Abstract
5′-isomiRs expand the repertoire of miRNA targets. However, how they are generated is not well understood. Previously, we showed that for some miRNAs in mammalian cells, Drosha cleaves at multiple sites to generate multiple pre-miRNAs that give rise to multiple 5′-isomiRs. Here, we showed that for some other miRNAs, 5′-isomiRs are generated by alternative Dicer processing. In addition, we showed that in miR-203, alternative Dicer processing is regulated by a conserved sliding-bulge structure at the Dicer processing site, which allows the pre-miRNA molecule to fold into two different structures that are processed differently by Dicer. So far no RNA motif that slides to change conformation and alter a protein–RNA interaction has been reported. Thus, our study revealed a novel RNA motif that regulates 5′-isomiR generation in some miRNAs. It might also contribute to regulating protein–RNA interactions in other biological processes, since it takes only one point mutation to generate the sliding bulge, and there are a large number of different RNAs in the cell.
Keywords: Biological sciences, Genetics, Cell biology
1. Introduction
miRNA genes are transcribed as long primary miRNAs (pri-miRNAs), which are processed by the Drosha–DGCR8 complex into pre-miRNAs, which are, in turn, processed by Dicer into mature miRNA duplexes. One or both strands of the duplexes are selected by Ago proteins to become mature miRNAs (Ha and Kim, 2014). Since the discovery of miRNAs, it has been observed that the ends of the mature miRNAs are not fixed but subject to variation (Lau et al., 2001). We and others have previously described the variation pattern at miRNA ends. For most miRNAs, the 5′ ends are homogeneous, while the 3′ ends are highly heterogeneous. However, for some miRNAs, the 5′ ends are also highly heterogeneous (Ruby et al., 2007; Wu et al., 2007). These miRNAs with 5′-end variations have been called “5′-isomiRs”, which are of particular interest because their 5′ seed sequences (ranging from positions 2–7 (Lewis et al., 2005)) are changed, which affects target specificity. In fact, multiple studies have shown recently that 5′ isomiRs appears to be a common mechanism that expands the pool of target genes regulated by miRNAs (Fukunaga et al., 2012; Humphreys et al., 2012; Llorens et al., 2013; Manzano et al., 2015; Tan et al., 2014).
An increasing number of 5′-isomiRs have been discovered recently (Gottwein et al., 2011; Humphreys et al., 2012; Llorens et al., 2013; Morin et al., 2008; Ruby et al., 2007; Seitz et al., 2008; Tan et al., 2014; Wu et al., 2007; Wu et al., 2009). However, how 5′-isomiRs are generated is not well understood. It is intriguing that they occur only in some miRNAs, while most miRNAs have highly homogeneous 5′ ends. It has been suspected that Drosha/Dicer cleavage is generally imprecise and thereby generates the heterogeneity of miRNA ends. However, downstream constraints, such as Ago protein loading, may limit selection of isomiRs, accounting for the homogeneous 5′ ends of most miRNAs (Seitz et al., 2008). We previously showed that, in most cases, Drosha cleavage is precise in generating one pre-miRNA that gives rise to miRNAs with homogeneous 5′ ends, however, for some miRNAs, Drosha cleaves at multiple sites to generate multiple pre-miRNAs that give rise to multiple 5′-isomiRs (Ma et al., 2013; Wu et al., 2009). Recently, Fukunaga et al. reported another mechanism in Drosophila in which binding of Dicer partner proteins, Loqs-PB, changes Dicer cleavage of miR-307a to generate 5′-isomiRs (Fukunaga et al., 2012). The mammalian Dicer partner TRBP, a Loqs-PB homolog, has also been reported to regulate Dicer processing of pre-miRNAs both in vitro (Lee et al., 2013) and in vivo (Kim et al., 2014; Wilson et al., 2015). It was also reported that nucleotide sequences at the Dicer cleavage site contribute to cleavage site selection by Dicer in vitro (Starega-Roslan et al., 2015). Here we showed that some miRNAs in mammals can be alternatively processed by Dicer to generate 5′-isomiRs in mammalian cells in vivo. In addition, we showed that a sliding-bulge structure can cause alternative Dicer processing to generate 5′-isomiRs, suggesting that the secondary structure of miRNAs is an alternative mechanism for generating these molecules.
2. Results
2.1. Alternative Dicer processing is a common mechanism for generating 5′-isomiRs
5′-isomiRs are commonly observed in the small RNA deep-sequencing data in publicly available databases and are also found in our own data. To understand how these molecules are generated, we selected miRNAs that showed heterogeneous 5′ ends according to miRBase and also our own deep-sequencing data. To determine how these isomers are generated, we cloned these pri-miRNAs in the pLL3.7 vector and transfected the constructs into 293FT cells. To avoid interference from endogenous miRNAs, we chose miRNAs that are not expressed or are poorly expressed in 293FT cells. The small RNAs from transfected 293 FT cells were cloned and sequenced to investigate the processing of these miRNAs by Drosha and Dicer.
