Significance
Seed desiccation tolerance (DT) is one of the most fascinating processes of higher plants, and has played a fundamental role in the evolution of land plants. DT allows plant seeds to remain viable in the dry state for years and even centuries. What the key transcription factors (TFs) are that activate the mechanisms that allow plant seeds to maintain cellular and DNA integrity for centuries remains largely unknown. In this paper, we report the identification of the TFs that act as major nodes of the transcriptional networks that regulate the acquisition of seed DT. We also report the functional validation of several of the major regulators of seed DT in plants.
Keywords: regulatory network, desiccation tolerance, drought tolerance, seed development, oligosaccharides
Abstract
Desiccation tolerance (DT) is a remarkable process that allows seeds in the dry state to remain viable for long periods of time that in some instances exceed 1,000 y. It has been postulated that seed DT evolved by rewiring the regulatory and signaling networks that controlled vegetative DT, which itself emerged as a crucial adaptive trait of early land plants. Understanding the networks that regulate seed desiccation tolerance in model plant systems would provide the tools to understand an evolutionary process that played a crucial role in the diversification of flowering plants. In this work, we used an integrated approach that included genomics, bioinformatics, metabolomics, and molecular genetics to identify and validate molecular networks that control the acquisition of DT in Arabidopsis seeds. Two DT-specific transcriptional subnetworks were identified related to storage of reserve compounds and cellular protection mechanisms that act downstream of the embryo development master regulators LEAFY COTYLEDON 1 and 2, FUSCA 3, and ABSCICIC ACID INSENSITIVE 3. Among the transcription factors identified as major nodes in the DT regulatory subnetworks, PLATZ1, PLATZ2, and AGL67 were confirmed by knockout mutants and overexpression in a desiccation-intolerant mutant background to play an important role in seed DT. Additionally, we found that constitutive expression of PLATZ1 in WT plants confers partial DT in vegetative tissues.
Desiccation tolerance (DT) can be operationally defined as the ability of an organism to dry to equilibrium with moderately dry air (50 to 70% relative humidity at 20 to 30 °C) and then resume normal function when rehydrated (1). DT organisms orchestrate a complex number of responses to protect cellular structures and prevent damage to proteins and nucleic acids. Early land plants evolved mechanisms to survive harsh drying environments to successfully exploit different ecosystems on land. Therefore, it has been postulated that the initial evolution of vegetative DT, in both vegetative and reproductive stages, was a crucial step required for the colonization of land by primitive plants of a fresh water origin (2).
Seed DT, a trait that allows terrestrial plants to survive long periods of sparse water until favorable conditions are present for germination, is probably part of the answer to Darwin’s “abominable mystery,” the sudden appearance of great angiosperm diversity in the fossil record. In angiosperms, DT is acquired at the seed maturation stage, which involves a complex regulatory network (3, 4) that activates a large subset of genes involved in a number of mechanisms that influence seed survival in the dry state. The set of genes required for seed DT includes genes encoding protective proteins such as late embryogenesis abundant (LEA) (5, 6) and heat shock proteins (HSPs) (7), enzymes involved in scavenging reactive oxygen species (8) and the biosynthesis of protective compounds such as oligosaccharides (3, 9), and antioxidants such as tocopherols and flavonoids (10, 11).
In Arabidopsis, embryo development and seed maturation, including the acquisition of DT, is orchestrated by a set of four master regulators: LEAFY COTYLEDON 1 (LEC1), a CCAAT box-binding factor, and three B3 domain-containing proteins (12), ABSCISIC ACID INSENSITIVE 3 (ABI3), FUSCA 3 (FUS3), and LEC2. In addition to controlling embryo formation and seed maturation, these master regulators also repress the expression of genes required for the transition from embryonic to vegetative development (13–16). Although the role of these master regulators during seed maturation is globally similar, some of their functions are very specific. For example, in contrast to mutations in LEC1, ABI3, or FUS3 that drastically affect DT (17, 18), lec2 mutants do not present this effect (18, 19). Interestingly, ectopic expression of LEC1, FUS3, or ABI3 in single- or double-mutant backgrounds of the other two regulators activated some processes of seed maturation, such as lipid and seed storage protein accumulation, but not DT, suggesting that all three regulators are required to activate DT. Genetic evidence suggests that downstream of LEC1, FUS3, ABI3, and LEC2, other transcription factors (TFs) play important roles in the network that regulates specific aspects of embryo development and seed maturation and, in particular, seed DT (20).
At least 13 independent cases of evolution (or reevolution) of vegetative DT occurred in the angiosperms and 1 in gymnosperms (21). The independent reevolution of vegetative DT in different clades of the angiosperms suggests that despite being a quite complex process, both vegetative and seed DT might be controlled by one or a few regulatory networks composed of a discrete number of TFs. The availability of seed desiccation-intolerant mutants allows a comparative analysis between desiccation-intolerant mutants, such as lec1-1, fus3-3, and abi3-5, with their corresponding DT wild-type ecotypes and other mutants (lec2-1) or alleles of the same gene (abi3-1), with similar phenotypes but that are desiccation-tolerant, to determine the transcriptional and metabolic differences between desiccation-tolerant and -intolerant lines during seed development. This type of comparative analysis should allow the identification of genes for which transcriptional activation is required to acquire seed DT but which fail to activate during seed maturation in desiccation-intolerant mutants. Understanding the networks that regulate seed DT in model plant systems will provide the tools to understand an evolutionary process that played a crucial role in the diversification of flowering plants. Although several processes involved in seed maturation and their regulatory mechanisms have been studied in Arabidopsis (22) and Medicago (3, 4), the regulatory interactions activating DT remain largely unknown. In this work, we used an integrated approach that included genomics, bioinformatics, metabolomics, and molecular genetics to identify and validate molecular networks that control the acquisition of DT in Arabidopsis seeds.
Results
Overview of the Experimental Comparative Strategy.
Our hypothesis was that there is a common subset of genes that fail to be activated in desiccation-intolerant mutants compared with their expression patterns in desiccation-tolerant lines, which should include most of the genes that are essential for the acquisition of seed DT, and that their activation patterns should allow the construction of regulatory networks to identify TFs that constitute the main nodes orchestrating the acquisition of DT. To identify the gene regulatory networks involved in the acquisition of DT downstream of LEC1, FUS3, and ABI3, we first designed a comparative transcriptomic analysis between the seed desiccation-intolerant lines lec1-1, fus3-3, and abi3-5 and the desiccation-tolerant lines lec2-1 and abi3-1. All analyses included the respective DT wild-type ecotype for each mutant. lec1 and lec2 have similar phenotypes, including morphological alterations during embryo development and reduced accumulation of storage compounds, and only differ in DT (SI Appendix, Table S1). Similarly, abi3-1 and abi3-5 are two alleles of ABI3 that are both insensitive to abscisic acid (ABA) but differ in their seed DT. Comparative transcriptomic analysis between desiccation-tolerant and -intolerant seed should allow the identification of genes differentially expressed during the seed maturation process between these two sets of lines. Among these differentially expressed genes, those that fail to be activated in all desiccation-intolerant mutants should contain those that are relevant for the acquisition of seed DT (SI Appendix, Fig. S1 and Table S1).
Global Transcriptional Analysis of Desiccation-Tolerant and -Intolerant Arabidopsis Seeds.
To obtain a global view of the transcriptional differences between desiccation-tolerant and -intolerant lines during seed maturation, we constructed RNA-sequencing (RNA-seq) libraries from RNA extracted from each mutant and their corresponding wild type at three different developmental stages (SI Appendix, Table S2): (i) 15 d after flowering (DAF), a developmental stage prior to drastic water loss; (ii) 17 DAF, when rapid water loss starts; and (iii) 21 DAF, when the seed is completely dry (SI Appendix, Fig. S1). To determine differentially expressed genes (DEGs), RNA-seq data were analyzed using two types of model analysis. First, we used a generalized linear model (GLM) based on an interaction model, which comprises genes for DT-specific mutant differences (SI Appendix, Table S3 and Datasets S1–S6). Using GLM and a stringency level of false discovery rate (FDR) of <0.05 and log2 fold change (log2FC) <1 and >−1, we identified 3,781 DEGs between tolerant and intolerant lines in at least one of the three developmental stages sampled for this analysis (Datasets S1–S6). Among the DEGs, 2,320 were up-regulated (Datasets S1–S3) and 1,461 were down-regulated (Datasets S4–S6).
