Abstract
The ATP-dependent DExH/D-box helicase DHX9 is a key participant in a number of gene regulatory steps, including transcriptional, translational, microRNA-mediated control, DNA replication, and maintenance of genomic stability. DHX9 has also been implicated in tumor cell maintenance and drug response. Here, we report that inhibition of DHX9 expression is lethal to human cancer cell lines and murine Eµ−Myc lymphomas. Using a novel conditional shDHX9 mouse model, we demonstrate that sustained and prolonged (6 months) suppression of DHX9 does not result in any deleterious effects at the organismal level. Body weight, blood biochemistry, and histology of various tissues were comparable to control mice. Global gene expression profiling revealed that although reduction of DHX9 expression resulted in multiple transcriptome changes, these were relatively benign and did not lead to any discernible phenotype. Our results demonstrate a robust tolerance for systemic DHX9 suppression in vivo and support the targeting of DHX9 as an effective and specific chemotherapeutic approach.
Keywords: DHX9, helicase, conditional mouse model, shRNA, drug target
INTRODUCTION
DHX9 (also known as Nuclear DNA Helicase II (NDH II) and RNA helicase A (RHA)) is an ATP-dependent DExH/D-box helicase capable of unwinding both RNA and DNA1, as well as aberrant polynucleotide structures2. Initially purified from bovine thymus3, homologs have been subsequently identified in human, mouse, Drosophila, and C. elegans4–6. DHX9 is comprised of two RNA-binding domains at the N-terminus, a core helicase region consisting of seven conserved motifs, and a DNA-binding domain and nuclear localization signal at the C-terminus7. The presence of numerous functional domains likely contributes to the multifunctional nature of DHX9, which has been implicated in a variety of biological processes. It participates at multiple levels of gene regulation, including transcriptional regulation via interaction with a number of transcription factors and complexes (e.g. CREB-binding protein, EGFR, BRCA1, NF-κB, and RNA polymerase II)8–11, translational regulation of specific mRNAs12, 13, miRNA processing14, and RNA transport15. DHX9 is also an important factor in DNA repair16 and maintenance of genome stability2, 17, 18.
We previously uncovered DHX9 as a synthetic lethal hit from an shRNA screen for modifiers of sensitivity to ABT-737 (an inhibitor of BCL-2 family pro-survival factors)19. Suppression of DHX9 acted in concert with MYC to sensitize lymphoma cells overexpressing BCL-2 to ABT-737. Subsequent examination of the effects of DHX9 suppression in primary human diploid, non-transformed fibroblasts revealed a pronounced growth arrest and premature senescence phenotype, but not cell death20. This was attributed to inhibition of DNA replication which activated a p53-dependent stress response to protect cells from aberrant DNA replication and genomic instability20. Hence, DHX9 appears to play an important role in DNA replication and normal cell cycle progression.
Although we initially discovered that inhibiting DHX9 had therapeutic properties in combination with ABT-737 in lymphomas overexpressing BCL-2, we also noted that long-term suppression of DHX9 in tumors with reduced BCL-2 overexpression was lethal on its own [see Suppl. Figs. 10 and 13 in Ref. 19]. In this study, we explore the possibility of DHX9 as a potential single-agent anti-neoplastic target and assess whether its suppression at the organismal level would be tolerated through the use of an inducible RNAi platform enabling DHX9 suppression in vivo in the mouse21, 22. Despite having detrimental effects on cellular fitness of tumor cells ex vivo and in vivo, we observed no adverse consequences resulting from reduced DHX9 expression at the organismal level. Our results support the notion of inhibiting DHX9 as a potential chemotherapeutic target with tolerable side effects.
RESULTS
DHX9 suppression reduces human cancer cell fitness
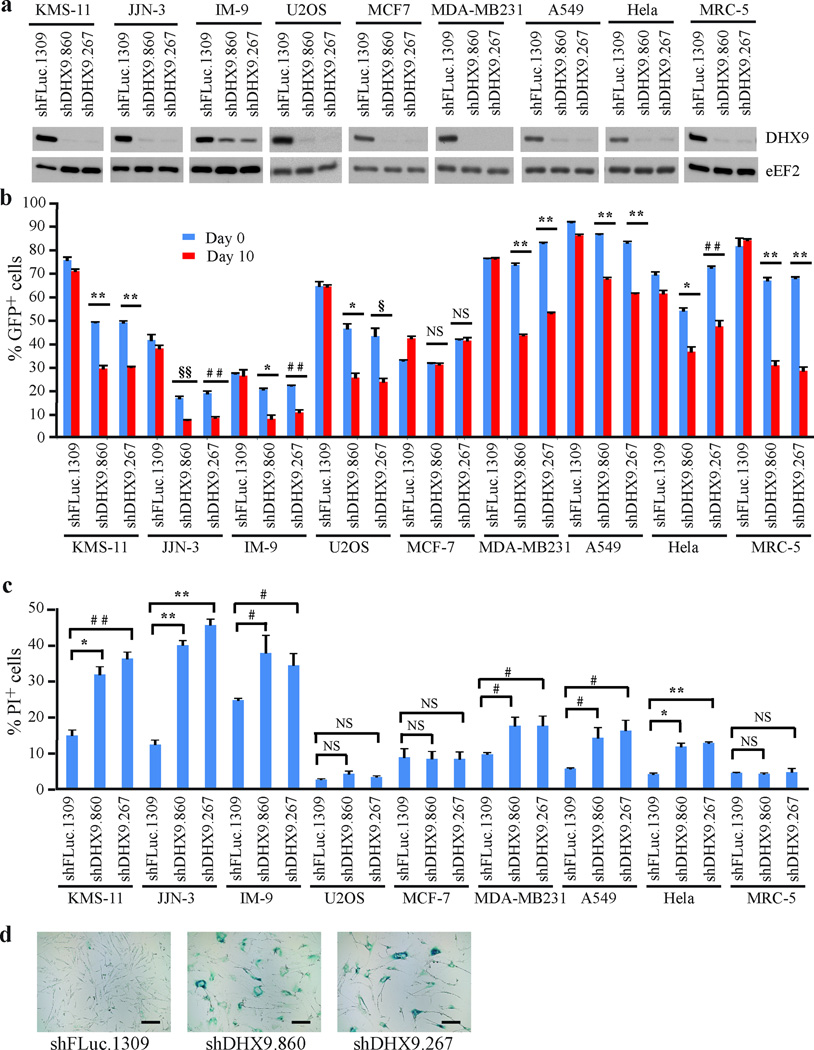
Whereas short-term suppression of DHX9 is synthetic lethal in combination with ABT-737 in Arf−/−Eµ-Myc/Bcl-2 lymphomas, we previously noted that DHX9 suppression is not well tolerated if BCL-2 is not supra-elevated19. To further document this latter effect on transformed cells, we suppressed DHX9 in different human tumor cell lines as well as in the non-immortalized MRC-5 line (Figure 1a). Initially, a representative panel of cell lines derived from different types of cancer was tested, including multiple myeloma (KMS-11, JJN-3, and IM-9), osteosarcoma (U2OS), breast (MCF-7 and MDA-MB231), lung (A549), and cervical (Hela) cancers. Infected cells (GFP+) were co-cultured with non-infected cells (GFP−) and the %GFP+ cells determined at t=0 and 10 days (Figure 1b). Suppression of DHX9 in all cell lines except MCF-7 led to a decrease in the GFP+ population over time (Figure 1b). To understand the molecular basis of this depletion, we quantified the extent of cell death that ensued following DHX9 suppression and found elevated apoptosis in all tumor lines (Figure 1c; 1.4 to 3.7-fold increase), except MCF-7 and U2OS. As previously reported20, MRC-5 cells did not show evidence of cell death but rather senesced (Figure 1c and d). We carried out cell cycle analysis on the tumor cell lines at day 10 after transduction with control or DHX9 shRNAs (Supplementary Figure 1). Upon DHX9 suppression, U2OS cells exhibited a pronounced (~15%) increase in the cells in the G0/G1 phase, and a 7–9% decrease in the number of cells in both the S and G2/M phases (Supplementary Figure 1). This demonstrates that U2OS cells were arresting in the G0/G1 phase, and that this correlated with depletion of shDHX9-expressing cells shown in Figure 1b. The IM-9 cells also showed a small G0/G1 arrest (~5% increase in G0/G1 cells). The remaining cell lines (KMS-11, JJN-3, MDA-MB231, A549, and Hela) did not show any significant changes in cell cycle distribution upon DHX9 knockdown, suggesting that apoptosis was the main mechanism of the depletion of shDHX9-expressing cells in these lines. The striking difference in phenotype obtained upon DHX9 suppression in the majority of transformed cells versus non-transformed cells prompted us to investigate DHX9 suppression as a potential anti-neoplastic approach.
Figure 1. DHX9 suppression leads to reduced fitness in human cancer cell lines.
(a) Western blot probing for DHX9 levels in the indicated cell lines infected with lentivirus expressing shRNAs targeting DHX9 or a neutral control, FLuc. (b) Competition assays showing the percentage of GFP+ cells over time, following infection of cell lines with the indicated shRNAs. T=Day 0 represents 48h following the final infection. N=3 biological replicates, each with 2 technical replicates, ±SEM. (c) Propidium Iodide (PI) staining of the indicated cell lines expressing the indicated shRNAs 7 days post-transduction. N=3 biological replicates, each with 2 technical replicates, ±SEM. (d) Senescence-associated β−galactosidase staining of MRC-5 cells transduced with lentivirus expressing the indicated shRNAs, 14 days post-infection. Bars represent 200 µm. # p≤0.05, § p≤0.01, * p≤0.005, ## p≤0.001, §§ p≤0.0005, ** p≤0.0001, NS – not significant.
