Abstract
RNF is a redox-driven ion (Na+ and in one case possibly H+) transporter present in many prokaryotes. It has been proposed that RNF performs a variety of reactions in different organisms, delivering low-potential reducing equivalents for specific cellular processes. RNF shares strong homology with the Na+-pumping respiratory enzyme Na+-NQR, although there are significant differences in subunit and redox cofactor composition. Here we report a topological analysis of the six subunits of RNF from Vibrio cholerae. Although individual subunits from other organisms have previously been studied, this is the first complete, experimentally derived, analysis of RNF from any one source. This has allowed us to identify and confirm key properties of RNF. The putative NADH binding site in RnfC is located on the cytoplasmic side of the membrane. FeS centers in RnfB and RnfC are also located on the cytoplasmic side. However, covalently attached FMNs in RnfD and RnfG are both located in the periplasm. RNF also contains a number of acidic residues that correspond to functionally important groups in Na+-NQR. The acidic residues involved in Na+ uptake and many of those implicated in Na+ translocation are topologically conserved. The topology of RNF closely matches the topology represented in the newly published structure of Na+-NQR, consistent with the close relation between the two enzymes. The topology of RNF is discussed in the context of the current structural model of Na+-NQR, and the proposed functionality of the RNF complex itself.
Graphical abstract
RNF is an energy-transducing membrane complex, found in a variety of bacteria and archaea, that links translocation of Na+ and, in one case, possibly H+ across the cell membrane to redox reactions in the cell.1 Although RNF is highly conserved throughout prokaryotes, RNF has been proposed to catalyze several different redox reactions in different organisms. In Acetobacterium woodii, RNF has been shown to oxidize ferredoxin and reduce NAD+, using the energy released to pump Na+ out of the cell.2 In Rhodobacter capsulatus, the enzyme has been proposed to operate in the opposite sense, driving electron flow from NADH to ferredoxin. This provides low-potential reducing equivalents for fixation of N2 using energy obtained by inward translocation of Na+.3,4 In Methanosarcina acetivorans, RNF oxidizes ferredoxin and has been proposed to reduce methanophenazine, a quinone-like substrate; it is believed to be a Na+ pump.5 In Clostridium ljungdahlii, RNF has been proposed to conduct the same redox reaction as A. woodii but to be a proton pump.6 In Escherichia coli7 and probably Vibrio cholerae, RNF is thought to oxidize NADH, providing electrons to keep the oxygen-sensing protein SoxR in its reduced state.
RNF is closely related to Na+-NQR, the Na+-pumping respiratory complex, though they do not share all subunits or redox cofactors. Sequence analysis indicates that RNF is closer to the primordial form.8 In contrast to the diversity among RNFs, all members of the Na+-NQR family appear to catalyze the same redox reaction, carrying electrons from NADH to ubiquinone.
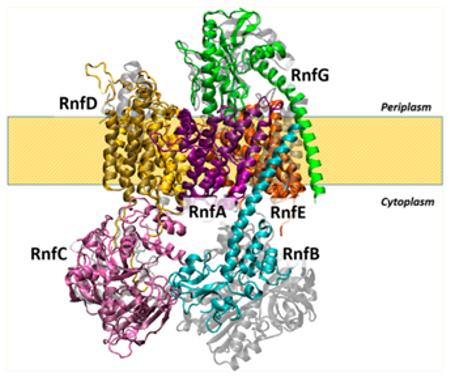
The RNF of V. cholerae, the subject of this paper, is composed of six subunits, RnfA, -B, -C, -D, -G, and -E (the out-of-order naming derives from the sequence of the corresponding ORFs in the operon). The substrate binding sites are located in RnfC and RnfB. RnfC contains a NADH binding motif, while RnfB is thought to be the proximal electron transfer subunit for the ferredoxin (or possibly SoxR) substrate.7 RNF contains two covalently bound FMN’s, one in RnfD and one in RnfG.9 Subunits RnfB and RnfC each contain two iron–sulfur centers. Spectroscopic results indicate that, in each case, at least one of the iron–sulfur centers has a 4Fe-4S structure.10,11 Many of the subunits and cofactors of RNF have clear homologues in Na+-NQR (see Table 1 and Figure 1). RnfA, -D, -G, and -E correspond to NqrE, -B, -C, and -D, respectively. RnfC is similar to NqrA in that both are soluble cytosolic proteins. RnfB has no corresponding subunit in Na+-NQR, and there is no subunit corresponding to NqrF in RNF.8,12 The NADH binding site of RNF is in RnfC, whereas in Na+-NQR, the NADH binding site is in NqrF. The FMN cofactors in RnfD and RnfG correspond to FMNs in NqrC and NqrB, binding with similar linkages at homologous sites. Na+-NQR contains riboflavin as a cofactor.13 In the case of RNF, riboflavin has thus far only been shown to present in the enzyme from Clostridium.14
Table 1.
RNF Subunits and Cofactors Compared to Na+-NQR
| subunit in RNF |
cofactors in RNF |
corresponding subunit in Na+-NQR |
cofactors in Na+- NQR |
|---|---|---|---|
| RnfA | none | NqrE | none |
| RnfB | FeS | None | |
| RnfC | NADH, FeS | NqrA | none |
| RnfD | FMN | NqrB | FMN, riboflavin |
| RnfG | FMN | NqrC | FMN |
| RnfE | none | NqrD | none |
| NqrF | NADH, FAD, FeS |
Figure 1.
Homology model representing the RNF subunits and cofactor localization with respect to the membrane and the proposed electron pathway. The two possible reactions, from ferredoxin to NAD + and from NADH to SoxR, are depicted.
The reaction catalyzed by RNF is “vectorial” in the sense that transport of ions across the cell membrane is defined as a forward or back reaction depending on whether Na+ is moving out of or into the cell. Moreover, RNF links together two different reactions (an oxidation–reduction process and ion translocation), and this coupling will also have a defined direction, or sense. For example, in the RNF from A. woodii, it has been shown that the flow of redox equivalents from ferredoxin to NAD is coupled to translocation of Na+ out of the cell,15,16 and thus, the translocation of Na+ into the cell will be coupled to the flow of redox equivalents in the opposite direction. If a different enzyme were to couple electron flow from NADH to ferredoxin for the translocation of Na+ into the cell, we would say that coupling in this enzyme has the opposite sense versus that in A. woodii.
One consequence of the reaction being “vectorial” is that the enzyme is expected to be asymmetrically organized in the membrane. This topology, i.e., how the polypeptide and various functionally important structures, such as substrate binding sites and redox cofactors, are organized with respect to the sides of the membrane, can give important insights into the mechanism. At the simplest level, sites that bind a water-soluble substrate like NADH or ferredoxin must be in contact with the aqueous compartment where the substrate is found. Similarly, to pump Na+ out of the cell, a site that functions in Na+ uptake must be in contact with the interior of the cell, while an exit site needs to be in contact with the periplasm. Finding a site with well-defined function in an apparently inappropriate location could indicate that the mechanism involves large conformational changes that relocate the site as part of the catalytic cycle. Furthermore, the locations of redox substrate binding sites and redox cofactors will determine whether the reaction involves electron transfer steps that cross the membrane and, thus, whether the redox reaction itself is electrogenic.
Several topological studies of RNF from different organisms have been published.1,9,11,17 However, some of these rely almost entirely on computer algorithms, without experimental data such as reporter gene fusion. Also, among the previous studies, there is a lack of consensus about the topology of almost every subunit. For example, in the case of RnfA, studies in E. coli and R. capsulatus, based on algorithm predictions and PhoA fusions, concluded that the subunit has six transmembrane helices with the N- and C-termini both located in the periplasm.18,11 However, a report conducted in A. woodii concluded that there could be seven to nine transmembrane helices,17 and a subsequent study conducted in M. acetivorans found six helices but located both termini in the cytosol;19 i.e., the subunit was predicted to have the opposite topology compated to that found in the earlier E. coli and R. capsulatus studies.10
In the case of RnfB, a study conducted in R. capsulatus concluded that the subunit could be removed by the membrane by high salt and is thus a peripheral membrane protein.11 A subsequent study, also in R. capsulatus, using similar methods concluded instead that RnfB is an integral membrane protein with at least one transmembrane helix.10 Later a study conducted in A. woodii1 based only on algorithm analysis concluded that RnfB is a peripheral membrane protein. A recent study in M. acetivorans used a histidine tag method to localize the subunit to the membrane fraction and, with this, together with an algorithm analysis that identified one transmembrane helix, concluded that RnfB is an integral membrane protein.19
In the case of RnfC, a study conducted in R. capsulatus concluded that the subunit is a peripheral membrane protein, without transmembrane helices.11 Another study conducted in the same organism reached the opposite conclusion.10 A study conducted in A. woodii concluded that RnfC is only peripherally associated with the membrane.17 None of these studies presented biochemical data to locate the RnfC on one side of the membrane or the other. These discrepancies point to the importance of biochemical methods and in particular reporter gene fusion data in providing experimental conformation for the results of algorithm analysis.
