ABSTRACT
The euryhaline cyanobacterium Synechococcus sp. strain PCC 7002 has an obligate requirement for exogenous vitamin B12 (cobalamin), but little is known about the roles of this compound in cyanobacteria. Bioinformatic analyses suggest that only the terminal enzyme in methionine biosynthesis, methionine synthase, requires cobalamin as a coenzyme in Synechococcus sp. strain PCC 7002. Methionine synthase (MetH) catalyzes the transfer of a methyl group from N5-methyl-5,6,7,8-tetrahydrofolate to l-homocysteine during l-methionine synthesis and uses methylcobalamin as an intermediate methyl donor. Numerous bacteria and plants alternatively employ a cobalamin-independent methionine synthase isozyme, MetE, that catalyzes the same methyl transfer reaction as MetH but uses N5-methyl-5,6,7,8-tetrahydrofolate directly as the methyl donor. The cobalamin auxotrophy of Synechococcus sp. strain PCC 7002 was complemented by using the metE gene from the closely related cyanobacterium Synechococcus sp. strain PCC 73109, which possesses genes for both methionine synthases. This result suggests that methionine biosynthesis is probably the sole use of cobalamin in Synechococcus sp. strain PCC 7002. Furthermore, a cobalamin-repressible gene expression system was developed in Synechococcus sp. strain PCC 7002 that was used to validate the presence of a cobalamin riboswitch in the promoter region of metE from Synechococcus sp. strain PCC 73109. This riboswitch acts as a cobalamin-dependent transcriptional attenuator for metE in that organism.
IMPORTANCE Synechococcus sp. strain PCC 7002 is a cobalamin auxotroph because, like eukaryotic marine algae, it uses a cobalamin-dependent methionine synthase (MetH) for the final step of l-methionine biosynthesis but cannot synthesize cobalamin de novo. Heterologous expression of metE, encoding cobalamin-independent methionine synthase, from Synechococcus sp. strain PCC 73109, relieved this auxotrophy and enabled the construction of a truly autotrophic Synechococcus sp. strain PCC 7002 more suitable for large-scale industrial applications. Characterization of a cobalamin riboswitch expands the genetic toolbox for Synechococcus sp. strain PCC 7002 by providing a cobalamin-repressible expression system.
INTRODUCTION
Synechococcus sp. strain PCC 7002 is a euryhaline, unicellular cyanobacterium that tolerates high light intensities and a wide range of sodium chloride concentrations (1, 2). This organism has one of the highest growth rates known among cyanobacteria (3), has a fully sequenced genome, and is naturally transformable (2, 4). A versatile system for genetic complementation and overexpression has been developed for this organism (5). Despite generally being considered to be photoautotrophic, Synechococcus sp. strain PCC 7002 has an obligate requirement for exogenous cobalamin (6), a large and structurally complex cobalt-chelating tetrapyrrole compound. Although cobalamin can only be synthesized de novo by some eubacteria and archaea, it is widely used as a coenzyme by many organisms, including eukaryotes, which cannot synthesize cobalamin and must obtain it from the environment (7, 8).
Similar to Synechococcus sp. strain PCC 7002, various cyanobacteria encompassing the genera Dermocarpa, Synechocystis, and Pleurocapsa, as well as other Synechococcus spp., also share this absolute requirement for exogenous cobalamin (9). In some prokaryotes, cobalamin acts as an essential coenzyme during the anaerobic fermentation of small molecules for growth, but it can also have roles in methionine synthesis and ribonucleotide reduction, functions that are common in aerobic organisms (8). Synechococcus sp. strain PCC 7002 performs oxygenic photosynthesis and can also perform aerobic respiration in the dark to provide energy for its metabolic processes (10). When aerobic respiration depletes the available oxygen, these photoautotrophs resort to fermentation to produce ATP for cellular processes (10). Only one annotated gene in Synechococcus sp. strain PCC 7002, that for cobalamin-dependent methionine synthase (metH), encodes an enzyme known to require cobalamin as a coenzyme.
Methionine synthases catalyze the transfer of a methyl group from N5-methyl-5,6,7,8-tetrahydrofolate to l-homocysteine in the final step in the biosynthesis of l-methionine (11). Two unrelated isozymes catalyze this methyl transfer: cobalamin-dependent methionine synthase (MetH, EC 2.1.1.13) and cobalamin-independent methionine synthase (MetE, EC 2.1.1.14). The distribution of the genes for these two enzymes varies widely. In plants, metE is common because these organisms do not synthesize cobalamin, but in mammals, metH is usually present because they can take up cobalamin from their food sources or acquire cobalamin from their intestinal microbiome. In bacteria and archaea, the distribution of metE and metH is less predictable, and although only metH is present in the Synechococcus sp. strain PCC 7002 genome, many eubacteria and some cyanobacteria have genes for both isozymes (12). Unlike the cyanobacterium Synechocystis sp. strain PCC 6803, which also solely utilizes MetH for methionine synthesis but does not require exogenous cobalamin as a growth factor (13, 14), Synechococcus sp. strain PCC 7002 is not capable of de novo cobalamin biosynthesis and thus is naturally a cobalamin auxotroph (6).
Bacteria with both metH and metE genes can take advantage of switching between the two isozymes to acclimate to changes in the supply of exogenous cobalamin (15). This regulation is usually modulated by cobalamin riboswitches (16). Riboswitches are highly structured elements in the 5′ untranslated leader regions of mRNAs that allow control of that gene by a metabolite, such as cobalamin, by inducing conformational changes in the RNA that result in attenuation at either the transcriptional or the translational level (17). Cobalamin riboswitches are widespread in prokaryotes (18) and are capable of sensing different vitamers of cobalamin with various affinities (19). The use of cobalamin riboswitches to regulate methionine synthase is not unique. Cobalamin is also known to regulate the btu genes responsible for its active transport (20), and the cob operon responsible for its de novo biosynthesis (21). Cobalamin riboswitches that control the transcription of the class Ia ribonucleotide reductase genes, nrdABS, are also known (e.g., in Streptomyces coelicolor [22]).
Because Synechococcus sp. strain PCC 7002 previously could not be studied in the complete absence of cobalamin, the effects of cobalamin limitation were little studied in this cyanobacterium until now. In this study, we show that the cobalamin auxotrophy of Synechococcus sp. strain PCC 7002 can be alleviated by complementation with the metE gene from a closely related cyanobacterium. The facile deletion of the native metH gene from metE-complemented Synechococcus sp. strain PCC 7002 with no evidence of any deleterious phenotype implies that cobalamin might solely be needed for the functioning of this single enzyme. Furthermore, a cobalamin riboswitch that potentially acts as a transcriptional attenuator was identified and functionally verified in the metE promoter of Synechococcus sp. strain PCC 73109. This riboswitch was used to create a cobalamin-repressible gene expression system in Synechococcus sp. strain PCC 7002.
MATERIALS AND METHODS
Strains, culture conditions and transformation procedure.
All of the cyanobacterial strains utilized in this study were obtained from the Pasteur Culture Collection, Institut Pasteur, Paris, France (9). A summary of the strains constructed and the plasmids utilized in this work can be found in Table 1. Wild-type (WT) Synechococcus sp. strain PCC 7002 was grown in medium A supplemented with 1 mg of NaNO3 ml–1 (designated medium A+), which additionally contains 4 μg of cobalamin (as cyanocobalamin) liter−1 (4, 23, 24). In order to evaluate if selected strains could grow in the absence of cobalamin, medium A+ was made without cobalamin (medium A+ B12–). WT Synechococcus sp. strain PCC 7002, Synechococcus sp. strain PCC 73109, and Synechococcus sp. strain PCC 7002 mutants (described below) were grown photoautotrophically in 20-mm culture tubes containing medium A+ or A+ B12– at 38°C with continuous cool white fluorescent illumination at 250 μmol of photons m–2 s–1 and sparging with 1% (vol/vol) CO2 in air, otherwise referred to as “standard conditions” (2). For mutant strains, the following antibiotic concentrations were supplied when appropriate: 50 μg of spectinomycin ml–1; 100 μg of kanamycin ml–1; 20 μg of erythromycin ml–1; and 20 μg of gentamicin ml–1. Transformation of Synechococcus sp. strain PCC 7002 was performed as described previously (5).
