Abstract
Chromatin-based DNA damage response (DDR) pathways are fundamental for preventing genome and epigenome instability, which are prevalent in cancer. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) catalyze the addition and removal of acetyl groups on lysine residues, a post-translational modification important for the DDR. Acetylation can alter chromatin structure as well as function by providing binding signals for reader proteins containing acetyl-lysine recognition domains, including the bromodomain (BRD). Acetylation dynamics occur upon DNA damage in part to regulate chromatin and BRD protein interactions that mediate key DDR activities. In cancer, DDR and acetylation pathways are often mutated or abnormally expressed. DNA damaging agents and drugs targeting epigenetic regulators, including HATs, HDACs, and BRD proteins, are used or are being developed to treat cancer. Here, we discuss how histone acetylation pathways, with a focus on acetylation reader proteins, promote genome stability and the DDR. We analyze how acetylation signaling impacts the DDR in the context of cancer and its treatments. Understanding the relationship between epigenetic regulators, the DDR, and chromatin is integral for obtaining a mechanistic understanding of genome and epigenome maintenance pathways, information that can be leveraged for targeting acetylation signaling, and/or the DDR to treat diseases, including cancer.
Introduction
Genome maintenance relies on the precise replication and repair of our genetic information. This is a daunting task given the 3 X 109 bp size of the human genome and the presence of thousands of DNA lesions that are generated per cell per genome every day by various genotoxic processes and agents. To maintain genome integrity in these adverse conditions, cells contain DNA damage response (DDR) pathways that detect, signal, and repair DNA lesions [1,2]. Multiple DNA repair pathways are present to accommodate diverse DNA lesions. A particularly cytotoxic DNA lesion is the DNA double-strand break (DSB). DSBs can promote mutations, DNA degradation, or ligation to other DNA ends, resulting in genome instability. DSBs are repaired mainly by non-homologous end-joining (NHEJ) and homologous recombination (HR) in mammalian cells [3]. NHEJ ligates the two broken DNA ends together [4], whereas HR uses a homologous DNA template to accurately copy and repair the DSB [5,6]. The importance of DDR pathways is highlighted by the various diseases associated with DDR defects, including neurodegenerative disorders, immune deficiencies, and cancer [1,7].
Eukaryotic nuclear DNA is organized into chromatin [8]. The basic unit of chromatin is the nucleosome, consisting of DNA wrapped around histone proteins [8,9]. Chromatin organizes the genome and controls its accessibility, making chromatin integral for DNA-based processes. Chromatin is highly modified by posttranslational modifications (PTMs), including phosphorylation, methylation, and acetylation [10–13]. PTMs regulate chromatin structure as well as modulate chromatin interactions of “reader” proteins that contain PTM binding domains [11,14–16]. Epigenetic changes and mutations within chromatin regulatory proteins are observed in different diseases, including cancer [17]. As many DDR activities occur within chromatin, understanding the interplay between chromatin and the DDR is fundamental for obtaining mechanistic details of DDR activities in both normal and diseased contexts.
Chromatin PTMs are dynamically regulated in response to DNA damage both locally at the lesion site and globally where they perform several functions [18–22]. These include modulating chromatin structure at DNA damage sites and across the genome to facilitate the DDR, including signaling [22–24], repair [25], and transcriptional responses [26,27]. Another fundamental role of PTMs is to provide docking sites for the recognition and accumulation of DDR factors at damage sites where they orchestrate DDR functions. For example, DNA damage-induced phosphorylation of histone variant H2AX (γH2AX) is recognized by the BRCT domains of MDC1, which mediate the recruitment of downstream signaling and repair proteins to damage sites [28]. 53BP1 represents another histone PTM DDR factor reader [29,30], which recognizes the bivalent marks H4K20me2 and H2AK13/15ub at damage sites [31,32]. Histone PTMs and their DDR functions have been comprehensively reviewed [18–21,33]. Here, we discuss acetylation signaling with a focus on acetylation readers that are involved in mammalian genome maintenance and DNA repair. We also consider DDR-related acetylation signaling in cancer and its potential impact on cancer therapies.
Acetylation Signaling in the DDR
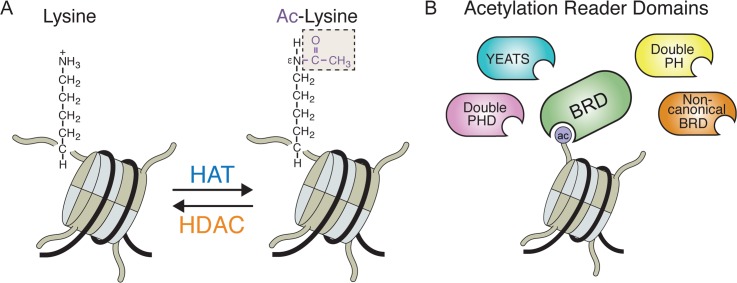
Acetylation is the covalent attachment of an acetyl-group (-COCH3) to the ε-amino groups of a lysine residue on histones and non-histone proteins by histone acetyltransferases (HATs) [34,35], which can be removed by histone deacetylases (HDACs) [36] (Fig 1A). Acetylation levels are regulated by the concerted activities of HATs and HDACs [21,35,36]. The importance of acetylation signaling is well established in many cellular processes, including transcription and the DDR [14]. One salient function of acetylation is to regulate chromatin structure. This is exemplified by H4K16ac, which blocks inter- and intra-chromosomal folding to promote an open chromatin structure [37]. Acetylated proteins are also bound by acetyl-lysine reader proteins. The principal acetylation recognition proteins contain the acetyl-lysine binding bromodomain (BRD), although other domains have acetyl-lysine interaction capabilities (Fig 1B) [16,38,39]. Since acetylation changes upon UV damage were observed over 30 years ago, numerous DNA damage responsive acetylations have been reported [21,40]. The involvement of acetylation reader proteins in deciphering these signals to promote the DDR is less clear. Recent studies have identified over one-third of human BRD proteins directly responding to DNA damage, suggesting that, collectively with HATs and HDACs, the entire acetylation signaling machinery orchestrates DDR activities within chromatin (Table 1) [41–44].
Fig 1. Acetylation signaling.
(A) HATs add an acetyl (Ac) moiety on the ε-amino group of a lysine residue, and HDACs reverse this reaction. (B) Acetylated lysines on histones are recognized and bound by proteins containing BRD domains and other acetyl-lysine binding domains. Abbreviations: ac, acetylation; HAT, histone acetyltransferase; HDAC, histone deacetylase; BRD, bromodomain; YEATS, Yaf9, ENL, AF9, Taf14, and Sas5; PHD, plant homeodomain; PH, pleckstrin-homology.
Table 1. DDR functions of mammalian acetylation readers.
| Domain | Protein | Complex | Histone Binding Ac targets | Damage recruitment | DDR Ac binding | DDR functions | Reference |
|---|---|---|---|---|---|---|---|
| BRD | ATAD2 | - | K3K14, H4K5 | - | - | - | [181,182] |
| ATAD2B | - | H4K5 | - | - | - | [38] | |
| ACF1 (BAZ1A) | CHRAC, ACF | - | Laser, R.E., UV | N | NHEJ, HR, NER, Checkpoint | [41,94,95,98] | |
| WSTF (BAZ1B) | WICH, B-WICH | - | Laser, UV | N | NER, DDR signaling, Checkpoint | [41,95,97,98] | |
| BAZ2B | - | H3K14 | - | - | - | [38] | |
| BPTF | NURF | H4K5/16 | Laser | - | - | [41] | |
| BRD2 | - | H3K14, H4K5/8/12/16/20 | - | - | - | [55] | |
| BRD3 | - | H3K14/18, H4K5/8/12/16/20 | - | - | - | [55] | |
| BRD4 | - | H3K9/14, H4K5/8/12/16 | - | Y | Regulate chromatin | [55,71] | |
| BRD7 | - | H3K9/14, H4K8/12/16 | - | - | Regulate p53 activity | [91,92,183] | |
| BRDT | - | H2BK12/15, H3K9/14/18/23, H4K5/8/12/16 | - | - | - | [55] | |
| BRPF1 | - | H2AK5, H3K14, H4K12 | - | - | - | [184] | |
| CECR2 | CERF | H3K9/14 | - | Y | DDR signaling | [38,185] | |
| CBP (CREBBP) | - | H2BK85, H3K9/14/36/56, H4K12/20/44 | Laser, R.E. | - | NHEJ, HR, NER | [44,55,76,186,187] | |
| GCN5 | SAGA, ATAC | H2AK5, H3K9/14, H4K8/14/16 | Laser, UV | - | NER, Regulate chromatin | [27,41,55,77,83] | |
| BAF180 (PBRM1) | PBAF | H3K4/9/14/18/23 | Laser | Y | Transcription repression, Regulate p53 activity, Promote cohesion | [42,55,89,91] | |
| p300 | - | H3K36/56, H4K12/20/44 | Laser, R.E., UV | - | NHEJ, HR, NER, DDR signaling, Checkpoint, Regulate p53 activity | [41,44,55,76,176,186–192] | |
| PCAF | ATAC | H3K9/14/36, H4K8/16/20 | Laser | - | Regulate p53 activity | [41,55,78,191] | |
| BRM (SMARCA2) | BAF | H3K9/14, H4K5/8/12/16 | Laser | Y | NHEJ, DDR signaling | [41,44,55,193] | |
| BRG1 (SMARCA4) | BAF, PBAF | H2BK5, H3K9/14, H4K8/K12/K16 | Laser, UV | Y | NER, Regulate chromatin, Transcription repression, Checkpoint, DDR signaling | [41,42,44,55,85–87,193,194] | |
| TAF1 | - | H3K9/14, H4K5/8/12/16 | - | - | Checkpoint | [195] | |
| TRIM24 | - | H3K23, H4K16 | Laser | - | Regulate p53 activity | [41,69,196] | |
| KAP1 (TRIM28) | - | No binding | IR, Laser | N | DDR signaling, Heterochromatin repair, Transcription repression | [64,66,68,113,197] | |
| TRIM33 | - | H3K18/23 | Laser | - | Regulate ALC1 activity | [41,43,70] | |
| ZMYND8 | NuRD | H4K5/8/12/16 | Laser | Y | Transcription repression, HR | [41] | |
| ZMYND11 | - | No binding | - | - | - | [160] | |
| Uncharacterized acetylation interactions: ASH1L, BAZ2A, BRD1, BRD8, BRD9, BRPF3, BRWD1, BRWD3, MLL, PHIP, SP100, SP110, SP140, SP140L, TAF1L, TRIM66 | |||||||
| Double PHD | DPF3 | PBAF | H3K14 | - | - | - | [114] |
| MORF | - | H3K9/14 | - | - | - | [115] | |
| MOZ | - | H3K9/14 | - | - | - | [115,116] | |
| YEATS | AF9 | - | H3K9/18/27 | - | - | Transcription repression | [39,119,120] |
| ENL | - | H3K9/27 | - | - | Transcription repression, NHEJ | [119,120] | |
| GAS41 | NuA4, SRCAP | H3K9/27 | - | - | - | [119] | |
| Uncharacterized acetylation interactions: YETS2 | |||||||
| Non-canonical BRD | DNA-PKcs | - | H2AXK5 | IR, Laser | Y | DDR signaling, NHEJ, Transcription repression | [112,122,198] |
Acetylation readers and their histone targets are provided. The reported damage recruitment, acetylation binding in the DDR, and DDR functions of acetylation readers are listed. Abbreviations: DDR, DNA damage response; NHEJ, nonhomologous end-joining; HR, homologous recombination; NER, nucleotide excision repair; IR, ionizing radiation; R.E., restriction enzyme; CHRAC, chromatin accessibility complex; WICH, WSTF-ISWI chromatin remodeling complex; NURF, nucleosome-remodeling factor; CERF, CECR2-containing remodeling factor; SAGA, Spt-Ada-Gcn5-acetyltransferase; ATAC, Ada2-containing chromatin-modifying complex; BAF, BRG1- or HBRM-associated factor; PBAF, polybromo-associated BAF; NuRD, nucleosome remodeling and histone deacetylase complex; NuA4, nucleosome acetyltransferase of H4 (or TIP60-p400 complex); SRCAP, Snf-2-related CREB-binding protein activator protein.
