Abstract
Identification of causative factors for common, chronic disorders is a major focus of current human health science research. These disorders are likely to be caused by multiple etiological agents. Available evidence also suggests that interactions between the risk factors may explain some of their pathogenic effects. While progress in genomics and allied biological research has brought forth powerful analytic techniques, the predicted complexity poses daunting analytic challenges. The search for pathogenesis of schizophrenia shares most of these challenges. We have reviewed the analytic and logistic problems associated with the search for pathogenesis. Evidence for pathogenic interactions is presented for selected diseases and for schizophrenia. We end by suggesting ‘recursive analyses’ as a potential design to address these challenges. This scheme involves initial focused searches for interactions motivated by available evidence, typically involving identified individual risk factors, such as candidate gene variants. Putative interactions are tested rigorously for replication and for biological plausibility. Support for the interactions from statistical and functional analyses motivates a progressively larger array of interactants that are evaluated recursively. The risk explained by the interactions is assessed concurrently and further elaborate searches may be guided by the results of such analyses. By way of example, we summarize our ongoing analyses of dopaminergic polymorphisms, as well as infectious etiological factors in schizophrenia genesis to exemplify this approach.
Keywords: Schizophrenia, interactions, etiology, epistasis, gene-environment, gene-gene
1. Introduction
Gene mapping research has illuminated the causes of rare monogenic conditions in humans [1] [2]. The challenge now is the search for prevalent chronic diseases such as cardiovascular diseases and obesity. The availability of the complete human genome sequence, its ongoing annotation, the public availability of DNA polymorphism data and the successful implementation of rapid, highly accurate and economical genotyping assays have all contributed to an explosion in our understanding of genetically complex traits and common, multi-factorial diseases. Nevertheless, daunting challenges remain.
The principal challenge is the evident complexity of the diseases and disorders of interest. Though many show familial segregation, simple Mendelian models appear insufficient to explain their inheritance. Though Mendelian laws can explain the inheritance of discrete traits elegantly, it has been debated whether they are applicable to quantitative variation in traits, as well as so called complexly inherited traits. Fisher [3] resolved this dilemma by suggesting that the correlation between related individuals for a quantitative trait could be explained by the cumulative, small-yet-discrete effects at a large number of genetic variants (loci). The resultant trait or phenotype imparted might be dichotomized by imposing an arbitrary threshold. This has evolved into a multi-factorial / polygenic threshold (MFPT) model for causation [4] [5]. The MFPT model proposes the presence of individual genetic risk factors of variable effect that may act discretely or interactively; environmental factors increase the variability of their expression. While the MFPT model has great explanatory power, defining the individual components and setting a threshold for a phenotype of interest can be difficult.
In the following sections, we initially review the analytic and logistical challenges in the search for etiology. Next, we provide selected examples of identified interactions in non-psychiatric disorders. The evidence necessarily has to involve not only statistical evidence such as gene mapping studies, but also functional evidence from biological models. A summary of challenges posed by schizophrenia (SZ) follows. Published examples of interactions identified in SZ are then discussed. We end by arguing for an approach that we call ‘recursive analysis’ and provide examples from our ongoing work.
2. Analytic and logistical challenges in the search for etiology
If more than one etiological factor appears to cause a disease, two key questions arise in relation to pathogenesis. Foremost, do such factors act in isolation? If not, do they interact and in what manner? As the risk conferred by individual risk factors for common, etiologically complex disorders generally appears to be modest (odds ratios ~ 1.1–2.0), it seems unlikely that such factors act in isolation. Therefore, interactions between risk factors envisaged in the MFPT model seem plausible. Such interactions can be conceptualized in the context of interactions between genetic risk factors (also called epistasis) and / or interactions between genetic and non-genetic or environmental risk factors (referred to here as G/E interactions). Further, the interactions need not be restricted to pairs of factors. Higher order interactions between numerous risk factors are likely.
The biological underpinnings for such interactions may be diverse and could occur at different levels. Thus, the interactions may reflect the impact of several risk variants within the same gene (e.g., variants in exons that impact the protein product may act in conjunction with variants that alter transcription), or they may occur across different genes (e.g., see chapter by Chang-Gyu Han and colleagues in this issue). The impact of such interactions may be reflected as alterations in activities of different cell types, by effects on different metabolic pathways or across different regions in the brain for neuropsychiatric disorders. Environmental factors may directly impact such processes or lead to epigenetic changes [6].
Regardless of the biological mechanisms, it is reasonable to assume that the interactions would be demonstrable statistically in appropriate samples of adequate size. Indeed, the statistical evidence is typically the starting point for delineation of the biological interactions. However, proving the interactions statistically using an agnostic approach is dogged by false positive (type I) and false negative (type II) errors. The former can be tested using appropriate corrections for multiple testing, and true positives validated by replicate studies. However, as the number of risk factors increase, the potential interactions and the analytic space also increase exponentially, posing challenges for detection. This is the so called ‘curse of dimensionality’. It imposes significant concerns related to type II errors given finite sample sizes. There is therefore a recurrent tradeoff between the difficulties of controlling type I errors and minimizing the type II error rate. When one considers interactions at a genome-wide level, this task can be daunting. Another concern is whether the individual risk factors have detectable statistical effects when analyzed in isolation. If such main effects are modified by another environmental or genetic variant, the power to detect the main effect may be reduced [7]. Furthermore, the interactions hamper efforts at replication, if the ascertainment schemes for replicate samples alter the impact or frequency of individual risk factors. Conventional statistical approaches that depend on hierarchical model building may fail to detect interaction effects in the absence of main effects [8].
