Abstract
In several preclinical tumor models, antitumor effects occur after intratumoral electroporation, also known as electrotransfer, of plasmid DNA devoid of a therapeutic gene. In mouse melanomas, these effects are preceded by significant elevation of several proinflammatory cytokines. These observations implicate the binding and activation of intracellular DNA-specific pattern recognition receptors or DNA sensors in response to DNA electrotransfer. In tumors, IFNβ mRNA and protein levels significantly increased. The mRNAs of several DNA sensors were detected, and DAI, DDX60, and p204 tended to be upregulated. These effects were accompanied with reduced tumor growth and increased tumor necrosis. In B16.F10 cells in culture, IFNβ mRNA and protein levels were significantly upregulated. The mRNAs for several DNA sensors were present in these cells; DNA-dependent activator of interferon regulatory factor (DAI), DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 (DDX60), and p204 were significantly upregulated while DDX60 protein levels were coordinately upregulated. Upregulation of DNA sensors in tumors could be masked by the lower transfection efficiency compared to in vitro or to dilution by other tumor cell types. Mirroring the observation of tumor necrosis, cells underwent a significant DNA concentration-dependent decrease in proliferation and survival. Taken together, these results indicate that DNA electrotransfer may cause the upregulation of several intracellular DNA sensors in B16.F10 cells, inducing effects in vitro and potentially in vivo.
Keywords: DNA sensor, electroporation, melanoma cells, melanoma tumors, plasmid DNA, pattern recognition receptor
Introduction
In vivo electroporation or electrotransfer, the application of controlled electric pulses, enhances delivery of plasmid DNA (pDNA) to a wide variety of healthy tissues as well as many tumor types.1,2,3,4 Electrotransfer of pDNA encoding therapeutic genes substantially increases gene expression, enhancing subsequent therapeutic effects. This gene delivery technique has reached clinical trials for cancer therapies, cancer vaccines, and infectious disease vaccines.5
In studies of cancer therapies in preclinical models, several groups have observed inhibition of tumor growth, increased survival time, and complete tumor regression after intratumor electrotransfer of oligonucleotides, plasmids devoid of encoded therapeutic genes, or plasmids encoding reporter genes. Antitumor effects have been described in melanomas,6,7,8,9,10,11,12 lung carcinomas,13,14 fibrosarcomas,15 pancreatic carcinomas,16 mammary tumors,17 and colorectal carcinomas.18,19,20,21 After electrotransfer of pDNA devoid of a therapeutic gene, increased expression of several proinflammatory cytokine and chemokine proteins, particularly CCL3, CCL4, IL-1β, and IL-6, was observed in B16.F10 melanoma tumors and preceded tumor regression.10 Subsequent tumor-localized inflammation might contribute to the observed tumor regression.7,11
During the process of electrotransfer, pDNA theoretically enters the cell via endocytosis.22,23 This theory was supported in vivo by the observation that the inhibition of endocytosis also inhibits gene expression in skeletal muscle.24 The observations that DNA enters cell via endocytosis during electrotransfer and that proinflammatory molecule expression was upregulated implicated the activation of the endosomal CpG motif DNA binding receptor toll-like receptor 9 (TLR9).25 However, regression was induced by electrotransfer of calf thymus DNA or non-CpG containing control oligonucleotides,11 which are not classic TLR9 ligands.
Electrotransfer also delivers pDNA to the cytosol, which is probably a dead-end pathway with respect to transgene expression.26,27 The presence and activity of several DNA-specific cytosolic pattern recognition receptors, also known as DNA sensors, has been demonstrated in a variety of cell types, including fibroblasts, tumor cells, and immune cells.28,29,30,31 pDNA electrotransfer may enhance the availability of pDNA to cytosolic DNA sensor binding, inducing the production of proinflammatory cytokine and chemokines, particularly type I interferons.28,29,31 Therefore, all cell types residing in the tumor could potentially respond to pDNA electrotransfer. However, the tumor cells themselves are universally present. The purpose of this study was to investigate whether B16.F10 mouse melanoma tumors and cells express cytosolic DNA sensors and whether these sensors respond to pDNA electrotransfer.
Results
Tumor growth delay and complete tumor regression induced by pDNA electrotransfer of empty vector plasmid is preceded by increased expression of interferon-β
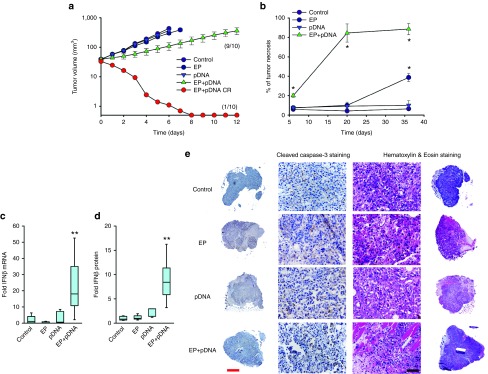
A single intratumor pDNA delivery by electrotransfer produced a significant growth delay in treated tumors (Figure 1a). In this experimental group, doubling time was decreased 3.2-fold; tripling time was decreased 2.8-fold. In addition, pDNA electrotransfer induced complete tumor regression in 1 out of 10 mice (Figure 1a). Hematoxylin & eosin (H&E) staining of tumor sections 6, 20, and 36 hours after pDNA electrotransfer demonstrated a statistically significant increased proportion of necrosis after pDNA electrotransfer at all three time points compared to pDNA injection alone, electrotransfer alone, and in unmanipulated control tumors. The control tumors had approximately 4–6% necrosis, while the proportion of necrosis in the experimental groups increased with time (Figure 1b,e) and reached 84% 20 hours after pDNA electrotransfer. The presence of inflammatory immune cells was observed at the tumor borders in the pDNA electrotransfer group (Supplementary Figure S1). Due to the early onset of necrotic cell death, the proportion of apoptotic cells as indicated by cleaved caspase 3 was evaluated at 6 hours after pDNA electrotransfer. No statistically significant difference in proportion of apoptotic cells between the groups was observed. At 6 hours post-treatment, necrosis was evenly distributed throughout the tumor tissue and no clear sharp boundary was observed between necrotic and apoptotic areas and viable tissue (Figure 1e). These results indicate that necrosis is more likely to occur after pDNA electrotransfer than apoptosis.
Figure 1.
Effect of pDNA electrotransfer on tumor growth, necrosis, and expression of IFNβ. Commercially prepared vector plasmid (gWiz Blank) was electrotransferred into palpable B16.F10 melanoma tumors in the flanks of C57Bl/6 mice. Control, no tumor manipulation; EP, 50 μl saline injection followed by the delivery of eight 5 ms pulses with a voltage-to-distance ratio of 800 V/cm with a caliper electrode; pDNA, injection of 50 μl 2 mg/ml gWiz Blank plasmid; EP+pDNA, injection of 50 μl 2 mg/ml gWiz Blank plasmid followed by the delivery of eight 5 ms pulses with a voltage-to-distance ratio of 800 V/cm with a caliper electrode. (a) Tumor growth was monitored as described. A complete response (CR, EP+pDNA CR) was observed in 1 out of 10 mice. Results represent two independent experiments, n = 5 for each experimental group in each experiment, and are expressed as mean ± SEM. (b) Necrosis was significantly increased in EP+pDNA group at all three time points compared to all other experimental groups. *P < 0.05 statistically significantly increased necrosis. (c) IFNβ mRNA levels as measured by real-time RT-PCR, n = 7–8. (d) IFNβ protein levels as measured by ELISA, n = 5–7. (e) Comparison of cleaved caspase-3 and hematoxylin & eosin staining of representative tumors sections 6 hours after pDNA electrotransfer. Results indicating that necrosis is more likely to occur after pDNA electrotransfer than apoptosis. Representative images, red scale bar = 1 mm and black scale bar = 50 µm.