We first characterized the Drosha cleavage sites in these miRNAs. While the 5′ end of a mature miRNA generated from the 5′ arm of the pre-miRNA (5p-miRNA) reliably indicates a Drosha cleavage site, some cleavage sites cannot be predicted by the 5′ end of the mature miRNAs, because after Drosha cleavage, (1) the pre-miRNAs might be degraded quickly or not be transported efficiently to the cytosol to be processed into the miRNA duplex or (2) one or both strands of the miRNA duplex may not be loaded efficiently into Ago proteins and, as a result, are degraded quickly. On the other hand, we have previously shown that miRNA offset RNAs (moRs), especially 5′-moRs, can be used to determine the Drosha cleavage site more reliably than the 5′ end of 5p-miRNAs (Ma et al., 2013). Thus, we mainly used 5′-moR sequences to determine Drosha cleavage sites.
Analysis of the moR sequences for miR-136, miR-383, and miR-411 suggested that Drosha cleaves at multiple sites, which leads to multiple pre-miRNAs generated from a single pri-miRNA (Fig. 1A, Table S1, Dataset S1 and S2). To confirm these results, we performed northern blotting. Fig. 1B shows that two distinct bands were observed for pre-miR-383 and pre-miR-411, indicating that Drosha cleaves at two sites and generates two different pre-miRNAs. However, for miR-136, although moR sequences indicated that Drosha cleaves at two sites to a similar degree (Fig. 1A and Table S1), only one band was visualized using a short-exposure autoradiogram (Fig. 1B), and a faint band at around 4 nt above the main pre-miRNA band was visualized using a long exposure (Fig. 1C). The lower level of the larger pre-miRNA shown by northern blot suggests that the larger pre-miRNA is degraded quickly. Consistent with this hypothesis, a mature miRNA corresponding to this pre-miRNA was cloned at a frequency of only 0.6% of total miR-136-5p reads (Dataset S2). And therefore, in light of 5′-moR sequences, pri-miR-136 was likely alternatively processed by Drosha. Thus, the 5′-isomiRs of miR-136, as well as of miR-383 and 411, were likely generated by alternative Drosha processing.
Fig. 1.
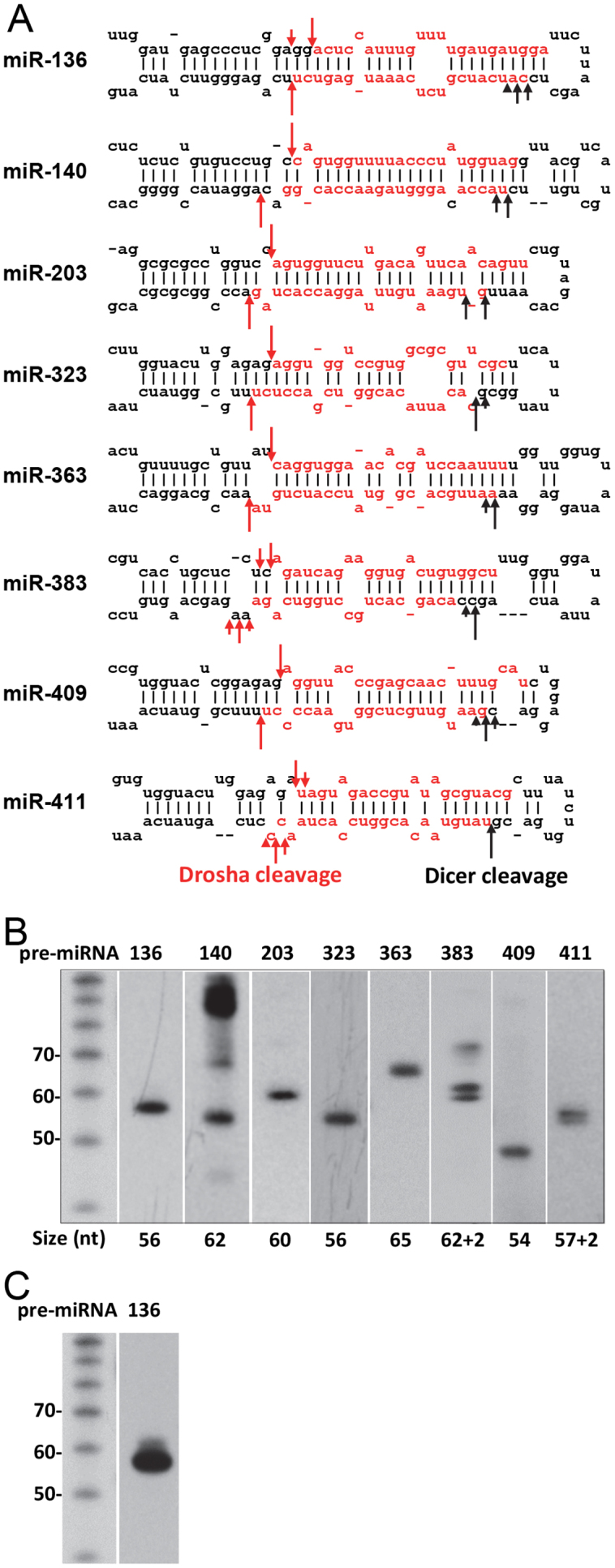
5′-isomiRs can be generated by alternative Drosha or Dicer processing. (A) The indicated pri-miRNAs were transfected into 293FT cells and the small RNAs cloned to determine the Drosha or Dicer cleavage sites. Mature miRNA sequences are highlighted in red. Representative reads of cloned small RNAs are found in Table S1. Red arrows indicate the Drosha cleavage sites and black arrows indicate the Dicer cleavage sites. Drosha cleavage sites were determined according to the moR sequences, while Dicer cleavage sites were determined according to the 5′ end of the mature miRNAs generated from the 3′ arm of the pre-miRNA. Therefore, only half of the Dicer cleavage sites are shown. The length of the arrows represents roughly the frequency of moRs for Drosha and the mature miRNA generated from the 3′ arm of the pre-miRNA for Dicer. (B) Northern blot analysis of the indicated pre-miRNAs in transfected cells. The numbers below the gel panels indicate the size of pre-miRNA predicted according to the moR sequences. (C) Long-exposure autoradiogram for pre-miR-136 northern blot. Please see Figs. 1S–5S in the Supplementary Material for full blot images.
