Abstract
Photonic crystal surfaces that are designed to function as wavelength-selective optical resonators have become a widely adopted platform for label-free biosensing, and for enhancement of the output of photon-emitting tags used throughout life science research and in vitro diagnostics. While some applications, such as analysis of drug-protein interactions, require extremely high resolution and the ability to accurately correct for measurement artifacts, others require sensitivity that is high enough for detection of disease biomarkers in serum with concentrations less than 1 pg/ml. As the analysis of cells becomes increasingly important for studying the behavior of stem cells, cancer cells, and biofilms under a variety of conditions, approaches that enable high resolution imaging of live cells without cytotoxic stains or photobleachable fluorescent dyes are providing new tools to biologists who seek to observe individual cells over extended time periods. This paper will review several recent advances in photonic crystal biosensor detection instrumentation and device structures that are being applied towards direct detection of small molecules in the context of high throughput drug screening, photonic crystal fluorescence enhancement as utilized for high sensitivity multiplexed cancer biomarker detection, and label-free high resolution imaging of cells and individual nanoparticles as a new tool for life science research and single-molecule diagnostics.
I. Introduction
Since the first description of photonic crystal (PC) surfaces coupled with biological recognition molecules that allowed them to function as highly specific and sensitive biosensors [1–3], the properties of PC biosensors have enabled them to become a broadly used and intensely researched technology platform. In contrast to the broad resonances produced by lossy metallic materials that comprise surface plasmon resonant (SPR) sensors, the low-loss dielectric materials of a PC optical resonator enable narrow (< 1 nm) bandwidth, high (~95–100%) reflectivity resonances that in turn provide the ability to measure small changes in the resonant wavelength with greater precision. Importantly, the resonant characteristics of a PC surface can be excited at normal incidence without the need for contacting the sensor with a coupling prism, which facilitates measurement of the resonant wavelength in either reflectance or transmittance configurations using low-intensity broadband light sources [4–6], light emitting diodes [4], and tunable lasers [7, 8]. While the first PC biosensors were designed to operate in the near-infrared portion of the spectrum [1, 3, 5, 6, 8, 9], the resonant wavelength of a PC biosensor may be designed to occur at wavelengths ranging from ultraviolet [10] through the visible parts of the spectrum [11–13] through selection of the PC period and dielectric materials to suit the need for specific applications. PC biosensor structures have been demonstrated using a wide variety of materials, including devices built upon glass [14–18], silicon [19–21], and polymers using fabrication approaches that include nanoreplica molding [3, 22, 23], roll-to-roll mass production [8], holographic lithography [1], optical lithography [21], and electron beam lithography [19, 20, 24–26]. For several of the fabrication approaches, PC biosensors have been fabricated with costs that are compatible with single-use disposable applications in formats that include microtiter plates [8, 23, 27], microscope slides [12, 13, 28–30], and microfluidic devices [31–34]. A wide variety of biomolecular assay applications have been demonstrated in these platforms that include small molecule high throughput screening [27, 35, 36], protein-protein interaction detection [37], inhibition of protein-DNA interactions [38], small molecule aggregation [27], molecular diagnostics [16, 21, 39–43], and direct detection of virus particles [44–46] at physiologically relevant concentrations. One of the most important application areas for PC biosensors has been for cell-based assays. Since the first demonstrations of cell adsorption sensing using a PC biosensor [5, 8], the platform has been utilized to study the interaction of cells with immobilized coatings of extracellular matrix (ECM) and a variety of immobilized antibodies and antigens [12, 28, 47, 48]. Exposure of PC-adsorbed cells is routinely used to monitor the effects of drugs upon surface-exposed cell receptors as a tool for measuring cytotoxicity [49–51], drug target specificity [52–61], and even the response of immobilized cells to apoptotic cells in the media [62].
In addition to the ability of PC surfaces to perform label-free detection of biological analytes, illumination of PCs with light that matches its resonant coupling condition creates high intensity electric field standing waves that can excite a wide variety of photon emitters with greater intensity than they would experience on an unstructured surface [63, 64], using an approach called “photonic crystal enhanced excitation.” PC surfaces have also been demonstrated to be capable of channeling the photon emission of sources that are embedded within the structure preferentially along the dispersion curve of the PC, which can be used to more efficiently extract photons towards detection optics, and to thus increase photon collection efficiency. The “PC enhanced extraction” effect operates independently of the PC enhanced excitation effect, and thus their contributions multiply, resulting in the ability to achieve overall PC enhanced fluorescence [65–67] (PCEF) effects as high as 7500× [17, 68, 69]. PCEF has been applied to boost the signal-to-noise ratio for fluorescent dye molecules [65, 70–72], quantum dots [65, 67], and Raman scattering [73–75] for applications in biosensing and high efficiency lighting [76, 77]. Within the field of biosensors, PCEF has been used to reduce the limits of detection for gene expression microarrays [30, 71], detection of cancer biomarkers in serum [16, 21, 39], and low-concentration transcription factors expressed by plant cells [43]. Label-free PC biosensing and PCEF have been implemented on PC structures and detection instruments that are capable of supporting both sensing modalities for applications that include DNA microarrays [29, 78] and cell-surface attachment imaging [47].
This review will focus upon the most recent advances in sensing structures, detection instruments, and applications for label-free PC biosensors and PCEF developed in the Cunningham Group at the University of Illinois, as summarized in an invited presentation at a special session on “Photonic and Phononic Crystal Sensors” at the 2014 IEEE Sensors Conference in Valencia, Spain. First, we will describe how the detection resolution of PC label-free biosensing can be extended to characterizing the interaction of small molecules (such as drugs) with immobilized protein targets by a new instrumentation approach in which the PC forms one mirror of an external cavity laser (ECL). Combined with PC biosensors embedded within a microplate format, the PC-ECL biosensing method provides an approach for extremely accurate referencing of the most common sources of biosensor noise, thus enabling small molecule screening of drug-protein interactions to be performed robustly. Next, we summarize the development and demonstration of a new form of microscopy, called “PC Enhanced Microscopy” (PCEM) that is used for label-free, kinetic imaging of cell-surface interactions. PCEM was developed to provide a new tool for biologists who seek a more clear understanding of how cells interact with basement membranes during stem cell differentiation, cancer cell metastasis, wound healing, and biofilm growth. PCEM has also proven capable of sensing individual dielectric and metallic nanoparticles when they are attached to a PC biosensor surface, opening potential application in “digital” diagnostics with single-analyte molecule resolution. Finally, we summarize recent development of the PCEF platform, in which the electromagnetic enhancement factor and extraction efficiency has recently been further improved through incorporation of a Fabry-Perot optical cavity underneath the PC, which serves to couple two independent cavities together.
II. External Cavity Laser (ECL) Biosensors: An Active Optical Resonator Instrument for Ultrahigh Resolution for Sensing Small Wavelength Shifts
PC structures have been utilized as label-free sensors for a wide variety of biomolecular assay applications [36–38, 79]. PC sensors detect refractive index changes induced by adsorption of biomolecules and allow direct measurement of the binding affinities and binding kinetics to characterize molecular interactions in real time [2, 3, 8]. The narrow spectral linewidth enables PC sensors to resolve small wavelength shifts associated with the detection of analytes at low concentration or detection of biomolecules with low molecular weight, such as small molecule drug compounds. To further extend the detection resolution of PC sensors, we have proposed the idea of laser biosensors, and have demonstrated various types of laser biosensors as a new route for high-precision, label-free detection [22, 35, 80–83]. Recent successful demonstrations include distributed feedback laser biosensors [22], PC-ECL biosensors [81], and plasmonic ECL biosensors [83]. These systems generate extremely narrow resonant linewidth and high intensity output beams through stimulated emission while maintaining high detection sensitivity and utilization of a simple detection instrument, enabling direct detection of small molecule binding to immobilized protein targets [35].
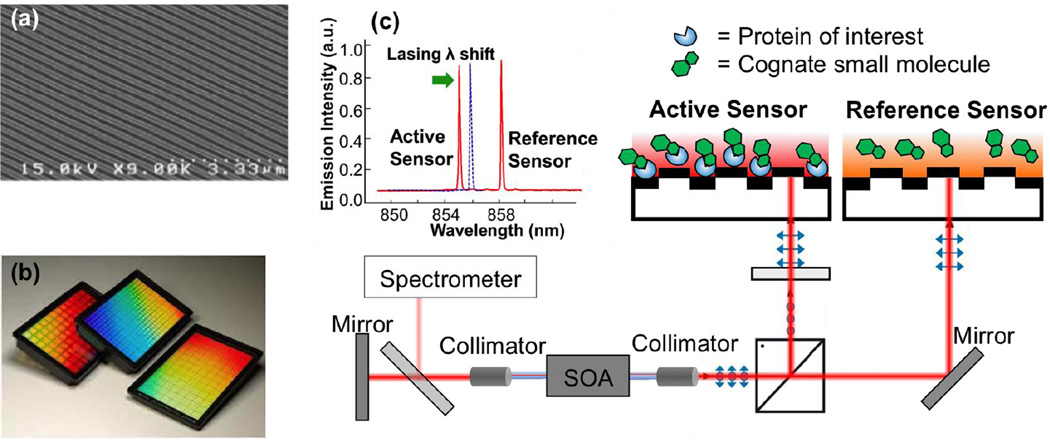
In the ECL biosensor, a PC resonant reflector surface is used as the transducer upon which biological material is adsorbed and serves as the wavelength selective element of the ECL cavity [81]. The PC sensor contains a one-dimensional surface grating structure with a period of 550 nm, as shown in Fig. 1a. It is produced via a room-temperature replica-molding process using a UV-curable polymer on a transparent polyester sheet [3]. The low refractive index polymer grating structure is subsequently coated with a film of high refractive index TiO2 to achieve the final sensor structure, which exhibits guided mode resonance with narrow bandwidth and high reflectivity at 850 nm. Adsorption of biomolecules modulates the resonant wavelength of the PC, which subsequently tunes the lasing wavelength of the ECL. Compared to simply measuring the passive resonant reflection spectrum of the PC, the ECL achieves high spectral resolution through stimulated emission and allows the measurement of small wavelength shifts. Importantly, since the optical gain is externally provided by a semiconductor optical amplifier (SOA) in the ECL, the PC sensor surface can be inexpensively manufactured over large surface areas to produce biosensors in a standard microplate format, as shown in Fig. 1b, for compatibility with automated liquid handling formats used for high-throughput applications.
Fig. 1.
Dual-wavelength self-referencing ECL detection approach using PC biosensors in 384-well microplates. A single SOA pumps two sensors (active + reference) using opposing polarizations separated by a beam splitting cube. (a) Top view of linear grating PC biosensor surface structure. (b) Photo of biosensor microplates, in which sensors on flexible plastic PC coupons are attached to bottomless microplates with adhesive. (c) Lasing spectra from active and reference sensors.
To improve the ability of the ECL biosensor to more easily distinguish true signals caused by biomolecular binding from a variety of sources of background noise, a self-referencing approach was developed based upon a dual-mode ECL [35, 82]. In the configuration of the dual-mode ECL system described in [35], two PC sensors in adjacent wells of a 384-well microplate were concurrently utilized as wavelength selective elements in the ECL cavity. Each sensor selects its own resonant wavelength, so the ECL system can lase at two independent wavelengths simultaneously. A schematic diagram of the instrument is shown in Fig. 1 with a dual-mode lasing spectrum shown as an inset in Fig. 1c. Self-referencing is achieved by designating one well as the reference well and the other as the active well, where both sensors were fabricated identically on the same substrate and were prepared identically with exception of the immobilized protein in the active well. Due to the close proximity of the active and reference sensors, accurate referencing is achieved to compensate for thermal variations and nonspecific binding. Kinetic monitoring of the lasing wavelength value (LWV) is achieved by directing a portion of the lasing emission energy with a beam splitter to a detection instrument such as a spectrometer or interferometer-based laser wavelength meter.
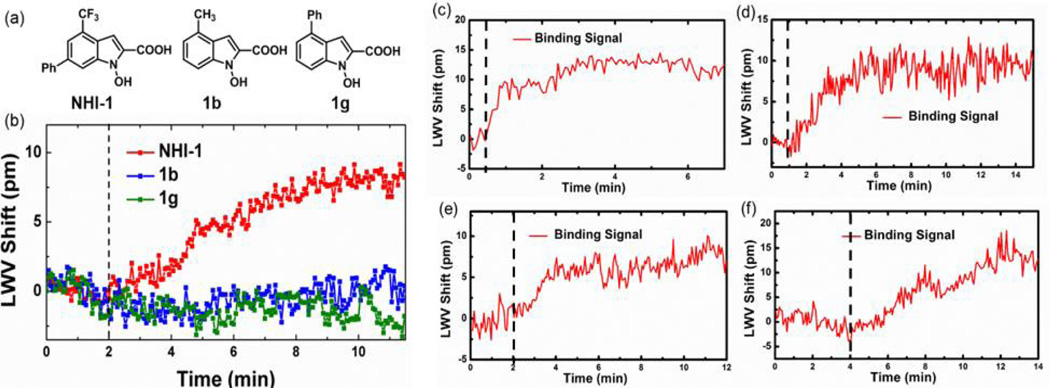
The self-referencing ECL biosensor instrument was used to study five well-characterized protein-small molecule binding interactions: CA II-dorzolamide (KD = 1.1 nM) [84], NQO1-dicoumarol (Ki = 1–10 nM), [85] XIAP-SM-164 (Ki = 0.56 nM) [86], caspase-3-Q-VD-OPh (IC50 < 25 nM) [87] and N-hydroxyindole-1 (NHI-1)-hLDH-A (Ki = 10.8 µM) [88]. These protein-small molecule interactions have binding affinities or inhibition constants ranging from sub-nanomolar to low-micromolar, which are typical of most protein-small molecule interactions. As shown in Fig. 2, the system can detect the interaction of the five protein-small molecule pairs. To further demonstrate the high throughput screening (HTS) capability of the ECL biosensor assay, a “needle-in-the-haystack” screen for inhibitors against CA II was performed. A 47-member compound collection obtained from an in-house screening library, plus dorzolamide, was screened at 50 µM with CA II immobilized to the sensor surface. Dorzolamide could be clearly differentiated from the other non-binding compounds, validating the HTS capability of the assay in identifying protein-small molecule binders.
Fig. 2.
(a) Structure of NIH-1 and two inactive variants 1b and 1g. Observed LWV shift from the binding of 50 µM (b) NHI-1, 1b and 1g to immobilized hLDH-A, (c) SM-164 to immobilized GST-XIAP, (d) dorzolamide to immobilized CA-II, (e) dicoumarol to immobilized NQO1, and (f) Q-VD-OPh to immobilized caspase-3. Dashed line represents the time at which small molecule was added to the active well. Self-referencing was applied to each. Reprinted with permission from [82], © 2014 American Chemical Society.
