Abstract
 The α-l-threofuranosyl nucleoside triphosphates of T, G, and D (tTTP, tGTP, and tDTP) were synthesized from the described 2‘-O-DMT-protected derivatives using the Eckstein method, while the corresponding C derivative (tCTP) was prepared from the 2‘-O-acetyl derivative.
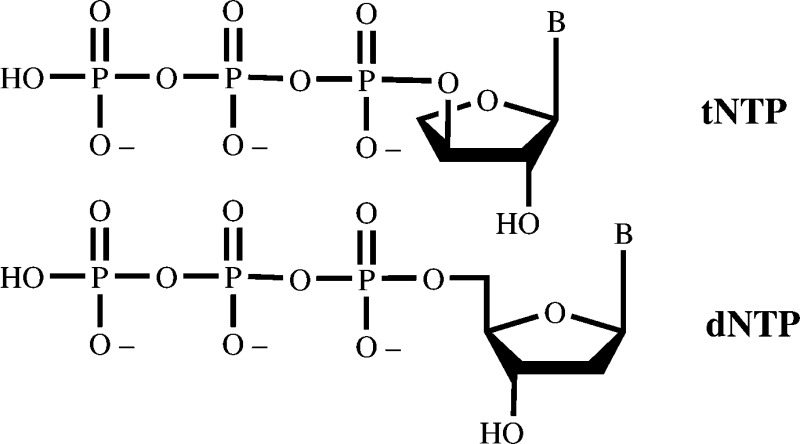
The prepared α-l-threofuranosyl nucleoside triphosphates, despite being one carbon shorter than the native 2‘-deoxyfuranosyl nucleoside
triphosphates, are effective substrates for selected DNA polymerases.
The α-l-threofuranosyl nucleoside triphosphates of T, G, and D (tTTP, tGTP, and tDTP) were synthesized from the described 2‘-O-DMT-protected derivatives using the Eckstein method, while the corresponding C derivative (tCTP) was prepared from the 2‘-O-acetyl derivative.
The prepared α-l-threofuranosyl nucleoside triphosphates, despite being one carbon shorter than the native 2‘-deoxyfuranosyl nucleoside
triphosphates, are effective substrates for selected DNA polymerases.
Unnatural (3‘ → 2‘)-α-l-threose oligonucleotides (TNA oligos) are linked together through their 3‘- and 2‘-hydroxyls and create a nucleic acid polymer with a backbone consisting of five bonds rather than the six-bond backbone of DNA and RNA. Even with the shorter backbone, TNA oligos were observed to cross-hybridize with complementary DNA, RNA, and TNA itself during a systematic study of alternative carbohydrate-based nucleic acids.500811b00001,500811b00002-d These findings have suggested new questions regarding the prebiotic development of these fundamental biological molecules. The property of intersystem base-paring, together with the structural simplicity of the TNA-based oligomers relative to natural RNA oligomers, suggests that TNA might be a candidate for an evolutionary progenitor of the proposed “RNA world”.500811b00002-c,500811b00003 Nucleic acids such as the TNAs would have carried out fundamental receptor−ligand binding as well as catalytic reactions in such an early stage in the evolution of life. The ability to select functional TNA molecules in vitro(500811b00004-c) from random pools of TNA sequences would provide some evidence for the plausibility of such a proposal. The ability to select functional TNA molecules requires the availability of the TNA building blocks, the nucleoside-3‘-triphosphates (tNTPs), and their ability to function with modern protein-based polymerization catalysts. Recently, we and others have observed that certain DNA polymerases can incorporate tTTP or tUTP to extend a DNA or RNA primer in response to a dA or A template.500811b00005-c Synthetic studies on these materials include the preparation of the 3‘-O-DMT 2‘-phosphoramidites necessary for the chemical synthesis of TNA oligomers.500811b00001 Here we describe the synthesis of the 3‘-tNTP analogues and their substrate activity.