To gain insight into the possible mechanisms contributing to the differences in response to DHX9 suppression among tumor cells, we compared the expression level of various cell cycle and apoptotic proteins (Supplementary Figure 2). Of all the cell lines tested, only U2OS and MRC-5 demonstrated a significant increase in CDKN1A levels, which may explain why DHX9 suppression elicited a growth arrest response rather than an apoptotic one. We observed a robust increase in p53 expression in JJN-3 and KMS-11 and a moderate increase in U2OS and MRC-5 cells. MDA-MB231 exhibited high basal levels of p53 but no upregulation upon DHX9 suppression, whereas Hela cells had almost non-existent p53 levels – these results are consistent with the former harboring mutated p53 (Ref. 23) and the latter overexpressing the E6 protein from human papillomavirus type 16, which induces the degradation of p53 (Ref. 24). While p53 activation may have contributed to the deleterious effect of DHX9 suppression in some of the cancer lines, it is not the only determinant, since both MDA-MB231 and Hela cells were susceptible to DHX9 inhibition. c-MYC expression was relatively high in A549 and Hela cells. Expression of the anti-apoptotic proteins MCL-1 and BCL-2 was highest in KMS-11, JJN-3 and MCF-7 cells. All cell lines expressed similar levels of the pro-apoptotic protein BAX, except for MCF-7, which expressed lower levels. Expression of Bim, another pro-apoptotic protein, was elevated in the three multiple myeloma lines compared to the other cancer lines. High levels of MCL-1 and BCL-2, combined with low levels of BAX and Bim, may have contributed to the resistance of MCF-7 cells to DHX9 suppression. These results indicate that the response of this set of cell lines to DHX9 suppression is not easily attributed to a single cell cycle or apoptotic modulator.
Given that all three multiple myeloma-derived lines showed robust depletion of GFP+ cells upon loss of DHX9 in the competition assay (Figure 1b), we were interested in extending these results. To this end, we assessed the effect of DHX9 suppression in 5 additional multiple myeloma lines: RPMI8226, U266B1, H929, OPM1.1, and OPM2. Of these, RPMI8226, H929, and OPM1.1 showed GFP depletion upon DHX9 knockdown in a competition assay (Supplementary Figure 3b). In total, 6 out of 8 multiple myeloma cell lines exhibited sensitivity to DHX9 suppression (Figure 1b and Supplementary Figure 3b). We also examined whether DHX9 would synergize with dexamethasone, a glucocorticoid (GC) used to treat multiple myeloma. Here we tested KMS-11 cells which are responsive to GC (IC50 = 50 nM following a 48 h exposure)25, JJN-3 cells which are resistant to GC with the defect occurring downstream of the GC receptor (IC50 >> 3 µM following a 48 h exposure)25, and IM-9 cells – a GC-resistant lymphoblastoid cell line. Loss of DHX9 was found to sensitize KMS-11 cells to dexamethasone by ~ 1.4–1.5 fold and JJN-3 cells by ~1.7 fold, but had little effect on IM-9 cells (Supplementary Figure 3c). These results indicate that DHX9 suppression is not well tolerated by a number of tumor cells, with multiple myeloma being a particularly susceptible cancer type.
Modeling DHX9 suppression in Eµ-Myc lymphomas
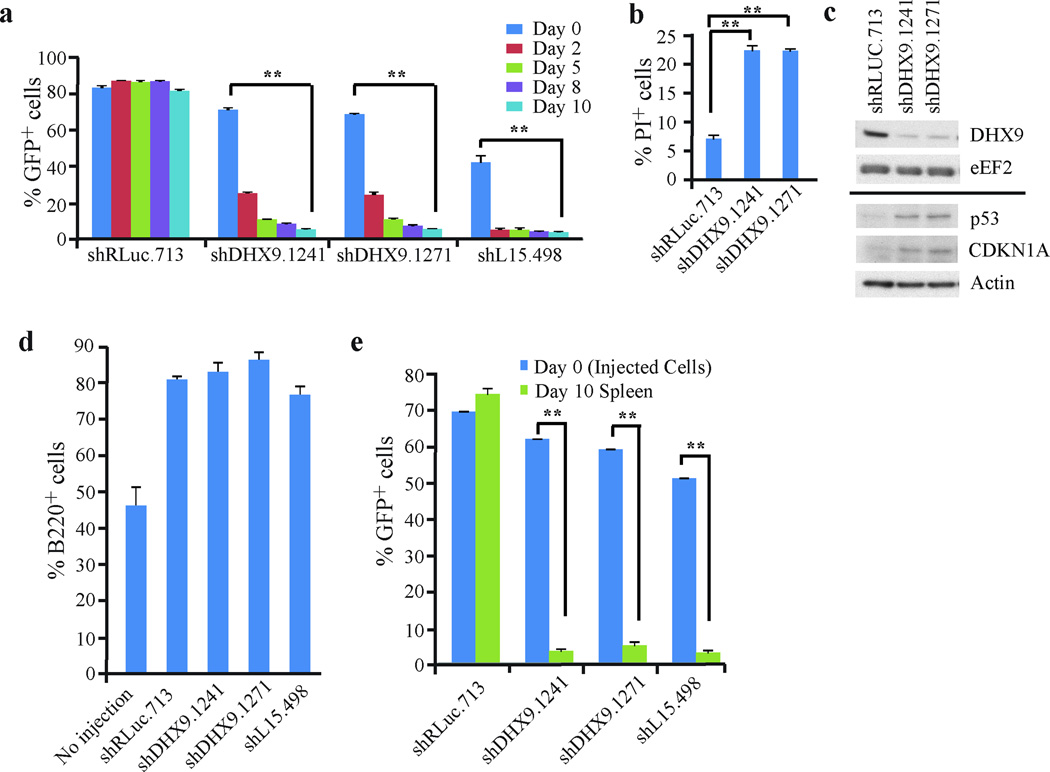
Given the above results, we sought to model the consequences of DHX9 knockdown in a more tractable murine model. First, we recapitulated the results described above using Tsc2+/−Eµ-Myc lymphomas, a Myc-driven tumor model26. Suppression of DHX9 in these cells was not well tolerated, with significant depletion of GFP+ cells occurring within 2 days post-infection using two independent DHX9 shRNAs (Figure 2a). This was comparable to what was observed with cells expressing shrpL15.498, which suppresses expression of the essential ribosomal protein, rpL15 (Ref. 27) (Figure 2a). Tsc2+/−Eµ-Myc lymphomas infected with MLS/shRLuc.713, a neutral shRNA targeting Renilla luciferase, were unaffected. A significant increase in apoptotic events was observed in DHX9 shRNA-expressing cells, compared to shRLuc.713-expressing cells, and was associated with elevated p53 and CDKN1A levels (Figures 2b and 2c).
Figure 2. DHX9 suppression is lethal in Tsc2+/−Eµ-Myc lymphoma cells.
(a) Ex vivo competition assay documenting %GFP+ cells over time following infection of Tsc2+/−Eµ-Myc cells with shRNAs targeting DHX9, RLuc (neutral control), or rpL15 (lethal positive control). The experiment was started 48 hours after the final infection (t=Day 0). N=5 biological replicates ±SEM. (b) PI staining of Tsc2+/−Eµ-Myc cells expressing the indicated shRNAs 7 days post-infection. N=5 biological replicates ±SEM. (c) Western blot analysis of extracts from Tsc2+/−Eµ-Myc cells expressing the indicated shRNAs. (d) Quantitation of the %B220+ cells in spleens harvested from mice which had been injected with Tsc2+/−Eµ-Myc cells expressing the indicated shRNAs. Spleens were harvested 10 days after injection. N=5 biological replicates ±SEM. (e) In vivo competition assay with Tsc2+/−Eµ-Myc cells. Tsc2+/−Eµ-Myc cells expressing the indicated shRNAs were introduced into mice via tail-vein injection 24h after the final infection (t=Day 0). Spleens were harvested 10 days post-injection and the %GFP+ tumor cells assessed. N=5 biological replicates ±SEM. ** p≤0.0001.
To determine whether the lethal effect of DHX9 could be recapitulated in vivo, shRNA-expressing Tsc2+/−Eµ-Myc lymphomas were introduced into C57BL/6 mice via tail-vein injection. Injected mice showed an increase in the percentage of splenic B-cells (~75–85%) compared to non-injected controls (45%), consistent with onset of lymphomagenesis (Figure 2d). Ten days later, spleens were harvested from the mice and the %GFP+ Tsc2+/−Eµ-Myc tumor cells determined (Figure 2e). Tsc2+/−Eµ-Myc tumor cells expressing the neutral shRLuc.713 control comprised the majority of the splenic cell population and showed a similar GFP+/GFP− ratio as the initial injected cell population (Figure 2e). In contrast, mice that had received Tsc2+/−Eµ-Myc tumor cells expressing shDHX9.1241, shDHX9.1271, or shL15.498 showed a profound (12–18 fold) depletion of GFP+ tumor cells 10 days following injection, with the majority of tumor cells exhibiting a GFP− phenotype, likely representing an outgrowth of non-infected tumor cells (~40–50%) arising from the initial population (Figure 2e). Taken together, these results demonstrate that DHX9 suppression is lethal in Tsc2+/−Eµ-Myc lymphomas ex vivo and in vivo.
