Abstract
Simultaneous multiplex mutation of large gene families using Cas9 has the potential to revolutionize agriculture and plant sciences. The targeting of multiple genomic sites at once raises concerns about the efficiency and specificity in targeting. The model Arabidopsis thaliana is widely used in basic plant research. Previous work has suggested that the Cas9 off-target rate in Arabidopsis is undetectable. Here we use deep sequencing on pooled plants simultaneously targeting 14 distinct genomic loci to demonstrate that multiplex targeting in Arabidopsis is highly specific to on-target sites with no detectable off-target events. In addition, chromosomal translocations are extremely rare. The high specificity of Cas9 in Arabidopsis makes this a reliable method for clean mutant generation with no need to enhance specificity or adopt alternate Cas9 variants.
Introduction
Many gene families that regulate key processes are highly redundant and spread across diverse chromosomal locations in plants. To understand gene function this necessitates the ability to simultaneously target and mutate distinct loci in a highly specific manner without affecting other genes. The CRISPR/Cas9 system has emerged as a powerful tool to create targeted mutations in plants [1]. By co-expressing multiple guide RNAs (gRNAs) with the Cas9 enzyme in plants efficient multiplex mutation of gene families can be achieved. Off-targets can be predicted computationally based on divergence from the desired on-target site [2]. Current data suggests that off-target mutagenesis rates in plants are low [3, 4] or undetectable, however this assessment is based on data from small scale targeting events. In many non-plant systems the use of Cas9 in targeted mutagenesis is off-set by considerable rates of off-target events. Several approaches targeting either Cas9 or the gRNA have been created by different groups that lead to reduced off-target rates in non-plant systems [5–7]. It is unclear if these approaches would be necessary in plant based systems. In higher order multiplex experiments the potential for off-target effects and chromosomal translocations increases significantly and has implications for the interpretation and confirmation of phenotypes. To explore these issues we performed whole genome re-sequencing of Arabidopsis plants targeting 14 genomic loci at once and assessed on-target and off-target Cas9 efficiencies and translocation events.
Results
Our lab is interested in the function of redundant peptide encoding genes that contribute to development. To facilitate the higher order multiplex mutation of plant peptide encoding genes (see methods for details), we assembled gRNA expression units into “stackable arrays” targeting up to 14 sites at once. We used our stackable array approach to target peptide families from the 11 member GOLVEN (GLV/RGF/CLEL) gene family which encode peptides that regulate several aspects of root stem cell development, but have not been studied in above ground tissues [8–10]. We targeted the six GLV genes that are expressed in aerial tissues, and CLE18, (S1 Table), which gives rise to a GLV-like peptide in aerial tissue [9, 11]. For each of the seven genes we selected two gRNA targets sites per gene using standard Cas9 targeting requirements. gRNA units, each consisting of a gRNA and Arabidopsis U6 promoter, were synthesized in clusters of four (see methods), and then stacked together to generate a vector harboring a total of fourteen gRNA units. This stacked array was cloned into the custom pCUT3 binary vector and transformed into wild-type Col-0 plants. We obtained multiple T1 plants, and first screened plants for editing of GLV1 by analyzing Sanger sequenced PCR fragments from T1 leaves to confirm Cas9 function in these lines (S2 Table). Although we typically observe high numbers of plants with editing activity using our system, we have also noted a bias in many cases against recovery of T1 plants containing editing activity when targeting genes that regulate embryogenesis or essential functions (Nimchuk, unpublished). In this case we observed evidence of GLV1 editing in only one plant from the six we surveyed, considerably lower than the near 100% efficiency we typically observe when targeting single non-essential genes. Details on the phenotypes arising from the stable septuple mutant plants will be presented elsewhere.
Leaves are composed of a large number of multiple cells types. In somatic Cas9 expressing plants each leaf cell is capable of having a distinct editing event. Thus a single leaf contains an array of editing events. Therefore, to assess genome wide editing efficiency we sampled whole leaf tissue pooled from 48 T2 plants to maximize the spectrum of detectable mutation types. We then purified DNA from the pooled leaves and subjected them to resequencing on the Illumnia HiSeq 2000 platform. Reads were mapped to the TAIR10 reference genome using BWA v 5.7 [12] and variants were called using FreeBayes v0.9.10 [13] and confirmed visually using the Integrated Genome Viewer v2.3.60 [14] (S1 Fig). Sequencing reads were generated across two lanes of Illumina. Editing events were only called if they occurred in both lanes and average coverage for on-target and off-target sites was 100X per lane (split pane in S1 Fig indicates each lane).