Interferon-β (IFNβ) is a marker of cytosolic DNA sensor activation.32,33,34,35,36,37,38,39,40 The effect of pDNA electrotransfer on intratumoral IFNβ mRNA and protein levels in tumors was determined. Real-time reverse transcriptase polymerase chain reaction (RT-PCR) demonstrated that while no change in IFNβ mRNA levels was detected in control tumors, tumors injected with pDNA, or tumors subjected to electrotransfer alone, the combination of pDNA and electrotransfer produced an increase of approximately 13-fold (P < 0.05) (Figure 1c). IFNβ protein levels correlated with the mRNA levels (Figure 1d); an 11-fold increase was observed only in the tumors subjected to the combination of pDNA injection and electrotransfer (P < 0.01).
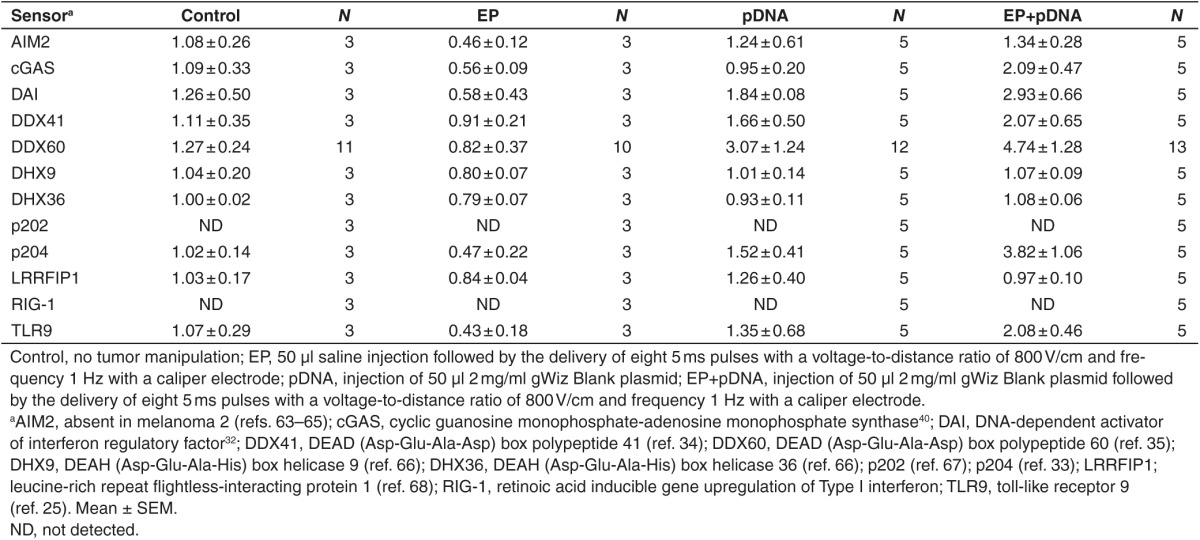
Tumor mRNA levels for several DNA sensors were determined by real-time RT-PCR 4 hours after the procedure (Table 1). While the all mRNAs tested except RIG-1 (retinoic acid inducible gene upregulation of type I interferon) and p202 were detected in the tumors, none were significantly upregulated in any experimental group. Either pDNA injection or pDNA electrotransfer tended to increase DDX60 (DEAD (Asp-Glu-Ala-Asp) box polypeptide 60) levels, while pDNA electrotransfer alone tended to increase DAI (DNA-dependent activator of interferon regulator factors) and p204 mRNA levels. Notably, the mRNA levels for the endosomal DNA sensor TLR9 did not change.
Table 1. Fold changes in mRNA levels of DNA sensors in B16.F10 tumors four hours after pDNA electrotransfer.
pDNA electrotransfer directly effects B16.F10 melanoma cells
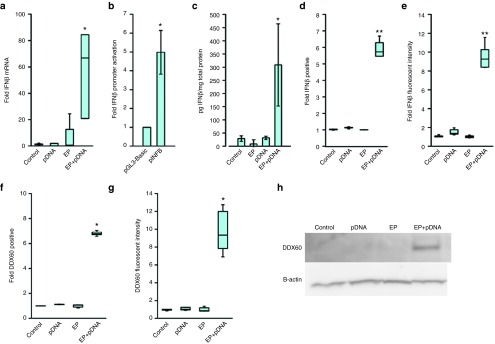
In vitro experiments were performed to determine if IFNβ was produced by B16.F10 cells subjected to pDNA electrotransfer. An increase in IFNβ mRNA of nearly 60-fold (P < 0.01) was detected in cells subjected to pDNA electrotransfer when compared to pDNA exposure alone, electrotransfer alone, and unmanipulated control cells (Figure 2a). This observation was supported by a fivefold increase in IFNβ promoter activation as measured by a reporter assay (Figure 2b, P < 0.05), indicating that transcriptional upregulation may be partially responsible. A significant increase in IFNβ protein levels was detected in B16.F10 cells by enzyme-linked immunosorbent assay (ELISA) (Figure 2c, P < 0.01). Flow cytometry demonstrated that the number of cells expressing IFNβ protein increased nearly sixfold (Figure 2d, P < 0.01) with an increase in median fluorescent intensity of nearly 10-fold (Figure 2e, P < 0.01) after pDNA electrotransfer.
Figure 2.
Levels of IFNβ and DDX-60 mRNA and protein after pDNA electrotransfer in B16.F10 mouse melanoma cells. (a) IFNβ mRNA levels as measured by real-time RT-PCR, n = 5–9. (b) IFNβ promotor activity as measured by luciferase expression, n = 4. (c) IFNβ protein levels as measured by ELISA, n = 3. (d) Fold cells positive for IFNβ as measured by flow cytometry normalized to the control, n = 6–8. (e) Median IFNβ fluorescent intensity as measured by flow cytometry normalized to the control, n = 6–8. (f) Fold cells positive for DDX60 as measured by flow cytometry normalized to the control, n = 7. (g) Median DDX60 fluorescent intensity as measured by flow cytometry normalized to the control, n = 7. (h) Western blot of cell lysate demonstrating an increased expression of DDX60 9 hours after pDNA electrotransfer. Control, 40 µl of cells and 10 µl of saline; EP, 40 µl of cells and 10 µl of saline were electroporated by the delivery of eight 5 ms pulses with a voltage-to-distance ratio of 600 V/cm with a plate electrode; pDNA, 40 µl of cells and 10 µl of 2 mg/ml gWiz Blank plasmid; EP+pDNA, 40 µl of cells and 10 µl of 2 mg/ml gWiz Blank plasmid were lectroporated by the delivery of eight 5 ms pulses with a voltage-to-distance ratio of 600 V/cm with a plate electrode.
