Abstract
Immunoaffinity enrichment of peptides coupled to multiple reaction monitoring-mass spectrometry (immuno-MRM) enables highly specific, sensitive, and precise quantification of peptides and post-translational modifications. Major obstacles to developing a large number of immuno-MRM assays are the poor availability of monoclonal antibodies (mAbs) validated for immunoaffinity enrichment of peptides and the cost and lead time of developing the antibodies de novo. Although many thousands of mAbs are commercially offered, few have been tested for application to immunoaffinity enrichment of peptides. In this study we tested the success rate of using commercially available mAbs for peptide immuno-MRM assays. We selected 105 commercial mAbs (76 targeting non-modified “pan” epitopes, 29 targeting phosphorylation) to proteins associated with the DNA damage response network. We found that 8 of the 76 pan (11%) and 5 of the 29 phospho-specific mAbs (17%) captured tryptic peptides (detected by LC-MS/MS) of their protein targets from human cell lysates. Seven of these mAbs were successfully used to configure and analytically characterize immuno-MRM assays. By applying selection criteria upfront, the results indicate that a screening success rate of up to 24% is possible, establishing the feasibility of screening a large number of catalog antibodies to provide readily-available assay reagents.
Keywords: antibody validation, DNA damage response, immuno-MRM, phosphorylation, protein quantification
The incorporation of an immunoaffinity enrichment step with mass spectrometry can produce highly reliable quantitative assays [1–4]. Major obstacles to developing a large number of peptide immuno-MRM assays are the poor availability of monoclonal antibodies (mAbs) validated for immunoaffinity enrichment of proteotypic peptides and the cost and lead time of developing the antibodies de novo. mAbs are especially attractive for use in immuno-MRM assays, since they are a renewable reagent that can be standardized [5]. Although there are thousands of commercially available mAbs (www.antibodypedia.com), few have been characterized for their performance in immuno-MRM assays. In a prior study, we demonstrated a high crossover success rate for using mAbs developed for peptide immuno-MRM in conventional protein detection platforms (e.g. Western blotting) [5]. In this study, we examine the converse by determining the success rate for using mAbs commercially developed for conventional protein detection platforms (e.g. Western blotting) in immuno-MRM assays. If successful, this approach has the potential to enable the development of a wide array of assays using existing antibodies, improving biological research by providing reproducible, multiplex, and highly specific quantification of proteins and post-translational modifications in a manner that can be standardized across laboratories [6–8].
For this study, we identified a panel of 105 commercially available mAbs for testing, based on two selection criteria: i) protein targets are involved in the cellular DNA damage response (DDR) network (as proof-of-concept), and ii) protein targets have been empirically observed in LC-MS/MS experiments (determined by searching publicly available mass spectrometry datasets [9, 10]). All mAbs were generated against peptide immunogens; however, we did not know the specific antigen sequences, and the antibody epitopes were not mapped. Two types of mAbs were chosen (Supporting Information Table 1): 76 that recognize non-modified epitopes (i.e. pan mAbs) and 29 that recognize post-translationally modified targets (i.e. phosphorylation mAbs).
Each mAb was tested for its ability to enrich peptides from trypsin-digested protein lysates from human cell lines (Figure 1A–B). MCF10A, HeLa, Hep G2, and HEK293 cell lines were selected based on confirmed expression data for the targeted proteins in the antibody vendor catalog (see Supporting Information). Whole cell lysates were used for enrichment. Pan antibodies (that had been coupled to Protein G magnetic beads) were applied to untreated cells, whereas antibodies for the modified targets (also bead-coupled) were applied to cells harvested two hours following exposure to 10 Gy ionizing radiation (to elicit a DNA damage phospho-signaling response). As a negative control, a “beads only” capture was performed (i.e., not containing antibodies). All immunoaffinity-enriched samples were analyzed by LC-MS/MS, and the data were deposited to the ProteomeXchange Consortium (see Supporting Information for details).
Figure 1. Evaluation and characterization of multiplex immuno-MRM assays using commercial mAbs.
