Abstract
Understanding factors that cause species’ geographic range limits is a major focus in ecology and evolution. The central marginal hypothesis (CMH) predicts that species cannot adapt to conditions beyond current geographic range edges because genetic diversity decreases from core to edge due to smaller, more isolated edge populations. We employed a population genomics framework using 24,235-33,112 SNP loci to test major predictions of the CMH in the ongoing invasion of the cane toad (Rhinella marina) in Australia. Cane toad tissue samples were collected along broad-scale, core-to-edge transects across their invasive range. Geographic and ecological core areas were identified using GIS and habitat suitability indices from ecological niche modeling. Bayesian clustering analyses revealed three genetic clusters, in the northwest invasion-front region, northeast precipitation-limited region, and southeast cold temperature-limited region. Core-to-edge patterns of genetic diversity and differentiation were consistent with the CMH in the southeast, but were not supported in the northeast and showed mixed support in the northwest. Results suggest cold temperatures are a likely contributor to southeastern range limits, consistent with CMH predictions. In the northeast and northwest, ecological processes consisting of a steep physiological barrier and ongoing invasion dynamics, respectively, are more likely explanations for population genomic patterns than the CMH.
Keywords: central marginal hypothesis, species range limits, population genomics, invasive species, amphibian, ecological niche model
Introduction
Understanding the factors that govern geographic range limits of species is a major focus in ecology and evolution (Darwin 1859, Haldane 1956, Mayr 1963, Macarthur 1972). The majority of species’ range edges occur in areas without major physiographic barriers to dispersal (Hoffman and Blows 1994, Parmesan et al. 2005, Sexton et al. 2009). Therefore, it is often unclear what causes the geographic range limits of many species, and why edge populations do not evolve traits that would allow them to expand their ranges (Bridle & Vines 2006, Kawecki 2008). Extensive theoretical work has been devoted to this topic, but empirical testing has lagged behind (Sexton et al. 2009). Testing species range limit hypotheses empirically in natural systems has become an urgent priority because global warming, exotic species invasions, and habitat alteration are currently changing the distributions of many species around the world (Parmesan et al. 2005).
The central marginal hypothesis (CMH) is one of the major evolutionary hypotheses for species’ range limits (Eckert et al. 2008, Sexton et al. 2009). The CMH is an extension of the abundant center hypothesis, which predicts that habitats in the center of a species’ range will be the most favorable for supporting high population densities (Brown 1984, Sagarin and Gaines 2002). Habitat quality, population densities, and connectivity among populations are consequently expected to decline towards the range periphery. The CMH describes the genetic consequences of the abundant center hypothesis (Eckert et al. 2008). That is, reduced habitat quality near range edges should lead to declines in effective population sizes (Ne), which should cause higher levels of genetic drift within populations and lower gene flow rates among populations. Therefore, reduced genetic diversity and increased genetic differentiation are expected within and among edge populations relative to core populations. In turn, edge populations may lack the genetic diversity necessary to adapt to habitat conditions beyond the range edge, leading to stable range limits (Hoffman and Blows 1994, Gomulkiewicz et al. 1999). Recent empirical studies testing the predictions of the CMH have been equivocal, so the generality of the CMH is unclear, particularly for invasive species (Sagarin and Gaines 2002, Garner et al. 2004, Eckert et al. 2008, Munwes et al. 2010, Dixon et al. 2013, Johansson et al. 2013, Micheletti and Storfer 2015, Ursenbacher et al. 2015). Serial founder effects and allele surfing are expected to cause reduced levels of genetic diversity at the expanding edge of an invasion wave, provided effective population sizes are small enough for genetic drift to dominate (Klopfstein et al. 2006, Excoffier and Ray 2008).
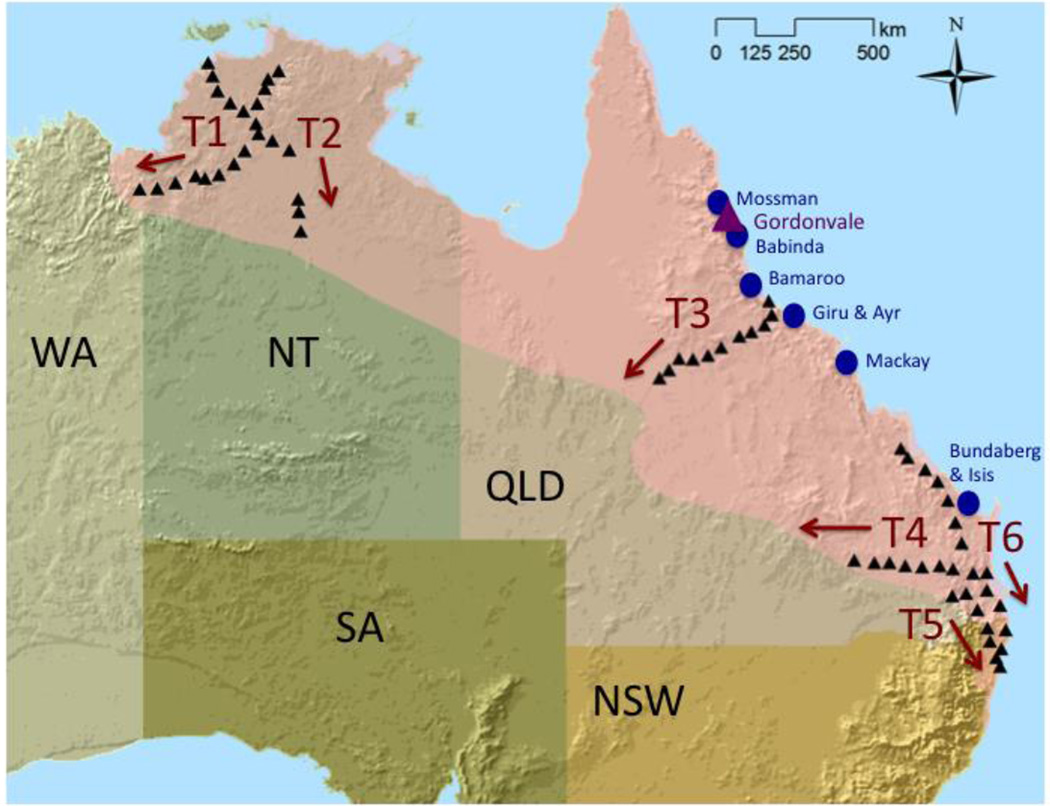
Species invasions provide unique, albeit unfortunate, opportunities to test evolutionary hypotheses for species’ geographic range limits (Sexton et al. 2009, Guo 2014). The cane toad (Rhinella [Bufo] marina) (Linnaeus, 1758; Pramuk et al. 2007), native to tropical and sub-tropical habitats in the Americas, has become a notorious worldwide invader (Lever 2001, Kraus 2009; www.issg.org). From the mid-1800’s to early-1900’s, cane toads were intentionally introduced to control insect crop pests on several Caribbean and tropical Pacific islands, including Bermuda, Martinique, Barbados, Jamaica, Puerto Rico, and Hawaii (Easteal 1981, Lever 2001). In 1935, 101 cane toads were collected from Hawaii and introduced to Gordonvale, Queensland, Australia as a biocontrol agent aimed at sugar cane beetles. From 1935–1937, offspring from this initial Australian introduction were subsequently released and established in six sugar cane growing regions along the east coast of Queensland (Sabath et al. 1981; Figure 1). Cane toads were not an effective biocontrol agent but instead became remarkably successful invaders, spreading continuously across a range of over 1.2 million square kilometers in northern and eastern Australia, and still spreading in the northwest (Urban et al. 2007, Kearney et al. 2008, Kolbe et al. 2010; Figure 1). Cane toads occupy a broader range of habitats in Australia than they do in their native range, possibly due to the lack of native Bufonid competitors in Australia (Lever 2001, Tingley et al. 2014). It is unclear where their highest habitat suitability, or ecological core areas (Martinez-Meyer et al. 2012, Lira-Noriega and Manthey 2014), occur in Australia, although arid habitats lacking suitable breeding ponds and cold temperatures currently limit their distributions in inland and southern portions of their range, respectively (Sutherst et al. 1996, Kearney et al. 2010, Kolbe et al. 2010, Tingley et al. 2012, McCann et al. 2014). In contrast, the northwestern edge of the cane toad’s range is an active invasion front, rapidly expanding at up to 55 km per year (Urban et al. 2008). Cane toads are of high conservation concern in Australia, as they negatively impact native biodiversity as novel toxic prey items, predators, and competitors (Kraus 2009, Llywellen et al. 2010, Shine 2010).
Figure 1.
Study area in Australia, including the cane toad’s range (pink), initial introduction site in 1935 (purple triangle), subsequent introduction sites from 1935–1937 (blue circles), sampling sites in 2010 and 2011 (black triangles), six core to edge transects (T1-T6), and states (WA=Western Australia, NT= the Northern Territory, QLD=Queensland, NSW=New South Wales, SA=Southern Australia).
Herein, we test key predictions of the CMH, using a population genomics framework to study the ongoing invasion of Australia by cane toads. To provide context for testing the CMH we first assess overall population structure across the range. We predict population structure to be low due to high dispersal and gene flow rates (Schwarzkopf and Alford 2002, Brown et al. 2006, Leblois et al. 2000, Estoup et al. 2001, 2004, 2010), or alternatively high due to serial founder effects and consequent genetic drift during their rapid range expansion (Klopfstein et al. 2006, Excoffier and Ray 2008). Second, the CMH predicts that reduced habitat suitability and smaller effective population sizes at the range edge will cause genetic diversity to decline from core to edge due to drift (Sagarin and Gaines 2002, Eckert et al. 2008). However, recent empirical findings in other systems have shown mixed support (Garner et al. 2004, Munwes et al. 2010, Dixon et al. 2013, Johansson et al. 2013, Micheletti and Storfer 2015, Ursenbacher et al. 2015). Third, the CMH predicts that reduced habitat suitability at the range edge will cause decreased gene flow and greater genetic differentiation among edge populations relative to those in the core (Sagarin and Gaines 2002, Eckert et al. 2008). Additionally, higher habitat suitability in the core will result in asymmetric gene flow from core to edge populations (Kirkpatrick and Barton 1997, Sexton et al. 2009). Alternatively, habitats may have high suitability until the edge of the range is reached, with a steep physiographic barrier at the range boundary (Hastings et al. 1997, Holt et al. 2005, Sexton et al. 2009). In this case, there would be no change in the levels of genetic diversity, or the rate and symmetry of gene flow, at the range edge relative to the core.