Here we present the first experimental topological analysis of the RNF from V. cholerae, including all six subunits. In each case, the sequence was first analyzed using a stronger multialgorithm prediction method to obtain a consensus model. Then reporter gene fusions were made and analyzed to verify the predictions. Two reporter genes were used: PhoA, which leads to alkaline phosphatase activity when the expressed protein is outside the cell, and a green fluorescent protein gene, which leads to fluorescence only when the expressed protein is inside the cell. For each subunit, at least one fusion construct was made, at the C-terminus, to confirm the overall orientation of the polypeptide in the membrane. Additional pairs of fusions were made to resolve ambiguities and confirm the topological localization of functionally important points in the polypeptide. The resulting complete topological model of RNF from V. cholerae is discussed in the context of earlier partial results for RNFs from other species and our topological model of Na+-NQR.
MATERIALS AND METHODS
Membrane Topology Prediction Methods
Eight web-based topology prediction services were used in this study: TopPred 1.10,20 MEMSAT,21 TMPred (http://www.ch.embnet.org/software/TMPRED_form.html), TMHMM 2.0,22 HMMTOP 2.0,23 Split4 (D. Juretic, University of Split, Split, Croatia), Consensus,24 and TOPCONS.25 Consensus and TOPCONS combine multiple algorithms (X and Y, respectively) to produce a consensus model, while the others each use a single algorithm. In all cases, the default settings were used. The topologies pictured in Figures 3, 4, and 7–9 are from the TOPCONS prediction method (Table 3) because it combines several algorithms to create a single prediction and had predictions that consistently fit the experimental data.
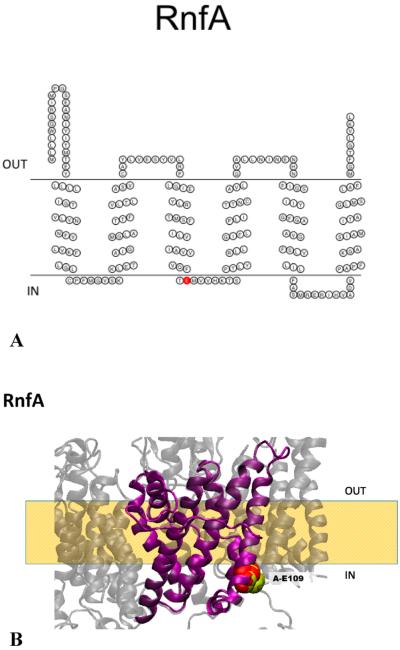
Figure 3.
(A) Membrane topology model of RnfA. RnfA-E109 is colored red. (B) Homology model of RnfA superimposed on the NqrE structure. RnfA-E109 (red) and the corresponding residue in Na+-NQR (NqrE-E95, yellow) are highlighted. The model created using RaptorX was selected as the best model; this selection was made on the basis of comparison to our RnfA topology data and the crystallographic structure of Na+-NQR.15
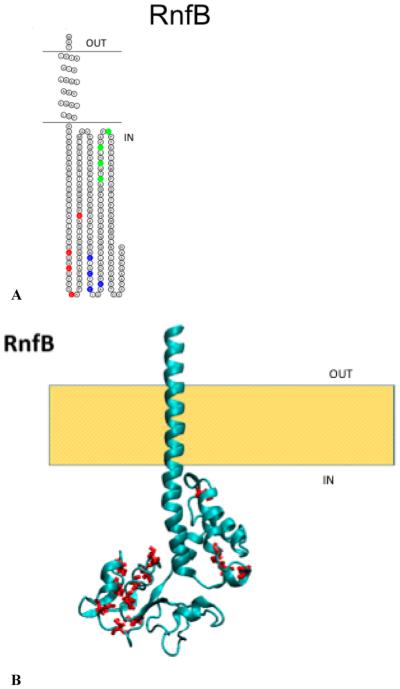
Figure 4.
(A) Membrane topology model of RnfB. Cysteine ligands for the three FeS centers are highlighted: red for C49, C52, C57, and C73; blue for C114, C117, C120, and C124; and green for C144, 147, C150, and C154. (B) Structural model of RnfB with the corresponding cysteine residues colored red. The model was created using ROBETTA.
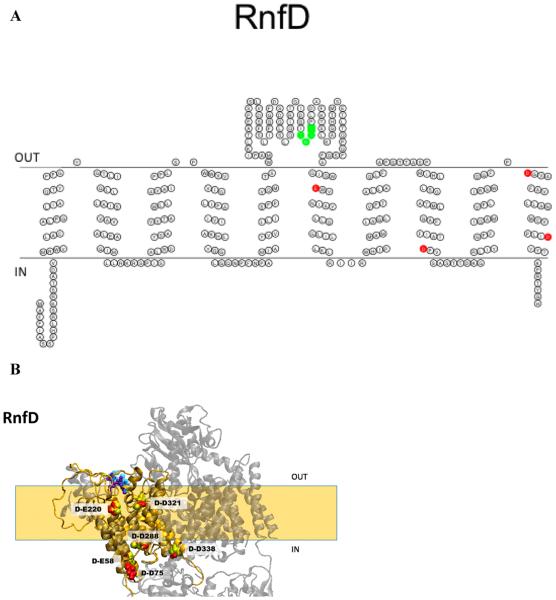
Figure 7.
(A) Membrane topology model of RnfD. Acidic residues implicated in sodium binding and release are colored red (E220, D288, D321, and D338). The FMN binding site is colored green (T184, M185, A186, and T187). (B) Homology model of RnfD superimposed on the NqrB structure. The acidic residues implicated in sodium transport in Na+-NQR are labeled in RNF (red) and Na+-NQR (yellow) structures [RnfD-E220 (NqrB-E274), RnfD-D288 (NqrB-D346), RnfD-D321 (NqrB-D380), and RnfD-D388 (NqrB-D397)]. The FMN binding site is also shown [RnfD-T184 (NqrB-S233), RnfD-M185 (NqrB-G234), RnfD-A186 (NqrB-A235), and RnfD-T187 (NqrB-T236)]. The model created using RaptorX was selected as the best model; this selection was made on the basis of comparison to our RnfD topology data and the crystallographic structure of Na+-NQR.
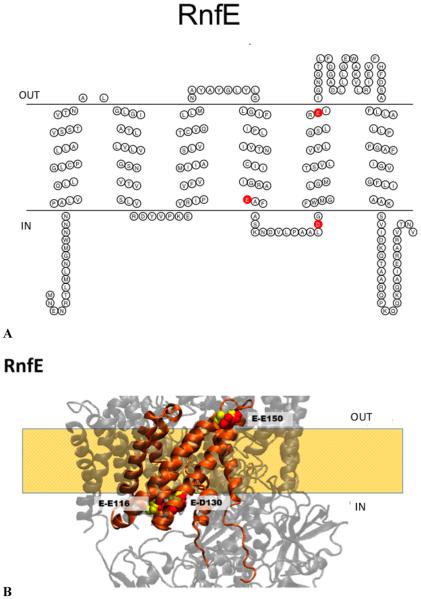
Figure 9.