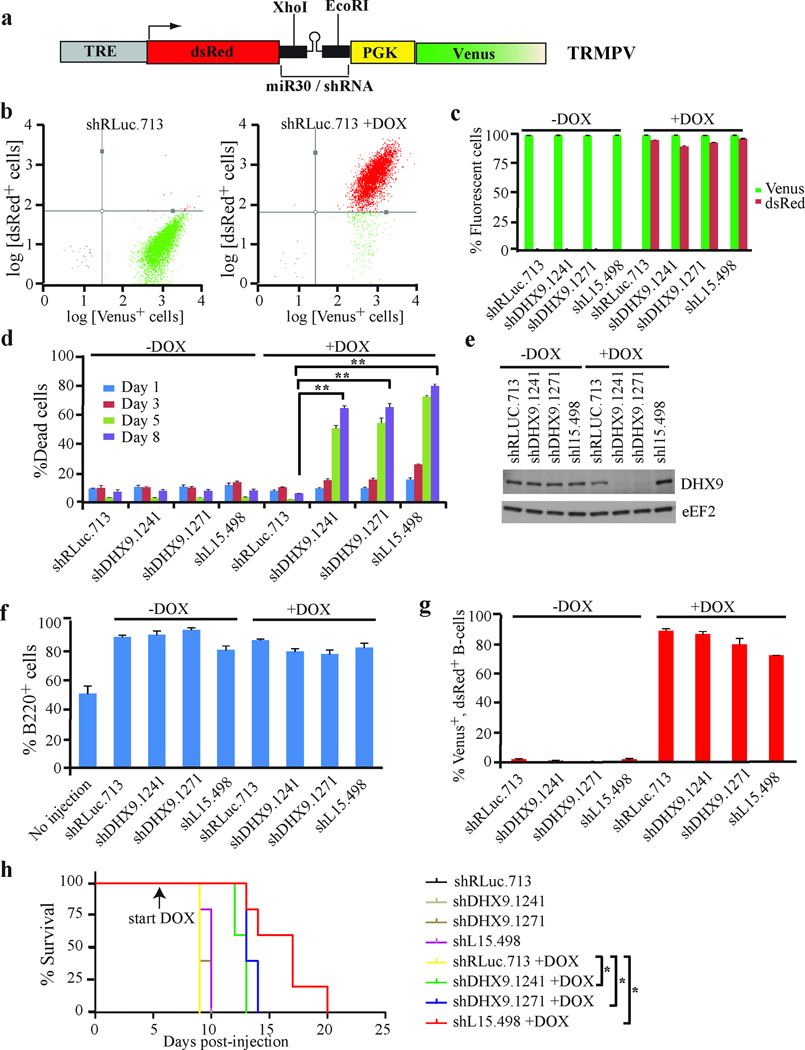
To determine whether DHX9 suppression had an effect on survival of mice harboring Tsc2+/−Eµ-Myc lymphomas, we took advantage of an shRNA doxycycline-inducible expression system utilizing the TRMPV vector28 (Figure 3a). Here, constitutive expression of Venus facilitates monitoring of infection efficiency whereas dsRed and miR30 expression are dependent on doxycycline and co-expression of rtTA. Tsc2+/−Eµ-Myc lymphomas expressing rtTA were generated by crossing Tsc2+/−Eµ-Myc mice with Rosa26-M2rtTA mice and harvesting the resulting tumors from triple-transgenic progeny22. Tsc2+/−Eµ-Myc/R26-M2rtTA tumors were transduced with TRMPV retroviruses expressing RLuc, DHX9 or L15 shRNAs. Ex vivo, addition of DOX resulted in robust induction of shRNA expression, as assessed by the percentage of Venus and dsRed double positive cells (Figures 3b and c). Conditional suppression of DHX9 resulted in a >6-fold increase in cell death (8 days post-induction) relative to shRLuc.713-infected controls (Figure 3d). Tsc2+/−Eµ-Myc/R26-M2rtTA lymphomas infected with TRMPV-shRNAs were then introduced into C57BL/6 mice via tail-vein injection with 50% of each cohort receiving doxycycline (DOX) 6 days post-injection. Spleens harvested at terminal disease stage were enlarged, showed an increased percentage of B-cells compared to non-injected controls (Figure 3f), and the majority of splenic B-cells were both Venus and dsRed positive in the DOX-treated mice (Figure 3g), demonstrating successful transplantation of the tumor cells and induction by DOX. Untreated and DOX-treated mice harboring tumors expressing shRLuc.713 reached terminal disease 9–10 days following tumor cell transplantation (Figure 3h). In contrast, DOX-treated mice harboring tumors expressing shDHX9.1241 and shDHX9.1271 reached terminal disease stage 12–14 days post-injection. Survival was extended to 13–20 days for the shL15.498 +DOX mice (Figure 3h). These results are consistent with suppression of DHX9 in vivo delaying lymphomagenesis and conferring a survival advantage.
Figure 3. DHX9 suppression enhances the survival of mice harboring Tsc2+/−Eµ-Myc lymphomas.
(a) Schematic diagram of the doxycyline-inducible vector, TRMPV. (b) Representative flow cytometry plots of Tsc2+/−Eµ-Myc/R26-M2rtTA cells transduced with TRMPV-shRLUC.713. Following infection, cells were sorted for a pure Venus-expressing population and then exposed to 1 µg/ml DOX. The %Venus+ and %dsRed+ cells were assessed 24h after DOX induction. (c) Quantification of Venus+ and dsRed+Tsc2+/−Eµ-Myc/R26-M2rtTA cells transduced with DOX-responsive retroviruses expressing the indicated shRNAs. Cells were sorted and treated with DOX as in (b), and the %Venus+ and %dsRed+ cells were assessed 24h after DOX induction. N=6 biological replicates ±SEM. (d) Assessment of cell death in Tsc2+/−Eµ-Myc/R26-M2rtTA cells transduced with DOX-responsive retroviruses expressing the indicated shRNAs. Dead cells were stained with the blue-fluorescent viability dye eFluor450 and the %dead cells was determined at the indicated time points post-DOX induction. N=5 biological replicates ±SEM, ** p≤0.0001. (e) Western blot analysis of extracts from Tsc2+/−Eµ-Myc/R26-M2rtTA cells expressing the indicated shRNAs. (f) Quantification of the %B220+ cells from spleens harvested from C57BL/6 mice injected with Tsc2+/−Eµ-Myc/R26-M2rtTA cells expressing the indicated shRNAs (treated with vehicle or 1 mg/ml DOX) at terminal disease stage. N=5 mice ±SEM. (g) Quantification of the %Venus+ dsRed+ B220+ spleen cells harvested from C57BL/6 mice injected with Tsc2+/−Eµ-Myc/R26-M2rtTA cells expressing the indicated shRNAs (treated with vehicle or 1 mg/ml DOX) at terminal disease stage. N=5 mice ±SEM. (h) Kaplan-Meier survival curve of C57BL/6 mice injected with Tsc2+/−Eµ-Myc/R26-M2rtTA cells expressing the indicated shRNAs. Mice were treated with vehicle or 1 mg/ml DOX at day 6 following injection of the tumor cells. * p<0.005 for comparisons between shRLuc.713+DOX and each of the shDHX9.1241+DOX, shDHX9.1271+DOX and shL15.498 cohorts. N=5 mice per condition.
Modeling conditional DHX9 suppression in the mouse
Whereas intriguing, the aforementioned results do not address whether DHX9 suppression would be tolerated at the organismal level. To this end, we took advantage of a previously described FLP/FRT-mediated site-specific recombination approach to introduce DHX9 shRNAs into the mouse germline22 (Supplementary Figure 4a). As a prelude to these studies, we tested several DHX9 shRNAs for suppression potency (Supplementary Table 1 and Supplementary Figure 4b). Several shRNAs (DHX9.1241, DHX9.1271 and DHX9.837) showed potent DHX9 suppression and two of these (DHX9.1271 and DHX9.837) were chosen for generating transgenic mice. Two mouse strains were generated, DHX9.837/rtTA and DHX9.1271/rtTA, containing DHX9 shRNA expression under TRE regulation at the Col1A1 locus and rtTA expression driven from the Rosa26 locus. Transgenic mice expressing a neutral shRNA targeting Firefly luciferase (FLuc.1309/rtTA) were used as controls22.