HATs, HDACs, and the DDR
The involvement of mammalian HATs and HDACs in the DDR is exemplified by their common localization to damage sites [21]. The HAT TIP60 (KAT5), a key component of the NuA4 complex, is involved in DSB repair from early signaling events to downstream repair pathway choices [45]. Upon DNA damage, TIP60 acetylates histones and the Ataxia telangiectasia mutated (ATM) kinase to enhance its activity [46–48]. Acetylated H4K16 by TIP60 promotes HR repair, whereas deacetylation facilitates NHEJ [47,49]. The DSB pathway choice regulated by TIP60 occurs in part by its ability to acetylate H2AK15, which impedes RNF168 ubiquitylation to block 53BP1 recruitment, thereby promoting HR [48]. The HAT Males absent on the first (MOF) regulates global H4K16Ac levels, and its loss impacts the DDR. Indeed, cells deficient of MOF exhibit impaired recruitment of DDR factors and a reduced capacity for DSB repair by HR and NHEJ [50–52]. Among HDACs, HDAC1 and HDAC2 play particularly important roles in DSB repair by deacetylating histones, including H3K56ac and H4K16ac [27,49]. HDACs can also target both histone and non-histone proteins. For example, SIRT6 deacetylates H3K56 and also the DDR factor CtIP, which regulates both the rapid recruitment of the remodeling factor SNF2H and the activity of CtIP at damage sites to promote DNA repair [53,54]. These studies highlight important functions of HATs and HDACs in the DDR (reviewed in [21]).
Readers of Acetylated Lysines in the DDR: Bromodomain Proteins
Emerging evidence has highlighted the importance of acetylation readers, including BRD proteins, in the DDR. One or more BRDs are encoded in 42 human proteins [38,55]. BRD domains consist of several α-helices linked by loops that form a hydrophobic cavity that specifically recognizes acetyl-lysines. BRD proteins can be broadly classified as HATs, components of ATP-dependent chromatin remodeling complexes, and/or transcriptional regulators [38,41,56]. Dysfunction of BRD proteins has been identified in diseases, including cancer [57]. The demonstrated druggability of the BRD by small molecule inhibitors has motivated targeting BRD proteins as cancer therapies [58–61].
Regulation of chromatin states by BRD proteins in the DDR
Chromatin exists between open and condensed states that impact its functionality. In heterochromatin, DSBs require specific factors as well as chromatin remodeling complexes to overcome the chromatin barrier to allow damage recognition and signaling to promote repair [62,63]. The BRD protein KAP1 (TRIM28) functions in DSB repair within heterochromatin. ATM phosphorylates KAP1 in response to DSBs [64], which disperses the nucleosome remodeler CHD3 from chromatin to trigger chromatin relaxation, allowing repair to occur within heterochromatin [64–66]. Three KAP1 tripartite motif-containing (TRIM) family paralogs (TRIM24, TRIM33, and TRIM66) also contain BRDs [67]. TRIM24 and TRIM33 are recruited to DNA damage and are involved in the DDR [41,43]. Although KAP1 BRD lacks acetyl-lysine binding [68], the plant homeodomain (PHD)-BRD tandem domains of TRIM24 and TRIM33 can read methylated or acetylated histones [69,70]. Whether these TRIM BRD proteins act interdependently or independently in the DDR awaits further investigation.
An isoform of BRD4, a member of the bromodomain and extra-terminal (BET) family of BRD proteins, has been reported to insulate chromatin to modulate γH2AX spreading around damaged DNA [71]. BRD4 contains two BRD domains and functions as a regulator of various transcriptional processes [72]. Successful targeting of the BRDs of BRD4 by small molecule inhibitors has gained widespread attention for its potential as an anticancer therapy and as a proof of concept for the “druggability” of BRD proteins [58,59]. In the DDR, inhibition of BRD4 with the BRD targeting inhibitor JQ1 resulted in increased DNA damage signaling of γH2AX formation, while survival from DNA damage was improved [71]. These data implicate the BRD of BRD4 as a key domain involved in regulating the cellular response to DNA damage. The BRDs of BRD4 have been shown to bind acetylated histone H3 and H4 [55]. BRD4 has also been reported to act as a HAT, acetylating H3 and H4 [73]. It is unclear how BRD4 recognizes damaged chromatin and if its HAT activity participates in its DDR functions. It will be important to distinguish the specific acetylated residues read and/or acetylated by BRD4 in cells upon DNA damage compared to those targeted during normal transcription to understand mechanistically how BRD4 promotes both transcriptional regulation in unperturbed cells and DNA damage signaling in response to DNA damage.
BRD-containing HATs in the DDR
The BRD-containing HATs GCN5, PCAF, p300, and CBP exist in multiple complexes from yeast to human that regulate transcription both through their enzymatic HAT activity and also through interacting with chromatin [74,75]. The BRD in HATs can broadly recognize acetyl-lysines mainly on H3 and H4 tails but also on H2A/H2B and non-histone factors [55]. For example, GCN5 and PCAF primarily acetylate H3 tails, but their BRDs bind H3ac and H4ac. This suggests the BRD may promote HAT activity by binding certain acetylated residues that would then promote the acetylation of other targets. Interestingly, these HATs are all recruited to DNA damage, where they participate in various aspects of the DDR including NER, DSB repair, and checkpoint regulation [21,41,44,76–78]. The DDR function of BRDs within these HATs is uncharacterized. Future studies identifying both HAT targets and BRD recognition signals will be essential to understand how HATs coordinate their catalytic activities with BRD reader capabilities to promote the DDR.
BRD protein components of chromatin remodeling complexes in the DDR
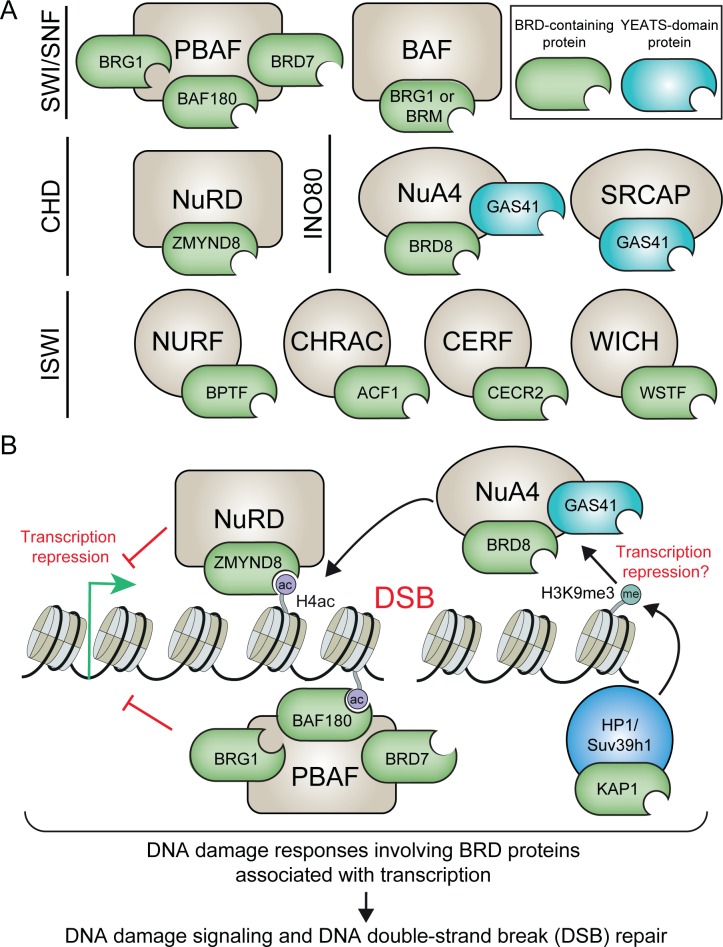
ATP-dependent chromatin remodeling complexes control the accessibility of chromatin factors to DNA by disrupting DNA–histone interaction, sliding/evicting nucleosome, or altering nucleosome composition, thereby regulating chromatin-based processes including transcription and DNA repair [79,80]. Chromatin remodeling complexes are organized into four principal families: switching defective/sucrose nonfermenting (SWI/SNF), imitation switch (ISWI), chromodomain, helicase, DNA binding (CHD), and inositol requiring 80 (INO80) [79]. In mammalian cells, ten BRD proteins have been identified within these four chromatin remodeling families (Fig 2A) [38,41,79]. Acetylation reader activities appear to represent a universal component of these complexes, and several studies are now defining their DDR functions within chromatin remodeling complexes.