(A) Membrane topology model of RnfE. Acidic residues implicated in sodium binding and release are colored red (E116, D130, and E150). (B) Homology model of RnfE superimposed on the NqrD structure. The acidic residues implicated in sodium transport in Na+-NQR are labeled in RNF (red) and Na+-NQR (yellow) structures [RnfE-E116 (NqrD-E119), RnfE-D130 (NqrB-D133), and RnfE-E150 (NqrD-E153)]. The model created using RaptorX was selected as the best model; this selection was made on the basis of comparison to our RnfE topology data and the crystallographic structure of Na+-NQR.
Table 3.
Topology Predictions for Subunits A-E from the Rnf Complex from V. choleraea
| TopPred 1.10 | MEMSAT | TMPrep | TMHMM 2.0 | HMMTOP 2.0 | TOPCONS | Split4 | consensus | |
|---|---|---|---|---|---|---|---|---|
| RnfA | 6P | 6C | 6P | 6C | 6P | 6P | 6P | 6P |
| RnfB | 1P | 1C | 1C | 1P | 1C | 1C | 1P | 1C |
| RnfC | 2C | 1P | 0 | 0 | 1C | 1C | 0 | 0 |
| RnfD | 7P | 10C | 7P | 10C | 10C | 10C | 10C | 10C |
| RnfG | 1P | 1C | 2C | 1P | 1P | 1P | 1P | 1P |
| RnfE | 6C | 6P | 6C | 6C | 5C | 6C | 5C | 5C |
Numbers indicate the number of predicted transmembrane helices. P and C indicate the predicted periplasm and cytoplasm C-terminal locations, respectively.
Molecular Genetic Techniques
Strains and plasmids used in this study are listed in Table 2A; primers are listed in Table 2B. Each primer sequence was designed to include restriction sites for insertion into pBAD vectors containing the reporter genes (KpnI for pBAD-PhoA and BmtI/NheI for pBAD-GFP). RnfB from E. coli, together with a sequence encoding a histidine tag, was cloned into the HindIII–EcoRI site of pBAD-HisA, together with an initial stop codon that prevented incorporation of an N-terminal histidine tag. The result was RnfB with a C-terminal six-histidine tag.
Table 2.
| strain or plasmid | (A) Strains and Plasmids Used in This Study genotype or description |
source or reference |
|---|---|---|
| strain E. coli Top 10 |
F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZ M15 ΔlacX74 recAl araD139 A(ara-leu)7697 galU galK rpsL (Strr) endAl nupG |
Invitrogen |
| E. coli Top 10 Δrnf | this study | |
| E. coli LMG194 | F− AlacX74 galE thi rpsL ΔphoA (Pvull) Aara714 leu::TnlO | Invitrogen |
| V. cholerae 0395N1 | Smr | 32 |
| V. cholerae 0395 INI Δnqr | Smr | 26 |
| plasmid | ||
| pBAD-PhoA | pBAD-22 derivative containing signal sequence-less phoA with a 5′ Kpnl site | 33 |
| pBAD-GFP | pBAD plasmid with cycle 3 grp | 34 |
| ARANC | pBAD-PhoA rnfA; full-length C-terminal fusion | this study |
| ARBNC | pBAD-PhoA rnfB; full-length C-terminal fusion | this study |
| ARDNC | pBAD-PhoA rnfD; full-length C-terminal fusion | this study |
| ARDN92 | pBAD-PhoA rnfD; C-terminal fusion at residue 92 | this study |
| ARDN266 | pBAD-PhoA rnfD; C-terminal fusion at residue 266 | this study |
| ARGNC | pBAD-PhoA rnfG; full-length C-terminal fusion | this study |
| GRCNC | pBAD-GFP rnfC; full-length C-terminal fusion | this study |
| GRDNC | pBAD-GFP rnfD; full-length C-terminal fusion | this study |
| GRENC | pBAD-GFP rnfE; fiall-length C-terminal fusion | this study |
| GRGNC | pBAD-GFP rnfG; full-length C-terminal fusion | this study |
| (B) Primers Used in This Study | ||
|---|---|---|
| fusion construct | forward 5′ → 3′' | reverse 5′ → 3′ |
| ARANC | GGTACCGTTATTGCTTTGGCAAAGCAGG | GGTACCTTCAGTTTCACCAATCCGGTAAAG |
| ARBNC | GGTACCGAGTACCATAGTCATCGCTG | GGTACCTTATTCGCACTTTTCTGCGC |
| ARDNC | GGTACCGGCCTTCTTTATTGCTAGCTC | GGTACCTTGTGTCCGTAAGTTCTCGG |
| ARDN92 | GGTACCGGCCTTCTTTATTGCTAGCTC | GGTACCAAGAGGGGCGGGATAG |
| ARDN266 | GGTACCGGCCTTCTTTATTGCTAGCTC | GGTACCAAGGTTGTTCCCGGTGC |
| ARGNC | GGTACCGCTGACAGCAATTCGAAAAAATG | GGTACCTTTTGACCCTCACACGGATTC |
| GRCNC | GCTAGCTTGTCATTAATCGAACAAATCAAATC | GCTAGCGTCTTTCTCCTCAGCGTTTAA |
| GRDNC | ACTAGTGCCTTCTTTATTGCTAGCTCCC | ACTAGTGTGTCCGTAAGTTCTCGGTTTG |
| GRENC | GCTAGCAATGAAAACCGAACTCTGATGTTAAATGGC | GCTAGCGACATTGGTCACGCGTGC |
| GRGNC RnfB |
GTAGCCTGACAGCAATTCGAAAAAATGG | GCTAGCTTGACCCTCACACGGATTC |
| RnfBEc | GCTAGAATTCGAGGAAATAATAAATGAATGCTATCTGGATTGCCG | AGTCAAGCTTTCAATGGTGATGGTGATGACCTTCAAGCTCGCCCTTAGCATGGTGTTCCACGG |
V. cholerae O395N1 genomic DNA was used as the template for polymerase chain reaction (PCR) cloning of all RNF subunits for reporter fusion constructs. In the case of RnfB, an additional construct was created, where the rnf B gene was amplified from E. coli TOP10 genomic DNA. Genomic DNA was extracted from overnight cultures with the PureLink Genomic DNA kit (Invitrogen), according to the manufacturer’s instructions. Complete or partial subunits were amplified with Q5 high-fidelity polymerase [New England Biolabs (NEB)] and subjected to agarose gel electrophoresis, and single bands of the correct size were purified from the agarose gel (QIAGEN). Purified DNA fragments were first cloned into pCR2.1-TOPO TA vectors (Invitrogen) and transformed into E. coli TOP10 cells (Invitrogen). Fragments were excised from the vector with appropriate restriction enzymes and purified from agarose gels. Fusion expression vectors were digested with corresponding restriction enzymes, dephosphorylated with Antarctic phosphatase (NEB), and heat-inactivated (65 °C for 15 min). Vectors and fragments were ligated with T4 ligase (NEB) overnight at 16 °C. The resulting plasmids were transformed into E. coli TOP10 cells, and the orientation was checked by restriction digestion. Plasmids with the rnf genes in the correct orientation relative to the reporter gene were transformed by electroporation (Bio-Rad) into E. coli LMG194 for PhoA fusions and V. cholerae O305N1 Δnqr for GFP fusions.
For the E. coli RnfB construct, the purified PCR fragment was digested with the appropriate restriction enzymes and cloned into the pBAD HisA expression vector. The resulting plasmid was transformed into E. coli Xl-1 Gold cells; the sequence of the rnf gene was verified by DNA sequencing. Vectors with the correct sequence were transformed by electroporation into E. coli TOP10 Δrnf cells.
Overnight cultures of strains containing pBAD-phoA/GFP fusions were re-inoculated into 50 mL cultures of Luria broth (Miller) with antibiotics (100 μg/mL ampicillin, and 50 μg/mL streptomycin for V. cholerae strains). Cells were grown to early log phase, and fusion expression was induced with l-(+)-arabinose (Sigma) to a final concentration of 0.2% (w/v). Cells were harvested at late log phase for analysis of PhoA or GFP activity.