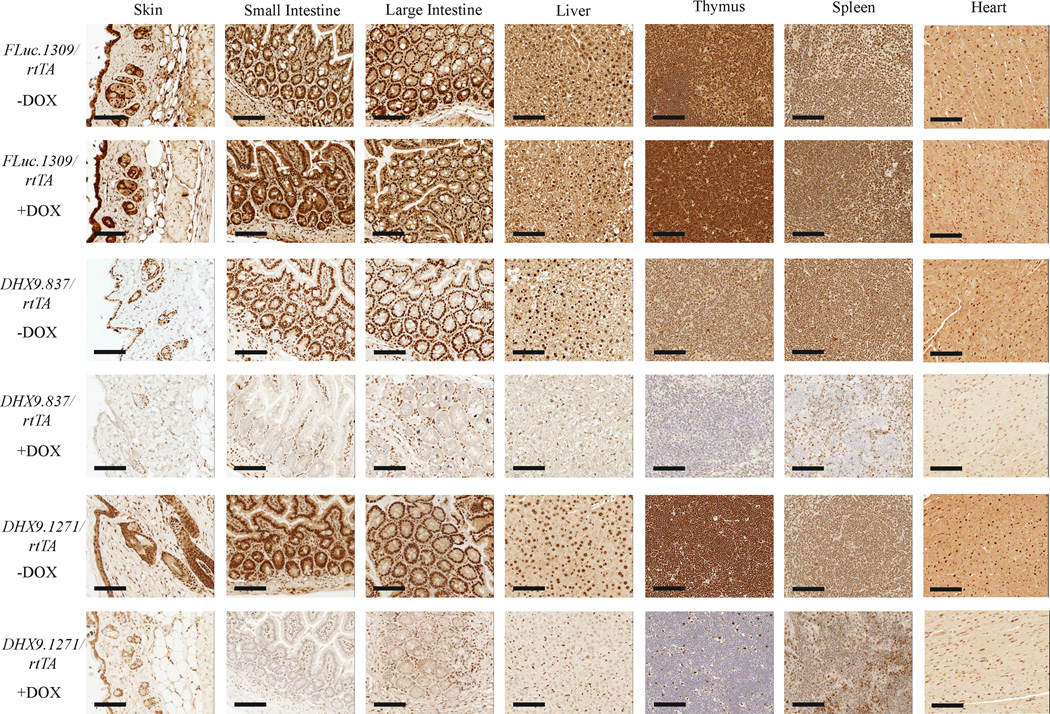
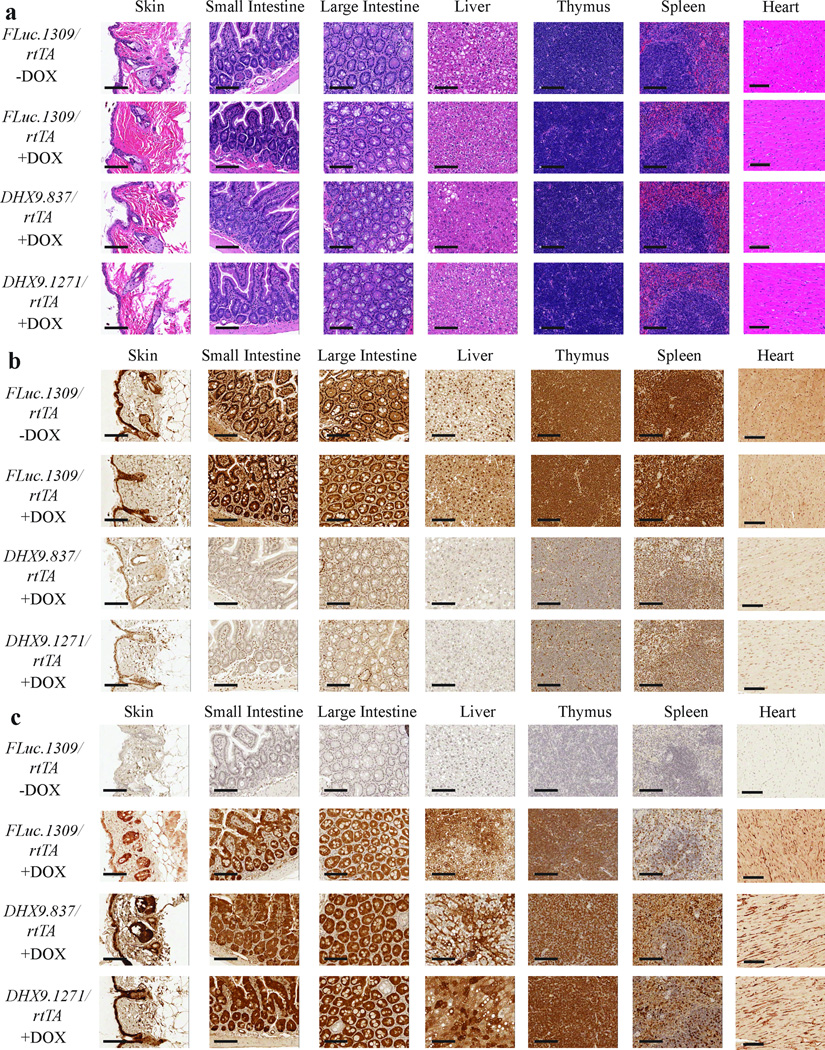
Examination of DHX9 expression in various tissues from FLuc.1309/rtTA, DHX9.837/rtTA, and DHX9.1271/rtTA mice treated with either vehicle or 1 mg/ml DOX for 14 days revealed suppression in all tissues examined, although the extent varied (Figure 4). The small intestine, large intestine, and thymus exhibited potent knockdown of DHX9 in both shDHX9 strains. In the skin, liver and heart, knockdown was moderate and the spleen exhibited weaker, mosaic suppression (Figure 4). Expression of DHX9 was predominantly nuclear (Supplementary Figure 5), consistent with previous studies29. TurboGFP, as expected, was DOX-inducible (Supplementary Figure 6). These results were confirmed by Western blot analysis of DHX9 knockdown and turboGFP expression in the aforementioned tissues (Supplementary Figure 7).
Figure 4. DOX-mediated DHX9 suppression in DHX9/rtTA mice.
Immunohistochemical analysis of representative tissues from FLuc.1309/rtTADHX9.837/rtTA, and DHX9.1271/rtTA mice treated with vehicle or 1 mg/ml DOX for 14 days. Sections were processed as described in the Supplementary Materials and Methods, probed with anti-DHX9 antibody, and counterstained with hematoxylin. Bars represent 100 µm.
Previous studies have shown that DHX9 knockdown increases p53 protein levels and activity in human fibroblasts, Arf−/−Eµ-Myc/Bcl-2 lymphomas, and U2OS cells19, 20. To determine whether these changes were recapitulated in our murine models, we examined RB1, p53, and CDKN1A status following DHX9 suppression (Supplementary Figure 7). We observed no consistent, robust induction of p53 in tissues from the two shDHX9 mouse strains either by Western blotting (Supplementary Figure 7) or immunohistochemistry (Supplementary Figure 8). There was also no consistent activation of the p53 target, CDKN1A. RB1 was expressed at very low levels in the skin and liver, and was unchanged upon DHX9 knockdown in all tissues analyzed (Supplementary Figure 7). These results indicate that under the conditions analyzed, we find no evidence for robust activation of p53, CDKN1A, or RB1 upon DHX9 suppression at the organismal level.
DHX9 knockdown in adult mice is well tolerated
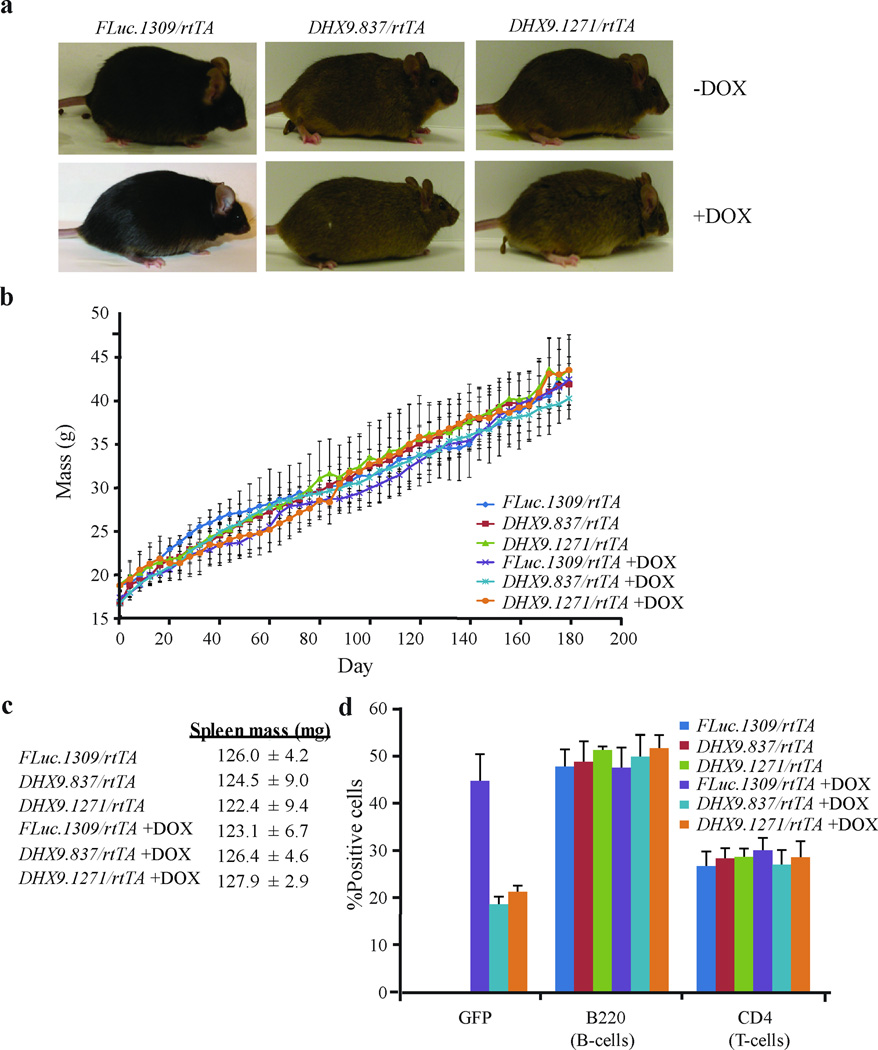
We next examined the consequences of DHX9 suppression to the general health and physiology of adult mice. Mice (4 week old) were treated with DOX for 6 months. Efficient long-term knockdown of DHX9 and induction of GFP was verified in various tissues (Figure 5). In this chronic DHX9-suppressed cohort, the weight, appearance, and behavior of DHX9.837/rtTA and DHX9.1271/rtTA mice on DOX were similar to those of FLuc.1309/rtTA (+DOX) and untreated mice (Figure 6a and b). We observed no evidence of weight loss, lowered activity levels, lack of grooming, hunched appearance, dehydration, or infections. Blood biochemical and hematological analyses of the treated cohorts revealed no significant differences from FLuc.1309/rtTA (+DOX) or untreated controls (Supplementary Tables 2 – 4), implying normal physiology and bone marrow function. Histopathological analysis of tissues from mice treated with DOX for 6 months did not reveal any pathological changes in the skin, small and large intestines, spleen, thymus, and heart specific to the DHX9/rtTA cohort (Table 1 and Figure 6c) nor in the % of splenic B and T cells (Figure 6d). In addition, Ki-67 and TUNEL staining was performed on the more proliferative tissues - namely skin, spleen, and small and large intestines. DHX9 suppression had no deleterious effects on proliferation in the skin and spleen, as determined by Ki-67 staining (Supplementary Figure 9). A slight decrease in Ki-67-positive cells was observed in the small and large intestines upon treatment with DOX, however this appeared to be due to the DOX itself, as it was seen in the FLuc.1309/rtTA +DOX mice as well. There appeared to be no significant difference between the FLuc.1309/rtTA and DHX9/rtTA DOX-treated samples in these tissues (Supplementary Figure 9). TUNEL staining revealed no increase in apoptotic cells upon DHX9 suppression in any of the tissues tested (Supplementary Figure 10).
