Abstract
This review discusses next-generation antibacterial agents developed using rational, or targeted, drug design strategies. The focus of this review is on small-molecule compounds that have been designed to bypass developing bacterial resistance, improve the antibacterial spectrum of activity, and/or to optimize other properties, including physicochemical and pharmacokinetic properties. Agents are discussed that affect known antibacterial targets, such as the bacterial ribosome, nucleic acid binding proteins, and proteins involved in cell-wall biosynthesis; as well as some affecting novel bacterial targets which do not have currently marketed agents. The discussion of the agents focuses on the rational design strategies employed and the synthetic medicinal chemistry and structure-based design techniques utilized by the scientists involved in the discoveries, including such methods as ligand- and structure-based strategies, structure-activity relationship (SAR) expansion strategies, and novel synthetic organic chemistry methods. As such, the discussion is limited to small-molecule therapeutics that have confirmed macromolecular targets and encompasses only a fraction of all antibacterial agents recently approved or in late-stage clinical trials. The antibacterial agents selected have been recently approved for use on the U.S. or European markets or have shown promising results in phase 2 or phase 3 U.S. clinical trials.
Keywords: Antibacterial, optimization, rational design, resistance, semi-synthesis, targeted
Graphical Abstract
Long past the historical “golden era” of antibacterial drug discovery, the modern “resistance era” is being countered by new legislation and advances in the rational design of antibacterial agents.
Introduction
Beginning with the discovery of penicillin in 1928, the early twentieth century ushered in an unprecedented “golden era” of antibacterial drug discovery. Phenotypic whole-cell screening of natural products predominated this era and carried with it a success that is yet to be reproduced. Effective drug discovery and development later transitioned into the mid- to late-twentieth century in the form of a strong “medicinal chemistry era.” This period was mainly oriented around synthetic modifications of known compounds, high-throughput screening (HTS), and library design.1 It was initially hoped that this subsequent age of antibacterial discovery and development would be a triumph over the long reign of bacterial infections, but instead it has more recently proven to be a simple prelude to the modern antibacterial era—an age marked by the looming threat of expanding bacterial resistance coupled with the stark reality of an inversely diminishing antibacterial pipeline.2-4 As opposed to those bygone golden and medicinal chemistry eras, the contemporary “resistance era” is marked by a drug discovery climate that is comparatively less fruitful and geared around more time-consuming rationalized design modalities featuring target-based HTS campaigns that make use of commonly exhaustive tools like combinatorial libraries.5
Continuing along the antibacterial timeline, the commonly forecasted “post-antibiotic era” showcases a bleak future in which it is estimated that by the year 2050 the number of human deaths attributed to resistant microorganisms will surpass 10 million per year, up from the current 700,000 per year.2 To combat this issue, a multi-faceted approach has commenced. The U.S. has recently launched infectious disease initiatives including legislation such as the Generating Antibiotics Incentives Now Act (GAIN Act) aimed at fast-tracking Qualified Infectious Disease Products (QIDPs). Europe has also responded with its own countermeasures, such as Innovative Medicines Initiative’s (IMI's) ENABLE project under the “New Drugs 4 Bad Bugs” (ND4BB) program aimed at battling Gram-negative bacteria.
To meet the antibacterial drug discovery and development challenges at hand, modern research efforts have further expanded upon rational design and optimization strategies—such efforts addressing specific targets or problems, such as cell penetration.5 These efforts revolve around improved activity, physicochemical properties, pharmacokinetics, adverse effects, off-target activities, and so forth. Moreover, researchers have recently appeared to “re-discover” natural products chemistry, additionally employing novel approaches like modern semi-synthetic techniques.6, 7 With newly implemented incentivization and modern advances in research methods, it is hoped that the threat of antibacterial resistance will be met with appropriate countermeasures. It is therefore the objective of this review to discuss the latest progress and developments in the rational design and optimization of antibacterial agents recently approved or in late-phase trials.
Discussion
Advanced generation cephalosporins
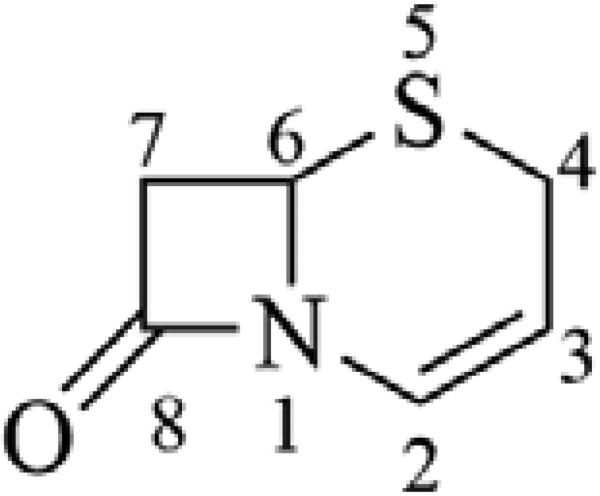
Recent advances in the chemistry of the β-lactam antibiotics have resulted in the approval or near approval of several advanced (or 5th) generation cephalosporins with enhanced spectra of activity, including methicillin-resistant Staphylococcus aureus (MRSA), and Pseudomonas aeruginosa (PA). While the general structure-activity relationship (SAR) of the cephalosporins has been covered well in previous communications,8 there has been some notable medicinal chemistry applied to the advanced generation cephalosporins that merits discussion here. The classical cephalosporin scaffold (Figure 1) includes a four-membered β-lactam ring fused with a dihydrothiazine ring (cephem) that contains a double bond between the 2- and 3-position carbons, and a 2-position carboxylate. Essential to the activity of the cephalosporin antibiotics is a functional group substitution at the 3-position that can stabilize a negative charge resulting from β-lactam ring opening and increase the overall reactivity of the β-lactam ring when interacting with target transpeptidase enzymes.
Figure 1.
Cephem ring, structure and positional numbering.
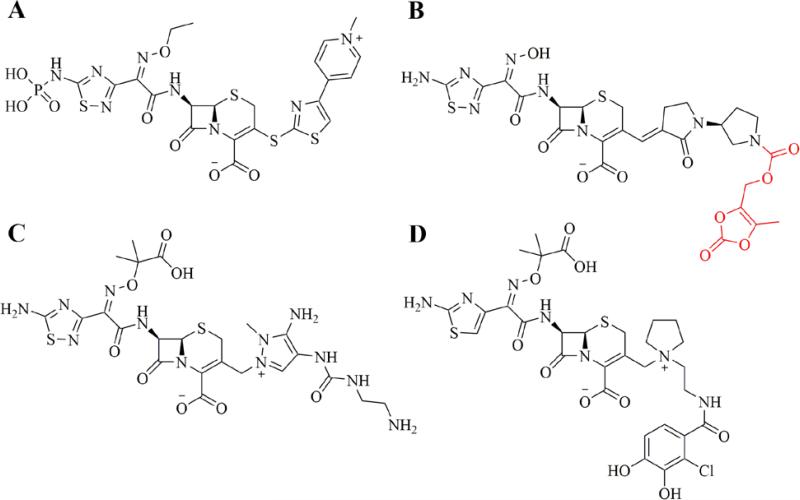
Ceftaroline (Teflaro® in the U.S., Zinforo® in Europe, Forest Laboratories) was approved for use in the U.S. for treatment of community-acquired bacterial pneumonia (CABP) and acute bacterial skin and skin structure infections (ABSSSI) in 2010. Ceftaroline is marketed as the prodrug Ceftaroline fosamil, which incorporates a hydrolysable phosphono group to improve aqueous solubility (Figure 2A).9 The drug has excellent Gram positive coverage, including activity against resistant strains of Streptococcus pneumoniae and MRSA, and moderate Gram negative coverage, but is less active against Pseudomonas aeruginosa, enterococci, or clinically relevant anaerobic organisms.10-12 Key to the drug's activity is a high binding affinity for the penicillin-binding protein 2a (PBP2a), which is known to play a significant role in resistance to anti-staphylococcal β-lactams, and PBP2x, found in β-lactam-resistant S. pneumoniae. As can be seen from the chemical structure (Figure 2A), ceftaroline retains an oxime group as part of the 7-position substituent, in this case an ethyl oxime. This is also seen in the third- and fourth-generation cephalosporins, and is known to confer resistance to some β-lactamase enzymes. The 1,2,4-thiadiazine ring component of the 7-position substituent is believed to improve the drug's penetration into Gram-negative organisms, as well as contribute to overall target affinity.13 The 1,3-thiazole ring, connected to the cephem ring at the 3-position by a unique sulfide bond, has been reported to play a key role in the improved affinity of ceftaroline for PBP2a and the resulting anti-MRSA activity.13 Finally, the quaternary nitrogen of the pyridine ring in the 3-position substituent contributes a positive charge to the overall zwitterion ceftaroline and improves drug penetration into Gram-negative organisms.
Figure 2.