The alkaline phosphatase activity was measured spectropho-tometrically as reported previously.26
The GFP fluorescence of the fusions was observed from 5 mL aliquots of cells after arabinose induction for 2 h. Cells were resuspended in 0.1 mL of 10 mM Tris-HCl (pH 8.0), mounted on 2% agarose pads, and covered with a coverslip. Images of live bacteria for the RnfC fusion were captured by a Zeiss confocal laser scanning LSM 510 META instrument (Carl Zeiss, Thornwood, NY) at CBIS/RPI core facility using a Plan-Apochromat 63X, 1.4 oil DIC objective lens with 488 nm excitation. Image analysis was accomplished using the LSM ExpertMode software (Carl Zeiss). Images of live bacteria for RnfD and RnfE fusions were captured by a Nikon eclipse Ti-U inverted microscope with a scan module using a Nikon Plan Flour 60X, 1.30 oil DIC objective lens, and an infrared pulsed laser with a 530/43 nm emission filter. The acquisition time was 40 μs, and the image size was 20 μm × 20 μm (256 pixels × 256 pixels). Images were analyzed using VistaVision (ISS, Colorado Springs, CO).
Membrane Isolation and Fluorescence Analysis
The GFP construct for RnfC was grown in LB medium to early log phase and induced with 0.2% l-(+)-arabinose. The culture was grown until early stationary phase and harvested. The cell pellets were washed and resuspended with 10 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 5 mM MgSO4 and broken with a microfluidizer (model 110S, Microfluidics) in the presence of DNase (Sigma) and the protease inhibitor AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride]. Cell debris and unbroken cells were removed by centrifugation at 4000 rpm (xx g) for 20 min, and the supernatant containing the membrane fraction was centrifuged at 100000g overnight. The resulting membrane pellet was separated from the supernatant and incubated for 30 min with the Tris buffer described above with 1 M NaCl. The supernatants from the overnight spin and wash steps were combined and concentrated. The membrane pellet was resuspended in Tris buffer and homogenized using a glass–Teflon homogenizer. The homogenized membranes were incubated at 4 °C in the presence of 2% Triton X-100 (Sigma) to solubilize membrane proteins. This mix was spun at 100000g for 30 min, and the solubilized membranes were kept for further analysis.
Solubilized membranes and the concentrated supernatant were analyzed with a Horiba Jobin Yvon Fluorolog Tau3A fluorimeter. The excitation wavelength was 395 nm, and the emission monochromator was scanned from 450 to 600 nm. The emission wavelength for GFP expression was 509 nm. The resulting spectrum was corrected for protein concentration. Protein was measured using the BCA method (Pierce).
Analysis of RnfB
To determine whether RnfB is strongly attached to the cell membrane, i.e., whether it has transmembrane helices or is simply peripherally associated with the membrane, we used an E. coli construct of RnfB. Cells were grown, and RnfB expression was induced with arabinose as described above. The membrane fraction and cell supernatant were run on a 12% SDS gel. The separated proteins were transferred to a PVDF membrane via wet transfer, and a Western blot was conducted following the protocol from the Western Breeze Kit (Invitrogen) with a mouse anti-His C-terminal primary antibody at a 1:2000 dilution. An alkaline phosphatase-conjugated anti-mouse secondary antibody was used, and the Western blot was resolved using a colorimetric substrate.
Structure Prediction
The three-dimensional structures of the RNF subunits were predicted on the basis of the recently published crystallographic structure of Na+-NQR using the following software tools: RnfA, RnfD, and RnfE, RaptorX; RnfC, Phyre; RnfG, I-TASSER. Each subunit was aligned to the Na+-NQR structure using VMD. In the case of RnfB, which does not have a corresponding subunit in Na+-NQR, the structure was predicted using ROBET.
RESULTS
The topological organization of each subunit of RNF was first analyzed using several different computer-based prediction methods, as described in Materials and Methods. These programs take the amino acid sequence of a polypeptide, and from this information, they predict the number and locations of transmembrane helices. In many cases, they also predict the locations of the N- and C-termini, thus defining the overall orientation of a subunit with respect to the sides of the membrane. The results of these analyses are summarized in Table 3. Although the predictions of the different algorithms are in general agreement, some significant discrepancies remained to be resolved.
The predictions were then tested experimentally using reporter–gene fusions. The overall orientation of a polypeptide in the membrane is likely the weakest point of the computer analysis. Thus, these predictions are tested by means of “full-length” fusions, in which the reporter gene is attached at the end of the gene encoding the subunit in question, so that the reporter protein is fused to the C-terminus of the complete subunit. “Partial-length” fusions, in which the reporter gene is attached to a truncated version of the target gene, can be used to resolve discrepancies of the number and locations of helices in the sequence and to confirm whether positions in the polypeptide sequence with functional significance are located on the inside or outside of the cell. For most fusion locations, we made two constructs. One was made with PhoA, a gene whose expression product has alkaline phosphatase activity, which can be detected by a simple colorimetric test, but only when located in the periplasm (Table 4). The other construct was made with GFP, whose product, the green fluorescent protein, glows under UV illumination only when located in the cytosol (Figure 2). With these complementary tools, we were able to avoid relying on negative results for any important conclusions. A full listing of fusion constructs can be found in Table 2A.
Table 4.
Alkaline Phosphatase Activitya
| fusion | alkaline phosphatase activity | location |
|---|---|---|
| ARANC | 548.83 ± 55.9 | periplasm |
| ARBNC | 1.847 ± 8.53 | cytoplasm |
| ARDNC | −1.65 ± 5.01 | cytoplasm |
| ARDN92 | 57.23 ± 5.55 | periplasm |
| ARDN266 | 58.51 ± 6.03 | periplasm |
| ARGNC | 263.63 ± 17.06 | periplasm |
Alkaline phosphatase activity was measured as pNPP hydrolyzed per minute per OD600 of cells.
Figure 2.
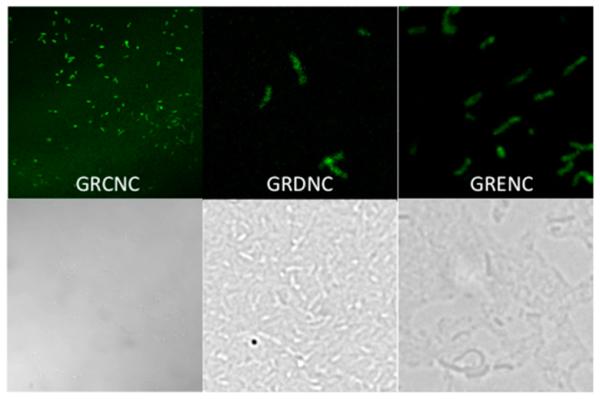
Fluorescence micrographs of GFP fusions. Top panels show fluorescence micrographs and bottom panels visible light micrographs. GRCNC indicates GFP fused to the complete rnfC gene in V. cholerae Δnqr cells. GRDNC indicates GFP fused to the complete rnfD gene in V. cholerae Δnqr cells. GRENC indicates GFP fused to the complete rnf E gene in V. cholerae Δnqr cells.
In describing the results of our topology studies on RNF, we will follow the ordering of ORFs in the rnf operon, which is alphabetical except that G follows D, and there is no RnfF.
RnfA is a relatively hydrophobic subunit consisting of 213 residues. The consensus topology algorithm predicts that this subunit consists of a cluster of six transmembrane helices, with small soluble domains at the N- and C-termini. Analysis of ful-llength fusions with PhoA (Table 4) and GFP (data not shown) showed that the C-terminus of RnfA is located on the periplasmic side of the membrane. This places the N-terminus on the periplasmic side (Figure 3A).
The RnfA subunit contains no cofactor binding sites, but the sequence does include at least six residues that are candidates to be ligands in cation binding sites (Table 5). Most notable is RnfA-E109, which corresponds to E95 in the E subunit of Na+-NQR (NqrE-95) (Figure 3A), one of a key group of acidic residues, close to the cytoplasmic side of the membrane, that are essential for Na+ uptake.27 This residue is fully conserved across all RNFs and Na+-NQRs. Our topological analysis and homology model using the Na+-NQR structure of RnfA place E109 close to the cytoplasmic face of the membrane, the same location as the corresponding residue in Na+-NQR (Figure 3B).
Table 5.
