Abstract
Silver nanoparticles (AgNPs) have been used as antimicrobials in a number of applications, including topical wound dressings and coatings for consumer products & biomedical devices. Ingestion is a relevant route of exposure for AgNPs, whether occurring unintentionally via Ag dissolution from consumer products, or intentionally from dietary supplements. AgNP have also been proposed as substitutes for antibiotics in animal feeds. While oral antibiotics are known to have significant effects on gut bacteria, the antimicrobial effects of ingested AgNPs on the indigenous microbiome or on gut pathogens are unknown. Additionally, AgNP size and coating have been postulated as significantly influential towards their biochemical properties and the influence of these properties on antimicrobial efficacy is unknown. We evaluated murine gut microbial communities using culture-independent sequencing of 16S rRNA gene fragments following 28 days of repeated oral dosing of well-characterized AgNPs of two different sizes (20 and 110 nm) and coatings (PVP and Citrate). Irrespective of size or coating, oral administration of AgNPs at 10 mg/kg body weight/day did not alter the membership, structure, or diversity of the murine gut microbiome. Thus, in contrast to effects of broad-spectrum antibiotics, repeat dosing of AgNP, at doses equivalent to 2000 times the oral reference dose and 100–400 times the effective in vitro anti-microbial concentration, does not affect the indigenous murine gut microbiome.
Keywords: nanomaterials, microbiome, antibiotics, toxicology, in vivo, pyrosequencing, mothur, mouse, Metastats
Introduction
Silver nanoparticle (AgNP) incorporation into consumer products or dietary supplements has become common as a purported means of antimicrobial protection (Volker et al., 2013). In vitro and topically, silver is demonstrably cytotoxic to bacteria, viruses, and fungal organisms (Pineda et al., 2012, Zarei et al., 2014). Historically, colloidal silver suspensions were used as anti-infectives before the development of modern-day antibiotics (Drake and Hazelwood, 2005), (El-Ansary and Al-Daihan, 2009, Varner et al., 2010). This application has potential for resurgence through the use of AgNP-coated food containers, as dietary supplements, (Bergin and Witzmann, 2013, Volker et al., 2013, Echegoyen and Nerin, 2013)and as an alternative to growth-promoting antibiotics in agricultural animals (Fondevila et al., 2009), (Ahmadi and Rahimi, 2011, Ahmadi, 2009). Thus, gastrointestinal exposure to AgNP may increase for many consumers, either directly from the aforementioned products, or indirectly from water contamination due to run-off or accumulation in aquatic organisms (Shaw and Handy, 2011, Volker et al., 2013).
The intestinal microbiome has been a major focus of research in the fields of microbiology and medicine(Wikoff et al., 2009, Young, 2012) but has only recently been considered in the context of potential toxicologic effects of ingested metals (Williams et al., 2014, Hadrup et al., 2012). Gut microbes influence digestion, metabolism, and the host immune system through the induction of both pro-inflammatory cytokines and immune tolerance(Atarashi et al., 2010, Atarashi et al., 2011). Since the majority of intestinal microbes cannot be cultured using standard microbiological techniques, culture-independent methods of microbial profiling, such as next generation sequencing and metagenomics, have been employed to further our understanding of intestinal microbial communities and the effects of their perturbation on human and animal health (Zoetendal et al., 2004). Alterations in gut microbiota have been associated with many gastro-intestinal and extra-digestive diseases (reviewed in (Kalliokoski et al., 2013, Morones-Ramirez et al., 2013). Additionally, a vast array of different substances may alter the gut microbiome. For instance, despite the fact that antibiotics are an essential treatment modality for infectious disease, many antibiotics also impair the normal, non-pathogenic gut microbiome after administration (Maneewattanapinyo et al., 2011).
The effects of ingested AgNPs on the indigenous gut microbiome have undergone only limited characterization (Hadrup et al., 2012, Williams et al., 2014). Moreover, AgNPs under current or proposed use differ widely in size, coating, or other physicochemical properties and the relative influence of these differences on their antimicrobial activity is unknown. The purpose of the current study was to evaluate culture-independent gut microbial community profiles in a mouse model following repeat dosing of well characterized AgNPs of two different sizes and coatings. Ionic silver (silver acetate) was used as a control to determine whether any effects seen were unique to nanoparticulate silver. Given the broad in vitro antimicrobial effects of AgNP (Morrill et al., 2013), we hypothesized that AgNP-associated changes to the gut microbiome, if present, would be similar to those associated with administration of a broad spectrum antibiotic.
Materials and Methods
Animal Studies
Male, C57BL/6NCrl mice (10–12 wk; Charles River Laboratories) were housed 5 per cage in static microisolation cages in an SPF barrier facility at the University of Michigan, which is an AAALAC-accredited institution. Mice were SPF for common murine viruses. Mice were fed an irradiated diet (PicoLab Laboratory Rodent Diet 5LOD, LabDiet, St Louis, MO) and water, both provided ad libitum. Animals were maintained on a 12:12-h light:dark cycle at 72 ± 2 °F. After a 7d acclimation period, animals were housed individually for the study duration. At study termination, animals were euthanized by CO2 inhalation. All procedures were approved by the University of Michigan’s IACUC.
NanoMaterials
AgNPs used in this study were manufactured by NanoComposix (San Diego, CA) and supplied by the NIEHS Centers for Nanotechnology Health Implications Research (NCNHIR) consortium. Particles consisted of colloidal suspensions of 20 and 110 nm polyvinylpyrrolidone (PVP) or citrate-stabilized particles, synthesized over a 5 nm Au core. Particles were at a concentration of 1 mg/mL in 2 mM citrate buffer or in 100 ug/ml PVP in water.
Characterization of materials
Physicochemical characterization of the particles used in this study was originally performed by the supplier and by the Nano Characterization Laboratory (NCL) at the National Cancer Institute as previously described (Wang et al., 2014). As assessed by kinetic turbidity and gel-clot Limulus Amoebocyte Lysate (LAL) assays, 20 nm Ag-Citrate, 110 Ag-Citrate, and 110 nm Ag-PVP particles were found to have endotoxin levels <0.5 EU/mL and 20 nm Ag-PVP particles had an average endotoxin level of 1.1 EU/mL. Particle composition was confirmed in-house using inductively coupled plasma mass spectrometry (ICPMS) and size/size distribution by dynamic light scattering, transmission electron microscopy, and Nanoparticle Tracking Analysis.
