Abstract
Airway epithelial cell responses are critical to the outcome of lung infection. In this study, we aimed to identify unique contributions of epithelial cells during lung infection. To differentiate genes induced selectively in epithelial cells during pneumonia, we compared genome-wide expression profiles from three sorted cell populations: epithelial cells from uninfected mouse lungs, epithelial cells from mouse lungs with pneumococcal pneumonia, and nonepithelial cells from those same infected lungs. Of 1,166 transcripts that were more abundant in epithelial cells from infected lungs compared with nonepithelial cells from the same lungs or from epithelial cells of uninfected lungs, 32 genes were identified as highly expressed secreted products. Especially strong signals included two related secreted and transmembrane (Sectm) 1 genes, Sectm1a and Sectm1b. Refinement of sorting strategies suggested that both Sectm1 products were induced predominantly in conducting airway epithelial cells. Sectm1 was induced during the early stages of pneumococcal pneumonia, and mutation of NF-κB RelA in epithelial cells did not diminish its expression. Instead, type I IFN signaling was necessary and sufficient for Sectm1 induction in lung epithelial cells, mediated by signal transducer and activator of transcription 1. For target cells, Sectm1a bound to myeloid cells preferentially, in particular Ly6GbrightCD11bbright neutrophils in the infected lung. In contrast, Sectm1a did not bind to neutrophils from uninfected lungs. Sectm1a increased expression of the neutrophil-attracting chemokine CXCL2 by neutrophils from the infected lung. We propose that Sectm1a is an epithelial product that sustains a positive feedback loop amplifying neutrophilic inflammation during pneumococcal pneumonia.
Keyword: pneumococcal pneumonia, lung epithelium, type I IFNs, neutrophil activation, CXCL2
Clinical Relevance
Pneumonia results from failures of the host to respond appropriately to microbes in the lung. The current studies identify immune signaling roles that are unique to the epithelial cells that line the surface of the lung.
Lung infections are one of the most prominent burdens of disease worldwide (1). Despite the development of antibiotics and vaccines, morbidity caused by pneumonia is still considerable. Severe pneumonias are difficult to treat and have a high mortality rate due to lung injury (2). They are responsible for the greatest number of infectious disease deaths in the United States (3). The bacteria causing pneumonia most frequently is the pneumococcus, Streptococcus pneumoniae (4, 5).
Epithelial cells constitute the entire surface of the respiratory tract, and several lines of evidence demonstrate that epithelial cells have special roles during pneumonia (6, 7). Interruptions of NF-κB and signal transducer and activator of transcription (STAT) 3 pathways in epithelial cells exacerbate lung infection and lung injury during pneumonia, implicating transcriptional remodeling in these cells as a critical determinant of tissue homeostasis (8, 9). Pulmonary epithelial cells are predominant sources of the cytokines, CXCL5, granulocyte/macrophage colony–stimulating factor (GM-CSF), and CCL20, all of which are induced in an NF-κB RelA–dependent mechanism (10, 11). CXCL5 and GM-CSF promote neutrophil recruitment during pneumococcal pneumonia (11). A purpose of the current study was to reveal additional unique and essential immune roles that epithelial cells perform during pneumonia.
Here, we introduce secreted and transmembrane (Sectm) 1 protein induction as a previously unrecognized, but immunologically relevant epithelial response to lung infection. Human Sectm1 and the mouse orthologs, Sectm1a and Sectm1b, are closely related Sectm proteins with a single extracellular domain containing two regions resembling Ig domains (12–14). Mouse Sectm1a and Sectm1b genes are located adjacent to one another on chromosome 11 and encode similar products (13, 14). Sectm1 proteins appear to stimulate diverse receptors, cells, and actions, but their functions are still only beginning to be defined. Sectm1 can serve as a costimulatory ligand for T cell proliferation (14, 15) and as a chemoattractant for monocytes (16). There are multiple receptors, including CD7, glucocorticoid-induced TNF receptor (GITR), and additional ones yet to be identified (13–17). To our knowledge, Sectm1 proteins have not previously been connected with pneumonia.
Materials and Methods
Mice
Experiments were performed using C57BL/6 mice, NK2 homeobox 1 (NKX2-1)-Cre Rela-floxed mice (11), surfactant protein (SP) C promoter–driven green fluorescent protein (GFP) transgenic mice (18) (kindly provided by John K. Heath, University of Birmingham, Birmingham, UK), and IFN-α/β receptor (IFNAR)-deficient mice (19). The SPC-GFP and IFNAR colonies were backcrossed at least eight generations onto the C57BL/6 background. For RelA targeting studies, all mice were homozygous for loxP insertions in the Rela gene, and Cre-positive mutants were compared with sex-matched wild-type (WT) control littermates not expressing Cre. For IFNAR-deficient mice, homozygous mutants were compared with sex-matched and age-matched nonlittermate C57BL/6 controls that were bred within the same mouse room as the mutants. SPC-GFP mice were used to allow comparisons of cell subsets and were not compared with mice of other genotypes. The age range throughout the studies was 7–19 weeks. All animal protocols were approved by Boston University Institutional Animal Care and Use Committee (Boston, MA) or the University of California, Los Angeles (Los Angeles, CA) Animal Research Committee.
Pneumonia
Mice were anesthetized by intraperitoneal administration of ketamine (50 mg/kg) and xylazine (5 mg/kg). An angiocatheter was inserted into the left bronchus and mice received intrabronchial delivery of 50 μl saline containing 106 CFU of S. pneumoniae serotype 19F EF3030 (11, 20). In IFNAR-deficient mouse experiments, mice were instilled with EF3030 into the trachea, as previously described (19). The EF3030 isolate of pneumococcus causes a self-limiting and nonbacteremic pneumonia in mice (8, 21, 22).
Sectm1a Binding Assay for Lung Leukocytes
The binding of Sectm1a to leukocytes was determined using flow cytometry (LSRII; BD Biosciences, San Jose, CA), with modification of methods previously described (17, 23). Lungs of C57BL/6 mice were perfused with 10 ml cold Hanks’ balanced salt solution (Life Technologies, Waltham, MA) via the right ventricle through the pulmonary artery. Perfused left lungs were harvested and digested into single-cell suspensions using collagenase (Worthington, Lakewood, NJ) (24). The lung cells were suspended in PBS and incubated with vehicle, recombinant mouse IgG2a Fc (stoichiometrically matched to recombinant mouse Sectm1a-Fc), or recombinant mouse Sectm1a-Fc chimera protein (with Sectm1a fused to mouse IgG2a Fc; R&D Systems, Minneapolis, MN) for 1 hour on ice in the presence of purified rat anti-mouse CD16/CD32 monoclonal antibody (BD Pharmingen, San Jose, CA). After incubation, cells were washed and stained with phycoerythrin (PE)-conjugated rat anti-mouse IgG2a (R&D systems), followed by staining with multiple surface marker antibodies consisting of Pacific Blue–conjugated anti-mouse CD45, FITC-conjugated anti-mouse Ly6g lymphocyte antigen 6 complex, locus G (Ly6G), PE/Cy7–conjugated anti-mouse/human CD11b, and APC-conjugated anti-mouse CD19 (Biolegend, San Diego, CA) to differentiate leukocytes. Dead cells were excluded using 7-aminoactinomycin D (BD Pharmingen).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism (GraphPad Software Inc., La Jolla, CA). Data are presented as means (±SEM). Single-group comparisons were done using Student’s t test. Multiple group comparisons were made using either a one-way or two-way ANOVA followed by Sidak’s post hoc test.
Please see the online supplement for descriptions of additional materials and methods, including experimental reagent, antibodies, lung epithelial cell sorting, microarray analysis, gene set enrichment analysis (GSEA), quantitative RT-PCR, ELISA, Cell culture, STAT1 knockdown by small interfering RNA, immunoblot, Sectm1a binding assay for blood leukocytes, and ex vivo neutrophil stimulation.