Location of Functionally Important Residues in Rnf
| subunit | |
|---|---|
| RnfA | conserved acidic residues |
| E109, end of helix 3 | |
| RnfB | three 4Fe-4S centers |
| C49-C52-C57-C73 | |
| C114-C117-C120-C124 | |
| C144-C147-C150-C154 | |
| RnfC | NADH binding site |
| G18, G19, I20, I21, A174, E175, C176, E177, P178 | |
| three 4Fe-4S centers | |
| C405-C408-C411-C415 | |
| C378-C381-C384-C388 | |
| C417-C420-C423-C427 | |
| RnfD | FMN binding site |
| T184, M185, A186, T187,a periplasmic region between helices 5 and 6 |
|
| conserved acidic residues | |
| E220, helix 6 | |
| D288, helix 8 | |
| D321, helix 10 | |
| D338, helix 10 | |
| RnfG | FMN binding site |
| T172, G173, A174, T175,a periplasmic region after helix 1 | |
| RnfE | conserved acidic residues |
| E116, helix 4 | |
| D130, cytoplasmic region between helices 4 and 5 | |
| E150, helix 5 |
Ligand for the covalently bound FMNs.
RnfA-E56 in the V. cholerae sequence is conserved as either Glu or Asp in all known RNFs, except in C. ljungdahlii, where there is an Asn, and M. acetivorans, where there is a Lys at this position (Figure 3A). Our topological analysis localizes this residue within the membrane, on the cytoplasmic side of helix II (Figure 3A). This residue is not conserved in Na+-NQR.
RnfA-E176, which corresponds to NqrE-E162, is fully conserved in all known RNF and Na+-NQR sequences (Figure 2B). Our topological analysis of RNF places this residue in a cytoplasmic loop between helices V and VI (Figure 3A), consistent with its location in Na+-NQR, as shown in the homology model shown in Figure 3B.
RnfA-D182 is only partially conserved in RNF sequences (Figure 3A) but corresponds to the fully conserved NqrE-D168. Our topological analysis places this residue in the same cytoplasmic loop as RnfA-E176 (Figure 3B), consistent with its location in Na+-NQR.
The overall organization of RnfA in the membrane is the same as that of the corresponding subunit in Na+-NQR (NqrE) (Figure 3B), consistent with its predicted role in the mechanism of ion pumping.27 The homologous subunit in Na+-NQR is NqrE, where Asp-95 likely has a role in Na+ uptake.27 This residue is conserved in RnfA (RnfA-E109 in V. cholerae, RnfA-E89 in R. capsulatus, and RnfA-E88) (Figure 3B) and on the same side of the membrane, suggesting a role in ion uptake. In the case of RnfA from M. acetivorans, RnfA-93 lies on the opposite side of the membrane.
The RnfB subunit is relatively hydrophilic. All of the topology models predicted the presence of only one transmembrane helix, with a small soluble domain at the N-terminus and a large soluble domain at the C-terminus. PhoA fused to the full-length RnfB subunit from V. cholerae showed no alkaline phosphatase activity (Table 4), indicating that the C-terminus is located in the cytosol (Figure 4A,B). Because the sequences of the RnfB proteins from V. cholerae and E. coli are 64% identical and 77% similar, we used E. coli cells expressing a histidine-tagged version of RnfB (pBAD-Topo RnfB) to verify that this subunit is in fact attached to the membrane. These cells were cultivated; cell membranes were isolated and washed with high salt to remove proteins peripherally associated with the membrane, and the resulting fraction was analyzed by SDS–PAGE and Western blotting with anti-His tag antibodies. The blot showed a clear band corresponding to RnfB, indicating that this subunit remained in the membrane fraction and is a true integral membrane protein (Figure 5).
Figure 5.
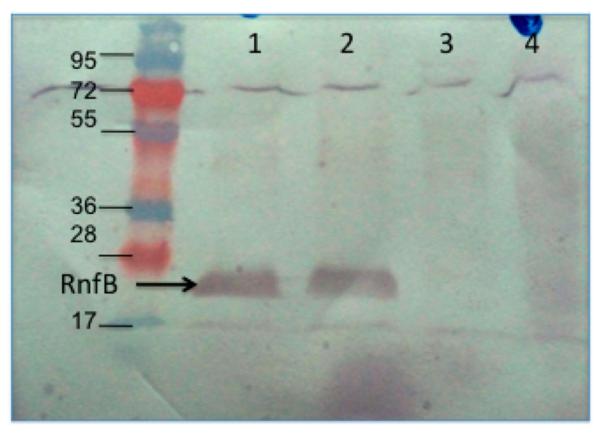
Western blot analysis of membrane and soluble fractions of E. coli cells expressing C-terminal His-tagged RnfB, using a six-His tag antibody: lane 1, 30 μg of membrane protein; lane 2, 10 μg of membrane protein; lane 3, 30 μg of protein from the soluble cell fraction; lane 4, 10 μg of protein from the soluble cell fraction. The arrow points to bands at approximately 20 kDa, the expected molecular mass of RnfB.
The RnfB subunit includes three binding motifs for iron–sulfur centers, located in the cytoplasmic domain (Figure 4A); as described above, EPR spectra consistent with at least one 4Fe-4S center have been observed in preparations from the RnfB subunit from R. capsulatus and M. acetivorans, expressed individually,10,11 and the RnfB subunit from E. coli has a UV–vis absorbance spectrum consistent with the presence of an iron–sulfur center (data not shown). The subunit does not include any conserved acidic residues (Table 5). However, the RnfB subunit from A. woodii is significantly longer, with 333 amino acids, and is predicted to have at least one additional iron–sulfur center.1 There is no corresponding subunit in Na+-NQR.
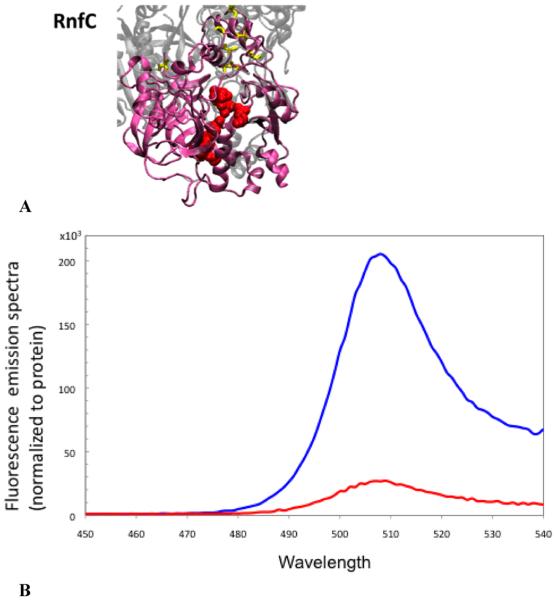
The RnfC subunit includes two binding motifs for iron–sulfur centers, as well as a GGIH motif indicative of an NADH binding site (Table 5). The RnfC subunit from R. capsulatus, expressed on its own, exhibits EPR signals consistent with at least one 4Fe-4S center.10 Our analysis with topology algorithms gave inconsistent results. Some predicted a single transmembrane helix, while others predicted none (Table 3). In our reporter gene fusion analysis, cells expressing a fusion of GFP to the full-length RnfC glowed, indicating that the C-terminus is located on the cytoplasmic side of the membrane. To resolve the question of whether RnfC is anchored to the membrane by a transmembrane helix, cell membranes from the fusion strain were isolated and then washed in high salt. After the membranes had been washed, significant fluorescence was detected in the supernatant, showing that the high-salt buffer was able to strip RnfC from the membrane (Figure 6B). This would not be expected for an integral membrane protein. Thus, RnfC is a peripheral protein, associated with the RNF complex on the cytoplasmic side of the membrane. This model is in agreement with the homology model shown in Figure 6A.
Figure 6.
(A) Structural model of RnfC showing the NADH binding site (G174, G176, G177, A178, F180, P181, A183, K185, L186, N199, C204, and P205) and the cysteine ligands for three FeS centers (yellow) (C405, C408, C411, C415, C444, C447, C450, and C454). The model obtained using Phyre was selected on the basis of comparison to the crystallographic structure of Na+-NQR (ref). (B) Fluorescence emission spectra of the RnfC–GFP fusion protein: membrane fraction (red) and soluble fractions (blue). Spectra were normalized by protein concentration.
