Summary
Many efforts are being devoted to the design and engineering of metalloenzymes with catalytic properties fulfilling the needs of practical applications. Progress in this field has recently been accelerated by advances in computational, molecular and structural biology. This review article focuses on recent examples of oxygen-activating metalloenzymes, developed through the strategies of de novo design, miniaturization process and protein redesign. The considerable progress in these diverse design approaches have produced many metal-containing biocatalysts able to adopt functions of native enzymes or even novel functions beyond those found in Nature.
Graphical abstract
Several tools are nowadays available for the design and engineering of artificial oxygen-activating metalloenzymes, with efficiency comparable to natural enzymes.
1. Introduction
Enzymes are known to catalyze a wide variety of challenging chemical and biological reactions with high efficiency and selectivity, and these traits highlight how far Nature has evolved enzymes towards catalyst optimization. A primary example is selective activation of dioxygen, which is far from simple and still represents a major challenge for chemists.1 Natural enzymes manage this task in a very efficient way, by using cofactors, such as transition metal ions, highly conjugated organic molecules, or both.2,3 Oxygen-activating metalloenzymes predominately employ iron or copper ions within their active sites. For example, efficient dioxygen activation and substrate hydroxylation is carried out by the heme monoxygenases cytochromes P450 (CYPs), where the heme iron is coordinated by a thiolate of Cys residue.4 A variant of P450, OleTJE, is able to perform oxidative decarboxylation chemistry, through hydrogen peroxide activation.5,6 A wide range of reactions involving dioxygen are also catalyzed by non-heme iron enzymes. Those enzymes include mononuclear nonheme iron oxygenases containing a 2-His-1-carboxylate facial triad motif, such as the intradiol-cleaving catechol dioxygenases,7,8 and carboxylate-bridged diiron proteins, such as the soluble methane monoxygenases, which hydroxylate a variety of organic substrates,9-11 and the stearoyl-acyl carrier protein (ACP) Δ9-desaturase, which introduces a double bond into saturated fatty acids.12,13 In addition to iron, copper is also utilized in enzymes involved in O2 binding, activation and subsequent substrate oxidation.14-16 The O2-reactive centers can be mononuclear (type 2 copper), dinuclear (type 3 copper) or trinuclear (type 2 and type 3 copper), with no direct correlation between the type of copper centers and reactivity.14-16 The type 2 copper sites are found in enzymes such as Cu,Zn superoxide dismutase and in galactose oxidase. The simplest of the metalloproteins containing type 3 copper center are hemocyanin (Hc), tyrosinase (Tyr), and catechol oxidase (CatO). The type 3 sites are also found in the multicopper oxidases (MCOs), which include fungal laccase (Lc), ceruloplasmin and ascorbase oxidase.17 More recently it was discovered that cellulose and chitin degrading polysaccharide monooxygenase enzymes (PMO) use only a single Cu center for catalysis.18-22 The selected examples, mentioned above, are intended to emphasize the relevance of metal centers in biology for the activation of dioxygen and subsequent oxidative chemistry. This fascinating chemistry has received growing interest over the years, and continuous efforts are devoted in preparing models of different size and complexity, to mimic the structures and functions of the natural counterparts.
Extensive studies on natural proteins and small molecule synthetic catalysts have formed the basis of our current understanding of dioxygen activation at various metal centers. A comprehensive description of small molecule mimics is beyond the scope of this review, and readers can refer to several excellent reviews, which collect the main results in this area.23-33 It should be outlined that these systems, which mainly reproduce the metal-site structure of their biological counterpart, have been and still represent a benchmark for understanding structure-function relationship in metalloproteins. They have deeply contributed in unravelling the role of the metal ion in the catalytic pathway, thus allowing a better interpretation of the chemistry supported by the natural systems. Despite the important progress made, most synthetic models developed so far are not able to reproduce some critical features of biocatalysts, such as high turn-over numbers under physiological conditions and high selectivity. The studies on small-molecule catalysts clearly demonstrated that enzymatic catalysis requires not only information encoded in the metal-ligand complex, but also those embodied within the protein framework (e.g. the secondary coordination sphere),34 which is difficult to reproduce in small-molecule.
The desire to develop artificial metalloenzymes that match or even exceed the performance of natural enzymes has stimulated researchers toward the construction of functional metal sites into more elaborate architectures. Several strategies have been employed to construct peptide/protein-based artificial metalloenzymes.35-47 In particular, three different design approaches, namely miniaturization process,36,37, de novo design,35,38-41 and protein redesign,38,40-47 have been extensively and successfully used to develop metalloenzyme mimics, and will be discussed in this review. The increasing number of structural data nowadays available, together with advanced computational methods for structure and sequence prediction, allows for the design of uniquely packed structures and metal-binding sites within either de novo or native protein scaffolds. Computational algorithms have been developed for designing metal binding site, by searching protein structures for backbone geometries suitable to accommodate the amino acid ligands in the appropriate conformation for metal binding.48-52
Before examining specific cases, we wish to outline the critical issues to be addressed and optimized simultaneously, in designing and engineering functional metalloenzymes: 1) the correct fold of the protein scaffold; 2) the coordination requirements of the metal ion; 3) the stability/activity trade-off, which is essential for structure and function. First, the interactions that contribute to protein folding should be preserved when designing a metal binding site into a protein scaffold, mainly hydrophobic interactions that provide a powerful driving force for folding, polar interactions that are essential for conformational specificity, H-bonds that stabilize secondary and tertiary structure.41,53,54 Secondly, for the construction of the metal binding site, an appropriate number of coordinating amino acid residues should be placed around the metal center, in the proper position and conformation, in order to satisfy the coordination geometry dictated by enzyme activity. More importantly, secondary coordination interactions, which play an important role in positioning the ligand in the correct orientation for binding and in modulating the properties of the metal center, need to be carefully considered.34 Finally, successful design of a metalloenzyme requires a critical balance between conformational stability and catalytic activity of the enzyme. Structural metal-binding sites in proteins frequently achieve stability by binding metal ions in coordinately saturated ligand environments, with idealized ligand-metal bond geometries.55,56 On the other hand, catalytic centers require coordinately unsaturated metal ions, sometimes with unusual geometries, to properly position and bind the substrates, to lower the activation energy for catalysis, and undergo conformational changes to perform catalysis.55,57 Thus, a successful construction of a metalloenzyme reflects a delicate interplay between opposing requirements, that is tight binding of the metal cofactor versus function, as has been clearly highlighted through the analysis of artificial systems.57
Key lessons on how to construct a functional metalloenzyme came not only from those involved in oxidation reactions, but also other activities. They represent major breakthroughs in protein design and redesign, and are instructive for researchers in the field. Leading examples include the artificial metalloenzymes mimicking the catalytic activity of the natural enzyme carbonic anhydrase (CA), reported by Pecoraro and coworkers.58,59 Structurally much simpler than natural hydrolases, the artificial enzymes are made up by de novo designed three-stranded coiled-coils.60,61 The most active model comprises a Hg(II) ion for structural stability, and an active-site Zn(II) ion and catalyzes CO2 hydration with an efficiency comparable to some naturally occurring CAs.58 These studies proved that a functional site can be engineered into a complete different scaffold, respect to the native enzyme. The designed coiled coils demonstrated well suited to accommodate the threefold symmetric histidine binding site of CA, thus allowing to reproduce the core elements necessary for function.
Computational and directed evolution methods also represent key tools in protein design: their combitation has proven a valuable strategy for developing function metalloenzymes and offers a general approach to explore new reactivities. Milestone results were reported by Baker and coworkers.62 They used RosettaMatch and RosettaDesign methodologies50,51 to identify a set of mutations in an adenosine deaminase that allowed it to catalyze the hydrolysis of a model organophosphate, by the insertion of a zinc-binding site. The selected scaffold was further improved by directed evolution, affording a redesigned enzyme with a catalytic efficiency (kcat/KM) of ~104 M−1 s−1. Crystal structure of the enzyme confirmed that all but one of the designed residues adopt the designed conformation. These results show the power of the approach, which can be readily applied to the generation of biocatalysts for different reactions. They highlight the impact of joining computational design with directed evolution, in order to optimize the desired activities, by introducing key elements, missed in the first stage.
Engineering desired functionalities into native protein scaffold offers the advantage of bypassing the design of a stable protein structure. Lu and co-workers demonstrated that native myoglobin (Mb) scaffold is well suited for the construction of additional metal-binding sites.63-65 The engineering of a nonheme iron binding center, through the incorporation of three histidines and one glutamate in the distal pocket of myoglobin, afforded a nitric oxide reductase (NOR) mimic.63-65 This NOR mimic not only reproduced the heme-nonheme diiron center of NORs structurally but also functionally, by selectively reducing nitric oxide to nitrous oxide as in NORs.
All of the above examples demonstrate tremendous success achieved in protein design as a tool for understanding and mimicking native metalloenzymes. Success is measured not only by how closely the designed metalloenzymes resembles the native protein, but also by the insights gained from the design and engineering process. Numerous excellent reviews recently published report the milestones reached in the design of functional metalloenzymes.35,39,44,66-81 The aim of this review is to discuss the current research in the development of metalloenzymes active in oxidation chemistry, namely oxygenases, peroxidases and oxidases. Dioxygen activation and its transformation are fundamental for both chemistry and biology, as they play key roles in designing new catalysts for oxygen reduction reaction in fuel cells with high efficiency and for functionalization of organic substrates.1 The triplet ground state and strong O-O bond of molecular oxygen makes it kinetically inert and quite stable towards reaction at room temperature. Metalloenzymes utilize paramagnetic metal ions like copper and iron to bind, activate and cleave the O-O bond of dioxygen and generate reactive high-valent species capable of oxidizing various substrates. We will highlight the efforts in constructing catalysts able to promote selective oxidations, with high regio- and stereo-selectivity, using “clean” oxidants, such as dioxygen and its two proton, two electron reduced form - hydrogen peroxide. In the process, we will point out the challenges that have been faced and the successes achieved, by analyzing the results from the following perspectives: i) capability of breaking dioxygen bond and/or reacting with hydrogen peroxide to form reactive species; ii) regio-/chemo-selectivity of the catalyzed reactions; iii) turnover frequency/turnover number of the designed catalysts; iv) adaptability of the catalyst to be tuned to oxidize a different substrate; and v) potential of the catalysts to be used in vivo via whole-cell biotransformation for industrial purposes. The artificial enzymes herein examined have been grouped according to the design strategy adopted, namely miniaturization, de novo design, and protein redesign. Using selected examples, we will try to highlight the potential of each design strategy in expanding the repertoire of protein functionalities and to provide perspectives for future development.
2. Artificial oxygen-activating metalloenzymes designed through miniaturization
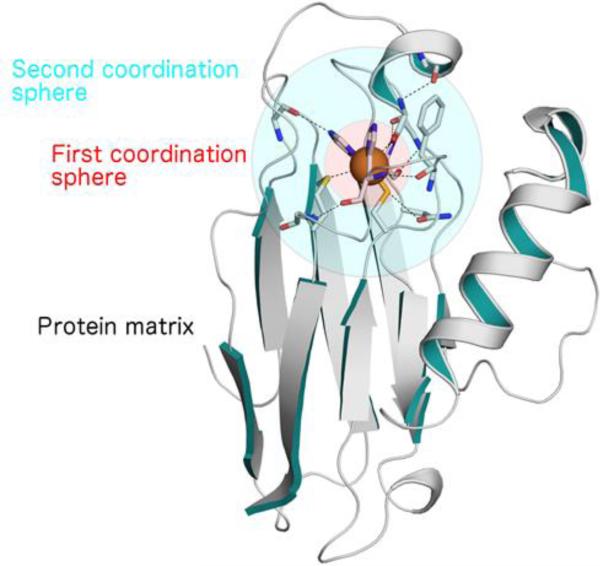
Many natural proteins have their functional properties located in discrete domains inside a large protein framework.82 In many cases, relatively few residues within these large surfaces actually participate in the catalytic activities. The approach of protein design through miniaturization aims to reduce the protein size to obtain small structured polypeptides or ‘mini-proteins’, still retaining the protein's functions. The goal of a miniaturization process is to design the minimal peptide sequence that contains sufficient information for proper folding and for an accurate reconstruction of the active site structure.36,37 This approach holds the advantage that the designed systems are generally simple enough, and therefore can be easily synthesized and characterized. Simultaneously, the polypeptide sequences are of sufficient size and chemical diversity to accommodate metal- binding centers, if any. An essential requirement for the miniaturization process is the structural knowledge of the parent system. More specifically, it is necessary to define: i) the type and number of constituents to be assembled; ii) the structure to be reconstructed and iii) the function to be reproduced. Metalloproteins, in particular, are well suited for miniaturization. The metal center represents a pivot point where spheres of variable diameters that circumscribe part of the protein can be centered.37 The larger the diameter of the sphere, the larger the number of constituents that must be included in the model. Figure 1 highlights this concept on the azurin crystal structure.83 Several strategies can be conceived to obtain a miniaturized model: for example, two or more parts pulled out from the entire protein could spontaneously associate to give folded non-covalent self-assembled oligomers, or two or more parts could be covalently connected through properly designed linkers to give folded monomers. Moreover, starting with the simplest molecular model, one can iteratively introduce further levels of complexity, as guided by experimental results, thus allowing selecting the minimal set of components dictating the desired structure, and finally housing the required function.
Fig. 1.
The concept of the miniaturization process highlighted on the azurin crystal structure (PDB code 4AZU). The metal center represents a pivot point where spheres of variable diameters that circumscribe part of the protein can be centered. The pink sphere includes all the elements of the first coordination sphere; the cyan sphere includes the elements of the secondary coordination sphere. The structure was generated with PyMol.335
One of the most studied family of oxidative enzymes that has been the subject of different miniaturization approaches is that of heme-containing enzymes.84-86 In peroxidases, catalases and monooxygenases, such as CYPs,87 catalysis take place at the heme iron center through the formation of a high-valent iron-oxo intermediate, whose fate depends on the specific environment created by the surrounding protein matrix.
A notable class of “mini-hemeproteins” with catalytic activity are the microperoxidases (MPs), derived from the proteolytic digestion of cytochrome c.88-91 The framework of MPs consists of a small peptide fragment (generally containing from 8 to 11 residues), containing the Cys-(Xaa)2-Cys-His motif, covalently linked to the heme c through thioether linkages with the two Cys residues.
The amino acid sequence of the polypeptide chain in MPs is numbered according to that of the parent cytochrome c, i.e., the two Cys residues occupy positions 14 and 17 of the peptide chain. Selective proteolysis of cytochrome c affords various MPs, differing in the peptide chain length.89 The best known among MPs is MP8, which is obtained from the tryptic digestion of horse heart cytochrome c; it retains the amino acid residues 14-21 of the starting protein. MP11 contains residues 11-21, and MP9 containing residues 14-22 have also been studied extensively. These MPs contain the minimal requirements for a heme-protein mimic, by retaining a His residue at position 18, which coordinates the heme iron and acts as proximal ligand. In addition, the sixth coordination site to the heme is open, occupied by a H2O molecule, which is readily displaced by an entering exogenous ligand.
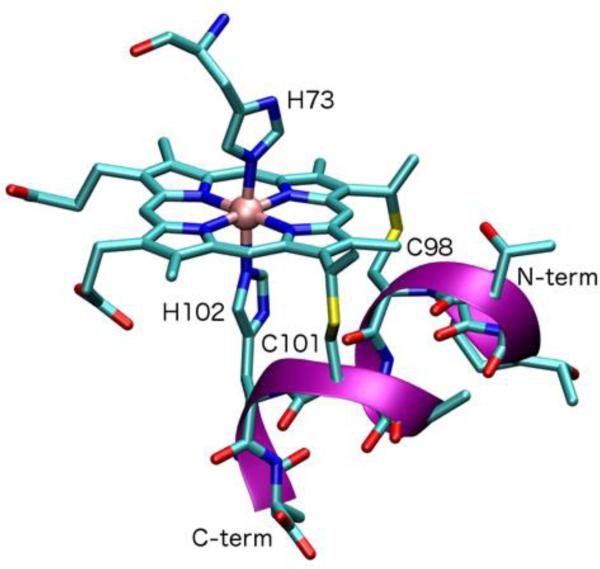
Despite their extensive use, no full atomic-resolution structure for any MP is available owing to the conformational flexibility of the peptide portion. Recently, a new MP analog, MP9cb562, was obtained from tryptic digestion of cytochrome cb562. The immobilization of MP9cb562 inside a protein lattice cage allowed the first crystallographic structure determination for any member of the MP family (Fig 2).92 MP9cb562 is anchored firmly to the protein surface through the coordination of His73 (coming from the protein cage) to the iron, affording a bis-His heme. The structure of MP9cb562 confirms that c-type linkages have little effect on the heme conformation and therefore may primarily serve other functions. For example such a linkage may affect the orientation of the coordinating His imidazole with respect to the porphyrin ring, which in turn may contribute to heme electronic asymmetry, and modulate electronic coupling to the heme iron.92
Fig. 2.
Crystal structure of MP9cb562, obtained from tryptic digestion of cytochrome cb562, distal position is occupied by the solvent exposed His 73 from the co-crystallized cytochrome cb562 (PDP ID: 3m4c).
One of the most interesting aspects of the MPs chemistry is that, despite their small size, they are able to selectively oxidize a variety of organic substrates, including ABTS, anilines, naphthols, phenols.93-99 MP-11 has been shown to oxidize sulfides enantioselectively with modest enantiomeric excess (ee) values (16-25%).98 However, the application of MPs as-catalysts are limited by their low stability under catalytic conditions. Accessibility of the distal side causes degradation of the porphyrin ring during catalysis, either by the direct action of H2O2 or by intermolecular reactions with another active iron-oxo species. Despite these limitations, MPs are still much more stable than simple protoporphyrin systems, indicating that the presence of the small peptide chain can play an important protective role and make the catalyst more robust. To minimize porphyrin degradation and to improve the stability of MPs for practical applications, several approaches have been undertaken. One approach deals with the encapsulation of these enzymes in mesoporous metal-organic framework (MOF).100-102 In a different approach, Casella and coworkers investigated the effect of distal residues on the formation of the active high-valent iron-oxo intermediate upon reaction with H2O2.90,91 Several MP8 derivatives were synthesized by covalent linkage to the N-terminal amino group of one (Pro-MP8) or two (Pro2-MP8) proline residues, or a single proline N-protected by an aromatic fluorenyl group (Fmoc-Pro-MP8).99 The proline-modified MP-8 showed a faster rate of oxidation of p-cresol with hydrogen peroxide relative to MP-8. Further studies revealed that the reactivity of MP8 derivatives is controlled by the polarity around the heme iron. In Fmoc-Pro-MP8, the amine protecting group reduces the polarity and introduces some steric hindrance and as a consequence the catalytic rate constant drops down with respect to the value observed for MP8 (k1=390 M−1 s−1 and k1=680 M−1 s−1, for Fmoc-Pro-MP8 and MP8, respectively). In Pro-MP8, the protonated proline -NH2+ group acts as an internal acid-base catalyst in the heterolytic cleavage of the heme-bound peroxide, thus increasing the activity of the catalyst (k1=1100 M−1 s−1). Moreover, the addition of a second Pro residue further reduces the distance between the proline N atom and the peroxide oxygen atom, increasing the k1 value for Pro2-MP8 (k1=2000 M−1 s−1).99
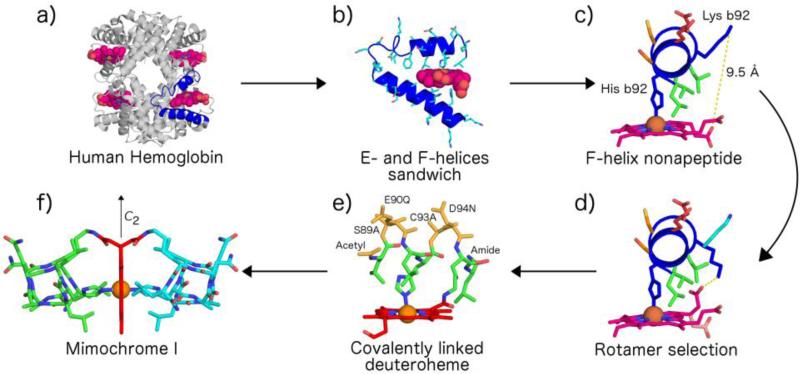
Minimal heme-proteins able to promote oxidation reaction (named mimocromes) have been successfully obtained by Lombardi, Pavone and co-workers through miniaturization.84,85,103-113 This approach allowed to design the shortest peptide sequences that could serve the same functions of the peptide chain in the natural proteins (Fig. 3a and 3b). A detailed analysis of heme-protein structures revealed that the prosthetic group in natural heme-proteins is strongly kept inside the protein structure by a large number of interactions, which were replaced in mimochromes by a few strong local constraints. The prototype molecule, mimochrome I, was patterned on the F helix of hemoglobin β-chain. Figure 3c shows the X-ray structure of β-chain Leu88-Leu96 F helix fragment facing the heme group in human deoxyhemoglobin.114 Simple molecular modeling studies showed that a change of conformation of the heme propionyl group and of Lys95 sidechain from a folded state to an extended state was sufficient to bring the propionyl carboxyl moiety and the lysine ε-amino functional group within bonding distance (Fig. 3d). This covalent bond appeared as a minimum requirement for positioning the helical scaffold in close proximity of the heme iron center and to drive the peptide chain to cover the heme face upon His axial coordination. N-terminal acetylation and C-terminal amidation were then added to avoid the presence of end charges that might affect the helix stability. Residues Ser89, Glu90, Cys93 and Asp94 were replaced with Ala, Gln, Ala and Asn, respectively, in order to simplify the synthetic procedure (Fig. 3e). The resulting sequence appeared to have a high propensity to assume the desired helical folding, due to the presence of five helix inducing residues (Leu1,4,9, Ala2,6) in a peptide of nine residues and to the N- and C-terminal protections.115-117 Deuteroporphyrin was preferred as a cofactor over the more common protoporphyrin IX to avoid degradation of the sensitive vinyl substituents during the synthesis. Finally, the helix-heme-helix sandwich mimochrome I, shown in Figure 3f was obtained by applying a C2 symmetry operation.103
Fig. 3.
Design through miniaturization: from hemoglobin (PDB ID: 2hhb) to mimochrome I. a), b) Active site identification and extraction. c) Isolation of heme covering nonapeptide. d) Mutual approach of heme propionate towards Lys92 by proper rotamer selection. e) Selection of solvent exposed residues and protoporphyrin IX to deuteroporphyrin mutation. f) Symmetry-generated hexacoordinated deuteroheme. The structure was generated with PyMol.335
The spectroscopic and structural features of mimochrome I indicated that it binds cobalt and iron in a low-spin bis-His–ligation, as envisioned in the design, even though some unexpected features were observed.103-105 The iron complex showed quite low solubility in water (in the μM range), thus limiting its possible application as a functional heme protein mimetic, whereas the cobalt complex was highly soluble (up to mM concentration). This different behavior has been ascribed to the stronger cobalt-histidine coordination, that stabilizes the sandwich structure. Therefore, aggregation by stacking of the porphyrin ring occurs in the iron complex, causing a lower water solubility.
Starting from mimochrome I prototype, numerous compounds have been gradually produced in order to favor the formation of a well-defined structure and to improve the water solubility.106-109 In particular, mimochrome II and IV feature a stable structure, with increased solubility in aqueous solution (up to mM concentration), either as free-base form, or as iron and cobalt complexes. To achieve this aim, two re-design strategies were used. The first strategy focused on peptides with longer (14 residue) sequences, modeled in both α-helical (residues 1-10) and extended (C-terminal part) conformations, leading to mimochrome II.106 The second strategy consisted in the introduction, into the mimochrome I sequence, of amino acid substitutions able provide intra-molecular, inter-helical interactions, leading to mimochrome IV.107,108
Taking into account the lesson learned on the bis-His-ligated complexes, attempts were made to stabilize a five-coordinate heme complex, apt to perform catalysis. A proximal and a distal site environment of the heme were designed, such that: i) a tetradecapetide (TD) bearing a His residue at position 6 as the axial ligand to the heme forms the proximal face and ii) a decapeptide (D) devoid of any heme-coordinating residue, and creating a cavity around the metal ion forms the distal face. As in mimochrome I, the two peptide chains embrace the metalloporphyrin in a fashion that the helical segments run antiparallel to each other and the helix axes are about parallel to the porphyrin plane. Stabilization of the tertiary structure was contributed by inter-chain ion-pairs between the carboxylate side chains of glutamate residues (Glu2) on one helix and the guanidine groups of arginines (Arg10) on the other helix, strategy proven to be effective in mimochrome IV design.107 Moreover, the positively charged Arg10 and the negatively charged Glu2 at the C-terminal and N-terminal ends respectively (with opposite sign relative to the helix dipole) may also provide stabilization to the secondary structure. Finally, several glutamines (Gln3,4,8) and a serine (Ser7) were introduced in the solvent exposed positions to improve water solubility. The analogue mimochrome VI thus designed (Fig. 4) embodies some of the key elements for functioning as peroxidase-like catalyst.110,111 The spectroscopic and functional characterization indicated that it functions as an efficient heme protein model, with a peroxidase-like catalytic activity. In the presence of hydrogen peroxide, it efficiently catalyzes the oxidation of several substrates exhibiting multiple turnovers. Mimochrome VI peptide framework, despite its small structure (a total of 24 amino acid residues), confers high efficiency to the porphyrin cofactor. Three important outcomes deserve highlighting: i) FeIII-mimochrome VI efficiently catalyzes the oxidation of different substrates, such as ABTS and guaiacol, by activating H2O2, and efficiently catalyzes the nitration of phenols; ii) FeIII-mimochrome VI displays a very high specific activity (104 mol mg−1 s−1 for ABTS oxidation), with respect to highly purified horse radish peroxidases (HRPs) (91 mol mg−1 s−1 for ABTS oxidation at pH 4.6) iii) FeIII-mimochrome VI exhibits multiple turnover kinetics: more than 4000 turnovers were observed in the ABTS oxidation, without bleaching. The evidence that both peptide chains modulate the properties of FeIII-mimochrome VI is a significant result, in terms of peroxidase activity. In fact, the derivative lacking the distal D chain showed higher bleaching susceptibility, as indicated by the much lower turnover number, suggesting a protective role of the D chain.