Materials and Methods
Study Area and field sampling
The cane toad’s invasive range in Australia lies between −11 and −31 degrees of latitude in northern and eastern Australia (Figure 1). It currently occupies four Australian states; Queensland (QLD), New South Wales (NSW), the Northern Territory (NT), and Western Australia (WA). Cane toads utilize a broad range of habitats in Australia; including grasslands, savannahs, dry broadleaf forests, and tropical rainforests (Lever et al. 2001). In general, precipitation decreases and aridity increases from coastal to inland sites in Australia. Vegetation tends to be denser with more tree cover along the coast, becoming sparser in inland areas. Broadly, temperature decreases along a north-south latitudinal gradient, and becomes more seasonal from coastal to inland sites. Most of the cane toad’s range is characterized by a summer monsoonal climate, with a warm wet season and a cooler dry season. However, the southeastern edge of the range is colder, more temperate, and receives most of its rainfall in the winter. Northern Australia has little topographic relief, whereas the Great Dividing Range runs along the east coast of Australia, providing some steep elevational gradients. The highest elevations are found in the northeastern and southeastern portions of the range, with some peaks exceeding 2,000 m.
Cane toad tissue samples were collected between January and April, 2010 and 2011. Population sampling was organized along core to edge transects, crossing key environmental gradients hypothesized to affect the cane toad’s distribution in Australia (e.g., temperature, precipitation, vegetation) at 50 km intervals. This sampling interval was chosen based on prior microsatellite work on cane toads from a small portion of their range, which showed little to no isolation by distance (IBD) up to 50 km (Leblois et al. 2000, Estoup et al. 2001, 2004, 2010). A total of 1,123 individuals were sequenced from 62 populations, with a mean of 18.1 individuals per population (Table S1, Supporting Information). Most individuals sampled were adults collected at breeding ponds, or from a localized area within approximately 3 km if a breeding pond could not be located. Tadpoles were collected at 2 locations and metamorphs at 1 location where few adults could be found (Table S1, Supporting Information). Here tadpoles and metamorphs were sampled from several, distant portions of the breeding pond to reduce the chances of collecting siblings.
Ecological Niche Modeling
We used ecological niche models (ENMs) to develop an index of habitat suitability across the Australian range of the cane toad to facilitate identification of geographic versus ecological core and edge areas (Phillips et al. 2006, Micheletti and Storfer 2015). ENMs were developed using Maxent (Phillips et al. 2006), which uses a machine-learning algorithm that maximizes the amount of entropy in the model, or minimizes the number of constraints, to create a continuous index of habitat suitability across space. It is accurate compared to other ecological niche modeling methods (Elith et al. 2006). Maxent utilizes presence-only locality data and continuous environmental data to calculate a habitat suitability index between 0 and 1. We gathered 3,384 non-repetitive, locality records from online biodiversity databases (Global Biodiversity Information Facility, Herpnet.org, Atlas of Living Australia), museums (Brisbane and Sydney Natural History Museums), and our own collections. Locality data spanned the cane toad’s Australian range, but were highly biased to the northwestern and southeastern edges of the range. Therefore, we created a 50 km grid and intersected it with our locality points using ArcGIS 10 (ESRI, Redlands, CA). We then randomly sampled one locality point per grid cell. This resulted in a less-biased dataset of 264 locality points from across the cane toad’s range to be used for ENM (Figure 2).
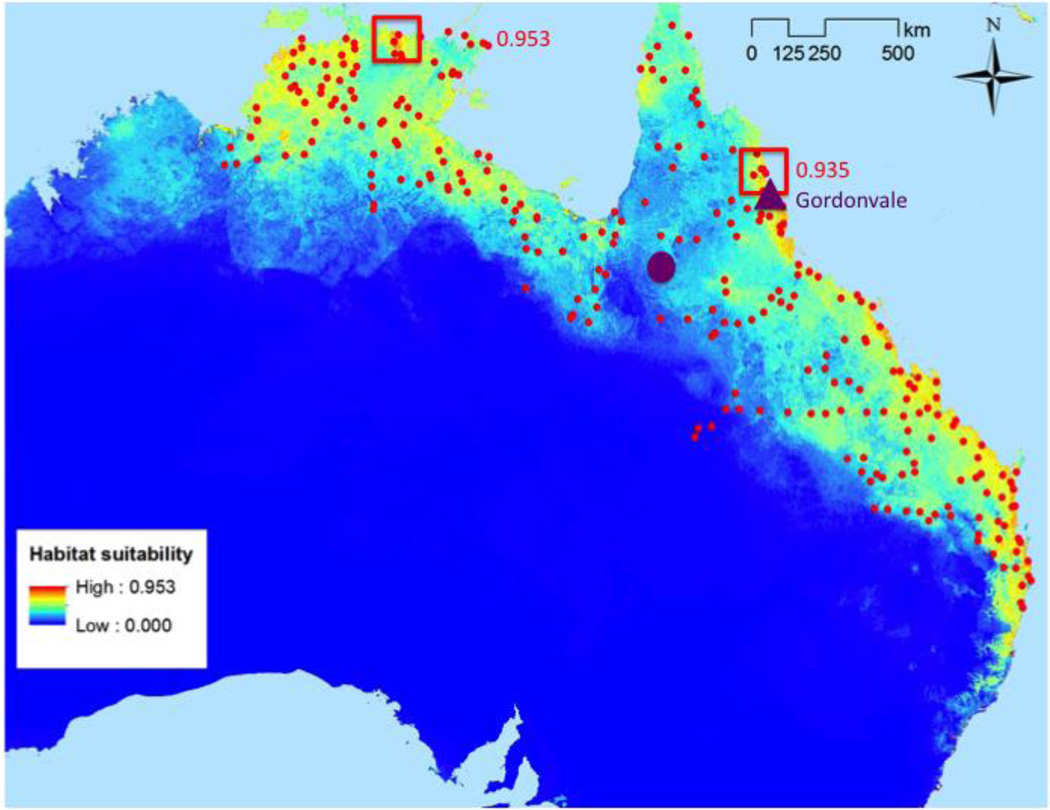
Figure 2.
Maxent model of habitat suitability (blue to red=low to high suitability); including 264 localities used to build (75%) and test (25%) the model (red circles); original introduction site in Gordonvale, QLD in 1935 (purple triangle); geographic center of the range (purple circle), and the highest habitat suitability values overall (0.953) and in the eastern portion of the range (0.935) (red boxes). Map shows median Maxent habitat suitability from 100 bootstrap replicates.
Twenty-six continuous environmental variables were collected for ENM (Table S2, Supporting Information). These variables were hypothesized to affect cane toad distributions across their range, and consisted of 19 temperature and precipitation layers, vegetation (enhanced vegetation index, leaf area index, tree cover), heat load index, moisture (compound topographic index), elevation, and topographic roughness (Gessler et al. 1995, McCune and Keon 2002, Hijmans et al. 2005; http://www.ga.gov.au, www.tern.org.au). However, some of these variables were likely to be correlated across Australia, which can reduce model accuracy (Elith et al. 2006). Therefore, we ran correlation tests on the variables using ENMTools (Warren et al. 2010) and removed strongly correlated variables (r > 0.9) for our final ENM dataset (Table S2, Supporting Information).
Maxent models were run for 100 bootstrapped replicates. We used 75% of the locality data to train the models and 25% of the data to test the models, with a regularization multiplier of 1. Area under the curve (AUC) scores of the receiver operating characteristic were calculated to assess the accuracy of the models (Swets 1988). Jackknife tests were used to determine individual variable contributions to the final models. Finally, we calculated an ecological core and a geographic core of the cane toad range in Australia (Martinez-Meyer et al. 2012, Lira-Noriega and Manthey 2014). The ecological core was defined as the highest median habitat suitability scores from 100 bootstrapped replicates, while the geographic core was the mean latitude and longitude of the final locality dataset of 264 individuals (Figure 2).
ddRAD sequencing
We followed the laboratory protocol of Peterson et al. (2012) to build ddRADseq libraries for 1,123 individuals. ddRADseq was chosen to reduce the total number of SNP loci, thereby increasing the average depth of coverage per locus, given that the cane toad has a relatively large genome size of approximately 4.1 Gb (Vinogradov 1998, Peterson et al. 2012; www.genomesize.com). Tissue samples were stored in 80% ethanol in −80° Celcius freezers. DNA was extracted using Qiagen DNEasy kits. We started the ddRADseq protocol with 500 ng of DNA. We digested the DNA using PstI and EcoRI restriction enzymes, which each recognize a different 6 bp cut site. We developed 96 P1 adapters with unique 6 bp barcodes for the PstI cut sites, and non-barcoded P2 adapters for the EcoRI cut sites. After ligating the adapters and pooling fragments with unique barcodes, we used a Sage Science Pippen Prep to size select 400–600 bp fragments. This size range was confirmed using an Agilent Technologies BioAnalyzer. We then ran 12 polymerase chain reaction (PCR) cycles using P1 primers and three uniquely-indexed P2 primers, using Phusion PCR master mix, for our final ddRADseq libraries. Using a combinatorial indexing technique with unique P1 adapter barcodes and unique P2 primer indices, we multiplexed approximately 162 individuals per library, for a total of eight libraries containing 1,123 individuals.
Each library was sequenced separately on a single Illumina HiSeq 2000 lane at the University of Oregon Genomics Core Facility (gc3f.uoregon.edu) using single-end 100 bp reads. We sequenced a total of 1.51 billion fragments, with 173–203 million reads per lane. We filtered out low quality reads with the process radtags program, and performed a de novo assembly and called SNPs using the denovo_map pipeline in Stacks Version 1.21 (Catchen et al. 2013). Stacks parameter values consisted of a minimum stack depth of 2 to report a locus (-m argument to the ustacks program), 5 mismatches allowed between loci when processing an individual (-M argument to ustacks), and 3 mismatches allowed between loci when building the SNP catalog (-n argument to cstacks). This resulted in a total of 1.29 million SNP loci, with a mean depth of coverage of 13x (range 2–192x). We removed singletons (i.e., alleles present in only 1 individual), as these could be due to sequencing error. We also removed RAD loci that had >30x depth of coverage (i.e., > 3 standard deviations from the mean), as well as loci with observed heterozygosity > 0.5, as these loci are likely to come from paralogous regions of the genome (e.g., Hohenlohe et al. 2011). This filtering resulted in a dataset of 337,394 putative SNP loci. Finally, we filtered our dataset individually by transect and region. For this step, we removed SNPs with a minor allele frequency (MAF)<0.01 for each transect (i.e., transects 1–6, 6–14 localities per transect), MAF<0.0075 within regions (i.e., regions 1 and 3, 24 and 27 localities, respectively), and MAF<0.005 across the range (62 localities). We also removed SNPs that were present in less than 1/3 of the individuals in each transect or region. This resulted in a final dataset of 24,235-33,112 SNPs per transect or region, with approximately 2 SNPs per 95 bp RAD locus on average (Table S3, Supporting Information). For analyses requiring unlinked or loosely-linked SNPs, we further filtered the dataset to one random SNP per RAD locus, resulting in a final dataset of 13,076–15,389 SNPs per transect or region (Table S3, Supporting Information).