Fig 2.
Acety-lysine readers within (A) chromatin remodeling complexes and (B) BRD protein pathways involving DNA damage within transcriptionally active chromatin. (A and B). Abbreviations: BRD, bromodomain; DSB, DNA double-strand break; ac, acetylation; me, methylation. Common and alternative names include: BRG1 (SMARCA4), BRM (SMARCA2), BAF180 (PBRM1), ACF1 (BAZ1A), WSTF (BAZ1B), KAP1 (TRIM28).
The mammalian SWI/SNF complexes BAF and PBAF contain BRD proteins including BRG1 (SMARCA4) and BRM (SMARCA2) that are involved in the DDR [81]. The ATPase subunit BRG1 functions in both complexes, whereas the ATPase BRM is exclusive to BAF. BRG1 is recruited to DNA damage and functions in the DDR, including through the binding of DSB-associated nucleosomes containing γH2AX, which requires the BRD to recognize GCN5-dependent H3ac [82–87]. BRM associates with DNA damage in a p300/CBP-dependent manner, which facilitates the recruitment of KU70 for NHEJ [41,44]. The role of BRM BRD in the DDR is unknown. PBAF contains two additional BRD proteins: BAF180 (PBRM1) and BRD7. BAF180 encodes six BRDs and is linked to the DDR [42,88,89]. With BRG1, BAF180 regulates PBAF in damage-induced transcription repression (see below) [42]. BAF180 also promotes cohesion, a function impaired upon mutations within its two BRDs, which leads to genome instability [89]. BRD7 interacts with BRCA1 and regulates p53, suggesting its involvement in the DDR [90–92].
The chromatin remodeler ISWI exists in many complexes that contain multiple BRD proteins [93]. The BRD protein ACF1 (BAZ1A) is a non-catalytic subunit of CHRAC and ACF ISWI complexes. ACF1 is recruited to damage, where it helps recruit KU for NHEJ [94]. ACF1 also regulates the G2/M DNA damage checkpoint [95]. Although ACF1 damage-recruitment is independent from its BRD, its chromatin binding domains could still promote DDR functions. WSTF (BAZ1B) is specific for the WICH ISWI complex [96]. WSTF is recruited to DNA damage where it regulates γH2AX as a kinase that targets H2AX Y142 [41,95,97]. The potential interplay between remodeling, acetylation binding, and kinase activity of this BRD DDR factor is worth investigating. ACF1 and WSTF are also recruited to UV-C laser irradiation and function in NER [98]. Currently, acetylation targets of ACF1 or WSTF BRDs remain unidentified, making mechanistic interpretations of these results challenging. The ISWI complex NURF also contains Bromodomain PHD Finger Transcription Factor (BPTF) that is recruited to DNA damage [41]. The BRD of BPTF recognizes H4K16ac to regulate transcription [99]. It will be interesting to determine whether BPTF participates in an acetylation-dependent DDR activity involving H4K16ac.
The CHD member nucleosome remodeling and histone deacetylase (NuRD) plays well-established roles in the DDR [100–103]. Although the canonical NuRD complex lacks any BRD protein, multiple studies have identified BRD proteins associated with this complex at DNA damage sites. For example, ZNF827 can specifically recruit NuRD to ALT telomere to regulate HR [104], whereas the BRD protein ZMYND8 recruits NuRD to DSBs within actively transcribing chromatin [41]. The INO80 chromatin remodeling family includes the NuA4 complex, which displays various DDR functions [45,63,79]. In addition to remodeling activity by p400, NuA4 also acetylates histones through its associated HAT activity by TIP60. Both chromatin remodeling and HAT activities play critical roles in DNA damage signaling and repair, including promoting HR and suppressing alternative-NHEJ [47,105–109]. The NuA4 complex contains the BRD protein BRD8 as well as GAS41, a YEATS protein with acetyl-lysine recognition capabilities [79,110]. These acetylation readers have the potential to coordinate the DDR functions of NuA4, although further studies are needed to elucidate the mechanistic details of how these large multi-subunit complexes promote the DDR in the context of acetylation. Thus, these studies highlight the involvement of BRD proteins within chromatin remodelers that have the potential to link acetylation signaling with the DDR.
Transcription, DNA damage, and BRD proteins
Transcription can be hindered by DNA damage, requiring molecular retooling of the chromatin environment to avoid conflicts between DNA repair and active transcription. Indeed, transcriptionally active genes located nearby DSBs are repressed by the DDR kinases ATM [111] and DNA-PK [112] to facilitate DSB repair. Acetylation readers have been identified that coordinate the DDR within transcriptionally active chromatin. The chromatin remodeling PBAF complex (SWI/SNF-B), which contains two BRD proteins BAF180 and BRG1, silences transcription upon DSBs and promotes NHEJ [42]. Although acetylation targets for the complex were not identified, point mutations identified in cancer genomes in BRDs of BAF180 rendered this complex defective for transcription silencing. The BRD protein ZMYND8 plays key roles in damage-induced transcriptional repression [41]. Upon DNA damage specifically within actively transcribing chromatin, ZMYND8 is recruited through its BRD to TIP60-mediated H4 acetylations. ZMYND8 associates with the NuRD complex and facilitates its accumulation at damage sites to mediate transcriptional repression and promote HR repair [41]. KAP1 participates in damage-induced formation of repressive chromatin. KAP1 is found in a methyltransferase complex with HP1 and Suv39h1, which promote H3K9me3 upon DSBs to form repressive chromatin transiently at break sites [113]. How KAP1-dependent H3K9me3 affects transcription within these damaged regions is unclear. These examples provide key examples for how BRD proteins participate in transcription responses to DNA damage (Fig 2B). Whether these pathways are coordinated or act independently at DSBs across varied chromatin landscapes needs further investigation. Interestingly, NuRD and KAP1 complexes are associated with gene repression, whereas PBAF is associated with active transcription. Identifying DDR-specific regulatory cues that switch these transcriptional regulators into DNA damage factors is needed to create a full view of the interplay between transcription and the DDR. It seems clear that transcription-associated DDR complexes must perform their normal functions in gene regulation while also being able to promote DNA repair of damaged DNA within transcriptionally active chromatin.
Non-BRD Acetylation Binding Proteins in the DDR
Several domains in addition to the BRD can bind acetylated lysines, including double PHD fingers [114], tandem PHD fingers [115,116], double pleckstrin homology (PH) domain [117], and the YEATS domain [39]. Four YEATS-containing proteins have been identified in humans and their functions in various cellular processes identified [118]. Several YEATS proteins participate in the DDR. In yeast, the YEATS domain of Taf14 binds H3K9ac, a mark that is responsive to DNA damage in human cells [27]. Disruption of Taf14 acetylation interactions impairs the DDR and sensitizes cells to DNA damaging agents [119]. The human YEATS protein ENL, a transcriptional elongation factor, is phosphorylated by ATM in response to DSBs. Phosphorylated ENL interacts with Polycomb Repressive Complex 1 specifically at DSBs to promote lesion-induced transcriptional repression [120]. Loss of ENL and transcriptional repression at DSB sites reduces the association of the NHEJ factor KU with damage sites, suggesting that this pathway acts to repress transcription in the presence of a DSB to allow its repair. Yeast Yaf9 and its close human homolog GAS41 are conserved components of the NuA4, a complex involved in the DDR (Fig 2) [105,107,118,121]. The DNA-dependent protein kinase, catalytic subunit, DNA-PKcs, contains a bromodomain-like module that recognizes TIP60-dependent H2AX K5-Ac to promote the formation of γH2AX at DSBs [122,123]. Thus, acetylation readers in addition to BRD proteins are also critical facilitators of the DDR.
Cancer Epigenetics: Acetylation Signaling
Acetylation modifiers and readers in cancer
Changes in acetylation signaling resulting from misregulated HATs or HDACs can cause abnormal gene expression patterns, including activation of proto-oncogenes and silencing of tumor suppressor genes [74,124,125] as well as impair DNA damage responses [21], which collectively can impact genome–epigenome stability (Fig 3). Altered acetylation signaling pathways have been identified in numerous cancers. Various genetic alterations in HATs have been found in hematological and solid cancers [124]. HATs can exhibit altered substrate targeting through mislocalization, aberrant protein interactions, or remodeled activities that can disrupt normal cell function. For instance, inactivating or truncated mutations in CBP/p300 HAT catalytic domains, which impair acetylation of H3K18 and non-histone substrates BCL6 and p53, have been identified in cancers [126–128]. BCL6 and p53 are transcriptional regulators involved in the DDR, and decreased acetylated BCL6 and p53 may enhance DNA damage tolerances, which can favor cancer survival [129–131]. Of note, no significant gene expression differences were found in mutant versus wt CBP/p300 containing small cell lung cancers, suggesting that mechanisms other than altered gene expression may contribute to these cancer-associated HAT mutations [128]. Given the broad involvement of CBP/p300 in gene regulation and the DDR, it remains unclear the primary cellular targets of these HATs in cancer. Analysis of genome stability pathways and acetylation reader associations with chromatin in CBP/p300 mutant cancers could provide answers to these outstanding questions.