3. Current approaches to identify epistasis
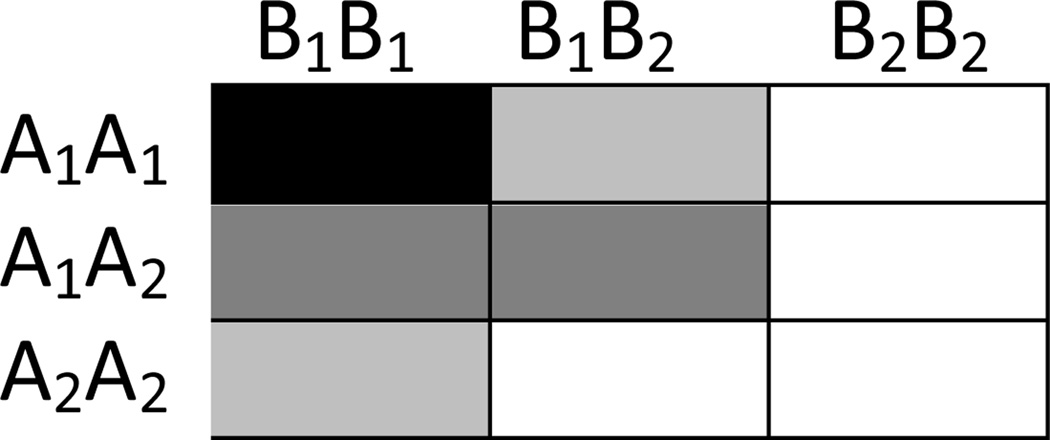
Traditional approaches include logistic regression, analysis of variance (ANOVA) and likelihood tests. Several novel approaches have been developed, such as multi-factorial dimensionality reduction (MDR), pattern recognition, neural network, cellular automata and genetic algorithm [9]. If two loci interact, then the interacting genotypes could be represented in a 3 × 3 matrix as shown in Figure 1. The cells are shaded to indicate the degree of risk contributed by the interactions between the genotypes. MDR utilizes a cross-validation approach to evaluate the classification. It takes account of empty cells or reduced cell sizes that can occur with relatively small samples. This approach was initially introduced for balanced case-control or balanced discordant sib pairs. Other investigators have extended this approach to family based designs [10] and to unbalanced samples [11].
Figure 1. Genotypes at two epistatic loci.
A, B indicate loci that are not in linkage disequilibrium. The subscripts indicate alleles at these loci. The shades indicate the differing risks imposed by combinations of genotypes, with darker shades referring to higher risk.
Several modifications of this approach have been developed. One of them combines the strength of logistic regression and the MDR. The two genotype interactions are partitioned into two to three genotype classes based on presumed risk level using a fixed classification scheme. The number of cases and controls in each classification scheme at different risk levels are then calculated. Interactions are then reported using Pearson’s χ2 statistic [12].
Additional approaches may prove useful on larger datasets such as genome-wide association studies (GWAS). For example, MDR identifies multiple order interactions through an exhaustive search and evaluates the association between each interaction and the disease by cross-validations. The exhaustive nature of the approach may be more appropriate for smaller datasets. Similarly, logic regression infers a tree-based relationship between the disease status and a set of markers, and evaluates the detected associations by permutation tests. A Bayesian approach may be more appropriate in the GWAS setting [13]. Zhang and Liu propose a Bayesian Epistasis Association Mapping algorithm (BEAM), which uses a Markov chain Monte Carlo method to evaluate individual markers based on the current status of other markers iteratively, providing a posterior probability that a marker is individually or epistatically associated with disease. Simulations under different models suggest that the method may be particularly useful when the marginal effect of an individual locus is small. The Bayesian method can also detect epistasis when no marginal effect is present, unlike the stepwise logistic procedures [13].
It should be noted that none of these methods are likely to be successful if the risk variant/s, or highly correlated ‘surrogates’ are not included in the analyses. (The surrogates may be available due to linkage disequilibrium (LD), the non-random association between polymorphisms at the population level that occurs throughout the genome). The power of all these methods understandably declines if the LD between the risk loci and the surrogate or measured polymorphisms is modest. In addition, discrepancies between the frequency of the true risk allele and the frequency of the linked or correlated allele at the measured loci can have a substantial impact on the power to detect interactions. Increases in sample size can help improve power in these circumstances, but this may be a limiting factor.
Most epistatic interactions are examined in case-control sample setting. These models are powerful and provide comparative odds for risk for a given trait. However, careful selection of control sample of adequate power is essential. Further, variations in genetic backgrounds between cases and controls could alter the magnitude of the interactions. Therefore, some innovative methods focus on case-only designs. One such approach utilizes entropy-based statistics [14] [15]. The entropy is used to measure the uncertainly of random variables. Such a measure represents non-linear transformation of variables of interest. Non-linear transformation of genotype frequencies amplifies the difference between the equilibrium (independence) and non-equilibrium (interaction) states of the genetic locus system. The difference between genotype combination frequencies between the observed data and the one assuming no interactions reflect the change in the entropy measure for a given trait. The entropy-based statistic asymptotically follows a chi square distribution. This method was recently used to suggest an interaction model for schizophrenia involving SNPs of NRG1, RGS4 and G72 [14]. A sequential forward selection procedure, where one SNP is added at a time may also be used to construct a genetic interaction network that shows the relative importance of a set of genetic loci on a clinical phenotype.
