Abstract
In all eukaryotes, the highly repeated 35S ribosomal DNA (rDNA) sequences encoding 18S-5.8S-26S ribosomal RNA (rRNA) typically show high levels of intragenomic uniformity due to homogenisation processes, leading to concerted evolution of 35S rDNA repeats. Here, we compared 35S rDNA divergence in several seed plants using next generation sequencing and a range of molecular and cytogenetic approaches. Most species showed similar 35S rDNA homogeneity indicating concerted evolution. However, Cycas revoluta exhibits an extraordinary diversity of rDNA repeats (nucleotide sequence divergence of different copies averaging 12 %), influencing both the coding and non-coding rDNA regions nearly equally. In contrast, its rRNA transcriptome was highly homogeneous suggesting that only a minority of genes (<20 %) encode functional rRNA. The most common SNPs were C > T substitutions located in symmetrical CG and CHG contexts which were also highly methylated. Both functional genes and pseudogenes appear to cluster on chromosomes. The extraordinary high levels of 35S rDNA diversity in C. revoluta, and probably other species of cycads, indicate that the frequency of repeat homogenisation has been much lower in this lineage, compared with all other land plant lineages studied. This has led to the accumulation of methylation-driven mutations and pseudogenisation. Potentially, the reduced homology between paralogs prevented their elimination by homologous recombination, resulting in long-term retention of rDNA pseudogenes in the genome.
Electronic supplementary material
The online version of this article (doi:10.1007/s00412-015-0556-3) contains supplementary material, which is available to authorized users.
Keywords: Cycadales, Living fossil, rDNA, Concerted evolution, Cytosine methylation
Introduction
In eukaryotes, rDNA encodes four types of ribosomal RNAs (rRNA), i.e. 18S, 5.8S, 26S and 5S rRNA. The 18S, 5.8S and 26S genes (referred to as 35S rDNA in plants) are transcribed as a single operon and are separated by internal transcribed spacers (ITS) and intergenic spacers (IGS). Cytogenetically, 35S rDNA are clustered forming distinct chromosomal domains called nucleolar organizer regions (NORs) or dispersed (Neves et al. 2005). Despite high copy number (hundreds to thousands of units in plants), the genes are nearly identical which are explained by their concerted evolution. Under such an evolutionary process, if the entire 35S rDNA unit is equally affected by concerted evolution, then we expect to observe (i) little or no differences in the degree of intragenomic homogeneity between coding and non-coding regions and (ii) that non-coding regions (i.e. the internal transcribed spacers (ITS1 and ITS2) and the intergenic spacer (IGS)) have lower selection constraints than the coding regions, with the latter evolving under strong purifying selection (Eickbush and Eickbush 2007). However, available data show there can be considerable intragenomic heterogeneity of ITS (Harpke and Peterson 2006; Kovarik et al. 2008; Zheng et al. 2008; Fehrer et al. 2009) and IGS (Matyasek et al. 2012; Galián et al. 2014) sequences, suggesting incomplete unit homogenisation.
Detection of intragenomic rDNA sequence diversity using conventional methods is particularly challenging given the large size of the rDNA unit (several kbs) and their repetitive nature. However, with the advent of next generation sequencing (NGS), it is now possible to quantify variations in the large repeat arrays, including rDNA (Buggs et al. 2012; Straub et al. 2012). Another potential advantage of NGS is the ability to obtain similar sequence coverage for regions with low and high sequence identity, avoiding potential ascertainment bias generated by classical PCR and cloning methods. NGS has already been used to determine intergenomic rDNA variation in the insect Drosophila (Stage and Eickbush 2007) and several fungi (Ganley and Kobayashi 2011; West et al. 2014). These studies found little or no variation in either the coding or non-coding regions, confirming classical studies and supporting concerted evolution models. In contrast, a few highly polymorphic sites within the coding regions were detected in angiosperm taxa (Zozomova-Lihova et al. 2014; Weitemier et al. 2015) suggesting that rDNA may be more polymorphic in some plants than in other species. Nevertheless, the frequency of highly polymorphic sites was low generally and not exceeding a few polymorphic sites per ~5 kb of the 18S-5.8S-26S operon.
Despite the ecological and economic importance of gymnosperms, their genomes have not been investigated comprehensively, especially in species outside of the conifers (Pinales) (Leitch and Leitch 2012). Recently, whole genome sequencing (WGS) was carried out in two conifers, Picea abies (Nystedt et al. 2013) and Pinus taeda (Neale et al. 2014). The genomes of both species showed similar organisation and were highly enriched in transposons and divergent repeated elements of unknown structure and function. However, unlike many repeats in angiosperms, they showed evidence of relatively slow evolutionary divergence. Other studies have shown that rDNA in gymnosperms is also unusual compared with many angiosperms. For example, (i) gymnosperms tend to have more 35S rDNA loci (Hizume et al. 1992a; Hizume et al. 1992b; Kondo et al. 1996; Lubaretz et al. 1996; Murray et al. 2002; Siljak-Yakovlev et al. 2002; Cai et al. 2006; Islam-Faridi et al. 2007 and reviewed in Roa and Guerra (2012)); (ii) the ITS1 region is longer, with considerable length heterogeneity between species (Maggini and Baldassini 1995; Liston et al. 1996; Won and Renner 2005). (iii) the size of the gymnosperm 35S rDNA unit is longer (exceeding 25 kb) than in studied angiosperms (Bobola et al. 1992; Ribeiro et al. 2008). but similar in length to those of early diverged land plants, such as Funaria hygrometrica (moss, Bryopsida) (Capesius 1997) and Marchantia polymorpha (liverwort, Marchantiopsida) (Sone et al. 1999). (iv) the 5S rRNA gene is frequently found embedded in the IGS (Garcia and Kovařík 2013) which is rarely observed in angiosperms (Garcia et al. 2009). (v) intragenomic ITS paralogs are often heterogeneous in Pinales (Wei et al. 2003; Campbell et al. 2005). Gnetales (Won and Renner 2005) and Cycadales (Bogler and Francisco-Ortega 2004; Xiao et al. 2010; Feng et al. 2014; Xiao and Moller 2015), a variability thought to be due to incomplete ITS concerted evolution linked to the high number of NORs in Cycas genomes and incomplete lineage sorting due to recent species divergence (Xiao et al. 2010).
Given these observations, we have applied NGS approaches to examine the genomic variation, unit structure and levels of sequence heterogeneity across both coding and non-coding regions of the 35S rDNA loci, in representatives of the major gymnosperm groups, focusing on key species, namely (i) Abies sibirica Ledeb. from Pinales, (ii) Ginkgo biloba L. from Ginkgoales, (iii) Cycas revoluta Thunb. from Cycadales and (iv) Gnetum gnemon L. from Gnetales. Considerable heterogeneity of C. revoluta rDNA contrasts with high homogeneity of its rRNA transcriptome. We also determined the number of rDNA loci in Gnetum gnemon for the first time. In addition, because many gymnosperms also have large genome sizes compared with most angiosperms (Leitch and Leitch 2012). we studied 35S rDNA heterogeneity in an angiosperm, Fritillaria imperialis L., with one of the largest genome sizes among seed plants (for the phylogenetic relationships between the genera, see Supplementary Fig. 1).
Material and methods
Plant material
Fresh leaf fragments from C. revoluta (accession number: 1990-0350/1) were obtained from Chelsea Physic Garden, London, UK. Cycas circinalis L. and Dioon edule Lindl. were obtained from Brno Botanical Garden, Czech Republic.
Fresh leaves of G. biloba (accession number: 1969–13276), G. gnemon (accession number: 1998–514), F. imperialis (accession number: s.n.) were collected from the Royal Botanic Gardens, Kew, UK. Nicotiana tabacum var. SR-1 was from a local collection of the Institute of Biophysics, Brno, CZ.
Genomic DNA extraction
Fresh leaf fragments were dried in silica gel for later use. Genomic DNA was extracted using the modified CTAB method (Kovarik et al. 1997). The quality and quantity of total genomic DNA were evaluated by NanoDrop (Thermo Scientific, USA) and a Qubit 2.0 fluorometer (Life Technologies, USA).
Illumina HiSeq sequencing
Total genomic DNAs from C. revoluta, G. biloba and G. gnemon were sent to BGI (Shenzhen, China) for paired-end sequencing (170-bp library insert size, Illumina HiSeq 2000 platform) to obtain NGS reads equivalent to 10 % of the genome size. Paired-end sequencing of F. imperialis (300–500 bp insert size) was carried out at the Centre for Genomic Research at the University of Liverpool on an Illumina HiSeq 2000 platform. Fastq format paired-end sequencing reads were supplied with the adaptor sequences removed. Reads with a phred score of <20 for >10 % of their bases were also removed. Genomic Illumina sequence reads of A. sibirica (run accession number: ERR268415) were downloaded from the Sequence Read Archive (SRA). The left-handed genomic reads of all the above species were imported into RepeatExplorer (Novak et al. 2013) for further analysis. Transcriptomic Illumina reads of C. revoluta were downloaded from EBI sequence archive (SRR1525778).
RNA extraction, genomic PCR and RT-PCR analysis
Fresh leaf tissues of C. revoluta, C. circinalis, G. gnemon, G. biloba and P. abies were collected and stored in RNAlater (Life Technologies, USA) at −20 °C until use. Total RNA was isolated using a RNeasy Plant Mini kit (Qiagen, Germany) following manufacturer’s instructions. RNA was measured using a spectrophotometer and quality checked by gel electrophoresis. DNA contamination was removed using DNase (Turbo DNA free, Ambion, Austin, TX). The reverse transcription mixture (20 μl) typically contained 1 μg RNA, 2 pmol of random nonamer primers, 10 nmol of each dNTP and 200 U of the Superscript reverse transcriptase II (Life Technologies, USA). The complementary DNA (cDNA) and gDNA were amplified with a low error-prone KAPA Taq DNA polymerase (Kapabiosystems, USA) using primers specific for rDNA. The 26S gene was amplified with primers derived from the tobacco sequence: 26S_2901_f, 5′-GAATTCACCAAGTGTTGGAT-3′ and 26S_3121_r, 5′-AGAGGCGTTCAGTCATAATC-3′. The PCR conditions were as described in Lim et al. (2000). The ~220 bp product was gel purified, extracted (PCR purification kit, Macherey Nagel, Germany) and ligated into the pDrive vector (Qiagen, Germany). Ligation was carried out overnight at 4 °C. A 1.0-μl aliquot was used to transform Escherichia coli competent cells (EZ strain, Qiagen, Germany). The inserts were sequenced by Sanger technology using the SP6 and T7 primers. Sequences were aligned in a BioEdit sequence editor. One clone of C. revoluta was used as a 26S rDNA probe in Southern blot hybridisation.