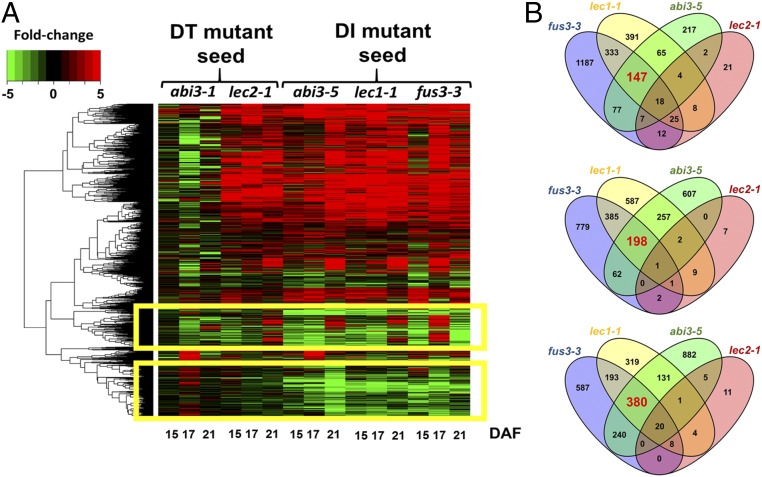
The second GLM analysis was based on pairwise comparisons between each mutant line and their corresponding wild-type ecotype (Fig. 1A and SI Appendix, Table S4). In this analysis, one large subset of genes was up-regulated in lec2-1, lec1-1, abi3-5, and fus3-3 with respect to abi3-1 and the WT controls, which likely represents genes that are activated as part of the direct transition from embryo to vegetative growth in these mutants as opposed to their inactivation during the entrance to dormancy and the acquisition of DT (see SI Appendix for details) (Fig. 1A and SI Appendix, Figs. S2–S4). A second subset of DEGs, which appeared as drastically repressed in all desiccation-intolerant mutants compared with desiccation-tolerant lines, could represent genes that are directly or indirectly relevant for the acquisition of DT in Arabidopsis seed (Fig. 1A). The number of down-regulated genes specific for desiccation-intolerant mutants increased as the level of water content decreased in the seed, from 147 genes at 15 DAF to 380 at 21 DAF (Fig. 1B).
Fig. 1.
Global gene expression profile in seed desiccation-intolerant mutants. (A) Heat map from hierarchical clustering of differentially expressed genes. Each mutant was compared with its corresponding wild type (lec1-1 and lec2-1 versus Ws; abi3-5 versus Ler; fus3-3 versus Col-0) at three developmental stages, 15, 17, and 21 DAF. Green indicates down-regulated values, red indicates up-regulated values, and black indicates unchanged values. The yellow rectangles indicate the common down-regulated genes between intolerant mutants. DI, desiccation-intolerant; DT, desiccation-tolerant. (B) Venn diagrams showing the number and distribution of differential genes across the comparison among mutants representing down-regulated genes at 15, 17, and 21 DAF. Genes potentially related to desiccation tolerance are indicated in red.
As predicted, desiccation-intolerant (DI)-specific down-regulated genes were enriched (FDR < 0.05) in the following Gene Ontology (GO) categories: molecular function: oxidoreductase activity and nutrient reservoir; and biological process: lipid and carbohydrate biosynthesis, seed development, and ABA and stress responses such as water, oxidation, and temperature (SI Appendix, Fig. S2B and Dataset S7). A more detailed analysis of the same set of genes using the MAPMAN system (23) showed enrichment in processes such as abiotic stress, LEA protein synthesis, and metabolic pathways including raffinose, stachyose, and trehalose biosynthesis (SI Appendix, Fig. S5). We observed that the number of genes for these stress categories increased at 17 and 21 DAF (SI Appendix, Fig. S5), when rapid water loss occurs. The finding that genes that are not activated in desiccation-intolerant mutants during seed maturation belong to water stress and cell protection mechanisms confirmed that desiccation-intolerant mutants fail to activate mechanisms required to acquire DT in the seed. lec2-1 and abi3-1 had a much lower number of down-regulated genes with respect to the WT controls than lec1-1, fus3-1, and abi3-5, which correlates well with the fact that these mutants are still capable of acquiring DT (SI Appendix, Fig. S7).
Carbohydrate Profiling of Seeds from Desiccation-Tolerant and -Intolerant Mutants.
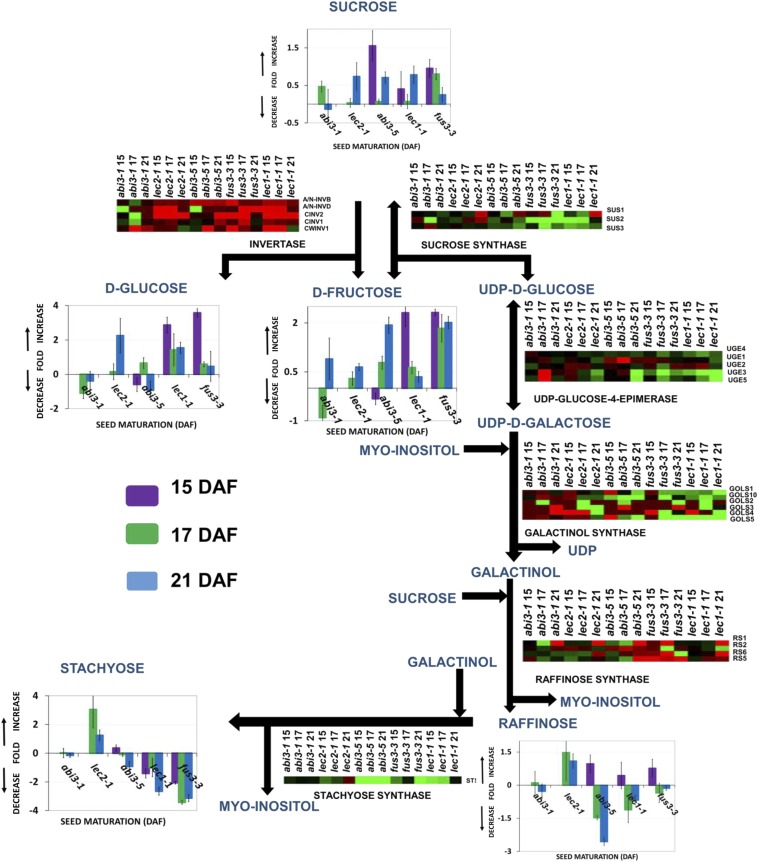
It has been reported that the raffinose family of oligosaccharides (RFOs), which accumulate during the last phase of seed maturation, can act as “water replacement” molecules that provide the hydrogen bonds required for membrane and protein stabilization as well as for protecting DNA against hydrolytic damage (24, 25). Therefore, to complement our transcriptional study, we performed carbohydrate profiling of desiccation-tolerant and -intolerant mutants using the same seed developmental stages chosen for RNA-seq analysis (15, 17, and 21 DAF). We specifically analyzed the sucrose-to-raffinose pathway (Fig. 2), as well as the invertase products d-glucose and d-fructose (Fig. 2). This analysis showed that in lec1-1, abi3-5, fus3-3, and lec2-1, sucrose levels at 15 DAF were 1-fold higher than in their wild-type controls and later decreased to almost wild-type levels at 21 DAF. Raffinose levels decreased 1.5- and 2.5-fold in abi3-5 and 1- and 0.7-fold in lec1-1 at 17 and 21 DAF, respectively, whereas in lec2-1, raffinose increased 2-fold at 21 DAF. Stachyose levels decreased 3-fold in lec1-1 and fus3-3, whereas for lec2-1 an increase of 3-fold with respect to the wild-type control was determined (Fig. 2). In agreement with the observed reduction in oligosaccharide accumulation, transcript levels of genes encoding key enzymes in the raffinose pathway, such as sucrose synthases (AT5G4919, SUS2 and AT4G02280, SUS3), UDP-d-galactose-4-epimerases (AT1G63180, UGE3 and AT4G10960, UGE5), galactinol synthases (AT2G4718, GOLS1 and AT1G09350, GOLS2), and stachyose synthase (AT4G01970, STS), were strongly repressed in desiccation-intolerant mutants (Fig. 2). Additionally, in agreement with the higher levels of d-glucose and d-fructose present in the seed of desiccation-intolerant mutants, invertase genes are up-regulated in these mutants. This suggests that sucrose is degraded to d-glucose and d-fructose via invertase enzymes and is not metabolized to stachyose and raffinose (Fig. 2), suggesting that the accumulation of stachyose and raffinose appears to be a key, common factor for the acquisition of DT in Arabidopsis seeds.