The RnfD subunit is very hydrophobic. Topology algorithms consistently predicted the presence of 10 transmembrane helices (Table 3), with a small soluble domain between helices V and VI containing the FMN binding motif, TMAT. Full-length fusions of RnfD to PhoA and GFP corroborated the finding that the C-terminus of RnfD is located in the cytoplasm9 (Figure 7A). Truncated fusions, at residues 92 and 266, showed that these sequence positions are both on the periplasmic side of the membrane (Table 4), confirming the topology shown in Figure 7A. This places the FMN binding site on the periplasmic side of the membrane, as is the case in the corresponding subunit of NqrB in the recent crystallographic structure (Figure 7B).28
RnfD also contains a number of conserved acidic amino acids, several of which correspond to residues in NqrB involved in Na+ translocation (see Table 5). Four acidic residues in the transmembrane helices of Na+-NQR have been specifically implicated in Na+ uptake,27 and two of these are located in NqrB (NqrB-E144 and NqrB-D397). NqrB-D397 occupies the same position in the sequence as RnfD-D338 (Figure 7B), and like its counterpart, RnfD-D338 is located close to the cytoplasmic side of the membrane, near the end of a transmembrane helix (Figure 7B). The sequence position corresponding to NqrB-E144 is occupied by nonacidic residues in RNF, but RNF sequences include two fully conserved acid groups close to that position (RnfD-E58 and RnfD-D75). Like NqrB-E144, these two residues are located at the ends of transmembrane helices, close to the cytoplasmic surface (Figure 7B). This location is consistent with a putative role in Na+ uptake for either or both of these RnfD residues.27
RnfD-D288 is homologous to NqrB-D346, one of three acidic residues in Na+-NQR that have been implicated in the transport of Na+ across the membrane because replacement with a nonacidic residue leads to inhibition of the transfer of an electron from FMNB to riboflavin and loss of the associated formation of ΔΨ.29 NqrB-D346 is located at the end of a transmembrane helix, close to the cytoplasmic side of the membrane. RnfD-D288 is located in a similar place in the corresponding helix (helix VIII) (Figure 7B).
Thus, in RnfD, two conserved acidic residues involved in Na+ uptake are located on the cytoplasmic side of the membrane, as are the corresponding residues in NqrB. The FMN binding site (T184, M185, A186, and T187) and the acidic group implicated in Na+ translocation (RnfD-D288) are located in the large loop between helices V and VI and in helix VIII, respectively, where they also have the same sidedness as in RnfD and NqrB (Figure 7B).
In M. acetivorans, the RnfD polypeptide is shorter than those in V. cholerae and other species. All parts of the sequence that correspond to transmembrane helices in V. cholerae are still present, but the topology prediction shows that only six helices are conserved.19 The missing part of the polypeptide is in the large loop between helices V and VI in V. cholerae RnfD, described above (Figure 7A), and includes the TMAT sequence where FMN would bind. Currently, there is no biochemical confirmation that this FMN is missing in the M. acetivorans RNF.
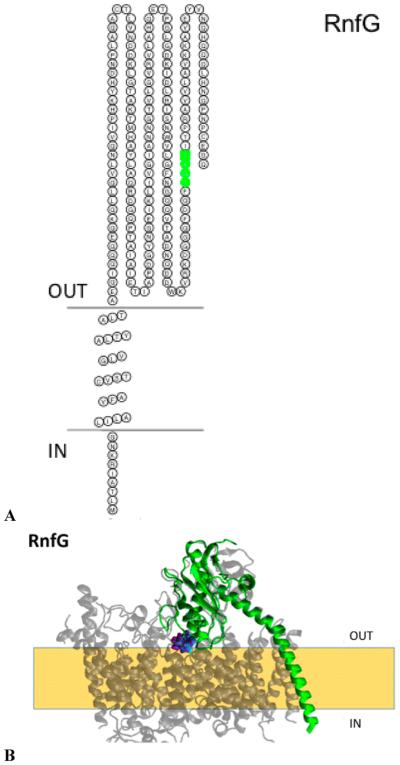
The RnfG subunit consists of a large hydrophilic domain at the C-terminus of the protein anchored to the membrane by a helix located near the N-terminus (Figure 8A). The subunit contains an S(T)GAT sequence (T172, G173, A174, and T175) where an FMN is covalently bound (Table 5). Cells expressing a fusion of the full-length RnfG to PhoA exhibited high alkaline phosphatase activity (Table 4),9 while cells with the corresponding GFP fusions were not fluorescent (data not shown), indicating that the C-terminus of RnfG is located in the periplasm. This topology places the FMN cofactor in the periplasmic domain, as does the topology prediction reported for M. acetivorans.19
Figure 8.
(A) Membrane topology model of RnfG. The FMN binding site is colored green (T172, G173, A174, and T175). (B) Homology model of RnfG (blue) superimposed on the NqrC (red) structure. The FMN binding site is also shown [RnfG-T1172 (NqrC-S222), RnfG-G173 (NqrC-G223), RnfC-A174 (NqrC-A224), and RnfC-T175 (NqrC-T225)]. The model created using RaptorX was selected as the best model; this selection was made on the basis of comparison to our RnfD topology data and the crystallographic structure of Na+-NQR.
This is also similar to the topological arrangement of the corresponding NqrC subunit in the crystallographic structure of Na+-NQR (Figure 8B), which places the FMN in the periplasm, in contrast to our earlier topology based on reporter–gene fusion analysis.28
The RnfE subunit is relatively hydrophobic, and some algorithms find six helices and others only five. This difference in the number of predicted helices occurs because some algorithms find a transmembrane helix within the first 36 residues from the N-terminus, where others find only a soluble domain. Thus, the first helix from the N-terminus in the five-helix models corresponds to the second helix in the six-helix models. Cells expressing a full-length fusion of RnfE from V. cholerae with GFP are fluorescent (Figure 2), showing that the C-terminus of the subunit is located in the cytoplasm.
Topology algorithms and the PhoA fusion for RnfE from E. coli suggest six transmembrane helices.18 The homologous subunit in Na+-NQR, NqrD, has six transmembrane helices; its C-terminus is located in the cytoplasm. The six transmembrane helices correspond to the six helices in RnfE from E. coli and M. acetivorans.18,19 The homology model with the Na+-NQR crystallographic model (Figure 9B) suggests a six-helix RnfE in V. cholerae, in agreement with the report about RnfE from E. coli.18
RnfE includes several residues that correspond to conserved acidic groups in Na+-NQR, shown to be important for Na+ transport.30 RnfE-Q85 in V. cholerae corresponds to NqrD-D88 (Figure 9A,B), which has been implicated in the generation of membrane potential via translocation of Na+, and possibly its release from the enzyme.30 In Na+-NQR, this sequence position is always occupied by an aspartate; in most RNFs, there is a glutamine, although in some sequences there is an aspartate or glutamate, and in a few cases another residue. Our analysis locates RnfE-Q85, like NqrD-D88, on the periplasmic side of the membrane (Figure 9A,B). The reported topology predictions for RnfE from R. capsulatus10 and M. acetivorans19 also locate the corresponding residues, in both cases aspartates, on the periplasmic side. RnfE-D130 corresponds to NqrD-D133, which is part of the Na+ uptake site. Consistent with this predicted function, our analysis locates RnfE-D130 on the cytoplasmic side of the membrane, like the corresponding residue in Na+-NQR. RnfE-E150 corresponds to NqrD-E153, a completely conserved residue whose replacement has a small but significant effect on the Na+ translocation step. Like NqrD-E153, RnfE150 is predicted to be on the periplasmic side of the membrane, a location consistent with the predicted function.