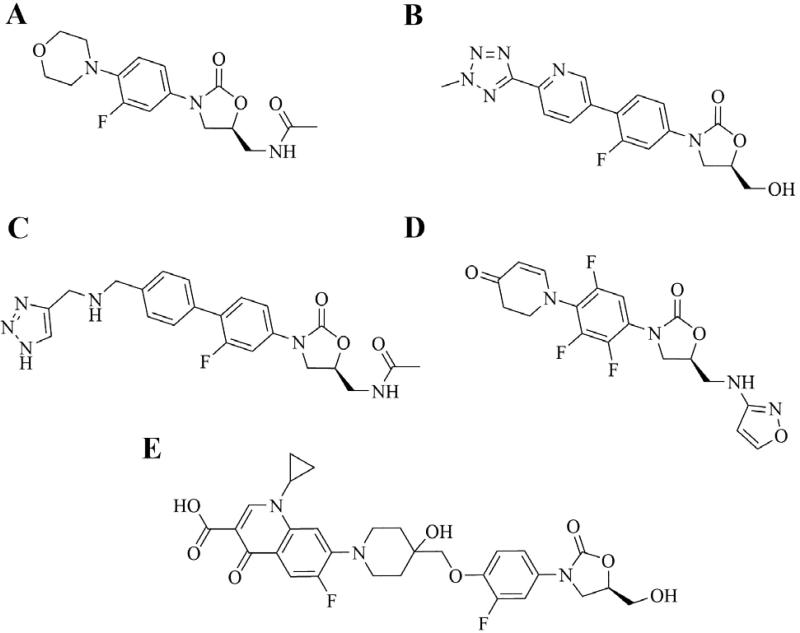
Advanced generation cephalosporins recently approved or in clinical trials. A. Ceftaroline fosamil. B. Ceftobiprole medocaril (prodrug moiety in red). C. Ceftolozane. D. S-649266
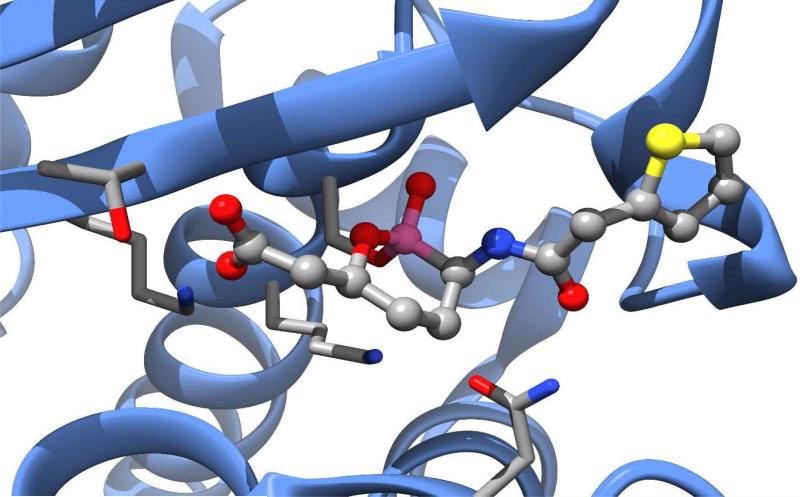
Ceftobiprole (Zevtera®, Basilea Pharmaceutica) was approved in Europe and Canada for the treatment of community-acquired pneumonia (CAP) and hospital-acquired pneumonia (HAP), excluding ventilator-associated pneumonia (VAP) in 2013. After initially being postponed in Europe and discontinued in Canada in 2010 due to recommendations from regulatory agencies in Europe and the U.S. regarding clinical trial concerns, Ceftobiprole is now marketed as ceftobiprole medocaril sodium, a prodrug incorporating a solubilizing group attached to the pyrrolidine nitrogen (Figure 2B) by a hydrolysable carbamate ester.14, 15 The drug has reported broad-spectrum activity against many Gram positive and Gram negative organisms, including resistant strains of S. aureus (MRSA & VRSA), S. pneumoniae (PRSP & CRSP), P. aeruginosa, and Enterobacteriaceae.16, 17 It is less active against clinically relevant anaerobic organisms, including Prevotella species and B. fragilis group species.18 The presence of the oxime functionality in the 7-position substituent confers a degree of β-lactamase resistance to ceftobiprole similar to that seen in the 3rd and 4th generation cephalosporins, however the drug is not resistant to the extended-spectrum β-lactamases (ESBL) produced by resistant strains of Enterobacteriaceae.19 Similar to ceftaroline, the activity of ceftobiprole is attributed to an increased affinity for PBP2a (S. aureus) and PBP2x (S. pneumoniae). The co-crystal structure of the complex of ceftobiprole bound to PBP2a has provided key insights in the nature of the binding interactions of the drug to its target (PDB accession code 4DKI).20 Key to the increased binding affinity of ceftobiprole for PBP2a appears to be the vinyl-pyrrolidinone component of the 3-position substituent, which interacts with a narrow groove of the PBP2a active site, forming pi-stacking interactions with a nearby tyrosine residue (Tyr-446) and general hydrophobic interactions with other nearby residues, resulting in a favorable positioning of the β-lactam ring for acylation by the key serine residue (Ser-403). Binding-induced conformational changes in secondary structure elements near the active site also appear to facilitate the acylation reaction.20
Another advanced generation cephalosporin, ceftolozane, was approved in the U.S. for use in combination with the β-lactamase inhibitor, tazobactam, for treatment of complicated intra-abdominal infections (in combination with metronidazole) and complicated urinary tract infections in 2014 (Zerbaxa™, Merck). Ceftolozane lacks activity against many clinically relevant Gram positive organisms, with no appreciable activity against MRSA. However, the addition of tazobactam gives the combination product excellent Gram negative activity including potent activity against many resistant P. aeruginosa strains and most ESBL-producing Enterobacteriaceae.21, 22 The combination (ceftolozane/tazobactam) shows highly variable activity against anaerobic organisms, particularly against B. fragilis species, which necessitates the use in combination with metronidazole for complicated intra-abdominal infections (cIAIs).23 Structurally, ceftolozane possesses the 7-substituent oxime functionality conferring a degree of β-lactamase resistance similar to the 3rd and 4th generation cephalosporins. Similar to ceftazidime, the oxime functionality incorporates a dimethyl acetic acid moiety that is believed to enhance activity against P. aeruginosa (Figure 2C).24 At the 3-position, the bulky, substituted pyrazole ring prevents hydrolysis of the β-lactam ring by steric hindrance and confers some stability to AmpC β-lactamases commonly seen in P. aeruginosa.25, 26 Unlike ceftaroline and ceftobiprole, the 3-position substituent of ceftolozane does not appear to improve the drug's affinity for PBP2a, which explains the compounds lack of activity against MRSA, and the drug has low overall affinity for PBP4, similar to ceftazidime, which explains it's very weak induction of AmpC expression.27 However, the ceftolozane has particularly strong affinity for PBP3, at least 2-fold higher than ceftazidime, which explains its potent P. aeruginosa activity.27 Indeed, the 3-substituent pyrazole substituents were intentionally designed to maximize this affinity and hence activity against P. aeruginosa.24, 26
Compound S-649266 (Shionogi, Inc.) is an unapproved, advanced generation cephalosporin currently in phase 2 clinical trials in the U.S for treatment of complicated urinary tract infections (cUTI). The compound is notable for its activity against bacterial strains producing ‘K. pneumoniae Carbapenemase’ (KPC) and ‘New Delhi Metallo-β-lactamase’ (NDM-1), particularly worrisome β-lactamases with the ability to inactivate most β-lactams, including carbapenems, and resistance to most marketed β-lactamase inhibitors.28, 29 The structure of S-649266 (Figure 2D) incorporates a catechol moiety in the 3-position substituent, which acts as a siderophore, and employs a ‘Trojan Horse’ strategy to increase cell penetration of the agent.30 In this case, the catechol group acts as an iron chelator and enables the active transport of S-649266 into the bacterial cell by way of the cell's iron transport systems. S-649266 possesses the same dimethyl acetic acid substituted oxime group at the 7-position seen in ceftolozane and ceftazidime, which confers a high degree of β-lactamase resistance and activity against P. aeruginosa.29 In fact, the compound has been reported to possess intrinsic resistance to the β-lactamases produced by many problematic strains of Enterobacteriaceae, including ESBL's, KPC, and NDM-1.31 Clinical trials currently underway are comparing S-649266 with imipenem/cilastatin for treatment of cUTI and are expected to be complete in September, 2016.32
Next Generation Beta-Lactamase Inhibitors
Bacteria-produced β-lactamase enzymes hydrolytically cleave the cyclic amide bond of the β-lactam antibacterial agents before they are able to bind to and inhibit their target PBPs, representing a challenging bacterial drug resistance strategy. There are a large and diverse number of β-lactamase enzymes produced by bacteria (Table 1). The development of β-lactam antibacterials with intrinsic resistance to β-lactamases (i.e., methicillin, higher generation cephalosporins, and the carbapenems) and mechanism-based inhibitors of the β-lactamase enzymes that can be co-administered with β-lactam antibacterials (Figure 3), initially helped to overcome this drug resistance mechanism.8 Unfortunately, the recent emergence of inhibitor-resistant β-lactamases with activity against the previously intrinsically resistant β-lactams, has led to a critical lack of antibacterials with the capability of treating resistant infections. Particularly problematic β-lactamases that have seen recent emergence in Gram negative organisms include the extended-spectrum β-lactamases (ESBLs) such as TEM, SHV & CTX-M (Class A), and OXA (Class D), that have the ability to hydrolyze extended-spectrum cephalosporins with the oxime (oxyimino) functional group.33-36 The AmpC-type β-lactamases (Class C), seen in Enterobacteriaceae and P. aeruginosa, have the ability to hydrolyze broad and extended spectrum cephalosporins (cephamycins & oxime cephalosporins) and are not inhibited by classical β-lactamase inhibitors (i.e., clavulanic acid).37, 38 Lastly, the very recent emergence of carbapenemases, including KPC (Class A), VIM, IMP, and NDM-1 (Class B), is of great concern as these enzymes have the ability to hydrolyze the carbapenems that have been used as a last resort for treatment of organisms expressing ESBLs and AmpC.39-42 The recent, or near approvals of two new classes of β-lactamase inhibitors has provided hope of some reprieve from pan-drug resistant infections. The novel chemistry and mechanisms of these agents is discussed below.
Table 1.
Ambler Classification of β-lactamases, substrate affinity, and selected examples. (Adapted with permission from Mandell's Principles & Practice of Infectious Disease, Chapter 18, Table 18-2, p. 240)
| Ambler Class1 | Enzyme Activity | Substrates | Examples |
|---|---|---|---|
| A | Broad-Spectrum (Penicillinases) | benzylpenicillins, aminopenicillins, carboxypenicillins, ureidopenicillins, narrow-spectrum cephalosporins | PCI (S. aureus); TEM-1, SHV-1 (E. coli, K. pneumoniae & other G-ve bacteria |
| A | Extended-Spectrum (ESBL) | Substrates of broad-spectrum, plus: oxyimino-β-lactams, aztreonam | TEM-derived, SHV-derived, CTX-M-derived (Enterobacteriaceae); PER-1, VEB-1, VEB-2, GES-1, GES-2, IBC-2 (P. aeruginosa) |
| A | Carbapenemases | Substrates of ESBLs, plus: cephamycins & carbapenems | KPC-1, KPC-2, KPC-3 (K. pneumoniae); NMC/IMI, SME family |
| B | Carbapenemases | Substrates of ESBLs, plus: cephamycins & carbapenems | NDM-1 (Enterobacteriaceae); IMP, VIM, GIM, SPM, SIM (P. aeruginosa, Acinetobacter spp. |
| C | Cephalosporinases | Substrates of ESBLs, plus: cephamycins | AmpC-type (Enterobacteriaceae, Acinetobacter spp.) |
| D | Broad-Spectrum (Oxacillinases) | Aminopenicillins, ureidopenicillins, cloxacillin, methicillin, oxacillin, and some narrow-spectrum cephalosporins | OXA-family (P. aeruginosa) |
| D | Extended-spectrum (ESBL) | Substrates of broad-spectrum, plus: oxyimino-β-lactams, aztreonam | OXA-derived (P. aeruginosa) |
| D | Carbapenemases | Substrates of ESBLs, plus: cephamycins & carbapenems | OXA-derived (Acinetobacter spp.) |
Ambler Class A, C, and D β-lactamases possess a catalytic active site serine; Ambler Class B β-lactamases are Zn2+ metallo-β-lactamases.
Figure 3.
Marketed, mechanism-based β-lactamase inhibitors. A. Clavulanic Acid. B. Tazobactam. C. Sulbactam.
Avibactam (formerly NLX104), a member of the diazabicyclo[3.2.1]octanone (DBO) series, is a novel β-lactamase inhibitor that does not possess the typical β-lactam ring seen in the previously marketed agents clavulanic acid, tazobactam, and sulbactam (Figure 4A).43, 44 Avibactam, in combination with the 3rd generation anti-pseudomonal cephalosporin, ceftazidime (Avycaz, Allergan, Inc.), was approved in the U.S. in 2015 for the treatment of cIAI (in combination with metronidazole), and cUTI caused by multidrug-resistant Gram negative bacteria. It is also in phase 2 clinical trials in Europe in combination with aztreonam for cIAIs and phase 3 trials in Europe for serious infections like complicated UTIs, acute pyelonephritis, and hospital acquired bacterial pneumonia. Avibactam has an extended spectrum of activity against β-lactamase enzymes compared to the classical β-lactamase inhibitors, including activity against ESBLs, AmpC, and KPC β-lactamases.45, 46 The combination of avibactam with ceftazidime extends the coverage of this agent to include activity against many resistant Gram-negative organisms, including ESBL-producing and carbapenem-resistant Enterobacteriaceae. The activity of the ceftazidime/avibactam combination against P. aeruginosa is modestly improved over ceftazidime alone, while Acinetobacter species remain mostly resistant to the combination drug.47, 48 The activity of the combination product against anaerobic and Gram positive organisms is limited and similar to that of ceftazidime alone, which necessitates the combination with metronidazole for the treatment of cIAI.49, 50 The structures of avibactam bound to Class A (CTX-M), Class C (AmpC), and Class D (OXA) β-lactamases have been solved and provide key insights into the drug's mechanism of β-lactamase inhibition.51-53 The classical β-lactamase inhibitors exert their activity by either transient or irreversible inhibition of the enzymes via the formation of an acyl-enzyme covalent bond associated with ring-opening. Hydrolysis of this bond can restore the active enzyme, but the β-lactamase inhibitor, once freed from the enzyme, is inactivated. Avibactam differs from the classical β-lactamase inhibitors by forming a carbamate linkage to the β-lactamase enzyme with opening of the diaza-bicyclo ring structure.52 The crystal structures indicate that the drug is more tightly bound within the β-lactamase active site compared with the classical inhibitors, with a large degree of rigidity along the carbamate linkage, and multiple hydrogen bonds in the active site.54 Avibactam's displacement of a key water molecule in the active site that is presumed to be involved in the hydrolytic release of the classical β-lactamase inhibitors has been proposed to contribute to the drug's longer residence time in the active site compared with the classical inhibitors; i.e., 50% of clavulanic acid is released from TEM-1 within 7 minutes, versus 7 days for avibactam.52, 55 Lastly, upon release from the β-lactamase active site, avibactam has been shown to return to its original active form, unlike the classical β-lactamase inhibitors, which are inactive upon release.56 Taken together, these factors contribute to the enhanced activity of avibactam over classical β-lactamase inhibitors against class A KPC β-lactamases, Class C β-lactamases, and some Class D β-lactamases.55, 57
Figure 4.
