ABSTRACT
In eukaryotic organisms, gene regulation occurs in the context of chromatin. In the interphase nucleus, euchromatin and heterochromatin occupy distinct space during cell differentiation, with heterochromatin becoming enriched at the nuclear and nucleolar peripheries. This organization is thought to fine-tune gene expression. To elucidate the mechanisms that govern this level of genome organization, screens were carried out in C. elegans which monitored the loss of heterochromatin sequestration at the nuclear periphery. This led to the identification of a novel chromodomain protein, CEC-4 (Caenorhabditis elegans chromodomain protein 4) that mediates the anchoring of H3K9 methylation-bearing chromatin at the nuclear periphery in early to mid-stage embryos. Surprisingly, the loss of CEC-4 does not derepress genes found in heterochromatic domains, nor does it affect differentiation under standard laboratory conditions. On the other hand, CEC-4 contributes to the efficiency with which muscle differentiation is induced following ectopic expression of the master regulator, HLH-1. This is one of the first phenotypes specifically attributed to the ablation of heterochromatin anchoring.
KEYWORDS: cell differentiation, heterochromatin, histone H3K9 methylation, nuclear organization, nuclear periphery
Tethering chromatin at the nuclear lamina
Studies in organisms including yeast, worms, flies and mammals have begun to shed light on the mechanisms that sequester chromatin at the nuclear periphery. Such studies generally rely on live imaging of genetic loci, tagged with a GFP-fusion protein. Not only did these studies show that silent genes are enriched at the nuclear periphery, but they found that repressed genes were also more constrained in their dynamics than active genes.1-3 This suggested that they might be molecularly tethered at the inner nuclear membrane (INM).
The radial distribution of chromatin changes during cell differentiation. A significant increase in chromatin positioned at the INM has been documented during mammalian hematopoiesis,4-7 C. elegans development,8,9 and upon the induction of neuronal cell types from pluripotent ES cells.10-12 This coincides with the restriction of gene transcription to cell-type specific profiles, which generally accompanies cell-fate determination. Gene activity is dependent on transcription factor availability, histone modification and local chromatin structure. It is likely that all 3 of these are affected by sequestration of chromatin, particularly in the form of heterochromatin, at the INM.
Heterochromatin is enriched for repressive histone modifications, in particular the di- and tri-methylation of histone H3K9. Moreover, the distribution of H3K9me2 and me3 changes during the establishment of differentiated cell states.13-16 Consistent with the notion that this modification regulates gene expression, stem cell differentiation in mammals was shown to be dependent on G9a, the methyltransferase responsible for H3K9me1 and me2.7 There also appears to be an increase in levels of H3K9me2 during ESC differentiation, and the concurrent formation of “large domains of chromatin bearing H3K9-modifications” or LOCKs.17 Conversely, the reduction of H3K9 methylation levels correlates with enhanced efficiency in cell fate reprogramming.18-21 Finally, down-regulation of both PRDM3 and PRDM16 (two H3K9-specific mono-methyltransferases) led to loss of H3K9me1, and the preclusion of higher levels of H3K9 methylation. This alteration resulted in the dispersal of centromeric foci, an accumulation of major satellite transcripts, and perturbation of the ultrastructure of the nuclear lamina.22 Although centromeric foci are not necessarily lamin-associated, secondary effects could arise from the release of satellite chromatin, disrupting general heterochromatin organization.
The major ligand of H3K9 methylation is HP1, which has at least 3 isoforms in mammalian cells (HP1α, HP1β and HP1γ), 2 in C. elegans (HPL-1 and HPL-2) and 2 in S. pombe (Chp2 and Swi6). All HP1 proteins contain an N-terminal chromodomain and a C-terminal chromo-shadow domain. The chromodomain specifically recognizes both H3K9me2 and me3,23,24 while the chromo-shadow domain mediates interaction with other proteins. Intriguingly, the different HP1 variants have very distinct roles in the ESC-to-differentiated cell transition, and not all HP1 binding correlates with heterochromatic gene repression.25 In S. pombe, the RNA binding functions of HP1 which are associated with the linker domain, actually restrict the spread of silencing.26,27 Thus, although HP1 might seem a good candidate for bridging from heterochromatin to the INM, direct involvement has not been shown.