Population genomics
We ran two Bayesian clustering programs to estimate population structure across the range of the cane toad in Australia; Admixture (Alexander et al. 2009) and fastStructure (Raj et al. 2014). These analyses were individual-based, as no a priori collection location information was included in either program. Both Admixture and fastStructure utilize a similar statistical model as Structure (Pritchard et al. 2000), but estimate ancestries using a faster numerical optimization algorithm. This allows them to process thousands of SNP loci in a reasonable timeframe. The change in log likelihood was used to assign the number of population clusters (Evanno et al. 2005).
We used custom software based on libsequence (Thornton 2003) to estimate genetic differentiation between populations by calculating pairwise FST for each locus. Mantel tests were performed in R using the ade4 package along each transect, comparing genetic distance (FST) to geographic distance (km) to determine if there were significant IBD patterns. We then calculated FST per km to control for varying distances between sites when examining core to edge patterns along transects (Micheletti et al. 2015). We divided FST by the ln(distance), as FST does not increase linearly with distance indefinitely, but reaches an asymptote.
Genetic diversity within populations was estimated by Watterson’s theta (θw; Watterson 1975) and theta pi (θπ; Nei 1987) for each locus. θw represents the total number of segregating sites observed (i.e., SNPs), corrected for the total number of sequences because the number of SNPs detected increases with sample size. θπ is the average number of pairwise difference across all sites. These two genetic diversity measures are proportional to the effective population size by the formula θ=4Neµ. We also calculated Tajima’s D (Tajima 1989), which is a comparison of the total number of segregating sites to the average pairwise difference. A D of 0 occurs when the total number of segregating sites equals the average pairwise difference, which is expected under neutral mutation-drift equilibrium. A negative D occurs when there is an excess of low frequency polymorphisms, causing there to be more segregating sites relative to the average pairwise difference. This suggests the population may have experienced strong positive or purifying selection, or a recent population expansion. A positive D occurs when there are few low frequency polymorphisms relative to intermediate frequency polymorphisms, causing the average pairwise difference to be higher than the number of segregating sites. This suggests the population may have experienced balancing selection or recent population bottlenecks.
Finally, we used a Bayesian assignment test BayesAss (Wilson and Rannala 2003) to estimate the symmetry of gene flow among populations, as well as NeEstimator v.2 (Do et al. 2014) to estimate effective population sizes (Ne). Asymmetric gene flow was calculated as the proportion of migrants moving from core to edge, as well as from edge to core. BayesAss was run for 10,000,000 Markov chain Monte Carlo (MCMC) iterations, discarding the first 1,000,000 iteration as burnin and sampling every 100 iterations. A migration rate mixing parameter (m) of 0.1 was used to optimize the acceptance rate. Ne estimates are more accurate with unlinked or loosely-linked SNPs (Do et al. 2014), so we first filtered the SNP datasets down to a single SNP per 95 bp RAD locus. (Table S3, Supporting Information). These datasets still exceeded the computer’s memory requirements when computing r2 between more than approximately 4,500 SNPs. Therefore, we filtered out SNPs covered in less than 75% of the individuals to get a high coverage SNP dataset, and then randomly selected half of these high coverage SNPs resulting in a final dataset of 3,173 – 4,217 SNPs per transect.
Results
Ecological niche modeling
Nine of the 26 environmental variables we tested were strongly correlated with other variables (r > 0.9), so we removed them from the niche models. This left 17 environmental variables in our models related to elevation and topography; vegetation density; moisture and heat load; precipitation minimum, maximum, and variation; and temperature minimum, maximum, and variation (Table 1). The mean AUC score across 100 bootstrapped Maxent models was 0.951, indicating high model sensitivity and specificity in predicting cane toad presence across the range.
Table 1.
Contributions of 17 environmental variables to Maxent habitat suitability models based on jackknife tests.
| Environmental Variable | AUC with only variable |
AUC without variable |
|---|---|---|
| Precipitation of the wettest quarter | 0.871 | 0.922 |
| Annual precipitation | 0.865 | 0.921 |
| Temperature annual range | 0.819 | 0.921 |
| Enhanced vegetation index | 0.813 | 0.920 |
| Isothermality | 0.801 | 0.921 |
| Minimum temperature of the coldest month | 0.781 | 0.921 |
| Precipitation seasonality | 0.748 | 0.919 |
| Precipitation of the coldest quarter | 0.737 | 0.921 |
| Precipitation of the driest quarter | 0.735 | 0.921 |
| Mean diurnal range temperature | 0.732 | 0.921 |
| Mean temperature of the wettest quarter | 0.721 | 0.920 |
| Elevation | 0.667 | 0.919 |
| Topographic roughness | 0.642 | 0.920 |
| Mean temperature of the warmest quarter | 0.624 | 0.921 |
| Mean temperature of the driest quarter | 0.611 | 0.921 |
| Heat load index | 0.598 | 0.921 |
| Compound topographic index of wetness | 0.583 | 0.919 |
Overall, Maxent predicted the highest habitat suitability for cane toads along the northern and northeastern coasts of Australia (Figure 2). The highest overall habitat suitability was located at the northern coast of the Northern Territory, in a large aboriginal area called Arnhem Land (Figure 2). This maximum habitat suitability area was approximately 1,100 km northwest of the geographic mean center of the range and 1,300 km northwest of the initial point of introduction. We also calculated the highest median habitat suitability in eastern Australia, due to the high degree of geographic and genetic separation between northwestern and eastern toads. It was located only 100 km north of the initial point of introduction at Gordonvale, Queensland and 400 km northeast of the geographic mean center point (Figure 2). At the individual transect scale, Maxent habitat suitability scores were generally poor predictors of genetic differentiation and diversity patterns (Figures S1 and S2, Supporting Information), Therefore, we used the maximum habitat suitability indices in the northwest and east (Figure 2) as a guide to designate more coastal and northerly sites as ecological core areas, and more inland and southerly sites as edge, for testing of CMH predictions (Figure 1).
Population genomics
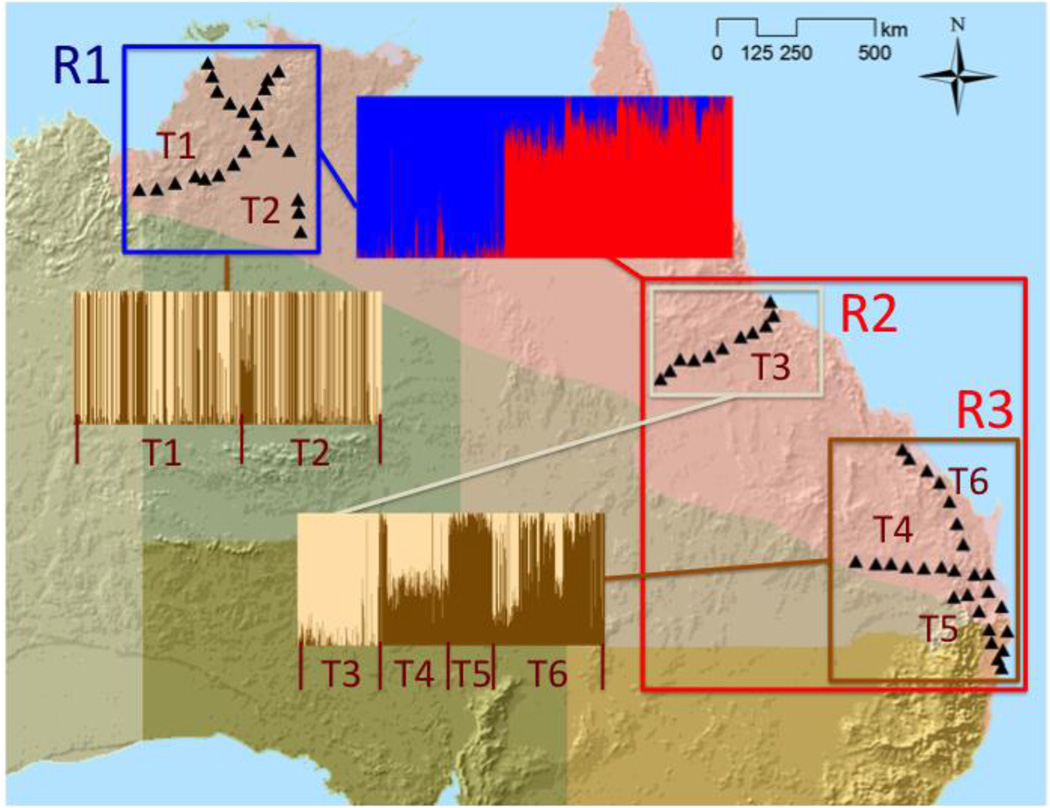
Bayesian clustering tests revealed little population structure across the cane toad’s invasive range in Australia (Figure 3). Admixture and fastStructure runs using all 1,123 individuals and 15,389 SNPs (i.e., filtered to 1 SNP per 95 bp RAD locus; Table S3, Supporting Information) separated the northwestern invasion front toads (transects 1 and 2) from all of the eastern toads. We then ran Admixture and fastStructure on these clusters separately. The northwestern invasion front toads showed no further population substructuring. Individuals were assigned randomly to K=2 populations with no spatial pattern, suggesting the real K value was 1. The eastern toads were further divided into K=2 clusters. The northeastern toads (transect 3) clustered together. The southeastern toads (transects 4–6) formed the second cluster, which had more population substructure and introgression than the other two regions (Figure 3). This suggests southeastern toad populations have more restricted gene flow, and/or more genetic drift, relative to the other two regions.
Figure 3.