The PC-ECL biosensor system achieves high detection sensitivity and high spectral resolution while being capable of eliminating thermal noise through accurate referencing. Protein-small molecule binding can be accurately detected in this robust detection system without temperature control of the reagents and the sensor, in an HTS compatible microplate format. In light of previous biodetection approaches based upon optical resonators [89–92], the ECL-based optical biosensing approach represents a substantial technical advancement by simultaneously achieving high resolution through stimulated emission and maintaining high sensitivity of PC sensors, offering a sensing platform for the identification and validation of small molecule binders to protein targets in a broad range of biologically significant applications.
III. Photonic Crystal Enhanced Microscopy (PCEM): A New Mode Of Optical Microscopy for Label-Free, High-Resolution, Time-Resolved Imaging of Cell/Surface Interactions and Sensing of Single Nanoparticles
Because the PC surface structure does not enable lateral propagation of light at the resonant wavelength, it is possible to create spatial maps of the resonant wavelength and the resonant damping that allow high resolution imaging of biomaterials distributed across the surface. Because the evanescent electric field is confined close (~200 nm) to the PC surface, the resonant characteristics only samples material that has a close association with the PC surface, such as membrane components of surface-adsorbed cells, or surface-attached nanoparticle tags. To date, PCEM has been used to image the surface attachment of live cells [12, 28], nanoparticles, and protein-protein binding events [13]. In addition, PCEM can be combined with a fluorescence imaging system (addressed in section 4) to enhance the intensity of fluorescenct tags applied to cell membrane or cell nucleus components, which serves as an approach to estimate the proximity of fluorescent emitters to the PC surface [47].
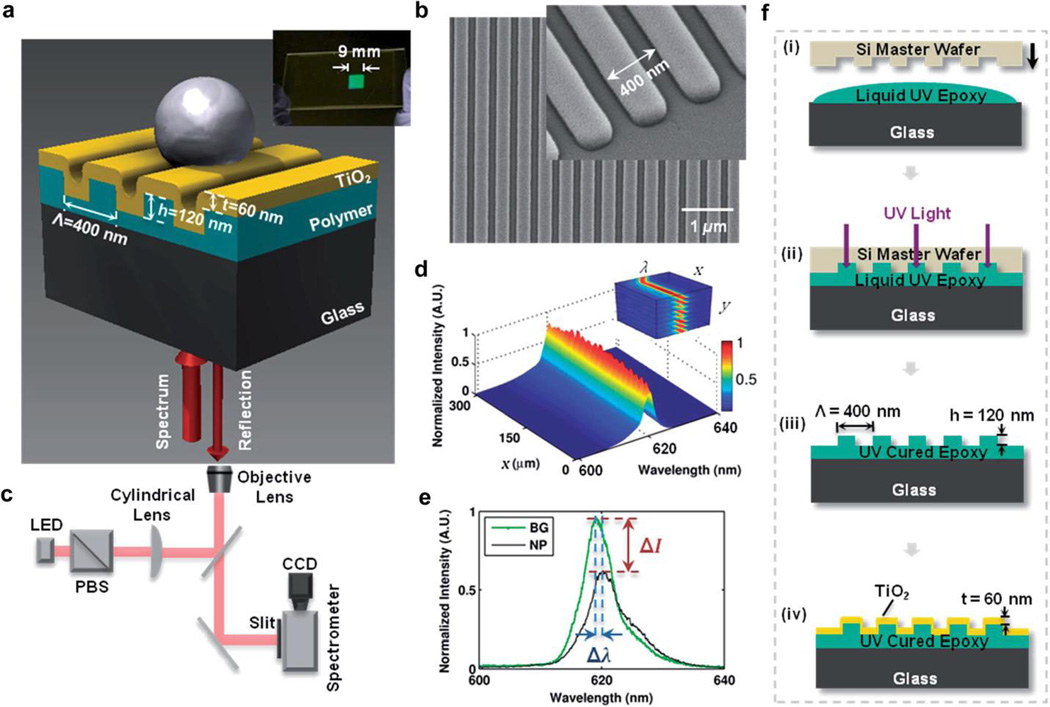
In the PCEM system (Fig. 3a–b) [12, 13, 28], a one-dimensional PC biosensor is fabricated of a periodic grating structure of low refractive index material (e.g. UV curable polymer) coated with a high refractive index layer (e.g. TiO2). The grating (period ~400 nm) enables phase matching of external radiation to the modes supported by the PC surface and symmetrically gives rise to the leaky nature of these modes by enabling re-radiation of the confined light. For a broadband light source with resonant incident angle and wavelength, the re-radiated modes couple light into and out of the high refractive index layer, and constructively and destructively interfere with the zeroth-order transmitted light in the sub-wavelength grating structure. Consequently, a sharp peak in reflection (with nearly 100% efficiency) can be observed in far field when illumination wavelengths (e.g., λ = 620 nm) and angles (e.g., θ = 0°) satisfy the resonance condition, indicating that complete interference occurs and no light is transmitted.
Fig. 3.
Principal of Photonic Crystal Enhanced Microscopy (PCEM). (a) Schematic diagram of a nanoparticle attached to a photonic crystal (PC) surface. Inset: photo of a PC fabricated on a glass cover slip. (b) Scanning electron micrograph of the PC surface. Inset: Zoomed in image. (c) Instrument schematic of the PCEM. (d) Normalized spectrum image (surface plot). Inset: PCEM-acquired 3D spectrum data. (e) Example spectrum with a peak wavelength value (PWV) shift and a peak intensity value (PIV) change with/without one detected nanoparticle (BG-background, NP-nanoparticle) on the PC surface. (f) Schematic diagram of the PC fabrication procedure: (i) the process begins with depositing a thin layer of liquid UV epoxy polymer between a Si wafer template and a glass substrate. (ii) The epoxy is converted to a solid with UV light exposure. (iii) The template is peeled away and the grating pattern is transferred to the glass (iv) A thin layer of sputter-deposited TiO2 film is applied over the grating structure. Reprinted with permission from [13], © 2014 RSC Publishing.
A linear scanning approach is used for PCEM imaging, as shown in Fig 3c. A standard inverted-microscope (with ordinary bright-field imaging components) was integrated into the PCEM system with an additional illumination path beneath the PC from a fiber-coupled broadband light-emitting diode (LED). The output light from the fiber was first polarized with a polarizing beamsplitter cube to preseve only the transverse magnetic (TM) mode of the PC, and then focused by a cylindrical lens to be formed as a linear beam at the back focal plane of the objective lens. After interacting with the material attached on PC surface, the reflected light was projected through a narrow slit aperture to an imaging spectrometer and then the spectograph was collected by a CCD camera. By measuring the peak wavelength value (PWV) or peak intensity value (PIV) at resonance of the optical spectrum on a pixel-by-pixel basis (Fig. 3d), an image of the attached material density within the evanescent field on a PC surface may be recorded.
Recently, PCEM has been successfully demonstrated as a label-free imaging system with many advantages. Measurement of cell attachment or characterization of protein-protein interactions can be achieved through locally tuned reflection spectra due to light-matter interaction on the PC within the evanescent electromagnetic field (Fig. 3e). As a label-free imaging modality, biomaterial attachment can be detected using PCEM without dyes or stains, which avoids photobleaching that is typically associated with fluorescent stains, and can hence be scanned repeatedly for long term experiments (hours or days). In the PCEM system, a PC can be illuminated in reflection mode from the bottom, efficiently avoiding the scattering from the cell body, which can happen in transmission mode when the light passes through the cell body, cell media, and the liquid-air meniscus. The use of broadband, noncoherent, low intensity light from an LED with normal incidence illumination allows the optical setup to be further simplified and avoids pre-tuning to a particular resonance condition, as typically required for SPR imaging. Furthermore, lateral light propagation in the PC surface structure is strictly limited, which enables sufficient imaging resolution for measuring subtle variations in biomaterial adhesion. In addition, a PC sensor can be fabricated (Fig. 3f) with inexpensive methods (e.g. replica molding) with UV-curable polymer and prepared with a variety of surface chemistry functionalizations. PCEM imaging provides novel information about cell/surface interactions that is fundamentally different from what is obtainable with conventional optical microscopy.
Live cells are integrated both structurally and functionally with their surrounding extracellular matrices (ECMs) in highly organized processes, which may be dynamically involved with thousands of proteins. The interaction between the cell membrane and the ECM is essential to regulate and mediate various cellular tasks, including cell adhesion, cell migration, stem cell differentiation, etc [93, 94]. However, due to the complexity of the cell–ECM adhesion process, the underlying mechanisms remain a topic of intense research interest. Conventional methods to characterize these interactions involve histological stains/dyes or fluorescent labeling, which may be cytotoxic or difficult to use for long term imaging due to photobleaching of dyes and labels. PCEM, as a label-free biosensor-based microscopy system with sufficient spatial resolution (0.6µm), temporal resolution (10s), and an integrated incubator, is an excellent approach for measuring cell-ECM interactions over extended time periods while still preserving the natural conformation of the cellular materials. Here we summarize PCEM imaging of cell-ECM interactions over time during chemotaxis, apoptosis, and differentiation.
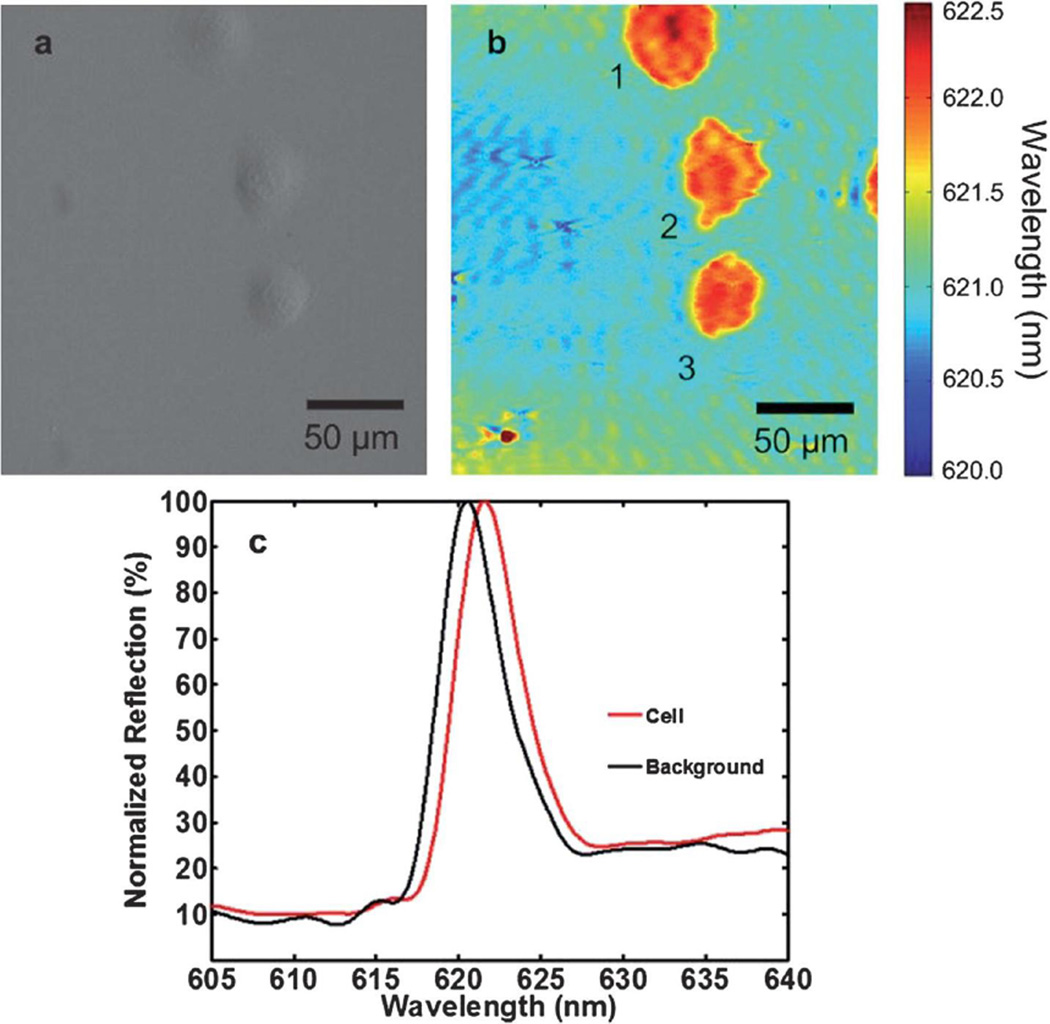
Using PCEM, PWV images are acquired and peak resonance shifts can be observed during live-cell attaching and spreading, indicating an increase in the concentration of cellular material on the PC sensor surface within the penetration depth of evanescent field [12]. Morphological surface profiles (Fig. 4b) and representative spectra (black curve: background, red curve: cell, Fig. 4c) are shown for surface-attached human pancreatic cancer cells (Panc-1). Compared with conventional brightfield images (Fig. 4a), PWV images (Fig. 4b) provide more subtle information for the non-uniformity of the attachment profile resulting from high axial resolution (~200 nm) of PCEM imaging. For instance, a local PWV shift demonstrates the presence of localized cellular adhesion while a sub-variation of PWV indicates modulation of attachment strength. A greater PWV shift along the cell edge suggests higher concentration of intracellular matter, which could be the formation of lamellipodia or actin filament bundles at the focal adhesion sites. A filopodial extension on a single cell during movement is easily identifiable and quantifiable from the acquired PWV images.
Fig. 4.
(a) Bright field and (b) PWV imaging of Panc-1 cells attached to the PC surface. Cells were seeded onto a fibronectin-coated sensor and allowed to incubate for 2 hours before imaging. Lamellipodial extensions are visible, especially from cell 2, demonstrating the ability of PCEM to resolve regional differences in single-cell attachment. Darker shading indicates regions of higher protein concentration, and is present in regions near the boundary of lamellipodia formation, consistent with the creation of actin bundles. (c) Representative regions of cellular attachment. Selected areas of the PWV image from beneath a cell show the PWV shift of a typical Panc-1 cell is ~1.0 nm, and consistent throughout the entire spectrum at those locations. Reprinted with permission from [12], © 2013 RSC Publishing.