Fig. 2.
Metabolic pathway of stachyose biosynthesis in Arabidopsis seeds. Graphs represent changes in the contents of sugars and oligosaccharides for each mutant during the desiccation period. Bar graphs indicate the log2 fold change-relative content with respect to the wild-type controls. Values are from two independently grown sets of plants with three technical replicates each. Heat maps represent expression profiles of genes encoding putative enzymes involved in raffinose and stachyose synthesis as well as sucrose degradation pathways. Red and green represent up-regulated and down-regulated genes, respectively. The biochemical and physiological pathways were classified according to the KEGG database (www.genome.jp/kegg/). ALKALINE/NEUTRAL INVERTASE (A/N-INVB, AT4G34860; A/N-INVD, AT1G22650), CYTOSOLIC INVERTASE (CINV2, AT4G09510; CINV1, AT1G35580), CELL WALL INVERTASE 1 (CWINV1, AT3G13790), SUCROSE SYNTHASE (SUS1, AT5G20830; SUS2, AT5G49190; SUS3, AT4G02280), UDP-d-GLUCOSE/UDP-d-GALACTOSE 4-EPIMERASE (UGE1, AT1G12780; UGE2, AT4G23920; UGE3, AT1G63180; UGE4, AT1G64440; UGE5, AT4G10960), GALACTINOL SYNTHASE (GOLS1, AT2G47180; GOLS2, AT1G56600; GOLS3, AT1G09350; GOLS4, AT1G60470; GOLS5, AT5G23790; GOLS10, AT5G30500), RAFFINOSE SYNTHASE (RS1, AT1G55740; RS2, AT3G57520; RS5, AT5G40390; RS6, AT5G20250), STACHYOSE SYNTHASE (STS, AT4G01970). Data are the means ± SD of two biological replicates and three technical replicates.
Inferring Novel Transcription Factors Involved in Desiccation Tolerance Acquisition by Arabidopsis Seeds.
Our transcriptomics data provided information on the genes possibly involved in DT acquisition. However, transcriptomics data by themselves do not unveil the regulatory networks that control complex processes, nor the key TFs that coordinate the expression of genes involved in such regulatory networks. To identify these regulatory pathways and predict novel TF genes involved in DT, we constructed two ARACNe-based (26, 27) coexpression networks using two carefully curated datasets obtained from 169 seed-specific CEL format files from 24 Affymetrix ATH1 microarray experiments (SI Appendix, Table S5). One of the coexpression networks was a global coexpression network containing all genes expressed in the seed that are represented in the ATH1 microarrays, which we named FullSeedNet. The second coexpression network contained only the TFs known to be expressed in the seed, which we named TFsSeedNet (26) (SI Appendix, Fig. S8A and Dataset S10). To identify TFs potentially involved in regulating the establishment of seed DT in Arabidopsis, we integrated into the TFsSeedNet TF genes repressed in desiccation-intolerant mutants (Datasets S1–S6), which represent TF genes that are activated by LEC1, FUS3, and ABI3 for the acquisition of DT but fail to be activated in lec1, fus3, and abi3.
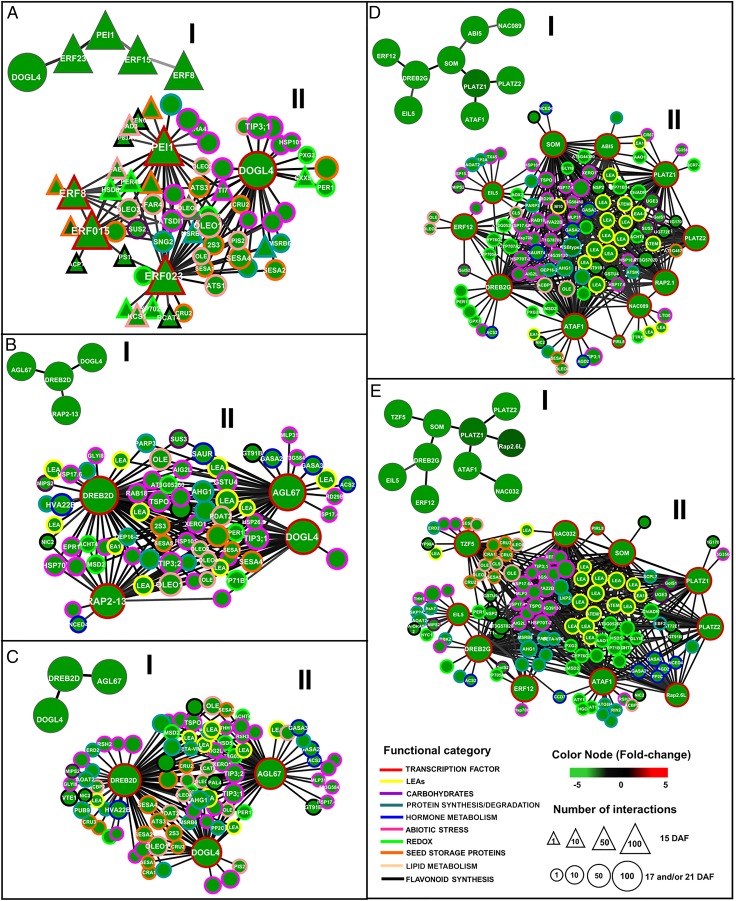
When TF genes that fail to be activated in desiccation-intolerant mutants were mapped to TFsSeedNet at 15, 17, and 21 DAF, we found that they formed two major coexpression subnetworks, which we termed TFsSeed-subNetDT1 and TFsSeed-subNetDT2 (Fig. 3 and SI Appendix, Fig. S8 B and C). TFsSeed-subNetDT1 was mainly composed of members of the APETALA2/ethylene responsive factor (AP2-ERF) TF gene family (DREB2D, AT1G75490; RAP2-13, AT1G22190; ERF15, AT4G31060; ERF23, AT1G01250; and ERF8, AT1G53170), but also contained members of the basic leucine zipper (bZIP12, AT2G41070), MADS-BOX (AGL67, AT1G77950), C2H2 zinc-finger (PEI1; AT5G07500), and delay of germination-like (DOG-like) (AT4G18650, which we named DOGL4) TF families (Fig. 3 A–C and SI Appendix, Fig. S8B). TFsSeed-subNetDT2 includes three members of the NAC family (ATAF1, AT1G01720; ANAC032, AT1G77450; and ANAC089, AT5G22290), two from the AP2-ERP family (DREB2G, AT5G18450 and ERF12, AT1G28360), two from the C3H zinc-finger family (SOM, AT1G03790 and TZF5, AT1G03790), two from the plant AT-rich sequence- and zinc-binding protein family (PLATZ) (AT1G21000, which we named PLATZ1, and AT1G76590, which we named PLATZ2), one from the bZIP family (ABI5, AT2G36270), and one from the ethylene insensitive 3 family (EIL5, AT5G65100) (Fig. 3 D and E and SI Appendix, Fig. S8C).
Fig. 3.