DNA sensor mRNA levels in B16.F10 cells were determined (Table 2). The mRNAs for TLR9 and AIM2 (absent in melanoma 2) were not detected. TLR9 is expressed in immune cells, while AIM2 is not normally expressed in melanoma cells. As with B16.F10 tumors, RIG-1 and p202 mRNAs were not detected. Interestingly, mRNAs for cGAS (cyclic guanosine monophosphate-adenosine monophosphate synthase), DDX41 (DEAD (Asp-Glu-Ala-Asp) box polypeptide 41), and LRRFIP1 (leucine-rich repeat flightless-interacting protein 1) as well as the CpG motif-specific sensors DHX9 (DexD/H-box helicase family 9) and DHX36 (DexD/H-box helicase family 36) were detected, although they were not upregulated in any group. The mRNAs of three cytosolic DNA sensors significantly increased after pDNA electrotransfer in B16.F10 cells. DAI mRNA was upregulated by approximately 355-fold, DDX60 mRNA was upregulated by more than 6-fold, and p204 mRNA was upregulated by approximately 18-fold. Upregulation of cytosolic DNA sensors after pDNA electrotransfer was confirmed using a different electroporation protocol (EP1). After EP1, similar mRNAs were upregulated as shown in Table 2. Although these pulse protocols demonstrated similar transfection efficiencies (Figure 3b), different levels of upregulation of DAI (P < 0.001) and p204 (P < 0.01) mRNAs were detected. Therefore, transfection efficiency may not relate to DNA sensor upregulation. Furthermore, the upregulation of cytosolic DNA sensors was confirmed also with an alternate plasmid, pEGFP-N1, which encodes enhanced GFP (pEGFP-N1) in another backbone. Electrotransfer of pEGFP-N1 resulted in DDX60 mRNA upregulation by 3.98 ± 1.15, DAI mRNA by 7.88 ± 0.15, and p204 mRNA by 4.46 ± 3.18-fold.
Table 2. Fold changes in mRNA levels of DNA sensors in B16.F10 tumor cells 4 hours after pDNA electrotransfer.
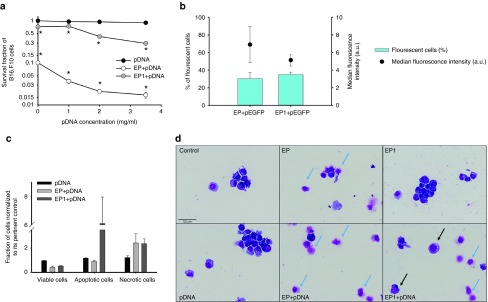
Figure 3.
Cell survival, transfection, cell death mechanism and morphology of B16.F10 cells after pDNA electrotransfer. Commercially prepared vector plasmid (gWiz Blank) was electrotransferred into B16.F10 melanoma cells. Control, 40 µl of cells and 10 µl of saline; pDNA 1 mg/ml, 40 µl of cells and 10 µl of 1 mg/ml gWiz Blank plasmid; pDNA 2 mg/ml, 40 µl of cells and 10 µl of 2 mg/ml gWiz Blank plasmid; pDNA 3.5 mg/ml, 40 µl of cells and 10 µl of 3.5 mg/ml gWiz Blank plasmid; EP, 40 µl of cells and 10 µl of saline were electroporated by the delivery of eight 5 ms pulses with a voltage-to-distance ratio of 600 V/cm with a plate electrode; EP+pDNA 1 mg/ml, 40 µl of cells and 10 µl of 1 mg/ml gWiz Blank plasmid were electroporated as described in EP group; EP+pDNA 2 mg/ml, 40 µl of cells and 10 µl of 2 mg/ml gWiz Blank plasmid were electroporated as described in EP group; EP+pDNA 3.5 mg/ml, 40 µl of cells and 10 µl of 3.5 mg/ml gWiz Blank plasmid were electroporated as described in EP group; EP1, 40 µl of cells and 10 µl of saline were electroporated by the delivery of six 100 µs pulses with a voltage-to-distance ratio of 1300 V/cm and frequency 4 Hz with a plate electrode; EP1+pDNA 1 mg/ml, 40 µl of cells and 10 µl of 1 mg/ml gWiz Blank plasmid were electroporated as described in EP1 group; EP1+pDNA 2 mg/ml, 40 µl of cells and 10 µl of 2 mg/ml gWiz Blank plasmid were electroporated as described in EP1 group; EP1+ pDNA 3.5 mg/ml, 40 µl of cells and 10 µl of 3.5 mg/ml gWiz Blank plasmid were electroporated as described in EP1 group. For image B, 40 µl of cells and 10 µl of 2 mg/ml pEGFP-N1 plasmid were electroporated as previously described. (a) Cell survival was measured 72 hours after electrotransfer and was normalized to the control group. (b) Transfection efficiency of B16.F10 cells after pEGFP-N1 electrotransfer after pDNA delivery with EP and EP1. *P < 0.05, statistically significant difference compared to all control groups and EP only. a.u., arbitrary units. (c) Cell death mechanism as quantified by flow cytometery and (d) cell morphology after pDNA electrotransfer. Blue arrows indicate necrotic cells displaying fragments of cytoplasm and only an outline of the nucleus. Black arrow indicates apoptotic cells with vacuolization of the cytoplasm and formation of apoptotic bodies.
An increase in DDX60 protein detected by two methods confirmed the mRNA results. A sevenfold increase in cells expressing DDX60 (Figure 2f, P < 0.05) and a 10-fold increase in the median fluorescent intensity in these cells was detected by flow cytometric after pDNA electrotransfer (Figure 2g, P < 0.05). On western blots, the DDX60 protein was detected in cell lysate only after pDNA electrotransfer as predicted by the 198 kDa band. No DDX60 protein was detected in any other experimental group (Figure 2h).
We next determined if pDNA electrotransfer using different protocols had a direct effect on melanoma cell survival. The exposure of cells to pDNA had no effect on cell survival, while the application of electric pulses alone decreased viability by 80% after EP and 30% after EP1 72 hours post-treatment. A significant pDNA concentration-dependent decrease in cell survival was observed after pDNA electrotransfer using both protocols (Figure 3a, P < 0.01); however, the decrease of survival after EP1 was smaller than after EP. The transfection efficiency with these electroporation protocols was identical (Figure 3b).
The mechanisms of cell death were investigated in vitro to confirm our previous findings in vivo. Using a flow cytometric assay, relative changes in annexin V and 7AAD detection are characteristic of apoptotic or necrotic cell death. Electrotransfer of pDNA with EP pulses caused necrotic cell death as indicated by a higher percentage of annexin V and 7AAD-positive cells (Supplementary Figure S2), while apoptosis after pDNA electrotransfer with these pulses was not increased as indicated by no change in percentage of cells positive for annexin V (Supplementary Figure S2). In contrast, electrotransfer of pDNA with EP1 pulses caused predominantly apoptosis, while necrosis was also increased (Figure 3c). These findings indicate that not only plasmid DNA but also pulse parameters are responsible for triggering cell death mechanisms. Moreover, morphological changes confirmed the flow cytometry data; necrosis as well as apoptosis were clearly seen after pDNA electrotransfer (Figure 3d).
Theoretically, the inhibition of endocytosis could modulate DNA sensor upregulation. As expected, exposure to the general inhibitor of endocytosis MβCD reduced transgene expression from approximately 30% of cells to 3% with reduced fluorescent intensity. MβCD pretreatment increased the levels of DDX60 and IFNβ mRNAs approximately twofold after pDNA electrotransfer (P < 0.05, P < 0.01 respectively), whereas DAI mRNA levels strongly decreased (Tables 2 and 3) (P < 0.05). TLR9 mRNA remained undetectable.
Table 3. Effect of the endocytosis inhibitor MβCD on fold changes in mRNA levels of IFNβ and DNA sensors in B16.F10 tumor cells 4 hours after DNA electrotransfer.
Discussion
The results of our study demonstrate that B16.F10 melanoma tumors secrete IFNβ after electrotransfer of pDNA (plasmid devoid of therapeutic genes) and that necrosis is produced. B16.F10 cells in culture express cytosolic DNA sensors and the expression of DAI, DDX60 and p204 mRNAs, DDX60 protein, and IFNβ m RNA and protein are increased after pDNA electrotransfer. These changes in mRNA and protein levels are accompanied by DNA concentration-dependent cell death. These effects may be due to the activation of signaling pathways mediated by the upregulated cytosolic DNA sensors.