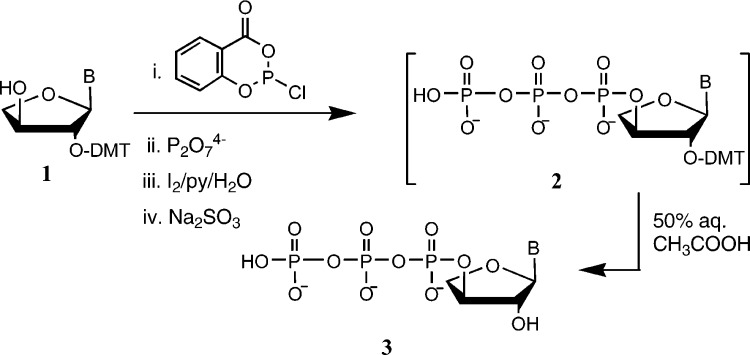
In the described syntheses of the TNA phosphoramidites,500811b00002 a DMT group was used as a temporary protecting group for the secondary 3‘-OH, but since the remaining 2‘-OH is also a secondary hydroxyl, this protection reaction typically resulted in a mixture of regioisomeric products. In most cases, the two DMT isomers (2‘- and 3‘-) could be chromatographically resolved, but in the case of α-l-threofuranosyl cytosine, the 3‘-isomer was preferentially obtained.500811b00002 The 3‘-DMT isomers were subsequently converted to the corresponding 2-phosphoramidites for use in the synthesis of TNA sequences. Here we explore whether the late-stage intermediate side products, the 2‘-DMT isomers (1, Scheme 1), can be used to prepare the 3‘-triphosphates (3).
Figure .
Syntheses of nucleoside-5‘-triphosphates using POCl3 in trimethyl phosphate rely upon the greater reactivity and lesser steric hindrance of the primary 5-hydroxyl. That approach was potentially problematic for the TNA building blocks with two secondary hydroxyls. Additionally, the acid-labile DMT group would be lost during such procedures. Eckstein's method500811b00006 (Scheme 1) can be more generally used, but it requires protection of the nontarget hydroxyl groups.
For three of the tNTPs, this strategy could work using the 2‘-DMT-protected TNA derivatives, providing that the acid conditions necessary for DMT removal after completion of the reaction sequence (2 → 3, Scheme 1) do not damage the 3‘-triphosphate. Using these late-stage intermediates for T, G, and α-l-threofuranosyl 2,6-diaminopurine (DAP), we first removed the nucleobase protecting groups in 8 M methylamine−ethanol/12 M methylamine−water 1:1. We then prepared the desired triphosphates using Eckstein's method and subsequently removed the 2‘-DMT group under mild acidic conditions with no observable cleavage of the triphosphate (3, Scheme 1).
The triphosphate products were purified by anion exchange HPLC chromatography (DEAE Sephadex), and after the fractions containing product were pooled and reduced in volume, desalting was performed using a 20 × 200 mm column of polydivinyl-benzene eluting with aqueous triethylammonium acetate. The products were lyophilized to dryness and stored at −20 °C.
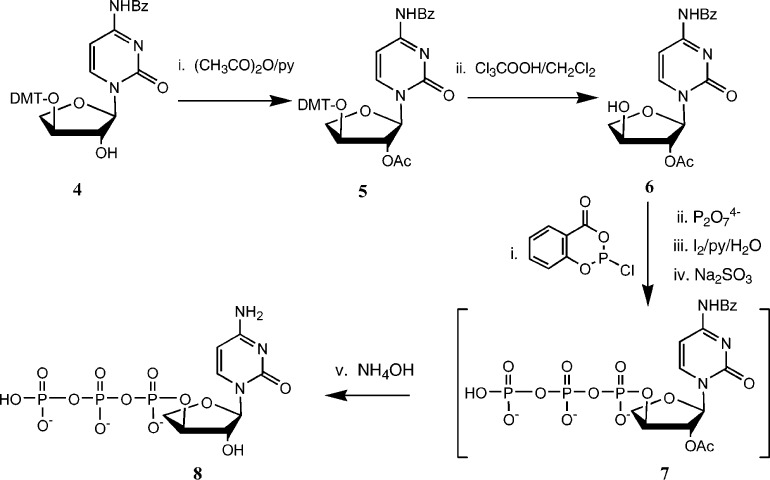
This strategy failed in the case of tCTP, where the 3‘-DMT protected nucleoside (4, Scheme 2) was the major product2b and the presence of the 2‘-regioisomer was negligible. We therefore prepared the 3‘-DMT derivative of the nucleobase-protected α-l-threofuranosyl cytosine and then acylated the 2‘-hydroxyl group (→ 5). Removal of the DMT group generated the 2‘-acetate (6). This derivative was used to prepare the corresponding triphosphate using Eckstein's method (tTTP has also been prepared5b using POCl3), after which concentrated aqueous ammonia was used to deprotect the nucleobase and generate the final product 8, which could be purified and desalted as described above.
