Summary
Post-translational protein modification by ubiquitin (Ub) and ubiquitin-like (Ubl) proteins such as small ubiquitin like modifier (SUMO) regulates processes including protein homeostasis, the DNA damage response, and the cell cycle. Proliferating cell nuclear antigen (PCNA) is modified by Ub or poly-Ub at Lys164 after DNA damage to recruit repair factors. Yeast PCNA is modified by SUMO on Lys164 and Lys127 during S-phase to recruit the anti-recombinogenic helicase Srs2. Lys164 modification requires specialized E2/E3 enzyme pairs for SUMO or Ub conjugation. For SUMO, Lys164 modification is strictly dependent on the E3 ligase Siz1, suggesting the E3 alters E2 specificity to promote Lys164 modification. The structural basis for substrate interactions in activated E3/E2-Ub/Ubl complexes remains unclear. Here, we report an engineered E2 protein and cross-linking strategies that trap an E3/E2-Ubl/substrate complex for structure determination, illustrating how an E3 can bypass E2 specificity to force-feed a substrate lysine into the E2 active site.
Keywords: E3 ligase, E2 conjugating enzyme, PCNA, SUMO, Signal transduction, DNA damage, Replication
Introduction
Ub and Ubl proteins are conjugated to substrate proteins by dedicated three-enzyme cascades involving E1 activating enzymes, E2 conjugating enzymes, and E3 ligases (reviewed in refs. 1–4). E1s catalyze Ubl activation and thioester transfer to E2s, and E3 Ubl isopeptide ligases often complete the cascade by co-localizing substrates and E2-Ubls to promote bond formation between the Ubl C-terminus and substrate (typically lysines).
E2s can exhibit substrate specificity. The E2Ubc9 catalyzes SUMO conjugation to lysine residues in SUMO consensus motifs (first exemplified for Ψ-K-X-E, where Ψ is a hydrophobic residue and K is lysine)5. Structures of a consensus site lysine bound to E2Ubc9 revealed E2 residues that contribute to lysine recognition and pKa suppression to promote catalysis6–8. Similarly important residues within other E2 active sites also contribute to particular E2-lysine specificities9–14.
Really Interesting New Gene (RING) domains, and their structural homologs, are found in several hundred proteins with E3 activity for Ub, SUMO and Nedd8 conjugation15. RING and some non-RING E3s bind the E2-Ubl thioester, stimulating conjugation by organizing the E2-Ubl into a closed activated conformation, first illustrated for a non-RING E3 (ref. 7) and subsequently shown for a variety of RING E3s16–22. PIAS (protein inhibitor of activated STATs) proteins23, known as Siz proteins in yeast24, were discovered in humans as inhibitors of STAT signaling and function in immune and cytokine signaling and cellular regulation. The Siz/PIAS-RING (SP-RING) proteins comprise the largest family of SUMO E3s, yet available structures lack E2 or substrate25.
PCNA is modified by Ub on Lys164 after DNA damage by the E2Rad6/E3Rad18 pair and sometimes extended into polyUb chains by E2Ubc13/UEVMms2 to recruit repair factors26–28. Yeast PCNA is modified by SUMO on Lys164 and Lys127 during S-phase to recruit Srs2 (refs. 29–31). Lys164 is a non-consensus lysine that requires the SUMO E3Siz1 for SUMO modification by E2Ubc9. SUMO modified PCNA enhances Ub modification32, suggesting pathway cross-talk. As such, PCNA represents a model system for understanding specificity determinants in Ub and SUMO pathways27.
Here, we present reconstitution of an E2Ubc9-SUMO thioester mimetic, an active E3Siz1 fragment, and substrate PCNA. The techniques used to engineer the E2-Ubl thioester mimetic leave the E2 active site cysteine available for cross-linking, generating a bridge between the E2 and PCNA at the endogenous site of Ubl modification with the same number of atoms as the predicted tetrahedral intermediate. We report the crystal structure of this complex at 2.85 Å resolution. The structure and biochemical data reveal molecular determinants of this E3 ligase complex that bypasses E2 specificity to promote modification at PCNA Lys164.
Reconstituting E2Ubc9-SUMO/E3Siz1/PCNA
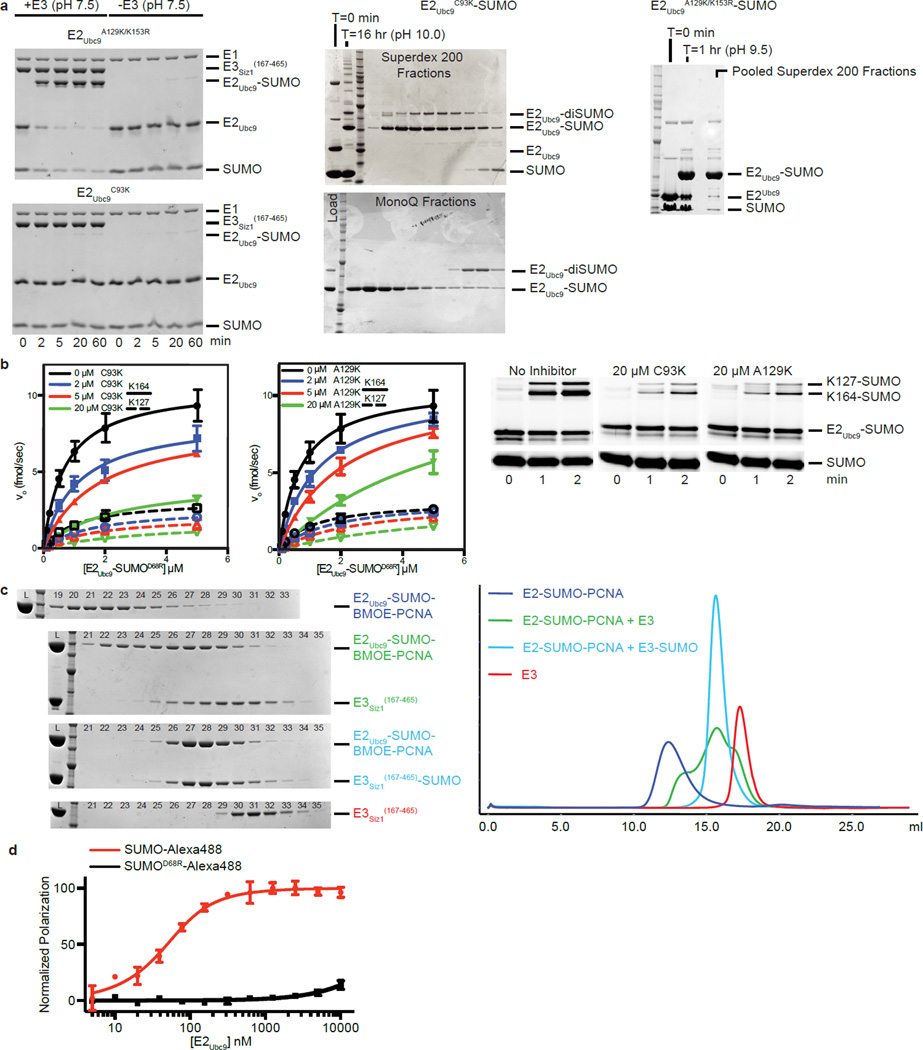
E2-Ubl thioester mimetics with stable E2-Ubl linkages are needed for structural studies because the E2-Ubl thioester is labile. E2 active site Cys to Lys generates a stable peptide bond16, but its side chain is longer than the native linkage and it could interfere with substrate interactions. In contrast, E2 Cys to Ser generates an ester linkage, but it is labile when combined with E3s9,13,17. As an alternative, an E2-Ubl thioester mimetic was engineered by substituting lysine for Ala129 in E2Ubc9 near the active site Cys93 (E2Ubc9A129K). At physiological pH, E3Siz1 stimulates SUMO conjugation to E2Ubc9A129K, but not E2Ubc9C93K, consistent with E1-catalyzed E2Ubc9A129K-SUMO thioester formation followed by Lys129 nucleophilic attack (Extended Data Fig. 1a). E2Ubc9A129K-SUMO and E2Ubc9C93K-SUMO are competitive inhibitors, with values of the dissociation constant for the mimetic binding, Ki, close to Km for E2-SUMO thioester interactions with E3Siz1 (Extended Data Fig. 1b; Extended Data Tables 1a,2). Importantly, Cys93 remains available for cross-linking in the E2Ubc9A129K-SUMO mimetic.
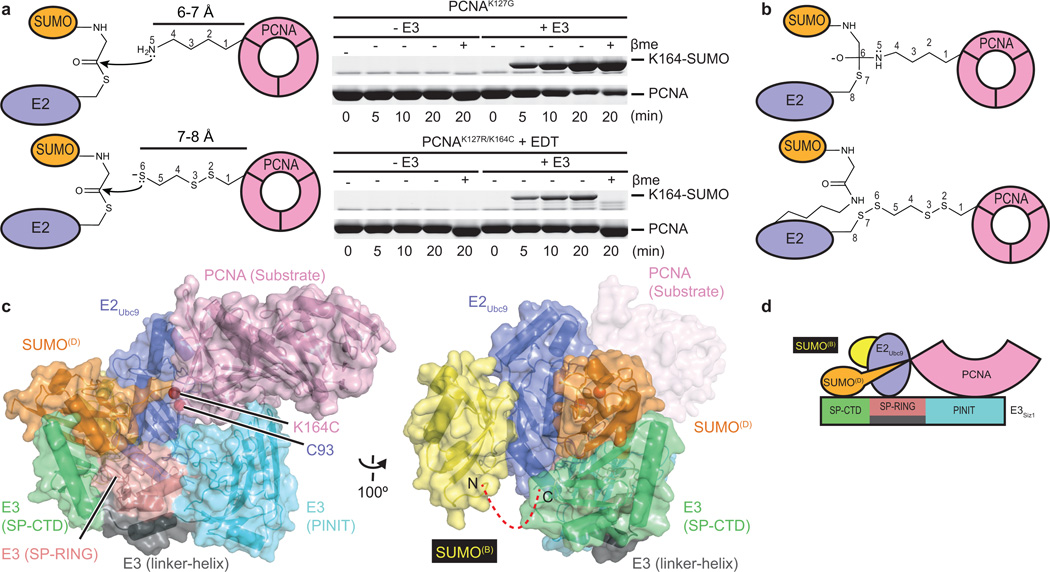
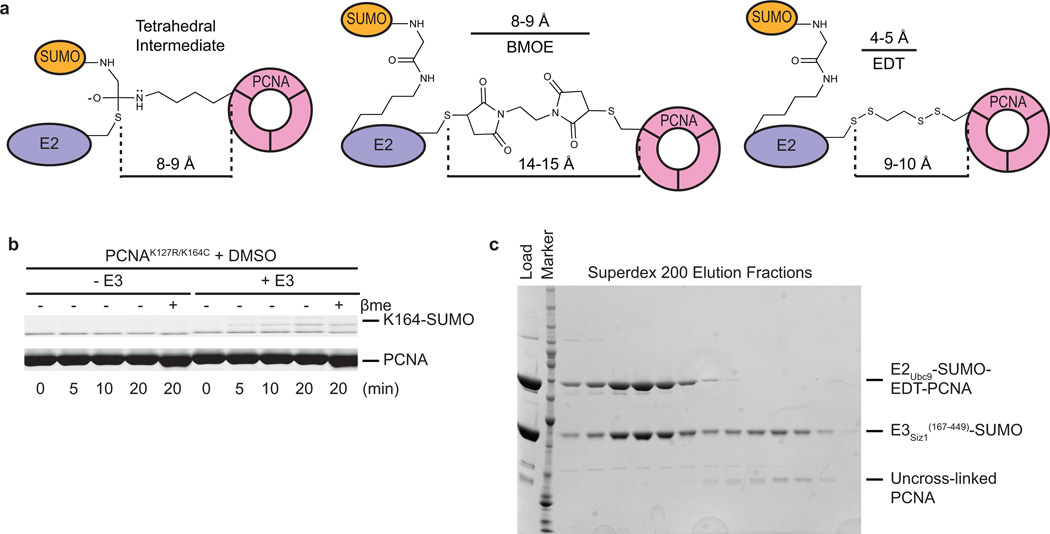
Bismaleimidoethane (BMOE) was used first to cross-link Cys93 in E2Ubc9A129K-SUMO to PCNA by replacing Lys164 with cysteine. E2-SUMO-BMOE-PCNA was combined with E3Siz1(167–465), and interactions were observed for E3/E2-SUMO-BMOE-PCNA by gel filtration, albeit in multiple peaks (Extended Data Fig. 1c). Our own efforts and prior studies33,34 suggested a second SUMO molecule (SUMOB) might stabilize the complex through non-covalent interactions with the E2Ubc9 backside given the high affinity measured between SUMO and E2Ubc9 (Kd of 25 nM ± 4 nM (Extended Data Fig. 1d; Extended Data Table 1b)). A second SUMO was provided by fusing SUMOB to the E3Siz1(167–465) C-terminus where its position relative to E2Ubc9 and E3Siz1 appeared ideal25. Using E3Siz1(167–465)-SUMOB, a stoichiometric complex with E2-SUMO-BMOE-PCNA was observed (Extended Date Fig. 1c). Although BMOE cross-linking trapped the complex, BMOE includes bulky maleimide groups at its ends and it is 4–5 Å longer than the estimated distance spanned by the tetrahedral intermediate (Extended Data Fig. 2a).
