Abstract
Microbial rhodopsins are light-activated, seven-α-helical, retinylidene transmembrane proteins that have been identified in thousands of organisms across archaea, bacteria, fungi, and algae. Although they share a high degree of sequence identity and thus similarity in structure, many unique functions have been discovered and characterized among them. Some function as outward proton pumps, some as inward chloride pumps, whereas others function as light sensors or ion channels. Unique among the microbial rhodopsins characterized thus far, Anabaena sensory rhodopsin (ASR) is a photochromic sensor that interacts with a soluble 14-kDa cytoplasmic transducer that is encoded on the same operon. The sensor itself stably interconverts between all-trans-15-anti and 13-cis-15-syn retinal forms depending on the wavelength of illumination, although only the former participates in a photocycle with a signaling M intermediate. A mutation in the cytoplasmic half-channel of the protein, replacing Asp217 with Glu (D217E), results in the creation of a light-driven, single-photon, inward proton transporter. We present the 2.3 Å structure of dark-adapted D217E ASR, which reveals significant changes in the water network surrounding Glu217, as well as a shift in the carbon backbone near retinal-binding Lys210, illustrating a possible pathway leading to the protonation of Glu217 in the cytoplasmic half-channel, located 15 Å from the Schiff base. Crystallographic evidence for the protonation of nearby Glu36 is also discussed, which was described previously by Fourier transform infrared spectroscopy analysis. Finally, two histidine residues near the extracellular surface and their possible role in proton uptake are discussed.
Introduction
Microbial rhodopsins have been identified in thousands of organisms (1, 2), spanning archaea, bacteria, fungi, and algae. Despite differences in protein function, they share structural similarities. Each possesses seven transmembrane helices, as well as a retinylidene chromophore covalently bound to a conserved lysine in the seventh transmembrane helix. The best-characterized are the haloarchaeal light-driven ion pumps bacteriorhodopsin (BR) (3, 4, 5, 6, 7, 8, 9, 10, 11) and halorhodopsin (HR) (12, 13, 14), as well as the signal transducers, sensory rhodopsin I (SRI) (15, 16, 17) and sensory rhodopsin II (SRII) (17, 18, 19, 20). BR functions as a light-driven ion pump, utilizing light energy to pump protons outward against an electrochemical gradient through a well-defined pathway (6), although the possibility that BR is a net inward OH- pump has been raised (21, 22, 23). BR’s ability to enforce vectorial ion transport can be explained partly by the lack of internal waters in its cytoplasmic half-channel during parts of its photocycle; whereas the extracellular half-channel of BR contains eight ordered water molecules, many charged residues, and a well-defined hydrogen-bonding network, the cytoplasmic half-channel only contains two waters and is predominantly hydrophobic, with the critical exception of Asp96 that is protonated in the ground state. During the course of the elucidation of its proton transfer mechanism, many mutational studies have been carried out, successfully converting BR into an inward chloride pump (24), as well as into a sensory rhodopsin (25). However, no successful attempts have been reported that reverse the direction of proton transport in BR, although two-photon, inward, net proton transfer activity has been documented (26), which perhaps highlights the importance of the hydrophobic cytoplasmic domain in preserving the integrity of BR as an outward-only pump.
Notably, Anabaena sensory rhodopsin’s (ASR) cytoplasmic half-channel is much more hydrophilic than that of the majority of known microbial rhodopsins, containing five ordered waters that interact with numerous polar residues. First identified in the freshwater cyanobacterium Anabaena (Nostoc) sp. PCC7120 (27), ASR is comprised of 261 residues and is structurally similar to BR (28). However, rather than pumping ions, it has been shown to regulate the expression of phycocyanin and phycoerythrocyanin, part of the phycobilisome light-harvesting complex, via a soluble 14-kDa cytoplasmic transducer (ASRT) (29, 30, 31, 32, 33, 34), found on a single operon (27). Unlike the haloarchaeal sensory rhodopsins, which interact with their cognate transmembrane transducers to mediate light-stimulated phototaxis (35, 36), it has been proposed that ASR’s photosignal is relayed to ASRT, which then translocates to the nucleus where it can bind specifically to the promoter regions of multiple genes (33, 34). Other studies have shown evidence that ASR alone in E. coli can bind DNA and regulate gene activity upon photostimulation (31).
Unlike any of the other known microbial rhodopsins, ASR is unique in that it stably photoconverts between an all-trans,15-anti retinal (ATR) form and a 13-cis,15-syn retinal (13C,15S) form, depending on the color of ambient light (28). In the dark, ASR is predominantly populated with ATR (∼97%), with an absorption maxima of 549 nm (37). Upon orange-light illumination (>560 nm), 13C,15S predominates (∼78%), with an accompanying absorption maximum of 537 nm. Illumination with blue light (∼470 nm) converts the 13C,15S configuration to ATR, resulting in a higher population of ATR (28, 37, 38). This wavelength-dependent interconversion between the ASR-ATR and ASR-13C,15S states suggests an involvement of the protein in response to environmental variables as a photochromic sensor, utilizing the color of ambient light to control the expression of phycobilisome proteins (27). It has been shown that upon photon absorption, the Schiff base proton is transferred to Asp217 in the cytoplasmic half (39), ∼15 Å from the retinal. However, because there is no significant net proton transfer through the protein, as determined by the monitoring of light-induced pH changes in nonbuffered solutions containing ASR-imbedded spheroplasts (40), it has been postulated that the Schiff base is reprotonated from the same residue (41, 42). As in most microbial rhodopsins, with the exception of proteorhodopsin (43), there is a tightly coordinated water molecule (Wat402) in the extracellular domain that is strongly hydrogen-bonded to the retinal Schiff base in the protonated state, while it is only weakly hydrogen-bonded to its counterion Asp75 (Asp85 in BR), which does not participate in reprotonation (41).
Previously, an inward proton transporter was engineered from ASR (40). Although the engineered D217E mutant protein is not technically a transporter, in the sense that it has not been shown to actively pump protons through the membrane against an electrochemical gradient, its role as a pump also cannot be discounted, as experimental data has not been provided to the contrary. Thus for the purposes of this manuscript, the mutant is designated a transporter, a term we use throughout this article.
In a study involving site-directed mutants of ASR, Kawanabe et al. replaced Asp217 with Glu217 and inadvertently created an inward proton transporter (40). In D217E ASR-containing spheroplast vesicles resuspended in a nonbuffered solution, illumination with yellow light (>500 nm) resulted in a net inward transport of protons, demonstrated by an increasingly alkaline medium. This effect was abolished by the addition of the proton uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP) and was not observed for protein expressed in the absence of retinal, showing that D217E ASR itself was responsible for an inward movement of protons. A linear relationship between illumination light intensity and a pH increase in the medium also demonstrated that the proton transport was driven by a single-photon reaction (40), setting it apart from a previously reported D85N BR mutant that demonstrated light-driven, two-photon inward proton transport (26). Whereas wild-type (WT) ASR also shows trace amounts of light-driven inward proton transport, the activity of D217E ASR is 10-fold higher, and has been shown to be pH-independent between pH 6.5 and 8.0. Subsequent Fourier transform infrared spectroscopy (FTIR) studies also showed that during formation of the ASR M intermediate, the Schiff base proton is transferred to Glu217, with 10-times higher levels of protonation than those of WT Asp217, correlating with activity assay results. It thus seems likely that protonation of residue 217 is instrumental in determining inward proton transport activity. Given the robust protonation of Glu217 compared with Asp217, and the significantly larger proton transport activity, it is likely that for D217E ASR the Schiff base is reprotonated from the extracellular side (40).