Next generation β-lactamase inhibitors recently approved or in clinical trials. A. Avibactam. B. Relebactam. C. Vaborbactam.
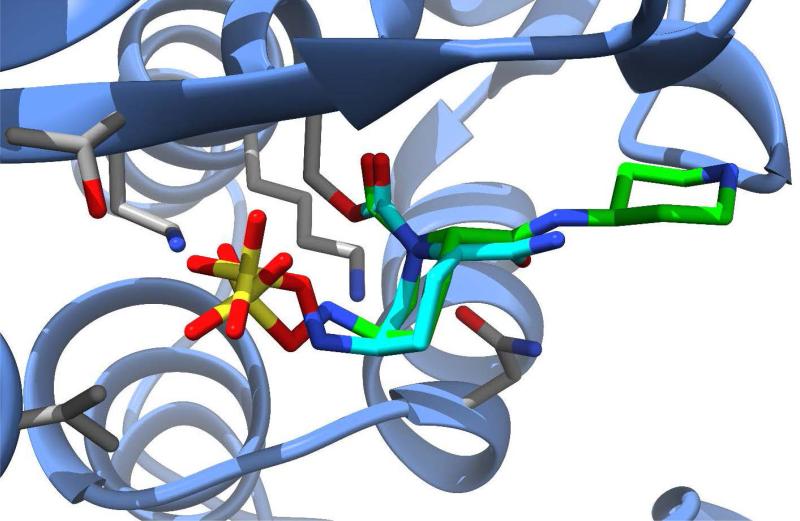
Relebactam (formerly MK-7655, Merck) is another new β-lactamase inhibitor in the DBO class that is currently undergoing phase 3 clinical trials in Europe and the U.S. (Figure 4B). The compound is being investigated for use in combination with imipenem-cilastatin (a carbapenem combined with a dehydropeptidase inhibitor that prevents degradation of imipenem in the kidney) for treatment of HAP, VAP, cIAI, and cUTI. Similar to avibactam, relebactam shows activity against class A and class C β-lactamases, including ESBLs.58 When combined with imipenem-cilastatin, relebactam improved the activity of imipenem against carbapenemase-producing (KPC) Enterobacteriaceae and AmpC-producing P. aeruginosa.59, 60 The combination is not effective against organisms expressing Class B metallo-β-lactamases or Class D carbapenemases and does not appear to add activity against Acinetobacter baumannii.61, 62 Structurally similar to avibactam, relebactam has a piperidine ring added to the carbamoyl moiety at the 2-position. This compound was synthesized in an effort to test the effects of positively and negatively charged side chain substitutions to the carbamoyl group.63 It is believed that a positively charged group at this position plays a key role in preventing the efflux of the drug from the bacterial cell, thereby facilitating synergy with imipenem. An x-ray crystal structure of relebactam bound in the active site of AmpC from P. aeruginosa (PDB entry 4NK3) shows the compound binds in a manner very similar to avibactam, with the piperidine substituent extending into an empty pocket of the active site and interacting primarily with water molecules (Figure 5).63
Figure 5.
Avibactam (cyan carbons) and relebactam (green carbons) shown bound into the active site of beta-lactamases, SHV-1 from K. pneumoniae [PDB ID 4ZAM] and AmpC from P. aeruginosa [PDB ID 4NK3), respectively.
In contrast to avibactam and relebactam, which possess a novel diaza-bicyclic-octanone scaffold, the novel β-lactamase inhibitor, vaborbactam (formerly RPX7009), possesses a unique, boronic acid-based cyclic scaffold (Figure 4C). Vaborbactam is currently undergoing phase 3 clinical trials in Europe for use in combination with meropenem (Carbavance™, Rempex Pharmaceuticals) for treatment of cUTI, HAP, VAP, & bacteremia caused by carbapenem-resistant Enterobacteriaceae.64, 65 Vaborbactam has been extensively studied in combination with the investigational carbapenem RPX2003 (biapenem).66, 67 The combination showed excellent activity against class A carbapenemase-producing Enterobacteriaceae, including KPC-producing strains of K. pneumoniae. The boronic acid pharmacophore has been known for many years to have affinity for serine proteases,68, 69 and boronic acid compounds have been explored as inhibitors of serine β-lactamases since the early 1970's.70 A structure of a boronic acid-based compound covalently bound to a TEM β-lactamase was published in the early 90's,71 and led to over a decade of structure-guided design of boronic acid-based β-lactamase inhibitors.72-76 Vaborbactam differs from previously reported boronic acid inhibitors of β-lactamase by the rational design of a cyclic boronate ester, which was hypothesized to provide selectivity toward β-lactamase enzymes over other serine proteases, the latter showing preference for acyclic substrates due to a more sterically restricted active site.77 Solved structures of vaborbactam bound to β-lactamases CTX-M-15 and AmpC show that the compound binds to the β-lactamase enzymes in a similar manner as avibactam and relebactam, with the exception that the catalytic serine residue forms a covalent bond with the boronic ester moiety (Figure 6).77
Figure 6.
RPX7009 bound to AmpC from E. cloacae [PDB ID 4XUX].
Next-Generation Oxazolidinone Antibacterials
The oxazolidinone class of antibacterials, a fully synthetic class of agents, has been considered a major breakthrough in antimicrobial drug development.78 The class is represented by the U.S. marketed agents linezolid (Zyvox®, Pfizer, approved 2000) and tedizolid (Syvextro®, Merck, approved 2014), and several other promising agents currently in clinical development (Figure 7). The history of the discovery and development of the oxazolidinone class as antibacterial agents have been previously reviewed,79-81 as well as early SAR studies.82-94 Oxazolidinones exert their antibacterial activity by binding to the bacterial 70S ribosome and inhibiting protein synthesis. The mechanism of this effect is unique from other classes of protein synthesis inhibitors in that they bind to the 50S ribosomal subunit, thereby preventing the binding of aminoacyl-tRNA to the peptidyl transferase center at the A-site.95-97 The antibacterial spectrum of the oxazolidinone class is excellent with respect to most clinically relevant Gram positive pathogens, including methicillin-resistant S. aureus (MRSA), penicillin-resistant S. pneumoniae (PRSP), macrolide-resistant streptococci, and vancomycin-resistant enterococci (VRE).78, 98 The class in general is clinically ineffective against most Gram negative pathogens, primarily due to an efflux mechanism of resistance.79, 99-101 Linezolid is currently approved for use in the U.S. for treatment of nosocomial and community-acquired pneumonia (CAP), uncomplicated and complicated skin and skin structure infections (cSSSI), and infections caused by vancomycin-resistant enterococci. While a very promising agent for treatment of infections caused by resistant pathogens, linezolid is, unfortunately, not without its drawbacks. The drug is known to cause reversible thrombocytopenia and bone marrow suppression, neuropathies with prolonged use, and has been associated with adverse serotonergic effects due to its inhibition of monoamine oxidase (MAO).102 Further, the recent emergence of linezolid resistance has driven the investigation and advancement of next-generation oxazolidinones with improved antibacterial activities and decreased adverse effect profiles.103-105
Figure 7.
Oxazolidinones currently marketed or in clinical trials. A. Linezolid. B. Tedizolid. C. Radezolid. D. MRX-I. E. Cadezolid.
Tedizolid (formerly torezolid and TR-701), is a second-generation oxazolidinone that was approved in the U.S. in 2014 for treatment of acute bacterial skin and skin structure infections (ABSSSI) caused by susceptible organisms (Figure 7B).96, 102, 106-109 The drug is marketed as tedizolid phosphate, a prodrug formulation of tedizolid that is phosphorylated at the 5-position hydroxymethyl group.110-112 Structurally similar to linezolid, tedizolid possesses the core oxazolidinone ring (A-ring) as well as the fluoro-phenyl B-ring. As discussed in previous communications, the 5-R configuration of the A-ring and the N-aryl B-ring are minimally required for antibacterial activity.81, 113 Fluorination of the B-ring improves antibacterial activity and this substituent is seen in all approved oxazolidinones as well as those in clinical development (Figure 7). In tedizolid, a C-5 hydroxymethyl group replaces the acetamide moiety seen in linezolid. This group serves as an attachment point for the prodrug phosphate which improves water solubility and may decrease MAO inhibition. Additionally, the substitution of hydroxymethyl at this position decreases steric bulk which has been proposed to allow for retained activity against bacterial species with resistance mediated by active site methylation (cfr gene).113, 114 Tedizolid also substitutes a bi-aryl system (C-ring pyridine and D-rings) for the morpholine C-ring seen in linezolid. This substitution has been suggested to be responsible for improved potency of tedizolid over linezolid, as the new ring system is able to form additional binding interactions in the 50S ribosomal subunit.115, 116 With respect to the spectrum of activity, tedizolid is up to 8-fold more potent than linezolid against most Gram-positive staphylococci, streptococci, and enterococci, including MRSA and VRE species.117-121 It possesses moderately improved activity over linezolid against the Gram-negative M. catarrhalis and H. influenzae, an 8- to 16-fold improvement in activity against Gram-positive anaerobes, and 2-fold improvement in activity against Gram-negative anaerobes.117, 120 Tedizolid also retains reasonable activity against linezolid-resistant Gram-positive cocci with cfr-mediated resistance.122 Additional favorable properties of tedizolid include a long half-life that allows once-daily administration (versus twice daily linezolid), low serotonergic effect due to decreased MAO interaction, and a significantly improved adverse event profile.96, 102, 106
Radezolid (formerly RX-1741 and Rx-01), Melinta Therapeutics) (Figure 7C) completed phase 2 clinical trials in the U.S. in 2009 for treatment of community-acquired pneumonia and uncomplicated skin and skin-structure infections.123 The compound was rationally designed using experimental ribosome structures and computational models to have enhanced Gram-negative activity and improved oral bioavailability.124-127 Structurally, the drug replaces the D-ring of tedizolid with a (triazolyl-methyl)-aminoyl-methyl moiety attached para to a phenyl C-ring and possesses the same acetamide group at the 5-position of the oxazolidinone A-ring seen with linezolid.113, 126-129 The antibacterial activity of radezolid has been evaluated in comparison to linezolid and the drug has been shown to have moderately improved activity against streptococci (S. pneumoniae and S. pyogenes), staphylococci (MSSA and MRSA), and enterococci, including some activity against linezolid-resistant staphylococci, enterococci, and S. pneumoniae.128 The safety profile and any advantages of radezolid compared to linezolid and tedizolid is not clear based on the current literature. To date, no phase 3 trials have been announced.