Admixture population clustering plots, showing the three regions that cluster together (R1-R3), six transects within regions (T1-T6), and collection localities in 2010 and 2011 (black triangles). Each line in a barplot represents an individual toad, and the colors represent the proportion of ancestry of the individual’s genotype (Q) assigned to each population cluster (K).
Genetic differentiation and diversity measures across the range were consistent with Bayesian clustering results, showing the highest levels of population structure in the southeast (Table 2). Genetic differentiation, as measured by pairwise FST, was low overall, suggesting high gene flow rates and/or insufficient time for genetic drift and differentiation to occur since their introduction. There was a cline of increasing genetic differentiation from the northwest to the southeast portion of the range (Table 2). This suggests gene flow is most restricted and/or genetic drift is highest in the southeast region, followed by the northeast region and the northwest invasion front. Genetic diversity measures, θw and θπ, were low and relatively stable across the range (Table 2), consistent with a single, recent introduction of a small founder population (Sabath et al. 1981). Tajima’s D was slightly positive across the cane toad’s range (Table 2), which is indicative of either balancing selection or a recent population bottleneck. Mantel tests revealed significant IBD patterns along all transects (Table 2), confirming the spatial scale of sampling was appropriate to detect patterns of genetic differentiation and diversity.
Table 2.
Genetic diversity measures, consisting of Watterson’s theta (θw) and theta pi (θπ); Tajima’s D; average pairwise FST; and Mantel tests for isolation by distance (IBD); for each transect and region.
| Transect or region |
Watterson’s theta (standard error) |
Theta pi (standard error) |
Tajima’s D (standard error) |
Pairwise FST (standard error) |
Mantel test for IBD R2 (p-value) |
|---|---|---|---|---|---|
| T1 | 0.00487 (0.00010) | 0.00549 (0.00011) | 0.438 (0.036) | 0.0136 (0.0006) | 0.363 (0.005) |
| T2 | 0.00434 (0.00007) | 0.00515 (0.00009) | 0.432 (0.029) | 0.0184 (0.0008) | 0.382 (0.008) |
| T3 | 0.00456 (0.00006) | 0.00541 (0.00007) | 0.434 (0.021) | 0.0372 (0.0017) | 0.704 (<0.001) |
| T4 | 0.00377 (0.00005) | 0.00471 (0.00007) | 0.454 (0.143) | 0.0485 (0.0031) | 0.572 (0.001) |
| T5 | 0.00354 (0.00008) | 0.00431 (0.00010) | 0.387 (0.143) | 0.0708 (0.0077) | 0.566 (0.040) |
| T6 | 0.00441 (0.00018) | 0.00518 (0.00018) | 0.371 (0.022) | 0.0712 (0.0020) | 0.471 (<0.001) |
| R1 | 0.00462 (0.00008) | 0.00532 (0.00008) | 0.424 (0.021) | 0.0159 (0.0004) | 0.484 (<0.001) |