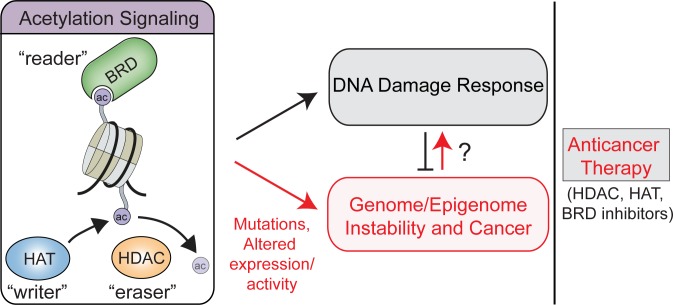
Fig 3. Model for the contribution of acetylation signaling in the DDR, cancer, and anticancer therapies that target epigenetic acetylation pathways.
Abbreviations: ac, acetylation; BRD, bromodomain; HAT, histone acetyltransferase; HDAC, histone deacetylase.
Decreased histone acetylation levels are most frequently observed in cancer [132], which, in addition to defective HAT activity, can also be a consequence of hyper-active HDACs. Altered HDAC levels, activities, and recruitment occur in cancer and all provide potential mechanisms to facilitate tumorigenesis [125,133]. For example, HDAC1 and HDAC2 expression is often enhanced in many cancers and has been correlated with transcriptional repression of tumor suppressors, including p21 [134,135]. Global reductions of H4K16ac have been identified in various cancer cell lines and tumors as being associated with tumor progression [136]. Because HDAC1 and HDAC2 deacetylate H4K16 to promote DSB repair [49], it is tempting to speculate that the observed HDAC1/HDAC2 overexpression and hypoacetylated H4K16 in cancers could deregulate the DDR, resulting in genetic and epigenetic instability that would promote tumorigenesis [137,138]. Although many HATs and HDACs are altered in cancer and concomitant histone modification changes are observed, mechanistic studies are needed to provide molecular insights into the relationship between epigenetic changes and the DDR that are involved in cancer.
Mutated or misregulated readers of acetylation, including BRD proteins, are involved in several different cancer types. The Mixed Lineage Leukemia (MLL) protein, a methyltransferase containing a BRD, is commonly fused with other chromatin proteins in haematological cancers and is able to induce leukemic transformation [139]. A non-MLL BRD contained within the MLL fusion or the loss of the BRD of MLL, a common event in MLL onco-fusion proteins, may also contribute to leukemogenesis [140–142]. For example, domain-swapping experiments identified a specific function of the CBP BRD in MLL–CBP-induced acute myeloid leukemia (AML) [143]. Recently, the development of inhibitors that block BRD acetyl-lysine interactions has uncovered the role of BRDs in several cancers [17,72]. In nuclear protein of the testis (NUT) midline carcinoma (NMC), BRD4 can be fused with the NUT protein [144,145], and these fusions are tethered to acetylated chromatin by BRDs. Inhibition of BRD-NUT by small molecule BET BRD inhibitors promoted differentiation and decreased MYC expression to inhibit proliferation [58,146]. In leukemia with MLL fusions, BRD4 can interact with MLL fusions and associated proteins, including the super elongation complex (SEC) and the polymerase-associated factor complex (PAFc), two critical regulators of transcriptional elongation [59]. Blocking BRD4 chromatin recruitment can constrain MYC and the expression of its target genes to inhibit proliferation and promote apoptosis [59,147–149]. TRIM BRD proteins have been linked to cancer [67], and the chromatin binding module including the BRD of TRIM24 has been implicated in transcription regulation of cancer-specific genes in breast [69] and prostate cancers [150]. Thus, BRD proteins are involved in several cancers, but whether their DDR activities also play a role in tumorigenesis remains an open question.
Components of several chromatin-remodeling complexes are highly mutated in cancer, including BRG1 and BAF180 in SWI/SNF and CHD4 in NuRD [110,151]. Mutations within these complexes have been associated with aberrant gene expression and genome instability [152,153], although it is not fully understood if DDR functions of these mutated complexes impact cancer. The BRD protein ZMYND8 is a component of the NuRD complex that, along with CHD4, displays mutations or altered expression in various cancers [154–158]. Aberrant regulation of the ZMYND8-NuRD DDR pathway could disrupt its transcription and DNA repair functions, resulting in genome instability [41]. In addition, enhancer-bound ZMYND8 has been shown to act as a controller for expression of enhancer RNA (eRNA), which are short, non-coding RNAs transcribed from enhancers [159]. Loss of ZMYND8 resulted in increased eRNA expression and hyper-enhancer activity that promoted cancer phenotypes [158]. The BRD protein ZMYND11, a paralog of ZMYND8, is an H3.3K36me3 reader that represses gene expression by regulating transcriptional elongation [160] and mRNA splicing [161]. Although ZMYND11 DDR functions are unknown, it appears to play a central role in cancer suppression. Indeed, ZMYND11 depletion caused up-regulation of MYC, and enhanced proliferation in cancer cells and mutations of ZMYND11 were identified in several cancers [160]. Interestingly, some mutations in the histone H3.3 variant, including at K36, have been identified in cancer, including chondroblastomas, in which high levels of mutations in H3.3 K36 and defects in HR repair are observed [162]. It is tempting to speculate that the tumor suppression of ZMYND11 could be linked to the DDR. Additional work is needed to understand the relationship between H3K36 methylation, the DDR, and cancer, information that could be clinically relevant for drugs targeting these pathways [163].
These studies highlight the involvement of acetylation “writers,” “erasers,” and “readers” in cancer. Although regulation of gene expression is most commonly associated with acetylation signaling, these pathways also play crucial roles in the DDR that promote genome and epigenome maintenance (Fig 3). How alterations in these pathways relate to DDR defects and their contributions to tumorigenesis is poorly understood. Defective DNA repair pathways are prevalent in many cancers, and genome instability is considered a hallmark of cancer. Although defects in acetylation signaling pathways could promote mutations and genome instability, these dysfunctional pathways might also provide opportunities to treat these cancers with epigenetic drugs, targeting acetylation signaling pathways as well as therapies that target cancer cells with defective DDR capacities (Fig 3). For example, PARP inhibitors can cause synthetic lethality to kill HR-defective tumors (i.e., BRCA1 and BRCA2 mutant cancers). Cancers exhibiting impaired acetylation signaling pathways resulting in defective DNA repair could be similarly treated. Understanding how DDR pathways are deregulated in acetylation signaling defective cancers can provide insights into the development and use of therapeutic strategies targeting the DDR [164].
Targeting acetylation signaling for cancer therapy
Deregulation of acetylation signaling in cancer has been intensively investigated and led to the development of a wide variety of small molecule inhibitors targeting acetylation signaling pathways (Table 2). The recent emergence of the importance of HATs, HDACs, and BRD proteins as key mediators of the DDR and genome maintenance (Table 1) requires an understanding of how these small molecule inhibitors impact the genome integrity functions of these pathways [21]. This information can reveal new insights into their drug mechanisms, which could provide a framework for considering opportunities for combinatorial treatment using these inhibitors with traditional cancer therapies. So far, HDAC inhibitors are one of the most well-characterized epigenome-targeting drugs and show promising therapeutic efficacy toward some cancers. HDAC inhibitors are known in some cases to directly modulate the cancer epigenome, leading to changes in gene expression profiles, an effect that is proposed to promote cell cycle arrest and cell death. HDAC inhibitors can also suppress DNA damage repair capacity in cancer cells. HDAC inhibitors reduced the expression level of key repair proteins such as KU proteins in NHEJ and RAD50 in HR in various cancer cell lines [165–169]. HDAC inhibitors also elevated reactive oxygen species (ROS) levels, a potential source for DNA damage [170,171]. Because HDACs regulate the DDR, including HDAC1/2, inhibition of these HDACs resulted in hyperacetyled H4K16 and H3K56 along with defective NHEJ, thereby impairing the DDR [49]. Thus, HDAC inhibitors impair the DDR in several ways, causing sensitization of cancer cells to DNA damaging agents including radiation (Table 2) [165–167]. HAT inhibitors including curcumin [172,173] also sensitize cells to DNA damaging agents (Table 2). However, in contrast to HDAC inhibitors, the progress of HAT inhibitors in cancer treatment has been slower due to their pleiotropic effects and poor bioavailability [174]. The potent and selective HAT inhibitor C646 has displayed antitumor efficacy in several cancer types and inhibition of DNA repair (Table 2) [175,176]. Additional studies identifying the anticancer drug mechanism of these acetylation signaling inhibitors are important to aid in the further development of acetylation signaling inhibitors for the treatment of cancer.