Association studies are designed to identify main effects of alleles across a potentially wide range of genetic backgrounds. To control for spurious associations, effects of the genetic background itself are often incorporated into the linear model, either in the form of sub-population effects in the case of structure or in the form of genetic relationship matrices in the case of complex pedigrees. In this context, epistatic interactions between loci can be captured as interaction effects between the associated locus and the genetic background. Recently, Jannink developed genetic and statistical models to align the locus by genetic background interaction concept with more standard concepts of epistasis, when genetic background is modeled using an additive relationship matrix [16].
Epistatic interactions in quantitative traits
Logistic regression may be used to estimate epistatic variance for quantitative traits. In this situation, epistatic variance is partitioned into four orthogonal components, namely additive × additive, additive × dominant, dominant × dominant, and dominant × additive [17]. Another approach to test potentially non-additive multi-locus genotypes as predictors of quantitative trait is called the restricted partition method (RPM). RPM is a partitioning algorithm for examining multi-locus genotypes as (potentially non-additive) predictors of a quantitative trait. RPM is designed to detect qualitative genetic and environmental factors contributing to a quantitative trait. This method takes a multi-locus measured genotype approach and assesses the mean trait values for different multi-locus genotypes, thus examining the potential contributors to quantitative traits. Different mean trait values of multi-locus genotypes indicate that a locus or a combination of loci contribute to trait variation. This method may identify loci that contribute epistatically to a quantitative trait even when no single locus effects are observed [18]. The same approach may be used to examine gene-environment interactions, and in case-control datasets where quantitative trait values are replaced with 0's or 1's indicating control or case status [18]. A computationally more intense approach is the combinatorial partitioning method (CPM) that attempts to identify partitions of multi-locus genotypes that predict variations in quantitative traits. Each partition is examined for phenotypic similarity and the dissimilarity of partition means. This method can help identify the effect of combination of loci even when the main effect of individual loci cannot be detected.
Analytic software
Several freewares are available. The reader is directed to a dedicated web site for analytic tools and for comprehensive discussion of other analytic methods (http://www.epistasis-list.org). For example, Genetic Association Interaction Analysis (GAIA) is a web-based application for testing for statistical interactions between loci. This tool is based on the widely used case-control study design for genetic association analysis and is designed so that non-specialists may routinely apply tests for interaction. GAIA allows simple testing of both additive and additive plus dominance interaction models and includes permutation testing to appropriately correct for multiple testing. GAIA also helps in prioritizing the loci in large scale studies before epistatic interactions are examined (http://www.bbu.cf.ac.uk/html/research/biostats.htm) [19].
4. Epistasis in genetically complex diseases
Several examples of persuasive interactions are available in the literature. An update list of published epistatic and G/E interactions is maintained by Drs Motsinger-Reif, and David M. Reif (http://www.epistasis-list.org). For example, an epistatic interaction between IL-13 and IL4Ralpha gene alleles for asthma has been noted in Dutch, as well as Chinese samples [20] [21]. Variations in triglyceride levels may be partly explained based on epistatic interactions. Interestingly, these interactions appear to be gender specific. In females, an interaction between ApoB and ApoE has been shown to be associated with triglyceride levels whereas in males it occurs between the ApoAI/CIII/AIV and the LDLR [22].
5. Epistasis in schizophrenia
A genetic etiology for SZ is widely accepted, but environmental factors necessarily need to be invoked [4]. The impact of genetic factors is likely to be substantial, with heritability estimates of 60–70% [23, 24] and sibling recurrence risk ratio, λs, estimated at 8–10 [25]. Segregation analyses, as well as linkage and association studies strongly suggest that the genetic liability may not be due to a single locus [26] [27]. Though earlier simulation studies suggested that the variation in liability could be explained by 3 – 4 loci [25], current analyses suggest a much larger number. The magnitude of risk conferred varies widely, from relatively modest odds ratios (OR) for common variants (~1.2) [28] to substantial risks due to relatively rare variants, such as copy number variations (CNVs) (~ 13) [29]; [30].
Complex behavioral traits such as hallucinations and delusions that are the hallmarks of schizophrenia can be construed as having their roots in higher order interactions of neural networks in the brain. Several neurotransmitters, neuromodulators, their receptors and transporters along with the enzymes that synthesize and catabolize them could interact at different levels, enabling ‘cross-talk’ between different neural networks. Genetic variations that quantitatively alter the expression or qualitatively alter the chemical structure of the products that may affect the function could conceivably be important from an etiological perspective. Interactions among these genetic variations could either accentuate or attenuate the impact of genetic factors on the biological systems that underlie the psychopathology of schizophrenia. Some of the epistatic interactions observed in schizophrenia are described below.
5.1. Dysbindin
The DTNBP1 gene that encodes dysbindin has been implicated in schizophrenia susceptibility by a series of independent genetic association and gene expression studies. Dysbindin is part of a protein complex, termed the biogenesis of lysosome-related organelles complex 1 (BLOC-1), the molecular components of which might be involved in the regulation of vesicular trafficking and dendrite branching. Using canonical correlation analysis (CCA) to perform gene-based tests of epistasis in schizophrenia, Morris et al [31] examined other BLOC-1 genes (MUTED, PLDN, CNO, SNAPAP, BLOC1S1, BLOC1S2, and BLOC1S3). They observed a main effect of BLOC1S3. Epistatic interactions were also observed between DTNBP1 and MUTED, though the latter did not have a main effect.