Fig. 4.
FeIII-Mimochrome VI and its analogues. Left: molecular model of FeIII-mimochrome VI, highlighting the R10-E2 ion pair interactions. Right: Peptide sequences of Mimochrome VI and its analogues: acid, basic and non-polar residues, in position 2 and 10, are indicated in red, blue and green, respectively. Reprinted with permission from ref. 113. Copyright (2015) Royal Society of Chemistry.
The success of the initial design demonstrates that it is possible to incorporate a functional heme site within a minimal scaffold. Therefore, modification of the structure, through specific amino acid substitutions in the first and/or in the secondary coordination shell, allowed for the structure–activity relationship studies of mimochromes. To explore the role of charges towards the peroxidase activity, Glu2 and Arg10 residues were individually substituted in both the peptide chains with an uncharged residue.113 These substitutions were expected to partly destabilize the molecules due to the consequent removal of inter-chain and/intra-chain interactions. To compensate this structure destabilization, leucine (Leu) was selected as the substituting residue, because of its high α-helical propensity, and for the potential hydrophobic interaction with the porphyrin. As a result of these substitutions, four analogues were obtained: Glu2Leu(TD), Glu2Leu(D) and Arg10Leu(TD), Arg10Leu(D). Figure 4 reports the sequence modifications of the new analogues respect to mimochrome VI, by using single letter code for amino acids, and the acronym MC6 for FeIII-mimochrome VI.
Table 1 lists the catalytic parameters for all the analogues. When compared to MC6 catalytic properties, kcat values are about 2-fold higher in the analogues substituted at the (TD) chain. The best performances were obtained for E2L(TD)-MC6, which displays improvement in the apparent catalytic constant (kcat = 7.8 × 102 s−1), and efficiency for both H2O2 and ABTS (kcat/KM = 25 mM−1 s−1, and 16 × 103 mM−1 s−1, respectively). In contrast, the analogues with the altered D chain showed an almost unmodified reactivity, with respect to MC6. The E2L(D)-MC6 analogue was found to be the worst catalyst. Its kcat is 3.8 × 102 s−1, and the kcat/KM values are 4.0 mM−1 s−1 for H2O2 and 3.3 × 103 mM−1 s−1 for ABTS.
Table 1.
MC6 and its analogues steady-state kinetic parameters. HRP parameters are provided for comparison.a
| Enzyme | pH | H2O2 | ABTS | H2O2 | ABTS | ||
|---|---|---|---|---|---|---|---|
| KM (mM) | KM 102 (mM) | kcat 10−2 (s−1) | kcat/KM (mM s−1) | kcat/KM 10−3 (mM s−1) | T.O.N. 10−3 | ||
| MC6 | 6.5 | 44 ± 2 | 8.4 ± 0.2 | 3.7 ± 0.1 | 8.4 ± 0.6 | 4.4 ± 0.2 | 4.0 |
| E2L(TD)-MC6 | 6.5 | 31 ± 2 | 5.0 ± 0.4 | 7.8 ± 0.6 | 25 ± 3 | 16 ± 2 | 5.9 |
| R10L(TD)-MC6 | 6.5 | 54 ± 2 | 3.8 ± 0.1 | 6.8 ± 0.3 | 13 ± 1 | 18 ± 1 | 5.6 |
| E2L(D)-MC6 | 6.5 | 96 ± 7 | 11 ± 1 | 3.8 ± 0.3 | 4.0 ± 0.6 | 3.4 ± 0.6 | 3.6 |
| R10L(D)-MC6 | 6.5 | 18 ± 1 | 3.0 ± 0.2 | 1.7 ± 0.1 | 9 ± 1 | 5.7 ± 0.7 | 3.3 |
| HRP | 4.6 | 0.85 ± 0.01 | 107 ± 1 | 62 ± 1 | (7.3 ± 0.2) 103 | 5.8 ± 0.1 | 50 |
| HRP | 7.0 | (1.15 ± 0.01) 10−2 | (5.1 ± 1.2) 102 | 0.52 ± 0.04 | (4.6 ± 0.5) 103 | (1.0 ± 0.3) 10−2 | 0.3 |
Adapted from ref. [113]
All MC6 analogues were able to perform several thousands of turnovers, without degradation. The turnover numbers (T.O.N.) reflect the trend of the catalytic efficiency, being higher in E2L(TD)-MC6 and lower in R10L(TD)-MC6. Based on the E2L(TD)-MC6 model, it is possible to hypothesize a catalytic role of R10 on the (D) chain. Its side chain may approach the ferric ion and interact, by hydrogen bonding, with the sixth ligand to the heme. Hence, R10 is supposed to mimic R38 in the HRP distal site and to enhance the efficiency of the compound I formation and ligand binding.118 E2L(TD)-MC6 behaves like natural peroxidases, whose reactivity is similarly influenced by the proximal and distal heme environments. Moreover, the E2L(TD)-MC6 catalytic cycle occurs with a peroxidase-like mechanism, through the formation of compound I, containing a FeIV=O center and a porphyrin radical cation, as evidenced by spectroscopic analysis.
Collectively, the experimental results allowed to point out the principle in heme-protein reactivity: slight structural modifications in the heme-distal pocket may greatly influence the peroxidase-like activity. Moreover, the characterization of this class of minienzymes demonstrates that their miniaturized structure holds essential elements to finely tune the reactivity. This represents a very important checkpoint for the rational design of new and improved bio-mimetic catalysts, tailored for specific applications. The overall results demonstrate that particular attention in future design should be paid in order to: i) stabilize both the helical and the sandwiched structures by introducing additional structural constraints; ii) favor the His-H2O coordination over a wider pH range; iii) introduce amino acid residues assisting the acid–base catalytic cleavage of the O–O bond, as His42 acts in HRP.119
3. Artificial oxygen-activating metalloenzymes by de novo design
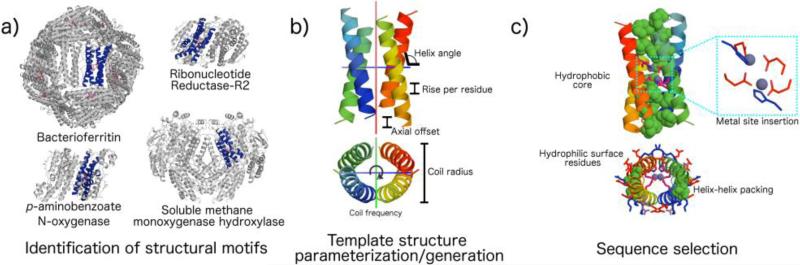
De novo metalloprotein design combines the fundamental knowledge of protein design and biomimetic chemistry, and can be defined as design “from first principles”. The strategy involves the construction of a peptide sequence that is not directly related to any natural protein and is able to fold into a unique, well-defined three-dimensional structure, and to incorporate a metal cofactor.39-41 De novo design is a valuable approach to analyze the factors that tune the structural and functional properties of metal binding sites in proteins. The possibility to isolate and investigate the active site of functional metalloproteins in a smaller, well-defined, model system allow researchers to accurately evaluate the role of first and secondary shell interactions in modulating metalloproteins activity. Figure 5 illustrates the principal steps in the process of de novo protein design, applied by DeGrado, Lombardi and coworkers to the development of diiron oxo protein models, DFs.120-122 The choice of the structural motif to be reproduced is a critical first step in de novo design process (Fig. 5a). A simple structural motif, very common among natural, functionally diverse proteins, including many metalloproteins, is the four-helix bundle.53,123 This motif is compact, thermodynamically stable and able to tolerate residue substitutions, deletions and insertions without perturbing the global folding. As a consequence, it represents a designable template in de novo design. The second step of de novo design involves the generation of the backbone coordinates from scratch (Fig. 5b), in order to gain high quality backbone templates, that are crucial for the success of the design process. Two main approaches are currently adopted to generate backbone coordinates.119 One approach (referred to as ideal backbone parameterization) consists in modelling folded structures through a few effective parameters, out of the very large number of parameters required to exactly define the geometry of a protein.124 It has been demonstrated, for some selected folds, that the approach can describe the structural variability, producing deviations between ideal and real structures within 1 Å. Examples of this include Crick parameterization of coiled coils,124-126 mathematical description of β-barrel structures,127 statistical parameterization of the structure of collagen,128 and parameterization of di-iron helical bundles129,130 and of transmembrane helix interaction geometry.131 The second approach consists in the fragment-based tertiary structure composition. Fragmentation of protein structures reduces the continuous protein structural space to a “parts list” of polypeptide building blocks.132 Since the pioneer study of Baker and colleagues,133 several groups adopted peptide fragment databases132 or atomistic tertiary fragment search,134-137 substantially mining the Protein Database for robust and conserved backbone arrangements apt to design.
Fig. 5.
Crucial steps in protein de novo design: from diiron proteins to DFs. a) The four-helix bundle structural motifs has been identified as a designable template, for its wide occurrence in natural diiron oxo proteins (bacterioferritin, PDB ID: 4am5, ribonucleotide reductase-R2, PDB ID: 1syy, p-aminobenzoate N-oxygenase, PDB ID: 3chh, and soluble methane monoxygenase hydroxylase, PDB ID: 1mty are reported). b) Geometrical parametrization has been used to generate backbone coordinates from scratch. c) The proper sequence has been selected, in order to stabilize the desired tertiary structure, and to include the metal-binding residues. The DF1 structure and details of the diiron site are depicted (PDB ID: 1ec5). The structures were generated with PyMol.335
The final step in de novo protein design (Fig. 5c), points to carefully identify the sequences and their properties in order to stabilize the selected structure and to include the desired function. This step requires the introduction and optimization of a large number of short- and long-range non-covalent interactions, which all together contribute to the stability of a protein.41,53,54,138,139 This includes the correct placement of hydrophobic and hydrophilic amino acids into the sequence, H-bonds and electrostatic interactions and the intrinsic conformational propensity of each amino acid. In addition, for a de novo metalloprotein design, metal-binding residues need to be included, in the proper position and orientation for metal ion coordination (Fig. 5c). Finally, the sequence should also contain elements of negative design, which aim to stabilize the target fold and destabilize any other alternative folds, thus providing conformational specificity. For example, inclusion of a large number of hydrophobic residues in a designed protein can lead to multiple, stable conformations; therefore, a correct balance between non-polar and polar interactions are required for correct folding and function.41,53,140,141 Examples of negative design include i) the placement of hydrophilic groups that are solvent exposed in the desired fold but buried in alternatively folded structures; ii) inclusion of buried H-bonds and salt bridges that are less stabilizing than hydrophobic interactions, but can be formed only in the desired structure; iii) solvent-accessible electrostatic interactions that are weakly stabilizing of the desired fold but strongly destabilize alternatives. Other elements of negative design include conformational restraints from residues such as Gly and Pro that are often used to interrupt elements of secondary structure. Shape complementarity and, more important, a lack thereof provide a third medium for negative design.
Because of the complexity of protein folding, an iterative process is often necessary for de novo designing a metalloprotein with a native-like metal-binding structure. It has been demonstrated that, through several cycles of design, synthesis, characterization and redesign, it is now possible to fine tune the structural properties of initial model, and to tailor functional metal-site into the interior.122 Outstanding reports have shown that α-helical bundles can be designed with the highest degree of confidence. In particular, the “rules” that control stability, oligomerization, helix-helix orientation and metal coordination are now well established and a variety of de novo designed α-helical coiled coils and bundles, with native-like structures, have been reported.34,35,40,41,54 Since the first examples of designed proteins and metalloproteins,41 much progress has been made in the construction of functional metalloproteins, with efficiency approaching that of their natural counterparts. Some successful examples of de novo designed metalloproteins with oxygen-activation activity are summarized in the next paragraphs, with main focus on de novo design of diiron oxo proteins and heme-proteins.
3.1 Artificial oxygen-activating metalloenzymes by de novo design of diiron oxo proteins
The ‘Due Ferri’ (two-iron; DF) family of proteins is a product of de novo protein design, successfully applied by DeGrado, Lombardi and coworkers for the development of artificial oxygen-activating metalloenzymes.120-122 The overall results obtained on DF family demonstrated that the DF structure represents an excellent scaffold for hosting different activities. The earliest developed models have contributed to our understanding of the principles governing protein folding, stabilization, as well as metal coordination and substrate binding. Through redesign processes, functional metalloproteins were successful produced. The progress in DF models, described in the following paragraphs, clearly illustrate that, starting from a very stable de novo designed scaffold, a relatively small number of amino acid mutations allowed to shift from inactive to active proteins, or even to produce a remarkable switch of activity.
Although inspired by highly complex natural diiron proteins,9,142-147 the original DF1 protein was designed using mathematical parameterization of the backbone (Fig. 5).129,130 DF1 is an antiparallel dimer of helix-loop-helix motif (α2), capable of binding di-metal cofactor close to the center of the structure. The metal-binding site comprises four glutamates and two histidines as first-shell ligands, which are positioned in the core of the protein by hydrogen bonded interactions with two aspartate, tyrosine and lysine residues.
Through an iterative process of design and characterization, catalytic diiron-oxo sites were successfully engineered into the prototype DF1 structure. However, adding functionality in this stable, uniquely folded DF1 protein required expansion of the active site to accommodate substrates. Towards this aim, several DF1 analogues were designed, with changes in both the sequence and loop conformation.148-155 The original DF1 protein had a pair of Leu residues at positions 9 and 13 (and 9’ and 13’ of neighboring helices), which entirely blocked access to the metal-binding site. By decreasing the bulk of Leu13 with Ala or Gly, the formation of a cavity just above the di-metal site was obtained, as indicated by crystal structure analysis.151-153 However, to allow rapid and efficient substrate access and binding, it appeared that it would be necessary to simultaneously mutate Leu9 to a smaller residue.
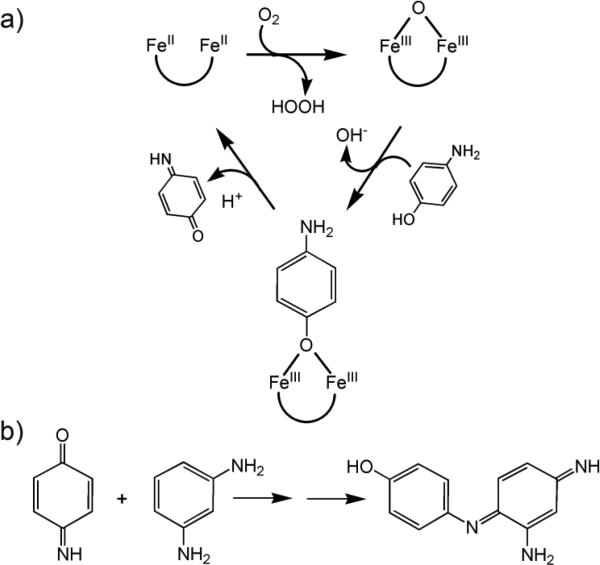
The sequence requirements for the catalytic activity were first evaluated in a combinatorial manner through the design of a four-chain four-helix bundle assembly composed of four unconnected helical peptides (DFtet).156-158 A series of asymmetrical variants were synthesized and mutations were performed at positions 15 and 19 (corresponding to the positions 9 and 13, respectively, of DF1). By mixing and matching different DFtet peptides, it was possible to produce catalysts for the oxidation of 4-amino-phenol (4-AP).158 The reaction involves the use of O2 to oxidize the diferrous protein to a diferric species (see Scheme 1). The diferric protein then reacts with the substrate 4-AP, producing benzoquinone monoimine. The reduced diferrous form is then oxidized by O2, thereby initiating another catalytic cycle. The released quinone monoimine product is quenched and spectroscopically detected by reacting with m-phenylen-diamine.159,160 The designated G4-DFtet analogue, in which both Leu-15 and Ala-19 in two peptide chains were substituted with Gly, enhanced the reaction rate by ≈1000-fold relative to the background reaction. The G4-DFtet exhibited at least 100 turnovers with a catalytic efficiency of kcat/K = 1,540 M−1 min−1 (see Table 1). Mutating either of the Gly residues at positions 19 or 15 to Ala decreased the rate by approximately 2.5- to 5-fold.158 However, the complex stoichiometry, marginal stability (in the absence of metal ions), and tendency to undergo ligand-exchange reactions of these assemblies hampered any attempts to fully characterize their three-dimensional structure and catalytic properties.
Scheme 1.
a) Proposed mechanism for the oxidation of 4-AP. b) Detection of the produced quinone monoimine by reaction with m-phenylen-diamine.
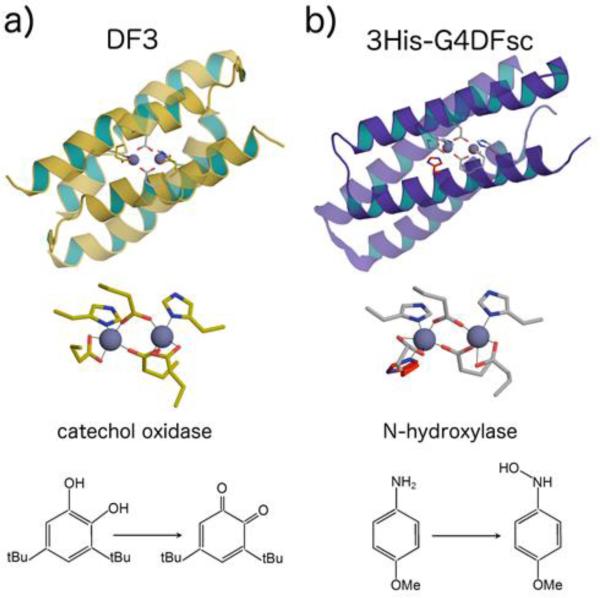
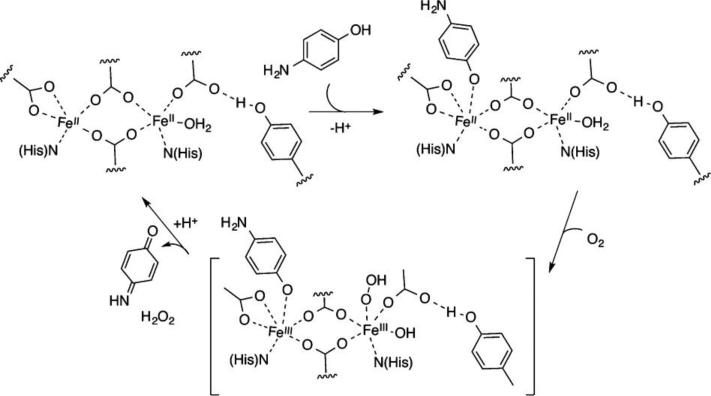
Following this work, the authors sought to introduce a phenol-oxidase activity into the original DF1 helix-loop-helix scaffold, similar to the naturally occurring alternative oxidase (AOX) and plastid terminal oxidase (PTOX). These catalysts cycle between di-FeII and di-FeIII states as they reduce O2, and then use the oxidizing equivalents to convert quinols to quinones.161 To make an enzyme to reach an activity similar to PTOX, it was essential to design a protein that is capable of: 1) generating a di-FeII cofactor ready for dioxygen oxidation; 2) stabilizing the resulting di-FeIII cofactor, which otherwise would be highly insoluble in aqueous solution; 3) binding phenols to allow two-electron oxidation of the substrate and regeneration of the di-ferrous cofactor; 4) releasing product, with concomitant entry into a new catalytic cycle. Furthermore, it was essential to engineer the system so that each reaction occurs in a two-electron process, thereby avoiding Fenton chemistry, destruction of the catalyst, and accumulation of toxic radicals and by-products. Thus, it appeared that the design strategy should consider both the functional requirements already highlighted through the DFtet protein subset as well as a careful redesign of the structural framework. It was therefore necessary to redesign the loop to compensate for stabilizing interactions lost during the incorporation of four glycines into the hydrophobic core (a single mutation of Leu13 to Gly destabilized DF1 by 10.8 kcal mol−1 dimer−1).57 To increase the conformational stability of the DF scaffold, the sequence of the inter-helical turn was modified, affording the helix-loop-helix dimer DF3 (Fig. 6a).162,163 This analogue in the apo- form is only slightly less α-helical than the holo form, but its thermodynamic stability outperforms DF1 mutants. DF3 retained the catalytic activity already observed for G4-DFtet. In the presence of ambient oxygen, di-FeIII-DF3 followed Michaelis-Menten kinetics in the oxidation of 4AP, with values of 1.97 ± 0.27 mM and 2.72 ± 0.19 min−1, respectively, for KM and kcat (Table 1, kcat/KM = 1,380 M−1 min−1). Measurement of the reaction over the course of an hour indicated that the protein was capable of at least 50 turnovers. As intended for this particular design, the active site cleft in DF3 could accommodate a larger substrate, such as 3,5-ditert-butyl-catechol (3,5-DTBC). Di-FeIII-DF3 catalyzed the oxidation of this catechol derivative to the corresponding quinone (3,5-DTBQ), with a fivefold higher value of kcat/KM than that for 4-AP, reflecting an increase in kcat (Table 2). Amino-aniline substrates were expected to bind more weakly to the diferric center. Indeed, kcat/KM for the two-electron oxidation of para-phenylenediamine (PPD) was 75-fold lower than the value for 3,5-DTBC, and no catalysis was observed for ortho-phenylenediamine (OPD).
Fig. 6.
Catalytically active DF compounds. a) Solution structure of Zn-DF3 (PDB ID: 2kik), and substrate conversion catalyzed by its iron derivative. b) Solution structure of Zn-3His-DFsc (PDB ID: 2lfd), and substrate conversion catalyzed by its iron derivative. The structure was generated with PyMol.335
Table 2.
Kinetics parameters obtained for the oxidation of the different substrates.a
| Protein | Substrate | KM (mM) | kcat (min−1) | kcat/KM (M−1min−1) |
|---|---|---|---|---|
| di-Fe(III)-DF3 | 3,5-DTBC | 2.09±0.31 | 13.2 | 6,315 |
| 4-AP | 1.97±0.27 | 2.72 | 1,380 | |
| PPD | 8.87±2.58 | 0.73 | 83 | |
| OPD | Not detected | |||
| G4-DFtet | 4-AP | 0.83±0.06 | 1.30 | 1,540 |
Adapted from ref. [162]
The encouraging results obtained with DFtet and DF3 analogues inspired the redesign of DFsc, the single-chain version of DF1,164 The DFsc scaffold is made up by a single polypeptide chain, in which the four helices are linked by three computationally designed loops. Four glycine residues were incorporated along the substrate access channel, affording the G4DFsc variant.165 Functional and spectroscopic analyses of this protein provides a detailed understanding of how solvent exposure and active site ligation impact O2 reactivity. G4DFsc, as for the above mentioned members of the DF family with unencumbered substrate access channels, successfully catalyzed the two-electron oxidation of 4-aminophenol to the corresponding quinone immine. However, some drawbacks arose: substitutions of four Gly residues into the DFsc scaffold resulted in an apo form slightly less α-helical than the holo form. In addition, better active-site accessibility increases the exposure of the iron atoms to the aqueous solvent, which renders them prone to hydrolysis. This reduces the stability of the Fe-protein complex and leads to the precipitation of iron oxides. Further spectroscopic analyses of G4DFsc showed that O2 binds to one iron and undergoes a 2-electron reduction to form an hemerythrin-like end-on hydroperoxo ferric intermediate. A larger access channel to the active site induces solvent coordination at the di-iron center, thus preventing O2 from binding to both iron ions to form a bridged peroxy species, as observed in previous DFs. The coordinative saturation of the iron ions may account for the lack of oxidase activity of the the diferric species. As a consequence, G4DFsc oxidase activity is observed when substrate binding to the biferrous site preceded O2 binding (Scheme 2).166,167
Scheme 2.
Proposed mechanism for the oxidation of 4-AP by G4DFsc. Reprinted with permission from ref. 166. Copyright (2015) American Chemical Society.