Table 2. Effects on DDR by small molecule inhibitors targeting acetylation signaling.
| Family | Target | Inhibitors | Phase | Tested cancer type | Effects on the DDR | References |
|---|---|---|---|---|---|---|
| HAT | CBP/p300 | Curcumin | Clinical | multiple | PARPi, CPT and HU sensitivity; (-) BRCA1 mRNA; (-) ATR activity; (-) Ku70/Ku80 recruitment | [172] |
| p300 | C646 | Preclinical | melanoma | Cisplatin sensitivity; (-) DNA repair genes mRNA i.e., Rad51; (-) γH2AX | [176] | |
| lung | IR sensitivity; (-) IR-p-CHK1 | [199] | ||||
| HDAC | Class I/II | Vorinostat (SAHA) | FDA approved in 2006 | multiple | IR sensitivity; (-) HR & NHEJ proteins, i.e., Rad50, Ku70; (+) γH2AX IRIF; (-) IR-induced HR & NHEJ proteins, i.e., Rad51, Ku80 | [165–167,200,201] |
| Class I | Romidepsin (FK228) | FDA approved in 2009 | ovarian | (+) γH2AX/Rad51/53BP1 foci with cisplatin c.t., and in s.c. | [202] | |
| lung | IR sensitivity; (+) IR-γH2AX | [203] | ||||
| thyroid | (+)γH2AX/ROS; (-) Ku70/80 and Rad51; (+) γH2AX in s.c. | [168] | ||||
| renal cell | (+) γH2AX/ROS with 5-FU c.t. and in s.c. | [204] | ||||
| Class I/II | Panobinostat (LBH589) | FDA approved in 2015 | lung, bladder | IR sensitivity; (+) IR-γH2AX; (-) Mre11/Nbs1/ Rad51 protein | [205,206] | |
| leukemia | (+) γH2AX; (-) DNA repair protein/signaling, i.e., Chk1/p-Chk1 with TOPIIi c.t. | [207,208] | ||||
| BRD | BET family | JQ1 | Preclinical | NMC, myeloma, prostate | - | [58,147,209] |
| glioma | (-) IR-γH2AX | [71] | ||||
| leukemia | (+) γH2AX & 53BP1 foci | [210] | ||||
| I-BET151 | Preclinical | leukemia, myeloma | - | [59,179] | ||
| I-BET762 | Clinical | myeloma | - | [177,179] | ||
| CBP/p300 | I-CBP112 | Preclinical | leukemia | Dox sensitivity; (+) γH2AX foci with JQ1 c.t. | [180] |
The inhibitor, clinical phase, cancer type, and effects on the DDR are provided for drug targeting of acetylation signaling factors. Abbreviations: CPT, camptothecin; c.t., co-treatment; Dox, doxorubicin; HU, hydroxyurea; IR, ionizing radiation; IRIF, ionizing radiation induced foci; PARPi, PARP inhibitor; ROS, reactive oxygen species; s.c., subcutaneous xenograft tumor model; TOPIIi, Topoisomerase II inhibitor; 5-FU, 5-fluorouracil. (-) indicates decreased levels; (+) indicates increased levels.
Small molecule inhibitors targeting the BRD have drawn significant attention for new classes of anticancer drugs. For example, JQ1, a potent BRD4 inhibitor, has displayed anti-tumor activities in preclinical studies [58]. In addition to JQ1, a variety of other BET family BRD inhibitors, including I-BET151 [59] and I-BET762 [177], show therapeutic promise in hematological malignancies [178]. These inhibitors have been shown to inhibit BRD4 chromatin interactions, thereby blocking MYC-mediated tumor growth and survival [59,147,179]. Reduced BRD4 function by JQ1 also alters the cellular response to ionizing radiation [71], indicating that BRD4 inhibition also impacts the DDR. A p300/CBP BRD inhibitor I-CBP112 showed enhanced topoisomerase inhibitor doxorubicin induced cytotoxicity effect in leukemic cell lines [180]. I-CBP112 also displayed a synergistic cytotoxic effect with JQ1, as more γH2AX foci were observed in co-treated cells (Table 2). These results suggest that CBP/p300 and BRD4 may function in different DDR pathways, thus providing selectivity for these inhibitors in either CBP/p300 mutant cells with BRD4 inhibitors or vice versa. These studies emphasize the promise of drugging acetylation signaling in cancer. As numerous BRD proteins are involved in the DDR, it will be vital to understand whether these BRD inhibitors impact DNA damage signaling/repair and whether these effects are advantageous or inhibitory towards the use of these compounds at therapeutic agents in cancer (Fig 3).
Summary
Mounting evidence highlights the crucial function of acetylation signaling in regulating the DDR and maintaining genome integrity. Acetylation reader proteins, including BRD proteins, are vital effector proteins for acetylated lysines that recognize and read these signals to orchestrate the DDR. Understanding the mechanisms by which acetylation reader proteins promote chromatin-based responses to DNA damage can provide critical insights into understanding how genome–epigenome maintenance is achieved. Genome instability is common in cancer as is defective acetylation signaling pathways. Whether altered acetylation signaling is causal or merely correlative for genome instability in cancer remains an important question. How acetylation signaling impacts cancer epigenetics and its contributions to the DDR and cancer treatments warrants further investigations. Given the rapid development of epigenetic drugs targeting acetylation signaling, including BRD inhibitors, it is critical to further decipher how inhibitors of acetylation signaling affect the DDR and whether this information can be leveraged to improve the use of these drugs in cancer treatment. We envision that a deeper understanding of how acetylation signaling is involved in the DDR and in cancer will help develop targeted therapies using epigenetic drugs either alone or in combination with DNA damaging agents to improve cancer treatments.
Funding Statement
This work in the K.M.M. laboratory was supported by the Cancer Prevention Research Institute of Texas (CPRIT, grant no. R1116) and the NIH National Cancer Institute grants CA198279 and CA201268. K.M.M. is a CPRIT scholar. The funders had no role in the preparation of the article.
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–8. 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40(2):179–204. 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Molecular cell. 2012;47(4):497–510. 10.1016/j.molcel.2012.07.029 [DOI] [PubMed] [Google Scholar]
- 4.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual review of biochemistry. 2010;79:181–211. 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nature structural & molecular biology. 2010;17(1):11–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Symington LS. Mechanism and regulation of DNA end resection in eukaryotes. Critical reviews in biochemistry and molecular biology. 2016:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability—an evolving hallmark of cancer. Nature reviews Molecular cell biology. 2010;11(3):220–8. 10.1038/nrm2858 [DOI] [PubMed] [Google Scholar]
- 8.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nature reviews Genetics. 2010;11(4):285–96. 10.1038/nrg2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184(4139):868–71. [DOI] [PubMed] [Google Scholar]
- 10.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annual review of biochemistry. 2011;80:473–99. 10.1146/annurev-biochem-061809-175347 [DOI] [PubMed] [Google Scholar]
- 11.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. [DOI] [PubMed] [Google Scholar]
- 12.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Molecular cell. 2008;29(6):653–63. 10.1016/j.molcel.2008.02.014 [DOI] [PubMed] [Google Scholar]
- 13.Campos EI, Reinberg D. Histones: annotating chromatin. Annual review of genetics. 2009;43:559–99. 10.1146/annurev.genet.032608.103928 [DOI] [PubMed] [Google Scholar]
- 14.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annual review of biochemistry. 2007;76:75–100. [DOI] [PubMed] [Google Scholar]
- 15.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nature reviews Molecular cell biology. 2007;8(12):983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nature structural & molecular biology. 2012;19(12):1218–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. The New England journal of medicine. 2012;367(7):647–57. 10.1056/NEJMra1112635 [DOI] [PubMed] [Google Scholar]
- 18.Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nature cell biology. 2011;13(10):1161–9. 10.1038/ncb2344 [DOI] [PubMed] [Google Scholar]
- 19.Miller KM, Jackson SP. Histone marks: repairing DNA breaks within the context of chromatin. Biochemical Society transactions. 2012;40(2):370–6. 10.1042/BST20110747 [DOI] [PubMed] [Google Scholar]
- 20.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Molecular cell. 2013;49(5):795–807. 10.1016/j.molcel.2013.01.017 [DOI] [PubMed] [Google Scholar]
- 21.Gong F, Miller KM. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutation research. 2013;750(1–2):23–30. 10.1016/j.mrfmmm.2013.07.002 [DOI] [PubMed] [Google Scholar]
- 22.Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, et al. Global chromatin compaction limits the strength of the DNA damage response. The Journal of cell biology. 2007;178(7):1101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess RC, Burman B, Kruhlak MJ, Misteli T. Activation of DNA damage response signaling by condensed chromatin. Cell reports. 2014;9(5):1703–17. 10.1016/j.celrep.2014.10.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, et al. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell reports. 2014;8(4):1049–62. 10.1016/j.celrep.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes & development. 2011;25(5):409–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, et al. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 2008;132(2):221–32. 10.1016/j.cell.2007.12.013 [DOI] [PubMed] [Google Scholar]
- 27.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. The EMBO journal. 2009;28(13):1878–89. 10.1038/emboj.2009.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123(7):1213–26. [DOI] [PubMed] [Google Scholar]
- 29.Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends in cell biology. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nature reviews Molecular cell biology. 2014;15(1):7–18. 10.1038/nrm3719 [DOI] [PubMed] [Google Scholar]
- 31.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127(7):1361–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499(7456):50–4. 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends in cell biology. 2009;19(5):207–17. 10.1016/j.tcb.2009.03.001 [DOI] [PubMed] [Google Scholar]
- 34.Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nature reviews Molecular cell biology. 2015;16(4):258–64. 10.1038/nrm3931 [DOI] [PubMed] [Google Scholar]
- 35.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nature reviews Molecular cell biology. 2007;8(4):284–95. [DOI] [PubMed] [Google Scholar]
- 36.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nature reviews Molecular cell biology. 2008;9(3):206–18. 10.1038/nrm2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311(5762):844–7. [DOI] [PubMed] [Google Scholar]
- 38.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149(1):214–31. 10.1016/j.cell.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell. 2014;159(3):558–71. 10.1016/j.cell.2014.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramanathan B, Smerdon MJ. Changes in nuclear protein acetylation in u.v.-damaged human cells. Carcinogenesis. 1986;7(7):1087–94. [DOI] [PubMed] [Google Scholar]