Results
Genes Induced Selectively in Epithelial Cells during Pneumococcal Pneumonia
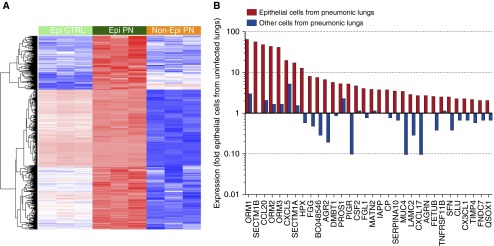
We have previously developed digestion and sorting strategies so that single-cell suspensions from the mouse lung can be separated into three distinct groups on the basis of relative expression of CD45 and CD326, also known as epithelial cell adhesion molecule (EpCAM), with epithelial cells identified as CD45−EpCAM+, leukocytes as CD45+, and other cell types being CD45−EpCAM− (11). To expand our understanding of epithelial-specific roles in pneumonia, we transcriptionally profiled sorted epithelial and nonepithelial cells using microarray to identify genes induced by pneumonia selectively in epithelial cells. Mice were instilled with pneumococcus, and left lungs were collected 15 hours after infection. Single-cell suspensions were generated and sorted into two separate populations, epithelial cells (CD45−EpCAM+) and others (all nonepithelial cells) (see Figure E1 in the online supplement). We also collected epithelial cells from the lungs of uninfected mice. Genome-wide expression profiling revealed 5,546 genes differentially expressed (false discovery rate [FDR] q < 0.05) in epithelial cells from pneumonic lungs compared with epithelial cells from uninfected lungs. To begin dissecting molecular and intercellular pathways that were coordinately regulated in epithelial cells due to pneumonia, we used GSEA to analyze all gene changes observed in epithelial cells due to pneumococcal pneumonia (25). GSEA indicated 94 gene sets being significantly coordinately up-regulated (FDR q < 0.05), confirming anticipated roles for some biological pathways, such as NF-κB and implicating diverse other pathways as well (Table 1 and Excel [Microsoft Corp., Redmond, WA] spreadsheet in the online supplement). After NF-κB, type I IFN signaling was the second most highly enriched gene set in epithelial cell responses to pneumococcal pneumonia (Table 1). Of the 5,546 genes induced by infection in the epithelial cells, 1,166 genes were significantly (FDR q < 0.05) more highly expressed in epithelial cells compared with nonepithelial cells from the same lungs (Figure 1A). The expression patterns in Figure 1A suggested that the majority of these genes may be constitutive epithelial products that were boosted by infection, as they tended to appear higher in epithelial cells from even noninfected lungs compared with nonepithelial cells from infected lungs. Approximately one-quarter of these transcripts (326 genes, upper cluster of Figure 1A) showed a different pattern, perhaps reflecting little epithelial-specific expression in the absence of infection, but strong induction during pneumonia. To interrogate our datasets for candidate genes relevant to signaling from epithelial cells to leukocytes, we filtered the 1,166 genes further to require a greater than twofold change in both directions (compared with uninfected epithelial cells and with infected nonepithelial cells) and used Database for Annotation, Visualization and Integrated Discovery (http://david.ncifcrf.gov) (26) to identify those genes that were annotated as encoding secreted proteins (SP_PIR_Secretion, FDR q < 0.044), retrieving a list of 32 genes (Figure 1B). The results confirmed expectations from prior studies, returning each of the three cytokines previously identified as epithelial in origin during pneumococcal pneumonia (CCL20, CXCL5, and CSF2), and none of the many cytokines previously identified as having prominent nonepithelial sources in the lung (such as CXCL1, CXCL2, and CSF3) (11). The regulation and function of the genes here identified as induced selectively in epithelial cells are of special interest. We chose to focus our first studies on Sectm1 proteins, because they are closely related and poorly understood proteins that represent two of the top seven signals in these analyses.
Table 1.
Gene Sets with Highest Enrichment of Up-Regulated Genes in Epithelial Cells
| Group | Gene Set Name | NES |
|---|---|---|
| Reactome pathway | ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS | 2.50 |
| Reactome pathway | INTERFERON_ALPHA_BETA_SIGNALING | 2.48 |
| Reactome pathway | INTERFERON_SIGNALING | 2.46 |
| Reactome pathway | ER_PHAGOSOME_PATHWAY | 2.45 |
| KEGG pathway | PROTEASOME | 2.42 |
| Reactome pathway | AUTODEGRADATION_OF_THE_E3_UBIQUITIN_LIGASE_COP1 | 2.39 |
| Reactome pathway | INTERFERON_GAMMA_SIGNALING | 2.36 |
| Reactome pathway | CDT1_ASSOCIATION_WITH_THE_CDC6_ORC_ORIGIN_COMPLEX | 2.36 |
| Reactome pathway | ANTIGEN_PROCESSING_CROSS_PRESENTATION | 2.34 |
| Reactome pathway | VIF_MEDIATED_DEGRADATION_OF_APOBEC3G | 2.33 |
| Reactome pathway | RP53_INDEPENDENT_G1_S_DNA_DAMAGE_CHECKPOINT | 2.33 |
| Reactome pathway | CYTOKINE_SIGNALING_IN_IMMUNE_SYSTEM | 2.33 |
| Reactome pathway | CDK_MEDIATED_PHOSPHORYLATION_AND_REMOVAL_OF_CDC6 | 2.32 |
| Reactome pathway | DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | 2.32 |
| GO term | INFLAMMATORY_RESPONSE | 2.30 |
Definition of abbreviations: GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; NES, Normalized Enrichment Score.
NES indicates skewness of each gene set with respect to pneumonia. All gene sets listed above have a false discovery rate q < 0.001.
Figure 1.
Genes induced by pneumonia selectively in epithelial cells. (A) Heatmap of 1,166 genes with significantly higher expression in epithelial cells from pneumonic lung compared with both other groups during pneumonia (false discovery rate [FDR] q < 0.05). Epi CTRL, epithelial cells from uninfected mouse lungs; Epi PN, epithelial cells from pneumonic mouse lungs; Non-Epi PN, nonepithelial cells from pneumonic mouse lungs (n = 3 mice per group, each column from an independent mouse). Red intensity represents the degree of higher level of expression, and blue intensity the degree of lower level of expression. (B) To identify secreted proteins induced especially in epithelial cells owing to pneumonia, the genes with significant (FDR q < 0.05) and strong (>twofold) up-regulation in epithelial cells from mice with pneumonia compared with both other groups (epithelial cells from uninfected mice and nonepithelial cells from pneumonic mice) were probed using Database for Annotation, Visualization and Integrated Discovery Bioinformatics. A total of 32 genes were identified as secreted proteins based on the SP_PIR_Secretion annotation. Expression of each transcript in the indicated cell population is shown normalized to the relative expression of that transcript in epithelial cells sorted from uninfected mouse lungs.