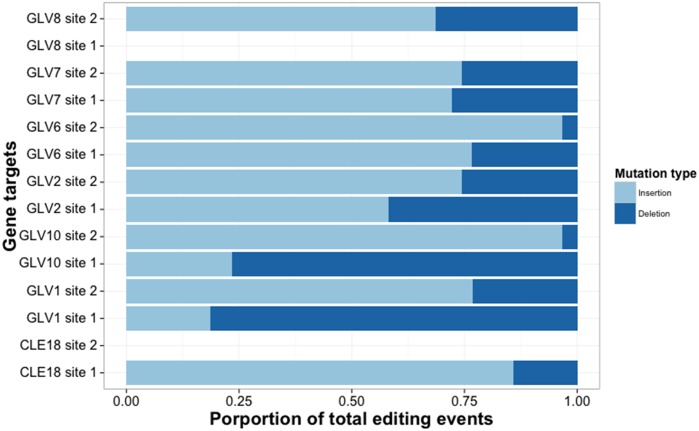
Cas9 preferentially cuts at the -3 position relative to the PAM sequence in a target site resulting in insertion/deletion events [15]. We therefore considered indel events at or near this location, rather than base substitutions, as being clear evidence of Cas9 activity rather than sequencing errors. We note that large deletion events created by dsDNA breaks at the -3 position followed by DNA deletion prior to repair could render individual reads un-mappable and may be selected against in our assembly. However these are expected to be infrequent as previous work has shown that Cas9 induced events are strongly biased to smaller indel events, with larger indels being increasingly rare [3]. As such if anything, our method will underrepresent on-target editing events. We first assessed the frequency of on-target editing efficiency in our multiplex system. Out of the 14 gRNAs transformed, two did not show any evidence of cutting (Table 1). This is comparable with frequencies of single gRNA expression in our lab (Nimchuk lab, unpublished). Of the remaining 12 gRNAs, on-target efficiencies ranged from 33% of sequence reads containing Cas9-consistent editing, to 92%. There was no clear correlation between editing efficiency and strand bias in the dataset. Both gRNAs that failed to edit targeted the minus strand, however several gRNAs that targeted the minus strand promoted editing at a rate equal to or greater than some of the gRNAs that targeted plus strands. We observed a significant bias in insertions over deletions (Fig 1, S3 and S4 Tables) which was not explained by strand, site, or distance from PAM (S4 Table), however, two genes (GLV1 and GLV10) showed the opposite effect.
Table 1. On-Target Rates.
| Site | Gene | Target Sequence | Total Eventsa | Total Reads | Percentb |
|---|---|---|---|---|---|
| CLE18_1 | AT1G66145.1 | tcgaggaaaacaatatcgac | 113 | 217 | 52.1 |
| CLE18_2 | AT1G66145.1 | aaaacaatatcgaccggtct | 0 | 0 | 0 |
| GLV1_1 | AT4G16515.1 | cttctttggacgtcttcaa | 124 | 181 | 68.5 |
| GLV1_2 | AT4G16515.1 | gagaaaggaggcgggcgtt | 121 | 183 | 66.1 |
| GLV2_1 | AT5G64770.1 | tgcgacgaggaaagacttg | 86 | 231 | 37.2 |
| GLV2_2 | AT5G64770.1 | aatagaacgctattggttg | 121 | 219 | 55.3 |
| GLV6_1 | AT2G03830.1 | gctagtgcacaaagaaaga | 219 | 251 | 87.3 |
| GLV6_2 | AT2G03830.1 | tggagttacaagcaataaaa | 90 | 267 | 33.7 |
| GLV7_1 | AT2G04025.1 | tgatgaaaaagatgatacg | 90 | 271 | 33.2 |
| GLV7_2 | AT2G04025.1 | ctgaaacgaagatgaagag | 149 | 198 | 75.3 |
| GLV8_1 | AT3G02242.1 | atcaatcccaaaaagaagaa | 0 | 0 | 0 |
| GLV8_2 | AT3G02242.1 | aagacaagcaataaagctg | 185 | 243 | 76.1 |
| GLV10_1 | AT5G51451.1 | aaggatcattgaagcaaca | 167 | 250 | 66.8 |
| GLV10_2 | AT5G51451.1 | gataatctgcaaataagag | 239 | 259 | 92.3 |
| Total | 1704 | 2770 | 61.5 |
a) Total number of editing events, including both insertions and deletions.
b) Number of editing events per number of reads analyzed.