Another study examined the interaction between DTNBP1, RGS4 and IL3 [32]. This study utilized a multi-factorial dimensionality reduction Pedigree Disequilibrium test (MDR-PDT) in an Irish family based sample, and MDR in an independent Irish case-control sample. Associations with single SNPs had been noted earlier at each of the three genes of interest in the same samples [33]. In the family based sample, a 3-locus interaction between IL3 SNP rs2069803, DTNBP1 SNP rs2619539, and RGS4 SNP rs2661319 was observed. In the case-control sample, a 2-locus interaction was observed between IL3 SNP rs31400 and DTNBP1 SNP rs760761. The different patterns of interactions were attributed to lower power in the case-control sample.
5.2. Dopaminergic genes
Tan and colleagues examined interactional effects of single nucleotide polymorphisms (SNPs) at Akt1 and catechol-O-methyl transferase (COMT) polymorphisms on prefrontal cortical function in schizophrenia [34]. They observed a main effect of Akt1 rs1130233 on a wide range of cognitive functions and fronto-striatal grey matter volume. In addition, an epistatic interaction of Akt1 with an exonic SNP of COMT (rs4680; val/met polymorphism) was observed on the prefrontal cortical activation. An epistatic interaction between allele A of rs1130233 and the val allele of COMT rs4580 was also observed on disproportionately inefficient prefrontal activation and reduced gray-matter volume of the prefrontal cortex.
5.3. DISC 1
Disrupted in schizophrenia 1 gene (DISC1) is disrupted in a t(1;11))q42.1;q14.3) translocation. In a large Scottish family, this gene segregates with schizophrenia, schizoaffective disorder and other psychiatric disorders [35]. DISC 1 is known to interact with several proteins such as NDEL1 and NDE1. Burdick et al reported an association between schizophrenia and a single haplotype block within NDEL1, but no significant association with individual SNPs at NDE1. They further found an epistatic interaction between NDEL1 SNP rs1391768 and DISC1 Ser704Cys. Further, an epistatic interaction was reported between DISC1 Ser704Cys and NDE1 rs3784859. These observations suggest epistatic interactions between DISC1, NDEL1 and NDE1 influences risk for SZ [36].
5.4. GABA & Dopamine
Postmortem brain studies have demonstrated reduced expression of glutamic acid decarboxylase 67 (GAD67), a key enzyme involved in the synthesis of γ-amino butyric acid (GABA). GAD-67 is encoded by GAD1. Straub et al reported an association of 8 of the 19 SNPs on GAD1 were associated with schizophrenia only against the background of COMT Val/Val genotypes at rs4680, but not the other genotypes among patients [37]. Further, they observed statistical epistasis using unconditional logistic regression between two SNPs in COMT and SNPs in GAD1, suggesting a potential biological synergism leading to increased risk.
5.5. Glutamatergic genes
The D-amino acid oxidase (DAO) signaling pathway has been implicated in the modulation of NMDA function in schizophrenia. Its catalytic activity depends on DAO activator (DAOA, formerly G72). Chumakov and colleagues first reported the association of DAOA/G30 with schizophrenia and a number of independent studies have since reported evidence of association between the DAOA and DAO genes and schizophrenia [38]. Though these associations have been questioned [39], an epistatic interaction was observed between the associated SNPs at DAOA (DAOA-M12, rs3916965) and DAO (DAO-M5, rs3918346) for schizophrenia risk (OR = 9.3) [40].
5.6. Glutamate and Dopamine
Functional MRI studies have pointed to the association between variants of COMT and the metabotropic glutamate receptor gene mgluR3 (GRM3) in regulating prefrontal activity. It was initially found that allele A of rs6465084 at GRM3 was associated with inefficient prefrontal processing of working memory and reduced NAA on 1H magnetic resonance spectroscopy [41]; [42] [43]. The combined effects of COMT and GRM3 were more pronounced on the prefrontal working memory processing. The GRM3 genotype earlier associated with suboptimal glutamatergic signaling was significantly associated with inefficient prefrontal engagement and altered prefrontal-parietal coupling against the background of COMT rs4680 (Val-homozygous genotype). Interestingly, a COMT rs4680 Met-homozygous background appeared to ameliorate the ‘deleterious’ effects of the GRM3 genotype on prefrontal processing [41].
The same group later reported epistatic interactions of certain COMT SNPs, including Val/Met (rs4680), rs2097603 and rs165599 with SNPs at RGS4, G72, GRM3, and DISC1 on prefrontal cortex processing efficiency. Three of five RGS4 SNPs (rs90387, rs951436 and rs2661319) did not have significant main effects, yet they showed a significant increase in risk in interaction with COMT SNPs. In addition, three SNPs on G72/G30 also showed significant interaction in the background of COMT variations. Similar observations were made on some SNPs on GRM3 and DISC1 [44].
5.7. Neuregulin
Neuregulin-1 (NRG1) was identified as a potential risk for schizophrenia in an Icelandic genome-wide linkage analysis [45]. Benzel et al, studied eight genes from NRG and erbB family of genes [46]. Out of the 365 tested single polymorphisms (SNPs) that spanned eight genes, significant epistasis was found with 42 SNPs (p< 0.05) among 396 schizophrenia cases and 1,342 blood bank controls. Gene-gene interactions in this study point towards three additional genes (NRG2, NRG3, and erbB1), which are expressed in the central nervous system, that play a role together in their association in schizophrenia.