Sectm1 Induction during Pneumococcal Pneumonia
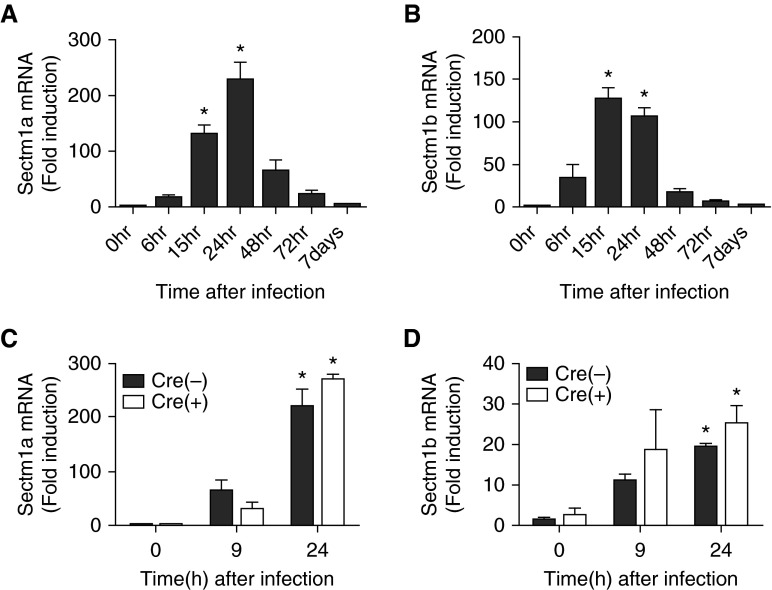
To verify that Sectm1 genes were indeed induced during pneumococcal pneumonia, we performed additional analyses of left-lung mRNA throughout a time course of infection. Both Sectm1a and Sectm1b transcripts were induced (Figures 2A and 2B). Expression of each transcript peaked 1 day after infection, and thereafter declined (Figures 2A and 2B). To investigate Sectm1 induction in epithelial cells specifically, mRNA was collected from CD45−EpCAM+ cells after 0, 9, or 24 hours of infection. In addition, results were compared between WT mice and those lacking functional RelA throughout the pulmonary epithelium as an assessment of potential activation mechanism. Sectm1a and Sectm1b were equivalently induced in sorted WT and RelA-deficient epithelial cells (Figures 2C and 2D). This contrasts with the three epithelial-specific cytokines that we previously studied, CXCL5, CSF2 (GM-CSF), and CCL20, all of which required NF-κB RelA in epithelial cells (11), as measured in the same samples here analyzed for Sectm1. Thus, pneumococcal pneumonia can induce Sectm1 transcription in epithelial cells by mechanisms other than RelA.
Figure 2.
Pneumonia can induce secreted and transmembrane (Sectm) 1 transcription in epithelial cells by mechanisms other than RelA. (A and B) C57BL/6 mice were infected with Streptococcus pneumoniae serotype 19F. mRNAs were measured in left-lung homogenates using quantitative RT-PCR (qRT-PCR) at indicated time points. Values represent fold induction relative to 0 hours and were expressed as means (±SEM). Significance was determined by one-way ANOVA with Sidak’s post hoc test (n = 4–8 per group). *P < 0.05 versus 0 hours. (C and D) NK2 homeobox 1–Cre Rela-floxed mice in which RelA was mutated throughout the lung epithelium, or wild-type (WT) control littermates with no Cre expression, were infected with pneumococcus and killed 9 or 24 hours after infection, after which left-lung epithelial cells, identified as CD45−EpCAM+, were isolated by FACS. Values represent fold induction relative to epithelial cells isolated from 0-hour Cre-negative mice and were expressed as mean (±SEM). Significance was determined by two-way ANOVA with Sidak’s post hoc test (n = 3–4 per group). *P < 0.05 versus 0 hour.
Type I IFN Signaling Mediates Sectm1 Induction
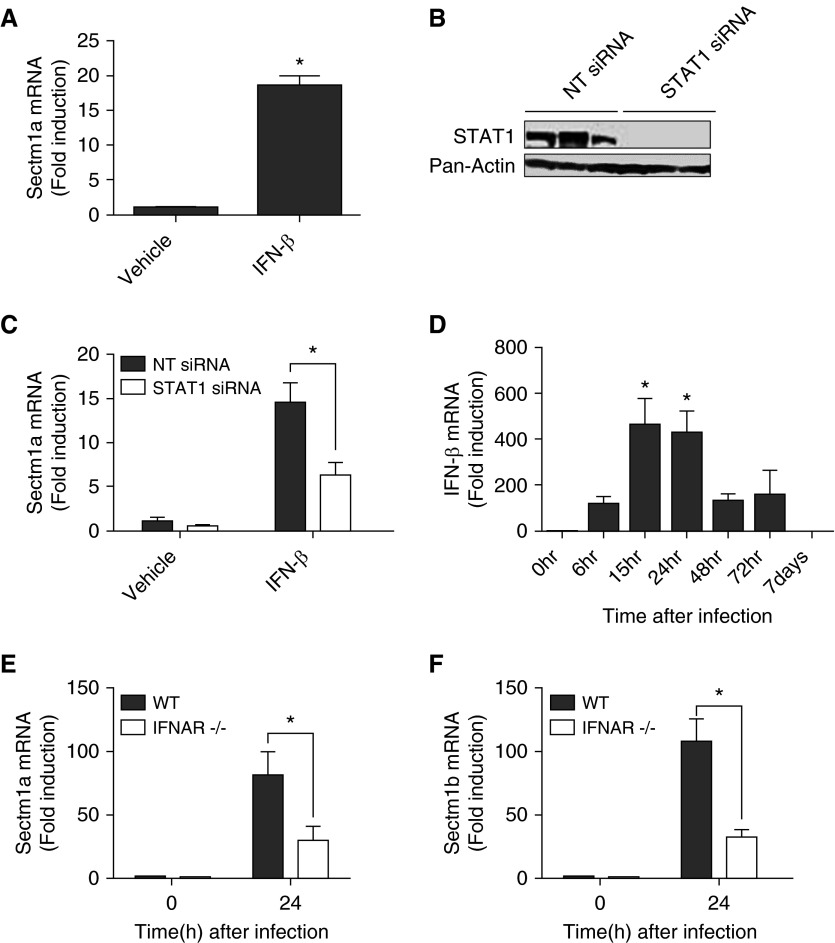
To gain a first insight into potential transcriptional signaling pathways that may mediate RelA-independent expression of Sectm1 in epithelial cells during pneumonia, we used the Gene2Promoter and Frameworker programs from the Genomatix software suite (Munich, Germany). In so doing, we identified transcription factor binding site modules that are present in the Sectm1a promoter and are also significantly overrepresented across the promoters of genes induced at least 10-fold in epithelial cells during pneumonia. The IFN regulatory factor family, including binding sites for 12 related transcription factors (IFN regulatory factors 1–9, Ring finger protein 31, Stat1, and Stat2), was the most significant (P = 3 × 10−12). This family of transcription factors is activated by type I IFNs and by cytoplasmic DNA sensing (27, 28), each of which was independently identified by GSEA as responses in epithelial cells during pneumonia (Table 1 and Excel spreadsheet in the online supplement). Using alveolar epithelial cell–like E10 cells, we observed that Sectm1a was induced by type I IFN signaling (Figure 3A). Type I IFNs activate multiple transcriptional regulatory pathways, most, but not all, of which involve STAT1 (29, 30). We used small interfering RNA to effectively knock down STAT1 in E10 cells (Figure 3B). Sectm1a induction by IFN-β was significantly decreased by STAT1 knockdown (Figure 3C), demonstrating a role for this specific transcription factor in epithelial expression of Sectm1a. During pneumococcal pneumonia, IFN-β was expressed in the lungs at times relevant to Sectm1 induction (Figure 3D). Signaling from all type I IFNs, including multiple IFN-α proteins as well as IFN-β, requires a common IFNAR (29). We infected IFNAR-deficient mice with pneumococcus and quantified Sectm1 mRNA expression. The induction of Sectm1a and Sectm1b by pneumococcal pneumonia was significantly diminished in mice lacking IFNAR (Figures 3E and 3F). Taken together, these data suggest that Sectm1 induction during pneumococcal pneumonia occurs in epithelial cells specifically, and is induced by type I IFN signaling through the IFNAR and likely using the STAT1 transcription factor.
Figure 3.