| R2 | 0.00456 (0.00006) | 0.00541 (0.00007) | 0.434 (0.021) | 0.0372 (0.0017) | 0.704 (<0.001) |
| R3 | 0.00407 (0.00012) | 0.00489 (0.00012) | 0.400 (0.015) | 0.0706 (0.0010) | 0.479 (<0.001) |
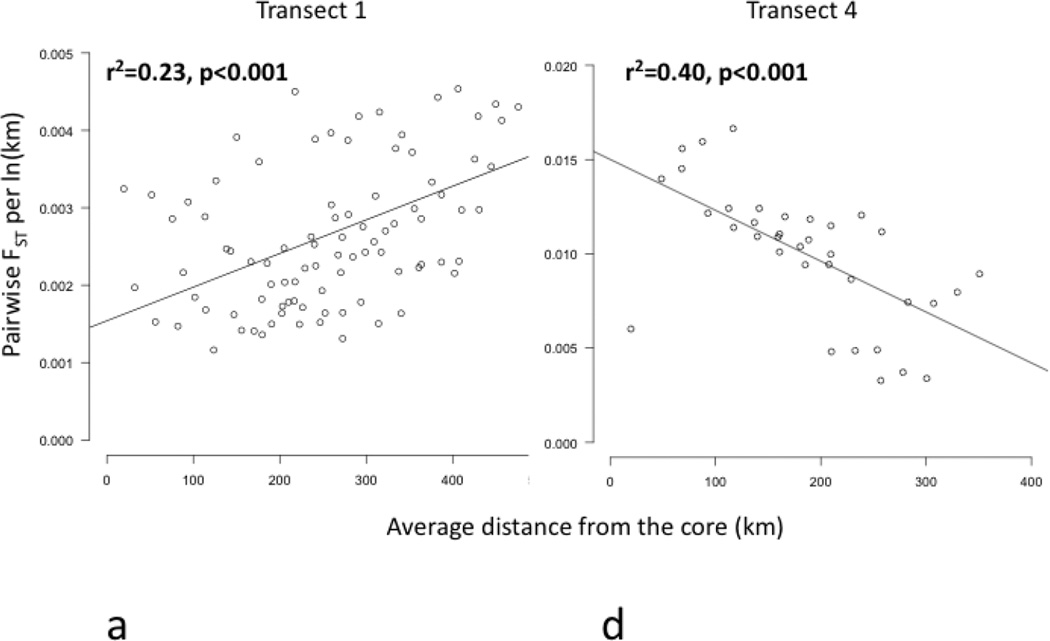
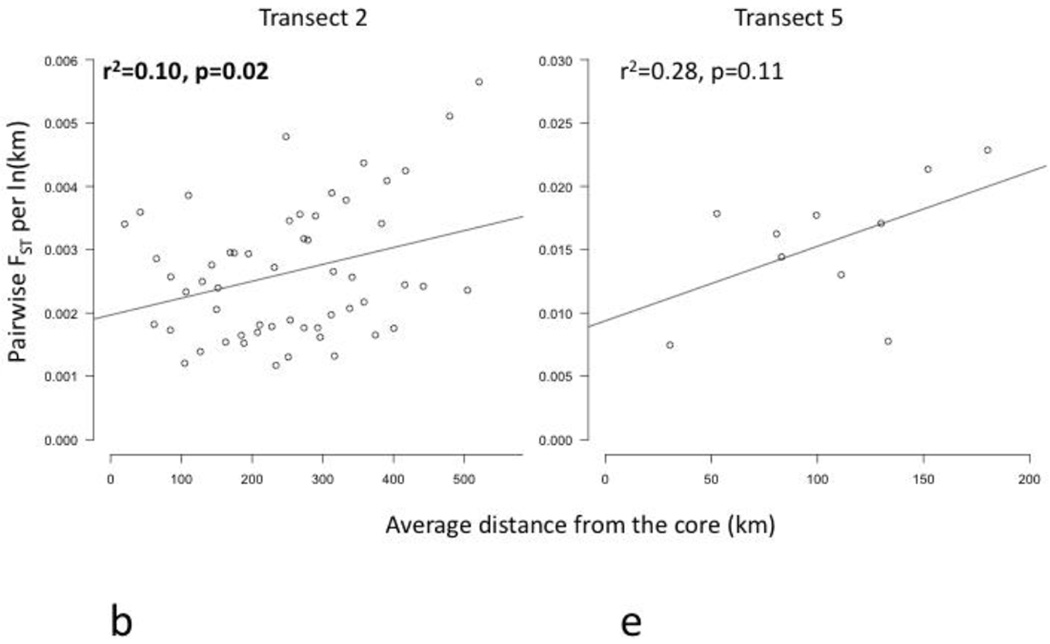
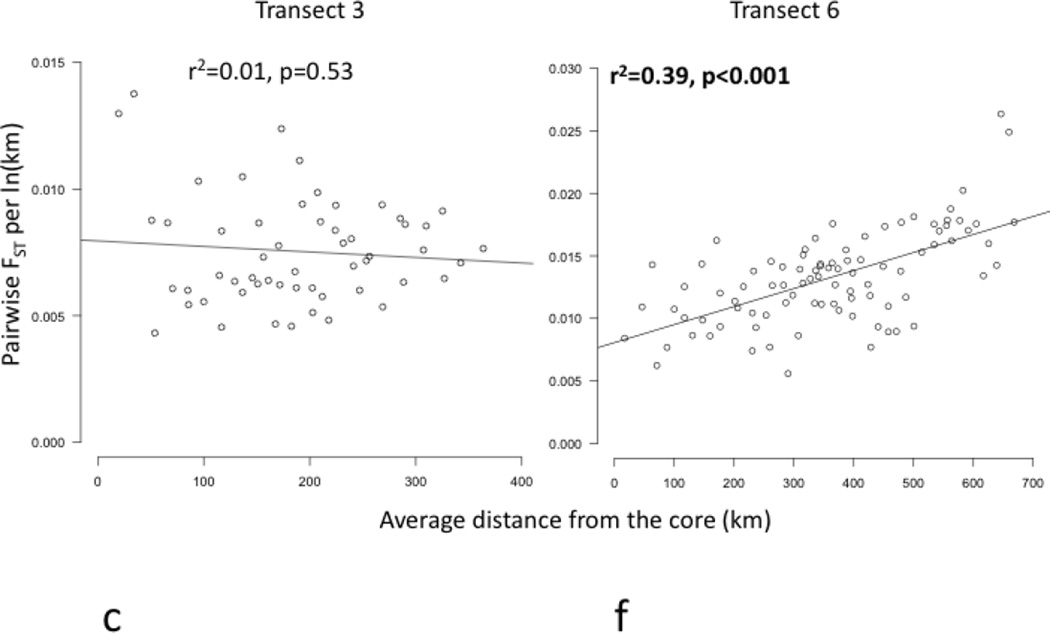
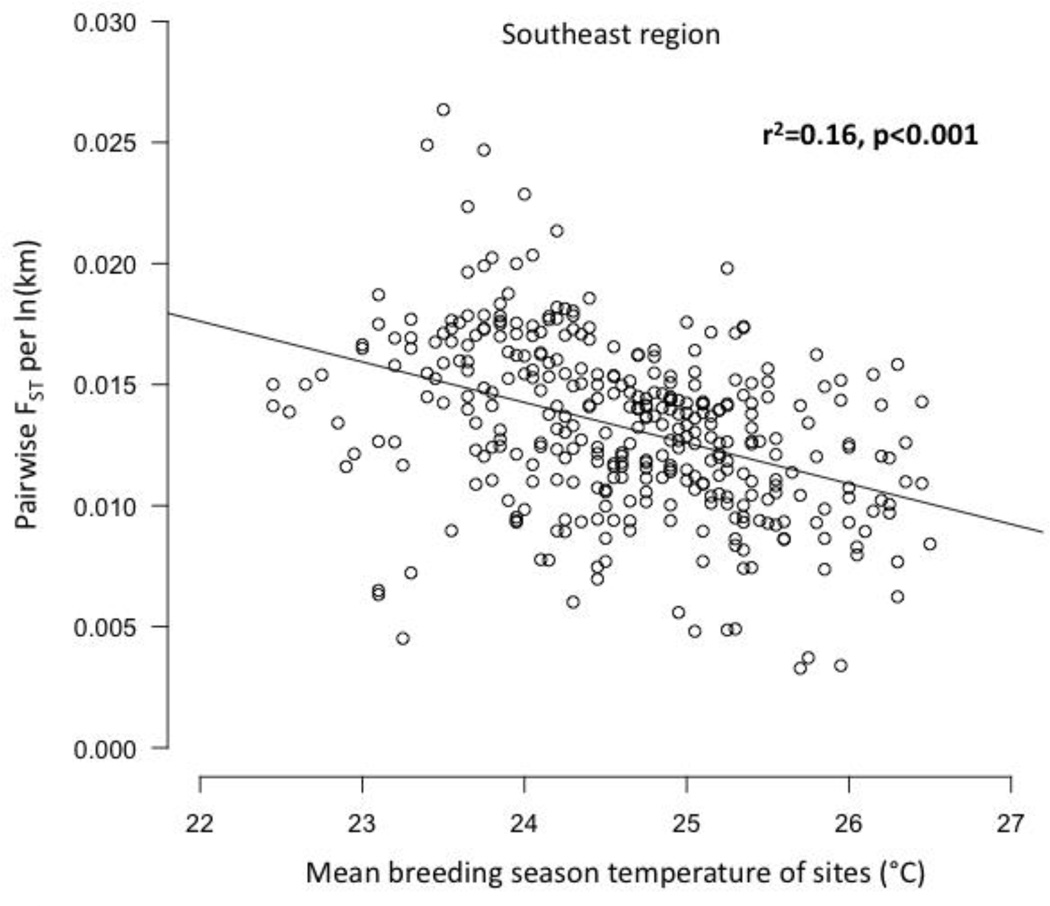
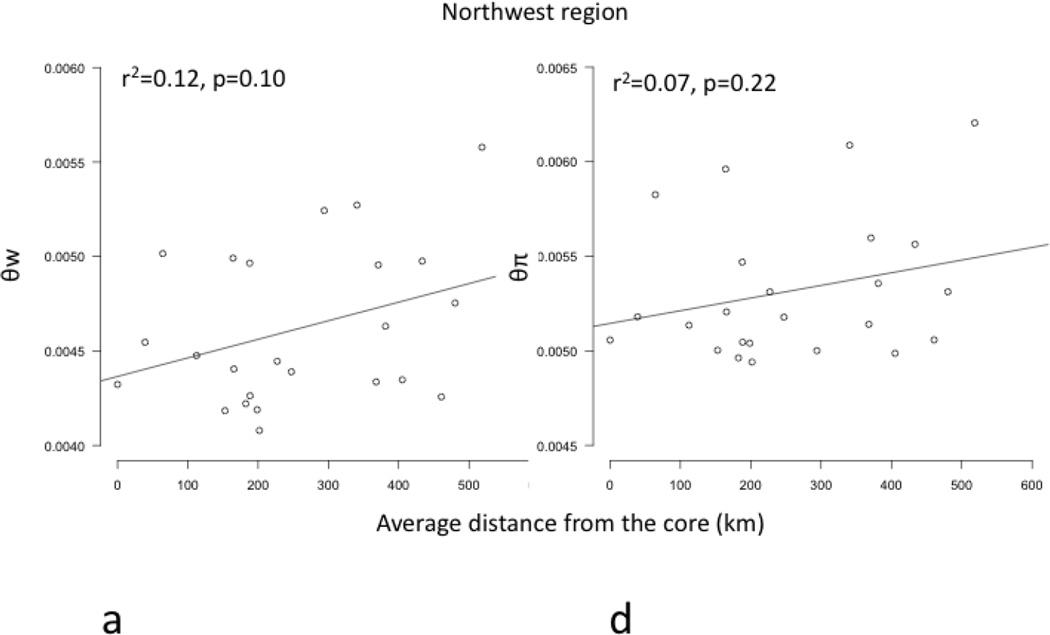
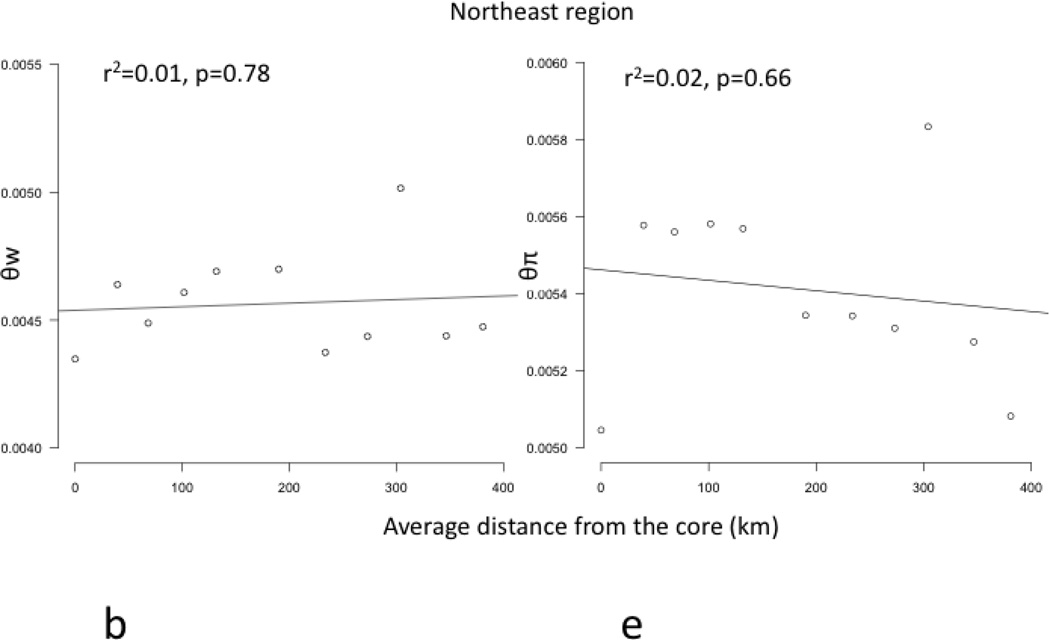
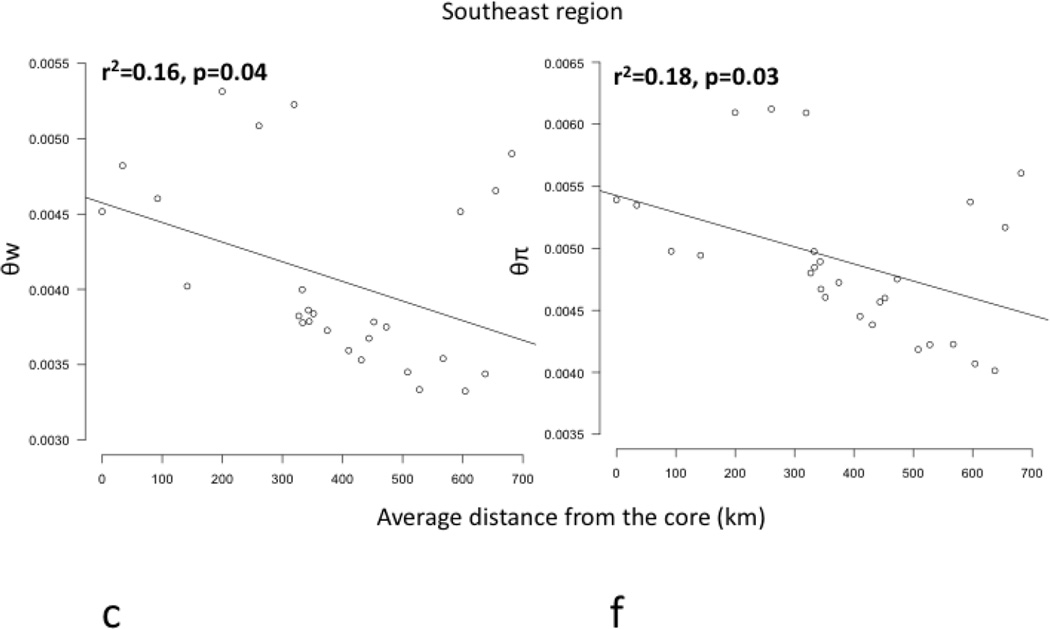
Core to edge transects showed markedly different patterns of genetic differentiation and genetic diversity across the range. Genetic differentiation increased significantly along core to edge transects 1, 2, and 6 (northwestern and southeastern regions; Figure 4a, b, f), suggesting gene flow becomes more restricted at these range edges. There was an increasing but non-significant trend in genetic differentiation along transect 5 in the southeast (Figure 4e), likely due to small sample size. Genetic differentiation did not increase or decrease from core to edge along transect 3 in the northeast (Figure 4c), suggesting no reduction in gene flow and habitat connectivity at this range edge. Unexpectedly, genetic differentiation decreased significantly from core to edge along transect 4 (Figure 4d). Since inland sites tended to be warmer than coastal sites in the southeast, we further investigated the environmental factor most likely to be limiting the cane toad distributions in the south: cold temperatures. We examined the relationship between mean temperature of the breeding season and genetic differentiation. We found a significant increase in genetic differentiation from warmer to colder sites, suggesting gene flow is more restricted between cold pairs of sites than warm pairs of sites, irrespective of spatial location (Figure 5). Genetic diversity, θw and θπ, showed no significant patterns along any transect, so we examined core to edge patterns by region. The southeast region showed significant declines in both θw and θπ from core to edge (Figure 6c, f). The northeast region (transect 3) showed no pattern in θw and θπ from core to edge (Figure 6b, e). The northwest region showed slightly increasing, non-significant trends in θw and θπ (Figure 6a, d).
Figure 4.
Regression plots of genetic differentiation, normalized by distance between populations, represented by pairwise FST per ln(km), versus average distance of each pair of sites to the core (km) along transects 1–6 (a–f). Transects 1, 2, and 6 (a, b, and f) showed significant increases in FST per ln(km) (bolded). Transect 4 (d) showed a significant decrease in FST per ln(km) (bolded). Transects 3 and 5 (c and e) showed no significant pattern in FST per ln(km).
