ABSTRACT
Expression of the human cytomegalovirus (HCMV) IE1 and IE2 proteins is critical for the establishment of lytic infection and reactivation from viral latency. Defining the mechanisms controlling IE1 and IE2 expression is therefore important for understanding how HCMV regulates its replicative cycle. Here we identify several novel transcripts encoding full-length IE1 and IE2 proteins during HCMV lytic replication. Two of the alternative major immediate early (MIE) transcripts initiate in the first intron, intron A, of the previously defined MIE transcript, while others extend the 5′ untranslated region. Each of the MIE transcripts associates with polyribosomes in infected cells and therefore contributes to IE1 and IE2 protein levels. Surprisingly, deletion of the core promoter region of the major immediate early promoter (MIEP) from a plasmid containing the MIE genomic locus did not completely abrogate IE1 and IE2 expression. Instead, deletion of the MIEP core promoter resulted in increased expression of alternative MIE transcripts, suggesting that the MIEP suppresses the activity of the alternative MIE promoters. While the canonical MIE mRNA was the most abundant transcript at immediate early times, the novel MIE transcripts accumulated to levels equivalent to that of the known MIE transcript later in infection. Using two HCMV recombinants, we found that sequences in intron A of the previously defined MIE transcript are required for efficient IE1 and IE2 expression and viral replication. Together, our results identify new regulatory sequences controlling IE1 and IE2 expression and suggest that multiple transcription units act in concert to regulate IE1 and IE2 expression during lytic infection.
IMPORTANCE The HCMV IE1 and IE2 proteins are critical regulators of HCMV replication, both during primary infection and reactivation from viral latency. This study expands our understanding of the sequences controlling IE1 and IE2 expression by defining novel transcriptional units controlling the expression of full-length IE1 and IE2 proteins. Our results suggest that alternative promoters may allow for IE1 and IE2 expression when MIEP activity is limiting, as occurs in latently infected cells.
INTRODUCTION
The human cytomegalovirus (HCMV) IE1 and IE2 proteins are critical regulators of the viral replicative cycle. Both proteins are immediately expressed upon infection and together stimulate the expression of host and viral genes necessary for virus replication (1). IE2 acts as a general transcription factor that broadly transactivates host genes and viral early and late genes to facilitate virus replication (2–12). IE1 promotes transcription from the HCMV genome by inhibiting histone deactylases (HDACs) (13–15), which otherwise limit virus transcription by forming inhibitory chromatin structures on the viral genome. Reexpression of IE1 and IE2 is also thought to be critical for the reactivation of quiescent HCMV genomes from latent infection. Thus, understanding the regulatory mechanisms controlling IE1 and IE2 expression is important to understand how HCMV regulates its replicative cycle.
The mRNAs encoding the 72-kDa IE1 and 86-kDa IE2 proteins are derived from a shared precursor RNA through alternative splicing (16). The mature IE1 and IE2 transcripts share the same first three exons (exons 1 through 3) but differ in their final exon. Transcripts that include exon 4 encode the 72-kDa IE1 protein, while transcripts that skip exon 4 and retain exon 5 encode the 86-kDa IE2 protein (17). Several IE1 and IE2 splice variants have been identified. For example, IE1 transcript variants arise from alternative splice donor and acceptor usage in exons 3 and 4, respectively (16–19). Similarly, IE2 transcript variants arise from alternative splicing within exon 5 (17). The proteins encoded by these alternatively spliced transcripts differ in function from full-length IE1 and IE2 proteins, highlighting the role of alternative splicing in expanding the functions of proteins encoded by the major immediate early (MIE) locus.
In addition to alternative splicing, multiple promoters direct the transcription of several distinct RNAs from the MIE locus. The most studied promoter in this region is the major immediate early promoter (MIEP), whose activity is regulated by multiple cellular and viral factors such as CREB/ATF, NF-κB, SP1, pp71, pp65, and pTRS1 (20–32). In addition, the IE1 and IE2 proteins themselves regulate MIEP activity. While IE1 stimulates MIEP activity, IE2 binds a wild-type cis repression sequence (crs) located adjacent to the MIEP transcription start site, inhibiting transcription initiation (33–37). Alternative promoters in the MIE locus also control expression of IE1 and IE2 transcript variants in specific contexts. A promoter proximal to the IE2-specific exon 5 drives the expression of several truncated IE2 isoforms during the late stage of infection (38–40). An additional promoter located 5′ of the MIEP is active in latently infected human monocytes (41), suggesting that MIE transcription may be driven by different promoters during lytic and latent infections.
In this study, we identified several previously unrecognized MIE transcripts encoding full-length IE1 and IE2 proteins that are expressed during HCMV lytic infection. The novel IE1 and IE2 transcripts differed from the known MIE transcript in their transcription start sites (TSSs), and thus their 5′ untranslated regions (5′UTRs). Sequences surrounding each transcription start site possessed promoter activity that increased during HCMV infection. Each transcript was associated with polyribosomes (polysomes) in infected cells, suggesting that each contributes to IE1 and IE2 protein expression during infection. While the canonical MIE transcript is the most abundant transcript encoding IE1 or IE2 during the immediate early stage of infection, the novel MIE transcripts accumulated as infection progressed. Two of the newly identified transcripts initiated in the first intron (intron A) of the known MIE transcript. Recombinant viruses lacking intron A or the region surrounding one of the “intronic” promoters expressed lower levels of IE1 and IE2 mRNA and protein and replicated less efficiently than the parental virus did. Together, our data show that multiple transcripts encode full-length IE1 and IE2 proteins during HCMV infection and suggest that multiple transcription units act in concert to regulate IE1 and IE2 expression during lytic replication.
MATERIALS AND METHODS
Cells and viruses.
Primary human foreskin fibroblasts (HFFs), MRC-5 fibroblasts, and HeLa cells were grown in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and penicillin and streptomycin. Unless otherwise indicated, bacterial artificial chromosome (BAC)-derived HCMV AD169 strain containing a green fluorescent protein (GFP) reporter driven by the simian virus 40 (SV40) promoter was used for all infections (42). The titers of cell-free virus were determined by the 50% tissue culture infective dose (TCID50) method on MRC-5 fibroblasts.
Construction of recombinant viruses.
Recombinant viruses were constructed in the AD169 genomic background by BAC-mediated recombineering as previously described (43–46). Briefly, the HCMVΔUTR378 BAC was constructed by amplifying a kanamycin cassette flanked by FLP recombination target (FRT) sites with primers HCMVΔUTR378-FRTKanF (Kan stands for kanamycin, and F stands for forward) (5′-TGGTGACGATACTTTCCATTACTAATCCATAACATGGCTCTTTGCCACAAACCACGTCGTGGAATGCC-3′) and HCMVΔUTR378-FRTKanR (R stands for reverse) (5′-ATCGTGCTGTGCCTAAGTCTGGCCTCCACTGTTAGGAGCAAGGAGCTGCCTCCCATGTGCAGGTGCTG-3′), which contained 50-nucleotide homology arms flanking the desired insertion site in the HCMV genome. The PCR product was electroporated into the recombination-competent E. coli EL250 strain, and recombinants were selected by growth on LB containing kanamycin and chloramphenicol. The kanamycin cassette was removed by induction of Flp recombinase with arabinose, leaving a 34-bp FRT sequence in place of the 265 bp surrounding the UTR378 transcription start site (nucleotides 174085 to 174349 deleted, according to the numbering in reference genome FJ527563.1). The deletion of intron A was accomplished using a two-step recombination approach. In the first step, the KanSacB gene was amplified with primers HCMVΔIntronA – KanSacBF (5′-GCGGCCGGGAACGGTGCATTGGAACGCGGATTCCCCGTGCCAAGAGTGACAATTCGAGCTCGGTACCCGG-3′) and HCMVΔIntronA – KanSacBR (5′-AGGGTCCATCTTTCTCTTGGCAGAGGACTCCATCGTGTCAAGGACGGTGACATCCCGGGAAAAGTGCCACC-3′), which contained 50-nucleotide homology arms flanking intron A. Recombination with the AD169 genome resulted in the replacement of intron A with the KanSacB cassette. Recombinants were selected by growth on LB plates containing kanamycin and chloramphenicol. For the second step, the primers Exon1/2Fusion F and R (F/R) (5′-CGGCCGGGAACGGTGCATTGGAACGCGGATTCCCCGTGCCAAGAGTGACTCACCGTCCTTGACACGATGGAGTCCTCTGCCAAGAGAAAGATGGACCCT-3′ and 5′-AGGGTCCATCTTTCTCTTGGCAGAGGACTCCATCGTGTCAAGGACGGTGAGTCACTCTTGGCACGGGGAATCCGCGTTCCAATGCACCGTTCCCGGCCG-3′, respectively) were used to amplify a region spanning the exon 1/exon 2 splice junction using IE2 cDNA as a template. The KanSacB cassette was then removed by a second round of recombineering using the above PCR product. The recombinants were grown on LB agar with chloramphenicol and 6% sucrose to select against colonies that retained the SacB gene. Sucrose-resistant colonies were then screened for kanamycin sensitivity to ensure loss of the kanamycin cassette. The resulting recombinant, HCMVΔIntronA contained a seamless fusion of exons 1 and 2. (nucleotides 173738 to 174564 deleted, according to numbering in reference genome FJ527563.1). The absence of genomic rearrangements was confirmed by restriction digestion of the recombinant BAC DNAs at each step. No errors were detected when the 500 bp on either side of each mutation were sequenced. To further confirm the absence of additional errors, the entire genome of the parental strain and both recombinants was sequenced. Next-generation sequencing libraries were prepared from BAC DNA using the Nextera library preparation kit (Illumina), and the libraries were sequenced on an Illumina MiSeq instrument. No changes other than the intended mutations were detected in the recombinant genomes.