Study Design
Animals were dosed by oral gavage once daily for 28 consecutive days followed by sacrifice on day 29 (i.e. 24 hours after the last dose). Animals were dosed between 9AM and 12 noon. Animals were randomly assigned to the following dose groups at n=6 per group: 1) Sterile, endotoxin-free water (Gibco Laboratories, cat# 15230), 2) silver acetate (AgOAc, Sigma Aldrich, #216674, St. Louis, MO), 3) 110 nm PVP AgNP, 4) 20 nm PVP AgNP, 5) 110 nm Citrate AgNP, 6) 20 nm Citrate AgNP. For groups 2 through 6 (AgOAc or AgNP), materials were dosed at 10 mg/kg bw/day, at the stock concentration of 1 mg/ml and a volume of 0.01 ml/g bw. This dose is equivalent to 2000x the EPA’s oral reference dose for colloidal silver (0.005 mg/kg bw/day) (CASRN, 1988). At study termination, animals were euthanized by CO2 inhalation. Cecal tips were removed at necropsy and stored at −80 for microbial sequencing. One experimental trial had an additional control group of animals (n=5) receiving the broad spectrum antibiotic cefoperazone (cefoperazone sodium salt, Alpha Aesar, #J65185), dosed in sterile drinking water (Gibco Laboratories, cat# 15230) at 0.5 mg/ml for 12 days (replaced every two days). After a 48 hour washout period these animals were sacrificed by CO2 inhalation on day 14.
DNA Extraction
Total DNA was extracted from cecal tips using a PowerSoil–htp 96 Well Soil DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) on the epMotion 5075 automated pipetting system (Eppendorf AG, Hamburg, Germany) according to the manufacturer’s instructions.
Amplification and sequencing of 16S rRNA encoding gene sequences
Gene amplicons representing the V3–V5 hypervariable region of the bacterial 16S rRNA encoding gene were generated and analyzed as previously described (Hashway et al., 2014) per protocols developed for the NIH Human Microbiome Project (http://www.hmpdacc.org/doc/16S_Sequencing_SOP_4.2.2.pdf). Pyrosequencing was performed on the Roche 454 GS Junior Titanium platform according to the manufacturer’s instructions (Roche 454 Life Sciences, Branford, CT, USA).
Sequence processing and analysis
Bacterial 16S rRNA gene sequences were processed using the software program mothur following a standard operating procedure for 454 pyrosequencing data (Schloss, 2009, Schloss, 2010). Sequences with <200 bases, ambiguous bases, homopolymers > 8 bases, or with missing or erroneous barcodes were removed. Sequences were aligned to a SILVA alignment database using the Needleman–Wunsch and NAST algorithms, and sequences that did not share a defined alignment space were trimmed (Schloss et al., 2011, Schloss, 2010, Schloss, 2009). Chimeras were removed using the UCHIME algorithm (Walters et al., 2014). The Ribosomal Database Project (RDP) naive Bayesian classifier within mothur was used to taxonomically assign 16S rRNA gene sequences, using a confidence cutoff of 80%. Sequences classified as ‘Chloroplast’, ‘Mitochondria’, or ‘unknown’ were removed (Cole et al., 2014). Sequences were grouped into operational taxonomic units (OTUs) based on ≥97% sequence similarity using the average neighbor algorithm (Cole et al., 2014). Sequences generated in this study were deposited in the NCBI Sequence Read Archive under Bioproject number XXXXXX. (AUTHOR NOTE: sequences entered 6/8/15; waiting for Project number)
θYC distance matrices were constructed to compare microbial communities and the θYC distance was calculated (Yue and Clayton, 2005). This data was visually represented by principal coordinates analysis (PCoA). Diversity scores at each timepoint were calculated within mothur using the inverse Simpson (1/D) index.
Statistical Analysis
θYC distances were statistically compared by analysis of molecular variance (AMOVA) within mothur, a non-parametric test useful for the comparison of calculated molecular distances (which by their nature have a non-normal distribution) (Anderson, 2001). Significance was defined as P < 0.05 after Bonferroni correction for multiple comparisons (included in mothur algorithm for AMOVA) (Anderson, 2001). Relative abundance of specific phyla, diversity indices (Shannon index and Inverse Simpson index), and measures of richness (Chao1) and evenness (Shannon evenness) were evaluated by one-way ANOVA and Tukey’s multiple comparisons test within Graph pad Prism (version 4.0, GraphPad Software, San Diego, CA) with significance defined as p<0.05 after multiple comparisons correction. Differences in specific OTUs were evaluated using Metastats within mothur, using q-values to represent the false discovery rate (White et al., 2009).
Results
Effects of repeated oral dosing of 110 nm PVP-coated AgNPs on gut microbial communities
110 nm AgNP-PVP was evaluated initially based on the hypothesis that the relatively large size and stable coating would minimize dissolution and absorption from the intestinal tract, leaving more of the material in contact with intraluminal intestinal microbes (Jiang et al., 2009, Li et al., 2010). Mouse cecal microbial communities were evaluated following 28 consecutive days’ of dosing at 10 mg/kg bw daily, in comparison to water and silver acetate (Figure 1A).
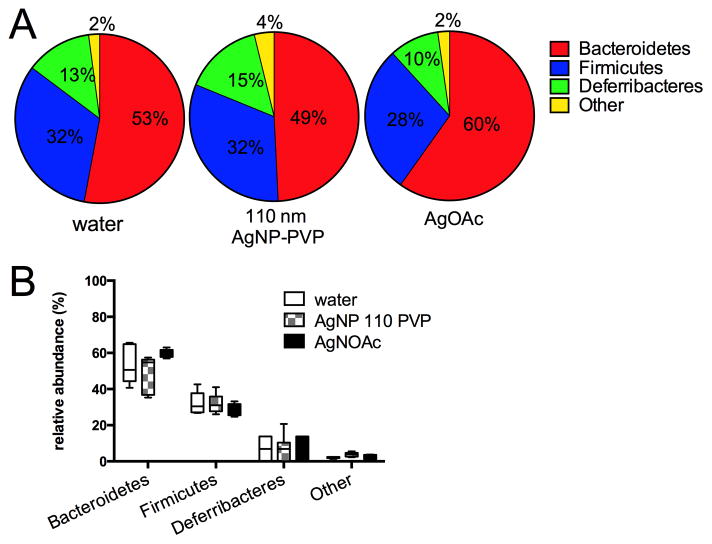
Figure 1.
Relative abundance of bacterial phyla from mouse cecal contents after 28 days of oral gavage with 10 mg/kg bw/day of 110 nm PVP-coated AgNP in comparison to water and silver acetate. A. Mean % relative abundance of all phyla for mice from each group, with mean percentage of each phylum indicated. For all groups, Bacteroidetes was the major phylum present, followed by Firmicutes and the murine-specific phylum Deferribacteres. Other phyla (combined) were present at <4% in each group and consisted of unclassified phyla (most abundant, 1–2%), followed by Actinobacteria, Proteobacteria, Tenericutes, Verrucomicrobia, and TM7, each present at <1%. B. Median (horizontal bar), interquartile range (box) and minimum/maximum for major phyla in each group. There were no significant differences between groups with respect to microbial community composition and structure at the phylum level (ANOVA, p=0.861).