ARACNe-inferred coexpression networks for genes involved in the acquisition of desiccation tolerance. “I” graphs represent TFsSeed-subNetDT1 or TFsSeed-subNetDT2 at data processing inequality (DPI) 0.0, and “II” graphs represent the interactions of transcription factors in II and nontranscription factors from FullSeedNet. (A–C) Graphs correspond to genes for TFsSeed-subNetDT1 involved in nutrient storage and stress tolerance at 15 (A), 17 (B), and 21 DAF (C). (D and E) Graphs correspond to genes for TFsSeed-subNetDT2 involved mainly in cellular protection mechanisms at 17 and 21 DAF. TFsSeed-subNetDT2 is not yet present at 15 DAF. Genes are represented as nodes and inferred interactions are represented as edges. Network attributes are presented in the information box. Nodes with a triangular shape represent genes that are expressed only at 15 DAF, and circles represent genes that are expressed at least at two of the sampled stages. Node borders represent functional categories (see Datasets S13 and S15 for details). Edge width and color intensity are proportional to the mutual information value of the interaction, with higher MI values corresponding to thicker and darker edges. I represent the TFs that act as major nodes at each stage of the integration of the regulatory networks and II represents the same TFs with all the putative target genes of each node.
Coexpression Subnetworks of Differentially Expressed Genes and Potential Targets.
We then searched for genes that are coexpressed with the TFs in TFsSeed-subNetDT1 and TFsSeed-subNetDT2 in FullSeedNet (SI Appendix, Fig. S9); these coexpression subnetworks were denoted FullSeed-subNetDT1 and FullSeed-subNetDT2. These genes represent the potential effector genes activated by the TFs that are part of the two regulatory subnetworks that failed to be activated in desiccation-intolerant mutants. FullSeed-subNetDT1 was composed of a total of 280 genes (Fig. 3 A–C, SI Appendix, Fig. S10, and Dataset S12), for which the most significantly enriched categories included nutrient reservoir activity and lipid storage (SI Appendix, Fig. S12 and Dataset S13). When FullSeed-subNetDT1 was analyzed at each developmental stage (15, 17, and 21 DAF), we found that at 15 DAF it is composed of five TF nodes, DOGL4, PEI1, ERF15, ERF8, and ERF23, that interact [mutual information (MI) > 0.5 and P < 1e-35; Fig. 3A and Dataset S10] with effector genes mainly involved in fatty acid, TAG biosynthesis, and storage protein genes such as those encoding the albumin proteins SESA1, SESA2, SESA3, SESA4, and SESA5, cruciferin proteins CRU2 and CRU3, and oleosin proteins OLE1, OLE2, and OLE4. At 17 DAF, TFsSeed-subNetDT1 significantly changed, maintaining only DOGL4 as a major node, and three other TFs (DREB2D, AGL67, and RAP2-13) were incorporated as nodes in this subnetwork. DREB2D appears to be a key TF in TFsSeed-subNetDT1 because it has the largest number of interactions, and it is strongly coexpressed (MI > 0.6 and P < 1e-40) with AGL67 and DOGL4. These three TFs have as predicted targets genes related to stress tolerance, including LEA genes (LEA4-1, LEA14), heat shock proteins (HSP17.4, HSP101, HSP70), dehydrins (XERO1, RAB18), and aquaporins (TIP3 and TIP1) (Fig. 3 B and C and Datasets S10 and S12). The evolution of FullSeed-subNetDT1 during seed maturation suggests that this subnetwork is initially involved in regulating genes involved in the accumulation of reserve compounds and then in DT acquisition.
FullSeed-subNetDT2 was composed of 317 genes, which represent 17% of all down-regulated genes from tolerance-specific mutant differences (Fig. 3 D and E and SI Appendix, Fig. S11). This subnetwork is enriched in effector genes in the following categories: redox activity, LEA protein, and response to abiotic stimulus (SI Appendix, Fig. S12 and Datasets S14 and S15). In FullSeed-subNetDT2, we identified ATAF1 as a TF with an important number of interactions (81), and is strongly coexpressed (MI > 0.5 and P < 1e-35) with other TFs in the subnetwork such as PLATZ1, SOM, and DREB2G. These four TFs are central in FullSeed-subNetDT2 because each has a large number of interactions and they share common nodes that correspond to genes involved in cellular protection. For example, among the genes involved in stress resistance that are predicted to be activated by these TFs, we found three LEA group 4 genes (LEA4-1, LEA4-5, and LEA4-2), two LEA group 1 genes (ATEM1 and ATEM6), three genes involved in raffinose synthesis (UGE3, SUS3, and GOLS1), two catalase genes (CAT2 and CAT3), one superoxide dismutase gene (SOD), two small heat shock protein genes (HSP17.4 and HSP17.6), and two genes involved in ABA signaling (PP2C and ABI5) (Fig. 3 D and E, SI Appendix, Fig. S11, and Datasets S10 and S14).
Interestingly, FullSeed-subNetDT2 was specifically activated at 17 DAF and became more complex at 21 DAF, which corresponds to the developmental stages at which rapid and total water loss occurs (Fig. 3 D and E). ATAF1, a NAC TF, was the node with the highest number of interactions in FullSeed-subNetDT2, and its potential targets are stress protection genes. In FullSeed-subNetDT2, we identified four other TFs with a high number of interactions, namely SOM, DREB2G, PLATZ1, and PLATZ2, which also preferentially interacted with genes associated with stress protection processes (Fig. 3 D and E). In summary, our network analysis supports a model in which at early stages FullSeed-subNetDT1 regulates seed-filling genes and at later stages FullSeed-subNetDT1 and FullSeed-subNetDT2 regulate DT genes.
We then searched for enriched cis-regulatory elements in the promoters of target genes in each subnetwork. In general, the enriched motifs were ABA signaling-related (ABF binding site motif, ABRE binding site motif, and ACGT ABRE motif A2OSE). The seed-specific motif RY-repeat was found in the three stages of FullSeed-subNetDT1, whereas dehydration and drought responses (ABRE-like binding site motif, DRE core motif, CBF1 BS in cor15a, and AtMYC2 BS in RD22) were enriched in FullSeed-subNetDT2 (SI Appendix, Table S6). This finding also supports our model for distinct but complementary roles for FullSeed-subNetDT1 and FullSeed-subNetDT2 in the acquisition of seed DT (SI Appendix, Fig. S20).
Mutants of the Major Nodes of FullSeed-SubNetDT1 and FullSeed-SubNetDT2 Have Reduced Desiccation Tolerance.
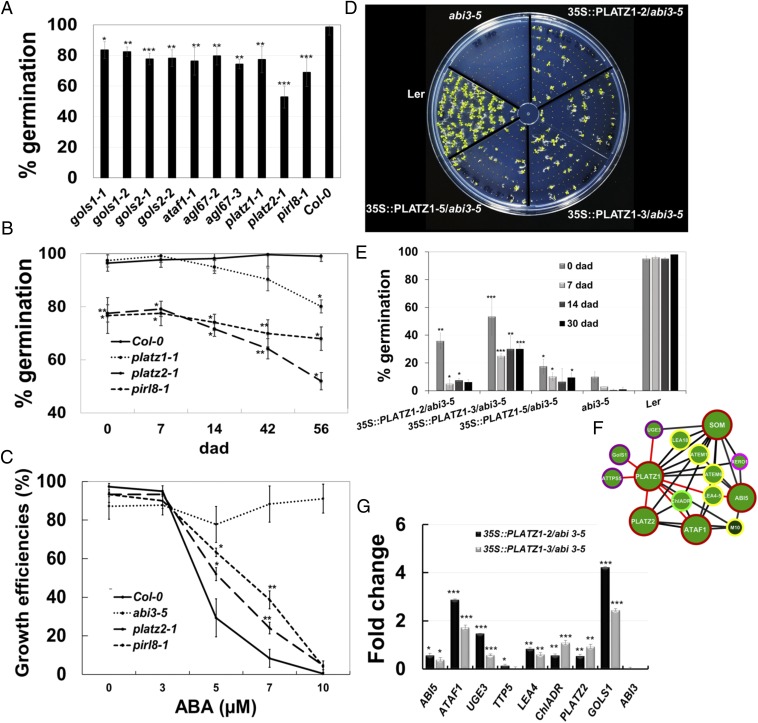
If the genes identified as major nodes in FullSeed-subNetDT2 indeed play roles in DT, it would be expected that the corresponding mutants should present some degree of desiccation intolerance. We therefore used T-DNA insertion mutants for the TFs PLATZ1, PLATZ2, AGL67, DREB2D, ERF23, and ATAF1 to determine whether they had a reduced germination percentage as a consequence of decreased DT. We observed that the germination percentage of dried ataf1, agl67, platz1, and platz2 seeds with respect to their WT was reduced to 76, 75, 77, and 53%, respectively (Fig. 4A). platz1-1 and platz2-1 showed a significantly reduced germination percentage when stored for 7 and 8 wk at 10% relative humidity (RH) (Fig. 4B), suggesting that PLATZ1 and PLATZ2 are important for DT in the dry seed stage.