Clustering genomic reads
RepeatExplorer (Novak et al. 2013). implemented in the Galaxy web-based server, is a pipeline employed to analyse repetitive elements based on sequencing reads from NGS platforms (e.g., Illumina HiSeq). Using RepeatExplorer, all the left-handed single reads of A. sibirica, C. revoluta, G. biloba, G. gnemon and F. imperialis were compared in pairwise BLAST comparisons and grouped into clusters that share at least 90 % similarity over 55 bp (61 %) of their sequence length. Read length for running clustering was 90 bp. Reads in each cluster are then displayed graphically using a Fruchterman-Reingold algorithm as described in Novak et al. (2010). Individual reads are represented as nodes (dots) on the graph, and the nodes are connected by “edges” (lines), the length of which increases with decreasing sequence similarity. In addition, reads within the clusters are assembled into one or multiple contigs based on their sequence similarities and can be compared with known repeat databases to identify the repeats (e.g. rRNA genes). Nucleotide composition of reads was determined using FastQC program implemented within the Galaxy Server (Goecks et al. 2010).
rDNA sequence assembly
Regions (18S, ITS, 26S and partial of IGS) of 35S rDNA of A. sibirica, C. revoluta, G. biloba, G. gnemon and F. imperialis were assembled into contigs by RepeatExplorer.
For those species whose 35S rDNA clusters did not cover all coding regions (i.e. the ITS2-26S region of G. biloba), missing sequences were downloaded through GenBank (G. biloba accession numbers: Y16892, AY095475). Different regions of 35S rDNA sequences (contigs) were ultimately assembled into one single consensus sequence for each species using Geneious 5.5 (Biomatters Limited, NZ) via default settings. The 35S rDNA sequence of Arabidopsis thaliana (accession number: X52322) was used as a reference to aid assembly. The boundaries of each region were determined by aligning all five studied species plus the 35S rDNA sequence of A. thaliana.
Intragenomic variation estimation
CLC Genomics Workbench 6.5.1 (CLCbio, Germany) was used to estimate intragenomic variation among 35S rDNA units in A. sibirica, C. revoluta, G. biloba, G. gnemon and F. imperialis. Illumina reads used for clustering were first mapped to their corresponding 35S rDNA consensus sequences with the mapping settings: mismatch cost value 2, insertion cost value 3, deletion cost value 3, with both the length fraction value and the similarity fraction value set at 0.8. Variations were then detected via the Probabilistic Variant Detection function tool in CLCbio, using default settings. Single nucleotide variants (SNVs), also called single nucleotide polymorphisms (SNPs), were filtered as follows: count (the number of countable reads supporting the allele) ≥10, frequency (the ratio of “the number of ‘countable’ reads supporting the allele” to “the number of 'countable' reads covering the position of the variant”) ≥20 %. Other variation types, i.e. deletion, insertion and multiple nucleotide variant (MNV), were produced by default settings.
Pi diversity values evaluation
The Pi diversity values (expressed as the number of substitutions per 100 bp of DNA sequence) were calculated for aligned reads using the DNASp software (Rozas et al. 2003). Data sets were obtained from the following whole genome shotgun archives using BLAST searches: A. thaliana (ERR171440_1), Oryza sativa (ERR605264), G. gnemon 2 (454 GS FLX, ERR268419), G. gnemon 1 (Illumina, ERR845260), Glycine max (SRR273385), A. sibirica (ERR268414), Taxus baccata (ERR26824), P. taeda (SRR1049544), C. revoluta (ERR845258), G. biloba (ERR845259), P. abies (ERP002565), Pinus sylvestris (ERP002572), F. imperialis (ERR845263), Cardamine amara (Zozomova-Lihova et al. 2014), Drosophila lowei (SRR330415), Bubalus bubalis (SRR032564) and Ornithorhynchus anatinus (SRR287787). As queries, we selected short 42–52-bp-long sequences derived from conserved subregions (Supplementary Table 1 and green bars in Supplementary Fig. 2). The phylogenetic trees were constructed from aligned 18S (region between V3 and V4 domains) and 26S (close to 3´end) reads using Neighbour Joining method (Jukes-Cantor model) with 1000 replications. Because of the lack of NGS data for C. circinalis and D. edule, we sequenced ~20 Sanger clones containing the 18S subregion (see blue bar in Supplementary Fig. 2a) from each species.
Bisulfite sequencing
Modification of C. revoluta genomic DNA with bisulfite was carried out with an EpiTect Bisulfite kit (Qiagen, Germany) using 1.3 μg of genomic DNA from leaves. The primers amplified a coding strand of the conserved part of the 18S gene (see blue bar in Supplementary Fig. 2a), and they did not discriminate between methylated and non-methylated templates. The sequences of primers were as follows: 18SBIS forward: 5′-TATGAGTYTGGTAATTGGAATG-3′; 18SBIS reverse: 5′-TTTAARCACTCTAATTTCTTCAAA-3′. PCR (25 μl) amplification used 1.0 μl of bisulfite-converted DNA as the template, 4 nmol of each dNTP, 8 pmol of each primer and 0.8 U of KAPA Taq DNA polymerase (Kapabiosystems, USA). Cycling conditions were as follows: initial denaturation (94 °C/3 min); 35 cycles of (94 °C/20 s; 55 °C/20 s; 72 °C/20 s) and a final extension (72 °C/10 min). The resulting ~300 bp products were separated by gel electrophoresis, purified and cloned into a TA vector (pDrive, Qiagen). After trimming of primers, the 241-bp-long sequences were aligned and statistically evaluated using a CyMATE software (Hetzl et al. 2007). One clone obtained from native DNA was used as the 18S probe in Southern blot hybridisation.
Southern blot hybridisation
Purified genomic DNA (~4 μg/sample) of G. biloba, C. revoluta, C. circinalis, D. edule, G. gnemon, N. tabacum and F. imperialis was digested with BstNI and TaqI restriction enzymes and hybridized with the 26S and 18S rDNA probe, respectively, (for the probe positions see black lines in Supplementary Fig. 2). In addition, C. revoluta and N. tabacum DNAs were digested with a further three methylation-sensitive restriction enzymes MspI, HpaII and ScrFI and hybridised with 18S rDNA probe. After the digestion, genomic DNA of those species was separated by gel electrophoresis on a 0.9 % (w/v) agarose gel. The gels were then alkali blotted onto Hybond-XL membranes (GE Healthcare, Little Chalfont, UK) and hybridised with a 32P-labelled DNA probe (DekaLabel kit, MBI, Fermentas, Vilnius, Lithuania) for the 26S and 18S rRNA gene, according to protocols described in Kovarik et al. (1997). After washing (2 × 5 min with 2x SSC, 0.1 % SDS at room temperature followed by 2 × 15 min with 0.6 × SSC, 0.1 % SDS, 65 °C), the hybridisation bands were visualised with a PhosphorImager (Typhoon 9410, GE Healthcare, PA, USA) and the data quantified by ImageQuant software (GE Healthcare, PA, USA).
Fluorescent in situ hybridisation (FISH) and immunostaining staining
To accumulate metaphases, fresh root tips from adult plants of C. revoluta, G. gnemon and F. imperialis were collected and immersed into 2 % 8-hydroxyquinoline for 4–6 h at room temperature. All root tips were subsequently fixed in 3:1 (v/v) ethanol:glacial acetic acid overnight or 24 h at room temperature. Finally, roots were transferred to 70 % ethanol and stored at −20 °C until use. Fixed root tips were digested for 90–120 min at 37 °C in a mixture of 1 % (v/v) pectinase and 2 % (v/v) cellulase in citrate buffer and spread on a slide in 3:1 (v/v) acetic acid:methanol.
18S rDNA probe was labelled with Alexa Fluor 488 (green fluorescence) and used in the hybridisation mix as described in Becher et al. (2014). Fluorescence in situ hybridisation was carried out as follows: Briefly, chromosome spreads were post-fixed in 4 % (v/v) formaldehyde in 2 × SSC (0.3 M sodium chloride and 0.03 M tri-sodium citrate) for 10 min, washed again in 2 × SSC, before dehydration through an ethanol series (70, 95 and 100 % ethanol). The slides and hybridization mix were denaturated together at 80 °C for 2 min using a DyadTM DNA engine carrying a PRINS block (Bio-Rad Laboratories, Inc., USA). After denaturation, the slides were incubated overnight at 37 °C in a well-sealed plastic box. Coverslips were removed in 2 × SSC at room temperature and the slides were incubated at 60 °C in 2 × SSC for 20 min and at room temperature for 10 min, dehydrated again in the ethanol series mentioned above and air-dried in the dark. Slides were mounted in Vector Shield TM mounting medium containing DAPI (4,6-diamidino-2-phenylindole) (Vector Laboratories, USA). Slides were visualised with a DM RA2 fluorescence microscope (Leica Microsystems GmbH, Germany) equipped with an Orca ERTM monochrome camera (Hamamatsu, Japan). Images were edited using OpenLabTM (Improvision, UK).
For detection of global DNA methylation, we followed the procedures described in Jasencakova et al. (2003). Briefly, nuclei were post−fixed and denatured as described for FISH and then incubated with a mouse antibody against 5-methylcytosine (5mC) (Eurogentec, cat-No.MMS-900P-A), followed by an Alexa 488-conjugated antimouse (1:100) antibody.
Results
Identifying 35S rDNA in Illumina HiSeq data
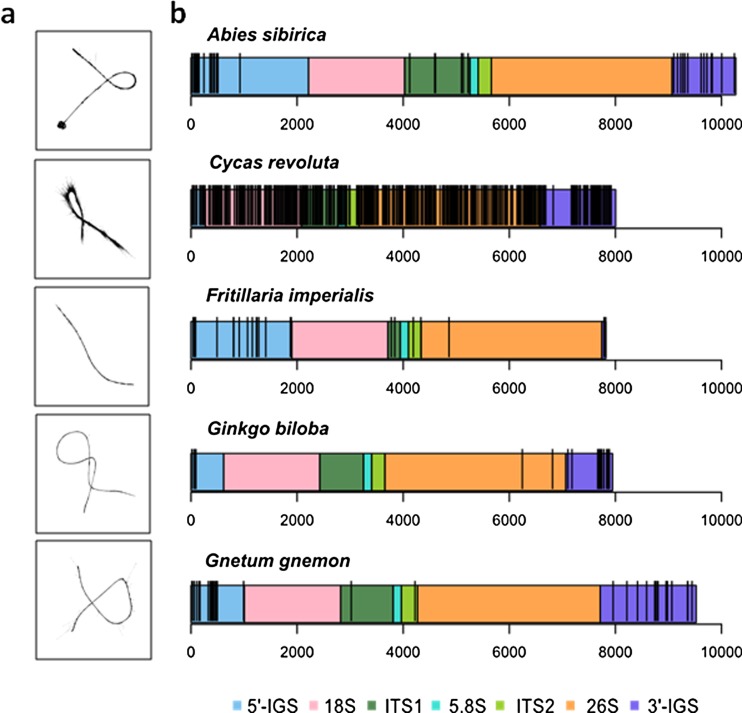
Illumina HiSeq data, amounting to approximately 5 % of the genome size of four gymnosperm species (A. sibirica, C. revoluta, G. gnemon and G. biloba) and 2 % of the genome of the angiosperm F. imperialis (Table 1) was analysed using RepeatExplorer (Novak et al. 2013). Those clusters containing reads from 18S rDNA are displayed graphically (Fig. 1a). The reads in clusters from A. sibirica, G. biloba, G. gnemon and F. imperialis form smooth lines in graphical displays because there is little sequence variation between the multiple 35S rDNA reads analysed. In contrast, the graphs reconstructed from C. revoluta reads are much fuzzier, indicating high levels of variation between reads from different 35S rDNA units.