Animals tolerated the dose well and all animals survived to endpoint with the exception of one animal from the silver acetate group that was euthanized due to gavage injury. All other animals were euthanized at 24 hours after the last dose and microbial community profiles were constructed by pyrosequencing of amplified bacterial 16S rRNA gene fragments from cecal tip DNA. After sequence processing, a total of 47,461 high-quality sequence reads were obtained, with mean number of sequences ± S.D per group of 2822 ± 601 (water group), 2726 ± 1112 (110 nm AgNP-PVP), and 2834 ± 350 (AgOAc). Sequences were subsampled to 1965 (the minimum number of sequences generated per sample) for analysis. Sequences were classified into operational taxonomic units (OTUs) on the basis of 3% dissimilarity (roughly equivalent to the taxonomic level of species). OTUs thus defined were then categorized into phyla by comparison to a standard database (Ribosomal Database Project classifier). A total of 9 phyla were represented in the intestinal microbial communities for these mice but 97.3% of the sequences present in all mice across groups were within three phyla, namely Bacteroidetes (54.0% mean total abundance), Firmicutes (30.9% mean total abundance), and Deferribacteres (12.4% mean total abundance) (Figure 1A). This is similar to intestinal microbial communities that have been previously described for mice (Krych et al., 2013, Antonopoulos et al., 2009, Bassis et al., 2014)}. There were no significant differences between any groups in the relative abundance of any phylum. (Figure 1B)
Microbial community diversity within each group, as indicated by the Inverse Simpson diversity index or the Shannon diversity index, was similar for the 110 PVP-AgNP, AgOAc and water groups (Table 1). Additionally, species richness (# of OTUs) and species evenness (equality of OTU distribution), evaluated by Chao1 richness and Shannon evenness scores, also did not differ between dose groups (Table 1).
Table 1.
Diversity parameters after 28 days administration of 110 nm PVP-AgNP or AgOAca
| Groupb | Shannon diversity index | Inverse Simpson index | Shannon evenness score | Chao1 (richness) |
|---|---|---|---|---|
| 110 nm AgNP- | 3.73 +/− | 19.77 +/− 4.40 | 0.72 +/− | 258.39 +/− 32.91 |
| sterile water | 3.67 +/− | 17.93 +/− 2.93 | 0.72 +/− | 241.92 +/− 51.23 |
| silver acetate | 3.65 +/− | 21.14 +/− 3.55 | 0.72 +/− | 226.71 +/− 14.82 |
No significant differences between groups for any parameter (ANOVA, p=0.396)
n=6 per group for AgNP and water, 5 per group for silver acetate
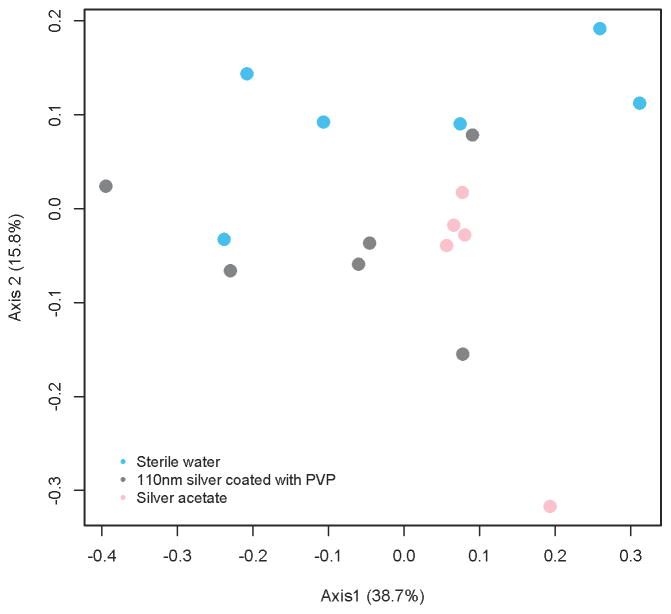
Microbial community structures did not differ between 110 nm AgNP-PVP, water, or AgOAc groups, as compared on the basis of θYC distances (AMOVA, p=0.111) (Figure 2).
Figure 2.
Cecal microbial communities in animals gavaged with 110 nm AgNP-PVP, AgOAc, or water for 28 days. Data are depicted as a principal components analysis (PCoA) plot representing the Yue Clayton (ØYC) distance metric. Microbial communities were not significantly different between groups. (AMOVA, p=0.111).
Effects of repeated oral dosing of 20 nm PVP-coated AgNP, 20 nm citrate-capped AgNP, and 110 nm citrate-capped AgNPs on gut microbial communities
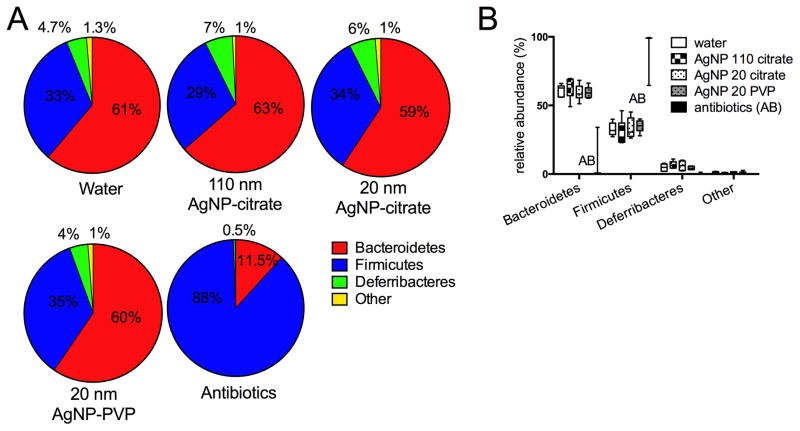
Microbial community alterations were next assessed in mice fed 20 nm AgNP-PVP, 20 nm AgNP-Citrate, or 110 nm AgNP-Citrate for 28 days, in comparison to water. In this trial, an additional positive control group for bacterial alterations was included that consisted of animals receiving a broad-spectrum antibiotic, cefoperazone, in the drinking water for 12 days. All animals tolerated the treatment well and survived to endpoint, with the exception of one mouse in the 110 nm AgNP-citrate group, which was euthanized due to gavage injury. Additionally, sterilized drinking water was utilized for all groups in an attempt to decrease inter and intra-group variation due to potential bacterial acquisition in the drinking water. Cecal microbial community profiles were constructed as for the previous trial. After sequence processing, a total of 46,826 high-quality sequence reads were obtained, with mean number of sequences ± S.D. per group of 2163 ± 598 (sterile water), 1750 ± 698 (110 ng AgNP-citrate), 1972 ± 665 (20 nm AgNP-citrate), 1894 ± 361 (20 nm AgNP-PVP), and 945 ± 277 (cefoperazone). Sequences were subsampled to 687 (the minimum number of sequences generated per sample) for analysis. Three of the antibiotic-dosed animals failed to generate sufficient sequence for analysis. A similar bacterial phyla distribution as in the previous analysis was present, with a total of 9 phyla represented and the majority (98.9%) distributed within three phyla- Bacteroidetes (56.2% mean total abundance), Firmicutes (37.7% mean total abundance), and Deferribacteres (5.04% mean total abundance). (Figure 3A). Only the cefoperazone treated animals had significant differences in phyla abundance, with markedly decreased Bacteroidetes and a shift to predominance of Firmicutes. (Figure 3B)
Figure 3.