Type I IFN signaling mediates Sectm1 induction through signal transducer and activator of transcription (STAT) 1. (A) Alveolar epithelial cell–like E10 cells were stimulated with IFN-β (1,000 U/ml) for 6 hours. Expression of Sectm1a was measured using qRT-PCR. Values represent fold induction relative to vehicle group and were expressed as means (±SEM). Significance was determined by Student’s t test. Results reflect data from three independent experiments. *P < 0.05 versus vehicle. (B) Immunoblot analysis of E10 cells 48 hours after transfection with either nontargeting (NT) or STAT1 small interfering RNA (siRNA). (C) qRT-PCR of Sectm1a expression in siRNA transfected cells treated with vehicle or IFN-β (1,000 U/ml) for 6 hours. Values represent fold induction compared with nontargeting and vehicle-treated group and were expressed as means (±SEM). Significance was determined by two-way ANOVA with Sidak’s post hoc test. Results reflect data from three independent experiments. *P < 0.05 versus nontargeting siRNA. (D) C57BL/6 mice were infected with S. pneumoniae serotype 19F. Expressions of IFN-β were measured in left-lung homogenates using qRT-PCR at indicated time points. Values represent fold induction relative to 0 hours and are expressed as means (±SEM). Significance was determined by one-way ANOVA with Sidak’s post hoc test (n = 4–8 per group). *P < 0.05 versus 0 hour. (E and F) WT and IFN-α/β receptor (IFNAR)-deficient mice were infected with S. pneumoniae serotype 19F and killed 24 hours after infection. mRNAs were measured in lung homogenates using qRT-PCR. Values represent fold induction relative to 0 hour, WT, and are expressed as means (±SEM). Significance was determined by two-way ANOVA with Sidak’s post hoc test (n = 4–5 in each experimental group). *P < 0.05 versus WT.
Conducting Airway Epithelial Cell Sources of Sectm1 during Pneumococcal Pneumonia
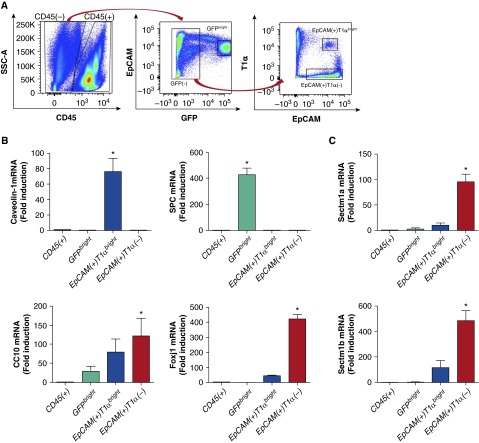
To discriminate Sectm1 expression among different types of pulmonary epithelial cells, we refined sorting strategies to distinguish subsets of cells within the CD45−EpCAM+ population (Figure 4A). The combination of EpCAM, T1α (also known as podoplanin), and an SPC-driven GFP transgene allowed us to differentiate lung epithelial cell populations into sets of GFPbright cells, CD45−GFP−EpCAM+T1αbright cells, and CD45−GFP−EpCAM+T1α− cells (Figure 4A). Using mRNAs for SPC, caveolin-1, and Forkhead box protein J1 (Foxj1) as indicative of the presence of type II cells, type I cells, and ciliated cells, respectively, we conclude that these three cell populations were effectively separated from each other using this sorting strategy (Figure 4B). CC10, a marker of club cells, was most abundantly expressed in CD45−GFP−EpCAM+T1α− cells, where the ciliated cells were also found, but was also expressed in both GFPbright cells and CD45−GFP−EpCAM+T1αbright cells (Figure 4B). This may be because club cells get collected in multiple sorted cell populations. Alternatively, this may result from cells other than club cells expressing CC10, such as cells expressing both CC10 and SPC that exist in bronchoalveolar duct junctions or alveolar regions (31–33). We measured Sectm1 transcripts across these four sorted cell populations, which revealed that Sectm1 transcripts were predominantly induced in the CD45−GFP−EpCAM+T1α− group containing both types of conducting airway epithelial cells (ciliated cells and club cells), but neither type I nor type II alveolar epithelial cells (Figure 4C). Induction was observed in the CD45−GFP−EpCAM+T1αbright cells as well, consistent with expression by type I cells, but this was modest in comparison to the CD45−GFP−EpCAM+T1α− cells (Figure 4C). Therefore, we conclude that conducting airway epithelial cells are predominant sites of Sectm1 induction during pneumococcal pneumonia.
Figure 4.
Conducting airway epithelial cells produce Sectm1 during pneumonia. (A and B) Surfactant protein (SP) C–green fluorescent protein (GFP) transgenic mice were infected with S. pneumoniae serotype 19F. Left lungs were harvested 24 hours after infection and digested to single-cell suspensions with elastase. Four cell populations consisting of CD45+, CD45−GFPbright, CD45−GFP−EpCAM+T1αbright, and CD45−GFP−EpCAM+T1α− were sorted by flow cytometry. Expression of SPC, caveolin-1, CC10, and Forkhead box protein J1 (Foxj1) were measured across these four cell populations using qRT-PCR. Values represent fold induction relative to CD45+ cells and are expressed as means (±SEM). Significance was determined by one-way ANOVA with Sidak’s post hoc test (n = 4 in each group). *P < 0.05 versus CD45+. (C) Sectm1 transcripts were measured using qRT-PCR across the four cell populations. Values represent fold induction relative to CD45+ cells and are expressed as means (±SEM). Significance was determined by one-way ANOVA with Sidak’s post hoc test (n = 4 in each group). *P < 0.05 versus CD45+. EpCAM, epithelial cell adhesion molecule; SSC, side scatter.
Sectm1a Interacts with Myeloid Cells in the Pneumonic Lung
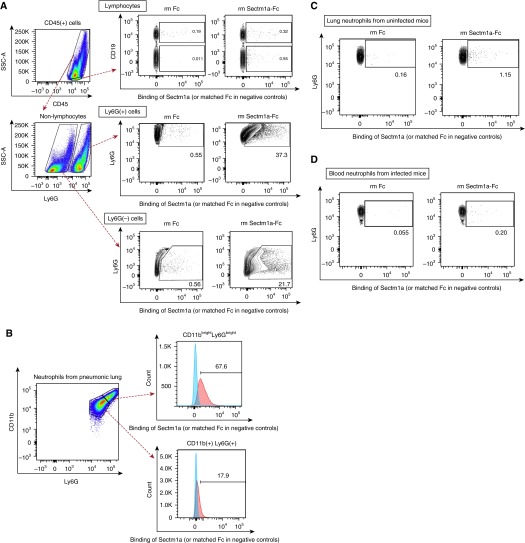
In an effort to identify cells potentially responsive to Sectm1a, we investigated cell-specific binding capacity. Lacking an antibody that recognized mouse Sectm1a, a recombinant Sectm1a-Fc fusion protein was used to identify Sectm1a-binding cells with flow cytometry. The Fc portion from mouse IgG2a, attached to mouse Sectm1a, was detected using a secondary PE-conjugated anti-mouse IgG2a antibody. By applying this method to single-cell suspensions from lungs of mice with pneumococcal pneumonia, we found that Sectm1a mainly interacts with myeloid cells, including both Ly6G-positive cells, considered to be neutrophils, and Ly6G-negative cells that likely included monocytes, macrophages, and dendritic cells (Figure 5A). The negative control, recombinant murine Fc protein without the Sectm1a fusion, consistently failed to bind these cells (Figure 5). To determine the binding of Sectm1a to lymphocytes more precisely, we differentiated T cells (CD45+CD3+), NK cells (CD45+CD3−NK1.1+), and B cells (CD45+CD19+) individually in independent experiments (Figure E2A), and found that Sectm1a could bind to T and NK cells (Figures E2B–E2D), but only a small fraction compared with myeloid cell binding. Intriguingly, the Sectm1a appeared to preferentially bind to the neutrophils that were brightest for Ly6G and CD11b in the pneumonic lung (Figure 5B), and neutrophils from uninfected lungs did not bind Sectm1a (Figure 5C). Furthermore, circulating blood neutrophils collected from either uninfected or infected mice also failed to bind Sectm1a (data not shown and Figure 5D). These data suggest that neutrophils become activated in the lung during pneumonia, and consequently acquire the ability to bind Sectm1a. Altogether, these data suggest that Sectm1a is elaborated by epithelial cells in the infected lung and interacts with highly activated neutrophils recruited there.