In this study, a reproducible negative effect on tumor growth was observed using vector pDNA electrotransfer, confirming previous studies with other pulse protocols.6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21 However, while some tumor regression was observed, in this case the effect manifested itself primarily as a tumor growth delay (Figure 1), which is consistent with the histological observation that 16% of the tumor cells in vivo and 20% in vitro may retain viability. Although the identical plasmid (gWiz Blank) produced a higher level of complete regressions in a previous study,11 the decreased antitumor effect observed in this study may be due to the differences between the electroporation protocols employed.
Some unknown characteristic of the DNA composition is an important variable in this antitumor effect. Using a similar pulse protocol (ten 5 ms pulses at a voltage-to-distance ratio of 800 V/cm), three deliveries of pUC18 in 1 week produced complete, long-term regression in 70% of mice,7 while three deliveries of gWiz Blank produced complete, long-term regression in only 25% of mice.11 Delivery of calf thymus DNA with the same pulse protocol induces regression in 15% of tumors.11 Clearly, the composition of the DNA itself is an important variable.
This antitumor effect was independent of caspase-3 and tumor necrosis was observed in the treated tumors (Figure 1b,c). A direct cytotoxic effect against melanoma cells subjected to pDNA electrotransfer in culture was also demonstrated (Figure 3a). Here, the mechanism of cell death was predominantly necrosis as observed in tumors in vivo. Interestingly, the apoptotic cell death of melanoma cells in culture was increased after EP1, which was also described previously in tumors in vivo.41 The level of cell death did not correlate with the transfection efficiency. Although these two electroporation protocols produced similar levels of transgene expression (Figure 3b), subsequent cell death varied greatly (Figure 3a). Therefore, it seems that besides the composition of the DNA itself, the electrical parameters contribute to the mechanism of cell death following the pDNA electrotransfer.
After pDNA electrotransfer, increased mRNA levels of several cytosolic DNA sensors associated with cell death were observed. For example, DAI is essential for programmed necrosis after murine cytomegalovirus infection.42 IFI16, the human ortholog of p204, is associated with p53-mediated apoptosis43 and caspase-1-dependent pyroptosis after CD4 cells are exposed to HIV-1 transcripts.44 Activation of DNA sensors associated with electrotransfer may cause cell death.
In this study, we found that the mRNA of the proinflammatory molecule IFNβ was significantly upregulated in B16.F10 tumors subjected to pDNA electrotransfer (Figure 1c). Many putative intracellular DNA sensors inducing IFNβ expression have been described, and one of these, an unknown sensor, or an unrelated mechanism might be responsible for the observed inflammatory protein production.28,29,30,31 While the mRNAs of several cytosolic sensors and the endosomal sensor TLR9 remained unchanged, the mRNAs of the cytosolic DNA sensors DDX60, p204, and DAI tended to be upregulated in tumors after pDNA electrotransfer (Table 2). The low transfection efficiency may have masked more pronounced responses, although we demonstrate in vitro that pulse protocols with similar transfection efficiencies (Figure 3b) can correlate to different levels of DNA sensor mRNA upregulation (Table 2). Using similar electrotransfer parameters, more than 50% of B16F1 cells were transfected in vitro, while only 3% of tumor area was transfected in vivo.45 Another possibility is that this upregulation is masked by the inherent cellular heterogeneity of tumors. All nucleated cells can respond to type I interferons46,47 and both IFNβ mRNA and protein were significantly upregulated. Immune cells, fibroblasts, and endothelial cells residing in the tumor could exhibit differential DNA sensor upregulation, masking the DNA sensors activated in the tumor cells themselves.
In B16.F10 cells in vitro, IFNβ transcription, mRNA levels, and protein levels were significantly elevated after pDNA electrotransfer but not after exposure to pDNA or electrotransfer individually (Figure 3). The mRNAs for DDX60, p204, and DAI were also significantly upregulated (Table 2). Other nonviral transfections also activate these sensors; several groups used lipid transfection reagent in the original descriptions of these DNA sensors32,33,39,40,48 and these sensors are activated in other cell types by pDNA electrotransfer, such as C2C12 mouse myoblasts (data not shown). Each of these DNA sensors may induce the production of IFNβ when activated.32,33,35 DDX60 and DAI are exclusively cytosolic; IFI16, the human ortholog of p204, is a nuclear sensor that moves to the cytosol after activation.49 The interesting observation that p204 mRNA is upregulated after pDNA electrotransfer may reflect the quantity of pDNA that reaches the nucleus. Since pDNA delivery to the nucleus is theoretically driven by endocytosis,22,23 this sensor may be activated by any successful electrotransfer protocol to any cell type that expresses it. A significant increase in DDX60 protein confirmed the mRNA upregulation (Figure 2).
Theoretically, the inhibition of endocytosis during electrotransfer could reduce gene expression by preventing entry of pDNA into the cell or alternatively by driving pDNA delivery to the cytosol. MβCD pretreatment doubled the upregulation of IFNβ and DDX60 mRNA levels; however, DAI mRNA levels significantly decreased (P < 0.05. Tables 2 and 3). These discordant observations may support the possibility of differential distribution of DDX60 and DAI within the cytosol; after electrotransfer accompanied by reduced endocytosis, pDNA may be available to DDX60 but unavailable to DAI.
The cytosolic DNA sensors upregulated in our study are activated by a variety of nucleic acids and possess overlapping downstream signaling pathways.28,29,30,31 DDX60 may bind both RNA and DNA35 and play a cell type- and/or ligand-specific role.50,51 When stimulated by mammalian, bacterial, viral, and synthetic dsDNA, DAI mediates the induction of proinflammatory molecules in a DNA dose-dependent manner.32 Similar to this study, DAI protein was upregulated after challenge with herpes simplex virus type 1 in glial cells52 and transcription was increased in hepatocellular carcinomas.53 After DNA electrotransfer to muscle, mRNA levels increased.54 In vivo pDNA electrotransfer of a plasmid encoding DAI acted as an adjuvant to a cancer vaccine delivery in mice.55
Even though human IFI16 and mouse p204 share only 37% amino acid identity, both induce the production of IFNβ, but not IL-1β, in several cell types in response to DNA.33 Interestingly, a fourfold increase in tumor IL-1β mRNA levels after pDNA electrotransfer (P < 0.05, data not shown) was not reflected in the B16.F10 cells in culture. This indicates that other cell types found in tumors, possibly immune cells, are also responding in vivo to pDNA electrotransfer. AIM2 and TLR9 mRNAs are detected in melanoma tumors but not melanoma cells. This is not unexpected, since these proteins are probably expressed in other cell types present in tumors.
Combined, these results indicate that the B16.F10 melanoma cells themselves produce IFNβ after pDNA electrotransfer, associated with the upregulation of multiple cytosolic DNA sensors. Programmed necrosis may contribute to the cell death observed in vitro, while both inflammation and programmed necrosis may contribute to the in vivo antitumor effects in our study. DNA delivery by electroporation is a common in vitro laboratory technique, and activation of DNA sensors may be important to consider in specific research areas. Since research utilizing in vivo pDNA electrotransfer is often directed toward clinical applications, it is important and significant to completely understand the underlying mechanisms involved.
The induction of inflammation may contribute to cancer-targeted immune therapies. In a previous study of the antitumor effects of vector electrotransfer, 70% of B16.F10 tumors completely regressed, and 70% of these tumor-free mice were resistant to challenge with B16.F10 cells,7 indicating the generation of an adaptive antitumor response. In a study of intratumor electrotransfer of a plasmid encoding IL-12 in the same tumor model, complete, long-term regression was produced in 89% of tumors.56 In this study, 100% of these mice were resistant to challenge with the same tumor cell line. A direct comparison is not possible since different plasmid backbones and differing tumor-directed pulse parameters were used in these studies; however, DNA sensor activation may contribute to the effectiveness of IL-12 as an anticancer therapy. Indeed, type I interferon production is required for the antitumor efficacy of a Semliki Forest virus encoding IL-12 (ref. 57).