1,2-ethanedithiol (EDT) was identified as a candidate to replace BMOE because it is only one atom longer than lysine when attached to PCNAK164C (Fig. 1a,b). Indeed, PCNAK164C-EDT was a substrate for E3-dependent conjugation by transthioesterification, suggesting EDT can mimic lysine (Fig. 1a; Extended Data Fig. 2b). Furthermore, EDT cross-linking of the E2Ubc9 active site cysteine and PCNAK164C yielded a bridge with the same number of atoms between PCNA and the E2 when compared to the tetrahedral intermediate (Fig. 1b). E2-SUMO-EDT-PCNA was reconstituted with E3Siz1(167–449)-SUMOB, yielding a monodisperse complex (Extended Data Fig. 2c).
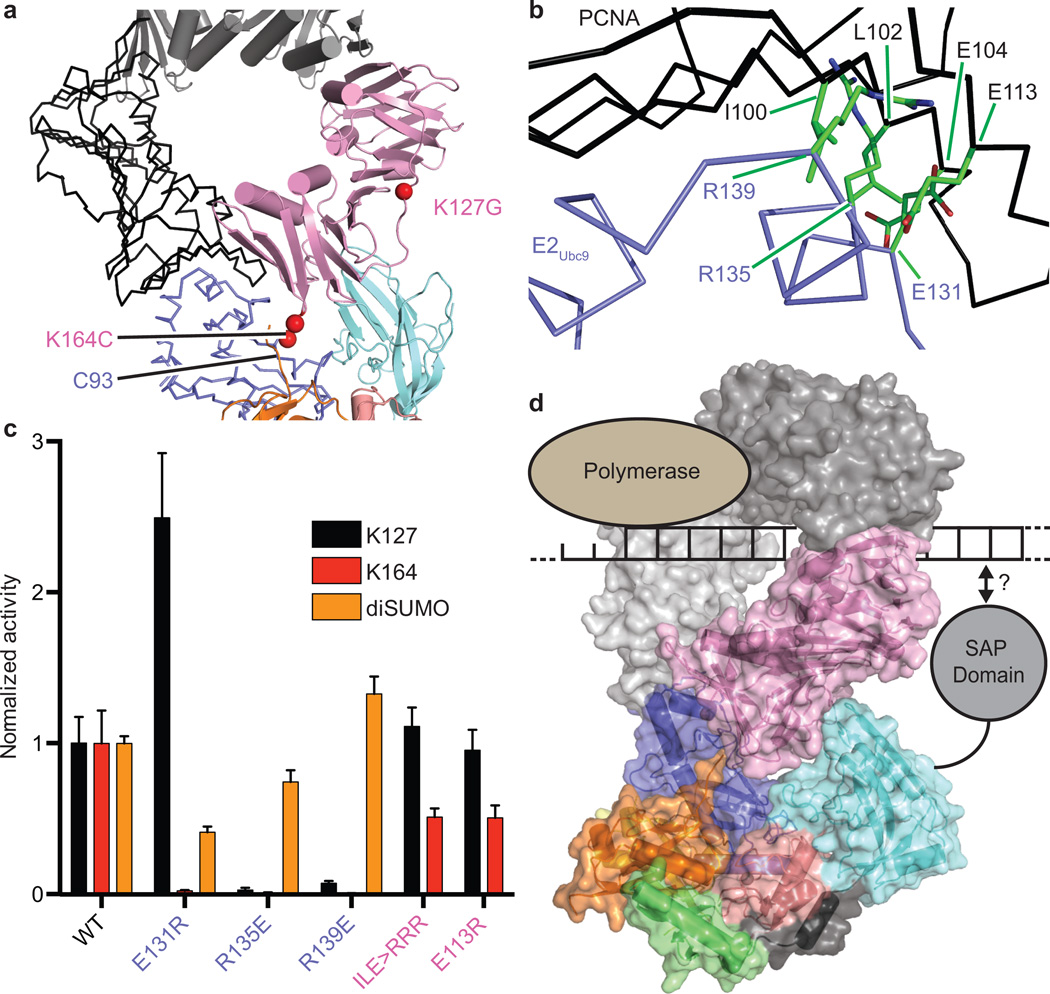
Figure 1. Reconstituting E2Ubc9-SUMOD/E3Siz1-SUMOB/PCNA.
a, Schematic for nucleophilic attack of the E2-SUMO thioester by lysine or EDT modified cysteine with in vitro SUMO modification of PCNA. b, Schematic of the tetrahedral intermediate during transthioesterification and EDT cross-linking of E2Ubc9A129K-SUMO and PCNA. c, Structure of the complex. d, Cartoon model of the complex. For gel source data, see Supplementary Figure 1.
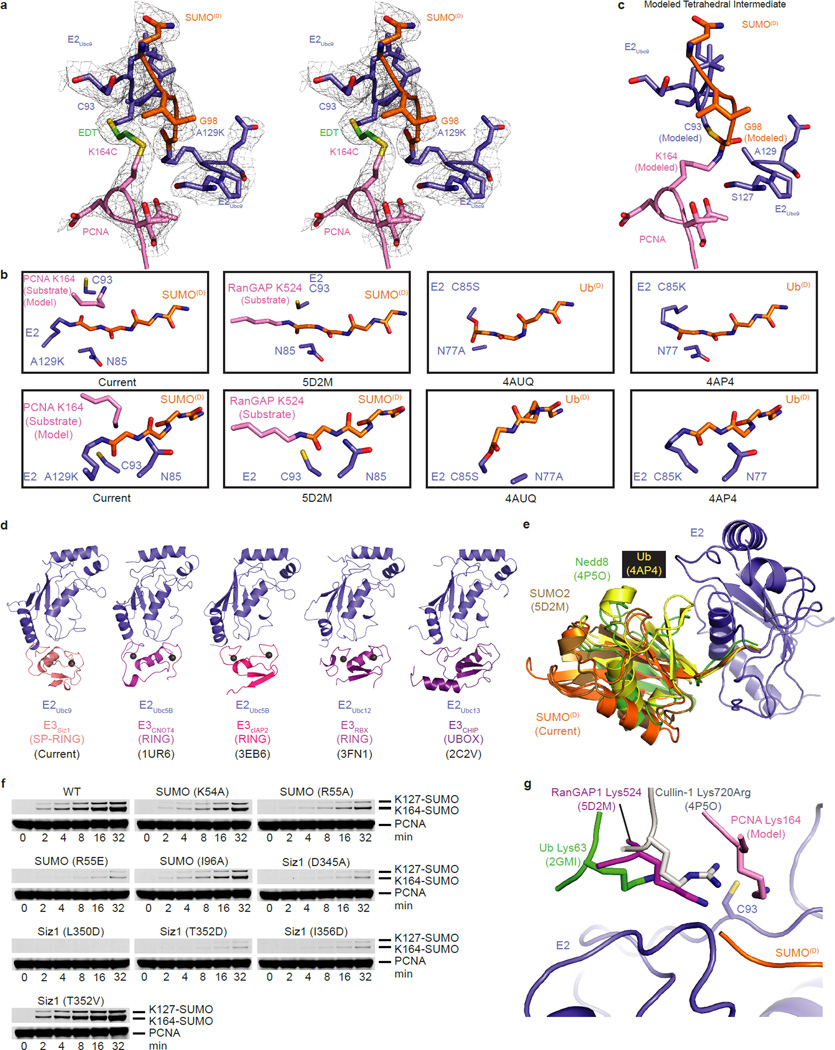
Reconstitutions used trimeric and monomeric PCNA, as both remain dependent on E3Siz1 for SUMO modification at Lys164 (ref. 31). Crystals containing trimeric PCNA did not diffract, however crystals containing monomeric PCNA diffracted to 2.85 Å. The structure was determined and contained two complexes in the asymmetric unit (Fig. 1c,d; Extended Data Table 3). Each complex includes an E2Ubc9A129K-SUMOD thioester mimetic with donor SUMOD bound to E3Siz1 in an activated closed conformation, SUMOB from E3Siz1(167–465)-SUMOB is bound to the E2Ubc9 backside, and EDT bridges Cys93 in E2Ubc9A129K and PCNAK164C above the SUMOD C-terminus which is linked to Lys129 in E2Ubc9A129K (Extended Data Fig. 3a). The SUMOD C-terminus superposes well onto other structures, but its C-terminal carbonyl oxygen points away from E2 Asn85, pushing Cys93 away from the active site (Extended Data Fig. 3a,b). A model of the predicted tetrahedral intermediate requires minimal side chain movements and no alterations in positions of PCNA relative to E2-SUMOD (Extended Data Fig. 3c).
E3Siz1 SP-RING/SP C-terminal Domain Activates E2-SUMOD
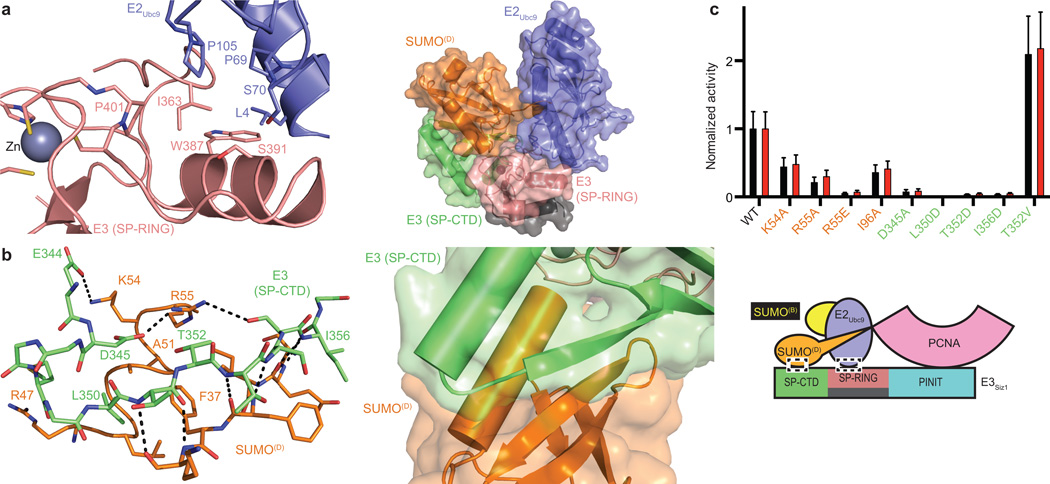
The SP-RING domain binds E2Ubc9 in a manner similar to E2 interactions with Ub RING domains (Fig. 2a; Extended Data Fig. 3d). Consistent with its function in general activation of E2-SUMOD, mutations in the E2/E3 interface including Siz1I363A, Siz1W387A and Siz1S391D, diminished conjugation to consensus and non-consensus lysine residues25,33.