5.8. Intermediate phenotypes
COMT and PRODH SNPs were examined for their associations with MRI morphometric measures in young patients with schizophrenia or schizoaffective disorder. A main effect of two non-synonymous SNPs at PRODH (rs2008720, rs450046 and rs372055) on frontal white matter reduction and one SNP on COMT (rs2097603) on superior temporal gyrus grey matter was observed. Epistatic interactions were observed on the inferior frontal lobe white matter when COMT Val allele was indexed with PRODH (rs20086720) alleles (GT or TT) and compared with the rest of the patients [47]. Quantitative phenotypes such as pre-pulse inhibition (PPI) and morphometric measurements on MRI scans have also been examined for epistatic interactions. In a small sample of female patients, the magnitude of eye-blink response did not show either a main effect or interaction effects with DRD2 Taq Ia and the COMT Val158Met polymorphisms [48].
5.9. Interactions between genetic and environmental risk factors
A ‘geneenvironment’ interaction model was examined in relation to obstetric complications (OCs), a putative environmental risk factor for SZ and a set of selected schizophrenia candidate genes (AKT1, BDNF, CAPON, CHRNA7, COMT, DTNBP1, GAD1, GRM3, NOTCH4, NRG1, PRODH, RGS4, TNF-alpha). Following multivariate analyses, variants of four genes, namely AKT1 (three SNPs), BDNF (two SNPs), DTNBP1 (one SNP) and GRM3 (one SNP) showed significant evidence for interactions with OCs [49].
6. A proposed systematic strategy for investigating interactions between risk factors
The studies reviewed above in relation to schizophrenia have largely examined interactions between carefully selected sets of genes, motivated by current concepts of the neurobiology of schizophrenia. They have to be considered tentative at present, as systematic replication has not been attempted for the majority. Furthermore, corrections for multiple comparisons have not been applied consistently. Thus, it is quite possible that some of these interactions reflect stochastic variation. In the face of such daunting challenges, is there a reasonable expectation of detecting meaningful interactions, or demonstrating that epistasis explains a greater proportion of disease risk than the main effects of individual risk alleles? Two general approaches are available at present. The first involves agnostic searches of GWAS data. The second involves focused searches based on prior evidence.
Statistical evidence for interactions can be garnered from GWAS datasets. These analyses are particularly persuasive when the interacting loci themselves have significant main effects. If main effects are undetectable initially, ‘true’ espistatic effects may still be present between these loci, or unmeasured risk alleles in LD with them. However, the investigator may wish to keep in mind the possibility that risk alleles or appropriate surrogates were not analyzed in the initial dataset. Replicate analyses are necessary to validate the epistatic effects. Replication of initial results from large GWAS datasets may thus require even larger samples for validation. Novel designs have been suggested to modify the classical independent replication model, such as a multi-stage design followed by joint analyses for single SNPs [50]. Such designs may eventually be applied for analysis of epistatic interactions.
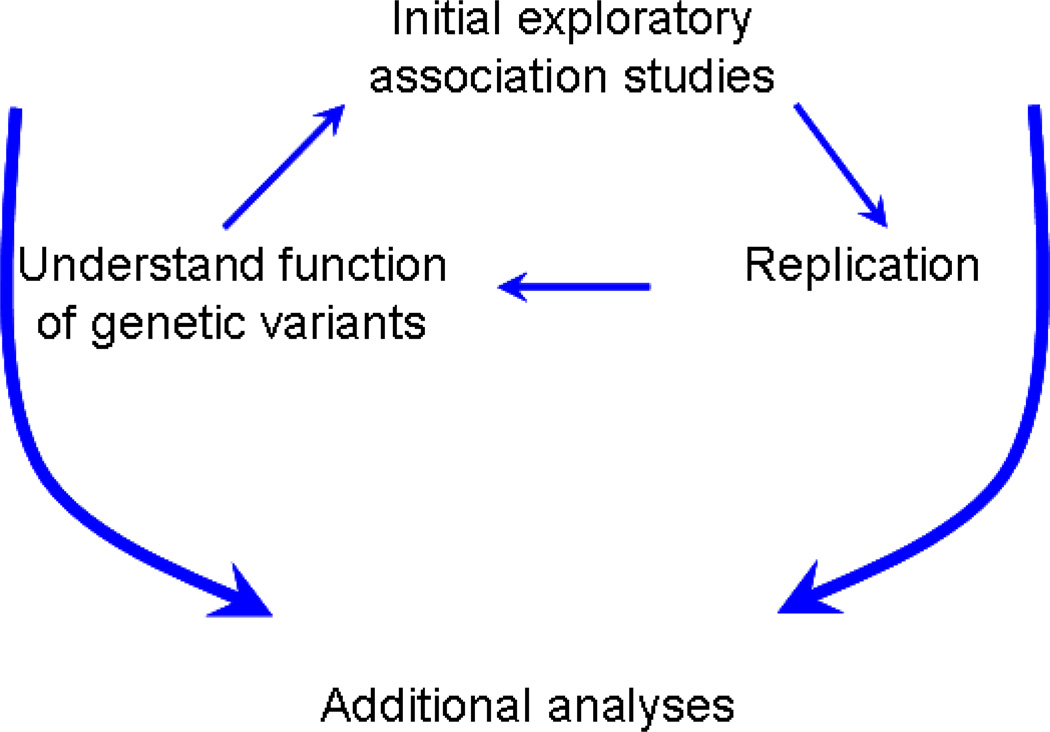
Alternatively, an incremental approach that we denote ‘recursive analysis’ may be used (Figure 2). In this approach, initial analyses are motivated by plausible interactions between plausible risk factors that individually appear to confer risk. This choice reflects an attempt to increase likelihood of detecting interactions a priori. Replication is attempted only if statistically meaningful interactions are noted. If replicable interactions are noted, a search for plausible biological function follows. Satisfactory functional evidence for the interactions then motivates searches for further interactions based on available data. This approach will enable a solid foundation for a MFPT model for SZ. While appealing, this approach is critically dependent on convincing identification of risk factor/s. It has been difficult to validate particular candidate gene risk alleles for schizophrenia. Furthermore, such an approach may fail to detect interactions in which individual risk factors or alleles themselves do not have main effects.