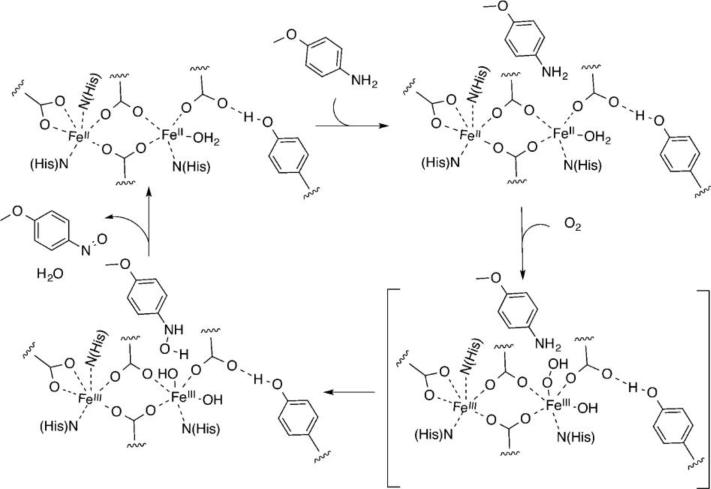
The reactivity potential achieved in the DFsc scaffold has been recently demonstrated by the switching of its activity from phenol oxidase to N-hydroxylase, through the incorporation of four mutations at different levels: one first-shell, two secondary shell and one third-shell.165 An additional iron-binding histidine was introduced in the active site to mimic the active site of the natural diiron oxo protein p-aminobenzoate N-oxygenase (AurF).168 The steric clashes caused by the insertion of an additional ligand in the active site were overcome by insertion of computationally screened mutations. The resulting 3His-G4DFsc (Fig. 6b) catalyst showed oxygenase activity, efficiently converting p-anisidine to the corresponding hydroxylamine, with loss of hydroquinone oxidation activity under steady state conditions. Spectroscopic studies demonstrated that the substrate is bound in the active site, which is able to cycle between bisferrous and ferric states, allowing for several turnovers to take place (Scheme 3).166 Furthermore, the loss of oxidase activity was attributed to the coordinative saturation of the metal center by addition of the third histidine leading to a very slow substrate binding kinetics, which may be rate limiting.166
Scheme 3.
Proposed mechanism for the oxidation of p-anisidine by 3HisG4DFsc. Reprinted with permission from ref. 166. Copyright (2015) American Chemical Society.
The overall work done on DF proteins highlights the power in using de novo design for the development of simplified metalloproteins as tools for future investigations on the geometric and electronic aspects that tune the diiron-oxo site properties, and for the design of novel catalysts with a wide variety of reactivities.
3.2 Artificial oxygen-activating metalloenzymes by de novo design of heme-proteins
Efforts to develop heme protein models by de novo design has greatly increased over the years, due to the important roles played by heme, which includes dioxygen transport and storage, electron transfer, and dioxygen activation. The high level of confidence reached in the design of structured four-helix bundles allowed the construction of several heme protein mimetics, in which the four-helix bundles serve as scaffolds for mono-heme and multi-heme binding.35 First and secondary shell interactions were accurately introduced in order to accomplish specific functions, i.e. electron transfer,169-172 oxygen binding,173 hydroxylase,174 oxygenase,175 peroxydase176-178 activities in de novo designed proteins.
Several approaches have been undertaken to develop functional heme-proteins by de novo design, and examples include the template-assembled synthetic four-helix bundle proteins,175,179-183 and the construction of variable gene libraries.176-178,184-186
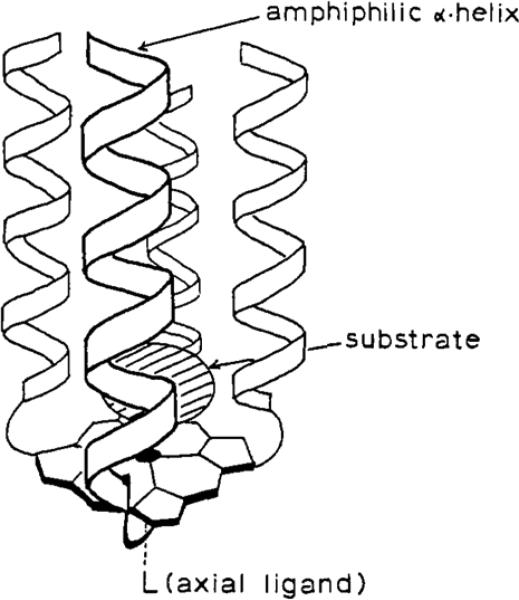
The first example of a functional heme protein mimetics, Helichrome, has been reported by Sasaki and Kaiser.174 They used the Template-Assembled Synthetic Proteins (TASP) approach, introduced by Mutter and coworkers, that consists in selecting a proper scaffold for the covalent assembly of a variety of tertiary structures.187-189 Helichrome represents a pioneering example of a catalytic heme-containing four-α-helix bundle, in which the porphyrin is also used as a template to generate a C4-symmetric four-α-helix bundle. This molecule is made up by four identical helical 15-residue peptide chains, N-terminally linked to the four propionic groups of coproporphyrin I. Helichrome was designed to mimic the hydroxylase activity of CYPs, whose structure is characterized by the presence of several α-helices surrounding the heme group forming a hydrophobic substrate recognition site. To this aim, the peptide sequence in helichrome was chosen such that it could stabilize an amphiphilic helix and create, in the folded state, a substrate binding hydrophobic pocket distal to the porphyrin ring, as schematically depicted in Fig. 7. The FeIII complex, similar to CYPs, catalyzed the hydroxylation of aniline to form p-aminophenol, using O2 as oxidant, in the presence of 7-acetylflavin and NADPH as sacrificial reductants, with kcat and KM values of 0.02 min−1 and 5.0 mM, respectively. Several hemeproteins, such as hemoglobin, indoleamine 2,3-dioxygenase, and L-tryptophane 2,3-dioxygenase have been reported to catalyze the hydroxylation of aniline in the presence of oxygen and appropriate reducing agents with kcat and KM ranging from 0.02 to 0.65 min−1 and from 3.7 to 5.4 mM, respectively.174 Comparison of kcat and KM values clearly demonstrate that helichrome has a hydroxylase activity quite similar to that of native hemeproteins. Interestingly, the rigid hydrophobic pocket formed by the peptide chains plays a key role towards helichrome activity (potentially by bringing the substrate in close proximity of the heme iron) as the iron(III) complex of the isolated coproporphyrin showed negligible catalytic activity under similar conditions.
Fig. 7.
Proposed structure of helichrome after folding of the peptide chains. Reprinted with permission from ref. 174. Copyright (1989) American Chemical Society.
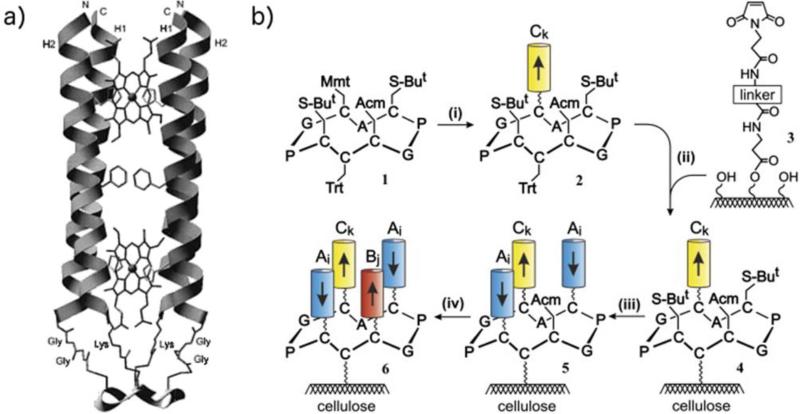
Following a similar approach, Haehnel and coworkers developed Modular Organized Proteins, MOPs, as suitable scaffolds for the incorporation of a variety of functions.175,179-182 The first molecule capable of binding heme, MOP1, was engineered using four α-helical peptides covalently connected to a cyclic decapeptide template.179 Subsequently, a bis-heme-binding protein was constructed on the basis of the cytochrome b subunit of the mitochondrial cytochrome bc1 complex. Two helices, H1 and H2, were designed in an attempt to reproduce the main features of the cytochrome b A-D helices: H1 constitutes the heme-binding helices and were covalently linked to the template in a parallel orientation. They run antiparallel to a parallel H2 helix couple, which plays the role of shielding the heme-binding pocket against the solvent (Fig. 8a). The spectroscopic properties of the bound heme resembled those of the natural protein suggesting the successful assembly of the model heme protein. Further improvement in the design of MOPs resulted in the development of artificial heme proteins with oxygenase activity, supported on cellulose membranes.175 To this aim, a library of mono-His-ligated hemeproteins was prepared using the peptide sequences derived from MOP1. A suitable chemo-selective synthetic strategy was adopted in order to control the directionality of the helical segments (Fig. 8b). Amino acid residues presumed to be in contact with the heme group were varied, in order to optimize heme binding and heme oxygenase activity. Heme oxygenase is an unusual enzyme in that the substrate heme serves also as cofactor. It catalyzes the degradation of heme to verdoheme, which can be followed in vitro in the presence of ascorbate as electron donor. A total of 352 synthetic heme proteins were screened on the cellulose support, by monitoring the kinetics of verdoheme formation, which showed first-order rate constant k1 values ranging by more than two orders of magnitude, between 0.1×10−3 and 50·10−3 ×min−1. Four out of 352 proteins were selected to cover a broad range of enzymatic activity (k1 in the range 2.6×10−2 to 0.01×10−2 min−1) for full characterization in solution. Faster reaction rates k1 of the soluble as compared to the immobilized proteins were observed. Moreover, two proteins showed reaction rates comparable to those of myoglobin mutants.175 Solution characterization revealed that the heme oxygenase activity was strictly related to the heme coordination and redox potential. In conclusion, the TASP approach was effective in the construction of stable four-helix bundles housing a functional heme, and showed very promising for engineering different scaffolds housing a variety of metal-binding sites.
Fig. 8.
De novo designed MOPs. a) Model of MOP1 bis-heme complex. The backbone is shown as ribbon. Amino acid side chains are shown to illustrate the thioether linkage of the four Cys of the template with the N-terminal two Gly of helix H1 and the C-terminal Lys of helix H2. The His residues which bind the two heme groups and the Phe residue in the center of helix H1 are presented as stick structures. N and C indicate the N- and C-terminus of helices H2 and H1, respectively. Reprinted with permission from ref. 179, Copyright (1998), American Chemical Society. b) Modular protein synthesis on peptide-templates linked to cellulose. The four-helix bundle protein consists of three different helix types, Ai (used twice in the assembly), Bj, and Ck, of which helix Bj provides the His residue as ligand to the heme. Reprinted with permission from ref. 175, Copyright (2007), Elsevier.
The screening of a library of variants was also used by Hecht and co-workers for the design of heme binding proteins based on the four-helix bundle scaffold.176-178,184-186 These authors developed a method for designing libraries of novel proteins that relies on the “binary patterning” of polar and nonpolar amino acids.190,191 Binary patterning incorporates polar and nonpolar amino acids in accordance with the structural periodicity of the target secondary structure. This binary code does not require to explicitly specify the residue identity, therefore the method has great potential for combinatorial diversity.192 Several libraries of sequences were generated and tested. Screening of hundred arbitrarily chosen sequences afforded many sequences able to bind heme; the resulting proteins had activities ranging from electron transfer184,185 to carbon monoxide binding.186 Furthermore, several designed proteins exhibited peroxidase activity at rates rivaling natural peroxidases.176 More recently, the authors sought to mimic natural selection by introducing random mutations to produce variants with improved activity.178 Directed molecular evolution on two previously characterized proteins, S824 and S836, yielded novel mutated sequences. The authors developed two different screens for peroxidase activity: a 96-well format that allowed for screening several hundred sequences, and a colony-based assay that allowed screening of thousands of clones. Several rounds of screening and selection allowed identifying in the progeny two mutants that were nearly three-fold more active than the parental sequence S836. The overall results demonstrate that the binary-patterned sequences, even though not related to any natural protein and not specifically designed for any catalytic activity, are useful starting materials for directed evolution designs. The peculiarity of the overall approach lies on the application of evolution to novel sequences that can be specified toward a desired biological function.
A further example of a de novo designed four-helix bundle heme protein with peroxidase activity is the MiniPeroxidase 3 (Fig. 9).193 The design was inspired by the bacterioferritin and HRP structures. By covalently linking deuteroporphyrin to two peptide chains of different compositions, an asymmetric α2/heme/α2 sandwich was obtained. The protein holds: i) a His residue on one chain that acts as an axial ligand to the iron ion; ii) a vacant distal site able to accommodate exogenous ligands or substrates; and iii) an Arg residue in the distal site that should assist hydrogen peroxide activation as in HRP. The MiniPeroxidase 3 was synthesized and characterized as its iron complex, and its spectroscopic and catalytic properties were compared with those of natural systems. FeIII-MiniPeroxidase 3 shows high catalytic turnover (kcat=535 s−1) and efficiency (kcat/KM=3.1 mM−1s−1) in the oxidation of ABTS by H2O2. However, the observed activity is lower respect to natural HRP, probably due to the mobility of the catalytic Arg in the distal site.
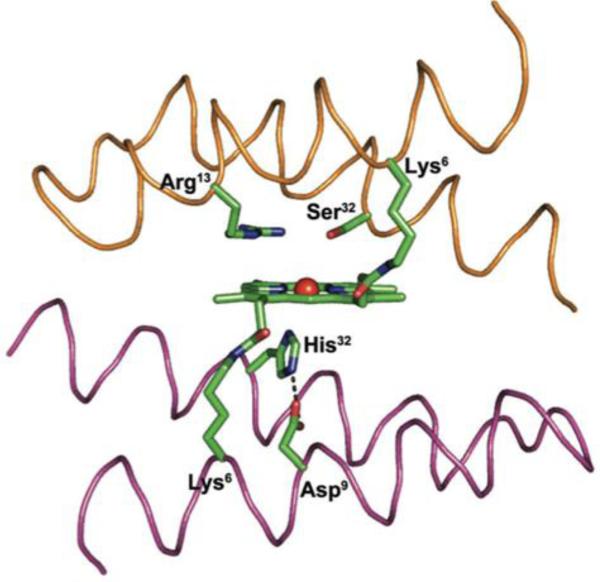
Fig. 9.
Computer model of MiniPeroxidase 3. Trace representation of the helices with key residues depicted as sticks: the His32-coordinating residue and the hydrogen-bonded Asp9 residue on the proximal site, the catalytic Arg13 residue on the distal site, and the heme group, which is covalently linked to the peptide chains through the side chain of two lysine residues. Adapted with permission from ref. 193, Copyright 2012 Wiley.
In conclusion, all the examples discussed above demonstrate the successful use and implementation of de novo design strategy to obtain heme-enzymes with increased catalytic efficiency, turnover numbers, and robustness.
4. Artificial oxygen-activating metalloenzymes by protein redesign
As outlined in the previous section, de novo protein design has been quite successful in developing artificial metalloenzymes. Despite the success, this approach is limited by the types of de novo designed scaffold (mainly based on α-helical bundles), thus by the range of protein environment available for controlling activity and selectivity of the metal-binding sites. Since the types of native protein scaffolds are much more than that of de novo designed scaffolds, designing artificial enzymes using native scaffolds can compensate for this particular limitation of de novo design approach, but a careful choice of the scaffold must be performed.38,42-44 Fortunately, there are several natural protein scaffolds available with high stability and therefore much more tolerant to mutations for the construction of a metal center, making it more likely for industrial applications. Nature is a master at taking a stable scaffold that has been evolved for millions of years and engineering different active sites into the same scaffold for different functions. Learning this “trick” from nature is an important component of artificial enzyme designs.
Using this approach, artificial oxygen-activating enzymes have been produced by modification of native protein scaffolds, by introducing nonnative cofactors, and by incorporation of metal cofactors into native scaffolds that do not contain metal ion.
4.1. Artificial oxygen-activating enzymes by modification of native protein scaffolds
Fantastic progresses has been made in modification of native protein scaffolds to either improve the enzyme activity, broaden the substrate scope, or introduce new functions by engineering new metal-binding sites.42-45,47 There are generally two approaches for protein design and engineering by modifications of native protein scaffolds.44 One is rational design based on knowledge of the desired chemical reaction, the original protein scaffold, and structure–function relationships from either previous experiments or computational modeling.42,50 Although this approach has been successful in designing new biocatalysts, it cannot be applied in the absence of structural information on the proteins. In this case, an alternative powerful strategy is the combinatorial design and directed evolution of new metalloenzymes.194-197 Directed evolution is a mimic of natural “Darwin evolution,” in which desired properties of proteins are obtained by in vitro or in vivo screening of mutant libraries constructed by random mutations, saturation mutagenesis at certain sites or gene shuffling.194-197 For many artificial enzyme designs, a combination of both approaches is often necessary in order to achieve the goal. In this section, we will report relevant results on iron- and copper-containing metalloenzymes, in which oxygen activation function of the native protein has been either improved or completely altered, or where new functionality has been engineered by modifying the protein scaffold. Since the field is rapidly moving forward, recent advances exploiting directed molecular evolution for the purposes of the design of oxygen-activating enzymes will also be described.
4.1.1 Heme proteins
One of the most successful artificial oxygen-activating enzymes produced by modification of native protein scaffolds are engineered CYPs, which are known to catalyze efficient and selective C-H bond functionalization using O2 as an oxidant, a major challenge in both chemical and biological catalysis.198 The use of oxygen as the oxidant also makes these enzymes very useful for chemical transformation in whole cells through fermentation.199-201 More than 11,500 distinct CYPs have been identified. Despite variations in amino acid sequences, different CYPs utilize the same conserved structural fold to hydroxylate a wide range of substrates. This generalizability of CYPs, along with the potential of their use in whole cell production of chemicals, have led to highly focused efforts in tuning their substrate binding pocket, thus expanding their substrate scope.202,203 The overall goal is to develop CYPs capable of performing highly selective C-H bond functionalization on any substrate of interest.
The resting state of CYP is the unreactive H2O-ligated low-spin ferric state; the binding of substrates, typically hydrocarbons, causes the transition of heme iron from the low-spin to the high-spin state and excludes water molecules from the site. These changes result in raising the reduction potential of the heme iron, making it possible for the resting Fe(III) heme to be reduced by its redox partner, thus allow oxygen binding and activation to as the initial step of oxidative catalysis.199-201 As a result of this requirement of electron transfer, almost all CYPs require a reductase to deliver electrons in order to function, which makes it difficult for industrial applications, because a separate reductase has to be produced in order to couple the electron transfer step with O2 activation. To overcome this limitation, a soluble, catalytically self-sufficient monooxygenase from bacillus megaterium called P450BM3 have been engineered by fusing an N-terminal heme domain (BMP) to a C-terminal FAD/FMN containing reductase domain (BMR) in a single polypeptide chain.204 The resulting enzyme complex performs hydroxylation of long chain (C12–C20) fatty acids, and their alcohols and amides, and the epoxydation of unsaturated fatty acids with the highest catalytic activity determined for a P450 monooxygenase (17,000 turnovers/min),205 due to efficient electron transfer between the reductase and the heme.204 Because the crystal structure of P450BM3 in complex with the substrate is available, and since the enzyme complex expresses extremely well in E. Coli, it represents an excellent platform for biocatalysis. Therefore, huge progress has been made by protein engineering techniques, such as site-directed mutagenesis, site-saturation mutagenesis, directed evolution or a combination of the three approaches, in enabling the creation of a wide range of P450BM3 variants with novel substrate selectivity.202-204,206-210
Directed evolution on P450BM3 has been used by Arnold and coworkers to increase substrate promiscuity of CYPs toward short-chain alkanes, such as ethane and propane, with strong C-H bonds.198,206,207 With the ultimate aim of engineering an ethane hydroxylase, the authors adopted an evolution strategy where they accumulated multiple generation of random as well as site-saturated mutagenesis and screening.206 The authors concentrated on the high-resolution crystal structure of enzyme bound to palmitoglycine substrate and chose amino acid residues that directly interact and bind either the heme or the substrate (Fig. 10). One of the mutants selected from this approach, called 53-5H, was shown to catalyze 5000 turnovers of propane hydroxylation, at a rate of 370 min−1, and at least 8000 turnovers of octane hydroxylation, at a rate of 660 min−1. Notably, the mutant 53-5H hydroxylated ethane to form ethanol as the sole product, albeit with a very slow rate of 0.4 min−1.Interestingly, 53-5H contained three active site mutations A78F, A82S and A328F, all of which replace alanine with a larger side chain and presumably reduce the volume of the active site and position small alkanes above the heme during catalysis.206
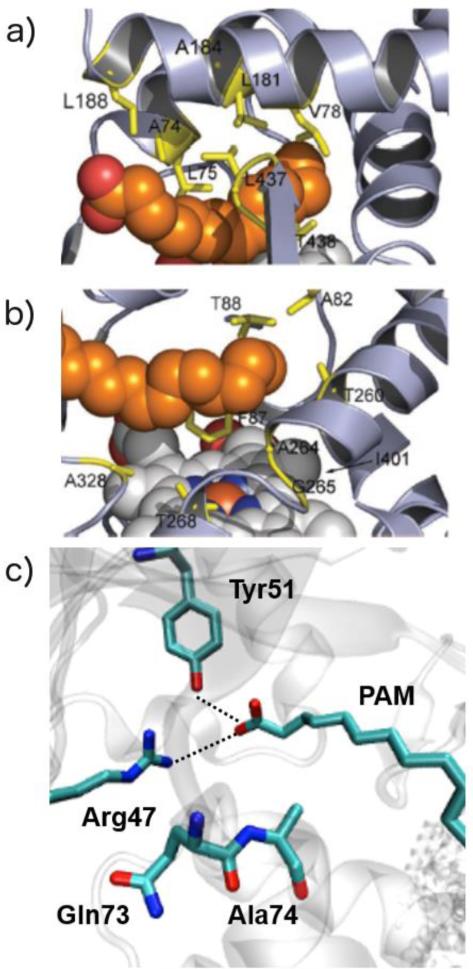
Fig. 10.
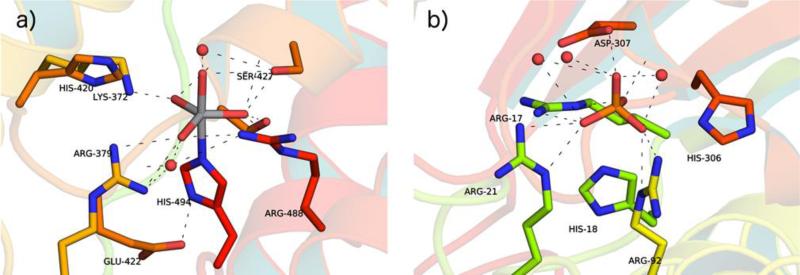
a) Substrate channel and b) active-site residues targeted for saturation mutagenesis mapped on the palmitate-bound structure of P450BM3 heme domain (PDB 1FAG). Heme (white) and fatty acid (orange) are shown in space -filling mode. c) H-bonding interactions binding the palmitoleic acid substrate in P450BM3 (PDB ID: 1FAG). Adapted from ref. 207, Copyright 2007 Wiley-VCH.
Even though the 53-5H attained thousands of turnovers in vitro, the utility of this catalyst remained limited because of its poor performance to produce products in whole cells, which was mostly due to the low efficiencies for coupling the product formation to cofactor consumption (17.4% for propane and 0.01% for ethane oxidation). To overcome this limitation, the authors used a domain-based protein-engineering strategy, in which the heme, flavin mononucleotide (FMN), and flavin adenine dinucleotide (FAD) domains of the P450BM3 variant were evolved separately in the context of the holoenzyme, and beneficial mutations were recombined in a final step. The enzyme P450PMO R2 obtained through the above approach had 11 mutations, most of which were clustered around the FAD domain and the nearby linker of the FMN. Previous chemical and thermal denaturation studies have shown that FMN is the most weakly bound cofactors in P450BM3, thus the mutations improved the coupling between various cofactors, and, in turn, improved the overall enzyme activity and robustness. The enzyme P450PMO R2, with improved activity and stability, not only supported more than 45,000 turnovers for propane hydroxylation in vitro, but also performed whole-cell biohydroxylation of propane at room temperature and pressure with air as oxidant.207
To exploit the potential of P450BM3 towards hydroxylation of drugs or polycyclic aromatic hydrocarbons (PAHs), Gilardi and coworkers used random mutagenesis to generate a library of mutants with novel activities.208-210 A double mutant, called A2, containing the Asp251Gly/Gln307His mutations, was found to catalyze specific hydroxylations of diclofenac, ibuprofen and tolbutamide.208 A2 was able to bind and turn over tolbutamide with rate constants higher than those measured for ibuprofen and diclofenac (kcat=0.94 min−1, kcat=0.10 min−1, kcat=0.048 min−1, for tolbutamide, ibuprofen and diclofenac, respectively). Even though the catalytic activity for A2 was low when compared to the values calculated for the known substrate lauric acid (kcat=1810 min−1 for WT P450BM3, and kcat=697 min−1 for A2, respectively), new binding and catalytic abilities have been introduced in the enzyme by mutating only two amino acids in positions not directly involved in substrate binding or turnover.
In subsequent studies, the authors used directed evolution of P450BM3 for the recognition and oxidation of chrysene and pyrene.209 Three different mutants, named M3, P2 and K4, showed higher affinity and coupling efficiency for both substrates with faster rates of product formation compared with the wild type enzyme. DNA sequencing of the mutants revealed the presence of 6 mutations in M3 (Asp251Gly, His266Ala, Glu267Arg, Thr269Asn, Ser270Glu, and Gln307His), 4 mutations in P2 (Asp208Arg, Ile209Tyr, Asp251Gly, and Gln307His), and only one single mutation, Val317Cys for K4. Measurement of the performance in terms of the reaction rate and relative coupling efficiency of the mutants and WT was carried out in reference to the production of 6-hydroxychrysene and 1-hydroxypyrene, respectively. Comparison between the data obtained from the mutants M3, P2 and K4 and chrysene and pyrene showed that both the rate of product formation and relative coupling efficiencies (calculated from the ratio between hydroxyl-product formed and NADPH consumed) were higher for pyrene than chrysene. These results are impressive demonstrations of how directed evolution may provide enzymes with new biocatalytic capabilities. This approach represent a powerful tool for producing protein variants with mutations in random positions, even far from the active site, otherwise unpredicted to affect the enzyme functionality by using a rational approach.