Fig 1. Insertion bias in editing events.
Bias in insertions relative to deletions within editing events (likelihood ratio test, X2 df = 1, p <<0.001). Light blue bars indicate insertion events, dark bars indicate deletion events. No editing activity was detected for either CLE18_2 or GLV8_1.
We next computed the most probable off-target sites for each gRNA using the CRISPR-P webtool interface (http://cbi.hzau.edu.cn/crispr/ [16]). This site uses algorithms based on the Hsu et al assessment of Cas9 specificity and gRNA-target mismatch tolerances in over 700 gRNAs tested in human cell lines [17], and allows for sites encoding a non-canonical NAG PAM site as well as the prototypical NGG PAM site for Cas9. We limited our analysis to the top 20 scoring off-target candidates for each on-target site but see below for other sites). For some on-target sites there were less than 20 off-target sites that matched the required criteria. A total of 178 off-target sites were assessed which were collectively covered by nearly 43500 reads in our dataset with an average read coverage of 100X (S5 Table). Surprisingly we found no evidence of Cas9-induced indel events at any site in the dataset. To screen for indel events across the genome at sites not predicted to be potential off-target site we first filtered our Freebayes derived indel set by quality and depth (to remove highly repetitive regions). From this data set we recovered 4 additional sites (not including the intentional Cas9 mutations) that had a minor allele frequency greater than 3%. Sequencing of three of these sites in different non-transgenic parental Col-0 plants lines revealed similar indels to our Cas9 containing line in some of our Col-0 plants for two of the sites. For the third we found a slightly larger deletion among the six sequenced plants that were at the same location (S7 Table) suggesting that this location is a hotspot for deletion events in Arabidopsis. We were unable to get quality sequencing data back for the 4th site among our non-transgenic plants despite several attempts. These data indicate that the deletions events we noted in the non-predicted off-target sites likely arose by natural divergence from the reference Col-0 in our local stocks prior to the introduction of Cas9.
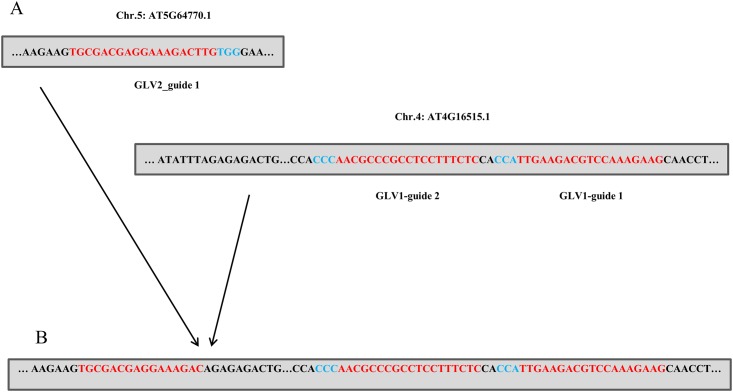
We next queried our data for evidence of translocation events using IGV v2.3.60. In multiplex gene targeting the prevalence of multiple dsDNA breaks in the same cell has the potential to generate translocation events. In mammalian systems this property has been exploited to create cell lines that mimic cancer causing chromosomal aberrations and has been noted in several studies [7, 18, 19]. To avoid sequencing errors we considered reads that originated in one on-target site and were contiguous with another on-target site and were supported by multiple reads on both sequencing lanes. Considering just the 7 genes targeted, and not the individual gRNAs, there is the potential for 42 different translocation events to arise between different targeted genes for simple translocation scenarios. Despite this, out of all the sites monitored we found only two independent events, both of which generated translocation events that occurred between GLV2 gRNA target site 1 and GLV1 gRNA sites 1 or 2 (Fig 2). These events arose in 1–2% of the total reads spanning both areas. Therefore Cas9 induced translocation events arise at a very low frequency in Arabidopsis.
Fig 2. Translocation Events Translocation between GLV2 and GLV1.
Target sites prior to (A) and after (B) CRISPR induced translocation. Red sequences indicate target sites while blue sequences indicate PAM sites.