Figure .
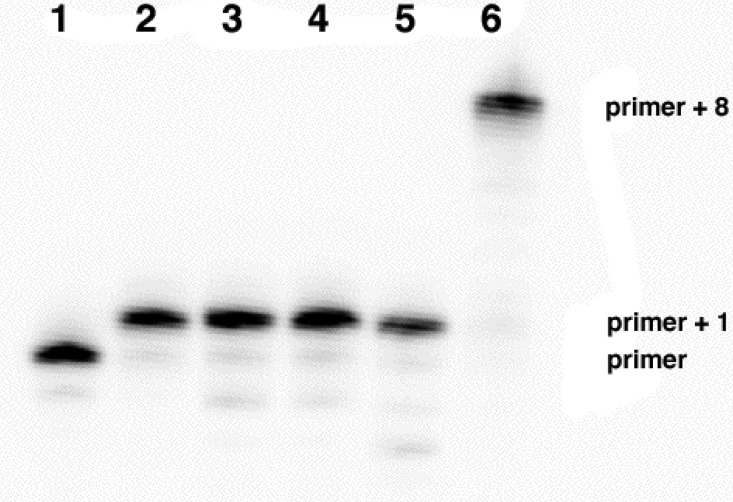
To probe the activity of these derivatives with a selected DNA polymerase, we prepared a primer sequence and a series of five templates. Four of the templates were used for single-nucleotide incorporation studies of each of the four tNTPs, and the fifth template was used with all four tNTPs (Figure 1). To date, the polymerase that most effectively uses the tNTPs as substrates is the thermophilic Therminator DNA polymerase, a site-specifically engineered (A485L) exonuclease-deficient form of “9°N” DNA polymerase. Incubation of a radiolabeled DNA primer with each appropriate templatepermitted an examination of single-nucleotide extension reactions using the tNTP substrates. In all cases, the single-elongation reactions were observed to occur essentially quantitatively within 15 min. Although most reactions functioned best in the presence of the divalent metal ions Mg2+ and Mn2+, tGTP only required the former. In the presence of all four tNTPS, the primer could be fully elongated by eight residues within 120 min.
Figure 1.
Radiolabeled annealed primer/template (250 nM). Position 1 in each template (lanes 2−5) provided the corresponding Watson−Crick complementary deoxynucleoside to test elongation of the DNA primer by one residue. Each reaction included 20 mM Tris-HCl, 10 mM KCl, 10 mM (NH4)2SO4, 20 mM MgSO4, 0.1% Triton X-100, 0.2 μg/μL BSA, 100 μM DTT, 0.2 units (37 nM) of Therminator Polymerase, 1.25 mM Mn2+, and 0.5 μM tNTP and were incubated at 55 °C for 15 min. Lane 1: Radiolabeled primer. Lane 2: tGTP (no Mn2+). Lane 3: tDTP. Lane 4: tTTP. Lane 5: tCTP. Lane 6: tGTP, tDTP, tTTP, and tCTP with an incubation time of 120 min.