In this study, we present the 2.3 Å structure of the D217E single-site mutant of Anabaena sensory rhodopsin, and discuss its structural differences from WT ASR, as well as examine the structural implications of the mutation as they pertain to the novel inward proton transport activity of the mutant.
Materials and Methods
Materials
The mutagenic oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) and Terrific Broth was from Fisher BioReagents (Hampton, NH). Anatrace (Maumee, OH) supplied n-dodecyl β-D-maltoside (DDM), and Clontech (Mountain View, CA) provided Co2+-affinity TALON resin. 100-kDa cutoff Amicon filter was from Millipore (Billerica, MA) and Sephacryl S-200 HR, Tricorn 10/300 GL size-exclusion column was from GE Healthcare (Chicago, IL). Monoolein was purchased from Sigma and the interconnected gas-tight syringes were from Hamilton.
Expression and purification
D217E ASR was constructed using site-directed mutagenesis, based on C-terminally 6x His-tagged WT ASR with a C-terminal truncation of 35 residues as the template. The ASR gene, and the resulting mutant, is located in the plasmid pKJ900, which also carries a gene for mouse 15-15′-β-dioxygenase (44). Polymerase chain reaction for the site-directed mutagenesis was performed using the oligonucleotide 5′-CTCCAAGGTTGGATTTAGTTTTCTGGAATTACACGGCTTACGTAA-3′ and its antisense.
D217E ASR-containing pKJ900 was transformed into an E. coli strain UT5600 previously transformed with a β-carotene expressing plasmid βORANGE. Five milliliters of overnight culture was transferred into 4 L of Terrific Broth, supplemented with 100 μg/ml ampicillin and 35μg/ml cloramphenicol, and grown at 30°C until the O.D.600 reached 0.4, at which point the temperature was lowered to 16°C and transcription was induced with 1 mM IPTG and 0.2% ʟ-arabinose. Cells were harvested after 20 h using centrifugation at 6,500 × g for 10 min at 4°C, and resuspended in a buffer containing 50 mM Tris-HCl pH 8.0, 300 mM NaCl, and 1 mg/mL lysozyme. Cells were lysed using sonication, intact cells removed by centrifugation at 6,500 × g for 10 min at 4°C, and the membrane fraction pelleted using centrifugation at 100,000 × g for 1 h at 4°C. The membrane fraction was resuspended in a buffer containing 50 mM Tris-HCl pH 8.0, 300 mM NaCl, and 1% n-dodecyl β-D-maltoside (DDM) using a dounce homogenizer, and allowed to solubilize overnight at 4°C with continuous stirring. The soluble fraction containing solubilized D217E ASR was separated after centrifugation at 100,000 × g for 1 h at 4°C and applied directly to a column packed with Co2+-affinity TALON resin; washed with a buffer containing 30 mM Tris-HCl pH 8.0, 300 mM NaCl, 50 mM imidazole, and 0.05% DDM; and eluted with a similar buffer containing 30 mM Tris-HCl pH 8.0, 300 mM NaCl, 200 mM imidazole, and 0.05% DDM. The fraction containing D217E ASR was collected, concentrated using a 100-kDa cutoff Amicon filter, and purified further using a Sephacryl S-200 HR, Tricorn 10/300 GL size-exclusion column and a buffer containing only 20 mM Tris-HCl pH 8.0 and 0.05% DDM.
Crystallization
D217E ASR was concentrated to 10 mg/mL using a 100-kDa cutoff Amicon filter. The protein was combined at a 60:40 lipid:protein ratio with molten monoolein melted at a temperature of 40°C. We mixed the components manually into a homogenous solution using two interconnected gas-tight syringes (Hamilton) until lipidic cubic phase was formed (45). We overlaid 200 nL of the lipid cubic phase mixture with 1 μL of crystallization solution, containing 6% MPD, 200 mM sodium citrate pH 5.6, 200 mM MgCl2, and 0.008% (w/v) of the following: 3-indolebutyric acid, hexadecanedioic acid, oxamic acid, pyromellitic acid, sebacic acid, and suberic acid (Silver Bullets HT screen reagent G4). Crystals appeared after 3 days as pink-colored diamonds and grew to ∼100 μm in length. Individual crystals were mechanically extracted directly from the lipidic cubic phase using Mitegen MicroMount loops and cryo-cooled with liquid nitrogen.
Data collection
Diffraction data to 2.3 Å were collected at 100 K from one dark-adapted crystal at beamline 11-1 at the Stanford Synchrotron Radiation Lightsource on a PILATUS 6M PAD detector. Images were indexed, integrated, and scaled using the program HKL2000 (46). Molecular replacement (MR) and refinement were performed using the CCP4 software suite (47); MR was performed using the program PHASER (48) with WT ASR (PDB: 1XIO) as the search model. Waters and retinal were removed, and Lys210 and Asp217 were reduced to Ala to avoid model bias. During later stages of refinement, the retinal, water molecules, and lipid tails were added manually using the program Coot (49) with 2Fo-Fc electron density maps contoured at 1.0 σ and Fo-Fc maps contoured at +/−3.0 σ. Multiple rounds of refinement were performed using the program REFMAC5 (50), with the final R factor (Rfree) of 20.9% (25.5%). All peptides fall into the allowed regions of the Ramachandran plot (Table 1).
Table 1.
X-ray Data Reduction and Refinement Statistics
| Data Collection | |
|---|---|
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 55.3, b = 104.3, c = 111.4 |
| X-ray wavelength (Å) | 0.97945 |
| Resolution range (Å) | 33.19–2.30 (2.34–2.30) |
| Total/unique reflections | 51,969/28,486 |
| Completeness (%) | 96.7 (98.7) |
| <I>/<σ(I)> | 11.4 (2.0) |
| Rmerge (%) | 11.3 (53.3) |
| Mosaicity (°) | 0.77 |
| Model Refinement | |
| Resolution range (Å) | 33.19–2.30 |
| Rwork/Rfree (%) | 21.98/24.95 |
| Monomers per asymmetric unit | 2 |
| No. of Non-H Atoms/Average B Factor (Å2) | |
| Protein | 3,582/24.64 |
| Retinal | 40/18.80 |
| Water | 109/34.15 |
| Lipids | 435/51.11 |
| RMSD from Ideal Geometry | |
| Bond lengths (Å) | 0.0081 |
| Bond angles (°) | 1.2032 |
| Ramachandran Plot | |
| Favored (%) | 423 (98.83) |
| Allowed (%) | 5 (1.17) |
| Outliers (%) | 0 (0.00) |
Results and Discussion
Crystallization, crystal packing, and overall structure of D217E Anabaena sensory rhodopsin
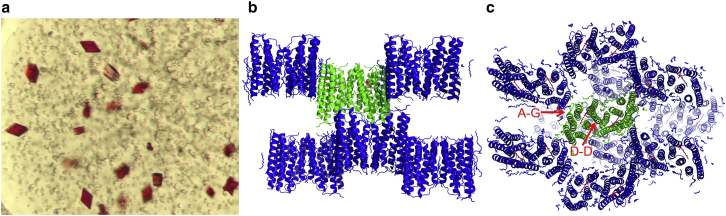
The expression, purification, and crystallization of D217E ASR are described in Materials and Methods. Pink-colored diamond-shaped crystals were grown in a lipidic cubic phase (45) (Fig. 1 a) and harvested directly into liquid nitrogen in the dark. Diffraction data were collected at BL11-1 at the Stanford Synchrotron Radiation Lightsource at a wavelength of 0.97945 Å to a resolution of 2.3 Å. The crystal belongs to orthorhombic space group P212121 with unit cell dimensions a = 55.3 Å, b = 104.3 Å, and c = 111.4 Å and two monomers per asymmetric unit. Initial phases were determined using molecular replacement with WT ASR (PDB: 1XIO) as the search model. After several rounds of refinement, the refined structure has an Rwork of 20.97% and an Rfree of 25.49% (Table 1).
