Abstract
The MEP (Methyl Erythritol Phosphate) isoprenoids biosynthesis pathway is an attractive drug target to combat malaria, due to its uniqueness and indispensability for the parasite. It is functional in the apicoplast of Plasmodium and its products get transported to the cytoplasm, where they participate in glycoprotein synthesis, electron transport chain, tRNA modification and several other biological processes. Several compounds have been tested against the enzymes involved in this pathway and amongst them Fosmidomycin, targeted against IspC (DXP reductoisomerase) enzyme and MMV008138 targeted against IspD enzyme have shown good anti-malarial activity in parasite cultures. Fosmidomycin is now-a-days prescribed clinically, however, less absorption, shorter half-life, and toxicity at higher doses, limits its use as an anti-malarial. The potential of other enzymes of the pathway as candidate drug targets has also been determined. This review details the various drug molecules tested against these targets with special emphasis to Plasmodium. We corroborate that MEP pathway functional within the apicoplast of Plasmodium is a major drug target, especially during erythrocytic stages. However, the major bottlenecks, bioavailability and toxicity of the new molecules needs to be addressed, before considering any new molecule as a potent antimalarial.
Keywords: Plasmodium, apicoplast, isoprenoids, MEP pathway, anti-malarial
Introduction
Isoprenoids are structurally and functionally the most diverse group of natural metabolites found in all three domains i.e., eubacteria, archaebacteria and eukarya. Depending on the number of precursor units [Isopentenyl Pyrophosphate (IPP) and Dimethylallyl Pyrophosphate (DMAPP)], they vary in structure and functions. Isoprenoids are known to play a key role in all aspects of life; e.g., in regulation of gene expression (prenylation of proteins), as membrane constituents (prenyl lipids in archaebacteria and sterol in eubacteria and eukaryotes), as vitamins, plant hormones (gibberellins, brassinosteroids, abscisic acid), photosynthetic pigments (carotenoids, side chain of chlorophyll), quinones in electron transport chain, and plant defense compounds (monoterpenes, sesquiterpenes, diterpenes) (Sacchettini and Poulter, 1997; Bach et al., 1999; Hunter, 2007). Two distinct pathways synthesize IPP and DMAPP, (a) the Mevalonate dependent pathway (MVA) functional in archaea and most eukaryotes (including all mammals and higher plants), and (b) the Methyl Erythritol Phosphate/1-deoxy-D-xylulose-5-phosphate (MEP/DOXP) pathway functional in bacteria, plant plastids, members of chlorophyta and pathogenic microorganisms (Banerjee and Sharkey, 2014). These pathways are significantly different from each other (Figure S1) in terms of preliminary substrates, mevalonate as an intermediate in MVA pathway only, and higher yield of IPP and DMAPP in MEP pathway.
Isoprenoids biosynthesis in Plasmodium
Plasmodium belongs to the phylum Apicomplexa, known to harbor a non-photosynthetic plastid like organelle of prokaryotic origin known as apicoplast (McFadden et al., 1996; Foth and McFadden, 2003). This organelle is indispensable for the survival of the parasite and is the functional site for four major metabolic pathways. The MEP/DOXP pathway is one of these pathways which is the only source for isoprenoids in the parasite and is absent in the human host. The first evidence for the presence of the MEP pathway in Plasmodium was given by Jomaa et al. (1999) who identified the presence of DOXP reductoisomerase (IspC) gene in preliminary staged Plasmodium falciparum whole genome database. Following this, few other enzymes of this pathway, IspD (Rohdich et al., 1999), IspF (Rohdich et al., 2001), IspG (Altincicek et al., 2001a), and IspH (Altincicek et al., 2001b) were characterized mainly from prokaryotes and were shown to be present in P. falciparum as well. Studies have detailed the import of initial substrates of the pathway, DHAP (Dihydroxy acetone phosphate) and PEP (Phosphoenol pyruvate) inside the Plasmodium apicoplast with the help of transporter molecules TPT (triose phosphate transporter) and PPT (phosphoenol pyruvate transporter) respectively, localized in the apicoplast membrane (Mullin et al., 2006) suggesting apicoplast as its functional site.
In Plasmodium, the MEP pathway is reported to be indispensable for both hepatic (Sparr et al., 2013) and erythrocytic stages (Cassera et al., 2004) of the parasite asexual life cycle. Recent reports have suggested that the “apicoplast less” Plasmodium parasite can divide indefinitely in culture if supplemented exogenously with IPP. This proves that during the erythrocytic stages, the only essential function of apicoplast is the synthesis of isoprene unit precursors, IPP and DMAPP (Yeh and DeRisi, 2011). Recent reports have also proven that the products of MEP pathway are required in the early stages of parasite gamete development (Wiley et al., 2015). All these studies suggest the importance of this pathway at different phases of the parasite's life cycle.
MEP pathway enzymes
The MEP pathway consists of seven enzymes, encoded by the parasite nuclear genome and targeted to apicoplast with the help of N-terminal bipartite leader sequence (van Dooren et al., 2002). While all these enzymes are well characterized in prokaryotes like Escherichia coli, only a few have been detailed from pathogenic organisms including Plasmodium. Here, we have detailed a comparative analysis of each enzyme of the pathway from different organisms and discussed the various natural and chemically derived molecules used as their inhibitors in recent years with special emphasis to Plasmodium (Table 1).