Discussion
Considerable effort has been made to improve the on-target efficiency of Cas9 in genome editing systems in non-plant systems. Our data demonstrate that the original Cas9 system based on the wild type Streptococcus pyrogenes Cas9 is already specific enough to make off-targeting events undetectable in Arabidopsis. Our work supports previous data using smaller scale targeting and suggests that there is no inherent need to adopt different Cas9 variants in Arabidopsis genome targeting systems. While it is formally possible that there may be some sites for which off-targets may be efficiently cleaved for some gRNAs, our data suggests that this will be very rare. Consistent with this off-target rates at computationally predicted sites were undetectable and the other indels surveyed in our system likely arose in our stock line of Columbia-0 due to spontaneous mutations rather than Cas9 editing activity [20]. In theory, if the average off-target cleavage rate of Cas9 in Arabidopsis were to be a mere 0.01% among our computationally predicted off-target sites we would still expect to find 4–5 such events in our total off-target dataset, and at 0.1% we would expect 43 events. The lack of detection of these events strongly suggests that Cas9 off-target effects are negligible in Arabidopsis. This contrasts with transcription activator-like effector nuclease (TALEN) and designer transcription activator-like effector (dTALE) systems which have been shown to result in off-target editing in plants at low rates.[21–23] To our knowledge no off-target editing has been reported using zinc finger nucleases (ZFN) [24]. Despite this, the the CRISPR/Cas9 system still offers advantages in ease of design compared to ZFNs. In addition, we find that Cas9-induced translocation events are extremely rare in Arabidopsis. This observation suggests that higher order multiplex genome editing will not likely be hindered by chromosomal aberrations. Clark and Krysan showed in a small scale survey of T-DNA mutants that chromosomal translocation events could be detected in 11/64 T-DNA insertion lines [25]. Our data suggest that translocation events induced by Cas9 are lower than the frequency of translocations that arise during T-DNA transformation in Arabidopsis [25]. In addition combining multiple T-DNA insertion events with common sequence similarity in the T-DNAs can stimulate recombination upon crossing [26]. As such higher order mutant creation with Cas9 might be a cleaner genetic system than T-DNA based crossing. It is likely that some translocation events are deleterious and are selected against and might not be identified in our analysis. However some translocation events are tolerated in Arabidopsis and we view it likely that dsDNA breaks induced by Cas9 are efficiently coupled to repair to select against translocation events. On-target mutant screening strategies based on PCR amplification using primer sets flanking gRNA on-target sites will further counter-select against rare chromosomal events.
While our findings indicate that Cas9 is highly specific in Arabidopsis it is not clear how these results would relate to other plant species. Detailed studies on the specificity of Cas9 in other plant species will require further research. It is not clear why Cas9 is highly specific in some systems and not others. Our study of the GLV gene family in Arabidopsis is likely to be representative of Arabidopsis, and not an effect of the GLV family specifically. Our gRNAs are diverse in sequence and are computationally predicted to have potential off-target sites throughout the genomes. As such, factors such as off-target chromosome location, methylation status, and nucleotide mismatch number and position would be expected to be variable and represented our dataset. Despite this we find no off target events. We have noticed that Cas9 accumulates to very low levels in both Arabidopsis and tobacco with our system despite being codon optimized for plants and being expressed from the UBIQUITIN10 promoter (ZN unpublished). High levels of Cas9 are correlated with the formation of energetically less favorable off-target events in others systems [27]. As such, our favored hypothesis is that low Cas9 levels in Arabidopsis limit off-target events to highly specific target pairing events. This hypothesis will require further experimentation to fully test. Our data presented here demonstrate that Cas9 is an extremely precise enzyme for genome editing in Arabidopsis and that off-targeting and chromosomal aberrations are not likely to be complications in multiplexing experiments.
Materials and Methods
CRISPR/Cas9 vector system construction and design
A dual maize and Arabidopsis optimized Cas9 gene was gene synthesized with a Hemagglutinin (HA) tag linker at the C-terminus fused to the N7 localization tag, which directs nuclear localization of expressed fusion protein [28], by GeneArts. This was subcloned into a modified pENTR3C vector (Invitrogen) containing an upstream omega translational enhancer sequence and then recombined into a pMOA vector series behind the Arabidopsis UBQ10 promoter sequence [29] to create the pCUT binary series. To create the pCUT-G gateway compatible series, Gateway cassettes (Invitrogen cat# 11828–029, rfB) were released from pBluescript with EcoRV and ligated into pCUT vectors linearized with PmeI. The resulting plasmids’ full-length sequences were confirmed via HTS.