Figure 2.
Model for recursive analysis
It is worthwhile to emphasize the need to correct for multiple testing. Such corrections can be complex when one is testing epistatic interactions, and particularly when such tests are being conducted recursively on the same sample/s. Simulations may be helpful in this context (see ref 52). However, replication in an independent sample remains the most convincing way to ensure that ‘true’ associations are reported. On the other hand, it needs to be remembered that failure to replicate can be due to factors other than a false positive initial result, such as sample heterogeneity.
The recursive approach is not merely a variant of the ‘candidate gene’ scheme. For example, the initial association tests may stem from a prior linkage or GWAS analysis. Furthermore, our scheme extends beyond a simplistic candidate gene approach as it implicitly seeks to build on initial associations with single polymorphisms.
We have adopted the recursive approach for systematic studies of dopamine (DA) gene variation in SZ. Though a large number of studies have already been published without consistent results, our review suggested that most published studies were inadequate either because an insufficient number of polymorphisms were analyzed or because the samples used were underpowered to detect relatively small effects (OR ~ 1.5) [51]. We systematically investigated 18 DA genes in two independent samples and detected replicable interactions between four DA genes. Simulations suggested that the observed interactions were unlikely to occur by chance [52].
We have also used the recursive analysis approach to investigate G/E interactions. A substantial number of studies have suggested that infections, particularly during the prenatal period may confer risk for SZ [53]. Many of these studies have been inconsistent, presumably due to difficulties in documenting remote exposure to infectious agents using a cross-sectional design. We reasoned that the infectious risk factor hypothesis could be tested indirectly by comparing exposure among families with single affected and multiply affected individuals. Consistent with our predictions, exposure was more frequent among the latter [54]. It should be noted that these results do not prove causality per se. Intriguingly, our association studies of the HLA region revealed associations with SZ; the associations were more prominent among individuals with prior exposure to cytomegalovirus (CMV) [55]. Further fine mapping analyses revealed plausible pleiotropic associations at exonic polymorphisms of the MHC Class I polypeptide-related sequence B (MICB) gene [55]. Replicate studies are now in progress.
7. Conclusions
The etiology of common disorders like schizophrenia may be meaningfully explained on the basis of interactive risk conferred by an unknown number of risk factors. The identification and analyses of such factors poses difficult challenges, particularly when the identity and impact of individual factors is uncertain. Current agnostic searches based on GWAS datasets may provide important insights. We propose another rational method, called ‘recursive analyses’. This approach relies on systematic, step-wise interrogation of interactions between individual factors for which reliable a priori information is available. Both approaches have relative strengths and weaknesses. A combined approach that utilizes their relative strengths may ultimately provide the most convincing evidence.
Acknowledgments
Supported by NIH (MH56242, MH63480 to VLN, MH72995 to KVP and MH080582 to MT) and the Stanley Medical Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
None
References
- 1.Collins FS. Positional cloning moves from perditional to traditional. Nature Genetics. 1995;9(4):347–350. doi: 10.1038/ng0495-347. [DOI] [PubMed] [Google Scholar]
- 2.Collins A, Ennis S, Taillon-Miller P, Kwok PY, Morton NE. Allelic association with SNPs: metrics, populations, and the linkage disequilibrium map. Hum Mutat. 2001;17(4):255–262. doi: 10.1002/humu.21. [DOI] [PubMed] [Google Scholar]
- 3.Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Transactions of the Royal Society of Edinburgh. 1918;52:399–433. [Google Scholar]
- 4.Gottesman I. Schizophrenia Genesis: The Origins of Madness. New York: WH Freeman; 1991. [Google Scholar]
- 5.Zerba KE, Kessling AM, Davignon J, Sing CF. Genetic structure and the search for genotype-phenotype relationships: an example from disequilibrium in the Apo B gene region. Genetics. 1991 Oct;129(2):525–533. doi: 10.1093/genetics/129.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petronis A. The origin of schizophrenia: genetic thesis, epigenetic antithesis, and resolving synthesis. Biol Psychiatry. 2004;55(10):965–970. doi: 10.1016/j.biopsych.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Cordell HJ, Clayton DG. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am J Hum Genet. 2002;70(1):124–141. doi: 10.1086/338007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Culverhouse R, Suarez BK, Lin J, Reich T. A perspective on epistasis: limits of models displaying no main effect. Am J Hum Genet. 2002 Feb;70(2):461–471. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24(2):150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- 10.Martin ER, Ritchie MD, Hahn L, Kang S, Moore JH. A novel method to identify gene-gene effects in nuclear families: the MDR-PDT. Genet Epidemiol. 2006;30(2):111–123. doi: 10.1002/gepi.20128. [DOI] [PubMed] [Google Scholar]