Figure 5.
Sectm1a interacts with myeloid cells in the pneumonic lung. (A) C57BL/6 mice were infected with S. pneumoniae serotype 19F, and left lungs were harvested 24 hours after infection. Single-cell suspensions from collagenase-digested left lung were incubated with vehicle (data not shown), recombinant Fc (25 μg/ml, stoichiometrically matched to recombinant mouse Sectm1a-Fc), or recombinant Sectm1a-Fc (40 μg/ml). The cells were washed and stained with phycoerythrin (PE)-conjugated anti-mouse IgG2a, followed by staining with anti-mouse CD45, Ly6G, CD11b, and CD19 antibodies. Binding of Sectm1a to cells was detected as PE signal. Results are representative of three independent experiments. Gates were determined based on cells incubated with vehicle (data not shown). (B) Neutrophils (CD45+Ly6G+) of the pneumonic lungs were further divided into two subpopulations based on the relative expression of CD11b and Ly6G, and binding of Sectm1a was determined as in (A). Cells represented in blue or red histograms reflect those incubated with Fc or Sectm1a-Fc, respectively. (C) Uninfected lungs were harvested and Sectm1a binding assay was performed using the same doses of Fc (25 μg/ml) and Sectm1a-Fc (40 μg/ml) as in (A) to determine the binding of Sectm1a-Fc to neutrophils (CD45+Ly6G+). Gates were determined based on vehicle group (data not shown). Results are representative of three independent experiments. (D) Blood was collected 20 hours after intrabronchial instillation of S. pneumoniae serotype 19F. Sectm1a binding assay was performed using the same doses of Fc (25 μg/ml) and Sectm1a-Fc (40 μg/ml) to determine the binding of Sectm1a-Fc to blood neutrophils (CD45+Ly6G+). Gates were determined based on vehicle group (data not shown). Results are representative of three independent experiments. Ly6G, Ly6g lymphocyte antigen 6 complex, locus G; rm, recombinant murine.
Sectm1a Promotes CXCL2 Production by Neutrophils from the Pneumonic Lung
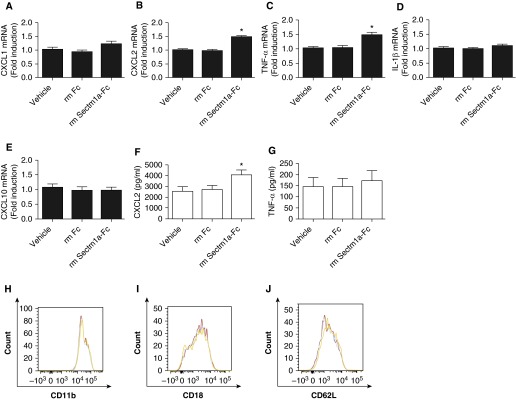
Based on preferential binding of Sectm1a to neutrophils from infected, but not uninfected, lungs, we endeavored to test the immunological influence of Sectm1a on highly activated neutrophils. We collected neutrophils from bronchoalveolar lavage (BAL) fluids of infected mice 20 hours after infection. BAL neutrophils were cultured with Sectm1a-Fc or negative controls (recombinant mouse Fc or vehicle). Transcript levels were measured by quantitative RT-PCR for cytokines including CXCL1, CXCL2, TNF-α, IL1-β, and CXCL10. Sectm1a significantly increased expression of CXCL2 and TNF-α, but not the other cytokines measured (Figures 6A–6E). CXCL2 protein levels were consistent with the mRNA data, indicating a significant increase in CXCL2 protein, but not in TNF-α protein from BAL neutrophils stimulated with Sectm1a (Figures 6F and 6G). Surface markers associated with neutrophil activation, including CD11b, CD18, and CD62L, were not further altered by Sectm1a stimulation (Figures 6H–6J). Collectively, these data suggest that epithelial-derived Sectm1a engages neutrophils to promote the expression and release of proinflammatory mediators, including CXCL2.
Figure 6.
Sectm1a promotes chemokine (C-X-C) motif ligand (CXCL) 2 production by neutrophils from the pneumonic lung. C57BL/6 mice were infected with S. pneumoniae serotype 19F, and bronchoalveolar lavage fluid was collected 20 hours after infection, from which neutrophils were isolated. Isolated neutrophils were cultured and stimulated with vehicle, Fc (39 μg/ml, stoichiometrically matched to recombinant mouse Sectm1a-Fc), or Sectm1a-Fc (60 μg/ml) for 2 hours (A–E), 8 hours (F and G), or 4 hours (H–J). (A–E) qRT-PCR was performed on total cellular RNA to determine the gene expressions of CXCL1, CXCL2, TNF-α, IL1-β, and CXCL10. Values represent fold induction relative to vehicle group and were expressed as means (±SEM). Significance was determined by one-way ANOVA with Sidak’s post hoc test (n = 9 per group). *P < 0.05 versus vehicle. (F and G) CXCL2 and TNF-α levels were measured by ELISA from culture supernatants of neutrophils. Values represent fold induction relative to vehicle and were expressed as means (±SEM). Significance was determined by one-way ANOVA with Sidak’s post hoc test (n = 9 per group). *P < 0.05 versus vehicle. (H–J) After stimulation, neutrophils were washed and stained with anti-mouse CD45, Ly6G, CD11b, CD18, and CD62L antibodies. The expression of surface markers, including CD11b, CD18, and CD62L, was determined in each group using flow cytometry. Cells represented in red, blue, or orange histograms reflect those stimulated with vehicle, Fc, or Sectm1a-Fc, respectively. Results are representative of four samples.
In Vivo Effect of Recombinant Sectm1a-Fc on Pneumococcal Pneumonia
In addition to ex vivo experiments, we examined the in vivo effect of exogenous Sectm1a on recruitment of leukocytes into the airspace and bacterial clearance during pneumococcal pneumonia. Sectm1a-Fc or negative controls (recombinant mouse Fc or vehicle) were coinstilled with pneumococcus. Sectm1a did not significantly change these outcomes compared with negative control groups (Table 2). These results suggest that the amount Sectm1 expressed during pneumonia was not present in limiting concentrations, that the timing of exogenous Sectm1 administration (with the initial bacterial infection, before any neutrophil recruitment) may be inadequate for observing Sectm1 activities, that Sectm1 might not contribute to the outcomes measured, or that the outcomes of interest may have already been at maximal levels.
Table 2.
In Vivo Effect of Recombinant Murine Secreted and Transmembrane 1a–Fc on Pneumococcal Pneumonia
| Vehicle | rm Fc | rm Sectm1a-Fc | |
|---|---|---|---|
| Cell counts (×106) in BALF from pneumonic lung | |||
| Total cells | 1.57 ± 0.03 | 1.12 ± 0.19 | 1.41 ± 0.20 |
| Macrophages and monocytes | 0.33 ± 0.02 | 0.28 ± 0.02 | 0.36 ± 0.04 |
| Neutrophils | 1.24 ± 0.04 | 0.84 ± 0.16 | 1.06 ± 0.17 |
| CFU (×106) in pneumonic lung | 1.17 ± 1.12 | 0.20 ± 0.16 | 0.52 ± 0.32 |
Definition of abbreviations: BALF, bronchoalveolar lavage fluid; rm, recombinant murine; Sectm1a, secreted and transmembrane 1a.