Quantitative real-time PCR analysis.
Total RNA was extracted using TRIzol according to the manufacturer's directions as described previously (47, 48). Briefly, cell pellets were resuspended in TRIzol and extracted with chloroform, and RNA was precipitated with isopropanol. The RNA was treated with Turbo DNase (Applied Biosystems) and quantified on a NanoDrop spectrophotometer. For quantitative reverse transcriptase PCR (qRT-PCR), cDNA was generated from 0.5 μg total RNA using the High Capacity cDNA reverse transcription kit (ThermoFisher) and random hexamer primers. The abundance of each transcript was determined using a real-time PCR machine (Bio-Rad) and SYBR Select master mix (ThermoFisher). The abundance of each product was determined by comparison to a standard curve generated from qPCR analysis of 10-fold serial dilutions of a DNA standard specific for each primer pair. For comparative analysis of MIE transcript abundance, each of the UTR-specific primer pairs were confirmed to (i) amplify only the specific UTR sequence, and not amplify the other MIE UTRs or HCMV BAC DNA (data not shown), (ii) have similar real-time PCR efficiencies (>95% efficiency for each primer pair), and (iii) have a similar linear range of detection of the appropriate DNA standard (linear between 108 and 102 copies; R2 >0.97 for all experiments included in this work). Where indicated, phosphonoacetic acid (PAA) (Sigma) was used at a final concentration of 200 μg/ml. The primers used for qRT-PCR were IE1 qPCR F/R (5′-CAAGTGACCGAGGATTGCAA-3′ and 5′-CACCATGTCCACTCGAACCTT-3′, respectively), IE2 qPCR F/R (5′-TGACCGAGGATTGCAACGA-3′ and 5′-CGGCATGATTGACAGCCTG-3′, respectively), GAPDH qPCR F/R (5′-CTGTTGCTGTAGCCAAATTCGT-3′ and 5′-ACCCACTCCTCCACCTTTGAC-3′, respectively), UTR70 qPCR (5′-TAGCTGACAGACTAACAGAC-3′), exon2-3 qPCR (5′-GGTCACGGGTGTCTCGGGCCGT-3′), UTR136 qPCR (5′-TTGACCTCCATAGAAGACAC-3′), UTR378 qPCR (5′-CGCATTTGGAAGACTTAAGG-3′), UTR136/378 qPCR Reverse (5′-CCTTGACACGATGGAGTCCT-3′), and UTR487 qPCR F/R (5′-GCATTATGCCCAGTACATGACC-3′ and 5′-GAAATCCCCGTGAGTCAAACC-3′, respectively).
For gene-specific reverse transcription reactions, primers specific for IE1 (exon 4) or IE2 (exon 5) were used in the cDNA synthesis step: exon 4 RT (5′-TCCTTTTTAGCACGGGCCTT-3′) and exon 5 RT (5′-CGCATCCACCTCACTCTTCA-3′). The cDNA was then amplified by PCR using the following reaction conditions: 94°C for 5 min, 30 cycles (with 1 cycle consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 5 min), followed by a 7-min extension at 72°C. The following PCR primers were used: exon 4 Out (5′-TGAATTTCTCTTCCGTCTGG-3′), exon 4 In (5′-AACCTTAATCTGTTTGACGA-3′), exon 5 Out (5′-CTGCAAGAGTGGGTTGTCAG-3′), exon 5 In (5′-CTGGGCGAGGATGTCACCGA-3′), UTR70 Out (5′-ACAGACTAACAGACTGTTCC-3′), UTR70 In (5′-TCCATGGGTCTTTTCTGCAG-3′), UTR136 Out (5′-TGACCTCCATAGAAGACACC-3′), UTR136 In (5′-TTCCCCGTGCCAAGAGTGAC-3′), UTR378 Out (5′-CGCTGACGCATTTGGAAGAC-3′), UTR378 In (5′-TGTTCTGATAAGAGTCAGAG-3′), UTR487 Out (5′-AAGTACGCCCCCTATTGACG-3′), and UTR487 In (5′-CCCCTATTGACGTCAATGAC-3′). PCR products were analyzed on agarose gels and visualized with ethidium bromide on an imager (Bio-Rad). PCR products were also sequenced to confirm the specificity of the PCR.
HCMV DNA was quantified essentially as described previously (49). Briefly, infected-cell pellets were lysed in DNA extraction buffer (400 mM NaCl, 10 mM Tris-HCl [pH 8.0], 10 mM EDTA) and digested with proteinase K (10 mg/ml) overnight at 37°C. The lysates were extracted with phenol-chloroform, digested with RNase A (10 mg/ml) for 1 h at 37°C, and again extracted with phenol-chloroform. DNA was precipitated with 100% ethanol and resuspended in 10 mM Tris-HCl (pH 8.0). The number of HCMV genomes was determined by comparing the threshold values to a series of HCMV BAC DNA standards containing from 108 to 101 HCMV genomes. Results were normalized to the total amount of DNA in the sample using qPCR primers specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primers used were UL99 qPCR F/R (5′-GTGTCCCATTCCCGACTCG-3′ and 5′-TTCACAACGTCCACCCACC-3′, respectively) and GAPDH qPCR F/R (listed above).
Plasmid construction.
To generate the promoter reporter constructs, the 500 nucleotides from −450 to +50 bp flanking each transcription start site were amplified by PCR and cloned into the HindIII and NotI sites of the pGL3-Basic vector (Promega) using Gibson cloning (NEB). The primers used were pGL3 Basic UTR70 F/R (5′-TCTTACGCGTGCTAGCCCGGCTATCGCCGATAGAGGCGACATCAAG-3′ and 5′-TACCAACAGTACCGGAATGCCAAGGCGGGCCATTTACCGTCATTG-3′, respectively), pGL3 Basic UTR136 F/R (5′-TCTACGCGTGCTAGCCCGGGCCGTATGTTCCCATAGTAACGCC-3′ and 5′-TACCAACAGTACCGGAATGCCAGTGTCTTCTATGGAGGTCAAAACAG-3′, respectively), pGL3 Basic UTR378 F/R (5′-TCTTACGCGTGCTAGCCCGGGCATCGCCTGGAGACGCCATCCAC-3′ and 5′-TACCAACAGTACGGAATGCCAATTCGCGTGGAGATCCCACGTTATG-3′, respectively), and pGL3 Basic UTR487 F/R (5′-TCTTACGCGTGCTAGCCCGGGCACCACCGTCCCCAGTGCCCGCAG-3′ and 5′-TACCAACAGTACCGGAATGCCACAGAAAAGACCCATGGAAAGGAAC-3′, respectively). The inserted region was sequenced to confirm the absence of mutations. To generate the plasmid pSVHΔMIEP, the nucleotides −94 to +64 relative to the canonical transcription start site in the MIEP were deleted from the pSVH vector (50) (generously provided by Q. Tang) using PCR-mediated mutagenesis and Gibson cloning. Primers pSVHΔMIEP F/R (5′-GGCACCAAAATCAACGGGACTTTCCATAGAAGACACCGGGACCGATCA-3′ and 5′-GGAAAGTCCCGTTGATTTTGGTGCC-3′, respectively) were designed to (i) flank the region to be deleted and (ii) contain 20 nucleotides of sequence complementary to the other primer. The plasmid was amplified by PCR, and the PCR product was circularized in a subsequent Gibson reaction. The clones were sequenced to confirm the absence of spurious mutations.
Luciferase assays.
Luciferase assays were performed as described previously (51). Briefly, 200,000 HeLa cells were transfected with the indicated plasmids using polyethylenimine (PEI) as the transfection reagent. Forty-eight hours after transfection, the cells were lysed in 150 μl of passive lysis buffer (Promega). Twenty microliters of lysate was mixed with 100 μl of luciferase reagent (Promega), and luciferase was measured on a luminometer (Molecular Devices). The luciferase results were normalized to the amount of protein in each sample as determined by the Bradford protein assay kit (VWR). Luciferase reactions were performed in duplicate, and the graphs show the mean values of at least three biological replicates performed on different days. In experiments using primary fibroblasts, 4 × 106 cells were electroporated with the reporter constructs and replated into six-well dishes 2 days prior to infection with HCMV. At the indicated time after infection, cells were harvested and analyzed as described above.