Figure 2. E3 Activation of E2Ubc9-SUMOD.
a, E2/SP-RING interactions (left) and the structure with PINIT removed (right). b, SP-CTD/SUMOD interactions (left) and overview (right). c, Quantification of multiple turnover assays of SUMO modification of PCNA with coupled E1, E2, and E3 activities. Quantified rate data show mean ± s.d. (n=3 technical replicates).
The SP C-terminal domain (SP-CTD) was required for SP-RING domain activity, but it was unclear how it worked25. Unexpectedly, a SUMO interaction motif (SIM)-like element embedded within the SP-CTD supports binding of SUMO in its activated conformation (Fig. 2b), similar to other activated E2-Ubls (Extended Data Fig. 3e). The SP-CTD is integral to the catalytic module as mutations that disrupt the interface diminished conjugation to consensus and non-consensus lysines (Fig. 2c; Extended Data Fig. 3f) (ref. 25). SIMs usually include three or four hydrophobic amino acids bordered by acidic residues, with the hydrophobic amino acids centered in a β-strand contacting SUMO (reviewed in ref. 2). A hydrophobic substitution (T352V) that makes the SP-CTD more SIM-like increased activity. Mutations with no measureable effect included SUMOF37A, SUMOA51I, Siz1Y337A, Siz1Y337E and Siz1Q431A.
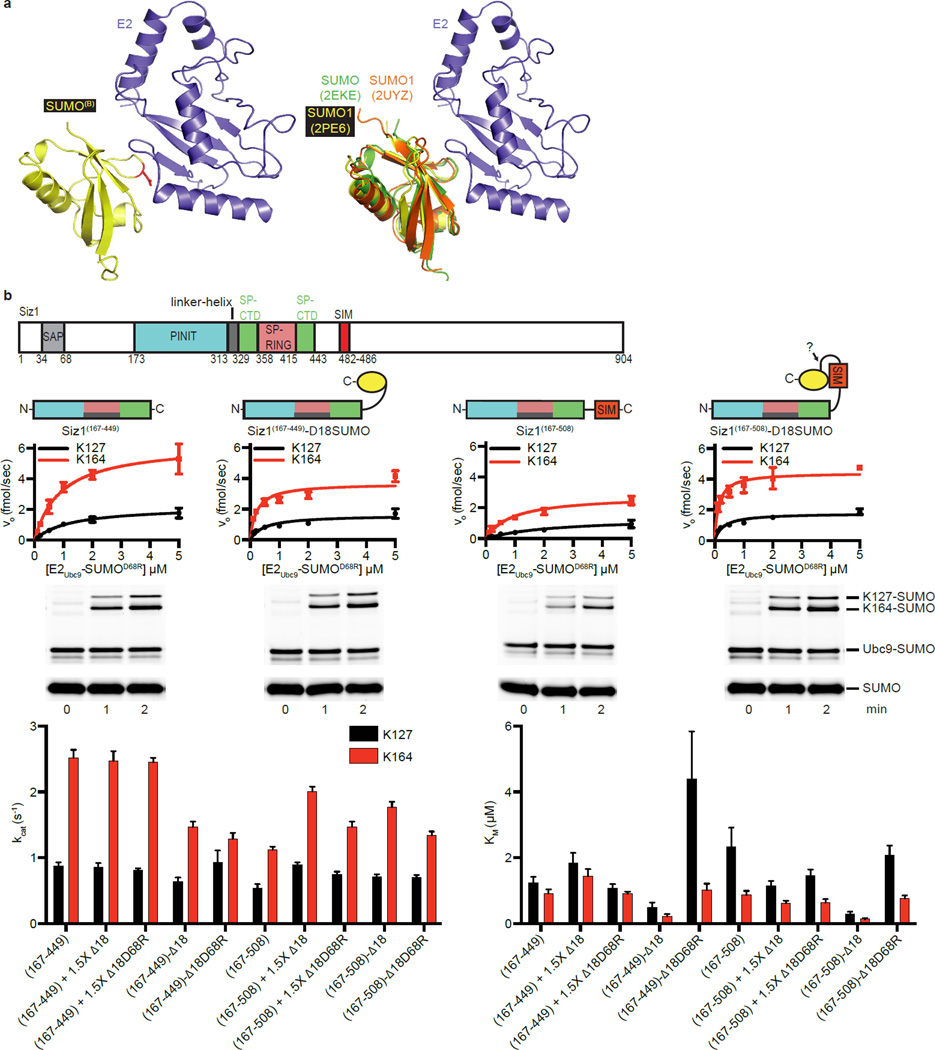
SUMOB Aids in E2-SUMOD Recruitment
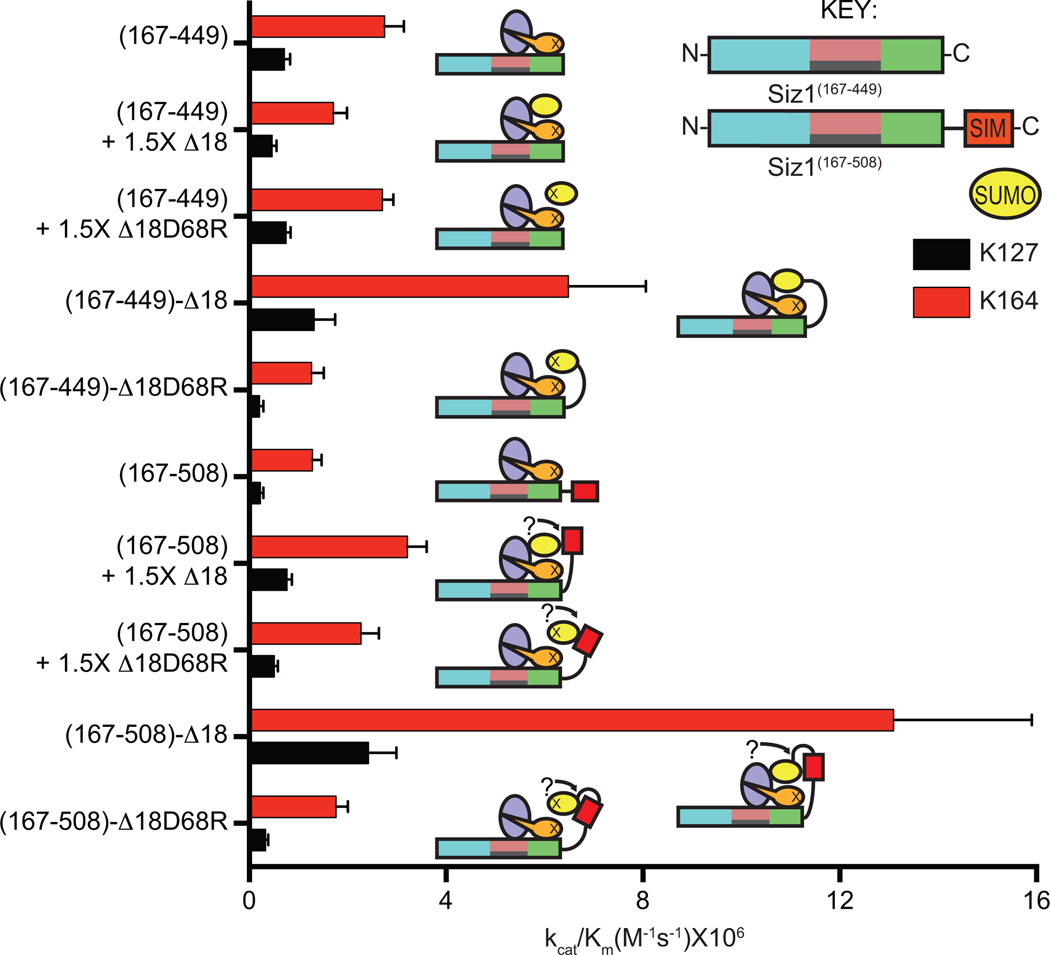
SUMOB adopts a similar configuration as observed in other noncovalent E2Ubc9/SUMOB complexes34–37 (Extended Data Fig. 4a). Several observations suggested that SUMOB could facilitate conjugation. In addition to the high affinity measured between E2Ubc9/SUMOB (Extended Data Fig. 1d; Extended Data Table 1b), Siz1 includes a SIM C-terminal to the SP-CTD38 that could interact with SUMOB. Finally, previous studies implicated E2/SUMOB interactions as important for PIAS and non-RING SUMO E3 activities33,34. To evaluate the E3Siz1 SIM and/or SUMOB in conjugation reactions, E3Siz1(167–449) (no SIM) was compared to E3Siz1(167–508) (plus SIM, residues 482–486) in the absence or presence of non-conjugatable SUMO or SUMOD68R at 1.5-fold molar excess, or as E3 C-terminal fusions.
Specific activity for E3Siz1(167–449) (no SIM) decreased slightly with exogenous SUMO and was unaffected by SUMOD68R, however specific activity for E3Siz1(167–508) (plus SIM) increased 2.6-fold with SUMO or 1.8-fold with SUMOD68R (Fig. 3; Extended Data Fig. 4b; Extended Data Table 2). Effects were most evident with Siz1-SUMOB fusions where kinetic data suggested that SUMO fusions increased activity by decreasing Km rather than increasing the rate constant Kcat, mirroring trends observed in the ubiquitin system21. These data suggest that SUMOB can enhance activity.
Figure 3. SUMOB Aids in E2Ubc9-SUMOD Recruitment.
Specific activities for E3 and E3-SUMO fusion construct-catalyzed multiple turnover reactions with E2-SUMOD68R thioester titrations with and without 1.5-fold excess SUMO. Quantified rate data show mean ± s.d. (n=3 technical replicates).