Fig. 4.
Transcriptomic network inference reveals novel genes involved in desiccation tolerance. (A) TF node mutants with reduced DT. Germination percentage of seeds 6 d after sowing. Values are the means ± SD of four biological replicates; n = 100 seeds per replicate. (B) Germination percentage of Col-0, platz1-1, platz2-1, and pirl8-1 seeds stored for 0, 7, 14, 43, and 56 d after desiccation (dad). Values are the means ± SD of three biological replicates. (C) Effect of ABA treatment on seedling viability or growth efficiency (scored as the % of seedlings with green cotyledons in each treatment); note that the highest percentage is detected for abi3-5 (reported as ABA-insensitive) and the lowest is for the WT control. Seedlings were grown on Murashige and Skoog (MS) medium with 0, 3, 5, 7, and 10 µM ABA and their phenotypes were scored 15 d after sowing. Values are the means ± SD of three replicates; n = 50 per replicate. (D) PLATZ1 overexpression partially rescues the abi3-5 seed desiccation intolerance phenotype. Seeds were grown on MS medium, and survival was scored 10 d after sowing. (E) Germination percentage of WT (Ler), abi3-5, and 35S::PLATZ1/abi3-5 seeds stored for 0, 7, 14, and 30 d after desiccation. Values are the means ± SD of three biological replicates. (F) PLATZ1 interactor genes (FullSeed-subNetDT2) related to cellular protection at DPI 0.1. (G) Real-time PCR of ABI5, ATAF1, UGE3, TTP5, LEA4, ChIADR, PLATZ2, GOLS1 (neighboring genes of PLATZ1 in F represented with red edges), and ABI3 in 35S::PLATZ1/abi3-5 line seeds at 21 DAF; abi3-5 was used as the reference gene. UBQ10 and TIP4L served as internal controls. Data are the means ± SD of two biological replicates and three technical replicates. Values are means ± SD of three biological replicates. Bars with asterisks are significantly different from the control (Student's t test, *P < 0.05, **P < 0.01, ***P < 0.0001).
TFs identified as major nodes in our DT subnetworks should regulate the expression of target genes directly involved in DT, such as those involved in raffinose biosynthesis or encoding LEA proteins. To determine whether FullSeed-subNetDT2 effector genes play a role in DT, we determined the germination capacity of T-DNA insertion mutants for some of these effector genes (SI Appendix, Table S9). A mutant for the non-TF gene PIRL8 had decreased germination and ABA sensitivity (Fig. 4C). PIRL8 belongs to a plant-specific class of intracellular leucine-rich repeats that likely mediate protein interaction, possibly in the context of signal transduction (28). PIRL8 is a neighbor of SOM, DREB2G, and ANAC089, and these TFs are coexpressed with an important number of genes involved in cellular protection mechanisms (Fig. 3E). We also found that mutants for the GOLS1 and GOLS2 genes (which encode enzymes involved in raffinose biosynthesis) had a reduction in germination of 20 to 30% with respect to their wild-type control (Fig. 4A). These results suggest that target genes of the major nodes of FullSeed-subNetDT2 do indeed play important roles in DT. A lower germination efficiency in these mutants with respect to their WT could be due to the formation of defective embryos rather than to a defect in seed DT. Therefore, to determine whether the lower germination phenotype observed in TF and effector mutants is due to developmental defects during embryo formation or to a reduced DT, the morphology of green and dry seeds for each mutant was evaluated. We did not detect any embryo defects in any of the TF or effector gene mutants that were tested in this work (SI Appendix, Fig. S13).
Overexpression of PLATZ1, AGL67, or DREB2D Partially Rescues the Desiccation Intolerance Phenotype of abi3-5.
If the TFs identified as major nodes in FullSeed-subNetDT1 and FullSeed-subNetDT2 act downstream of ABI3, FUS3, and LEC1 and play important roles in activating effector genes involved in DT, then overexpression of some these TFs in a desiccation-intolerant background, for example abi3-5, should partially revert the desiccation intolerance phenotypes of these mutants. To test this, we expressed AGL67, DREB2D, and ERF23 from TFsSeed-subNetDT1 and PLATZ1, PLATZ2, and DREB2G from TFsSeed-subNetDT2 under the control of the 35S promoter in the abi3-5 background (Fig. 4 D and F and SI Appendix, Fig. S14). Dry seeds were collected and stored for 0, 1, 2, and 4 wk at 10% RH and then tested for germination efficiency. As previously reported (29) (Fig. 4E), abi3-5 seeds rapidly lose viability after desiccation and, after 1 wk of storage, germination was reduced to less than 5%. In contrast, seeds from the abi3-5/35S::PLATZ1 (Fig. 4 D and E), abi3-5/35S::AGL67 (SI Appendix, Fig. S14 A and B), and abi3-5/35S::DREB2G (SI Appendix, Fig. S14 C and D) lines showed a germination rate of 25, 30, and 12%, respectively, after 4 wk of storage. These results demonstrate that overexpression of some of the TFs identified as major nodes in TFsSeed-subNetDT1 and TFsSeed-subNetDT2 can partially rescue DT in abi3-5 seeds.
The capacity of PLATZ1, AGL67, and DREBG2 to partially complement the desiccation intolerance phenotype of abi3-5 seeds should, in principle, involve the activation of the effector target genes with which these transcription factors interact in TFsSeed-subNetDT1 and TFsSeed-subNetDT2. To confirm that the corresponding predicted target genes are indeed transcriptionally activated by these TFs, we evaluated the effect of the expression of the 35S::PLATZ1 gene construct (SI Appendix, Fig. S15) in the abi3-5 genetic background on the expression of some of the putative PLATZ1 target genes with a high MI value, such as LEA-related, LEA4 and XERO1; antioxidants, NRS; oligosaccharide synthesis, UGE3, GOLS1, and TTP5; oleosins, OLE3; and TFs, ABI5, PLATZ2, and ATAF1 (Fig. 4 F and G). Using two independent PLATZ1-overexpressing lines (SI Appendix, Fig. S15), we found that in most cases the putative target genes had a 1.5- to 4-fold higher expression in the 35::PLATZ1-overexpressing abi3-5 lines than in the abi3-5 control line. These results suggest that the expression of the genes predicted to be regulated by PLATZ1 is indeed directly or indirectly activated by this TF.
Constitutive Expression of PLATZ1 Confers Tolerance to Low Water Availability in Vegetative Tissues.