The procedures described here provide a straightforward approach to the preparation of the 3‘-nucleoside triphosphate derivatives of the α-l-threofuranosyl sugars. These building blocks will be important for the study of polymerase activity as well as for the selection of functional TNA sequences.
Acknowledgments
We thank R. Krishnamurthy and A. Eschenmoser for the generous gift of advanced intermediates for the synthesis of D, G, and T tNTPs and Dr. Meena for helpful discussions. This work was supported by grants from the NIH (R01 GM053936) and the NASA Astrobiology Institute (NNA04CC12A) to J.W.S. and a grant from the NSF (MCB0077667) to L.W.M. J.W.S. is an Investigator of the Howard Hughes Medical Institute.
Supporting Information Available
Supporting Information Available
Experimental procedures and 1H and 13C NMR for compound 5 and 6 and 31P NMR for 8 and compounds 3. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Eschenmoser A. Science 1999, 284, 2118–2124. 10.1126/science.284.5423.2118. [DOI] [PubMed] [Google Scholar]
- a Schöning K.-U.; Scholz P.; Guntha S.; Wu X.; Krishnamurthy R.; Eschenmoser A. Science 2000, 290, 1347–1351. 10.1126/science.290.5495.1347. [DOI] [PubMed] [Google Scholar]; b Schöning K.-U.; Scholz P.; Wu X.; Guntha S.; Delgado G.; Krishnamurthy R.; Eschenmoser A. Helv. Chim. Acta 2002, 85, 4111–4153. 10.1002/hlca.200290000. [DOI] [Google Scholar]; c Wu X.; Guntha S.; Ferencic M.; Krishnamurthy R.; Eschenmoser A. Org. Lett. 2002, 4, 1279–1282. 10.1021/ol020015x. [DOI] [PubMed] [Google Scholar]; d Wu X.; Degado G.; Krishnamurthy R.; Eschenmoser A. Org. Lett. 2002, 4, 1283–1286. 10.1021/ol020016p. [DOI] [PubMed] [Google Scholar]
- a Orgel L. E. Science 2000, 290, 1306–1307. 10.1126/science.290.5495.1306. [DOI] [PubMed] [Google Scholar]; b Joyce G. F. Nature 2002, 418, 214–221. 10.1038/418214a. [DOI] [PubMed] [Google Scholar]; c Herdewijn P. Angew. Chem., Int. Ed. 2001, 40, 2249–2051. 10.1002/1521-3773(20010618)40:12%3C2249::AID-ANIE2249%3E3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- a Carothers J. M.; Oestreich S. C.; Davis J. H.; Szostak J. W. J. Am. Chem. Soc. 2004, 126, 5130–5137. 10.1021/ja031504a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tuerk C.; Gold L. Science 1990, 249, 505–510. 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]; c Ellington A. D.; Szostak J. W. Nature 1990, 346, 818–822. 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]; d Famulok M. Curr. Opin. Struct. Biol. 1999, 9, 324–329. 10.1016/S0959-440X(99)80043-8. [DOI] [PubMed] [Google Scholar]; e Wilson D. S.; Szostak J. W. Annu. Rev. Biochem. 1999, 68, 611–647. 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]; f Breaker R. R. Chem. Rev. 1997, 97, 371–390. 10.1021/cr960008k. [DOI] [PubMed] [Google Scholar]
- a Chaput J. C.; Szostak J. W. J. Am. Chem. Soc. 2003, 125, 9274–9275. 10.1021/ja035917n. [DOI] [PubMed] [Google Scholar]; b Kempeneers V.; Vastmans K; Rozenski J.; Herdewijn P. Nucleic Acids Res. 2003, 31, 6221–6226. 10.1093/nar/gkg833. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Horhota A.; Zou K.; Ichida J. K.; Biao Y.; McLaughlin L. W.; Szostak J. W.; Chaput J. C. J. Am. Chem. Soc. 2005, submitted. [DOI] [PMC free article] [PubMed]
- Ludwig J.; Eckstein F. J. Org. Chem. 1989, 54, 631–635. 10.1021/jo00264a024. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.