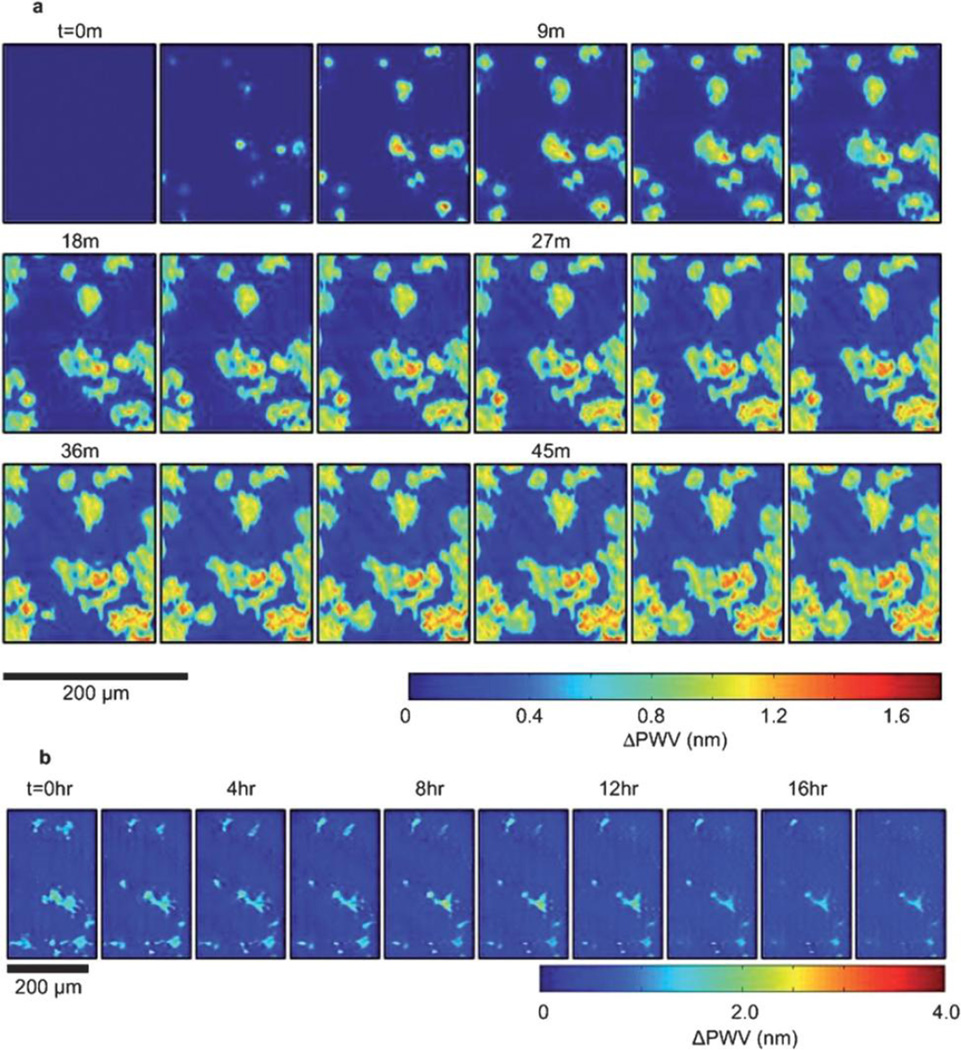
Time sequences of PWV images can also be acquired in PCEM to capture the dynamic movement of cellular processes. Fig. 5a demonstrates sequential frames from a PWV movie taken with three minute intervals for a dynamic adhesion procedure of murine dental stem cells (mHAT9) seeded on a fibronectin-coated PC surface. Stem cells initially exhibit small, round attachment areas in PWV images and progressively increase their contact area with PC while the cell edge becomes more irregular. Cell detachment and apoptosis, induced via protein kinase inhibition (by adding staurosporine), were detected and recorded as movies using the PCEM imaging system (Fig. 5b). Furthermore, stem cell chemotaxis experiments were performed on the PC surface and imaged using PCEM. The random detachment or locomotion of cells is observable and shows a full recovery of initial local PWV in the absence of cells at the original location. In summary, compared with conventional live-cell imaging methods, PCEM demonstrates unique imaging performance, such as directly and dynamically monitoring surface profiles of cell movement at the single-cell level over an extended period of time. These capabilities offer a novel technique with significant potential for the future investigation of numerous cellular processes, including cancer cell metastasis and stem cell differentiation.
Fig. 5.
(a) Time lapse PWV images of cellular attachment of mHAT9a cells. Cells were seeded at 20 000 cells per mL on a fibronectin-coated sensor surface. After 3 minutes, regions initial cell attachment appear as small, round regions, consistent with spheroid, trypsinized cells coming out of suspension and attaching to a surface. As time progresses, both the size of the cells and intensity of the PWV shift induced by them increases, indicating a higher localization of cellularmaterial at the sensor surface, which can be expected during cell spreading. Finally, once cells are sufficiently attached, cellular processes can be observed sensing the cells’ microenvironment in all directions. The outer irregular boundaries of the cells have a relatively low PWV, consistent with thin, exploratory filopodia, accompanied by a more heavily attached region slightly immediately adjacent in the cell interior, likely a result of actin bundle formation. (b) Time lapse PWV images of mHAT9a apoptosis and detachment. Cells were seeded at 8000 cells per ml onto a fibronectin-coated sensor surface. Cells that detach can be observed by the gradual retraction of filopodia and overall cell rounding before the PWV shift disappears entirely. Some cells appear to undergo apoptosiswhile still attached, leaving remnants of cellmembranes and protein on the sensor surface. DPWV datawas attained via background subtraction from an initial image taken before cell attachment (t = 0). Reprinted with permission from [12], © 2013 RSC Publishing.
As mentioned earlier, fluorescent dyes provide enhanced contrast to bioimaging modalities, but suffer from photo-bleaching. Alternatively, nanoparticles (NPs) may hold great potential when functionalized as biosensing tags for biomolecule detection [95]. This is due to their size on the nanometer scale (which is on the order of the target biomolecules) and ability to be prepared with ligands (which can enable specific binding with target biomolecules). Therefore, nanoparticle-based detection has become an important element of biosensing technology. However, single nanoparticle detection over large areas remains challenging due to the difficulties in directing the targeting molecules to specific locations [20, 96]. Nevertheless, imaging-based optical biosensing approaches, including SPR-based sensors [97, 98], dielectric thin film interference sensors [99, 100], and photonic crystal sensors [1–4, 39], demonstrate advantages since they may detect highly localized variations of dielectric permittivity produced by target molecules. Here we summarize the detection of dielectric and metallic nanoparticles (smaller than the pixel size) with highly localized effects observed by two distinct mechanisms using PCEM [13].
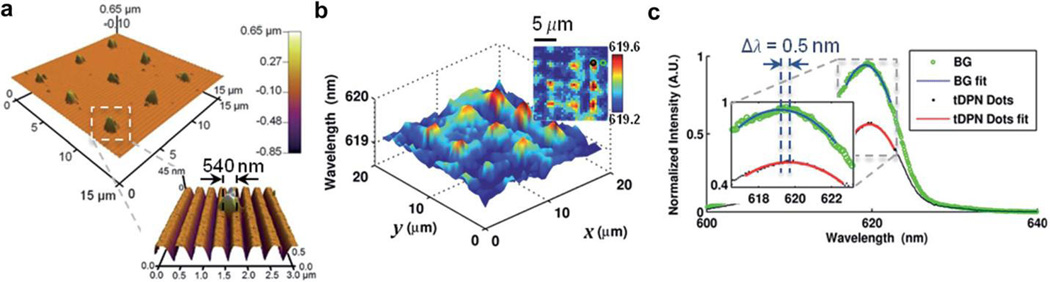
For dielectric nanoparticle detection, both mechanisms can be applied: an increase in the dielectric permittivity from the surrounding medium (e.g. nwater = 1.33) to the nanoparticle (e.g. nNP = 1.5) results in a local shift of resonance wavelength, while light intensity output from the PC exhibits a local reduction of peak intensity in the reflection spectrum. Fig. 6a shows a 3×3 array of polystyrene nanodots (dimensions of ~500× 500x45 nm3) deposited using thermal Dip-Pen Nanolithography (tDPN) with a heated AFM tip [101, 102]. The PWV image (Fig. 6b) demonstrates the PCEM detection of the nanodots, which yields results consistent with the AFM images. One pair of representative spectra (Fig. 6c) between a nanoparticle and background pixel reveal local variation in the peak resonance of wavelength and intensity. Randomly distributed TiO2 nanoparticles of similar dimensions can also be detected using PCEM with a sufficiently large peak-wavelength shift and peak-intensity reduction [13]. The mechanism is slightly different for metallic nanoparticle detection, as the reduction of resonance intensity may be caused by highly localized optical absorption of metallic nanoparticles (resulting from the imaginary part of the refractive index in metal). Gold nanoparticles (AuNP, diameter of ~100 nm) randomly attached on a PC surface had only a small shift in PWV yet had detectable variation in PIV images.
Fig. 6.
PCEM detection of nano-dots printed by thermal Dip-Pen Nanolithography (tDPN). (a) Atomic force microscopy (AFM) images of tDPNprinted 3×3 arrays of nano-dots. Inset: Zoomed in AFM images of one 540×540×40 nm3 tDPN nano-dot. (b) PCEM reflected PWV image of the tDPN nano-dots displayed in a 3D surface plot within a 20×20 µm2 field of view. Inset: 2D PWV image, demonstrating the ability of PCEM to resolve PWV differences caused by single nano-dot attachment to the PC surface. (c) Normalized spectrum of a representative tDPN nano-dot (black line) and a background pixel (green line). Inset: Zoomed in image of the spectrum with 2D polynomial fitting (tDPN nano-dot fitting in red line, background fitting in blue line), indicating a PWV shift of Δλ = 0.5 nm. Reprinted with permission from [13], © 2014 RSC Publishing.
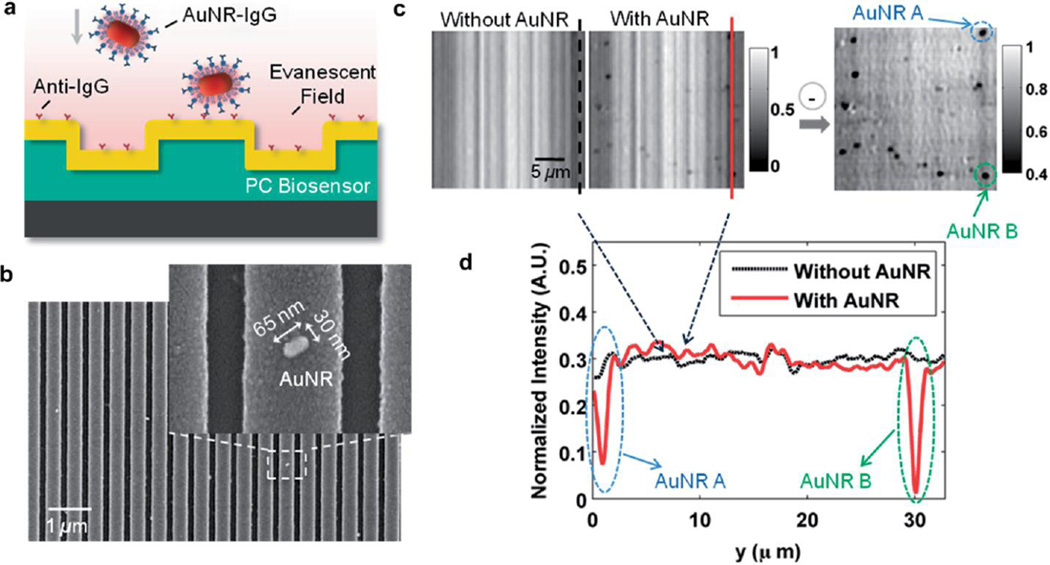
One application of nanoparticle detection using PCEM is demonstrated to detect antibody-antigen binding with nanoparticles as tags [13]. The detection performance of the imaging system can be further optimized through tuning either the resonance wavelength of the PC sensor or the localized surface plasmon resonance (LSPR) of the nanoparticles to closely match the other. Described by Mie-Gans theory [103], the depolarization factor along the longitudinal direction of a nanoparticle exhibits a strong dependence on the aspect ratio, and thus the longitudinal plasmon resonance wavelength is linearly proportional to the aspect ratio [104]. Therefore, we designed and synthesized gold nanorods (AuNRs) to improve the imaging contrast by matching the nanoparticle’s resonant wavelength (λAuNR = 620nm) with that of the PC. Fig. 7 demonstrates PCEM detection of antibody-antigen binding with nanoparticles as tags. AuNRs with dimensions of 65×65×30 nm3 were successfully synthesized with an LSPR around 620 nm, which matches the PC resonance. Then, the AuNRs were conjugated with target molecules (e.g. Rabbit Immunoglobulin G (IgG)) via flexible linkers (e.g. thiolterminated polyethylene glycol (SH-PEG)) and exposed to the PC surface immobilized with capture antibodies (e.g. anti-Rabbit IgG). After rinsing with phosphate buffered saline to remove weakly adsorbed antibodies, the PC was measured with PCEM and the PIV images were recorded. Fig. 7c (Right) plots the background-removed PIV image. The attached nanoparticles can be observed clearly in the image, which demonstrates the detection capability of PCEM for protein-protein binding using nanoparticle as tags.
Fig. 7.
PCEM detection of protein–protein binding. (a) Schematic illustration of the attachment of AuNR–IgG (AuNR conjugated with SH–PEG–IgG) on the PC biosensor surface. (b) SEM images of AuNR–IgG attached to the PC biosensor surface. Inset: Zoomed in image. (c) PCEM detected PIV images (in gray scale) and the difference between without and with AuNR–IgG on the PC surface. (d) Two representative cross-section lines of the normalized intensity images with/without two AuNRs-IgG on the PC surface. Reprinted with permission from [13], © 2014 RSC Publishing.
Label-free detection of nanoparticles using PCEM offers an attractive tool in biosensing as it avoids photobleaching of fluorescent dyes, utilizes a simple optical setup, and provides long-term imaging capability as well as versatile applications for any type of nanoparticle tags.
IV. Photonic Crystal Enhanced Fluorescence (PCEF): An Approach for High Sensitivity Assays Using Enhanced Excitation of Photon-Emitting Tags Combined With Enhanced Collection Efficiency
While the first demonstrations of enhanced fluorescence appeared shortly after the discovery of surface enhanced Raman scattering almost three decades ago [105–107], the application of this method to improving bioassays has only occurred recently, as the use of fluorescence for standard protocols in life sciences research has become increasingly popular in the past decade. Enhancing fluorescence typically relies on an interaction between a fluorophore and a resonant optical structure, the most common of which are metal nanoparticles with surface plasmon resonances. These resonances can affect fluorophores in a variety of ways: they can amplify excitation light [108], alter the spatial distribution of the fluorophore emission [109], modify the radiative lifetime of the fluorophore [110], or simultaneously perform more than one of these functions [111–113]. Metal nanostructures have been demonstrated to enhance fluorescence for applications such as immunoassays [114] and cell imaging [115] in addition to microarrays. Previous demonstrations of metalinduced enhancement of microarray dyes have shown a signal increase of more than one order of magnitude [116, 117]. However, fluorophores in close proximity to metals (within 10 nm) often transfer their energy non-radiatively as well [118]. Fluorescence enhancement using metal surfaces or metal nanoparticles suffers from quenching if the fluorophore is too close to the metal, while electromagnetic fields associated with localized surface plasmons decay rapidly with distance as one moves away from the metal – resulting in very stringent requirements for surface-fluorophore spacing [119]. The low Quality-factor of metal-based resonances, due to optical absorption, further reduces the achievable amplification factor for metal-enhanced fluorescence [120, 121].