For multiplex stackable array construction, a digital template of 4 gRNA units was created in LaserGene and saved as a.gb file with 19Ns replacing the putative target sequence in each gRNA unit. For each gene of interest to be targeted, Cas9 target sites were selected using standard bioinformatics criteria [30]. Our template contains a transcriptional start site G in each gRNA unit between the U6 promoter sequence and the gRNA. We have found that this renders the requirement for an upstream G in the standard G-19/20N-NGG consensus site not strictly necessary and that N-19/20N-NGG sites will also be targeted efficiently (S2 Fig). To be conservative, where possible G-19/20N-NGG sites were selected. Target sites were then pasted into each gRNA unit to create a digital array that was then synthesized (GeneArts; Regensburg, Germany). Stacked arrays were cloned together by restriction enzyme digestion of one array (donor) into a pMA gRNA array linearized with EcoRV (recipient), thus destroying the PmeI sites in the donor but preserving the flanking PmeI sites in the recipient (S3 Fig). Stacked arrays were then cloned into pCUT3 using the Mighty Mix ligation system (Takara).
Transformation of Arabidopsis
Plant expression vectors were transformed into an Agrobacterium tumefacians recA- strain via triparental mating (strain C58, gift from Stephen Farrand). Wild type Arabidopsis thaliana Col-0 plants were grown in a growth chamber with constant light at 23°C. Plants were transformed using the floral dip method [31]. T1 seed derived from plants transformed with binary pCUT3 were selected on B5 media lacking sucrose and containing 100mg/L kanamycin (S6 Table).
Sanger sequencing analysis of CRISPR events
Plant and bacterial DNA were extracted as previously published [29]. Forward and reverse primers were designed to anneal to genomic DNA sequences at approximately 250 bp flanking the putative clipping sites (S2 Table). PCR was performed using PfuUltra II Hotstart 2X Master Mix (Agilent Technologies, Inc). After amplification the PCR products were purified using the Zymo Clean and Concentrator Kit (Zymoresearch, USA). For direct sequencing of PCR products, the same primer set for amplifications were used. For sequencing of cloned PCR products, PCR fragments were cloned directly into pCR2.1 using the manufacturer’s recommendations for the Zero Blunt Topo cloning kit (Life Technologies), and sequenced using M13F and M13R primers. In both cases DNA traces were compared to wild type sequences using the SeqMan function from LaserGene analysis package. For directly sequenced PCR products, deviations from a consensus trace that originated near the -3 position relative to the NGG PAM site, the known site of Cas9 cutting, were scored as evidence of genome editing in somatic tissues relative to wild type traces.
Illumina DNA preparation, sequencing and bioinformatics
A total of 150 ng of each plasmid was used to prepare libraries following the TruSeq DNA prep protocol (Illumina). In brief, DNA was sheared to a mean size of 400 bp by ultrasonication (Covaris), end-repaired, adenylated and ligated to barcoded adapters. Adapter-ligated molecules were then submitted to 14 cycles of PCR using the KAPA HiFi DNA polymerase (KAPA Biosystems). The amplified libraries were pooled and sequenced on an Illumina HiSeq2500 instrument to generate 48 bp paired-end reads. Approximately 230,000 paired-reads were obtained for each library. The Velvet software package [32] was used to assemble the plasmids into single contigs using k-mers that ranged from 44 to 46 nt. Sequencing data has been deposited in the SRA database under project SRP073942.
Statistical analysis
We used the glmer function in the lme4 package developed for the R computing platform to generate logistic regression models to test for bias in event types. All fitted models included total editing events as the response variable and gene as a random effect. Models with differing fixed effects (insertions, deletions, site, and distance from PAM) were used for multi-model inference (MMI). The best fit models were chosen based on the change in AICc (Akikiae information criterion corrected for small sample size) and likelihood ratio test (S4 Table).
Supporting Information
Sequencing lanes were kept separate and are displayed in different panels (upper panel is lane 1, lower panel is lane 2). Blue symbols represent insertions and dashes represent deletions in the mapped reads. In this view, the editing events can be seen for both site 1 and site 2. See https://www.broadinstitute.org/igv/ for a complete description and guide for the use of the IGV.