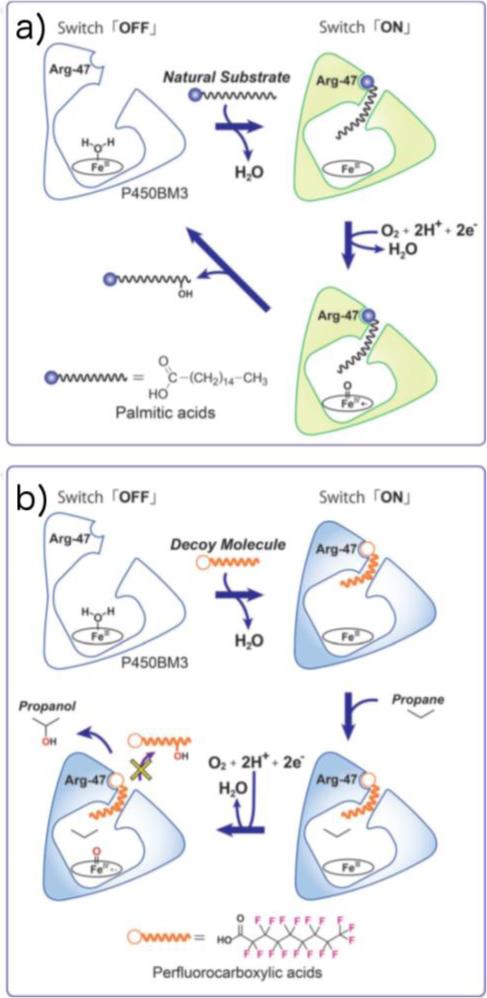
Site-directed mutagenesis is an excellent strategy to increase the substrate scope of CYPs; however, it often results in a lower yield and stability than those of the native enzyme. As an alternate strategy, Reetz211 and Watanabe212-214 groups have used small decoy molecules into “fooling” CYPs to catalyze C-H bond functionalization on non-native substrates. The crystal structure of palmitoleic acid bound P450BM3 shows that the substrate is fixed by two major interactions (1) hydrophobic interaction of the alkyl chain with amino acids at the substrate binding site and (2) interaction of the substrate carboxylate group with Tyr51 and Arg47 (Fig. 10c). Both of these interactions were required for the proper placement of the substrate in the active site of enzyme such that the enzyme is triggered ‘on’, reduced and then binds O2 at the heme iron to perform the catalytic C-H bond oxidation. Building on this observation, Watanabe and coworkers designed perfluorinated versions of long chain fatty acid substrates (called PFCs) as they would “trick” CYPs into believing that the substrate was recognized and hence remain switched on for catalytic hydroxylation of non-native substrates (Fig. 11). The PFCs were expected to initiate the activation of molecular O2 in the same manner as do the long-alkyl-chain fatty acids and induce the generation of Compound I (the key oxidizing species in CYPs) to oxidize non-native substrates like propane, while PFCs will never be oxidized due to their stronger C-F bonds (~116 kcal/mol). The first generation PFCs, thus designed, contained 9-13 carbon atoms giving enough space for the binding of propane at the catalytic heme center. As was expected, the product formation rates were highly dependent on the alkyl-chain length of PFCs, with the PFC10 displaying the highest rate of product formation (67 min−1 for propane).213 Building upon previous substrate occupancy studies on CYPs, the authors designed decoy molecules with increased H-bonding interactions at the substrate binding site. These second generation PFCs not only showed at least 100-fold improved binding affinity to P450BM3 but also increased the turnover rates to 256 min−1 for propane and 45 min−1 for ethane.214 Most importantly, there were no over-oxidation products for these transformations, which encouraged the authors to investigate the hydroxylation of aromatic compounds using a series of PFCs (PFC8 – PFC12). PFC9, in particular, afforded the largest turnover rate of 120 min−1 indicating that the active site provided by PFC9 was suitable for the accommodation of benzene. Interestingly, the o-position of monosubstituted benzenes were selectively hydroxylated, regardless of the substituents, suggesting that the structural recognition is responsible for the selectivity of CYP reaction.212 At the same time, PFCs of different chain lengths were screened to obtain a suitable activator for selective hydroxylation of propane and butane isomers. Remarkably, the PFC of formula CF3(CF2)7COOH could hydroxylate methane to methanol, the “holy grail” of C-H bond functionalization through dioxygen activation, displaying more than 2000 turnovers.211 These results on CYPs suggest that the use of decoy molecules is an excellent technique to alleviate enzymes from their dependence on substrate cofeeds and expand their substrate scope.
Fig. 11.
Schematic of the reaction mechanisms of a) the natural reaction system and b) the decoy molecule system of P450BM3. a) Subterminal carbons of fatty acids are hydroxylated in the natural reaction. b) By simple addition of a decoy molecule such as PFC9, the hydroxylation reaction of a small alkane (for example, propane) is catalyzed by P450BM3 because of substrate misrecognition of P450BM3. Reprinted with permission from ref. 212, Copyright 2015 Wiley.
Moreover, decoy molecules can eventually be combined with traditional mutagenesis on the target enzyme to increase its catalytic efficiency. The CYPs are extremely versatile in that they not only perform selective C-H bond hydroxylation but also other transformations including epoxidation, oxidative deformylation, dehydrogenation, rearrangements, Baeyer-Villiger oxygenation, and oxidative decarboxylation.215 Recently, a novel class of nitrating CYP TxtE have also been discovered that catalyze the direct and regioselective aromatic nitration using oxygen and nitric oxide as substrates.216 This generalizability in the CYPs reactivity has encouraged researchers to expand its reactivity (through directed evolution and replacement of axial cysteine residue) to include different transformations like halogenation, cyclopropanation, N-H insertion, C-H amination and sulfimination.217,218 However, these reactions are beyond the scope of this review and readers are referred to a recent review focusing on non-natural reactions by CYPs.202,219
The example of CYPs discussed above shows how a protein can be engineered to perform chemo- and site-selective catalysis on different substrates. Engineering and redesigning a protein to model structural units of another protein also helps to understand the structure-function relationship in the latter in greater detail and result in different applications, such as alternative energy productions. A notable example of that is the redesign of myoglobin to mimic the catalytic center of cytochrome c oxidase (CcO). The CcO is a trans-membrane protein, which catalyzes the four-electron reduction of oxygen to water during aerobic respiration. The catalytic center of CcOs consists of a heme-copper center, where the copper is coordinated to three histidines with one of the histidine crosslinked to a tyrosine residue (Fig. 12).220,221 Despite decades of investigation of oxygen reduction in CcOs, structural features responsible for the efficient reaction are not well understood. This gap in understanding is primarily due to the features of the membranous CcO protein as well as to the presence of multiple metal cofactors (for example a CuA and a heme center that deliver electrons to the heme-copper active site where O2 is reduced) in the enzyme complex, which makes its spectroscopic investigation very challenging. Furthermore, CcO is also a model catalyst for oxygen reduction reaction (ORR) in fuel cells, as it uses earth abundant metal ions and has a much lower overpotential than the current state-of-the-art platimum-based ORR catalysts. However, the large size and low stability of the CcO makes it difficult for its application as ORR catalysts. In order to overcome these challenges, a number of small molecule-based synthetic models of CcO have been developed and these systems have provided tremendous knowledge about the structure and functions of CcOs.222,223 even though most of the models have low activity and turnover numbers.
Fig. 12.
a) The heme-copper catalytic center of cbb3 oxidase with the CuB coordinated to three histidines with one of the histidines crosslinked to tyrosine (PDB: 3mk7). b) The designed heme-copper center in the CcO mimic in myoglobin (F33Y-CuBMb) containing the CuB coordinated to three histidines. A Tyr33 has also been incorporated in the active site to mimic the conserved Tyr in CcOs. (PDB: 4fwy).
To complement the synthetic modeling approach, Lu and coworkers have developed an alternative method to study the oxygen reduction catalysis in CcOs by designing a heme-copper center in a small, easy-to-purify heme-containing soluble protein, myoglobin, that is free of other metal cofactors (Fig. 12). The first generation models focused on designing a copper center distal to the histidine coordinated high-spin heme. To engineer a copper binding center, two histidines (L29H, F43H) were introduced in the distal pocket of myoglobin that, along with the third histidine already present (H64), could potentially bind copper similar to CcOs; this myoglobin mutant was named CuBMb.224-226 The copper binding in the distal pocket of the heme center was confirmed by several spectroscopic and crystallographic methods.224 The introduction of the copper binding center, while increased the product selectivity of the oxygen reduction (the ratio of water produced with respect to reactive oxygen species such as superoxide and peroxide) with respect to WTMb, also lowered the total rate of oxygen reduction. To improve the activity and product selectivity of the CcO model, tyrosines were engineered close to the heme-copper catalytic center, thus affording F33Y-CuBMb and G65Y-CuBMb, which not only improved the activity and product selectivity (~80% water production), but also performed more than a thousand turnovers (Fig. 13).227 In fact, by increasing the electron transfer rates to the heme cofactor using the native electron transfer partner of Mb (cyt. b5), the G65Y-CuBMb variant was shown to reduce oxygen with rates (52 s−1) comparable to a native CcO (50 s−1).228 Moreover, when the designed protein was immobilized onto electrodes, the electrocatalytic reduction of oxygen using G65Y-CuBMb not only resulted in very selective oxygen reduction (~ 96% water) but also exhibited rates (5000 s−1) exceeding the fastest CcO (bovine CcO with rates of 500 s−1).229 These results demonstrated that it is possible to redesign the metal-binding sites in native scaffolds that display new functions (e.g., from reversible O2 binding in WTMb to catalytic O2 reduction to H2O) that can meet or even exceed the catalytic efficiency of native enzymes.
Fig. 13.
a) Correlation of the oxidase activity of Phe33Tyr-CuBMb, Phe33ClY-CuBMb, Phe33F2Y-CuBMb, and Phe33F3Y-CuBMb vs pKa of phenols on the Tyr and its analogs. b) Correlation of water produced in oxygen reduction reaction performed by these proteins vs the pKa of phenols on the Tyr and its analogs. c) Correlation of oxidase activity by these proteins vs peak potential at pH 13 (Ep) of the corresponding Tyr and Tyr analogs. The structures of various Tyr analogues used in the study are displayed left. Adapted with permission from ref. 228, Copyright 2015 American Chemical Society.
The rationally designed functional mimics of CcO thus provided an efficient method to probe and understand the role of various structural units, such as the cross-linked tyrosine conserved in all CcOs. To elucidate the role of tyrosine towards oxygen reduction, unnatural tyrosine variants, such as mono chloro-, dichloro-, fluoro- and methoxy- tyrosines, were incorporated genetically at the 33 position of the functional CcO mimic, F33Y-CuBMb (Fig. 13).230 The oxygen reduction activity measurements showed a systematic increase in the oxidase activity of the CcO mimics with an increase in Tyr redox potential and decrease in Tyr pKa, strongly suggesting the role of a tyrosine as an H radical donor to oxygen for its reduction. To prove the role of Tyr and observe oxygen reduction intermediates in the CcO mimic, the reduced F33Y-CuBMb was reacted with oxygen and the reaction mixture, freeze-quenched at a short time scale (~ 20 ms), and probed via continuous wave-EPR. The experiments revealed the formation of a Tyr radical during reaction conditions, which for the first time, established the role of Tyr as an H radical donor to oxygen in CcOs.231
Another unique structural feature of CcOs is the presence of different heme types at its catalytic center. While, A- and B-types oxidases contain heme a and o, the C- type oxidases contain heme b (Fig. 14).232 An efficient method to understand the role of different heme types towards the function and stability of CcOs would be to replace different heme types and studying their impact on enzymatic activity. However, the presence of different heme cofactors (e.g. the heme that delivers electrons to the heme-copper center) and the membranous nature of CcO makes such a study very difficult, if not impossible. The Lu group addressed this issue, by incorporating mimics of heme a and heme o in the designed CcO mimic.233 The incorporation of a heme variant with hydroxyl group attached to the porphyrin core (similar to heme o) increased the stability of CuBMb by decreasing the rate of heme degradation by approximately 19-fold.233 Furthermore, the incorporation of heme a mimics (with one or two formyl groups conjugated to porphyrin core) increased the heme redox potential and in turn the overall rate of oxygen reduction by more than six-fold (Fig. 14g).234 Overall, this study reveals that the CcO uses different heme types such as heme a and heme o to control the heme redox potential, oxygen reduction activity and stability of the enzyme.
Fig. 14.
a) Protein scaffold of F33Y-CuBMb. b) Heme b cofactor present in F33Y-CuBMb. c) Heme a present in the catalytic site of bovine CcO. d) Diacetyl heme. e) Monofomyl heme. f) Diformyl heme incorporated in F33YCuBMb apo-protein. g) Variation of oxygen reduction activity with heme E° for F33Y-CuBMb variants. Dotted blue line indicates E° of WTMb and bovine CcO. Reprinted with permission from ref. 234, Copyright 2014 American Chemical Society.
Apart from designing the Cu binding site in Mb, Lu and coworkers have also designed a Mn binding site in cytochrome c peroxidase (CcP), to mimic the function of manganese peroxidase (MnP).235 The MnP from the white rot fungus P. chrysosporium plays a vital role in lignin degradation. MnP binds two CaII, one MnII and a heme FeIII that oxidize MnII to MnIII via hydrogen peroxide activation. The enzyme-generated MnIII is subsequently used to oxidize organic substrates. To test the current understanding of the structure and function of MnP and to find an alternative catalyst for oxidative delignification, the authors designed and engineered a MnII binding site similar to MnP in easy-to-work-with peroxidase, CcP, that can be readily expressed in E. coli in high yields.236 The CcP mutant (MnCcP), thus created, based on structural comparisons and computer modeling, binds MnII in a manner similar to the native enzyme and shows that the incorporation of the MnII binding site facilitates MnII oxidation. Further mutations of the residues in heme distal pocket (W191 and W51) in MnCcP to the corresponding Phe in MnP conferred even higher MnP activity of the protein model. Interestingly, the two mutations do not contribute equally to the activity increase. A much larger increase arises from the W51F mutation because W51 stabilizes the heme compound II (oxo-ferryl heme species). Since, the reaction of compound II with MnII is rate-limiting, a more reactive compound II increases MnP activity.237
Despite the above progress made in redesigning CcP into MnP, most of the designed enzymes still suffer from relatively lower activity and moderate to low binding affinities for the metal than those in native MnP. It was hypothesized that one reason for such low activity and metal-binding affinity is our lack of understanding about the subtle secondary coordination sphere interactions around the active site that are responsible for fine-tuning the activity.34 While these interactions, such as hydrogen bonding, salt bridges to the metal ion ligands, hydrophobicity of the site, and overall rigidity or flexibility of the site, have been recognized to be important, few studies have demonstrated engineering these interactions to improve enzyme activity and metal-binding affinity.110,162,165,170 To address the above issue, specific secondary sphere residues around the Mn(II)-binding site of the engineered MnCcP (MnCcP1) were rationally mutated. This resulted in improved binding affinity to Mn(II) and/or catalytic efficiency toward Mn(II) oxidation and degradation of lignin (Fig. 15).238 First, removing a hydrogen bond to Glu45 through Tyr36Phe mutation, enhanced Mn(II)-binding affinity, as evidenced by a decrease of KM of Mn(II) oxidation 2.8 fold. Second, introducing a salt bridge through Lys179Arg improved Glu35 and Glu181 coordination of Mn(II), decreasing KM 2.6 fold. Third, eliminating a steric clash that prevented Glu37 from orienting towards Mn(II) resulted in an 8.6 fold increase in kcat/KM arising primarily from a 3.6 fold decrease in KM. These effort has resulted in one variant (Ile40Gly) displaying a KM values that is comparable to that of native MnPs. (0.28 mM vs. 0.2 mM for Pleurotus eryngii MnP PS3). This work further demonstrated that while the effects of Tyr36Phe and Lys179Arg mutations are additive, other combinations of mutations were antagonistic. Using both alkali treated lignin and a lignin model compound, the variants were also shown to be functional mimics of MnPs. This study not only provides a better functional model of MnP, but more importantly it confirmed the importance of the secondary coordination interactions to enhance metal binding affinities and overall activity.
Fig. 15.
Secondary shell residues rationally mutated in MnCcP.1 and their effect on activity and metal affinity. Reprinted with permission from ref. 238, Copyright 2016 American Chemical Society.
4.1.2 Copper proteins
Copper proteins fulfill important roles in nature, including catalytic oxygen activation. Therefore it is not surprising that they have been the subject of many protein engineering studies, wherein the native proteins are altered in an attempt to improve their functionality. Several approaches have been employed depending on the protein family under study. For example, in the case of MCOs, directed evolution has been preferred over rational design approaches, due to the lack of detailed structural information on these enzymes.239
Tyrosinases are copper-containing enzymes widely distributed in Nature, which utilize dioxygen to catalyze two successive enzymatic reactions: (i) the ortho-hydroxylation of monophenols to orthodiphenols (monophenolase activity) and (ii) the oxidation of ortho-diphenols to ortho-quinones (diphenolase activity).240 These enzymes have a strong preference for phenolic and diphenolic substrates and are somewhat limited in their reaction scope, always producing an activated quinone as product. Despite this fact, they can be potentially utilized in several biotechnological applications, including the production of L-DOPA, phenol and novel mixed melanins, biosensors of phenols, and removal of dye and other compounds.241 Therefore, many efforts have been devoted to the applications of directed evolution methods to engineer improved tyrosinases. Fishman and coworkers used directed evolution on tyrosinase from Bacillus megaterium (TyrBm) in an attempt to improve its monophenolase/diphenolase activity ratio.242 Through this approach, they discovered one variant, R209H, displaying a 1.7 fold improvement in monophenolase activity and a 1.5 decrease in diphenolase activity, with an overall 2.6-fold improvement in the monophenolase/diphenolase activity ratio. The authors hypothesized that the imidazole group of the newly introduced histidine residue obstructs the entrance to the active site, thus interfering with the binding of L-DOPA to one of the two copper ions. Figure 16 illustrates the active site structures for both wild-type TyrBm (Fig. 16a) and variant R209H (Fig. 16b).243 Comparison of wild-type structures with the structure of the site-specific variant R209H, which possesses a higher monophenolase/diphenolase activity ratio, lent further support to the hypothesized mechanism by which monophenolic substrates dock mainly to CuA. The Arg209 residue is positioned in proximity to the entrance of the active site, on helix α6 adjacent to His208, which coordinates CuB. In subunit 1 of TyrBm, two conformations of this residue were observed, indicating its flexibility (Fig. 16a). In the R209H TyrBm mutant structure, His209 can be seen to shield CuB and to obstruct the entrance to the active site (Fig. 16b). This structural evidence further supports the hydroxylation mechanism in which a monophenol substrate docks to CuA. This work clearly highlight the potential of directed evolution approach to modulate the function of native proteins, and it is of great relevance since the hypothetical mechanism has been then confirmed by structural data.243
Fig. 16.
The active sites of wild-type TyrBm and variant R209H. (a) The active site of wild-type TyrBm (TyrBm1) is presented in blue, and the two conformations of Arg209 identified in the electron density maps are presented in yellow. (b) The active site of variant R209H (structure of TyrBm1_mut) and residue His209 are shown in light blue and yellow, respectively. Reprinted with permission from ref. 243, Copyright (2011) Elsevier.
Among MCO family, laccases represents the most attractive enzymes for their possible application in bioremediation, organic synthesis, biosensors and other industrial applications. They show broad substrate specificity and relative autonomy, since they use molecular oxygen from air as an electron acceptor and they only produce water as the by-product.17 They couple the four single-electron oxidations of the reducing substrate to the four electron reductive cleavage of the dioxygen bond, using four Cu atoms distributed over three sites.15,16 Typical metal content of laccases includes one type 1 copper (T1Cu), and one type 2 copper (T2Cu) and two type 3 copper ions (T3Cu), with T2Cu and T3Cu arranged in a trinuclear cluster (TNC).244 The range of functions exhibited by laccases is broadly divided into three categories: (1) cross-linking of monomers, (2) degradation of polymers, and (3) ring cleavage of aromatic compounds. In order to highlight the power of directed evolution approaches in the development of copper-based oxygen activating enzymes, several selected examples of engineered fungal laccases are described in the following.245,246 However, for a more detailed description of laccases, their properties and their applications, the reader can refer to previous excellent reviews on the topic.244,247,248
The first successful example of directed evolution to modify a fungal laccase was reported by Arnold and coworkers.249 After 10 rounds of selection, they obtained a laccase variant with 14 mutations accounting for a 170-fold improvement in total activity, 8-fold increase in the expression levels and 22-fold in the kcat for ABTS. Among laccases, those possessing high redox potentials at the T1 copper site (ranging from +430 mV in bacterial and plant laccases, to +790 mV in some fungal laccases) are classified as high redox potential laccases (HRPLs). They aretypically secreted by ligninolytic basidiomycetes (white-rot fungi), and are excellent candidates for industrial applications, since their high redox-potential values allows them to oxidize a wide variety of substrates.245,250 Thermostable laccases with a high-redox potential have been engineered by Matè and coworkers,251 through a strategy that combines directed evolution with rational approaches. The starting point was the basidiomycete PM1 HRPL functionally expressed in S. cerevisiae, which exhibits a remarkable stability and activity, including thermal activation. After eight rounds of evolution, the total laccase activity was enhanced 34,000-fold, culminating in the OB-1 mutant as the last variant of the evolution process, displaying a highly active and stable enzyme in terms of temperature, pH range, and organic cosolvents.251 Similarly, Camarero and coworkers described a directed-evolution platform for the development of Pycnoporus cinnabarinus laccase (PcL) with different properties.252 After six rounds of evolution coupled to high-throughput (HTP) assays based on the oxidation of natural and synthetic mediators, the total laccase activity was improved 8000 times. The final mutant of this study (the 3PO variant) accumulated a total of 15 mutations in the fusion gene. Five mutations were responsible for a 40-fold enhancement in secretion by S. cerevisiae (~2 mg/L), while the ten beneficial mutations in the mature protein led to a 13.7-fold increase in the kcat for ABTS.252 Semi-rational mutagenesis of the laccase POXA1b from Pleurotus ostreatus was performed through a combination of directed evolution with elements of rational enzyme modification, by Sannia and coworkers.253-255 The final evolved mutant, containing five non-synonymous mutations, increased 3.5-fold in its activity towards ABTS and two-fold towards 2,6-dimethoxyphenol (DMP), with higher affinity towards DMP. A significant changes in the laccase selectivity towards the oxidation of phenolic substrates has been obtained by Wang and coworkers, who prepared several mutants of Klebsiella sp. 601 MCO.256 They found that theα-helix (from Leu 351 to Gly 378) covering the substrate binding pocket strongly influence substrate selectivity, probably by modulating the electron transfer between the substrate and the T1 copper site. Seven mutations were introduced in this helical fragment, affording the α351-380M mutant. These mutations seems to disrupt the helix secondary structure, thus enhancing specificity toward the phenolic substrate DMP. This finding make the mutant very attractive for applications in biotechnology and green chemistry.
Azurin from pseudomonas aeruginosa is a type 1 copper protein involved in biological electron transfer. It has been a subject of extensive protein design and engineering to understand structural features responsible for efficient electron transfer.257,258 Recently, it has been used by the Lu group to understand post-translational modification of cysteinyl thiolate to sulfonate, which plays important roles in biology, such as redox signalling, enzyme and gene regulation, by designing an azurin cavity mutant (M121G-Az) that makes the copper center accesible to small molecules like hydrogen peroxide and oxygen.259 The reaction of hydrogen peroxide with Cu(I)– M121G-Az resulted in the first copper-sulfenate characterized in a protein environment, as supported by resonance Raman spectroscopy, electrospray mass spectrometry using isotopically enriched hydrogen peroxide, and density functional theory calculations correlated to the experimental data. Further, structural and computational studies suggested that in the secondary coordination sphere noncovalent interactions played an important role in stabilizing this highly reactive species, which can further react with oxygen to form a sulfinate and then a sulfonate species, as demonstrated by mass spectrometry. Engineering the electron transfer protein azurin into an active copper enzyme that forms a copper–sulfenate center and demonstrating the importance of noncovalent secondary sphere interactions in stabilizing it are significant in understanding the metal-sulfenate species in biological systems.