Figure 5.
Regression plot of genetic differentiation, normalized by distance between populations, represented by pairwise FST per ln(km), versus mean temperature of the warmest quarter (i.e., cane toad breeding season in the southeast) of each pair of sites. FST per ln(km) decreases significantly from colder to warmer pairs of sites in the southeastern region.
Figure 6.
Regression plots of genetic diversity, represented by θw (a, b, c) and θπ (d, e, f), versus distance from the core (km) in 3 regions (northwest=a, d; northeast =b, e; southeast=c, f). θw and θπ decline significantly in the southeastern region (c, f) (bolded). The northwestern region (a, d) and northeastern region (b, e) showed no significant patterns in θw or θπ.
Finally, we found limited evidence of asymmetric, core-to-edge gene flow using BayesAss, as well as strong regional differences in effective population sizes using NeEstimator. Transects 2, 4, 5 had a higher proportion of migrants assigned moving from core to edge than from edge to core, whereas transects 1, 3, and 6 had approximately equal proportions of core to edge migrants as edge to core migrants (Table 3). Estimates of Ne showed no significant patterns along core to edge transects, but overall Nes were largest in the northwest invasion front (123.5–172.1), intermediate in the northeast (103.0), and smallest in the cold southeast region (39.4–61.2) (Table S4, Supporting Information). This marked decline in Ne from northwest to southeast provides further support for small, isolated populations in the cold southeast region, as well as surprisingly large populations at the actively expanding edge of the northwest invasion front.
Table 3.
Asymmetric gene flow estimation using the Bayesian assignment test BayesAss. Mean migrant proportions and standard deviation across model runs were calculated from 5 independent BayesAss runs for each transect.
| Transect | Mean proportion migrants moving in (edge-to-core) |
Mean proportion migrants moving out (core-to-edge) |
Standard deviation across model runs |
|---|---|---|---|
| 1 | 0.499 | 0.501 | 0.009 |
| 2 | 0.438 | 0.562 | 0.006 |
| 3 | 0.507 | 0.493 | 0.001 |
| 4 | 0.400 | 0.600 | 0.003 |
| 5 | 0.469 | 0.531 | <0.001 |
| 6 | 0.502 | 0.493 | 0.006 |
Discussion
Geographic range limits of species can be caused by a combination of interacting ecological and evolutionary factors, such as demography and genetic diversity or habitat heterogeneity and gene flow (Hoffman and Blows 1994, Parmesan et al. 2005, Sexton et al. 2009). The CMH is an evolutionary hypothesis that predicts decreased habitat suitability at the range edge will result in smaller, more isolated populations with decreased genetic diversity and gene flow relative to the core (Sagarin and Gaines 2002, Eckert et al. 2008). Edge populations may lack sufficient genetic diversity to adapt to environmental conditions at the range edge, which can cause either stable range limits or punctuated range expansions over time (Hoffman and Blows 1994, Gomulkiewicz et al. 1999, Holt et al 2005). For the invasive cane toad in Australia, patterns of genomic diversity and differentiation followed the predictions of the CMH in the southeastern portion of the range, but there was mixed support in the northwest and no support in the northeast (Figure 3). In the southeast, effective population sizes were the smallest, genetic diversity decreased, and genetic differentiation increased from core to edge, consistent with the CMH (Figures 4 and 6, Table S4, Supporting Information). Additionally, asymmetric, core-to-edge gene flow was found along some transects in the southeast (Table 3), potentially caused by higher quality habitats in the core relative to the edge (Mayr 1963, Kirkpatrick and Barton 1997). Cold temperatures appear to be a major limiting factor to further range expansions in the southeast (Figure 5; Sutherst et al. 1996, Kolbe et al. 2012, McCann et al. 2014). While allele surfing can also cause patterns of reduced genetic diversity at expanding range edges (Klopfstein et al. 2006, Excoffier and Ray 2008), this explanation is less likely than the CMH in eastern Australia given that cane toads have been there the longest (Figure 1), range limits are stable, and effective population sizes are smaller than those at the northwest invasion front where serial founder effects should result in the highest magnitude of genetic drift (Table S4, Supporting Information).
The CMH was not supported in the northeast and showed mixed support in the northwest. Here, simpler ecological explanations of genomic patterns are more likely than the CMH. In the northeast, the rate and symmetry of gene flow and levels of genetic diversity did not change from core to edge, in spite of a strong precipitation gradient from the tropical coast to semi-desert inland habitats (Table 3, Figures 4 and 6; Tingley et al. 2012, 2014). The inland northeast is characterized by an extreme physiological barrier, drought, combined with patchily distributed habitats that contain enough water resources for breeding and desiccation avoidance, including large rivers, cattle ponds, and irrigated human developments. With sufficient habitat patch sizes and connectivity, these ecological conditions can cause abrupt species range limits with large population sizes and high connectivity all the way out to the range edge (Hastings et al. 1997, Holt et al. 2005, Sexton et al. 2009). At the northwest invasion front, increased genetic differentiation and asymmetric gene flow were detected from core to edge (Table 3, Figure 4). However, effective population sizes were the largest in this region, overall levels of genetic differentiation were lowest, and genetic diversity did not decline from core to edge, but actually showed a slightly increasing, non-significant trend (Table 2, Figure 6, Table S4, Supporting Information). Here, the ecological dynamics of an active, ongoing species invasion are more likely to explain these genomic patterns than the CMH (Sakai et al. 2001, Klopfstein et al. 2006, Excoffier and Ray 2008, Sexton et al. 2009, Shine et al. 2012, Rollins et al. 2015). These mixed results highlight the value of assessing multiple transects across a species’ geographic range to detect varying ecological and evolutionary processes operating at different range edges (Sexton et al. 2009).
Tests of the CMH
Although unfortunate, species invasions provide unique opportunities to test hypotheses for the evolution of species’ range limits (Sexton et al. 2009, Guo 2014). However, one challenge in testing species’ range limit theory is the identification of core and edge regions, particularly in invasive species that have large, non-circular geographic ranges that still expanding (Hoffman and Blows 1994, Sexton et al. 2009, Micheletti and Storfer 2015). Using an ENM-based habitat suitability index, we found that the highest habitat suitability in the cane toad’s Australian range was located at the northern coast of the Northern Territory, over 1,000 km west of both the geographic center of the range and the initial point of introduction (Figure 2). Restricting ENMs to populations in eastern Australia (Figure 2), the highest habitat suitability was only 100 km north of the site of the initial introduction in tropical, northeast Queensland (Figure 2). The geographic mean center of the range had a relatively low habitat suitability index score (0.283; Figure 2). This low score highlights the importance of using a habitat-based suitability measure to define the ecological core of the range, rather than a simple geographic approximation of the range center as most empirical range limit studies have done in the past (Garner et al. 2004, Munwes et al. 2010, Martinez-Meyer et al. 2012, Dixon et al. 2013, Johansson et al. 2013, Lira-Noriega and Manthey 2014, Micheletti and Storfer 2015, Ursenbacher et al. 2015). ENM-based estimates of the core largely matched our intuitive predictions that more coastal and northerly regions would have higher habitat suitability (Figure 2). However, it was not apparent which of the northwestern coastal populations should be assigned as core in analyses that combined transects 1 and 2 until ENM was performed (Figures 3, 6).
The southeastern region had the smallest effective population sizes, and it was the only portion of the cane toad’s range where we found significant declines in genetic diversity from core to edge (Figure 6, Table S4, Supporting Information). Cold temperatures are a likely contributing factor, as colder sites were significantly more genetically isolated than warmer sites (Figure 5). Therefore, small population sizes and lack of genetic diversity at cold edge sites likely limit cane toad range expansion to the south. However, ongoing global climate change may allow for future cane toad range expansions in the south by increasing local temperatures, resulting in higher habitat suitabilities and thus potentially a shallower environmental gradient to adapt (Kawecki 2008, Sexton et al. 2009).
In the northeast, there was no evidence of increased genetic differentiation, decreased genetic diversity, or asymmetric gene flow, as predicted by the CMH (Figures 4 and 6, Table 3; Eckert et al. 2008). Here, simpler ecological explanations appear more likely than the CMH. Cane toads are limited by a steep physiological barrier at this range edge, consisting of extreme aridity and drought resulting in a lack of breeding ponds and high desiccation risk (Tingley et al. 2012, 2014). However, there are also large inland rivers (e.g., Flinders River), permanent cattle ponds, and irrigated human developments located near this range edge. Arid habitats that contain permanent moisture to avoid desiccation, and at least occasional standing water for breeding, likely allow dispersal and gene flow to remain high within otherwise inhospitable edge environments. When suitable habitat patches are of sufficient size and connectivity, steep ecological barriers to dispersal can result in stable levels of gene flow and genetic diversity all the way out to the range edge (Hastings et al. 1997, Holt et al. 2005, Sexton et al. 2009).
The northwest region is still an expanding invasion front (Kearney et al. 2008, Urban et al. 2008), so the CMH is an unlikely explanation for genomic patterns of diversity and gene flow. Instead, continual establishment of founder populations at the invasion front can lead to scattered populations with low gene flow among them (Sakai et al. 2001, Klopfstein et al. 2006, Excoffier and Ray 2008, Shine et al. 2012). Therefore, edge populations are expected to be more genetically isolated, with asymmetric gene flow from larger, more contiguous core populations (Figure 4, Table 3; Sakai et al. 2001, Kirkpatrick and Barton 1997, Eckert et al. 2008). Surprisingly, effective population sizes were not smaller than the core in this recently invaded region, and genetic diversity did not decline but rather increased slightly from core to edge (Figure 6a, d, Table S4, Supporting Information). Abundance of cane toads appears to be highest in newly-colonized areas and decreases in long-colonized areas, perhaps due to reduced food availability or increased parasite loads (Freeland 1986, Shine 2010). High habitat suitability in the northwest (Figure 2), and potentially the evolution of increased dispersal phenotypes in an expanding invasion front (Alford et al. 2009, Phillips et al. 2010, Shine et al. 2012, Rollins et al. 2015), may also help explain the high overall levels of gene flow, genetic diversity, and large effective population sizes found in this region (Table 1, Figure 3, Table S4, Supporting Information).