Table 1.
Inhibitors reported for different enzyme involved in the MEP pathway with their chemical properties.
| Gene/Protein | Enzyme | Inhibitor molecules | Inhibition mechanism | Organism with IC50 value | References | |
|---|---|---|---|---|---|---|
| DXS | 1-deoxy-D-xylose-5-phosphate (DXP) Synthase | *6-benzyl-3-(4-chlorophenyl)-5-methyl-2-(trifluoromethyl)pyrazolo[1,5-a]pyrimidin-7(4H)-one | 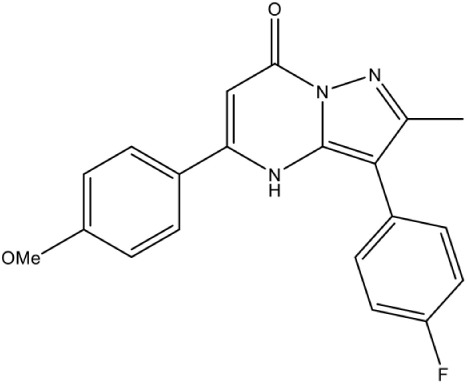 |
Competitive inhibition | M. tuberculosis (IC5010.6 μM) | Mao et al., 2008 |
| Ketoclomazone PubChem CID: 12811046 | 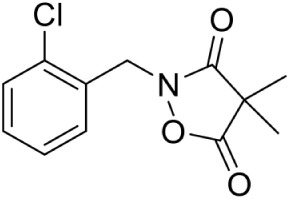 |
Non competitive inhibition | E. coli (IC50800 μg/mL) and H. influenzae (IC5012.5 μg/mL) | Matsue et al., 2010 | ||
| β-fluoropyruvate PubChem CID: 67946 | 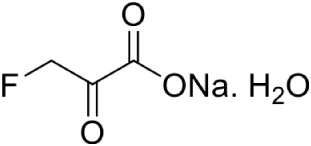 |
Competitive inhibition | P. vivax (IC5035 ± 1.7 μM) P. falciparum (IC5043 ± 3.8 μM) | Battistini et al., 2016 | ||
| Methylacetylphosphonate PubChem CID: 23674726 | 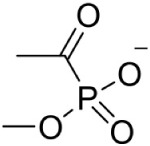 |
Competitive inhibition | P. vivax (IC5080 μM) P. falciparum (IC5046 ± 3.8 μM) | |||
| IspC | DXP reductoisomerase | Fosmidomycin PubChem CID: 572 | 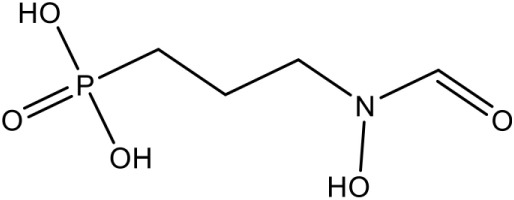 |
Competitive inhibition | P. falciparum (IC50350 ± 170 nM) | Jomaa et al., 1999; Lell et al., 2003; Umeda et al., 2011 |
| FR900098 PubChem CID: 162204 | 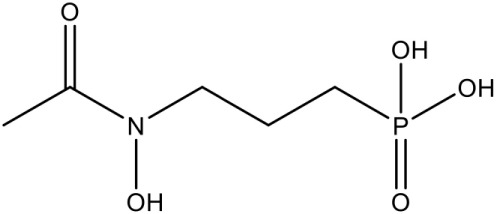 |
Competitive inhibition | P. falciparum (IC50170 ± 100 nM) | Jomaa et al., 1999 | ||
| *[1-(3,4-Difluorophenyl)-4-(hydroxylamino)-4-oxobutyl] phosphonic acid (Fosmidomycin reverse derivative) | 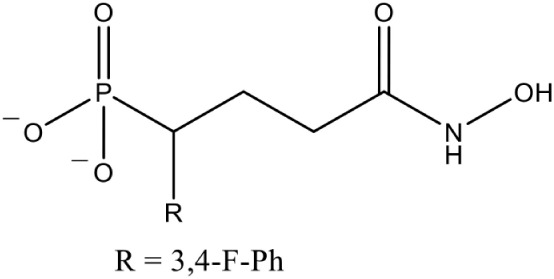 |
Competitive inhibition | P. falciparum (IC503 nM) | Behrendt et al., 2011 | ||
| *((3,4-Difluorophenyl)(2-(hydroxy(methyl)amino)-2-oxoethoxy)methyl) phosphonic acid | 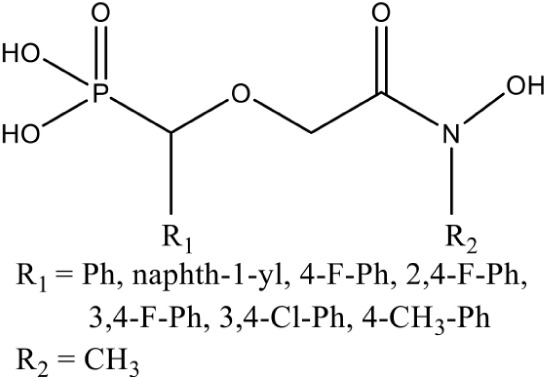 |
Competitive inhibition | P. falciparum (IC5012 nM) | Brücher et al., 2012 | ||
| *4-[Hydroxy(methyl)amino]-1-(4-methoxyphenyl)-4-oxobutylphosphonic acid (Fosmidomycin reverse analog | 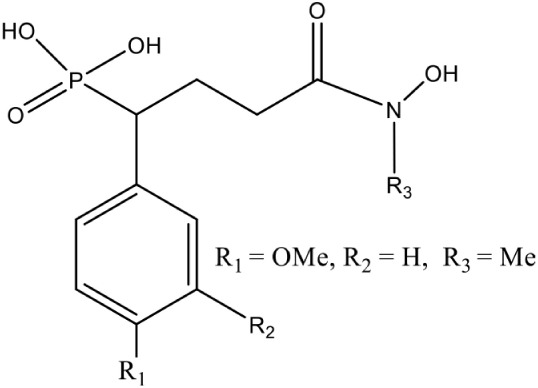 |
Competitive inhibition | P. falciparum (IC5020 nM) | Konzuch et al., 2014 | ||
| IspD | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | *L-erythritol-4-phosphate | 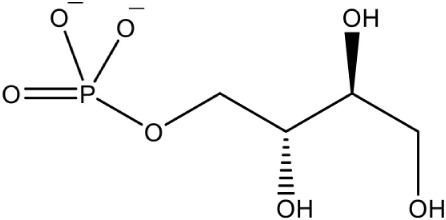 |
Competitive inhibition | B. abortus (IC501.36 mM) | Lillo et al., 2003 |
| 7-hydroxy-[1,2,4] triazolo [1,5-a] pyrimidine PubChem CID: 75629 (2503-56-2) | 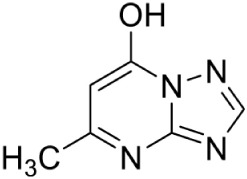 |
Allosteric inhibition | A. thaliana (IC50140 ± 10 nM) | Witschel et al., 2011 | ||
| *6-Amino-7-(1H-benzo[d]imidazol-2-yl)-5-[5-(diethylamino)-1-methylbut-1-yl)-5H-pyrrolo [3,2-b] pyrazine-2,3-dicarbonitrile | 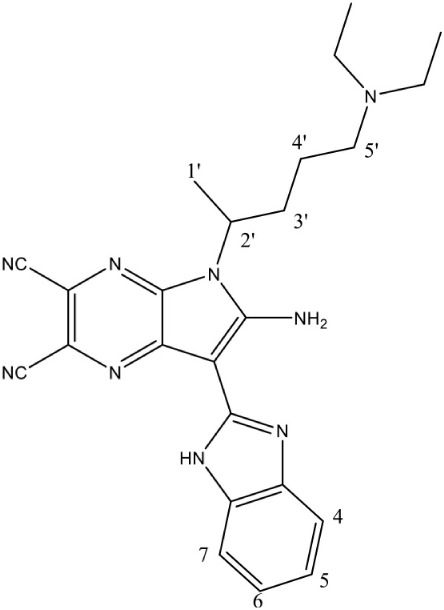 |
Non competitive inhibition | P. falciparum (EC50 50 nM) | Reker et al., 2014 | ||
| Pyrroloquinoxaline | 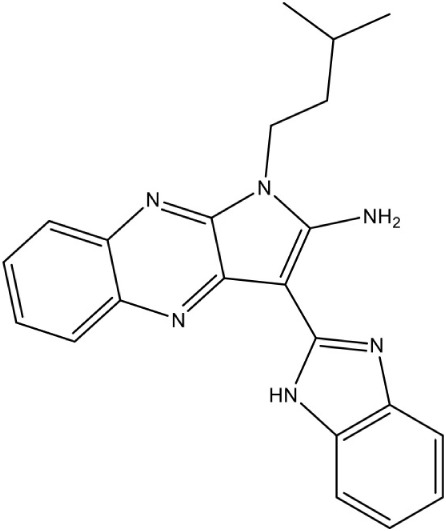 |
Non competitive inhibition | A. thaliana (IC50 1.6 μM) | Reker et al., 2014 | ||
| MMV008138 PubChem CID: 2829106 | 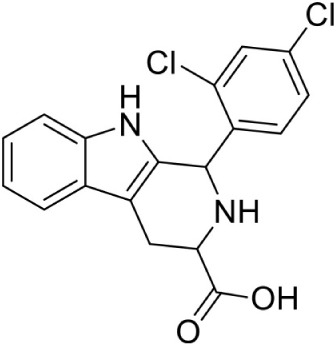 |
Competitive inhibition | P. falciparum (IC5047 nM) and P. vivax (IC50310 nM) | Imlay et al., 2015 | ||
| *Ethyl {3-[4-amino-5-{3-[(cyclopropylsulfonyl) amino] prop-1-yn-1-yl}-2-oxopyrimidin-1(2H)-yl] oxetan-3-yl} acetate | 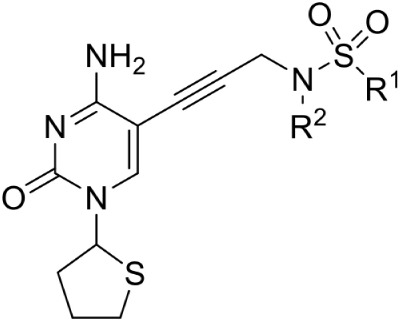 |
Competitive inhibition | A. aeolicus (IC50590 ± 10 nM) | Hirsch et al., 2008 | ||
| IspE | 4-(cytidine-5-diphospho)-2-C-methyl-D-erythritol kinase | 6-(benzylthio)-2-(2-hydroxyphenyl)-4-oxo-3,4-dihydro-2H-1,3-thiazine-5-carbonitrile PubChem CID: 3768522 | 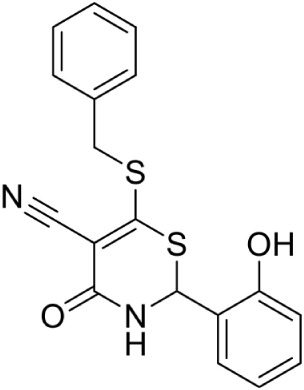 |
Competitive inhibition | E. coli (IC505.5 μM) | Tang et al., 2011 |
| *Diammonium 5′-O-{[({[2-({[5-(Dimethylamino) naphthalene-1- yl]sulfonyl}amino) ethyl] oxy}phosphinato)oxy] phosphinato} cytidine | 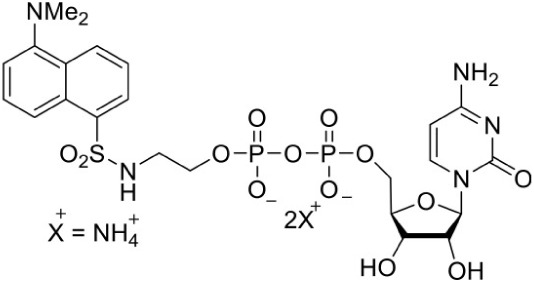 |
Competitive inhibition | E. coli (IC503.0 μM) | Crane et al., 2006 | ||
| IspF | 2C-Methyl-D-erythritol-2, 4-cyclodiphosphate synthase | Thiazolopyrimidine PubChem CID: 330031 | 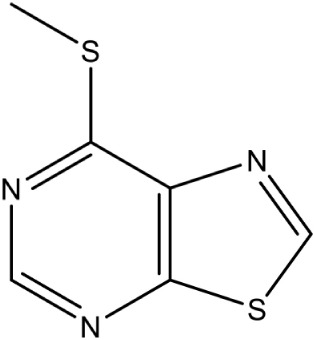 |
Competitive inhibition | P. falciparum (IC509.6 μM) and M. tuberculosis (IC506.1 μM) | Geist et al., 2010 |
| Aryl bis sulphonamide PubChem CID: 5333 | 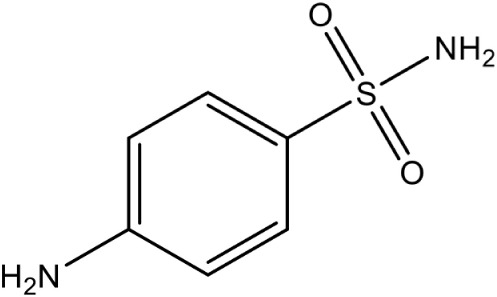 |
Competitive inhibition | P. falciparum (IC501.4 μM) and A. thaliana (IC50240 nM) | Thelemann et al., 2015 | ||
| Propargyl diphosphate PubChem CID: 46236597 | 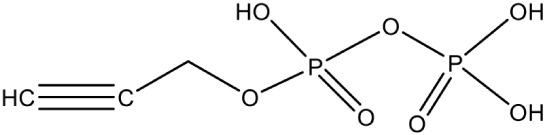 |
Competitive inhibition | E. coli (IC50750 nM) | Wang et al., 2010 | ||
| IspG | 4-Hydroxy-3-methyl-2-(E)-butenyl-4-diphosphate synthase | Prop-2-yn-1-yl trihydrogen diphosphate PubChem CID: 448670 | 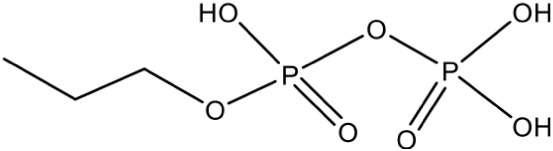 |
Competitive inhibition | T. thermophilus (IC50770 nM) | Quitterer et al., 2015 |
| But-3-yn-1-yl trihydrogen diphosphate PubChem CID: 46236598 | 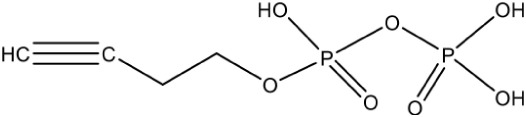 |
Competitive inhibition | T. thermophilus (IC50580 nM) | Wang et al., 2010 | ||
| But-3-yn-1-yl trihydrogen diphosphate PubChem CID: 46236598 | 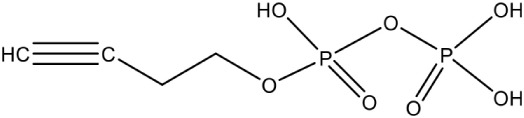 |
A. aeolicus (IC50450 nM) | ||||
| IspH | 4-Hydroxy-3-methyl-2-(E)-butenyl-4-diphosphate reductase | Pyridine phosphate PubChem CID: 10866885 | 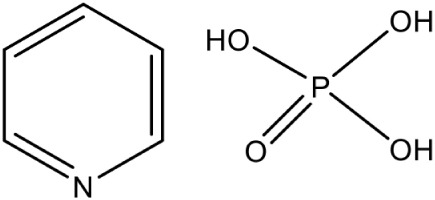 |
Competitive inhibition | A. aeolicus (IC5035 μM) | Wang et al., 2011 |
| *(E)-4-mercapto-3-methyl but-2 enyl diphosphate (Alkyne diphosphate derivative) | 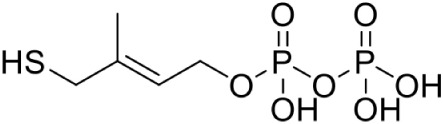 |
Competitive inhibition | E. coli (IC50210 nM) | Janthawornpong et al., 2013 | ||
PubChem Id not available.