By contrast, for miR-140, 203, 323, 363, and 409, Drosha cleaves at only one site, according to their moR sequences, to generate only one pre-miRNA (Fig. 1A and Table S1), which was also confirmed by northern analysis (Fig. 1B). Thus, the 5′-isomiRs observed in these miRNAs appear to be the result of alternative Dicer processing, suggesting that it might be a common mechanism for generating 5′-isomiRs.
2.2. The mismatches in the miRNA duplex are not responsible for the alternative Dicer processing of miR-203
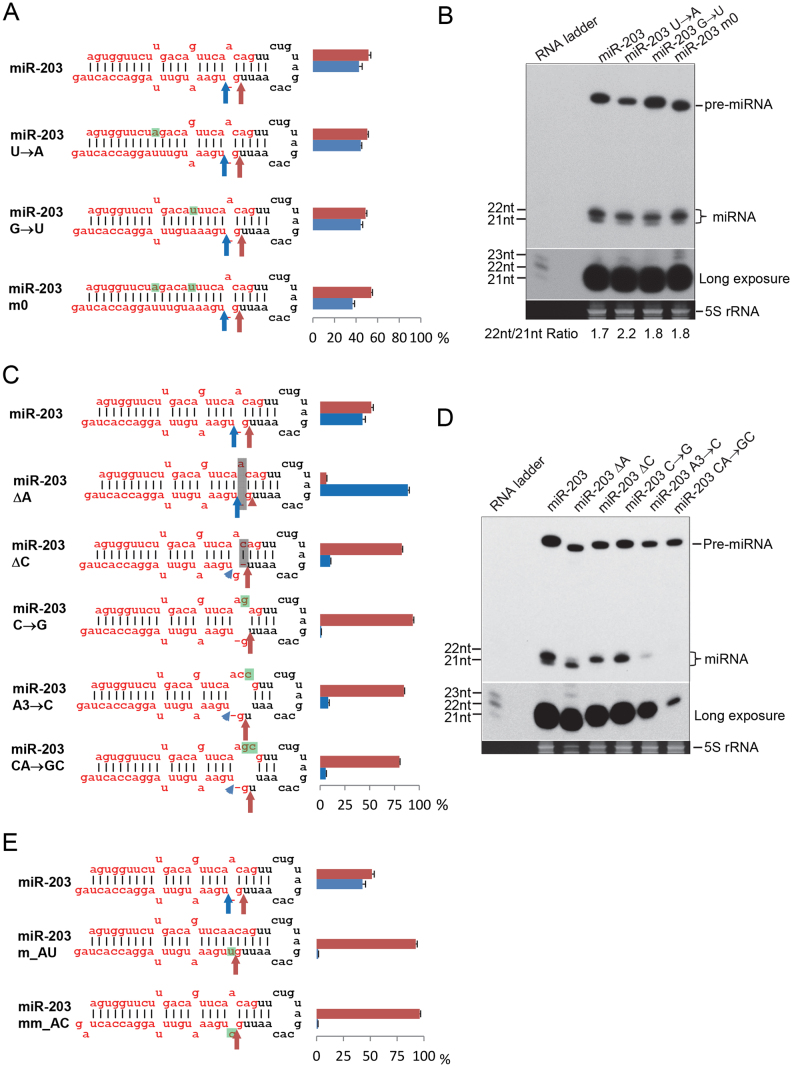
To determine why alternative Dicer processing occurred in these miRNAs, we chose miR-203 as a model miRNA to investigate the underlying mechanism. There are two mismatches in the pre-miR-203 stem, which might affect the stem length of the pre-miRNA and therefore change Dicer cleavage site selection, since Dicer acts as a molecular ruler that measures the stem length and cleaves ∼22 nt from the end generated by Drosha cleavage (Macrae et al., 2006). To test this hypothesis, we introduced mutations to remove the mismatches, as shown in Fig. 2A. The moR sequences suggested that the Drosha processing site was not affected (Table S1), which was confirmed by cloning the pre-miRNAs (Table S1 and Dataset S3). Removing mismatches did not change the overall isomiR pattern of miR-203, according to the mature miR-203 sequences. Two dominant isomiRs were generated in all mutants, although the ratio of 21-nt/22-nt isomiRs was slightly changed (Fig. 2A and Table S1). Northern blot results also showed that the mature miR-203 has two bands at 21 and 22 nts. In contrast to the small RNA cloning results, in which two forms of mature miR-203 were almost same, the intensity of the 21-nt band is less than the 22-nt band; however, this pattern appears to be similar between mutant and wild type miR-203 (Fig. 2B). Thus, the mismatches in the miRNA duplex are unlikely to have caused the alternative Dicer processing observed in miR-203.