- 41.Gong F, Chiu LY, Cox B, Aymard F, Clouaire T, Leung JW, et al. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes & development. 2015;29(2):197–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Kunzel J, et al. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Molecular cell. 2014;55(5):723–32. 10.1016/j.molcel.2014.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kulkarni A, Oza J, Yao M, Sohail H, Ginjala V, Tomas-Loba A, et al. Tripartite Motif-containing 33 (TRIM33) Protein Functions in the Poly(ADP-ribose) Polymerase (PARP)-dependent DNA Damage Response through Interaction with Amplified in Liver Cancer 1 (ALC1) Protein. The Journal of biological chemistry. 2013;288(45):32357–69. 10.1074/jbc.M113.459164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S, et al. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 2011;30(18):2135–46. 10.1038/onc.2010.592; [DOI] [PubMed] [Google Scholar]
- 45.Gursoy-Yuzugullu O, House N, Price BD. Patching Broken DNA: Nucleosome Dynamics and the Repair of DNA Breaks. Journal of molecular biology. 2016;428(9 Pt B):1846–60. 10.1016/j.jmb.2015.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Molecular and cellular biology. 2007;27(24):8502–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nature structural & molecular biology. 2013;20(3):317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert JP, Roques C, Pandita RK, et al. The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Molecular cell. 2016;62(3):409–21. 10.1016/j.molcel.2016.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nature structural & molecular biology. 2010;17(9):1144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Molecular and cellular biology. 2010;30(14):3582–95. 10.1128/MCB.01476-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, et al. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Molecular and cellular biology. 2010;30(22):5335–47. 10.1128/MCB.00350-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta A, Hunt CR, Hegde ML, Chakraborty S, Chakraborty S, Udayakumar D, et al. MOF phosphorylation by ATM regulates 53BP1-mediated double-strand break repair pathway choice. Cell reports. 2014;8(1):177–89. 10.1016/j.celrep.2014.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Molecular cell. 2013;51(4):454–68. 10.1016/j.molcel.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329(5997):1348–53. 10.1126/science.1192049 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS letters. 2012;586(17):2692–704. 10.1016/j.febslet.2012.04.045 [DOI] [PubMed] [Google Scholar]
- 56.Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harbor perspectives in biology. 2014;6(7):a018762 10.1101/cshperspect.a018762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muller S, Filippakopoulos P, Knapp S. Bromodomains as therapeutic targets. Expert reviews in molecular medicine. 2011;13:e29 10.1017/S1462399411001992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–73. 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–33. 10.1038/nature10509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barbieri I, Cannizzaro E, Dawson MA. Bromodomains as therapeutic targets in cancer. Briefings in functional genomics. 2013;12(3):219–30. 10.1093/bfgp/elt007 [DOI] [PubMed] [Google Scholar]
- 61.Zhang G, Smith SG, Zhou MM. Discovery of Chemical Inhibitors of Human Bromodomains. Chemical reviews. 2015;115(21):11625–68. 10.1021/acs.chemrev.5b00205 [DOI] [PubMed] [Google Scholar]
- 62.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Molecular cell. 2012;46(6):722–34. 10.1016/j.molcel.2012.06.002 [DOI] [PubMed] [Google Scholar]
- 63.Price BD, D'Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152(6):1344–54. 10.1016/j.cell.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nature cell biology. 2006;8(8):870–6. [DOI] [PubMed] [Google Scholar]
- 65.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Molecular cell. 2008;31(2):167–77. 10.1016/j.molcel.2008.05.017 [DOI] [PubMed] [Google Scholar]
- 66.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nature structural & molecular biology. 2011;18(7):831–9. [DOI] [PubMed] [Google Scholar]
- 67.Hatakeyama S. TRIM proteins and cancer. Nature reviews Cancer. 2011;11(11):792–804. 10.1038/nrc3139 [DOI] [PubMed] [Google Scholar]
- 68.Zeng L, Yap KL, Ivanov AV, Wang X, Mujtaba S, Plotnikova O, et al. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nature structural & molecular biology. 2008;15(6):626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468(7326):927–32. 10.1038/nature09542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Agricola E, Randall RA, Gaarenstroom T, Dupont S, Hill CS. Recruitment of TIF1gamma to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Molecular cell. 2011;43(1):85–96. 10.1016/j.molcel.2011.05.020 [DOI] [PubMed] [Google Scholar]
- 71.Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature. 2013;498(7453):246–50. 10.1038/nature12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Molecular cell. 2014;54(5):728–36. 10.1016/j.molcel.2014.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Devaiah BN, Case-Borden C, Gegonne A, Hsu CH, Chen Q, Meerzaman D, et al. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nature structural & molecular biology. 2016;23(6):540–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Farria A, Li W, Dent SY. KATs in cancer: functions and therapies. Oncogene. 2015;34(38):4901–13. 10.1038/onc.2014.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5(1):9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cazzalini O, Sommatis S, Tillhon M, Dutto I, Bachi A, Rapp A, et al. CBP and p300 acetylate PCNA to link its degradation with nucleotide excision repair synthesis. Nucleic acids research. 2014;42(13):8433–48. 10.1093/nar/gku533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo R, Chen J, Mitchell DL, Johnson DG. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic acids research. 2011;39(4):1390–7. 10.1093/nar/gkq983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Love IM, Sekaric P, Shi D, Grossman SR, Androphy EJ. The histone acetyltransferase PCAF regulates p21 transcription through stress-induced acetylation of histone H3. Cell Cycle. 2012;11(13):2458–66. 10.4161/cc.20864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annual review of biochemistry. 2009;78:273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- 80.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nature reviews Molecular cell biology. 2006;7(6):437–47. [DOI] [PubMed] [Google Scholar]
- 81.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nature reviews Cancer. 2011;11(7):481–92. 10.1038/nrc3068 [DOI] [PubMed] [Google Scholar]
- 82.Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. The EMBO journal. 2006;25(17):3986–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. The EMBO journal. 2010;29(8):1434–45. 10.1038/emboj.2010.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lans H, Marteijn JA, Vermeulen W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics & chromatin. 2012;5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang L, Zhang Q, Jones K, Patel M, Gong F. The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage. Cell Cycle. 2009;8(23):3953–9. [DOI] [PubMed] [Google Scholar]
- 86.Zhao Q, Wang QE, Ray A, Wani G, Han C, Milum K, et al. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. The Journal of biological chemistry. 2009;284(44):30424–32. 10.1074/jbc.M109.044982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gong F, Fahy D, Liu H, Wang W, Smerdon MJ. Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle. 2008;7(8):1067–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brownlee PM, Chambers AL, Oliver AW, Downs JA. Cancer and the bromodomains of BAF180. Biochemical Society transactions. 2012;40(2):364–9. 10.1042/BST20110754 [DOI] [PubMed] [Google Scholar]
- 89.Brownlee PM, Chambers AL, Cloney R, Bianchi A, Downs JA. BAF180 promotes cohesion and prevents genome instability and aneuploidy. Cell reports. 2014;6(6):973–81. 10.1016/j.celrep.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harte MT, O'Brien GJ, Ryan NM, Gorski JJ, Savage KI, Crawford NT, et al. BRD7, a subunit of SWI/SNF complexes, binds directly to BRCA1 and regulates BRCA1-dependent transcription. Cancer research. 2010;70(6):2538–47. 10.1158/0008-5472.CAN-09-2089 [DOI] [PubMed] [Google Scholar]
- 91.Burrows AE, Smogorzewska A, Elledge SJ. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(32):14280–5. 10.1073/pnas.1009559107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Drost J, Mantovani F, Tocco F, Elkon R, Comel A, Holstege H, et al. BRD7 is a candidate tumour suppressor gene required for p53 function. Nature cell biology. 2010;12(4):380–9. 10.1038/ncb2038 [DOI] [PubMed] [Google Scholar]
- 93.Aydin OZ, Vermeulen W, Lans H. ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle. 2014;13(19):3016–25. 10.4161/15384101.2014.956551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, et al. The ACF1 complex is required for DNA double-strand break repair in human cells. Molecular cell. 2010;40(6):976–87. 10.1016/j.molcel.2010.12.003 [DOI] [PubMed] [Google Scholar]
- 95.Sanchez-Molina S, Mortusewicz O, Bieber B, Auer S, Eckey M, Leonhardt H, et al. Role for hACF1 in the G2/M damage checkpoint. Nucleic acids research. 2011;39(19):8445–56. 10.1093/nar/gkr435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu X, Meng X, Morris CA, Keating MT. A novel human gene, WSTF, is deleted in Williams syndrome. Genomics. 1998;54(2):241–9. [DOI] [PubMed] [Google Scholar]
- 97.Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457(7225):57–62. 10.1038/nature07668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aydin OZ, Marteijn JA, Ribeiro-Silva C, Rodriguez Lopez A, Wijgers N, Smeenk G, et al. Human ISWI complexes are targeted by SMARCA5 ATPase and SLIDE domains to help resolve lesion-stalled transcription. Nucleic acids research. 2014;42(13):8473–85. 10.1093/nar/gku565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145(5):692–706. 10.1016/j.cell.2011.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(43):18475–80. 10.1073/pnas.1012946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. The Journal of cell biology. 2010;190(5):731–40. 10.1083/jcb.200912135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. The EMBO journal. 2010;29(18):3130–9. 10.1038/emboj.2010.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. The Journal of cell biology. 2010;190(5):741–9. 10.1083/jcb.201001048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Conomos D, Reddel RR, Pickett HA. NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nature structural & molecular biology. 2014;21(9):760–70. [DOI] [PubMed] [Google Scholar]