DXS (DXP synthase)
The initial substrates pyruvate and Glyceraldehyde-3-Phosphate are acted upon by a thiamine pyrophosphate dependent enzyme DXS (EC 4.1.3.37) to generate 1-deoxy-D-xylose-5-phosphate (DOXP/DXP) (Sprenger et al., 1997). This first step is believed to be rate limiting in some organisms and represents a branch point for Vitamin B and isoprenoids synthesis in bacteria (Hahn et al., 2001). The X-ray crystal structure of DXS enzyme has been elucidated from E. coli (PDB Id: 2O1S) and Deinococcus radiodurans (PDB Id: 2O1X). This enzyme consists of three functional domains: Thiamine Pyrophosphate (TPP/ThDP) binding domain, Pyrimidine (PYR) binding domain and transketolase C domain. In E. coli, the interaction of ThDP and Mg2+ has been observed with amino acid residues present in the TPP DXS domain (Xiang et al., 2007). However, in Plasmodium, the characterization of DXS from both P. falciparum and P. vivax suggests its existence as a homodimer (Handa et al., 2013) which contains one-bound Mg (II) per enzyme molecule, having both a catalytic and structural role in the enzyme. All ThDP dependent enzymes catalyze two successive half reactions. The first step involves the attack of an activated ThDP ylide on the first substrate GA3P. The next step can occur via three distinct mechanisms: (i) the most common classical ping-pong mechanism; (ii) through an ordered sequential kinetic mechanism or (iii) through an alternate random sequential mechanism (Brammer et al., 2011). Enzyme kinetics of DXS in Plasmodium points toward a random sequential kinetic mechanism, an unusual finding for ThDP-dependent enzymes similar to Rhodobacter capsulatus (Battistini et al., 2016).
In Mycobacterium tuberculosis, transketolase inhibitors like derivatives of Pyrimidinone (IC50 = 10.6 μM) have been shown to inhibit DXS enzyme (Mao et al., 2008). Later, a herbicide Ketoclomazone was shown to inhibit the DXS enzyme in both E. coli and Haemophilus influenzae by binding to a site, which differs from both the pyruvate and GA3P binding sites, thus suggesting non-competitive inhibition (Matsue et al., 2010). However, yet, the binding site of the drug is not elucidated. In another study, Smith et al. (2014) has demonstrated the selective inhibition of ThDP dependent enzymes in E. coli, Yersinia pestis, and M. tuberculosis by unnatural by-substrate analog butylacetylphosphonate (BAP) and its synergistic action with established anti-microbial agents like Fosmidomycin (Fos). However, BAP exhibits weak anti-microbial activity, possibly due to poor cellular uptake. Recently, analogs of pyruvate, β-Fluoropyruvate (F-Pyr) and methylacetylphosphonate (MAP) were demonstrated as competitive inhibitors of the DXS enzyme in both P. falciparum and P. vivax (Handa et al., 2013; Battistini et al., 2016).
IspC (DXP reductoisomerase/DXR)
IspC (EC 1.1.1.267) is the first enzyme committed to the isoprenoids biosynthesis in the MEP pathway. In a rate limiting step, it is responsible for the conversion of DOXP to 2-C-methyl-D-erythritol-4-phosphate (MEP) by using the NADPH pro-S hydride (Brammer et al., 2011). Its structure has been well defined in E. coli (PDB Id: 1Q0L; Mac Sweeney et al., 2005), P. falciparum (PDB Id: 3AU9; Umeda et al., 2011, 2015) and various other pathogens, including M. tuberculosis (PDB Id: 2Y1C; Andaloussi et al., 2011) and Zymomonas mobilis (PDB Id: 1ROK; Ricagno et al., 2004; Henriksson et al., 2006). It is a class B dehydrogenase enzyme that exists as a homodimer where each subunit is composed of three domains; an N-terminal domain for cofactor binding (DXP_reductoisom), central domain having active site residues (DXP_redisom_C) and a C-terminal helical domain (DXPR_C). The N-terminal domain is a member of dinucleotide binding fold and serves for binding of NADPH. The central catalytic domain harbors the binding sites for divalent cations (like Mn2+ or Mg2+), phosphate of the substrate and the catalytic loop. The C-terminal domain is connected to the catalytic domain by a linker region that spans entire monomer and appears to have a structural role in supporting the catalytic domain. Normally, the active site has two different conformations, open and closed. The open conformation allows the substrate, DXP, to enter and bind to the active site while in the closed conformation, a flap covers the active site and catalytic function is activated (Mac Sweeney et al., 2005). In P. falciparum IspC, NADPH molecule bind to the cavity composed of conserved residues D231, E233, S269, S270, W296, M298, S306, N311, K312, and E315 present toward the C-terminal of the enzyme. These residues are conserved in all human malaria parasites (Kunfermann et al., 2013).
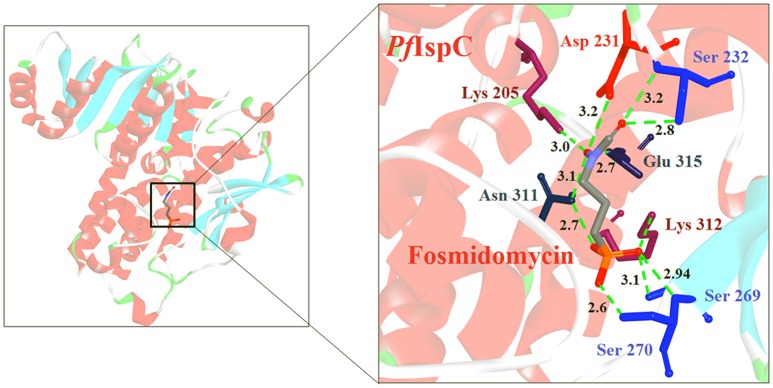
In E. coli and P. falciparum, Fos has been characterized as an inhibitor (IC50 = 0.032 μM) of IspC enzyme. It behaves as a substrate analog of DOXP and competes for its binding site on IspC (Jomaa et al., 1999; Steinbacher et al., 2003). In P. falciparum, Fos interacts with K205, D231, S232, S269, S270, N311, K312, and E315 residues of the catalytic domain (Figure 1; Umeda et al., 2011). The effect of Fos has been reported to be varied amongst different stages of Plasmodium development as well as amongst different apicomplexans. It was found that Fos is only effective on the erythrocytic stages of Plasmodium due to the formation of new permeability pathways (minute anionic selective channels formed due to entry of parasite in erythrocytes), but has no or minimal effect on the liver stages of P. berghei. In addition, Fos is not effective on many apicomplexans including Toxoplasma, Eimeria even at higher concentrations, probably due to its impermeability to the parasite plasma membrane (Baumeister et al., 2011; Nair et al., 2011).
Figure 1.
X-ray crystal structure (PDB Id: 3AU9; Umeda et al., 2011) of P. falciparum IspC enzyme interacting with Fosmidomycin (represented as stick). The drug interacts with K205, D231, S232, S269, S270, N311, K312, and E315 residues of enzyme. CPK color scheme followed and distance represented in Å.
Fos is one of those few drugs which was able to qualify the Phase–II clinical trials (in combination with Clindamycin), however, its treatment faces several drawbacks, including low bioavailability, rapid clearance from the parasite, less absorption (16.6 μg/mL after an oral dose of 40 mg/kg) and shorter half–life (1.14 h) leading to incidences of recurrence. Due to its toxicity at higher doses, this drug alone could not act as potential antimalarial (Murakawa et al., 1982; Kuemmerle et al., 1987; Lell et al., 2003). To overcome the bottlenecks in the use of Fos as an anti-malarial, different groups have synthesized analogs of Fos. Jansson et al. (2013) reported various analogs of Fos but only one among these, a disubstituted Fos (1-(3,4-Dichlorophenyl)-3-(N-hydroxyphenylamido) propylphosphonic acid) was shown to have equivalent activity (IC50 = 0.04 μM) as Fos. Recently, a Fos derivative (phosphonohydroxamates) was observed to inhibit parasite growth in the in-vitro culture of P. falciparum Dd2 strain, resistant to chloroquine and mefloquine (Faísca Phillips et al., 2015). In search of better derivatives of Fos, reverse analogs of Fos were also synthesized. Behrendt et al. (2011) synthesized the reverse hydroxamate based inhibitors amongst which [1-(3,4-Difluorophenyl)-4-(hydroxylamino)-4-oxobutyl] phosphonic acid is considered most effective with an IC50 of 3 nM. Later Brücher et al. (2012) synthesized α-Aryl-substituted β-oxa isosteres of Fos with a reverse orientation of the hydroxamic acid group and tested them against recombinant P. falciparum IspC and chloroquine-sensitive and resistant strains of P. falciparum. They found an inhibitory activity of these derivatives against P. falciparum IspC, with the most active derivative ((3,4-Difluorophenyl) (2-(hydroxy(methyl)amino)-2-oxoethoxy) methyl) phosphonic acid showing an IC50 value of 12 nM and potent in-vitro anti-plasmodial activity. Following similar approach for synthesis of reverse analogs, Konzuch et al. (2014) showed a new compound 4-[Hydroxy(methyl)amino]-1-(4-methoxyphenyl)-4-oxobutylphosphonic acid, to exhibits more than one order of magnitude of activity in comparison to Fos.
Another approach has been tried by Haymond et al. (2014), where several compounds from resolved crystal structures of M. tuberculosis MEP synthase in complex with Fos were designed, containing an amide-linked or O-linked functional group. The strategy was to target two major binding sites in MEP synthase; the Fos/DXP site and the NADPH site, bridging these adjacent sites to yield a highly specific inhibitor ligand. Amongst the compounds tested, the most effective inhibitor (diethyl 3-(benzyloxyamino) propylphosphonate) binds to both the NADPH and DXP sites, acting as a potent tight binding inhibitor of the enzyme in both M. tuberculosis and Y. pestis. However, a growth inhibition secondary screen revealed that the whole-cell inhibitory activity of this compound is relatively poor, indicating the need for additional structure-activity relationship studies to elucidate the underlying etiology.
Apart from the synthesis of Fos derivatives, various other trials are underway to increase the efficacy of Fos. Nair et al. (2011) had shown the increase in permeability and uptake of Fos in T. gondii in presence of recombinant GA3P transporter (GlpT) protein of E. coli. Similarly, Sparr et al. (2013) has recently reported a mechanism of tagging Fos with cell penetrating peptides consisting of octa-arginine which could ultimately block the hepatic stages of the parasite.