4.2. Artificial oxygen-activation enzymes by cofactor replacement
To expand the repertoire of metalloenzyme functions and to design new artificial molecules, the strategy of replacing the natural cofactor housed in a native protein has been reported since the 1980s.260 Towards this aim, small molecule-based chemical catalysts show some interesting properties.261 They can be produced in high quantity and at low cost through chemical synthesis, possess high stability and resistance to severe conditions of temperatures and pressures. However, compared to natural biocatalysts, chemical catalysts often show low selectivity and turnover numbers, due to the lack of a proper molecular environment, which controls both the first and secondary coordination spheres. This disadvantage can be overcome by inserting a chemical catalyst within a protein environment.261,262
Heme proteins are particularly well suited for cofactor replacement, since the heme pocket represents a proper protein cavity for inserting an unnatural cofactor. An intricate set of different interactions allows heme to be hold into the protein matrix. These interactions create a molecular architecture around the cofactor, which contribute to a fine tuning of the heme properties. The number, type and donor properties of the axial ligands directly contribute to heme properties.34 Hydrophobic, ion pairing and hydrogen bonding interactions, dominate the secondary coordination sphere and control several features of the heme environment, such as the local dielectric constant, heme exposure, the ligand orientation, etc. Furthermore, the protein regulates the accessibility of solvent and substrates into the active site, thus allowing to selectively discriminate between different ligands and/or substrates. Replacing the heme with an unnatural cofactor can be facilitated by the enormous amount of structural data available on natural and mutated heme proteins: a careful inspection of the pocket structure at a molecular level can suggest that the cofactor can be inserted and mutations to be performed for housing that cofactor.
Through this approach, Hayashi and coworkers reported the engineering of new functions inside myoglobin (Mb) (Fig. 17a).263,264 Myoglobin is a dioxygen binding protein and lacks a recognition site of substrates different from dioxygen in the heme proximity. Thus, the first aim was to provide myoglobin with a substrate binding pocket. Aryl substituents were introduced on the propionate groups of protoheme IX, thus giving the un-natural cofactor 2, shown in Figure 17b. Mb reconstitution with this un-natural cofactor afforded a peroxidase-like catalyst, which was able to activate hydrogen peroxide and catalyze guaiacol oxidation with an efficiency 13-fold higher than native myoglobin. Next, in order to increase the catalytic properties of this molecule, some key amino acid mutations were introduced in the myoglobin structure.265 In particular, mutation of the distal His64 to Asp, aimed at expanding the substrate binding pocket, into myoglobin resulted into an 430-fold enhancement of the catalytic efficiency (kcat/KM) toward guaiacol oxidation with respect to WT myoglobin. In an effort to increase the kcat value, modified hemes substituted at only one of the two propionates (cofactors 3a and 3b, Fig. 17b) were prepared. Reconstitution of MbH64D with these cofactors resulted in an artificial enzyme with an increased kcat value respect to MbH64D-1 (MbH64D-3 kcat=24 s−1; MbH64D-1 kcat=1.2 s−1) and with a catalytic efficiency comparable to that of native HRP (MbH64D-3 kcat/KM=85000 M−1s−1; HRP kcat/KM=72000 M−1s−1).118
Fig. 17.
Myoglobin reconstitution with an artificial cofactor for inserting new functions : a) General strategy for myoglobin and its mutants reconstitution. Adapted from ref. 265, Copyright (2004) American Chemical Society. b) Molecular structures of the modified hemes. c) Molecular structure of porphycene (Pc).
These results demonstrated that the replacement of the native heme with an artificially synthesized heme is an attractive strategy to regulate the myoglobin function. In subsequent works, the authors tried to increase the peroxidase and peroxygenase activity of myoglobin by modifying the tetrapyrrolic framework of the prosthetic group, using porphyrinoids. Towards this aim, porphycene (Pc), a constitutional porphyrin isomer coordinated by FeIII, was used (Fig. 17c).266 As revealed from the analysis of the crystal structure, the FeIIIPc cofactor was bound to the protein scaffold situated between the E and F helices, and with the His93 of the F helix as a fifth ligand. Spectroscopic analysis of the FeIIIPc reconstituted myoglobin revealed that the un-natural cofactor is more tightly coordinated to His93 with respect to WT myoglobin. The authors suggested that the strong coordination of the Fe-His93 bond in the reconstituted FeIIIPc-myoglobin contribute to the enhancement of the catalytic activity toward the oxidation of substrates such as guaiacol, thioanisole, and styrene, respect to native myoglobin. At pH 7.0 and 20 °C, the initial rate of the guaiacol oxidation was found to be 11-fold faster than that observed for the native myoglobin. Because manganese porphyrins and porphyrinoids such as manganese corrole and corrolazine have been studied as catalysts of C–H bond activation, more recently the authors used myoglobin reconstituted with MnIIIPc cofactor to obtain an hydroxylation catalyst.267 As revealed from the solution of the crystal structure of the reconstituted protein, obtained at 2.2 Å resolution (Fig. 18), and as already observed for the iron complex, MnIIIPc is coordinated by His-93, thus being accommodated into the natural heme binding site. Upon addition of H2O2 at pH 8.5, rMb(MnPc) promotes hydroxylation of ethylbenzene with a total turnover number (TON) of 13 and initial turnover frequency (TOF) of 33 h, yielding 1-phenylethanol as a single product.267 The finding that myoglobin, only when reconstituted with MnPc, is able to catalyze hydroxylation reactions is clearly related to the presence of MnIII. In addition, the direct involvement of the metal ion-center into the hydroxylation reaction mechanism was demonstrated by using O18 labeled hydrogen peroxide. The results of this work clearly demonstrate that by choosing the correct un-natural cofactor, artificial metalloenzymes with un-precedent functional properties can be designed.
Figure 18.
Cofactor replacement in myoglobin as adopted in the work by Hayashi et al.267 Crystal structures of WT-myoglobin (left PDP ID: 1myo) and MnIIIPc substituted myoglobin (PDP ID: 3wi8).
The heme pocket of hemeproteins can also accommodate non-tetrapyrrolic cofactors. Ueno and Watanabe first reported a detailed screening of the insertion of Schiff-base metal complexes into protein environment by non-covalent conjugation.268 The interest in reconstituting myoglobin with Schiff- base metal complexes derived from the finding that these complexes, containing MnIII and CrIII, have been reported as oxidation catalysts in organic solvents.269,270 In addition, their molecular size and coordination geometry are similar to heme, and it is easy to modify the ligand size and hydrophobicity. The authors reported the incorporation of several Fe, Mn and Cr salen and salophen complexes (Fig. 19) into myoglobin and some mutants, to analyze the stereoselective oxidation of thioanisole. Detailed screening of metal binding ability and stability, together with resolution of the crystal structures of some of the reconstituted proteins allowed for the redesign of metal complex/protein composites with improved catalytic activity and stereoselectivity.288,271-274 From these studies, they conclude that A71G mutation seemed to be necessary to make myoglobin suitable for accommodating the Schiff-base complexes without steric bumps. Replacement of His64 to Asp (H64D mutation) was expected to increase access of substrates and oxidants to the vacant distal site above the metal ion center.
Fig. 19.
Schiff base complexes inserted into myoglobin: salophen ligand series (a) and salen ligand series (b) screened for the activity.
The crystal structure of Fe·2·salophen apo-A71GMb confirmed that the iron complex Fe·2 is fixed in the heme cavity of apo-Mb (Fig. 20a), and axially coordinated by His93.273 Specific interactions with Phe43, Leu89, His97 and Ile99 influence the orientation of the Fe·2 complex in the cavity of apo-A71GMb. The side chain of Ile107 is located between the 3- and 3’-methyl groups of Fe·2. Inspection of the crystal structure of Mn·2·apo-A71GMb revealed a narrow channel between His64 and the phenylenediamine unit of 2. Replacement of His64 to Asp (H64D mutation) was expected to increase access of substrates and oxidants to the vacant distal site above the metal ion center. In addition, to further enlarge the active site access, phenylendiammine unit was replaced by ethylendiammine affording the salen ligands series (Fig. 19 b). The rate with Mn·4·apo-H64D/A71GMb was found to be threefold higher than that with Mn·2·apo-H64D/A71GMb, thus confirming that the enlargement of the substrate cavity increase the accessibility of the substrate.233 Substitutions at the 3- 3’- positions strongly affect the enantioselectivity of the sulfoxidation reaction. Interestingly, while Mn·4·apo-H64D/A71GMb showed 32% ee (R) selectivity, introduction of bulkier groups at the 3,3’-positions induces relative S-selectivity to end up 13% (S) for 6. All the results demonstrate that the enantioselectivity and catalytic activity of unnatural metal cofactors in protein cavities can be regulated by rational design of the cofactors.
Fig. 20.
a) Crystal structure of apo-A71GMb reconstituted with the Fe 2 salophen ligand. Reprinted with permission from ref. 273, Copyright (2004) American Chemical Society. b) (top) Salen ligand modified by methyl thiosulfonate linking arms. (bottom) Computer model of the Mb cavity with MnSalen (green) positioned to overlap the space occupied by the native heme cofactor. The residues selected as possible anchor positions are visible. Left anchors T39 and Y103 are purple, and right anchors L72 and S108 are orange. Reprinted with permission from ref. 278, Copyright (2011) American Chemical Society.
An alternative approach for site-specific attachment of a non-natural cofactor into a protein scaffold is by covalently linking the two. Towards this aim, manganese salen with methyl thiosulfonate linking arms (Fig. 20b) was covalently attached to Cys residues inside the Mb cavity by the Lu group, who demonstrate that incorporation of MnSalen into a protein scaffold enhances the chemoselectivity in sulfoxidation of thioanisole 275 and that both the polarity and hydrogen bonding of the protein scaffold play an important role in tuning the chemoselectivity.276 Furthermore, a comparison of the effect of different pH on sulfoxidation and ABTS oxidation indicates that, while the intermediate produced at low pH conditions could only perform sulfoxidation, the intermediate at high pH could oxidize both sulfoxides and ABTS. Such a fine-control of reactivity through hydrogen-bonding interactions by the distal ligand to bind, orient and activate H2O2 is very important for designing artificial enzymes with dramatic different and tunable reactivity from catalysts without protein scaffolds.277 Importantly, the effect of different anchoring positions on reactivity and selectivity were investigated.278 This metalloenzyme design started by modeling the salen ligand into the myoglobin structure and searching for the best position for covalent anchoring the cofactor in a location similar to that of the heme in the protein binding pocket. The alignment of the metals and donor atoms of the heme and the model salen complex allowed the positioning of Mn-salen in the heme-binding site. The calculation suggested T39, L72, Y103 and S108 as proper anchoring groups to link the salen ligand in a position similar to that of the native cofactor (see Fig. 20b). Four myoglobin mutants Y103C/S108C, T39C/S108C, Y103C/L72C, and T39C/L72C carrying two Cys residues necessary for the covalent linkage of the salen ligand arms to the protein, were expressed and purified. Next, the effects of the different anchoring position on the reactivity of the reconstituted proteins toward thioanisole sulfoxidation were analyzed. In order to distinguish between the impact of the different anchoring positions, anchor residues were clustered as belonging to right side (L72 or S108) or left side (T39 or Y103) with respect to the salen ligand (see Fig. 20b). Interestingly, the anchor position of the salen ligand strongly affects both the reaction rate and selectivity, and the effects are additive in nature and independent. For example, changing the anchor positions, from 1 Mb(Y103C/S108C) to 1·Mb(T39C/L72C), result in total rate enhancements of 7-fold and an 80% increase in selectivity for the S enantiomer. To gain more insight into the molecular basis for the observed selectivity, the reactivity of the reconstituted Mbs were assayed toward a series of substituted aryl methyl sulfides, with different substituents. The right anchor site was observed to have a greater influence than the left anchor site on the reactivity and selectivity in sulfoxidation, and 1·Mb(T39C/L72C) showed the highest reactivity (TON up to 2.32 min−1) and selectivity (ee % up to 83%) among the different anchoring positions examined. Molecular dynamic simulations revealed differences in substrate access path for the L72C anchor position; steric effects inside the protein cavity forces the substrates to enter in a different way limiting the conformational mobility thus enhancing stereoselectivity. Overall, this work highlights that attachment strategies may have considerable influence on the activities of non-native cofactors.
Simple replacement of the metal ion has also been shown to be effective in providing new enzymes with oxygen-activating activity. Several examples deal with the replacement of the Zn(II) ion with redox active ions, such as MnIII and CuII.279-281 Substitution of manganese for zinc in carbonic anhydrase (CA) converted this hydrolase to an enantioselective peroxidase.280 Manganese-substituted carbonic anhydrase (CA[Mn]) shows peroxidase activity with a bicarbonate-dependent mechanism towards the oxidation of o-dianisidine with kcat/KM=1.4 × 106 M−1s−1. In addition, CA[Mn] also catalyzed the moderately enantioselective epoxidation of olefins to epoxides.281 Enantiomeric excesses (ee's) (up to 67% were obtained, although higher ee's were associated with low conversion.
4.3 Artificial oxygen-activating enzymes by incorporation of metal cofactors into native scaffolds that do not contain metal ion
While native enzymes catalyze many reactions with high efficiency and selectivity, the scope of the reactions are still limited in comparison to chemical catalysts, partially due to the limitation of utilizing only physiological available metal ions such as iron and copper, and metal-containing cofactors such as heme. To expand the scope the reactions for more diverse chemical transformations, chemical catalysts have been inserted in a non-metal containing protein scaffolds. An initial example of this approach came from Whitesides and coworkers, as early as in 1978, which utilized the extremely high affinity of protein avidin with its substrate biotin and its variants (K ~ 10−15 M), to obtain an asymetric hydrogenation catalyst. More specifically, a biotin conjugated variant of hydrogenation catalyst Rh(I)-norbodiene was constructed and inserted into avidin.282 The tertiary structure of the enzyme avidin was expected to provide the chirality for enantioselective hydrogenation. Recently, Ward and coworkers have made significant progress in exploring, developing and realizing the full potential of this strategy and much beyond (see Fig. 21a) by designing several biotin/streptavidin-based metalloenzymes to catalyze various enantioselective transformations.283-286 They showed that changes in the biotin-spacer-ligand moiety (see Fig. 21b), site-directed mutation on avidin, and metal ion had strong implications on the activity of the biotin-based catalyst. For example, an artificial metalloenzyme for the reduction of prochiral imines was obtained through the insertion of a biotinylated cyclopentadienyl rhodium complex into a mutated streptavidin (Fig. 21a). Further, an aminosulfonamide ruthenium complex (Fig. 21b), when incorporated in avidin, oxidized secondary alcohols (using tert-butylhydroperoxide as the oxidizing agent) with over 90% yield.287
Fig. 21.
Biotinylated complex inserted into streptavidin. a) Crystal structure of a biotynilated cyclopentadienyl rhodium complex, inserted into the K121H streptavidine (PDB ID: 4gjs). b) Molecular structure of biotinylated aminosulfonamide ruthenium complex.
Additionally, the authors also developed a vanadium-dependent artificial peroxidase for enantioselective sulfoxidation by inserting a vanadyl ion into streptavidin.288 The vanadium-oxo complex [VO]2+ was inserted into the protein scaffold by the using VOSO4 as vanadium source and the resulting hybrid protein catalyzed sulfoxidation of thioanisole giving 94% yield and 46% ee (R). EPR spectroscopy on the vanadium reconstituted protein yielded a spectrum very similar to that of [VO(H2O)5]2+. This finding together with docking experiments suggested that the interaction between vanadium and the protein was mainly dictated by hydrogen bonds in the second coordination sphere of the metal ion (Fig. 22). To improve upon the stereo-selectivity of the enzyme, a screening on different prochiral sulfide substrates was performed with enzyme displaying highest enantioselectivity for dialkyl and alkyl-aryl substrates (up to 93% ee). Interestingly, increasing the steric bulk of the aromatic moiety of the sulfide led to an increase in selectivity showing the importance of proper substrate positioning in the active site for efficient catalysis.
Fig. 22.
Putative binding interaction between vanadium oxo specie and streptavidin-biotin binding site. The interacting substrate is also showed for clarity. Adapted from ref. 288 Copyright (2008) American Chemical Society.
Artificial metalloenzymes with peroxidase activity were also developed through the so-called “Trojan horse” strategy, by Mahy and coworkers.289 They took advantage of the remarkable affinity of a monoclonal antibody for its estradiol antigen (KD = 9.5 ×10−10 M) to design an artificial hemoproteins by inserting Fe- or Mn-porphyrin–estradiol conjugates into the anti-estradiol antibody (Fig. 23).289 These biocatalysts catalyzed selective oxidations, such as the enantioselective sulfoxidation of thioanisole by H2O2, with a 10% ee and the chemoselective epoxidation of styrene by potassium peroxymonosulfate (KHSO5). Differences in enantioselectivity were rationalized by docking experiments which suggested specific interactions of the substrate with residues in the active site to be critical for enantioselectivity. Interestingly, the antibody still retains good affinity for its antigen, even when linked to such a bulky molecule as a porphyrin, and interacts closely enough with the catalytic center to influence its activity.
Fig. 23.
Fe- porphyrin–estradiol conjugates
The same authors developed a different class of artificial hemoproteins, named “hemoabzymes”, by raising monoclonal antibodies against metalloporphyrins and microperoxidases.290-295 In their first study, using iron(III)-meso-tetrakis(ortho-carboxyphenyl (Fe(ToCPP) porphyrin as the hapten, three monoclonal anti-porphyrin antibodies were found to recognize the porphyrin. Two of them, 13G10 and 14H7, bound Fe(ToCPP) with nanomolar Kd values and catalyzed the oxidation of ABTS by H2O2, exhibiting catalytic efficiencies at least fivefold higher than free Fe(ToCPP).291-293 This result clearly highlighted the importance of the protein matrix in modulating the activity of the cofactor. However, the catalytic efficiency of these enzymes were far lower than that of the native peroxidases. This finding was attributed to the lack of an axial His ligand on catalytic iron. To overcome this limitation, the authors used MP8 as the hapten (for details on MP8 see paragraph 1).290 Seven antibodies recognizing MP8 were produced, and the best of them, 3A3, was found to bind MP8 with a moderate KD of 1×10−7 M. Despite the modest affinity, the 3A3-MP8 complex displayed good peroxidase activity toward o-dianisidine characterized by a kcat/KM value of 2×106 M−1 min−1. In addition, the 3A3-MP8 complex catalyzed the regioselective nitration of phenol in the presence of H2O2 with 2-nitrophenol as a preferred product over 4-nitrophenol.294 Moreover, the 3A3–MP8 complex was shown to catalyze the oxidation of thioanisole by H2O2 in the presence of 5% tert-butyl alcohol, with a 45% ee in favour of the R isomer.295
Chemical catalysts can also be incorporated in protein via ionic interactions. One such examples comes from the Mahy lab, where Xylanase A (Xln10A, possessing a globally positive charge) was reconstituted with anionic porphyrins.296,297 Thus, Xln10A, which in its native form possesses no metal center, bound FeIII-meso-tetra-p-carboxy-phenylporphyrin (Fe-TpCPP) and in the presence of imidazole as co-catalyst catalyzed the chemo- and stereoselective oxidation of thioanisole to sulfoxide, with 85% yield and 40% ee.296 Replacement of iron with manganese allowed for the selective oxidation of aromatic alkenes. Indeed, the Mn-based complex catalyzed the oxidation of para-methoxystyrene by KHSO5 with a 16% yield and a very high enantioselectivity (80% in favour of the R isomer ).297
The approach of constructing hybrid catalysts requires the selection of a protein capable of binding the catalyst and of modulating its activity in order to drive the targeted reaction. Given these requirements, several examples focused on the use of serum albumin as protein scaffold as the enzyme has a unique ability to bind a variety of hydrophobic molecules, including steroids, fatty acids and heme.298 A simple noncovalent adduct of albumin-conjugated manganese corroles was reported by Gross and coworkers as a catalyst for asymmetric oxidations.299 The MnIII corrole reconstituted serum albumin catalyzed the sulfoxidation of asymmetric aryl-sulfide in the presence of hydrogen peroxide with up to 74% ee. Further experiments revealed direct involvement of the metal cofactor in the reaction mechanism and supported that the ee was due to the chiral environment surrounding the albumin-conjugated catalyst. Similarly, human serum albumin (HSA) was also functionalized by MnIII-salen complexes yielding MnIII–salen⊂HSA hybrid proteins as Mn monooxygenase mimics (see Fig. 24 for MnIII-salen complexes structures).300 These hybrids proven to be very efficient (almost complete conversion of substrate), stable (Kd values in the micromolar range) and soluble in aqueous medium. Notably, comparison of the catalytic efficiency of the complexes and the hybrids shows that the polarity of the protein environment is crucial for efficiency and selectivity. In fact, the hybrids selectively catalyzed the oxidation of thioanisole, by sodium hypochlorite, to the corresponding sulfoxide, without further oxidation to sulfone.
Fig. 24.
Mn salen complexes inserted into HAS.
All the examples discussed above were built upon visual inspection of proteins and its physico-chemical properties to insert a chemical catalyst in it. As an alternate strategy, Ward and coworkers developed a computer search algorithm, called “Search for Three dimensional Atom Motifs in Protein Structure” (STAMPS), which help identify protein structures with a given topological motif similar to the functional motif in a reference protein, through a systematic search in the protein databank (PDB).301,302 Using this approach, the authors demonstrated that it is possible, by in silico study, to create an artificial metallo-peroxidase upon addition of a metal cofactor to a nonmetal containing protein, that harbor a potential metal binding site. A search within the PDB allowed to identify pre-organized two histidine and one carboxylate triads in proteins, that appeared suitable for metal ion coordination.303 Several scaffolds bearing an HHD/E motif, located within a pocket predisposed to bind metals, were selected. The search was further expanded to include HHN/Q motifs, revealing six additional potential metal binders following a single point mutation. Out of the thirteen cloned proteins, six were overexpressed in E. coli. One of these proteins, 6-phosphogluconolactonase, bearing a N131D mutation (6-PGLac) bound CuII and showed peroxidase activity toward o-dianisidine. Structural insights into the metalloenzyme activity were gained by x-ray crystallography that confirmed copper ion to be coordinated by two His residues. This strategy looks very promising since it is possible to generate novel functionality within a protein scaffold, and can be valuable for mechanistic enzymology and evolutionary analysis of enzymatic activity.
Another successful example of redesigning protein activity is the work carried out by Sheldon and coworkers, who developed semi-synthetic peroxidase through the incorporation of vanadate ions into acid phosphatases and phytases.304-307 Towards this goal, the authors exploited the structural similarity between vanadium haloperoxidases and the acid phosphatases. To begin with, vanadate (VO43−) and phosphate ions (PO43−) are very similar in their structures. In addition, in the apo-form, vanadium haloperoxidases (which contains a vanadate ion) exhibits phosphatase-like activity, while vanadate ions are potent inhibitors of the phytases and sulfatases activity. Thus, Vanadium haloperoxidases and phosphatases (which bonds and hydroxylates phosphate) were considered to have similar active sites. (Fig. 25).308 Based on these structural and functional similarities, the authors incorporated vanadate ion into phosphatases to produce a novel, semi-synthetic peroxidases. The resulting vanadium phosphatase catalyzed the enantioselective oxidation of several sulfides, with slight ee. Furthermore, the semi-synthetic peroxidase was found to be more stable at relatively high H2O2 concentrations (as compared to Vanadium haloperoxidase) displaying catalytic activity approaching that of heme peroxidases (see Table 3).
Fig. 25.
Active site crystal structure of a) vanadium chloroperoxidase (PDB ID: 3w36) and b) phytase (PDB ID: 4aro)
Table 3.
Comparison of the catalytic properties of three different peroxidases based on the oxidation of thioanisolea
| Heme CPO C. fumago | Phytase/VO43− | VBPO A. nodosum | |
|---|---|---|---|
| ee (%) | 99 (R) | 68 (S) | 83 (R) |
| TOF (min−1) | 900 | 5.5 | 0.78 |
| TTN (-) | 108000 | 25000 | 750 |
Adapted from ref. [307]
Another method to incorporate metal cofactors artificially in protein scaffold is by addition of metal-coordinating unnatural amino acids (UAAs).77,309 UAAs are now routinely being incorporated into peptides and proteins to enable functions beyond those accessible using natural amino acids.310-314 UAAs have been used to enable new modes of metal coordination or covalent attachment, which has led to construction of several new artificial metalloenzymes and metallopeptides not possible using natural amino acids. In particular, Lewis and coworkers have recently used protein scaffolds, selectively modified with the UAA residue p-azido-l-phenylalanine (Az), to covalently link catalytically active alkyne-substituted metal complexes, via strain-promoted azide-alkyne cycloaddition.315 For example, an alkyne-substituted dirhodium catalyst, when incorporated into prolyl oligopeptidase (POP), performed highly selective (92% ee) olefin cyclopropanation.316 Even though this strategy has not yet used for developing oxygen-activating metalloenzymes, it deserves great attention since it can be used, in principle, to conjugate any desired metal complex to various scaffolds, targeting different functions.
5. Future targets for metalloprotein design
Many oxygen-activating metalloenzymes have recently been discovered and characterized, offering exciting opportunities for the design and engineering of novel artificial enzymes. In this section, we give few examples of iron and copper-based oxygen activating enzymes, which represent important future targets for metalloprotein design. Reproducing their intriguing features in model systems will not only expand the repertoire of artificial metalloenzymes but it will also be valuable in biotechnological applications.