Fig. 2.
A single-nucleotide bulge at the Dicer processing site regulates alternative Dicer processing of miR-203. (A) Dicer cleavage sites determined by mature miR-203 ends. Mismatches in the duplex were mutated, and the mutated nucleotides are highlighted in green. Dicer processing sites were determined as in Fig. 1A. Dicer cleavage sites are indicated with color-coded arrows corresponding to bars indicating the proportion of each isomiR (n = 3). Representative reads of cloned small RNAs are found in Table S1. (B) Northern blot to visualize the intermediate processing products of the indicated mutants. The number below the image is the 22-nt/21-nt ratio. (C) Various mutations introduced to change the bulge at the Dicer processing site. Dicer cleavage sites were determined by cloned reads of mature miR-203 ends (see Table S1). Deletions are highlighted in gray, while mutations are highlighted in green. Dicer processing sites were determined as in Fig. 1A. (D) Northern blot to visualize the intermediate processing products of the indicated mutants. (E) Introducing mutations in guide strand also eliminates alternative Dicer processing. A nucleotide was introduced into guide strand to create a matched (miR-203 m_AU) or mismatched (miR-203 mm_AC) pair. Dicer processing sites were determined as in Fig. 1A. Please see Figs. 6S–7S in the Supplementary Material for full blot images.
The inconsistency in which 22-nt bands were more frequent in northern blots than in the small-RNA cloning data can be explained partially by the fact that the 21-nt isomiR-203 (UGAAAUGUUUAGGACC ACUAG) tended to be modified with an additional U more often than the 22-nt isomiR (GUGAAAUGUUUAGGACCACUAG) (Table S2 and Dataset S1). The 21-nt isomiR with an additional U has a length of 22 nt, which would appear as a 22-nt isomiR in a northern blot. Thus, small-RNA cloning followed by deep sequencing is a more reliable way to determine Drosha/Dicer processing sites in vivo than northern blot.
2.3. The single-nucleotide bulge at the Dicer processing site regulates alternative Dicer processing of miR-203
There is a single-nucleotide bulge at the Dicer processing site of pre-miR-203. We suspected that the bulge caused the alternative Dicer processing, so we introduced different mutations to remove the bulge (Fig. 2C). As expected, after removing the bulge, Dicer cleaved predominantly at one site and generated one predominant mature miR-203 (Fig. 2C, D). Interestingly, removing the bulge by deleting the bulge nucleotide (miR-203 ΔA) or deleting a neighboring nucleotide to make a mismatch (miR-203 ΔC) caused Dicer to cut predominantly at different sites (Fig. 2C, D). A similar effect was observed when the bulge size was increased to 2 or 3 nt (miR-203C→G, A3→C, or CA→GC mutants, Fig. 2C, D), suggesting that the single-nucleotide bulge regulates the alternative Dicer processing.
It is noteworthy that mutants miR-203 A3→C and CA→GC have much less mature miRNA product according to both northern blot (Fig. 2D) and small RNA cloning results (Table S1), suggesting that structures around the Dicer processing site can significantly affect mature miRNA production.
So far, we have made all the mutants by modifying the passenger strand while leaving the guide strand intact. To test whether changing the bulge to a match or a mismatch by modifying the guide strand would eliminate alternative Dicer processing as well, we made mutants as shown Fig. 2E. In both cases, only one predominant mature miRNA was generated (Fig. 2E and Table S1), suggesting that the bulge is indeed important for alternative Dicer processing.
2.4. The conserved, flexible, single-nucleotide bulge regulates Dicer processing
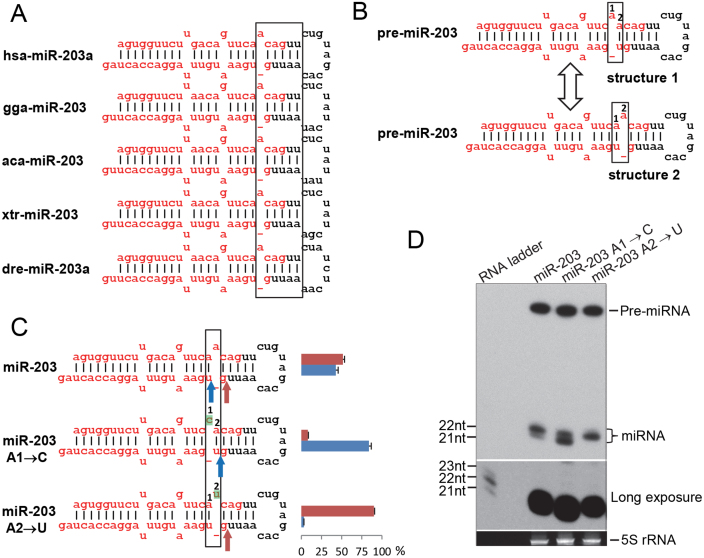
Next, we investigated the pre-miR-203s from different species and found that the single-bulge structure is highly conserved—all of the species examined that express miR-203 have the same bulge plus a 5-bp extension into the terminal loop (Fig. 3A), suggesting that this structure is important. Interestingly, it appears that two neighboring A residues at the bulge site compete for pairing with the same U to form two different structures. When A1 pairs with U, the pre-miR-203 forms the pre-miR-203-1 structure, and when A2 pairs with U, it converts the molecule to the pre-mir-203-2 structure (Fig. 3B). The single-nucleotide bulge can therefore slide one nucleotide to form two different structures. We suspected that this conserved sliding bulge at the Dicer processing site causes Dicer to cleave at different sites, because it can slide one nucleotide to change the pre-miR-203 structures at the Dicer processing site. To test this hypothesis, we introduced mutations to make the sliding bulge fixed in place to yield either structure 1 (miR-203 A1→C mutant) or structure 2 (miR-203 A2→U mutant), as shown in Fig. 3C. Exactly as predicted, Dicer cleaved predominantly at site 1 for the miR-203 A1→C mutant and at site 2 for the miR-203 A2→U mutant, judging from the small RNA reads (Fig. 3C and Table S1). Northern blotting results were consistent with the small RNA cloning results (Fig. 3D). Thus, the alternative Dicer processing in miR-203 is regulated by the sliding bulge, which generates two different pre-miRNA structures, depending on which A was chosen to pair with the U.