Western blotting.
Western blotting was performed essentially as described previously (52). Briefly, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate) with cOmplete protease inhibitors (Roche) on ice for 15 min, and insoluble material was removed by centrifugation at 21,000 × g for 10 min. Protein concentration was measured by the Bradford assay, and equal amounts of protein (20 μg) were loaded in each lane. Sample buffer (45 mM Tris-HCl [pH 6.8], 30% glycerol, 2% sodium dodecyl sulfate [SDS], 150 mM dithiothreitol [DTT], and 180 μM bromophenol blue) was added to each sample prior to boiling at 95°C for 10 min. Insoluble material was removed by centrifugation for 5 min in a microcentrifuge at 21,000 × g, and the samples were then resolved on SDS-polyacrylamide gels. Proteins were transferred to nitrocellulose membranes (Amersham) and blocked in Tris-buffered saline with Tween 20 (TBST) (20 mM Tris-HCl [pH 7.6], 140 mM NaCl, 0.1% Tween 20) with 5% nonfat milk for at least 1 h at room temperature. For monoclonal antibodies, the membranes were incubated with the indicated dilution of antibody in TBST with 1% bovine serum albumin (BSA) for 1 h at room temperature. For polyclonal antibodies, the membranes were incubated with the antibody in TBST with 5% BSA overnight at 4°C. Following incubation with primary antibody, the membranes were washed thrice with TBST and then incubated with horseradish peroxidase-coupled secondary antibodies for 1 h at room temperature in TBST with 1% BSA. The membranes were then incubated with WesternBright enhanced chemiluminescence (ECL) reagent (BioExpress) and imaged on a chemiluminescence detection system (Bio-Rad). The following antibodies were used at the indicated dilution: ICP36 (UL44; Virusys), 1:1,000; tubulin (Sigma), 1:10,000; GFP (Roche), 1:2,000. The IE1 (53) (1:100), IE2 (54) (1:100), and pp28 (55) (1:100) antibodies were a generous gift from the Shenk lab.
5′ rapid amplification of cDNA ends.
5′ rapid amplification of cDNA ends (5′ RACE) was performed using the RLM-RACE (Ambion) kit according to the manufacturer's protocol as previously described (47). Briefly, total or polysome-associated RNA was treated with calf intestinal phosphatase (CIP) to dephosphorylate uncapped or degraded RNAs. The 5′ 7-methylguanosine cap was then removed by incubation with tobacco acid pyrophosphatase (TAP). An RNA linker was ligated to the newly exposed phosphate on the 5′ end of the RNA and was then reverse transcribed with random hexamer primers. Nested PCR was performed using primers specific for the linker sequence ligated to the mRNA 5′ end and exon 2 of the MIE transcript. The primers used were 5′ RACE Outer exon 2 (5′-TTCGGCCAACTCTGGAAACAGC-3′) and 5′ RACE Inner exon 2 (5′-ATCAGGGTCCATCTTTCTCTTGG-3′). PCR products were visualized by ethidium bromide staining on agarose gels, shotgun cloned into the pCR-Blunt vector (Invitrogen), and sequenced by Sanger sequencing at the University of North Carolina (UNC) Genome Analysis Facility.
Polysome isolation.
Polysomes and polysome-associated RNAs were isolated as described previously (47, 48). Briefly, at the time of harvest, cells were incubated in normal growth medium containing 0.1 mg/ml cycloheximide (CHX) for 10 min. The cells were then washed three times with phosphate-buffered saline (PBS) containing CHX and lysed in 1 ml of polysome buffer (20 mM Tris-HCl [pH 7.4], 140 mM KCl, 5 mM MgCl2) containing 0.1% Triton X-100, 10 mM DTT, and 100 μM CHX. The lysate was passed five times through a 27-gauge needle to disrupt the cells, and nuclei were pelleted by centrifugation for 5 min at 1,150 × g. The cytoplasmic lysate was then centrifuged for 10 min at 21,000 × g in a microcentrifuge to remove mitochondria and insoluble debris. The supernatant was layered onto a linear 10 to 50% sucrose gradient prepared in polysome buffer. The gradients were spun for 2 h in an SW40 rotor at 32,000 rpm without interruption, and the gradients were fractionated through a gradient fractionation system (Brandel) with continuous monitoring of the optical density at 254 nm (OD254) (UA-6; Isco). Gradient fractions containing polysomes were extracted with TRIzol and treated with Turbo DNase (Applied Biosystems), and the RNA was converted to cDNA as described above.
RESULTS
Multiple mRNAs with different transcription start sites encode full-length IE1 and IE2 proteins.
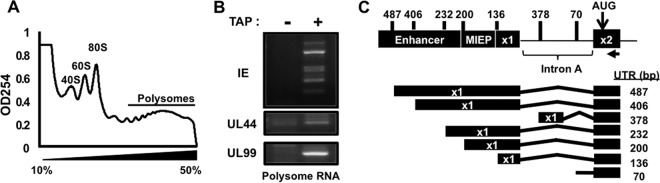
In the course of identifying splice junctions in HCMV transcripts using next-generation sequencing, we observed novel spliced transcripts in the major immediate early (MIE) locus encoded on the same strand as the IE1 and IE2 reading frames (data not shown). These results suggested the potential existence of undescribed IE1 and IE2 transcripts with alternative 5′ termini. We therefore performed 5′ rapid amplification of cDNA ends (5′ RACE) analysis to identify transcription start sites (TSSs) for mRNAs containing exon 2 of the MIE locus. To focus on transcripts that were translated during infection, we used polysome-associated RNA in the 5′ RACE reactions. Cytoplasmic lysates of HCMV-infected cells were resolved through 10 to 50% linear sucrose gradients, and RNA was extracted from fractions of the gradient containing polysomes. A representative absorbance profile showing the location of polysomes in the gradient is shown in Fig. 1A. For 5′ RACE analysis, we used the RLM-RACE procedure, wherein the mRNA cap is cleaved from mature transcripts by tobacco acid pyrophosphatase (TAP) and replaced with an RNA oligonucleotide containing a known sequence. Following random-hexamer-primed reverse transcription, the cDNA was amplified using primers specific to the RNA oligonucleotide sequence and exon 2 of the MIE transcript. Multiple bands were present when the PCR products were visualized on agarose gels (Fig. 1B), demonstrating that transcripts containing exon 2 are associated with multiple 5′ ends. No products were obtained in the absence of TAP, demonstrating the specificity of the PCR for mature, capped mRNAs.
FIG 1.
Multiple transcription start sites give rise to mRNAs containing MIE exon 2. (A) HFFs were infected with HCMV (MOI of 3) and harvested at 72 h after infection. Cytoplasmic lysates were resolved through a 10 to 50% linear sucrose gradient to separate ribosomal subunits (40S and 60S), monosomes (80S), and polysomes. (B) 5′ RACE was performed on RNA extracted from gradient fractions containing polysomes in the presence (+) or absence (−) of tobacco acid pyrophosphatase (TAP). 5′ RACE PCR products were visualized on agarose gels, cloned, and sequenced. (C) The cartoon shows the structures of the identified transcripts (not to scale). The number to the right of each transcript indicates the length of the 5′ untranslated region (UTR) of each transcript relative to the translation start site (AUG) located in exon 2.
To identify the 5′ ends of the exon 2-containing transcripts, we cloned and sequenced the PCR products of the 5′ RACE reaction. Herein we refer to each MIE transcript by the length of their 5′ untranslated region (UTR). In addition to the previously identified MIE TSS (referred to as UTR136) (56), multiple clones containing additional 5′ ends were found (Fig. 1C). One of the novel transcription start sites (UTR487) was located 351 nucleotides 5′ of the start site of UTR136. Several clones identified a TSS previously observed in latently infected monocytes isolated from HCMV-infected individuals (UTR406) (41). Transcripts initiating at this site have a 270-nucleotide extension of the 5′ end of exon 1 compared to the UTR136 start site. Two additional start sites, UTR232 and UTR200, were located between the UTR406 TSS and the UTR136 TSS. Each of the above transcripts used the previously described splice donor in exon 1 and splice acceptor in exon 2 of the MIE transcript. The directionality of the splice donor and acceptor sites confirmed that each transcript is encoded on the same strand of the viral genome as the IE1 and IE2 open reading frames (ORFs).
We also identified two transcripts that initiated in intron A of the MIE locus. The first of these “intronic” transcripts, UTR378, initiated in the middle of intron A, 350 nucleotides 3′ of the canonical exon 1 splice donor site. This transcript used a unique splice donor site with a canonical splice donor sequence (57) and was spliced to the known splice acceptor in MIE exon 2. We also found several clones containing transcripts that initiated 54 nucleotides 5′ of the splice acceptor site of MIE exon 2 (UTR70). These results show that multiple transcription start sites give rise to a collection of transcripts containing MIE exon 2. The association of each transcript with polysomes suggests that the transcripts are both trafficked to the cytoplasm and translated into protein during HCMV infection.