Table 1.
The volume of Illumina HiSeq reads analysed with RepeatExplorer
| Species | 1C Genome size (Gb)a | Number of reads analysed | Genome proportion of analysed reads (%) | Genome proportion of 35S rDNAb (%) |
|---|---|---|---|---|
| Abies sibirica | 15.452 | 8,212,026 | 4.8 | 0.235 |
| Cycas revoluta | 13.399 | 6,927,500 | 4.7 | 1.095 |
| Ginkgo biloba | 11.492 | 5,402,020 | 4.2 | 0.468 |
| Gnetum gnemon | 3.785 | 2,102,778 | 5.0 | 0.845 |
| Fritillaria imperialis | 42.005 | 9,200,000 | 2.0 | 0.116 |
aThe values are from Plant DNA C-values database (Bennett and Leitch 2012)
bCoding region plus internal transcribed spacers (i.e. 18S-ITS1-5.8S-ITS2-26S)
Fig. 1.
Intragenomic variation of 35S rDNA NGS Illumina reads in five species. a Graphical outputs from RepeatExplorer showing repeat clusters containing 18S rDNA sequences. Smooth line graphs indicate little sequence variation between reads (the pattern in most species), the more diffuse graph for C. revoluta indicates considerable heterogeneity between read sequences. b Distribution of SNPs across the 35S rDNA sequences. Each vertical black bar represents a single highly confident SNP
Assembly of 35S rDNA sequence
To assemble the 18S-5.8S-26S coding regions, we used RepeatExplorer to identify clusters containing rDNA sequences (as for the 18S gene domains, Fig. 1a). Based on the contigs generated by RepeatExplorer, we generated a single 35S rDNA consensus sequence for each species (Fig. 1b). The correct order of domains was confirmed by Southern blot hybridisation (not shown) and alignment of the 35S rDNA consensus sequences of all the studied species and A. thaliana. The total length of the recovered 18S-5.8S-26S regions (including ITS sequences) varied from 5844 bp in F. imperialis to 6854 bp in A. sibirica (Table 2). The length variation was mostly accounted for by differences in length of ITS1 (Supplementary Fig. 3), as the length of ITS2 did not differ substantially between species, in line with previous reports (Maggini and Baldassini 1995). The ITS1 of A. siberica and G. biloba contained short tandem duplications (Supplementary Fig. 4). We were unable to reconstruct the whole 35S rDNA unit from any of the species analysed because the repetitive IGS region cannot be assembled from short reads. The genome proportion that each 35S rDNA cluster comprised was calculated from the number of reads in each 35S rDNA cluster compared with the total number of reads analysed by RepeatExplorer. From the genome proportion, the copy numbers of 35S rDNA sequences were estimated using the formula: (genome size × genome proportion of 35S rDNA clusters (Table 1)) / (35S rDNA sequence length (Table 2)). Copy number ranged from c. 3300 in G. gnemon to c. 17,000 copies in C. revoluta distributed over two (G. biloba and F. imperialis) to twelve loci (C. revoluta). Because of the known underestimation of repeated families among NGS libraries (Emadzade et al. 2014). we validated in silico results experimentally by Southern blot hybridisation (Table 2 and Supplementary Fig. 5d). In general, the values calculated from NGS were in a good agreement with those experimentally assessed.
Table 2.
Chromosomal distribution, length and number of rDNA gene domains and spacer sequences in the analysed species
| Species | Number of chromosomes (2n) | Number of 18S rDNA loci (sites at metaphase) | Length of 35S rDNA including ITS sequences (bp) | 35S rDNA copy numberd(1C) | |
|---|---|---|---|---|---|
| NGS | S.blot hyb. | ||||
| Abies sibirica | 24a | – | 6854 | c. 4300 | n.d. |
| Cycas revoluta | 22 | 12 (24) | 6306 | c. 17,000 | c. 18,300 |
| Ginkgo biloba | 24b | 2 (4) | 6448 | c. 6300 | c. 5800 |
| Gnetum gnemon | 48c | 5 (10) | 6707 | c. 3300 | c. 3400 |
| Fritillaria imperialis | 24 | 2 (4) | 5844 | c. 6200 | c. 4000 |
aSedelnikova and Pimenov (2005)
bGalián et al. (2012)
cThe estimated chromosome number (2n = 48) is consistent with Pearson (1908) while it differs from the other report (2n = 44, (Fagerlind 1941)). The same number was obtained from counting of >20 G. gnemon metaphases
dCopies calculated from the genome representation (NGS) and a Southern blot experiment (Supplementary Fig. 4d). Genome sizes are given in Table 1.
Distribution of single nucleotide polymorphisms
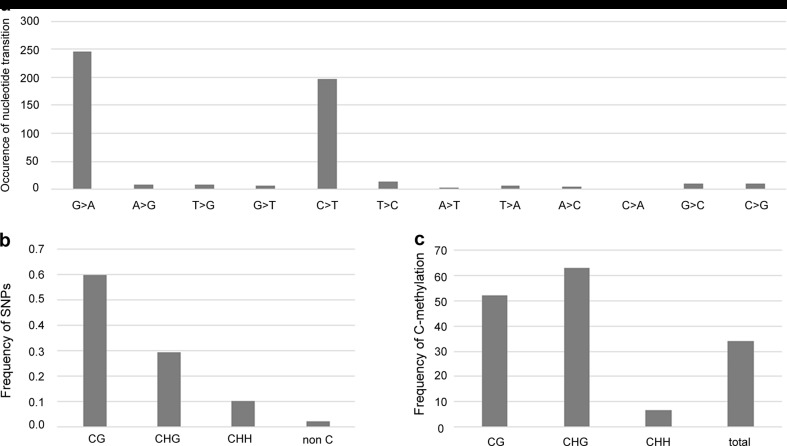
The distribution of SNPs across the 35S rDNA sequences was determined using CLCbio software, with a threshold of at least two SNPs and ten reads. For A. sibirica, G. biloba, G. gnemon and F. imperialis, SNPs were found predominantly in the non-coding sequences of 35S rDNA as expected (Fig. 1b and Supplementary Fig. 6). However, in C. revoluta, there was an astonishing density of SNPs across both coding and non-coding regions with the highest SNPs/kb values out of all species analysed. In this species, the distribution of mutations was almost uniform, without significant differences between individual 35S rDNA subregions. We also determined the nature of the SNPs. The vast majority of SNPs were C > T and G > A transitions (Fig. 2a) located almost exclusively in symmetrical CG and CHG contexts (Fig. 2b). These mutations apparently influenced overall nucleotide composition of rDNA since C. revoluta 18S genes had significantly lower G + C content compared to other species (Supplementary Fig. 7a). Indels were far less abundant than substitutions (Supplementary Fig. 7b).
Fig. 2.
Characterization of SNPs and methylation analysis in C. revoluta 35S rDNA. a Occurrence and distribution of SNPs across the 35S rDNA sequence obtained from Illumina reads. b Frequency of mutations in different dinucleotide and trinucleotide motifs in a 300 bp subregion of 18S genes (Supplementary Fig. 2a). c Frequency of methylation in the same region as (b) analysed by bisulfite sequencing of 23 Sanger-sequenced clones
Southern blot hybridisation
As noted above, the complete IGS region could not be reconstructed in any species from our NGS data due to its long length and highly repetitive structure (Supplementary Fig. 4). To estimate the number of IGS length variants, we carried out Southern blot hybridisation to BstNI-restricted DNA using a 26S rDNA probe (Supplementary Fig. 5a). Digestion of genomic DNAs with this enzyme yielded 1–3 strong hybridisation bands in all species analysed, indicating a similar number of amplified variants.
To confirm the data obtained by SNP analysis (see above), we examined the degree of conservation of restriction sites within the 18S genes. For this, we selected TaqI, an enzyme that digests the TCGA motif, and which is methylation-insensitive and has six target sites in the 18S gene. The C > T/G > A mutations would result in the loss of the restriction site and result in larger hybridisation fragments than expected. The 18S probe hybridised predominantly to a single band in G. gnemon and N. tabacum (Supplementary Fig. 5b) (and other species, data not shown) indicating conservation of restriction sites. However, in C. revoluta, the probe hybridised to multiple bands in a smeared high molecular weight region indicating extensive site polymorphisms.
Comparison of 35S rDNA intragenomic homogeneity between species
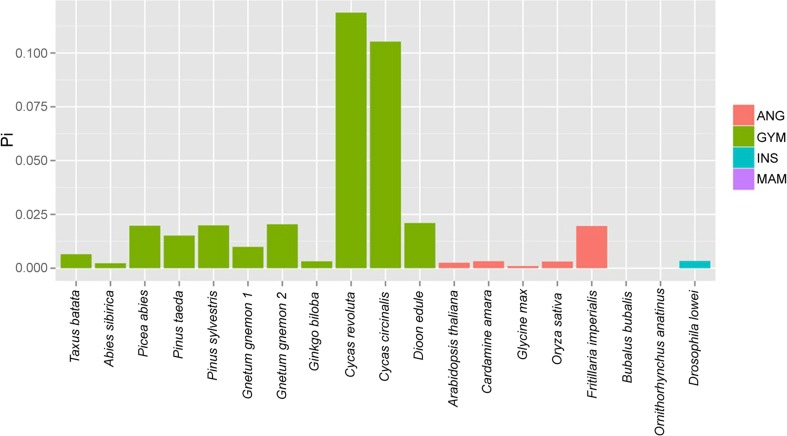
To determine whether gymnosperms have particularly diverse 35S rDNA sequences, we calculated the number of substitutions per nucleotide (Pi) in a 42-bp subregion of the 18S rRNA gene (Fig. 3). The subregion comprises a core region occurring between the V3 and V4 domains (see green bar in Supplementary Fig. 2a), which is thought to be highly conserved between plants and animals. The data analysed included the Illumina data generated here, together with data from a further four gymnosperm species, four angiosperm species, one insect (D. lowei) and two mammals (the water buffalo, B. bubalis, and the duck-billed platypus, O. anatinus) which are available in public databases. Average Pi diversity values of the 18S rDNA subregion ranged from zero in the two mammals to almost 12 % in C. revoluta and C. circinalis. Most angiosperms and animals displayed lower intragenomic variation than most gymnosperms. The exceptions were A. sibirica and G. biloba which had a low Pi values compared with other gymnosperms.