TABLE 1.
Bacterial strains and plasmids used in this study
| Plasmid or strain | Relevant characteristics | Reference(s) or source |
|---|---|---|
| Plasmids | ||
| pRL409 | Positive-selection cloning vector, ermC (Emr) | 30, 31 |
| pSRA81 | pUC19 containing aadA from pHP45Ω lacking transcription terminators (Smr Spr) | 27, 28 |
| pRL161 | Positive-selection cloning vector, aphAII (Kmr) | 30 |
| pAQ1Ex | pGEM-7zf pMB1 vector backbone with Synechococcus sp. strain PCC 7002 pAQ1 flanking sites, Spr | 5 |
| pAQ1Ex-PcpcBA[metE] | pAQ1Ex metE-expressing vector under control of PcpcBA, Spr | This study |
| pAQ1Ex-PpsbA2[metE] | pAQ1Ex metE-expressing vector under control of PpsbA2, Spr | This study |
| pAQ3Ex | pGEM-7zf pMB1 vector backbone with Synechococcus sp. strain PCC 7002 pAQ3 endogenous plasmid flanking sites, Kmr | 5 |
| pAQ3Ex-PmetE[yfp] | pAQ3Ex yfp-expressing vector under control of PmetE, Kmr | This study |
| pAQ3Ex-Pfused[yfp] | pAQ3Ex yfp-expressing vector under control of Φ(PcpcBA-PmetE), Kmr | This study |
| pAQ3Ex-PmetE-1[yfp] | pAQ3Ex yfp-expressing vector with mutated PmetE (PmetE-1), Kmr | This study |
| Strains | ||
| Synechococcus sp. strain PCC 73109 | 9 | |
| Synechococcus sp. strain PCC 7002 | 9 | |
| Modified forms of Synechococcus sp. strain PCC 7002 | ||
| AAP001 | pAQ1Ex-PcpcBA[metE] | This study |
| AAP002 | pAQ1Ex-PcpcBA[metE] ΔmetH::ermC | This study |
| AAP003 | pAQ1Ex-PcpcBA[metE]/pAQ3Ex-PmetE[yfp] | This study |
| AAP004 | pAQ1Ex-PpsbA2[metE] | This study |
| AAP005 | pAQ1Ex-PpsbA2[metE]/pAQ3Ex-Pfused[yfp] | This study |
| AAP006 | pAQ1Ex-PcpcBA[metE]/pAQ3Ex-PmetE[yfp] ΔmetH::ermC | This study |
| AAP007 | pAQ1Ex-PcpcBA[metE]/pAQ3Ex-PmetE-1[yfp] ΔmetH::ermC | |
| E. coli Top10F′ | F′ [lacIq Tn10(Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
PCR amplification, digestions, and ligations.
The PCR primers utilized in this study are listed in Table S1 in the supplemental material. The PCR amplifications in this work were performed with Phusion High-Fidelity DNA polymerase (catalog number M0530S) from New England BioLabs Inc. (Ipswich, MA). Purification of the PCR products prior to digestion was performed as described previously (5). Genomic DNA was isolated as described previously (25). Digested fragments were purified by electrophoresis on 0.8% (wt/vol) agarose gels with the EZ-10 Spin Column DNA Extraction kit from Bio Basic Inc. (catalog number BS353). Ligations were carried out with T4 DNA ligase in an equimolar reaction mixture (volume of 20 μl) for 8 h at 16°C. Recombinant DNA procedures were performed with chemically competent Escherichia coli strain TOP10F′ cells (Invitrogen, ThermoFisher Scientific, Waltham, MA).
Construction of metE-complemented Synechococcus sp. strain PCC 7002.
The BLASTP program (26) was utilized to identify the cobalamin-independent methionine synthase (MetE) gene (http://www.ncbi.nlm.nih.gov/protein/975878911) in Synechococcus sp. strain PCC 73109 by comparing it to other cyanobacterial metE genes (see Fig. S1 in the supplemental material). The metE gene of Synechococcus sp. strain PCC 73109 was amplified from isolated genomic DNA with primers metEF and metER (see Table S1 in the supplemental material). The metE amplicon was cloned into the endogenous plasmid-based pAQ1Ex_PcpcBA expression system (5) under the control of the strong constitutive phycocyanin promoter (PcpcBA) of Synechocystis sp. strain PCC 6803 (Fig. 1A) to generate plasmid pAQ1Ex-PcpcBA[metE]. This plasmid contains an aadA cassette from pSRA81 (27, 28) that confers spectinomycin resistance and an N-terminal His10 tag between the NcoI and NdeI sites. pAQ1Ex-PcpcBA[metE] was linearized by ScaI digestion and transformed as described previously (5) into WT Synechococcus sp. strain PCC 7002 to generate strain AAP001 (Table 1). Spectinomycin resistance was used for selection of the mutant strain. MetE synthesis was verified by immunoblotting of whole-cell extracts using commercial rabbit anti-His6 epitope tag antibodies with conjugated horseradish peroxidase (Rockland Antibodies & Assays, Gilbertsville, PA; product code 600-403-382). Immunoblotting was performed as previously described (29).
FIG 1.
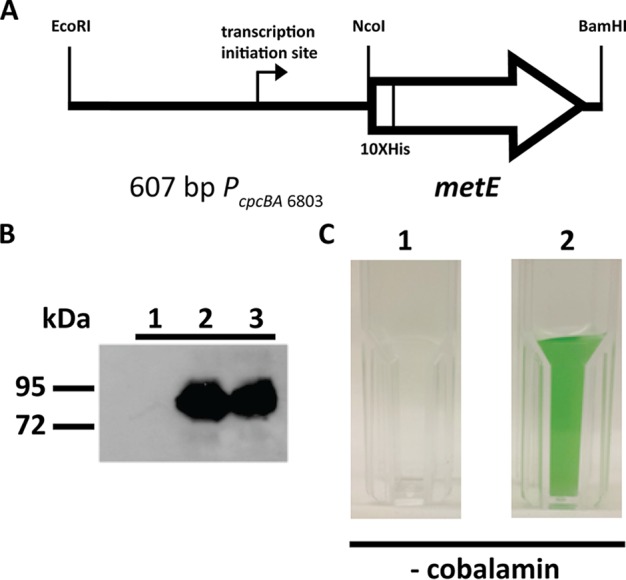
The metE gene of Synechococcus sp. strain PCC 73109 can complement the cobalamin auxotrophy of Synechococcus sp. strain PCC 7002. (A) Representation of pAQ1Ex-PcpcBA[metE]. (B) Immunoblot assay of whole-cell extracts with rabbit anti-His6 antibodies to detect N-His10.MetE. MetE synthesis in strain AAP001 in the presence (lane 2) or absence (lane 3) of cobalamin is shown. No MetE was detected in WT Synechococcus sp. strain PCC 7002 (lane 1). (C) Strain AAP001 (panel 2) grows in medium A+ B12– lacking cyanocobalamin, whereas a WT strain does not (panel 1).
Deletion of the native metH gene from metE-complemented Synechococcus sp. strain PCC 7002.
Upstream and downstream fragments of the metH gene (GenBank locus tag SYNPCC7002_A2466) were obtained by PCR with Synechococcus sp. strain PCC 7002 genomic DNA as the template (see Fig. S2 in the supplemental material). The upstream fragment was amplified with primers dmetHA_F and dmetHA_R, and the downstream fragment was amplified with primers dmetHB_F and dmetHB_R (see Table S1 in the supplemental material). The ermC erythromycin resistance cassette from plasmid pRL409 (30, 31) was obtained through sequential digestion with BamHI and EcoRI and was ligated with the metH upstream and downstream fragments. The reaction product was used as the template for nested PCR with nested primers Nest_metHF and Nest_metHR (see Table S1). The nested PCR product was used to transform strain AAP001 to obtain strain AAP002. Segregation of the metH and ΔmetH::ermC alleles was performed on plates prepared with medium A+ B12– supplemented with erythromycin and spectinomycin. The segregation status was assessed by PCR with primers Nest_metHF and Nest_metHR (see Table S1).