Figure 1.
Crystallization of D217E ASR. (a) Pink-colored crystals were grown from the cubic lipid phase and belong to orthorhombic space group P212121, with two molecules in the asymmetric unit. (b and c) Crystal packing of D217E ASR is shown. Crystal contacts between neighboring dimers cause an undulation in the lipid bilayer. To see this figure in color, go online.
D217E ASR crystals belong to a different spacegroup than the previously reported WT ASR crystals (spacegroup C2221) (28), possibly because of the presence of further additives required for crystallization, or a necessary second purification step, but there are similarities in crystal packing. D217E ASR and WT ASR share similar interfaces between protomers, including the hydrogen-bonded contacts between pairs of helix D that allow both proteins to crystallize as a dimer; the WT ASR interface contains six hydrogen-bonded contacts with a dimer interface surface of 682 Å2, whereas D217E ASR has seven contacts with an interface area of 701 Å2, as calculated with the program PISA (51). Crystal contacts between helix A and helix G (Fig. 1 b) cause an undulation in the type I protein-lipid layer, resulting in a bilayer that is nonplanar. This is in contrast to the majority of previously reported microbial rhodopsin structures crystallized from the lipid cubic phase, such as BR (5, 45) and HR (13), which contain planar bilayers; however, similar undulations have been observed in SRII (18) due to interactions between helices A and G. Structural studies of WT ASR using solid-state NMR suggest that this undulation is specific to the arrangement in crystals; the same study shows that WT ASR forms a trimer (52).
D217E ASR is a seven-transmembrane helical retinylidene protein with a backbone structure nearly identical to that of WT ASR, with a root mean square deviation (RMSD) of the backbone calculated as 0.313 Å using the SSM Superpose (53) function in the program Coot (49), with the largest deviations at C-terminal residues Asn223 to Asp226, which may be attributed to differences in crystal packing. Similar to WT ASR, D217E ASR does not show electron density for residues that comprise the extracellular BC loop. Of the seven helices, helix G shows the largest perturbations from an ideal α helix due to the presence of a proline residue at Pro206 (φ = −69.2°, ψ = −22.8°), as well as a π bulge at Ser209, Lys210, and Val211 (54); notably, α helix G of WT ASR is not disturbed by the proline at position 206, which in BR is Asp212 (28). In D217E ASR, minor changes in the main chain at Pro206 in comparison with WT ASR (∼0.24 Å) bring the Pro206 carbonyl closer to the neighboring Ser209 N-H, creating a 3.15 Å intrahelical hydrogen bond.
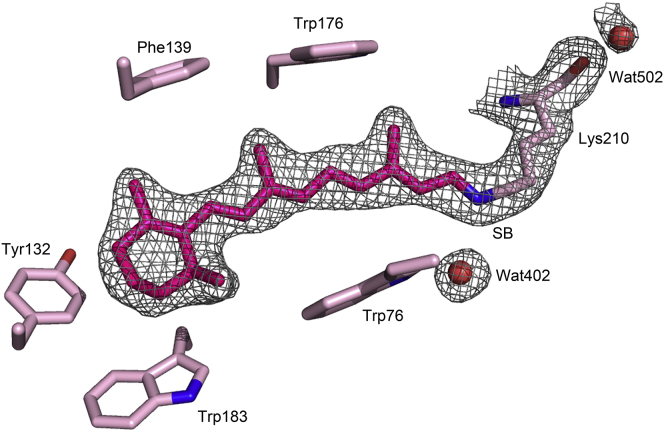
The dark-adapted D217E ASR structure contains only all-trans retinal, in contrast to the previously reported WT ASR structure (28), which showed a mixture of all-trans and 13-cis retinal due to white-light illumination before crystal harvesting (Fig. 2). A previous report based on the extraction of retinal isomers after different light illumination schemes showed that in dark-adapted ASR, the all-trans form of retinal predominates (28). This is supported by the electron density of dark-adapted D217E ASR, which does not appear to contain significant quantities of 13-cis retinal. This photochromism, the stable conversion between all-trans,15-anti retinal and 13-cis,15-syn retinal, appears unique to ASR among the known microbial rhodopsins. Unlike other microbial rhodopsins, ASR maintains different retinal configurations in its ground state depending on the wavelength of ambient light, requiring orange illumination (>560 nm) to stably convert the retinal chromophore to its 13-cis,15-syn configuration (55). Conversion of the retinal back to the all-trans configuration requires dark-adaptation or illumination with blue light (<470 nm) (56).
Figure 2.
Retinal binding pocket of D217E ASR. This simulated annealed omit map contoured at 1σ supports all-trans retinal as the predominant chromophore. Dark-adapted ASR has populations of all-trans retinal near unity, whereas orange light illumination (>580 nm) causes stable interconversion to 13-cis retinal (∼78%). To see this figure in color, go online.
Hydrogen-bond network in cytoplasmic half of D217E ASR