Historical demography
Overall, cane toads in Australia showed low levels of population genomic structure and genetic diversity across their vast invaded range (Table 2, Figure 3). These genomic patterns are consistent with numerous ecological studies documenting the toad’s extreme dispersal abilities relative to other amphibians (Schwarzkopf and Alford 2002, Brown et al. 2006, Shine 2010) and a well-documented history of founder effects during their invasion (Easteal 1981, Sabbath et al. 1981, Lever 2001). Mean Tajima’s D values across loci were slightly positive (Table 2). Whereas range expansions can cause Tajima’s D values to be negative by increasing the number of low frequency mutations through allele surfing and genetic drift (Klopfstein et al. 2006, Excoffier and Ray 2008), strong population bottlenecks can cause Tajima’s D values to be positive by eliminating low frequency mutations. Low θ values across the cane toad’s geographic range can be caused by strong population bottlenecks as well (Table 1), as θ is proportional to past effective populations sizes and mutation rates (θ=4Neµ). Prior studies using 12 microsatellites from a smaller portion of the cane toad’s Australian range have also shown evidence of population bottlenecks (Estoup et al. 2001, 2004, 2010).
Cane toads in Australia have a well-documented invasion history of multiple population bottlenecks and founder effects (Easteal 1981, Sabbath et al. 1981, Lever 2001). In the mid-1800s, cane toads were introduced from their native South American countries of Guyana and French Guiana to the Caribbean islands of Bermuda, Martinique, Barbados, and Jamaica. In 1920 and 1923, they were introduced from Barbados and Jamaica to Puerto Rico. In 1932, they were introduced from Puerto Rico to the Hawaiian Islands. Finally in 1935, 101 cane toads were collected from just a few localities on the island of Oahu and introduced to Gordonvale, Queensland, Australia. Offspring from this initial Australian introduction were subsequently released and established in six sugar cane growing regions along the east coast of Queensland from 1935–1937 (Figure 1; Sabath et al. 1981). Thus cane toads have only been present in Australia for approximately 80 generations, assuming a minimum 1 year generation time (Phillips and Shine 2005). Our population genomic results suggest that high dispersal and gene flow, as well as strong and relatively recent population bottlenecks, are dominating the effects of isolation and genetic drift due to serial founder effects and allele surfing (Klopfstein et al. 2006, Excoffier and Ray 2008) during the cane toad’s rapid invasion of Australia.
Conclusion
Cane toads have been extremely successful invaders in Australia, quickly becoming one of the largest and most damaging amphibian invasions in the world (Lever 2001, Kraus 2009, Shine 2010; www.issg.org). This vast invasion is ongoing, as the northwestern edge of their range is still an actively expanding invasion front. ENMs predict cane toad spread along the northern and western coasts into the southern part of Western Australia (Kearney et al. 2008, Kolbe et al. 2010). Moreover, the southeastern range edge is likely to expand further with warming temperatures due to global climate change. We found that the CMH and cold temperatures are likely contributors to the toad’s current range limits in the southeast, but not in the other two regions. Therefore, future management efforts in the southeast should consider targeting populations with high levels of adaptive genetic diversity. In arid, interior portions of the range, management measures should limit toad access to moisture and standing water wherever possible, such as cattle ponds and irrigated developed areas. Overall we found mixed support for the role of the CMH in evolving new range limits in this infamous and ongoing species invasion.
Supplementary Material
Acknowledgments
We thank the editor and two anonymous reviewers for their constrctive comments on this manuscript. For help with field research, conceptual advice, and funding in Australia, we thank Mathew Vickers, John Llewelyn, Richard Duffy, Lexie Edwards, Joost Kunst, Reid Tingley, Sharon Lehman, Jordy Groffen, Lee Scott-Virtue, Jeremy VanDerWal, Ben Phillips, Richard Shine, and an Australian Research Council grant. For help with lab research, conceptual advice, and funding in the USA, we thank Tamara Max, Sarah Emel, Steven Micheletti, Cody Wiench, Rose Marie Larios, Patricia Frias, Matt Settles, Stephen Spear, Jon Eastman, Richard Gomulkiewicz, Lisette Waits, the National Science Foundation (NSF) Doctoral Dissertation Improvement Grant (DDIG) Award Number 1407335, NSF Integrative Graduate Education and Research Traineeship (IGERT) Program in Evolutionary Modeling (IPEM) fellowship, a National Institute of Health (NIH) grant P30 GM103324, and Washington State University Elling Trust Travel Awards and Brislawn Award.
Footnotes
Data Accessibility
ddRADseq and locality data used in genomic and niche modeling analyses are available at Dryad (doi:10.5061/dryad.5ps15).
Author Contributions
D.R.T. collected field data, performed laboratory work, performed population genomic and ecological niche modeling analyses, and wrote the manuscript; B.E. performed population genomic analyses; L.S. designed research and directed field work; P.A.H. designed laboratory and population genomics research and directed laboratory work; R.A.A. directed field work; A.S. directed the project, designed research, contributed to data analyses, and contributed to writing the manuscript; and all authors contributed input to draft and final versions of the manuscript.
Table S1: Field samples collected from 62 cane toad populations; including sample size, transect membership, and life stage.
Table S2: Correlation matrix of 26 environmental variables considered for ecological niche modeling with Maxent.
Table S3: Number of SNP loci per transect or region retained after filtering, as well as the number of sampling sites and number of individuals sequenced per transect or region.
Table S4: Effective population sizes (Ne), as well as means and standard errors across each transect, estimated using NeEstimator v.2 (linkage disequilibrium method).
Figure S1: Regression plots of genetic differentiation, normalized by distance between populations, represented by pairwise FST per ln(km), versus average Maxent habitat suitability index of each pair of sites along transects 1–6 (a–f). Transect 1 showed a significant decrease in FST per ln(km) (bolded). Transects 2–6 showed no significant pattern in FST per ln(km).
Figure S2: Regression plots of genetic diversity, represented by θw (a, b, c) and θπ (d, e, f), versus Maxent habitat suitability index in 3 regions (northwest=a, d; northeast =b, e; southeast=c, f). θw and θπ showed no significant patterns in any region.
References
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Research. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford RA, Brown GP, Schwarzkopf L, Phillips BL, Shine R. Comparisons through time and space suggest rapid evolution of dispersal behaviour in an invasive species. Wildlife Research. 2009;36:23–28. [Google Scholar]
- Blows MW, Hoffmann AA. A reassessment of genetic limits to evolutionary change. Ecology. 2005;86:1371–1384. [Google Scholar]
- Bonin A, Nicole F, Pompanon F, Miaud C, Taberlet P. Population adaptive index: a new method to help measure intraspecific genetic diversity and prioritize populations for conservation. Conservation Biology. 2007;3:697–708. doi: 10.1111/j.1523-1739.2007.00685.x. [DOI] [PubMed] [Google Scholar]
- Bridle JR, Vines TH. Limits to evolution at range margins: When and why does adaptation fail? Trends in Ecology and Evolution. 2006;22:140–147. doi: 10.1016/j.tree.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Brown JH. On the relationship between abundance and distribution of species. American Naturalist. 1984;124:255–279. [Google Scholar]
- Brown GP, Phillips BL, Webb JK, Shine R. Toad on the road: use of roads as dispersal corridors by cane toads (Bufo marinus) at an invasion front in tropical Australia. Biological Conservation. 2006;133:88–94. [Google Scholar]
- Catchen JM, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Molecular Ecology. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin CR. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London, England: John Murray; 1859. p. 362. [Google Scholar]
- Dixon AL, Herlihy CR, Busch JW. Demographic and population-genetic tests provide mixed support for the abundant centre hypothesis in the endemic plant Leavenworthia stylosa . Molecular Ecology. 2013;22:1777–1791. doi: 10.1111/mec.12207. [DOI] [PubMed] [Google Scholar]
- Do C, Wapels RS, Peel D, et al. NeEstimator v2; re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Molecular Ecology Resources. 2014;14:209–214. doi: 10.1111/1755-0998.12157. [DOI] [PubMed] [Google Scholar]
- Easteal S. The history of introductions of Bufo marinus (Amphibia: Anura); a natural experiment in evolution. Biological Journal of the Linnean Society. 1981;16:93–113. [Google Scholar]
- Eckert CG, Samis KE, Lougheed SC. Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Molecular Ecology. 2008;17:1170–1188. doi: 10.1111/j.1365-294X.2007.03659.x. [DOI] [PubMed] [Google Scholar]
- Elith J, Graham CH, Anderson RP, et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 2006;29:129–151. [Google Scholar]
- Estoup A, Wilson IJ, Sullivan C, Cornuet JM, Moritz C. Inferring population history from microsatellite and enzyme data in serially introduced cane toads, Bufo marinus . Genetics. 2001;159:1671–1687. doi: 10.1093/genetics/159.4.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estoup A, Beaumont E, Sennedot F, Moritz C, Cornuet JM. Genetic analysis of complex demographic scenarios: spatially expanding populations of the cane toad, Bufo marinus . Evolution. 2004;58(9):2021–2036. doi: 10.1111/j.0014-3820.2004.tb00487.x. [DOI] [PubMed] [Google Scholar]
- Estoup A, Ray N, Cornuet MJM, Santos F, Beaumont MA, Excoffier L. Combining genetic, historical and geographical data to reconstruct the dynamics of bioinvasions: application to the cane toad Bufo marinus . Molecular Ecology Resources. 2010;10:886–901. doi: 10.1111/j.1755-0998.2010.02882.x. [DOI] [PubMed] [Google Scholar]
- Evanno GS, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Ray N. Surfing during population expansions promotes genetic revolutions and structuration. Trends in Ecology & Evolution. 2008;23:347–351. doi: 10.1016/j.tree.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Freeland WJ. Populations of cane toad, Bufo marinus, in relation to time since colonisation. Australian Wildlife Research. 1986;13:321–329. [Google Scholar]