Figure 5. Characterization of long-term DHX9 suppression.
(a) H/E staining of representative tissues from vehicle and DOX-treated (6 months) FLuc.1309/rtTADHX9.837/rtTA, and DHX9.1271/rtTA mice. Bars represent 100 µm. (b) Immunohistochemical analysis of representative tissues from untreated and DOX-treated (6 months) FLuc.1309/rtTADHX9.837/rtTA, and DHX9.1271/rtTA mice. Sections were probed with anti-DHX9 antibody and counterstained with hematoxylin. Bars represent 100 µm. (c) Immunohistochemical analysis of representative tissues from untreated and DOX-treated (6 months) FLuc.1309/rtTADHX9.837/rtTA, and DHX9.1271/rtTA mice. Sections were probed with anti-GFP (for FLuc.1309/rtTA) or anti-turboGFP (for DHX9.837/rtTA and DHX9.1271/rtTA) antibodies and counterstained with hematoxylin. Bars represent 100 µm.
Figure 6. Long-term suppression of DHX9 is well-tolerated in mice.
(a) Photographs of FLuc.1309/rtTADHX9.837/rtTA, and DHX9.1271/rtTA mice after treatment with vehicle or 1 mg/ml DOX for 6 months. (b) Weight of FLuc.1309/rtTADHX9.837/rtTA, and DHX9.1271/rtTA mice treated with vehicle or DOX over a 6 month period. Mice were weighed every 4 days starting at 4 weeks of age (t=Day 0). N=3 mice ±SEM. (c) Weight of spleens extracted from FLuc.1309/rtTADHX9.837/rtTA, and DHX9.1271/rtTA mice treated with vehicle or DOX for 6 months. Spleen weights were normalized to mouse weights. N=3 mice ±SEM. (d) Flow cytometry analysis of splenocytes extracted from FLuc.1309/rtTADHX9.837/rtTA, and DHX9.1271/rtTA mice after treatment with vehicle or DOX for 6 months. Single-cell suspensions were prepared from mouse spleen and stained with PE-conjugated antibodies against B220 and CD4, and the percentage of stained cells determined by FACS analysis. N=3 mice ±SEM.
Table 1. Histopathological analysis following long-term DHX9 suppression in DHX9/rtTA mice.
Histopathological analysis was performed on representative tissues extracted from FLuc.1309/rtTADHX9.837/rtTA, and DHX9.1271/rtTA mice treated with 1 mg/ml DOX for 6 months. Multifocal, random microgranulomas (Grade 1) were present in the liver of all mice analyzed and are likely a consequence of non-specific bacterial or viral infection. One DHX9.837/rtTA (+DOX) liver sample displayed moderate hepatic lipidosis which was not observed in the DHX9.1271/rtTA liver sample.
| FLuc.1309/ rtTA |
FLuc.1309/ rtTA |
DHX9.837/ rtTA |
DHX9.1271/ rtTA |
||
|---|---|---|---|---|---|
| DOX: | − | + | + | + | |
| Skin | N | N | N | N | |
| Small Intestine | N | N | N | N | |
| Large Intestine | N | N | N | N | |
| Liver | A | A | A | A | |
| Microgranuloma, multifocal, random |
1 | 1 | 1 | 1 | |
| Hepatic lipidosis, microvacuolar, centrolubular, diffuse |
3 | ||||
| Thymus | N | N | N | N | |
| Spleen | N | N | N | N | |
| Heart | N | N | N | N |
Key: N = no significant lesion, A = lesion observed, 0 = no tissue; Grade 1 = modest, rare, Grade 2 = mild, infrequent, Grade 3 = moderate, frequent, Grade 4 = severe, diffuse.
Assessing DHX9 suppression on global gene expression
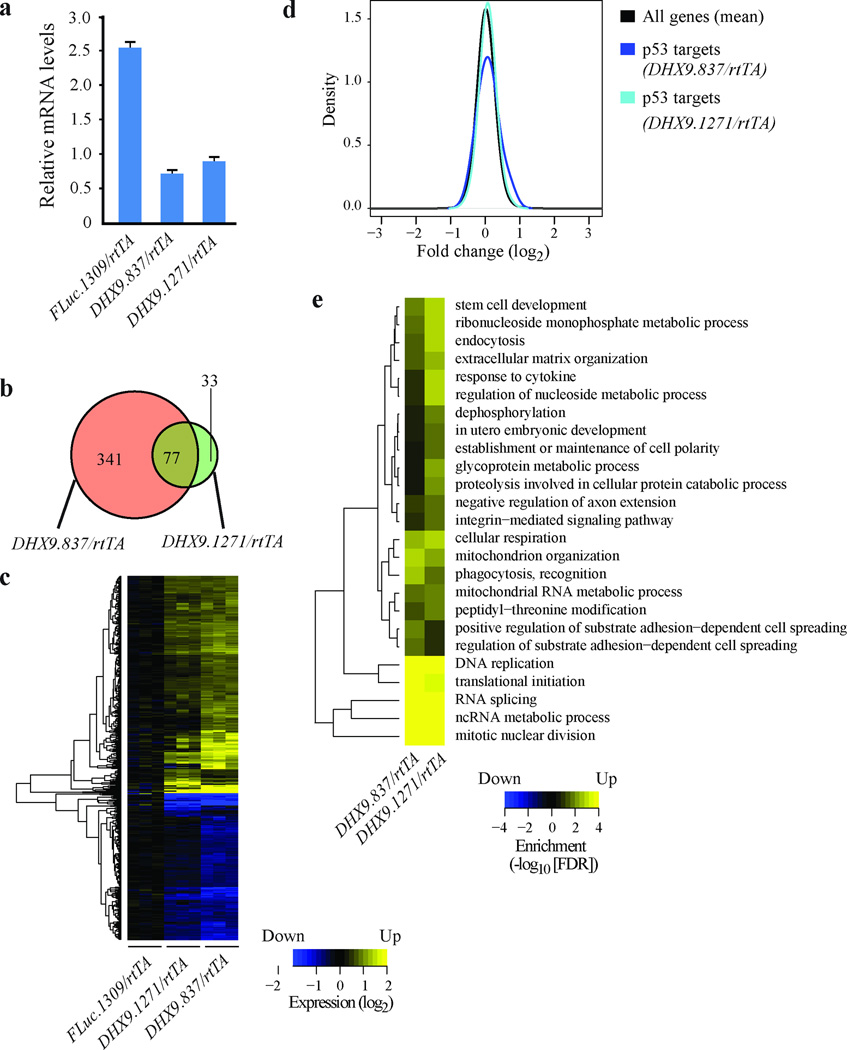
Having observed no apparent negative physiological effects of DHX9 suppression in mice, we asked whether reduced DHX9 levels induced any changes in global gene expression in vivo. To address this, we conducted gene expression analysis on the large intestine isolated from FLuc.1309/rtTA, DHX9.837/rtTA and DHX9.1271/rtTA mice treated with DOX for 14 days. The large intestine was chosen because it was one of the tissues showing potent suppression of DHX9, which was validated in samples isolated for global gene expression studies (Figure 7a). A comparison of the expression patterns between DHX9/rtTA and FLuc.1309/rtTA samples identified 451 transcripts as differentially expressed in at least one of the DHX9/rtTA mice (fold-change>1.5 and false discovery rate (FDR)<0.05; Figure 7b and Supplementary Table 5). Although only 77 transcripts were significantly altered in both shDHX9 transgenic samples, the fold-changes observed for the combined set of genes were largely consistent for both DHX9 shRNAs (Figure 7c). Our previous transcriptome-wide analysis of DHX9 suppression in cell lines showed activation of the p53 signaling pathway in ex vivo contexts19, 20. This phenomenon was not observed in the large intestine of DOX-treated DHX9.837/rtTA and DHX9.1271/rtTA mice (Figure 7d). Moreover, there were only minor changes in levels of p53 mRNA itself (1.2 or 1.4 fold for DHX9.837/rtTA and DHX9.1271/rtTA, respectively) without concomitant changes in CDKN1A, consistent with Western blot results (Supplementary Figure 7). Gene set enrichment analysis was applied to identify functions that were enriched among genes showing differential expression. The identified biological processes found to be most affected by DHX9 suppression were DNA replication, translation, RNA splicing, non-coding RNA processes, and nuclear division (Figure 7e and Supplementary Tables 6 and 7). Taken together, these results indicate significant transcriptome changes as a consequence of DHX9 suppression in the large intestine, but these do not lead to any overt pathological perturbations.
Figure 7. Gene expression analysis on large intestine of DHX9/rtTA mice.
(a) Quantitative RT-PCR analysis showing DHX9 knockdown in large intestines of DHX9.837/rtTA and DHX9.1271/rtTA mice treated with DOX for 14 days. mRNA levels were normalized to GAPDH. N=3 biological replicates ±SEM. (b) Venn diagram highlighting the number of common and distinct differentially expressed genes from DHX9.837/rtTA and DHX9.1271/rtTA mice. (c) Heatmap showing genes from FLuc.1309/rtTADHX9.837/rtTA, and DHX9.1271/rtTA mice up-or down-regulated at least 1.5 fold, FDR<0.05. (d) Densities of fold-changes (DHX9/rtTA vs FLuc.1309/rtTA) for all genes and a subset of p53 target genes. (e) Heatmap showing highly significant enrichment of biological processes among genes differentially expressed upon reduced DHX9 expression, FDR<0.01. Non-redundant biological processes were defined by the Gene Ontology Consortium.