On the side of the INM, one finds the nuclear lamina, which is composed of integral INM proteins including LAP2, Emerin and MAN1 (the so-called LEM proteins) and nuclear lamins. In mouse differentiated tissues heterochromatin sequestration is dependent on both Lamin A/C and LBR (Lamin B Receptor) in a partially redundant manner.16 Lamins interact directly with histones and DNA in vitro,28,29 yet it is unclear whether or not these low affinity interactions are relevant in vivo. Indeed, there are still no lamin mutations identified that interfere with the recognition of specific chromatin motifs. Alternatively, a loss of lamin function could affect the stability or spatial distribution of different lamin-associated proteins, such as LEM domain proteins or the Lamin-B receptor (LBR).30 LBR contains a Tudor domain that was reported to bind histone H4K20me2 in vitro,31 yet this histone modification is distributed broadly across the genome without strong enrichment in lamin associated heterochromatin.32 LBR was also reported to bind HP1α and HP1γ,33 although the ablation of HP1α or HP1β in pluripotent or differentiated embryonic stem cells failed to alter pericentric heterochromatin organization.25 Thus the link between the INM and chromatin was unclear.
Perinuclear chromatin segregation via H3K9 methylation in C. elegans
Several studies have used C. elegans to examine the impact of nuclear organization on gene expression during differentiation (for reviews see refs. 34, 35). In worms, large integrated gene arrays become transcriptionally silenced and bear histone modifications that are typical for heterochromatin, notably histone H3K9 and H3K27 methylation. Moreover, these heterochromatic arrays are sequestered at the INM.36 In a genome-wide RNAi screen that exploited such heterochromatic arrays tagged with lacO/LacI-GFP, factors were identified that silence the array and link it to the INM. This screen looked first for RNAi targets that led to the derepression of a constitutive promoter on the array. Among the 29 hits were many histone modifiers, yet only one of the RNAi constructs lead to release of the array from the nuclear periphery. This one identified a pair of closely related genes encoding S-adenosyl methionine synthetase (SAMS – sams-3 and sams-4). Loss of these enzymes reduced histone methylation globally, provoking both transcriptional up-regulation and displacement of the array from the nuclear periphery. Given that the other RNAi clones allowed derepression without affecting localization, one concluded that peripheral anchoring does not silence per se. In other words, gene expression is compatible with perinuclear localization.
Based upon this result, a second, targeted screen was performed to identify the relevant histone methyltransferases (HMTs) necessary for array sequestration at the nuclear envelope. Beyond the HMT genes tested in the genome-wide RNAi screen, 12 single and double mutant combinations in a range of putative C. elegans HMTs were tested. The majority of the different mutants were defective in array silencing, but not in array anchoring. Only the combined elimination of 2 HMTs, MET-2 and SET-25, showed a release of heterochromatic arrays that was as efficient as the SAMS-3/-4 knockdown. MET-2 is the worm homolog of human SetDB1/ESET, while the catalytic domain of SET-25 is similar to human G9a. These experiments, together with mass spectroscopy analysis of histone methylation in single and double mutants, revealed that (1) both MET-2 and SET-25 target lysine 9 of histone H3, (2) H3K9me1 and -me2 are sufficient for array anchoring, while H3K9me3 also mediates INM contact, and (3) that H3K9me3 was needed for transcriptional silencing of the reporter array.37 Since chromatin bearing H3K9me1 and me2 was anchored but not repressed, one can conclude that different levels of H3K9 methylation distinguish anchoring signals from transcriptional repression, even though they concern modification of the same lysine residue.
Interestingly, the SET-25 enzyme (the sole specific for H3K9me3 in worms) was shown to colocalize with peripheral heterochromatin, although its binding did not mediate anchoring. Indeed, at this point it was unclear what component of the INM might recognize the H3K9me1, me2 or me3 marks, and whether it required one or several proteins. The binding of HP1 homolog (HPL-2) and/or LIN-61, an MBT (Malignant Brain Tumor) domain protein, which both recognize H3K9me2/me3, were able to mediate transcriptional repression, but even the double mutant did not alter chromatin anchorage significantly.37,38 Similarly downregulation of HPL-1 affected the transcription of endogenous loci, but did not affect INM anchoring.37,39
Importantly, we note that although H3K9 methylation is essential for chromatin anchoring in early embryos, this pathway is not the only INM anchoring mechanism in adult worms. As early as L1 larvae, when most cells have acquired a cell-type specific pattern of gene expression and morphology, sequences that were released from the INM in H3K9-methylation deficient embryos became re-anchored, despite the persistent absence of the H3K9 methylation mark.37 This led to the conclusion that alternative pathways for heterochromatin anchoring are induced during terminal differentiation. This may account for the unexpected finding that C. elegans embryos lacking all histone H3K9 methylation still develop into adult worms with the full range of differentiated tissues.