MRX-I (MicuRx Pharmaceuticals, Inc.) is another next-generation oxazolidinone currently in early phase 2 clinical trials in the U.S. for the treatment ABSSSI (Figure 7D). This compound is reported to have been rationally designed to possess significantly decreased myelosuppression and inhibition of MAO resulting in an improved safety profile over linezolid.130 MRX-I deviates from traditional SAR of the oxazolidinones by incorporating a novel 2,4,6-trifluorophenyl B-ring, a diydropyridone C-ring, and a unique isoxazolyl-aminomethyl substitution at the A-ring 5-position. The trifluorophenyl B-ring is believed to be responsible for the improvement in the safety profile of MRX-I possibly due to a distortion of A-ring, B-ring planarity forced by the ortho-fluoro substituent that affects off-target binding to MAO and the mitochondrial ribosome.130 With respect to the drug's spectrum of activity, MRX-I has similar antibacterial range to other oxazolidinones, including MRSA, PRSP, and VRE, and is comparable or slightly better than that of linezolid.131 The literature is not clear on the activity of MRX-I against linezolid-resistant bacterial strains.
The last agent of interest within the developing oxazolidinone class is cadazolid (ACT-179811, Actelion Pharmaceuticals, Ltd.).132 Structurally, cadazolid is almost a complete departure from molecules previously discussed. It is a bifunctional molecule containing the oxazolidinone core scaffold on one half tethered to a fluoroquinolone via a phenoxyether linkage (Figure 7E).133-135 The common structural feature that serves as a bridge between the two molecules is a piperidine ring. This positioning of this ring is analogous to the morpholine seen in linezolid and the piperazine ring found in ciprofloxacin. Most reports seem to indicate that the primary mechanism of action for cadazolid is protein synthesis with very little effect on DNA synthesis as a possible secondary mechanism.136 This would suggest that this agent acts more like an oxazolidinone with very little fluoroquinolone activity. It is being studied for the treatment of C. difficile-associated diarrhea (CDAD), and has progressed to phase 3 clinical trials in Europe and in the U.S. in comparison with vancomycin.137, 138 It has very limited oral bioavailability due to its acidic and lipophilic nature, which means its activity is primarily localized to the gut.139, 140 The drug has received QIDP and Fast Track development status from the U.S. FDA.
Next Generation Bacterial Topoisomerase Inhibitors
Currently, there are four independent drug classes that target the bacterial type II topoisomerases DNA Gyrase and topoisomerase IV. Chronologically, Novobiocin is the classic representative of the first class of such antibacterial agents. No longer commercially available, Novobiocin operates via the competitive inhibition of ATP on DNA Gyrase. Next, while technically not the first class to exploit type II topoisomerases, the outstanding success of the quinolone class has almost single-handedly validated the target via the formation of DNA-quinolone-enzyme ternary complexes. Since their discovery in the 1960s, wide use of quinolones has resulted in high resistance levels. As such, rational improvements are continually being made to these indispensable agents, some of which are discussed below. Thirdly, a novel class of bacterial type II topoisomerase inhibitor has arisen that occupies a topoisomerase binding site distinctly separate from the quinolones and, therefore, possesses a unique MOI. These agents, known as novel bacterial topoisomerase inhibitors (NBTIs), consequently lack cross-resistance among pathogens possessing quinolone resistance. Lastly, the newer spiropyrimidinetrione class has recently arisen with yet another unique mechanism of topoisomerase II inhibition. Though discovered via whole-cell activity screens, spiropyrimidinetriones have since been rationally optimized.141 As one would expect, the unique MOIs of these newer classes imply accordingly unique structures, scaffolds, and SARs.
Zoliflodacin (previously known as ETX0914/AZD0914, Entasis Therapeutics, Inc.) is an oral antibacterial agent with QIDP fast-track status currently in phase 2 clinical trials in the U.S. for the treatment of uncomplicated gonorrhea. It is a spiropyrimidinetrione, a novel class of topoisomerase II inhibitors that uniquely inhibits re-ligation and consequently causes a bactericidal buildup of DNA double-strand cleavages.142, 143 It shows no cross-resistance with other type II topoisomerase inhibitor classes and exhibits an antibacterial spectrum of activity (SOA) that includes Gram-positive organisms; fastidious Gram-negative organisms such as Neissaria gonorrhoeae, Haemophilus influenzae, and Moraxella catarrhalis; as well as anaerobes.142-144 While much of the SAR was previously optimized or intolerable to manipulation, the benzisoxazole 3-position on the spiropyrimidinetrione scaffold proved to be modifiable. Thorough SAR optimization at this position resulted in the minimization of toxicity issues seen with fluoroquinolones (Figure 8A).142
Figure 8.
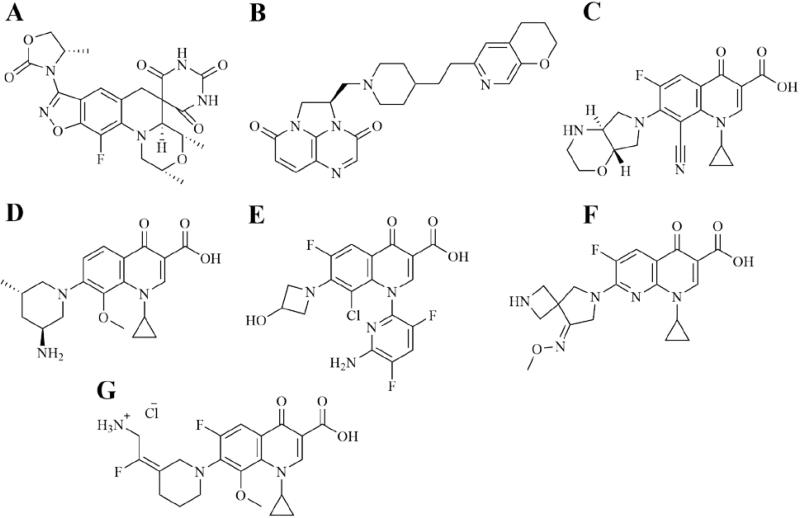
Next generation bacterial topoisomerase inhibitors. A. Zoliflodacin. B. Gepotidacin. C. Finafloxacin. D. Nemonoxacin. E. Delafloxacin. F. Zabofloxacin. G. Avarofloxacin.
Gepotidacin (also known as GSK2140944, GlaxoSmithKline plc) is a novel NBTI currently in phase 2 clinical trials in Europe for uncomplicated urogenital gonorrhea and the U.S. for Gram-positive ABSSSI, respiratory tract infections, and uncomplicated gonorrhea. It is a triazaacenaphthylene that binds to both gyrA and parC subunits at sites distinctly different from the fluoroquinolones and aminocoumarin-binding sites.145, 146 Early NBTIs arose from whole cell antibacterial screens and have since been structurally characterized.147, 148 Structural motifs include a bicyclic aromatic heterocycle connected to another aromatic heterocycle by a variable linker that must contain a basic nitrogen at position-7 in order to form a requisite salt bridge with Asp83 as seen via X-ray crystallographic structural data (Figure 8B).146, 147 NBTIs commonly exhibit high potencies and rather broad-spectrums of activity, but are generally plagued by unacceptable hERG activity that appears to be determined by overall compound polarity.146 Gepotidacin, however, has shown relatively low hERG activity with an IC50 of 1.4 mM.146
Finafloxacin (MerLion Pharmaceuticals Pte Ltd., Singapore; licensed as Xtoro® under Alcon Pharmaceuticals in the U.S.) is an 8-cyano fluoroquinolone with QIDP status that, while already approved in the U.S. in the form of an otic suspension for the treatment of acute otitis externa, is currently in phase 2 clinical trials in the U.S. for the treatment of ventilator-associated bacterial pneumonia (VABP) due to pseudomonas, acute bacterial skin and skin structure infections (ABSSSI), and complicated intra-abdominal infections. Unlike other fluoroquinolones, which exhibit reduced activity under acidic conditions, finafloxacin shows improved activity under slightly acidic conditions (pH 5.0 – 6.0) due to a unique 7-substituent chiral base component that confers a distinctive base capacity (Figure 8C).149, 150 It is presumed that the stereoconfiguration of the 7a hydrogen on the base component is responsible for this characteristic. Finafloxacin's ability to act under harsh acidic environments grants it a number of potential applications, such as Helicobacter pylori infections, urinary tract infections (UTIs), and more.
Nemonoxacin (TaiGen Biotechnology Co., Ltd.) is a non-fluorinated C-8-methoxy quinolone with QIDP status that has entered phase 3 clinical trials for the treatment of community acquired pneumonia (CAP), ABSSSI, and diabetic foot infection. The C-8-methoxy group grants nemonoxacin activity against both DNA Gyrase and topoisomerase IV, thereby decreasing resistant mutant selection, while the removal of the C-6-fluorine is presumed to decrease the incidence of toxic side effects (Figure 8D).151, 152 As compared to standard fluoroquinolones, it has shown increased activity against certain Gram-positive pathogens, including MSSA, MRSA, and multi-drug resistant S. pneumonia, but generally shows less activity against Gram-negative pathogens, including similar or inferior results as compared to standard fluoroquinolones.152, 153 Nemonoxacin has also shown varying degrees of activity against different quinolone-resistant pathogens.154
Delafloxacin (Baxdela™, Melinta Therapeutics) is a fluoroquinolone with QIDP status in phase 3 clinical trials for the treatment of ABSSSI and at the time of this review, is soon set to begin phase 3 trials for the treatment of CAP. Unlike other fluoroquinolones, delafloxacin exhibits a similar affinity for both DNA Gyrase and topoisomerase IV in both Gram-positive and Gram-negative pathogens and, therefore, is expected to be less selective for resistance mutants.155, 156 Delafloxacin has not been found to be a more potent topoisomerase inhibitor than other fluoroquinolones, but has been found to have greater potency, particularly against Gram-positive bacteria.156 Structural characteristics implicated for this attribute include a bulky heteroaromatic substituent (6-amino-3,5-difluoropyridine) at the 1 position, a C-8-chlorine that grants weak polarity, and the lack of a basic group at the 7 position (3-hydroxyazetidine) (Figure 8E).156 This last characteristic grants delafloxacin properties particularly unique among quinolones. Lacking a basic group available for protonation at the 7 position results in an anionic charge at neutral pH values (whereas most quinolone antibacterials form zwitterions at a physiological pH) and a mainly neutral charge at a slightly acidic pH.156 As such, delafloxacin has shown improved activity under slightly acidic conditions, which as mentioned above, is a rare characteristic for quinolones, and results in improved MICs against different bacteria as compared to moxifloxacin and ciprofloxacin.157-159
While there is less rational design information readily available at the time of this review, two more quinolones are being developed and in late-phase trials. Zabofloxacin (Dong Wha Pharmaceutical Co., Ltd.) is a fluoroquinolone that has completed phase 3 trials for the treatment of acute bacterial exacerbations of chronic obstructive pulmonary disease (COPD) (Figure 8F). Avarofloxacin (previously known as JNJ-Q2 and JNJ-32729463, Janssen Pharmaceutica, licensed to Furiex Pharmaceuticals) is a wide-spectrum fluoroquinolone with QIDP status that has completed phase 2 trials for the treatment of complicated skin and skin structure infections (CSSSIs) (Figure 8G).