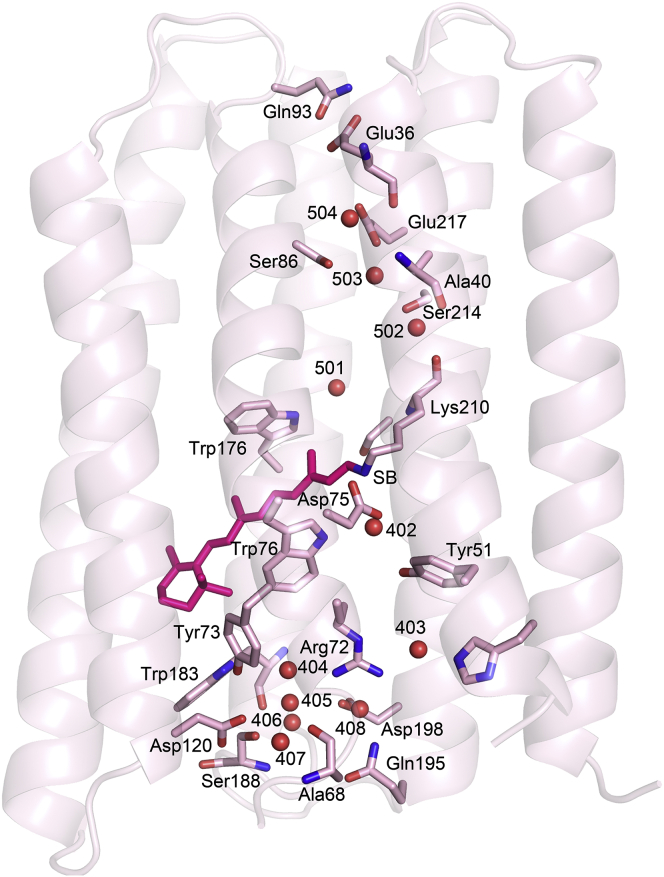
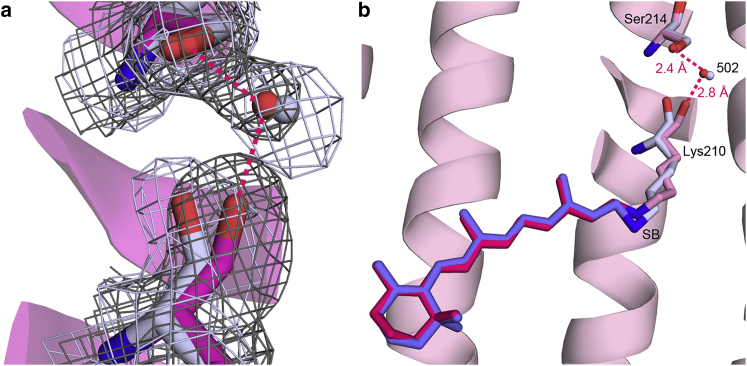
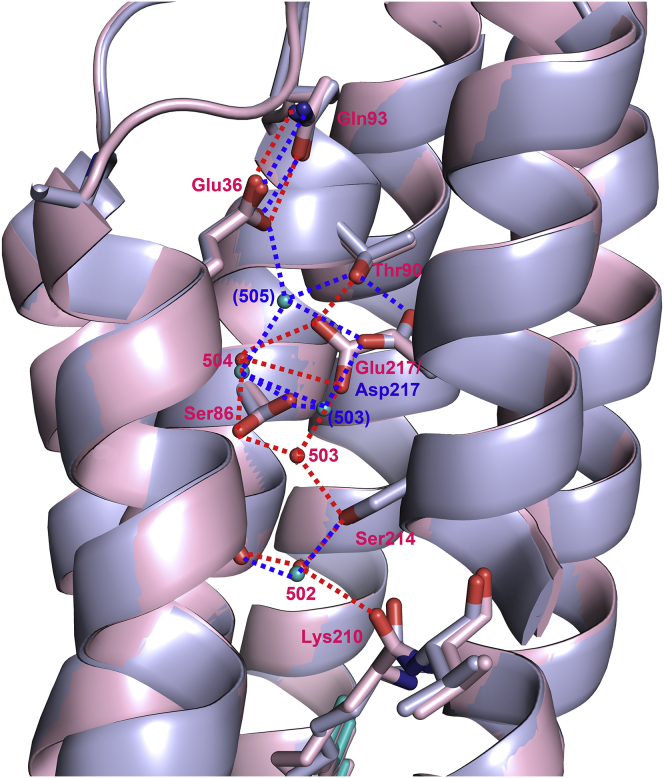
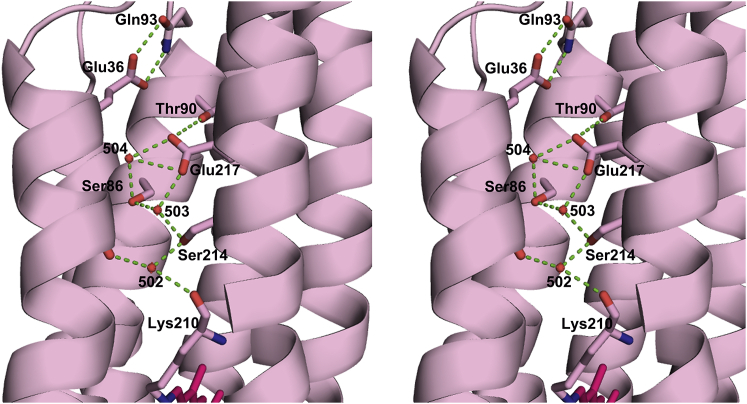
There are networks of ordered water molecules in both the cytoplasmic and extracellular halves of the protein (Fig. 3), the former of which is significantly rearranged due to the mutation of Asp217 to Glu. In D217E ASR, the peptide plane between Lys210 and Val211 is tilted from its orientation in WT ASR (Fig. 4 a) such that the Lys210 carbonyl is within hydrogen-bonding distance (2.8 Å) of nearby Wat502, which is part of a hydrogen-bonding network involving Ser214, Ser86, Glu217, Thr90, and two more ordered waters (Wat503, Wat504). The electron density strongly supports only one placement of the D217E ASR Lys210 C = O. In contrast, in examining the previously deposited structure (PDB: 1XIO) and electron density map for WT ASR (28), the conformation of WT ASR Lys210 associated with the all-trans retinal places the C = O outside the range of hydrogen-bond distances to Wat502, with a distance of 3.4 Å (Fig. 4 b). The difference between the D217E and WT ASR Lys210 carbonyl vectors is ∼68°.
Figure 3.
Internal waters in D217E ASR and their hydrogen-bonding partners. D217E ASR contains seven ordered waters in its extracellular domain, and four ordered waters in its cytoplasmic domain, which form a hydrogen-bonding network with several polar residues. The cytoplasmic region is shown at the top of the figure, with the extracellular surface at the bottom. The retinal is shown in magenta. To see this figure in color, go online.
Figure 4.
Rotation of the Lys210 peptide backbone creates a hydrogen bond between the carbonyl oxygen and Wat502. (a) An examination of the electron densities (2Fo-Fc contoured at 1σ) of WT ASR (light gray) and D217E ASR (pink) unambiguously supports a repositioning by ∼68° of the Lys210 C = O. (b) The newly positioned Lys210 C = O now accepts a hydrogen bond from Wat502, which is also hydrogen-bonded to Ser214. To see this figure in color, go online.
The region immediately surrounding the mutated residue 217 shows structural changes from WT ASR (Fig. 5). The Glu217 carboxyl replaces Wat505, which is no longer present in the mutant structure. For the WT, Wat505WT shared a hydrogen bond with Glu36WT (2.95 Å), as well as one of the side-chain oxygens of Asp217WT (2.95 Å). Glu217D217E also displaces Wat503, which is shifted by 1.8 Å in comparison to WT ASR, bringing it within hydrogen-bonding distance to Ser214D217E (2.7 Å, Fig. 6). In WT ASR, Ser214WT and Wat503WT are separated by 3.6 Å, which is outside the typical range for hydrogen bonds. Although there are two alternate conformations of the Ser86 side chain in WT ASR supported by electron density (not shown), there is only one conformation for Ser86D217E, which places the hydroxyl side chain away from the water network. The hydroxyl receives a hydrogen bond from Wat503D217E, which in turn is hydrogen-bonded to Wat504D217E; both waters donate hydrogen bonds to Glu217. Among the ordered waters, Wat502 and Wat504 are in nearly identical positions compared with WT, which is expected, as both are distant from the site of the D217E mutation.
Figure 5.
Comparison of WT and D217E ASR cytoplasmic hydrogen-bonding networks. WT ASR and its residues are drawn in blue with blue waters; D217E ASR is drawn in pink with red waters. The replacement of Asp217 with Glu extends the carboxyl into the interior of the protein, which replaces Wat505 in D217E ASR. The carboxylate of Glu217 also displaces Wat503 by 1.8 Å, such that it is now 2.7 Å from Ser214, within hydrogen-bonding distance. To see this figure in color, go online.
Figure 6.
Stereo view of the cytoplasmic half-channel of D217E ASR. A network of ordered waters and polar residues connects the Lys210 carbonyl with Glu217, which is protonated upon formation of the M intermediate of D21E ASR. The placement of the Glu217 carboxyl has replaced Wat505. Furthermore, Wat503 is displaced, forming a hydrogen bond with the Ser214 hydroxyl. Glu36, protonated in the L intermediate, is located near the cytoplasmic surface along with Gln93, and may serve as a proton gate, as well as the site of proton release into the cytoplasm. To see this figure in color, go online.