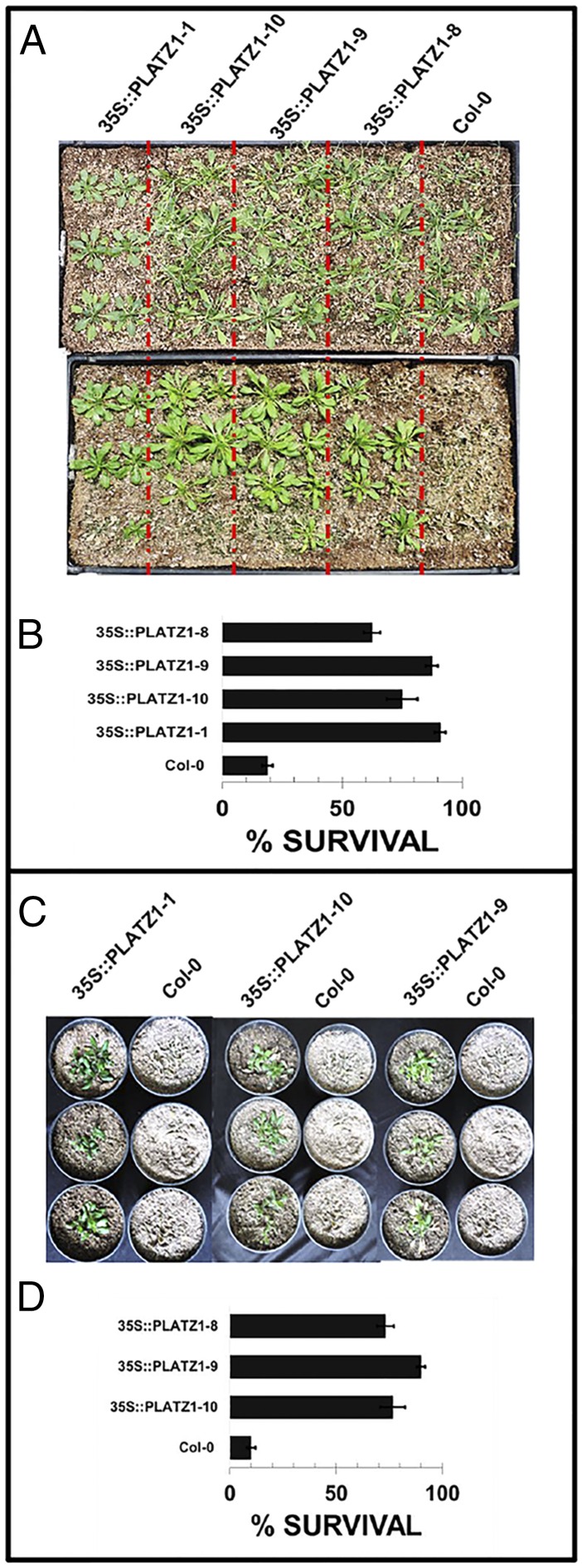
Our results suggest that PLATZ1 is capable of activating a subset of TFsSeed-subNetDT2 genes that appears to be important for the acquisition of DT in Arabidopsis seeds. To test whether PLATZ1 indeed plays a relevant role in DT, we introduced the 35S::PLATZ1 gene construct into the WT Arabidopsis Col-0 ecotype and tested whether its constitutive overexpression had any effect on the phenotype of vegetative tissue subjected to water stress. With this aim, seedlings from four independent 35S::PLATZ1 transgenic lines were grown under full irrigation in the greenhouse and subjected to a period of 7 d without irrigation, and then the number of surviving plants 4 d after a recovery irrigation treatment was scored. The four tested lines showed a 70 to 80% plant survival rate compared with 10% survival recorded for WT Col-0 (Fig. 5 A and B). Similar results were obtained in experiments carried out in growth chambers in which temperature and light conditions were controlled (Fig. 5 C and D). These results show that transgenic plants ectopically expressing PLATZ1 have an enhanced tolerance to low water availability compared with the WT. Although additional experiments are required to determine whether enhance tolerance to low water availability is due to the activation of protective mechanisms, a reduced cell metabolism, or a reduced growth rate, the finding that plants expressing PLATZ1 under the 35S promoter are capable of better surviving low water availability in the soil suggests that the DT regulatory subnetworks are relevant to seed DT, as activation of any of these mechanisms or their combinations could play an important role in allowing cells in the Arabidopsis embryo to enter into a low metabolic dormant state in which cells are protected against water stress and become more tolerant to desiccation.
Fig. 5.
Constitutive expression of PLATZ1 confers tolerance to low water availability in vegetative tissues. 35S::PLATZ1/Col-0-overexpressing lines display increased tolerance to low water availability under both greenhouse (A and B) and growth chamber (C and D) conditions. Plants were grown under greenhouse conditions for 20 d and then subjected to a low water availability treatment by withholding water irrigation for 15 d; after the drought treatment, plants were evaluated 5 d after being irrigated once to field capacity. (A, Upper) Transgenic and WT plants before the low water availability treatment. (A, Lower) 35S::PLATZ1 and control plants 5 d after the low water availability treatment (flowering stems were cut to show the rosette leaves). (B) Percentage of surviving plants after the low water availability treatment and the recovery irrigation. Values are the means ± SD of three biological replicates (n ≥ 18). (C) Independent pots with Arabidopsis plants grown under growth chamber conditions were subjected to a low water availability stress in which water was withheld for 15 d and then subjected to a recovery irrigation treatment; images of representative Col-0 and 35S::PLATZ1 plants 5 d after the recovery irrigation are shown (flowering stems were cut to show the rosette leaves). (D) Percentage of surviving plants after the low water availability treatment and the recovery irrigation. Values are means ± SD of three biological replicates (n ≥ 18). Growth chamber conditions were 22 °C with a 16-h photoperiod of 200 μmol⋅m−2⋅s−1.
Discussion
Desiccation tolerance has allowed seed plants to conquer ecosystems with long periods of limited water availability by providing a state in which the embryo is maintained as viable until favorable conditions for germination are encountered. This adaptive feature allows seeds to remain dried for hundreds or even thousands of years without losing their ability to germinate (30–32). Significant advances have been achieved in our understanding of the processes involved in seed maturation and their regulatory mechanisms in Arabidopsis (22) and Medicago (3, 4). However, the molecular networks that regulate the DT process remain largely unknown. Our results provide novel clues on the molecular networks acting downstream of the master genes LEC1, ABI3, FUS3, and LEC2 that modulate seed development, including DT, during seed maturation.
ABI3, FUS3, LEC1, and LEC2 have been shown to control most aspects of seed maturation and to repress the transition from embryo to vegetative development (13, 17, 18). Although some aspects of their function are specific, namely chlorophyll breakdown controlled by ABI3 (33) and production of somatic embryos controlled by LEC1 and LEC2 (12, 16), their role in seed maturation is globally similar. A genetic analysis of single and double mutants suggests that these genes integrate an important regulatory network with overlapping functions, which could be explained by synergism or redundancy (19). Interestingly, ectopic expression of LEC2, FUS3, or ABI3 in the single- or double-mutant backgrounds of the other two regulators was able to initiate seed lipid and storage protein accumulation but not DT, suggesting that all three regulators are required to activate the expression of TFs directly involved in DT (20). Our comparative transcriptomic analysis confirms that ABI3, FUS3, LEC1, and LEC2 repress the transition from embryo to vegetative development, because up-regulated genes in the corresponding null mutants were mainly associated with high metabolic activity, including genes involved in photosynthesis and anthocyanin synthesis that are probably responsible for the green and purple seed phenotypes observed in these mutants (SI Appendix, Table S8) as well as genes involved in leaf development and trichrome formation (SI Appendix, Table S8). These results are in agreement with the notion that ABI3, FUS3, LEC1, and LEC2 are key genes determining seed identity and that mutations in any of these genes transit directly from embryo to vegetative development.
Our comparative transcriptional global analysis from the interaction model allowed us to identify DT-specific differentially expressed genes independent of other genes whose altered expression contributes to the pleiotropic phenotype of lec1-1, lec2-1, fus3-3, and abi3-5 (SI Appendix, Tables S1 and S3). To remove the background related to the development transition, we used the comparison of lec1-1, fus3-3, and abi3-5, which have similar phenotypes, and lec2-1 and abi3-1 to identify specific genes of DT (SI Appendix, Table S1). Therefore, in our model, down-regulated genes represent specific genes that fail to be activated in desiccation-intolerant mutants, which should include TF genes that coordinate the establishment of DT and effector genes that are directly responsible for the different components that result in DT.