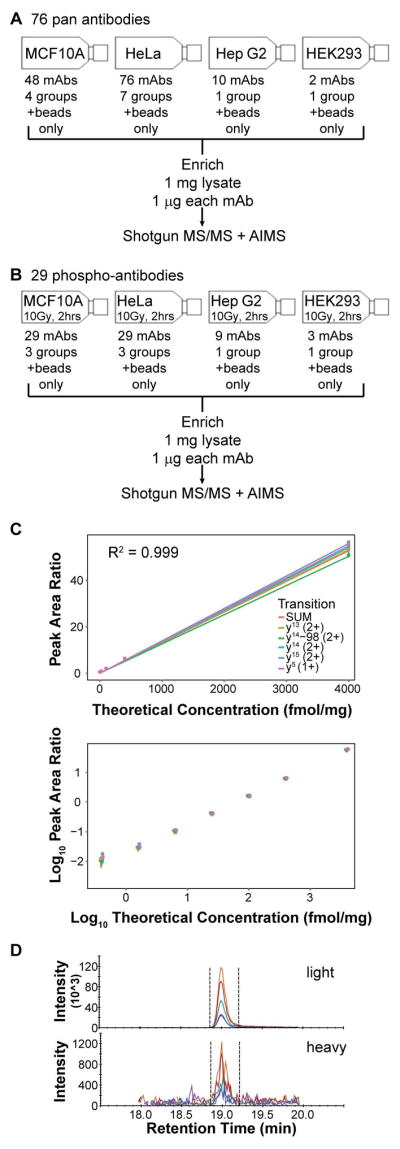
(A) Overview of evaluation of 76 pan antibodies, and (B) evaluation of 29 anti-phospho antibodies. (C) Response curve for TTTPGPSLS[PO4]QGVSVDEK (pS343) to Nibrin (NBN), plotted in linear and log space. Error bars indicate the range of the peak area ratios of the three capture and LC-MRM-MS replicates. (D) Chromatogram for light and heavy peptides for TTTPGPSLS[PO4]QGVSVDEK (pS343) to Nibrin (NBN) in 0.5 mg MCF10A + HeLa 10Gy IR lysate. Heavy peptide is added at a concentration of 0.4 fmol/mg. AIMS, accurate inclusion mass screening.
A total of 4884 unique peptides corresponding to 1703 proteins were identified across all immunoaffinity enrichment experiments. Overall, 27 targets of the mAbs were identified; however, a large number of identifications were due to non-specific binding to the antibodies, Protein-G, or the magnetic beads. To evaluate the specificity of the pull-downs, we plotted a heatmap for the number of identifications in each pull-down and a histogram of the fraction of total identifications across the pull-downs (Supporting Information Figures 1 and 2, respectively). Several filters were applied to the dataset to classify confidently identified peptides associated with the antibody’s reported specificity. First, using a strategy applied in identifying proteins ubiquitous in immunoprecipitation (IP) experiments (i.e. the CRAPome) [11, 12], we eliminated peptides identified in greater than 8% (n=5) of total capture experiments and any peptide identified in the bead-only control capture. Next, we required that the identified peptide was present in a capture in which the corresponding target antibody was applied, and that matching peptides were proteotypic (i.e. unique to the protein of interest). We also eliminated all identifications for which only 1 spectral match was observed. Using these conservative criteria, peptides corresponding to 8 of the 76 pan mAbs (11%) and to 5 of the 29 phosphorylation mAbs (17%) were confidently detected in the LC-MS/MS data (see Supporting Information Table 2). Success rates were higher for antibodies that were previously validated for IP of the full length protein (i.e., amongst the 105 mAbs, 40 were previously validated for protein IP, and 8 (20%) of these were amongst the 13 successful mAbs; conversely, 65 mAbs were not previously validated for protein IP, and only 5 (8%) of these were amongst the 13 successful mAbs).