IspD [2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (YgbP)]
IspD (EC 2.7.7.60) enzyme, catalyzes the cytidylation process and participates in the third step of the pathway where its activity is highly dependent upon divalent cations Mg2+ or Mn2+ (Richard et al., 2001). Nucleotide derivatives, i.e., Cytosine-5′-triphosphate (CTP) and phosphate groups i.e., 2-C-methyl-D-erythritol-4-phosphate (MEP) are introduced directly as substrates in the IspD catalyzed reaction and produces 4-diphosphocytidyl-2C-methyl-D-erythritol (CDP-ME) with pyrophosphate as a bi-product. The catalytic mechanism of IspD enzyme is unique and comes under the category of associative mechanism. According to this process, when there is a nucleophile attack on the α-phosphate of CTP by the 4-phosphate of MEP, a negatively charged penta-coordinate transition state is formed. The collapse of this charged transition state finally leads to pyrophosphate release and CDP-ME formation. Two lysine residues (Lys27 and Lys213 in E. coli IspD) are critical to stabilize this pentavalent transition state (Richard et al., 2004). Amino acid alignments show that the basic residues in the IspD active site, in particular the E. coli IspD Lys27/Lys213 are also conserved in P. falciparum (Hunter, 2011). The X-ray crystal structure of IspD protein is known from several organisms, such as M. tuberculosis (PDB Id: 2WXN; Björkelid et al., 2011), Thermotoga maritima (PDB Id: 1VPA; unpublished), Neisseria gonorrhoeae (PDB Id: 1VGZ; Badger et al., 2005), Thermus thermophilus (PDB Id: 2PX7; unpublished), Listeria monocytogenes (PDB Id: 3F1C; unpublished) and Arabidopsis thaliana (PDB Id: 1W77; Gabrielsen et al., 2006). The presence of a conserved CDP-ME synthase domain (GT-A superfamily) is important for the functionality of IspD enzyme and follows a Rossmann fold arrangement which only allows the sequestration of pyrimidine nucleotide i.e., CTP. The presence of two signature motifs GXG and [IVT] -[LIVMC] -[IVT] -[HS] -D-[SGAV] -[AV] -R is another important feature of IspD enzyme where the Glycine rich GXG motif at N- terminal provides a proper orientation for the binding of substrate near to the other conserved signature motif of the enzyme (Shi et al., 2007).
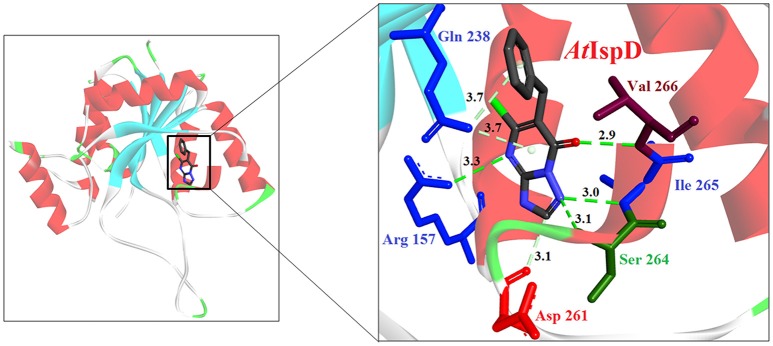
The first inhibitor of IspD enzyme was reported from Brucella abortus, where a compound named L-erythritol-4-phosphate (E4P) showed an IC50 of 1.36 mM (Lillo et al., 2003). Later, Witschel et al. (2011) showed the inhibition of IspD in A. thaliana by a synthetic compound 7-hydroxy [1,2,4] triazolo [1,5-a] pyrimidine (Azolopyrimidine). Crystallization studies of this compound with IspD showed interactions at R157, Q238, D261, S264, I265, and V266 residues (Figure 2) suggesting an allosteric inhibition. Further, direct enzyme based assays by Reker et al. (2014) suggested Pyrrolopyrazines derivatives as potent inhibitor of A. thaliana IspD. They have also shown the potential of these pyrrolopyrazines for inhibiting the growth of malaria parasite in cell based assays, where, 6-Amino-7-(1H-benzo[d]imidazol-2-yl)-5-[5-(diethylamino)-1-methylbut-1-yl)-5H-pyrrolo [3,2-b] pyrazine-2,3-dicarbonitrile was shown to be effective at nano molar concentration. However, the authors have also suggested an additional mechanism of action for the compound in Plasmodium. In another study, highly halogenated marine alkaloid of class pseudilins isolated from the marine bacterium Pseudomonas bromoutilis exhibited both herbicidal and anti-malarial activity (EC50 of 1–12 μM in cell based assays). However, cytotoxicity of these pseudilins in mammalian cell also suggests additional molecular targets apart from IspD (Kunfermann et al., 2014). Recently, another inhibitor MMV008138 (shortlisted from Malaria Box) was found effective against Pf IspD enzyme showing competitive inhibition with CTP substrate (Wu et al., 2015; Yao et al., 2015), whereas, in P. vivax, it is effective only at a lower concentration of CTP substrate (Imlay et al., 2015). In addition, MMV008138 does not exhibit activity against liver stages of P. yoelli nor does it have activity against sexual stages of P. falciparum (Bowman et al., 2014). Thus, further investigation is required for using MMV008138 as a common inhibitor for the IspD enzyme of both P. falciparum and P. vivax.
Figure 2.
X-ray crystal strucuture (PDB Id: 2YC3; Witschel et al., 2011) of A. thaliana IspD interacting with Azolopyrimidines (represented as stick). The inhibitory molecule 7-hydroxy-[1,2,4] triazolo [1,5-a] pyrimidine interact with R157, Q238, D261, S264, I265, and V266 residues of enzyme. CPK color scheme followed and distance represented in Å.
IspE [4-(cytidine-5-diphospho)-2-C-methyl-D-erythritol kinase (CMK)]
The fourth step of the pathway is catalyzed by IspE (EC 2.7.1.148) enzyme, an ATP and Mg2+ dependent enzyme belonging to the ATP–dependent GHMP kinase super-family (Andreassi and Leyh, 2004). It catalyzes the conversion of CDP-ME to 4-diphosphocytidyl-2-C-methyl-D-erythritol-2-phosphate (CDP-ME2P) in an ATP-dependent manner. The X-Ray crystal structure of IspE has been elucidated from E. coli, T. thermophilus HB8 (PDB Ids: 2WW4 and 1UEK; Wada et al., 2003), Aquifex aeolicus (PDB Id: 2VF3; Sgraja et al., 2008) and M. tuberculosis (PDB Id: 3PYD; Shan et al., 2011). The IspE enzyme exists as a monomer and displays the characteristic two-domain fold of the GHMP kinase superfamily. The active site of IspE is enclosed in a deep cleft between these two domains (Rohdich et al., 2000) where three known conserved motifs are present. Motif A (Lys13 to Leu18) forms the substrate binding site, Motif B (Gly102 to Ser107) forms a glycine-rich phosphate binding loop that interacts with the ATP and Motif C (Val254 to Gly258) helps to stabilize the conformation of motifs A and B rather than interacting with ligands directly (Sgraja et al., 2008; Eoh et al., 2009; Shan et al., 2011). In addition, a small, hydrophobic pocket lies adjacent to the CDP-binding site lined by amino acids Leu14, Ile27, Tyr175, and Leu208 (Hirsch et al., 2007).
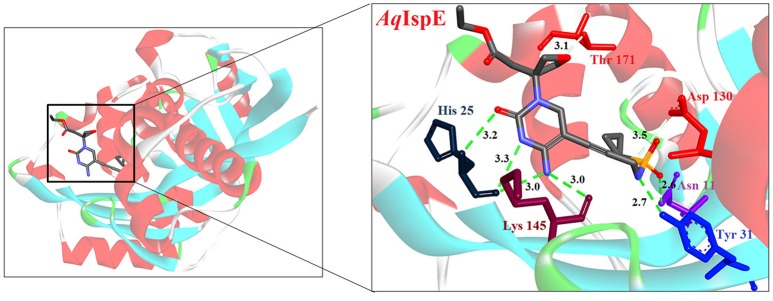
Hirsch et al. (2008) used the structure-based design approach and developed various inhibitors to target the substrate instead of enzyme. On screening of these compounds, Ethyl {3-[4-amino-5-{3-[(cyclopropylsulfonyl) amino] prop-1-yn-1-yl}-2-oxopyrimidin-1(2H) -yl] oxetan-3-yl} acetate was found to be the most promising candidate with an IC50 of 590 ± 10 nM. Further, co-crystallization studies of IspE enzyme along with this inhibitor in E. coli and A. aeolicus confirmed that the inhibitor fits properly in the cytidine-binding pocket of the enzyme, where the cyclopropyl substituent of the sulfone moiety occupies the small cavity not used by the substrate (Figure 3). The highly hydrophobic Phe185 in E. coli IspE is replaced with a more hydrophilic tyrosine (Tyr) residue in M. tuberculosis, P. falciparum and A. aeolicus, affecting the binding characteristics of this pocket, by strongly reducing its hydrophobic character. Thus, the cyclopropyl ring may not locate to this sub-pocket and indeed, the cyclopropyl ring prevents any solvation of the phenolic hydroxyl group of Tyr175. This explains the reduced inhibitory activity of the above compound against A. aeolicus and P. falciparum IspE (Hirsch et al., 2007, 2008).
Figure 3.
X-ray crystal strucuture (PDB Id: 2VF3; Hirsch et al., 2008) of A. aeolicus IspE interacting with ethyl {3-[4-amino-5-{3-[(cyclopropylsulfonyl) amino] prop-1-yn-1-yl}-2-oxopyrimidin-1(2H)-yl] oxetan-3-yl} acetate (represented as stick). The inhibitory molecule interacts with N11, H25, Y31, D130, K145, and T171 residues of enzyme. CPK color scheme followed and distance represented in Å.
In another study, from a list of existing small molecule inhibitors of GHMP kinases, Tang et al. (2011) identified and confirmed two novel scaffold classes of CDP-ME kinase inhibitors in E. coli; 6-(benzylthio)-2-(2-hydroxyphenyl)-4-oxo-3,4-dihydro-2H-1,3-thiazine-5-carbonitrile and (Z)-3-methyl-4-((5-phenylfuran-2-yl) methylene) isoxazol-5(4H)-one. Selection of these compounds was based on the predicted binding mode where 6-benzylthio and 5-phenylfuran ring moieties present in these compounds show a strong π-π interaction with cytidine binding pocket of CDP-ME created by three critical residues, viz. Tyr25, His26, and Phe185. In addition, binding of first compound show that its central core dihydro-2H-1,3-thiazine-5-carbonitrile –C = O mimics the α-, β-phosphates of substrate CDP-ME and participates in H-bonding interaction with Asp141 –NH…O-, whereas the 2-hydroxy-aryl ring position toward the binding site of the D-erythritol moiety of CDP-ME. IC50 values of these compounds were 18 and 5.5 μM, respectively. Substructure search and docking experiments based on these two scaffolds further identified 23 analogs for structure-activity relationship (SAR) studies. Three new compounds from the isoxazol-5(4H)-one series have shown inhibitory activities against E. coli and Y. pestis CDP-ME kinases with the IC50 values ranging from 7 to 13 μM (Tang et al., 2011).
Another compound which has shown inhibitory activity against Pf IspE is 1,3-diiminoisoindoline carbohydrazide with an IC50 value <100 nM in cell based assay. Synthesis of a variety of derivatives allowed an improvement of the initial antimalarial activity down to IC50 = 18 nM for the most potent compound (Mombelli et al., 2012). However, till date, none of the tested derivatives have shown any inhibitory activity against P. falciparum IspE in-vitro below 100 μM (Masini and Hirsch, 2014), which demands further investigation into this enzyme.