DISCUSSION
One of the major challenges in chemotherapy is finding a therapeutic window in which drugs can efficiently eliminate tumor cells with minimal damage to normal tissues. Traditional strategies rely on the administration of cytotoxic agents (e.g. paclitaxel, etoposide, doxorubicin, etc.), which act by killing rapidly dividing cells, a property shared by both cancer cells and highly proliferative normal cells. With the advent of targeted molecular therapeutics, the development of drugs that inhibit specific gene products involved in tumor maintenance offer greater selectivity in eliminating cancer cells. However, the challenge of finding targets with suitable therapeutic indices remains, as targeting these same gene products in normal tissues can have deleterious effects. Hence, the search for novel molecular targets for chemotherapeutic use is a critical ongoing endeavor.
In the present study, we examined the outcome of DHX9 suppression in several human cancer cell lines, mouse engineered lymphomas, and at the organismal level in the mouse. DHX9 suppression resulted in a reduction in proliferative fitness in most (10 out of 13) human cancer cell lines representing five different types of cancers. This demonstrates that targeting DHX9 can be potentially effective against many, but not all, cancers. What makes a cancer a good candidate may be dependent on the relative expression levels of genes involved in proliferation, cell cycle progression, or apoptotic pathways. What makes MCF-7 breast cancer cells resistant to DHX9 suppression (Figure 1b) awaits further investigation but may be due to the fact that these express high levels of the anti-apoptotic proteins BCL-2 and MCL-1, and low levels of pro-apoptotic proteins c-MYC, Bim, and Bax (Supplementary Figure 2 and Ref. 30). Although the multiple myeloma lines JJN-3 and KMS-11 express BCL-2, the fact that they also express the pro-apoptotic proteins Bim, Bax, and Bak at relatively high levels (Supplementary Figure 2 and Ref. 31) may contribute to their inability to tolerate DHX9 suppression. All cell lines tested herein (Figure 1) have been reported to contain wildtype p53, with the exception of MDA-MB231 (Ref. 23). Hela cells contain wildtype p53 but overexpress the E6 protein from human papillomavirus type 16, which induces the degradation of p53 (Ref. 24). KMS-11 and JJN-3 exhibited the highest fold-increase in p53 levels, which may be a contributing factor to the robust depletion of DHX9 shRNA-expressing cells observed in the competition assay for these cell lines. However, the level of p53 activation was not the only determinant of susceptibility to DHX9. Both MDA-MB231 and Hela cells were sensitive to DHX9 suppression, despite harboring mutated p53 or low p53 levels, while MCF-7 cells, which contain wild-type p53, were resistant. Thus, it is likely that susceptibility of a cancer cell line to DHX9 inhibition is not dependent on a single factor, a topic that will require further investigation. Of the cancer cell lines which showed GFP depletion upon DHX9 suppression in the competition assay (Figure 1b), increased cell death was documented in all cell lines except the U2OS osteosarcoma line, which underwent a G0/G1 arrest instead (Supplementary Figure 1). We note that only the U2OS and MRC-5 cells demonstrated a significant increase in CDKN1A levels, an inhibitor of proliferation. Activation of CDKN1A has been shown to lead to cell cycle arrest and senescence rather than apoptosis32, 33. In fact, studies have shown that in some cases it may actually inhibit apoptosis34 and may explain why loss of DHX9 does not result in increased cell death in U2OS and MRC-5 cells but causes growth arrest and/or senescence19, 20, 35. In all the other cell lines, which did not show CDKN1A induction, it appeared that apoptosis rather than cell cycle arrest was primarily responsible for the decrease in proliferative fitness observed upon DHX9 knockdown (Figure 1c and Supplementary Figure 1). These results illustrate that the precise effects of DHX9 suppression may depend on biological context. The lethal effect observed in several human cancer lines with DHX9 suppression was recapitulated in murine Eµ-Myc lymphomas ex vivo and in vivo.
Having demonstrated that DHX9 suppression is not tolerated by most tumor cells, we investigated the consequences on normal tissues in vivo. To this end, we generated inducible RNAi-based transgenic mice to study the consequences of DHX9 loss at the organismal level. This inducible model allowed us to achieve conditional and reversible suppression of DHX9, which would not have been possible using a straight knockout model. Indeed, Lee and colleagues previously generated a DHX9 knockout mouse and observed that homozygous loss leads to embryonic lethality36. Their results suggest that DHX9 is essential for the differentiation of the embryonic ectoderm36. Consistent with this, the C. elegans homologue, rha-1, is necessary for germline transcriptional control and proliferation37. In this report, we find that partial suppression of DHX9 is well tolerated at the organismal level in adult mice, a situation that reflects the scenario one would expect with a small molecule inhibitor of DHX9. Indeed, chronic suppression of DHX9 in the mouse for 6 months had no noticeable effect, indicating that the organism appears well buffered to tolerate profound changes in DHX9 levels.
Although the mice appeared to suffer no ill effects resulting from DHX9 suppression, analysis of global gene expression in the large intestine identified numerous genes whose expression was affected upon DHX9 loss (Figure 7). Our enrichment analysis revealed that genes involved in metabolism, DNA replication, translational initiation, mitotic nuclear division, RNA splicing, amongst others, were upregulated. It should be noted this analysis included genes which negatively regulate the aforementioned processes. An example is the increased expression of eIF4EBP1, a translational inhibitor. However, eIF4G1, eIF2A, and DHX29, all known to stimulate translation38, 39, were also upregulated (Supplementary Tables 6 and 7); thus the net result on translation is difficult to predict. Similarly, genes involved in mitotic nuclear division found to be upregulated included promoters of this process (e.g. SMC2, CDK1, CDC20, CDC25C, and Plk1) as well as checkpoint proteins (e.g. Mad2/1, CHEK1, CDC27) (Supplementary Tables 6 and 7). Hence, there may be little net effect on cell cycle progression. Interestingly, there appeared to be no activation of a p53 transcriptional program upon DHX9 suppression, in contrast to previous genome-wide expression studies in Arf−/−Eµ-Myc/Bcl-2 lymphomas and MRC-5 human fibroblasts, where p53 pathway activation elicited an apoptotic or senescence response respectively19, 20. On a more general level, comparison of our present data with previous data generated in Arf−/−Eµ-Myc/Bcl-2 and MRC-5 cells revealed no significant overlap with the previous studies (data not shown). In MRC-5 cells, DHX9 suppression resulted in downregulation of many genes involved in promoting DNA replication, mitosis, and cell cycle progression20. This was not recapitulated in vivo. The fact that we did not observe activation of the p53 pathway or global downregulation of proliferative and replication genes at the organismal level is consistent with the lack of detectable phenotype resulting from reduced DHX9 expression in mice. Conversely, the upregulation of many biological processes may represent a compensatory response to the cellular perturbations caused by DHX9 suppression – a mechanism that may occur in vivo but not ex vivo. In any event, the changes in global gene expression were small compared to those sustained ex vivo, and appear to be relatively benign, as they resulted in no drastic consequences in the mice.
While the two DHX9 shRNAs exhibited similar levels of knockdown ex vivo and in most tissues in vivo (Figures 4, 5 and 7a, and Supplementary Figures 4b, 6, and 7), histopathological analysis revealed that the DHX9.837/rtTA +DOX mice exhibited moderate hepatic lipidosis, which was not found in the DHX9.1271/rtTA +DOX mice (Table 1). Although we have not investigated the potential reasons for this additional phenotype, we do not feel it can be explained by differences in knockdown potency (which appear quite similar). We cannot rule out that it may represent a possible off-target effect of shDHX9.837 at this time.
Why loss of DHX9 has such detrimental effects on tumor cells but no negative impact on normal adult tissues is not entirely clear, but this finding has been documented in a number of previous settings with key regulatory proteins. DDX5 is a DEAD-box RNA helicase with many similarities to DHX9. It is multifunctional, with important roles in transcriptional regulation, microRNA processing, RNA splicing, and ribosome biogenesis40–43. Furthermore, loss of DDX5 is embryonic lethal42. In a study by Mazurek et al.44, DDX5 knockdown induced apoptosis in acute myeloid leukemia (AML) cells but was well tolerated in adult mice. Mice expressing DDX5 shRNAs were healthy, gained weight normally, suffered no physiological defects, and had no deleterious effects on organ morphology44. Similarly, Soucek and colleagues modeled the therapeutic impact of systemic Myc inhibition in a Kras-induced lung cancer mouse model using a dominant-negative Myc allele45. Conditional expression of the mutant (termed Omomyc) allele impeded initiation and maintenance of Kras-induced lung adenomas by eliciting apoptosis in the tumors. On the other hand, adult mice expressing Omomyc exhibited no significant differences compared to control mice in terms of body weight, blood chemistry, and general health and activity45. Organs with low proliferative indices were unaffected by MYC inhibition; however, highly proliferative tissues such as skin and intestines did suffer from increased apoptosis, degeneration, and decreased proliferation, effects that were reversible upon restoration of Myc function. Another example illustrating the differential effects of suppressing a protein in tumor cells versus normal tissues involves suppression of the translation initiation factor eIF4E, which catalyzes the ribosome recruitment step of translation initiation. Suppression of eIF4E in the Eµ-Myc lymphoma model significantly delayed MYC-dependent tumor initiation by augmenting apoptosis and impairing cell division in premalignant B-cells21. eIF4E suppression in adult mice resulted in a reduction in body weight and increased apoptosis and loss of differentiation in the intestines, but these effects were reversible upon DOX withdrawal21. These mouse models, like the shDHX9 mice described herein, demonstrate that although a gene product may be essential, its partial suppression (as would be expected from a small molecule inhibitor) may be well tolerated at the organismal level.