One alternative anchorage pathway that had already been characterized tethers C. elegans telomeric repeats.40 The terminal repeats are anchored at the INM, becoming increasingly peripheral in terminally differentiated cells. Their anchorage is not dependent on H3K9 methylation, but requires a factor that recognizes the ss telomeric overhang (POT-1) and an inner nuclear membrane SUN domain protein, SUN-1. The interaction is also affected by sumoylation, reminiscent of telomere anchoring in budding yeast.40,41
A novel perinuclear anchor: CEC-4
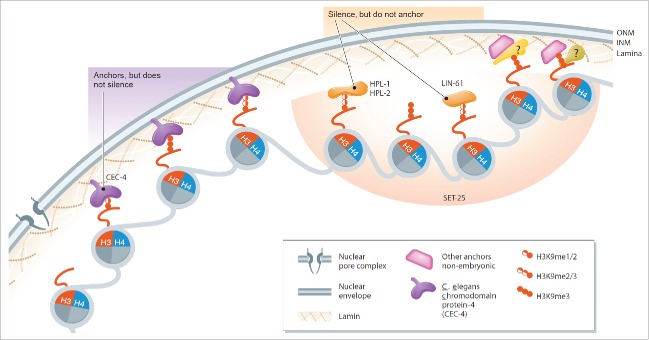
Knowing that H3K9 methylation is required for anchoring of C. elegans heterochromatin,37 another genetic screen was implemented to identify H3K9 methylation readers that might function as perinuclear tethers. In a screen of all potential histone methylation mark readers, a previously uncharacterized C. elegans chromodomain protein, CEC-4, was identified as a perinuclear heterochromatin anchor.42 CEC-4 binds preferentially mono-, di-, or tri-methylated H3K9 and localizes at the nuclear envelope independently of H3K9 methylation and nuclear lamin. CEC-4 is necessary for the perinuclear anchoring of endogenous heterochromatin, but does not affect transcriptional repression. This is in clear contrast to other known worm H3K9 methyl-binders, which mediate gene repression but not perinuclear anchoring (Fig. 1).
Figure 1.
Anchoring chromatin to the nuclear periphery in C. elegans. In early embryos, CEC-4 is a H3K9 me1, me2 or me3 ligand that mediates anchoring to the nuclear periphery, without necessarily repressing transcription. The H3K9me-ligands, HPL-1, HPL-2 and LIN-61 mediate transcriptional repression by binding H3K9 methylation, but do not anchor chromatin. SET-25 recognizes the H3K9me3-containing chromatin that it creates and together with HP1 homologues and LIN-61, leads to repression. In differentiated cells alternative anchors may be present. Reprinted from Harr et al.48 © SM Gasser. Reproduced by permission of the authors. Permission to reuse must be obtained from the rightsholder.
Although CEC-4 was identified using the transgene-based heterochromatic reporter, the majority of the effects seen with the heterochromatic reporter also held true for endogenous chromatin. Chromatin immunoprecipitation confirmed that CEC-4 contributes to the perinuclear sequestration of endogenous H3K9me3 marked chromatin, yet the distribution of H3K9me2 or me3 along the chromosome arms was independent of CEC-4. Finally, CEC-4 localized constitutively to the INM, which enabled it to carry out its anchoring function. Intriguingly, in differentiated tissues of the L1 larvae, CEC-4 function is redundant with other anchoring pathways, meaning that CEC-4 is only essential for anchoring in embryos. Nonetheless, as described below, further experiments identified a functional role for CEC-4-mediated perinuclear chromatin anchoring in an induced muscle differentiation program.