IspF [2C-methyl-D-erythritol-2, 4-cyclodiphosphate synthase (ygbB)]
IspF (EC 4.6.1.12) catalyzes the conversion of CDP-ME-2P into 2-C-methyl-D-erythritol-2,4-cyclodiphosphate (MECP), and like IspD, is dependent on divalent cations Zn2+ or Mn2+ for its activity. X-ray crystal structures of IspF from E. coli (PDB Id: 2GZL; Crane et al., 2006); (PDB Id: 1KNJ; Richard et al., 2002); (PDB Id: 1U3L; Steinbacher et al., 2002), A. thaliana (PDB Id: 2PMP; Calisto et al., 2007), H. influenzae (PDB Id: 1VH8; Lehmann et al., 2002), M. smegmatis (PDB Id: 2UZH; Buetow et al., 2007), P. vivax (PDB Id: 3BN6) and P. falciparum (PDB Id: 4C81; O'Rourke et al., 2014) have been determined, showing formation of a homo-trimeric quaternary structure. The active sites are located at the interface of two monomer units where the pocket involved in binding the phosphate moiety of the substrate is capped with a flexible loop that becomes completely ordered when the reaction product is bound. In E. coli, the downstream products of Isoprenoids pathway like geranyl- or farnesyl-pyrophosphate have been observed to bind IspF trimers at the central hydrophobic cavity, which suggests a possible feedback inhibition of the enzyme (Kemp et al., 2005). However, in Plasmodium, reports of geranyl pyrophosphate interacting with IPP in the cytoplasm (Jordão et al., 2013) indicates that though IspF is present as homo-trimer in P. falciparum apicoplast, it may not be affected by any feedback mechanism of geranyl pyrophosphate localized in cytoplasm.
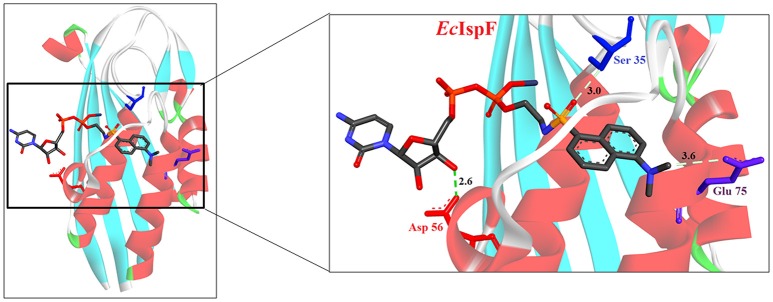
Despite the fact that the active site of IspF is considered the most druggable, based on the presence of high apolar amino acid residues, very few inhibitors have been reported. Crane et al. (2006) tested various anthranilate compounds with cytidine moiety, amongst which fluorescent diammonium 5′-O-{[({[2-({[5-(Dimethylamino) naphthalene-1- yl] sulfonyl} amino) ethyl] oxy} phosphinato) oxy] phosphinato} cytidine was found to be effective against E. coli IspF. In in-vitro studies, this inhibitor was reported to interact with S35, D56 and E77 residues of E. coli IspF enzyme (Figure 4). The most successful inhibitors identified till date for IspF are non-cytidine-like thiazolopyrimidine derivatives, with high activity against both P. falciparum (IC50 = 9.6 μM) and M. tuberculosis IspF (IC50 = 6.1 μM) (Geist et al., 2010), and the aryl bis-sulphonamide inhibitors showing inhibition of P. falciparum IspF and A. thaliana IspF with IC50 values as low as 1.4 μM and 240 nM, respectively (Thelemann et al., 2015). However, their binding mode and optimization is yet to be reported.
Figure 4.
X-ray crystal strucuture (PDB Id: 2GZL; Crane et al., 2006) of E. coli IspF interacting with diammonium 5′-O-{[({[2-({[5-(Dimethylamino) naphthalene-1- yl]sulfonyl}amino) ethyl] oxy}phosphinato)oxy] phosphinato} cytidine (represented as stick). The fluorescent inhibitory molecule interacts with S35, D56, and E77 residues of enzyme. CPK color scheme followed and distance represented in Å.
IspG [4-hydroxy-3-methyl-2-(E)-butenyl-4-diphosphate synthase (gcpE)] and IspH [4-hydroxy-3-methyl-2-(E)-butenyl-4-diphosphate reductase (lytB)]
IspG and IspH enzymes participate in the last two steps of the pathway, respectively. Initially, IspG catalyzes the reduction of MECP through a multistep reaction and converts it into 4-hydroxy-3-methyl-2-(E)-butenyl-4-diphosphate (HMBDP) and then IspH converts it to IPP and DMAPP (Rohdich et al., 2002). The X-ray crystal structure of IspG has been derived from A. aeolicus (PDB Id: 3NOY; Lee et al., 2010) and T. thermophilus (PDB Id: 2YOF; Rekittke et al., 2011). The IspG enzyme consists of two conserved domains: an N-terminal TIM barrel domain for the binding of MECP substrate and a C-terminal domain for binding of Fe-S clusters. In P. falciparum, the electrons required for the binding of MECP substrate to the IspG enzyme are provided by ferredoxin/ferredoxin NADP+ reductase system through [4Fe-4S] clusters (Röhrich et al., 2005). This binding of the MECP substrate is responsible for the formation of a double bond that converts cyclic form of the MECP molecule to aliphatic HMBDP.
The HMBDP formed by IspG is then converted to the IPP and DMAPP by IspH enzyme and this conversion consists of three steps: (i) removal of a hydroxyl group, (ii) transfer of two electrons from the [4Fe-4S] cluster, and (iii) the protonation of an intermediate allylic anion (Laupitz et al., 2004). The X-ray crystal structure of IspH has been derived from E. coli (PDB Id: 3KE8; Gräwert et al., 2010), A. aeolicus (PDB Id: 3DNF; Rekittke et al., 2008) and P. falciparum (PDB Id: 4N7B; Rekittke et al., 2013). IspH consists of two domains, a LytB domain for the binding of HMBDP and a Fe-S cluster binding domain similar to IspG. Specific requirements for the activity of IspG and IspH enzyme was proven by a study in Saccharomyces cerevisiae, where the reconstruction of the complete MEP pathway was aimed, however, even after incorporation of all the necessary components including all the enzymes, substrates and cofactors, the pathway remained non-functional. It was concluded that a reducing environment and compartmentalization is required for the functionality of these enzymes (Partow et al., 2012).
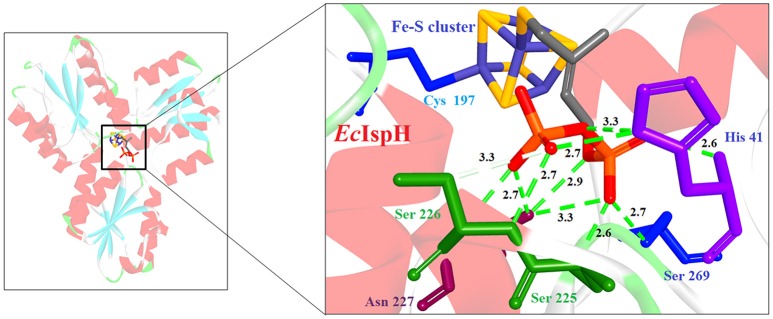
Most of the inhibitors developed initially against IspG and IspH targeted the Fe-S clusters, especially the unique fourth iron site, but as most of the metallo-proteins in mammals, including humans require these Fe-S clusters for their activity, these inhibitors have selectivity issues. Substrate analogoues that bind to IspG and IspH have been tried, but they could not turn out to be potent inhibitors. Replacing the diphosphate group with another moiety like carbamates or aminosulfonyl carbamates showed only very weak inhibitory activity (Van Hoof et al., 2008). In search of inhibitor, amongst various chemically synthesized compounds, (E)-4-mercapto-3-methyl but-2 enyl diphosphate (Alkyne diphosphate derivative) was shown to inhibit E. coli IspH enzyme (Figure 5) where interaction was observed with H41, S225, S226, N227, and S269 (Xiao et al., 2011; Span et al., 2013). E126 residue is reported to be important for the activity of IspH enzyme in E. coli and A. aeolicus, and a mutation at this residue can cause a significant effect on its activity (Wang et al., 2010). This residue is conserved in other organisms, including Apicomplexans and plants.
Figure 5.
X-ray crystal strucuture (PDB Id: 4H4E; Span et al., 2013) of E. coli IspH interacting with (E)-4-mercapto-3-methyl but-2 enyl diphosphate (represented as stick). The inhibitory molecule interacts with H41, S225, S226, N227, and S269 residues of enzyme. CPK color scheme followed and distance represented in Å.
Bhuyan et al. (2015) screened a large ligand data-set containing diphospate group against the P. falciparum IspH structure, and based on Goldscore and Chemscore identified 17 lead compounds amongst which 5-((hydroxymethyl)-O-pyrophosphoryl) uracil shows best binding affinities with Plasmodium IspH, and thus can be considered as its potential inhibitor. Recently, the derivatives of diphosphonate such as alkyl phosphate have been identified as potential inhibitors for IspG and IspH enzymes of A. aeolicus, E. coli, and P. falciparum (Guerra et al., 2014), however their exact mechanism for inhibition is unknown.
MEP pathway as a potential drug target in Plasmodium
MEP pathway and its enzymes present attractive new targets for the development of broad-spectrum novel anti-microbial and anti-malarial drugs. In the last decade, various researchers have extensively explored these enzymes and their active sites to identify/design probable inhibitors in prokaryotes (Hale et al., 2012). In-silico studies and certain in-vitro assays performed on the malaria parasite culture using drugs tested on prokaryotic homologs have detailed few compounds that can block the parasite growth by specifically binding to the enzymes of MEP pathway. These compounds can be further analyzed for their pharmacokinetics and pharmacological efficacy to establish them as probable anti-malarials.
The natural antibiotic Fos, an inhibitor of IspC/DXR enzyme has been widely used as an anti-bacterial and as an anti-malarial to block in-vitro parasite cultures. Despite the apparent merits of Fos, the drawbacks associated with it like low absorption and shorter half-life, hampered its market introduction. Several research groups attempted to improve its structural activity by chemical modification, but none has been able to achieve activity similar to Fos in vivo. However, the natural acetyl derivative of Fos, FR900098, when tested for its anti-malarial property has shown better activity (IC50 = 0.018 μM) than Fos. Thus, an extensive research could help in the development of new chemically synthesized derivatives of Fos and similar compounds, to be used as potential anti-malarial.
With the studies performed on the initial enzymes of the Plasmodium MEP pathway, showing the essentiality and the uniqueness of the pathway for the survival of the parasite, it becomes imperative to study the remaining enzymes of the pathway. With the availability of the X-ray crystal structure of almost half of them (IspC/DXR, IspF & IspH) from P. falciparum, showing the conserved functional domains and substrate binding sites, extrapolating the prokaryotic MEP inhibitors data to Plasmodium is a promising way of identifying novel drugs. Another approach can be to test the efficacy of clinically approved and commercially established anti-bacterials targeting these enzymes, as initially Fosmidomycin was also one amongst those compounds.