The electric field standing waves that form when a PC is illuminated with light that matches its resonant coupling condition are capable of enhancing fluorescence in a similar fashion to surface plasmon resonances, taking advantage of two phenomena: enhanced excitation and enhanced extraction. Enhanced excitation is the result of incident radiation coupling to a PC resonance, which increases the local electric field intensity throughout the structure. These fields decay exponentially as one moves way from the substrate surface, in a similar fashion to total internal reflection fluorescence (TIRF) microscopy, but the resonance coupling provides a constructive interference effect that amplifies the incident wave [122]. Thus enhanced excitation provides the benefit of localized surface-bound fluorophore excitation observed with TIRF, but with increased performance due to field enhancement. Multiplied with this enhancement effect is enhanced extraction, whereby fluorophore emission couples to the PC and is redirected along the PC dispersion [65]. This mechanism helps to claim emitted light that otherwise may have been lost to guided modes within the substrate or to emission at oblique angles not collected by the detection optics. This process provides an optically active surface capable of providing uniform fluorescence enhancement over large areas without the quenching effects that limit metal enhanced fluorescence approaches.
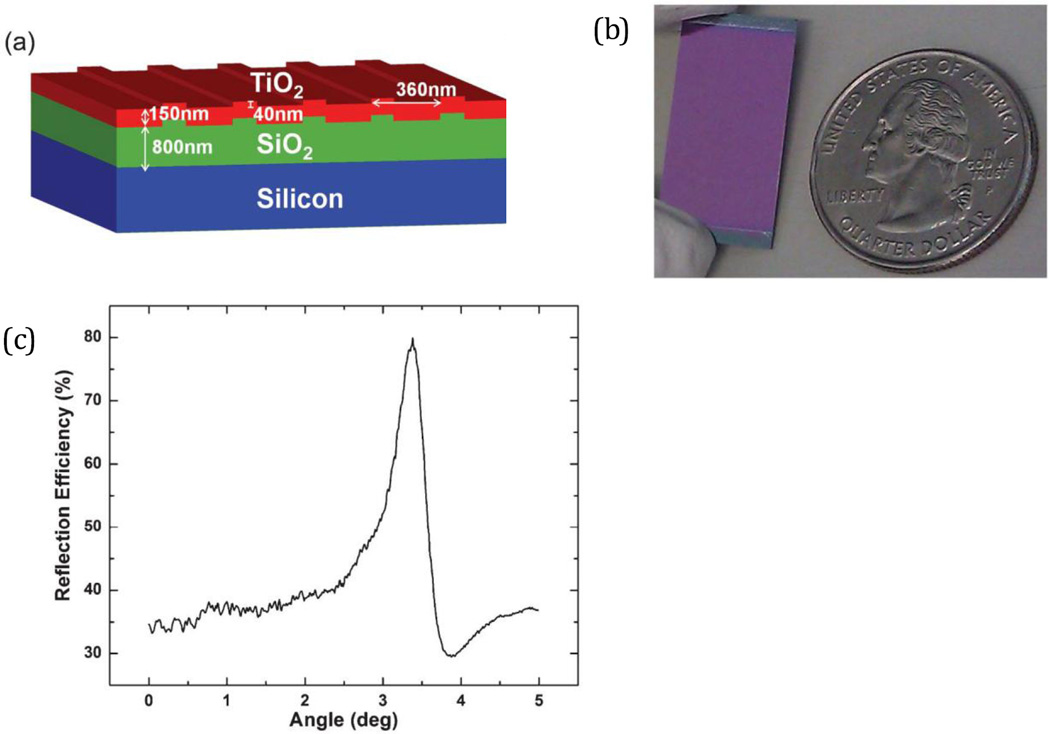
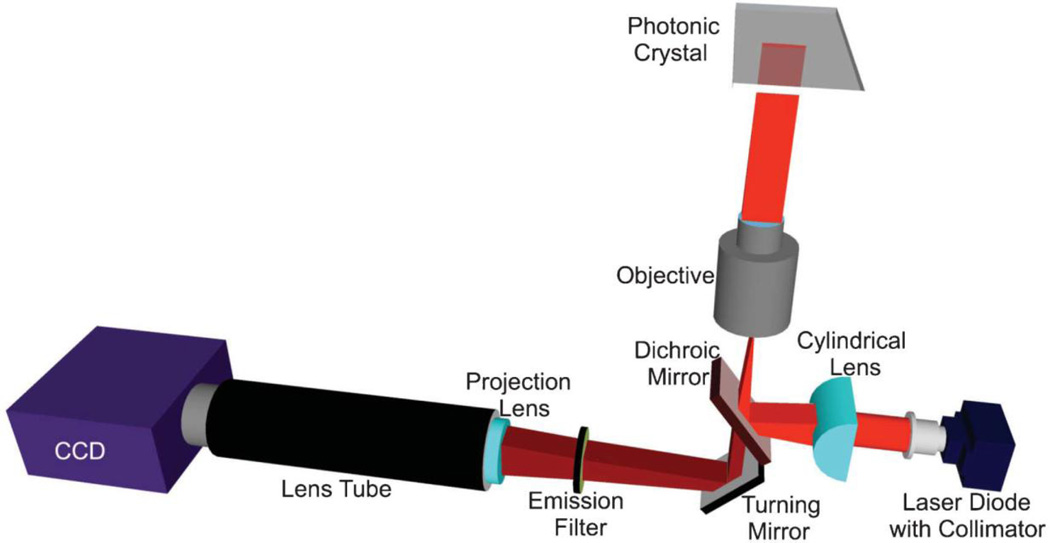
We recently demonstrated PCEF to amplify the output of surface-based fluorescence assays for multiplexed blood-based biomarker detection with sensitivity in the pg/mL range using silicon-based PCs and an objective-coupled line scanning (OCLS) instrument. A one-dimensional nanostructured grating was patterned on a silicon substrate, in a thermally grown layer of low refractive index silicon oxide (SiO2) beneath a coating of high index titanium oxide (TiO2), as shown in Fig. 8. Optimum fluorescence enhancement occurs at a specific excitation wavelength and incident angle combination, controlled by the OCLS instrument shown in Fig. 9. In brief, a 6 µm-wide focused laser line (637 nm) shines along the PC grating at a corresponding angle exhibiting peak reflectance, facilitating enhanced excitation and extraction of fluorescence.
Fig. 8.
(a) Schematic of the silicon PC (Si-PC) design. (b) Image of fabricated Si-PC. (c) Reflection spectra of Si-PC showing optimum incidence angle of 3.5 degrees using an excitation source of 637 nm. Reprinted with permission from [21], © 2013 RSC Publishing.
Fig. 9.
Overview of the components of the objective-coupled line scanning (OCLS) instrument. The solid-state laser diode emits 637 nm light which exits the objective as a 6 µm × 1 mm line onto the PC surface. Reflected light is directed toward a CCD. The PC is translated in 2 um steps in one direction for each scan. Angle tuning is achieved by translating the cylindrical lens and dichroic mirror via a motorized stage. Reprinted with permission from [21], © 2013 RSC Publishing.
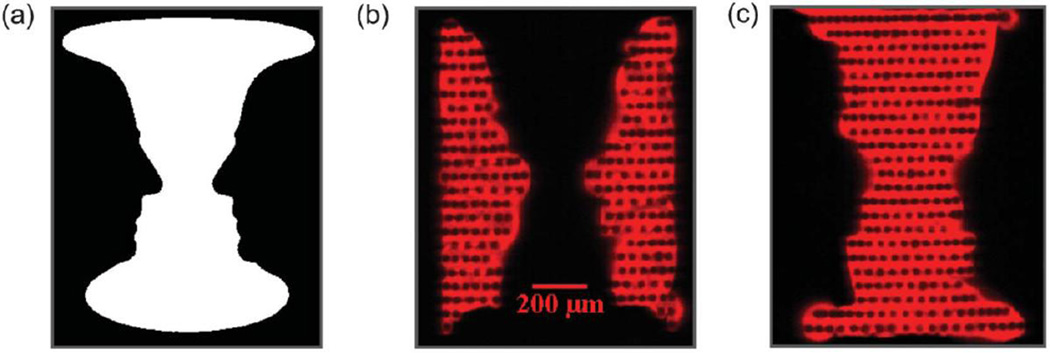
Adsorption of biomaterial (such as surface chemical functionalizaiton and application of capture molecules) on a PC surface increases the incidence angle at which resonance occurs, as demonstrated in Fig. 10. Cyanine-5 (Cy5)-labeled streptavidin (Cy5-SA) and Cy5-labeled oligonucleotide (Cy5-oligo) were used to print, via electrohydrodynamic jet (e-jet) deposition [123, 124], the two regions of the grayscale image of Rubin’s Vase onto a silanized PC surface. The protein-printed spots had higher surface density, and therefore a ~0.1 degree higher resonant coupling angle than the oligonucleotide-printed region. The PC was scanned at two different incident angles corresponding to the resonant conditions of the Cy5-SA and Cy5-oligo regions, resulting in the images seen in Fig. 10. Thus, we demonstrated the ability of the OCLS system to obtain uniform, high-resolution images of fluorescence intensity and the importance of illuminating the PC at the proper resonance condition.
Fig. 10.
(a) Rubin’s Vase optical illusion, depicting either (b) two faces or (c) a vase. Fluorescent images were obtained by printing Cy5-labeled oligonucleotide (Cy5-oligo) and Cy5-labeled protein (Cy5-SA) on the two different regions (b and c, respectively) of the same PC, then scanning at the angle tuned to the resonant condition of each. Cy5-SA has a greater surface density when printed, resulting in a ~0.1 degree greater incident angle, thus demonstrating the importance of tuning the angle of incidence. Reprinted with permission from [21], © 2013 RSC Publishing.
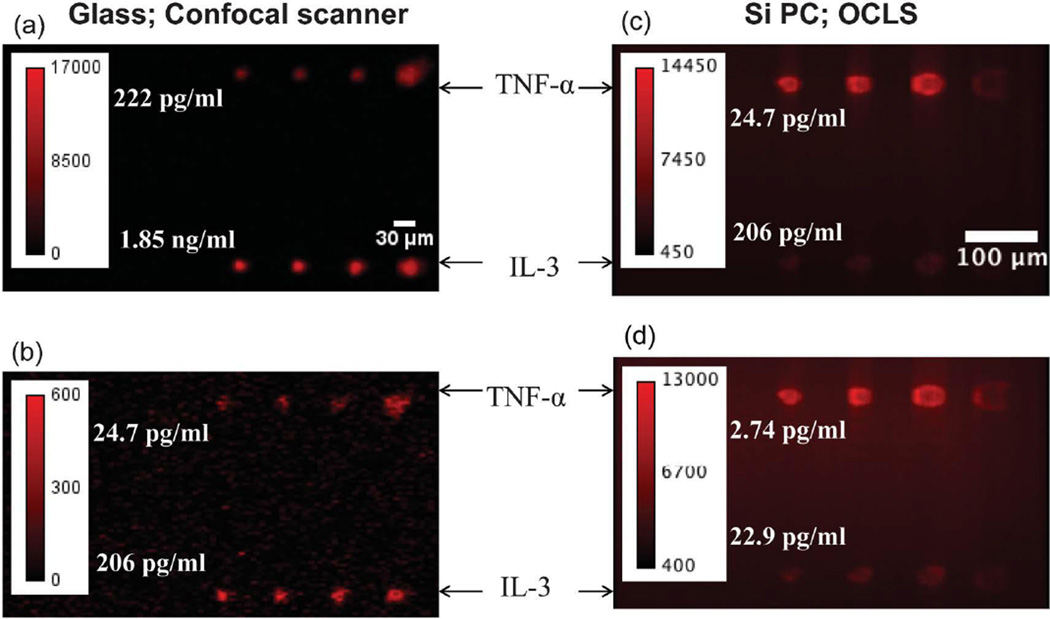
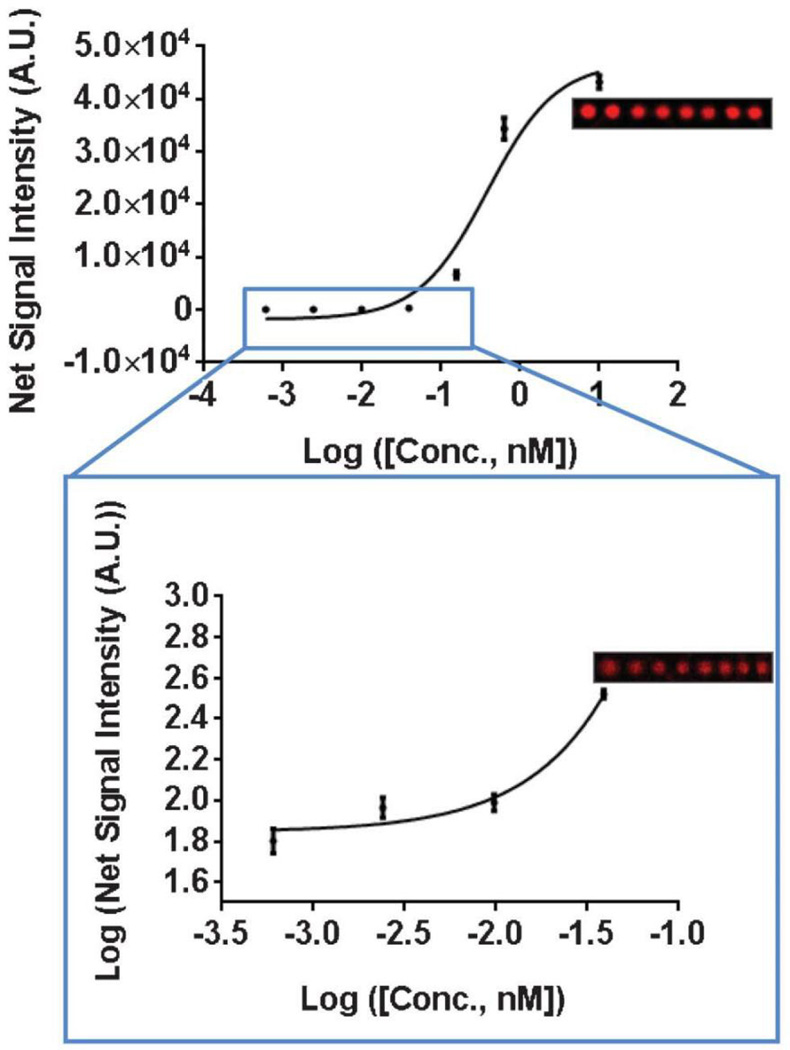
The Si-PC was also used to perform a sandwich immunoassay for two biomarkers, TNF-α and IL-3. Capture antibodies for TNF-α and IL-3 were printed on silanized Si-PC surfaces and glass slides (negative control) using dip-pen lithography [125, 126]. A mix of TNF-α and IL-3 was assayed over a range of 7 concentrations in a 3-fold dilution series with starting concentrations of 2.0 ng/mL and 17 ng/mL, respectively. The lowest concentrations detectable using spots printed on the glass slides and measured using a commercially available confocal microarray scanner (Tecan-LS), were 25 pg/mL for TNF-α and 0.21 ng/mL for IL-3. Meanwhile, all seven assayed concentrations were detectable using the Si-PC and OCLS system, the lowest concentrations being 2.7 pg/mL and 23 pg/mL for TNF-α and IL-3, respectively (Fig. 11). Lastly, a microspot assay of miR-21, a miRNA sequence implicated in the progression of breast cancer [127], was performed on the Si-PC. All seven concentrations of the miR-21 target sequence, ranging from 10 nM to 0.6 pM, were detectable. The obtained dose-response curve is shown in Fig. 12.