C57BL/6 mice were coinstilled with vehicle, rm Fc (50 μg/ml), or rm Sectm1a-Fc (80 μg/ml) into left lungs in addition to Streptococcus pneumoniae serotype 19F. BALF was collected 20 hours after coinstillation, and total and differential cell counts were determined. Bilateral lungs were harvested at 24 hours after coinstillation, and bacterial burden was measured in whole-lung homogenates using CFU analysis. There were no statistically significant differences among three experimental groups in both experiments. Values are expressed as means (±SEM). Significance was determined by one-way ANOVA with Sidak’s post hoc test (n = 3–8 per group and 6–7 per group, respectively).
Discussion
In this study, we sought to identify epithelial-specific immunomodulators during pneumonia. Our genome-wide expression profiling revealed that more than 5,000 genes are differentially expressed in epithelial cells after infection, indicating that these cells undergo diverse and considerable transcriptional remodeling during pneumococcal pneumonia. We have extensively studied the role of NF-κB RelA in pulmonary epithelial cells (8, 10, 11), and NF-κB was prominently reflected in these transcriptional responses of epithelial cells. In addition, GSEA identified IFN signaling, particularly from type I IFNs (IFN-α and -β), as the next most up-regulated gene set in epithelial cells due to pneumococcal pneumonia. A fourth of the genes induced in epithelial cells were expressed at higher levels compared with other cells from the same infected lungs. A limitation inherent to these analyses is that the pool of all nonepithelial cells was compared with the epithelial cells. It is conceivable that a subset within the nonepithelial cell population (such as neutrophils alone or smooth muscle cells alone) may have a signal that is diluted by the RNA from other cell types not expressing that gene, and hence expression by subsets of nonepithelial cells may be underestimated. Another limitation is that only lung cells were examined, and some of these genes may also be expressed by cells outside of the lungs. For instance, orosomucoids (ORMs; ORM1, ORM2, ORM3), also known α1-acid glycoproteins, are immunomodulatory proteins (34, 35) that were some of the most highly induced epithelial-specific products in the infected lung in the present studies. ORMs are better known as acute-phase proteins that are synthesized by hepatocytes in the liver, including during pneumococcal pneumonia (36). Although ORMs and other proteins may be induced outside the lungs also, we interpret these data as suggesting that local expression within the lung is epithelial cell predominant for several hundred genes. The set of 32 genes that we identified as pneumococcal pneumonia–induced secreted products from epithelial cells likely represents an underestimation. However, perfect matching, to our knowledge base of whether cytokines derive from epithelial or nonepithelial cells during pneumococcal pneumonia (11), provides confidence in ascribing these as epithelial-specific secreted proteins. We selected the Sectm1 proteins as deserving of further study, because they were such prominent signals and are so poorly understood, but other genes identified in Figure 1B as pneumococcal pneumonia–induced secreted products from epithelial cells demand further investigation as potential regulators of lung immunity.
To our knowledge, this is the first study to report connections of Sectm1 with pulmonary infection. There is limited prior knowledge about Sectm1 and disease, other than an association with melanoma (16). During pneumococcal pneumonia, Sectm1 transcripts were induced during early stages of amplifying inflammation. Mouse Sectm1a and Sectm1b were reported to be similar proteins in terms of their chromosomal location and amino acid sequences (13, 14). Here, we find that they are similarly regulated in the inflamed lung. Sectm1 transcripts were induced by a RelA-independent mechanism, which was unexpected. NF-κB RelA is a central transcription factor that regulates many inflammatory and immune-modulatory reactions during acute bacterial infection (8, 37). All three cytokines previously identified as epithelial cell derived during pneumonia (CCL20, CXCL5, and GM-CSF) require NF-κB RelA for their induction (11). In contrast to these, our data suggest that induction of both Sectm1a and Sectm1b is stimulated by type I IFNs and mediated by IFNAR and STAT1.
Type I IFNs likely have multiple influences on host defense against bacterial infection (38), with functions that are more complex and heterogeneous than those observed in the setting of viral infection (38, 39). In postinfluenza pneumococcal pneumonia, type I IFNs are deleterious for impairing production of neutrophil chemoattractants, including CXCL1 and CXCL2 (19). On the other hand, in primary pneumococcal pneumonia, type I IFNs can enhance host defense, decrease bacterial burdens in the lungs and blood, and improve survival rates (40, 41). Differential roles of type I IFNs during pneumococcal pneumonia may vary according to characteristics of the host (including genetic and environmental variations in immune pathways) and the microbe (including virulence factors), and demand further investigation. Sectm1 is induced by type I IFNs during pneumococcal pneumonia, and may contribute to some of the phenotypes mediated by IFNAR signaling.
The current results indicate that type I IFNs contribute to Sectm1 expression. However, these are probably not the only inflammatory mediators driving Sectm1 expression during pneumonia. Both Sectm1a and Sectm1b were significantly reduced in IFNAR-deficient mice with pneumococcal pneumonia, but some residual induction of each remained. It has been reported that human Sectm1 can be induced by IFN-γ (15), and IFN-γ is increased in the lung during S. pneumoniae infection (42); thus, IFN-γ may be an additional contributor to Sectm1 induction in the lungs. We observed here that Sectm1 induction by IFN-β is mediated by the STAT1 transcription factor, which also mediates signaling from IFN-γ (29), so other STAT1-activating cytokines that are expressed during pneumonia may be additional activators of Sectm1 in the infected lung. The STAT1 knockdown was performed in E10 cells, which resemble type I alveolar epithelial cells, so asserting that STAT1 mediates induction in columnar epithelial cells and in the lungs is based on extrapolation. Roles of IFNAR were determined in whole lungs in vivo. The present data suggest that the majority, but not all, of the Sectm1 induced during pneumonia depends on type I IFN signaling through IFNAR.
Our results indicate that Sectm1a exhibits greater binding to myeloid cells than to lymphocytes. Prior reports identified an interaction between Sectm1 and T cells or NK cells (13–15), which was detectable with such cells from the pneumonic lung, but only with a modest signal. In in vitro studies, human Sectm1 and mouse Sectm1a costimulate T cells, inducing their proliferation as well as cytokine production (14, 15), and human Sectm1 increases expression of activation markers of NK cells, such as CD69 (13). Our results do not disagree with the prior studies of Sectm1 interactions with lymphocytes, as our assay system (relying on stable binding of recombinant tagged Sectm1 to the cell surface, followed by detection of the recombinant fusion protein with a fluorescent anti-tag antibody) was very different from assays in prior studies. For example, this seeming disparity may be explained if Sectm1 interacts with receptors on neutrophil surfaces with greater avidity than with the receptors on T cell or NKT cell surfaces. To the best of our knowledge, this is the first report that Sectm1a binds to neutrophils, and, using this assay, this interaction was more prominent than its binding to lymphocytes. Interestingly, it appears that Sectm1a preferentially binds to subsets of neutrophils. Sectm1a does not bind to neutrophils from uninfected lungs or from the blood of either infected or uninfected mice, but it does bind to some of the neutrophils within pneumonic lungs, notably those with the greatest expression of both CD11b and Ly6G. Although neutrophils are defined by common elements (such as their distinctive-shaped nucleus and their characteristic granules), it is becoming increasingly clear that this is a remarkably heterogeneous population of cells, even at a healthy baseline (43). Changes in neutrophils during the maturation stages combined with their brief lifespans add to this complexity. Furthermore, acute and chronic inflammatory stimuli additionally alter neutrophil phenotypes. Generally, surface levels of CD11b and of Ly6G are increased in the most mature neutrophils and with activation by proinflammatory stimuli (43–46). The facts that Sectm1a binds to CD11bbrightLy6Gbright neutrophils from an inflammatory site, but not neutrophils from other sites or times, suggest to us that neutrophils must be both mature and activated to interact with this ligand. The neutrophil receptor(s) for Sectm1a are unknown. Human and mouse Sectm1 proteins interact with T cells via multiple receptors, including CD7 and GITR, but other Sectm1 receptors on T cells are implicated as well, and are yet to be identified (13, 14). We find that neutrophils collected from infected mouse lungs express vanishingly low levels of CD7 and GITR, far less than mouse T cells (data not shown), making these unlikely candidates for Sectm1 receptors on activated neutrophils.