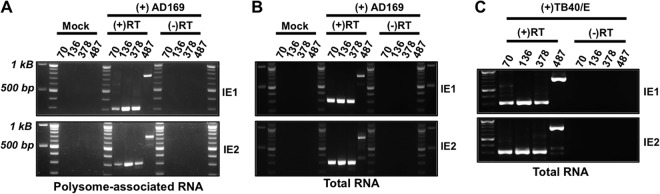
Each of the new MIE transcripts included exon 2, which contains the initiator methionine for both the IE1 and IE2 proteins (16). To determine whether the novel MIE transcripts encoded full-length IE1 or IE2 protein, we used reverse transcriptase PCR (RT-PCR) to determine whether IE1- or IE2-specific sequence (exon 4 or exon 5, respectively) was found on mRNAs originating from each of the newly identified TSSs. We focused our analysis on four transcript isoforms: (i) the longest transcript (UTR487), (ii) the previously described MIE transcript (UTR136), and (iii) the two transcripts initiating in intron A (UTR378 and UTR70). Total or polysome-associated RNA was extracted from HCMV-infected cells and reverse transcribed using primers specific for either IE1 (exon 4) or IE2 (exon 5). The resulting cDNA was amplified using primers specific to exon 4 or exon 5 together with a primer recognizing the individual transcript isoforms. No PCR product was obtained from cDNA from uninfected cells or in the absence of reverse transcriptase, demonstrating the specificity of each reaction (Fig. 2A and B). In the presence of reverse transcriptase, PCR products were obtained from both polysome-associated (Fig. 2A) and total (Fig. 2B) cDNA for each 5′ UTR linked to either IE1- or IE2-specific exons. Each MIE transcript was also detected in total RNA from cells infected with the HCMV TB40/E strain (Fig. 2C). Sequence analysis of the amplicons confirmed that each transcript contained exons 2 and 3 and was properly spliced to either exon 4 or exon 5 (data not shown). We conclude that transcripts arising from each MIE transcription start site give rise to mature cytoplasmic transcripts encoding full-length IE1 or IE2 and that each transcript has the potential to contribute to IE1 and IE2 protein expression during infection.
FIG 2.
Transcripts originating from each MIE transcription start site give rise to mature IE1 and IE2 mRNAs. Mock-infected (Mock) or HCMV-infected MRC-5 fibroblasts were harvested at 72 h after infection. (A) Polysome-associated RNA was extracted from AD169-infected cells and reverse transcribed using a primer specific for either exon 4 (IE1) or exon 5 (IE2). The resulting cDNA was amplified using a primer specific for either IE1 or IE2 together with a primer specific for each UTR (the primers are indicated by the length [in base pairs] of their their 5′ UTR [UTR70, UTR136, UTR378, and UTR487]), and the PCR products were visualized on agarose gels. Reverse transcriptase was omitted in a set of samples [(-)RT] to ensure the absence of contaminating DNA. (B) Total RNA was extracted and analyzed as in panel A. (C) As in panel B, except cells were infected with the HCMV TB40/E strain (MOI of 3). The PCR products in panels A, B, and C were cloned and sequenced to ensure their specificity.
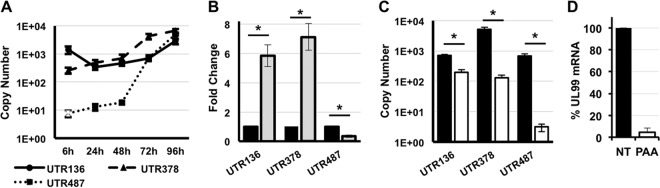
To measure the temporal accumulation of each MIE transcript, we used quantitative reverse transcriptase PCR (qRT-PCR) to measure the abundance of each transcript at different times after infection. RNA was harvested at various times after infection, and the abundance of each MIE transcript was determined by comparison to a standard curve specific for each amplicon using qRT-PCR (Fig. 3A). Each primer pair had a similar range of detection and amplified its target with similar efficiency. The abundance of the previously described MIE 5′ UTR (UTR136) decreased between 6 and 24 h after infection (16) (Fig. 3A). UTR136 abundance did not return to initial levels until 96 h after infection. The abundance of the UTR378 transcript increased fivefold between 6 and 48 h after infection, with a maximal increase at 96 h. The longest transcript, UTR487, displayed a similar trend, but it was even more abundant at 96 h than at the 6-h time point.
FIG 3.
Temporal analysis of MIE transcript abundance. (A) Primary fibroblasts were infected with HCMV (MOI of 3). The abundance of the indicated MIE transcript at each time point was measured by qRT-PCR using the absolute abundance method as described in Materials and Methods. (B) Cells were infected as described above for panel A in the presence of cycloheximide (gray bars) or absence of cycloheximide (black bars). The relative abundance of the indicated transcript was measured by qRT-PCR at 6 h after infection. Transcript abundance in untreated cells was set at one. (C) Cells were infected as in panel A in the presence (open bars) or absence (black bars) of PAA. Transcript abundance was measured by qRT-PCR using the absolute abundance method. Values that are significantly different (P value < 0.001) are indicated by a bar and asterisk. (D) Cells were infected and treated as in panel C. The reduction of UL99 mRNA expression in the presence of PAA is shown for comparison. NT, not treated.
Temporal regulation of abundance and promoter activity of alternative MIE transcripts.
In comparing the abundance of the different transcripts, the known MIE transcript (UTR136) was by far the most abundant MIE transcript expressed 6 h after infection. However, by 72 h after infection, the UTR378 and UTR487 transcripts were expressed to at least the level of the UTR136 transcript. Thus, the UTR136 transcript is the most abundant MIE transcript at immediate early times after infection. However, transcription of the novel MIE transcript isoforms increases as infection progresses, eventually rivaling expression of the previously described MIE transcript, UTR136. Like the UTR136 transcript, UTR378 transcript abundance was increased in the presence of cycloheximide. In contrast, cycloheximide significantly reduced UTR487 abundance (Fig. 3B). To measure the role of viral DNA replication in the accumulation of each MIE transcript, we measured the effect of phosphonoacetic acid (PAA), an inhibitor of viral DNA replication, on MIE transcript abundance. PAA significantly inhibited accumulation of the novel MIE transcripts, with a more pronounced effect on accumulation of the UTR378 and UTR487 mRNAs (Fig. 3C). These data show that the newly identified MIE transcripts have a complex expression profile, which together with the UTR136 transcript allows for continued IE1/2 transcription throughout the HCMV lytic cycle.
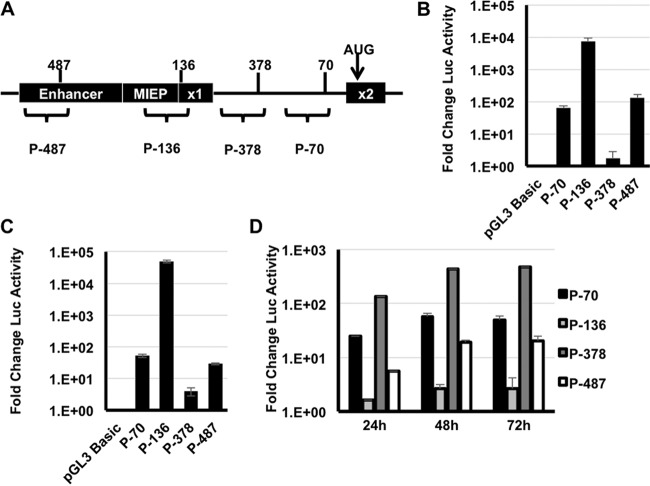
Sequences surrounding transcription start sites act as promoters that recruit RNA polymerase to initiate transcription. To determine whether the sequences surrounding the novel MIE transcription start sites possessed promoter activity, we cloned the region from −450 to +50 bp flanking each transcription start site into a promoterless luciferase reporter vector (Fig. 4A) and measured luciferase expression after transfection into HeLa cells (Fig. 4B) or primary human fibroblasts (Fig. 4C). Each promoter region increased the amount of luciferase activity in both cell types compared to the empty luciferase vector lacking a promoter, although to various degrees. The reporter containing the MIEP was the most active promoter, increasing luciferase expression by >49,000-fold compared to the empty vector (Fig. 4C). The least active promoter segment was the region surrounding the UTR378 TSS, which resulted in a modest but consistent >3-fold increase in luciferase activity. The regions surrounding the UTR487 and UTR70 transcription start sites had greater activity, increasing luciferase activity by 28- and 51-fold, respectively, compared to the empty vector.
FIG 4.