DISCUSSION
In this paper, we present a topological analysis of the RNF enzyme from V. cholerae. Initially, each subunit was analyzed by computer-based topology prediction algorithms. Then reporter–gene fusions were constructed and expressed, to verify the overall orientation of the subunits in the membrane, to resolve discrepancies between the results of the different prediction algorithms, and to definitively localize important points in the sequence. Two reporter groups were used: PhoA/alkaline phosphatase, which has activity only in the periplasm, and green fluorescent protein (GFP), which glows only in the cytoplasm. Finally, for two subunits, RnfB and RnfC, there was a question of whether the protein is actually anchored to the membrane by a transmembrane helix.10,11 In these cases, cells were separated into cytosolic and membrane fractions for analysis. A Western blot showed that a histidine tag on RnfB remained in the membrane fraction showing that this subunit is a true membrane protein. Fluorescence spectroscopy showed that GFP fused to RnfC was removed from the membrane fraction by high salt, showing that this subunit is peripherally associated with the RNF complex on the cytoplasmic side. The topology data were supported by a homology model, in which the RNF sequence was independently superimposed on the crystal structure for the homologous subunits in Na+-NQR.
Although some subunits of RNF have been analyzed previously,9,11,18,19 this is the first complete example, experimentally derived from any one source via topological analysis of RNF (see Table 5). These results make it possible to determine the sidedness of all predicted substrate binding sites, redox cofactor binding sites, and other parts of the sequence predicted to be important for function, such as conserved acidic residues that could provide ligands for Na+ binding. They also provide additional information for the comparison of RNF sequences from different species, especially in cases where topological analyses for subunits from other species are available. Importantly, they also make it possible to compare the topological organization of RNF to that of Na+-NQR, from our earlier work and the recently published crystallographic structure. Na+-NQR is far better understood than RNF; its electron transfer pathway has been thoroughly investigated,28,31 and the roles of many acidic residues in Na+ uptake and transport have been analyzed.27 There are significant homologies between Na+-NQR and RNF, and having complete topologies for both enzymes adds an important layer to the sequence comparisons.
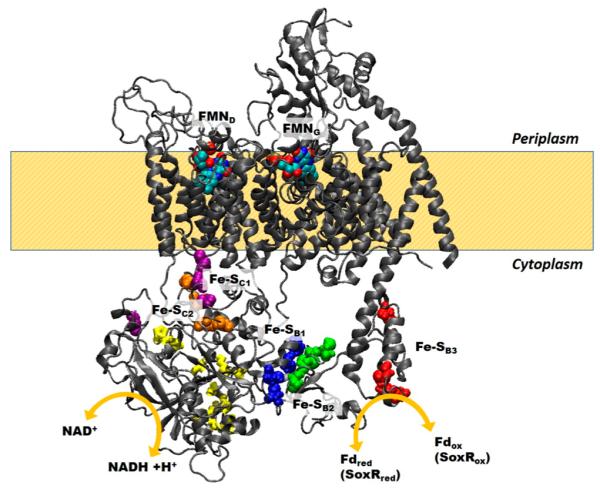
RNF is a redox-coupled ion (Na+ and in one case possibly H+) transporter, and topological locations of redox substrate binding sites and redox cofactors are expected to be crucial for mechanism. On the basis of motifs in the sequence, RNF is predicted to have a NADH binding site in RnfC, covalently bound FMN cofactors in RnfD and RnfG, and two iron–sulfur centers each in RnfB and RnfC. The FMN cofactors in RnfD and RnfG have been confirmed in biochemical studies,9 and RnfB and RnfC have each been shown to contain at least one 4Fe-4S center by EPR.10,11 Our topological analysis and homology modeling data locate the NADH binding site on the cytoplasmic side of the membrane, consistent with its predicted function. The iron–sulfur centers are also predicted to be on the cytoplasmic side. The two FMN cofactors are predicted to be on the periplasmic side, similar to their location in the recent crystallographic structure of Na+-NQR.
Thus, RnfC is a peripheral membrane protein on the cytoplasmic side of the membrane that apparently mediates redox interactions with NADH. In Na+-NQR, this is the role of the NqrF subunit, and integral membrane protein that does not have a homologue in RNF. NqrF contains the NADH binding site, the FAD cofactor that serves as an initial electron acceptor, and a 2Fe-2S center. RnfC appears to contain a NADH binding site and one pair of 4Fe-4S centers.11 RnfC shares homology with NqrA, which is also not an integral membrane protein, but NqrA does not include the regions containing the NADH binding site or the iron–sulfur centers.
RnfB has been proposed to mediate redox interactions with ferredoxin. This subunit contains the other two iron–sulfur centers thus far identified in RNF, both on the cytoplasmic side of the membrane. There is no homologous subunit in Na+-NQR.
RnfD and RnfG, in which the two covalently attached FMN cofactors (FMND and FMNG) are located, are strongly homologous to NqrC and NqrB, respectively.9,28 In Na+-NQR, these two subunits bind the FMNC and FMNB cofactors, which are involved in the redox steps coupled to Na+ uptake and its electrogenic transport across the membrane, respectively.29 The current topological analysis and the homology modeling of RNF place both FMNs in the periplasm. This is in contrast to our earlier topological model of Na+-NQR based on reporter–gene fusion but is consistent with the recent crystallographic structure of Na+-NQR.14,15 If, as proposed, electrons enter RNF from cytoplasmic ferrodoxin at RnfC and exit to cytoplasmic NAD+ at RnfB, and if the two FMNs are part of the redox pathway, electrons must traverse the membrane twice. The recent crystallographic structure of Na+-NQR identified what is believed to be an iron, attached to cysteines in subunits NqrD and NqrE, close to the center of the membrane, and it was proposed that this could be a center that facilitates electron transfer through the membrane span. Homologous cysteine residues are present in RNF. It is worth noting that this would still account for only one of two transits of electrons across the membrane.
Our topological data also shed light on possible structures involved in ion transport in RNF. Three of the four key cytoplasmic residues involved in Na+ uptake in Na+-NQR are present and fully conserved in RNF. NqrB-D397 corresponds to RnfD-D338, NqrD-D133 to RnfE-D131, and NqrE-E95 to RnfA-E109.27 The fourth residue, NqrB-E144, corresponds to a nonconserved residue at position 63 in the RnfD sequence, but there is a conserved Glu five residues away (RnfD-E58) or a conserved Asp 12 residues away (RnfD-D75) that could fill the same functional role. Our topological analysis places all four of these residues on the cytoplasmic side of the membrane, like their counterparts in Na+-NQR, consistent with their proposed function.
Three residues in Na+-NQR have been shown to be important for Na+ translocation, and formation of ΔΨ (NqrB-E28, NqrD-D88, and NqrB-D346). The first of these, NqrB-E28, is completely missing in RNF. The fact that the entire first helix of the NqrB subunit, where NqrB-E28 is located, is not present in the corresponding RnfD subunit suggests the possibility that this functionality has moved elsewhere in the enzyme structure in RNF. The second residue, NqrD-D88, is replaced by a glutamine in RNF (RnfE-Q85). Like its counterpart in Na+-NQR, RnfE-Q85 is on the periplasmic side of the membrane, which would be consistent with a role in Na+ efflux. Although in Na+-NQR this sequence position is always occupied by an aspartate, the residue at the corresponding location in RNF is not fully conserved. In most RNF sequences, there is a glutamine, although aspartate, glutamate, and sometimes other residues are also seen. It is worth noting that although glutamine is not an exact functional replacement for aspartate, it could potentially serve as a ligand in a Na+ binding site.
The third residue, NqrB-D346, corresponds to RnfD-D288. The current analysis places RnfD-D288 on the cytoplasmic side of the membrane, in contrast with our earlier analysis of Na+-NQR that located NqrB-D346 on the periplasmic side,26 but consistent with its location in the recent crystallographic structure.28 This location raises interesting questions; like the other two residues, replacement of NqrB-D346 by a nonacidic residue leads to inhibition of the same FMNB to the riboflavin electron transfer step and suppression of the related formation of ΔΨ. However, given this location, neither RnfD-D288 nor NqrB-D346 is likely to participate in a Na+ exit site, although they could play a direct or indirect role in the electrogenic movement of Na+ across the membrane.
The complete topology of RNF coincides closely with that of the homologous regions of Na+-NQR (see Figures 3, 4, and 6–9). Although the earlier parts of the electron transfer pathway in Na+-NQR are very different with essentially no cofactors in common with RNF, the two FMN cofactors are present in both enzymes and in both cases are located on the periplasmic side of the membrane. This indicates that at some point the electron transfer pathway crosses from one side of the membrane to the other, which means that some electron transfer steps are expected to be electrogenic, although the entire redox reaction is not. The acidic residues involved in Na+ uptake and many of those implicated in Na+ translocation are topologically conserved. The significance of these findings should become clearer as the mechanisms of RNF and Na+-NQR emerge more fully and begin to be compared on a detailed level.