Studies of metE-complemented Synechococcus sp. strain PCC 7002 growth.
Strain AAP002 was grown in parallel with WT Synechococcus sp. strain PCC 7002. Starter cultures of the strains were grown for 2 to 3 days under standard conditions on medium A+ or A+ B12– without the use of antibiotics for the WT and mutant strains. Aliquots were subsequently transferred to 20-mm culture tubes in triplicate. Samples were diluted to an optical density at 730 nm (OD730) of 0.05 with medium, and the OD730 was recorded every 2 h for the first 18 h to follow growth during exponential phase; additional measurements were made until 110 h.
Prediction of a cobalamin riboswitch in the promoter region of metE.
The previously identified B12 riboswitch regulons in cyanobacterial genomes were extracted from the RegPrecise database (http://regprecise.lbl.gov) (32). The comparative genomic analysis of metE genes in Synechococcus sp. genomes was performed with the SEED genomic platform (http://pubseed.theseed.org), which supports the encoding and projection of a metabolic subsystem across prokaryotic genomes (33). Identification of orthologs in closely related genomes and genome neighborhood analysis were performed with the MicrobesOnline tool (http://www.microbesonline.org) (34). Computational identification of B12 riboswitches was performed with the RibEx Riboswitch Explorer tool (35). The RNA secondary structures of the B12 riboswitches and potential antiterminators and antisequestors were predicted with Zuker's algorithm of free-energy minimization (36) implemented in the Mfold program (http://www.bioinfo.rpi.edu/applications/mfold/). Multiple-sequence alignments of DNA regions upstream of the metE gene were constructed with ClustalX (37).
Construction of a cobalamin-regulated expression system.
Synechococcus sp. strain PCC 73109 genomic DNA was used as the template for the amplification of a 612-bp metE promoter region containing the putative cobalamin riboswitch with primers PmetE73109F and PmetE73109R (see Table S1 in the supplemental material). The amplified metE promoter (PmetE) was cloned into the endogenous plasmid-based expression system pAQ3Ex_PcpcBA[yfp] (Kmr) (R. Alvey and D. Bryant, unpublished results), a variant version of the pAQ1Ex_PcpcBA expression platform (5), with flanking sites adapted for homologous recombination with the native pAQ3 plasmid. This construct is identified as pAQ3Ex-PmetE[yfp] (Table 1). pAQ3Ex_PcpcBA[yfp] (Kmr) contains the gene for enhanced yellow fluorescent protein (YFP), the strong phycocyanin promoter, PcpcBA, of Synechocystis sp. strain PCC 6803, and the aphAII kanamycin resistance cassette from pRL161 (Table 1) (30). Both pAQ3Ex-PcpcBA[yfp] (Kmr) and PmetE were digested with SacII and NcoI, and resulting fragments were electrophoretically and column purified prior to the ligation reaction. Transformed E. coli cells were grown on Luria-Bertani medium and plates supplemented with 30 mg of kanamycin ml–1. Plasmid pAQ3Ex-PmetE[yfp] was linearized with ScaI and transformed into strain AAP001 grown in medium A+ B12–. Transformants were selected on plates prepared with medium A+ B12– supplemented with kanamycin and spectinomycin, and a resultant transformant was selected and designated strain AAP003 (Table 1), which is also identified as the YFP fluorescence reporter system.
In vivo evaluation of cobalamin-regulated yfp expression.
A starter culture of strain AAP003 was grown under standard conditions in medium A+ B12– supplemented with kanamycin and spectinomycin to an OD730 of 1.0. A WT Synechococcus sp. strain PCC 7002 starter culture was grown in medium A+ under standard conditions. Subcultures of the mutant strain were inoculated at an OD730 of 0.05 and grown under standard conditions in the presence (A+ B12+) or absence (A+ B12–) of cobalamin to an OD730 of 1.0. The YFP fluorescence amplitude per OD730 unit was determined with an SLM-Aminco 8100C fluorometer modernized for computerized data acquisition by On-Line Instrument Systems (Bogart, GA) (5). Cultures were standardized to an OD730 of 1.0 for data acquisition, and the excitation wavelength was 488 nm. The emission wavelength was scanned from 500 to 600 nm, and the emission maximum of YFP at 527 nm was monitored.
Construction of a strong promoter regulated by cobalamin.
By using an introduced NheI site, the metE promoter region from Synechococcus sp. strain PCC 73109, which contains the putative cobalamin riboswitch element, was transcriptionally fused downstream from the strong cpcBA promoter of Synechocystis sp. strain PCC 6803 (38). Primers cpcBA6803_fusF and cpcBA6803_fusR (see Table S1 in the supplemental material) were used to amplify the cpcBA promoter. An intrinsic SacII restriction site near the 5′ end of the PcpcBA amplified region was used for further cloning purposes. Primers metE73109_fusF and metE73109_fusR (see Table S1) were used with genomic DNA from Synechococcus sp. strain PCC 73109 as the template to amplify the metE promoter with the cobalamin riboswitch elements. Digested fragments were gel purified and then ligated. Nested PCR with primers cpcBA6803_fusF and metE73109_fusR (see Table S1) was performed with the ligation mixture as the template, and the nested PCR product was purified and digested. Plasmid pAQ3Ex-PcpcBA[yfp] was digested with SacII and NcoI to replace the existing PcpcBA promoter with the fused promoter construct to generate plasmid pAQ3Ex-Pfused[yfp]. E. coli strains containing pAQ3Ex-Pfused[yfp] were grown in Luria-Bertani liquid or solid medium supplemented with 30 mg of kanamycin ml–1. To avoid the possibility of homologous recombination between the PcpcBA promoter controlling metE expression and the homologous sequence of PcpcBA in the fused promoter, the psbA2 promoter from Synechocystis sp. strain PCC 6803 (5) was used to drive metE expression. Primers psbA26803_F and psbA28603_R (see Table S1) were used to amplify PpsbA2 from isolated Synechocystis sp. strain PCC 6803 genomic DNA, which was then introduced into the pAQ1Ex plasmid as described above to obtain plasmid pAQ1Ex-PpsbA2[metE]. Strain AAP004 was constructed from WT Synechococcus sp. strain PCC 7002 by introducing plasmid pAQ1Ex-PpsbA2[metE] (Table 1; results not shown). Strain AAP005 was then constructed by transforming strain AAP004 with plasmid pAQ3Ex-Pfused[yfp] that had been linearized with ScaI. Strain AAP005 contains the enhanced YFP fluorescence reporter system. YFP levels in cells containing the fused promoter construct were fluorometrically evaluated in vivo as described above.
Evaluation of the putative cobalamin riboswitch in the metE leader sequence.
Plasmid pAQ3-PmetE[yfp] was transformed into strain AAP002 to obtain strain AAP006 as a WT control for the assay to study mutations in the PmetE cobalamin riboswitch mutant. A variant promoter, PmetE-1, was synthesized (GenScript, Piscataway, NJ) by introducing a cytosine-to-thymine transition double mutation (CC to TT in the DNA; CC to UU in the RNA) at the P1 helix-B12 box interface of the putative metE cobalamin riboswitch. This construct was transcriptionally fused to YFP in pAQ3Ex to obtain plasmid pAQ3-PmetE-1[yfp], and the resulting construction was transformed into strain AAP002 to obtain strain AAP007. The in vivo fluorescence amplitudes of YFP controlled by PmetE and PmetE-1 in strains AAP006 and AAP007 were measured as described above.
Total RNA extraction.
Synechococcus sp. strain PCC 73109 cells that had been grown continuously in white light were inoculated into three tubes containing fresh medium A+ or A+ B12– at an OD730 of 0.05 and incubated under standard growth conditions. When cells grew to an OD730 of ∼0.7, 20-ml aliquots of culture were collected from all three tubes and combined. The combined culture sample was centrifuged for 5 min at 5000 × g at 4°C, and the resulting cell pellet was quickly frozen with liquid nitrogen and stored at −80°C until processing. For RNA isolation, cell pellets were suspended in 50 mM Tris-HCl, pH 8.0. An equal volume of glass beads was mixed with the cell suspension, which was then subjected to a brief bead beating (4,200 rpm for 10 s) with a Mini-Beadbeater (Biospec Products, Bartlesville, OK). Total RNA was extracted with phenol and further purified with the High Pure RNA Isolation kit (Roche, Indianapolis, IN) as described previously (2).
