological and Mathematical Sciences, National Science Foundation grant No. DMS-1463657, National Science Foundation grant No. DMS-1206772, and the San Diego Center for Systems Biology, National Institutes of Health grant No. P50-GM085764.
Editor: Richard Bertram.
Footnotes
Supporting Materials and Methods and 12 figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30616-6.
Contributor Information
Ruth J. Williams, Email: williams@math.ucsd.edu.
Jeff Hasty, Email: hasty@ucsd.edu.
Lev S. Tsimring, Email: ltsimring@ucsd.edu.
Supporting Material
References
- 1.Rao C.V., Wolf D.M., Arkin A.P. Control, exploitation and tolerance of intracellular noise. Nature. 2002;420:231–237. doi: 10.1038/nature01258. [DOI] [PubMed] [Google Scholar]
- 2.Eldar A., Elowitz M.B. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsimring L.S. Noise in biology. Rep. Prog. Phys. 2014;77:026601. doi: 10.1088/0034-4885/77/2/026601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S.Y., Ferrell J.E., Jr. Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell. 2007;128:1133–1145. doi: 10.1016/j.cell.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 5.Buchler N.E., Louis M. Molecular titration and ultrasensitivity in regulatory networks. J. Mol. Biol. 2008;384:1106–1119. doi: 10.1016/j.jmb.2008.09.079. [DOI] [PubMed] [Google Scholar]
- 6.Ray J.C.J., Wickersheim M.L., Balázsi G. Cellular growth arrest and persistence from enzyme saturation. PLOS Comput. Biol. 2016;12:e1004825. doi: 10.1371/journal.pcbi.1004825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bak P., Tang C. Earthquakes as a self-organized critical phenomenon. J. Geophys. Res. 1989;94:15635–15637. [Google Scholar]
- 8.Sneppen K., Bak P., Jensen M.H. Evolution as a self-organized critical phenomenon. Proc. Natl. Acad. Sci. USA. 1995;92:5209–5213. doi: 10.1073/pnas.92.11.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elf J., Paulsson J., Ehrenberg M. Near-critical phenomena in intracellular metabolite pools. Biophys. J. 2003;84:154–170. doi: 10.1016/S0006-3495(03)74839-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krotov D., Dubuis J.O., Bialek W. Morphogenesis at criticality. Proc. Natl. Acad. Sci. USA. 2014;111:3683–3688. doi: 10.1073/pnas.1324186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauro H.M. Future Skill Software; Seattle, WA: 2012. Enzyme Kinetics for Systems Biology. [Google Scholar]
- 12.Levine E., Hwa T. Stochastic fluctuations in metabolic pathways. Proc. Natl. Acad. Sci. USA. 2007;104:9224–9229. doi: 10.1073/pnas.0610987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mather W.H., Cookson N.A., Williams R.J. Correlation resonance generated by coupled enzymatic processing. Biophys. J. 2010;99:3172–3181. doi: 10.1016/j.bpj.2010.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cookson N.A., Mather W.H., Hasty J. Queueing up for enzymatic processing: correlated signaling through coupled degradation. Mol. Syst. Biol. 2011;7:561. doi: 10.1038/msb.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mather W.H., Hasty J., Williams R.J. Translational cross talk in gene networks. Biophys. J. 2013;104:2564–2572. doi: 10.1016/j.bpj.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hochendoner P., Ogle C., Mather W.H. A queueing approach to multi-site enzyme kinetics. Interface Focus. 2014;4:20130077. doi: 10.1098/rsfs.2013.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vind J., Sørensen M.A., Pedersen S. Synthesis of proteins in Escherichia coli is limited by the concentration of free ribosomes. Expression from reporter genes does not always reflect functional mRNA levels. J. Mol. Biol. 1993;231:678–688. doi: 10.1006/jmbi.1993.1319. [DOI] [PubMed] [Google Scholar]