Next Generation Tetracyclines
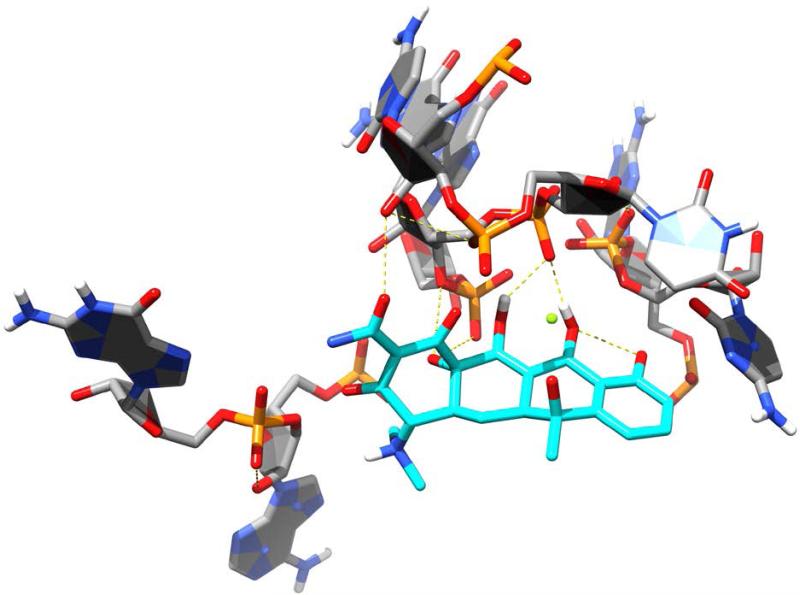
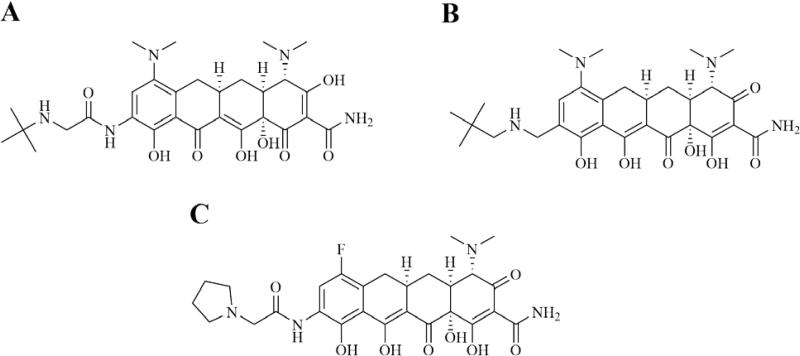
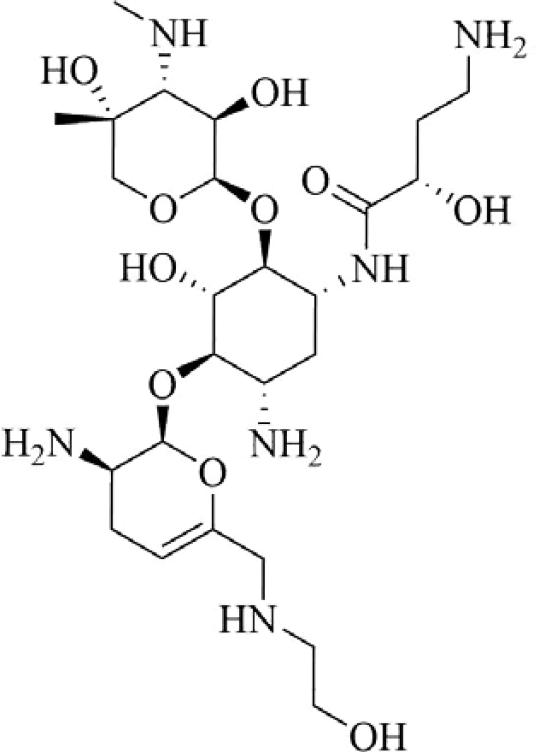
Since their discovery in the 1940s, tetracyclines have been widely used to treat both Gram-positive and Gram-negative pathogens. SAR studies of tetracycline analogues and the cocrystallization of tetracycline with the 30S subunit revealed significant interactions between several functional groups in the drug and nucleotides in the ribosomal structure (Figure 9).160 The same studies showed a lack of significant interactions between ring D and ribosomal nucleotides. The lack of significant binding at the C-7 through C-9 positions spurred the search for ring D-modified analogs, with the resulting development of tigecycline. Tigecycline (Tygacil®; Wyeth Pharmaceuticals Inc., a subsidiary of Pfizer Inc., initial U.S. approval in 2005) is the first semi-synthetic representative of a class of antimicrobials called glycylcyclines, which are 9-t-butylglycylamido derivatives of minocycline, a second generation tetracycline (Figure 10A).161 The design and synthesis of glycylcyclines were intended to overcome the major problems of resistance associated with the tetracyclines, namely ribosomal protection and efflux-pump-mediated resistance.162, 163 Tigecycline consequently demonstrates good in vitro and in vivo activity against a wide range of mutant bacterial strains enabled with these resistance mechanisms, as well as potent activity against MRSA, penicillin-resistant streptococci (PRS), and VRE.160, 162, 164 SAR studies determined that the t-butylglycylamido moiety of tigecycline confers the highest activity, and replacement of glycine with other amino acids does not further enhance it.165 The 7-deaminated analog of tigecycline showed comparable antimicrobial activity.166 The binding mode of tigecycline to the 70S ribosome has been elucidated as well, showing similar interactions as observed with tetracycline, but with some significant differences.167 In particular, although both the 7-dimethylamino and the 9-tbutylglycylamido moieties of tigecycline do not seem to directly interact with any ribosomal nucleotide, they may enhance binding by increasing electron density of ring D, thereby enhancing the π-stacking interaction with C1054. The rigidity of the 9-t-butylglycylamido moiety also seems to induce conformational changes that allow stacking between C1054 and U1196, similar to the ribosomal conformation when tRNA is bound to the A site. The success and utility of tigecycline, coupled with climbing resistance rates against other major antibacterial classes, have recently prompted the development of several more novel tetracycline analogues.
Figure 9.
Tetracycline (cyan carbons) shown binding to the bacterial ribosome, 30S subunit [PDB ID 1HNW]. Hydrogen bonds are shown as dashed, yellow lines.
Figure 10.
Next generation tetracyclines & glycylcyclines marketed or in clinical trials. A. Tigecycline. B. Omadacycline. C. Eravacycline.
Omadacycline (Paratek Pharmaceuticals) is a novel tetracycline derivative with QIDP status in phase 3 trials for the treatment of ABSSSIs and CAP. Specifically, omadacycline is a C-9-substituted semi-synthetic minocycline derivative known as an aminomethylcycline (AMC) (Figure 10B).168, 169 It is a broad-spectrum antibacterial that shows strong activity against various tetracycline-resistant Gram-positive and Gram-negative organisms.168, 169 It has shown in vitro activity against additional organisms as well, including anaerobes, atypical pathogens, and organisms resistant to other classes of antibacterials, such as methicillin, vancomycin and ciprofloxacin.168 During the development of omadacycline, diversity sets based around a 9-aminomethylminocycline intermediate were analyzed to find and explore hits against tetracycline-resistant Gram-positive organisms.169 Initially increasing alkyl group size at this C-9 position was found to substantially increase activity against these organisms, while the side chain seen at this position on omadacycline was found to be optimal for activity as further increased methyl branching and different alkyl chain length were both found to decrease activity.169 Omadacycline is being formulated in both oral tablet form and intravenous form.168
Eravacycline (Tetraphase Pharmaceuticals) is another next-generation tetracycline, known as a fluorocycline. It has QIDP status and is in phase 3 trials for the treatment of cUTIs and cIAIs. While semi-synthetic development of tetracyclines has previously been hindered by a limited ability to manipulate C-7 and C-9 substituents, total synthesis approaches have recently been developed that allow for improved diversity at these positions (as well as C-8), which has allowed for the development of new 7,9-disubstituted tetracycline analogues, including eravacycline.170 As electronegativity at the C-7 substituent has been shown to effect the antibacterial potency of tetracyclines, a fluoro substituent at this position is found on eravacycline (Figure 10C).170, 171 Furthermore, SAR studies showed a small alkylamine substituent on the C-9 side chain to be the optimum amino group for antibacterial activity.170 Eravacycline has shown broad-spectrum in vitro activity against aerobic and anaerobic Gram-positive and Gram-negative pathogens, as well as MDR pathogens including ESBL- and carbapenemase-producing pathogens.172 It also maintains activity against tetracycline efflux pumps and ribosomal protection.173 Additionally, while results are still being analyzed, it is reported that eravacycline did not achieve its primary endpoint of statistical non-inferiority compared to levofloxacin for cUTI in the phase 3 trial IGNITE2.174
Next Generation Macrolides
Macrolide antibiotics are natural product-based antibacterials that are composed of a large, macrocyclic lactone core, to which a variety of sugar substituents may be attached. Erythromycin, which was discovered in soil samples collected in the Philippine islands, entered the U.S. market in 1952 and was followed by a variety of other macrolides over the following decades, notably clarithromycin (Biaxin, approved 1995) and azithromycin (Zithromax, approved 1996). Macrolide antibiotics exert their antibacterial effect by binding to and inhibiting the action of the bacterial 70S ribosome, specifically by binding to the 23S rRNA component of the 50S subunit and subsequently inhibiting cellular protein synthesis. The macrolide antibiotics are notable for their Gram-positive activity and are typically prescribed to treat respiratory tract infections caused by Gram-positive organisms (Streptococci, Staphylococci, and Enterococci) as well as certain “atypical” organism, including Legionella pneumophila, Mycoplasma spp., Chlamydia spp. and some Rickettsia. Disadvantages of the macrolide antibiotics include limited Gram-negative activity, acid instability limiting the oral effectiveness of some compounds in the class, and interaction with human drug metabolizing enzymes in the cytochrome P-450 system that can result in significant drug interactions.175 Further, emerging resistance, typically due to alterations of the macrolide binding site on the ribosome, has limited the clinical utility of older compounds in the class.
Telithromycin (Ketek, Sanofi-Aventis) was the first ‘next-generation’ macrolide to be approved for use in the U.S. in 2004 (Figure 11A). The compound, the first in the ‘ketolide’ subclass, is a semi-synthetic erythromycin derivative that substitutes a keto functionality at the 3-position for an L-cladinose. Additionally, a novel carbamate ring has been attached to the central lactone system at the 11- and 12-positions with a linked aryl-alkyl moiety.176 The 3-keto functionality is believed to be responsible for several desirable properties of the ketolides, including a higher degree of acid stability (aided by the 6-position methyl group), a lack of induction of macrolide resistance (MLSB phenotype), and the ability to overcome resistance caused by methylation of the 23S subunit (a component of the 50S subunit).176, 177 Further, the carbamate substituent plays a role in the expanded activity of telithromycin compared to erythromycin, and has been suggested to mediate the drug's resistance to bacterial efflux and play a role in improved pharmacokinetic properties.100, 101, 176, 178 Classical macrolides interact with domain V of the 23S rRNA subunit. The increased binding affinity of telithromycin has been shown to be due to an additional interaction between the carbamate functionality of telithromycin and residues in domain II of the 23S subunit.179 This increased binding affinity presumably allows the ketolide antibiotics to overcome resistance due to active site methylation and confers activity of telithromycin against erythromycin-resistant Gram-positive organisms. The drug shows excellent activity against most Gram-positive aerobic organisms, including macrolide-resistant strains of S. pneumoniae, excellent activity against atypical pathogens including C. pneumoniae, L. pneumophila, and M. pneumoniae, and good activity against some Gram-negative aerobes including M. catarrhalis and H. influenzae.177, 180-182 Although initially showing great promise and receiving U.S. approval for use in treatment of community-acquired pneumonia, acute bacterial sinusitis, and bacterial exacerbations of chronic bronchitis, telithromycin was subsequently discovered to cause irreversible hepatotoxicity in some patients and the sinusitis and bronchitis indications were removed by the FDA in 2007 and a black box warning added to the package literature.183 Clinical trial safety and data integrity concerns with this drug have been raised and have somewhat overshadowed the promise of the ketolide class.184, 185
Figure 11.