Although our study does not identify a single major pathway affected by DHX9 suppression in tumor cells, its role in a multitude of cellular processes (see Introduction) may indicate a higher dependency of tumor cells for this gene product. In the mouse, since the majority of cells are not actively proliferating, but are instead quiescent46, these cells may have a lower requirement for DHX9. Interestingly, DHX9 knockdown had no deleterious consequences in tissues with high proliferative indices, such as the intestines and skin. This may be due to the fact that although suppression is significant in these tissues, levels of remaining DHX9 are sufficient to maintain normal cell function and prevent activation of a p53 response. It should be noted that ex vivo, both tumor cells and normal diploid fibroblasts were susceptible to DHX9 knockdown, although the specific effect elicited was different (apoptosis versus senescence). The differential effects of DHX9 suppression observed in vivo versus ex vivo in terms of cellular fitness and p53 pathway activation may be partly attributed to cell culture stress. In a physiological environment, reduction in DHX9 levels may be well tolerated. However, the added stress of being cultured ex vivo, at higher oxygen levels and separated from their native extracellular matrix, may sensitize cells sufficiently to activate a p53 stress response. The knockdown of DHX9 in vivo may not be as potent as what can be achieved ex vivo, and may not reach a high enough threshold to activate p53. Another possible explanation is that there may be some form of compensation for the suppression of DHX9 in vivo – for example, upregulation of other replication factors. In sum, our study demonstrates that DHX9 is dispensable for normal tissue homeostasis in the adult mouse and warrants exploration of DHX9 as an anti-neoplastic target. What makes DHX9 particularly attractive is that unlike other potential therapeutic targets that have been explored using conditional shRNA mice21, 27, 45, its suppression does not have negative effects on tissues harboring highly proliferative cells.
MATERIALS AND METHODS
Cell Lines
NIH3T3, Hela, and HEK293T/17 cells (ATCC, Manassas, VA, USA), and MRC-5 cells (a kind gift from Dr. Nahum Sonenberg, McGill University, Canada) were maintained in DMEM (Multicell, St-Bruno, QC, Canada). U2OS cells (ATCC) were maintained in McCoy 5A (Multicell). MCF-7 (ATCC), MDA-MB231 (ATCC), and JJN-3, KMS-11, IM-9, RPMI8226, U266B1, H929, OPM1.1, and OPM2 cells (kindly provided by Dr. Michael Sebag, McGill University, Canada) were maintained in RPMI-1640 media (Multicell). A549 cells (ATCC) were maintained in F12K (Multicell). All media was supplemented with 10% fetal bovine serum (Multicell). ATCC authenticates all their cell lines using Short Tandem Repeat profiling. Cells were not tested for mycoplasma contamination. Tsc2+/−Eµ−Myc lymphoma cells were derived from tumors in Tsc2+/− mice crossed with Eµ−Myc mice and inbred on the C57BL/6 (Charles River Laboratories) background for over ten generations. Tsc2+/−Eµ-Myc/R26-M2rtTA lymphomas were generated by crossing Tsc2+/−Eµ−Myc mice with mice expressing the M2rtTA transgene at the Rosa26 locus and harvesting tumors from the resultant triple transgenic progeny. Lymphomas were cultured in B-cell media (45% DMEM, 45% Iscove’s media, 55 mM β-mercaptoethanol, 10% fetal bovine serum) on irradiated Ink4A−/− MEF feeder layers.
Virus Generation and Transductions
For suppression of DHX9 in murine cell lines (NIH3T3 and Eµ-Myc lymphomas), shRNAs targeting mouse DHX9 (DHX9.1241 and DHX9.1271) and a control shRNA targeting renilla luciferase (RLuc.713) were transduced into cells using the MSCV/LTR/miR30/PuroR-IRES-GFP (MLP) or MSCV/LTR/miR30/SV40-GFP (MLS) retroviral vectors (Supplementary Table 1). Retroviral infections were performed using ecotropic Phoenix packaging cells following established protocols (http://www.stanford.edu/group/nolan/retroviral_systems/retsys.html). Briefly, 20 µg of plasmid were transfected into Phoenix ecotropic cells in a 10 cm dish by calcium phosphate-mediated delivery. The media was changed 12 hours later and 48 hours post-transfection, retroviral supernatant was collected every 8 hours up to 72 hours and added to target cells. For infections using MLP, forty-eight hours after the last transduction, stable integrants were selected using 2 µg/ml puromycin for at least 3 days. For suppression of DHX9 in human cell lines, two shRNAs targeting human DHX9 (DHX9.860 and DHX9.267) and a control shRNA targeting firefly luciferase (FLuc.1309) were transduced into cells using pPrime-PGK-Puro (Addgene, Cambridge, MA, USA) (Supplementary Table 1). Lentiviral transduction was performed following published procedures47. Briefly, 15 µg of pPrime-PGK-Puro-shRNA, 7.5 µg of packaging plasmid pSPAX2, and 3.75 µg of envelope-encoding vector, CMV-VSVG, were mixed and transfected into HEK293T/17 cells in a 10 cm dish by calcium phosphate-mediated delivery.
Generation, genotyping, and induction of transgenic mice
Transgenic mice harboring a Rosa26 (R26)-m2rtTA allele and DHX9.837 or DHX9.1271 shRNAs under the control of a tetracycline-inducible promoter (TRE) were generated through FLP/FRT-mediated site specific recombination at the Col1A1 locus on chromosome 11 by Mirimus (Cold Spring Harbor, NY, USA). PCR was used for genotyping using shRNA-specific forward primers [FLuc.1309: 5’AAGCCACAGATGTATTAATCAGAGA3’, DHX9.837: 5’AAGCCACAGATGTATTTAGATCCAT3’, and DHX9.1271: 5’AAGCCACAGATGTATAAATTATGAT3’], combined with a common reverse primer, Col1A1 Rev43 [5’GAAAGAACAATCAAGGGTCC3’]. Genotyping for the R26-m2rtTA allele was performed using Mutant For: 5’AAAGTCGCTCTGAGTTGTTAT3’, Rev: 5’GCGAAGAGTTTGTCCTCAACC3’, and WT For: 5’GGAGCGGGAGAAATGGATATG3’ primers.
All mice strains were maintained on a C57BL/6 background. Induction of shRNA expression was performed in 4-week-old shRNA/rtTA mice by supplying doxycycline (DOX) at 1 mg/ml in the drinking water (plus 5% sucrose) for 14 days (for short-term experiments) or 6 months (for long-term experiments). The DOX-supplemented water was changed every 4 days. All animal studies were approved by the McGill University Faculty of Medicine Animal Care Committee.
Immunohistochemical analysis and TUNEL staining of mouse tissues
Tissues from shRNA/rtTA mice were harvested, fixed in 10% formalin for 48h, and embedded in paraffin. Sections (4 µm) were deparaffinized in xylene, and rehydrated through a series of decreasing ethanol washes (100%, 95%, and 75%), followed by washing in water for 2 × 5 minutes. Antigen retrieval was performed by boiling the slides in 10 mM citric acid buffer [pH 6.0] for 15 minutes. Immunohistochemistry was performed using the HRP/DAB Detection Kit (ab64261; Abcam, Cambridge, MA, USA) according to the manufacturer’s instructions and as previously described21. The following primary antibodies and dilutions were used: DHX9 (ab26271; Abcam, Cambridge, MA, USA) (1:100 dilution), GFP (#2555; Cell Signaling, Danvers, MA, USA) (1:800 dilution), turboGFP (AB514; Evrogen, Moscow, Russia) (1:5000 dilution) and Ki-67 (Sp6; Neomarkers, Fremont, Ca, USA). After visualizing the signal using DAB chromogen and substrate, sections were counterstained with hematoxylin, destained with 0.5% HCl in 70% ethanol, incubated with 0.2% lithium carbonate, and rinsed with tap water. The sections were then dehydrated through an ethanol gradient (75%, 95%, and 100% ethanol), followed by xylene washes (2 × 5 minutes), and mounted using Permount (Fisher, Ottawa, ON, Canada). Sections were scanned using an Aperio ScanScope XT (Aperio Technologies, Vista, CA, USA). TUNEL staining was performed using the In situ Cell Death Detection Kit, TMR-Red (#12156792910; Roche, Mannheim, Germany) according to the manufacturer’s instructions. Images were taken with a LSM 510 Meta Confocor2 Confocal Microscope (Zeiss) at 40X magnification and the percentage of apoptotic cells quantitated using ImageJ (National Institutes of Health, Bethesda, Maryland, USA).
Ex vivo competition assays
Ex vivo competition assays were performed by transducing cells with MLS-based (for mouse cells) or pPrime-PGK-puro-based (for human cells) shRNAs. The percentage of GFP-positive cells was measured 48h after the final infection (t=0) using a GUAVA EasyCyte HT flow cytometer (Millipore, Billerica, MA, USA). Cell death was assessed by staining cells with 4 µg/ml propidium iodide (PI) and measuring the percentage of PI-positive cells. For competition assays using the DOX-inducible vector TRMPV, Tsc2+/−Eµ-Myc/R26-M2rtTA cells were transduced with TRMPV-shRNA and FAC-sorted to obtain a pure Venus+ population. shRNA expression was induced by treatment with vehicle or 1 µg/ml DOX and cell death assessed by staining with Fixable Viability Dye eFluor450 (eBioscience) and measuring the percentage of stained cells using a LSRII cytometer (BD Biosciences).
In vivo competition assays
Tsc2+/−Eµ-Myc cells were transduced with MLS-based shRNAs and two days post-infection, 106 cells were injected into the tail vein of C57BL/6 recipients (not randomized or blinded to the investigator). Mice were monitored until they reached terminal disease stage (characterized by the appearance of tumors, decreased activity, hunched posture, dehydration, paralysis, and weight loss), at which point the spleen was harvested and analyzed for GFP-positive B-cells using flow cytometry.