- 11.Velez DR, White BC, Motsinger AA, Bush WS, Ritchie MD, Williams SM, et al. A balanced accuracy function for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction. Genet Epidemiol. 2007 May;31(4):306–315. doi: 10.1002/gepi.20211. [DOI] [PubMed] [Google Scholar]
- 12.DeWan A, Klein RJ, Hoh J. Linkage disequilibrium mapping for complex disease genes. Methods in molecular biology (Clifton, NJ. 2007;376:85–107. doi: 10.1007/978-1-59745-389-9_7. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Liu JS. Bayesian inference of epistatic interactions in case-control studies. Nat Genet. 2007 Sep;39(9):1167–1173. doi: 10.1038/ng2110. [DOI] [PubMed] [Google Scholar]
- 14.Kang G, Yue W, Zhang J, Huebner M, Zhang H, Ruan Y, et al. Two-stage designs to identify the effects of SNP combinations on complex diseases. J Hum Genet. 2008;53(8):739–746. doi: 10.1007/s10038-008-0307-x. [DOI] [PubMed] [Google Scholar]
- 15.Cui Y, Kang G, Sun K, Qian M, Romero R, Fu W. Gene-centric genomewide association study via entropy. Genetics. 2008 May;179(1):637–650. doi: 10.1534/genetics.107.082370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jannink JL. Identifying quantitative trait locus by genetic background interactions in association studies. Genetics. 2007 May;176(1):553–561. doi: 10.1534/genetics.106.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cockerham CC, Weir BS. Covariances of relatives stemming from a population undergoing mixed self and random mating. Biometrics. 1984 Mar;40(1):157–164. [PubMed] [Google Scholar]
- 18.Culverhouse R, Klein T, Shannon W. Detecting epistatic interactions contributing to quantitative traits. Genet Epidemiol. 2004 Sep;27(2):141–152. doi: 10.1002/gepi.20006. [DOI] [PubMed] [Google Scholar]
- 19.Macgregor S, Khan IA. GAIA: An easy-to-use web-based application for interaction analysis of case-control data. BMC Med Genet. 2006;7(1):34. doi: 10.1186/1471-2350-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard LM, Kumar C, Leese M, Thornicroft G. The general fertility rate in women with psychotic disorders. Am J Psychiatry. 2002;159(6):991–997. doi: 10.1176/appi.ajp.159.6.991. [DOI] [PubMed] [Google Scholar]
- 21.Chan IH, Leung TF, Tang NL, Li CY, Sung YM, Wong GW, et al. Gene-gene interactions for asthma and plasma total IgE concentration in Chinese children. The Journal of allergy and clinical immunology. 2006 Jan;117(1):127–133. doi: 10.1016/j.jaci.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 22.Nelson MR, Kardia SL, Ferrell RE, Sing CF. A combinatorial partitioning method to identify multilocus genotypic partitions that predict quantitative trait variation. Genome Res. 2001 Mar;11(3):458–470. doi: 10.1101/gr.172901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao DC, Morton NE, Gottesman II, Lew R. Path analysis of qualitative data on pairs of relatives: application to schizophrenia. Human Heredity. 1981;31(6):325–333. doi: 10.1159/000153233. [DOI] [PubMed] [Google Scholar]
- 24.McGue M, Gottesman II, Rao DC. The transmission of schizophrenia under a multifactorial threshold model. American Journal of Human Genetics. 1983;35(6):1161–1178. [PMC free article] [PubMed] [Google Scholar]
- 25.Risch N. Genetic linkage and complex diseases, with special reference to psychiatric disorders. Genet Epidemiol. 1990;7(1):3–16. doi: 10.1002/gepi.1370070103. discussion 7–45. [DOI] [PubMed] [Google Scholar]
- 26.Rao DC, Morton NE, Gottesman II, Lew R. Path analysis of qualitative data on pairs of relatives: application to schizophrenia. Hum Hered. 1981;31(6):325–333. doi: 10.1159/000153233. [DOI] [PubMed] [Google Scholar]
- 27.Carter CL, Chung CS. Segregation analysis of schizophrenia under a mixed genetic model. Human Heredity. 1980;30(6):350–356. doi: 10.1159/000153156. [DOI] [PubMed] [Google Scholar]
- 28.Shirts BH, Nimgaonkar V. The genes for schizophrenia: finally a breakthrough? Curr Psychiatry Rep. 2004;6(4):303–312. doi: 10.1007/s11920-004-0081-1. [DOI] [PubMed] [Google Scholar]
- 29.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008 Sep 11;455(7210):232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Consortium TIS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008 Sep 11;455(7210):237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris DW, Murphy K, Kenny N, Purcell SM, McGhee KA, Schwaiger S, et al. Dysbindin (DTNBP1) and the biogenesis of lysosome-related organelles complex 1 (BLOC-1): main and epistatic gene effects are potential contributors to schizophrenia susceptibility. Biol Psychiatry. 2008 Jan 1;63(1):24–31. doi: 10.1016/j.biopsych.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 32.Edwards TL, Wang X, Chen Q, Wormly B, Riley B, O'Neill FA, et al. Interaction between interleukin 3 and dystrobrevin-binding protein 1 in schizophrenia. Schizophr Res. 2008 Dec;106(2–3):208–217. doi: 10.1016/j.schres.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen SF, Chen CH, Chen JY, Wang YC, Lai IC, Liou YJ, et al. Support for association of the A277C single nucleotide polymorphism in human vesicular monoamine transporter 1 gene with schizophrenia. Schizophr Res. 2007 Feb;90(1–3):363–365. doi: 10.1016/j.schres.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 34.Tan HY, Nicodemus KK, Chen Q, Li Z, Brooke JK, Honea R, et al. Genetic variation in AKT1 is linked to dopamine-associated prefrontal cortical structure and function in humans. J Clin Invest. 2008 Jun;118(6):2200–2208. doi: 10.1172/JCI34725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336(8706):13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 36.Burdick KE, Kamiya A, Hodgkinson CA, Lencz T, DeRosse P, Ishizuka K, et al. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Hum Mol Genet. 2008 Aug 15;17(16):2462–2473. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Straub RE, Lipska BK, Egan MF, Goldberg TE, Callicott JH, Mayhew MB, et al. Allelic variation in GAD1 (GAD67) is associated with schizophrenia and influences cortical function and gene expression. Mol Psychiatry. 2007 Sep;12(9):854–869. doi: 10.1038/sj.mp.4001988. [DOI] [PubMed] [Google Scholar]