Relative abundance of bacterial phyla from mouse cecal contents after 28 days of oral gavage with 10 mg/kg bw/day of 110 nm, 20 nm citrate-coated, or 20 nm PVP-coated AgNP in comparison to 28 days oral gavage with water or 12 days of antibiotic (cefoperazone) administration in the drinking water. A. Mean % relative abundance of predominant phyla for mice from each group. All AgNP and water- dosed groups had a preponderance of Bacteroidetes and Firmicutes, with lesser numbers of Deferribacteres. Other phyla (combined) were present at <2% in each group and consisted of unclassified phyla, Actinobacteria, Proteobacteria, Tenericutes, Verrucomicrobia, and TM7. B. Median (horizontal bar), interquartile range (box), and minimum/maximum for major phyla identified in all groups. Only the antibiotics-treated group had significant differences from other groups, consisting of increased Firmicutes and decreased Bacteroidetes (ANOVA, Tukey’s multiple comparisons test, p<0.0001). N=5–6 for water or AgNP groups (see text). Only two points are shown for the antibiotics group since only two animals had sufficient recovery of gut bacterial DNA for sequencing remaining after antibiotic dosing, despite the 24 hour washout period.
Microbial diversity was not significantly different in groups receiving 110 PVP-AgNP, 20 nm citrate-AgNP, 110 nm citrate-AgNP, or water, as shown in Table 2. Evenness and richness were likewise not significantly different (Table 2). Animals dosed with the positive control antibiotic (cefoperazone), had significant decreases in diversity parameters in comparison to the AgNP and water-treated groups (Table 2). The pronounced effects of cefoperazone on microbial communities was also clear since 3 of 5 mice failed to generate sufficient sequence numbers for analysis, even after a 24 hour washout period.
Table 2.
Alpha diversity parameters after 28 days administration of 20 nm PVP AgNPs, 20 nm citrate AgNPs, 110 nm citrate AgNPs, or cefoperazone (antibiotic)a
| Groupb | Shannon diversity index | Inverse Simpson index | Shannon evenness score | Chao1 (richness) |
|---|---|---|---|---|
| AgNP 20 PVP | 3.99 +/− 0.14 | 29.26 +/− 6.11 | 0.78 +/− 0.02 | 281.79 +/− 35.48 |
| AgNP 20 Citrate | 4.01 +/− 0.10 | 31.44 +/− 2.13 | 0.78 +/− 0.03 | 292.28 +/− 76.03 |
| AgNP 110 Citrate | 3.86 +/− 0.12. | 27.54 +/− 4.08 | 0.77 +/− 0.02 | 282.41 +/− 30.64 |
| sterile water | 3.93 +/− 0.17 | 28.10 +/− 5.57 | 0.76 +/− 0.01 | 297.19 +/− 40.78 |
| cefoperazonea | 0.16 +/− 0.04 | 1.05 +/− 0.01 | 0.06 +/− 0.01 | 59.5 +/− 33.50 |
There were no significant differences between water and AgNP groups for any parameter. The cefoperazone-treated group was significantly different from all other groups (ANOVA, Tukey’s multiple comparisons test, p<0.0001).
n=6 per group for AgNP 20 PVP and citrate, n=5 for 110 citrate and n=3 for cefoperazone (only 3 animals had sufficient recovery of microbial sequences for analysis)
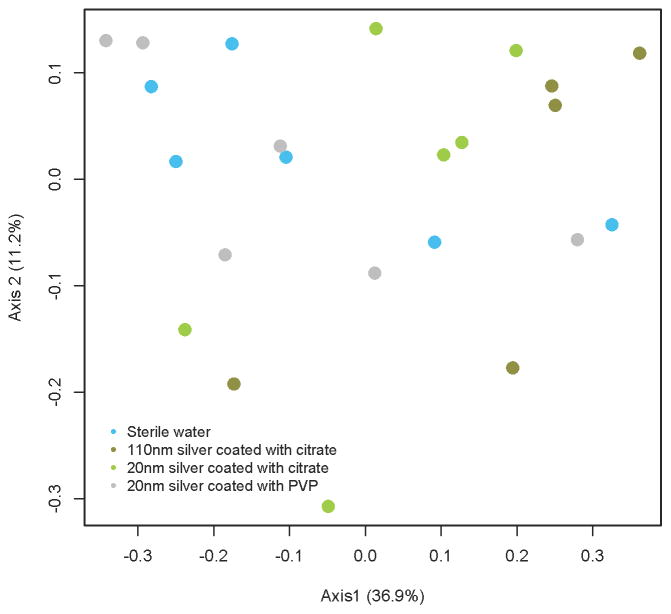
Microbial community structures were not significantly different when comparing AgNP-dosed groups to one another or to water (AMOVA, p=0.113) (Figure 4). In contrast, microbial communities of cefoperazone-dosed animals were distinct from the AgNP-dosed or water-dosed groups (Supplemental Figure 1).
Figure 4.
Cecal microbial community differences between groups gavaged with AgNP of varying size and coating. Data are depicted as a PCoA plot representing the ØYC distance metric. Microbial communities were not significantly different between groups. (AMOVA, p=0.114).
Effects of AgNP dosing on specific OTUs
There were no significant differences in individual OTUs between the AgNP groups and the water or AgOAc groups, as indicated by a Metastats q-value (indication of false discovery rate) of 1 for all OTU comparisons between groups. This included no difference in relative abundance between groups for OTU 19 (Lactobacillus), OTUs 6,8, and 20 (Bacteroides), and OUT 164 (Bifidobacterium), which represented genera previously described as reduced in rats dosed with AgNP for 13 wks (Williams et al., 2014). Of note, Bifidobacterium was only present in 3 mice in the study (1 each in the water, 110 nm AgNP-PVP, and silver acetate groups) at relative abundances of only 0.08–0.17%, thus this genus may not represent a relevant target for evaluating antimicrobial effects in all host species. Lactobacillus was present in all animals, but only at a mean relative abundance of 1.25% (range 0.08–4.92%).