One of the most intriguing questions about how seeds in the desiccated state can remain viable for periods of time that can exceed centuries is particularly how the integrity of DNA is preserved to prevent permanent damage making the seed unviable. Three types of DNA damage under physiological conditions have been reported: hydrolysis of the N-glycosyl bond, hydrolytic deamination of cytosine to form uracil, and DNA damage by oxidation. The first two types of DNA damage are catalyzed by water, and therefore the last layers of water that interact with DNA need to be removed or decreased to prevent damage, and the third is mediated mainly by reactive oxygen species (ROS) (34, 35). In Arabidopsis, some oligosaccharides accumulate very late during seed development and have been proposed as possible key factors in the acquisition of DT, acting as protective agents via a water replacement mechanism (9). This hypothesis suggests that hydroxyl groups of these sugars substitute for water and provide the required hydrophilic interaction for membrane and protein stabilization, as well as DNA protection against hydrolytic damage. In agreement with the notion that oligosaccharides play an important role in preserving protein, membranes, and DNA from damage during seed desiccation and the dry seed stage, we found that among the DT-specific gene categories unveiled by our transcriptome analysis one of the most affected was oligosaccharide synthesis (mainly raffinose, stachyose, and trehalose synthesis). In transcriptome analysis, we also found a significant number of genes related to the control of cellular damage by free radicals. These ROS detoxification enzymes such as superoxide dismutases, catalases, ascorbate peroxidases, glutathione peroxidases, and glutathione reductase (SI Appendix, Table S8) could play an important role in the context of dry quiescent seeds in which reactive oxygen species are generated. Therefore, accumulation of antioxidant components during the late maturation stage contributes to controlling their storage potential, and helps to prevent damage from accumulated ROS during seed maturation. Accumulation of RFOs and activation of mechanisms that prevent damage by ROS allow maximum metabolism reduction to decrease the production of toxic compounds and to prevent membrane, DNA, RNA, and protein damage.
Some studies have proposed that the accumulation of oligosaccharides is not sufficient for the acquisition of DT in Arabidopsis seeds (29, 36). However, the fact that the level of oligosaccharides was found to be reduced in desiccation-intolerant mutants, but not in lec2 and the WT, suggests a clear correlation between DT and raffinose accumulation. Moreover, the reduction in germination displayed by gols1 and gols2 T-DNA insertion mutants provides direct evidence supporting the role of RFOs in the acquisition of DT.
Our comparative transcriptomic analysis of desiccation-tolerant and -intolerant mutants identified biological processes important for DT acquisition in seeds and coincides with findings from previous reports (3, 37). However, there is no information available on TFs that regulate the genetic networks involved in the acquisition of seed DT. In this work, we identified TF genes that are major nodes in the subnetworks related to DT. The importance of these TFs in the acquisition of seed DT was confirmed by the observation that seed viability was reduced in T-DNA insertion mutants of some of these TFs, such as PLATZ1, PLATZ2, AGL67, and ATAF1. Moreover, network prediction of the potential targets of the TFs identified as major regulatory nodes was confirmed by the observation that the expression of several of the putative targets of PLATZ1 was indeed up-regulated in PLATZ1-overexpressing lines. All these data support the notion that TFsSeed-subNetDT1 and TFsSeed-subNetDT2 downstream of LEC1, FUS3, and ABI3 are essential for the acquisition of DT in seeds. It remains to be determined whether the regulatory subnetworks that our data strongly suggest regulate, at least in part, DT in Arabidopsis seeds are or are not conserved in other angiosperms. In this regard, it is quite suggestive that several of the nodes in TFsSeed-subNetDT1 and TFsSeed-subNetDT2 have seed-specific expression in different plant species. For example, PLATZ1 orthologs are specifically expressed during seed maturation in rice, soybean, and maize. Interestingly, maize PLATZ1 is orthologous; in addition to being expressed during seed maturation, it is also strongly induced by drought stress (SI Appendix, Fig. S17).
A number of previously published reports further support our results on the prediction of regulatory subnetworks involved in seed DT: (i) Some of the TFs that are major nodes in these subnetworks seem to be directly activated by LEC1; for example, it has been shown that PEI1 and ERF23 are activated by ectopic expression of LEC1 (38); (ii) overexpression in Arabidopsis of ATAF1, one of the TFsSeed-subNetDT2 nodes with a high number of interactions with effector genes, was previously reported to enhance drought tolerance in Arabidopsis (39); (iii) SOM is a TF belonging to the CCCH-type zinc-finger family that has been reported to negatively regulate seed germination by activating ABA biosynthesis and inhibiting gibberellic acid biosynthesis (40); and (iv) although the precise functions of DREB2G and DREB2D are still unknown, they belong to a TF family that is generally involved in abiotic stress tolerance (41).
Because LEC1 and ABI3 are highly conserved from bryophytes to angiosperms (42), an important question is whether vegetative DT was originally controlled by these TFs, which were then recruited to control seed development, or whether the networks responsible for DT were controlled by other master regulators responsible for the activation of DT genes in an inducible manner in vegetative tissues. The study of basal plants that have vegetative DT would allow us to answer this question, which has significant importance for understanding the evolution of land plants. Moreover, vegetative DT is an ancestral feature of plants and has reevolved at least eight times in angiosperms, suggesting that a conserved set of regulatory genes has been recruited several times to activate vegetative desiccation during plant evolution. Thus, another interesting question is whether the regulatory subnetworks that control vegetative DT in basal plants are similar to those controlling DT in seeds. We propose that recruitment of the DT regulators that act downstream of LEC1 and ABI3 for expression in vegetative tissues might be responsible for the independent reevolution of vegetative DT during plant evolution (SI Appendix, Fig. S20). As a first attempt to answer this question, we performed a phylogenetic analysis of the conservation of the key nodes of the TFsSeed-subNetDT1 and TFsSeed-subNetDT2 networks. We found that PLATZ1, PLATZ2, AGL67, DREB2D, and DREB2G are conserved in the bryophyte Physcomitrella patents, vascular DT basal plants such as Selaginella moellendorffii, the basal angiosperm Amborella trichopoda, the monocotyledonous species Oryza sativa (rice) and Zea mays (corn), the DT plant Oropetium thomaeum, and the dicotyledonous species Glycine max (soybean) and Solanum lycopersicum (tomato) (SI Appendix, Figs. S16–S19).
This opens up the possibility that a core DT regulatory network could indeed have been conserved throughout plant evolution. Further research will be needed to explore whether DT is orchestrated by regulatory networks in which at least a common core of TFs has been conserved during plant evolution and to determine how it has been rewired several times to be activated in seeds and in vegetative tissues.
Materials and Methods
Plants were grown in a sterile mix of vermiculite and soil in a growth chamber at 22 °C with a 16-h photoperiod at 200 μmol⋅m−2⋅s−1. For RNA-seq and carbohydrate profiles, flowers were marked and, at specific times after flowering (15, 17, and 21 DAF), siliques were harvested from 24 plants and seeds were collected and immediately frozen in liquid nitrogen and stored at −80 °C.
Statistical Analyses.
Gene counts were normalized using edgeR’s (version 2.9.16) (4) TMM [trimmed mean M (1/4 log fold-change gene expression)] algorithm. For the analysis of differentially expressed genes, only genes with at least five reads across all samples were included. For tolerance designation, we tested for differential expression of genes using the multifactor generalized linear model (glms) approach in edgeR.
Categorization and Functional Analysis.
GO annotation analysis on gene clusters was performed using the BiNGO 2.3 plugin tool in Cytoscape version 2.6 with GO_full and GO_slim categories (www.cytoscape.org).
Gene Expression Analysis.
Gene-specific primer pairs (SI Appendix, Table S10) designed using the National Center for Biotechnology Information Primer-BLAST tool (10) were used for real-time PCR. A total of 10 μg of RNA was reverse-transcribed using SuperScript III Reverse Transcriptase (Life Technologies) according to the manufacturer’s instructions.
A more detailed description of all material and methods is provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank G. Corona-Armenta for support with the transcriptome experiments. We also thank M. J. Ortega-Estrada for support with the quantitative RT-PCR experiments and J. I. Cervantes-Luevano for support with bioinformatics tools. S.I.G.-M. is indebted to CONACyT (México) for a PhD fellowship. This research was supported by Consejo Nacional de Ciencia y Tecnología (México) Grant Fronteras 137 and Howard Hughes Medical Institute Grant 4367 (to L.H.-E.).
Footnotes
The authors declare no conflict of interest.