DNA sequences surrounding the novel MIE transcription start sites have promoter activity. (A) Cartoon showing the location of the 500 nucleotide regions tested for promoter activity. (B) HeLa cells were transfected with the indicated reporter plasmids, and luciferase activity was measured at 24 h after transfection. The graph shows the fold change in luciferase (Luc) activity relative to cells transfected with the empty pGL3 Basic vector, which lacks a promoter for the luciferase gene. (C) The indicated reporters were electroporated into MRC-5 fibroblasts, and luciferase activity was measured at 24 h after electroporation. (D) MRC-5 fibroblasts were electroporated with the indicated reporters and then infected with HCMV (MOI of 3). Luciferase activity was measured at the indicated times after HCMV infection. The graph shows the fold change in luciferase activity relative to the activity of each reporter at 6 h after infection. P-, promoter.
We next measured the effect of HCMV infection on the activity of each promoter. Fibroblasts were electroporated with the reporter constructs and infected with HCMV, and luciferase activity was measured over a time course of infection (Fig. 4D). Infection had minimal effect on the activity of the UTR136 promoter, increasing its activity by twofold throughout infection. Consistent with our qRT-PCR analysis, each of the novel promoters was induced by HCMV infection. Compared to uninfected cells, the UTR70 promoter was induced 25-, 58-, and 50-fold at 24, 48, or 72 h after infection, respectively. At 24, 48, or 72 h after infection, the UTR378 promoter was similarly induced 130-, 435-, and 470-fold, respectively, and UTR487 was induced 5-, 20-, and 20-fold, respectively (Fig. 4D). We conclude that the regions surrounding the novel MIE transcription start sites have promoter activity that is induced during HCMV infection.
The core promoter region of the MIEP is not necessary for IE1 and IE2 expression outside the context of HCMV infection.
We next investigated whether the novel MIE promoters were active outside of infection in the context of the MIE genomic locus. The pSVH plasmid contains the region of the HCMV genome from 840 nucleotides upstream of the MIEP transcription start site to 1,413 nucleotides downstream of the polyadenylation signal following exon 5 (17). 5′ RACE analysis of polysome-associated RNA from pSVH-transfected HeLa cells using a gene-specific primer recognizing MIE exon 2 produced a single prominent band (Fig. 5D). When the PCR product was sequenced, it was found to correspond to the previously defined MIEP transcription start site, UTR136. Thus, the UTR136 transcript is the predominant MIE transcript expressed from the MIE locus in uninfected cells.
FIG 5.
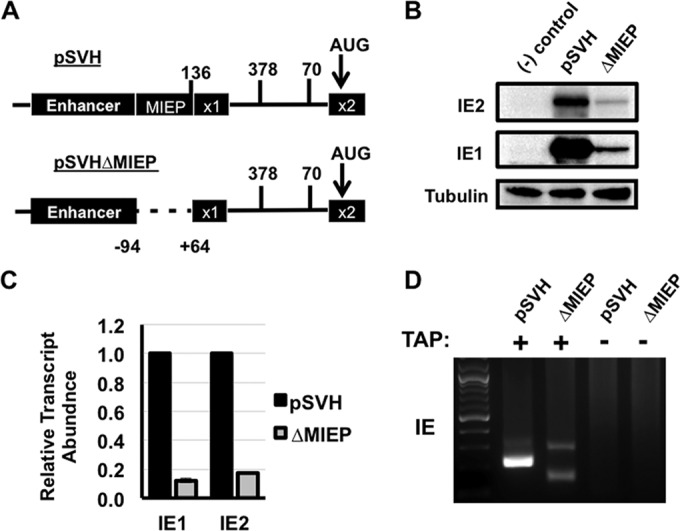
The core promoter region of the MIEP is not necessary for IE1 and IE2 expression outside the context of HCMV infection. (A) Cartoon showing a portion of the MIE locus in pSVH. The numbers show the locations of the MIE transcription start sites for the indicated MIE transcripts shown in Fig. 1. pSVHΔMIEP was created by removing a 158-bp region containing the MIEP core promoter (−94 to +64 relative to the transcription start site). (B) HeLa cells were left untransfected {negative control [(-) control]} or transfected with pSVH or pSVHΔMIEP (ΔMIEP). IE1 and IE2 protein levels were measured by Western blotting at 24 h after transfection. (C) HeLa cells were transfected and harvested as in panel B. The relative abundance of the IE1 and IE2 mRNAs in pSVHΔMIEP-transfected cells compared to pSVH-transfected cells was determined by qRT-PCR using the ΔΔCT method. IE1 and IE2 abundance in pSVH-transfected cells is set at one. (D) HeLa cells were transfected as in panel B. 5′ RACE analysis of polysome-associated RNA was performed using gene-specific primers located in exon 2. 5′ RACE PCR products were visualized on agarose gels and subsequently cloned and sequenced. No PCR products were obtained in cells where tobacco acid pyrophosphatase (TAP) was omitted (−), demonstrating that the PCR products were derived from mRNAs containing a 5′ m7G cap.
Previous studies found that the highly active MIEP can suppress the activity of weaker promoters by competing for RNA polymerase II recruitment (58, 59). To determine whether promoter competition might explain our failure to observe transcripts arising from the alternative promoters in pSVH-transfected cells, we generated a mutated version of the pSVH plasmid (pSVHΔMIEP) in which the core promoter region of the MIEP (nucleotides −94 to +64 bp surrounding the transcription start site) was deleted (Fig. 5A). Both IE1 and IE2 protein and mRNA were expressed following pSVHΔMIEP transfection, albeit at significantly reduced levels compared to the level in pSVH-transfected cells (Fig. 5B and C, respectively). This could reflect increased alternative promoter activity driven by a change in proximity to the MIE enhancer. In any case, these data show that the core promoter region of the MIEP is not required for IE1 and IE2 expression from the MIE genomic locus outside the context of infection.
5′ RACE analysis of polysome-associated RNAs from pSVHΔMIEP-transfected cells generated two PCR products, neither of which matched the size of the UTR136 5′ UTR found in cells transfected with wild-type pSVH. When the PCR products were cloned and sequenced, they were found to correspond to the UTR378 and UTR70 transcription start sites in intron A (Fig. 5D). Thus, the regions flanking the alternative transcription start sites in intron A have promoter activity that is sufficient to drive IE1 and IE2 expression in the context of the MIE genomic locus in the absence of the MIEP. These data also suggest that promoter suppression by the MIEP limits the activity of the intronic MIE promoters.
Intron A of the MIE locus is important for efficient IE1 and IE2 expression and HCMV replication.
Our results suggested that sequences in intron A contributed to the expression of IE1 and IE2. We therefore constructed an HCMV recombinant, HCMVΔIntronA, wherein the entire intron A was deleted, leaving a seamless fusion of exon 1 to exon 2 of the canonical MIE transcript. We also constructed a second recombinant, HCMVΔUTR378, lacking the 250 nucleotides surrounding the transcription start site for the UTR378 transcript (Fig. 6A). The entire genome of each recombinant and the parental virus was resequenced to confirm the absence of spurious mutations. Infectious virus was recovered following electroporation of the recombinant genomes into MRC-5 fibroblasts, demonstrating that neither intron A nor the sequences surrounding the UTR378 promoter are essential for virus replication (Fig. 6B).
FIG 6.
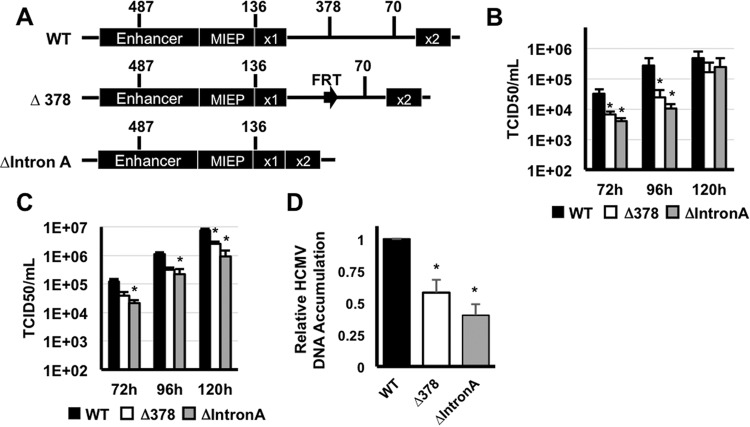
Removal of MIE intron A delays HCMV replication. (A) Diagram depicting the genomic regions removed from each recombinant HCMV BAC. (B) MRC-5 fibroblasts were infected with wild-type (WT) HCMV, HCMVΔIntron A (ΔIntron A), or HCMVΔUTR378 (ΔUTR378) (MOI of 0.5). Cell-free virus was quantified at the indicated times after infection by the TCID50 assay. (C) Fibroblasts were infected with HCMV (MOI of 3), and cell-free virus was quantified as in panel B. Values that are significantly different (P < 0.05) from the WT value are indicated by an asterisk. (D) Fibroblasts were infected as in panel B, and the relative abundance of viral DNA was quantified by real-time PCR at 72 h after infection. The amount of viral DNA in cells infected with wild-type virus was set at one. Values that are significantly different (P < 0.001) from the WT value are indicated by an asterisk.