From the structure (Fig. 6), we obtain a more complete glimpse of the water network in the cytoplasmic half-channel, along with its hydrogen-bonding partners, beginning from the tilted Lys210D217E carbonyl. From there, a hydrogen-bond network can be traced to Glu217, 15 Å away, via two possible pathways (Lys210 C = O — Wat502 — Ser214 — Wat503 — Glu217) or (Lys210 C = O — Wat502 — Ser214 — Wat503— Ser86 — Wat504 — Glu217). The establishment of this hydrogen-bond network, caused by both the rotation of the Lys210 peptide backbone and the reorganization of ordered waters in the cytoplasmic half-channel, offers insight into the mechanism of proton-pumping of D217E, compared with that of the similarly hydrophilic WT ASR. We know from FTIR studies that upon formation of the M intermediate, the Schiff base is deprotonated, whereas Glu217 is protonated (40). Further rearrangement of hydrogen bonds upon light illumination may account for the eventual expulsion of a proton into the cytoplasm, perhaps via Glu36.
Recent studies have shown that at least one other microbial rhodopsin also possesses several ordered water molecules in the cytoplasmic half. The crystal structure of the sodium pump Krokinobacter eikastus rhodopsin 2 (KR2) reveals a hydrophilic cavity that protrudes from the protein surface, in which water molecules coordinate with interior polar side chains and backbone atoms (57, 58). Further experimentation showed that the cavity in KR2 is important in ion uptake, and in the absence of sodium and potassium ions, KR2 maintains a stable stationary current (58). This not only strengthens the importance of ordered cytoplasmic water molecules in microbial rhodopsins as it pertains to directional ion and proton movement, but indicates a structural importance in determining protein function, as seen with D217E ASR.
Protonated Glu36 at cytoplasmic face may act as gateway
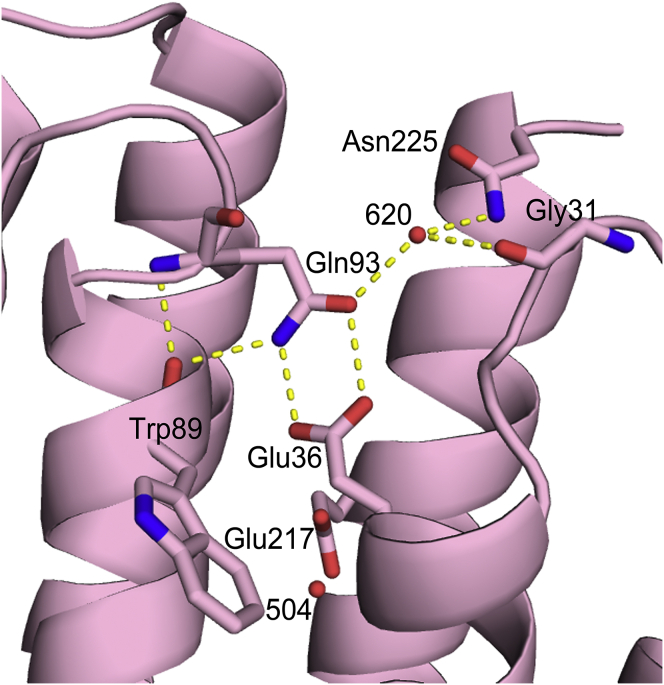
Near the cytoplasmic surface of the protein, residue Gln93 donates a hydrogen bond from its amide Nε to nearby Glu36 Oε2, while it receives a hydrogen bond from Glu36 Oε1 to its amide carbonyl Oε, indicating that in the crystal structures for both WT ASR and D217E ASR, Glu36 is protonated (Fig. 7). The carboxyl of Glu36 and the amide of Gln93 are in plane, with the distances being 2.99 Å (chain B: 3.02 Å) between Gln93 Oε and Glu36 Oε1, and 2.75 Å (chain B: 2.56 Å) between Gln93 Nε and Glu36 Oε2. After careful analysis of the Gln93 side-chain B factors for D217E ASR, we decided to flip the Gln93 side-chain amide by 180° around the χ3 bond compared with the WT model. Examination of the resulting hydrogen-bond pattern also supports this change, which is summarized in Table 2. Such a B factor analysis was performed for WT ASR as well and it was determined that the placement of the Gln93 side chain for the deposited structure is correct. It is worth noting that Gln93 also shares a hydrogen bond with a neighboring water molecule that is exposed to the bulk solvent. Although both the crystal structures of D217E and WT ASR suggest that Glu36 is protonated due its hydrogen bonding with Gln93, there are inherent limitations in discussing water-exposed hydrogen bonds based on crystallographic structures solved at cryogenic temperatures.
Figure 7.
Evidence of Glu36 protonation. Residues within typical hydrogen-bonding distance (< 3.5 Å) are connected with yellow dashed lines. The carboxyl of Glu36 and the amide of Gln93 are in plane. The distance between Gln93 Oε and Glu36 Oε1 is 2.99 Å, whereas the distance between Gln93 Nε and Glu36 Oε2 is 2.75 Å. Based on crystallographic evidence, Glu36 appears to be protonated in the structure of dark-adapted D217E ASR. The possibility arises that upon photon absorption in D217E ASR, the Glu36 proton is released into the cytoplasm, whereupon it is reprotonated by the now-protonated Glu217. To see this figure in color, go online.
Table 2.
Summary of Residues Located within Typical Hydrogen-Bonding Distance with Ordered Waters in the Extracellular Domain for Both WT ASR and D217E ASR
| Residue (H-Bond Distance (Å)) WT | Water | Water RMSD (Å) | Residue (H-Bond Distance (Å)) D217E |
|---|---|---|---|
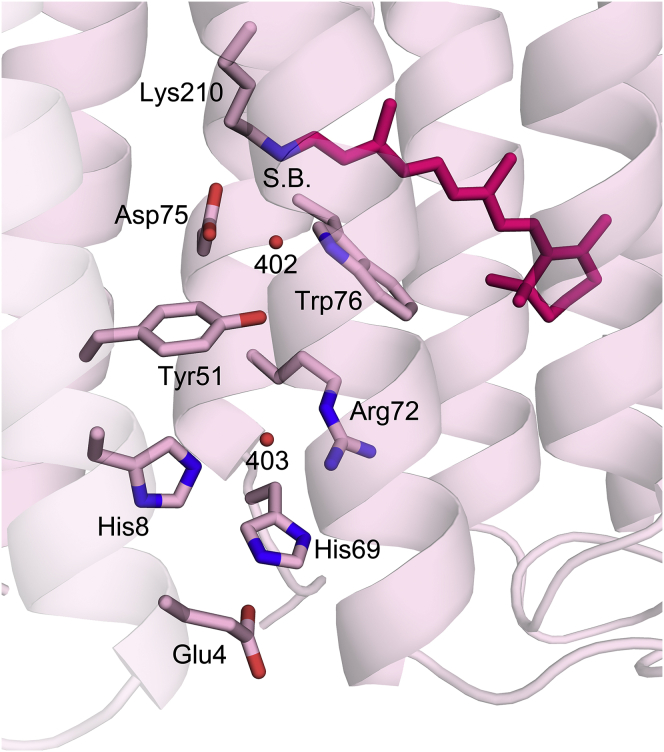
| ATR Schiff base (2.57), Tyr51 (2.92), Asp75 (2.74), Trp76 (2.94) | 402 | 0.22 | ATR Schiff base (3.13), Tyr51 (2.99), Asp75 (2.91), Trp76 (2.88) |
| Arg72 NH1 (3.14) | 403 | 0.91 | His8 (2.74), Arg72 NH1 (2.74) |
| Arg72 (3.14), Tyr73 (3.18), Wat405 (2.96) | 404 | 0.87 | Arg72 (2.67), Tyr73 (3.02), Wat405 (2.50) |
| Ser188 (2.81), Wat406 (2.81) | 405 | 0.49 | Ser188 (2.71), Wat406 (2.88) |
| Trp183 C = O (2.79), Ser188 NH (2.96), Asp198 (3.16) | 406 | 0.15 | Trp183 C = O (2.72), Ser188 NH (2.98), Asp198 (3.10) |
| Ala68 C = O (2.96), Asp120 (2.59) | 407 | 0.43 | Ala68 C = O (2.96), Asp120 (2.48) |
| Arg72 NH2 (2.98), Gln195 (2.58), Asp198 (2.56) | 408 | 1.72 | Arg72 NH1 (3.14), Arg72 NH2 (3.06), Gln195 (2.32), Asp198 (2.25) |
Previous FTIR studies with WT ASR not only showed an alteration of hydrogen bonding of a water but also of Glu36, ∼20 Å from the Schiff base, as early as during the formation of the L state at 170K (42). This work also demonstrated that Glu36WT displays an elevated pKa (between 6 and 7). The latter finding supports a protonated Glu36 since ASR crystals were grown at pH 5.6. Furthermore, pKa calculations for the dark-adapted, ground-state of D217E ASR suggest a pKa of 7.0 for the Glu36 carboxylate (59). And although in the published white-illuminated WT ASR structure, the positioning of the Glu36 carboxylate and the Gln93 amide carbonyl implies that Glu36 is protonated, it is possible that in a dark-adapted ground state, for which a crystal structure is not available, conformational and resulting pKa changes may lead to a deprotonated Glu36.