Fig. 3.
A sliding-bulge structure caused alternative Dicer processing in miR-203. (A) The sliding-bulge structure is conserved across species. The box indicates the sliding bulge with the extended duplex that helps to maintain the bulge structure. (B) pre-miR-203 can have two different structures, depending on which A (at positions 1 or 2) pairs with the U nucleotide. The box indicates the sliding-bulge structure. (C) Dicer cleavage sites in miR-203 mutants with a fixed bulge. The A at position 1 was mutated to C so that the miR-203 A1→C mutant had the fixed bulge of structure 1, while the A at position 2 was mutated to U so that the miR-203 A2→U mutant had the fixed bulge of structure 2, as shown in (B). Dicer processing sites were determined as in Fig. 1A. (D) Northern blot to visualize the mature miR-203 derived from the indicated mutants. Please see Fig. 8S in the Supplementary Material for full blot images.
2.5. The sliding bulge is a general mechanism for regulating Dicer processing
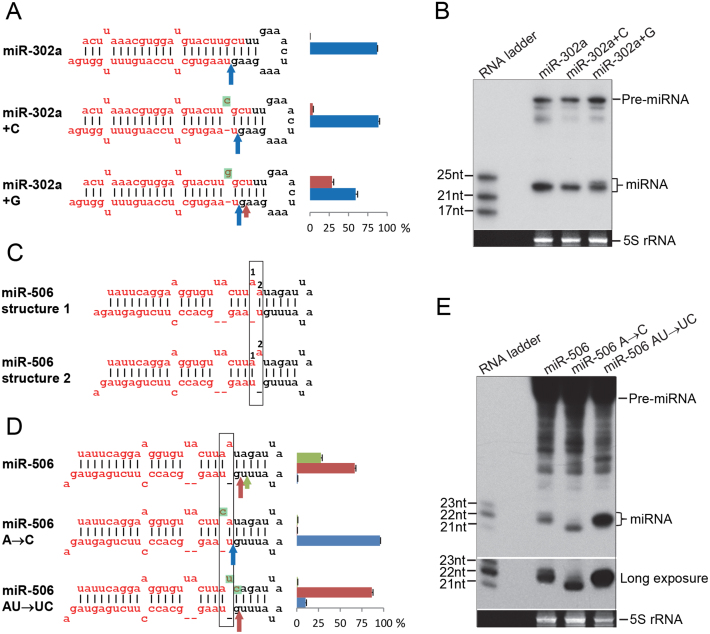
To investigate whether the sliding bulge is a general mechanism regulating alternative Dicer processing, we introduced a sliding bulge into miR-302a, which doesn't have such a structure originally. We inserted either a C to create a fixed bulge (miR-302a + C) or a G to create a sliding bulge (miR-302a + G), in which the inserted G competes with the neighboring G for pairing to the U (Fig. 4A). The mutant with the artificial sliding bulge (miR-302a + G) generates two isomiRs that are different from those generated by the wild type miR-302a or the mutant with the fixed bulge, which generates only one predominant mature miRNA (Fig. 4A, B). The insertion of a bulge appeared to affect the accuracy of Drosha processing in both mutants similarly (Table S1). However, it should not interfere with the interpretation of Dicer processing data, since both sliding or fixed bulges affect Drosha processing similarly in this case (Table S1). Therefore, the artificial sliding bulge indeed caused alternative Dicer processing.
Fig. 4.