The interactions that properly localize CEC-4 are unclear. While CEC-4 does not have a putative transmembrane domain, attempts to identify an additional protein interactor that would hold CEC-4 at the nuclear periphery were fruitless. Indeed, CEC-4 was able to locate at the INM independently of lamin and other INM proteins when expressed in budding yeast. Either CEC-4 interacts directly with the membrane (e.g. through a lipid anchor) or it may interact redundantly with a range of INM proteins, such that single knockouts were insufficient to trigger its delocalization.
Perinuclear localization of chromatin is not necessary for gene silencing nor for differentiation under normal developmental conditions
CEC-4 recognizes the methylation of H3K9 on chromatin for anchoring purposes,42 yet gene expression profiles are not altered in embryos that lose this anchor. Thus, stable INM anchoring does not drive transcriptional silencing. Although many studies have reported correlations between INM association and gene repression, they have not resolved whether the INM sequestration of chromatin results from or is causal for gene repression. The Gonzalez-Sandoval study clearly demonstrates that in C. elegans early embryonic development, chromatin anchoring is not needed for proper gene expression under normal growth conditions. Nonetheless, since perinuclear chromatin accumulates over time, one cannot exclude that at later stages of development, anchoring might play a role in maintaining proper gene expression under specific conditions.
CEC-4 is not required for the proper timing of embryonic development under standard laboratory conditions. Brood-size and embryonic viability are unchanged in the cec-4 mutant, and there is only a very slight increase in male induction in mutant worms at 26°C. A similar observation applies to the anchoring of H3K9 methylated chromatin during cell differentiation in mice, i.e., Lamin A/C and the Lamin B Receptor or LBR. In mice that lack both of Lamin A/C and LBR proteins, there are major issues in cell structure and nuclear organization, and the animals die immediately after birth, yet ablation of these 2 pathways of chromatin anchoring does not prevent cell differentiation.16
Other aspects of genome biology may be influenced by chromatin positioning. Intriguingly, genome-wide analysis of early and late DNA replication domains shows that these correlate with the 3D organization of chromatin domains.43,44 Chromatin associated with the nuclear lamina (LADs) correlate with late replicating domains.45 Upon neuronal differentiation, 20% of the genome changes replication timing concomitant with changes in gene expression of intermediate/low CpG-containing promoters and a shift in radial position for the loci tested.46 Perhaps perinuclear localization of chromatin plays a more important role in relation to replication, than in the control of gene expression per se. However, in order to prove a causal link between subnuclear position and an altered function, it will be necessary to show genetically that the mechanism that mediates positioning also mediates late replication.
Chromatin anchoring supports commitment to an induced differentiation program
At later stages of worm development additional chromatin anchorage mechanisms are induced, which compensate for the null cec-4 mutation and restore anchoring of heterochromatic arrays.42 These compensatory mechanisms may help maintain cell-type identity, and likely mediate the residual anchoring observed for endogenous sequences in cec-4 mutant embryos. They are likely to involve chromatin modifications other than histone H3K9 methylation, given that the absence of SET-25 and MET-2, the 2 H3K9me histone methyltransferases, also is permissive to cell differentiation under normal laboratory conditions.37
In order to probe more deeply for the function of perinuclear anchoring in embryos, muscle cell fate was induced during an early embryonic stage, either in the absence or presence of CEC-4. Efficient muscle cell programming was triggered by the heat-shock induced expression of the master regulator HLH-1 (known as MyoD in mammals).47 In wild-type embryos, the induction of HLH-1 forces cells into a muscle specification pathway, resulting in the 100% conversion of embryonic cells into muscle. In contrast, in the cec-4 mutant about 25% of the embryos continued to develop over the next 24 hours to the point of hatching from the eggshell. One interpretation of this result is that the loss of heterochromatin anchoring impaired the proper commitment to muscle, allowing some cells to pursue other developmental programs. Unfortunately, the mechanism is not yet clear, as the resulting larva-like embryos do not survive. Under this forced differentiation condition, heterochromatin anchoring by CEC-4 may indeed support stable gene silencing, or else anchoring may influence events that prepare genes for tissue-restricted patterns of expression, such as replication timing.