In the dark-adapted D217E ASR structure, Glu36 Oε2 is located ∼3.7 Å from the nearest side-chain oxygen of Glu217. Although the details of the proton release mechanism are still unclear, it is possible that in D217E ASR, upon photon absorption, a proton is expelled into the cytoplasm from Glu36, which is subsequently reprotonated by protonated Glu217. Incidentally, in comparison with the arrangement in archaeal rhodopsins such as BR and SRII, Gln93 is part of a four-residue insertion in the C-D loop, which in concert with the A-B and E-F loops may interact with ASR’s cytoplasmic transducer, ASRT (29).
Extracellular half-channel of D217E ASR
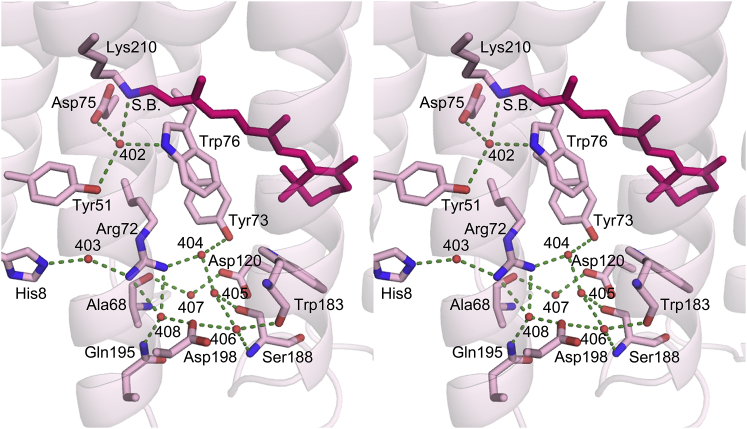
In the extracellular portion of both D217E and WT ASR, there are seven water molecules, the positions of which are generally similar in both structures, although the repositioning of some of the waters leads to the formation of new hydrogen bonds. Wat403D217E is displaced 0.91 Å from Wat403WT, resulting in a new hydrogen bond between Wat403D217E and His8D217E (2.74 Å) and a shortened bond between the water and Arg72 (2.74 Å versus 3.14 Å in WT). Furthermore, a difference in the side-chain rotamer of Asp198 displaces Wat408 by 1.72 Å. And whereas Wat408WT is hydrogen-bonded to Gln195 Nε, Asp198 Oδ2, Thr199 Oγ, and Arg72 NH2, Wat408D217E forms hydrogen bonds with Gln195 Nε, Asp198 Oδ2, and both Arg72 NH1 and NH2 (Fig. 8).
Figure 8.
Stereo view of the extracellular domain of D217E ASR. Ordered internal waters (Wat 402–408) are shown as red spheres. Hydrogen bonds have been drawn between waters and their corresponding binding partners. Distances are listed in Table 2. The number of waters is conserved between D217E ASR and WT ASR, although Wat403 and Wat408 are displaced by 0.91 Å and 1.72 Å, respectively. The movement of Wat403 potentially creates a new hydrogen bond with His8. To see this figure in color, go online.
In the region of the retinal, there is a water molecule (Wat402) that is strongly hydrogen-bonded to the Schiff base in dark-adapted ASR-ATR, but is only weakly hydrogen-bonded to the Schiff base counterion Asp75 (41). In BR, it is possible that the counter ion (Asp85BR) accepts the Schiff base proton via the analogous Wat402BR during the L-to-M transition, but in contrast to the BR photocycle, Asp75 is never protonated during the ASR photocycle (41). This supports the published theory that for ASR, although the Schiff base proton is transferred to Asp217 in the cytoplasmic half-channel, the Schiff base is also reprotonated from the same residue, resulting in no net proton transport across the membrane (39). Given the robust inward proton transporting nature of D217E ASR, in the mutant the Schiff base is likely reprotonated from the extracellular side, from a residue other than Asp75.
Interestingly, there are two histidines near the extracellular surface (His8 and His69), one or both of which may be involved in vectorial reprotonation of the Schiff base. The residues are located on adjacent helices, with His8 on helix A and His69 in the turn immediately preceding helix C (Fig. 9). Experimentally determined pKa values for either His8 or His69 are not available, but are predicted to be 3.62 and 4.22, respectively, using the program PROPKA (59). In contrast, BR does not possess any histidine residues at all, and an alignment of the primary sequences for ASR, BR, HR, SRI, SRII, as well as xanthorhodopsin and proteorhodopsin (Polaribacter sp MED152) (60) shows no conservation of either histidine in those positions (not shown). Therefore, either His8 or His69 could be a potential proton donor to the Schiff base, while subsequently taking up a proton from the extracellular surface or via the hydrogen-bonding network comprised of waters 403–408, and residues Ala68, Arg72, Tyr73, Asp120, Trp183, Ser188, Gln195, and Asp198 (Fig. 8). One possible mechanism of reprotonation from the extracellular side thus entails the movement of the Arg72 guanidinium moiety in response to Schiff base deprotonation (analogous to the role of Arg82 in BR (61), located between the Schiff base and the pair of histidines that connect to the extracellular bulk via exposed Glu4.
Figure 9.
Histidines near the extracellular surface may serve as potential proton donors for the Schiff base. Located on helices A and C, neither His8 nor His69 are conserved among the prototypic ion pumping or sensory rhodopsins. Their proximity to the solvent-accessible surface, as well as their innate ability to be protonated at physiological pH make them an intriguing possibility as the extracellular proton donor in D217E ASR. Although the WT ASR Schiff base is likely reprotonated from the cytoplasm, the strong proton transport activity in D217E ASR makes it likely that reprotonation originates from the extracellular domain. To see this figure in color, go online.