The sliding bulge is a general mechanism for regulating Dicer processing. (A) Introducing a sliding bulge causes alternative Dicer processing in miR-302a. A fixed bulge was created for miR-302a + C by inserting a C, while a sliding bulge was created for miR-302a + G by inserting a G, which competes with the neighboring G for the same U and therefore, can slide one nucleotide. Dicer processing sites were determined as in Fig. 1A. (B) Northern blot to visualize the products of pre-miR-302a mutants. (C) pre-miR-506 has a similar sliding structure, as observed in miR-203, in which two As at positions 1 and 2 compete for pairing with U. The box indicates the sliding-bulge structure. The Dicer cleavage sites were determined by mature miR-506 sequences, and the Drosha processing site was determined by moR sequences. The reads of mature miR-506 and moRs are found in Table S1. (D) Dicer cleavage sites in miR-506 mutants with a fixed bulge. Dicer processing sites were determined as in Fig. 1A. (E) Northern blot to visualize the mature miR-506 of the indicated mutants. Please see Fig. 9S-10S in the Supplementary Material for full blot images.
Thus, it is extremely easy to generate the sliding bulge structure, as a single point mutation is sufficient. We searched miRBase to see whether other miRNAs have similar structures to miR-203. miR-506 appears to have a similar sliding-bulge structure around the Dicer processing site, which allows it to fold into two different structures (Fig. 4C), although it is noteworthy that the sliding bulge in miR-506 is not conserved. To determine whether the sliding bulge also causes alternative Dicer processing similar to what we observed in miR-203, we made mutants with fixed (not sliding) bulges (Fig. 4D). According to the small RNAs generated from these constructs, pre-miR-506 was alternatively processed by Dicer at two sites (Fig. 4D). Fixing the bulge in place caused Dicer to cleave predominantly at one site (Fig. 4D). Northern blot results were consistent with the small RNA cloning results (Fig. 4E). Thus, the sliding bulge also regulates alternative Dicer processing in miR-506. It is noteworthy that the mutant cleavage sites did not correlate precisely with the WT cleavage sites in miR-506, as we observed for miR-203. The reason might be that the sliding bulge in WT miR-506 is not exactly localized at the Dicer cleavage site, which is 1 nt away from the Dicer processing site, while the sliding bulge in miR-203 is exactly localized at the Dicer processing site.
3. Discussion
Our study revealed a new mechanism in which isomiRs are generated by alternative Dicer processing that is regulated by secondary structure. A conserved sliding-bulge structure in miR-203 affects protein processing of RNA and regulates alternative Dicer processing and therefore the generation of 5′-isomiRs, since disrupting the bulge structure or fixing the bulge causes Dicer to cut predominantly at different sites (Fig. 2 and Fig. 3). This kind of sliding bulge allows the RNAs to fold into two different structures, which interact with protein differently. Thus, our study has revealed a novel RNA structure that can regulate RNA–protein interactions and is the mechanism behind 5′-isomiR generation in some miRNAs.
Considering the large number of RNAs in cells and that it is extremely easy to generate this kind of structure—a single point mutation is sufficient—we believe that the sliding bulge structure might also contribute to regulating other protein–RNA interactions and should be taken into consideration generally when studying protein–RNA interactions.
Furthermore, we found more 5′-isomiRs that were generated by alternative Dicer processing in mammalian cells, such as miR-140, 323, 363, and 409 (Fig. 1). Thus, alternative Dicer processing appears to be a common mechanism for generating 5′-isomiRs, although we did not find any sliding-bulge structures around the Dicer processing site in these miRNAs. The reason for alternative Dicer processing of these miRNAs therefore remains to be determined.
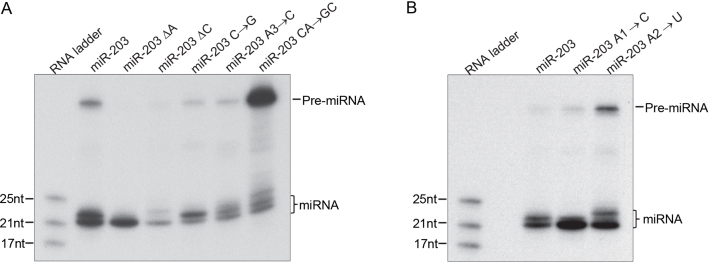
The effect of the sliding-bulge structure on Dicer processing can be visualized using an in vitro Dicer-processing assay, which avoids the complication introduced by in vivo processing steps, such as duplex unwinding and loading. Therefore, we performed an in vitro Dicer cleavage assay for pre-miR-203 mutants disrupting the single-nucleotide bulge and mutants with a fixed sliding bulge (Fig. 5A, B). Wild type pre-miR-203 was processed to produce two predominant isomiRs of length 22 nt and 21 nt. Mutant miR-203 ΔA and miR-203 A1→C produced predominantly the 21-nt isomiR, consistent with the in vivo result. However, the processing of the other mutants appears to be quite different from the in vivo results (Fig. 5A, B). Some other studies also suggested that in vitro Dicer processing assays using purified Dicer might introduce a serious bias, such as Dicer cleavage at multiple sites to generate multiple bands for many miRNAs (Lee and Doudna, 2012; Starega-Roslan et al., 2011), which are likely cleaved by Dicer at one site, as indicated by the mature miRNA sequences cloned from cells or animals. Thus, in vitro Dicer processing might not always recapitulate Dicer processing in vivo. Although our in vitro data was not fully consistent with the in vivo results, it does suggest that the sliding bulge regulates Dicer processing.
Fig. 5.