This study suggests that gene or promoter positioning at the nuclear lamina does help fine-tune gene regulation under conditions of developmental perturbation or environmental stress. Anchoring is not essential for muscle-fate induction, but it appears to contribute to the repression of alternative developmental pathways. While there is no clear mammalian homolog of CEC-4 identified to date, the function may be covered by 2 or more proteins working together. These results suggest that the repression of differentiation pathways is distinct from pathway induction, and that both can be necessary under some conditions of growth.
C. elegans has proven to be an invaluable tool for the study of nuclear organization. Further experiments may reveal other situations in which INM anchoring contributes to cell-type specific gene repression. The identification and ablation of other anchors that function at later stages of development, will help the field elucidate the gene regulatory function of this universally conserved nuclear subcompartment.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 2002; 12:439-45; PMID:11909528; http://dx.doi.org/ 10.1016/S0960-9822(02)00695-4 [DOI] [PubMed] [Google Scholar]
- [2].Hediger F, Dubrana K, Gasser SM. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J Struct Biol 2002; 140:79-91; PMID:12490156; http://dx.doi.org/ 10.1016/S1047-8477(02)00533-6 [DOI] [PubMed] [Google Scholar]
- [3].Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome Dynamics in the Yeast Interphase Nucleus. Science 2001; 294:2181-6; PMID:11739961; http://dx.doi.org/ 10.1126/science.1065366 [DOI] [PubMed] [Google Scholar]
- [4].Brown KE, Amoils S, Horn JM, Buckle VJ, Higgs DR, Merkenschlager M, Fisher AG. Expression of [alpha]- and [beta]-globin genes occurs within different nuclear domains in haemopoietic cells. Nat Cell Biol 2001; 3:602; PMID:11389446; http://dx.doi.org/ 10.1038/35078577 [DOI] [PubMed] [Google Scholar]
- [5].Hubner B, Lomiento M, Mammoli F, Illner D, Markaki Y, Ferrari S, Cremer M, Cremer T. Remodeling of nuclear landscapes during human myelopoietic cell differentiation maintains co-aligned active and inactive nuclear compartments. Epigenetics Chromatin 2015; 8:47; PMID:26579212; http://dx.doi.org/ 10.1186/s13072-015-0038-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kosak ST, Scalzo D, Alworth SV, Li F, Palmer S, Enver T, Lee JSJ, Groudine M. Coordinate Gene Regulation during Hematopoiesis Is Related to Genomic Organization. PLoS Biology 2007; 5:e309; PMID:18031200; http://dx.doi.org/ 10.1371/journal.pbio.0050309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ugarte F, Sousae R, Cinquin B, Martin EW, Krietsch J, Sanchez G, Inman M, Tsang H, Warr M, Passegue E, et al.. Progressive Chromatin Condensation and H3K9 Methylation Regulate the Differentiation of Embryonic and Hematopoietic Stem Cells. Stem Cell Reports 2015; 5:728-40; PMID:26489895; http://dx.doi.org/ 10.1016/j.stemcr.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fakhouri THI, Stevenson J, Chisholm AD, Mango SE. Dynamic Chromatin Organization during Foregut Development Mediated by the Organ Selector Gene PHA-4/FoxA. PLoS Genet 6:e1001060; PMID:20714352; http://dx.doi.org/ 10.1371/journal.pgen.1001060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes & Development 2010; 24:766-82; PMID:20395364; http://dx.doi.org/ 10.1101/gad.559610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al.. Chromatin architecture reorganization during stem cell differentiation. Nature 2015; 518:331-6; PMID:25693564; http://dx.doi.org/ 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al.. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 2010; 38:603-13; PMID:20513434; http://dx.doi.org/ 10.1016/j.molcel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Williams RRE, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, et al.. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci 2006; 119:132-40; PMID:16371653; http://dx.doi.org/ 10.1242/jcs.02727 [DOI] [PubMed] [Google Scholar]