The induction of inflammation may also aid in the induction of immune responses to vaccines58 and can be beneficial in cancer gene therapies based on stimulation of immune responses. However, therapies requiring simple transgene expression may be inhibited when an immune response is not desirable. To our knowledge, no pDNA electrotransfer gene therapy has been inhibited by the innate immune response. However, this inhibition is a potential concern for certain gene therapies.59 DNA sensor upregulation and subsequent activation of downstream signaling pathways should be fully understood in order to harness or control it for therapeutic applications.
Materials and methods
Plasmids. gWiz Blank, an empty vector, and gWiz Luc, which encodes the firefly luciferase gene driven by the CMV promoter, were commercially prepared (Aldevron, Fargo, ND). The promoterless pGL3-Basic (Promega, Madison, WI) and pGL3-IFNβ-prom, which encodes firefly luciferase driven by the mouse IFNβ promoter, were kind gifts of Judith A. Smith.60 These plasmids and pEGFP-N1 (BD Biosciences Clontech, Palo Alto, CA), which encodes the enhanced green fluorescent protein gene driven by the CMV promoter, were prepared using Qiagen Maxi-Endo-Free Kits according to the manufacturer's instructions. All plasmids were suspended at 2 µg/µl in physiological saline unless otherwise noted.
Cells and tumors. All procedures were approved by the Veterinary Administration of The Ministry of Agriculture and the Environment of the Republic of Slovenia (#34401–12/2009/6). B16F.10 mouse melanoma cells (ATCC CRL-6475, American Type Culture Collection, Manassas, VA) in the exponential growth phase were used in experiments in vitro and in vivo. For tumor induction, 1 × 106 B16.F10 melanoma cells in 50 µl phosphate-buffered saline (PBS) were injected subcutaneously in the left flank of female 7–8-week-old C57Bl/6 mice (Envigo, Udine, Italy). Tumors were allowed to grow approximately 8 days to a mean tumor diameter of 4 mm before experiments were performed.
In vivo DNA electrotransfer. Mice were anesthetized using a mixture of 2.5% isoflurane and 97.5% O2. Tumors were injected with 50 µl of pDNA solution and eight square electric pulses with a voltage-to-distance ratio of 800 V/cm, a pulse duration of 5 ms, and a frequency 1 Hz were delivered through two parallel stainless steel electrodes using an Electro Cell B10 electric pulse generator (LEROY biotech, L'Union, France). Tumors were measured twice to three times weekly using a digital caliper. Tumor volume was calculated by the formula v = ab2π/6, where a is the longest diameter, and b is the next longest diameter perpendicular to a. Mice were humanely euthanized when the tumor volume reached 1,000 mm3 or when the animal's behavior indicated discomfort. Animals with tumors in regression were followed up to 100 days and were considered to be in complete regression if no tumor regrowth was observed during this time. For postmortem tumor analysis, mice were humanely sacrificed, tumors removed, and snap frozen on dry ice.
Real-time reverse transcription PCR. RNA was extracted from tumors or cells 4 hours after electrotransfer using Trizol Reagent (Invitrogen, Carlsbad, CA) then purified using RNeasy columns (Qiagen, Valencia, CA). After extraction, 250 ng of total RNA was transcribed into cDNA using the SuperScript VILO cDNA Synthesis Kit (Invitrogen, Thermo Fisher Scientific,Waltham, MA). according to manufacturer's instructions and diluted 10-fold. Messenger RNA was quantified on a CFX96 Real Time PCR Detection System (Bio-Rad, Hercules, CA) or a Primus 25 advanced thermal cycler (Peqlab, Erlangen, Germany) using custom primers (Integrated DNA Technologies, Coralville, IA) in SYBR Green Master Mix (Applied Biosystems, Thermo Fisher Scientific Waltham, MA). See Supplementary Table S1 for primer sequences. Spleen RNA acted as a positive control. Relative quantification was performed by comparison to the housekeeping genes β-actin and glyceraldehyde 3-phosphate dehydrogenase using the ΔΔCt method.61
Protein quantification by ELISA. Tumors were homogenized in PBS containing protease inhibitors and homogenates were centrifuged to remove cell debris. bicinchoninic acid (BCA) assays (Pierce Biotechnology, Rockford, IL) were performed and samples normalized to 1 mg/ml total protein. ELISAs were performed on the normalized homogenates per manufacturer's instructions (PBL Assay Science, Piscataway, NJ).
Immunohistochemistry. Histological analysis was performed at three different time points, 6, 20, and 36 hours. Three mice from each experimental group were sacrificed in two independent experiments. The tumors were excised at selected time point, fixed in immunohistochemistry (IHC) zinc fixative (BD Pharmingen, BD Biosciences, San Jose, CA) overnight, and embedded in paraffin. Consecutive 2-μm thick tumor sections were cut from each paraffin block. The first section of each tumor sample was stained with hematoxylin & eosin (H&E) and analyzed. Based on H&E staining, the appropriate time point for analysis of apoptosis was determined and following tumor sections were stained with rabbit monoclonal antibodies against cleaved caspase-3 (Ca-3, Cell signaling Technology, Danvers, MA, dilution 1:1,500). A peroxidase-conjugated streptavidin–biotin system (Rabbit-specific HRP/DAB detection IHC kit, ab64261, Abcam, Cambridge, England, UK) was used as a colorogenic reagent followed by hematoxylin counterstaining. The images of stained tumor sections were captured with a DP72 CCD camera connected to a BX-51 microscope (Olympus, Hamburg, Germany). Whole tumor sections were captured at ×4 magnification for H&E staining and 5 images at ×60 magnification for Ca-3 staining. The percentage of tumor necrosis and the number of Ca-3-positive cells were determined by two independent observers.
In vitro DNA electrotransfer. Trypsinized B16.F10 mouse melanoma cells were suspended in electroporation buffer and pDNA added as described previously.62 Aliquots were transferred to cuvettes or pipetted between 2 mm gap steel electrodes. Two different pulse types were used, eight square wave electric pulses at an amplitude over distance ratio 600 V/cm, a pulse duration of 5 ms, and a frequency 1 Hz (EP), and six square wave electric pulses at an amplitude over distance ratio 1,300 V/cm, a pulse duration of 100 µs, and a frequency of 4 Hz (EP1) electric pulses were generated by an in-house built electroporator (GT-01, Faculty of Electrical Engineering, University of Ljubljana, Slovenia) or with a T820 Electrosquare porator (BTX Molecular Delivery Systems, Holliston, MA). Cells were immediately diluted in medium.
IFNβ transcription assay. Cells were transfected by DNA electrotransfer with pGL3-Basic, pGL3-IFNβ-prom, or gWiz Luc. After transfection, 2.5 × 105 cells in 100 µl medium per well were incubated 16 hours in white opaque 96-well plates. The medium was replaced with fresh medium containing 250 µg/ml luciferin and luciferase expression was immediately quantified (Lumistar Omega, BMG Labtech, Cary, NC). The medium was again replaced with 100 µl medium containing PrestoBlue (Invitrogen, Thermo Fisher Scientific), incubated for 2 hours, and viability quantified as determined by reducing ability (Gemini XPS, Molecular Devices, Sunnyvale, CA). Luciferase expression was normalized to viability.