Sufficient antibody was available for quantitative assay development for 7 of the mAbs showing activity for immunopeptide enrichment (Table 1). For these targets, stable isotope-labeled peptide standards were used to develop MRM methods (see Supporting Information Table 3 for transitions selected). The antibodies were configured into one multiplex immuno-MRM group, and were characterized by response curves and the ability to detect endogenous analyte from a digest of whole cell lysate. For phosphorylated targets, we attempted to develop assays to the modified and non-modified versions of the peptides, as previously demonstrated [13].
Table 1. Immuno-MRM assay performance and characterization data.
The amount of mAb used per capture depended on the mAb amount received from the vendor. “Endo” refers to detection of endogenous analyte. The cost of mAb per microgram was based on advertised price and amount delivered. [PO4] = phosphorylation on the preceding amino acid; LOD, limit of detection; LLOQ, lower limit of quantification; ULOQ, upper limit of quantification.
| Catalog # (Abcam) |
Gene symbol (phos- site) |
Protein Description |
Peptide Sequence | mAb amt./ capture (μg) |
Endo | LOD (fmol / mg) |
LLOQ (fmol / mg) |
ULOQ (fmol / mg) |
%CV at LLOQ |
mAb cost per μg |
|---|---|---|---|---|---|---|---|---|---|---|
| ab92312 | MLH1 | DNA mismatch repair protein Mlh1 | LDETVVNR | 1.0 | No | $ 7.13 | ||||
| ab92471 | MSH6 | DNA mismatch repair protein Msh6 | QSTLYSFFPK | 0.2 | Yes | 0.02 | 0.4 | ≥4,000 | 1.9 | $ 50.13 |
| ab109214 | UNG | Uracil-DNA glycosylase | APAGQEEPGTPPSSPLSAEQLDR | 1.0 | Yes | 0.2 | 0.4 | ≥4,000 | 9.1 | $ 12.83 |
| ab81292 | ATM (S1981) | Serine-protein kinase ATM | SLAFEEGS[PO4]QSTTISSLSEK SLAFEEGSQSTTISSLSEK |
1.2 | Yes No |
13.9 | 25 | ≥4,000 | 3.9 | $ 20.20 |
| ab109453 | NBN (S343) | Nibrin | TTTPGPSLS[PO4]QGVSVDEK TTTPGPSLSQGVSVDEK |
1.0 | Yes No |
0.2 | 0.4 | ≥4,000 | 16.2 | $ 5.19 |
| ab133441 | PRKDC (S2612) | DNA-dependent protein kinase catalytic subunit | STVLTPMFVETQAS[PO4]QGTLQTR STVLTPMFVETQASQGTLQTR |
1.0 | Yes No |
2.4 | 6.3 | 400 | 7.5 | $ 9.21 |
| ab32385 | JUN (S63) | Transcription factor AP-1 | NSDLLTS[PO4]PDVGLLK NSDLLTSPDVGLLK |
0.8 | Yes Yes |
0.6 1.0 |
1.6 1.6 |
≥4,000 ≥4,000 |
12.1 13.9 |
$ 38.50 |
Response curves were used to determine the limits of detection (LODs), lower and upper limits of quantification (LLOQs and ULOQs), precision, and linearity of the multiplexed assay. Each concentration point was measured in technical triplicate (including replicates of the immunoaffinity enrichment and mass spectrometry steps). Endogenous peptide analyte and a linear response were observed for 6 of the 7 antibodies (including 7 peptides, since one of the antibodies targeting pJUNSer63 was capable of enriching the phosphorylated and non-modified peptides). The failure of MLH1 (LDETVVNR) could be due to the matrix used, as it was detected in non-irradiated lysates in the shotgun studies described above. An example response curve is shown in Figure 1C, and figures of merit are reported in Table 1 (response curves for the 7 assays are shown in Supporting Information Figure 3, and the raw data are given as Supporting Information in a Skyline document). The LLOQs ranged from 0.4 to 25 fmol/mg digest, with a median of 1.6 fmol/mg. An example chromatogram showing detection of the NBNpS343 peptide at the LLOQ is shown in Figure 1D. The %CVs at the LLOQs ranged from 1.9 to 16.2%, with a median of 9.1%. The assays were linear for ≥3 orders of magnitude for 5 of the 7 assays. (One assay with less than 3 orders was still linear at the highest concentration point, thus underestimating the true linear range.) These figures of merit are comparable to those seen previously using monoclonal antibodies specifically generated for immuno-MRM [5].