Author contributions
GS, SG, and VS conceptualized and designed the manuscript. GS and VS drafted the manuscript. GS, ZP, SG, and VS read and critically revised the contents of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
GS acknowledges Senior Research fellowship from the University Grant Commission, Govt. of India, New Delhi, India. ZP acknowledges Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR), Govt. of India, New Delhi, India. VS acknowledges University Grant Commission, Govt. of India, New Delhi, India for the financial support through the grant [F.No.42-182/2013 (SR)]. The authors wish to thank Birla Institute of Technology and Science, Pilani, India for providing the required infrastructural facilities during this study.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01421
References
- Altincicek B., Kollas A. K., Eberl M., Wiesner J., Sanderbrand S., Hintz M., et al. (2001b). LytB, a novel gene of the 2-C-methyl-D-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. FEBS Lett. 499, 37–40. 10.1016/S0014-5793(01)02516-9 [DOI] [PubMed] [Google Scholar]
- Altincicek B., Kollas A. K., Sanderbrand S., Wiesner J., Hintz M., Beck E., et al. (2001a). GcpE is involved in the 2-C-methyl-Derythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. J. Bacteriol. 183, 2411–2416. 10.1128/JB.183.8.2411-2416.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andaloussi M., Henriksson L. M., Wieckowska A., Lindh M., Björkelid C., Larsson A. M., et al. (2011). Design, synthesis, and X-ray crystallographic studies of α-aryl substituted fosmidomycin analogues as inhibitors of Mycobacterium tuberculosis 1-deoxy-d-xylulose 5-phosphate reductoisomerase. J. Med. Chem. 54, 4964–4976. 10.1021/jm2000085 [DOI] [PubMed] [Google Scholar]
- Andreassi J. L., II, Leyh T. S. (2004). Molecular functions of conserved aspects of the GHMP kinase family. Biochemistry 43, 14594–14601. 10.1021/bi048963o [DOI] [PubMed] [Google Scholar]
- Bach T. J., Boronat A., Campos N., Ferrer A., Vollack K. U. (1999). Mevalonate biosynthesis in plants. Crit. Rev. Biochem. Mol. 34, 107–122. 10.1080/10409239991209237 [DOI] [PubMed] [Google Scholar]
- Badger J., Sauder J. M., Adams J. M., Antonysamy S., Bain K., Bergseid M. G., et al. (2005). Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins 60, 787–796. 10.1002/prot.20541 [DOI] [PubMed] [Google Scholar]
- Banerjee A., Sharkey T. D. (2014). Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat. Prod. Rep. 31, 1043–1055. 10.1039/C3NP70124G [DOI] [PubMed] [Google Scholar]
- Battistini M. R., Shoji C., Handa S., Breydo L., Merkler D. J. (2016). Mechanistic binding insights for 1-deoxy-d-Xylulose-5-Phosphate synthase, the enzyme catalyzing the first reaction of isoprenoid biosynthesis in the malaria-causing protists, Plasmodium falciparum and Plasmodium vivax. Protein Express. Purif. 120, 16–27. 10.1016/j.pep.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister S., Wiesner J., Reichenberg A., Hintz M., Bietz S., Harb O. S., et al. (2011). Fosmidomycin uptake into Plasmodium and Babesia-infected erythrocytes is facilitated by parasite-induced new permeability pathways. PLoS ONE 6:19334. 10.1371/journal.pone.0019334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendt C. T., Kunfermann A., Illarionova V., Matheeussen A., Pein M. K., Grãwert T., et al. (2011). Reverse fosmidomycin derivatives against the antimalarial drug target IspC (Dxr). J. Med. Chem. 54, 6796–6802. 10.1021/jm200694q [DOI] [PubMed] [Google Scholar]
- Bhuyan R., Nandy S. K., Seal A. (2015). An in silico structural insights into Plasmodium LytB protein and its inhibition. J. Biomol. Struct. Dyn. 33, 1198–1210. 10.1080/07391102.2014.938248 [DOI] [PubMed] [Google Scholar]
- Björkelid C., Bergfors T., Henriksson L. M., Stern A. L., Unge T., Mowbray S. L., et al. (2011). Structural and functional studies of mycobacterial IspD enzymes. Acta Crystallogr. Sect. D Biol. Crystallogr. 67, 403–414. 10.1107/S0907444911006160 [DOI] [PubMed] [Google Scholar]
- Bowman J. D., Merino E. F., Brooks C. F., Striepen B., Carlier P. R., Cassera M. B. (2014). Antiapicoplast and gametocytocidal screening to identify the mechanisms of action of compounds within the malaria box. Antimicrob. Agents Chemother. 58, 811–819. 10.1128/AAC.01500-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brammer L. A., Smith J. M., Wade H., Meyers C. F. (2011). 1-Deoxy-D-xylulose-5-phosphate synthase catalyzes a novel random sequential mechanism. J. Biol. Chem. 286, 36522–36531. 10.1074/jbc.M111.259747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brücher K., Illarionov B., Held J., Tschan S., Kunfermann A., Pein M. K., et al. (2012). α-Substituted β-oxa isosteres of fosmidomycin: synthesis and biological evaluation. J. Med. Chem. 55, 6566–6575. 10.1021/jm300652f [DOI] [PubMed] [Google Scholar]
- Buetow L., Brown A. C., Parish T., Hunter W. N. (2007). The structure of Mycobacteria 2 C-methyl-D-erythritol-2, 4-cyclodiphosphate synthase, an essential enzyme, provides a platform for drug discovery. BMC Struct. Biol. 7:68 10.1186/1472-6807-7-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisto B. M., Perez-Gil J., Bergua M., Querol-Audi J., Fita I., Imperial S. (2007). Biosynthesis of isoprenoids in plants: structure of the 2C-methyl-d-erithrytol 2, 4-cyclodiphosphate synthase from Arabidopsis thaliana. Comparison with the bacterial enzymes. Prot. Sci. 16, 2082–2088. 10.1110/ps.072972807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassera M. B., Gozzo F. C., D'Alexandri F. L., Merino E. F., del Portillo H. A., Peres V. J., et al. (2004). The methylerythritol phosphate pathway is functionally active in all intraerythrocytic stages of Plasmodium falciparum. J. Biol. Chem. 279, 749–759. 10.1074/jbc.M408360200 [DOI] [PubMed] [Google Scholar]
- Crane C. M., Kaiser J., Ramsden N. L., Lauw S., Rohdich F., Eisenreich W., et al. (2006). Fluorescent inhibitors for IspF, an enzyme in the non-mevalonate pathway for isoprenoid biosynthesis and a potential target for antimalarial therapy. Angew. Chem. 118, 1082–1087. 10.1002/ange.200503003 [DOI] [PubMed] [Google Scholar]
- Eoh H., Narayanasamy P., Brown A. C., Parish T., Brennan P. J., Crick D. C. (2009). Expression and characterization of soluble 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase from bacterial pathogens. Chem. Biol. 16, 1230–1239. 10.1016/j.chembiol.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faísca Phillips A. M., Nogueira F., Murtinheira F., Barros M. T. (2015). Synthesis and antimalarial evaluation of prodrugs of novel fosmidomycin analogues. Bioorg. Med. Chem. Lett. 25, 2112–2116. 10.1016/j.bmcl.2015.03.077 [DOI] [PubMed] [Google Scholar]
- Foth B. J., McFadden G. I. (2003). The apicoplast: a plastid in Plasmodium falciparum and other Apicomplexan parasites. Int. Rev. Cytol. 224, 57–110. 10.1016/S0074-7696(05)24003-2 [DOI] [PubMed] [Google Scholar]
- Gabrielsen M., Kaiser J., Rohdich F., Eisenreich W., Laupitz R., Bacher A., et al. (2006). The crystal structure of a plant 2C-methyl-D-erythritol 4-phosphate cytidylyltransferase exhibits a distinct quaternary structure compared to bacterial homologues and a possible role in feedback regulation for cytidine monophosphate. FEBS J. 273, 1065–1073. 10.1111/j.1742-4658.2006.05133.x [DOI] [PubMed] [Google Scholar]
- Geist J. G., Lauw S., Illarionova V., Illarionov B., Fischer M., Gräwert T., et al. (2010). Thiazolopyrimidine inhibitors of 2- methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium tuberculosis and Plasmodium falciparum. Chem. Med. Chem. 5, 1092–1101. 10.1002/cmdc.201000083 [DOI] [PubMed] [Google Scholar]
- Gräwert T., Span I., Eisenreich W., Rohdich F., Eppinger J., Bacher A., et al. (2010). Probing the reaction mechanism of IspH protein by X-ray structure analysis. Proc. Natl. Acad. Sci. U.S.A. 107, 1077–1081. 10.1073/pnas.0913045107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra F., Wang K., Li J., Wang W., Liu Y. L., Amin S., et al. (2014). Inhibition of the 4Fe–4S proteins IspG and IspH: an EPR, ENDOR and HYSCORE investigation. Chem. Sci. 5, 1642–1649. 10.1039/c3sc53301h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn F. M., Eubanks L. M., Testa C. A., Blagg B. S., Baker J. A., Poulter C. D. (2001). 1-Deoxy-d-Xylulose 5-Phosphate synthase, the gene product of open reading frame (ORF) 2816 and ORF 2895 in Rhodobacter capsulatus. J. Bacteriol. 183, 1–11. 10.1128/JB.183.1.1-11.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale I., O'Neill P. M., Berry N. G., Odom A., Sharma R. (2012). The MEP pathway and the development of inhibitors as potential anti-infective agents. Med. Chem. Commun. 3, 418–433. 10.1039/c2md00298a [DOI] [Google Scholar]
- Handa S., Ramamoorthy D., Spradling T. J., Guida W. C., Adams J. H., Bendinskas K. G., et al. (2013). Production of recombinant 1-deoxy-d-xylulose-5-phosphate synthase from Plasmodium vivax in Escherichia coli. FEBS Open Biol. 3, 124–129. 10.1016/j.fob.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haymond A., Johny C., Dowdy T., Schweibenz B., Villarroel K., Young R., et al. (2014). Kinetic characterization and allosteric inhibition of the Yersinia pestis 1-deoxy-d-xylulose 5-phosphate reductoisomerase (MEP synthase). PLoS ONE 9:e106243. 10.1371/journal.pone.0106243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson L. M., Björkelid C., Mowbray S. L., Unge T. (2006). The 1.9Å resolution structure of Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate reductoisomerase, a potential drug target. Acta Crystallogr. D. Biol. Crystallogr. 62, 807–813. 10.1107/S0907444906019196 [DOI] [PubMed] [Google Scholar]