Protein detection by flow cytometry. Immunofluorescence staining and subsequent flow cytometry analysis was performed for protein level quantification. Cells were collected from 24-well ultra-low attachment plates 6 hours after electrotransfer. Cells were fixed in 4% paraformaldehyde (Alfa Aesar, A Johnson Matthey Company, Ward Hill, MA) for 15 minutes, permeabilized with 0.5% Tween 20 (TWEEN 20, Sigma-Aldrich, Steinheim, DE) for 10 minutes and incubated in 10% donkey serum (Sigma-Aldrich) for 30 minutes. Samples were incubated overnight at 4 °C with primary rabbit anti-mouse polyclonal antibodies (Anti-DDX60, ab139807, Abcam, dilution 1:100). Donkey anti-rabbit secondary antibodies (Cy3 AffiniPure Donkey Anti-Rabbit IgG (H+L), 711-165-152, Jackson ImmunoResearch Laboratories, West Grove, PA, dilution 1:150) were added and incubated for 1 hour. Cells were washed with PBS between each step. Measurements were performed with FACSCanto II flow cytometer (BD Biosciences, San Jose, CA) with appropriate filters (excitation: 488 nm, emission: 530 nm). A histogram of cells gated to eliminate debris against their fluorescence intensity was recorded and the number of fluorescent cells and their median fluorescence intensity were determined (software: BD FACSDiva V6.1.2, BD Biosciences, San Jose, CA).
Protein detection by western blot. After performing assays at 4, 6, 9, and 20 hours after pDNA electrotransfer, the highest expression of DDX60 was observed after 9 hours. Cells were collected and washed with ice-cold PBS from 24-well ultra-low attachment plates 9 hours after electrotransfer or trypsinyzed from 6 cm Petri-dish plate 20 hours after electrotransfer. Cell lysis was performed in radioimmuno precipitation assay lysis buffer supplemented with protease and phosphatase inhibitors (Santa Cruz Biotechnology, Santa Cruz, CA) and incubated on ice for 30 minutes with constant mixing. Whole-cell extracts were centrifuged and supernatants were collected. BCA assays (Pierce Biotechnology, Rockford, IL) were used for protein concentrations determination. A total of 25 µg of total proteins from each sample was separated on NuPAGE 3–8% Tris-Acetate Midi-Gels for 35 minutes at 200 V using NuPAGE Tris Acetate Sample Buffer and dry transferred to a polyvinylidene fluoride membrane using iBlot Gel Transfer Stack (Invitrogen, Thermo Fisher Scientific, Waltham). The membranes were blocked with 5% low-fat dry milk (Pomurske mlekarne, Murska Sobota, Slovenia) in Tris-buffered saline (pH 7.4) containing 0.1% Tween-20 for 2 hours at room temperature and then incubated with primary rabbit anti-mouse polyclonal antibodies, as listed in the section Protein detection by flow cytometry and β-actin (ab 75186, Abcam, dilution 1:1,000) as a loading control overnight at 4 °C. The next day washed membranes were incubated with horseradish peroxide-conjugated donkey anti-rabbit secondary antibody for 45 minutes at room temperature. Protein bands were detected with Image Quant LAS 4000 (GE Healthcare, Little Chalfont, UK) after 5 minutes incubation of the membrane in SuperSignal West Pico Chemiluminescent Substrate (Invitrogen, Thermo Fischer Scientific).
Cell survival assay. After electrotransfer of 1, 2, and 3.5 mg/ml gWiz Blank plasmid, 1 × 103 cells were cultured in 100 µl of medium in 96-well plates and incubated for 72 hours. Fresh medium containing Presto Blue (Invitrogen, Carlsbad, CA) was added to the cells and the fluorescence intensity was measured by microplate reader (Infinite 200, Tecan, Männedorf, Switzerland) 30 minutes thereafter. Cell viability was normalized to control group.
Proliferation assay. After electrotransfer, cells were cultured in 6 cm Petri dishes for 16 hours to recover. Viable B16F10 cells (2.5 × 102) were cultured in 100 µl of medium in 96-well plates for the proliferation assay. A Presto Blue assay was performed 2, 48, and 96 hours after viable cell culture according to manufacturer's instructions. Cell proliferation in each experimental group was normalized to day 0 for that group. A second normalization to the untreated control group at day 4 was performed to determine percentage of reduced proliferation.
Determination of cell death mechanism. Cell death mechanisms were determined 20 hours after pDNA electrotransfer using a FITC Annexin V Apoptosis Detection Kit with 7-AAD (7-amino-actinomycin D, BioLegend, San Diego, CA) according to manufacturer's instructions. A FACSCanto II flow cytometer (BD Biosciences, San Jose, CA), with a 488-nm laser (air-cooled, 20 mW solid state) was used for the excitation and both 530 and 650-nm band-pass filter were used for detection of green and red fluorescence.
Cell morphology. For morphological observation of cells, cytospins were prepared. For each sample, a labeled slide, chamber and blotter were prepared and assembled. Six hours after pDNA electrotranfer, 80 µl of 1 × 103 B16.F10 cells were added to a slide chamber and spun at 1,000 rpm for 4 minutes in a cytocentrifuge (Cytospin 2, Thermo Shandon, Runcorn, UK). The slides were air dried then stained with Giemsa's Azure methylene blue solution (Merck, Germany) according to the manufacturer's protocol. Cell images were captured with a DP72 CCD camera connected to a BX-51 microscope.
Endocytosis inhibition. Cells were trypsinized and counted. 4.5 × 106 B16F10 cells were incubated 30 minutes in 3.75 ml of medium containing 7.5 mmol/l methyl-β cyclodextrin (MβCD). Fifteen milliliters of electroporation buffer were added then cells were centrifuged and resuspended in 44 µl of ice-cold electroporation buffer. 11 µl of 2 µg/µl gWiz Blank were added. 50 µl of the mixture was electroporated and total RNA was extracted for further analysis as described above.
Transfection efficiency. One day after pDNA electrotransfer, transfection efficiency was determined by fluorescent microscopy and quantified by flow cytometry. Three different observation fields of bright-field and fluorescent (exposure 400 ms) images of the cells were captured at ×100 objective magnification with Olympus IX-70 (Hamburg, Germany) and appropriate filters (excitation: 460–490 nm, emission: 505 nm). The same samples were later trypsinized and resuspended in 400 µl of phosphate-buffered saline for flow cytometry analysis. Flow cytometry was performed as previously described.
Statistical analysis. For graphical representation and statistical analysis, SigmaPlot Software (Systat Software, Chicago, IL) was used. The data were first tested for normality of distribution with the Shapiro-Wilk test. The differences between the experimental groups were statistically evaluated by one-way analysis of variance followed by a Holm-Sidak test for multiple comparison. A P value of less than 0.05 was considered to be statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. The presence of inflammatory immune cells in melanoma tumor after pDNA electrotransfer. Figure S2. Cell death mechanisms asdetermined by flow cytometry(FITC Annexin V Apoptosis Detection Kit with 7-AAD, BioLegend, San Diego, CA, USA). Table S1. Primers used in this study.
Acknowledgments
The authors acknowledge the financial support from the state budget by the Slovenian Research Agency (programs no. P3-0003, bilateral projects BI-US/13-14-007 and BI-US/15-16-019). Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number R01CA196796. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The research is a result of networking efforts within COST TD1104 Action. We would like to thank Mira Lavric, Andreja Brozic (Institute of Oncology Ljubljana, Ljubljana, Slovenia), Petra Hudler (Institute of Biochemistry, Medical Center for Molecular Biology, Faculty of Medicine, University of Ljubljana, Slovenia) and Andrej Coer (Faculty of Health Sciences, University of Primorska, Slovenia) for all the valuable help they contributed to this paper. Plasmids pGL3-Basic (Promega, Madison, WI, USA) and pGL3-IFNβ-prom were kind gifts of Judith A. Smith, Department of Pediatrics, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA.