Advanced generation macrolides. A. Telithromycin. B. Cethromycin. C. Solithromycin.
Two new ketolide antibiotics that are currently in or have completed U.S. clinical trials include cethromycin (Figure 11B) (Restanza, ABT-773, Advanced Life Sciences, Inc.) and solithromycin (Figure 11C) (CEM-101, Cempra, Inc.). Cethromycin completed phase 3 clinical trials in 2007 for treatment of community-acquired pneumonia. However, while it was found to be safe, the drug was denied U.S. approval by the FDA in 2009 for reasons related to clinical trial design.186 Structurally, cethromycin possesses the 3-position keto group characteristic of the ketolide class as well as the 11, 12 carbamate moiety. Similar to telithromycin, cethromycin possesses an aryl-alkyl side chain, but in this case it is a quinolylallyl side chain attached to the C6 position versus the carbamate nitrogen in telithromycin. These structural features impart pharmacology similar to telithromycin, with two points of interaction with the 23S rRNA subunit, at domains II and V, affording the drug a spectrum of activity similar to telithromycin including activity against erythromycin resistant organisms and some resistance to efflux.187, 188 Solithromycin (CEM-101, Cempra, Inc.) is a novel fluoroketolide in phase 2 clinical trials in Europe for COPD and has recently completed phase 3 clinical trials in the U.S. comparing it to moxifloxacin for the treatment of community-acquired pneumonia.189, 190 Solithromycin is structurally similar to telithromycin, with a 3-keto moiety, an 11, 12 carbamate with an aryl-alkyl substitution (in this case a butyl-[1,2,3]-triazolyl-aminophenyl), and a unique fluorine substitution at the 2-position of the lactone (Figure 11C). With respect to its spectrum of activity, solithromycin shows a similar profile to telithromycin, with enhanced activity against some telithromycin intermediate and resistant organisms.191 The enhanced activity has been proposed to be due to improved binding of solithromycin to the bacterial ribosome over telithromycin, including Erm-methylated ribosomes.192 The x-ray crystal structure of solithromycin bound to the E. coli ribosome provides insight into possible reasons for this improved binding.192 First, the aryl-alkyl side chain forms interactions with rRNA base pairs in domain II of the 23S subunit, including a possible hydrogen bond mediated by the amino group. Second, weak electrostatic interactions of the 2-fluorine with an rRNA base near domain V may enhance the activity of solithromycin over telithromycin. Lastly, as with other ketolides, the substitution of the keto group at the 3-position for the cladinose sugar seen in earlier macrolide generations contributes to the binding affinity of the drug for Erm-methylated bacterial ribosomes.
Next Generation Aminoglycosides
Plazomicin (ACHN-490, Achaogen, Inc.) is a next generation, semi-synthetic aminoglycoside (neoglycoside) with reported activity against a broad range of Gram-positive and Gram-negative organisms, including carbapenem-resistant Enterobacteriaceae, when used in combination therapy with other antibacterial agents.193 Derived by synthetic modification of sisomicin, a hydroxy-aminbutyric acid substituent at position 1 and a hydroxyethyl substituent at position 6’ are reported to protect the compound against aminoglycoside-modifying enzymes in clinically relevant organisms (Figure 12).194 Plazomicin is not active against organisms with aminoglycoside resistance due to the expression of ribosomal methyltransferases. This agent is currently undergoing phase 3 clinical trials in Europe for blood stream infections and nosocomial pneumonia due to carbapenem-resistant Enterobacteriacea as well as cUTIs and acute pyelonephritis. It is also in phase 3 trials in the U.S. for treatment of complicated urinary tract infections, hospital- and ventilator-associated bacterial pneumonia, and complicated intraabdominal infection.
Figure 12.
Plazomicin.
Novel Bacterial Folate Synthesis Inhibitors
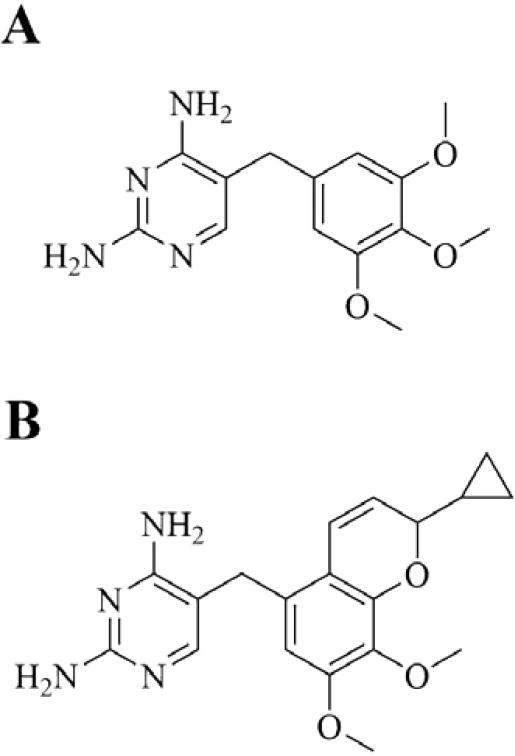
Iclaprim (Motif Bio plc) is a next generation dihydrofolate reductase (DHFR) inhibitor with QIDP status in phase 3 clinical trials for ABSSSI and cSSSI. It is a novel 2,4-diaminopyrimidine trimethoprim analogue that was rationally designed and synthesized by F. Hoffman-La Roche Ltd. using structural data and molecular modeling (Figure 13).195 Studies show a single point mutation on DHFR to be responsible for trimethoprim resistance, illustrated by an F98Y mutation in S. aureus strain B71 that causes the loss of a single H-bond between trimethoprim's 4-amino group and the enzyme, resulting in a 64-fold increase in MIC.196 It is posited that increased hydrophobic interactions between DHFR and iclaprim increase affinity, thereby overcoming trimethoprim resistance. Structural data shows that within the hydrophobic channel of DHFR, positioning of the p-aminobenzoic acid moiety of folate is similar to that of the chromene moiety of iclaprim.197 It has potent Gram-positive bactericidal activity, including activity against MSSA, MRSA, and trimethoprim-resistant F98Y mutant strains.197 Activity against Gram-negative and atypical organisms is reported to be similar to that of trimethoprim.198 The drug has reported synergy with the sulfonamide inhibitors of dihydropteroate synthase (DHPS), another key enzyme in the bacterial folate pathway.199, 200 In clinical trials, iclaprim has been compared with linezolid as monotherapy for cSSSI (ASSIST-1 and ASSIST-2 trials) and further phase 3 trials are underway comparing the drug, again as monotherapy, with vancomycin for ABSSSI (REVIVE-1 and REVIVE-2 trials).32 With respect to selectivity over mammalian DHFR, iclaprim has been found to inhibit bacterial DHFR at submicromolar concentrations and lack inhibition of human DHFR at more than 5 orders of magnitude higher concentrations, showcasing favorable selectivity.195
Figure 13.
Bacterial DHFR inhibitors. A. Trimethoprim. B. Iclaprim.
Novel Inhibitors of the Bacterial FAS-2 Pathway
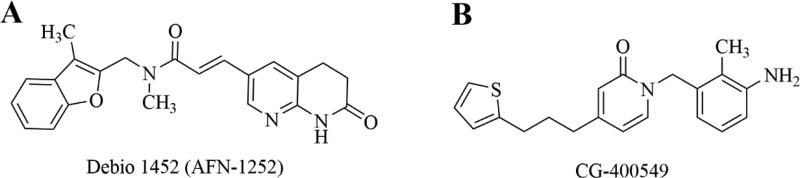
The type II bacterial fatty acid synthesis pathway, FAS II, provides fatty acid precursors for membrane phospholipids that are essential to bacterial cells. The pathway is distinct from the mammalian FAS I pathway which is composed of a single, multifunctional synthase, thus the FAS II enzymes represent novel and selective antibacterial targets that remain relatively unexploited. The structure and function of the enzymes in the bacterial FAS II pathway have been previously reviewed.201-204 As a metabolic pathway there is some concern about the ability of bacteria to bypass FAS II inhibition using exogenous fatty acids and this has been the subject of intense debate.205-207 Recent work, however, has demonstrated that certain bacteria, including S. aureus, remain susceptible to FAS II inhibition even in the presence of exogenous fatty acids due to differences in their regulation of genetic expression and feedback regulatory systems.208, 209 As a rate-limiting enzyme in the FAS-II pathway, FabI, enoyl-[acyl-carrier protein (ACP)] reductase, represents a particularly attractive drug target and a number of inhibitors of this enzyme have been characterized.210-221 Triclosan, a stereotypical FabI inhibitor with a diphenyl ether scaffold, has been marketed for a number of years and is used in a variety of over-the-counter household products as a sterilizing agent, however it has low utility for systemic use because of poor bioavailability.222, 223 Two promising FabI inhibitors, Debio 1452 and CG400549, with activity against S. aureus, have progressed sufficiently in clinical trials to merit discussion here.
Debio 1452 (previously AFN-1252, Debiopharm Group) is a FabI inhibitor with specific activity against staphylococci, including MRSA.219, 224 The drug has little to no activity against non-staphylococci Gram-positive organisms, including streptococci, due to the ability of these organisms to bypass the FAS II pathway with exogenous fatty acids or the absence of the FabI target enzyme in some species such as the streptococci.208 Further, Debio 1452 has negligible Gram-negative activity, making the drug quite selective for S. aureus and coagulase-negative staphylococci. The drug has recently completed U.S. phase 2 clinical trials for ABSSSI with favorable outcomes.225 A prodrug formulation, Debio 1450, allowing for oral and IV formulations is also currently in phase 2 clinical trials for ABSSSI.226 Structurally, Debio 1452 consists of a 3-methyl-benzofuran and an oxo-tetrahydro-naphthyridine linked by an N-methylpropenamide (Figure 14A).227 The compound was initially discovered using an iterative, structure-guided strategy, and has an interesting mechanism of inhibition involving the drug binding to a binary complex of enzyme and oxidized cofactor, NADP+ 219, 227, 228.
Figure 14.
Inhibitors of the bacterial FAS2 pathway. A. Debio 1452. Debio 1450 is a reported prodrug of Debio 1452, with undisclosed structure. B. CG-400549.