Discussion
This study investigated the effects of AgNPs of differing size and coating on the cecal microbiome of mice, as evaluated by culture-independent microbial sequencing. Here, we demonstrated that repeated dose administration of 20 nm or 110 nm AgNP, with either PVP or citrate coatings, for 28 days did not significantly alter indigenous gut microbial communities of mice. The dose of AgNP used in this study corresponds to 2000x the oral reference dose (i.e. daily intake considered “safe” over a lifetime) for colloidal silver in humans (CASRN, 1988).
Although AgNPs have demonstrable antimicrobial effects in vitro (Sondi and Salopek-Sondi, 2004, Kim et al., 2007, Percival et al., 2005, Loh et al., 2009, Morrill et al., 2013) there is currently minimal evidence for in vivo efficacy of ingested AgNP, either for pathogens or the indigenous gut microbiome. In pigs administered AgNP of between 60–100 nm in a sepiolite carrier at concentrations up to 40 mg Ag per kg feed (~ 11.3 mg Ag/kg body weight/day) for 14 days, there was a non-significant trend towards decreased coliforms, with no effects on lactobacilli, as determined by bacterial culture, and a significant but mild decrease in overall ileal bacteria, as determined by fluorescence in situ hybridization (Fondevila et al., 2009). Rats gavaged with 14 nm PVP-coated AgNP at a dose of 9 mg/kg bw/day for 28 days showed no difference in the ratio of the two main bacterial phyla of the gastrointestinal tract, Firmicutes and Bacteroidetes, as determined by quantitative PCR (Hadrup et al., 2012). In contrast, (Williams et al., 2014) recently reported alterations in indigenous microbial populations in Sprague Dawley rats gavaged with PVP-coated AgNPs of 10, 75, and 110 nm diameter at doses of 9, 18, and 36 mg/kg bw/day for 13 weeks. Specifically, there were significantly decreased colony-forming units (CFU) recovered upon culture of ileal microbes from AgNP-dosed groups. The most pronounced effects on cultivable bacteria were for lower doses and smaller diameter particles (no differences were reported for 110 nm particles at the highest dose). (Williams et al., 2014) also compared the ratio of Bacteroidetes to Firmicutes phyla, as assessed by quantitative PCR from ileal microbial populations, Here the most pronounced effect was seen for 110 nm particles at the highest dose, which was associated with a decrease in Firmicutes. Conversely, an increase in Firmicutes in comparison to vehicle control was reported for the 75 and 110 nm particles at the low dose (9 mg/kg bw/day). Female rats had less pronounced changes. Statistical significance and inter-animal variation were not reported for Firmicutes/Bacteroidetes ratios. For any AgNP-associated alteration, there was no indication of a physiologic effect, either detrimental or beneficial, for the rats. In contrast, (Williams et al., 2014)found that high doses of AgOAc (100–400 mg/kg bw/day) delivered by gavage to rats in the same study resulted in significant gastrointestinal ulceration.
There are several potential explanations for the lack of microbial alterations reported in our study and by (Hadrup et al., 2012) and the differences in cultivable bacteria and Firmicutes/Bacteroidetes ratio reported by (Williams et al., 2014). The latter study used a longer duration (13 weeks) and higher dose range (up to 36 mg/kg bw/day) in comparison to our study. However, (Williams et al., 2014) observed no differences between any dosed group when 16S rRNA gene sequence-based evaluation of microbial communities was performed, consistent with the lack of differences seen in our study and in (Hadrup et al., 2012). The ability of PCR-based methods to detect both live and dead bacteria was cited in (Williams et al., 2014) in explanation of this discrepancy. An additional explanation could be that AgNP administration affects cultivation efficacy. Whether this indicates a methodological issue or a true AgNP-dependent impediment to vegetative growth (ie. dormancy) is not known. Further, it should be remembered that the cultivable bacterial fraction represents only a portion of the total gut microbial community, with some estimates of the non-cultivable fraction as high as 60–70% (Hayashi et al., 2002). Alterations in the cultivable fraction will thus have to be assessed in terms of their functional significance to the whole gut microbial community. Multiple factors, ranging from diet to intestinal microbiota, can affect gut microbial composition and stochastic effects can generate significant inter-individual variation, even in genetically identical animals and even with identical starting microbial populations (Kalliokoski et al., 2013). Thus AgNP-associated alterations, such as a decrease in cultivable bacteria will have to be assessed in light of inter-animal variation and functional significance to host health, before concluding that this alteration represents an adverse effect.
There are several potential reasons for the lack of in vivo antimicrobial AgNP effects seen in our study. These include AgNP concentration at the site (cecum) of highest microbial concentration. AgNP ≤ 5 nm have been demonstrated as having in vitro growth inhibitory effects for a variety of microbes at a concentration of 2.5 ppm (for the particles), albeit only with multiple dosing applications (Morrill et al., 2013). It is difficult to extrapolate the administered dose in our study (10 mg/kg bw/day), which was selected as a high multiple (2000x) of the current regulatory limits for silver ingestion (CASRN, 1988), to the true ppm exposure for the gut microbes. The administered dose had a concentration of 1000 ppm (1 mg/ml), equivalent to 400x the concentration (2.5 ppm) observed to have antimicrobial effects in vitro (Morrill et al., 2013). It is unlikely, however, that this represented the AgNP concentration at the site of bacterial exposure. The average water content of the murine gastrointestinal tract has been estimated as ~1.0 ml (McConnell et al., 2008). Assuming even dispersion (unlikely), the concentration of AgNP in a 0.2 ml dose for a 20 g mouse, would be diluted to ~200 ppm (~0.2 mg/ml), which is still ~100x the concentration reported as having antimicrobial effects in vitro. However the delivered dose (to intestinal microbes) may be considerably less than these extrapolations, which do not account for dissolution to Ag ions or physicochemical interaction of AgNPs or Ag ions with the molecularly diverse intestinal contents. Nevertheless, the discrepancy observed is illustrative of the difficulties in extrapolating doses which demonstrate in vitro antimicrobial effects to similar effects in vivo, particularly for the oral route. It is possible that higher AgNP oral doses would result in antimicrobial effects for either indigenous microbes or for intestinal pathogens. (Bhol and Schechter, 2007) reported that a polydisperse suspension of AgNP in polyvinyl alcohol administered orally or intracolonically at doses of 40 mg/kg bw/day, but not 0.4 or 4 mg/kg bw/day, was anti-inflammatory in a rat model of dinitrobenzene sulfonic acid-induced ulcerative colitis. This was not specifically tied to antimicrobial alterations, but shows that higher AgNP doses may have physiological effects. From an environmental exposure perspective, the AgNP concentration typical of current consumer products could not result in doses this high without egregious and intentional over-consumption. Nevertheless, further evaluation for antimicrobial effects may be warranted using AgNP specifically formulated at high concentrations for this purpose.