PCNA Binding and Substrate Specificity
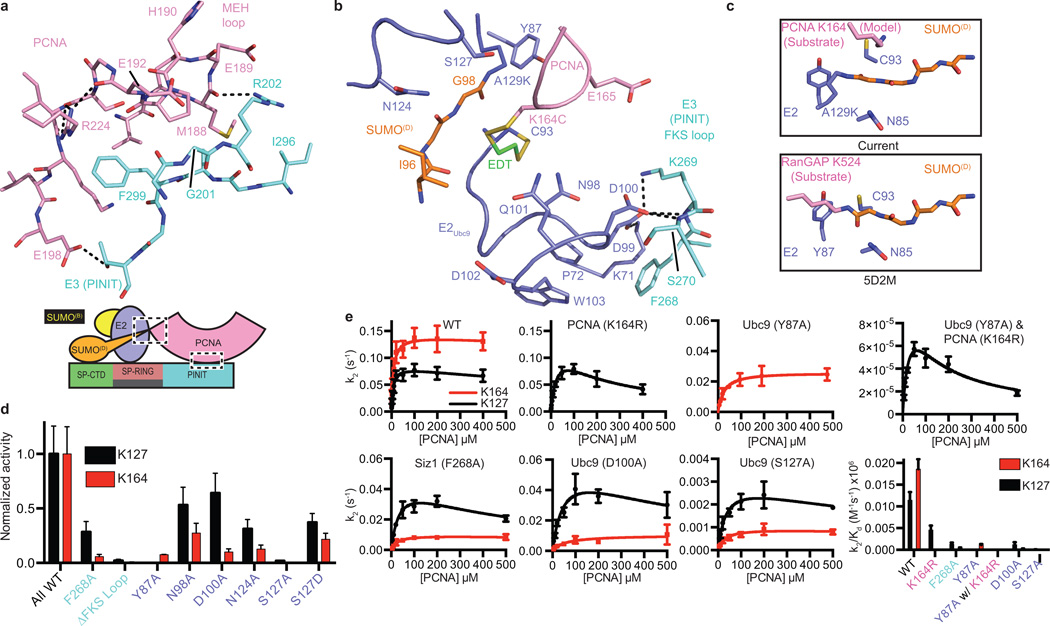
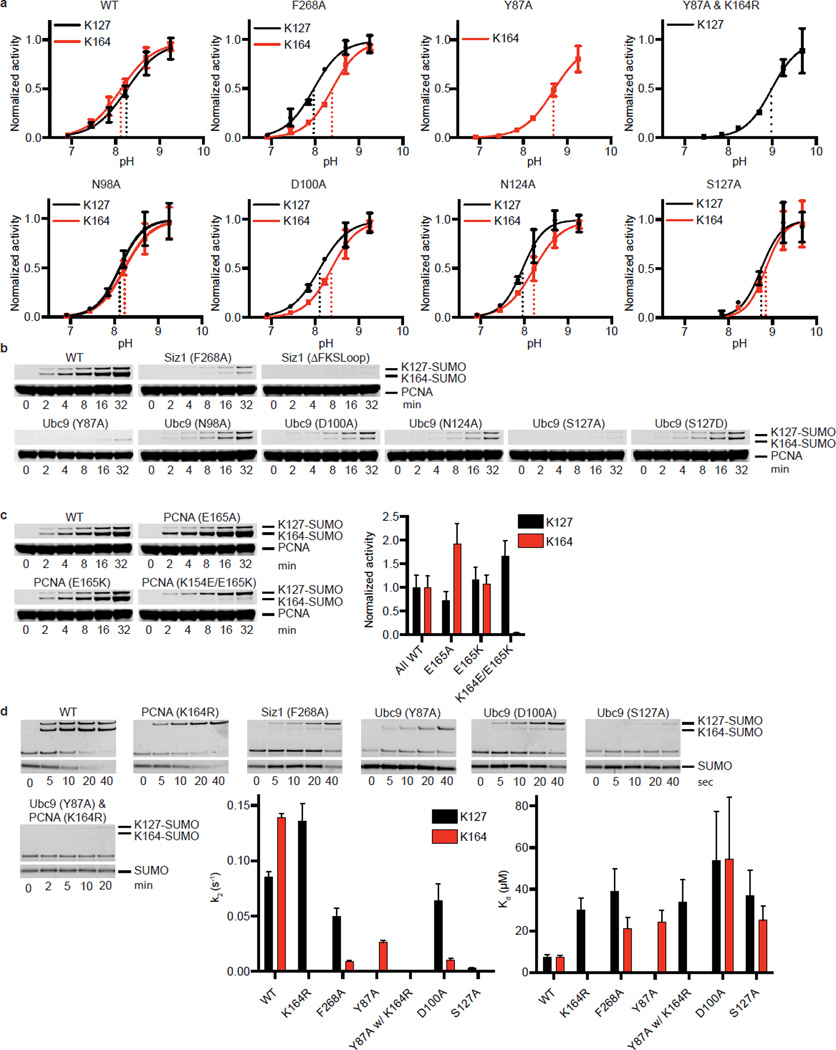
The E3Siz1 PINIT domain forms an interface between the E3 and substrate (Fig. 4a). Consistent with our structure and studies showing that the PINIT domain was required for PCNA Lys164 modification25, mutation of Siz1 Phe299 or Arg202, the PCNA MEH loop (Met188/Glu189/His190), or a combination, reduced or eliminated detectable modification at Lys164, but not Lys127. Because E2Ubc9 cannot modify PCNA Lys164, we posit that PINIT/PCNA interactions are required to force Lys164 into the E2 active site. Indeed, the modeled conformation for PCNA Lys164 differs from those observed for a SUMO consensus site lysine6–8,34, Lys63 from Ub9, or Arg720 from Cullin-1 (ref. 20) (Fig. 4b,c; Extended Data Fig. 3g).
Figure 4. E3/PCNA interactions and Lysine Specificity.
a, E3Siz1 PINIT/PCNA interactions. b, E2 active site interactions with the FKS loop from the E3Siz1 PINIT domain (EDT in green). c, Comparison of the E2Ubc9 active sites with PCNA or RanGAP1. d, Quantification of multiple turnover assays for SUMO modification of PCNA with coupled E1, E2 and E3 activities. e, Kinetics of single turnover assays with E2Ubc9-SUMOD68R thioester, E3 and PCNA. For panels d and e, quantified data show mean ± s.d. (n=3 technical replicates).
E2Ubc9 side chains near the active site coordinate the lysine nucleophile while lowering its pKa (ref. 8). We examined if E3Siz1 makes Lys164 a better nucleophile than Lys127 by differential effects on pKa suppression, however single turnover assays revealed no differences (Extended Data Fig. 5a). We next analyzed E2 mutations previously implicated in coordinating consensus lysine residues such as Lys127. As anticipated, Y87A disrupted Lys127 modification to below detection, however Lys164 modification was still evident, albeit diminished (Fig. 4d,e; Extended Data Fig. 5b). In contrast, S127A, S127D, N98A and N124A, selectively reduced activity toward Lys164 when compared to Lys127.
Unanticipated interactions were observed between the PINIT FKS loop (residues 268–270) and a loop containing E2Ubc9 Asp100 (Fig. 4b). Specifically, Siz1 Phe268 occupies a hydrophobic pocket on the E2 while backbone nitrogen atoms from Ser270 and Lys269 interact with backbone and side chain atoms of E2Ubc9 Asp100. Siz1F268A or deletion of the FKS loop reduced modification at both lysine residues, however defects were 6 to 7-fold greater for Lys164 (Fig. 4d). E2Ubc9D100A also decreased modification of Lys164 relative to Lys127. These mutations did not show differential pKa suppression for respective lysine nucleophiles (Extended Data Fig. 5a). Due to its proximity to Lys164, PCNA Glu165 was tested and eliminated as a potential catalytic residue for Lys164 modification (Extended Data Fig. 5c). No measureable defects were observed for E2Ubc9Q101A and E2Ubc9Q101K.
Catalytic defects described in preceding sections were explored using single turnover assays to calculate kinetics of modification at each lysine. Apparent Kd values were similar for each lysine using WT proteins (~7.5 µM), while the k2 was 1.6-fold higher for modification of Lys164 over Lys127 (Fig. 4e; Extended Data Fig. 5d; Extended Data Table 4). Selective defects for Lys164 modification by Siz1F268A, E2Ubc9D100A and E2Ubc9S127A were attributed to impaired turnover rather than binding. For instance, Siz1F268A reduced k2 by 1.7-fold for Lys127 versus 15.1-fold for Lys164, while apparent affinities increased 5-fold for Lys127 and 2.8-fold for Lys164. Lys127 modification of WT PCNA by E2Ubc9Y87A and E3Siz1 was below detection, necessitating use of PCNAK164R. While the apparent Kd increased 1.1-fold, a 1162-fold defect in k2 was observed for Lys127 modification (E2Ubc9 versus E2Ubc9Y87A with PCNAK164R). For comparison, the k2 for Lys164 modification decreased 5.3-fold (E2Ubc9 versus E2Ubc9Y87A with WT PCNA) while the apparent Kd increased 3.2-fold. Binding defects exist, but mutations altering lysine preference are best explained by differential rate defects, perhaps because these mutants fail to stabilize lysine in productive conformations for catalysis.
Kinetics for Lys127 modification was better modeled by accounting for substrate inhibition while Lys164 modification was better modeled without substrate inhibition (Fig. 4e; Extended Data Table 4). The apparent Ki for WT PCNA was 1430 µM, while the Ki for PCNAK164R was 190 µM. Mutations detrimental for Lys164 modification (E3Siz1F268A and E2Ubc9D100A) also resulted in substrate inhibition (Ki of ~400–500 µM). These results suggest that Lys164 is the preferred substrate. When Lys164 modification is impaired, these non-productive complexes behave as inhibitors for Lys127 modification. The high Ki value suggests inhibition is not a factor in vivo; however, these results illustrate a complex kinetic relationship between the two modifications.
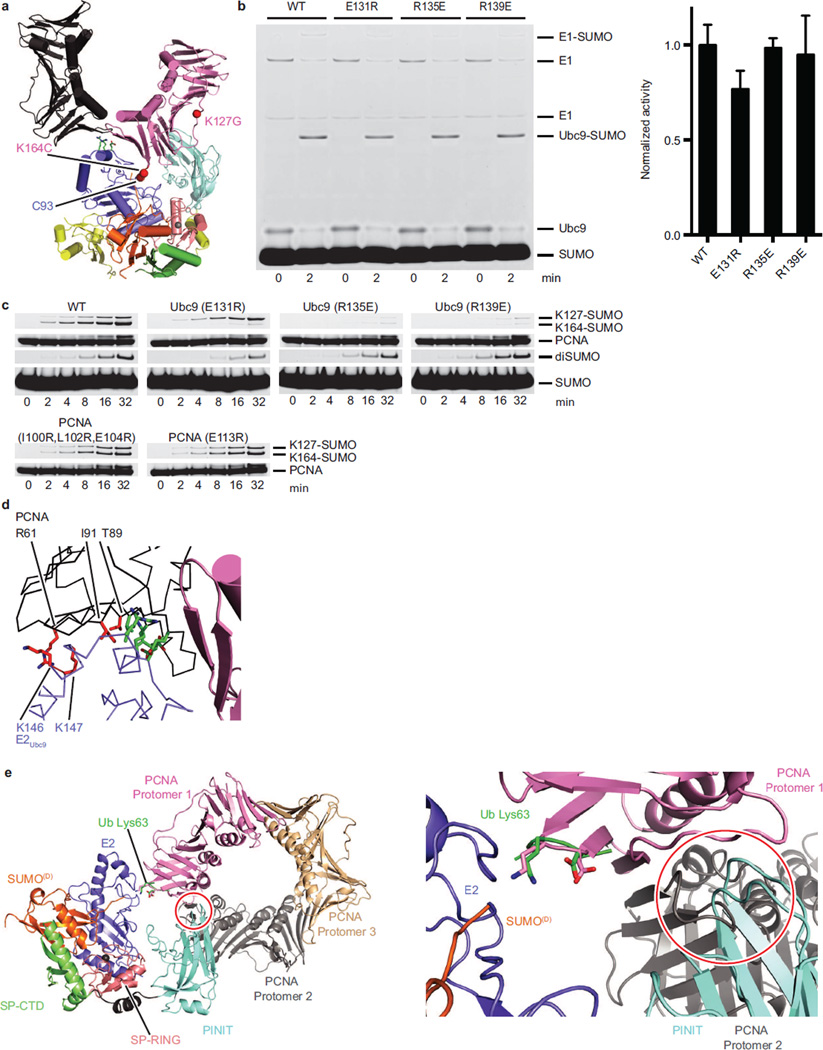
Surface Complementarity between E2-PCNA
PCNA exists as a trimer but our structure includes a monomer of PCNA. Lattice contacts in the crystal reveal PCNA/PCNA contacts reminiscent of the PCNA ring (Extended Data Fig. 6a) and docking trimeric PCNA onto PCNA in our structure revealed no backbone clashes and few side chain clashes, with E2Ubc9 accommodated in the concave surface at the PCNA/PCNA interface (Fig. 5a).