Fig. 3.
Intragenomic variation within the 18S gene in selected plant and animal species. The sequences were extracted from NGS data either obtained in the present work or downloaded from public databases, aligned and pairwise distance calculated as number of substitutions per 100 bp (Pi). A 42-bp core region (green bar in Supplementary Fig. 2a) was used for alignments. The data for C. circinalis and D. edule were obtained by conventional cloning of PCR products amplified from the corresponding 18S subregion (green bar in Supplementary Fig. 2a). ANG angiosperms, GYM gymnosperms, INS insects, MAM mammals
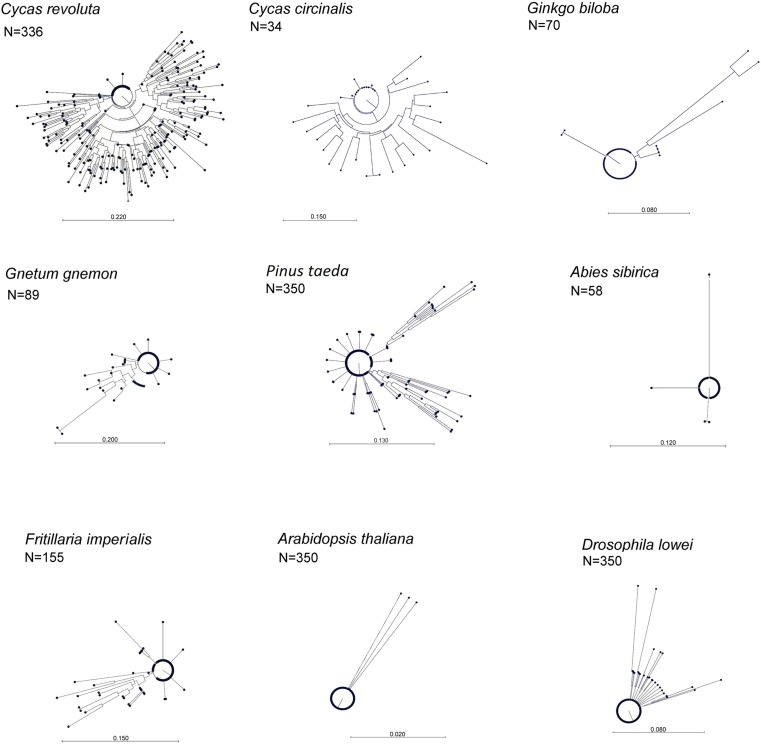
To compare the level of intragenomic sequence divergence within the 35S rDNA between species, we analysed the divergence of individual sequence reads for a selection of six gymnosperms, two angiosperms and one insect (Fig. 4). Each phylogram comprised 50–350 aligned 18S reads. It is evident that in A. thaliana and D. lowei, the majority of sequences grouped at the base of the tree indicating their homogeneity. The few reads emanating from the base comprise haplotypes differing in a single mutation and most likely represent sequencing artefacts. However, in C. revoluta, the homogeneous population of unresolved reads comprised a relatively minor fraction of the reads analysed, most formed highly divergent branches. The most divergent sequences differed from the homogeneous inner cluster by more than 20 % of nucleotides. In addition, G. gnemon, P. taeda and F. imperialis contained a small fraction of units separating into diverging groups. In contrast, C. revoluta (and C. circinalis, from Sanger sequencing of clones) had a large fraction of highly divergent units. Trees constructed from 26S reads showed essentially similar topologies as the 18S trees (Supplementary Fig. 8).
Fig. 4.
Phylograms showing the contrasting levels of intragenomic sequence diversity in the 18S rDNA subregion (green bar in Supplementary Fig. 2a) for a selection of six gymnosperms, two angiosperms and one insect species. Phylogenetic relationships between the individual 18S reads were constructed from aligned sequences using a NJ method. Note the large scale bar for C. revoluta and C. circinalis indicating high divergence between sequences in these species. Because the data sets from D. lowei, A. thaliana and P. taeda were too large to compare with others, we reduced the number of sequence reads analysed to 350. Note, there is no correlation between the number of reads and degree of branching of the trees (N = number of reads analysed)
Expression of 35S rDNA
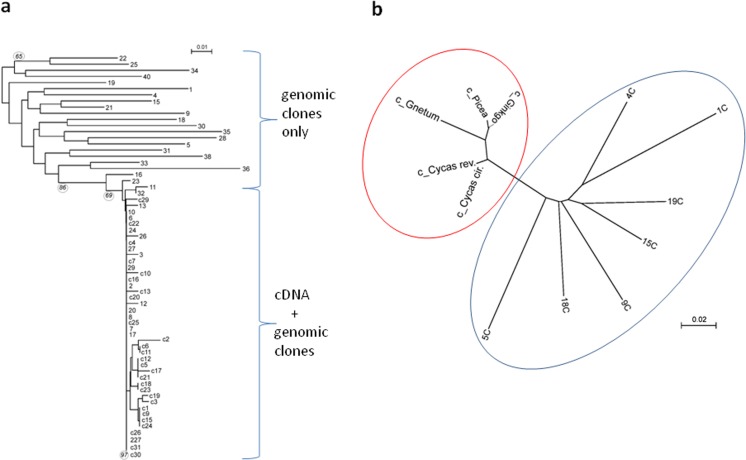
To examine whether the diversity of 35S rDNA sequences observed in the genomic DNA was expressed in C. revoluta, we analysed the homogeneity of genomic and transcriptomic (SRR1525778) reads across the rDNA unit (Fig. 5a and Supplementary Table S1). It is evident that the diversity (Pi, expressed as the number of substitutions per 100 bp) of genomic rDNA reads was at least 10-fold higher than that of transcriptomic rDNA reads. To validate these results, we sequenced 28 Sanger clones from each cDNA and genomic (gDNA) amplicons of 26S rDNA (for the position, see Supplementary Fig. 2b) for seven gymnosperm species. The Pi values were highly congruent with the NGS analysis (not shown). Sequences of the cDNA clones from C. revoluta were not identical, and several mutation hot spots were identified. Of these, the most prominent one was at position 2922 (with respect to the first 26S nucleotide): 19 (68 %) cDNA clones had a T, 6 (21 %) had an A and 3 had a G (11 %). The same position was polymorphic in the closely related C. circinalis but not in the distantly related cycad D. edule (not shown). The phylogram constructed from the cDNA and genomic 26S sequences is shown in Fig. 6a. There were two branches supporting two distinct groups of sequences: group (i) containing highly divergent genomic clones and group (ii) containing cDNA and genomic clones with little sequence divergence. The latter group also clustered with sequences obtained from the cDNA clones of the other gymnosperms analysed (Fig. 6b).
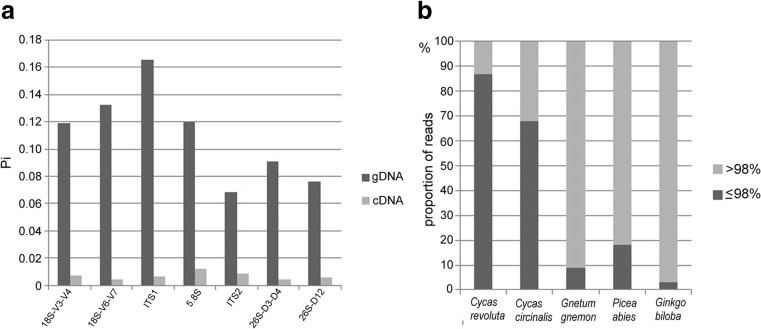
Fig. 5.
a Bar charts showing intragenomic diversity of genomic (gDNA) and transcriptomic (cDNA) reads in Cycas revoluta. The Pi values were calculated from aligned reads. The alignments comprised >150 reads for each subregion. b Bar charts showing the proportion of functional genes (light grey > 98 % sequence identity to consensus cDNA sequence) to pseudogenes (dark grey ≤ 98 % sequence identity) in the genomes of five gymnosperm species
Fig. 6.
a Sequence homogeneity at the rRNA expression level contrasts with sequence heterogeneity at the genomic DNA level in C. revoluta. The phylogram was constructed from the genomic DNA (number) and cDNA (c + number) clones of 26S rDNA subregion (see blue double-headed arrow in Supplementary Fig. 2b). Bootstrap support of branching (>60) are indicated. (b) Phylogram constructed from cDNA (red oval) sequences from G. biloba, C. revoluta, C. circinalis, G. gnemon and Picea abies and randomly selected pseudogene (blue oval) clones from C. revoluta
The highly diverse and heavily mutated genomic clones were not found amongst the cDNA clones indicating they are pseudogenes. To determine the proportion of functional genes to pseudogenes in individual genomes, we BLASTed the NGS rDNA genomic reads of C. revoluta, C. circinalis, G. gnemon and G. biloba against their consensus sequences of 26S rRNA (for the position, see Supplementary Fig. 2b) obtained from >20 cDNA Sanger clones. The cDNA consensus for C. revoluta and Picea abies was obtained from the NGS transcriptome libraries (SRR1525778 and ERR260407, respectively). The recovered sequences were filtered and separated into two groups according to their similarities. The “high (>98 %) identity” group was considered to originate from mostly functional genes while the “low (≤98 %) identity” group was likely to have been derived from pseudogenes. In contrast to the two Cycas species analyzed (C. revoluta and C. circinalis) had high proportions of “low identity” sequences, the other gymnosperm genomes analyzed contained only a low proportion of these types of sequences (Fig. 5b). In both Cycadales, the G + C content of functional genes and pseudogene fraction was 50 and 41 %, respectively.
Comparison of methylation levels between pseudogenes and functional copies of 35S rRNA genes
To study the occurrence of cytosine methylation in genomic DNA, we carried out bisulfite sequencing and restriction enzyme analysis of 18S rDNA in C. revoluta.