The work was performed in Norfolk, Virginia, USA and Ljubljana, Slovenia.
Supplementary Material
References
- Cemazar, M, Golzio, M, Sersa, G, Rols, MP and Teissié, J (2006). Electrically-assisted nucleic acids delivery to tissues in vivo: where do we stand? Curr Pharm Des 12: 3817–3825. [DOI] [PubMed] [Google Scholar]
- Heller, LC and Heller, R (2006). In vivo electroporation for gene therapy. Hum Gene Ther 17: 890–897. [DOI] [PubMed] [Google Scholar]
- Favard, C, Dean, DS and Rols, MP (2007). Electrotransfer as a non viral method of gene delivery. Curr Gene Ther 7: 67–77. [DOI] [PubMed] [Google Scholar]
- Heller, LC and Heller, R. Translation of electroporation mediated DNA delivery to the clinic. In: Markov M, Miklavcic D and Pakhomov A (eds.). Advanced Electroporation Techniques in Biology and Medicine. CRC Press: Boca Raton, FL; 2010. pp. 19-11-13. [Google Scholar]
- Heller, R and Heller, LC (2015). Gene electrotransfer clinical trials. Adv Genet 89: 235–262. [DOI] [PubMed] [Google Scholar]
- Niu, G, Heller, R, Catlett-Falcone, R, Coppola, D, Jaroszeski, M, Dalton, W et al. (1999). Gene therapy with dominant-negative Stat3 suppresses growth of the murine melanoma B16 tumor in vivo. Cancer Res 59: 5059–5063. [PubMed] [Google Scholar]
- Heller, LC and Coppola, D (2002). Electrically mediated delivery of vector plasmid DNA elicits an antitumor effect. Gene Ther 9: 1321–1325. [DOI] [PubMed] [Google Scholar]
- McCray, AN, Ugen, KE, Muthumani, K, Kim, JJ, Weiner, DB and Heller, R (2006). Complete regression of established subcutaneous B16 murine melanoma tumors after delivery of an HIV-1 Vpr-expressing plasmid by in vivo electroporation. Mol Ther 14: 647–655. [DOI] [PubMed] [Google Scholar]
- Ugen, KE, Kutzler, MA, Marrero, B, Westover, J, Coppola, D, Weiner, DB et al. (2006). Regression of subcutaneous B16 melanoma tumors after intratumoral delivery of an IL-15-expressing plasmid followed by in vivo electroporation. Cancer Gene Ther 13: 969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller, LC, Cruz, YL, Ferraro, B, Yang, H and Heller, R (2010). Plasmid injection and application of electric pulses alter endogenous mRNA and protein expression in B16.F10 mouse melanomas. Cancer Gene Ther 17: 864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller, L, Todorovic, V and Cemazar, M (2013). Electrotransfer of single-stranded or double-stranded DNA induces complete regression of palpable B16.F10 mouse melanomas. Cancer Gene Ther 20: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero, B, Shirley, S and Heller, R (2014). Delivery of interleukin-15 to B16 melanoma by electroporation leads to tumor regression and long-term survival. Technol Cancer Res Treat 13: 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack, A, Bovenzi, V, Bigey, P, Ivanov, MA, Ramchandani, S, Bhattacharya, S et al. (2002). Antisense MBD2 gene therapy inhibits tumorigenesis. J Gene Med 4: 381–389. [DOI] [PubMed] [Google Scholar]
- Prud'homme, GJ, Glinka, Y, Khan, AS and Draghia-Akli, R (2006). Electroporation-enhanced nonviral gene transfer for the prevention or treatment of immunological, endocrine and neoplastic diseases. Curr Gene Ther 6: 243–273. [DOI] [PubMed] [Google Scholar]
- Grosel, A, Sersa, G, Kranjc, S and Cemazar, M (2006). Electrogene therapy with p53 of murine sarcomas alone or combined with electrochemotherapy using cisplatin. DNA Cell Biol 25: 674–683. [DOI] [PubMed] [Google Scholar]
- Deharvengt, S, Rejiba, S, Wack, S, Aprahamian, M and Hajri, A (2007). Efficient electrogene therapy for pancreatic adenocarcinoma treatment using the bacterial purine nucleoside phosphorylase suicide gene with fludarabine. Int J Oncol 30: 1397–1406. [PubMed] [Google Scholar]
- Radkevich-Brown, O, Piechocki, MP, Back, JB, Weise, AM, Pilon-Thomas, S and Wei, WZ (2010). Intratumoral DNA electroporation induces anti-tumor immunity and tumor regression. Cancer Immunol Immunother 59: 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, YS, Tsang, YW, Chi, CH, Chang, CC, Chu, RM and Chi, KH (2008). Synergistic anti-tumor effect of combination radio- and immunotherapy by electro-gene therapy plus intra-tumor injection of dendritic cells. Cancer Lett 266: 275–285. [DOI] [PubMed] [Google Scholar]
- Vidic, S, Markelc, B, Sersa, G, Coer, A, Kamensek, U, Tevz, G et al. (2010). MicroRNAs targeting mutant K-ras by electrotransfer inhibit human colorectal adenocarcinoma cell growth in vitro and in vivo. Cancer Gene Ther 17: 409–419. [DOI] [PubMed] [Google Scholar]
- Forde, PF, Sadadcharam, M, Hall, LJ, O' Donovan, TR, de Kruijf, M, Byrne, WL et al. (2014). Enhancement of electroporation facilitated immunogene therapy via T-reg depletion. Cancer Gene Ther 21: 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde, PF, Hall, LJ, de Kruijf, M, Bourke, MG, Doddy, T, Sadadcharam, M et al. (2015). Non-viral immune electrogene therapy induces potent antitumour responses and has a curative effect in murine colon adenocarcinoma and melanoma cancer models. Gene Ther 22: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M and Yuan, F (2011). Membrane binding of plasmid DNA and endocytic pathways are involved in electrotransfection of mammalian cells. PLoS One 6: e20923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosazza, C, Phez, E, Escoffre, JM, Cézanne, L, Zumbusch, A and Rols, MP (2012). Cholesterol implications in plasmid DNA electrotransfer: Evidence for the involvement of endocytotic pathways. Int J Pharm 423: 134–143. [DOI] [PubMed] [Google Scholar]
- Markelc, B, Skvarca, E, Dolinsek, T, Kloboves, VP, Coer, A, Sersa, G et al. (2015). Inhibitor of endocytosis impairs gene electrotransfer to mouse muscle in vivo. Bioelectrochemistry 103: 111–119. [DOI] [PubMed] [Google Scholar]
- Hemmi, H, Takeuchi, O, Kawai, T, Kaisho, T, Sato, S, Sanjo, H et al. (2000). A Toll-like receptor recognizes bacterial DNA. Nature 408: 740–745. [DOI] [PubMed] [Google Scholar]
- Bertling, W, Hunger-Bertling, K and Cline, MJ (1987). Intranuclear uptake and persistence of biologically active DNA after electroporation of mammalian cells. J Biochem Biophys Methods 14: 223–232. [DOI] [PubMed] [Google Scholar]
- Golzio, M, Teissie, J and Rols, MP (2002). Direct visualization at the single-cell level of electrically mediated gene delivery. Proc Natl Acad Sci USA 99: 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz, P and Monack, DM (2013). Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol 13: 551–565. [DOI] [PubMed] [Google Scholar]
- Desmet, CJ and Ishii, KJ (2012). Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat Rev Immunol 12: 479–491. [DOI] [PubMed] [Google Scholar]
- Hornung, V and Latz, E (2010). Intracellular DNA recognition. Nat Rev Immunol 10: 123–130. [DOI] [PubMed] [Google Scholar]
- Keating, SE, Baran, M and Bowie, AG (2011). Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol 32: 574–581. [DOI] [PubMed] [Google Scholar]
- Takaoka, A, Wang, Z, Choi, MK, Yanai, H, Negishi, H, Ban, T et al. (2007). DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448: 501–505. [DOI] [PubMed] [Google Scholar]