Conclusions
Significant structural changes in the cytoplasmic hydrogen-bonding network in D217E ASR at 2.3 Å shed light on the D217E ASR’s unique activity as a single-photon-driven inward proton transporter. The Lys210 carbonyl is tilted 68° toward conserved Wat502, bringing its oxygen within hydrogen-bonding distance and completing a network of residues and ordered waters that trace a putative proton translocation pathway from the Schiff base to Glu217 located 15 Å in the direction of the cytoplasmic surface. Two of the cytoplasmic waters in WT ASR are displaced by the mutation of Asp217 to Glu217, bringing Wat503 within hydrogen-bonding distance to Ser214, while Wat505 is lost entirely. Meanwhile, the Ser86 hydroxyl is forced to adopt a single conformation away from the center of the water network, while maintaining its hydrogen bonds with Wat503 and Wat504. Using this structure, we can now envision how a proton might be transferred to Glu217 upon deprotonation of the Schiff base during formation of the M intermediate. Later in the proton transport cycle, based on analysis of the structure of the extracellular domain of D217E ASR, we have identified a pair of histidine residues (His8, His69) near the extracellular surface that has the potential to act as a proton donor for the reprotonation of the Schiff base; however, additional mutagenesis studies will be necessary.
We also examine crystallographic evidence for the protonation of Glu36, which is positioned near the cytoplasmic surface of D217E and WT ASR. Its protonation is inferred from the proximity between the Glu36 carboxyl and the Gln93 amide, which are coplanar and within typical hydrogen-bonding distance. The D217E ASR structure differs from WT ASR in this region in that the amide side chain of Gln93 has been rotated 180° around the χ3 bond, whereas analysis of side-chain B factors for the WT structure indicates that the original model does not need to be modified. The protonation of Glu36 and its position near the cytoplasmic surface suggest this residue as the site of proton release into the cytoplasm, as well as a switch for the interaction with the ASR transducer, ASRT.
Author Contributions
B.D. performed the majority of the research, analyzed data, and wrote the article. L.S.M. assisted with certain aspects of protein expression, purification, and crystallization. H.L. designed the research, analyzed the x-ray data, and wrote the article.
Acknowledgments
Plasmid pKJ900 with wild-type ASR, plasmid βORANGE, and E. coli strain UT5600 were a generous gift from Dr. Kwang-Hwan Jung at the Department of Life Science at Sogang University in Seoul, South Korea. We are grateful to Janos Lanyi for discussions, and we thank the beamline staff at the Stanford Synchrotron Radiation Laboratory (SSRL) for their support. The atomic coordinates and structure factors of D217E Anabaena sensory rhodopsin are available at the Protein Data Bank with code 4TL3.
This research was supported by NIH grants R01 GM067808 and AI078000, the UC Irvine Center for Biomembrane Systems, and a UCI Chancellor’s Fellowship. L.S.M. holds a JAE-Doc fellowship from CSIC, Spain. Additionally, we acknowledge support from the Ikerbasque Foundation and the Spanish Ministerio de Economía, grant BFU-2012-36241.
Editor: Leonid Brown.
References
- 1.Venter J.C., Remington K., Smith H.O. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 2.Zhao M., Chen F., Jiao N. Genetic diversity and abundance of flavobacterial proteorhodopsin in China seas. Appl. Environ. Microbiol. 2009;75:529–533. doi: 10.1128/AEM.01114-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathies R.A., Lin S.W., Pollard W.T. From femtoseconds to biology: mechanism of bacteriorhodopsin’s light-driven proton pump. Annu. Rev. Biophys. Biophys. Chem. 1991;20:491–518. doi: 10.1146/annurev.bb.20.060191.002423. [DOI] [PubMed] [Google Scholar]
- 4.Grigorieff N., Ceska T.A., Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J. Mol. Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 5.Luecke H., Schobert B., Lanyi J.K. Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 6.Luecke H., Richter H.T., Lanyi J.K. Proton transfer pathways in bacteriorhodopsin at 2.3 angstrom resolution. Science. 1998;280:1934–1937. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- 7.Kimura Y., Vassylyev D.G., Fujiyoshi Y. Surface of bacteriorhodopsin revealed by high-resolution electron crystallography. Nature. 1997;389:206–211. doi: 10.1038/38323. [DOI] [PubMed] [Google Scholar]
- 8.Takeda K., Sato H., Kouyama T. A novel three-dimensional crystal of bacteriorhodopsin obtained by successive fusion of the vesicular assemblies. J. Mol. Biol. 1998;283:463–474. doi: 10.1006/jmbi.1998.2103. [DOI] [PubMed] [Google Scholar]
- 9.Pebay-Peyroula E., Rummel G., Landau E.M. X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases. Science. 1997;277:1676–1681. doi: 10.1126/science.277.5332.1676. [DOI] [PubMed] [Google Scholar]
- 10.Essen L., Siegert R., Oesterhelt D. Lipid patches in membrane protein oligomers: crystal structure of the bacteriorhodopsin-lipid complex. Proc. Natl. Acad. Sci. USA. 1998;95:11673–11678. doi: 10.1073/pnas.95.20.11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitsuoka K., Hirai T., Fujiyoshi Y. The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution. J. Mol. Biol. 1999;286:861–882. doi: 10.1006/jmbi.1998.2529. [DOI] [PubMed] [Google Scholar]
- 12.Otomo J. Anion selectivity and pumping mechanism of halorhodopsin. Biophys. Chem. 1995;56:137–141. doi: 10.1016/0301-4622(95)00025-s. [DOI] [PubMed] [Google Scholar]
- 13.Kolbe M., Besir H., Oesterhelt D. Structure of the light-driven chloride pump halorhodopsin at 1.8 A resolution. Science. 2000;288:1390–1396. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- 14.Váró G., Brown L.S., Lanyi J.K. Proton transport by halorhodopsin. Biochemistry. 1996;35:6604–6611. doi: 10.1021/bi9601159. [DOI] [PubMed] [Google Scholar]
- 15.Bogomolni R.A., Spudich J.L. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc. Natl. Acad. Sci. USA. 1982;79:6250–6254. doi: 10.1073/pnas.79.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spudich J.L., Bogomolni R.A. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984;312:509–513. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoff W.D., Jung K.H., Spudich J.L. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu. Rev. Biophys. Biomol. Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 18.Royant A., Nollert P., Navarro J. X-ray structure of sensory rhodopsin II at 2.1-A resolution. Proc. Natl. Acad. Sci. USA. 2001;98:10131–10136. doi: 10.1073/pnas.181203898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolff E.K., Bogomolni R.A., Stoeckenius W. Color discrimination in halobacteria: spectroscopic characterization of a second sensory receptor covering the blue-green region of the spectrum. Proc. Natl. Acad. Sci. USA. 1986;83:7272–7276. doi: 10.1073/pnas.83.19.7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi T., Tomioka H., Kobatake Y. A photosystem other than PS370 also mediates the negative phototaxis of Halobacterium halobium. FEMS Microbiol. Lett. 1985;28:161–164. [Google Scholar]
- 21.Facciotti M.T., Rouhani S., Glaeser R.M. Crystal structures of bR(D85S) favor a model of bacteriorhodopsin as a hydroxyl-ion pump. FEBS Lett. 2004;564:301–306. doi: 10.1016/S0014-5793(04)00208-X. [DOI] [PubMed] [Google Scholar]