RT-PCR.
Reverse transcription (RT)-PCR analysis of total RNA isolated from WT Synechococcus sp. strain PCC 73109 was performed with a MyTaq One-Step RT-PCR kit (Bioline USA Inc., Taunton, MA). The concentration of RNA was determined by NanoDrop measurements (ThermoFisher Scientific, Waltham, MA). The absence of DNA in RNA samples was verified by RT-PCR assays without reverse transcriptase. Primers RT_metE_F and RT_metE_R (see Table S1 in the supplemental material) were designed to amplify specifically a region of the metE gene of Synechococcus sp. strain PCC 73109. Primers RT_16S_F and RT_16S_R (see Table S1) were designed to amplify a region of the 16S rRNA gene as a control. The amplicons from the RT-PCR analysis were analyzed by agarose gel electrophoresis.
Accession numbers.
The genome sequence of Synechococcus sp. strain PCC 73109 is available in GenBank under accession numbers CP013998 to CP014003.
RESULTS
Synechococcus sp. strain PCC 73109 contains both metE and metH genes.
Synechococcus sp. strain PCC 73109 is a marine cyanobacterium that is very closely related to Synechococcus sp. strain PCC 7002 (39). This cyanobacterium can grow in the presence or absence of exogenous vitamin B12 (cyanocobalamin) (9; Z. Liu, R. Alvey, Z. Li, and D. A. Bryant, unpublished results). With the BLASTP program (26), the metE gene, encoding cobalamin-independent methionine synthase in Synechococcus sp. strain PCC 73109, was identified on a plasmid (unnamed plasmid 5; GenBank accession number CP014003.1) by its high amino acid sequence identity (73%) and similarity (85%) to the metE-2 gene product of Synechococcus sp. strain JA-3-3Ab (CyanoBase ID CYA_0167) (see Fig. S1 in the supplemental material). The amino acid sequence of MetE is available in GenBank under accession number AMA10842.1. The availability of this gene from a very closely related Synechococcus sp. strain led us to question whether this metE gene could complement the cobalamin auxotrophy of Synechococcus sp. strain PCC 7002.
The metE gene from Synechococcus sp. strain PCC 73109 complements the cobalamin auxotrophy of Synechococcus sp. strain PCC 7002.
MetE synthesis driven by the strong PcpcBA promoter (5, 40) in strain AAP001 occurs in both the presence and the absence of exogenous cobalamin, and His10-tagged MetE was detected in cells under both conditions by immunoblotting (Fig. 1B). WT Synechococcus sp. strain PCC 7002 is initially able to grow in A+ B12– medium because of cellular reserves, residual MetH, and cobalamin carried over from the stock cultures derived from A+ medium plates or liquid cultures. For this reason, multiple serial transfers in medium A+ B12– were required to dilute the cobalamin and MetH to the point at which cells could no longer grow. After three serial transfers, WT Synechococcus sp. strain PCC 7002 is unable to grow in medium A+ B12–. Strain AAP001 was still able to grow after similar treatment (Fig. 1C). Complementation with metE is sufficient to eliminate the obligate requirement for exogenous cobalamin in Synechococcus sp. strain PCC 7002, although strain AAP001 had a slightly lower growth rate (see below). Consistent with the notion that strain AAP001 no longer requires cobalamin, the growth rate of this strain was the same in the presence or absence of cobalamin (data not shown).
Deletion of the native metH gene from metE-complemented Synechococcus sp. strain PCC 7002.
Synechococcus sp. strain PCC 7002 contains a cobalamin-dependent methionine synthase gene (CyanoBase ID SYNPCC7002_A2466 [metH]), and bioinformatic analyses have suggested that this is probably the only enzyme that requires cobalamin as a coenzyme. Thus, the inability of this strain to synthesize cobalamin might explain its obligate requirement for exogenous cobalamin (41). Preliminary growth experiments supported this interpretation. Cells were grown in the absence of cobalamin with or without added methionine (1 mM). In the absence of added methionine, cells were unable to grow in medium A+ B12–, as expected. Although Synechococcus sp. strain PCC 7002 apparently lacks a methionine transporter, cells were able to grow with supplied methionine even after multiple serial transfers to deplete cells of cobalamin. These cells grew in medium A+ B12– at approximately half the rate of the WT in medium A+ (data not shown). This result suggested that methionine synthase was likely to be the only cellular function absolutely dependent on cobalamin or, at minimum, was a cell growth rate-limiting factor under these conditions.
To study this behavior further, a metH deletion construct was made by cloning the upstream and downstream flanking regions of metH surrounding an erythromycin resistance cassette (ermC) from plasmid pRL409 (30, 31) (see Fig. S2A in the supplemental material). This construct was transformed into strain AAP001 to produce a metH null mutant, AAP002. Full segregation of the metH and ΔmetH::ermC alleles in the deletion strain was confirmed by PCR (see Fig. S2B). Compared to WT Synechococcus sp. strain PCC 7002, this strain grew ∼20% slower under photoautotrophic conditions (Fig. 2). The calculated doubling times in exponential phase were 4.22 ± 0.14 h for the WT strain and 5.08 ± 0.18 h for strain AAP002. In addition, strain AAP002 no longer required antibiotics to maintain plasmid pAQ1Ex-PcpcBA[metE], as the deletion of metH makes metE essential for viability (data not shown).
FIG 2.
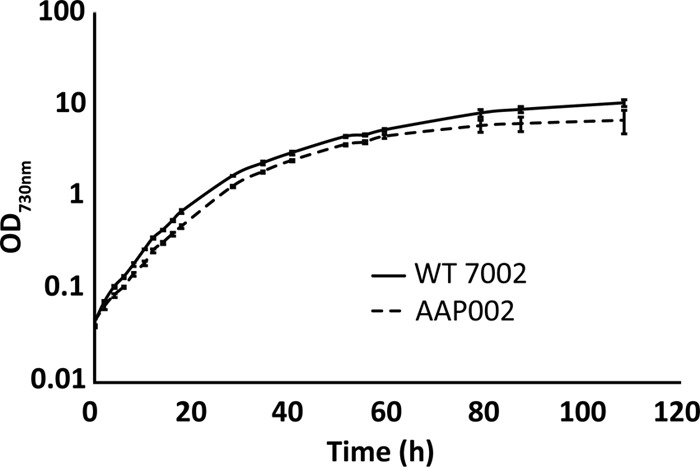
Comparison of the growth of strain AAP002 in medium A+ B12– and the WT strain in medium A+ containing cobalamin. The average values of three independent cultures of each strain are plotted, and the error bars show the standard deviations.
The 5′ untranslated region of the metE gene of Synechococcus sp. strain PCC 73109 contains a putative cobalamin riboswitch.
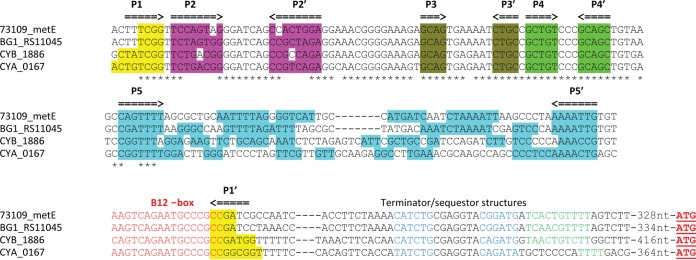
The Synechococcus sp. strain PCC 73109 metE locus, comprising the metE gene and ∼600 bp upstream of the metE start codon, was analyzed with the online riboswitch prediction tool RibEx (35). This tool identified a B12 box (21), a conserved motif commonly found in cobalamin riboswitches. Other elements were identified that correspond to a system that should control gene expression at the transcriptional level, including a putative terminator helix and a terminator-like poly-T (U) tract (22) (Fig. 3). These elements were identified 328 nucleotides upstream of the start codon of the metE gene. The function, if there is any, of the remainder of the leader sequence is unknown.
FIG 3.