(TIF)
A 4 gRNA unit array. Each unit contains an Arabidopsis U6 promoter (blue), target site pasting location (poly n streach), and gRNA backbone with poly T terminator (yellow). Each unit is separated by a unique blunt restriction sites (underlined) in the following order: SmaI, SnaBI, HpaI, NaeI, EcoRV (S3 Fig). The entire array is flanked by PmeI sites on either site (Red). Higher order arrays can be synthesized but take considerably longer than a 4 unit array. Note: the poly n stretch is proceeded in each case by a transcriptional G. Each poly N stretch is replaced with the desired genomic target site, both 19 and 20 nt target site will work.
(TIF)
A) The pCUT binary vector system. A plant codon optimized HA-tagged nuclear localized (N7) Cas9 is expressed from the Arabidopsis UBQ10 promoter. A transformer boosting sequence (TBS [1]) separates the Cas9 expression cassette from a unique PmeI site (red). OCS; octopine synthase terminator. B) Strategy for array stacking. Array gRNA units (A-L), are synthesized or cloned in groups of four containing flanking PmeI sites (red) and unique blunt cutting sites (blue). Arrays are stacked as PmeI fragments cloned into unique Sma1 or EcoRV blunt sites iteratively to preserve the flanking PmeI sites. Example shows stacking of three 4 unit arrays to create a 12 unit stacked array. Reference 1: Hily JM, Singer SD, Yang Y, Liu Z. A transformation booster sequence (TBS) from Petunia hybrida functions as an enhancer-blocking insulator in Arabidopsis thaliana. Plant cell reports. 2009;28(7):1095–104. doi: 10.1007/s00299-009-0700-8. PubMed PMID: 19373469.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
This work was supported by: start-up funds from Virginia Tech (DCH and ZLN) and UNC (ZLN), the Howard Hughes Medical Institute, supported by the HHMI and the Gordon and Betty Moore Foundation (GBMF3030, JLD), and The Pew Latin American Fellows Program in the Biomedical Sciences (00026198-PJPLT). Additional support was provided by the NSF-IOS (IOS-1455607) to ZLN.
Data Availability
All sequence read archive (SRA) files are available from the NCBI Sequence Read Archive database (accession number SRP073942). http://www.ncbi.nlm.nih.gov/sra.
Funding Statement
Funding was provided by start up funds from University of North Carolina at Chapel Hill to ZLN; start up funds from Virginia Tech to ZLN and DCH; the Pew Latin American Fellows Program in the Biomedical Sciences (00026198 to PJPLT), http://www.pewtrusts.org/en/projects/pew-latin-american-fellows; National Science Foundation-Division of Integrated Organismal Systems (IOS-1455607 to ZLN), http://www.nsf.gov/div/index.jsp?div=IOS; and Howard Hughes Medical Institute, supported by the HHMI and the Gordon and Betty Moore Foundation (GBMF3030 to JLD), https://www.hhmi.org/programs/biomedical-research/investigator-program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lozano-Juste J, Cutler SR. Plant genome engineering in full bloom. Trends Plant Sci. 2014;19(5):284–7. 10.1016/j.tplants.2014.02.014 . [DOI] [PubMed] [Google Scholar]
- 2.O'Geen H, Yu AS, Segal DJ. How specific is CRISPR/Cas9 really? Curr Opin Chem Biol. 2015;29:72–8. 10.1016/j.cbpa.2015.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(12):4632–7. 10.1073/pnas.1400822111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nature biotechnology. 2013;31(8):691–3. 10.1038/nbt.2655 . [DOI] [PubMed] [Google Scholar]
- 5.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–9. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–5. 10.1038/nature16526 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature biotechnology. 2014;32(3):279–84. 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez A, Hilson P, Beeckman T. GOLVEN peptides as important regulatory signalling molecules of plant development. Journal of experimental botany. 2013;64(17):5263–8. 10.1093/jxb/ert248 . [DOI] [PubMed] [Google Scholar]
- 9.Meng L, Buchanan BB, Feldman LJ, Luan S. CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(5):1760–5. 10.1073/pnas.1119864109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science. 2010;329(5995):1065–7. 10.1126/science.1191132 . [DOI] [PubMed] [Google Scholar]
- 11.Jun J, Fiume E, Roeder AH, Meng L, Sharma VK, Osmont KS, et al. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant physiology. 2010;154(4):1721–36. 10.1104/pp.110.163683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012;preprint arXiv:1207.3907 [q-bio.GN]
- 14.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–92. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21. 10.1126/science.1225829 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Molecular plant. 2014;7(9):1494–6. 10.1093/mp/ssu044 . [DOI] [PubMed] [Google Scholar]