The target region for bisulfite analysis is shown in Supplementary Fig. 2a (see blue bar), and it includes the V3 domain (part), core and part of the V4 domain. Cytosines at symmetrical CG and CHG motifs were shown to be highly methylated (>50 % of cytosines) in this region, while low (c. 7 %) methylation was found at non-symmetrical (CHH) sites (Fig. 2c). Since many C. revoluta genomic clones must have originated from mutated copies, we compared the methylation levels of pseudogenes with functional genes separately. In principle, bisulfite sequencing cannot discriminate C > T mutations from non-methylated Cs. To distinguish between these two possibilities, we took advantage of the fact that bisulfite clones always originate from one DNA strand only. Thus, every G > A substitution represents a true mutation in native DNA. Therefore, clones with no G > A transitions likely represent functional genes while clones with G > A transitions represent pseudogenes. The proportion of methylated Cs in each fraction is shown in Supplementary Fig. 9. It is evident that pseudogenes have less methylation (25 %) compared with functional genes (53 %). The difference is explained by a reduced number of symmetrical methylatable sites in pseudogenised copies.
Because bisulfite sequencing analysis of several clones may not be representative for the whole cluster comprising thousands of units, we carried out global analysis of cytosines at restriction sites using the methylation-sensitive restriction enzymes MspI, HpaII and ScrFI and hybridisation of restricted DNAs with the 18S probe (Supplementary Fig. 5c). We used the MspI/HpaII isoschizomers that cut at CCGG and are sensitive to CHG and CG methylation, respectively. BstNI and ScrFI are nearly isoschizomeric pairs cutting at CCWGG and CCNGG, respectively. BstNI is methylation-insensitive, ScrFI is sensitive to methylation of the inner C. There are more than five target restriction sites in each 18S gene. The probe hybridised to high molecular weight bands produced by digestion with methylation-sensitive MspI, HpaII and ScrFI enzymes. In contrast, the probe hybridised to low-molecular weight BstNI fragments. As a control, we used N. tabacum (tobacco) DNA which has one of the highest levels of rDNA methylation among angiosperms (Kovarik et al. 1997). The extent of tobacco rDNA digestion with methylation sensitive enzymes was comparable or even higher (MspI, HpaII) than that of C. revoluta indicating heavy methylation of C. revoluta genes.
Fluorescence in situ hybridisation (FISH) and immunostaining of chromatin
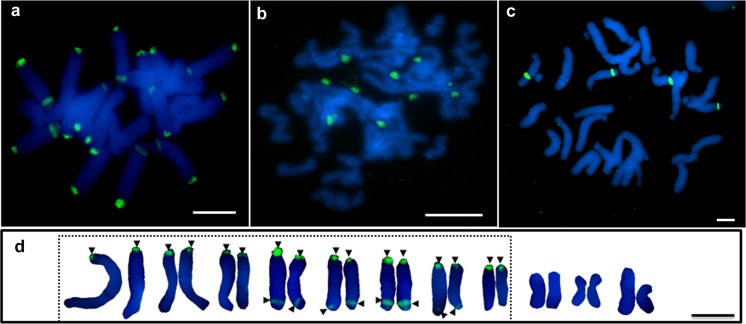
FISH using an 18S rDNA probe was conducted to determine the distribution of the 35S rDNA loci on metaphase chromosomes of C. revoluta, G. gnemon and F. imperialis (Fig. 7, Table 2). Metaphases of C. revoluta had 24 signals on 16 out of the 22 chromosomes. Of these, eight chromosomes had a strong terminal and a more defuse sub-terminal signal, whereas the other eight chromosomes carried only one terminal signal (Fig. 7a, d). This observation is inconsistent with a previous report (Hizume et al. 1992a) where only 16 terminally located 35S rDNA sites were detected. FISH to G. gnemon (Fig. 7b) was sub-optimal as high quality metaphases proved difficult to obtain. However, it represents the first attempt to cytogenetically analyse rDNA in this species. There were ten sub-terminal 18S rDNA signals of variable intensity and condensation. The probe hybridised to four strong sub-terminal sites in F. imperialis (Fig. 7c). Compared to C. revoluta and G. gnemon, all sites were highly condensed in F. imperialis.
Fig. 7.
Fluorescent in situ hybridisation of 18S rDNA (green signal) in a Cycas revoluta, b Fritillaria imperialis and c Gnetum gnemon. Karyotype ideograms of Cycas revoluta (d) showing chromosomes bearing 18S rDNA signals (dashed line boxed), rDNA locations indicated by arrowheads. Scale bar = 1 μm
In order to study the distribution of DNA methylation in C. revoluta chromatin, we immunostained nuclei with an antibody recognizing methylated cytosine residues (5mC) (Supplementary Fig. 10). The nuclei were counterstained with DAPI revealing several large and numerous small bright DAPI-positive spots apparently representing highly condensed chromocenters. Subsequent immunodetection with an antibody against 5mC resulted in diffuse signals relatively evenly distributed across the nuclei preventing discrimination between sparsely and heavily methylated loci. This may suggest that in C. revoluta (and perhaps other large genomes) densely methylated heterochromatin is not limited to classical heterochromatic sites (mostly centromeres) as in Arabidopsis (Jasencakova et al. 2003) and Citrus (Marques et al. 2011). but its distribution is rather global.
Discussion
Inefficient homogenisation of 35S rRNA genes in Cycas revoluta
In eukaryotes, genes encoding 18S-5.8S-26S rRNAs generally display little or no intragenomic sequence variability (Dvorak et al. 1987; Eickbush and Eickbush 2007; Nieto Feliner and Rossello 2012). In contrast to these observations, our data for C. revoluta are novel and striking, as evolution has converted most of these genes into heavily mutated and non-functional pseudogenes. Although some rRNA pseudogenes have been reported in various plant and animal species, those described here for C. revoluta show distinct features:
Their abundance is extremely high. We estimate that the pseudogenes account for c. 80 % of the rDNA (~14,000 copies) in C. revoluta. The number may actually be higher since many of them might have escaped BLAST searches due to reduced homology.
In contrast to previous reports showing that rRNA pseudogenes are frequently dispersed as orphan single or low copy repeats outside the NORs (Mentewab et al. 2011). in C. revoluta, the pseudogenes appear to be clustered on chromosomes (Fig. 7), although we do not know whether they split into different functional and non-functional loci. Given that up to 13 nucleoli were reported in C. revoluta nuclei (Hizume et al. 1992a). and provided that each locus forms its own nucleolus, it is likely that some loci may contain both functional genes and pseudogenes. Further work is needed, possibly employing antibodies to chromatin epigenetic marks (Neves et al. 2005; Marques et al. 2011). to determine the higher order organisation of rRNA genes.
The Cycas pseudogenes harbour extraordinary haplotypic diversity indicating that almost all units are different. This apparent absence of homogenisation is indicative of extremely high mutation rates and/or considerable periods of time since the arrays last homogenised. Such variability contrasts with the situation in some animals where pseudogenes (transposon-inserted) seem to undergo at least partial homogenisation (Eickbush et al. 1997; Glass et al. 2008; Martoni et al. 2015). The high homogeneity of 18S reads in D. lowei (Fig. 4) is consistent with this hypothesis. Interestingly, the high diversity of ITS pseudogenes has recently been used as outgroup sequences in a Cycadalean phylogeny (Xiao and Moller 2015). Potentially, 26S rRNA pseudogenes may be used as outgroup sequences in phylogenies of all four major gymnosperm groups (Cycadales, Ginkgoales, Gnetales and Coniferales) (Fig. 6b).
Point mutations occur at nearly the same frequency in coding and non-coding regions and between the core domains of 18S and 26S rRNA genes. In other organisms studied, it is more usual to observe higher SNP diversity in non-coding regions (Eickbush and Eickbush 2007). For example, heterogeneous ITS was reported in Laryx potaninii (Pinales) (Wei et al. 2003) and Mammillaria (Cactaceae) (Harpke and Peterson 2007). In the latter, quantification of ITS pseudogenes revealed that as few as 3 % of ITS repeats could be functional. It will be interesting to determine if coding sequences are also highly mutated in Mammillaria (as in C. revoluta) or if higher frequencies of homogenisation influence coding domains more than the non-coding domains as in most eukaryotes (Stage and Eickbush 2007; Matyasek et al. 2012; West et al. 2014; Weitemier et al. 2015).
Efficient silencing of 35S rRNA pseudogenes in Cycas revoluta
The rRNA pseudogenes of C. revoluta do not seem to be expressed despite their high copy number. By contrast, Podisma pedestris (a grasshopper), which also harbours a large proportion of mutated rDNA copies, shows some pseudogene expression (Keller et al. 2006). It is known that silencing of pseudogenes and transposons involves epigenetic factors, including cytosine methylation (Kim and Zilberman 2014). Perhaps, differences in levels of pseudogene expression between C. revoluta and P. pedestris reflect different efficiencies of their epigenetic silencing systems. In C. revoluta, 35S rDNA was highly methylated in symmetrical CG and CHG sites and to a lesser extent in non-symmetrical CHH sites. Such a pattern is similar to repeat sequence methylation described in the monocot F. imperialis (Becher et al. 2014) and other angiosperms (Kovarik et al. 1997) and gymnosperms (Ma et al. 2015). Methylation of cytosine may not be the only factor contributing to pseudogene silencing in C. revoluta. For example, regulatory elements and promoters not analysed in this study could be inactivated by mutations. Probably the absence of transcription of rRNA pseudogenes is likely to be explained by a combination of genetic and epigenetic factors. Immunostaining with an antibody against 5mC revealed strong signals across the nuclei (Supplementary Fig. 10) indicating relatively even distribution of heavily methylated loci in the C. revoluta genome.