Multiple-sequence alignment of the regions upstream of the metE gene of Synechococcus spp. and identification of conserved RNA secondary structures of the B12 riboswitches. The alignment shows the regions upstream of the metE gene of Synechococcus sp. strain PCC 73109 (73109_metE), Synechococcus sp. strain JA-3-3A (CYA_0167), Synechococcus sp. strain JA-3-3Ab (CYB_1886), and Synechococcus sp. strain NKBG15041c (BG1_RS11045). The arrows above the sequences indicate the complementary stems of the RNA secondary structure. Base-paired positions are highlighted in matching colors. The sequences of conserved B12 box elements are in red font. Secondary structures of putative terminators/ribosomal sequesters are in blue font; poly-T tracts in terminators are in green font. Start codons are in bold red font and underlined.
The putative riboswitch-containing metE promoter of Synechococcus sp. strain PCC 73109 represses yfp expression in the presence of exogenous cobalamin.
The yfp gene, encoding YFP, has been used as a versatile reporter that can be assayed in whole cells by spectrofluorometry (5). To assess the functionality of the potential cobalamin riboswitch in the metE promoter, strain AAP003 was grown in the absence or presence of cobalamin. The PmetE promoter that controls the expression of yfp in this strain contains the putative cobalamin riboswitch and all of its associated elements (Fig. 4A). YFP fluorescence was detected in AAP003 cells grown in medium A+ B12–; however, the YFP fluorescence signal was strongly repressed when cells were grown in A+ growth medium containing exogenous cobalamin (Fig. 4B). Compared to the fluorescence emission of WT Synechococcus sp. strain PCC 7002, a small amount of YFP fluorescence was detected even in the presence of cobalamin in AAP003 cells (Fig. 4B). These data support the in silico prediction of a cobalamin riboswitch in the metE promoter region that controls metE expression via repression in the presence of exogenous cobalamin.
FIG 4.
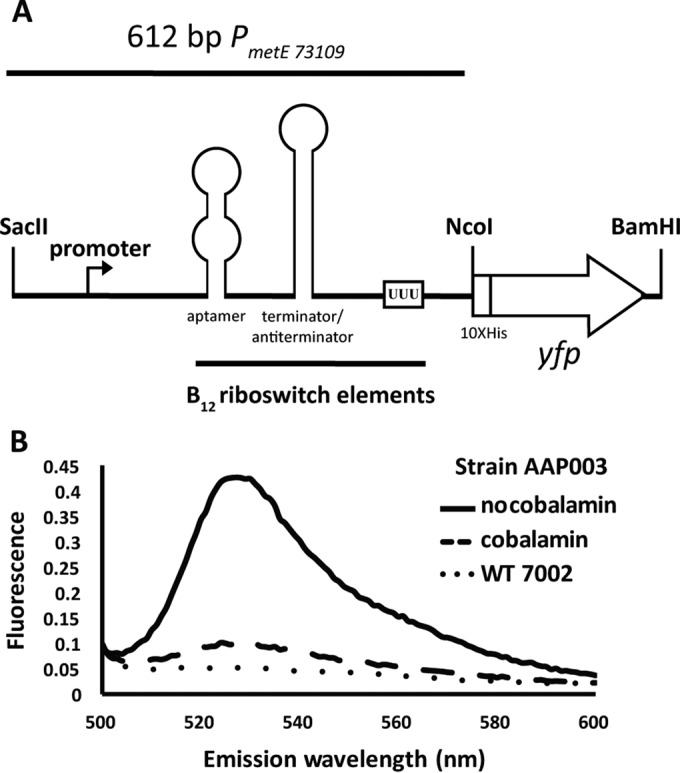
The metE promoter from Synechococcus sp. strain PCC 73109 contains a putative cobalamin riboswitch. (A) Representation of PmetE in plasmid pAQ3Ex-PmetE[yfp]. (B) Detection of YFP expression in strain AAP003. Fluorescence emission spectra of YFP in vivo with Synechococcus sp. strain PCC 7002 normalized at an OD730 of 1.0 in growth medium A+ with or without exogenous cobalamin. The dotted line shows fluorescence emission from WT Synechococcus sp. strain PCC 7002 grown in medium A+ as a negative control. The dashed line shows YFP fluorescence emission in strain AAP003 in the presence of cobalamin in A+ medium, and the solid line shows YFP fluorescence emission for the same strain grown in medium A+ B12–.
YFP expression is enhanced but still regulated by cobalamin when the metE promoter is fused with the cpcBA promoter from Synechocystis sp. strain PCC 6803.
The metE promoter (PmetE) of Synechococcus sp. strain PCC 73109 enables repression of YFP synthesis in the presence of cobalamin in strain AAP003. In the absence of cobalamin, only very low levels of YFP expression occurred (Fig. 4B). To enhance the expression of yfp, a fused promoter was constructed by using the strong cpcBA promoter from the cyanobacterium Synechocystis sp. strain PCC 6803 (5, 40). In plasmid pAQ3Ex-Pfused[yfp], the cpcBA promoter (PcpcBA) (38) was fused upstream of PmetE and cloned into pAQ3Ex to control yfp expression (Table 1; Fig. 5A). Plasmid pAQ3Ex-Pfused[yfp] was transformed into strain AAP004 to produce strain AAP005. In contrast to the results obtained with AAP003, strain AAP005 had much higher levels of YFP fluorescence in the absence of exogenous cobalamin (Fig. 5B). A low background level of YFP production still occurred in the presence of cobalamin, but the YFP fluorescence intensity increased >6-fold in the absence of cobalamin. The fused hybrid PcpcBA/metE promoter in pAQ3Ex-Pfused[yfp] includes the transcription start sites for both the PmetE (based on sequence alignment; results not shown) and PcpcBA promoters (40, 42, 43). Results in Fig. 5B demonstrate that this does not affect the strong expression directed by PcpcBA or the cobalamin-dependent repression directed by the PmetE promoter.
FIG 5.
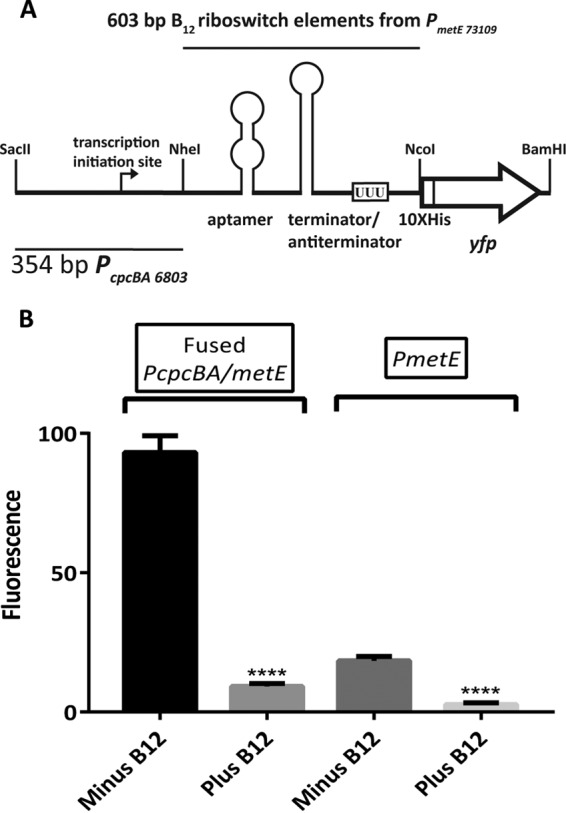
A fused promoter (Pfused) can control the expression of YFP in response to cobalamin. (A) Fused promoter Pfused construct (derived from PcpcBA and PmetE) in plasmid pAQ3Ex-Pfused[yfp]. (B) In vivo fluorescence measurements of YFP in strains AAP005 and AAP003. Synechococcus sp. strain PCC 7002 was standardized at an OD730 of 1.0 in growth medium A+ with or without exogenous cobalamin. Each bar represents the average emission amplitude of YFP at 527 nm of three biological replicates after normalization by using the highest fluorescence amplitude as 100%. The error bars represent the standard deviations of these measurements. The asterisks represent statistical significance with P values as follows: none, P > 0.05; ****, P ≤ 0.0001.