- Unterholzner, L, Keating, SE, Baran, M, Horan, KA, Jensen, SB, Sharma, S et al. (2010). IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11: 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z, Yuan, B, Bao, M, Lu, N, Kim, T and Liu, YJ (2011). The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 12: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita, M, Oshiumi, H, Matsumoto, M and Seya, T (2011). DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol 31: 3802–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser, A, Bauernfeind, F, Hartmann, G, Latz, E, Fitzgerald, KA and Hornung, V (2009). RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol 10: 1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, MK, Wang, Z, Ban, T, Yanai, H, Lu, Y, Koshiba, R et al. (2009). A selective contribution of the RIG-I-like receptor pathway to type I interferon responses activated by cytosolic DNA. Proc Natl Acad Sci USA 106: 17870–17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, BJ, Mansur, DS, Peters, NE, Ren, H and Smith, GL (2012). DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife 1: e00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, T, Kobayashi, J, Saitoh, T, Maruyama, K, Ishii, KJ, Barber, GN et al. (2013). DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci USA 110: 2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L, Wu, J, Du, F, Chen, X and Chen, ZJ (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339: 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, LH, Sen, A, Murphy, SP, Jahreis, GP, Fuji, H and Hui, SW (1999). Apoptosis induced by DNA uptake limits transfection efficiency. Exp Cell Res 253: 541–550. [DOI] [PubMed] [Google Scholar]
- Upton, JW, Kaiser, WJ and Mocarski, ES (2012). DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 11: 290–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aglipay, JA, Lee, SW, Okada, S, Fujiuchi, N, Ohtsuka, T, Kwak, JC et al. (2003). A member of the Pyrin family, IFI16, is a novel BRCA1-associated protein involved in the p53-mediated apoptosis pathway. Oncogene 22: 8931–8938. [DOI] [PubMed] [Google Scholar]
- Monroe, KM, Yang, Z, Johnson, JR, Geng, X, Doitsh, G, Krogan, NJ et al. (2014). IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343: 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cemazar, M, Sersa, G, Wilson, J, Tozer, GM, Hart, SL, Grosel, A et al. (2002). Effective gene transfer to solid tumors using different nonviral gene delivery techniques: electroporation, liposomes, and integrin-targeted vector. Cancer Gene Ther 9: 399–406. [DOI] [PubMed] [Google Scholar]
- Pichlmair, A and Reis e Sousa, C (2007). Innate recognition of viruses. Immunity 27: 370–383. [DOI] [PubMed] [Google Scholar]
- Randow, F, MacMicking, JD and James, LC (2013). Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science 340: 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X, Brann, TW, Zhou, M, Yang, J, Oguariri, RM, Lidie, KB et al. (2011). Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol 186: 4541–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur, N, Veettil, MV, Sharma-Walia, N, Bottero, V, Sadagopan, S, Otageri, P et al. (2011). IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe 9: 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi, H, Miyashita, M, Okamoto, M, Morioka, Y, Okabe, M, Matsumoto, M et al. (2015). DDX60 Is Involved in RIG-I-Dependent and Independent Antiviral Responses, and Its Function Is Attenuated by Virus-Induced EGFR Activation. Cell Rep 11: 1193–1207. [DOI] [PubMed] [Google Scholar]
- Goubau, D, van der Veen, AG, Chakravarty, P, Lin, R, Rogers, N, Rehwinkel, J et al. (2015). Mouse superkiller-2-like helicase DDX60 is dispensable for type I IFN induction and immunity to multiple viruses. Eur J Immunol 45: 3386–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr, SR, Chauhan, VS, Moerdyk-Schauwecker, MJ and Marriott, I (2011). A role for DNA-dependent activator of interferon regulatory factor in the recognition of herpes simplex virus type 1 by glial cells. J Neuroinflammation 8: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, TH, Kwon, KM, Kim, YE, Kim, KK and Ahn, JH (2013). DNA sensing-independent inhibition of herpes simplex virus 1 replication by DAI/ZBP1. J Virol 87: 3076–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, CJ, Anguela, XM, Montané, J, Obach, M, Roca, C, Ruzo, A et al. (2012). Molecular signature of the immune and tissue response to non-coding plasmid DNA in skeletal muscle after electrotransfer. Gene Ther 19: 1177–1186. [DOI] [PubMed] [Google Scholar]
- Lladser, A, Mougiakakos, D, Tufvesson, H, Ligtenberg, MA, Quest, AF, Kiessling, R et al. (2011). DAI (DLM-1/ZBP1) as a genetic adjuvant for DNA vaccines that promotes effective antitumor CTL immunity. Mol Ther 19: 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, ML and Heller, R (2003). IL-12 gene therapy using an electrically mediated nonviral approach reduces metastatic growth of melanoma. DNA Cell Biol 22: 755–763. [DOI] [PubMed] [Google Scholar]
- Melero, I, Quetglas, JI, Reboredo, M, Dubrot, J, Rodriguez-Madoz, JR, Mancheño, U et al. (2015). Strict requirement for vector-induced type I interferon in efficacious antitumor responses to virally encoded IL12. Cancer Res 75: 497–507. [DOI] [PubMed] [Google Scholar]
- Babiuk, S, Baca-Estrada, ME, Foldvari, M, Middleton, DM, Rabussay, D, Widera, G et al. (2004). Increased gene expression and inflammatory cell infiltration caused by electroporation are both important for improving the efficacy of DNA vaccines. J Biotechnol 110: 1–10. [DOI] [PubMed] [Google Scholar]
- Nayak, S and Herzog, RW (2010). Progress and prospects: immune responses to viral vectors. Gene Ther 17: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L, Liu, YP, Sha, H, Chen, H, Qi, L and Smith, JA (2010). XBP-1 couples endoplasmic reticulum stress to augmented IFN-beta induction via a cis-acting enhancer in macrophages. J Immunol 185: 2324–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, KJ and Schmittgen, TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Todorovic, V, Sersa, G, Mlakar, V, Glavac, D and Cemazar, M (2012). Assessment of the tumourigenic and metastatic properties of SK-MEL28 melanoma cells surviving electrochemotherapy with bleomycin. Radiol Oncol 46: 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer, T, Baumann, C, Blüml, S, Dixit, E, Dürnberger, G, Jahn, H et al. (2009). An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol 10: 266–272. [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri, T, Yu, JW, Datta, P, Wu, J and Alnemri, ES (2009). AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458: 509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung, V, Ablasser, A, Charrel-Dennis, M, Bauernfeind, F, Horvath, G, Caffrey, DR et al. (2009). AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458: 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T, Pazhoor, S, Bao, M, Zhang, Z, Hanabuchi, S, Facchinetti, V et al. (2010). Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci USA 107: 15181–15186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, TL, Idris, A, Dunn, JA, Kelly, GM, Burnton, CM, Hodgson, S et al. (2009). HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 323: 1057–1060. [DOI] [PubMed] [Google Scholar]
- Yang, P, An, H, Liu, X, Wen, M, Zheng, Y, Rui, Y et al. (2010). The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol 11: 487–494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.