In addition to the administered concentration, it is difficult to account for the in vivo effects of AgNP interaction with other gut intestinal contents as a possible explanation for the lack of antimicrobial activity. Most in vitro experimental evidence suggests that dissolution and local silver ion concentration play a significant role in the antimicrobial activity of AgNP (McQuillan et al., 2012, McQuillan and Shaw, 2014, Behra et al., 2013), although some aspects are not entirely explained by Ag ion availability (McQuillan and Shaw, 2014, Ivask et al., 2014). Assuming that antimicrobial effects of silver are dependent on direct interaction with gut microbes (Volker et al., 2013, Behra et al., 2013), particles present in higher luminal concentrations in the distal gut, which is the site of maximal bacterial concentration, may be more likely to have antimicrobial effects. Smaller nanoparticles are thought to dissolve more completely in the gastric environment (Mwilu et al., 2013)and thus may be more readily absorbed from the intestinal tract, decreasing the luminal concentration (Park et al., 2011). However, neither large nor small AgNP appeared to have antimicrobial activity against enteric bacteria in this study. Although subject to experimental confirmation, a possible explanation may involve physicochemical particle alterations affecting the available concentration of Ag ion in the distal tract. For example, interaction of AgNP with plasma proteins is known to generate a stable protein corona, that alters the surface properties of the AgNP (Monopoli et al., 2011). Given the molecular complexity and variability of the intestinal luminal contents, additional work in synthetic or ex vivo gastrointestinal environments would be helpful in defining the potential physicochemical transformations undergone by ingested AgNP, and the impact of these alterations on antimicrobial effects.
Conclusions
Ingested AgNPs of varied size and coating, administered in repeated doses equivalent to 2000x the oral reference dose for silver and ~100–400x the concentrations having in vitro antimicrobial activity, did not cause alterations in the overall community membership, structure, or diversity of the gut microbiome in mice as assessed using pyrosequencing. Thus, despite having in vitro antimicrobial properties (Morrill et al., 2013, McQuillan et al., 2012, McQuillan and Shaw, 2014, Ivask et al., 2014), ingested AgNPs do not appear to have a similar effect on the gut microbiome as do broad-spectrum antibiotics (Theriot et al., 2014, Theriot et al., 2011, Bassis et al., 2014). This finding is significant in assessing the potential for antimicrobial effects of ingested silver nanoparticles at exposure levels likely to be encountered in consumer products and dietary additives. Additional investigation of potential differences in the cultivable fraction of intestinal bacteria (Williams et al., 2014) may be useful with respect to the functional relevance of such alterations. Additionally, ex vivo or in vitro evaluation of the physicochemical alteration of AgNPs in the complex molecular environment of the intestine, may be informative to explain discrepancies between the lack of in vivo effects and previously reported in vitro antimicrobial effects.
Supplementary Material
Supplemental Figure 1. Cecal microbial communities from groups gavaged with AgNP of varying size and coating for 28 days in comparison to 12 days of dosing with cefoperazone, a broad-spectrum antibiotic. Data are depicted as a PCoA plot representing the ØYC distance metric. Only two animals treated with cefoperazone had sufficient gut microbial populations remaining after dosing and washout (24 hours) to generate enough sequence for analysis. Microbial communities from cefoperazone-treated animals were highly similar to one another and points representing these animals are superimposed on one another in the plot. There were no significant differences amongst the water or AgNP-treated groups.
Acknowledgments
We gratefully acknowledge Dr. Vincent Young and personnel of the Michigan Microbiome Initiative Sequencing core for assistance with pyrosequencing in this study. We additionally thank Mayu Uchihashi for assistance in sample collection, and Andy Ault, Jessica Axson, and Diana Stark for materials characterization. The silver engineered nanomaterials used in these studies have been procured, characterized supplied by the NIEHS as part of NCNHIR consortium efforts.
Footnotes
Declaration of Interests
The authors have no conflicts of interest to declare. This study was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under award number 1U01ES020128-01 as part of the NIEHS Centers for Nanotechnology Health Implications Research Consortium (NCNHIR). The manuscript was reviewed by the NCNHIR consortium prior to submission, however the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the NCNHIR.
References
- AHMADI F, RAHIMI F. The effect of different levels of nano silver and retention of silver in edible tissues of broilers. World Applied Sciences Journal. 2011;12:01–04. [Google Scholar]
- AHMADI J. Application of different levels of silver nanoparticles in food on the performance and some blood parameters of broiler chickens. World Applied Sciences Journal. 2009;7:24–27. [Google Scholar]
- ANDERSON MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- ANTONOPOULOS DA, HUSE SM, MORRISON HG, SCHMIDT TM, SOGIN ML, YOUNG VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–75. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ATARASHI K, TANOUE T, HONDA K. Induction of lamina propria Th17 cells by intestinal commensal bacteria. Vaccine. 2010;28:8036–8. doi: 10.1016/j.vaccine.2010.09.026. [DOI] [PubMed] [Google Scholar]