How Sectm1 alters immune and inflammatory processes within the lung is now proposed as an important new question, which only begins to be addressed here. Sectm1a enhanced CXCL2 production by neutrophils from the pneumonic lung. CXCL2 is a potent inducer of neutrophil chemotaxis (47), and it plays critical roles in neutrophil recruitment during lung infections (48, 49). Neutrophil-derived CXCL2 is central to a positive-feedback loop for neutrophil recruitment in the infected lung (50). Taken together, these data suggest that epithelial-derived Sectm1a is proinflammatory by amplifying a neutrophil-driven positive-feedback loop in the pneumonic lung. If Sectm1 is most highly expressed by columnar epithelial cells, such as club cells or ciliated cells, then this effect on neutrophil recruitment may be most prominent in the conducting airways. However, a major limitation of this study is the inability to inhibit Sectm1 function in vivo, due to the absence of effective tools at present. To our knowledge, reports of mice deficient in Sectm1a and/or Sectm1b have yet to be published, nor have reports of blocking antibodies against mouse Sectm1a and/or Sectm1b. Future studies in which Sectm1a and Sectm1b can be interrupted in vivo, independently as well as concurrently, will be essential to assessing the full significance of this newly recognized pathway in lung immunity.
In conclusion, a genome-wide approach demonstrates that expression of many genes is altered in the epithelium in response to pneumococcal infection, which involves diverse pathways, including some that had not previously been implicated in pneumonia. Sectm1a is a largely uncharacterized epithelial product that is induced by type I IFNs, mediated by STAT1. Sectm1a interacts with neutrophils in the pneumonic lung, enhancing their production of CXCL2, which we propose feeds into a positive-feedback loop amplifying neutrophilic inflammation. The full functional significance of Sectm1a in pneumococcal pneumonia is an important focus for further studies.
Supplementary Material
Acknowledgments
Acknowledgments
The authors thank Patrick Autissier (Flow Cytometry Core Facility, Boston University School of Medicine, Boston, MA) for outstanding technical help with these studies.
Footnotes
This work was supported by National Institutes of Health grants R01 HL0068153 (J.P.M.), R01 HL 079392 (J.P.M.), R00 HL 092956 (L.J.Q.), R01 HL 111449 (L.J.Q.), R01 HL 104053 (M.R.J.), T32 AI 899673 (G.A.W.), UL1-TR001012 (A.C.G), and by an American Lung Association Senior Research Fellowship (K.Y.)
Author Contributions: All authors participated in the design and interpretation of experiments and review of the manuscript; H.K. led the performance of all experiments, drafted the manuscript, and performed the statistical analyses; J.P.M. supervised the study, managed the research team, and helped draft the manuscript; K.Y. contributed to the genome-wide transcriptional profiling; G.A.W. contributed to the in vitro experiments; M.C.Z. assisted with many of the in vivo experiments; M.I.R. contributed to type II cell transgenic mouse experiments; A.C.G. contributed to analyses of microarray data; A.C.B. contributed to flow cytometry analyses; C.K.Y., W.Y.L., and J.C.D. contributed to IFN-α/β receptor–deficient mouse experiments; L.J.Q. and M.R.J. contributed to experimental design, data analysis, and scientific interpretation.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2015-0261OC on April 11, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Vos T, Flaxman AD, Naghavi M, Lozano R, Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, Aboyans V, et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2163–2196. doi: 10.1016/S0140-6736(12)61729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer TT, Ewig S, Rodloff AC, Müller EE. Acute respiratory distress syndrome and pneumonia: a comprehensive review of clinical data. Clin Infect Dis. 2006;43:748–756. doi: 10.1086/506430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong GL, Conn LA, Pinner RW. Trends in infectious disease mortality in the United States during the 20th century. JAMA. 1999;281:61–66. doi: 10.1001/jama.281.1.61. [DOI] [PubMed] [Google Scholar]
- 4.van der Poll T, Opal SM. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet. 2009;374:1543–1556. doi: 10.1016/S0140-6736(09)61114-4. [DOI] [PubMed] [Google Scholar]
- 5.Jain S, Self WH, Wunderink RG, Fakhran S, Balk R, Bramley AM, Reed C, Grijalva CG, Anderson EJ, Courtney DM, et al. CDC EPIC Study Team. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015;373:415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleaver JO, You D, Michaud DR, Pruneda FA, Juarez MM, Zhang J, Weill PM, Adachi R, Gong L, Moghaddam SJ, et al. Lung epithelial cells are essential effectors of inducible resistance to pneumonia. Mucosal Immunol. 2014;7:78–88. doi: 10.1038/mi.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker D, Prince A. Innate immunity in the respiratory epithelium. Am J Respir Cell Mol Biol. 2011;45:189–201. doi: 10.1165/rcmb.2011-0011RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinton LJ, Jones MR, Simms BT, Kogan MS, Robson BE, Skerrett SJ, Mizgerd JP. Functions and regulation of NF-κB RelA during pneumococcal pneumonia. J Immunol. 2007;178:1896–1903. doi: 10.4049/jimmunol.178.3.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinton LJ, Jones MR, Robson BE, Simms BT, Whitsett JA, Mizgerd JP. Alveolar epithelial STAT3, IL-6 family cytokines, and host defense during Escherichia coli pneumonia. Am J Respir Cell Mol Biol. 2008;38:699–706. doi: 10.1165/rcmb.2007-0365OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamoto K, Ferrari JD, Cao Y, Ramirez MI, Jones MR, Quinton LJ, Mizgerd JP. Type I alveolar epithelial cells mount innate immune responses during pneumococcal pneumonia. J Immunol. 2012;189:2450–2459. doi: 10.4049/jimmunol.1200634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto K, Ahyi AN, Pepper-Cunningham ZA, Ferrari JD, Wilson AA, Jones MR, Quinton LJ, Mizgerd JP. Roles of lung epithelium in neutrophil recruitment during pneumococcal pneumonia. Am J Respir Cell Mol Biol. 2014;50:253–262. doi: 10.1165/rcmb.2013-0114OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slentz-Kesler KA, Hale LP, Kaufman RE. Identification and characterization of K12 (SECTM1), a novel human gene that encodes a Golgi-associated protein with transmembrane and secreted isoforms. Genomics. 1998;47:327–340. doi: 10.1006/geno.1997.5151. [DOI] [PubMed] [Google Scholar]
- 13.Lyman SD, Escobar S, Rousseau AM, Armstrong A, Fanslow WC. Identification of CD7 as a cognate of the human K12 (SECTM1) protein. J Biol Chem. 2000;275:3431–3437. doi: 10.1074/jbc.275.5.3431. [DOI] [PubMed] [Google Scholar]