Survival analysis following tumor transplantation
Tsc2+/−Eµ-Myc/R26-M2rtTA lymphoma cells were transduced with TRMPV retroviruses expressing shRNAs to RLuc, DHX9 or L15, FAC-sorted to obtain a pure Venus+ population, and 106 cells were introduced into 10 C57BL/6 mice for each shRNA via tail-vein injection. 6 days post-injection, 5 mice for each shRNA was treated with 1 mg/ml DOX (in 5% sucrose) and the other 5 mice were treated with 5% sucrose. The mice were sacrificed when they reached terminal disease stage, at which point the spleen was harvested and the percentages of B220+, Venus+, and dsRed+ cells were analyzed by flow cytometry. The survival (number of days to reach terminal stage) of each mouse was plotted on a Kaplan-Meier curve using Graphpad Prism (v. 5.03, GraphPad Software Inc., La Jolla, CA, USA) and the p-values determined using the Log-rank (Mantel-Cox) test.
Statistical analysis
Statistical analysis was carried out using GraphPad Prism (v. 5.03, GraphPad Software Inc., La Jolla, CA, USA) and data is shown as mean ±SEM. For all analyses except the Kaplan-Meier analysis (see above), statistically significant differences were determined using the unpaired two-tailed t-test and represented as p-values.
Supplementary Material
Acknowledgments
We thank Francis Robert for performing tail-vein injections of lymphoma cells into mice. TL was supported by a Maysie MacSporran graduate studentship and fellowships from the CIHR-sponsored Chemical Biology and Systems Biology Training Programs. This work is supported by grants from the Canadian Cancer Society Research Institute [#701362] and National Institutes of Health [CA163291] to JP and grants from the Swedish Research Council and the Swedish Cancer Society to OL.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
TL performed all experiments under the guidance of JP. MP provided the histopathological analysis of tissue sections. OL analyzed the gene expression data. TL and JP wrote the manuscript.
REFERENCES
- 1.Zhang S, Grosse F. Nuclear DNA helicase II unwinds both DNA and RNA. Biochemistry. 1994;33:3906–3912. doi: 10.1021/bi00179a016. [DOI] [PubMed] [Google Scholar]
- 2.Jain A, Bacolla A, Chakraborty P, Grosse F, Vasquez KM. Human DHX9 helicase unwinds triple-helical DNA structures. Biochemistry. 2010;49:6992–6999. doi: 10.1021/bi100795m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang SS, Grosse F. Purification and characterization of two DNA helicases from calf thymus nuclei. J Biol Chem. 1991;266:20483–20490. [PubMed] [Google Scholar]
- 4.Lee CG, Hurwitz J. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3’ to 5’ direction. J Biol Chem. 1992;267:4398–4407. [PubMed] [Google Scholar]
- 5.Lee CG, Hurwitz J. Human RNA helicase A is homologous to the maleless protein of Drosophila. J Biol Chem. 1993;268:16822–16830. [PubMed] [Google Scholar]
- 6.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S, Grosse F. Domain structure of human nuclear DNA helicase II (RNA helicase A) J Biol Chem. 1997;272:11487–11494. doi: 10.1074/jbc.272.17.11487. [DOI] [PubMed] [Google Scholar]
- 8.Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, et al. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 9.Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nature Genetics. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 10.Huo L, Wang YN, Xia W, Hsu SC, Lai CC, Li LY, et al. RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. PNAS USA. 2010;107:16125–16130. doi: 10.1073/pnas.1000743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tetsuka T, Uranishi H, Sanda T, Asamitsu K, Yang JP, Wong-Staal F, et al. RNA helicase A interacts with nuclear factor kappaB p65 and functions as a transcriptional coactivator. European Journal of Biochemistry / FEBS. 2004;271:3741–3751. doi: 10.1111/j.1432-1033.2004.04314.x. [DOI] [PubMed] [Google Scholar]
- 12.Hartman T, Qian S, Bolinger C, Fernandez S, Schoenberg D, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat Struct Mol Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 13.Manojlovic Z, Stefanovic B. A novel role of RNA helicase A in regulation of translation of type I collagen mRNAs. RNA. 2012;18:321–334. doi: 10.1261/rna.030288.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 15.Tang H, Gaietta GM, Fischer WH, Ellisman MH, Wong-Staal F. A cellular cofactor for the constitutive transport element of type D retrovirus. Science. 1997;276:1412–1415. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Schlott B, Gorlach M, Grosse F. DNA-dependent protein kinase (DNA-PK) phosphorylates nuclear DNA helicase II/RNA helicase A and hnRNP proteins in an RNA-dependent manner. Nucleic Acids Res. 2004;32:1–10. doi: 10.1093/nar/gkg933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain A, Bacolla A, Del Mundo IM, Zhao J, Wang G, Vasquez KM. DHX9 helicase is involved in preventing genomic instability induced by alternatively structured DNA in human cells. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakraborty P, Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst) 2011;10:654–665. doi: 10.1016/j.dnarep.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 19.Mills JR, Malina A, Lee T, Di Paola D, Larsson O, Miething C, et al. RNAi screening uncovers Dhx9 as a modifier of ABT-737 resistance in an Eµ-myc/Bcl-2 mouse model. Blood. 2013;121:3402–3412. doi: 10.1182/blood-2012-06-434365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee T, Di Paola D, Malina A, Mills JR, Kreps A, Grosse F, et al. Suppression of the DHX9 helicase induces premature senescence in human diploid fibroblasts in a p53-dependent manner. J Biol Chem. 2014;289:22798–22814. doi: 10.1074/jbc.M114.568535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CJ, Nasr Z, Premsrirut PK, Porco JA, Jr, Hippo Y, Lowe SW, et al. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Reports. 2012;1:325–333. doi: 10.1016/j.celrep.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hui L, Zheng Y, Yan Y, Bargonetti J, Foster DA. Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D. Oncogene. 2006;25:7305–7310. doi: 10.1038/sj.onc.1209735. [DOI] [PubMed] [Google Scholar]
- 24.Hoppe-Seyler F, Butz K. Repression of endogenous p53 transactivation function in HeLa cervical carcinoma cells by human papillomavirus type 16 E6, human mdm-2, and mutant p53. J Virology. 1993;67:3111–3117. doi: 10.1128/jvi.67.6.3111-3117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robert F, Roman W, Bramoulle A, Fellmann C, Roulston A, Shustik C, et al. Translation initiation factor eIF4F modifies the dexamethasone response in multiple myeloma. PNAS USA. 2014;111:13421–13426. doi: 10.1073/pnas.1402650111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 27.McJunkin K, Mazurek A, Premsrirut PK, Zuber J, Dow LE, Simon J, et al. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. PNAS USA. 2011;108:7113–7118. doi: 10.1073/pnas.1104097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nature biotechnology. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang H, McDonald D, Middlesworth T, Hope TJ, Wong-Staal F. The carboxyl terminus of RNA helicase A contains a bidirectional nuclear transport domain. Mol Cell Biol. 1999;19:3540–3550. doi: 10.1128/mcb.19.5.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calcabrini A, Garcia-Martinez JM, Gonzalez L, Tendero MJ, Ortuno MT, Crateri P, et al. Inhibition of proliferation and induction of apoptosis in human breast cancer cells by lauryl gallate. Carcinogenesis. 2006;27:1699–1712. doi: 10.1093/carcin/bgl044. [DOI] [PubMed] [Google Scholar]
- 31.Trudel S, Stewart AK, Li Z, Shu Y, Liang SB, Trieu Y, et al. The Bcl-2 family protein inhibitor, ABT-737, has substantial antimyeloma activity and shows synergistic effect with dexamethasone and melphalan. Clinical Cancer Research. 2007;13:621–629. doi: 10.1158/1078-0432.CCR-06-1526. [DOI] [PubMed] [Google Scholar]
- 32.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 33.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nature reviews Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 34.Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Molecular Cancer Therapeutics. 2002;1:639–649. [PubMed] [Google Scholar]
- 35.Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. PNAS USA. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee CG, da Costa Soares V, Newberger C, Manova K, Lacy E, Hurwitz J. RNA helicase A is essential for normal gastrulation. PNAS USA. 1998;95:13709–13713. doi: 10.1073/pnas.95.23.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walstrom KM, Schmidt D, Bean CJ, Kelly WG. RNA helicase A is important for germline transcriptional control, proliferation, meiosis in C. elegans. Mechanisms of Development. 2005;122:707–720. doi: 10.1016/j.mod.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Parsyan A, Shahbazian D, Martineau Y, Petroulakis E, Alain T, Larsson O, et al. The helicase protein DHX29 promotes translation initiation, cell proliferation, and tumorigenesis. PNAS USA. 2009;106:22217–22222. doi: 10.1073/pnas.0909773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dardenne E, Pierredon S, Driouch K, Gratadou L, Lacroix-Triki M, Espinoza MP, et al. Splicing switch of an epigenetic regulator by RNA helicases promotes tumor-cell invasiveness. Nature Structural & Molecular Biology. 2012;19:1139–1146. doi: 10.1038/nsmb.2390. [DOI] [PubMed] [Google Scholar]
- 41.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nature Cell Biology. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 43.Saporita AJ, Chang HC, Winkeler CL, Apicelli AJ, Kladney RD, Wang J, et al. RNA helicase DDX5 is a p53-independent target of ARF that participates in ribosome biogenesis. Cancer Research. 2011;71:6708–6717. doi: 10.1158/0008-5472.CAN-11-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mazurek A, Park Y, Miething C, Wilkinson JE, Gillis J, Lowe SW, et al. Acquired dependence of acute myeloid leukemia on the DEAD-box RNA helicase DDX5. Cell Reports. 2014;7:1887–1899. doi: 10.1016/j.celrep.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nature Reviews Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 47.Barde I, Salmon P, Trono D. Production and titration of lentiviral vectors. Curr Protoc Neurosci. 2010 doi: 10.1002/0471142301.ns0421s53. Chapter 4: Unit 4.21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.