α-Ketoglutarate (αKG) dependent dioxygenase and halogenase
They are a class of mononuclear non-heme iron-dependent dioxygenases that require α-KG as cosubstrate to perform catalysis. In fact, these enzymes couple the oxidative decarboxylation of the cofactor α-KG to carbon dioxide and succinate to the oxidative transformations of a variety of substrates. Crystallographic data from several member of this class reveal a common structural arrangement of the active site that consists of a Fe(II) metal center, facially coordinated by two histidines and one carboxylate ligand, from either a glutamate or aspartate residue.5 This “facial triad motif” leaves empty three remaining sites, which are occupied by weakly bound solvent molecules in the resting-state enzyme, and thus are available for binding of additional exogenous ligands, as the α-KG cofactor or substrates. This flexibility in coordination environment at the metal accounts for the observed diversity in catalyzed oxidative transformations.8
Among (αKG) dependent dioxygenase, taurine/2-oxoglutyrate dioxygenase (TauD) catalyzes the hydroxylation of taurine TauD) enabling E. coli to use the aliphatic sulfonate taurine as a sulfur source during periods of sulfate starvation.317 The nonheme iron in TauD activates the oxygen molecule, cleaving the O-O bond to obtain a high-valent iron oxo intermediate, which is then able to abstract an H radical from the nearby C-H bond to perform hydroxylation. Analogous to TauD are another set of nonheme iron dependent enzymes, halogenases that catalyze the site-selective chlorination/bromination of saturated hydrocarbon (specifically the C-4 position of amino acid Thr).318 The crystal structure of SyrB2 halogenase in the resting state reveals that the nonheme iron exhibits a facial triad geometry similar to TauD, except that the Glu/Asp ligand of TauD is replaced by a halogenide (Cl− or Br−).319 Furthermore, the mechanism of oxygen/C-H bond activation and OO bond cleavage in SyrB2 are also similar to TauD except that the halogenase very selectively halogenates the C-H bond with almost no hydroxylated product.320 Studies into the structure-function relationship of SyrB2 have revealed that the hydrogen-bonding to the ferryl intermediate, as well as the positioning of substrate, play important role in controlling the chemo- and regio-selectivity of this reaction, However, exact details of this control is not understood especially because the SyrB2 is dependent on a conjugate enzyme SyrB1.321-323 These two examples highlight once again that similar active sites are able to perform very different functions, depending on the protein environment. Thus, they are interesting candidates for metalloprotein design, as controlling secondary coordination sphere interactions and substrate positioning is nowadays feasible using the tools of protein design and engineering.
Polysaccharide monooxygenases
An exciting target for future copper-protein design and engineering is the polysaccharide monooxygenases (PMOs), also known as lytic PMOs (LPMOs), which utilize molecular oxygen to cleave glycosidic bonds.18-21 The interests in this class of enzymes are growing considerably, since they enhance the depolymerization of recalcitrant polysaccharides, such as cellulose, chitin and diverse marine polysaccharides, by hydrolytic cleavage. In this respect, PMOs hold a commercial potential in the enhancement of biomass degradation, and could find application in industrial biofuel production. Several structural, spectroscopic and activity studies, have just started to shed light on the features of these powerful enzymes.20,22 PMOs are now known to be copper-dependent oxygenases with unusual active sites, since they perform O2 activation using only a single Cu center. The PMO active site contains a mononuclear type II copper center coordinated by three equatorial nitrogen ligands in a histidine brace motif. Solomon and coworkers have recently determined the coordination properties of the PMO from T. aurantiacus in solution.22 Using a combination of spectroscopic and computational analysis, they found a different geometry for Cu(II) and Cu(I): a four-coordinate tetragonal geometry in the oxidized state, and a three-coordinate T-shaped structure in the reduced state. Three protein-derived nitrogen ligands coordinate copper in both redox state, whereas a labile hydroxide ligand is lost upon reduction. The unique protein structure favors the formation of the T-shaped Cu(I) site, providing an open coordination position for strong O2 binding with very little reorganization energy, and enabling the thermodynamically difficult one-electron reduction of O2 by Cu(II)-superoxide formation. This work represents an important step toward elucidating the mechanism by which mononuclear Cu oxygenases activate the inert O2 molecule by one-electron reduction for subsequent degradation of polysaccharides. Reproducing this mechanism of O2 reduction by a single copper center in either de novo designed or engineered protein scaffolds will be challenging for protein designers, but will also aid in the development of more and more efficient enzymes.
Methane monoxygenases (MMOs)
MMOs are key enzymes in the aerobic bacterial degradation of hydrocarbons. Their catalytic chemistry is extraordinary, since they catalyze the activation of the extremely strong C-H bond of methane to methanol, under ambient conditions, without over-oxidation.324-327 There are two distinct types of MMO, an iron-dependent, soluble cytosolic protein (sMMO), and a copper-dependent membrane protein, particulate MMO (pMMO). Even though the two enzymes perform the same transformation, their structures, active sites, and chemical mechanism are completely different. A huge amount of structural and functional data are available for the sMMO, and in particular for its diiron active center. In contrast, structural features responsible for activity of copper-containg pMMO and its catalytic mechanism remain to be fully elucidated.324-328 The protein models of MMOs may help in understanding how the two different MMO metal centers efficiently accomplish this challenging chemistry under physiological conditions.
6. Conclusion and future perspectives
The field of design and engineering of artificial metalloenzymes has progressed spectacularly within the last decade. What started as minimal structural mimics that have no activity has evolved into functional metalloenzymes achieving rates close to that of native enzymes. Much of the progress has been made as a result of recent development in computational and molecular biology. At the same time, advances in structural biology, in terms of obtaining crystallographic and NMR structures, also help in better characterization of the resulting designed enzymes to further improve them. In this review, we have used design of oxygen-activating metalloenzymes as examples to illustrate progress made so far and to highlight approaches employed to achieve this progress. Significant advances has been made in designing many oxygen-activating metalloenzymes by miniaturization, by de novo design and by protein redesign using native protein scaffolds. For all three practices, both rational design using computer modeling and directed evolution or combinatorial selection have been employed. Suitable practices or approaches to be used depend on: i) the level of the understanding of the enzyme one aims to design; ii) the reaction one targets to achieve; iii)the technical difficulty inherent to the selected design. For instance, protein miniaturization is preferred if the geometric parameters of the metal cofactors are well defined, with minimal help from the protein scaffolds to maintain its stability and integrity.37,113 Different types of heme cofactors are excellent examples and that's why miniaturization of heme enzymes have enjoyed most success in this practice.85,113 When the geometric parameters of the metal-binding sites require substantial participation of protein scaffolds to define and maintain, such as diiron centers, one needs to consider applying either design using de novo scaffolds, or redesign using native scaffolds. Protein design using de novo scaffolds is an ultimate goal in engineering enzymes, as it is a testimony of complete understanding of the proteins. Only limited de novo designed scaffolds are available and most of success in de novo design of functional metalloproteins has been reached using helical bundles as scaffolds35,38,39,61 They are so stable to tolerate multiple mutations without disruption of the global folding. This is allowing one to explore how changes to the first- and second-shell ligands, as well as residues lining the substrate binding pocket, affect the reactivity and catalytic properties of the metal site. The design of metal sites that are natively inserted in helical bundles is relatively easy, thus allowing one to unravel each specific contribution to function. This also enables to clearly identify which feature could be responsible for switching towards alternative reactivity. In this perspective, various examples show how the initial designed function can be finely tuned, or even reprogrammed.162,165,173 Moreover, it is possible to engineer metal sites that are not natively found in helical bundles, by taking advantage of intrinsic symmetry of the natural coordination environment, which can be fitted well in a de novo designed coiled coil. An outstanding example is reported by Pecoraro and coworkers, who engineered the threefold symmetric site of carbonic anhydrase, an all beta enzyme, into a three-stranded coiled coil.58 Finally, it should be noted that the field is expanding, and other than α-folds are being investigated, in an attempt to fully de novo design protein structures beyond those found in Nature.329
For metal-binding sites that are not easily designable in helical bundles, design using native scaffolds becomes advantageous. In addition, the combination of gains in our knowledge and experience in protein design together with recent advances in modern computational biology and biophysical characterization techniques, rational design has enjoyed much success recently. However, there are still a number of gaps in our knowledge and computational methods, especially when we wish to design enzymes with very high activity. In this case, directed evolutions become very useful and can complement rational design well. In fact, some of the most successful examples of artificial enzymes are results of using the combined approach.202,330,331
Most reported designs of metalloenzymes aim to gain deeper insights in how metalloproteins are assembled. Given much smaller and more well-defined structures, these designed proteins have provided deeper insights into the structure, mechanism and function of the “target” protein. For example, in the design of N-oxygenase, DeGrado and coworkers contributed to clarify the role of the third histidine in the primary coordination sphere of a diiron center in the N-oxygenation reaction.165 Moreover, designing the CcO active site in myoglobin gave considerable inputs into the role of different structural features like conserved tyrosines and different heme types in CcOs.25,228,234 In some exceptional cases, a number of artificial metalloenzymes have been engineered to have an impact in chemical and biotechnological applications. For example, CYP mutants developed by Arnold and coworkers can now oxidize ethane to ethanol (an industrial raw material which is typically produced via high-energy thermochemical methods) with high selectivity and no over-oxidation products.206 This CYP mutant along with others are currently being utilized in a biotech startup companies to obtain fuel/raw materials from cellulose and lignin.
While the progress made so far is quite exciting, much more work is required to reach the full potential of the field. For example, most artificial metalloenzymes designed for oxygen-activation reviewed here use highly reactive and unstable H2O2 as the sacrificial oxidant. For further advancement of this field and to be able to use these designed metalloenzymes for whole cells biotransformation suitable for fermentation, metalloenzymes using bioavailable and inexpensive O2 as the oxidant need to be developed. This is especially important if we wish to scale up the reactions for industrial use. The issue with using oxygen is that the O-O bond is difficult to break efficiently, thus lessons should be learnt from native enzymes like CYPs332 to design oxygen utilizing artificial metalloenzymes.
For nonheme metal-binding sites, the metal-binding affinity of the designed site is normally much weaker than those of native enzymes, making the metal-binding site much less selective than those of native enzymes, and making the designed metalloenzyme more difficult to characterize and to carry out enzymatic assays at low concentrations. Such limitations could be overcome through site-specific incorporation of metal-chelating unnatural amino acids by means of recent genetic code expansion techniques. This strategy can in the same time recapitulate the functions of metalloproteins bearing uncommon metal binding sites and expand the native functions resulting in molecular systems with unprecedented efficiency and selectivity.314
More importantly, with a few exceptions,58,59,113,162,228 most designed metalloenzymes have very low activity and turnover numbers. A main contributing factor to both issues is the non-covalent secondary sphere interactions around the primary coordination sphere that influence both the metal-binding affinity and enzymatic efficiency. While more recent reports have begun to address this issue,113,162,165,170,238 more attention needs to be paid in designing non-covalent secondary sphere interactions.
Furthermore, only a very small number of the oxygen-activation enzymes described in the review perform in whole cells that are amenable to producing products by fermentation. To overcome this challenge, more emphasis should be given into using moderate oxidants like oxygen and chlorite for catalytic biotransformation. Moreover, light energy can be stored in form of chemical energy by converting light into energy for oxidation reaction. One such recent example is by Lewis and coworkers who utilized acridinium cofactor embedded in a protein to perform sulfoxidation.333 Rubipy conjugated heme has been incorporated in CYPs in attempts to perform light based C-H bond oxidation.334 While, this is an excellent idea, the photo-catalysts designed until now are not very selective and display low quantum efficiency. Thus, more impetus should be provided to develop light based-oxidation catalysts.
Overall, the metalloenzyme design has allowed chemists, biochemists and chemical engineers to answer challenging questions about metal biochemistry, the importance of the protein matrix, and ultimately be able to design new metalloproteins capable of performing desired functions not necessarily in the repertoire of biology. The examples herein discussed make significant progress toward these goals.
Supplementary Material
Acknowledgements
We wish to thank all co-workers for their contributions to the results described in this review, as well as Fabrizia Sibillo for help with editing the manuscript. The work from authors’ groups reported herein has been supported by US National Institutes of Health (GM062211), US National Science Foundation (CHE 14-13328), the European Union (EU) (Cost Action CM1003 - Biological Oxidation Reactions: Mechanisms and Design of New Catalysts) and the Scientific Research Department of Campania Region (BIP Project, POR FESR 2007/2013, grant number B25C13000290007, and the STRAIN Project, POR FSE 2007/2013, grant number B25B0900000000 for a postdoctoral fellowship to M. C.).
References
- 1.Doble MV, Ward ACC, Deuss PJ, Jarvis AG, Kamer PCJ. Bioorg. Med. Chem. 2014;22:5657–5677. doi: 10.1016/j.bmc.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Hess CR, Welford RWD, Klinman JP. In: Wiley Encyclopedia of Chemical Biology. Wiley John., editor. Vol. 3. 2009. pp. 529–540. [Google Scholar]
- 3.Decker A, Solomon EI. Curr. Opin. Chem. Biol. 2005;9:152–163. doi: 10.1016/j.cbpa.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Ortiz de Montellano PR. Chem. Rev. 2010;110:932–948. doi: 10.1021/cr9002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rude MA, Baron TS, Brubaker S, Alibhai M, Del Cardayre SB, Schirmer A. Appl. Environ. Microbiol. 2011;77:1718–1727. doi: 10.1128/AEM.02580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belcher J, McLean KJ, Matthews S, Woodward LS, Fisher K, Rigby SEJ, Nelson DR, Potts D, Baynham MT, Parker DA, Leys D, Munro AW. J. Biol. Chem. 2014;289:6535–6550. doi: 10.1074/jbc.M113.527325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koehntop KD, Emerson JP, Que Jr L. J. Biol. Inorg. Chem. 2005;10:87–93. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- 8.Bruijnincx PCA, van Koten G, Klein Gebbink RJM. Chem. Soc. Rev. 2008;37:2716–2744. doi: 10.1039/b707179p. [DOI] [PubMed] [Google Scholar]
- 9.Wallar BJ, Lipscomb JD. Chem. Rev. 1996;96:2625–2657. doi: 10.1021/cr9500489. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Liang AD, Lippard SJ. Acc. Chem. Res. 2015;48:2632–2639. doi: 10.1021/acs.accounts.5b00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tinberg CE, Lippard SJ. Acc. Chem. Res. 2011;44:280–288. doi: 10.1021/ar1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindqvist Y, Huang W, Schneider G, Shanklin J. EMBO J. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- 13.Fox BG, Shanklin J, Ai J, Loehr TM, Sanders-Loehr J. Biochemistry. 1994;33:12776–12786. doi: 10.1021/bi00209a008. [DOI] [PubMed] [Google Scholar]
- 14.Rosenzweig AC, Sazinsky MH. Curr. Opin. Struct. Biol. 2006;16:729–735. doi: 10.1016/j.sbi.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Solomon EI, Sundaram U, Machonkin T. Chem. Rev. 1996;96:2563–2606. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- 16.Solomon EI, Heppner DE, Johnston EM, Ginsbach JW, Cirera J, Qayyum M, Kieber-Emmons MT, Kjaergaard CH, Hadt RG, Tian L. Chem. Rev. 2014;114:3659–3853. doi: 10.1021/cr400327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dwivedi UN, Singh P, Pandey VP, Kumar A. J. Mol. Cat. B. 2011;68:117–128. [Google Scholar]
- 18.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sørlie M, Eijsink VGH. Science. 2010;330:219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 19.Quinlan RJ, Sweeney MD, Leggio LL, Otten H, Poulsen JCN, Salomon Johansen K, Krogh KBRM, Jørgensen CI, Tovborg M, Anthonsen A, Tryfona T, Walter CP, Dupree P, Xu F, Davies GJ, Walton PH. Proc. Nat. Acad. Sci. USA. 2011;108:15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beeson WT, Vu VV, Span EA, Phillips CM, Marletta MA. Annu. Rev. Biochem. 2015;84:923–946. doi: 10.1146/annurev-biochem-060614-034439. [DOI] [PubMed] [Google Scholar]
- 21.Hemsworth GR, Henrissat B, Davies GJ, Walton PH. Nat. Chem. Biol. 2014;10:122–126. doi: 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kjaergaard CH, Qayyum MF, Wong SD, Xu F, Hemsworth GR, Walton DJ, Young NA, Davies GJ, Walton PH, Salomon Johansen K, Hodgson,f KO, Hedman B, Solomon EI. Proc. Nat. Acad. Sci. USA. 2014;111:8797–8802. doi: 10.1073/pnas.1408115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleespies ST, Oloo WN, Mukherjee A, Lawrence Que., Jr. Inorg. Chem. 2015;54:5053–5064. doi: 10.1021/ic502786y. [DOI] [PubMed] [Google Scholar]
- 24.Puri M, Que L. Jr., Acc. Chem. Res. 2015;48:2443–2452. doi: 10.1021/acs.accounts.5b00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itoh S. Acc. Chem. Res. 2015;48:2066–2074. doi: 10.1021/acs.accounts.5b00140. [DOI] [PubMed] [Google Scholar]
- 26.Che CM, Huang JS. Chem. Commun. 2009;27:3996–4015. doi: 10.1039/b901221d. [DOI] [PubMed] [Google Scholar]
- 27.Costas M. Coord. Chem. Rev. 2011;255:2912–2932. [Google Scholar]
- 28.Serrano-Plana J, Garcia-Bosch I, Company A, Costas M. Acc. Chem. Res. 2015;48:2397–2406. doi: 10.1021/acs.accounts.5b00187. [DOI] [PubMed] [Google Scholar]
- 29.Friedle S, Reisner E, Lippard SJ. Chem. Soc. Rev. 2010;39:2768–2779. doi: 10.1039/c003079c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hematian S, Garcia-Bosch I, Karlin KD. Acc. Chem. Res. 2015;48:2462–2474. doi: 10.1021/acs.accounts.5b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rolff M, Schottenheim J, Decker H, Tuczek F. Chem. Soc. Rev. 2011;40:4077–4098. doi: 10.1039/c0cs00202j. [DOI] [PubMed] [Google Scholar]
- 32.Dalle KE, Meyer F. Eur. J. Inorg. Chem. 2015;21:3391–3405. [Google Scholar]
- 33.Abdolahzadeh S, de Boer JW, Browne WR. Eur. J. Inorg. Chem. 2015;21:3432–3456. [Google Scholar]
- 34.Maglio O, Nastri F, Lombardi A. In: Ionic Interactions in Natural and Synthetic Macromolecules. Ciferri A, Perico A, editors. John Wiley & Sons; Hoboken (New Jersey): 2012. pp. 361–450. [Google Scholar]
- 35.Nastri F, Bruni R, Maglio O, Lombardi A. In: Coordination Chemistry in Protein Cages: Principles, Design, and Applications. Ueno T, Watanabe Y, editors. John Wiley & Sons; Hoboken (New Jersey): 2013. pp. 45–85. [Google Scholar]
- 36.Xing G, DeRose VJ. Curr. Opin. Chem. Biol. 2001;5:196–200. doi: 10.1016/s1367-5931(00)00190-3. [DOI] [PubMed] [Google Scholar]
- 37.Lombardi A, Marasco D, Maglio O, Di Costanzo L, Nastri F, Pavone V. Proc. Nat. Acad. Sci. USA. 2000;97:11922–11927. doi: 10.1073/pnas.97.22.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zastrow ML, Pecoraro VL. Coord. Chem. Rev. 2013;257:2565–2588. doi: 10.1016/j.ccr.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu F, Cangelosi VM, Zastrow ML, Tegoni M, Plegaria JS, Tebo AG, Mocny CS, Ruckthong L, Qayyum H, Pecoraro VL. Chem. Rev. 2014;114:3495–3578. doi: 10.1021/cr400458x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tebo AG, Pecoraro VL. Curr. Opin. Chem. Biol. 2015;25:65–70. doi: 10.1016/j.cbpa.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. Annu. Rev. Biochem. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- 42.Lu Y, Yeung N, Sieracki N, Marshall NM. Nature. 2009;460:855–862. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Y, Berry SM, Pfister TD. Chem. Rev. 2001;101:3047–3080. doi: 10.1021/cr0000574. [DOI] [PubMed] [Google Scholar]