Figure 5. Surface Complementarity between E2Ubc9-SUMOD/E3 and PCNA.
a, Trimeric PCNA (black ribbon and grey cartoon) modeled onto the current structure (color). b, Location of E2 and PCNA residues with detrimental effects on activity. c, Multiple turnover assays of SUMO modification of PCNA with coupled E1, E2, and E3 activities. Quantified data show mean ± s.d. (n=3 technical replicates). d, Model of E2-SUMO/E3/Trimeric PCNA indicating the site for polymerase opposite the presumed position of the N-terminal E3Siz1 SAP domain.
Mutations were generated for E2Ubc9 on the penultimate alpha helix at positions predicted to point into the PCNA/PCNA interface (Fig. 5b). Each E2 mutant supported conjugation to a SUMO consensus site as evidenced by diSUMO formation (Fig. 5c; Extended Data Fig. 6b,c). In contrast, E2Ubc9E131R resulted in a 44-fold reduction for Lys164 modification and a 2.5-fold increase for Lys127 modification. E2Ubc9R135E and E2Ubc9R139E diminished modification at both sites, although defects were worse for Lys164. PCNAE104R/I100R/L104R or PCNAE113R also diminished conjugation to Lys164 relative to Lys127. In contrast, E2Ubc9K146E/K147E, PCNAR61E, PCNAT89R and PCNAI91R showed no measurable defect (Extended Data Fig. 6d). These data suggest that steric accommodation of E2Ubc9 within the PCNA/PCNA interface contributes to SUMO modification of PCNA at Lys164.
Conclusions
Strategies employed here to link an engineered E2-thioester mimetic and substrate at the endogenous site of modification with the same number of atoms as the predicted tetrahedral intermediate enabled structure determination of an E2-Ubl-Substrate/E3 ligase complex, providing a snapshot for otherwise transient states during conjugation. These methods could be used to trap other E2-Ubl/E3/Substrate complexes.
Our structure shows how the SP-RING domain interacts with E2Ubc9 and how the SP-CTD coordinates the closed E2-SUMO conformation via an embedded SIM-like motif, extending strategies observed for SIMs in two non-RING SUMO ligases, RanBP2 and ZNF451 (refs. 7,34). Our data expands the utility of a second Ubl (SUMOB) in promoting E2-Ubl/E3 complex formation, including a potential role for the Siz/PIAS SIM motif in recruiting activated SUMOB/E2Ubc9-SUMOD. The affinity measured for SUMOB/E2Ubc9 is ~1,000-fold better compared to other UbB/E2 interactions21, suggesting E2Ubc9 may constitutively bind SUMO in vivo. Other possible roles for SUMOB/E2Ubc9 interactions include feed forward mechanisms, SUMO modification of the E3 could enhance activity as proposed for ZNF451 (ref. 34), or SUMO modified proteins could amplify conjugation cascades by recruiting SUMO enzymes as proposed in the DNA damage response39.
E3-dependent modification of PCNA Lys164 does not result because the E3 alters E2 mediated pKa suppression of Lys164 or Lys127. Instead, specificity arises because the E3 binds the substrate to force-feed Lys164 into the E2 active site. This conformation appears particularly relevant for PCNA Lys164 modification as PCNA cannot be accommodated in the complex based on previously observed substrate lysine conformations (Extended Data Fig. 6e). Furthermore, residues surrounding the E2 active site play differential roles in coordinating Lys164 and Lys127, with a distinct subset of mutations that bias modification of consensus or non-consensus lysine residues (Fig. 4e). It is unclear if E2Rad6/E3Rad18 utilize a similar strategy to direct mono Ub modification of PCNA Lys164.
The three dimensional shape of the E2-Ubl/E3 complex must complement the shape of the substrate for selective modification of a particular lysine(s), a theme noted in other E3 ligase complexes9,20,40–42. In our structure, surface complementarity extends beyond E3 contacts to PCNA, and includes complementarity between the E2 and substrate. Although not required in vitro, SUMO modification of PCNA is enhanced when PCNA is loaded on DNA, while in vivo studies suggest loading of PCNA on DNA is a prerequisite for SUMO modification43. Our structure appears consistent with modification of PCNA on DNA as the predicted location of the N-terminal Siz1 SAP domain that binds duplex DNA is opposite from PCNA surfaces that interact with polymerase (Fig. 5d).
Methods
Cloning, expression and purification of recombinant proteins
Expression and purification of mature yeast SUMO (Smt3), Smt3K11C, ΔN18Smt3, yeast E1 (Aos1/Uba2ΔC-term1–554), yeast E2 (Ubc9, wt and K153R), yeast E3 active fragments (Siz1(167–465) & Siz1(167–508)) and tag free yeast PCNA (wt, K127G and K164R) have been described previously6,7,25,31,44,45. Point mutants of Smt3 (D68R, R55A, and R55E) were introduced into Smt3K11C by PCR mutagenesis and expressed and purified as above. Non-conjugatable ΔN18Smt3K19RΔGG97/98LeuGlyHis6Stop and ΔN18Smt3K19R/D68RΔGG97/98LeuGlyHis6Stop were generated by PCR amplification and insertion into pET28b with NcoI/XhoI without a stop codon, creating a C-terminal His6 extension. These were purified as the other Smt3 proteins without tag cleavage. The Ubc9C93K point mutant was generated by PCR mutagenesis and expressed and purified as Ubc9. Other Ubc9 point mutants (C5S/A129K, A129K, Y87A, N98A, D100A, D100E, S127A, S127D, N124A, E131R, R135E, and R139E) were introduced into Ubc9K153R (to minimize auto-conjugation46) by PCR mutagenesis and expressed and purified as Ubc9. The yeast Siz1C361D mutant was generated to minimize oxidative inactivation of the Siz1 SP-RING domain and has comparable activity to WT. Aspartic acid is found at the analogous position of Candida albicans and Caenorhabditis elegans Siz1 orthologs. Siz1(167–449) was cloned into the NdeI/XhoI sites of pET28b and purified as the Siz1(167–465). The Siz1(167–465) point mutants (D345A, F268A, I356D, L350D, T352D, and T352V) were generated by PCR mutagenesis and expressed and purified as Siz1(167–465). Siz1(167–465)ΔFKSLoop was generated by replacing Siz1(167–465) amino acid residues 267–274 with Gly-Ser-Gly by PCR mutagenesis and expressed and purified as Siz1(167–465). Siz1(167–449)C361D-ΔN18Smt3, Siz1(167–449)-ΔN18Smt3K19RΔGG97/98-L-E-His6, Siz1(167–449)-ΔN18Smt3K19R/D68RΔGG97/98-L-E-His6, Siz1(167–508)-ΔN18Smt3K19RΔGG97/98-L-E-His6, and Siz1(167–508)-ΔN18Smt3K19R/D68RΔGG97/98-L-E-His6 fusion proteins were generated by inserting the indicated Siz1 sequence into the pSmt3 (ref. 47) vector with BamHI/HindIII and subsequently the indicated ΔN18Smt3 sequences into the HindIII/XhoI sites 3’ of Siz1. These were expressed as N-terminal His6-Smt3 fusions that were cleaved of the N-terminal His6-Smt3 and purified as described previously for His6-Smt3-Siz1(167–465) (ref. 25). Yeast PCNAK77D/C81E/R110D and PCNAK77D/C81E/R110D/K127G/K164C were generated to create a monomeric versions of PCNA (residues were identified for ability to induce monomerization) with the latter capable of sulfhydryl based cross-linking at the K164 position. PCNA point mutants (I100R/L102R/E104R, E113R, K127R/K164C, E165A, E165K, E165A/E165K) were generated by PCR mutagenesis. All PCNA constructs were expressed and purified as WT PCNA.
The Ubc9C93K-Smt3 thioester mimetic was generated in a reaction containing 20 mM BIS-TRIS propane (pH 10.0), 50 mM NaCl, 10 mM MgCl2, 0.1% Tween-20, 2 mM ATP, 1 µM E1, 25 µM Ubc9C93K, and 200 µM Smt3 for 16 hours at 30°C and purified by Superdex75 and MonoQ. The Ubc9A129K/K153R-Smt3 thioester mimetic was generated in a reaction containing 20 mM BIS-TRIS propane (pH 9.5), 50 mM NaCl, 10 mM MgCl2, 0.1% Tween-20, 2 mM ATP, 1.0 µM E1, 200 µM Ubc9A129K/K153R, and 400 µM Smt3 for 1 hour at 30°C and purified by Superdex75. The Ubc9C5S/A129K/K153R-Δ18Smt3 thioester mimetic was generated in a reaction containing 20 mM BIS-TRIS propane (pH 9.5), 50 mM NaCl, 10 mM MgCl2, 0.1% Tween-20, 2 mM ATP, 0.5 µM E1, 100 µM Ubc9C5S/A129K/K153R, and 200 µM Smt3 for 1 hour at 30°C and purified by Superdex75.
Fluorescence Polarization
Smt3K11C was labeled with Alexa Fluor 488 (hereafter Alexa488) maleimide as recommended by the manufacturer. Smt3K11C-Alexa488, Smt3K11C/D68R-Alexa488, and Ubc9 were buffer exchanged into 20 mM HEPES (pH 7.5), 50 mM NaCl, 0.1% Tween-20, and 1 mM β-me. Fluorescence polarization was performed at 22°C with a SpectraMax M5 (Molecular Devices) microplate reader in 384-well microplates. The 20 µl incubations contained 50 nM Smt3K11C-Alexa488 or Smt3K11C/D68R-Alexa488 and buffer alone in the first well followed by a serial dilution of E2Ubc9 from 5 nM to 10 µM, performed in triplicate. Data was analyzed in Prism fitted to a single-site binding model accounting for receptor depletion, as described previously31.
Complex Reconstitution and Crystallization
Bismaleimidoethane (BMOE, Pierce) crosslinking: The purified Ubc9C5S/A129K/K153R-ΔN18Smt3 thioester mimetic (~800 µl at 493 µM) and yPCNAK77D/C81E/R110D/K127G/K164C (~480 µl at 3290 µM) were incubated with 1 mM TCEP (Soltec Ventures, reconstituted in 10 mM HEPES (pH 7.5)) for 15 minutes at 22°C. Each protein was desalted into cross-linking buffer (20 mM HEPES (pH 7.0), 200 mM NaCl and 5 mM EDTA). Twelve µl of 100 mM BMOE in DMSO was added to the E2-SUMO mimetic (~3 fold excess of BMOE) and incubated 3 min at 22°C and desalted to cross-linking buffer. The E2-Smt3-BMOE was mixed with desalted PCNA (~4 fold excess of PCNA) and incubated 15 min to form Ubc9C5S/A129K/K153R-ΔN18Smt3-BMOE-PCNAK77D/C81E/R110D/K127G/K164C and was quenched with 1 µl β-me. Complex was purified on Superdex200 in 20 mM TRIS-HCl (pH8.0), 350 mM NaCl, and 1 mM β-me, concentrated, adjusted to 200 mM NaCl and purified on MonoQ.
1,2-ethanedithiol (EDT, Sigma) crosslinking: The purified Ubc9C5S/A129K/K153R-ΔN18Smt3 thioester mimetic (~400 µl at 1238 µM) and yPCNAK77D/C81E/R110D/K127G/K164C (~825 µl at 1575 µM) were incubated with 1 mM TCEP for 15 minutes at 22°C. Both were exchanged to cross-linking buffer. Four µl of 0.3 M Aldrithiol-2 (AT2, Sigma) in DMSO was added to the desalted E2-ΔN18Smt3, incubated 15 min at 22°C, and desalted to cross-linking buffer. Five µl of 0.3 M EDT in DMSO was added to the desalted E2-ΔN18Smt3-AT2, incubated 15 min at 22°C, and desalted to cross-linking buffer. Five µl of 0.3 M AT2 was added to the desalted E2-ΔN18Smt3-EDT, incubated 15 min at 22°C, and desalted to cross-linking buffer. The desalted E2-ΔN18Smt3-EDT-AT2 was mixed with the desalted PCNA (~3 fold excess of PCNA) and incubated 20 min at 22°C to form Ubc9C5S/A129K/K153R-ΔN18Smt3-EDT-PCNAK77D/C81E/R110D/K127G/K164C. The complex was purified by Superdex200 and MonoQ chromatography as for BMOE complex, excluding β-me.