A cytosine-to-thymine double transition mutation in the conserved 3′ region of the B12 box causes loss of cobalamin regulation.
Mutations in conserved regions of cobalamin riboswitches are often employed to demonstrate that cobalamin can regulate gene expression through a riboswitch mechanism (16). A transition mutation of the conserved CC motif in the 3′ end of the B12 box-P1 helix interface to TT (UU in RNA) was introduced to obtain plasmid pAQ3Ex-PmetE-1[yfp] (Fig. 6A). Figure 6B shows that YFP expression no longer occurs in response to exogenous cobalamin when yfp expression is controlled by promoter PmetE-1 instead of WT promoter PmetE.
FIG 6.
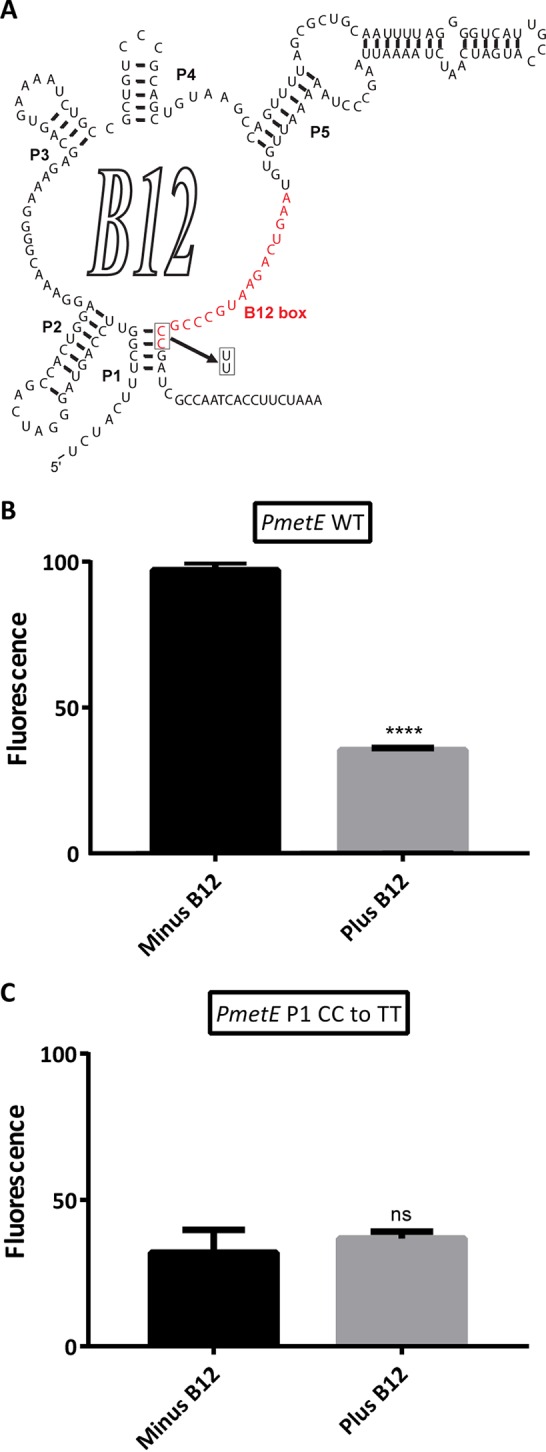
Mutational analysis of the putative cobalamin riboswitch in the PmetE promoter. (A) Putative secondary structure of the cobalamin riboswitch in the metE leader region of Synechococcus sp. strain PCC 73109 in plasmid pAQ3-PmetE-1[yfp]. The boxed bases represent the B12 box-P1 helix interface, where a CC-to-TT (UU in the RNA structure) transition mutation was introduced to generate PmetE-1. (B, C) Expression of YFP in the PmetE promoter variants in the presence or absence of cobalamin. (B) Fluorescence amplitudes of strain AP006 (PmetE promoter) in the presence or absence of exogenous cobalamin. (C) Fluorescence amplitudes of strain AP007 (PmetE-1, CC-to-UU mutation) in the presence or absence of exogenous cobalamin. Each bar represents the average emission amplitude of YFP at 527 nm of three biological replicates after normalization by using the highest fluorescence value as 100%. The error bars represent the standard deviations of these measurements. The asterisks represent statistical significance with P values as follows: none, P > 0.05; ****, P ≤ 0.0001; ns, not significant.
The cobalamin riboswitch in the metE promoter of Synechococcus sp. strain PCC 73109 probably acts as a transcription terminator in the presence of cobalamin.
It was of interest to determine whether the cobalamin riboswitch operates at the transcriptional or translational level. RT-PCR of total RNA from Synechococcus sp. strain PCC 73109 with specific primers for the metE gene of Synechococcus sp. strain PCC 73109 showed that metE transcripts were not detectable in cells grown in the presence of cobalamin (Fig. 7A). Conversely, metE transcripts were readily detectable when Synechococcus sp. strain PCC 73109 cells were grown in the absence of exogenous cobalamin (Fig. 7B). This result strongly implies, but further experiments are required to confirm definitively, that the cobalamin riboswitch in the metE untranslated leader sequence operates as a transcription terminator when cells are grown in the presence of cobalamin.
FIG 7.
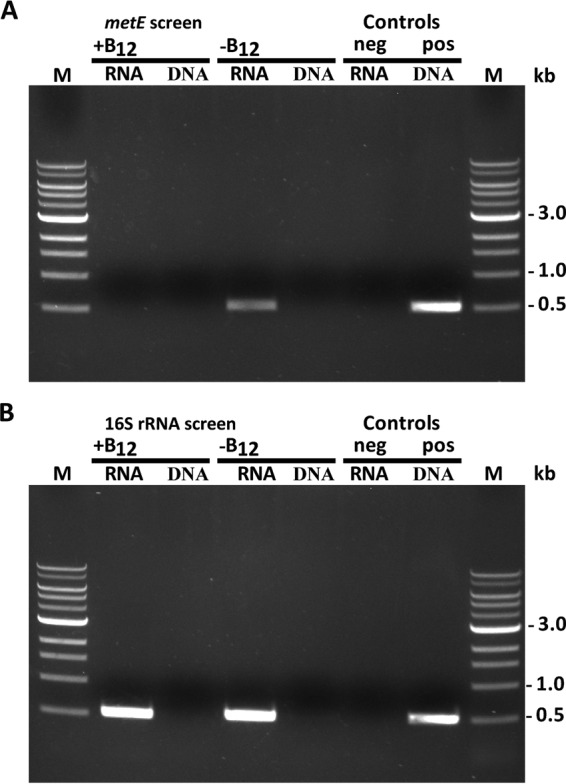
The cobalamin riboswitch represses the expression of yfp in the presence of cobalamin. (A) RT-PCR evaluation of the Synechococcus sp. strain PCC 73109 metE gene in which the total RNA template (RNA) was set to 1 ng/μl from cultures grown in the presence of 4 μg of cyanocobalamin liter−1 (+B12) or in medium lacking cobalamin (–B12). DNA lanes represent RT-PCRs lacking reverse transcriptase. The negative control (neg) refers to an RT-PCR mixture containing all of the reagents and amplified under standard RT-PCR conditions but lacking a template to survey for extraneous nucleic acid contamination (no-template control). The positive control (pos) refers to the use of metE RT-PCR primers in a PCR with Synechococcus sp. strain PCC 73109 genomic DNA as the template. (B) RT-PCR evaluation of 16S rRNA in the total RNA of Synechococcus sp. strain PCC 73109 grown in the presence or absence of cobalamin. Conditions are exactly the same as those for panel A, except that primers specific for the 16S rRNA were utilized. All PCR products are 504 bp in size. Lanes M contained molecular size markers.