- [13].Bian Q, Khanna N, Alvikas J, Belmont AS. beta-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J Cell Biol 2013; 203:767-83; PMID:24297746; http://dx.doi.org/ 10.1083/jcb.201305027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell 2013; 153:178-92; PMID:23523135; http://dx.doi.org/ 10.1016/j.cell.2013.02.028 [DOI] [PubMed] [Google Scholar]
- [15].Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 2009; 137:356-68; PMID:19379699; http://dx.doi.org/ 10.1016/j.cell.2009.01.052 [DOI] [PubMed] [Google Scholar]
- [16].Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al.. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 2013; 152:584-98; PMID:23374351; http://dx.doi.org/ 10.1016/j.cell.2013.01.009 [DOI] [PubMed] [Google Scholar]
- [17].Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nature genetics 2009; 41:246-50; PMID:19151716; http://dx.doi.org/ 10.1038/ng.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bao X, Wu H, Zhu X, Guo X, Hutchins AP, Luo Z, Song H, Chen Y, Lai K, Yin M, et al.. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell research 2015; 25:80-92; PMID:25512341; http://dx.doi.org/ 10.1038/cr.2014.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Baxter J, Sauer S, Peters A, John R, Williams R, Caparros M-L, Arney K, Otte A, Jenuwein T, Merkenschlager M, et al.. Histone hypomethylation is an indicator of epigenetic plasticity in quiescent lymphocytes. The EMBO journal 2004; 23:4462-72; PMID:15510223; http://dx.doi.org/ 10.1038/sj.emboj.7600414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, et al.. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013; 155:1479-91; PMID:24360272; http://dx.doi.org/ 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, et al.. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1$\gamma$ in reprogramming to pluripotency. Nature cell biology 2013; 15:872-82; PMID:23748610; http://dx.doi.org/ 10.1038/ncb2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pinheiro I, Margueron R, Shukeir N, Eisold M, Fritzsch C, Richter FM, Mittler G, Genoud C, Goyama S, Kurokawa M, et al.. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 2012; 150:948-60; PMID:22939622; http://dx.doi.org/ 10.1016/j.cell.2012.06.048 [DOI] [PubMed] [Google Scholar]
- [23].Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001; 410:116-20; PMID:11242053; http://dx.doi.org/ 10.1038/35065132 [DOI] [PubMed] [Google Scholar]
- [24].Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 2002; 416:103-7; PMID:11882902; http://dx.doi.org/ 10.1038/nature722 [DOI] [PubMed] [Google Scholar]
- [25].Mattout A, Aaronson Y, Sailaja BS, Raghu Ram EV, Harikumar A, Mallm JP, Sim KH, Nissim-Rafinia M, Supper E, Singh PB, et al.. Heterochromatin Protein 1beta (HP1beta) has distinct functions and distinct nuclear distribution in pluripotent versus differentiated cells. Genome biology 2015; 16:213; PMID:26415775; http://dx.doi.org/ 10.1186/s13059-015-0760-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Keller C, Kulasegaran-Shylini R, Shimada Y, Hotz H-R, Bhler M. Noncoding RNAs prevent spreading of a repressive histone mark. Nature structural \& molecular biology 2013; 20:994-1000; PMID:23872991; http://dx.doi.org/ 10.1038/nsmb.2619 [DOI] [PubMed] [Google Scholar]
- [27].Stunnenberg R, Kulasegaran-Shylini R, Keller C, Kirschmann MA, Gelman L, Bhler M. H3K9 methylation extends across natural boundaries of heterochromatin in the absence of an HP1 protein. EMBO J 2015; 34(22):2789-803; PMID:26438724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Goldberg M, Harel A, Brandeis M, Rechsteiner T, Richmond TJ, Weiss AM, Gruenbaum Y. The tail domain of lamin Dm0 binds histones H2A and H2B. Proc Natl Acad Sci U S A 1999; 96:2852-7; PMID:10077600; http://dx.doi.org/ 10.1073/pnas.96.6.2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Luderus ME, de Graaf A, Mattia E, den Blaauwen JL, Grande MA, de Jong L, van Driel R. Binding of matrix attachment regions to lamin B1. Cell 1992; 70:949-59; PMID:1525831; http://dx.doi.org/ 10.1016/0092-8674(92)90245-8 [DOI] [PubMed] [Google Scholar]