- ATARASHI K, TANOUE T, SHIMA T, IMAOKA A, KUNAWAHARA T, MOMOSE Y, CHENG G, YAMASAKI S, SAITO T, OHBA Y, TANIGUCHI T, TAKEDA K, HORI S, IV, ANOV II, UMESAKI Y, ITOH K, HONDA K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BASSIS CM, THERIOT CM, YOUNG VB. Alteration of the murine gastrointestinal microbiota by tigecycline leads to increased susceptibility to Clostridium difficile infection. Antimicrob Agents Chemother. 2014;58:2767–74. doi: 10.1128/AAC.02262-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEHRA R, SIGG L, CLIFT MJ, HERZOG F, MINGHETTI M, JOHNSTON B, PETRI-FINK A, ROTHEN-RUTISHAUSER B. Bioavailability of silver nanoparticles and ions: from a chemical and biochemical perspective. J R Soc Interface. 2013;10:20130396. doi: 10.1098/rsif.2013.0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERGIN IL, WITZMANN FA. Nanoparticle toxicity by the gastrointestinal route: evidence and knowledge gaps. Int J Biomed Nanosci Nanotechnol. 2013;3 doi: 10.1504/IJBNN.2013.054515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BHOL KC, SCHECHTER PJ. Effects of nanocrystalline silver (NPI 32101) in a rat model of ulcerative colitis. Digestive Diseases and Sciences. 2007;52:2732–2742. doi: 10.1007/s10620-006-9738-4. [DOI] [PubMed] [Google Scholar]
- CASRN; (IRIS), I. R. I. S, editor SIlver (CASRN 7440-22-4) 1988. [Google Scholar]
- COLE JR, WANG Q, FISH JA, CHAI B, MCGARRELL DM, SUN Y, BROWN CT, PORRAS-ALFARO A, KUSKE CR, TIEDJE JM. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DRAKE PL, HAZELWOOD KJ. Exposure-related health effects of silver and silver compounds: A review. Annals of Occupational Hygiene. 2005;49:575–585. doi: 10.1093/annhyg/mei019. [DOI] [PubMed] [Google Scholar]
- ECHEGOYEN Y, NERIN C. Nanoparticle release from nano-silver antimicrobial food containers. Food Chem Toxicol. 2013;62:16–22. doi: 10.1016/j.fct.2013.08.014. [DOI] [PubMed] [Google Scholar]
- EL-ANSARY A, AL-DAIHAN S. On the toxicity of therapeutically used nanoparticles: an overview. J Toxicol [Online] 2009;2009 doi: 10.1155/2009/754810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FLORENCE AT, HILLERY AM, HUSSAIN N, JANI PU. Nanoparticles as carriers for oral peptide absorption- studies on particle uptake and fate. Journal of Controlled Release. 1995;36:39–46. [Google Scholar]
- FONDEVILA M, HERRER R, CASALLAS MC, ABECIA L, DUCHA JJ. SIlver nanoparticles as a potential antimicrobial additive for weaned pigs. Animal Feed Science and Technology. 2009;150:259–269. [Google Scholar]
- HADRUP N, LOESCHNER K, BERGSTROM A, WILCKS A, GAO X, VOGEL U, FRANDSEN HL, LARSEN EH, LAM HR, MORTENSEN A. Subacute oral toxicity investigation of nanoparticulate and ionic silver in rats. Arch Toxicol. 2012;86:543–51. doi: 10.1007/s00204-011-0759-1. [DOI] [PubMed] [Google Scholar]
- HASHWAY SA, BERGIN IL, BASSIS CM, UCHIHASHI M, SCHMIDT K, YOUNG V, ARONOFF D, PATTON D, BELL J. Impact of a hormone-releasing intrauterine system on the vaginal microbiome: a prospective baboon model. Journal of Medical Primatology. 2014;43:89–99. doi: 10.1111/jmp.12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAYASHI H, SAKAMOTO M, BENNO Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46:535–48. doi: 10.1111/j.1348-0421.2002.tb02731.x. [DOI] [PubMed] [Google Scholar]
- IVASK A, ELBADAWY A, KAWEETEERAWAT C, BOREN D, FISCHER H, JI Z, CHANG CH, LIU R, TOLAYMAT T, TELESCA D, ZINK JI, COHEN Y, HOLDEN PA, GODWIN HA. Toxicity mechanisms in Escherichia coli vary for silver nanoparticles and differ from ionic silver. ACS Nano. 2014;8:374–86. doi: 10.1021/nn4044047. [DOI] [PubMed] [Google Scholar]
- JIANG JK, OBERDORSTER G, BISWAS P. Characterization of size, surface charge, and agglomeration state of nanoparticle dispersions for toxicological studies. Journal of Nanoparticle Research. 2009;11:77–89. [Google Scholar]
- KALLIOKOSKI O, JACOBSEN KR, DARUSMAN HS, HENRIKSEN T, WEIMANN A, POULSEN HE, HAU J, ABELSON KSP. Mice Do Not Habituate to Metabolism Cage Housing-A Three Week Study of Male BALB/c Mice. [Accessed Mar];Plos One [Online] 2013 8 doi: 10.1371/journal.pone.0058460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM JS, KUK E, YU KN, KIM JH, PARK SJ, LEE HJ, KIM SH, PARK YH, HWANG CY, KIM YK, LEE YS, JEONG DH, CHO MH. Antimicrobial effects of silver nanoparticles. Nanomedicine. 2007;3:95–101. doi: 10.1016/j.nano.2006.12.001. [DOI] [PubMed] [Google Scholar]
- KRYCH L, HANSEN CH, HANSEN AK, VAN DEN BERG FW, NIELSEN DS. Quantitatively different, yet qualitatively alike: a meta-analysis of the mouse core gut microbiome with a view towards the human gut microbiome. PLoS One. 2013;8:e62578. doi: 10.1371/journal.pone.0062578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI X, LENHART JJ, WALKER HW. Dissolution-accompanied aggregation kinetics of silver nanoparticles. Langmuir. 2010;26:16690–8. doi: 10.1021/la101768n. [DOI] [PubMed] [Google Scholar]
- LOH JV, PERCIVAL SL, WOODS EJ, WILLIAMS NJ, COCHRANE CA. SIlver resistance in MRSA isolated from wound and nasal sources in humans and animals. International Wound Journal. 2009;6:32–38. doi: 10.1111/j.1742-481X.2008.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANEEWATTANAPINYO P, BANLUNARA W, THAMMACHAROEN C, EKGASIT S, KAEWAMATAWONG T. An evaluation of acute toxicity of colloidal silver nanoparticles. J Vet Med Sci. 2011;73:1417–23. doi: 10.1292/jvms.11-0038. [DOI] [PubMed] [Google Scholar]
- MCCONNELL EL, BASIT AW, MURDAN S. Measurements of rat and mouse gastrointestinal pH, fluid, and lymphoid tissue, and implications for in vivo experiments. Journal of Pharmacy and Pharmacology. 2008;60:63–70. doi: 10.1211/jpp.60.1.0008. [DOI] [PubMed] [Google Scholar]
- MCQUILLAN JS, INFANTE HG, STOKES E, SHAW AM. Silver nanoparticle enhanced silver ion stress response in Escherichia coli K12. Nanotoxicology. 2012;6:857–66. doi: 10.3109/17435390.2011.626532. [DOI] [PubMed] [Google Scholar]
- MCQUILLAN JS, SHAW AM. Differential gene regulation in the Ag nanoparticle and Ag(+)-induced silver stress response in Escherichia coli: a full transcriptomic profile. Nanotoxicology. 2014;8(Suppl 1):177–84. doi: 10.3109/17435390.2013.870243. [DOI] [PubMed] [Google Scholar]