DISCUSSION
Few studies have examined the role of cobalamin in Synechococcus sp. strain PCC 7002, even though this compound is an essential nutrient for the growth of this organism and other cyanobacteria (6). In fermenting bacteria, cobalamin is associated with essential functions in the metabolism of certain small molecules (e.g., 1,3-propanediol) and is used as a coenzyme in some reactions of the Wood-Ljungdahl pathway (44), but cobalamin is also important in processes including methionine and ribonucleotide biosynthesis (8). With the advent of bioinformatics and the ability to sequence and study complete genomes, the role of cobalamin in Synechococcus sp. strain PCC 7002 was revisited with an emphasis on the cobalamin-dependent methionine synthase (metH) gene, which is apparently the only gene whose product requires this vitamin as a coenzyme in this cyanobacterium. The absence of the cobalamin-independent methionine synthase (metE) gene and the occurrence of metH in Synechococcus sp. strain PCC 7002 indicate that this cyanobacterium requires cobalamin for the synthesis of l-methionine (12, 45). The only other alternative would be to take up this amino acid from the surrounding milieu with a specialized transporter (46). The inability to obtain sufficient l-methionine from external sources would render cobalamin an essential component for the otherwise photoautotrophic lifestyle of Synechococcus sp. strain PCC 7002.
In a previous study, Wilhelm and Trick (6) investigated the role of cobalamin in Synechococcus sp. strain PCC 7002 by comparing cells that had been grown under cobalamin-deficient and cobalamin-replete conditions. The cell densities achieved correlated with the levels of cobalamin in the medium, and cells grown in limiting cobalamin had lower levels of chlorophyll a and exhibited changes in the integrity of the thylakoid membranes. Lower cobalamin levels also led to the appearance of several outer membrane proteins, which was the first evidence that a TonB-like active-transport uptake system for cobalamin transport might be regulated by cobalamin itself (47). Nevertheless, a direct correlation between these phenotypes and cobalamin was never demonstrated.
Cobalamin has been suggested to be an important molecule in aquatic systems where extensive exchange of nutrients occurs among diverse organisms and has been shown to play a limiting role in marine systems with cyanobacteria and eukaryotic phytoplankton (48, 49). Synechococcus sp. strain PCC 7002 is similar to other types of marine phytoplankton that are unable to produce cobalamin de novo and require external sources of this key nutrient (49). However, unlike some of its marine photoautotrophic eukaryotic counterparts that still retain auxotrophy for cobalamin when grown with exogenous l-methionine (50), Synechococcus sp. strain PCC 7002 is capable of slow cobalamin-independent growth if a relatively high level of methionine (1 mg ml–1) is added to the growth medium (data not shown). The genome of Synechococcus sp. strain PCC 7002 does not appear to encode a methionine transporter but does encode a predicted ABC transporter for branched-chain amino acids (KEGG Pathway entry syp02010), which might account for some methionine transport.
In this study, we confirmed that metH is responsible for the cobalamin auxotrophy of Synechococcus sp. strain PCC 7002, as it is the sole route for de novo methionine biosynthesis in this cyanobacterium. Providing an alternate route for methionine biosynthesis via the expression of metE allowed cells to grow in cobalamin-free medium; this rendered metH unnecessary, as evidenced by the facile deletion of the metH gene (see Fig. S2 in the supplemental material). Cobalamin-independent strain AAP002 grew a bit (∼20%) more slowly than its WT counterpart when both were grown under photoautotrophic conditions. There are several possible reasons for the lower growth rate, including the lower turnover rate of MetE than of MetH, which utilizes cobalamin as a coenzyme to facilitate the conversion of l-homocysteine into l-methionine (51). Alternatively, the constitutive overexpression of metE from the high-copy-number pAQ1Ex expression system and the strong cpcBA promoter in plasmid pAQ1Ex-PcpcBA[metE] could perturb cellular metabolites and siphon off important metabolic resources. The adoption of an alternate pathway for methionine biosynthesis could itself affect the global metabolic fluxes, thereby influencing the growth rate.
The identification and validation of a cobalamin riboswitch, with potential transcriptional regulation, in the 5′-upstream untranslated region of the metE gene from Synechococcus sp. strain PCC 73109 provide a novel genetic tool for controlling gene expression. They also represent an efficient system for characterizing putative cobalamin riboswitches from other organisms that lack available genetic systems by experimentally using YFP as a reporter system in cobalamin-independent Synechococcus sp. strain PCC 7002. Future experimentation could also utilize this system to identify genes responsible for uptake of the large cobalamin molecule (16) into Synechococcus sp. strain PCC 7002.
The development of a cobalamin-independent version of Synechococcus sp. strain PCC 7002 uncovered the unique role of cobalamin in this marine cyanobacterium that serves as a model organism. Development of this strain favors the use of Synechococcus sp. strain PCC 7002 for industrial large-scale culturing without the need to provide external cobalamin. The possible implementation of MetE as a selection marker that would simply require the removal of cobalamin is also more feasible than implementing commonly used selection methods based on antibiotic resistance and other methods that require expensive reagents and/or the development of mutant host strains (52). With this development, another step has been taken to establish Synechococcus sp. strain PCC 7002 as a versatile and cost-effective model organism for industrial applications.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge former lab member Richard Alvey, who first showed that Synechococcus sp. strain PCC 7002 can utilize exogenous l-methionine. We also acknowledge Alexander Yakhnin and Paul Babitzke for helpful discussions about the design of genetic fusions for promoter constructs.
This work was supported by NSF grant MCB-1021725 and DOE contract DE-FG-02-05-ER46222 to D.A.B. D.A.B. and D.A.R. were supported by the Genomic Science Program (GSP), Office of Biological and Environmental Research (OBER), U.S. Department of Energy (DOE). The NSF GK-12 CarbonEARTH grant (award number 0947962) provided full tuition and stipend support to A.A.P.
Footnotes
This study is a contribution of the Pacific Northwest National Laboratory (PNNL) Foundational Scientific Focus Area.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00475-16.
For a companion article on this topic, see doi:10.1128/JB.00476-16.
REFERENCES
- 1.Sakamoto T, Bryant DA. 2002. Synergistic effect of high-light and low temperature on cell growth of the Delta12 fatty acid desaturase mutant in Synechococcus sp. PCC 7002. Photosynth Res 72:231–242. [DOI] [PubMed] [Google Scholar]
- 2.Ludwig M, Bryant DA. 2011. Transcription profiling of the model cyanobacterium Synechococcus sp. strain PCC 7002 by Next-Gen (SOLiD™) sequencing of cDNA. Front Microbiol 2:41. doi: 10.3389/fmicb.2011.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein HC, Konopka A, Melnicki MR, Hill EA, Kucek LA, Zhang S, Shen G, Bryant DA, Beliaev AS. 2014. Effect of mono- and dichromatic light quality on growth rates and photosynthetic performance of Synechococcus sp. PCC 7002. Front Microbiol 5:488. doi: 10.3389/fmicb.2014.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevens SE, Porter RD. 1980. Transformation in Agmenellum quadruplicatum. Proc Natl Acad Sci U S A 77:6052–6056. doi: 10.1073/pnas.77.10.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y, Alvey RM, Byrne PO, Graham JE, Shen G, Bryant DA. 2011. Expression of genes in cyanobacteria: adaptation of endogenous plasmids as platforms for high-level gene expression in Synechococcus sp. PCC 7002. Methods Mol Biol 684:273–293. doi: 10.1007/978-1-60761-925-3_21. [DOI] [PubMed] [Google Scholar]
- 6.Wilhelm SW, Trick CG. 1995. Effects of vitamin B12 concentration on chemostat cultured Synechococcus sp. strain PCC 7002. Can J Microbiol 41:145–151. doi: 10.1139/m95-019. [DOI] [Google Scholar]
- 7.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. 2003. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem 278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- 8.Roth JR, Lawrence JG, Bobik TA. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol 50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 9.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111:1–61. [Google Scholar]