The results of this study demonstrate the feasibility of using commercially available mAbs in quantitative peptide immuno-MRM assays. Based on the results, we estimate an overall success rate in developing working assays of ~14% (combining success rates in detection and qualification of assays in MRM). This success rate is lower than a previous investigation using polyclonal antibodies generated against protein fragments for peptide immunoaffinity enrichment [14], possibly since use of polyclonals can increase the likelihood of a working antibody by enriching multiple peptides (i.e. multiple epitopes) for each protein target. Limitations of polyclonal antibodies (e.g., limited resource, batch-to-batch variation) make the use of mAbs more attractive for a distributable and standardizable assay reagent.
Several parameters could be taken into account to improve the likelihood of developing a successful assay. First, considering known epitopes of commercial antibodies will improve screening results by allowing selection of targets for which the peptide is amenable to LC-MS analysis (for example, targeting epitope-containing tryptic peptides that are neither too short nor too long, nor disrupted by a cleavage site). Parameters for suitable MRM-able peptides have been published [15]. Likewise, antibodies for phosphorylated targets could be considered for only phosphopeptides that yield an analyzable tryptic peptide. For example, 8 phosphosites in our study were contained within tryptic phosphopeptides that are not ideal for mass spectrometry (peptide length > 35). Removing these targets from the study increases the success rate of screening anti-phosphopeptide antibodies to 5/21 (24%). For some of these difficult targets, alternative peptides may be produced through digestion with enzymes other than trypsin. Finally, the higher success rate for antibodies previously characterized by protein IP indicates that these characterization data are useful in identifying antibodies most likely to work in peptide immuno-MRM. Publicly available antibody databases and resources, such as antibodypedia (antibodypedia.com), 1degreebio (1degreebio.org), CiteAb (citeab.com), Human Protein Atlas [16], or the National Cancer Institute’s Antibody Portal [5] (antibodies.cancer.gov), could be searched for additional validation methods, epitope information, and mAb uses, which would increase the chances of a mAb working in immuno-MRM assays.
The availability of commercial antibodies is a definite advantage in terms of time savings for assay development. However, the cost benefits are short-term given the current formulations of antibodies. For example, Table 1 shows the per sample cost of the antibodies as formulated and used in this study, ranging from $5 to over $50 per sample (by comparison, the cost of using an existing custom-made monoclonal is $0.50 per sample (assuming $1000–$3000 for production and purification of 6 mg) [17]). Even adding in the considerable costs associated with de novo generation of a custom monoclonal antibody, assays based on the custom antibody would be $2.33 – $3.50 per sample (assuming $14,000–$21,000 for antibody generation and production of 6 mg). Thus, it may be beneficial to use commercially-available antibodies for limited studies requiring small numbers of assays, in order to generate data supporting development of a less expensive antibody source for long-term or larger studies.
Regardless, the large number of commercially-available affinity reagents combined with the relatively good success rate for working antibodies makes a broad screening effort worthwhile. The potential for rapidly developing quantitative, multiplexed immuno-MRM assays to a wide array of targets has the potential to greatly expand the assay content and contribute immensely to quantitative biological studies.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number U24CA160034. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We acknowledge the helpful contributions of Xiuwen Liu and Meng Yun Chou in making available antibody reagents.
Abbreviations
- CV
coefficient of variation
- DDR
DNA damage response
- immuno-MRM
immunoaffinity enrichment of peptides coupled to multiple reaction monitoring-mass spectrometry
- IP
immunoprecipitation
- IR
ionizing radiation
- LC
liquid chromatography
- LLOQ
lower limit of quantification
- LOD
limit of detection
- mAb
monoclonal antibody
- ULOQ
upper limit of quantification
Footnotes
The authors have declared no conflict of interest.