- Hirsch A. K., Alphey M. S., Lauw S., Seet M., Barandun L., Eisenreich W., et al. (2008). Inhibitors of the kinase IspE: structure–activity relationships and co-crystal structure analysis. Org. Biomol. Chem. 6, 2719–2730. 10.1039/b804375b [DOI] [PubMed] [Google Scholar]
- Hirsch A. K. H., Lauw S., Gersbach P., Schweizer W. B., Rohdich F., Eisenreich W., et al. (2007). Nonphosphate inhibitors of IspE protein, a kinase in the non-mevalonate pathway for isoprenoid biosynthesis and a potential target for antimalarial therapy. Chem. Med. Chem. 2, 806–810. 10.1002/cmdc.200700014 [DOI] [PubMed] [Google Scholar]
- Hunter W. N. (2007). The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 282, 21573–21577. 10.1074/jbc.R700005200 [DOI] [PubMed] [Google Scholar]
- Hunter W. N. (2011). Isoprenoid precursor biosynthesis offers potential targets for drug discovery against diseases caused by apicomplexan parasites. Curr. Top. Med. Chem. 11, 2048–2059. 10.2174/156802611796575867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay L. S., Armstrong C. M., Masters M. C., Li T., Price K. E., Edwards R. L., et al. (2015). Plasmodium IspD (2-C-Methyl-D-erythritol 4-Phosphate Cytidyltransferase), an essential and druggable antimalarial target. ACS Infect. Dis. 1, 157–167. 10.1021/id500047s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson A. M., Wieckowska A., Björkelid C., Yahiaoui S., Sooriyaarachchi S., Lindh M, et al. (2013). DXR inhibition by potent mono-and disubstituted fosmidomycin analogues. J. Med. Chem. 56, 6190–6199. 10.1021/jm4006498 [DOI] [PubMed] [Google Scholar]
- Janthawornpong K., Krasutsky S., Chaignon P., Rohmer M., Poulter C. D., Seemann M. (2013). Inhibition of IspH, a [4Fe–4S] 2+ enzyme involved in the biosynthesis of isoprenoids via the methylerythritol phosphate pathway. J. Am. Chem. Soc. 135, 1816–1822. 10.1021/ja309557s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomaa H., Wiesner J., Sanderbrand S., Altincicek B., Weidemeyer C., Hintz M., et al. (1999). Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 285, 1573–1575. 10.1126/science.285.5433.1573 [DOI] [PubMed] [Google Scholar]
- Jordão F. M., Gabriel H. B., Alves J. M., Angeli C. B., Bifano T. D., Breda A., et al. (2013). Cloning and characterization of bifunctional enzyme farnesyl diphosphate/geranylgeranyl diphosphate synthase from Plasmodium falciparum. Malar. J. 12:184. 10.1186/1475-2875-12-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp L. E., Alphey M. S., Bond C. S., Ferguson M. A., Hecht S., Bacher A., et al. (2005). The identification of isoprenoids that bind in the intersubunit cavity of Escherichia coli 2C-methyl-D-erythritol-2, 4-cyclodiphosphate synthase by complementary biophysical methods. Acta Crystallogr. D Biol. Crystallogr. 61, 45–52. 10.1107/S0907444904025971 [DOI] [PubMed] [Google Scholar]
- Konzuch S., Umeda T., Held J., Hähn S., Brücher K., Lienau C, et al. (2014). Binding modes of reverse fosmidomycin analogs toward the antimalarial target IspC. J. Med. Chem. 57, 8827–8838. 10.1021/jm500850y [DOI] [PubMed] [Google Scholar]
- Kuemmerle H. P., Murakawa T., De Santis F. (1987). Pharmacokinetic evaluation of fosmidomycin, a new phosphonic acid antibiotic. Chemioterapia 6, 113–119. [PubMed] [Google Scholar]
- Kunfermann A., Lienau C., Illarionov B., Held J., Gräwert T., Behrendt C. T., et al. (2013). IspC as target for anti-infective drug discovery: synthesis, enantiomeric separation, and structural biology of fosmidomycin thia isosters. J. Med. Chem. 56, 8151–8162. 10.1021/jm4012559 [DOI] [PubMed] [Google Scholar]
- Kunfermann A., Witschel M., Illarionov B., Martin R., Rottmann M., Höffken H. W., et al. (2014). Pseudilins: Halogenated, Allosteric Inhibitors of the non-mevalonate pathway Enzyme IspD. Angew. Chem. 53 2235–2239. 10.1002/anie.201309557 [DOI] [PubMed] [Google Scholar]
- Laupitz R., Gräwert T., Rieder C., Zepeck F., Bacher A., Arigoni D., et al. (2004). Stereochemical studies on the making and unmaking of isopentenyl diphosphate in different biological systems. Chem. Biodiv. 1, 1367–1376. 10.1002/cbdv.200490099 [DOI] [PubMed] [Google Scholar]
- Lee M., Gräwert T., Quitterer F., Rohdich F., Eppinger J., Eisenreich W., et al. (2010). Biosynthesis of Isoprenoids: crystal structure of the [4Fe–4S] cluster Protein IspG. J. Mol. Biol. 404, 600–610. 10.1016/j.jmb.2010.09.050 [DOI] [PubMed] [Google Scholar]
- Lehmann C., Lim K., Toedt J., Krajewski W., Howard A., Eisenstein E., et al. (2002). Structure of 2C-methyl-D-erythrol-2, 4-cyclodiphosphate synthase from Haemophilus influenzae: activation by conformational transition. Proteins 49, 135–138. 10.1002/prot.10182 [DOI] [PubMed] [Google Scholar]
- Lell B., Ruangweerayut R., Wiesner J., Missinou M. A., Schindler A., Baranek T., et al. (2003). Fosmidomycin, a novel chemotherapeutic agent for malaria. Antimicrob. Agents Chemother. 47, 735–738. 10.1128/AAC.47.2.735-738.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo A. M., Tetzlaff C. N., Sangari F. J., Cane D. E. (2003). Functional expression and characterization of EryA, the erythritol kinase of Brucella abortus, and enzymatic synthesis of L-erythritol-4-phosphate. Bioorg. Med. Chem. Lett. 13, 737–739. 10.1016/S0960-894X(02)01032-6 [DOI] [PubMed] [Google Scholar]
- Mac Sweeney A., Lange R., Fernandes R. P., Schulz H., Dale G. E., Douangamath A., et al. (2005). The crystal structure of E. coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation. J. Mol. Biol. 345, 115–127. 10.1016/j.jmb.2004.10.030 [DOI] [PubMed] [Google Scholar]
- Mao J., Eoh H., He R., Wang Y., Wan B., Franzblau S. G., et al. (2008). Structure–activity relationships of compounds targeting Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate synthase. Bioorg. Med. Chem. Lett. 18, 5320–5323. 10.1016/j.bmcl.2008.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masini T., Hirsch A. K. H. (2014). Development of inhibitors of the 2C-Methyl-D- erythritol 4-Phosphate (MEP) pathway Enzymes as potential anti-infective agents. J. Med. Chem. 57, 9740–9763. 10.1021/jm5010978 [DOI] [PubMed] [Google Scholar]
- Matsue Y., Mizuno H., Tomita T., Asami T., Nishiyama M., Kuzuyama T. (2010). The herbicide ketoclomazone inhibits 1-deoxy-D-xylulose 5-phosphate synthase in the 2-C-methyl-D-erythritol 4-phosphate pathway and shows antibacterial activity against Haemophilus influenzae. J. Antibiot. 63, 583–588. 10.1038/ja.2010.100 [DOI] [PubMed] [Google Scholar]
- McFadden G. I., Reith M. E., Munholland J., Lang-Unnasch N. (1996). Plastid in human parasites. Nature 381, 482. 10.1038/381482a0 [DOI] [PubMed] [Google Scholar]
- Mombelli P., Witschel M. C., van Zijl A. W., Geist J. G., Rottmann M., Freymond C., et al. (2012). Identification of 1, 3-Diiminoisoindoline Carbohydrazides as potential antimalarial candidates. Chem. Med. Chem. 7, 151–158. 10.1002/cmdc.201100441 [DOI] [PubMed] [Google Scholar]
- Mullin K. A., Lim L., Ralph S. A., Spurck T. P., Handman E., McFadden G. I. (2006). Membrane transporters in the relict plastid of malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 103, 9572–9577. 10.1073/pnas.0602293103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakawa T., Sakamoto H., Fukada S., Konishi T., Nishida M. (1982). Pharmacokinetics of fosmidomycin, a new phosphonic acid antibiotic. Antimicrob. Agents Chemother. 21, 224–230. 10.1128/AAC.21.2.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S. C., Brooks C. F., Goodman C. D., Sturm A., McFadden G. I., Sundriyal S., et al. (2011). Apicoplast isoprenoid precursor synthesis and the molecular basis of fosmidomycin resistance in Toxoplasma gondii. J. Exp. Med. 208, 1547–1559. 10.1084/jem.20110039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke P. E., Kalinowska-Tłuścik J., Fyfe P. K., Dawson A., Hunter W. N. (2014). Crystal structures of IspF from Plasmodium falciparum and Burkholderia cenocepacia: comparisons inform antimicrobial drug target assessment. BMC Struct. Biol. 14:1. 10.1186/1472-6807-14-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partow S., Siewers V., Daviet L., Schalk M., Nielsen J. (2012). Reconstruction and evaluation of the synthetic bacterial MEP pathway in Saccharomyces cerevisiae. PLoS ONE 7:e52498. 10.1371/journal.pone.0052498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quitterer F., Frank A., Wang K., Rao G., O'Dowd B., Li J., et al. (2015). Atomic-resolution structures of discrete stages on the reaction coordinate of the [Fe4S4] Enzyme IspG (GcpE). J. Mol. Biol. 427, 2220–2228. 10.1016/j.jmb.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reker D., Seet M., Pillong M., Koch C. P., Schneider P., Witschel M. C., et al. (2014). Deorphaning pyrrolopyrazines as potent multi-target antimalarial agents. Angew. Chem. Int. Ed. 53, 7079–7084. 10.1002/anie.201311162 [DOI] [PubMed] [Google Scholar]
- Rekittke I., Nonaka T., Wiesner J., Demmer U., Warkentin E., Jomaa H., et al. (2011). Structure of the E-1-hydroxy-2-methyl-but-2-enyl-4-diphosphate synthase (GcpE) from Thermus thermophilus. FEBS Lett. 585, 447–451. 10.1016/j.febslet.2010.12.012 [DOI] [PubMed] [Google Scholar]
- Rekittke I., Olkhova E., Wiesner J., Demmer U., Warkentin E., Jomaa H., et al. (2013). Structure of the (E)-4-hydroxy-3-methyl-but-2-enyl-diphosphate reductase from Plasmodium falciparum. FEBS Lett. 587, 3968–3972. 10.1016/j.febslet.2013.10.029 [DOI] [PubMed] [Google Scholar]
- Rekittke I., Wiesner J., Röhrich R., Demmer U., Warkentin E., Xu W., et al. (2008). Structure of (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase, the terminal enzyme of the non-mevalonate pathway. J. Am. Chem. Soc. 130, 17206–17207. 10.1021/ja806668q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Grolle S., Bringer-Meyer S., Sahm H., Lindqvist Y. (2004). Crystal structure of 1-deoxy-D-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9Å resolution. Biochim. Biophys. Acta 1698, 37–44. 10.1016/j.bbapap.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Richard S. B., Bowman M. E., Kwiatkowski W., Kang I., Chow C., Lillo A. M., et al. (2001). Structure of 4-diphosphocytidyl-2-C-methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis. Nat. Struct. Biol. 8, 641–648. 10.1038/89691 [DOI] [PubMed] [Google Scholar]