Fig. 11.
Fluorescence images of the TNF-α and IL-3 immunoassay, run on spots printed on silanized (a,b) glass and (c,d) Si-PC substrates and imaged using Tecan microarray scanner and OCLS, respectively. Reprinted with permission from [21], © 2013 RSC Publishing.
Fig. 12.
Dose-response curve of miR-21, a breast cancer biomarker miRNA. Representative microspot images are shown for miR-21 concentrations of 2.5 nM and 39 pM. Reprinted with permission from [21], © 2013 RSC Publishing.
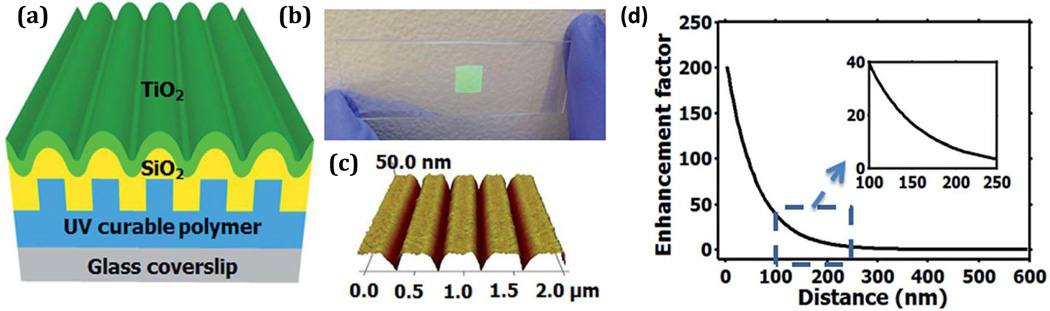
Characterization of cell-substrate interactions is useful for understanding the dynamics of cellular morphology, motion, function, and behavior throughout the cell cycle and in response to environmental cues, which have major implications in fields such as immunology and cancer biology. We recently published the first report of a PCEF microscopy approach comprising of a sensor structure, detection instrument, and image processing algorithm for imaging and quantifying cell-substrate distances. The PC used in this work is comprised of a grating structure formed in a low-refractive-index ultraviolet curable polymer (UVCP) upon a glass cover-slip. The polymer grating structure is coated with a ~200 nm SiO2 spacer layer followed by a ~95 nm high-refractive-index TiO2 thin film, as shown in Fig. 13a. The PC was fabricated via room-temperature nanoreplica molding as a 9×9 mm2 region on the surface of a standard microscope slide (Fig. 13b) and characterized using atomic force microscopy (Fig. 13c).
Fig. 13.
(a) Schematic showing the layers which comprise the PC substrate. (b) Image of the PC substrate on a microscope slide. (c) AFM image of the PC gratings. (d) Enhancement factor as a function of distance of fluorophore from PC substrate surface. Reprinted with permission from [47], © 2014 RSC Publishing.
The surface was designed to exhibit resonance at both the excitation and emission wavelengths of dyes in the spectral range of Cy5, specifically, λex = 637 nm and λem ~ 680 nm. The PC structure selectively provides enhanced excitation via the transverse magnetic (TM) mode at the on-resonance angle while always yielding enhanced emission via the transverse electric (TE) mode for emitters on the PC surface matching the TE mode wavelength. Finite Difference Time Domain (FDTD) Solutions (Lumerical Inc.) was used to simulate the optical field distribution and the distance-dependence of the enhancement effect for this PC design. The enhancement factor, or the ratio of averaged electric field intensity at on- and off-resonance conditions, was calculated at heights ranging from 2 to 600 nm, as shown in Fig. 13d.
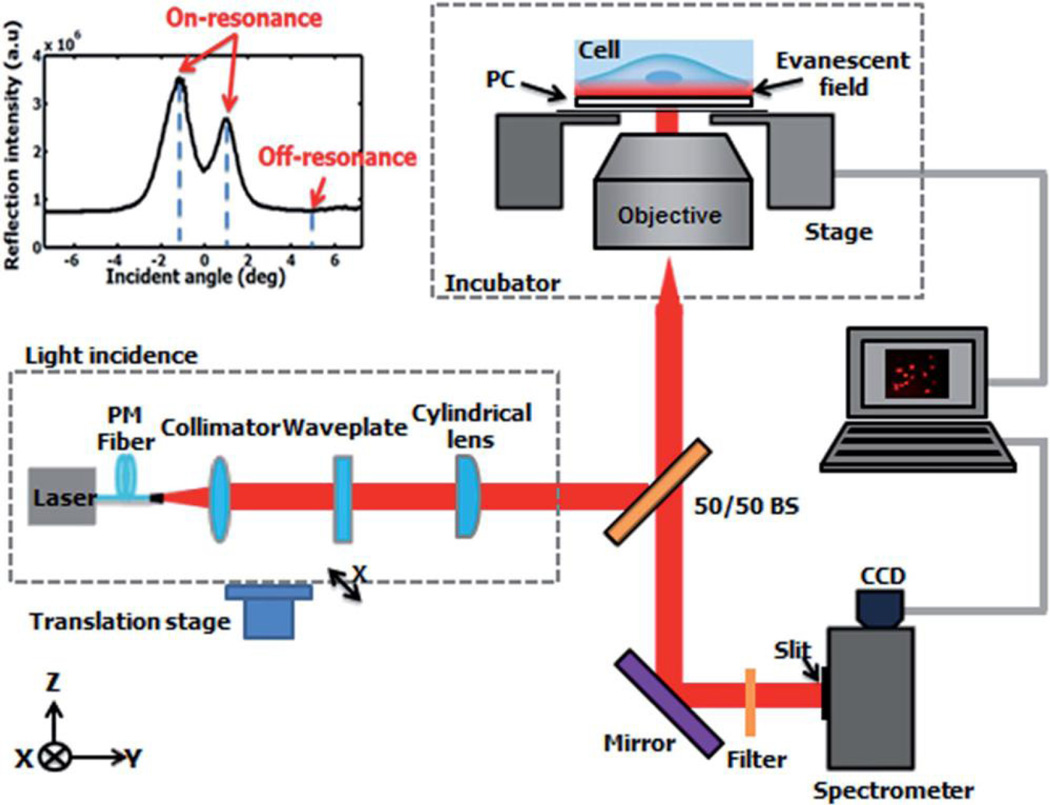
All images were acquired using a custom-built detection instrument (Fig. 14). In short, the instrumentation includes a fiber-coupled semiconductor laser excitation source (λ = 637 nm) that illuminates the cells from beneath the PC, perpendicular to the PC grating lines. Reflected light from the PC is captured by a CCD camera after passing through an emission filter. This set-up is built upon the body of a standard microscope (Carl Zeiss Axio Observer Z1) and includes an incubation chamber that maintains a constant 37°C temperature and 5% CO2 level.
Fig. 14.
Overview of the detection instrumentation, composed of a laser light source directed into a microscope objective enclosed in an incubation chamber. The incident angle is tuned via the translation stage. The sample stage holding the PC also translates along the axis perpendicular to the imaged line for each scan. Scans are performed in 0.6 µm increments, at a rate of 0.1s per line to form a whole image. Reprinted with permission from [47], © 2014 RSC Publishing.
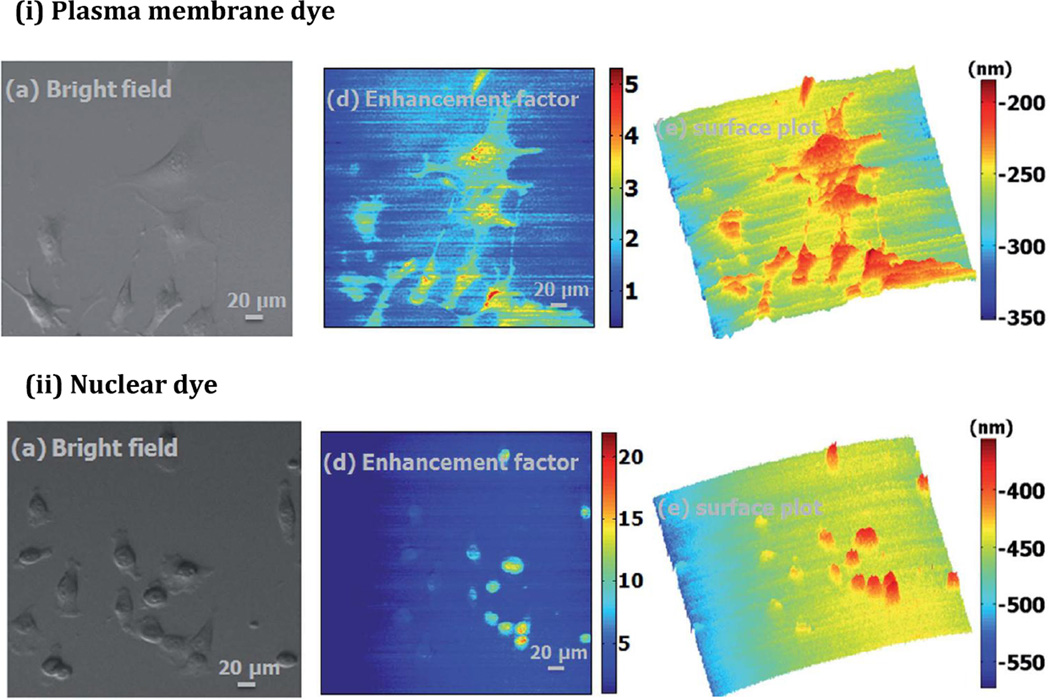
As a proof of concept, PCEF microscopy was performed on 3T3 fibroblast cells (ATCC) incubated on fibronectin-treated PC surfaces. The cells were stained with either Cell-Mask™ Deep Red plasma membrane dye (Life Technologies, λex/λem = 659/674 nm) or NucRed™ Live 647 nuclear dye (Life Technologies, λex/λem = 638/686 nm), the latter to explore the PCEF microscopy functionality when internal components of the cells are labeled. Each PC was first scanned at off-resonance—selected as 5 degrees away from the on-resonance angle—to avoid overestimation of the enhancement factor due to photobleaching. Consequently the fluorescence enhancement effect is underreported by a known factor, α, which was determined through a photobleaching test for each dye and accounted for in the analytical model. While the output fluorescence intensity of the off-resonance images only depends on the local dye concentration, that of the on-resonance images depends on both the concentration and the dye-substrate distance. This accounts for the variability in fluorescence intensity when comparing off- and on-resonance images. That is, regions of high intensity at off-resonance but low intensity at on-resonance indicates that the fluorescence is simply due to high local dye concentration, while regions of low intensity at off-resonance and high intensity at on-resonance indicates that the fluorescence is mainly due to the proximity of the stained cell component to the substrate. As such, the ratio of electric field intensities of the on- and off-resonance images is important and was obtained and termed the “enhancement factor image.” Cell-substrate distance is related to the enhancement factor by the exponential decay curve of the evanescent region of the PC and was depicted in 3D surface plots, shown in Fig. 15.
Fig. 15.
(a) Brightfield, (b) enhancement factor, and (c) surface plot images of 3T3 fibroblast cells with (i) plasma membrane dye and (ii) nuclear dye. Reprinted with permission from [47], © 2014 RSC Publishing.
We demonstrated the use of PCEF microscopy for characterizing the distance between cellular components (plasma membrane and the nucleus) of 3T3 fibroblast cells grown on fibronectin-covered PC substrates. This method can be utilized further to observe cell adhesion to gather important information regarding cell morphology, function, and behavior for the investigation of several biologically-relevant applications.