- 18.Baumgartner B.L., Bennett M.R., Hasty J. Antagonistic gene transcripts regulate adaptation to new growth environments. Proc. Natl. Acad. Sci. USA. 2011;108:21087–21092. doi: 10.1073/pnas.1111408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mauri M., Klumpp S. A model for sigma factor competition in bacterial cells. PLOS Comput. Biol. 2014;10:e1003845. doi: 10.1371/journal.pcbi.1003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo S.-H., Mohawk J.A., Takahashi J.S. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prindle A., Selimkhanov J., Hasty J. Rapid and tunable post-translational coupling of genetic circuits. Nature. 2014;508:387–391. doi: 10.1038/nature13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Figliuzzi M., Marinari E., De Martino A. MicroRNAs as a selective channel of communication between competing RNAs: a steady-state theory. Biophys. J. 2013;104:1203–1213. doi: 10.1016/j.bpj.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosia C., Pagnani A., Zecchina R. Modelling competing endogenous RNA networks. PLoS One. 2013;8:e66609. doi: 10.1371/journal.pone.0066609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babtie A., Tokuriki N., Hollfelder F. What makes an enzyme promiscuous? Curr. Opin. Chem. Biol. 2010;14:200–207. doi: 10.1016/j.cbpa.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 25.Cohen P. The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem. Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 26.Kelly F.P. Cambridge University Press; Cambridge, UK: 2011. Reversibility and Stochastic Networks. [Google Scholar]
- 27.Williams R.J. Diffusion approximations for open multiclass queueing networks: sufficient conditions involving state space collapse. Queueing Syst. 1998;30:27–88. [Google Scholar]
- 28.Bramson M. State space collapse with application to heavy traffic limits for multiclass queueing networks. Queueing Syst. 1998;30:89–140. [Google Scholar]
- 29.Yi T.-M., Huang Y., Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc. Natl. Acad. Sci. USA. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Detwiler P.B., Ramanathan S., Shraiman B.I. Engineering aspects of enzymatic signal transduction: photoreceptors in the retina. Biophys. J. 2000;79:2801–2817. doi: 10.1016/S0006-3495(00)76519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Samad H., Goff J.P., Khammash M. Calcium homeostasis and parturient hypocalcemia: an integral feedback perspective. J. Theor. Biol. 2002;214:17–29. doi: 10.1006/jtbi.2001.2422. [DOI] [PubMed] [Google Scholar]
- 32.Muzzey D., Gómez-Uribe C.A., van Oudenaarden A. A systems-level analysis of perfect adaptation in yeast osmoregulation. Cell. 2009;138:160–171. doi: 10.1016/j.cell.2009.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni X.Y., Drengstig T., Ruoff P. The control of the controller: molecular mechanisms for robust perfect adaptation and temperature compensation. Biophys. J. 2009;97:1244–1253. doi: 10.1016/j.bpj.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furusawa C., Kaneko K. Adaptation to optimal cell growth through self-organized criticality. Phys. Rev. Lett. 2012;108:208103. doi: 10.1103/PhysRevLett.108.208103. [DOI] [PubMed] [Google Scholar]
- 35.Hatakeyama T.S., Kaneko K. Kinetic memory based on the enzyme-limited competition. PLOS Comput. Biol. 2014;10:e1003784. doi: 10.1371/journal.pcbi.1003784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillespie D.T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977;81:2340–2361. [Google Scholar]
- 37.He F., Fromion V., Westerhoff H.V. (Im)Perfect robustness and adaptation of metabolic networks subject to metabolic and gene-expression regulation: marrying control engineering with metabolic control analysis. BMC Syst. Biol. 2013;7:131. doi: 10.1186/1752-0509-7-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.François P., Siggia E.D. A case study of evolutionary computation of biochemical adaptation. Phys. Biol. 2008;5:026009. doi: 10.1088/1478-3975/5/2/026009. [DOI] [PubMed] [Google Scholar]
- 39.Bak P., Tang C., Wiesenfeld K. Self-organized criticality. Phys. Rev. A Gen. Phys. 1988;38:364–374. doi: 10.1103/physreva.38.364. [DOI] [PubMed] [Google Scholar]
- 40.Awazu A., Kaneko K. Self-organized criticality of a catalytic reaction network under flow. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2009;80:010902. doi: 10.1103/PhysRevE.80.010902. [DOI] [PubMed] [Google Scholar]
- 41.Friedman N., Ito S., Butler T.C. Universal critical dynamics in high resolution neuronal avalanche data. Phys. Rev. Lett. 2012;108:208102. doi: 10.1103/PhysRevLett.108.208102. [DOI] [PubMed] [Google Scholar]
- 42.Xie Y., Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. USA. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.