To measure the effect of each mutation on virus replication, we compared the replication kinetics of the wild-type and recombinant viruses. Cells were infected at a multiplicity of infection (MOI) of 0.5 or 3, and the accumulation of cell-free virus was measured over a single round of virus replication. We observed reduced accumulation of cell-free virus with both mutants at 72 and 96 h after infection at either multiplicity of infection, though the effect was more pronounced at the lower multiplicity (Fig. 6B and C). In addition, both recombinant viruses had a modest, but reproducible, reduction in the accumulation of viral DNA (Fig. 6D). Thus, deletion of intron A or the region surrounding the UTR378 promoter resulted in delayed replication kinetics compared to wild-type virus.
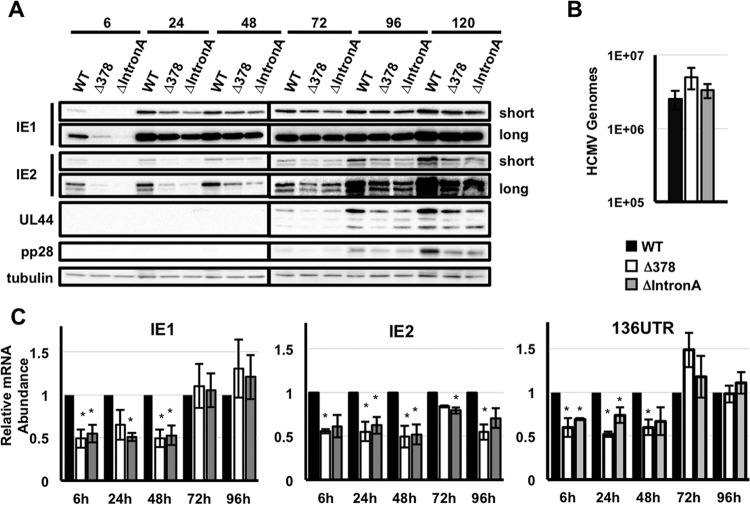
To better define the defect in viral replication after low-multiplicity infection, we measured the accumulation of representative immediate early, early, and late proteins after infection with the recombinant viruses. Both recombinants expressed decreased levels of the IE1 and E2 proteins compared to the wild-type virus. In addition, the levels of the early protein pUL44 and the late protein pp28 were expressed with delayed kinetics and to reduced levels (Fig. 7A). The delayed replication of the HCMVΔIntron A and HCMVΔUTR378 viruses thus correlated with decreased IE1 and IE2 protein expression.
FIG 7.
Deletion of MIE intron A results in decreased IE1 and IE2 mRNA and protein expression. (A) MRC-5 fibroblasts were infected with the indicated strains of HCMV (MOI of 0.5). The expression of representative HCMV immediate early (IE1 and IE2), early (UL44), and late (pp28) proteins was measured by Western blotting. (B) Fibroblasts were infected as in panel A. DNA was extracted from infected cells at 2 h after infection, and HCMV genome abundance was measured by qPCR. (C) MRC-5 fibroblasts were infected as in panel A. The relative abundance of IE1, IE2, and UTR136 mRNAs in cells infected with each recombinant virus was measured by qRT-PCR and compared to the levels in cells infected with wild-type HCMV, which was set at one. Values that are significantly different (P < 0.05) from the WT value are indicated by an asterisk.
To determine whether the reduction in IE1 and IE2 protein levels were the result of decreased IE1 and IE2 transcript levels, we measured the accumulation of IE1 and IE2 transcripts after infection with each recombinant virus. Despite the presence of equivalent numbers of viral genomes (Fig. 7B), IE1 and IE2 mRNA levels were reduced in cells infected with the recombinant viruses than in cells infected with the wild-type virus at early times of infection (Fig. 7C). At late times after infection, both recombinants expressed reduced levels of IE2 mRNA, but IE1 mRNA levels were similar to those in cells infected with the wild-type virus (Fig. 7C). Expression of the UTR136 transcript closely mirrored IE1 expression; less UTR136 mRNA was present early after infection with either mutant, but its expression reached the same level as in cells infected with wild-type virus by 72 h. Thus, the defect in IE1 and IE2 mRNA levels correlated with a decrease in IE1 and IE2 protein expression. We conclude that sequences in intron A, and the sequences surrounding the UTR378 TSS in MIE intron A, are important for maximal expression of IE1 and IE2 and efficient HCMV replication.
DISCUSSION
Proper regulation of IE1 and IE2 expression is critical for both lytic and latent HCMV infection. In this study, we describe novel IE1 and IE2 transcript isoforms arising from alternative transcription start site usage. The novel transcripts encode full-length IE1 and IE2 proteins, but they differ from the previously described MIE transcript in their 5′ UTRs (Fig. 2). Several of the new transcript isoforms extended exon 1 of the previously described MIE transcript, while two of the novel transcripts initiated within intron A (Fig. 1). The DNA sequences surrounding the new transcription start sites had promoter activity (Fig. 4B and C) that was induced upon HCMV infection (Fig. 4D), consistent with the accumulation of the novel transcript isoforms as infection progressed (Fig. 3A). Deletion of MIE intron A or the region surrounding the UTR378 TSS in intron A delayed the production of cell-free virus (Fig. 6B and C) and decreased IE1 and IE2 mRNA and protein accumulation (Fig. 7). Together, these results suggest that multiple transcription units within the MIE region control the expression of full-length IE1 and IE2 proteins during lytic virus replication.
Several spliced and alternatively spliced transcripts are transcribed from the MIE locus (16–19). Many of the alternative IE1 and IE2 mRNAs encode truncated versions of the IE1 and IE2 proteins that have distinct roles in regulating HCMV gene expression (18, 19). Our results suggest that in addition, the MIE locus gives rise to multiple transcripts encoding full-length IE1 and IE2 proteins. RT-PCR analysis of infected-cell RNA demonstrated that each of the MIE transcription start sites was linked to both exons 2 and 3, as well as the IE1- or IE2-specific exons (exon 4 or 5, respectively; Fig. 2A). The alternative transcripts were expressed from both laboratory (AD169) and clinical (TB40/E) HCMV strains (Fig. 2B and C) and thus likely are not the result of adaptation of the virus to in vitro passage. Each MIE transcript was associated with polysomes in HCMV-infected cells, suggesting that these transcripts contribute to IE1 and IE2 protein expression (Fig. 2A). In addition, our results show that the upstream MIE transcription start site previously found in latently infected monocytes (UTR406 [41]) is also utilized during lytic infection (Fig. 1). Thus, multiple transcription units contribute to the expression of full-length IE1 and IE2 during lytic HCMV infection.
We also found that deleting either intron A of the MIE locus or a portion of intron A containing one of the novel MIE promoters decreased IE1 and IE2 mRNA and protein levels (Fig. 7) and delayed virus replication (Fig. 6B and C) without affecting viral entry (Fig. 7B). These results are consistent with a role for the MIE transcripts that initiate in intron A in the expression of IE1 and IE2 during infection. We obtained similar results with the smaller deletion of sequences surrounding the UTR378 start site. Interestingly, while IE1 and UTR136 mRNA levels were similar at late times after infection with the wild-type and deletion viruses, IE2 expression remained reduced at all time points (Fig. 7). This may suggest that the alternative MIE transcripts are more important for IE2 expression, particularly during the late stage of infection. However, it is important to note that other undefined regulatory elements could also be affected by these mutations and thus could contribute to the observed decrease in IE1 and IE2 expression, as suggested by the decreased expression of the UTR136 transcript at early times after infection when intron A was deleted. Detailed mapping of transcription factor binding sites and regulatory sequences controlling the intronic promoters together with a series of subtle viral mutants will be needed to better define the factors that regulate intronic promoter activity.
Our results suggest that the transcriptional regulation of full-length IE1 and IE2 is more complex than previously appreciated. We found that the MIE locus contains multiple promoters, including two promoters located in intron A of the previously described MIE transcript. This could explain results showing that the presence of intron A modestly enhances the expression of downstream reporter genes (60–62). Perhaps this reflects a contribution of the UTR378 and UTR70 promoters to transgene expression. Our data also suggest that the coordinated regulation of the MIE promoters allows for appropriate levels of IE1 and IE2 expression throughout infection. The MIEP suppressed the weaker alternative MIE promoters in the context of the MIE locus (Fig. 5), which correlates with the accumulation of the alternative MIE transcripts later in infection when increased IE2 protein levels should limit MIEP activity (32, 35, 37, 63). We also find that the UTR378 transcript can be transcribed in the presence of cycloheximide, suggesting a potential role in the immediate early expression of IE1 and IE2 as well (Fig. 3B). However, the contribution of this transcript to IE1 and IE2 expression at IE times is likely to be minor, as it is approximately 10-fold less abundant than the UTR136 transcript at this time.