Hypothetical model of 35S rRNA pseudogene evolution
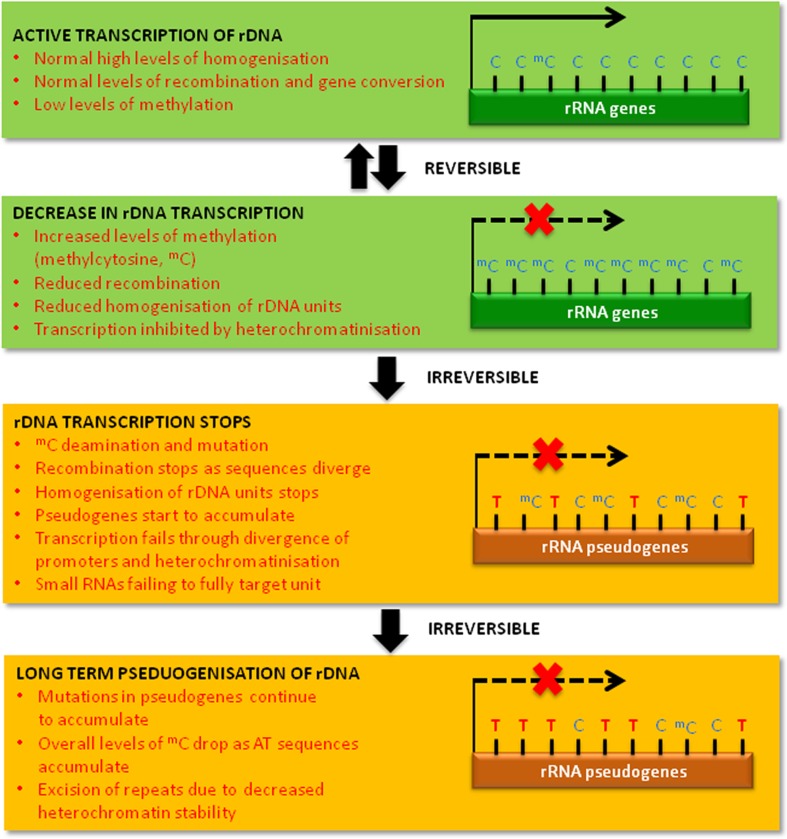
It has been proposed that epigenetic mechanisms regulating homologous recombination within transposable elements contribute to genome size variation (Fedoroff 2012). Here, we adopt this model to explain the formation and accumulation of 35S rRNA pseudogenes in plant genomes (Fig. 8):
Functional and transcriptionally active 35S rDNA units typically show low levels of cytosine methylation and epigenetic marks for heterochromatin. The copies of these units are usually highly homogenous in sequence through ill-defined processes that are likely to involve recombination and gene conversion events.
Some functional units may be inactive and carry epigenetic modifications, including DNA methylation. Potentially, methylation decreases the frequency of meiotic crossovers, as already reported in some other organisms, including plants (Melamed-Bessudo and Levy 2012). There is also evidence that inactive 35S rDNA units are less vulnerable to intragenomic homogenisation (Lim et al. 2000). However, methylation is reversible and could be reset following cell division. Hence, methylated units may still retain the potential to recombine and homogenise, although perhaps at a lower frequency.
Mutations arising from methylated cytosine (mC) deamination (mC > T) results in mC to T transitions (Poole et al. 2001). Strikingly, in rRNA pseudogenes, mutations have occurred almost exclusively in CG and CHG motifs, which are also heavily methylated. Methylated cytosine deamination is irreversible and increases sequence divergence among rDNA units. Potentially, as a consequence, rDNA may become non-functional forming pseudogenes. These will have reduced homology that could further restrict rDNA homogenisation and the targeting of small RNAs, the latter causing reduced cytosine methylation. Indeed, the intragenomic homogeneity of C. revoluta rRNA pseudogenes is low, the sequences divergent and cytosine methylation levels reduced. In such a scenario, pseudogenes would no longer recombine and would continue to accumulate mutations. A low turnover of repetitive sequences in gymnosperms (Nystedt et al. 2013; Cafasso and Chinali 2014) is consistent with this hypothesis.
Over time, with continued mC deamination, the sequences would also become ever more AT rich. This may also decrease gene conversion events known to be favoured by GC-rich rRNA genes (Escobar et al. 2011). The retention of low complexity A/T-rich genomic “graveyards” may contribute to reduced GC content observed in large plant genomes (Smarda et al. 2014).
Fig. 8.
A hypothetical model of 35S rDNA pseudogene evolution. In this model, from the second to the third stage, we observe four types of variation: (1) mC > T; (2) mC > C; (3) the originally unmethylated Cs; (4) mCs that keep their methylated status
Why is Cycas so unusual?
Although all the genomes analysed here, and elsewhere, contain some pseudogenised 35S rDNA copies, their abundance in Cycas is extraordinary. The question arises as to why the balance between mutation and mutation correction mechanisms has been disrupted in this species:
rDNAs may fulfil other functions in addition to ribosomal biosynthesis. One can assume that they help to stabilise these large chromosomes by binding heterochromatic proteins (Kobayashi 2011). Their strict localisation at subtelomeric positions of nearly all chromosomes may provide a protective role against degradation of chromosome ends. In this context, it is known that in some organisms telomeres are formed by rDNA and transposons rather than canonical simple repeats (Pich and Schubert 1998; Fajkus et al. 2005).
Large genomes could be associated with the accumulation of repeats with high sequence diversity, including rDNA, as observed in other animal and plants with large genomes (Martin and Gordon 1995; Kelly et al. 2015). C. revoluta certainly has a large genome (1C value = 13.40 Gb), compared with most angiosperms measured (modal 1C = 0.59 Gb, mean 1C = 5.77 Gb) (Leitch and Leitch 2012). However, many other gymnosperms also have similarly large genomes (e.g., P. abies, 1C = 19.57 Gb, P. taeda 1C = 21.61 Gb) but without such an exceptional diversity of SNPs in their rDNA. Furthermore, the angiosperm F. imperialis (1C = 42.0 Gb) has a genome nearly four times the size of C. revoluta, yet, its rDNA units do not have such a high SNP diversity either (Fig. 1b) (1C-values from Bennett and Leitch (2012)). Thus, there is no simple relationship between genome size, SNP diversity and frequency of rDNA unit homogenisation.
It has been proposed that subtelomeric rDNA loci tend to recombine more frequently than those at interstitial locations (Cronn et al. 1996; Pedrosa-Harand et al. 2006). However, 18S rDNAs (both functional genes and pseudogenes) are terminal in C. revoluta arguing against a role for chromosome position in the failure of rDNA sequences to homogenise.
The number of pseudogenes is proportional to the number of loci, which in C. revoluta is unusually high. Interlocus homogenisation is considered to be less frequent than intralocus homogenisation (Schlotterer et al. 1994). suggesting that species with multiple loci should have more diverged rDNA sequences than species with a single rDNA locus. However, this is at odds with the observation of relatively high rDNA homogeneity in P. taeda (Islam-Faridi et al. 2007), which has even more loci (10 major and up to 9 minor) than C. revoluta with 12 loci (8 major and 4 minor). Thus, a large number of copies or loci alone does not account for the exceptionally high abundance of pseudogenised rDNA in C. revoluta.
Cytosine methylation inhibits the homogenisation processes. If so then the high levels of methylation observed in C. revoluta are predicted to result in reduced or no homogenisation of the rDNA, leading to the accumulation of pseudogenes. In further support of such a link, Drosophila has negligible amounts of cytosine methylation, yet efficiently homogenises rDNA arrays (Stage and Eickbush 2007) while the methylated genome of Podisma pedestris with CG dinucleotide depletion has considerable rDNA sequence diversity (Keller et al. 2007).
Thus, C. revoluta may not be so different from other gymnosperm and angiosperm species in individual characteristics suspected to influence homogenisation processes. However, the large genome size, dense DNA methylation, frequent methylated cytosine deamination and multiple rDNA loci may act together to reduce the frequency of rDNA homogenisation. Furthermore, C. revoluta is known to exhibit very slow growth (2–3 inches a year) and a relatively long life span (more than 1000 years). Perhaps, methylation-induced mutations may accumulate during extended periods of vegetative growth (Dubrovina and Kiselev 2015). Potentially, the meiotic correction system is failing because the reduced homology between repeats inhibits rDNA recombination. Perhaps, some highly divergent rDNA paralogs could be as old as the genus itself, which is considered to be very ancient (>250 myr, (Chaw et al. 2000; Condamine et al. 2015). A long retention time of pseudogenes may not be happening, or only at low frequency, in relatively short-lived organisms, with more frequently occurring meiotic events, and lower levels of cytosine methylation. Indeed, coding regions of rRNA genes are highly homogeneous in Drosophila and Arabidopsis, both with generation times of just a few weeks and negligible (D. melanogaster) or low (A. thaliana) methylation levels. Thus, rDNA evolution in both these species is fully congruent with a concerted evolution model while concerted evolution largely fails in C. revoluta. Despite its overall paralog heterogeneity, a small subset of rRNA genes that are expressed has remained similar, perhaps because they have undergone homogenisation (via, e.g. unequal recombination) or because these copies have been maintained by selection.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PPTX 4.97 MB)
Acknowledgments
The Illumina data for F. imperialis was funded by NERC (NE/ G01724/1) and generated by the Centre of Genomic Research, based at the University of Liverpool, UK. The authors would like to thank Maite Guignard from Queen Mary, University of London for helping with R plot, Nick Bailey and Nell Jones from Chelsea Physic Garden (UK) for providing plant material. Two anonymous referees are acknowledged for their helpful comments. Wencai Wang is sponsored by China Scholarship Council (CSC) and Sònia Garcia by a Juan de la Cierva Postdoctoral Contract from the Government of Spain. The work was supported by the Czech Science Foundation (P501/13/10057S and P501/12/G090) and NERC (UK).
Author contributions
W.W. carried our most of the bioinformatic part, rDNA unit reconstruction, SNP analysis and FISH. H.B. analysed Fritillaria whole genomic data. L.M prepared probes for FISH. S.G. was involved in cloning and sequencing of rDNA. I.J.L. collected the material. A.K. and A.R.L. designed the study and W.W., A.K., A.R.L. and I.J.L wrote the paper.
Compliance with ethical standards
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
DNA deposition
Sequence data from this article can be found in the EMBL/GenBank data sequence read archives under accession numbers ERR845258-ERR845263.