V. Cavity-Coupled PCEF: An Approach for Further Increasing the PCEF Enhancement Factor
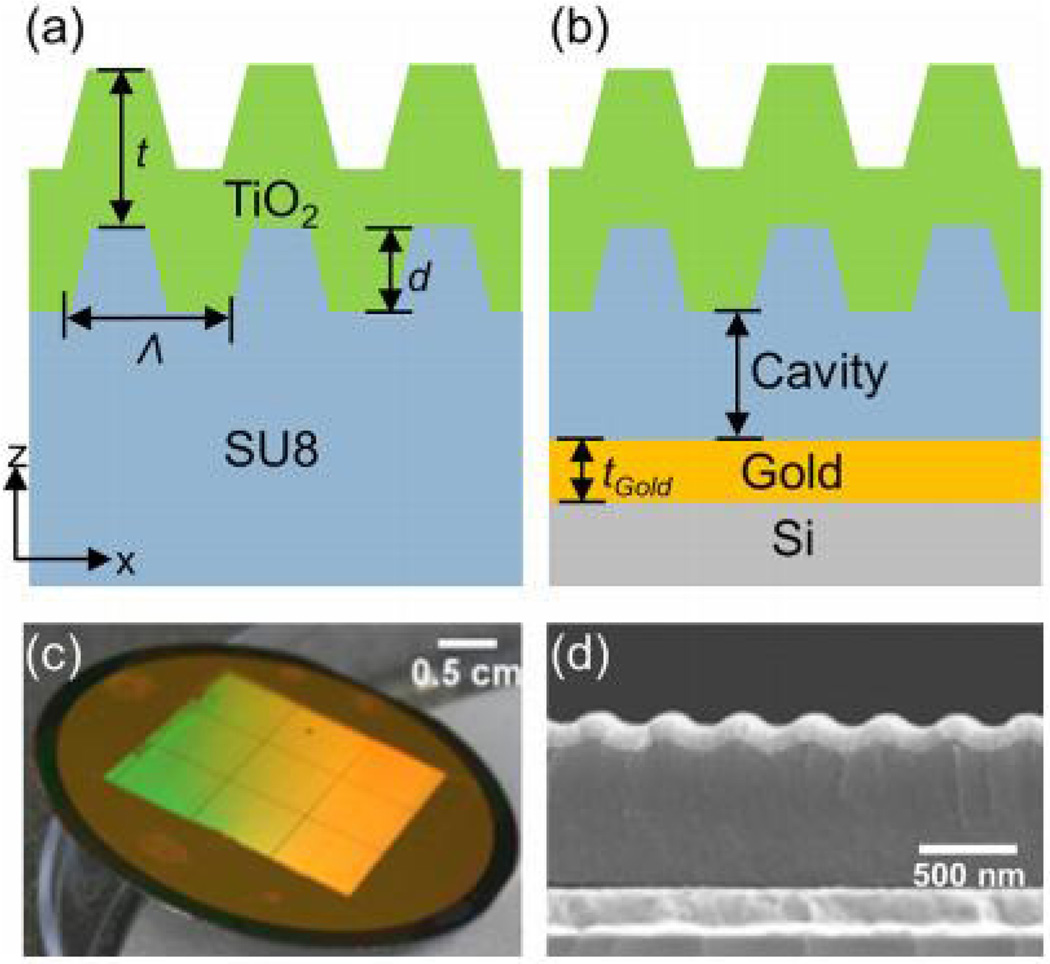
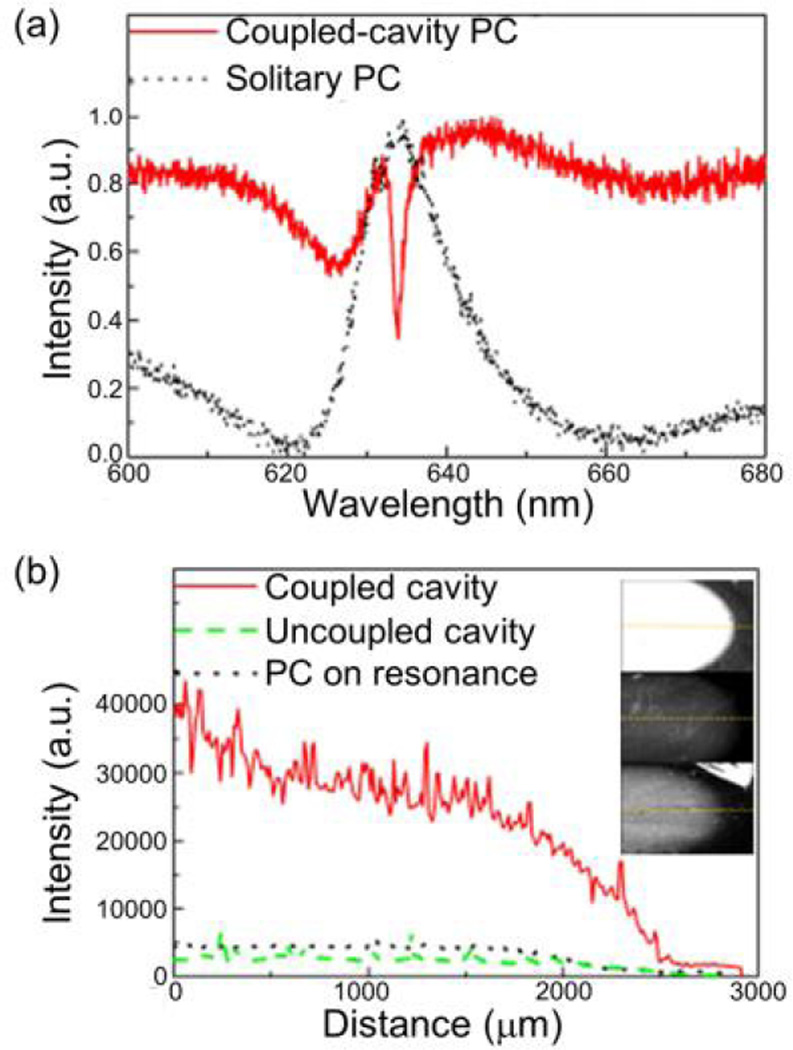
PCs have been extensively used to enhance the signal from small concentrations of biological materials [16, 21, 29, 30, 39, 43, 47, 71, 78]. As a way to both increase the quality factor of the resonator and to get additional signal enhancement, we have demonstrated coupling of a Fabry-Perot cavity with a photonic crystal resonator, as shown in Fig. 16 [128]. The gold layer, deposited at the correct cavity depth for coupling with the PC modes, increases the evanescent field strength on the PC surface, allowing for E-field enhancement. The cavity-coupled PC also has a transmission dip instead of a reflection peak at the resonance coupling condition, as shown in Fig. 17a. The cavity-coupled PC structure is fabricated on a gold-coated, silicon wafer. Before fabrication, simulations were done using an RWCA tool to find the optimal thickness for the cavity. The cavity is formed of SU8 resist, and a PDMS stamp is used to create the grating structure, followed by sputter deposition of a TiO2 thin film. It is important to note that, in order for coupling to occur, the cavity height has to satisfy the condition for constructive interference, where all the light at the resonance wavelength is transmitted through the device and not lost to destructive interference. For a range of cavity heights, this effect is periodic and can be satisfied for every λeff/2 in extra height, where λeff = λ/neff.
Fig. 16.
(a) Cross-section of regular PC. (b) Cross section of cavity-coupled PC. (c) Cavity-coupled PC fabricated on 2in gold-coated Si wafer. (d) SEM cross-section of cavity coupled PC. Reprinted with permission from [128]. © 2013 AIP Publishing.
Fig. 17.
(a) Far-field reflection spectrum for PC and cavity-coupled PC. (b) Intensity profile and image (inset) of PPL-Alexa 647 dye for coupled cavity, uncoupled cavity, and resonant PC. Reprinted with permission from [128]. © 2013 AIP Publishing.
In order to test the effectiveness of the cavity-coupled PC, the surfaces of three devices (cavity-coupled PC, PC with non-resonant cavity, and solitary PC) were prepared with a fluorescently-labeled polypeptide. The results, shown in Fig. 17b, demonstrate that cavity-coupled PC exhibits a 6x and 10x increase in fluorescence intensity over the uncoupled cavity and the solitary PC, respectively. Thus, we have shown that we can improve the sensitivity of a PC by coupling it to a Fabry-Perot cavity, enabling further work into lowering the limit of detection fluorescence-based biosensors.
VI. Conclusion
As life science researchers seek tools with ever-expanding capabilities for high sensitivity, high detection throughput, and robust operation for applications that range from drug discovery, disease diagnostics, environmental monitoring, or development of a more fundamental understanding of biological processes, there will continue to be robust research activity that can harness the capabilities of nanostructures that can interact with biological analytes. At the same time, detection instruments that are able to operate in concert with nanostructures for optimal light coupling, efficient collection of optical signals, and the ability to rapidly capture data will be a key part of any technology. This paper summarized several recent technological developments associated with photonic crystal nanostructures. For ECL biosensors, the PC can now be used in a new operating mode, where it serves as the tunable element of a laser cavity, enabling extremely narrow bandwidth optical output that can be easily measured for detection of sub-picometer wavelength shifts. Further, the ECL sensing approach incorporates a highly accurate self-referencing method that corrects for the most common label-free optical biosensor noise artifacts, resulting in a robust detection instrument for sensing small molecule interactions with much larger proteins. The PCEM represents another new detection instrument approach for PCs operating in the visible part of the spectrum that takes advantage of the lack of lateral light propagation within a PC surface. PCEM can create quantitative images of cell-surface interactions with sub-cellular resolution, and can gather images in short time intervals to generate “movies” of cell attachment processes over extended time periods. Finally PCEF represents a novel approach for coupling electric fields to fluorophores and quantum dots that are commonly used as tags for biomolecules and cells. We have recently demonstrated new objective-coupled laser scanning detection approachs for coupling a laser to a PC surface that results in enhancement factors large enough to enable multiplexed biomarker detection in a microarray format, and a cell microscopy approach that can derive enhancement factor images by modification of a conventional brightfield microscope.
Acknowledgments
The authors are grateful to support from the National Institutes of Health (R21EB009695, R01GM90220, R01GM086382, R33 CA177446, and R25CA154015) the National Science Foundation (CBET 07-54122, CBET 11-32225, and CBET 11-32301), and the US Army TATRC (grant W81XWH0810701).
Biographies

Brian T. Cunningham is the Willett Professor of Engineering in the Department of Electrical and Computer Engineering at the University of Illinois at Urbana-Champaign, where he also serves as the Director of the Micro and Nanotechnology Laboratory, and as Director of the NSF Center for Innovative Instrumentation Technology. His research is in the development of biosensors and detection instruments for pharmaceutical high throughput screening, disease diagnostics, point-of-care testing, life science research, and environmental monitoring. He has published 135 peer-reviewed journal articles, and is an inventor on 78 patents. Prof. Cunningham was a co-founder of SRU Biosystems in 2000, and founded Exalt Diagnostics in 2012 to commercialize photonic crystal enhanced fluorescence technology for disease biomarker detection. Acoustic MEMS biosensor technology that he developed at Draper Laboratory has been commercialized by Bioscale, Inc.. Prof Cunningham’s work was recognized with the IEEE Sensors Council Technical Achievement Award and the IEEE Engineering in Medicine and Biology Technical Achievement Award. He is a member of the National Academy of Inventors and a Fellow of IEEE, OSA, and AIMBE.

Meng Zhang received the B.S. in Physics from Nanjing University and the Ph.D. in Physics from University of Illinois at Urbana-Champaign. During her Ph.D. study, she worked in the Nano Sensors Group with Prof. Brian Cunningham. Her research focused on the development of laser biosensor for high sensitivity and high resolution label-free detection, and its application for drug screening.

Yue Zhuo received the B.S. and M.S. degrees in Electrical and Computer Engineering from the Northwestern University and the Chinese Academic of Science, and the Ph.D. degree in Bioengi-neering from the University of Illinois at Urbana-Champaign, respectively. She is currently working as a Post-doctoral Associate under the direction of Prof. Brian Cunningham in the Micro and Nanotechnology Laboratory at University of Illinois at Urbana-Champaign. She is also a research scientist at Exalt Diagnostics, Urbana. Her research focuses on the development of nano-optical biosensor and detection system, and their applications for biosensing and bioimaging.
Lydia Kwon received the B.S. degree in Biomedical Engineering from the University of California, Davis in 2012. She is interested in developing devices for point-of-care medical applications and is currently working on a Ph.D. degree in Bioengineering under the direction of Prof. Brian T. Cunningham at the University of Illinois at Urbana-Champaign.
Caitlin Race received the B.E.E. and M.S. degrees in electrical and computer engineering from the University of Minnesota - Twin Cities in 2011 and 2012, respectively. She is currently working on her Ph.D. degree as a Graduate Research Assistant under the direction of Prof. Brian T. Cunningham at the University of Illinois at Urbana-Champaign, Urbana. Her research focuses on the design and characterization of optical biosensors and device fabrication processes.