- 105.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nature cell biology. 2006;8(1):91–9. [DOI] [PubMed] [Google Scholar]
- 106.Sun Y, Jiang X, Price BD. Tip60: connecting chromatin to DNA damage signaling. Cell Cycle. 2010;9(5):930–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102(4):463–73. [DOI] [PubMed] [Google Scholar]
- 108.Courilleau C, Chailleux C, Jauneau A, Grimal F, Briois S, Boutet-Robinet E, et al. The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks. The Journal of cell biology. 2012;199(7):1067–81. 10.1083/jcb.201205059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taty-Taty GC, Chailleux C, Quaranta M, So A, Guirouilh-Barbat J, Lopez BS, et al. Control of alternative end joining by the chromatin remodeler p400 ATPase. Nucleic acids research. 2016;44(4):1657–68. 10.1093/nar/gkv1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mayes K, Qiu Z, Alhazmi A, Landry JW. ATP-dependent chromatin remodeling complexes as novel targets for cancer therapy. Advances in cancer research. 2014;121:183–233. 10.1016/B978-0-12-800249-0.00005-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141(6):970–81. 10.1016/j.cell.2010.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nature structural & molecular biology. 2012;19(3):276–82. [DOI] [PubMed] [Google Scholar]
- 113.Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(25):9169–74. 10.1073/pnas.1403565111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466(7303):258–62. 10.1038/nature09139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ali M, Yan K, Lalonde ME, Degerny C, Rothbart SB, Strahl BD, et al. Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. Journal of molecular biology. 2012;424(5):328–38. 10.1016/j.jmb.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Qiu Y, Liu L, Zhao C, Han C, Li F, Zhang J, et al. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes & development. 2012;26(12):1376–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Su D, Hu Q, Li Q, Thompson JR, Cui G, Fazly A, et al. Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106. Nature. 2012;483(7387):104–7. 10.1038/nature10861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schulze JM, Wang AY, Kobor MS. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2009;87(1):65–75. 10.1139/O08-111 [DOI] [PubMed] [Google Scholar]
- 119.Shanle EK, Andrews FH, Meriesh H, McDaniel SL, Dronamraju R, DiFiore JV, et al. Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response. Genes & development. 2015;29(17):1795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ui A, Nagaura Y, Yasui A. Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair. Molecular cell. 2015;58(3):468–82. 10.1016/j.molcel.2015.03.023 [DOI] [PubMed] [Google Scholar]
- 121.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419(6905):411–5. [DOI] [PubMed] [Google Scholar]
- 122.Wang L, Xie L, Ramachandran S, Lee Y, Yan Z, Zhou L, et al. Non-canonical Bromodomain within DNA-PKcs Promotes DNA Damage Response and Radioresistance through Recognizing an IR-Induced Acetyl-Lysine on H2AX. Chemistry & biology. 2015;22(7):849–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Molecular and cellular biology. 2007;27(20):7028–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Di Cerbo V, Schneider R. Cancers with wrong HATs: the impact of acetylation. Briefings in functional genomics. 2013;12(3):231–43. 10.1093/bfgp/els065 [DOI] [PubMed] [Google Scholar]
- 125.Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Molecular oncology. 2007;1(1):19–25. 10.1016/j.molonc.2007.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471(7337):235–9. 10.1038/nature09727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–95. 10.1038/nature09730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nature genetics. 2012;44(10):1104–10. 10.1038/ng.2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133(4):612–26. 10.1016/j.cell.2008.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nature genetics. 2002;32(4):606–13. [DOI] [PubMed] [Google Scholar]
- 131.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432(7017):635–9. [DOI] [PubMed] [Google Scholar]
- 132.Chervona Y, Costa M. Histone modifications and cancer: biomarkers of prognosis? American journal of cancer research. 2012;2(5):589–97. [PMC free article] [PubMed] [Google Scholar]
- 133.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. The Journal of clinical investigation. 2014;124(1):30–9. 10.1172/JCI69738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(5):1241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huang BH, Laban M, Leung CH, Lee L, Lee CK, Salto-Tellez M, et al. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell death and differentiation. 2005;12(4):395–404. [DOI] [PubMed] [Google Scholar]
- 136.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature genetics. 2005;37(4):391–400. [DOI] [PubMed] [Google Scholar]
- 137.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–70. [DOI] [PubMed] [Google Scholar]
- 138.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–13. [DOI] [PubMed] [Google Scholar]
- 139.Deshpande AJ, Bradner J, Armstrong SA. Chromatin modifications as therapeutic targets in MLL-rearranged leukemia. Trends in immunology. 2012;33(11):563–70. 10.1016/j.it.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang J, Muntean AG, Hess JL. ECSASB2 mediates MLL degradation during hematopoietic differentiation. Blood. 2012;119(5):1151–61. 10.1182/blood-2011-06-362079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bursen A, Schwabe K, Ruster B, Henschler R, Ruthardt M, Dingermann T, et al. The AF4.MLL fusion protein is capable of inducing ALL in mice without requirement of MLL.AF4. Blood. 2010;115(17):3570–9. 10.1182/blood-2009-06-229542 [DOI] [PubMed] [Google Scholar]
- 142.Meyer C, Hofmann J, Burmeister T, Groger D, Park TS, Emerenciano M, et al. The MLL recombinome of acute leukemias in 2013. Leukemia. 2013;27(11):2165–76. 10.1038/leu.2013.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Santillan DA, Theisler CM, Ryan AS, Popovic R, Stuart T, Zhou MM, et al. Bromodomain and histone acetyltransferase domain specificities control mixed lineage leukemia phenotype. Cancer research. 2006;66(20):10032–9. [DOI] [PubMed] [Google Scholar]
- 144.French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer research. 2003;63(2):304–7. [PubMed] [Google Scholar]
- 145.French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27(15):2237–42. [DOI] [PubMed] [Google Scholar]
- 146.Grayson AR, Walsh EM, Cameron MJ, Godec J, Ashworth T, Ambrose JM, et al. MYC, a downstream target of BRD-NUT, is necessary and sufficient for the blockade of differentiation in NUT midline carcinoma. Oncogene. 2014;33(13):1736–42. 10.1038/onc.2013.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–17. 10.1016/j.cell.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(40):16669–74. 10.1073/pnas.1108190108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–8. 10.1038/nature10334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Groner AC, Cato L, de Tribolet-Hardy J, Bernasocchi T, Janouskova H, Melchers D, et al. TRIM24 Is an Oncogenic Transcriptional Activator in Prostate Cancer. Cancer cell. 2016;29(6):846–58. 10.1016/j.ccell.2016.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Le Gallo M, O'Hara AJ, Rudd ML, Urick ME, Hansen NF, O'Neil NJ, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nature genetics. 2012;44(12):1310–5. 10.1038/ng.2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Helming KC, Wang X, Roberts CW. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer cell. 2014;26(3):309–17. 10.1016/j.ccr.2014.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nature reviews Cancer. 2011;11(8):588–96. 10.1038/nrc3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bierkens M, Krijgsman O, Wilting SM, Bosch L, Jaspers A, Meijer GA, et al. Focal aberrations indicate EYA2 and hsa-miR-375 as oncogene and tumor suppressor in cervical carcinogenesis. Genes, chromosomes & cancer. 2013;52(1):56–68. [DOI] [PubMed] [Google Scholar]
- 155.Panagopoulos I, Micci F, Thorsen J, Haugom L, Buechner J, Kerndrup G, et al. Fusion of ZMYND8 and RELA genes in acute erythroid leukemia. PLoS ONE. 2013;8(5):e63663 10.1371/journal.pone.0063663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Park J, Betel D, Gryfe R, Michalickova K, Di Nicola N, Gallinger S, et al. Mutation profiling of mismatch repair-deficient colorectal cncers using an in silico genome scan to identify coding microsatellites. Cancer research. 2002;62(5):1284–8. [PubMed] [Google Scholar]
- 157.Wada Y, Matsuura M, Sugawara M, Ushijima M, Miyata S, Nagasaki K, et al. Development of detection method for novel fusion gene using GeneChip exon array. Journal of clinical bioinformatics. 2014;4(1):3 10.1186/2043-9113-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shen H, Xu W, Guo R, Rong B, Gu L, Wang Z, et al. Suppression of Enhancer Overactivation by a RACK7-Histone Demethylase Complex. Cell. 2016;165(2):331–42. 10.1016/j.cell.2016.02.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–7. 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D, et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508(7495):263–8. 10.1038/nature13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Guo R, Zheng L, Park JW, Lv R, Chen H, Jiao F, et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Molecular cell. 2014;56(2):298–310. 10.1016/j.molcel.2014.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fang D, Gan H, Lee JH, Han J, Wang Z, Riester SM, et al. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science. 2016;352(6291):1344–8. 10.1126/science.aae0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pfister SX, Markkanen E, Jiang Y, Sarkar S, Woodcock M, Orlando G, et al. Inhibiting WEE1 Selectively Kills Histone H3K36me3-Deficient Cancers by dNTP Starvation. Cancer cell. 2015;28(5):557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nature reviews Cancer. 2008;8(3):193–204. 10.1038/nrc2342 [DOI] [PubMed] [Google Scholar]
- 165.Blattmann C, Oertel S, Ehemann V, Thiemann M, Huber PE, Bischof M, et al. Enhancement of radiation response in osteosarcoma and rhabdomyosarcoma cell lines by histone deacetylase inhibition. International journal of radiation oncology, biology, physics. 2010;78(1):237–45. 10.1016/j.ijrobp.2010.03.010 [DOI] [PubMed] [Google Scholar]