- 38.Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, et al. Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13675–23680. doi: 10.1073/pnas.182412499. Epub 2002 Oct 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kvajo M, Dhilla A, Swor DE, Karayiorgou M, Gogos JA. Evidence implicating the candidate schizophrenia/bipolar disorder susceptibility gene G72 in mitochondrial function. Mol Psychiatry. 2008 Jul;13(7):685–696. doi: 10.1038/sj.mp.4002052. [DOI] [PubMed] [Google Scholar]
- 40.Corvin A, McGhee KA, Murphy K, Donohoe G, Nangle JM, Schwaiger S, et al. Evidence for association and epistasis at the DAOA/G30 and D-amino acid oxidase loci in an Irish schizophrenia sample. Am J Med Genet B Neuropsychiatr Genet. 2007 Oct 5;144B(7):949–953. doi: 10.1002/ajmg.b.30452. [DOI] [PubMed] [Google Scholar]
- 41.Tan HY, Chen Q, Sust S, Buckholtz JW, Meyers JD, Egan MF, et al. Epistasis between catechol-O-methyltransferase and type II metabotropic glutamate receptor 3 genes on working memory brain function. Proc Natl Acad Sci U S A. 2007 Jul 24;104(30):12536–12541. doi: 10.1073/pnas.0610125104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marenco S, Steele SU, Egan MF, Goldberg TE, Straub RE, Sharrief AZ, et al. Effect of metabotropic glutamate receptor 3 genotype on N-acetylaspartate measures in the dorsolateral prefrontal cortex. Am J Psychiatry. 2006 Apr;163(4):740–742. doi: 10.1176/appi.ajp.163.4.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egan MF, Straub RE, Goldberg TE, Yakub I, Callicott JH, Hariri AR, et al. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc Natl Acad Sci U S A. 2004 Aug 24;101(34):12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicodemus KK, Kolachana BS, Vakkalanka R, Straub RE, Giegling I, Egan MF, et al. Evidence for statistical epistasis between catechol-O-methyltransferase (COMT) and polymorphisms in RGS4, G72 (DAOA), GRM3, and DISC1: influence on risk of schizophrenia. Hum Genet. 2007 Feb;120(6):889–906. doi: 10.1007/s00439-006-0257-3. [DOI] [PubMed] [Google Scholar]
- 45.Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71(4):877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benzel I, Bansal A, Browning BL, Galwey NW, Maycox PR, McGinnis R, et al. Interactions among genes in the ErbB-Neuregulin signalling network are associated with increased susceptibility to schizophrenia. Behav Brain Funct. 2007;3:31. doi: 10.1186/1744-9081-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zinkstok J, Schmitz N, van Amelsvoort T, Moeton M, Baas F, Linszen D. Genetic variation in COMT and PRODH is associated with brain anatomy in patients with schizophrenia. Genes Brain Behav. 2008 Feb;7(1):61–69. doi: 10.1111/j.1601-183X.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 48.Montag C, Hartmann P, Merz M, Burk C, Reuter M. D2 receptor density and prepulse inhibition in humans: negative findings from a molecular genetic approach. Behav Brain Res. 2008 Mar 5;187(2):428–432. doi: 10.1016/j.bbr.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 49.Nicodemus KK, Marenco S, Batten AJ, Vakkalanka R, Egan MF, Straub RE, et al. Serious obstetric complications interact with hypoxia-regulated/vascular-expression genes to influence schizophrenia risk. Mol Psychiatry. 2008 Sep;13(9):873–877. doi: 10.1038/sj.mp.4002153. [DOI] [PubMed] [Google Scholar]
- 50.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 51.Talkowski ME, Bamne M, Mansour H, Nimgaonkar VL. Dopamine genes and schizophrenia: case closed or evidence pending? Schizophr Bull. 2007 Sep;33(5):1071–1081. doi: 10.1093/schbul/sbm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talkowski ME, Kirov G, Bamne M, Georgieva L, Torres G, Mansour H, et al. A network of dopaminergic gene variations implicated as risk factors for schizophrenia. Hum Mol Genet. 2008 Mar 1;17(5):747–758. doi: 10.1093/hmg/ddm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yolken RH, Torrey EF. Are some cases of psychosis caused by microbial agents?A review of the evidence. Mol Psychiatry. 2008 Feb 12; doi: 10.1038/mp.2008.5. [DOI] [PubMed] [Google Scholar]
- 54.Kim JJ, Shirts BH, Dayal M, Bacanu S, Wood J, Xie W, et al. Are exposure to cytomegalovirus and genetic variation on chromosome 6p joint risk factors for schizophrenia? Annals of Medicine. 2007;39:145–153. doi: 10.1080/07853890601083808. [DOI] [PubMed] [Google Scholar]
- 55.Kim JJ, Dayal M, Bacanu S-A, Shirts BH, Wood J, Xie W, et al. Exposure to cytomegalovirus and polymorphisms in two genes on chromosome 6p21-23 as joint risk factors for schizophrenia. American Journal of Medical Genetics. 2004 Sep 15;130B(1):19. [Google Scholar]