- 22.Herzfeld J., Lansing J.C. Magnetic resonance studies of the bacteriorhodopsin pump cycle. Annu. Rev. Biophys. Biomol. Struct. 2002;31:73–95. doi: 10.1146/annurev.biophys.31.082901.134233. [DOI] [PubMed] [Google Scholar]
- 23.Luecke H. Atomic resolution structures of bacteriorhodopsin photocycle intermediates: the role of discrete water molecules in the function of this light-driven ion pump. Biochim. Biophys. Acta. 2000;1460:133–156. doi: 10.1016/s0005-2728(00)00135-3. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki J., Brown L.S., Lanyi J.K. Conversion of bacteriorhodopsin into a chloride ion pump. Science. 1995;269:73–75. doi: 10.1126/science.7604281. [DOI] [PubMed] [Google Scholar]
- 25.Spudich E.N., Ozorowski G., Luecke H. A transporter converted into a sensor, a phototaxis signaling mutant of bacteriorhodopsin at 3.0 Å. J. Mol. Biol. 2012;415:455–463. doi: 10.1016/j.jmb.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tittor J., Schweiger U., Bamberg E. Inversion of proton translocation in bacteriorhodopsin mutants D85N, D85T, and D85,96N. Biophys. J. 1994;67:1682–1690. doi: 10.1016/S0006-3495(94)80642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung K.H., Trivedi V.D., Spudich J.L. Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 2003;47:1513–1522. doi: 10.1046/j.1365-2958.2003.03395.x. [DOI] [PubMed] [Google Scholar]
- 28.Vogeley L., Sineshchekov O.A., Luecke H. Anabaena sensory rhodopsin: a photochromic color sensor at 2.0 A. Science. 2004;306:1390–1393. doi: 10.1126/science.1103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogeley L., Trivedi V.D., Luecke H. Crystal structure of the Anabaena sensory rhodopsin transducer. J. Mol. Biol. 2007;367:741–751. doi: 10.1016/j.jmb.2006.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S., Kim S.Y., Brown L.S. A eukaryotic-like interaction of soluble cyanobacterial sensory rhodopsin transducer with DNA. J. Mol. Biol. 2011;411:449–462. doi: 10.1016/j.jmb.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Irieda H., Morita T., Sudo Y. Photo-induced regulation of the chromatic adaptive gene expression by Anabaena sensory rhodopsin. J. Biol. Chem. 2012;287:32485–32493. doi: 10.1074/jbc.M112.390864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown L.S. Eubacterial rhodopsins—unique photosensors and diverse ion pumps. Biochim. Biophys. Acta. 2014;1837:553–561. doi: 10.1016/j.bbabio.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Kim S.Y., Yoon S.R., Jung K.H. A role of Anabaena sensory rhodopsin transducer (ASRT) in photosensory transduction. Mol. Microbiol. 2014;93:403–414. doi: 10.1111/mmi.12635. [DOI] [PubMed] [Google Scholar]
- 34.Kim S.H., Kim S.Y., Kim D. DNA binding activity of Anabaena sensory rhodopsin transducer probed by fluorescence correlation spectroscopy. Biosci. Biotechnol. Biochem. 2015;79:1070–1074. doi: 10.1080/09168451.2015.1015950. [DOI] [PubMed] [Google Scholar]
- 35.Luecke H., Schobert B., Spudich J.L. Crystal structure of sensory rhodopsin II at 2.4 angstroms: insights into color tuning and transducer interaction. Science. 2001;293:1499–1503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordeliy V.I., Labahn J., Engelhard M. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature. 2002;419:484–487. doi: 10.1038/nature01109. [DOI] [PubMed] [Google Scholar]
- 37.Sineshchekov O.A., Trivedi V.D., Spudich J.L. Photochromicity of Anabaena sensory rhodopsin, an atypical microbial receptor with a cis-retinal light-adapted form. J. Biol. Chem. 2005;280:14663–14668. doi: 10.1074/jbc.M501416200. [DOI] [PubMed] [Google Scholar]
- 38.Kawanabe A., Kandori H. Photoreactions and structural changes of Anabaena sensory rhodopsin. Sensors (Basel) 2009;9:9741–9804. doi: 10.3390/s91209741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi L., Yoon S.R., Brown L.S. Cytoplasmic shuttling of protons in Anabaena sensory rhodopsin: implications for signaling mechanism. J. Mol. Biol. 2006;358:686–700. doi: 10.1016/j.jmb.2006.02.036. [DOI] [PubMed] [Google Scholar]
- 40.Kawanabe A., Furutani Y., Kandori H. Engineering an inward proton transport from a bacterial sensor rhodopsin. J. Am. Chem. Soc. 2009;131:16439–16444. doi: 10.1021/ja904855g. [DOI] [PubMed] [Google Scholar]
- 41.Furutani Y., Kawanabe A., Kandori H. FTIR spectroscopy of the all-trans form of Anabaena sensory rhodopsin at 77 K: hydrogen bond of a water between the Schiff base and Asp75. Biochemistry. 2005;44:12287–12296. doi: 10.1021/bi050841o. [DOI] [PubMed] [Google Scholar]
- 42.Kawanabe A., Furutani Y., Kandori H. FTIR study of the L intermediate of Anabaena sensory rhodopsin: structural changes in the cytoplasmic region. Biochemistry. 2008;47:10033–10040. doi: 10.1021/bi800941a. [DOI] [PubMed] [Google Scholar]
- 43.Ran T., Ozorowski G., Luecke H. Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes. Acta Crystallogr. D Biol. Crystallogr. 2013;69:1965–1980. doi: 10.1107/S0907444913017575. [DOI] [PubMed] [Google Scholar]
- 44.Kim S.Y., Waschuk S.A., Jung K.H. Screening and characterization of proteorhodopsin color-tuning mutations in Escherichia coli with endogenous retinal synthesis. Biochim. Biophys. Acta. 2008;1777:504–513. doi: 10.1016/j.bbabio.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Landau E.M., Rosenbusch J.P. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. USA. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski Z., Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.Winn M.D., Ballard C.C., Wilson K.S. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy A.J., Grosse-Kunstleve R.W., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Vagin A.A., Steiner R.A., Murshudov G.N. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 51.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 52.Wang S., Munro R.A., Ladizhansky V. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nat. Methods. 2013;10:1007–1012. doi: 10.1038/nmeth.2635. [DOI] [PubMed] [Google Scholar]
- 53.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 54.Cartailler J.P., Luecke H. Structural and functional characterization of π bulges and other short intrahelical deformations. Structure. 2004;12:133–144. doi: 10.1016/j.str.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Kawanabe A., Furutani Y., Kandori H. Photochromism of Anabaena sensory rhodopsin. J. Am. Chem. Soc. 2007;129:8644–8649. doi: 10.1021/ja072085a. [DOI] [PubMed] [Google Scholar]
- 56.Tahara S., Kato Y., Ohtani H. pH-dependent photoreaction pathway of the all-trans form of Anabaena sensory rhodopsin. J. Phys. Chem. B. 2013;117:2053–2060. doi: 10.1021/jp311217a. [DOI] [PubMed] [Google Scholar]
- 57.Kato H.E., Inoue K., Nureki O. Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump. Nature. 2015;521:48–53. doi: 10.1038/nature14322. [DOI] [PubMed] [Google Scholar]
- 58.Gushchin I., Shevchenko V., Gordeliy V. Crystal structure of a light-driven sodium pump. Nat. Struct. Mol. Biol. 2015;22:390–395. doi: 10.1038/nsmb.3002. [DOI] [PubMed] [Google Scholar]
- 59.Olsson M.H., Søndergaard C.R., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 60.González J.M., Fernández-Gómez B., Pedrós-Alió C. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria) Proc. Natl. Acad. Sci. USA. 2008;105:8724–8729. doi: 10.1073/pnas.0712027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luecke H., Schobert B., Lanyi J.K. Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution. Science. 1999;286:255–260. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]