- 14.Howie D, Garcia Rueda H, Brown MH, Waldmann H. Secreted and transmembrane 1A is a novel co-stimulatory ligand. PLoS One. 2013;8:e73610. doi: 10.1371/journal.pone.0073610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang T, Huang C, Lopez-Coral A, Slentz-Kesler KA, Xiao M, Wherry EJ, Kaufman RE. K12/SECTM1, an interferon-γ regulated molecule, synergizes with CD28 to costimulate human T cell proliferation. J Leukoc Biol. 2012;91:449–459. doi: 10.1189/jlb.1011498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang T, Ge Y, Xiao M, Lopez-Coral A, Li L, Roesch A, Huang C, Alexander P, Vogt T, Xu X, et al. SECTM1 produced by tumor cells attracts human monocytes via CD7-mediated activation of the PI3K pathway. J Invest Dermatol. 2014;134:1108–1118. doi: 10.1038/jid.2013.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bade-Doding C, Gottmann W, Baigger A, Farren M, Lee KP, Blasczyk R, Huyton T. Autocrine GM-CSF transcription in the leukemic progenitor cell line KG1a is mediated by the transcription factor ETS1 and is negatively regulated through SECTM1 mediated ligation of CD7. Biochim Biophys Acta. 2014;1840:1004–1013. doi: 10.1016/j.bbagen.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 18.Lo B, Hansen S, Evans K, Heath JK, Wright JR. Alveolar epithelial type II cells induce T cell tolerance to specific antigen. J Immunol. 2008;180:881–888. doi: 10.4049/jimmunol.180.2.881. [DOI] [PubMed] [Google Scholar]
- 19.Shahangian A, Chow EK, Tian X, Kang JR, Ghaffari A, Liu SY, Belperio JA, Cheng G, Deng JC. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest. 2009;119:1910–1920. doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis KP, Yu J, Bellinger-Kawahara C, Joh D, Hawkinson MJ, Xiao G, Purchio TF, Caparon MG, Lipsitch M, Contag PR. Visualizing pneumococcal infections in the lungs of live mice using bioluminescent Streptococcus pneumoniae transformed with a novel gram-positive lux transposon. Infect Immun. 2001;69:3350–3358. doi: 10.1128/IAI.69.5.3350-3358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones MR, Quinton LJ, Simms BT, Lupa MM, Kogan MS, Mizgerd JP. Roles of interleukin-6 in activation of STAT proteins and recruitment of neutrophils during Escherichia coli pneumonia. J Infect Dis. 2006;193:360–369. doi: 10.1086/499312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Briles DE, Hollingshead SK, Paton JC, Ades EW, Novak L, van Ginkel FW, Benjamin WH., Jr Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J Infect Dis. 2003;188:339–348. doi: 10.1086/376571. [DOI] [PubMed] [Google Scholar]
- 23.Lefort CT, Rossaint J, Moser M, Petrich BG, Zarbock A, Monkley SJ, Critchley DR, Ginsberg MH, Fässler R, Ley K. Distinct roles for talin-1 and kindlin-3 in LFA-1 extension and affinity regulation. Blood. 2012;119:4275–4282. doi: 10.1182/blood-2011-08-373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauer KA, Scholtes P, Karwot R, Finotto S. Isolation of CD4+ T cells from murine lungs: a method to analyze ongoing immune responses in the lung. Nat Protoc. 2006;1:2870–2875. doi: 10.1038/nprot.2006.435. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 28.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Decker T, Müller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- 30.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 32.Rock JR, Gao X, Xue Y, Randell SH, Kong YY, Hogan BL. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 2011;8:639–648. doi: 10.1016/j.stem.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotton DN, Morrisey EE. Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med. 2014;20:822–832. doi: 10.1038/nm.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YS, Choi JW, Hwang I, Lee JW, Lee JH, Kim AY, Huh JY, Koh YJ, Koh GY, Son HJ, et al. Adipocytokine orosomucoid integrates inflammatory and metabolic signals to preserve energy homeostasis by resolving immoderate inflammation. J Biol Chem. 2010;285:22174–22185. doi: 10.1074/jbc.M109.085464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hochepied T, Berger FG, Baumann H, Libert C. Alpha1-acid glycoprotein: an acute phase protein with inflammatory and immunomodulating properties. Cytokine Growth Factor Rev. 2003;14:25–34. doi: 10.1016/s1359-6101(02)00054-0. [DOI] [PubMed] [Google Scholar]
- 36.Quinton LJ, Blahna MT, Jones MR, Allen E, Ferrari JD, Hilliard KL, Zhang X, Sabharwal V, Algül H, Akira S, et al. Hepatocyte-specific mutation of both NF-κB RelA and STAT3 abrogates the acute phase response in mice. J Clin Invest. 2012;122:1758–1763. doi: 10.1172/JCI59408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizgerd JP. Acute lower respiratory tract infection. N Engl J Med. 2008;358:716–727. doi: 10.1056/NEJMra074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrero JA. Confounding roles for type I interferons during bacterial and viral pathogenesis. Int Immunol. 2013;25:663–669. doi: 10.1093/intimm/dxt050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LeMessurier KS, Häcker H, Chi L, Tuomanen E, Redecke V. Type I interferon protects against pneumococcal invasive disease by inhibiting bacterial transmigration across the lung. PLoS Pathog. 2013;9:e1003727. doi: 10.1371/journal.ppat.1003727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Damjanovic D, Khera A, Medina MF, Ennis J, Turner JD, Gauldie J, Xing Z. Type 1 interferon gene transfer enhances host defense against pulmonary Streptococcus pneumoniae infection via activating innate leukocytes. Mol Ther Methods Clin Dev. 2014;1:5. doi: 10.1038/mtm.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada M, Gomez JC, Chugh PE, Lowell CA, Dinauer MC, Dittmer DP, Doerschuk CM. Interferon-γ production by neutrophils during bacterial pneumonia in mice. Am J Respir Crit Care Med. 2011;183:1391–1401. doi: 10.1164/rccm.201004-0592OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KW, Low D, Ruedl C, Riccardi-Castagnoli P, Poidinger M, et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol. 2014;15:1181–1189. doi: 10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- 44.Yong KL, Rowles PM, Patterson KG, Linch DC. Granulocyte-macrophage colony–stimulating factor induces neutrophil adhesion to pulmonary vascular endothelium in vivo: role of β2 integrins. Blood. 1992;80:1565–1575. [PubMed] [Google Scholar]
- 45.Satake S, Hirai H, Hayashi Y, Shime N, Tamura A, Yao H, Yoshioka S, Miura Y, Inaba T, Fujita N, et al. C/EBPβ is involved in the amplification of early granulocyte precursors during candidemia-induced “emergency” granulopoiesis. J Immunol. 2012;189:4546–4555. doi: 10.4049/jimmunol.1103007. [DOI] [PubMed] [Google Scholar]
- 46.Lee PY, Wang JX, Parisini E, Dascher CC, Nigrovic PA. Ly6 family proteins in neutrophil biology. J Leukoc Biol. 2013;94:585–594. doi: 10.1189/jlb.0113014. [DOI] [PubMed] [Google Scholar]
- 47.Miller MD, Krangel MS. Biology and biochemistry of the chemokines: a family of chemotactic and inflammatory cytokines. Crit Rev Immunol. 1992;12:17–46. [PubMed] [Google Scholar]
- 48.Greenberger MJ, Strieter RM, Kunkel SL, Danforth JM, Laichalk LL, McGillicuddy DC, Standiford TJ. Neutralization of macrophage inflammatory protein-2 attenuates neutrophil recruitment and bacterial clearance in murine Klebsiella pneumonia. J Infect Dis. 1996;173:159–165. doi: 10.1093/infdis/173.1.159. [DOI] [PubMed] [Google Scholar]
- 49.Standiford TJ, Kunkel SL, Greenberger MJ, Laichalk LL, Strieter RM. Expression and regulation of chemokines in bacterial pneumonia. J Leukoc Biol. 1996;59:24–28. doi: 10.1002/jlb.59.1.24. [DOI] [PubMed] [Google Scholar]
- 50.Brandes M, Klauschen F, Kuchen S, Germain RN. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154:197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.