ACKNOWLEDGMENTS
This work was supported by Grant MCB-1052234 from the National Science Foundation (B.B.). H.D.H. is supported by National Institutes of Health Grant GM086801 (to Dr. Angel Garcia). We thank Dr. Catherine Royer for her help with the microscopy experiments.
Footnotes
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Biegel E, Schmidt S, González JM, Müller V. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell. Mol. Life Sci. 2011;68:613–634. doi: 10.1007/s00018-010-0555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Muller V, Imkamp F, Biegel E, Schmidt S, Dilling S. Discovery of a Ferredoxin:NAD+-Oxidoreductase (Rnf) in Acetobacterium woodii: A Novel Potential Coupling Site in Acetogens. Ann. N.Y. Acad. Sci. 2008;1125:137–146. doi: 10.1196/annals.1419.011. [DOI] [PubMed] [Google Scholar]
- (3).Schmehl M, Jahn A, Meyer zu Vilsendorf A, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: A putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- (4).Jeong HS, Jouanneau Y. Enhanced nitrogenase activity in strains of Rhodobacter capsulatus that overexpress the rnf genes. J. Bacteriol. 2000;182:1208–1214. doi: 10.1128/jb.182.5.1208-1214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Schlegel K, Welte C, Deppenmeier U, Müller V. Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J. 2012;279:4444–44452. doi: 10.1111/febs.12031. [DOI] [PubMed] [Google Scholar]
- (6).Tremblay P, Zhang T, Dar SA, Leang C, Lovley DR. The Rnf complex of Clostridium ljungdahlii is a proton-translocating ferredoxin:NAD+ oxidoreductase essential for autotrophic growth. mBio. 2012;26:e00406–e00412. doi: 10.1128/mBio.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Koo MS, Lee JH, Rah SY, Yeo WS, Lee JW, Koh YS, Kang SO, Roe JH. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J. 2003;22:2614–2622. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Reyes-Prieto A, Barquera B, Juarez O. Origin and evolution of the sodium -pumping NADH: ubiquinone oxidoreductase. PLoS One. 2014;9:e96696. doi: 10.1371/journal.pone.0096696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Backiel J, Juarez O, Zagorevski DV, Wang Z, Nilges MJ, Barquera B. Covalent Binding of Flavins to RnfG and RnfD in the Rnf Complex from Vibrio cholerae. Biochemistry. 2008;47:11273–11284. doi: 10.1021/bi800920j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jouanneau Y, Jeong HS, Hugo N, Meyer C, Willison JC. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus: Characterization of two membrane-bound iron-sulfur proteins. Eur. J. Biochem. 1998;251:54–64. doi: 10.1046/j.1432-1327.1998.2510054.x. [DOI] [PubMed] [Google Scholar]
- (11).Kumagai H, Fujiwara T, Matsubara H, Saeki K. Membrane localization, topology, and mutual stabilization of the rnfABC gene products in Rhodobacter capsulatus and implications for a new family of energy-coupling NADH oxidoreductases. Biochemistry. 1997;36:5509–5521. doi: 10.1021/bi970014q. [DOI] [PubMed] [Google Scholar]
- (12).Hayashi M, Nakayama Y, Yasui M, Maeda M, Furuishi K, Unemoto T. FMN is covalently attached to a threonine residue in the NqrB and NqrC subunits of Na+-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 2001;488:5–8. doi: 10.1016/s0014-5793(00)02404-2. [DOI] [PubMed] [Google Scholar]
- (13).Barquera B, Zhou W, Morgan JE, Gennis RB. Riboflavin is a component of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10322–10324. doi: 10.1073/pnas.162361299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Jayamani E. Biologie. Philipps-Universität Marburg; Marburg, Germany: 2008. A unique way of energy conservation in glutamate fermienting clostridia; p. 104. [Google Scholar]
- (15).Biegel E, Muller V. Bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18138–18142. doi: 10.1073/pnas.1010318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Hess V, Schuchmann K, Müller V. The ferredoxin:NAD+ oxidoreductase (Rnf) from the acetogen Acetobacterium woodii requires Na+ and is reversibly coupled to the membrane potential. J. Biol. Chem. 2013;288:496–502. doi: 10.1074/jbc.M113.510255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Biegel E, Schmidt S, Muller V. Genetic, immunological and biochemical evidence for a Rnf complex in the acetogen Acetobacterium woodii. Environ. Microbiol. 2009;11:1438–1443. doi: 10.1111/j.1462-2920.2009.01871.x. [DOI] [PubMed] [Google Scholar]
- (18).Saaf A, Johansson M, Wallin E, von Heijne G. Divergent evolution of a membrane protein topology: The Escherichia coli RnfA and RnfE homologues. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8540–8544. doi: 10.1073/pnas.96.15.8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Suharti S, Wang M, de Vries S, Ferry JG. Characterization of the RnfB and RnfG subunits of the Rnf complex from the archaeon Methanosarcina acetivorans. PLoS One. 2014;9:e97966. doi: 10.1371/journal.pone.0097966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Claros MG, von Heijne G. ToPRed II: An imprived software for membrane protein structure predictions. Comput. Appl. Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- (21).Jones D. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bio-informatics. 2007;23:538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- (22).Melen K, Krogh A, von Heijne G. Reliability measures for membrane protein topology prediction algorithms. J. Mol. Biol. 2003;327:735–744. doi: 10.1016/s0022-2836(03)00182-7. [DOI] [PubMed] [Google Scholar]
- (23).Tusnady GE, Simon I. The HMMTOP transmembrane topology predicton server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- (24).Ikeda M, Arai M, Lao DM, Shimizu T. Transmembrane topology prediction methods: A re-assesssment and improvement by a consensus method using a data set of experimentally-characterized transmembrane topologies. In Silico Biol. 2002;2:19–33. [PubMed] [Google Scholar]
- (25).Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: Consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37:W465–W468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Duffy E, Barquera B. Membrane topology mapping of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae by PhoA-green fluorescent protein fusion analysis. J. Bacteriol. 2006;188:8343–8351. doi: 10.1128/JB.01383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Juarez O, Athearn K, Gillespie P, Barquera B. Acid residues in the transmembrane helices of the Na+-pumping NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae involved in sodium translocation. Biochemistry. 2009;48:9516–9524. doi: 10.1021/bi900845y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Steuber J, Vohl G, Casutt MS, Vorburger T, Diederichs K, Fritz G. Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase. Nature. 2014;516:62–67. doi: 10.1038/nature14003. [DOI] [PubMed] [Google Scholar]
- (29).Juarez O, Morgan JE, Nilges MJ, Barquera B. The energy transducing redox steps of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12505–12510. doi: 10.1073/pnas.1002866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Juárez O, Morgan JE, Barquera B. The electron transfer pathway of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 2009;284:8963–8972. doi: 10.1074/jbc.M809395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Neehaul Y, Juarez O, Barquera B, Hellwig P. Thermodynamic contribution to the regulation of electron transfer in the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry. 2012;51:4072–4077. doi: 10.1021/bi300343u. [DOI] [PubMed] [Google Scholar]
- (32).Mekalanos JJ, Swartz DJ, Pearson GD, Harford N, Groyne F, de Wilde M. Cholera toxin genes: Nucleotide sequence, deletion analysis and vaccine development. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- (33).Melchers K, Schuhmacher A, Buhmann A, Weitzenegger T, Belin D, Grau S, Ehrmann M. Membrane topology of CadA homologous P-type ATPase of Helicobacter pylori as determined by expression of phoA fusions in Escherichia coli and the positive inside rule. Res. Microbiol. 1999;150:507–520. doi: 10.1016/s0923-2508(99)00106-0. [DOI] [PubMed] [Google Scholar]
- (34).Lu C, Bentley WE, Rao G. A high-throughput approach to promoter study using green fluorescent protein. Biotechnol. Prog. 2004;20:1634–1640. doi: 10.1021/bp049751l. [DOI] [PubMed] [Google Scholar]