To reconstitute the final complex for crystallization, the purified Ubc9-ΔN18Smt3-EDT-PCNA complex was mixed with the purified Siz1(167–449)C361D-ΔN18Smt3 fusion in 1:1 ratio. This was dialyzed versus 20 mM TRIS-HCl (pH 8.0), 50 mM NaCl and 5 mM EDTA and resolved by Superdex200 in same buffer. Peak fractions were concentrated to ~7.5 mg/ml, supplemented with TCEP to 1 mM, aliquoted, and flash frozen at −80°C for later use. The complex was crystallized at 18°C by hanging-drop vapor diffusion by mixing 0.5 µl of the complex with 0.5 µl of the reservoir solution containing 0.1 M TRIS-HCl (pH 8.5), 5% PEG 10,000, 0.2 M NaCl, 10% glycerol, and 3% dioxane. Crystals were cryoprotected by gradually increasing the glycerol concentration in the drop by repeated additions of well solution supplemented with 30% glycerol. The crystal was removed from the drop and swiped through another drop of the well solution supplemented with 30% glycerol and then flash cooled in liquid nitrogen. Data was collected at 100K at the 24-IDE beamline at APS with an ADSC Q315 CCD detector at a 0.979 Å wavelength. Data was indexed, integrated, and scaled with HKL-2000 (ref. 48) to a 2.85 Å resolution. Molecular replacement was performed with Phenix49 using the crystal structures of Ubc9, Smt3, Siz1, and PCNA (2EKE, 2EKE, 3ID2 and 1PLQ, respectively) as search models. Refinement was performed with Phenix and model building was performed with Coot50 and CNS51,52. The geometry of the structure was analyzed with MolProbity53. 96.1% of residues are found in the favored configuration, with 0.06% Ramachandran outliers (1 residue). The structure has a clash score of 2.3 (100th percentile) and a MolProbity score of 1.28 (100th percentile). Figures were prepared with PyMol (http://www.pymol.org/).
Multiple turnover assay with purified Ubc9K153R-Smt3K11C/D68R-Alexa488 Thioester
Smt3K11C/D68R was labeled with Alexa488-maleimide (Life Technologies) as recommended by manufacturer. The Ubc9K153R-Smt3K11C/D68R-Alexa488 thioester was formed in a reaction mixture containing 20 mM HEPES (pH7.5), 50 mM NaCl, 10 mM MgCl2, 0.1% Tween-20, 2 mM ATP, 0.4 mM DTT, 11 µM E1, 200 µM Ubc9K153R, and 100 µM Smt3K11C/D68R-Alexa488 for 5 minutes at 30°C. The thioester was diluted and purified by Superdex75 in 50 mM NaCitrate (pH 5.5), 200 mM NaCl, 5% glycerol, concentrated, aliquoted and flash frozen at −80°C for later use. A serial dilution of the purified thioester was prepared in 20 mM NaCitrate (pH 5.5), 50 mM NaCl, and 5% glycerol. Ten µl of the thioester dilutions were delivered to a 50 µl reaction mixture containing 50 mM HEPES (pH 7.5), 50 mM NaCl, 0.1 % Tween-20, 1 nM of the indicated E3, and 32 µM PCNA, and incubated at 30°C. To determine the Ki for thioester mimetics, parallel reaction mixtures included the thioester mimetic at concentrations indicated. For the experiments investigating the role of backside bound SUMO, a 1.5-fold excess of the indicated nonconjugatable ΔN18Smt3 was included in the thioester serial dilution. Aliquots were removed at 1 and 2 minutes and quenched in equal volume of 4X LDS NuPAGE loading dye (Life Technologies), resolved by non-reducing 12% SDS-PAGE with MOPS running buffer (Life Technologies), and imaged on Typhoon FLA 9500 with a 473-nm laser and an LPB filter. All gels were imaged with a serial dilution Smt3Alexa488 reference gel to convert band intensity to pmol of conjugate with ImageJ (NIH). Experiments were performed in triplicate. Rates were determined by plotting the pmol of conjugates versus time in Excel. Rates were plotted versus E2-thioester concentration in Prism (GraphPad) and fitted to the equation v=Vmax[S]/(KM+[S]), where Vmax=kcat[E]t, [E]t is the E3 concentration, Km is the substrate concentration at the half-maximal velocity, and [S] is the substrate concentration. The Ki was measured by fitting the rate data for all the inhibitor concentrations [I] to the equation v=Vmax[S]/(KM(1+([I]/Ki))+[S]) in Prism.
Multiple turnover assay with coupled E1, E2, and E3 Activities
Reaction mixtures containing 20 mM HEPES (pH7.5), 50 mM NaCl, 10 mM MgCl2, 0.1% Tween-20, 2 mM ATP, 1 mM DTT, 200 nM E1, 100 nM of indicated E2Ubc9, 50 nM of the indicated Siz1(167–465), 80 µM of the indicated Smt3, and 4 µM of the indicated PCNA were incubated at 30°C. Aliquots were removed at the indicated times and quenched in equal volume of 4X LDS NuPAGE loading dye with 1 M β-mercaptoethanol (β-me), resolved by 12% SDS-PAGE with MOPS running buffer. Proteins were stained with SYPRO Ruby (Bio-Rad) and imaged on Typhoon FLA 9500 with a 473-nm laser and an LPG filter. Band intensities were integrated with ImageJ and plotted against time in Excel to determine rates relative to wild type. Experiments were performed in triplicate.
Single turnover assays with purified Ubc9K153R-Smt3K11C/D68R-Alexa488 and Ubc9 Mutant Thioesters and Siz1 Mutants25,45
Ubc9Y87A/K153R-Smt3K11C/D68R-Alexa488, Ubc9N98A/K153R-Smt3K11C/D68R-Alexa488, Ubc9D100A/K153R-Smt3K11C/D68R-Alexa488, Ubc9N124A/K153R-Smt3K11C/D68R-Alexa488, Ubc9S127A/K153R-Smt3K11C/D68R-Alexa488 and Ubc9S127D/K153R-Smt3K11C/D68R-Alexa488 thioesters were formed and purified as above. Ten µl of 25 nM of the indicated purified thioester (diluted in 20 mM NaCitrate (pH 5.5), 50 mM NaCl, and 5% glycerol) were delivered to a 50 µl reaction mixture containing 50 mM HEPES (pH 7.5), 50 mM NaCl, 0.1 % Tween-20, 50 nM of the indicated E3, and the indicated concentration of PCNA, and incubated at 4°C. Aliquots were removed at the indicated times, quenched with equal volume of 4X LDS NuPAGE loading dye, resolved by non-reducing 12% SDS-PAGE with MOPS running buffer, and imaged and quantitated as above. Experiments were performed in triplicate. Rates were determined by plotting pmol of conjugates versus time in Excel. Due to the speed of reaction, rates at the higher PCNA concentrations were estimated by the single 5 sec time point. Rates were plotted versus PCNA concentration in Prism (GraphPad) and fitted to the equation v=Vmax[S]/(Kd+[S]), where Vmax=k2[E]t for Lys164 data, where [E]t is E2-Smt3 thioester concentration, Kd is the apparent dissociation constant, and [S] is the substrate concentration. Rates plotted versus PCNA concentration were fitted to the equation v=Vmax[S]/(Kd + [S](1+[S]/Ki)) for Lys127 data, where Vmax=k2[E]t, [E]t is E2-Smt3 thioester concentration, Kd is the apparent dissociation constant, [S] is the substrate concentration, and Ki is the dissociation constant for substrate binding modeled by considering that two substrates can bind to one enzyme.
Multiple turnover assays with purified E2 thioester at various pH
These assays were performed similar to those described for the previous multiple turnover assays, except the reaction and dilution buffers contained BIS-TRIS propane (Sigma) with the pH adjusted from 6.35 to 9.75, measured at 4°C. The indicated purified E2-thioester was diluted to 700 nM (in 20 mM NaCitrate (pH 5.5), 50 mM NaCl, and 5% glycerol) and 10 µl was delivered to a 70 µl reaction containing 50 mM BIS-TRIS propane (pH as indicated), 50 mM NaCl, 0.1 % Tween-20, 5 nM of the indicated Siz1(167–465) and 4 µM PCNA and incubated at 4°C. Besides the E2-thioester dilution, each protein for each reaction was diluted immediately prior to initiation to minimize pH effects on protein stability. Aliquots were removed at the indicated times, quenched with equal volume of 4X LDS NuPAGE loading dye, resolved by non-reducing 12% SDS-PAGE with MOPS running buffer, and imaged and quantitated as above. Experiments were performed in triplicate.
Extended Data
Extended Data Figure 1. E2Ubc9-SUMO Thioester Mimic and Cross-Linking to Substrate PCNA for Reconstitution With E3Siz1.
a, SDS-PAGE analysis of in vitro E2Ubc9A129K or E2Ubc9C93K charging with SUMO in the presence and absence of E3Siz1(167–465) at pH (7.5) (left) and purification of the E2Ubc9C93K-SUMO (middle) and E2Ubc9A129K-SUMO (right) thioester mimetics. b, Plots of rates for in vitro SUMO modification of PCNA in assays utilizing various concentrations of purified E2Ubc9-SUMOD68R-Alexa488 labeled thioester, 1 nM E3Siz1(167–465) and 32 µM PCNA with 0, 2, 5 or 20 µM of the E2Ubc9C93K-SUMO or E2Ubc9A129K-SUMO thioester mimic (left) with exemplary non-reducing SDS-PAGE for the 0.5 µM E2Ubc9-SUMOD68R-Alexa488 reactions (right). The calculated Km and Ki from these fits are shown in Extended Data Tables 1a and 2 and the quantified data show mean ± s.d. (n=3 technical replicates). c, SDS-PAGE analysis (left) of numbered 0.5 ml fractions from Superose6 analytical gel-filtration analysis (right) of complex reconstitution between E2Ubc9-SUMO-BMOE-PCNA and E3Siz1(167–465) (green) or the E3Siz1(167–465)-SUMO fusion (blue). Elution profiles for E2Ubc9-SUMO-BMOE-PCNA (purple) and E3Siz1(167–465) (red) alone are shown. d, Plot of the normalized change in polarization observed upon addition of serially diluted E2Ubc9 with Alexa488 labeled SUMO or SUMOD68R. Data were fit to single-site binding model accounting for receptor depletion. Data show mean ± s.d. (n=3 technical replicates). For gel source data, see Supplementary Figure 1.
Extended Data Figure 2. Comparing Strategies for Crosslinking the E2-SUMO Thioester Mimic and Substrate PCNA.
a, Chemical structures of the proposed tetrahedral intermediate formed during PCNA Lys164 attack of E2Ubc9-SUMO thioester (left), a BMOE cross-link (middle) or an EDT cross-link (right) between E2Ubc9-SUMO C93 and PCNA K164C. Indicated distances were estimated with ChemDraw15 (PerkinElmer). b, Control non-reducing SDS-PAGE panel for Fig. 1a showing mock treated PCNA K127R/K164C (DMSO instead of EDT in DMSO) is unable to accept transthioesterification of SUMO at position 164. c, SDS-PAGE analysis of the 5 ml fractions from the final preparative Superdex200 gel-filtration purification of the E2Ubc9-SUMO-EDT-PCNA/ E3Siz1(167–449)-SUMO complex. For gel source data, see Supplementary Figure 1.