References
- 1.Ackermann BL, Berna MJ. Coupling immunoaffinity techniques with MS for quantitative analysis of low-abundance protein biomarkers. Expert Rev Proteomics. 2007;4:175–186. doi: 10.1586/14789450.4.2.175. [DOI] [PubMed] [Google Scholar]
- 2.Madian AG, Rochelle NS, Regnier FE. Mass-linked immuno-selective assays in targeted proteomics. Anal Chem. 2013;85:737–748. doi: 10.1021/ac302071k. [DOI] [PubMed] [Google Scholar]
- 3.Bostrom T, Takanen JO, Hober S. Antibodies as means for selective mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2015 doi: 10.1016/j.jchromb.2015.10.042. [DOI] [PubMed] [Google Scholar]
- 4.Whiteaker JR, Zhao L, Abbatiello SE, Burgess M, et al. Evaluation of large scale quantitative proteomic assay development using peptide affinity-based mass spectrometry. Molecular & cellular proteomics. 2011;10:M110 005645. doi: 10.1074/mcp.M110.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoenherr RM, Saul RG, Whiteaker JR, Yan P, et al. Anti-peptide monoclonal antibodies generated for immuno-multiple reaction monitoring-mass spectrometry assays have a high probability of supporting Western blot and ELISA. Molecular & cellular proteomics. 2015;14:382–398. doi: 10.1074/mcp.O114.043133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kennedy JJ, Abbatiello SE, Kim K, Yan P, et al. Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Nat Methods. 2014;11:149–155. doi: 10.1038/nmeth.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn E, Whiteaker JR, Mani DR, Jackson AM, et al. Interlaboratory evaluation of automated, multiplexed peptide immunoaffinity enrichment coupled to multiple reaction monitoring mass spectrometry for quantifying proteins in plasma. Molecular & cellular proteomics. 2012;11:M111 013854. doi: 10.1074/mcp.M111.013854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Addona TA, Abbatiello SE, Schilling B, Skates SJ, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mertins P, Yang F, Liu T, Mani DR, et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Molecular & cellular proteomics. 2014;13:1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcon E, Jain H, Bhattacharya A, Guo H, et al. Assessment of a method to characterize antibody selectivity and specificity for use in immunoprecipitation. Nat Methods. 2015;12:725–731. doi: 10.1038/nmeth.3472. [DOI] [PubMed] [Google Scholar]
- 13.Whiteaker JR, Zhao L, Yan P, Ivey RG, et al. Peptide Immunoaffinity Enrichment and Targeted Mass Spectrometry Enables Multiplex, Quantitative Pharmacodynamic Studies of Phospho-Signaling. Molecular & cellular proteomics. 2015;14:2261–2273. doi: 10.1074/mcp.O115.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edfors F, Bostrom T, Forsstrom B, Zeiler M, et al. Immunoproteomics using polyclonal antibodies and stable isotope-labeled affinity-purified recombinant proteins. Molecular & cellular proteomics. 2014;13:1611–1624. doi: 10.1074/mcp.M113.034140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoofnagle AN, Whiteaker JR, Carr SA, Kuhn E, et al. Recommendations for the Generation, Quantification, Storage, and Handling of Peptides Used for Mass Spectrometry-Based Assays. Clin Chem. 2016;62:48–69. doi: 10.1373/clinchem.2015.250563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas--a tool for pathology. J Pathol. 2008;216:387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 17.Zhao L, Whiteaker JR, Voytovich UJ, Ivey RG, Paulovich AG. Antibody-Coupled Magnetic Beads Can Be Reused in Immuno-MRM Assays To Reduce Cost and Extend Antibody Supply. J Proteome Res. 2015;14:4425–4431. doi: 10.1021/acs.jproteome.5b00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