In vitro Dicer processing assay. pre-miRNAs were treated with recombinant Dicer protein and visualized with northern blot. (A) miR-203 and its mutants from Fig. 2C. (B) miR-203 and its mutants from Fig. 3C. Please see Fig. 11S-12S in the Supplementary Material for full blot images.
For miRNAs generated from the 3′ arm of pre-miRNAs (3p-miRNAs), the 5′ ends of the mature miRNAs are generated by Dicer processing. Multiple 5′-isomiRs for 3p-miRNAs usually indicates that Dicer cleaves at multiple sites, which can be explained by two different mechanisms: (1) Alternative Drosha processing generates multiple pre-miRNAs, which leads to Dicer cutting at multiple sites, although Dicer might cut precisely at only one site for each pre-miRNA, as is the case for miR-142 (Ma et al., 2013; Wu et al., 2009). (2) For some miRNAs, such as miR-140, 203, 323, 363, and 409, multiple isomiRs were generated by alternative Dicer processing of a single pre-miRNA that was generated by precise Drosha cleavage. The key to distinguishing between these two mechanisms is finding a method that can reliably determine the Drosha cleavage site. The 5′ end of mature miRNAs generated from the 5′ arm of pre-miRNAs might be used to determine the Drosha cleavage site; however, this approach might be misleading in some cases. For example, there is only one mature miRNA for miR-136-5p, which might lead one to assume that Drosha precisely cleaves at one site; however, judging from 5′-moR sequences, which we showed previously are a more reliable resource for determining Drosha cleavage sites (Ma et al., 2013), Drosha cleaves at two sites. With this method using 5′-moR sequences to determine Drosha cleavage sites, we correctly identified several 5′-isomiRs that were generated by alternative Dicer processing.
It is interesting that the mature miRNA amount is significantly reduced in two mutants of miR-203 (miR-203 A3→C and CA→GC; Fig. 2D and Table S1), suggesting that the structures close to the Dicer processing site at the terminal region are important for Dicer processing efficiency, which is consistent with a previous study by Zhang et al., in which it was shown that the terminal loop region is important for Dicer processing efficiency for pre-miRNAs (Zhang and Zeng, 2010). Our data also showed that the structures around the Dicer processing site are important for Dicer processing accuracy, which is consistent with a previous study by Gu et al., in which it was shown that the structure around terminal loop region is important for the accuracy of Dicer processing of shRNAs (Gu et al., 2012). Thus, collectively, these results suggest that the structure around the Dicer processing site should be taken into consideration in designing shRNA for accurate and efficient Dicer processing.
4. Materials and methods
4.1. Plasmids
All constructs, including pri-miRNA serial mutations, were designed as oligos and inserted into pLL3.7 lentiviral vector at restriction sites HpaI and XhoI. The inserted sequences are listed in Table S3.
4.2. Cell culture and transfection
293FT cells (Invitrogen) were cultured according to the supplier’s instructions. Plasmid (0.8 μg) was transfected into 293FT cells that had been seeded into 24-well plates at 2 × 105 cells/well the day before transfection. The small-RNA fraction was extracted 28 h later with the miRNeasy kit (Qiagen), as instructed by the manufacturer.
4.3. Small-RNA cloning.
Small RNAs were cloned as described previously (Ma et al., 2013), in which we have made major modifications to improve adaptor ligation efficiency and thereby minimize cloning bias. Briefly, the small RNAs were purified with the miRNeasy kit (Qiagen). Small RNA (50 ng) was ligated with 3′ and 5′ linkers (with barcode) with a truncated form of T4 RNA ligase 2 (K227Q) at 25 °C for 3 h and T4 RNA ligase 1 (NEB) at 37 °C for 1 h, successively, in a ligation buffer containing 17.5% PEG and 10% DMSO. For pre-miRNA cloning, the incubation time for the second ligation step was 5 h. The ligated small RNAs were reverse-transcribed and amplified with the KAPA library amplification kit (KAPA Biosystems) for 10 cycles and the library sequenced using the Illumina HiSeq2000 or MiSeq instruments. The deep-sequencing data were processed with in-house-developed software. Note that for small RNAs to be efficiently cloned with the method described here, the 5' phosphate and 3' hydroxyl groups must be preserved.
4.4. Northern blot
Small RNAs was extracted with the miRNeasy kit (Qiagen) from 293FT cells transfected with the pLL3.7 vector harboring pri-miRNAs or mutants 28 h before. Total small RNA (1.5 μg) was separated on a 15% TBE-UREA PAGE gel (Invitrogen) and transferred onto a hybridization transfer membrane (GeneScreen Plus, Perkin Elmer) in a semi-dry transfer cell (Trans Blot SD, BioRad). The probes were synthesized as DNA or RNA oligos and labeled with γ-32P-ATP by T4 PNK. The probe sequences are listed in Table S4. Hybridization was carried out in 50 ml of hybridization solution at 50 °C overnight.
4.5. In vitro Dicer processing assay
The substrates for the in vitro Dicer cleavage assay were prepared according to Ferre-D'Amare and Doudna et al. (Ferre-D'Amare and Doudna, 1996). A hammerhead ribozyme and a VS ribozyme substrate were introduced to generate pre-miRNAs with homogeneous ends by PCR (see Table S4 for primer sequences). After transcribing with T7 polymerase (HiScribe, NEB), the 84-nt product was PAGE purified and digested with VS ribozyme, then treated with T4 PNK (NEB, USA) and again PAGE purified. The purified oligo (∼6 ng) was heated at 94 °C for 30 s and immediately chilled on ice to get a homogeneous solution of monomers before using in assays with recombinant human Dicer (Genlantis, USA). The products were visualized by northern blot.