- 44.Petrik ID, Liu J, Lu Y. Curr. Opin. Chem. Biol. 2014;19:67–75. doi: 10.1016/j.cbpa.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Durrenberger M, Ward TR. Curr. Opin. Chem. Biol. 2014;19:99–106. doi: 10.1016/j.cbpa.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 46.Ward TR. Acc. Chem. Res. 2011;44:47–57. doi: 10.1021/ar100099u. [DOI] [PubMed] [Google Scholar]
- 47.Matsuo T, Hirota S. Bioorg. Med. Chem. 2014;22:5638–5656. doi: 10.1016/j.bmc.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 48.Lazar GA, Handel TM. Curr. Opin. Chem. Biol. 1998;2:675–679. doi: 10.1016/s1367-5931(98)80102-6. [DOI] [PubMed] [Google Scholar]
- 49.Butterfoss GL, Kuhlman B. Annu. Rev. Biophys. Biomol. Struct. 2006;35:49–65. doi: 10.1146/annurev.biophys.35.040405.102046. [DOI] [PubMed] [Google Scholar]
- 50.Zanghellini A, Jiang L, Wollacott AM, Cheng G, Meiler J, Althoff EA, Röthlisberger D, Baker D. Protein Sci. 2006;15:2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuhlman B, Baker D. Proc. Nat. Acad. Sci. USA. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baker D. J. Am. Chem. Soc. 2013;135:13393–13399. doi: 10.1021/ja403503m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohn WD, Hodge RS. Tibtech. 1998;16:379–389. [Google Scholar]
- 54.Baltzer L, Nilsson H, Nilsson J. Chem. Rev. 2001;101:3153–3163. doi: 10.1021/cr0000473. [DOI] [PubMed] [Google Scholar]
- 55.Holm RH, Kennepohl P, Solomon EI. Chem. Rev. 1996;96:2239–2314. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]
- 56.Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. University Science Books; Mill Valley, CA: 1994. [Google Scholar]
- 57.Maglio O, Nastri F, Pavone V, Lombardi A, DeGrado WF. Proc. Nat. Acad. Sci. USA. 2003;100:3772–3777. doi: 10.1073/pnas.0730771100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zastrow ML, Peacock AF, Stuckey JA, Pecoraro VL. Nat. Chem. 2012;4:118–123. doi: 10.1038/nchem.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cangelosi VM, Deb A, Penner-Hahn JE, Pecoraro VL. Angew. Chem. Int. Ed. 2014;53:7900–7903. doi: 10.1002/anie.201404925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosh D, Pecoraro VL. Curr. Opin. Chem. Biol. 2005;9:97–103. doi: 10.1016/j.cbpa.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 61.Mocny CS, Pecoraro VL. Acc. Chem. Res. 2015;48:2388–2396. doi: 10.1021/acs.accounts.5b00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khare SD, Kipnis Y, Jr Greisen P, Takeuchi R, Ashani Y, Goldsmith M, Song Y, Gallaher JL, Silman I, Leader H, Sussman JL, Stoddard BL, Tawfik DS, Baker D. Nat. Chem. Biol. 2012;8:294–300. doi: 10.1038/nchembio.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeung N, Lin YW, Gao YG, Zhao X, Russell BS, Lei L, Miner KD, Robinson H, Lu Y. Nature. 2009;426:1079–1082. doi: 10.1038/nature08620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin YW, Yeung N, Gao YG, Miner KD, Tian S, Robinson H, Lu Y. Proc. Nat. Acad. Sci. USA. 2010;107:8581–8586. doi: 10.1073/pnas.1000526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin YW, Yeung N, Gao YG, Miner KD, Lei L, Robinson H, Lu Y. J. Am. Chem. Soc. 2010;132:9970–9972. doi: 10.1021/ja103516n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bos J, Roelfes G. Curr. Opin. Chem. Biol. 2014;19:135–143. doi: 10.1016/j.cbpa.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 67.Creus M, Ward TR. Prog. Inorg. Chem. 2012;57:203–253. [Google Scholar]
- 68.Deuss PJ, den Heeten R, Laan W, Kamer PCJ. Chem. Eur. J. 2011;17:4680–4698. doi: 10.1002/chem.201003646. [DOI] [PubMed] [Google Scholar]
- 69.Drienovska I, Rioz-Martinez A, Draksharapu A, Roelfes G. Chem. Sci. 2015;6(1):770–776. doi: 10.1039/c4sc01525h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heinisch T, Ward TR. Curr. Opin. Chem. Biol. 2010;14(2):184–199. doi: 10.1016/j.cbpa.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 71.Heinisch T, Ward TR. Eur. J. Inorg. Chem. 2015;2015(21):3406–3418. [Google Scholar]
- 72.Hoarau M, Hureau C, Gras E, Faller P. Coord. Chem. Rev. 2015 (Ahead of Print) [Google Scholar]
- 73.Ilie A, Reetz MT. Isr. J. Chem. 2015;55(1):51–60. [Google Scholar]
- 74.Koehler V, Wilson YM, Lo C, Sardo A, Ward TR. Curr. Opin. Biotechnol. 2010;21(6):744–752. doi: 10.1016/j.copbio.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 75.Kohler V, Turner NJ. Chem. Commun. 2015;51(3):450–464. doi: 10.1039/c4cc07277d. [DOI] [PubMed] [Google Scholar]
- 76.Lewis JC. ACS Catal. 2013;3(12):2954–2975. [Google Scholar]
- 77.Lewis JC. Curr. Opin. Chem. Biol. 2015;25:27–35. doi: 10.1016/j.cbpa.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marchi-Delapierre C, Rondot L, Cavazza C, Menage S. Isr. J. Chem. 2015;55(1):61–75. [Google Scholar]
- 79.Praneeth VK, Ward TR. John Wiley & Sons, Inc.; 2013. [Google Scholar]
- 80.Ringenberg MR, Ward TR. Chem. Commun. 2011;47(30):8470–8476. doi: 10.1039/c1cc11592h. [DOI] [PubMed] [Google Scholar]
- 81.Rosati F, Roelfes G. ChemCatChem. 2010;2(8):916–927. [Google Scholar]
- 82.Cunningham BC, Wells JA. Curr. Opin. Struct. Biol. 1997;7:457–462. doi: 10.1016/s0959-440x(97)80107-8. [DOI] [PubMed] [Google Scholar]
- 83.Nar H, Messerschmidt A, Huber R, van de Kamp M, Canters GW. J. Mol. Biol. 1991;221:765–772. doi: 10.1016/0022-2836(91)80173-r. [DOI] [PubMed] [Google Scholar]
- 84.Nastri F, Lombardi A, D'Andrea LD, Sanseverino M, Maglio O, Pavone V. Biopolymers. 1998;47:5–22. doi: 10.1002/(SICI)1097-0282(1998)47:1<5::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 85.Lombardi A, Nastri F, Pavone V. Chem. Rev. 2001;101:3165–3189. doi: 10.1021/cr000055j. [DOI] [PubMed] [Google Scholar]
- 86.Reedy CJ, Gibney BR. Chem. Rev. 2004;104:617–650. doi: 10.1021/cr0206115. [DOI] [PubMed] [Google Scholar]
- 87.Dolphin D. The Porphyrins. Vol. 7. Academic Press; New York: 1979. [Google Scholar]
- 88.Munro OQ, Marques HM. Inorg. Chem. 1996;35:3752–3767. doi: 10.1021/ic9502842. [DOI] [PubMed] [Google Scholar]
- 89.Marques HM. Dalton Trans. 2007;39:4371–4385. doi: 10.1039/b710940g. [DOI] [PubMed] [Google Scholar]
- 90.Nicolis S, Casella L, Roncone R, Dallacosta C, Monzani E. C. R. Chimie. 2007;10:380–391. [Google Scholar]
- 91.Dallacosta C, Casella L, Monzani E. ChemBioChem. 2004;5:1692–1699. doi: 10.1002/cbic.200400175. [DOI] [PubMed] [Google Scholar]
- 92.Ni TW, Tezcan FA. Angew. Chem. Int. Ed. 2010;49:7014–7018. doi: 10.1002/anie.201001487. [DOI] [PubMed] [Google Scholar]
- 93.Adams PA. J. Chem. Soc., Perkin Trans. 1990;2:1407–1414. [Google Scholar]
- 94.Cunningham ID, Bachelor J, Pratt J. J. Chem. Soc., Perkin Trans. 1991;2:1839–1843. [Google Scholar]
- 95.Cunnigham ID, Snare GR. J. Chem. Soc., Perkin Trans. 1992;2:2019–2023. [Google Scholar]
- 96.Cunningham ID, Bachelor J, Pratt J. J. Chem. Soc., Perkin Trans. 1994;2:1347–1350. [Google Scholar]
- 97.Osman AM, Koerts J, Boersma MG, Boeren S, Veeger C, Rietjens I. Eur. J. Biochem. 1996;240:232–238. doi: 10.1111/j.1432-1033.1996.0232h.x. [DOI] [PubMed] [Google Scholar]
- 98.Colonna S, Gaggero N, Carrea G, Pasta P. Tetrahedron Lett. 1994;35:9103–9104. [Google Scholar]
- 99.Casella L, De Gioia L, Frontoso Silvestri G, Monzani E, Redaelli C, Roncone R, Santagostini L. J. Inorg. Biochem. 2000;79:31–39. doi: 10.1016/s0162-0134(99)00243-3. [DOI] [PubMed] [Google Scholar]
- 100.Feng D, Liu T-F, Su J, Bosch M, Wei Z, Wan W, Yuan D, Chen Y-P, Wang X, Wang K, Lian X, Gu Z-Y, Park J, Zou X, Zhou H-C. Nature Comm. 2015:6. doi: 10.1038/ncomms6979. doi:10.1038/ncomms6979. [DOI] [PubMed] [Google Scholar]
- 101.Lykourinou V, Chen Y, Wang X-S, Meng L, Hoang T, Ming L-J, Musselman RL, Ma S. J. Am. Chem. Soc. 2011;133:10382–10385. doi: 10.1021/ja2038003. [DOI] [PubMed] [Google Scholar]
- 102.Wu X, Hou M, Ge J. Catal. Sci. Technol. 2015;5:5077–5085. [Google Scholar]
- 103.Nastri F, Lombardi A, Morelli G, Maglio O, D'Auria G, Pedone C, Pavone V. Chem. Eur. J. 1997;3:340–349. [Google Scholar]
- 104.D'Auria G, Maglio O, Nastri F, Lombardi A, Mazzeo M, Morelli G, Paolillo L, Pedone C, Pavone V. Chem. Eur. J. 1997;3:350–362. [Google Scholar]
- 105.Nastri F, Lombardi A, Morelli G, Pedone C, Pavone V, Chottard G, Battioni P, Mansuy D, Biol J. Inorg. Chem. 1998;3:671–681. [Google Scholar]
- 106.Lombardi A, Nastri F, Sanseverino M, Maglio O, Pedone C, Pavone V. Inorg. Chim. Acta. 1998;301:275–276. [Google Scholar]
- 107.Lombardi A, Nastri F, Marasco D, Maglio O, De Sanctis G, Sinibaldi F, Santucci R, Coletta M, Pavone V. Chem. Eur. J. 2003;9:5643–5654. doi: 10.1002/chem.200304831. [DOI] [PubMed] [Google Scholar]
- 108.Di Costanzo L, Geremia S, Randaccio L, Nastri F, Maglio O, Lombardi A, Pavone V. J. Biol. Inorg. Chem. 2004;9:1017–1027. doi: 10.1007/s00775-004-0600-x. [DOI] [PubMed] [Google Scholar]
- 109.Vicari C, Saraiva IH, Maglio O, Nastri F, Pavone V, Louro RO, Lombardi A. Chem. Commun. 2014;50:3852–3855. doi: 10.1039/c3cc49123d. [DOI] [PubMed] [Google Scholar]
- 110.Nastri F, Lista L, Ringhieri P, Vitale R, Faiella M, Andreozzi C, Travascio P, Maglio O, Lombardi A, Pavone V. Chem. Eur. J. 2011;17:4444–4453. doi: 10.1002/chem.201003485. [DOI] [PubMed] [Google Scholar]
- 111.Ranieri A, Monari S, Sola M, Borsari M, Battistuzzi G, Ringhieri P, Nastri F, Pavone V, Lombardi A. Langmuir. 2010;26:17831–17835. doi: 10.1021/la103744x. [DOI] [PubMed] [Google Scholar]
- 112.Vitale R, Lista L, Lau-Truong S, Tucker RT, Brett MJ, Limoges B, Pavone V, Lombardi A, Balland V. Chem. Commun. 2014;50:1894–1896. doi: 10.1039/c3cc48489k. [DOI] [PubMed] [Google Scholar]
- 113.Vitale R, Lista Liliana, Cerrone C, Caserta G, Chino M, Maglio O, Nastri F, Pavone V, Lombardi A. Org. Biomol. Chem. 2015;13:4859–4868. doi: 10.1039/c5ob00257e. [DOI] [PubMed] [Google Scholar]
- 114.Perutz MF, Hasnain SS, Duke PJ, Sessler JL, Hahn JE. Nature. 1982;295:535–538. doi: 10.1038/295535a0. [DOI] [PubMed] [Google Scholar]
- 115.O'Neil KT, DeGrado WF. Science. 1990;250:646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 116.Creamer TP, Rose GD. Proteins Struct. Funct. Genet. 1994;19:85–97. doi: 10.1002/prot.340190202. [DOI] [PubMed] [Google Scholar]
- 117.Serrano L, Fersht AR. Nature. 1989;342:296–299. doi: 10.1038/342296a0. [DOI] [PubMed] [Google Scholar]
- 118.Savenkova MI, Kuo JM, Ortiz de Montellano PR. Biochemistry. 1998;37:10828–10836. doi: 10.1021/bi9725780. [DOI] [PubMed] [Google Scholar]
- 119.Newmyer SL, de Montellano PR. J. Biol. Chem. 1996;271:14891–14896. doi: 10.1074/jbc.271.25.14891. [DOI] [PubMed] [Google Scholar]
- 120.Maglio O, Nastri F, Torres Martin de Rosales R, Faiella M, Pavone V, DeGrado WF, Lombardi A. C. R. Chimie. 2007;10:703–720. [Google Scholar]
- 121.Calhoun JR, Nastri F, Maglio O, Pavone V, Lombardi A, DeGrado WF. Biopolymers. 2005;80:264–278. doi: 10.1002/bip.20230. [DOI] [PubMed] [Google Scholar]
- 122.Chino M, Maglio O, Nastri F, Pavone V, DeGrado WF, Lombardi A. Eur. J. Inorg. Chem. 2015;21:3371–3390. doi: 10.1002/ejic.201500470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lombardi A. Nat. Chem. Biol. 2015;11:760–761. doi: 10.1038/nchembio.1918. [DOI] [PubMed] [Google Scholar]
- 124.Grigoryan G, DeGrado WF. J. Mol. Biol. 2011;405:1079–1100. doi: 10.1016/j.jmb.2010.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Crick FH. Acta Crystallogr. 1953;6:685–689. [Google Scholar]
- 126.Wood CW, Bruning M, Ibarra AA, Bartlett GJ, Thomson AR, Sessions RB, Brady RL, Woolfson DN. Bioinformatics. 2014;30:3029–3035. doi: 10.1093/bioinformatics/btu502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Murzin AG, Lesk AM, Chothia C. Principles determining the structure of β-sheet barrels in proteins. I. A theoretical analysis. J. Mol. Biol. 1994;236:1369–1381. doi: 10.1016/0022-2836(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 128.Rainey JK, Goh MC. A statistically derived parameterization for the collagen triple-helix. Protein Sci. 2002;11:2748–2754. doi: 10.1110/ps.0218502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Summa CM, Lombardi A, Lewis M, DeGrado WF. Curr. Opin. Struct. Biol. 1999;9:500–508. doi: 10.1016/S0959-440X(99)80071-2. [DOI] [PubMed] [Google Scholar]
- 130.Lombardi A, Summa C, Geremia S, Randaccio L, Pavone V, DeGrado WF. Proc. Nat. Acad. Sci. USA. 2000;97:6298–6305. doi: 10.1073/pnas.97.12.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goldberg SD, Soto CS, Waldburger CD, DeGrado WF. Determination of the physiological dimer interface of the PhoQ sensor domain. J. Mol. Biol. 2008;379:656–665. doi: 10.1016/j.jmb.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Verschueren E, Vanhee P, van der Sloot AM, Serrano L, Rousseau F, Schymkowitz J. Current Opinion in Structural Biology. 2011;21:452–459. doi: 10.1016/j.sbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 133.Simons KT, Kooperberg C, Huang E, Baker D. J. Mol. Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 134.Zhou J, Grigoryan G. Protein Science. 2015;24:508–524. doi: 10.1002/pro.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gonzalez G, Hannigan B, DeGrado WF. PLoS Compu.t Biol. 2014;10:e1003750. doi: 10.1371/journal.pcbi.1003750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moll M, Bryant DH, Kavraki LE. BMC Bioinform. 2010;11:555. doi: 10.1186/1471-2105-11-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.He L, Vandin F, Pandurangan G, Bailey-Kellogg C. J. Comput. Biol. 2013;20:137–151. doi: 10.1089/cmb.2012.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.DeGrado WF, Wasserman ZR, Lear JD. Science. 1989;243:622–628. doi: 10.1126/science.2464850. [DOI] [PubMed] [Google Scholar]
- 139.Bryson JW, Betz SF, Lu HS, Suich DJ, Zhou HX, O'Neil KT, DeGrado WF. Science. 1995;270:935–94. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- 140.Willis MA, Bishop B, Regan L, Brunger AT. Structure. 2000;8:1319–1328. doi: 10.1016/s0969-2126(00)00544-x. [DOI] [PubMed] [Google Scholar]
- 141.Hill B, Raleigh DP, Lombardi A, DeGrado WF. Acc. Chem. Res. 2000;33:745–754. doi: 10.1021/ar970004h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Feig AL, Lippard SJ. Chem. Rev. 1994;94:759–805. [Google Scholar]
- 143.Nordlund P, Sjöberg B-M, Eklund H. Nature. 1990;345:593–598. doi: 10.1038/345593a0. [DOI] [PubMed] [Google Scholar]
- 144.Lange SJ, Que L. Curr. Opin. Chem. Biol. 1998;2:159–172. doi: 10.1016/s1367-5931(98)80057-4. [DOI] [PubMed] [Google Scholar]
- 145.Andersson KK, Gräslund A. Adv. Inorg. Chem. 1995;43:359–408. [Google Scholar]
- 146.Nordlund P, Eklund H. Curr. Opin. Struct. Biol. 1995;5:758–766. doi: 10.1016/0959-440x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 147.Lipscomb JD. Annu. Rev. Microbiol. 1994;48:371–399. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- 148.Pasternak A, Kaplan J, Lear JD, DeGrado WF. Protein Sci. 2001;10:958–969. doi: 10.1110/ps.52101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Maglio O, Nastri F, Calhoun JR, Lahr S, Pavone V, DeGrado WF, Lombardi A. J. Biol. Inorg. Chem. 2005;10:539–549. doi: 10.1007/s00775-005-0002-8. [DOI] [PubMed] [Google Scholar]
- 150.Lahr SJ, Engel DE, Stayrook SE, Maglio O, North B, Geremia S, Lombardi A, DeGrado WF. J. Mol. Biol. 2005;346:1441–1454. doi: 10.1016/j.jmb.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 151.Geremia S, Di Costanzo L, Randaccio L, Engel DE, Lombardi A, Nastri F, DeGrado WF. J. Am. Chem. Soc. 2005;127:17266–17276. doi: 10.1021/ja054199x. [DOI] [PubMed] [Google Scholar]
- 152.DeGrado WF, Di Costanzo L, Geremia S, Lombardi A, Pavone V, Randaccio L. Angew. Chem., Int. Ed. 2003;42:417–420. doi: 10.1002/anie.200390127. [DOI] [PubMed] [Google Scholar]
- 153.Di Costanzo L, Wade H, Geremia S, Randaccio L, DeGrado WF, Pavone V, Lombardi A. J. Am. Chem. Soc. 2001;123:12749–12757. doi: 10.1021/ja010506x. [DOI] [PubMed] [Google Scholar]
- 154.Wade H, Stayrook SE, DeGrado WF. Angew. Chem., Int. E. 2006;45:4951–4954. doi: 10.1002/anie.200600042. [DOI] [PubMed] [Google Scholar]
- 155.Wei P, Skulan AJ, Wade H, DeGrado WF, Solomon EI. J. Am. Chem. Soc. 2005;127:16098–16106. doi: 10.1021/ja053661a. [DOI] [PubMed] [Google Scholar]
- 156.Summa CM, Rosenblatt MM, Hong JK, Lear JD, DeGrado WF. J. Mol. Biol. 2002;321:923–938. doi: 10.1016/s0022-2836(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 157.Marsh EN, DeGrado WF. Proc. Nat. Acad. Sci. USA. 2002;99:5150–5154. doi: 10.1073/pnas.052023199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kaplan J, DeGrado WF. Proc. Nat. Acad. Sci. USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Corbett JF. J. Chem. Soc. B. 1969:823–826. [Google Scholar]
- 160.Corbett JF, Gamson EP. J. Chem. Soc. Perkin Trans. 1972;2:1531–1537. [Google Scholar]
- 161.A Berthold D, Stenmark P. Annu. Rev. Plant Biol. 2003;54:497–517. doi: 10.1146/annurev.arplant.54.031902.134915. [DOI] [PubMed] [Google Scholar]
- 162.Faiella M, Andreozzi C, Torres Martin de Rosales R, Pavone V, Maglio O, Nastri F, DeGrado WF, Lombardi A. Nat. Chem. Biol. 2009;5:882–884. doi: 10.1038/nchembio.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Torres Martin de Rosales R, Faiella M, Farquhar E, Que L, Andreozzi C, Pavone V, Maglio O, Nastri F, Lombardi A. J. Biol. Inorg. Chem. 2010;15:717–728. doi: 10.1007/s00775-010-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Calhoun JR, Kono H, Lahr S, Wang W, DeGrado WF, Saven JG. J. Mol. Biol. 2003;334:1101–1115. doi: 10.1016/j.jmb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 165.Reig AJ, Pires MM, Snyder RA, Wu Y, Jo H, Kulp DW, Butch SE, Calhoun JR, Szyperski T, Solomon EI, DeGrado WF. Nat. Chem. 2012;4:900–906. doi: 10.1038/nchem.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Snyder RA, Butch SE, Reig AJ, DeGrado WF, Solomon EI. J. Am. Chem. Soc. 2015;137:9302–9314. doi: 10.1021/jacs.5b03524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Snyder RA, Betzu J, Butch SE, Reig AJ, DeGrado WF, Solomon EI. Biochemistry. 2015;54:4637–4651. doi: 10.1021/acs.biochem.5b00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chanco E, Choi YS, Sun N, Vu M, Zhao H. Bioorg. Med. Chem. 2014;22:5569–5577. doi: 10.1016/j.bmc.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 169.Huang SS, Koder RL, Lewis M, Wand AJ, Dutton PL. Proc. Nat. Acad. Sci. USA. 2004;101:5536–5541. doi: 10.1073/pnas.0306676101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Koder RL, Dutton PL. Dalt. Trans. 2006:3045–3051. doi: 10.1039/b514972j. [DOI] [PubMed] [Google Scholar]
- 171.Shifman JM, Moser CC, Kalsbeck WA, Bocian DF, Dutton PL. Biochemistry. 1998;37:16815–16827. doi: 10.1021/bi9816857. [DOI] [PubMed] [Google Scholar]
- 172.Lichtenstein BR, Farid TA, Kodali G, Solomon LA, Anderson JLR, Sheehan MM, Ennist NM, Fry BA, Chobot SE, Bialas C, Mancini JA, Armstrong CT, Zhao Z, Esipova TV, Snell D, Vinogradov SA, Discher BM, Moser C.r C., Dutton PL. Biochem Soc Trans. 2012;40:561–566. doi: 10.1042/BST20120067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Koder RL, Anderson JLR, Solomon LA, Reddy KS, Moser CC, Dutton PL. Nature. 2009;458:305–309. doi: 10.1038/nature07841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sasaki T, Kaiser ET. J. Am. Chem. Soc. 1989;111:380–381. [Google Scholar]
- 175.Monien BH, Drepper F, Sommerhalter M, Lubitz W, Haehnel W. J. Mol. Biol. 2007;371:739–753. doi: 10.1016/j.jmb.2007.05.047. [DOI] [PubMed] [Google Scholar]
- 176.Das A, Hecht MH. J. Inorg. Biochem. 2007;101:1820–1826. doi: 10.1016/j.jinorgbio.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Moffet DA, Certain LK, Smith AJ, Kessel AJ, Beckwith KA, Hecht MH. J. Am. Chem. Soc. 2000;122:7612–7613. [Google Scholar]
- 178.Patel SC, Hecht MH. Protein Eng. Des. Sel. 2012;25:445–451. doi: 10.1093/protein/gzs025. [DOI] [PubMed] [Google Scholar]
- 179.Rau HK, Haehnel W. J. Am. Chem. Soc. 1998;120:468–476. [Google Scholar]
- 180.Rau HK, DeJonge N, Haehnel W. Angew. Chem. Int. Ed. 2000;39:250–253. [PubMed] [Google Scholar]
- 181.Rau HK, DeJonge N, Haehnel W. Proc. Nat. Acad. Sci. USA. 1998;95:11526–11531. doi: 10.1073/pnas.95.20.11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Albrecht T, Li W, Ulstrup J, Haehnel W, Hildebrandt P. Chem. Phys. Chem. 2005;6:961–970. doi: 10.1002/cphc.200400597. [DOI] [PubMed] [Google Scholar]
- 183.Mutter M, Hersperger R, Gubernator K, Muller K. Proteins: Struct. Funct. Genet. 1989;5:13–21. doi: 10.1002/prot.340050104. [DOI] [PubMed] [Google Scholar]
- 184.Rojas NRL, Kamtekar S, Simons CT, Mclean JE, Vogel KM, Spiro TG, Farid RS, Hecht MH. Protein Sci. 1997;6:2512–2524. doi: 10.1002/pro.5560061204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wei Y, Liu T, Sazinsky SL, Moffet DA, Pelczer I, Hecht MH. Protein Sci. 2003;12:92–102. doi: 10.1110/ps.0228003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Moffet DA, Case MA, House JC, Vogel K, Williams RD, Spiro TG, McLendon GL, Hecht MH. J. Am. Chem. Soc. 2001;123:2109–2115. doi: 10.1021/ja0036007. [DOI] [PubMed] [Google Scholar]
- 187.Mutter M. CHIMIA International Journal for Chemistry. 2013;67:868–873. doi: 10.2533/chimia.2013.868. [DOI] [PubMed] [Google Scholar]
- 188.Mutter M, Altmann E, Altmann K-H, Hersperger R, Koziej P, Nebel K, et al. Helvetica Chimica Acta. 1988;71:835–847. [Google Scholar]
- 189.Mutter M, Tuchscherer G. Cellular and Molecular Life Sciences CMLS. 1997;53:851–863. doi: 10.1007/s000180050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190F1.Kamtekar S, Schiffer JM, Xiong H, Babik JM, Hecht MH. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 191F2.Beasley JR, Hecht MH. J. Biol. Chem. 1997;24:2031–2034. doi: 10.1074/jbc.272.4.2031. [DOI] [PubMed] [Google Scholar]
- 192F3.David A. Moffet and Michael H. Hecht* Chem. Rev. 2001;101:3191–3203. doi: 10.1021/cr000051e. [DOI] [PubMed] [Google Scholar]