Extended Data Figure 3. E2Ubc9 Active Site, Conformation of SUMOD and Comparison to Relevant Structures.
a, Stereo image of simulated annealing electron density map showing the EDT linkage and the SUMO Gly98 linkage to E2Ubc9 A129K. The 2Fo-Fc electron density map is contoured at 0.8σ (grey mesh). b, Alignment of the E2s from the current structure, SUMO modified RanGAP1 bound to E2Ubc9K14R and E3Znf451 (5D2M), E2Ubc5BS22R/N77A/C85S-Ub bound to the RING dimer from E3BIRC7 (4AUQ) and E2 E2Ubc5AS22R/C85K-Ub bound to the RING dimer from E3RNF4 (4AP4) showing two orientations of the E2 active site. c, Model of tetrahedral intermediate generated by comparing our structure to other structures of E2-Ubl/E3 complexes, particularly Protein Data Bank (PDB) 5DM2 and 4P5O. d, Alignment of the current structure and three E2/RING (1UR6, 3EB6, and 3FN1) complexes and one E2/UBox (2C2V) complex (aligned by the E2). e, Alignments of four E2-Ubls/E3 complexes (aligned by the E2) in the closed activated confirmation for the current structure, E2Ubc9 K14R -SUMO (5D2M), E2Ubc5AS22R/C85K-Ub (4AP4) and E2Ubc12N103S/C111S-Nedd8 (4P5O). f, SDS-PAGE analysis of multiple turnover assays of SUMO modification of PCNA utilizing in vitro reactions with coupled E1 (200 nM), E2 (100 nM), and E3 (50 nM) activities with 4 µM PCNA for the quantified data shown in Fig. 2c. g, Alignments of the E2 from relevant structures with lysine or arginine residues within or projecting toward the E2 active sites compared to the current structure. Lysine 63 from acceptor ubiquitin projecting toward the active site of the E2Ubc13C87S-Ub is shown in green (2GMI). Lysine 524 from SUMO modified RanGAP1 laying across the active site of E2Ubc9K14R is shown in magenta. The Lys720Arg from Cullin-1 projecting into the active site of E2Ubc12-Nedd8 is shown in grey (4P5O). For the current structure, EDT was removed from the model, Cys164 was mutated back to lysine and the side chain was fit to the electron density and is shown in pink in reference to the current E2 (blue) and donor SUMO (orange). For gel source data, see Supplementary Figure 1.
Extended Data Figure 4. SUMOB Bound to the E2 Backside enhances E2Ubc9-SUMO Recruitment.
a, Alignment of the current E2Ubc9/backside SUMOB (left) to previously observed E2Ubc9/backside SUMO complexes (right). The position of the D68R mutation is shown in red sticks (left). b, Primary E3Siz1 structure (top). Cartoons indicating the E3Siz1 or E3Siz1-SUMO fusion constructs utilized in the multiple turnover in vitro assays (middle) shown in Fig. 3 utilizing a titration of the purified E2Ubc9-SUMOD68R-Alexa488 thioester with or without 1.5-fold excess of the indicated additional molecule of non-conjugatable SUMO, 1 nM of the indicated E3 construct and 32 µM PCNA. Representative non-reducing SDS-PAGE showing the 0.5 µM E2Ubc9-SUMOD68R-Alexa488 thioester reactions below the plots of the rates of reaction for each E2Ubc9-SUMOD68R concentration (middle). The kinetics of SUMO modification of PCNA were calculated and the Km and kcat were determined (bottom) and are also shown in Extended Data Table 2. The quantified rate data show mean ± s.d. (n=3 technical replicates). For gel source data, see Supplementary Figure 1.
Extended Data Figure 5. E2Ubc9 and E3Siz1 Determinants of Lysine Specificity.
a. Plots of the rates observed at different pH values for multiple turnover in vitro assays of SUMO modification of PCNA utilizing 0.1 µM purified E2Ubc9-SUMOD68R-Alexa488 thioester (or E2Ubc9 mutant thioesters) with 5 nM E3Siz1 and 4 µM PCNA at 4°C. b, SDS-PAGE analysis of multiple turnover assays of SUMO modification of PCNA utilizing in vitro reactions with coupled E1 (200 nM), E2 (100 nM), and E3 (50 nM) activities with 4 µM PCNA for the quantified data shown in Fig. 4d. c, SDS-PAGE analysis of multiple turnover assays of SUMO modification of PCNA utilizing in vitro reactions with coupled E1 (200 nM), E2 (100 nM), and E3 (50 nM) activities with 4 µM PCNA and quantified. d, Representative non-reducing SDS-PAGE analysis of the single turnover in vitro assays of SUMO modification of PCNA shown in Fig. 4e. These assays utilize 5 nM of the E2Ubc9-SUMOD68R-Alexa488 thioester (or E2Ubc9 mutant thioesters) in reactions with 50 nM of the indicated E3Siz1 and a titration of PCNA. Shown are typical SDS-PAGE analyses from the 10 µM PCNA reactions. The data were used to extract the kinetic constants for the reactions shown as histograms and in Extended Data Table 4. For panels a,c and d the quantified rate data show mean ± s.d. (n=3 technical replicates). For gel source data, see Supplementary Figure 1.
Extended Data Figure 6. Shape Complementarity between the E2Ubc9-SUMO/E3 Complex and PCNA.
a, The current structure (color) with the crystallographic packing of a lattice mate PCNA molecule (black). b, Non-reducing SDS-PAGE analysis of 2 minute endpoint in vitro E2Ubc9-SUMO thioester formation reactions with 0.05 µM E1, 0.4 µM of the indicated E2Ubc9 and 22 µM SUMO (left) and the quantitated E2-SUMO band (right). The quantified band intensity shows mean ± s.d. (n=3 technical replicates). c, SDS-PAGE analysis of multiple turnover assays of SUMO modification of PCNA utilizing in vitro reactions with coupled E1 (200 nM), E2 (100 nM), and E3 (50 nM) activities with 4 µM PCNA or without PCNA (diSUMO formation) shown quantified in Fig. 5c. d, Location of E2Ubc9 and PCNA mutations that had no effect (red sticks) on activities observed for in vitro assays similar to those performed in Fig. 5c in relation to residues that did show effects (green sticks). e, The E2Ubc13C87S-Ub was aligned to E2Ubc9 in the current structure and subsequently the Lys164/Glu165 loop from trimeric PCNA (pink) was aligned onto the Lys63/Glu64 loop from acceptor ubiquitin (2GMI, green). Within this conformation the E3Siz1 PINIT domain (cyan) clashes with another protomer of the PCNA trimer (grey). For gel source data, see Supplementary Figure 1.
Extended Data Table 1.
Calculated Km for E2Ubc9-SUMOD68R-Alexa488 binding to E3Siz1(167–465) and calculated Ki values for competitive inhibition of this interaction by the indicated thioester mimetic using multiple turnover SUMO conjugation of PCNA at 30° C with 100 nM–5 µM purified E2Ubc9-SUMOD68R-Alexa488 thioester with 1 nM E3 and 32 µM PCNA in the absence and presence of 2–20 µM of the indicated mimetic. Data show mean ± s.d. (n=3 technical replicates).
| Extended Data Table 1a. Summary of Kinetic Constants or Inhibition Constants for E2Ubc9-SUMO Thioester and Thioester Mimetic Association to E3Siz1. | |||
|---|---|---|---|
| Thioester Mimetic | Km (µM) | Ki (µM) | |
| - | K127 | 1.07 ± 0.10 | - |
| C93K | K127 | 0.96 ± 0.08 | 2.05 ± 0.19 |
| A129K | K127 | 1.11 ± 0.07 | 4.33 ± 0.33 |
| - | K164 | 0.70 ± 0.07 | - |
| C93K | K164 | 0.72 ± 0.05 | 1.44 ± 0.13 |
| A129K | K164 | 0.63 ± 0.05 | 3.15 ± 0.27 |
| Extended Data Table 1b. Summary of Binding Curve Fits of Fluorescent Polarization Data for Ubc9 Binding Alexa488 Labeled SUMO or SUMOD68R. | ||
|---|---|---|
| FP (Receptor Depletion Model) | SUMO-Alexa488 | SUMOD68R-Alexa488 |
| Best Fit Values (+/− Std. Error) | ||
| Limiting anisotropy free ligand | 0.09 ± 2.77 | −0.02 ± 0.51 |
| Limiting anisotropy bound ligand | 99.94 ± 1.67 | 100.00 ± 236.90 |
| [Ligand] | = 50.00 nM | = 50.00 nM |
| Kd (nM) | 24.6 ± 4.4 | 62,819 ± 169,826 |
| 95% Confidence Intervals | ||
| Limiting anisotropy free ligand | −5.55 to 5.73 | −1.07 to 1.02 |
| Limiting anisotropy bound ligand | 96.55 to 103.3 | −381.9 to 581.9 |
| Kd (nM) | 15.57 to 33.62 | 0.0 to 408,332 |
| Goodness of Fit | ||
| Degrees of Freedom | 33 | 33 |
| R square | 0.97 | 0.75 |
| Absolute Sum of Squares | 1253 | 188.4 |
| Sy. x | 6.16 | 2.39 |
| Constraints | ||
| [Ligand] | = 50.00 nM | = 50.00 nM |
| Kd | Kd > 0.0 | Kd > 0.0 |
| Number of Points | ||
| Analyzed | 36 | 36 |
Extended Data Table 2. Summary of Kinetic Constants For Multiple Turnover Experiments with Purified E2Ubc9-SUMOD68R Thioester.
(top) Catalytic constants for multiple turnover SUMO conjugation of PCNA to K127 at 30° C with 100 nM–5 µM purified E2Ubc9-SUMOD68R-Alexa488 thioester with and without a 1.5-fold excess of an additional molecule of SUMO, 1 nM E3 and 32 µM PCNA. Data show mean ± s.d. (n=3 technical replicates). (bottom) Catalytic constants for multiple turnover SUMO conjugation of PCNA to K164 at 30° C with 100 nM–5 µM purified E2Ubc9-SUMOD68R-Alexa488 thioester with and without a 1.5-fold excess of an additional molecule of SUMO, 1 nM E3 and 32 µM PCNA. Data show mean ± s.d. (n=3 technical replicates).