- Richard S. B., Ferrer J. L., Bowman M. E., Lillo A. M., Tetzlaff C. N., Cane D. E., et al. (2002). Structure and mechanism of 2-C-methyl-D-erythritol 2, 4-cyclodiphosphate synthase an enzyme in the mevalonate-independent isoprenoid biosynthetic pathway. J. Biol. Chem. 277, 8667–8672. 10.1074/jbc.C100739200 [DOI] [PubMed] [Google Scholar]
- Richard S. B., Lillo A. M., Tetzlaff C. N., Bowman M. E., Noel J. P., Cane D. E. (2004). Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and mutants, reveals roles of active site amino acids. Biochemistry 43, 12189–12197. 10.1021/bi0487241 [DOI] [PubMed] [Google Scholar]
- Rohdich F., Eisenreich W., Wungsintaweekul J., Hecht S., Schuhr C. A., Bacher A. (2001). Biosynthesis of terpenoids. 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (IspF) from Plasmodium falciparum. Eur. J. Biochem. 268, 3190–3197. 10.1046/j.1432-1327.2001.02204.x [DOI] [PubMed] [Google Scholar]
- Rohdich F., Hecht S., Gärtner K., Adam P., Krieger C., Amslinger S., et al. (2002). Studies on the non-mevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc. Natl. Acad. Sci. U.S.A. 99, 1158–1163. 10.1073/pnas.032658999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdich F., Wungsintaweekul J., Fellermeier M., Sagner S., Herz S., Kis K., et al. (1999). Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc. Natl. Acad. Sci. U.S.A. 96, 11758–11763. 10.1073/pnas.96.21.11758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdich F., Wungsintaweekul J., Luttgen H., Fischer M., Eisenreich W., Schuhr C. A., et al. (2000). Biosynthesis of terpenoids: 4-Diphosphocytidyl-2-C-methyl-D-erythritol kinase from tomato. Proc. Natl. Acad. Sci. U.S.A. 97, 8251–8256. 10.1073/pnas.140209197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrich R. C., Englert N., Troschke K., Reichenberg A., Hintz M., Seeber F., et al. (2005). Reconstitution of an apicoplast-localised electron transfer pathway involved in the isoprenoid biosynthesis of Plasmodium falciparum. FEBS Lett. 579, 6433–6438. 10.1016/j.febslet.2005.10.037 [DOI] [PubMed] [Google Scholar]
- Sacchettini J. C., Poulter C. D. (1997). Creating isoprenoid diversity. Science 277, 1788–1789. 10.1126/science.277.5333.1788 [DOI] [PubMed] [Google Scholar]
- Sgraja T., Alphey M. S., Ghilagaber S., Marquez R., Robertson M. N., Hemmings J. L., et al. (2008). Characterization of Aquifex aeolicus 4-diphosphocytidyl-2C-methyl-d-erythritol kinase–ligand recognition in a template for antimicrobial drug discovery. FEBS J. 275, 2779–2794. 10.1111/j.1742-4658.2008.06418.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan S., Chen X., Liu T., Zhao H., Rao Z., Lou Z. (2011). Crystal structure of 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (IspE) from Mycobacterium tuberculosis. FASEB J. 25, 1577–1584. 10.1096/fj.10-175786 [DOI] [PubMed] [Google Scholar]
- Shi W., Feng J., Zhang M., Lai X., Xu S., Zhang X., et al. (2007). Biosynthesis of isoprenoids: characterization of a functionally active recombinant 2-C-methyl-D-erythritol 4-phosphate cytidyltransferase (IspD) from Mycobacterium tuberculosis H37Rv. BMB Rep. 40, 911–920. 10.5483/bmbrep.2007.40.6.911 [DOI] [PubMed] [Google Scholar]
- Smith J. M., Warrington N. V., Vierling R. J., Kuhn M. L., Anderson W. F., Koppisch A. T., et al. (2014). Targeting DXP synthase in human pathogens: enzyme inhibition and antimicrobial activity of butylacetylphosphonate. J. Antibiot. 67, 77–83. 10.1038/ja.2013.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Span I., Wang K., Wang W., Jauch J., Eisenreich W., Bacher A., et al. (2013). Structures of fluoro, amino, and thiol inhibitors bound to the [Fe4S4] protein IspH. Angew. Chem. 52, 2118–2121. 10.1002/anie.201208469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparr C., Purkayastha N., Kolesinska B., Gengenbacher M., Amulic B., Matuschewski K. (2013). Improved efficacy of fosmidomycin against Plasmodium and Mycobacterium species by combination with the cell-penetrating peptide octaarginine. Antimicrob. Agents Chemother. 57, 4689–4698. 10.1128/AAC.00427-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger G. A., Schörken U., Wiegert T., Grolle S., de Graff A. A., Taylor S. V., et al. (1997). Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. U.S.A. 94, 12857–12862. 10.1073/pnas.94.24.12857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S., Kaiser J., Eisenreich W., Huber R., Bacher A., Rohdich F. (2003). Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol 4-phosphate synthase (IspC). J. Biol. Chem. 278, 18401–18407. 10.1074/jbc.M300993200 [DOI] [PubMed] [Google Scholar]
- Steinbacher S., Kaiser J., Wungsintaweekul J., Hecht S., Eisenreich W., Gerhardt S., et al. (2002). Structure of 2C-methyl-D-erythritol-2, 4-cyclodiphosphate synthase involved in mevalonate-independent biosynthesis of isoprenoids. J. Mol. Biol. 316, 79–88. 10.1006/jmbi.2001.5341 [DOI] [PubMed] [Google Scholar]
- Tang M., Odejinmi S. I., Allette Y. M., Vankayalapati H., Lai K. (2011). Identification of novel small molecule inhibitors of 4-diphosphocytidyl-2-C-methyl-D-erythritol (CDP-ME) kinase of Gram-negative bacteria. Bioorg. Med. Chem. 19, 5886–5895. 10.1016/j.bmc.2011.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelemann J., Illarionov B., Barylyuk K., Geist J., Kirchmair J., Schneider P., et al. (2015). Aryl Bis-Sulfonamide Inhibitors of IspF from Arabidopsis thaliana and Plasmodium falciparum. Chem. Med. Chem. 10, 2090–2098. 10.1002/cmdc.201500382 [DOI] [PubMed] [Google Scholar]
- Umeda T., Kusakabe Y., Sakamoto Y., Odanaka Y., Matsubayashi S., Kitagawa Y., et al. (2015). Crystal structures of 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Plasmodium falciparum complexed with fosmidomycin analogs. Phot. Fact. Act. Rep. 33:BL17A/2007G025. [Google Scholar]
- Umeda T., Tanaka N., Kusakabe Y., Nakanishi M., Kitade Y., Nakamura K. T. (2011). Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum. Sci. Rep. 1:9. 10.1038/srep00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dooren G. G., Su V., D'Ombrain M. C., McFadden G. I. (2002). Processing of an apicoplast leader sequence in Plasmodium falciparum, and the identification of a putative leader cleavage enzyme. J. Biol. Chem. 277, 23612–23619. 10.1074/jbc.M201748200 [DOI] [PubMed] [Google Scholar]
- Van Hoof S., Lacey C. J., Röhrich R. C., Wiesner J., Jomaa H., Van Calenbergh S. (2008). Synthesis of analogues of (E)-1-hydroxy-2-methylbut-2-enyl 4-diphosphate, an isoprenoid precursor and human γδ T cell activator. J. Org. Chem. 73, 1365–1370. 10.1021/jo701873t [DOI] [PubMed] [Google Scholar]
- Wada T., Kuzuyama T., Satoh S., Kuramitsu S., Yokoyama S., Unzai S., et al. (2003). Crystal structure of 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis. J. Biol. Chem. 278, 30022–30027. 10.1074/jbc.M304339200 [DOI] [PubMed] [Google Scholar]
- Wang W., Li J., Wang K., Huang C., Zhang Y., Oldfield E. (2010). Organometallic mechanism of action and inhibition of the 4Fe-4S isoprenoid biosynthesis protein GcpE (IspG). Proc. Natl. Acad. Sci. U.S.A. 107, 11189–11193. 10.1073/pnas.1000264107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Li J., Wang K., Smirnova T. I., Oldfield E. (2011). Pyridine inhibitor binding to the 4Fe-4S protein A. aeolicus IspH (LytB): a HYSCORE Investigation. J. Am. Chem. Soc. 133, 6525–6528. 10.1021/ja2008455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley J. D., Merino E. F., Krai P. M., McLean K. J., Tripathi A. K., Vega-Rodríguez J., et al. (2015). Isoprenoid precursor biosynthesis is the essential metabolic role of the apicoplast during gametocytogenesis in Plasmodium falciparum. Eukaryot. Cell 14, 128–139. 10.1128/EC.00198-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witschel M. C., Höffken H. W., Seet M., Parra L., Mietzner T., Thater F., et al. (2011). Inhibitors of the herbicidal target IspD: allosteric site binding. Angew. Chem. 50, 7931–7935. 10.1002/anie.201102281 [DOI] [PubMed] [Google Scholar]
- Wu W., Herrera Z., Ebert D., Baska K., Cho S. H., DeRisi J. L., et al. (2015). A chemical rescue screen identifies a Plasmodium falciparum apicoplast inhibitor targeting MEP isoprenoid precursor biosynthesis. Antimicrob. Agents Chemother. 59, 356–364. 10.1128/AAC.03342-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S., Usunow G., Lange G., Busch M., Tong L. (2007). Crystal structure of 1-deoxy-D-xylulose 5-phosphate synthase, a crucial enzyme for isoprenoids biosynthesis. J. Biol. Chem. 282, 2676–2682. 10.1074/jbc.M610235200 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Chang W. C., Liu H. W., Liu P. (2011). Study of IspH, a key enzyme in the methylerythritol phosphate pathway using fluoro-substituted substrate analogues. Org. Lett. 13, 5912–5915. 10.1021/ol202559r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z. K., Krai P. M., Merino E. F., Simpson M. E., Slebodnick C., Cassera M. B., et al. (2015). Determination of the active stereoisomer of the MEP pathway-targeting antimalarial agent MMV008138, and initial structure–activity studies. Bioorg. Med. Chem. Lett. 25, 1515–1519. 10.1016/j.bmcl.2015.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., DeRisi J. L. (2011). Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. PLoS Biol. 9:e1001138. 10.1371/journal.pbio.1001138 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.