References
- Becher H, Ma L, Kelly LJ, Kovarik A, Leitch IJ, Leitch AR. Endogenous pararetrovirus sequences associated with 24 nt small RNAs at the centromeres of Fritillaria imperialis L. (Liliaceae), a species with a giant genome. Plant J. 2014;80:823–833. doi: 10.1111/tpj.12673. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ (2012) Angiosperm DNA C-values database (release 8.0, Dec. 2012) http://www.kew.org/cvalues/
- Bobola MS, Smith DE, Klein AS (1992) Five major nuclear ribosomal repeats represent a large and variable fraction of the genomic DNA of Picea rubens and P. mariana. Mol Biol Evol 9:125–137 [DOI] [PubMed]
- Bogler DJ, Francisco-Ortega J. Molecular systematic studies in cycads: Evidence from trnL intron and ITS2 rDNA sequences. Bot Rev. 2004;70:260–273. doi: 10.1663/0006-8101(2004)070[0260:MSSICE]2.0.CO;2. [DOI] [Google Scholar]
- Buggs RJ, Renny-Byfield S, Chester M, Jordon-Thaden IE, Viccini LF, Chamala S, Leitch AR, Schnable PS, Barbazuk WB, Soltis PS, Soltis DE. Next-generation sequencing and genome evolution in allopolyploids. Am J Bot. 2012;99:372–382. doi: 10.3732/ajb.1100395. [DOI] [PubMed] [Google Scholar]
- Cafasso D, Chinali G. An ancient satellite DNA has maintained repetitive units of the original structure in most species of the living fossil plant genus Zamia. Genome. 2014;57:125–135. doi: 10.1139/gen-2013-0133. [DOI] [PubMed] [Google Scholar]
- Cai Q, Zhang DM, Liu ZL, Wang XR. Chromosomal localization of 5S and 18S rDNA in five species of subgenus Strobus and their implications for genome evolution of Pinus. Ann Bot (Lond) 2006;97:715–722. doi: 10.1093/aob/mcl030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CS, Wright WA, Cox M, Vining TF, Major CS, Arsenault MP. Nuclear ribosomal DNA internal transcribed spacer 1 (ITS1) in Picea (Pinaceae): sequence divergence and structure. Mol Phylogenet Evol. 2005;35:165–185. doi: 10.1016/j.ympev.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Capesius I. Analysis of the ribosomal RNA gene repeat from the moss Funaria hygrometrica. Plant Mol Biol. 1997;33:559–564. doi: 10.1023/A:1005740031313. [DOI] [PubMed] [Google Scholar]
- Chaw SM, Parkinson CL, Cheng YC, Vincent TM, Palmer JD. Seed plant phylogeny inferred from all three plant genomes: Monophyly of extant gymnosperms and origin of Gnetales from conifers. Proc Natl Acad Sci U S A. 2000;97:4086–4091. doi: 10.1073/pnas.97.8.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condamine FL, Nagalingum NS, Marshall CR, Morlon H. Origin and diversification of living cycads: a cautionary tale on the impact of the branching process prior in Bayesian molecular dating. BMC Evol Biol. 2015;15:65. doi: 10.1186/s12862-015-0347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn RC, Zhao X, Paterson AH, Wendel JF. Polymorphism and concerted evolution in a tandemly repeated gene family: 5S ribosomal DNA in diploid and allopolyploid cottons. J Mol Evol. 1996;42:685–705. doi: 10.1007/BF02338802. [DOI] [PubMed] [Google Scholar]
- Dubrovina AS, Kiselev KV. Age-associated alterations in the somatic mutation and DNA methylation levels in plants. Plant Biol (Stuttg) 2015 doi: 10.1111/plb.12375. [DOI] [PubMed] [Google Scholar]
- Dvorak J, Jue D, Lassner M. Homogenization of tandemly repeated nucleotide-sequences by distance-dependent nucleotide-sequence conversion. Genetics. 1987;116:487–498. doi: 10.1093/genetics/116.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Eickbush DG. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Burke WD, Eickbush DG, Lathe WC. Evolution of R1 and R2 in the rDNA units of the genus Drosophila. Genetica. 1997;100:49–61. doi: 10.1023/A:1018396505115. [DOI] [PubMed] [Google Scholar]
- Emadzade K, Jang TS, Macas J, Kovařík A, Novák P, Parker J, Weiss-Schneeweiss H. Differential amplification of satellite PaB6 in chromosomally hypervariable Prospero autumnale complex (Hyacinthaceae) Ann Bot (London) 2014;114:1597–1608. doi: 10.1093/aob/mcu178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar JS, Glemin S, Galtier N. GC-Biased gene conversion impacts ribosomal DNA evolution in vertebrates, angiosperms, and other eukaryotes. Mol Biol Evol. 2011;28:2561–2575. doi: 10.1093/molbev/msr079. [DOI] [PubMed] [Google Scholar]
- Fagerlind F (1941) Bau und Entwicklung der Gnetum-Gametophyten, Stockholm
- Fajkus J, Sýkorová E, Leitch AR. Telomeres in evolution and evolution of telomeres. Chromosome Res. 2005;13:469–479. doi: 10.1007/s10577-005-0997-2. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV. Transposable elements, epigenetics, and genome evolution. Science. 2012;338:758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- Fehrer J, Krak K, Chrtek J., Jr Intra-individual polymorphism in diploid and apomictic polyploid hawkweeds (Hieracium, Lactuceae, Asteraceae): disentangling phylogenetic signal, reticulation, and noise. BMC Evol Biol. 2009;9:239. doi: 10.1186/1471-2148-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Wang Y, Gong X. Genetic diversity, genetic structure and demographic history of Cycas simplicipinna (Cycadaceae) assessed by DNA sequences and SSR markers. BMC Plant Biol. 2014;14:187. doi: 10.1186/1471-2229-14-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galián JA, Rosato M, Rosselló JA. Early evolutionary colocalization of the nuclear ribosomal 5S and 45S gene families in seed plants: evidence from the living fossil gymnosperm Ginkgo biloba. Heredity (Edinb) 2012;108:640–646. doi: 10.1038/hdy.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galián JA, Rosato M, Rosselló JA. Incomplete sequence homogenization in 45S rDNA multigene families: intermixed IGS heterogeneity within the single NOR locus of the polyploid species Medicago arborea (Fabaceae) Ann Bot (Lond) 2014;114:243–251. doi: 10.1093/aob/mcu115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley ARD, Kobayashi T. Monitoring the rate and dynamics of concerted evolution in the ribosomal DNA repeats of Saccharomyces cerevisiae using experimental evolution. Mol Biol Evol. 2011;28:2883–2891. doi: 10.1093/molbev/msr117. [DOI] [PubMed] [Google Scholar]
- Garcia S, Kovařík A. Dancing together and separate again: gymnosperms exhibit frequent changes of fundamental 5S and 35S rRNA gene (rDNA) organisation. Heredity. 2013;111:23–33. doi: 10.1038/hdy.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S, Lim KY, Chester M, Garnatje T, Pellicer J, Valles J, Leitch AR, Kovarik A. Linkage of 35S and 5S rRNA genes in Artemisia (family Asteraceae): first evidence from angiosperms. Chromosoma. 2009;118:85–97. doi: 10.1007/s00412-008-0179-z. [DOI] [PubMed] [Google Scholar]
- Glass SK, Moszczynska A, Crease TJ. The effect of transposon Pokey insertions on sequence variation in the 28S rRNA gene of Daphnia pulex. Genome. 2008;51:988–1000. doi: 10.1139/G08-092. [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. The Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;25:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpke D, Peterson A. Non-concerted ITS evolution in Mammillaria (Cactaceae) Mol Phylogenet Evol. 2006;41:579–593. doi: 10.1016/j.ympev.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Harpke D, Peterson A. Quantitative PCR revealed a minority of its copies to be functional in Mammillaria (Cactaceae) Int J Plant Sci. 2007;168:1157–1160. doi: 10.1086/520729. [DOI] [Google Scholar]
- Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007;51:526–536. doi: 10.1111/j.1365-313X.2007.03152.x. [DOI] [PubMed] [Google Scholar]
- Hizume M, Ishida F, Kondo K. Differential staining and in situ hybridization of nucleolar organizers and centromeres in Cycas revoluta chromosomes. Jpn J Genet. 1992;67:381–387. doi: 10.1266/jjg.67.381. [DOI] [Google Scholar]
- Hizume M, Ishida F, Murata M. Multiple locations of the ribosomal-RNA genes in chromosomes of pines, Pinus densiflora and P. thunbergii. Jpn J Genet. 1992;67:389–396. doi: 10.1266/jjg.67.389. [DOI] [Google Scholar]
- Islam-Faridi MN, Nelson CD, Kubisiak TL. Reference karyotype and cytomolecular map for loblolly pine (Pinus taeda L.) Genome. 2007;50:241–251. doi: 10.1139/G06-153. [DOI] [PubMed] [Google Scholar]
- Jasencakova Z, Soppe WJJ, Meister A, Gernand D, Turner BM, Schubert I. Histone modifications in Arabidopsis—high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 2003;33:471–480. doi: 10.1046/j.1365-313X.2003.01638.x. [DOI] [PubMed] [Google Scholar]
- Keller I, Chintauan-Marquier IC, Veltsos P, Nichols RA. Ribosomal DNA in the grasshopper Podisma pedestris: Escape from concerted evolution. Genetics. 2006;174:863–874. doi: 10.1534/genetics.106.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller I, Bensasson D, Nichols RA. Transition-transversion bias is not universal: a counter example from grasshopper pseudogenes. PLoS Genet. 2007;3:e22. doi: 10.1371/journal.pgen.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LJ, Renny-Byfield S, Pellicer J, Macas J, Novak P, Neumann P, Lysak MA, Day PD, Berger M, Fay MF, Nichols RA, Leitch AR, Leitch IJ. Analysis of the giant genomes of Fritillaria (Liliaceae) indicates that a lack of DNA removal characterizes extreme expansions in genome size. New Phytol. 2015;208:596–607. doi: 10.1111/nph.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Zilberman D. DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 2014;19:320–326. doi: 10.1016/j.tplants.2014.01.014. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell Mol Life Sci. 2011;68:1395–1403. doi: 10.1007/s00018-010-0613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Kokubugata G, Honda Y (1996) Marking and identification of certain chromosomes in wild Chrysanthemums and Cycads by fluorescence in situ hybridization using pTa71 rDNA probe. J Appl Genet 37:24–26
- Kovarik A, Matyasek R, Leitch A, Gazdova B, Fulnecek J, Bezdek M. Variability in CpNpG methylation in higher plant genomes. Gene. 1997;204:25–33. doi: 10.1016/S0378-1119(97)00503-9. [DOI] [PubMed] [Google Scholar]
- Kovarik A, Werlemark G, Leitch AR, Souckova-Skalicka K, Lim YK, Khaitova L, Koukalova B, Nybom H. The asymmetric meiosis in pentaploid dogroses (Rosa sect. Caninae) is associated with a skewed distribution of rRNA gene families in the gametes. Heredity. 2008;101:359–367. doi: 10.1038/hdy.2008.63. [DOI] [PubMed] [Google Scholar]