References
- 1.Cunningham BT, Li P, Lin B, Pepper J. Colorimetric resonant reflection as a direct biochemical assay technique. Sensors and Actuators B. 2002;81:316–328. [Google Scholar]
- 2.Cunningham B, Qiu J, Li P, Lin B. Enhancing the surface sensitivity of colorimetric resonant optical biosensors. Sensors and Actuators B-Chemical. 2002;87:365–370. [Google Scholar]
- 3.Cunningham BT, Qiu J, Li P, Pepper J, Hugh B. a plastic colorimetric resonant optical biosensor for multiparallel detection of label-free biochemical interactions. Sensors and Actuators B. 2002;85:219–226. [Google Scholar]
- 4.Cunningham BT, Li P, Schulz S, Lin B, Baird C, Gerstenmaier J, Genick C, Wang F, Fine E, Laing L. Label-Free Assays on the BIND System. Journal of Biomolecular Screening. 2004;9:481–490. doi: 10.1177/1087057104267604. [DOI] [PubMed] [Google Scholar]
- 5.Lin B, Li P, Cunningham BT. A label-free biosensor-based cell attachment assay for characterization of cell surface molecules. Sensors and Actuators B-Chemical. 2006;114:559–564. [Google Scholar]
- 6.Lin B, Pepper J, Li P, Pien H, Cunningham BT. A label-free high throughput optical technique for detecting small molecule interactions. Biosensors & Bioelectronics. 2002;17:827–834. doi: 10.1016/s0956-5663(02)00077-5. [DOI] [PubMed] [Google Scholar]
- 7.Mateus CFR, Huang MCY, Chang-Hasnain CJ, Foley JE, Beatty R, Li P, Cunningham BT. Ultra-sensitive immunoassay using VCSEL detection system. Electronics Letters. 2004;40:649–651. [Google Scholar]
- 8.Mateus CFR, Huang MCY, Cunningham BT, Chang-Hasnain CJ. Compact label-free biosensor using VCSEL-based measurement system. IEEE Photonics Technology Letters. 2004;16:1712–1714. [Google Scholar]
- 9.Li P, Lin B, Gerstenmaier J, Cunningham BT. A new method for label-free imaging of biomolecular interactions. Sensors and Actuators B. 2004;99:6–13. [Google Scholar]
- 10.Ganesh N, Block ID, Cunningham BT. Near ultraviolet-wavelength photonic-crystal biosensor with enhanced surface-to-bulk sensitivity ratio. Applied Physics Letters. 2006;89 [Google Scholar]
- 11.Gallegos D, Long KD, Yu HJ, Clark PP, Lin YX, George S, Nath P, Cunningham BT. Label-free biodetection using a smartphone. Lab on a Chip. 2013;13:2124–2132. doi: 10.1039/c3lc40991k. [DOI] [PubMed] [Google Scholar]
- 12.Chen WL, Long KD, Lu M, Chaudhery V, Yu H, Choi JS, Polans J, Zhuo Y, Harley BAC, Cunningham BT. Photonic crystal enhanced microscopy for imaging of live cell adhesion. Analyst. 2013;138:5886–5894. doi: 10.1039/c3an01541f. [DOI] [PubMed] [Google Scholar]
- 13.Zhuo Y, Hu H, Chen WL, Lu M, Tian LM, Yu HJ, Long KD, Chow E, King WP, Singamaneni S, Cunningham BT. Single nanoparticle detection using photonic crystal enhanced microscopy. Analyst. 2014;139:1007–1015. doi: 10.1039/c3an02295a. [DOI] [PubMed] [Google Scholar]
- 14.Block ID, Chan LL, Cunningham BT. Photonic crystal optical biosensor incorporating structured low-index porous dielectric. Sensors and Actuators B-Chemical. 2006;120:187–193. [Google Scholar]
- 15.Block ID, Chan LL, Cunningham BT. Large-area submicron replica molding of porous low-k dielectric films and application to photonic crystal biosensor fabrication. Microelectronic Engineering. 2007;84:603–608. [Google Scholar]
- 16.Huang CS, Chaudhery V, Pokhriyal A, George S, Polans J, Lu M, Tan RM, Zangar RC, Cunningham BT. Multiplexed Cancer Biomarker Detection Using Quartz-Based Photonic Crystal Surfaces. Analytical Chemistry. 2012;84:1126–1133. doi: 10.1021/ac202817q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pokhriyal A, Lu M, Chaudhery V, Huang CS, Schulz S, Cunningham BT. Photonic crystal enhanced fluorescence using a quartz substrate to reduce limits of detection. Optics Express. 2010;18:24793–24808. doi: 10.1364/OE.18.024793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George S, Pokhriyal A, Jones SI, Lu M, Vodkin LO, Cunningham BT. Using Photonic Crystal Enhanced Fluorescence on Quartz Substrates to Improve the Sensitivity of DNA Microarrays; 2011 Annual International Conference of the Ieee Engineering in Medicine and Biology Society (Embc); 2011. pp. 26–29. [DOI] [PubMed] [Google Scholar]
- 19.Lee M, Fauchet PM. Two-dimensional silicon photonic crystal based biosensing platform for protein detection. Optics Express. 2007;15:4530–4535. doi: 10.1364/oe.15.004530. [DOI] [PubMed] [Google Scholar]
- 20.Lee MR, Fauchet PM. Nanoscale microcavity sensor for single particle detection. Optics Letters. 2007;32:3284–3286. doi: 10.1364/ol.32.003284. [DOI] [PubMed] [Google Scholar]
- 21.George S, Chaudhery V, Lu M, Takagi M, Amro N, Pokhriyal A, Tan YF, Ferreira P, Cunningham BT. Sensitive detection of protein and miRNA cancer biomarkers using silicon-based photonic crystals and a resonance coupling laser scanning platform. Lab on a Chip. 2013;13:4053–4064. doi: 10.1039/c3lc50579k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu M, Choi SS, Irfan U, Cunningham BT. Plastic distributed feedback laser biosensor. Applied Physics Letters. 2008;93 [Google Scholar]
- 23.Tan YF, Ge C, Chu A, Lu M, Goldshlag W, Huang CS, Pokhriyal A, George S, Cunningham BT. Plastic-Based Distributed Feedback Laser Biosensors in Microplate Format. IEEE Sensors Journal. 2012;12:1174–1180. [Google Scholar]
- 24.Buswell SC, Wright VA, Buriak JM, Van V, Evoy S. Specific detection of proteins using photonic crystal waveguides. Optics Express. 2008;16:15949–15957. doi: 10.1364/oe.16.015949. [DOI] [PubMed] [Google Scholar]
- 25.Skivesen N, Tetu A, Kristensen M, Kjems J, Frandsen LH, Borel PI. Photonic-crystal waveguide biosensor. Optics Express. 2007;15:3169–3176. doi: 10.1364/oe.15.003169. [DOI] [PubMed] [Google Scholar]
- 26.Topol’ancik J, Bhattacharya P, Sabarinathan J, Yu PC. Fluid detection with photonic crystal-based multichannel waveguides. Applied Physics Letters. 2003;82:1143–1145. [Google Scholar]
- 27.Chan LL, Lidstone EA, Finch KE, Heeres JT, Hergenrother PJ, Cunningham BT. A Method for Identifying Small-Molecule Aggregators Using Photonic Crystal Biosensor Microplates. JALA. 2009;14:348–359. doi: 10.1016/j.jala.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lidstone EA, Chaudhery V, Kohl A, Chan V, Wolf-Jensen T, Schook LB, Bashir R, Cunningham BT. Label-free imaging of cell attachment with photonic crystal enhanced microscopy. Analyst. 2011;136:3608–3615. doi: 10.1039/c1an15171a. [DOI] [PubMed] [Google Scholar]
- 29.George S, Block ID, Jones SI, Mathias PC, Chaudhery V, Vuttipittayamongkol P, Wu HY, Vodkin LO, Cunningham BT. Label-Free Prehybridization DNA Microarray Imaging Using Photonic Crystals for Quantitative Spot Quality Analysis. Analytical Chemistry. 2010;82:8551–8557. doi: 10.1021/ac101551c. [DOI] [PubMed] [Google Scholar]
- 30.Mathias PC, Jones SI, Wu HY, Yang F, Ganesh N, Gonzalez DO, Bollero G, Vodkin LO, Cunningham BT. Improved Sensitivity of DNA Microarrays Using Photonic Crystal Enhanced Fluorescence. Analytical Chemistry. 2010;82:6854–6861. doi: 10.1021/ac100841d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi CJ, Belobraydich AR, Chan LL, Mathias PC, Cunningham BT. Comparison of label-free biosensing in microplate microfluidic and spot-based affinity capture assays. Anal Biochem. 2010;405:1–10. doi: 10.1016/j.ab.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 32.Tan Y, Sutanto E, Alleyne AG, Cunningham BT. Photonic crystal enhancement of a homogeneous fluorescent assay using sub-micron fluid channels fabricated by E-jet patterning. J Biophotonics. 2014;7:266–275. doi: 10.1002/jbio.201300158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi CJ, Cunningham BT. Single-step fabrication and characterization of photonic crystal biosensors with polymer microfluidic channels. Lab on a Chip. 2006;6:1373–1380. doi: 10.1039/b603514k. [DOI] [PubMed] [Google Scholar]
- 34.Choi CJ, Cunningham BT. A 96-well microplate incorporating a replica molded microfluidic network integrated with photonic crystal biosensors for high throughput kinetic biomolecular interaction analysis. Lab on a Chip. 2007;7:550–556. doi: 10.1039/b618584c. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M, Peh J, Hergenrother PJ, Cunningham BT. Detection of protein-small molecule binding using a self-referencing external cavity laser biosensor. J Am Chem Soc. 2014;136:5840–5843. doi: 10.1021/ja500636p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin B, Qiu J, Gerstenmeier J, Li P, Pien H, Pepper J, Cunningham B. A label-free optical technique for detecting small molecule interactions. Biosens Bioelectron. 2002;17:827–834. doi: 10.1016/s0956-5663(02)00077-5. [DOI] [PubMed] [Google Scholar]
- 37.Heeres JT, Kim SH, Leslie BJ, Lidstone EA, Cunningham BT, Hergenrother PJ. Identifying Modulators of Protein-Protein Interactions Using Photonic Crystal Biosensors. J Am Chem Soc. 2009;131:18202–18203. doi: 10.1021/ja907066r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan LL, Pineda M, Heeres JT, Hergenrother PJ, Cunningham BT. A general method for discovering inhibitors of protein-DNA interactions using photonic crystal biosensors. ACS Chem Biol. 2008;3:437–448. doi: 10.1021/cb800057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunningham BT, Zangar RC. Photonic crystal enhanced fluorescence for early breast cancer biomarker detection. J Biophotonics. 2012;5:617–628. doi: 10.1002/jbio.201200037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson R, Cunningham B, Andrade J. An enhanced photonic crystal biosensor-based immunodiagnostic assay for detection of soluble transferrin receptor as a biomarker of iron deficiency. Faseb Journal. 2014;28 [Google Scholar]
- 41.Peterson RD, Cunningham B, Andrade J, Engeseth N. A label-free photonic crystal biosensor for the assessment of iron status using ferritin. Faseb Journal. 2013;27 [Google Scholar]
- 42.Peterson RD, Cunningham BT, Andrade JE. A photonic crystal biosensor assay for ferritin utilizing iron-oxide nanoparticles. Biosensors & Bioelectronics. 2014;56:320–327. doi: 10.1016/j.bios.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 43.Jones SI, Tan Y, Shamimuzzaman M, Cunningham BT, Vodkin LO. Direct detection of transcription factors in cotyledons during seedling development using sensitive silicon photonic crystal protein arrays. Plant Physiology. 2015;167:639–649. doi: 10.1104/pp.114.253666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pineda MF, Chan LLY, Kuhlenschmidt T, Choi CJ, Kuhlenschmidt M, Cunningham BT. Rapid Specific and Label-Free Detection of Porcine Rotavirus Using Photonic Crystal Biosensors. IEEE Sensors Journal. 2009;9:470–477. [Google Scholar]
- 45.Huang MCY, Mateus CFR, Foley JE, Beatty R, Cunningham BT, Chang-Hasnain CJ. VCSEL optoelectronic biosensor for detection of infectious diseases. IEEE Photonics Technology Letters. 2008;20:443–445. [Google Scholar]
- 46.Shafiee H, Lidstone EA, Jahangir M, Inci F, Hanhauser E, Henrich TJ, Kuritzkes DR, Cunningham BT, Demirci U. Nanostructured Optical Photonic Crystal Biosensor for HIV Viral Load Measurement. Scientific Reports. 2014;4 doi: 10.1038/srep04116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen WL, Long KD, Yu HJ, Tan YF, Choi JS, Harley BA, Cunningham BT. Enhanced live cell imaging via photonic crystal enhanced fluorescence microscopy. Analyst. 2014;139:5954–5963. doi: 10.1039/c4an01508h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shamah SM, Cunningham BT. Label-free cell-based assays using photonic crystal optical biosensors. Analyst. 2011;136:1090–1102. doi: 10.1039/c0an00899k. [DOI] [PubMed] [Google Scholar]
- 49.Chan LL, George S, Ahmad I, Gosangari SL, Abbasi A, Cunningham BT, Watkin KL. Cytotoxicity Effects of Amoora rohituka and chittagonga on Breast and Pancreatic Cancer Cells. Evidence-Based Complementary and Alternative Medicine. 2011:1–8. doi: 10.1155/2011/860605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan LL, Gosangari SL, Watkin KL, Cunningham BT. A label-free photonic crystal biosensor imaging method for detection of cancer cell cytotoxicity and proliferation. Apoptosis. 2007;12:1061–1068. doi: 10.1007/s10495-006-0031-y. [DOI] [PubMed] [Google Scholar]
- 51.Chan LL, Gosangari SL, Watkin KL, Cunningham BT. Label-free imaging of cancer cells using photonic crystal biosensors and application to cytotoxicity screening of a natural compound library. Sensors and Actuators B-Chemical. 2008;132:418–425. [Google Scholar]
- 52.Fang Y, Ferrie A, Fontaine N, Ki Yuen P. Optical biosensors for monitoring dynamic mass redistribution in living cells mediated by epidermal growth factor receptor activation. Conf Proc IEEE Eng Med Biol Soc. 2005;1:666–669. doi: 10.1109/IEMBS.2005.1616501. [DOI] [PubMed] [Google Scholar]
- 53.Fang Y, Ferrie AM. Optical biosensor differentiates signaling of endogenous PAR1 and PAR2 in A431 cells. BMC Cell Biol. 2007;8 doi: 10.1186/1471-2121-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang Y, Ferrie AM. Label-free optical biosensor for ligand-directed functional selectivity acting on beta(2) adrenoceptor in living cells. FEBS Lett. 2008;582:558–564. doi: 10.1016/j.febslet.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 55.Fang Y, Ferrie AM, Fontaine NH, Mauro J, Balakrishnan J. Resonant waveguide grating biosensor for living cell sensing. Biophys J. 2006;91:1925–1940. doi: 10.1529/biophysj.105.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang Y, Ferrie AM, Fontaine NH, Yuen PK. Characteristics of dynamic mass redistribution of epidermal growth factor receptor signaling in living cells measured with label-free optical biosensors. Analytical Chemistry. 2005;77:5720–5725. doi: 10.1021/ac050887n. [DOI] [PubMed] [Google Scholar]
- 57.Fang Y, Ferrie AM, Li G. Probing cytoskeleton modulation by optical biosensors. FEBS Lett. 2005;579:4175–4180. doi: 10.1016/j.febslet.2005.06.050. [DOI] [PubMed] [Google Scholar]
- 58.Fang Y, Ferrie AM, Li G. Cellular functions of cholesterol probed with optical biosensors. Biochim Biophys Acta. 2006;1763:254–261. doi: 10.1016/j.bbamcr.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 59.Fang Y, Ferrie AM, Tran E. Resonant waveguide grating biosensor for whole-cell GPCR assays. Methods Mol Biol. 2009;552:239–252. doi: 10.1007/978-1-60327-317-6_17. [DOI] [PubMed] [Google Scholar]
- 60.Ferrie AM, Goral V, Wang C, Fang Y. Label-free functional selectivity assays. Methods Mol Biol. 2015;1272:227–246. doi: 10.1007/978-1-4939-2336-6_16. [DOI] [PubMed] [Google Scholar]
- 61.Pai S, Verrier F, Sun H, Hu H, Ferrie AM, Eshraghi A, Fang Y. Dynamic mass redistribution assay decodes differentiation of a neural progenitor stem cell. J Biomol Screen. 2012;17:1180–1191. doi: 10.1177/1087057112455059. [DOI] [PubMed] [Google Scholar]