- 193.Faiella M, Maglio O, Nastri F, Lombardi A, Lista L, Hagen WR, Pavone V. Chem. Eur., J. 2012;18:5960–15971. doi: 10.1002/chem.201201404. [DOI] [PubMed] [Google Scholar]
- 194.Brustad EM, Arnold FH. Curr. Opin. Chem. Biol. 2011;15:201–210. doi: 10.1016/j.cbpa.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Romero PA, Arnold FH. Nat. Rev. Mol. Cell. Bio. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jäckel C, Kast P, Hilvert D. Annu. Rev. Biophys. 2008;37:153–73. doi: 10.1146/annurev.biophys.37.032807.125832. [DOI] [PubMed] [Google Scholar]
- 197.Bornscheuer UT, Pohl Martina. Curr. Opin. Chem. Biol. 2001;5:137–143. doi: 10.1016/s1367-5931(00)00182-4. [DOI] [PubMed] [Google Scholar]
- 198.Lewis JC, Coelho PS, Arnold FH. Chem. Soc. Rev. 2011;40:2003–2021. doi: 10.1039/c0cs00067a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Murray RI, Fisher MT, Debrunner PG, Sligar SG. Top. Mol. Struct. Biol. 1985;6:157–206. [Google Scholar]
- 200.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 201.Shimada H, Sligar SG, Yeom H, Ishimura Y. Catal. Met. Complexes. 1997;1:195–221. [Google Scholar]
- 202.Renata H, Wang ZJ, Arnold FH. Angew. Chem., Int. Ed. 2015;54:3351–3367. doi: 10.1002/anie.201409470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Fasan R, Meharenna Y, Snow CD, Poulos TL, Arnold FH. J. Mol. Biol. 2008;383:1069–1080. doi: 10.1016/j.jmb.2008.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Munro AW, Leys DG, McLean KJ, Marshall KR, Ost TW, Daff S, Miles CS, Chapman SK, Lysek DA, Moser CC, Page CC, Dutton PL. Trends Biochem. Sci. 2002;27:250–257. doi: 10.1016/s0968-0004(02)02086-8. [DOI] [PubMed] [Google Scholar]
- 205.Noble MA, Miles CS, Chapman SK, Lysek DA, Mackay AC, Reid GA, Hanzlik RP, Munro AW. Biochem. J. 1999;339:371–379. [PMC free article] [PubMed] [Google Scholar]
- 206.Meinhold P, Peters MW, Chen MMY, Takahashi K, Arnold FH. ChemBioChem. 2005;6:1765–1768. doi: 10.1002/cbic.200500261. [DOI] [PubMed] [Google Scholar]
- 207.Fasan R, Chen MM, Crook NC, Arnold FH. Angew. Chem., Int. Ed. 2007;46:8414–8415. doi: 10.1002/anie.200702616. [DOI] [PubMed] [Google Scholar]
- 208.Tsotsou GE, Sideri A, Goyal A, Nardo GD, Gilardi G. Chem. Eur. J. 2012;18:3582–3588. doi: 10.1002/chem.201102470. [DOI] [PubMed] [Google Scholar]
- 209.Sideri A, Goyal A, Di Nardo G, Tsotsou GE, Gilardi G. J. .Inorg. Biochem. 2013;120:1–7. doi: 10.1016/j.jinorgbio.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 210.Di Nardo G, Gilardi G. Int. J. Mol. Sci. 2012;13:15901–15924. doi: 10.3390/ijms131215901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zilly FE, Acevedo JP, Augustyniak W, Deege A, Häusig UW, Reetz MT. Angew. Chem. 2011;50:2720–2724. doi: 10.1002/anie.201006587. [DOI] [PubMed] [Google Scholar]
- 212.Shoji O, Watanabe Y. Isr. J. Chem. 2015;55:32–39. [Google Scholar]
- 213.Kawakami N, Shoji O, Watanabe Y. Angew. Chem. Int. Ed. 2011;50:5315–5318. doi: 10.1002/anie.201007975. [DOI] [PubMed] [Google Scholar]
- 214.Cong Z, Shoji O, Kasai C, Kawakami N, Sugimoto H, Shiro Y, Watanabe Y. ACS Catal. 2015;5:150–156. [Google Scholar]
- 215.Sligar SG, Makris TM, Denisov IG. Biochem. Biophys. Res. Comm. 2005;338:346–354. doi: 10.1016/j.bbrc.2005.08.094. [DOI] [PubMed] [Google Scholar]
- 216.Barry SM, Kers JA, Johnson EG, Song L, Aston PR, Patel B, Krasnoff SB, Crane BR, Gibson DM, Loria R, Challis GL. Nat. Chem. Biol. 2012;8:814–816. doi: 10.1038/nchembio.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Coelho PS, Brustad EM, Kannan A, Arnold FH. Science. 2013;339:307–310. doi: 10.1126/science.1231434. [DOI] [PubMed] [Google Scholar]
- 218.Hyster TK, Farwell CC, Buller AR, McIntosh JA, Arnold FH. J. Am. Chem. Soc. 2014;136:15505–15508. doi: 10.1021/ja509308v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Prier CK, Arnold FH. J. Am. Chem. Soc. 2015;137:13992–14006. doi: 10.1021/jacs.5b09348. [DOI] [PubMed] [Google Scholar]
- 220.Kaila VRI, Verkhovsky MI, Wikström M. Chem. Rev. 2010;110:7062–7081. doi: 10.1021/cr1002003. [DOI] [PubMed] [Google Scholar]
- 221.Ferguson-Miller S, Babcock GT. Chem. Rev. 1996;96:2889–2907. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- 222.Karlin KD, Fox S, Nanthakumar A, Murthy NN, Wei N, Obias HV, Martes CF. Pure Appl. Chem. 2009;67:289–296. [Google Scholar]
- 223.Collman JP, Herrmann PC, Boitrel B, Zhang X, Eberspacher TA, Fu L, Wang J, Rosseau DL, Williams ER. J. Am. Chem. Soc. 1994;116:9783–9784. [Google Scholar]
- 224.Sigman JA, Kim HK, Zhao X, Carey JR, Lu Y. Proc. Nat. Acad. Sci. USA. 2003;100:3629–3634. doi: 10.1073/pnas.0737308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sigman JA, Kwok BC, Gengenbach A, Lu Y. J. Am. Chem. Soc. 1999;121:8949–8950. [Google Scholar]
- 226.Sigman JA, Kwok BC, Lu Y. J. Am. Chem. Soc. 2000;122:8192. [Google Scholar]
- 227.Miner KD, Mukherjee A, Gao Y-G, Null EL, Petrik ID, Zhao X, Yeung N, Robinson H, Lu Y. Angew. Chem. Intl. Ed. 2012;51:5589–5592. doi: 10.1002/anie.201201981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Yu Y, Chang C, Liu X, Petrik ID, Wang J, Lu Y. J. Am. Chem. Soc. 2015;137:11570–11573. doi: 10.1021/jacs.5b07119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mukherjee S, Mukherjee A, Bhagi-Damodaran A, Mukherjee M, Lu Y, Dey A. Nat. Commun. 2015 doi: 10.1038/ncomms9467. doi:10.1038/ncomms9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yu Y, Lv X, Li J, Zhou Q, Cui C, Hosseinzadeh P, Mukherjee A, Nilges MJ, Wang J, Lu Y. J. Am. Chem. Soc. 2015;137:4594–4597. doi: 10.1021/ja5109936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Yu Y, Mukherjee A, Nilges MJ, Hosseinzadeh P, Miner KD, Lu Y. J. Am. Chem. Soc. 2014;136:1174–1177. doi: 10.1021/ja4091885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.de Vries S, Schroder I. Biochem. Soc. Trans. 2002;30(4):662–667. doi: 10.1042/bst0300662. [DOI] [PubMed] [Google Scholar]
- 233.Wang NY, Zhao X, Lu Y. J. Am. Chem. Soc. 2005;127:16541–16547. doi: 10.1021/ja052659g. [DOI] [PubMed] [Google Scholar]
- 234.Bhagi-Damodaran A, Petrik ID, Marshall N, Robinson H, Lu Y. J. Am. Chem. Soc. 2014;136:11882–11885. doi: 10.1021/ja5054863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yeung BK, Wang X, Sigman JA, Petillo PA, Lu Y. Chemistry and Biology. 1997;4(3):215–221. doi: 10.1016/s1074-5521(97)90291-x. [DOI] [PubMed] [Google Scholar]
- 236.Pfister TD, Mirarefi AY, Gagenbach AJ, Zhao X, Danstrom C, Conaster N, Gao Y-G, Robinson H, Zukoski CF, Wang AHJ, Lu Y. J. Biol. Inorg. Chem. 2007;12(1):126–137. doi: 10.1007/s00775-006-0171-0. [DOI] [PubMed] [Google Scholar]
- 237.Wang X, Lu Y. Biochem. 1999;38:9146–9157. doi: 10.1021/bi990235r. [DOI] [PubMed] [Google Scholar]
- 238.Hosseinzadeh P, Mirts EN, Pfister TD, Gao YG, Mayne C, Robinson H, Tajkhorshid E, Lu Y. Biochemistry. 2016;55:1494–1502. doi: 10.1021/acs.biochem.5b01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Marshall NM, Miner KD, Wilson TD, Lu Y. In: Coordination Chemistry in Protein Cages: Principles, Design, and Applications. Ueno T, Watanabe Y, editors. John Wiley & Sons; Hoboken (New Jersey): 2013. pp. 111–147. [Google Scholar]
- 240.Selinheimo E, NiEidhin D, Steffensen C, Nielsen J, Lomascolo A, Halaouli S, Record E, O'Beirne D, Buchert J, Kruus K. J. Biotechnol. 2007;30:471–480. doi: 10.1016/j.jbiotec.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 241.Fairhead M, Thöny-Meyer L. New. Biotech. 2012;29:183–191. doi: 10.1016/j.nbt.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 242.Shuster Ben-Yosef V, Sendovski M, Fishman A. Enzyme Microb. Tech. 2010;47:372–376. [Google Scholar]
- 243.Sendovski M, Kanteev M, Shuster Ben-Yosef V, Adir N, Fishman A. J. Mol. Biol. 2011;405:227–237. doi: 10.1016/j.jmb.2010.10.048. [DOI] [PubMed] [Google Scholar]
- 244.Giardina P, Faraco V, Pezzella C, Piscitelli A, Vanhulle S, Sannia G. Cell. Mol. Life Sci. 2010;67:369–385. doi: 10.1007/s00018-009-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Mate DM, Alcalde M. Biotech. Advances. 2015;33:25–40. doi: 10.1016/j.biotechadv.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 246.Pardo I, Camarero S. Cell. Mol. Life Sci. 2015;72:897–910. doi: 10.1007/s00018-014-1824-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Alcalde M. In: Industrial enzymes. Structure, function and applications. Polaina J, MacCabe AP, editors. Springer Dordrecht; 2007. pp. 461–476. [Google Scholar]
- 248.Gianfreda L, Xu F, Bollag J. Bioremediat. J. 1999;3:1–25. [Google Scholar]
- 249.Bulter T, Alcalde M, Sieber V, Meinhold P, Schlachtbauer C, Arnold FH. Appl. Environ. Microbiol. 2003;69:987–995. doi: 10.1128/AEM.69.2.987-995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Rodgers CJ, Blanford CF, Giddens SR, Skamnioti P, Armstrong FA, Gurr SJ. Trends Biotechnol. 2010;28:63–72. doi: 10.1016/j.tibtech.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 251.Mate D, García-Burgos C, García-Ruiz E, Ballesteros AO, Camarero S, Alcalde M. Chemistry & Biology. 2010;17:1030–1041. doi: 10.1016/j.chembiol.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 252.Camarero S, Pardo I, Cañas I,AI, Molina P, Record E, Martinez AT, Martinez MJ, Alcade M. Appl. Environ. Microbiol. 2012;78:1370–1384. doi: 10.1128/AEM.07530-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Festa G, Autore F, Fraternali F, Giardina P, Sannia G. Proteins. 2008;72:25–34. doi: 10.1002/prot.21889. [DOI] [PubMed] [Google Scholar]
- 254.Miele A, Giardina P, Notomista E, Piscitelli A, Sannia G, Faraco V. Mol. Biotechnol. 2010;46:149–156. doi: 10.1007/s12033-010-9289-y. [DOI] [PubMed] [Google Scholar]
- 255.Miele A, Giardina P, Sannia G, Faraco V V. J. Appl. Microbiol. 2010;108:998–1006. doi: 10.1111/j.1365-2672.2009.04505.x. [DOI] [PubMed] [Google Scholar]
- 256.Li Y, Gong Z, Li X, Li Y, Wang XG. BMC Biochemistry. 2011;12:30. doi: 10.1186/1471-2091-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Shiga D, Nakane D, Inomata T, Funahashi Y, Masuda H, Kikuchi A, Oda M, Noda M, Uchiyama S, Fukui K, Kanaori K, Tajima K, Takano Y, Nakamura H, Tanaka T. J. Am. Chem. Soc. 2010;132:18191–18198. doi: 10.1021/ja106263y. [DOI] [PubMed] [Google Scholar]
- 258.Shiga D, Hamano Y, Kamei M, Funahashi Y, Masuda H, Sakaguchi M, Ogura T, Tanaka T. J. Biol. Inorg. Chem. 2012;17:1025–1031. doi: 10.1007/s00775-012-0916-x. [DOI] [PubMed] [Google Scholar]
- 259.Sieracki NA, Tian S, Hadt RG, Zhang JL, Woertink JS, Nilges MJ, Sun F, Solomon EI, Lu Y. Proc. Nat. Acad. Sci. 2014;111:924–929. doi: 10.1073/pnas.1316483111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kaiser ET, Lawrence DS. Science. 1984;226:505–511. doi: 10.1126/science.6238407. [DOI] [PubMed] [Google Scholar]
- 261.Hamels DR, Ward TR. Comprehensive Inorganic Chemistry II. 2013;6:737–761. [Google Scholar]
- 262.Lu Y. Angew. Chem. Int. Ed. 2006;45:5588–5601. doi: 10.1002/anie.200600168. [DOI] [PubMed] [Google Scholar]
- 263.Hayashi T. In: Coordination Chemistry in Protein Cages. Ueno T, Watanabe Y, editors. John Wiley & Sons; Hoboken (New Jersey): 2013. pp. 87–110. [Google Scholar]
- 264.Hayashi T, Hisaeda Y. Acc. Chem. Res. 2002;35:35–43. doi: 10.1021/ar000087t. [DOI] [PubMed] [Google Scholar]
- 265.Sato H, Hayashi T, Ando T, Hisaeda Y, Ueno T, Watanabe J Y. Am. Chem. Soc. 2004;126:436–437. doi: 10.1021/ja038798k. [DOI] [PubMed] [Google Scholar]
- 266.Hayashi T, Murata D, Makino M, Sugimoto H, Matsuo T, Sato H, Shiro Y, Hisaeda Y. Inorg. Chem. 2006;45:10530–10536. doi: 10.1021/ic061130x. [DOI] [PubMed] [Google Scholar]
- 267.Oohora K, Kihira Y, Mizohata E, Inoue T, Hayashi T. J. Am. Chem. Soc. 2013;135:17282–17285. doi: 10.1021/ja409404k. [DOI] [PubMed] [Google Scholar]
- 268.Ueno T, Koshiyama T, Abe S, Yokoi N, Ohashi M, Nakajima H, Watanabe Y. J. Organomet. Chem. 2007;692:142–147. [Google Scholar]
- 269.Jacobsen EN. Comprehensive Organometallic Chemistry II. Vol. 12. Pergamon; New York: 1995. [Google Scholar]
- 270.Katsuki T. Coord. Chem. Rev. 1995;140:189–214. [Google Scholar]
- 271.Ueno T, Abe S, Yokoi N, Watanabe Y. Coord. Chem. Rev. 2007;251:2717–2731. [Google Scholar]
- 272.Ueno T, Koshiyama T, Ohashi M, Kondo K, Kono M, Suzuki A, Yamane T, Watanabe Y. J. Am. Chem. Soc. 2005;127:6556–6562. doi: 10.1021/ja045995q. [DOI] [PubMed] [Google Scholar]
- 273.Ueno T, Ohashi M, Kono M, Kondo K, Suzuki A, Yamane T, Watanabe Y. Inorg. Chem. 2004;43:2852–2858. doi: 10.1021/ic0498539. [DOI] [PubMed] [Google Scholar]
- 274.Ohashi M, Koshiyama T, Ueno T, Yanase M, Fujii H, Watanabe Y. Angew. Chem., Int. Ed. 2003;42:1005–1008. doi: 10.1002/anie.200390256. [DOI] [PubMed] [Google Scholar]
- 275.Carey JR, Ma SK, Pfister TD, Garner DK, Kim HK, Abramite JA, Wang Z, Guo Z, Lu Y. J. Am. Chem. Soc. 2004;126:10812–10813. doi: 10.1021/ja046908x. [DOI] [PubMed] [Google Scholar]
- 276.Zhang JL, Garner DK, Liang L, Chen Q, Lu Y. Chem. Commun. 2008:1665–1667. doi: 10.1039/b718915j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zhang JL, K Garner D, Liang L, A Barrios D, Lu Y. Chem. Eur. J. 2009;15:7481–7489. doi: 10.1002/chem.200802449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Garner DK, Liang L, Barrios DA, Zhang JL, Lu Y. ACS Catal. 2011;1:1083–1089. doi: 10.1021/cs200258e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Yamamura K, Kaiser ET. J. Chem. Soc. Chem. Commun. 1976:830–831. [Google Scholar]
- 280.Okrasa K, Kazlauskas RJ. Chem. Eur. J. 2006;12:1587–1596. doi: 10.1002/chem.200501413. [DOI] [PubMed] [Google Scholar]
- 281.Fernandez-Gacio A, Codina A, Fastrez J, Riant O, Soumillion P. Chembiochem. 2006;7:1013–1016. doi: 10.1002/cbic.200600127. [DOI] [PubMed] [Google Scholar]
- 282.Wilson ME, Whitesides GM. J. Am. Chem. Soc. 1978;100:306–307. [Google Scholar]
- 283.Ward TR. Acc. Chem. Res. 2011;44:47–57. doi: 10.1021/ar100099u. [DOI] [PubMed] [Google Scholar]
- 284.Zimbron JM, Heinisch T, Schmid M, Hamels D, Nogueira ES, Schirmer T, Ward TR. J. Am. Chem. Soc. 2013;135:5384–5388. doi: 10.1021/ja309974s. [DOI] [PubMed] [Google Scholar]
- 285.Mallin H, Hestericová M, Reuter R, Ward TR. Nature Protocols. 2016;11:835–852. doi: 10.1038/nprot.2016.019. [DOI] [PubMed] [Google Scholar]
- 286.Hyster TK, Knörr L, Ward TR, Rovis T. Science. 2012;338:500–503. doi: 10.1126/science.1226132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Thomas CM, Letondor C, Humbert N, Ward TR. J. Organomet. Chem. 2005;690:4488–4491. [Google Scholar]
- 288.Pordea A, Creus M, Panek J, Duboc C, Mathis D, Novic M, Ward TR. J. Am. Chem. Soc. 2008;130:8085–8088. doi: 10.1021/ja8017219. [DOI] [PubMed] [Google Scholar]
- 289.Raffy Q, Ricoux R, Sansiaume E, Pethe S, Mahy J-P. J. Mol. Catal. A: Chem. 2010;317:19–26. [Google Scholar]
- 290.Ricoux R, Sauriat-Dorizon H, Girgenti E, Blanchard D, Mahy J-P. J. Immunol. Methods. 2002;269:39–57. doi: 10.1016/s0022-1759(02)00223-5. [DOI] [PubMed] [Google Scholar]
- 291.Quilez R, de Lauzon S, Desfosses B, Mansuy D, Mahy J-P. FEBS Lett. 1996;395:73–76. doi: 10.1016/0014-5793(96)01006-x. [DOI] [PubMed] [Google Scholar]
- 292.de Lauzon S, Desfosses B, Mansuy D, Mahy J-P. FEBS Lett. 1999;443:229–234. doi: 10.1016/s0014-5793(98)01703-7. [DOI] [PubMed] [Google Scholar]
- 293.de Lauzon S, Quilez R, Lion L, Desfosses B, Desfosses B, Lee I, Sari MA, Benkovic SJ, Mansuy D, Mahy J-P. Eur. J. Biochem. 1998;257:121–130. doi: 10.1046/j.1432-1327.1998.2570121.x. [DOI] [PubMed] [Google Scholar]
- 294.Ricoux R, Girgenti E, Sauriat-Dorizon H, Blanchard D, Mahy J-P. J. Protein Chem. 2002;21:473–477. doi: 10.1023/a:1021351120772. [DOI] [PubMed] [Google Scholar]
- 295.Ricoux R, Lukowska E, Pezzotti F, Mahy J-P. Eur. J. Biochem. 2004;271:1277–1283. doi: 10.1111/j.1432-1033.2004.04032.x. [DOI] [PubMed] [Google Scholar]
- 296.Ricoux R, Allard M, Dubuc R, Dupont C, Marećhal J-D, Mahy J-P. Org. Biomol. Chem. 2009;7:3208–3211. doi: 10.1039/b907534h. [DOI] [PubMed] [Google Scholar]
- 297.Allard M, Dupont C, Munoz Robles V, Doucet N, Lledos A, Marechal J-D, Urvoas A, Mahy J-P, Ricoux R. ChemBioChem. 2012;13:240–251. doi: 10.1002/cbic.201100659. [DOI] [PubMed] [Google Scholar]
- 298.Peters T. All About Albumin: Biochemistry, Genetics and Medical Applications. Academic Press; San Diego: 1995. [Google Scholar]
- 299.Mahammed A, Gross Z. J. Am. Chem. Soc. 2005;127:2883–2887. doi: 10.1021/ja045372c. [DOI] [PubMed] [Google Scholar]
- 300.Rousselot-Pailley P, Bochot C, Marchi-Delapierre C, Jorge-Robin A, Martin L, Fontecilla-Camps JC, Cavazza C, Menage S. ChemBioChem. 2009;10:545–552. doi: 10.1002/cbic.200800595. [DOI] [PubMed] [Google Scholar]
- 301.Debret G, Martel A, Cuniasse P. Nucl. Acids Res. 2009;37:W459–W464. doi: 10.1093/nar/gkp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Amrein B, Schmid M, Collet G, Cuniasse P, Gilardoni F, Seebeck FP, Ward TR. Metallomics. 2012;4:379–388. doi: 10.1039/c2mt20010d. [DOI] [PubMed] [Google Scholar]
- 303.Fujieda N, Schätti J, Stuttfeld E, Ohkubo K, Maier T, Fukuzumicde S, Ward TR. Chem. Sci. 2015;6:4060–4065. doi: 10.1039/c5sc01065a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.van de Velde F, Könemann L, van Rantwijk F, Sheldon RA. Chem. Commun. 1998:1891–1892. [Google Scholar]
- 305.van de Velde F, Könemann L, van Rantwijk F, Sheldon RA. Biotechnol. Bioeng. 2000;67:87–96. doi: 10.1002/(sici)1097-0290(20000105)67:1<87::aid-bit10>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- 306.van de Velde F, Arends IWCE, Sheldon RA. Top. Catal. 2000;13:259–265. [Google Scholar]
- 307.van de Velde F, Arends IWCE, Sheldon RA. J. Inorg. Biochem. 2000;80:81–89. doi: 10.1016/s0162-0134(00)00043-x. [DOI] [PubMed] [Google Scholar]
- 308.Littlechild J, Garcia-Rodriguez E, Dalby A, Isupov M. J. Mol. Recognit. 2002;15:291–296. doi: 10.1002/jmr.590. [DOI] [PubMed] [Google Scholar]
- 309.Lu Y. Curr. Opin. Chem. Biol. 2005;9:118–126. doi: 10.1016/j.cbpa.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 310.Berry SM, Ralle M, Low DW, Blackburn NJ, Lu Y. J. Am. Chem. Soc. 2003;125:8760–8768. doi: 10.1021/ja029699u. [DOI] [PubMed] [Google Scholar]
- 311.Garner DK, Vaughan MD, Hwang HJ, Savelieff MG, Berry SM, Honek JF, Lu Y. J. Am. Chem. Soc. 2006;128:15608–15617. doi: 10.1021/ja062732i. [DOI] [PubMed] [Google Scholar]
- 312.Clark KM, Yu Y, Marshall NM, Sieracki NA, Nilges MJ, Blackburn NJ, van der Donk WA, Yi Lu J. Am. Chem. Soc. 2010;132:10093–10101. doi: 10.1021/ja102632p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.M Clark K, Yu Y, van der Donk WA, Blackburn N, Lu Y. Inorg. Chem. Frontiers. 2014;1:153–158. doi: 10.1039/C3QI00096F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Hu C, Chan SI, Sawyer EB, Yu Y, Wang J. Chem. Soc. Rev. 2014;43:6498–6510. doi: 10.1039/c4cs00018h. [DOI] [PubMed] [Google Scholar]
- 315.Yang H, Srivastava P, Zhang C, Lewis JC. ChemBioChem. 2014;15:223–227. doi: 10.1002/cbic.201300661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Srivastava P, Yang H, Ellis-Guardiola K, Lewis JC. Nat. Commun. 2015;6:7789–7795. doi: 10.1038/ncomms8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- p[317.Costas M, Mehn MP, Jensen MP, Que L. Chem. Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 318.Vaillancourt FH, Yin J, Walsh CT. Proc. Nat. Acad. Sci. USA. 2005;102:10111–10116. doi: 10.1073/pnas.0504412102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Blasiak LC, Vaillancourt FH, Walsh CT, Drennan C. Nature. 2006;440:368–371. doi: 10.1038/nature04544. [DOI] [PubMed] [Google Scholar]
- 320.Wong SD, Srnec M, Matthews ML, Liu LV, Kwak Y, Park K, Bell III CB, Alp EE, Zhao J, Yoda Y, Kitao S, Seto M, Krebs C, Bollinger JM, Solomon EI. Nature. 2013;499:320–323. doi: 10.1038/nature12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.L Matthews M, Krest CM, W Barr E, Vaillancourt FH, Walsh CT, Green MT, Krebs C, Bollinger JM. Biochemistry. 2009;48:4331–4343. doi: 10.1021/bi900109z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.L Matthews M, Neumann CS, Miles LA, Grove TL, Booker SJ, Krebs C, Walsh CT, Bollinger JM. Proc. Nat. Acad. Sci. USA. 2009;106:17723–17728. doi: 10.1073/pnas.0909649106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Kulik HJ, Drennan CL. J. Biol. Chem. 2013;288:11233–11241. doi: 10.1074/jbc.M112.415570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Hakemian AS, Rosenzweig AC. Annu. Rev. Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- 325.Sirajuddin S, Rosenzweig AC. Biochemistry. 2015;54:2283–2294. doi: 10.1021/acs.biochem.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Sazinsky MH, Lippard SJ. Met. Ions Life Sci. 2015;15:205–256. doi: 10.1007/978-3-319-12415-5_6. [DOI] [PubMed] [Google Scholar]
- 327.Culpepper MA, Rosenzweig AC. Crit. Rev. Biochem. Mol. Biol. 2012;47:483–492. doi: 10.3109/10409238.2012.697865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Himes RA, Karlin KD. Curr. Opin. Chem. Biol. 2009;13:119–131. doi: 10.1016/j.cbpa.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Woolfson DN, Bartlett GJ, Burton AJ, Heal JW, Niitsu, Andrew A, Thomson R, Wood CW. Curr. Opin. Struct. Biol. 2015;33:16–26. doi: 10.1016/j.sbi.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 330.Karanicolas J, Corn JE, Chen I, Joachimiak LA, Dym O, Peck SH, Albeck S, Unger T, Hu W, Liu G, Delbecq S, Montelione GT, Spiegel CP, Liu DR, Baker D. Mol. Cell. 2011;42:250–260. doi: 10.1016/j.molcel.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Verma R, Schwaneberg U, Roccatano D. Comput. Struct. Biotechnol. J. 2012;2:1–12. doi: 10.5936/csbj.201209008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Namuswe F, Kasper GD, Sarjeant AAN, Hayashi T, Krest CM, Green MT, Moenne-Loccoz P, Goldberg David P. J. Am. Chem. Soc. 2008;130:14189–14200. doi: 10.1021/ja8031828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Gu Y, Ellis-Guardiola K, Srivastava P, Lewis JC. ChemBioChem. 2015;16:1880–1883. doi: 10.1002/cbic.201500165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Ener ME, Lee Y-T, Winkler JR, Gray HB, Cheruzel L. Proc. Nat. Acad. Sci. USA. 2010;107:18783–18786. doi: 10.1073/pnas.1012381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.The PyMOL Molecular Graphics System, version 1.7.4. Schrödinger, LLC; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.