- Leitch AR, Leitch IJ. Ecological and genetic factors linked to contrasting genome dynamics in seed plants. New Phytol. 2012;194:629–646. doi: 10.1111/j.1469-8137.2012.04105.x. [DOI] [PubMed] [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, Bezdek M, Lichtenstein CP, Leitch AR. Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma. 2000;109:161–172. doi: 10.1007/s004120000074. [DOI] [PubMed] [Google Scholar]
- Liston A, Robinson WA, Oliphant JM, AlvarezBuylla ER. Length variation in the nuclear ribosomal DNA internal transcribed spacer region of non-flowering seed plants. Syst Bot. 1996;21:109–120. doi: 10.2307/2419742. [DOI] [Google Scholar]
- Lubaretz O, Fuchs J, Ahne R, Meister A, Schubert I. Karyotyping of three Pinaceae species via fluorescent in situ hybridization and computer-aided chromosome analysis. Theor Appl Genet. 1996;92:411–416. doi: 10.1007/BF00223687. [DOI] [PubMed] [Google Scholar]
- Ma L, Hatlen A, Kelly LJ, Becher H, Wang W, Kovarik A, Leitch IJ, Leitch AR. Angiosperms are unique among land plant lineages in the occurrence of key genes in the RNA-dependent DNA methylation (RdDM) pathway. Genome Biol Evol. 2015;7:2648–2662. doi: 10.1093/gbe/evv171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggini F, Baldassini S. Ribosomal-RNA genes in the genus Pinus.1. Caryologia. 1995;48:17–25. doi: 10.1080/00087114.1995.10797314. [DOI] [Google Scholar]
- Marques A, Fuchs J, Ma L, Heckmann S, Guerra M, Houben A. Characterization of eu- and heterochromatin of Citrus with a focus on the condensation behavior of 45S rDNA chromatin. Cytogenet Genome Res. 2011;134:72–82. doi: 10.1159/000323971. [DOI] [PubMed] [Google Scholar]
- Martin CC, Gordon R. Differentiation trees, a junk DNA molecular clock, and the evolution of neoteny in salamanders. J Evol Biol. 1995;8:339–354. doi: 10.1046/j.1420-9101.1995.8030339.x. [DOI] [Google Scholar]
- Martoni F, Eickbush DG, Scavariello C, Luchetti A, Mantovani B. Dead element replicating: Degenerate R2 element replication and rDNA genomic turnover in the Bacillus rossius stick insect (Insecta: Phasmida) PLoS One. 2015;10:e0121831. doi: 10.1371/journal.pone.0121831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyasek R, Renny-Byfield S, Fulnecek J, Macas J, Grandbastien MA, Nichols R, Leitch A, Kovarik A. Next generation sequencing analysis reveals a relationship between rDNA unit diversity and locus number in Nicotiana diploids. BMC Genomics. 2012;13:722. doi: 10.1186/1471-2164-13-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed-Bessudo C, Levy AA. Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:E981–988. doi: 10.1073/pnas.1120742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentewab AB, Jacobsen MJ, Flowers RA. Incomplete homogenization of 18 S ribosomal DNA coding regions in Arabidopsis thaliana. BMC Res Notes. 2011;4:93. doi: 10.1186/1756-0500-4-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BG, Friesen N, Heslop-Harrison JS. Molecular cytogenetic analysis of Podocarpus and comparison with other gymnosperm species. Ann Bot (Lond) 2002;89:483–489. doi: 10.1093/aob/mcf047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale DB, Wegrzyn JL, Stevens KA, Zimin AV, Puiu D, Crepeau MW, Cardeno C, Koriabine M, Holtz-Morris AE, Liechty JD, Martinez-Garcia PJ, Vasquez-Gross HA, Lin BY, Zieve JJ, Dougherty WM, Fuentes-Soriano S, Wu LS, Gilbert D, Marcais G, Roberts M, Holt C, Yandell M, Davis JM, Smith KE, Dean JF, Lorenz WW, Whetten RW, Sederoff R, Wheeler N, McGuire PE, Main D, Loopstra CA, Mockaitis K, deJong PJ, Yorke JA, Salzberg SL, Langley CH. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 2014;15:R59. doi: 10.1186/gb-2014-15-3-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves N, Delgado M, Silva M, Carpeta A, Morais-Cecílio L, Viegas W. Ribosomal DNA heterochromatin in plants. Cytogenet Genome Res. 2005;109:104–111. doi: 10.1159/000082388. [DOI] [PubMed] [Google Scholar]
- Nieto Feliner G, Rossello JA. Concerted evolution of multigene families andhomeologous recombination. In: Wendel JF, editor. Plant genome diversity. Wien: Springer; 2012. pp. 171–194. [Google Scholar]
- Novak P, Neumann P, Macas J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics. 2010;11:378. doi: 10.1186/1471-2105-11-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P, Neumann P, Pech J, Steinhaisl J, Macas J. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics. 2013;29:792–793. doi: 10.1093/bioinformatics/btt054. [DOI] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, Vicedomini R, Sahlin K, Sherwood E, Elfstrand M, Gramzow L, Holmberg K, Hallman J, Keech O, Klasson L, Koriabine M, Kucukoglu M, Kaller M, Luthman J, Lysholm F, Niittyla T, Olson A, Rilakovic N, Ritland C, Rossello JA, Sena J, Svensson T, Talavera-Lopez C, Theissen G, Tuominen H, Vanneste K, Wu ZQ, Zhang B, Zerbe P, Arvestad L, Bhalerao R, Bohlmann J, Bousquet J, Gil RG, Hvidsten TR, de Jong P, MacKay J, Morgante M, Ritland K, Sundberg B, Thompson SL, Van de Peer Y, Andersson B, Nilsson O, Ingvarsson PK, Lundeberg J, Jansson S. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–584. doi: 10.1038/nature12211. [DOI] [PubMed] [Google Scholar]
- Pearson HHW. Further observations on Welwitschia. P R Soc Lond B-Conta. 1908;80:530–531. doi: 10.1098/rspb.1908.0048. [DOI] [Google Scholar]
- Pedrosa-Harand A, de Almeida CCS, Mosiolek M, Blair M, Schweizer D, Guerra M. Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Theor Appl Genet. 2006;112:924–933. doi: 10.1007/s00122-005-0196-8. [DOI] [PubMed] [Google Scholar]
- Pich U, Schubert I. Terminal heterochromatin and alternative telomeric sequences in Allium cepa. Chromosome Res. 1998;6:315–321. doi: 10.1023/A:1009227009121. [DOI] [PubMed] [Google Scholar]
- Poole A, Penny D, Sjoberg BM. Confounded cytosine! Tinkering and the evolution of DNA. Nat Rev Mol Cell Biol. 2001;2:147–151. doi: 10.1038/35052091. [DOI] [PubMed] [Google Scholar]
- Ribeiro T, Barao A, Viegas W, Morais-Cecilio L. Molecular cytogenetics of forest trees. Cytogenet Genome Res. 2008;120:220–227. doi: 10.1159/000121070. [DOI] [PubMed] [Google Scholar]
- Roa F, Guerra M. Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol Biol. 2012;12:225. doi: 10.1186/1471-2148-12-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Schlotterer C, Hauser MT, von Haeseler A, Tautz D. Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Mol Biol Evol. 1994;11:513–522. doi: 10.1093/oxfordjournals.molbev.a040131. [DOI] [PubMed] [Google Scholar]
- Sedelnikova T, Pimenov A. A karyological study of swamp and dry valley populations of Siberian fir (Abies sibirica Ledeb.) Biol Bull. 2005;32:16–21. doi: 10.1007/s10525-005-0004-7. [DOI] [PubMed] [Google Scholar]
- Siljak-Yakovlev S, Cerbah M, Coulaud J, Stoian V, Brown SC, Zoldos V, Jelenic S, Papes D. Nuclear DNA content, base composition, heterochromatin and rDNA in Picea omorika and Picea abies. Theor Appl Genet. 2002;104:505–512. doi: 10.1007/s001220100755. [DOI] [PubMed] [Google Scholar]
- Smarda P, Bures P, Horova L, Leitch IJ, Mucina L, Pacini E, Tichy L, Grulich V, Rotreklova O. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proc Natl Acad Sci U S A. 2014;111:E4096–4102. doi: 10.1073/pnas.1321152111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone T, Fujisawa M, Takenaka M, Nakagawa S, Yamaoka S, Sakaida M, Nishiyama R, Yamato KT, Ohmido N, Fukui K, Fukuzawa H, Ohyama K. Bryophyte 5S rDNA was inserted into 45S rDNA repeat units after the divergence from higher land plants. Plant Mol Biol. 1999;41:679–685. doi: 10.1023/A:1006398419556. [DOI] [PubMed] [Google Scholar]
- Stage DE, Eickbush TH. Sequence variation within the rRNA gene loci of 12 Drosophila species. Genome Res. 2007;17:1888–1897. doi: 10.1101/gr.6376807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub SCK, Parks M, Weitemier K, Fishbein M, Cronn RC, Liston A. Navigating the tip of the genomic iceberg: Next generation sequencing for plant systemtatics. Am J Bot. 2012;99:349–364. doi: 10.3732/ajb.1100335. [DOI] [PubMed] [Google Scholar]
- Wei XX, Wang XQ, Hong DY. Marked intragenomic heterogeneity and geographical differentiation of nrDNA ITS in Larix potaninii (Pinaceae) J Mol Evol. 2003;57:623–635. doi: 10.1007/s00239-003-2512-8. [DOI] [PubMed] [Google Scholar]
- Weitemier K, Straub SC, Fishbein M, Liston A. Intragenomic polymorphisms among high-copy loci: a genus-wide study of nuclear ribosomal DNA in Asclepias (Apocynaceae) PeerJ. 2015;3:e718. doi: 10.7717/peerj.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West C, James SA, Davey RP, Dicks J, Roberts IN. Ribosomal DNA sequence heterogeneity reflects intraspecies phylogenies and predicts genome structure in two contrasting yeast species. Syst Biol. 2014;63:543–554. doi: 10.1093/sysbio/syu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Renner SS. The internal transcribed spacer of nuclear ribosomal DNA in the gymnosperm Gnetum. Mol Phylogenet Evol. 2005;36:581–597. doi: 10.1016/j.ympev.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Xiao LQ, Moller M. Nuclear ribosomal ITS functional paralogs resolve the phylogenetic relationships of a late-miocene radiation Cycad Cycas (Cycadaceae) PLoS One. 2015;10:1. doi: 10.1371/journal.pone.0117971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao LQ, Moller M, Zhu H (2010) High nrDNA ITS polymorphism in the ancient extant seed plant Cycas: Incomplete concerted evolution and the origin of pseudogenes. Mol Phylogenet Evol 55:168–177 [DOI] [PubMed]
- Zheng XY, Cai DY, Yao LH, Teng YW. Non-concerted ITS evolution, early origin and phylogenetic utility of ITS pseudogenes in Pyrus. Mol Phylogenet Evol. 2008;48:892–903. doi: 10.1016/j.ympev.2008.05.039. [DOI] [PubMed] [Google Scholar]
- Zozomova-Lihova J, Mandakova T, Kovarikova A, Muhlhausen A, Mummenhoff K, Lysak MA, Kovarik A. When fathers are instant losers: homogenization of rDNA loci in recently formed Cardamine x schulzii trigenomic allopolyploid. New Phytol. 2014;203:1096–1108. doi: 10.1111/nph.12873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PPTX 4.97 MB)