- 62.Pattabiraman G, Lidstone EA, Palasiewicz K, Cunningham BT, Ucker DS. Recognition of apoptotic cells by viable cells is specific, ubiquitous, and species independent: analysis using photonic crystal biosensors. Molecular Biology of the Cell. 2014;25:1704–1714. doi: 10.1091/mbc.E13-11-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Budach W, Abel AP, Bruno AE, Neuschaefer D. Planar waveguides as high-performance sensing platforms for fluorescence-based multiplexed oligonucleotide hybridization assays. Analytical Chemistry. 1999;71:3347–3355. [Google Scholar]
- 64.Budach W, Neuschafer D, Wanke C, Chibout S-D. Generation of transducers for fluorescence-based microarrays with enhanced sensitivity and their application for gene expression profiling. Analytical Chemistry. 2003;75:2571–2577. doi: 10.1021/ac026390k. [DOI] [PubMed] [Google Scholar]
- 65.Ganesh N, Block ID, Mathias PC, Zhang W, Chow E, Malyarchuk V, Cunningham BT. Leaky-mode assisted fluorescence extraction: Application to fluorescence enhancement biosensors. Optics Express. 2008;16:21626–21640. doi: 10.1364/oe.16.021626. [DOI] [PubMed] [Google Scholar]
- 66.Ganesh N, Mathias PC, Zhang W, Cunningham BT. Distance dependence of fluorescence enhancement from photonic crystal surfaces. Journal of Applied Physics. 2008;103:083104. [Google Scholar]
- 67.Ganesh N, Zhang W, Mathias PC, Chow E, Soares JANT, Malyarchuk V, Smith AD, Cunningham BT. Enhanced fluorescence emission from quantum dots on a photonic crystal surface. Nature Nanotechnology. 2007;2:515–520. doi: 10.1038/nnano.2007.216. [DOI] [PubMed] [Google Scholar]
- 68.Pokhriyal A, Lu M, Huang C-S, Schulz SC, Cunning-ham BT. Multi-color fluorescence enhancement from a photonic crystal surface. Applied Physics Letters. 2010;97:121108–121110. doi: 10.1063/1.3485672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pokhriyal A, Lu M, Ge C, Cunningham BT. Coupled external cavity photonic crystal enhanced fluorescence. J Biophotonics. 2014;7:332–340. doi: 10.1002/jbio.201200173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mathias PC, Ganesh N, Zhang W, Cunningham BT. Graded wavelength one-dimensional photonic crystal reveals spectral characteristics of enhanced fluorescence. Journal of Applied Physics. 2008;103:094320. [Google Scholar]
- 71.Mathias PC, Wu HY, Cunningham BT. Employing two distinct photonic crystal resonances to improve fluorescence enhancement. Applied Physics Letters. 2009;95:021111. doi: 10.1063/1.3184573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu H-Y, Zhang W, Mathias PC, Cunningham BT. Magnification of photonic crystal fluorescence enhancement via TM resonance excitation and TE resonance extraction on a dielectric nanorod surface. Nanotechnology. 2009;21:125203–125210. doi: 10.1088/0957-4484/21/12/125203. [DOI] [PubMed] [Google Scholar]
- 73.Kim SM, Zhang W, Cunningham BT. Photonic crystals with SiO(2)-Ag “post-cap” nanostructure coatings for surface enhanced Raman spectroscopy. Applied Physics Letters. 2008;93 [Google Scholar]
- 74.Kim SM, Zhang W, Cunningham BT. Coupling discrete metal nanoparticles to photonic crystal surface resonant modes and application to Raman spectroscopy. Optics Express. 2010;18:4300–4309. doi: 10.1364/OE.18.004300. [DOI] [PubMed] [Google Scholar]
- 75.Wu HY, Cunningham BT. Plasmonic coupling of SiO2-Ag “post-cap” nanostructures and silver film for surface enhanced Raman scattering. Applied Physics Letters. 2011;98 [Google Scholar]
- 76.Yang FC, Cunningham BT. Enhanced quantum dot optical down-conversion using asymmetric 2D photonic crystals. Optics Express. 2011;19:3908–3918. doi: 10.1364/OE.19.003908. [DOI] [PubMed] [Google Scholar]
- 77.See GG, M.S N, Tang T, Bonita Y, Joo J, Trefonas P, Desh-pande K, Kenis PJA, Nuzzo RG, Cunningham BT. Region specific enhancement of quantum dot emission using interleaved two-dimensional photonic crystals. Applied Optics. 2015;54:2302–2308. doi: 10.1364/AO.54.002302. [DOI] [PubMed] [Google Scholar]
- 78.Block ID, Mathias PC, Ganesh N, Jones ID, Dorvel BR, Chaudhery V, Vodkin L, Bashir R, Cunningham BT. A detection instrument for enhanced fluorescence and label-free imaging on photonic crystal surfaces. Optics Express. 2009;17:13222–13235. doi: 10.1364/oe.17.013222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chan LL, Cunningham BT, Li PY, Puff D. A Self-Referencing Method for Microplate Label-Free Photonic-Crystal Biosensors. Sensors Journal, IEEE. 2006;6:1551–1556. [Google Scholar]
- 80.Tan Y, Chu A, Lu M, Cunningham BT. Distributed Feedback Laser Biosensor Noise Reduction. Sensors Journal, IEEE. 2013;13:1972–1978. [Google Scholar]
- 81.Ge C, Lu M, George S, Flood TA, Wagner C, Zheng J, Pokhriyal A, Eden JG, Hergenrother PJ, Cunningham BT. External cavity laser biosensor. Lab on a Chip. 2013;13:1247–1256. doi: 10.1039/c3lc41330f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang M, Ge C, Lu M, Zhang Z, Cunningham BT. A self-referencing biosensor based upon a dual-mode external cavity laser. Applied Physics Letters. 2013;102:213701. doi: 10.1063/1.4801427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang M, Lu M, Ge C, Cunningham BT. Plasmonic External Cavity Laser Refractometric Sensor. Optics Express. 2014;22:20347–20357. doi: 10.1364/OE.22.020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hasegawa T, Hara K, Hata S. Binding of dorzolamide its metabolite, N-deethylated dorzolamide to human erythrocytes in vitro. Drug Metabolism and Disposition. 1994;22:377–382. [PubMed] [Google Scholar]
- 85.Asher G, Dym O, Tsvetkov P, Adler J, Shaul Y. The Crystal Structure of NAD(P)H Quinone Oxidoreductase 1 in Complex with Its Potent Inhibitor Dicoumarol†. Biochemistry. 2006;45:6372–6378. doi: 10.1021/bi0600087. [DOI] [PubMed] [Google Scholar]
- 86.Lu J, Bai L, Sun H, Nikolovska-Coleska Z, McEachern D, Qiu S, Miller RS, Yi H, Shangary S, Sun Y, Meagher JL, Stuckey JA, Wang S. SM-164: A Novel, Bivalent Smac Mimetic That Induces Apoptosis and Tumor Regression by Concurrent Removal of the Blockade of cIAP-1/2 and XIAP. Cancer Research. 2008;68:9384–9393. doi: 10.1158/0008-5472.CAN-08-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Caserta TM, Smith AN, Gultice AD, Reedy MA, Brown TL. Q-VD-OPh, a broad spectrum caspase inhibitor with potent antiapoptotic properties. Apoptosis. 2003;8:345–352. doi: 10.1023/a:1024116916932. [DOI] [PubMed] [Google Scholar]
- 88.Calvaresi EC, Granchi C, Tuccinardi T, Di Bussolo V, Huigens RW, Lee HY, Palchaudhuri R, Macchia M, Martinelli A, Minutolo F, Hergenrother PJ. Dual Targeting of the Warburg Effect with a Glucose-Conjugated Lactate Dehydrogenase Inhibitor. ChemBioChem. 2013;14:2263–2267. doi: 10.1002/cbic.201300562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arnold S, Khoshsima M, Teraoka I, Holler S, Vollmer F. Shift of whispering-gallery modes in microspheres by protein adsorption. Optics Letters. 2003;28:272–274. doi: 10.1364/ol.28.000272. [DOI] [PubMed] [Google Scholar]
- 90.Vollmer F, Arnold S. Whispering-gallery-mode biosensing: label-free detection down to single molecules. Nat Meth. 2008;5:591–596. doi: 10.1038/nmeth.1221. [DOI] [PubMed] [Google Scholar]
- 91.White IM, Oveys H, Fan X. Liquid-core optical ring-resonator sensors. Optics Letters. 2006;31:1319–1321. doi: 10.1364/ol.31.001319. [DOI] [PubMed] [Google Scholar]
- 92.Fang W, Buchholz DB, Bailey RC, Hupp JT, Chang RPH, Cao H. Detection of chemical species using ultraviolet microdisk lasers. Applied Physics Letters. 2004;85:3666–3668. [Google Scholar]
- 93.Boudreau NJ, Jones PL. Extracellular matrix and integrin signalling: the shape of things to come. Biochem J. 1999;339(Pt 3):481–488. [PMC free article] [PubMed] [Google Scholar]
- 94.Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–80. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Biosensing with plasmonic nanosensors. Nat Mater. 2008;7:442–53. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 96.Zhu JG, Ozdemir SK, Xiao YF, Li L, He LN, Chen DR, Yang L. On-chip single nanoparticle detection and sizing by mode splitting in an ultrahigh-Q microresonator (vol 4, pg 46, 2010) Nature Photonics. 2010;4:122–122. [Google Scholar]
- 97.Rothenhäusler B, Knoll W. Surface-plasmon microscopy. Nature Nanotechnology. 1988;332:615–617. [Google Scholar]
- 98.Wang W, Tao N. Detection counting and imaging of single nanoparticles. Analytical Chemistry. 2014;86:2–14. doi: 10.1021/ac403890n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vedula R, Daaboul G, Reddington A, Ozkumur E, Bergstein DA, Unlu MS. Self-referencing substrates for optical interferometric biosensors. J Mod Opt. 2010;57:1564–1569. [Google Scholar]
- 100.Monroe MR, Daaboul GG, Tuysuzoglu A, Lopez CA, Little FF, Unlu MS. Single nanoparticle detection for multiplexed protein diagnostics with attomolar sensitivity in serum and unprocessed whole blood. Analytical Chemistry. 2013;85:3698–706. doi: 10.1021/ac4000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hu H, Mohseni PK, Pan L, Li X, Somnath S, Felts JR, Shannon MA, King WP. Fabrication of arbitrarily-shaped silicon and silicon oxide nanostructures using tip-based nanofabrication. J Vac Sci Technol B. 2013;31:06FJ01. [Google Scholar]
- 102.Sheehan PE, Whitman LJ, King WP, Nelson BA. Nanoscale deposition of solid inks via thermal dip pen nanolithography. Applied Physics Letters. 2004;85:1589–1591. [Google Scholar]
- 103.Gans R. Use the Form Ultramicroscopic Particles of Gold. Ann Phys. 1912;342:881–900. [Google Scholar]
- 104.Tian L, Chen E, Gandra N, Abbas A, Singamaneni S. Gold Nanorods as Plasmonic Nanotransducers: Distance-Dependent Refractive Index Sensitivity. Langmuir. 2012;28:17435–17442. doi: 10.1021/la3034534. [DOI] [PubMed] [Google Scholar]
- 105.Weber WH, Eagen CF. Energy transfer from an excited dye molecule to the surface plasmons of an adjacent metal. Optics Letters. 1979;4:236–238. doi: 10.1364/ol.4.000236. [DOI] [PubMed] [Google Scholar]
- 106.Glass AM, Liao PF, Bergman JG, Olson DH. Interaction of metal particles with adsorbed dye molecules: absorption and luminescence. Optics Letters. 1980;5:368–370. doi: 10.1364/ol.5.000368. [DOI] [PubMed] [Google Scholar]
- 107.Knoll W, Philpott MR, Swalen JD, Girlando A. Emission of light from Ag metal gratings coated with dye monolayer assemblies. Journal of Chemical Physics. 1981;75:4795–4799. [Google Scholar]
- 108.Hung Y-J, Smolyaninov II, Davis CC. Fluorescence enhancement by surface gratings. Optics Express. 2006;14:10825–10830. doi: 10.1364/oe.14.010825. [DOI] [PubMed] [Google Scholar]
- 109.Gryczynski I, Malicka J, Gryczynski Z, Lakowicz JR. Radiative decay engineering 4. Experimental studies of surface plasmon-coupled directional emission. Anal Biochem. 2004;324:170–182. doi: 10.1016/j.ab.2003.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Muskens OL, Giannini V, Sanchez-Gil JA, Rivas JG. Strong Enhancement of the Radiative Decay Rate of Emitters by Single Plasmonic Nanoantennas. Nano Lett. 2007;7:2871–2875. doi: 10.1021/nl0715847. [DOI] [PubMed] [Google Scholar]
- 111.Lakowicz JR. Radiative decay engineering: biophysical and biomedical applications. Anal Biochem. 2001;298:1–24. doi: 10.1006/abio.2001.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu Y, Blair S. Fluorescence enhancement from an array of subwavelength metal apertures. Optics Letters. 2003;28:507–509. doi: 10.1364/ol.28.000507. [DOI] [PubMed] [Google Scholar]
- 113.Chen Y, Munechika K, Ginger DS. Dependence of Fluorescence Intensity on the Spectral Overlap between Fluorophores and Plasmon Resonant Single Silver Nanoparticles. Nano Lett. 2007;7:690–696. doi: 10.1021/nl062795z. [DOI] [PubMed] [Google Scholar]
- 114.Zhang J, Matveeva E, Gryczynski I, Leonenko Z, Lakowicz JR. Metal-Enhanced Fluoroimmunoassay on a Silver Film by Vapor Deposition. Journal of Physical Chemistry B. 2005;109:7969–7975. doi: 10.1021/jp0456842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang J, Fu Y, Liang D, Zhao RY, Lakowicz JR. Enhanced Fluorescence Images for Labeled Cells on Silver Island Films. Langmuir. 2008;24:12542–12457. doi: 10.1021/la801749f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sabanyagam CR, Lakowicz JR. Increasing the sensitivity of DNA microarrays by metal-enahnced fluorescence using surface-bound silver nanoparticles. Nucleic Acids Research. 2007;35:e13. doi: 10.1093/nar/gkl1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moal EL, Leveque-Fort S, Potier M-C, Fort E. Nanoroughened plasmonic films for enhanced biosensing detection. Nanotechnology. 2009;20:225502. doi: 10.1088/0957-4484/20/22/225502. [DOI] [PubMed] [Google Scholar]
- 118.Pockrand I, Brillante A, Mobius D. Nonradiative Decay of Excited Molecules Near a Metal Surface. Chemical Physics Letters. 1980;69:499–504. [Google Scholar]
- 119.Zhang J, Fu Y, Chowdhury MH, Lakowicz JR. Metalenhanced signle-molecule fluorescence on silver particle monomer and dimer: coupling effect between metal particles. Nano Lett. 2007;7:2101–2107. doi: 10.1021/nl071084d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hayakawa T, Selvan ST, Nogami M. Field enhancement effect of small Ag particles on teh fluorescence from Eu(3+)-doped SiO2 glass. Applied Physics Letters. 1999;74:1513–1515. [Google Scholar]
- 121.Selvan ST, Hayakawa T, Nogami M. Remarkable influence of silver islands on the enhancement of fluorescence from Eu(3+) ion-doped silica gels. J. Phys. Chem B. 1999;103:7064–7067. [Google Scholar]
- 122.Ganesh N, Mathias PC, Zhang W, Cunningham BT. Distance dependence of fluorescence enhancement from photonic crystal surfaces. Journal of Applied Physics. 2008;103:083104. [Google Scholar]
- 123.Park JU, Hardy M, Kang SJ, Barton K, Adair K, Mukhopadhyay DK, Lee CY, Strano MS, Alleyne AG, Georgiadis JG, Ferreira PM, Rogers JA. High-resolution electrohydrodynamic jet printing. Nat Mater. 2007;6:782–789. doi: 10.1038/nmat1974. [DOI] [PubMed] [Google Scholar]
- 124.Park JU, Lee JH, Paik U, Lu Y, Rogers JA. Nanoscale patterns of oligonucleotides formed by electrohydrodynamic jet printing with applications in biosensing and nanomaterials assembly. Nano Lett. 2008;8:4210–4216. doi: 10.1021/nl801832v. [DOI] [PubMed] [Google Scholar]
- 125.Hong S, Zhu J, Mirkin CA. Multiple ink nanolithography: toward a multiple-Pen nano-plotter. Science. 1999;286:523–525. doi: 10.1126/science.286.5439.523. [DOI] [PubMed] [Google Scholar]
- 126.Salaita K, Wang Y, Mirkin CA. Applications of dip-pen nanolithography. Nat Nanotechnol. 2007;2:145–155. doi: 10.1038/nnano.2007.39. [DOI] [PubMed] [Google Scholar]
- 127.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 128.Pokhriyal A, Lu M, Chaudhery V, George S, Cunningham BT. Enhanced fluorescence emission using a photonic crystal coupled to an optical cavity. Applied Physics Letters. 2013;102:221114. doi: 10.1063/1.4809513. [DOI] [PMC free article] [PubMed] [Google Scholar]