We also found that maximal accumulation of the alternative MIE transcripts required viral DNA replication (Fig. 3C). The promoters of herpesvirus late genes contain TATT motifs, which are recognized by a complex of viral proteins that facilitate RNA polymerase II recruitment (64, 65). Interestingly, TATT elements are found approximately 30 nucleotides upstream of several of the alternative MIE transcription start sites (UTR487, UTR406, and UTR378; our unpublished observations), suggesting a role for HCMV proteins in regulating the accumulation of the alternative MIE transcripts. Thus, the specific sequence elements in the alternative MIE promoters may allow for the temporal or context-specific regulation of IE1 and IE2 transcription.
Together, our data suggest a model for the regulation of IE1 and IE2 expression during HCMV infection. During the immediate early stage of infection, IE1 and IE2 transcription is controlled primarily by the MIEP. As IE2 accumulates, it binds the crs sequence in the MIEP and represses MIEP promoter activity. Suppression of the MIEP by IE2 allows the weaker promoters controlling expression of the UTR70 and UTR378 transcripts to become active, which together account for the majority of the IE1 and IE2 transcripts made during the later stages of infection. The combined action of the multiple MIE promoters thus allows for continued transcription of IE1 and IE2 despite the accumulation of IE2 protein as infection progresses.
An interesting hypothesis arising from these results is that the existence of multiple regulatory regions controlling IE1 and IE2 transcription might also contribute to cell-type- or context-dependent expression. While our data suggest a role for the alternative MIE promoters in fibroblasts, perhaps the alternative MIE transcription units play a more pronounced role in controlling IE1 and IE2 expression in monocytes or macrophages where MIEP activity is limited. The alternative promoters could also respond to distinct environmental cues, similar to the situation for ORF50 of the gammaherpesviruses. The gammaherpesvirus Rta protein is functionally analogous to IE2, transactivating viral gene expression to drive lytic replication and reactivate from latency (66–68). Multiple transcripts arising from at least three distinct promoters regulate ORF50 expression in both Kaposi's sarcoma-associated herpesvirus (KSHV) and murid herpesvirus 68 (MHV68) (69), and each ORF50 promoter responds uniquely to temporal and cell-specific cues (70, 71). Together with our data, these results suggest that the use of alternative promoters may be a common herpesvirus strategy to allow for the precise regulation of herpesvirus IE gene expression. Future experiments will focus on identifying the roles of specific sequence elements that regulate the alternative MIE promoters and their roles in other cell types and during latent infection.
ACKNOWLEDGMENTS
We thank members of the Moorman, Heise, de Silva, and de Paris labs as well as the members of the UNC Virology community for helpful discussions and input. We also thank Westefer Sanders for help sequencing the recombinant HCMV strain and Qi Tang (University of Puerto Rico) for providing the pSVH plasmid.
This work was supported by NIH grants AI03311 and AI123811 to N.J.M., the North Carolina University Cancer Research Fund, and awards from the UNC Virology Training grant (T32 AI07419 to B.Z. and K.C.A.) and National Science Foundation Graduate Research Fellowship grant (DGE-1144081) to K.C.A.
REFERENCES
- 1.Stinski MF, Thomsen DR, Stenberg RM, Goldstein LC. 1983. Organization and expression of the immediate early genes of human cytomegalovirus. J Virol 46:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hermiston TW, Malone CL, Witte PR, Stinski MF. 1987. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol 61:3214–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwamoto GK, Monick MM, Clark BD, Auron PE, Stinski MF, Hunninghake GW. 1990. Modulation of interleukin 1 beta gene expression by the immediate early genes of human cytomegalovirus. J Clin Invest 85:1853–1857. doi: 10.1172/JCI114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geist LJ, Monick MM, Stinski MF, Hunninghake GW. 1991. The immediate early genes of human cytomegalovirus upregulate expression of the interleukin-2 and interleukin-2 receptor genes. Am J Respir Cell Mol Biol 5:292–296. doi: 10.1165/ajrcmb/5.3.292. [DOI] [PubMed] [Google Scholar]
- 5.Crump JW, Geist LJ, Auron PE, Webb AC, Stinski MF, Hunninghake GW. 1992. The immediate early genes of human cytomegalovirus require only proximal promoter elements to upregulate expression of interleukin-1 beta. Am J Respir Cell Mol Biol 6:674–677. doi: 10.1165/ajrcmb/6.6.674. [DOI] [PubMed] [Google Scholar]
- 6.Monick MM, Geist LJ, Stinski MF, Hunninghake GW. 1992. The immediate early genes of human cytomegalovirus upregulate expression of the cellular genes myc and fos. Am J Respir Cell Mol Biol 7:251–256. doi: 10.1165/ajrcmb/7.3.251. [DOI] [PubMed] [Google Scholar]
- 7.Geist LJ, Monick MM, Stinski MF, Hunninghake GW. 1994. The immediate early genes of human cytomegalovirus upregulate tumor necrosis factor-alpha gene expression. J Clin Invest 93:474–478. doi: 10.1172/JCI116995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caswell R, Bryant L, Sinclair J. 1996. Human cytomegalovirus immediate-early 2 (IE2) protein can transactivate the human hsp70 promoter by alleviation of Dr1-mediated repression. J Virol 70:4028–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yurochko AD, Huong SM, Huang ES. 1999. Identification of human cytomegalovirus target sequences in the human immunodeficiency virus long terminal repeat. Potential role of IE2-86 binding to sequences between −120 and −20 in promoter transactivation. J Hum Virol 2:81–90. [PubMed] [Google Scholar]
- 10.Lukac DM, Harel NY, Tanese N, Alwine JC. 1997. TAF-like functions of human cytomegalovirus immediate-early proteins. J Virol 71:7227–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrik DT, Schmitt KP, Stinski MF. 2007. The autoregulatory and transactivating functions of the human cytomegalovirus IE86 protein use independent mechanisms for promoter binding. J Virol 81:5807–5818. doi: 10.1128/JVI.02437-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagemeier C, Walker SM, Sissons PJ, Sinclair JH. 1992. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J Gen Virol 73(Part 9):2385–2393. doi: 10.1099/0022-1317-73-9-2385. [DOI] [PubMed] [Google Scholar]
- 13.Nevels M, Paulus C, Shenk T. 2004. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc Natl Acad Sci U S A 101:17234–17239. doi: 10.1073/pnas.0407933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saffert RT, Penkert RR, Kalejta RF. 2010. Cellular and viral control over the initial events of human cytomegalovirus experimental latency in CD34+ cells. J Virol 84:5594–5604. doi: 10.1128/JVI.00348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zalckvar E, Paulus C, Tillo D, Asbach-Nitzsche A, Lubling Y, Winterling C, Strieder N, Mucke K, Goodrum F, Segal E, Nevels M. 2013. Nucleosome maps of the human cytomegalovirus genome reveal a temporal switch in chromatin organization linked to a major IE protein. Proc Natl Acad Sci U S A 110:13126–13131. doi: 10.1073/pnas.1305548110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenberg RM, Witte PR, Stinski MF. 1985. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol 56:665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenberg RM, Depto AS, Fortney J, Nelson JA. 1989. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J Virol 63:2699–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Awasthi S, Isler JA, Alwine JC. 2004. Analysis of splice variants of the immediate-early 1 region of human cytomegalovirus. J Virol 78:8191–8200. doi: 10.1128/JVI.78.15.8191-8200.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirakata M, Terauchi M, Ablikim M, Imadome K, Hirai K, Aso T, Yamanashi Y. 2002. Novel immediate-early protein IE19 of human cytomegalovirus activates the origin recognition complex I promoter in a cooperative manner with IE72. J Virol 76:3158–3167. doi: 10.1128/JVI.76.7.3158-3167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bresnahan WA, Shenk TE. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc Natl Acad Sci U S A 97:14506–14511. doi: 10.1073/pnas.97.26.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantrell SR, Bresnahan WA. 2005. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J Virol 79:7792–7802. doi: 10.1128/JVI.79.12.7792-7802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantrell SR, Bresnahan WA. 2006. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J Virol 80:6188–6191. doi: 10.1128/JVI.02676-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O'Keefe ES, Rout MP, Chait BT, Shenk T. 2010. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol 84:7803–7814. doi: 10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meier JL, Keller MJ, McCoy JJ. 2002. Requirement of multiple wild-type cis-acting elements in the human cytomegalovirus major immediate-early distal enhancer for viral gene expression and replication. J Virol 76:313–326. doi: 10.1128/JVI.76.1.313-326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunninghake GW, Monick MM, Liu B, Stinski MF. 1989. The promoter-regulatory region of the major immediate-early gene of human cytomegalovirus responds to T-lymphocyte stimulation and contains functional cyclic AMP-response elements. J Virol 63:3026–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penkert RR, Kalejta RF. 2012. Tale of a tegument transactivator: the past, present and future of human CMV pp71. Future Virol 7:855–869. doi: 10.2217/fvl.12.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saffert RT, Kalejta RF. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J Virol 80:3863–3871. doi: 10.1128/JVI.80.8.3863-3871.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romanowski MJ, Garrido-Guerrero E, Shenk T. 1997. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J Virol 71:5703–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romanowski MJ, Shenk T. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J Virol 71:1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macias MP, Huang L, Lashmit PE, Stinski MF. 1996. Cellular or viral protein binding to a cytomegalovirus promoter transcription initiation site: effects on transcription. J Virol 70:3628–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isomura H, Tsurumi T, Stinski MF. 2004. Role of the proximal enhancer of the major immediate-early promoter in human cytomegalovirus replication. J Virol 78:12788–12799. doi: 10.1128/JVI.78.23.12788-12799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lashmit PE, Stinski MF, Murphy EA, Bullock GC. 1998. A wild-type repression sequence adjacent to the transcription start site of the human cytomegalovirus US3 gene is required to down regulate gene expression at early and late times after infection. J Virol 72:9575–9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu B, Hermiston TW, Stinski MF. 1991. A wild-type cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol 65:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pizzorno MC, Hayward GS. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J Virol 64:6154–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherrington JM, Khoury EL, Mocarski ES. 1991. Human cytomegalovirus ie2 negatively regulates alpha gene expression via a short target sequence near the transcription start site. J Virol 65:887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macias MP, Stinski MF. 1993. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc Natl Acad Sci U S A 90:707–711. doi: 10.1073/pnas.90.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reeves M, Murphy J, Greaves R, Fairley J, Brehm A, Sinclair J. 2006. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J Virol 80:9998–10009. doi: 10.1128/JVI.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders RL, Del Rosario CJ, White EA, Spector DH. 2008. Internal deletions of IE2 86 and loss of the late IE2 60 and IE2 40 proteins encoded by human cytomegalovirus affect the levels of UL84 protein but not the amount of UL84 mRNA or the loading and distribution of the mRNA on polysomes. J Virol 82:11383–11397. doi: 10.1128/JVI.01293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White EA, Del Rosario CJ, Sanders RL, Spector DH. 2007. The IE2 60-kilodalton and 40-kilodalton proteins are dispensable for human cytomegalovirus replication but are required for efficient delayed early and late gene expression and production of infectious virus. J Virol 81:2573–2583. doi: 10.1128/JVI.02454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez V, Clark CL, Yen JY, Dwarakanath R, Spector DH. 2002. Viable human cytomegalovirus recombinant virus with an internal deletion of the IE2 86 gene affects late stages of viral replication. J Virol 76:2973–2989. doi: 10.1128/JVI.76.6.2973-2989.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kondo K, Xu J, Mocarski ES. 1996. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc Natl Acad Sci U S A 93:11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang D, Bresnahan W, Shenk T. 2004. Human cytomegalovirus encodes a highly specific RANTES decoy receptor. Proc Natl Acad Sci U S A 101:16642–16647. doi: 10.1073/pnas.0407233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terhune S, Torigoi E, Moorman N, Silva M, Qian Z, Shenk T, Yu D. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J Virol 81:3109–3123. doi: 10.1128/JVI.02124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terhune SS, Moorman NJ, Cristea IM, Savaryn JP, Cuevas-Bennett C, Rout MP, Chait BT, Shenk T. 2010. Human cytomegalovirus UL29/28 protein interacts with components of the NuRD complex which promote accumulation of immediate-early RNA. PLoS Pathog 6:e1000965. doi: 10.1371/journal.ppat.1000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenarcic EM, Ziehr B, De Leon G, Mitchell D, Moorman NJ. 2014. Differential role for host translation factors in host and viral protein synthesis during human cytomegalovirus infection. J Virol 88:1473–1483. doi: 10.1128/JVI.02321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziehr B, Lenarcic E, Cecil C, Moorman NJ. 2016. The eIF4AIII RNA helicase is a critical determinant of human cytomegalovirus replication. Virology 489:194–201. doi: 10.1016/j.virol.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moorman NJ, Shenk T. 2010. Rapamycin-resistant mTORC1 kinase activity is required for herpesvirus replication. J Virol 84:5260–5269. doi: 10.1128/JVI.02733-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stenberg RM, Fortney J, Barlow SW, Magrane BP, Nelson JA, Ghazal P. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol 64:1556–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziehr B, Lenarcic E, Vincent HA, Cecil C, Garcia B, Shenk T, Moorman NJ. 2015. Human cytomegalovirus TRS1 protein associates with the 7-methylguanosine mRNA cap and facilitates translation. Proteomics 15:1983–1994. doi: 10.1002/pmic.201400616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ziehr B, Vincent HA, Moorman NJ. 2016. Human cytomegalovirus pTRS1 and pIRS1 antagonize protein kinase R to facilitate virus replication. J Virol 90:3839–3848. doi: 10.1128/JVI.02714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu H, Shen Y, Shenk T. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol 69:7960–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cuevas-Bennett C, Shenk T. 2008. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J Virol 82:9525–9536. doi: 10.1128/JVI.00946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silva MC, Yu QC, Enquist L, Shenk T. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J Virol 77:10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stenberg RM, Thomsen DR, Stinski MF. 1984. Structural analysis of the major immediate early gene of human cytomegalovirus. J Virol 49:190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharp PA. 1988. RNA splicing and genes. JAMA 260:3035–3041. doi: 10.1001/jama.1988.03410200091032. [DOI] [PubMed] [Google Scholar]
- 58.Lee T, Bradley ME, Walowitz JL. 1998. Influence of promoter potency on the transcriptional effects of YY1, SRF and Msx-1 in transient transfection analysis. Nucleic Acids Res 26:3215–3220. doi: 10.1093/nar/26.13.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reddy JC, Hosono S, Licht JD. 1995. The transcriptional effect of WT1 is modulated by choice of expression vector. J Biol Chem 270:29976–29982. doi: 10.1074/jbc.270.50.29976. [DOI] [PubMed] [Google Scholar]
- 60.Chapman BS, Thayer RM, Vincent KA, Haigwood NL. 1991. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res 19:3979–3986. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hennighausen L, Fleckenstein B. 1986. Nuclear factor 1 interacts with five DNA elements in the promoter region of the human cytomegalovirus major immediate early gene. EMBO J 5:1367–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeang KT, Rawlins DR, Rosenfeld PJ, Shero JH, Kelly TJ, Hayward GS. 1987. Multiple tandemly repeated binding sites for cellular nuclear factor 1 that surround the major immediate-early promoters of simian and human cytomegalovirus. J Virol 61:1559–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lang D, Stamminger T. 1993. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J Virol 67:323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perng YC, Qian Z, Fehr AR, Xuan B, Yu D. 2011. The human cytomegalovirus gene UL79 is required for the accumulation of late viral transcripts. J Virol 85:4841–4852. doi: 10.1128/JVI.02344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong-Ho E, Wu TT, Davis ZH, Zhang B, Huang J, Gong H, Deng H, Liu F, Glaunsinger B, Sun R. 2014. Unconventional sequence requirement for viral late gene core promoters of murine gammaherpesvirus 68. J Virol 88:3411–3422. doi: 10.1128/JVI.01374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moser JM, Farrell ML, Krug LT, Upton JW, Speck SH. 2006. A gammaherpesvirus 68 gene 50 null mutant establishes long-term latency in the lung but fails to vaccinate against a wild-type virus challenge. J Virol 80:1592–1598. doi: 10.1128/JVI.80.3.1592-1598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pavlova IV, Virgin HW IV, Speck SH. 2003. Disruption of gammaherpesvirus 68 gene 50 demonstrates that Rta is essential for virus replication. J Virol 77:5731–5739. doi: 10.1128/JVI.77.10.5731-5739.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu TT, Tong L, Rickabaugh T, Speck S, Sun R. 2001. Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J Virol 75:9262–9273. doi: 10.1128/JVI.75.19.9262-9273.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wakeman BS, Johnson LS, Paden CR, Gray KS, Virgin HW, Speck SH. 2014. Identification of alternative transcripts encoding the essential murine gammaherpesvirus lytic transactivator RTA. J Virol 88:5474–5490. doi: 10.1128/JVI.03110-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goodwin MM, Canny S, Steed A, Virgin HW. 2010. Murine gammaherpesvirus 68 has evolved gamma interferon and stat1-repressible promoters for the lytic switch gene 50. J Virol 84:3711–3717. doi: 10.1128/JVI.02099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gray KS, Allen RD III, Farrell ML, Forrest JC, Speck SH. 2009. Alternatively initiated gene 50/RTA transcripts expressed during murine and human gammaherpesvirus reactivation from latency. J Virol 83:314–328. doi: 10.1128/JVI.01444-08. [DOI] [PMC free article] [PubMed] [Google Scholar]