- 17.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31(9):827–32. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nature communications. 2014;5:3728 10.1038/ncomms4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nature communications. 2014;5:3964 10.1038/ncomms4964 . [DOI] [PubMed] [Google Scholar]
- 20.Ossowski S, Schneeberger K, Lucas-Lledo JI, Warthmann N, Clark RM, Shaw RG, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327(5961):92–4. 10.1126/science.1180677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du H, Zeng X, Zhao M, Cui X, Wang Q, Yang H, et al. Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol. 2016;217:90–7. 10.1016/j.jbiotec.2015.11.005 . [DOI] [PubMed] [Google Scholar]
- 22.Mahfouz MM, Piatek A, Stewart CN Jr., Genome engineering via TALENs and CRISPR/Cas9 systems: challenges and perspectives. Plant Biotechnol J. 2014;12(8):1006–14. 10.1111/pbi.12256 . [DOI] [PubMed] [Google Scholar]
- 23.Morbitzer R, Romer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(50):21617–22. 10.1073/pnas.1013133107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(26):12028–33. 10.1073/pnas.0914991107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark KA, Krysan PJ. Chromosomal translocations are a common phenomenon in Arabidopsis thaliana T-DNA insertion lines. The Plant journal: for cell and molecular biology. 2010;64(6):990–1001. 10.1111/j.1365-313X.2010.04386.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtis MJ, Belcram K, Bollmann SR, Tominey CM, Hoffman PD, Mercier R, et al. Reciprocal chromosome translocation associated with TDNA-insertion mutation in Arabidopsis: genetic and cytological analyses of consequences for gametophyte development and for construction of doubly mutant lines. Planta. 2009;229(4):731–45. 10.1007/s00425-008-0868-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids. 2015;4:e264 10.1038/mtna.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(7):3718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr Biol. 2011;21(5):345–52. 10.1016/j.cub.2011.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant journal: for cell and molecular biology. 1998;16(6):735–43. . [DOI] [PubMed] [Google Scholar]
- 32.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome research. 2008;18(5):821–9. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequencing lanes were kept separate and are displayed in different panels (upper panel is lane 1, lower panel is lane 2). Blue symbols represent insertions and dashes represent deletions in the mapped reads. In this view, the editing events can be seen for both site 1 and site 2. See https://www.broadinstitute.org/igv/ for a complete description and guide for the use of the IGV.
(TIF)
A 4 gRNA unit array. Each unit contains an Arabidopsis U6 promoter (blue), target site pasting location (poly n streach), and gRNA backbone with poly T terminator (yellow). Each unit is separated by a unique blunt restriction sites (underlined) in the following order: SmaI, SnaBI, HpaI, NaeI, EcoRV (S3 Fig). The entire array is flanked by PmeI sites on either site (Red). Higher order arrays can be synthesized but take considerably longer than a 4 unit array. Note: the poly n stretch is proceeded in each case by a transcriptional G. Each poly N stretch is replaced with the desired genomic target site, both 19 and 20 nt target site will work.
(TIF)
A) The pCUT binary vector system. A plant codon optimized HA-tagged nuclear localized (N7) Cas9 is expressed from the Arabidopsis UBQ10 promoter. A transformer boosting sequence (TBS [1]) separates the Cas9 expression cassette from a unique PmeI site (red). OCS; octopine synthase terminator. B) Strategy for array stacking. Array gRNA units (A-L), are synthesized or cloned in groups of four containing flanking PmeI sites (red) and unique blunt cutting sites (blue). Arrays are stacked as PmeI fragments cloned into unique Sma1 or EcoRV blunt sites iteratively to preserve the flanking PmeI sites. Example shows stacking of three 4 unit arrays to create a 12 unit stacked array. Reference 1: Hily JM, Singer SD, Yang Y, Liu Z. A transformation booster sequence (TBS) from Petunia hybrida functions as an enhancer-blocking insulator in Arabidopsis thaliana. Plant cell reports. 2009;28(7):1095–104. doi: 10.1007/s00299-009-0700-8. PubMed PMID: 19373469.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All sequence read archive (SRA) files are available from the NCBI Sequence Read Archive database (accession number SRP073942). http://www.ncbi.nlm.nih.gov/sra.