CG400549 (Crystal Genomics, Inc.) completed a phase 2 clinical trial in 2012 for treatment of cABSSSI caused by MRSA.32 Similar to Debio 1452, CG400549 has selective anti-staphylococcal activity, including activity against MRSA.229-231 The company announced the completion of the phase 2 trial and confirmed in vivo efficacy with minimal adverse events, however no plans for subsequent clinical trials have been reported.232 Structurally, the compound is a 2-pyridone analog (Figure 14B), having been rationally designed to improve on the poor pharmacokinetics of the diphenyl ether compounds represented by triclosan, specifically by replacing a metabolically unstable phenol group of triclosan with the pyridone ring system and the ether functionality of triclosan with a methylene group.215, 232 Recent x-ray structures have revealed the binding interactions of CG400549 in E. coli FabI and S. aureus FabI.232 The keto group of CG400549's pyridone ring, similar to the amide keto in Debio 1452, forms electrostatic interactions with both the protein (a key tyrosine residue) and the NADPH cofactor. Unlike Debio 1452, CG400549 is believed to bind to the protein-NADPH (reduced cofactor) binary complex, making the drug uncompetitive with respect to NADPH (vs. NADP+ for Debio 1452) and competitive with respect to the enoyl substrate. This structural information has guided recent efforts to expand the activity of CG400549 by iterative, structure-guided design strategies, to include Gram-negative pathogens and mycobacteria.232
Conclusions
The number of novel antibacterials recently approved or under late-stage development is highly encouraging, as is recent U.S. and European legislation facilitating the rapid development and approval of new antibacterials. At the time of this review, there are almost 60 antimicrobial drugs that have gained QIDP status in the U.S., 6 of which have already attained U.S. approval, and 37 that are in late-phase clinical trials.233 These results stand as evidence for the initial success of these new initiatives and their ability to drive the advancement of antibacterial development. A thorough analysis of the antibacterial pipeline, however, showcases the relatively low number of compounds under development with original targets, thereby highlighting the urgent need for the characterization and validation of such. Furthermore, it has been argued that ample attention should be given to the future development of selective antibacterial targets, giving way to what some predict will be the next phase of antibacterial discovery and development, the “narrow spectrum-era”.1 It is reasonable to believe that antibacterial agents geared toward such selective targets would have substantial benefits—such as the possibility of developing effective microbiome-sparing and resistance-mitigating therapeutic strategies. Characterizing and validating novel narrow-spectrum targets, of course, would require an understandably significant amount of resources, innovation, and effort—including fully rational drug design based around heretofore unconventional targets, the efficient use of modern techniques like diversity-oriented synthesis, as well as a certain degree of anticipated success.
Current techniques and methods being implemented in the rational design and optimization of antibacterial agents forecast a possible change in the state of affairs regarding infectious disease. As described above, recent advances in various techniques have helped gain ground in a struggle against bacterial infections where prolonged trends have demonstrated mostly the opposite. Promising examples include specific chiral configurations of new quinolones that grant improved activity under acidic conditions, fully-synthetic techniques used in the next-generation tetracyclines that allow for novel side-chains that grant activity against resistant pathogens, and semi-synthetic techniques used in the next-generation macrolides that allow for the modification of functional groups that yields subsequent binding improvements and, again, activity against resistant pathogens. Upon review, the recent advances in the rational design and optimization of antibacterial agents are, for the time being, highly encouraging.
Supplementary Material
Acknowledgments
KH gratefully acknowledges ITHS Rising Stars Career Development Program at University of Washington, which is supported by grants UL1TR000423, KL2TR000421, and TL1TR000422 from the NIH National Center for Advancing Translational Sciences through the Clinical and Translational Science Awards Program (CTSA). KH also acknowledges the support of the Mountain West IDeA Clinical and Translational Research – Infrastructure Network (CTR-IN), which is supported by a grant from the National Institute of General Medical Sciences (5 U54 GM104944). Support from the Idaho State University faculty development programs (Office of Research and College of Pharmacy) is also gratefully acknowledged.
Abbreviations
- ABSSSI
acute bacterial skin and skin structure infection
- AMC
aminomethylcycline
- CAP
community acquired pneumonia
- cIAI
complicated intra-abdominal infection
- COPD
chronic obstructive pulmonary disease
- cSSSI
complicated skin and skin structure infection
- DHFR
dihydrofolate reductase
- GAIN Act
Generating Antibiotics Incentives Now Act
- HTS
high-throughput screening
- IMI
Innovative Medicines Initiative
- MOI
mode of inhibition
- MRSA
methicillin-resistant Staphylococcus aureus
- MSSA
methicillin-sensitive Staphylococcus aureus
- NBTIs
Novel Bacterial Topoisomerase Inhibitors
- ND4BB
New Drugs 4 Bad Bugs
- QIDP
Qualified Infectious Disease Product
- SOA
spectrum of activity
- VABP
ventilator-associated bacterial pneumonia
Footnotes
The authors declare no competing interests.
Competing Interests
The authors declare no competing interests regarding the content of this manuscript.
References
- 1.Brown ED, Wright GD. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 2.Review on Antimicrobial Resistance: Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. 2014 [Google Scholar]
- 3.Centers_for_Disease_Control_and_Prevention [June 17, 2015];Antibiotic resistance threats in the United States. 2013 [Google Scholar]
- 4.Cooper MA, Shlaes D. Nature. 2011;472:32. doi: 10.1038/472032a. [DOI] [PubMed] [Google Scholar]
- 5.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Nat. Rev. Drug Discovery. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 6.Lewis K. Nat. Rev. Drug Discovery. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 7.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Testero SA, Fisher JF, Mobashery S. In: Burger's Medicinal Chemistry and Drug Discovery. 7th edn. Abraham DJ, Rotella DP, editors. Vol. 7. John Wiley and Sons, Inc.; Hoboken, NJ: 2010. pp. 257–402. [Google Scholar]
- 9.Ishikawa T, Matsunaga N, Tawada H, Kuroda N, Nakayama Y, Ishibashi Y, Tomimoto M, Ikeda Y, Tagawa Y, Iizawa Y, Okonogi K, Hashiguchi S, Miyake A. Bioorganic & medicinal chemistry. 2003;11:2427–2437. doi: 10.1016/s0968-0896(03)00126-3. [DOI] [PubMed] [Google Scholar]
- 10.Flamm RK, Sader HS, Farrell DJ, Jones RN. Antimicrobial agents and chemotherapy. 2012;56:2933–2940. doi: 10.1128/AAC.00330-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sader HS, Farrell DJ, Mendes RE, Flamm RK, Castanheira M, Jones RN. Diagn. Microbiol. Infect. Dis. 2015;82:78–84. doi: 10.1016/j.diagmicrobio.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein EJ, Citron DM, Merriam CV, Tyrrell KL. Diagn. Microbiol. Infect. Dis. 2013;76:347–351. doi: 10.1016/j.diagmicrobio.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 13.Zhanel GG, Sniezek G, Schweizer F, Zelenitsky S, Lagace-Wiens PR, Rubinstein E, Gin AS, Hoban DJ, Karlowsky JA. Drugs. 2009;69:809–831. doi: 10.2165/00003495-200969070-00003. [DOI] [PubMed] [Google Scholar]
- 14.Del Pozo JL, Patel R. Drugs of today. 2008;44:801–825. doi: 10.1358/dot.2008.44.11.1264007. [DOI] [PubMed] [Google Scholar]
- 15.Canada H. [June 14, 2016];Zeftera (ceftobiprole medocaril) for Injection - Discontinuation of Sale - Notice to Hospitals. http://www.healthycanadians.gc.ca/recall-alert-rappel-avis/hcsc/2010/14632a-eng.php.
- 16.Fritsche TR, Sader HS, Jones RN. Diagn. Microbiol. Infect. Dis. 2008;61:86–95. doi: 10.1016/j.diagmicrobio.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Bustos C, Del Pozo JL. Infect Drug Resist. 2010;3:5–14. doi: 10.2147/idr.s3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstein EJ, Citron DM, Merriam CV, Warren YA, Tyrrell KL, Fernandez HT. Antimicrobial agents and chemotherapy. 2006;50:3959–3962. doi: 10.1128/AAC.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Issa NC, Rouse MS, Piper KE, Wilson WR, Steckelberg JM, Patel R. Diagn. Microbiol. Infect. Dis. 2004;48:73–75. doi: 10.1016/j.diagmicrobio.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Lovering AL, Gretes MC, Safadi SS, Danel F, de Castro L, Page MG, Strynadka NC. J. Biol. Chem. 2012;287:32096–32102. doi: 10.1074/jbc.M112.355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sader HS, Rhomberg PR, Farrell DJ, Jones RN. Antimicrobial agents and chemotherapy. 2011;55:2390–2394. doi: 10.1128/AAC.01737-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livermore DM, Mushtaq S, Ge Y. The Journal of antimicrobial chemotherapy. 2010;65:1972–1974. doi: 10.1093/jac/dkq248. [DOI] [PubMed] [Google Scholar]
- 23.Armstrong ES, Farrell DJ, Palchak M, Steenbergen JN. Antimicrobial agents and chemotherapy. 2015;60:666–668. doi: 10.1128/AAC.01964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toda A, Ohki H, Yamanaka T, Murano K, Okuda S, Kawabata K, Hatano K, Matsuda K, Misumi K, Itoh K, Satoh K, Inoue S. Bioorg. Med. Chem. Lett. 2008;18:4849–4852. doi: 10.1016/j.bmcl.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 25.Murano K, Yamanaka T, Toda A, Ohki H, Okuda S, Kawabata K, Hatano K, Takeda S, Akamatsu H, Itoh K, Misumi K, Inoue S, Takagi T. Bioorganic & medicinal chemistry. 2008;16:2261–2275. doi: 10.1016/j.bmc.2007.11.074. [DOI] [PubMed] [Google Scholar]
- 26.Takeda S, Ishii Y, Hatano K, Tateda K, Yamaguchi K. Int. J. Antimicrob. Agents. 2007;30:443–445. doi: 10.1016/j.ijantimicag.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 27.Moya B, Zamorano L, Juan C, Ge Y, Oliver A. Antimicrobial agents and chemotherapy. 2010;54:3933–3937. doi: 10.1128/AAC.00296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohira N, West J, Ito A, Ito-Horiyama T, Nakamura R, Sato T, Rittenhouse S, Tsuji M, Yamano Y. Antimicrobial agents and chemotherapy. 2015;60:729–734. doi: 10.1128/AAC.01695-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito A, Kohira N, Bouchillon SK, West J, Rittenhouse S, Sader HS, Rhomberg PR, Jones RN, Yoshizawa H, Nakamura R, Tsuji M, Yamano Y. The Journal of antimicrobial chemotherapy. 2016;71:670–677. doi: 10.1093/jac/dkv402. [DOI] [PubMed] [Google Scholar]
- 30.Mollmann U, Heinisch L, Bauernfeind A, Kohler T, Ankel-Fuchs D. BioMetals. 2009;22:615–624. doi: 10.1007/s10534-009-9219-2. [DOI] [PubMed] [Google Scholar]
- 31.Ishii Y, Tateda K, Horiyama T, Tsuji M, Yamano Y, Shimada J, Yamaguchi K. presented in part at the 54th Interscience Conference on Antimibrobial Agents and Chemotherapy; Washington, DC: 2014. [Google Scholar]
- 32. [2/24/2016]; ClinicalTrials.gov.