Data deposition: All the data presented in this paper, including the raw data of RNA-sequencing experiments, have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE76015).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610985113/-/DCSupplemental.
References
- 1.Alpert P, Oliver MJ. 2002. Drying without dying. Desiccation and Survival in Plants: Drying Without Dying, eds Black M, Pritchard HW (CABI, Wallingford, Oxfordshire, UK), pp 3–46.
- 2.Mishler BD, Churchill SP. Transition to a land flora: Phylogenetic relationships of the green algae and bryophytes. Cladistics. 1985;1(4):305–328. doi: 10.1111/j.1096-0031.1985.tb00431.x. [DOI] [PubMed] [Google Scholar]
- 3.Verdier J, et al. A regulatory network-based approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiol. 2013;163(2):757–774. doi: 10.1104/pp.113.222380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Righetti K, et al. Inference of longevity-related genes from a robust coexpression network of seed maturation identifies regulators linking seed storability to biotic defense-related pathways. Plant Cell. 2015;27(10):2692–2708. doi: 10.1105/tpc.15.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manfre AJ, LaHatte GA, Climer CR, Marcotte WR., Jr Seed dehydration and the establishment of desiccation tolerance during seed maturation is altered in the Arabidopsis thaliana mutant atem6-1. Plant Cell Physiol. 2009;50(2):243–253. doi: 10.1093/pcp/pcn185. [DOI] [PubMed] [Google Scholar]
- 6.Delahaie J, et al. LEA polypeptide profiling of recalcitrant and orthodox legume seeds reveals ABI3-regulated LEA protein abundance linked to desiccation tolerance. J Exp Bot. 2013;64(14):4559–4573. doi: 10.1093/jxb/ert274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wehmeyer N, Vierling E. The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol. 2000;122(4):1099–1108. doi: 10.1104/pp.122.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailly C. Active oxygen species and antioxidants in seed biology. Seed Sci Res. 2004;14(2):93–107. [Google Scholar]
- 9.Baud S, Boutin J, Miquel M, Lepiniec L, Rochat C. An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem. 2002;40(2):151–160. [Google Scholar]
- 10.Mène-Saffrané L, Jones AD, DellaPenna D. Plastochromanol-8 and tocopherols are essential lipid-soluble antioxidants during seed desiccation and quiescence in Arabidopsis. Proc Natl Acad Sci USA. 2010;107(41):17815–17820. doi: 10.1073/pnas.1006971107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M, et al. The effect of transparent TESTA2 on seed fatty acid biosynthesis and tolerance to environmental stresses during young seedling establishment in Arabidopsis. Plant Physiol. 2012;160(2):1023–1036. doi: 10.1104/pp.112.202945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lotan T, et al. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93(7):1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 13.Nambara E, Naito S, McCourt P. A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele. Plant J. 1992;2(4):435–441. [Google Scholar]
- 14.Giraudat J, et al. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell. 1992;4(10):1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luerssen H, Kirik V, Herrmann P, Miséra S. FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 1998;15(6):755–764. doi: 10.1046/j.1365-313x.1998.00259.x. [DOI] [PubMed] [Google Scholar]
- 16.Stone SL, et al. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA. 2001;98(20):11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keith K, Kraml M, Dengler NG, McCourt P. fusca3: A heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell. 1994;6(5):589–600. doi: 10.1105/tpc.6.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meinke DW, Franzmann LH, Nickle TC, Yeung EC. Leafy cotyledon mutants of Arabidopsis. Plant Cell. 1994;6(8):1049–1064. doi: 10.1105/tpc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.To A, et al. A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell. 2006;18(7):1642–1651. doi: 10.1105/tpc.105.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roscoe TT, Guilleminot J, Bessoule JJ, Berger F, Devic M. Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in Arabidopsis. Plant Cell Physiol. 2015;56(6):1215–1228. doi: 10.1093/pcp/pcv049. [DOI] [PubMed] [Google Scholar]
- 21.Gaff DF, Oliver M. The evolution of desiccation tolerance in angiosperm plants: A rare yet common phenomenon. Funct Plant Biol. 2013;40(4):315–328. doi: 10.1071/FP12321. [DOI] [PubMed] [Google Scholar]
- 22.Fait A, et al. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 2006;142(3):839–854. doi: 10.1104/pp.106.086694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thimm O, et al. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37(6):914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 24.Crowe LM. Lessons from nature: The role of sugars in anhydrobiosis. Comp Biochem Physiol A Mol Integr Physiol. 2002;131(3):505–513. doi: 10.1016/s1095-6433(01)00503-7. [DOI] [PubMed] [Google Scholar]
- 25.Obendorf RL. Oligosaccharides and galactosyl cyclitols in seed desiccation tolerance. Seed Sci Res. 1997;7(2):63–74. [Google Scholar]
- 26.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37(4):382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 27.Chávez Montes RA, et al. ARACNe-based inference, using curated microarray data, of Arabidopsis thaliana root transcriptional regulatory networks. BMC Plant Biol. 2014;14:97. doi: 10.1186/1471-2229-14-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forsthoefel NR, Cutler K, Port MD, Yamamoto T, Vernon DM. PIRLs: A novel class of plant intracellular leucine-rich repeat proteins. Plant Cell Physiol. 2005;46(6):913–922. doi: 10.1093/pcp/pci097. [DOI] [PubMed] [Google Scholar]
- 29.Ooms J, Leon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM. Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana (a comparative study using abscisic acid-insensitive abi3 mutants) Plant Physiol. 1993;102(4):1185–1191. doi: 10.1104/pp.102.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver MJ, Tuba Z, Mishler BD. The evolution of vegetative desiccation tolerance in land plants. Plant Ecol. 2000;151(1):85–100. [Google Scholar]
- 31.Oliver MJ, Velten J, Mishler BD. Desiccation tolerance in bryophytes: A reflection of the primitive strategy for plant survival in dehydrating habitats? Integr Comp Biol. 2005;45(5):788–799. doi: 10.1093/icb/45.5.788. [DOI] [PubMed] [Google Scholar]
- 32.Illing N, Denby KJ, Collett H, Shen A, Farrant JM. The signature of seeds in resurrection plants: A molecular and physiological comparison of desiccation tolerance in seeds and vegetative tissues. Integr Comp Biol. 2005;45(5):771–787. doi: 10.1093/icb/45.5.771. [DOI] [PubMed] [Google Scholar]
- 33.Delmas F, et al. ABI3 controls embryo degreening through Mendel’s I locus. Proc Natl Acad Sci USA. 2013;110(40):E3888–E3894. doi: 10.1073/pnas.1308114110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kranner I, Birtic S. A modulating role for antioxidants in desiccation tolerance. Integr Comp Biol. 2005;45(5):734–740. doi: 10.1093/icb/45.5.734. [DOI] [PubMed] [Google Scholar]
- 35.Rajjou L, Debeaujon I. Seed longevity: Survival and maintenance of high germination ability of dry seeds. C R Biol. 2008;331(10):796–805. doi: 10.1016/j.crvi.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 36.Bentsink L, et al. Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol. 2000;124(4):1595–1604. doi: 10.1104/pp.124.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angelovici R, Galili G, Fernie AR, Fait A. Seed desiccation: A bridge between maturation and germination. Trends Plant Sci. 2010;15(4):211–218. doi: 10.1016/j.tplants.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Mu J, et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008;148(2):1042–1054. doi: 10.1104/pp.108.126342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y, et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009;19(11):1279–1290. doi: 10.1038/cr.2009.108. [DOI] [PubMed] [Google Scholar]
- 40.Kim DH, et al. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell. 2008;20(5):1260–1277. doi: 10.1105/tpc.108.058859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu H, Xiong L. Genetic engineering and breeding of drought-resistant crops. Annu Rev Plant Biol. 2014;65:715–741. doi: 10.1146/annurev-arplant-050213-040000. [DOI] [PubMed] [Google Scholar]
- 42.Cagliari A, et al. New insights on the evolution of Leafy Cotyledon1 (LEC1) type genes in vascular plants. Genomics. 2014;103(5–6):380–387. doi: 10.1016/j.ygeno.2014.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.