- 10.Peschek GA, Obinger C, Paumann M. 2004. The respiratory chain of blue-green algae (cyanobacteria). Physiol Plant 120:358–369. doi: 10.1111/j.1399-3054.2004.00274.x. [DOI] [PubMed] [Google Scholar]
- 11.McNeely K, Xu Y, Ananyev G, Bennette N, Bryant DA, Dismukes GC. 2011. Synechococcus sp. strain PCC 7002 nifJ mutant lacking pyruvate:ferredoxin oxidoreductase. Appl Environ Microbiol 77:2435–2444. doi: 10.1128/AEM.02792-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pejchal R, Ludwig ML. 2005. Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication. PLoS Biol 3:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. 2011. Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol 28:2921–2933. doi: 10.1093/molbev/msr124. [DOI] [PubMed] [Google Scholar]
- 14.Tanioka Y, Yabuta Y, Yamaji R, Shigeoka S, Nakano Y, Watanabe F, Inui H. 2009. Occurrence of pseudovitamin B12 and its possible function as the cofactor of cobalamin-dependent methionine synthase in a cyanobacterium Synechocystis sp. PCC6803. J Nutr Sci Vitaminol (Tokyo) 55:518–521. doi: 10.3177/jnsv.55.518. [DOI] [PubMed] [Google Scholar]
- 15.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. 2004. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res 32:3340–3353. doi: 10.1093/nar/gkh659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warner DF, Savvi S, Mizrahi V, Dawes SS. 2007. A riboswitch regulates expression of the coenzyme B12-independent methionine synthase in Mycobacterium tuberculosis: implications for differential methionine synthase function in strains H37Rv and CDC1551. J Bacteriol 189:3655–3659. doi: 10.1128/JB.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nahvi A, Barrick JE, Breaker RR. 2004. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res 32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. 2003. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA 9:1084–1097. doi: 10.1261/rna.5710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson JE Jr, Reyes FE, Polaski JT, Batey RT. 2012. B12 cofactors directly stabilize an mRNA regulatory switch. Nature 492:133–137. doi: 10.1038/nature11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nou X, Kadner RJ. 2000. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci U S A 97:7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravnum S, Andersson DI. 1997. Vitamin B12 repression of the btuB gene in Salmonella typhimurium is mediated via a translational control which requires leader and coding sequences. Mol Microbiol 23:35–42. doi: 10.1046/j.1365-2958.1997.1761543.x. [DOI] [PubMed] [Google Scholar]
- 22.Borovok I, Gorovitz B, Schreiber R, Aharonowitz Y, Cohen G. 2006. Coenzyme B12 controls transcription of the Streptomyces class Ia ribonucleotide reductase nrdABS operon via a riboswitch mechanism. J Bacteriol 188:2512–2520. doi: 10.1128/JB.188.7.2512-2520.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakuragi Y, Zybailov B, Shen G, Bryant DA, Golbeck JH, Diner BA, Karygina I, Pushkar Y, Stehlik D. 2005. Recruitment of a foreign quinone into the A1 site of photosystem I. Characterization of a menB rubA double deletion mutant in Synechococcus sp. PCC 7002 devoid of FX, FA, and FB and containing plastoquinone or exchanged 9,10-anthraquinone. J Biol Chem 280:12371–12381. [DOI] [PubMed] [Google Scholar]
- 24.Stevens SE, Patterson CO, Myers J. 1973. The production of hydrogen peroxide by blue-green algae: a survey. J Phycol 9:427–430. [Google Scholar]
- 25.Schluchter WM, Shen G, Zhao J, Bryant DA. 1996. Characterization of psal and psaL mutants of Synechococcus sp. strain PCC 7002: a new model for state transitions in cyanobacteria. Photochem Photobiol 64:53–66. doi: 10.1111/j.1751-1097.1996.tb02421.x. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Frigaard N-U, Sakuragi Y, Bryant DA. 2004. Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods Mol Biol 274:325–340. [DOI] [PubMed] [Google Scholar]
- 28.Frigaard N-U, Maresca JA, Yunker CE, Jones AD, Bryant DA. 2004. Genetic manipulation of carotenoid biosynthesis in the green sulfur bacterium Chlorobium tepidum. J Bacteriol 186:5210–5220. doi: 10.1128/JB.186.16.5210-5220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen G, Zhao J, Reimer SK, Antonkine ML, Cai Q, Weiland SM, Golbeck JH, Bryant DA. 2002. Assembly of photosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of photosystem I activity. J Biol Chem 277:20343–20354. [DOI] [PubMed] [Google Scholar]
- 30.Elhai J, Wolk CP. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 31.Frigaard N-U, Bryant DA. 2001. Chromosomal gene inactivation in the green sulfur bacterium Chlorobium tepidum by natural transformation. Appl Environ Microbiol 67:2538–2544. doi: 10.1128/AEM.67.6.2538-2544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, Kazanov MD, Riehl W, Arkin AP, Dubchak I. 2013. RegPrecise 3.0—a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 14:745. doi: 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS. 2010. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res 38:D396–D400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abreu-Goodger C, Merino E. 2005. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res 33:W690–W692. doi: 10.1093/nar/gki445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyngsø RB, Zuker M, Pedersen CN. 1999. Fast evaluation of internal loops in RNA secondary structure prediction. Bioinformatics 15:440–445. doi: 10.1093/bioinformatics/15.6.440. [DOI] [PubMed] [Google Scholar]
- 37.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshimura T, Imamura S, Tanaka K, Shirai M, Asayama M. 2007. Cooperation of group 2 σ factors, SigD and SigE for light-induced transcription in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett 581:1495–1500. [DOI] [PubMed] [Google Scholar]
- 39.Robertson BR, Tezuka N, Watanabe MM. 2001. Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content. Int J Syst Evol Microbiol 51:861–871. doi: 10.1099/00207713-51-3-861. [DOI] [PubMed] [Google Scholar]
- 40.Zhou J, Zhang H, Meng H, Zhu Y, Bao G, Zhang Y, Li Y, Ma Y. 2014. Discovery of a super-strong promoter enables efficient production of heterologous proteins in cyanobacteria. Sci Rep 4:4500. doi: 10.1038/srep04500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonnet S, Webb EA, Panzeca C, Karl DM, Capone DG, Wilhelmy SAS. 2010. Vitamin B12 excretion by cultures of the marine cyanobacteria Crocosphaera and Synechococcus. Limnol Oceanogr 55:1959–1964. doi: 10.4319/lo.2010.55.5.1959. [DOI] [Google Scholar]
- 42.Camsund D, Lindblad P. 2014. Engineered transcriptional systems for cyanobacterial biotechnology. Front Bioeng Biotechnol 2:40. doi: 10.3389/fbioe.2014.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, Hess WR. 2011. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci U S A 108:2124–2129. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bender G, Pierce E, Hill JA, Darty JE, Ragsdale SW. 2011. Metal centers in the anaerobic microbial metabolism of CO and CO2. Metallomics 3:797–815. doi: 10.1039/c1mt00042j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerjee RV, Matthews RG. 1990. Cobalamin-dependent methionine synthase. FASEB J 4:1450–1459. [DOI] [PubMed] [Google Scholar]
- 46.Kadaba NS, Kaiser JT, Johnson E, Lee A, Rees DC. 2008. The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science 321:250–253. doi: 10.1126/science.1157987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richter-Dahlfors AA, Ravnum S, Andersson DI. 1994. Vitamin B12 repression of the cob operon in Salmonella typhimurium: translational control of the cbiA gene. Mol Microbiol 13:541–553. doi: 10.1111/j.1365-2958.1994.tb00449.x. [DOI] [PubMed] [Google Scholar]
- 48.Chimento DP, Kadner RJ, Wiener MC. 2003. The Escherichia coli outer membrane cobalamin transporter BtuB: structural analysis of calcium and substrate binding, and identification of orthologous transporters by sequence/structure conservation. J Mol Biol 332:999–1014. doi: 10.1016/j.jmb.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 49.Swift D. 1981. Vitamin levels in the Gulf of Maine and ecological significance of vitamin B12 there. J Mar Res 39:375–403. [Google Scholar]
- 50.Croft MT, Warren MJ, Smith AG. 2006. Algae need their vitamins. Eukaryot Cell 5:1175–1183. doi: 10.1128/EC.00097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.González JC, Banerjee RV, Huang S, Sumner JS, Matthews RG. 1992. Comparison of cobalamin-independent and cobalamin-dependent methionine synthases from Escherichia coli: two solutions to the same chemical problem. Biochemistry 31:6045–6056. doi: 10.1021/bi00141a013. [DOI] [PubMed] [Google Scholar]
- 52.Goh S, Good L. 2008. Plasmid selection in Escherichia coli using an endogenous essential gene marker. BMC Biotech 8:61. doi: 10.1186/1472-6750-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.