- [30].Wilson KL, Foisner R. Lamin-binding Proteins. Cold Spring Harb Perspect Biol 2010; 2:a000554; PMID:20452940; http://dx.doi.org/ 10.1101/cshperspect.a000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hirano Y, Hizume K, Kimura H, Takeyasu K, Haraguchi T, Hiraoka Y. Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation. The Journal of biological chemistry 2012; 287:42654-63; PMID:23100253; http://dx.doi.org/ 10.1074/jbc.M112.397950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129:823-37; PMID:17512414; http://dx.doi.org/ 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- [33].Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. The Journal of biological chemistry 1996; 271:14653-6; PMID:8663349; http://dx.doi.org/ 10.1074/jbc.271.25.14653 [DOI] [PubMed] [Google Scholar]
- [34].Meister P, Mango SE, Gasser SM. Locking the genome: nuclear organization and cell fate. Current Opinion in Genetics & Development 2011; 21:167-74; PMID:21345665; http://dx.doi.org/ 10.1016/j.gde.2011.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Meister P, Taddei A. Building silent compartments at the nuclear periphery: a recurrent theme. Curr Opin Genet Dev 2013; 23(2):96-103; PMID:23312840 [DOI] [PubMed] [Google Scholar]
- [36].Towbin BD, Meister P, Pike BL, Gasser SM. Repetitive transgenes in C. elegans accumulate heterochromatic marks and are sequestered at the nuclear envelope in a copy-number- and lamin-dependent manner. Cold Spring Harb Symp Quant Biol 2010; 75:555-65; PMID:21467137; http://dx.doi.org/ 10.1101/sqb.2010.75.041 [DOI] [PubMed] [Google Scholar]
- [37].Towbin Benjamin D, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser Susan M. Step-Wise Methylation of Histone H3K9 Positions Heterochromatin at the Nuclear Periphery. Cell 2012; 150:934-47; PMID:22939621; http://dx.doi.org/ 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- [38].Koester-Eiserfunke N, Fischle W. H3K9me2/3 Binding of the MBT Domain Protein LIN-61 Is Essential for Caenorhabditis elegans Vulva Development. PLoS Genetics 2011; 7:e1002017; PMID:21437264; http://dx.doi.org/ 10.1371/journal.pgen.1002017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Studencka M, Konzer A, Moneron G, Wenzel D, Opitz L, Salinas-Riester G, Bedet C, Kruger M, Hell SW, Wisniewski JR, et al.. Novel roles of Caenorhabditis elegans heterochromatin protein HP1 and linker histone in the regulation of innate immune gene expression. Mol Cell Biol 2012; 32:251-65; PMID:22083954; http://dx.doi.org/ 10.1128/MCB.05229-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ferreira HC, Towbin BD, Jegou T, Gasser SM. The shelterin protein POT-1 anchors Caenorhabditis elegans telomeres through SUN-1 at the nuclear periphery. J Cell Biol 2013; 203:727-35; PMID:24297748; http://dx.doi.org/ 10.1083/jcb.201307181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ferreira HC, Luke B, Schober H, Kalck V, Lingner J, Gasser SM. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat Cell Biol 2011; 13:867-74; PMID:21666682; http://dx.doi.org/ 10.1038/ncb2263 [DOI] [PubMed] [Google Scholar]
- [42].Gonzalez-Sandoval A, Towbin BD, Kalck V, Cabianca DS, Gaidatzis D, Hauer MH, Geng L, Wang L, Yang T, Wang X, et al.. Perinuclear Anchoring of H3K9-Methylated Chromatin Stabilizes Induced Cell Fate in C. elegans Embryos. Cell 2015; 163:1333-47; PMID:26607792; http://dx.doi.org/ 10.1016/j.cell.2015.10.066 [DOI] [PubMed] [Google Scholar]
- [43].Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al.. Topologically associating domains are stable units of replication-timing regulation. Nature 2014; 515:402-5; PMID:25409831; http://dx.doi.org/ 10.1038/nature13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 2010; 20:761-70; PMID:20430782; http://dx.doi.org/ 10.1101/gr.099655.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014; 159:1665-80; PMID:25497547; http://dx.doi.org/ 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 2008; 6:e245; PMID:18842067; http://dx.doi.org/ 10.1371/journal.pbio.0060245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fukushige T, Krause M. The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development 2005; 132:1795-805; PMID:15772130; http://dx.doi.org/ 10.1242/dev.01774 [DOI] [PubMed] [Google Scholar]
- [48].Harr JC, Gonzalez-Sandoval A, Gasser SM. Histones and histone modifications in perinuclear chromatin anchoring: from yeast to man. EMBO Rep 2016; 17(2):139-55; PMID:2679293 [DOI] [PMC free article] [PubMed] [Google Scholar]