- Garner TWJ, Pearman PB, Angelone S. Genetic diversity across a vertebrate species’ range: a test of the central-peripheral hypothesis. Molecular Ecology. 2004;13:1047–1053. doi: 10.1111/j.1365-294X.2004.02119.x. [DOI] [PubMed] [Google Scholar]
- Gessler PE, Moore ID, McKenzie NJ, Ryan PJ. Soil-landscape modeling of plant species distribution. International Journal of GIS. 1995;9(4):421–432. [Google Scholar]
- Gomulkiewicz R, Holt RD, Barfield M. The effects of density dependence and immigration on local adaptation and niche evolution in a black-hole sink environment. Theoretical Population Biology. 1999;55:283–296. doi: 10.1006/tpbi.1998.1405. [DOI] [PubMed] [Google Scholar]
- Guo Q. Central-marginal population dynamics in species invasions. Frontiers in Ecology and Evolution. 2014;2(23):1–17. [Google Scholar]
- Haldane JBS. The relation between density regulation and natural selection. Proceedings of the Royal Society London Series B. 1956;145:306–308. doi: 10.1098/rspb.1956.0039. [DOI] [PubMed] [Google Scholar]
- Hastings A, Harrison S, McCann K. Unexpected spatial patterns in an insect outbreak match a predator diffusion model. Proceedings of the Royal Society B. 1997;264:1837–1840. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005;25:1965–1978. [Google Scholar]
- Hoffmann AA, Blows MW. Species borders: ecological and evolutionary perspectives. Trends in Ecology and Evolution. 1994;9:223–227. doi: 10.1016/0169-5347(94)90248-8. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Amish S, Catchen JM, Allendorf FW, Luikart G. Next-generation RAD sequencing identifies thousands of SNPs for assessing hybridization between rainbow and westslope cutthroat trout. Molecular Ecology Resources. 2011;11:117–122. doi: 10.1111/j.1755-0998.2010.02967.x. [DOI] [PubMed] [Google Scholar]
- Holt RD, Keitt TH, Lewis MA, Maurer BA, Taper ML. Theoretical models of species’ borders: single species approaches. Oikos. 2005;108:18–27. [Google Scholar]
- Johansson H, Stoks R, Nilsson-Ortman V, Ingvarsson PK, Johansson F. Large-scale patterns in genetic variation, gene flow and differentiation in five species of European Coenagrionid damselfly provide mixed support for the central-marginal hypothesis. Ecography. 2013;36:744–755. [Google Scholar]
- Kawecki TJ. Adaptation to marginal habitats. Annual Review of Ecology, Evolution, and Systematics. 2008;39:321–342. [Google Scholar]
- Kearney MR, Phillips BL, Tracy CR, Christian KA, Betts G, Porter WP. Modelling species distributions without using species distributions: the cane toad in Australia under current and future climates. Ecography. 2008;31:423–434. [Google Scholar]
- Kirkpatrick M, Barton NH. Evolution of a species’ range. American Naturalist. 1997;150:1–23. doi: 10.1086/286054. [DOI] [PubMed] [Google Scholar]
- Klopfstein S, Currat M, Excoffier l. The fate of mutations surfing on the wave of a range expansion. Molecular Biology and Evolution. 2006;23(3):482–490. doi: 10.1093/molbev/msj057. [DOI] [PubMed] [Google Scholar]
- Kolbe JJ, Kearney M, Shine R. Modeling the consequences of thermal trait variation for the cane toad invasion of Australia. Ecological Applications. 2010;20:2273–2285. doi: 10.1890/09-1973.1. [DOI] [PubMed] [Google Scholar]
- Kraus F. Alien reptiles and amphibians: a scientific compendium and analysis. Dordrecht, The Netherlands: Springer Science; 2009. [Google Scholar]
- Leblois R, Rousset F, Tikel D, Moritz C, Estoup A. Absence of evidence for isolation by distance in an expanding cane toad (Bufo marinus) population: an individual-based analysis of microsatellite genotypes. Molecular Ecology. 2000;9:1905–1909. doi: 10.1046/j.1365-294x.2000.01091.x. [DOI] [PubMed] [Google Scholar]
- Lever C. The cane toad: the history and ecology of a successful colonist. West Yorkshire, United Kingdom: Westbury Publishing; 2001. [Google Scholar]
- Lira-Noriega A, Manthey JD. Relationship of genetic diversity and niche centrality: a survey and analysis. Evolution. 2014;68(4):1082–1093. doi: 10.1111/evo.12343. [DOI] [PubMed] [Google Scholar]
- Llewelyn J, Phillips BL, Brown GP, Schwarzkopf L, Alford RA, Shine R. Adaptation or preadaptation: why are keelback snakes (Tropidonophis mairii) less vulnerable to invasive cane toads (Bufo marinus) than are other Australian snakes? Evolutionary Ecology. 2010;25:13–24. [Google Scholar]
- Luikart G, England PR, Tallmond D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nature Reviews Genetics. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- MacArthur RH. Geographical ecology. New York, USA: Harper and Row; 1972. p. 288. [Google Scholar]
- Martinez-Meyer E, Diaz-Porras D, Peterson AT, Yanez-Arenas C. Ecological niche structure and rangewide abundance patterns of species. Biology Letters. 2012;9:1–5. doi: 10.1098/rsbl.2012.0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Animal species and evolution. Massachusetts, USA: Harvard University Press, Cambridge; 1963. [Google Scholar]
- McCann S, Greenlees MJ, Newell D, Shine R. Rapid acclimation to cold allows the cane toad to invade montane areas within its Australian range. Functional Ecology. 2014;25(5):1166–1174. [Google Scholar]
- McCune B, Keon D. Equations for potential annual direct incident radiation and heat load. Journal of Vegetation Science. 2002;13:603–606. [Google Scholar]
- Micheletti SJ, Storfer A. A test of the central-marginal hypothesis using population genetics and ecological niche modelling in an endemic salamander (Ambystoma barbouri) Molecular Ecology. 2015;24:967–979. doi: 10.1111/mec.13083. [DOI] [PubMed] [Google Scholar]
- Munwes I, Geffen E, Roll U, et al. The change in genetic diversity down the core-edge gradient in the eastern spadefoot toad (Pelobates syriacus) Molecular Ecology. 2010;19:2675–2689. doi: 10.1111/j.1365-294X.2010.04712.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York, USA: Columbia University Press; 1987. [Google Scholar]
- Parmesan C, Gaines S, Gonzalez L, Kaufman DM, Kingsolver J, Peterson AT, Sagarin R. Empirical perspectives on species borders: from traditional biogeography to global change. Oikos. 2005;108:58–75. [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE. 2012;7(5):1–11. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramuk JB, Robertson T, Sites JW, Jr, Noonan BP. Around the world in 10 million years: biogeography of the nearly cosmopolitan true toads (Anura: Bufonidae) Global Ecology and Biogeography. 2007;17:72–83. [Google Scholar]
- Phillips BL, Shine R. The morphology, and hence impact, of an invasive species (the cane toad, Bufo marinus): changes with time since colonization. Animal Conservation. 2005;8:407–413. [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecological Modelling. 2006;190:231–259. [Google Scholar]
- Phillips BL, Brown GP, Shine R. Evolutionarily accelerated invasions: the rate of dispersal evolves upwards during the range advance of cane toads. Journal of Evolutionary Biology. 2010;23:2595–2601. doi: 10.1111/j.1420-9101.2010.02118.x. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Stephens M, Pritchard JK. FastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics. 2014;179(2):573–589. doi: 10.1534/genetics.114.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins LA, Richardson MF, Shine R. A genetic perspective on rapid evolution in cane toads (Rhinella marina) Molecular Ecology. 2015;24:2264–2276. doi: 10.1111/mec.13184. [DOI] [PubMed] [Google Scholar]
- Sabath MD, Boughton WC, Easteal S. Expansion of the range of the introduced toad Bufo marinus in Australia from 1935 to 1974. Copeia. 1981;3:676–680. [Google Scholar]
- Sagarin RD, Gaines SD. The abundant centre distribution: to what extent is it a biogeographical rule? Ecology Letters. 2002;5:137–147. [Google Scholar]
- Sakai AK, Allendorf FW, Holt JS, et al. The population biology of invasive species. Annual Review of Ecology and Systematics. 2001;32:305–332. [Google Scholar]
- Schwarzkopf L, Alford RA. Nomadic movement in tropical toads. Oikos. 2002;96(3):492–506. [Google Scholar]
- Sexton JP, McIntyre PJ, Angert AL, Rice KJ. Evolution and ecology of species range limits. Annual Review of Ecology, Evolution, and Systematics. 2009;40:415–436. [Google Scholar]
- Shine R. The ecological impact of invasive cane toads (Bufo Marinus) in Australia. The Quarterly Review of Biology. 2010;85(3):253–291. doi: 10.1086/655116. [DOI] [PubMed] [Google Scholar]
- Shine R, Brown GP, Phillips BL. An evolutionary process that assembles phenotypes through space rather than through time. Proceedings of the National Academy of Sciences. 2012;108(14):5708–5711. doi: 10.1073/pnas.1018989108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherst RW, Floyd RB, Maywald GF. The potential geographical distribution of the cane toad, Bufo marinus, in Australia. Conservation Biology. 1996;10:294–299. [Google Scholar]
- Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240:1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K. Libsequence, a C++ class library for evolutionary genetic analysis. Bioinformatics. 2003;19(17):2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- Tingley R, Phillips BL, Letnic M, Brown GP, Shine R, Baird SJE. Identifying optimal barriers to halt the invasion of cane toads Rhinella marina in arid Australia. Journal of Applied Ecology. 2012;50:129–137. [Google Scholar]
- Tingley R, Vallinoto M, Sequeirac F, Kearney MR. Realized niche shift during a global biological invasion. Proceedings of the National Academy of Sciences. 2014;111(28):10233–10238. doi: 10.1073/pnas.1405766111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban MC, Phillips BL, Skelly DK, Shine R. A toad more traveled: the heterogeneous invasion dynamics of cane toads in Australia. The American Naturalist. 2008;171(3):134–148. doi: 10.1086/527494. [DOI] [PubMed] [Google Scholar]
- Ursenbacher S, Guillon M, Cubizolle H, Dupoué A, Demers GB, Lourdais O. Postglacial recolonisation in a cold climate specialist in Western Europe: patterns of genetic diversity in the adder (Vipera berus) support the central-marginal hypothesis. Molecular Ecology. 2015 doi: 10.1111/mec.13259. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. Genome size and GC-percent in vertebrates as determined by flow cytometry: the triangular relationship. Cytometry. 1998;31:100–109. doi: 10.1002/(sici)1097-0320(19980201)31:2<100::aid-cyto5>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Warren DL, Glor RE, Turelli M. ENMTOOLS: a toolbox for comparative studies of environmental niche models. Ecography. 2010;33:607–611. [Google Scholar]
- Watterson G. On the number of segregating sites in genetical models without recombination. Theoretical Population Biology. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Wilson GA, Rannala B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics. 2003;163:1177–1191. doi: 10.1093/genetics/163.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.