Declarations
Author contribution statement
Hongming Ma: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Yonggan Wu, Qi Niu, Junli Zhang, Gengxiang Jia: Performed the experiments.
N. Manjunath: Analyzed and interpreted the data; Wrote the paper.
Haoquan Wu: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported partially by TTUHSC El Paso Seed Grant Program grant for H.W.
Competing interest statement
The authors declare the following conflict of interests: H.W. is a founder and stakeholder of KoBio LLC and Kanglin Biotech (Hangzhou) Co. Ltd.
Additional information
Data associated with this study has been deposited at the NCBI Gene Expression Omnibus database with accession numbers GSE80771 and GSE80772.
Acknowledgements
We thank Dr. Richard A. Collins for kindly providing the VS ribozyme expressing plasmids and technical suggestions.
Contributor Information
Hongming Ma, Email: hongmingma@wustl.edu.
Haoquan Wu, Email: haoquan.wu@ttuhsc.edu.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Ferre-D'Amare A.R., Doudna J.A. Use of cis- and trans-ribozymes to remove 5' and 3' heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R., Han B.W., Hung J.H., Xu J., Weng Z., Zamore P.D. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–546. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E., Corcoran D.L., Mukherjee N., Skalsky R.L., Hafner M., Nusbaum J.D., Shamulailatpam P., Love C.L., Dave S.S., Tuschl T. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe. 2011;10:515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S., Jin L., Zhang Y., Huang Y., Zhang F., Valdmanis P.N., Kay M.A. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Humphreys D.T., Hynes C.J., Patel H.R., Wei G.H., Cannon L., Fatkin D., Suter C.M., Clancy J.L., Preiss T. Complexity of murine cardiomyocyte miRNA biogenesis, sequence variant expression and function. PLoS One. 2012;7:e30933. doi: 10.1371/journal.pone.0030933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Yeo J., Lee J.H., Cho J., Seo D., Kim J.S., Kim V.N. Deletion of human tarbp2 reveals cellular microRNA targets and cell-cycle function of TRBP. Cell Rep. 2014;9:1061–1074. doi: 10.1016/j.celrep.2014.09.039. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee H.Y., Doudna J.A. TRBP alters human precursor microRNA processing in vitro. RNA. 2012;18:2012–2019. doi: 10.1261/rna.035501.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.Y., Zhou K., Smith A.M., Noland C.L., Doudna J.A. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013;41:6568–6576. doi: 10.1093/nar/gkt361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Llorens F., Banez-Coronel M., Pantano L., del Rio J.A., Ferrer I., Estivill X., Marti E. A highly expressed miR-101 isomiR is a functional silencing small RNA. BMC Genomics. 2013;14:104. doi: 10.1186/1471-2164-14-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Wu Y., Choi J.G., Wu H. Lower and upper stem-single-stranded RNA junctions together determine the Drosha cleavage site. Proc. Natl. Acad. Sci. U S A. 2013;110:20687–20692. doi: 10.1073/pnas.1311639110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Manzano M., Forte E., Raja A.N., Schipma M.J., Gottwein E. Divergent target recognition by coexpressed 5'-isomiRs of miR-142-3p and selective viral mimicry. RNA. 2015;21:1606–1620. doi: 10.1261/rna.048876.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R.D., O'Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Stark A., Johnston W.K., Kellis M., Bartel D.P., Lai E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H., Ghildiyal M., Zamore P.D. Argonaute loading improves the 5' precision of both MicroRNAs and their miRNA strands in flies. Curr. Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starega-Roslan J., Galka-Marciniak P., Krzyzosiak W.J. Nucleotide sequence of miRNA precursor contributes to cleavage site selection by Dicer. Nucleic Acids Res. 2015;43:10939–10951. doi: 10.1093/nar/gkv968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starega-Roslan J., Krol J., Koscianska E., Kozlowski P., Szlachcic W.J., Sobczak K., Krzyzosiak W.J. Structural basis of microRNA length variety. Nucleic Acids Res. 2011;39:257–268. doi: 10.1093/nar/gkq727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G.C., Chan E., Molnar A., Sarkar R., Alexieva D., Isa I.M., Robinson S., Zhang S., Ellis P., Langford C.F. 5' isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 2014;42:9424–9435. doi: 10.1093/nar/gku656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.C., Tambe A., Kidwell M.A., Noland C.L., Schneider C.P., Doudna J.A. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell. 2015;57:397–407. doi: 10.1016/j.molcel.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Neilson J.R., Kumar P., Manocha M., Shankar P., Sharp P.A., Manjunath N. miRNA Profiling of Naive, Effector and Memory CD8 T Cells. PLoS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Ye C., Ramirez D., Manjunath N. Alternative processing of primary microRNA transcripts by Drosha generates 5' end variation of mature microRNA. PLoS ONE. 2009;4:e7566. doi: 10.1371/journal.pone.0007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010;38:7689–7697. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.