- 166.Chen X, Wong P, Radany EH, Stark JM, Laulier C, Wong JY. Suberoylanilide hydroxamic acid as a radiosensitizer through modulation of RAD51 protein and inhibition of homology-directed repair in multiple myeloma. Molecular cancer research: MCR. 2012;10(8):1052–64. 10.1158/1541-7786.MCR-11-0587 [DOI] [PubMed] [Google Scholar]
- 167.Chinnaiyan P, Vallabhaneni G, Armstrong E, Huang SM, Harari PM. Modulation of radiation response by histone deacetylase inhibition. International journal of radiation oncology, biology, physics. 2005;62(1):223–9. [DOI] [PubMed] [Google Scholar]
- 168.Lin SF, Lin JD, Chou TC, Huang YY, Wong RJ. Utility of a histone deacetylase inhibitor (PXD101) for thyroid cancer treatment. PLoS ONE. 2013;8(10):e77684 10.1371/journal.pone.0077684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Munshi A, Kurland JF, Nishikawa T, Tanaka T, Hobbs ML, Tucker SL, et al. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11(13):4912–22. [DOI] [PubMed] [Google Scholar]
- 170.Butler LM, Zhou X, Xu WS, Scher HI, Rifkind RA, Marks PA, et al. The histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ungerstedt JS, Sowa Y, Xu WS, Shao Y, Dokmanovic M, Perez G, et al. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(3):673–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ogiwara H, Ui A, Shiotani B, Zou L, Yasui A, Kohno T. Curcumin suppresses multiple DNA damage response pathways and has potency as a sensitizer to PARP inhibitor. Carcinogenesis. 2013;34(11):2486–97. 10.1093/carcin/bgt240 [DOI] [PubMed] [Google Scholar]
- 173.Oike T, Ogiwara H, Amornwichet N, Nakano T, Kohno T. Chromatin-regulating proteins as targets for cancer therapy. Journal of radiation research. 2014;55(4):613–28. 10.1093/jrr/rrt227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Furdas SD, Kannan S, Sippl W, Jung M. Small molecule inhibitors of histone acetyltransferases as epigenetic tools and drug candidates. Archiv der Pharmazie. 2012;345(1):7–21. 10.1002/ardp.201100209 [DOI] [PubMed] [Google Scholar]
- 175.Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chemistry & biology. 2010;17(5):471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yan G, Eller MS, Elm C, Larocca CA, Ryu B, Panova IP, et al. Selective inhibition of p300 HAT blocks cell cycle progression, induces cellular senescence, and inhibits the DNA damage response in melanoma cells. The Journal of investigative dermatology. 2013;133(10):2444–52. 10.1038/jid.2013.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468(7327):1119–23. 10.1038/nature09589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chaidos A, Caputo V, Karadimitris A. Inhibition of bromodomain and extra-terminal proteins (BET) as a potential therapeutic approach in haematological malignancies: emerging preclinical and clinical evidence. Therapeutic advances in hematology. 2015;6(3):128–41. 10.1177/2040620715576662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chaidos A, Caputo V, Gouvedenou K, Liu B, Marigo I, Chaudhry MS, et al. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood. 2014;123(5):697–705. 10.1182/blood-2013-01-478420 [DOI] [PubMed] [Google Scholar]
- 180.Picaud S, Fedorov O, Thanasopoulou A, Leonards K, Jones K, Meier J, et al. Generation of a Selective Small Molecule Inhibitor of the CBP/p300 Bromodomain for Leukemia Therapy. Cancer research. 2015;75(23):5106–19. 10.1158/0008-5472.CAN-15-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Revenko AS, Kalashnikova EV, Gemo AT, Zou JX, Chen HW. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Molecular and cellular biology. 2010;30(22):5260–72. 10.1128/MCB.00484-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Caron C, Lestrat C, Marsal S, Escoffier E, Curtet S, Virolle V, et al. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene. 2010;29(37):5171–81. 10.1038/onc.2010.259 [DOI] [PubMed] [Google Scholar]
- 183.Sun H, Liu J, Zhang J, Shen W, Huang H, Xu C, et al. Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4. Biochemical and biophysical research communications. 2007;358(2):435–41. [DOI] [PubMed] [Google Scholar]
- 184.Poplawski A, Hu K, Lee W, Natesan S, Peng D, Carlson S, et al. Molecular insights into the recognition of N-terminal histone modifications by the BRPF1 bromodomain. Journal of molecular biology. 2014;426(8):1661–76. 10.1016/j.jmb.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lee SK, Park EJ, Lee HS, Lee YS, Kwon J. Genome-wide screen of human bromodomain-containing proteins identifies Cecr2 as a novel DNA damage response protein. Molecules and cells. 2012;34(1):85–91. 10.1007/s10059-012-0112-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Tillhon M, Cazzalini O, Nardo T, Necchi D, Sommatis S, Stivala LA, et al. p300/CBP acetyl transferases interact with and acetylate the nucleotide excision repair factor XPG. DNA repair. 2012;11(10):844–52. 10.1016/j.dnarep.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 187.Ogiwara H, Kohno T. CBP and p300 histone acetyltransferases contribute to homologous recombination by transcriptionally activating the BRCA1 and RAD51 genes. PLoS ONE. 2012;7(12):e52810 10.1371/journal.pone.0052810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jang ER, Choi JD, Lee JS. Acetyltransferase p300 regulates NBS1-mediated DNA damage response. FEBS letters. 2011;585(1):47–52. 10.1016/j.febslet.2010.11.034 [DOI] [PubMed] [Google Scholar]
- 189.Kim MK, Shin JM, Eun HC, Chung JH. The role of p300 histone acetyltransferase in UV-induced histone modifications and MMP-1 gene transcription. PLoS ONE. 2009;4(3):e4864 10.1371/journal.pone.0004864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rubbi CP, Milner J. p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. Embo Journal. 2003;22(4):975–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, et al. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Molecular and cellular biology. 1999;19(2):1202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Qi W, Chen H, Xiao T, Wang R, Li T, Han L, et al. Acetyltransferase p300 collaborates with chromodomain helicase DNA-binding protein 4 (CHD4) to facilitate DNA double-strand break repair. Mutagenesis. 2016;31(2):193–203. 10.1093/mutage/gev075 [DOI] [PubMed] [Google Scholar]
- 193.Smith-Roe SL, Nakamura J, Holley D, Chastain PD 2nd, Rosson GB, Simpson DA, et al. SWI/SNF complexes are required for full activation of the DNA-damage response. Oncotarget. 2015;6(2):732–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Husain A, Begum NA, Taniguchi T, Taniguchi H, Kobayashi M, Honjo T. Chromatin remodeller SMARCA4 recruits topoisomerase 1 and suppresses transcription-associated genomic instability. Nature communications. 2016;7:10549 10.1038/ncomms10549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Buchmann AM, Skaar JR, DeCaprio JA. Activation of a DNA damage checkpoint response in a TAF1-defective cell line. Molecular and cellular biology. 2004;24(12):5332–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jain AK, Allton K, Duncan AD, Barton MC. TRIM24 is a p53-induced E3-ubiquitin ligase that undergoes ATM-mediated phosphorylation and autodegradation during DNA damage. Molecular and cellular biology. 2014;34(14):2695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nature cell biology. 2010;12(2):177–84. 10.1038/ncb2017 [DOI] [PubMed] [Google Scholar]
- 198.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, et al. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. The Journal of cell biology. 2007;177(2):219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Oike T, Komachi M, Ogiwara H, Amornwichet N, Saitoh Y, Torikai K, et al. C646, a selective small molecule inhibitor of histone acetyltransferase p300, radiosensitizes lung cancer cells by enhancing mitotic catastrophe. Radiotherapy and oncology: journal of the European Society for Therapeutic Radiology and Oncology. 2014;111(2):222–7. [DOI] [PubMed] [Google Scholar]
- 200.Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(33):14639–44. 10.1073/pnas.1008522107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Munshi A, Tanaka T, Hobbs ML, Tucker SL, Richon VM, Meyn RE. Vorinostat, a histone deacetylase inhibitor, enhances the response of human tumor cells to ionizing radiation through prolongation of gamma-H2AX foci. Molecular cancer therapeutics. 2006;5(8):1967–74. [DOI] [PubMed] [Google Scholar]
- 202.Wilson AJ, Lalani AS, Wass E, Saskowski J, Khabele D. Romidepsin (FK228) combined with cisplatin stimulates DNA damage-induced cell death in ovarian cancer. Gynecologic oncology. 2012;127(3):579–86. 10.1016/j.ygyno.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhang Y, Adachi M, Zou H, Hareyama M, Imai K, Shinomura Y. Histone deacetylase inhibitors enhance phosphorylation of histone H2AX after ionizing radiation. International journal of radiation oncology, biology, physics. 2006;65(3):859–66. [DOI] [PubMed] [Google Scholar]
- 204.Kim MJ, Lee JS, Park SE, Yi HJ, Jeong IG, Kang JS, et al. Combination treatment of renal cell carcinoma with belinostat and 5-fluorouracil: a role for oxidative stress induced DNA damage and HSP90 regulated thymidine synthase. The Journal of urology. 2015;193(5):1660–8. 10.1016/j.juro.2014.11.091 [DOI] [PubMed] [Google Scholar]
- 205.Geng L, Cuneo KC, Fu A, Tu T, Atadja PW, Hallahan DE. Histone deacetylase (HDAC) inhibitor LBH589 increases duration of gamma-H2AX foci and confines HDAC4 to the cytoplasm in irradiated non-small cell lung cancer. Cancer research. 2006;66(23):11298–304. [DOI] [PubMed] [Google Scholar]
- 206.Groselj B, Kerr M, Kiltie AE. Radiosensitisation of bladder cancer cells by panobinostat is modulated by Ku80 expression. Radiotherapy and oncology: journal of the European Society for Therapeutic Radiology and Oncology. 2013;108(3):429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Maiso P, Colado E, Ocio EM, Garayoa M, Martin J, Atadja P, et al. The synergy of panobinostat plus doxorubicin in acute myeloid leukemia suggests a role for HDAC inhibitors in the control of DNA repair. Leukemia. 2009;23(12):2265–74. 10.1038/leu.2009.182 [DOI] [PubMed] [Google Scholar]
- 208.Xie C, Drenberg C, Edwards H, Caldwell JT, Chen W, Inaba H, et al. Panobinostat enhances cytarabine and daunorubicin sensitivities in AML cells through suppressing the expression of BRCA1, CHK1, and Rad51. PLoS ONE. 2013;8(11):e79106 10.1371/journal.pone.0079106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–82. 10.1038/nature13229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Stewart HJ, Horne GA, Bastow S, Chevassut TJ. BRD4 associates with p53 in DNMT3A-mutated leukemia cells and is implicated in apoptosis by the bromodomain inhibitor JQ1. Cancer medicine. 2013;2(6):826–35. 10.1002/cam4.146 [DOI] [PMC free article] [PubMed] [Google Scholar]