- MONOPOLI MP, WALCZYK D, CAMPBELL A, ELIA G, LYNCH I, BOMBELLI FB, DAWSON KA. Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J Am Chem Soc. 2011;133:2525–34. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- MORONES-RAMIREZ J, WINKLER J, SPINA C, COLLINS J. Silver enhances antibiotic activity against gram-negative bacteria. Sci Transl Med [Online] 2013;5 doi: 10.1126/scitranslmed.3006276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORRILL K, MAY K, LEEK D, LANGLAND N, JEANE LD, VENTURA J, SKUBISZ C, SCHERER S, LOPEZ E, CROCKER E, PETERS R, OERTLE J, NGUYEN K, JUST S, ORIAN M, HUMPHREY M, PAYNE D, JACOBS B, WATERS R, LANGLAND J. Spectrum of Antimicrobial Activity Associated with Ionic Colloidal Silver. Journal of Alternative and Complementary Medicine. 2013;19:224–231. doi: 10.1089/acm.2011.0681. [DOI] [PubMed] [Google Scholar]
- MWILU SK, EL BADAWY AM, BRADHAM K, NELSON C, THOMAS D, SCHECKEL KG, TOLAYMAT T, MA LZ, ROGERS KR. Changes in silver nanoparticles exposed to human synthetic stomach fluid: Effects of particle size and surface chemistry. Science of the Total Environment. 2013;447:90–98. doi: 10.1016/j.scitotenv.2012.12.036. [DOI] [PubMed] [Google Scholar]
- PARK K, PARK EJ, CHUN IK, CHOI K, LEE SH, YOON J, LEE BC. Bioavailability and toxicokinetics of citrate-coated silver nanoparticles in rats. Arch Pharm Res. 2011;34:153–8. doi: 10.1007/s12272-011-0118-z. [DOI] [PubMed] [Google Scholar]
- PERCIVAL SL, BOWLER PG, RUSSEL D. Bacterial resistance to silver in wound care. Journal of Hospital Infection. 2005;60:1–7. doi: 10.1016/j.jhin.2004.11.014. [DOI] [PubMed] [Google Scholar]
- PINEDA L, CHWALIBOG A, SAWOSZ E, LAURIDSEN C, ENGBERG R, ELNIF J, HOTOWY A, SAWOSZ F, GAO Y, ALI A, MOGHADDAM HS. Effect of silver nanoparticles on growth performance, metabolism and microbial profile of broiler chickens. Arch Anim Nutr. 2012;66:416–29. doi: 10.1080/1745039X.2012.710081. [DOI] [PubMed] [Google Scholar]
- SCHLOSS PD. [Accessed June 30, 2014 2014];Schloss_SOP [Online] 2009 Available: http://www.mothur.org/wiki/
- SCHLOSS PD. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput Biol. 2010;6:e1000844. doi: 10.1371/journal.pcbi.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHLOSS PD, GEVERS D, WESTCOTT SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PloS One. 2011;6 doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHAW BJ, HANDY RD. Physiological effects of nanoparticles on fish: A comparison of nanometals versus metal ions. Environment International. 2011;37:1083–1097. doi: 10.1016/j.envint.2011.03.009. [DOI] [PubMed] [Google Scholar]
- SONDI I, SALOPEK-SONDI B. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. J Colloid Interface Sci. 2004;275:177–182. doi: 10.1016/j.jcis.2004.02.012. [DOI] [PubMed] [Google Scholar]
- THERIOT CM, KOENIGSKNECHT MJ, CARLSON PE, JR, HATTON GE, NELSON AM, LI B, HUFFNAGLE GBJZL, YOUNG VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THERIOT CM, KOUMPOURAS CC, CARLSON PE, BERGIN II, ARONOFF DM, YOUNG VB. Cefoperazone-treated mice as an experimental platform to assess differential virulence of Clostridium difficile strains. Gut Microbes. 2011;2:326–34. doi: 10.4161/gmic.19142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VARNER KE, EL-BADAWY A, FELDHAKE D, VENKATAPATHY R. State of the science literature review: everything nanosilver and more. Washington, D.C: 2010. [Google Scholar]
- VOLKER C, OETKEN M, OEHLMANN J. The biological effects and possible modes of action of nanosilver. Rev Environ Contam Toxicol. 2013;223:81–106. doi: 10.1007/978-1-4614-5577-6_4. [DOI] [PubMed] [Google Scholar]
- WALTERS CR, POOL EJ, SOMERSET VS. Ecotoxicity of silver nanomaterials in the aquatic environment: A review of literature and gaps in nano-toxicological research. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2014;49:1588–601. doi: 10.1080/10934529.2014.938536. [DOI] [PubMed] [Google Scholar]
- WANG X, JI ZX, CHANG CH, ZHANG HY, WANG MY, LIAO YP, LIN SJ, MENG H, LI RB, SUN BB, WINKLE LV, PINKERTON KE, ZINK JI, XIA T, NEL AE. Use of Coated Silver Nanoparticles to Understand the Relationship of Particle Dissolution and Bioavailability to Cell and Lung Toxicological Potential. Small. 2014;10:385–398. doi: 10.1002/smll.201301597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITE JR, NAGARAJAN N, POP M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIKOFF WR, ANFORA AT, LIU J, SCHULTZ PG, LESLEY SA, PETERS EC, SIUZDAK G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMS K, MILNER J, BOUDREAU MD, GOKULAN K, CERNIGLIA CE, KHARE S. Effects of subchronic exposure of silver nanoparticles on intestinal microbiota and gut-associated immune responses in the ileum of Sprague-Dawley rats. Nanotoxicology. 2014:1–11. doi: 10.3109/17435390.2014.921346. [DOI] [PubMed] [Google Scholar]
- YOUNG VB. The intestinal microbiota in health and disease. Current Opinions in Gastroenterology. 2012;28:63–69. doi: 10.1097/MOG.0b013e32834d61e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YUE JC, CLAYTON MK. A similarity measure based on species proportions. Communications in Statistics-Theory and Methods. 2005;34:2123–2131. [Google Scholar]
- ZAREI M, JAMNEJAD A, KHAJEHALI E. Antibacterial effect of silver nanoparticles against four foodborne pathogens. Jundishapur J Microbiol. 2014;7:e8720. doi: 10.5812/jjm.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZOETENDAL EG, COLLIER CT, KOIKE S, MACKIE RI, GASKINS HR. Molecular ecological analysis of the gastrointestinal microbiota: A review. Journal of Nutrition. 2004;134:465–472. doi: 10.1093/jn/134.2.465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Cecal microbial communities from groups gavaged with AgNP of varying size and coating for 28 days in comparison to 12 days of dosing with cefoperazone, a broad-spectrum antibiotic. Data are depicted as a PCoA plot representing the ØYC distance metric. Only two animals treated with cefoperazone had sufficient gut microbial populations remaining after dosing and washout (24 hours) to generate enough sequence for analysis. Microbial communities from cefoperazone-treated animals were highly similar to one another and points representing these animals are superimposed on one another in the plot. There were no significant differences amongst the water or AgNP-treated groups.