| Sizl Isoform | Additional SUMO Isoform | Km (µM) | kcat (s−1) | kcat/ Km (M−1/s−1) |
| 167–465 | - | 1.07 ± 0.10 | 1.27 ± 0.05 | 1.18 × 106 ± 0.12 × 106 |
| 167–449 | - | 1.23 ± 0.19 | 0.88 ± 0.05 | 7.15 × 105 ± 0.12 × 105 |
| 167–449 | Δ18Smt3ΔGGHIS | 1.84 ± 0.31 | 0.86 ± 0.06 | 4.68 × 105 ± 0.09 × 105 |
| 167–449 | Δ18Smt3D68RΔGGHIS | 1.08 ± 0.12 | 0.81 ± 0.03 | 7.49 × 105 ± 0.09 × 105 |
| 167–449-Δ18Smt3ΔGGHIS | - | 0.48 ± 0.15 | 0.64 ± 0.06 | 1.32 × 106 ± 0.43 × 106 |
| 167–449-Δ18Smt3D68RΔGGHIS | - | 4.40 ± 1.44 | 0.93 ± 0.18 | 2.11 × 105 ± 0.08 × 105 |
| 167–508 | - | 2.34 ± 0.58 | 0.54 ± 0.06 | 2.31 × 105 ± 0.06 × 105 |
| 167–508 | Δ18Smt3ΔGGHIS | 1.15 ± 0.14 | 0.89 ± 0.04 | 7.71 × 105 ± 0.10 × 105 |
| 167–508 | Δ18Smt3D68RΔGGHIS | 1.46 ± 0.18 | 0.75 ± 0.04 | 5.13 × 105 ± 0.07 × 105 |
| 167–508-Δ18Smt3ΔGGHIS | - | 0.29 ± 0.07 | 0.71 ± 0.04 | 2.43 × 106 ± 0.57 × 106 |
| 167–508-Δ18Smt3D68RΔGGHIS | - | 2.07 ± 0.29 | 0.70 ± 0.04 | 3.38 × 105 ± 0.05 × 105 |
| Sizl Isoform | Additional SUMO Isoform | Km (µM) | kcat (s−1) | kcat/ Km (M−1/s−1) |
| 167–465 | - | 0.70 ± 0.07 | 4.26 ± 0.14 | 6.09 × 106 ± 0.69 × 106 |
| 167–449 | - | 0.91 ± 0.12 | 2.52 ± 0.12 | 2.76 × 106 ± 0.39 × 106 |
| 167–449 | Δ18Smt3ΔGGHIS | 1.44 ± 0.21 | 2.47 ± 0.15 | 1.71 × 106 ± 0.27 × 106 |
| 167–449 | Δ18Smt3D68RΔGGHIS | 0.90 ± 0.07 | 2.45 ± 0.07 | 2.71 × 106 ± 0.22 × 106 |
| 167–449-Δ18Smt3ΔGGHIS | - | 0.23 ± 0.05 | 1.47 ± 0.08 | 6.48 × 106 ± 1.58 × 106 |
| 167–449-Δ18Smt3D68RΔGGHIS | - | 1.02 ± 0.19 | 1.29 ± 0.09 | 1.26 × 106 ± 0.25 × 106 |
| 167–508 | - | 0.91 ± 0.12 | 1.12 ± 0.05 | 1.29 × 106 ± 0.18 × 106 |
| 167–508 | Δ18Smt3ΔGGHIS | 0.62 ± 0.07 | 2.01 ± 0.07 | 3.22 × 106 ± 0.39 × 106 |
| 167–508 | Δ18Smt3D68RΔGGHIS | 0.65 ± 0.10 | 1.47 ± 0.08 | 2.27 × 106 ± 0.36 × 106 |
| 167–508-Δ18Smt3ΔGGHIS | - | 0.14 ± 0.03 | 1.77 ± 0.08 | 13.09 × 106 ± 2.81 × 106 |
| 167–508-Δ18Smt3D68RΔGGHIS | - | 0.76 ± 0.10 | 1.34 ± 0.06 | 1.77 × 106 ± 0.24 × 106 |
Extended Data Table 3. Data collection and refinement statistics.
A single crystal was used.
| E2Ubc9-SUMO/E3sizl-SUMO/PCNA | |
|---|---|
| Data collection | |
| Space group | C121 |
| Cell dimensions | |
| a, b, c (Å) | 93.42, 205.88, 142.50 |
| α, β, γ (°) | 90.00, 95.30, 90.00 |
| Resolution (Å) | 48.4-2.85 (2.95-2.85)* |
| Rmerge | 10.7 (55.6) |
| I/σI | 9.1 (1.82) |
| Completeness (%) | 99.0 (98.0) |
| Redundancy | 3.6 (2.9) |
| Refinement | |
| Resolution (Å) | 47.3-2.85 |
| No. reflections | 61981 |
| Rwork / Rfree | 0.210/0.248 |
| No. atoms | 13744 |
| Protein | 13403 |
| Ligand/ion | 70 |
| Water | 271 |
| B-factors | |
| Protein | 58.3 |
| Ligand/ion | 64.5 |
| Water | 43.0 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.001 |
| Bond angles (°) | 0.42 |
Values in parentheses are for highest-resolution shell.
Extended Data Table 4. Summary of Kinetic Constants For Single Turnover Experiments with Purified E2Ubc9-SUMOD68R Thioester.
(Top) Catalytic constants for single turnover SUMO conjugation of PCNA to K127 at 4° C with 5 nM purified E2Ubc9-SUMOD68R-Alexa488 thioester, 50 nM E3 and 0.5–500 µM PCNA as indicated. Data show mean ± s.d. (n=3 technical replicates).
| Ubc9- Smt3D68RAlexa488 Isoform |
Sizl(167–465) Isoform |
PCNA Isoform |
Kd (µM) |
k2 (s−1) |
k2/Kd (M−1/s−1) |
Kinetic Model |
Ki (µM) |
| K154R (WT) | WT | WT | 7.46 ± 1.15 | 8.54×10−2 ± 0.47×10−2 | 1.14×104 ± 0.19×104 | S.I.* | 1430 ± 690 |
| K154R (WT) | WT | K164R | 29.96 ± 5.85 | 1.36×10−1 ± 0.16×10−1 | 4.54×103 ± 1.03×103 | S.I.* | 190 ± 50 |
| K154R (WT) | F268A | WT | 39.15 ± 10.83 | 4.99×10−2 ± 0.72×10−2 | 1.27×103 ± 0.40×103 | S.I.* | 410 ± 140 |
| Y87A/K154R | WT | WT | Below Detect. | Below Detect. | Below Detect. | - | - |
| Y87A/K154R | WT | K164R | 33.66 ± 10.88 | 1.17×10−4 ± 0.23×10−4 | 3 ± 1 | S.I.* | 110 ± 40 |
| D100A/K154R | WT | WT | 53.92 ± 23.58 | 6.38×10−2 ± 1.52×10−2 | 1.18×103 ± 0.59×103 | S.I.* | 490 ± 280 |
| S127A/K154R | WT | WT | 36.96 ± 12.19 | 3.22×10−3 ± 0.50×10−3 | 90 ± 30 | S.I.* | 840 ± 450 |
| Ubc9- Smt3D68RAlexa488 Isoform |
Sizl(167–465) Isoform |
PCNA Isoform |
Kd (µM) |
k2 (s−1) |
k2/Kd (M−1/s−1) |
Kinetic Model |
Ki (µM) |
| K154R (WT) | WT | WT | 7.47 ± 0.88 | 1.39×10−1 ± 0.04×10−1 | 1.86×104 ± 0.23×103 | M.M.† | - |
| K154R (WT) | WT | K164R | - | - | - | - | - |
| K154R (WT) | F268A | WT | 21.06 ± 5.26 | 9.18×10−3 ± 0.57×10−3 | 4.36×102 ± 1.12×102 | M.M.† | - |
| Y87A/K154R | WT | WT | 24.24 ± 5.65 | 2.63×10−2 ± 0.16×10−2 | 1.08×103 ± 0.26×103 | M.M.† | - |
| Y87A/K154R | WT | K164R | - | - | - | - | - |
| D100A/K154R | WT | WT | 54.40 ± 29.80 | 1.03×10−2 ± 0.18×10−2 | 190 ± 100 | M.M.† | - |
| S127A/K154R | WT | WT | 25.12 ± 6.93 | 8.96×10−4 ± 0.64×10−4 | 40 ± 10 | M.M.† | - |
Substrate Inhibition. (Bottom) Catalytic constants for single turnover SUMO conjugation of PCNA to K164 at 4° C with 5 nM purified E2Ubc9-SUMOD68R-Alexa488 thioester, 50 nM E3 and 0.5–500 µM PCNA as indicated.
Data show mean ± s.d. (n=3 technical replicates).
Michaelis-Menton.
Supplementary Material
Acknowledgments
Research at NE-CAT beamlines was funded by P41 GM103403 (National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS)), S10 RR029205 (NIH-ORIP (Office of Research Infrastructure Programs) High-End Shared Equipment grant) at the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory (DE-AC02-06CH11357). Research was supported in part by GM065872 and GM118080 (NIH/NIGMS, C.D.L) and P30CA008748 (NIH/NCI). Content is the authors’ responsibility and does not represent the official views of the NIH. C.D.L is a Howard Hughes Medical Institute Investigator.
Footnotes
Author Contributions F.C.S. and C.D.L executed experiments, data analysis, and manuscript preparation.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession number 5JNE.
The authors declare no competing financial interests.
Literature Cited
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Streich FC, Jr, Lima CD. Structural and functional insights to ubiquitin-like protein conjugation. Annu Rev Biophys. 2014;43:357–379. doi: 10.1146/annurev-biophys-051013-022958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 6.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 7.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 9.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 10.Bosanac I, et al. Modulation of K11-linkage formation by variable loop residues within UbcH5A. J Mol Biol. 2011;408:420–431. doi: 10.1016/j.jmb.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Page RC, Pruneda JN, Amick J, Klevit RE, Misra S. Structural insights into the conformation and oligomerization of E2~ubiquitin conjugates. Biochemistry. 2012;51:4175–4187. doi: 10.1021/bi300058m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigo-Brenni MC, Foster SA, Morgan DO. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol Cell. 2010;39:548–559. doi: 10.1016/j.molcel.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 16.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruneda JN, et al. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat Struct Mol Biol. 2013;20:982–986. doi: 10.1038/nsmb.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott DC, et al. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell. 2014;157:1671–1684. doi: 10.1016/j.cell.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buetow L, et al. Activation of a primed RING E3-E2-ubiquitin complex by non-covalent ubiquitin. Mol Cell. 2015;58:297–310. doi: 10.1016/j.molcel.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Wright JD, Mace PD, Day CL. Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity. Nat Struct Mol Biol. 2016;23:45–52. doi: 10.1038/nsmb.3142. [DOI] [PubMed] [Google Scholar]
- 23.Rytinki MM, Kaikkonen S, Pehkonen P, Jaaskelainen T, Palvimo JJ. PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol Life Sci. 2009;66:3029–3041. doi: 10.1007/s00018-009-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 25.Yunus AA, Lima CD. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol Cell. 2009;35:669–682. doi: 10.1016/j.molcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 27.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Parker JL, Ulrich HD. Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J. 2009;28:3657–3666. doi: 10.1038/emboj.2009.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 30.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Armstrong AA, Mohideen F, Lima CD. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature. 2012;483:59–63. doi: 10.1038/nature10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker JL, Ulrich HD. A SUMO-interacting motif activates budding yeast ubiquitin ligase Rad18 towards SUMO-modified PCNA. Nucleic Acids Res. 2012;40:11380–11388. doi: 10.1093/nar/gks892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mascle XH, et al. Identification of a non-covalent ternary complex formed by PIAS1, SUMO1, and UBC9 proteins involved in transcriptional regulation. J Biol Chem. 2013;288:36312–36327. doi: 10.1074/jbc.M113.486845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cappadocia L, Pichler A, Lima CD. Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat Struct Mol Biol. 2015;22:968–975. doi: 10.1038/nsmb.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 2007;26:2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capili AD, Lima CD. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J Mol Biol. 2007;369:608–618. doi: 10.1016/j.jmb.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duda DM, et al. Structure of a SUMO-binding-motif mimic bound to Smt3p-Ubc9p: conservation of a non-covalent ubiquitin-like protein-E2 complex as a platform for selective interactions within a SUMO pathway. J Mol Biol. 2007;369:619–630. doi: 10.1016/j.jmb.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stehmeier P, Muller S. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol Cell. 2009;33:400–409. doi: 10.1016/j.molcel.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Jentsch S, Psakhye I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genet. 2013;47:167–186. doi: 10.1146/annurev-genet-111212-133453. [DOI] [PubMed] [Google Scholar]
- 40.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattiroli F, Uckelmann M, Sahtoe DD, van Dijk WJ, Sixma TK. The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A. Nat Commun. 2014;5:3291. doi: 10.1038/ncomms4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parker JL, et al. SUMO modification of PCNA is controlled by DNA. EMBO J. 2008;27:2422–2431. doi: 10.1038/emboj.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 44.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. Embo Journal. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yunus AA, Lima CD. Purification of SUMO conjugating enzymes and kinetic analysis of substrate conjugation. Methods of Molecular Biology. 2009;497:167–186. doi: 10.1007/978-1-59745-566-4_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knipscheer P, et al. Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell. 2008;31:371–382. doi: 10.1016/j.molcel.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 47.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Molecular Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter CW Jr, Sweet RM, editors. Vol. 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 52.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 53.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.