- 33.Babic M, Hujer AM, Bonomo RA. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2006;9:142–156. doi: 10.1016/j.drup.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S. Infection. 1983;11:315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 35.Cornaglia G, Giamarellou H, Rossolini GM. Lancet Infect. Dis. 2011;11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 36.Philippon A, Labia R, Jacoby G. Antimicrobial agents and chemotherapy. 1989;33:1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers RA. Curr. Drug Targets. 2015;17:1051–1060. doi: 10.2174/1573399811666150615144707. [DOI] [PubMed] [Google Scholar]
- 38.Livermore DM. Eur. J. Clin. Microbiol. 1987;6:439–445. doi: 10.1007/BF02013107. [DOI] [PubMed] [Google Scholar]
- 39.Gupta N, Limbago BM, Patel JB, Kallen AJ. Clin. Infect. Dis. 2011;53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 40.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. Antimicrobial agents and chemotherapy. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh TR. Clin. Microbiol. Infect. 2005;11(Suppl 6):2–9. doi: 10.1111/j.1469-0691.2005.01264.x. [DOI] [PubMed] [Google Scholar]
- 42.Nordmann P, Poirel L, Walsh TR, Livermore DM. Trends Microbiol. 2011;19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Zasowski EJ, Rybak JM, Rybak MJ. Pharmacotherapy. 2015;35:755–770. doi: 10.1002/phar.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhanel GG, Lawson CD, Adam H, Schweizer F, Zelenitsky S, Lagace-Wiens PR, Denisuik A, Rubinstein E, Gin AS, Hoban DJ, Lynch JP, 3rd, Karlowsky JA. Drugs. 2013;73:159–177. doi: 10.1007/s40265-013-0013-7. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Estabrook M, Jacoby GA, Nichols WW, Testa RT, Bush K. Antimicrobial agents and chemotherapy. 2015;59:1789–1793. doi: 10.1128/AAC.04191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshizumi A, Ishii Y, Aoki K, Testa R, Nichols WW, Tateda K. J. Infect. Chemother. 2015;21:148–151. doi: 10.1016/j.jiac.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 47.Sader HS, Castanheira M, Mendes RE, Flamm RK, Farrell DJ, Jones RN. Antimicrobial agents and chemotherapy. 2015;59:3656–3659. doi: 10.1128/AAC.05024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sader HS, Castanheira M, Flamm RK, Farrell DJ, Jones RN. Antimicrobial agents and chemotherapy. 2014;58:1684–1692. doi: 10.1128/AAC.02429-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubreuil LJ, Mahieux S, Neut C, Miossec C, Pace J. Int. J. Antimicrob. Agents. 2012;39:500–504. doi: 10.1016/j.ijantimicag.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 50.Citron DM, Tyrrell KL, Merriam V, Goldstein EJ. Antimicrobial agents and chemotherapy. 2011;55:3616–3620. doi: 10.1128/AAC.01682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lahiri SD, Johnstone MR, Ross PL, McLaughlin RE, Olivier NB, Alm RA. Antimicrobial agents and chemotherapy. 2014;58:5704–5713. doi: 10.1128/AAC.03057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lahiri SD, Mangani S, Durand-Reville T, Benvenuti M, De Luca F, Sanyal G, Docquier JD. Antimicrobial agents and chemotherapy. 2013;57:2496–2505. doi: 10.1128/AAC.02247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lahiri SD, Mangani S, Jahic H, Benvenuti M, Durand-Reville TF, De Luca F, Ehmann DE, Rossolini GM, Alm RA, Docquier JD. ACS Chem. Biol. 2015;10:591–600. doi: 10.1021/cb500703p. [DOI] [PubMed] [Google Scholar]
- 54.Krishnan NP, Nguyen NQ, Papp-Wallace KM, Bonomo RA, van den Akker F. PLoS One. 2015;10:e0136813. doi: 10.1371/journal.pone.0136813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonnefoy A, Dupuis-Hamelin C, Steier V, Delachaume C, Seys C, Stachyra T, Fairley M, Guitton M, Lampilas M. The Journal of antimicrobial chemotherapy. 2004;54:410–417. doi: 10.1093/jac/dkh358. [DOI] [PubMed] [Google Scholar]
- 56.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aktas Z, Kayacan C, Oncul O. Int. J. Antimicrob. Agents. 2012;39:86–89. doi: 10.1016/j.ijantimicag.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 58.Young K, Raghoobar S, Hairston N, et al. presented in part at the 50th Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC); Boston, MA. September 12-15, 2010. [Google Scholar]
- 59.Livermore DM, Warner M, Mushtaq S. The Journal of antimicrobial chemotherapy. 2013;68:2286–2290. doi: 10.1093/jac/dkt178. [DOI] [PubMed] [Google Scholar]
- 60.Hirsch EB, Ledesma KR, Chang KT, Schwartz MS, Motyl MR, Tam VH. Antimicrobial agents and chemotherapy. 2012;56:3753–3757. doi: 10.1128/AAC.05927-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young K, Hackel M, Lascols C, et al. presented in part at the 52nd Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC); San Franscisco, CA. September 9-12, 2012. [Google Scholar]
- 62.Lapuebla A, Abdallah M, Olafisoye O, Cortes C, Urban C, Landman D, Quale J. Antimicrobial agents and chemotherapy. 2015;59:5029–5031. doi: 10.1128/AAC.00830-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blizzard TA, Chen H, Kim S, Wu J, Bodner R, Gude C, Imbriglio J, Young K, Park YW, Ogawa A, Raghoobar S, Hairston N, Painter RE, Wisniewski D, Scapin G, Fitzgerald P, Sharma N, Lu J, Ha S, Hermes J, Hammond ML. Bioorg. Med. Chem. Lett. 2014;24:780–785. doi: 10.1016/j.bmcl.2013.12.101. [DOI] [PubMed] [Google Scholar]
- 64.Wenzler E, Gotfried MH, Loutit JS, Durso S, Griffith DC, Dudley MN, Rodvold KA. Antimicrobial agents and chemotherapy. 2015;59:7232–7239. doi: 10.1128/AAC.01713-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lapuebla A, Abdallah M, Olafisoye O, Cortes C, Urban C, Quale J, Landman D. Antimicrobial agents and chemotherapy. 2015;59:4856–4860. doi: 10.1128/AAC.00843-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Livermore DM, Mushtaq S. The Journal of antimicrobial chemotherapy. 2013;68:1825–1831. doi: 10.1093/jac/dkt118. [DOI] [PubMed] [Google Scholar]
- 67.Goldstein EJ, Citron DM, Tyrrell KL, Merriam CV. Antimicrobial agents and chemotherapy. 2013;57:2620–2630. doi: 10.1128/AAC.02418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smoum R, Rubinstein A, Dembitsky VM, Srebnik M. Chem. Rev. 2012;112:4156–4220. doi: 10.1021/cr608202m. [DOI] [PubMed] [Google Scholar]
- 69.Kiener PA, Waley SG. Biochem. J. 1978;169:197–204. doi: 10.1042/bj1690197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dobozy O, Mile I, Ferencz I, Csanyi V. Acta Biochim. Biophys. Acad. Sci. Hung. 1971;6:97–105. [PubMed] [Google Scholar]
- 71.Strynadka NC, Adachi H, Jensen SE, Johns K, Sielecki A, Betzel C, Sutoh K, James MN. Nature. 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 72.Martin R, Jones JB. Tetrahedron Lett. 1995;36:8399–8402. [Google Scholar]
- 73.Strynadka NC, Martin R, Jensen SE, Gold M, Jones JB. Nat. Struct. Biol. 1996;3:688–695. doi: 10.1038/nsb0896-688. [DOI] [PubMed] [Google Scholar]
- 74.Ness S, Martin R, Kindler AM, Paetzel M, Gold M, Jensen SE, Jones JB, Strynadka NC. Biochemistry. 2000;39:5312–5321. doi: 10.1021/bi992505b. [DOI] [PubMed] [Google Scholar]
- 75.Morandi F, Caselli E, Morandi S, Focia PJ, Blazquez J, Shoichet BK, Prati F. J. Am. Chem. Soc. 2003;125:685–695. doi: 10.1021/ja0288338. [DOI] [PubMed] [Google Scholar]
- 76.Wang X, Minasov G, Blazquez J, Caselli E, Prati F, Shoichet BK. Biochemistry. 2003;42:8434–8444. doi: 10.1021/bi034242y. [DOI] [PubMed] [Google Scholar]
- 77.Hecker SJ, Reddy KR, Totrov M, Hirst GC, Lomovskaya O, Griffith DC, King P, Tsivkovski R, Sun D, Sabet M, Tarazi Z, Clifton MC, Atkins K, Raymond A, Potts KT, Abendroth J, Boyer SH, Loutit JS, Morgan EE, Durso S, Dudley MN. Journal of medicinal chemistry. 2015;58:3682–3692. doi: 10.1021/acs.jmedchem.5b00127. [DOI] [PubMed] [Google Scholar]
- 78.Moellering RC. Ann. Intern. Med. 2003;138:135–142. doi: 10.7326/0003-4819-138-2-200301210-00015. [DOI] [PubMed] [Google Scholar]
- 79.Ford CW, Zurenko GE, Barbachyn MR. Curr. Drug Targets Infect. Disord. 2001;1:181–199. doi: 10.2174/1568005014606099. [DOI] [PubMed] [Google Scholar]
- 80.Slee AM, Wuonola MA, McRipley RJ, Zajac I, Zawada MJ, Bartholomew PT, Gregory WA, Forbes M. Antimicrobial agents and chemotherapy. 1987;31:1791–1797. doi: 10.1128/aac.31.11.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barbachyn MR, Ford CW. Angew. Chem. Int. Ed. Engl. 2003;42:2010–2023. doi: 10.1002/anie.200200528. [DOI] [PubMed] [Google Scholar]
- 82.Gregory WA, Brittelli DR, Wang CL, Kezar HS, 3rd, Carlson RK, Park CH, Corless PF, Miller SJ, Rajagopalan P, Wuonola MA, et al. Journal of medicinal chemistry. 1990;33:2569–2578. doi: 10.1021/jm00171a035. [DOI] [PubMed] [Google Scholar]
- 83.Gregory WA, Brittelli DR, Wang CL, Wuonola MA, McRipley RJ, Eustice DC, Eberly VS, Bartholomew PT, Slee AM, Forbes M. Journal of medicinal chemistry. 1989;32:1673–1681. doi: 10.1021/jm00128a003. [DOI] [PubMed] [Google Scholar]
- 84.Park CH, Brittelli DR, Wang CL, Marsh FD, Gregory WA, Wuonola MA, McRipley RJ, Eberly VS, Slee AM, Forbes M. Journal of medicinal chemistry. 1992;35:1156–1165. doi: 10.1021/jm00084a022. [DOI] [PubMed] [Google Scholar]
- 85.Barbachyn MR, Cleek GJ, Dolak LA, Garmon SA, Morris J, Seest EP, Thomas RC, Toops DS, Watt W, Wishka DG, Ford CW, Zurenko GE, Hamel JC, Schaadt RD, Stapert D, Yagi BH, Adams WJ, Friis JM, Slatter JG, Sams JP, Oien NL, Zaya MJ, Wienkers LC, Wynalda MA. J. Med. Chem. 2003;46:284–302. doi: 10.1021/jm020248u. [DOI] [PubMed] [Google Scholar]
- 86.Borthwick ADB, K., Rocherolle V, Cox DM, Chung GAC. Med. Chem. Res. 1996;6:22–27. [Google Scholar]
- 87.Dennis A, Villette T. Bioorg. Med. Chem. Lett. 1994;4:1925–1930. [Google Scholar]
- 88.Quesnelle CA, Gill P, Roy S, Dodier M, Marinier A, Martel A, Snyder LB, D'Andrea SV, Bronson JJ, Frosco M, Beaulieu D, Warr GA, Denbleyker KL, Stickle TM, Yang H, Chaniewski SE, Ferraro CA, Taylor D, Russell JW, Santone KS, Clarke J, Drain RL, Knipe JO, Mosure K, Barrett JF. Bioorg. Med. Chem. Lett. 2005;15:2728–2733. doi: 10.1016/j.bmcl.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 89.Snyder LB, Meng Z, Mate R, D'Andrea SV, Marinier A, Quesnelle CA, Gill P, DenBleyker KL, Fung-Tomc JC, Frosco M, Martel A, Barrett JF, Bronson JJ. Bioorg. Med. Chem. Lett. 2004;14:4735–4739. doi: 10.1016/j.bmcl.2004.06.076. [DOI] [PubMed] [Google Scholar]
- 90.Brickner SJ. Curr. Pharm. Des. 1996;2:175–194. [Google Scholar]
- 91.Barbachyn MR, Hutchinson DK, Brickner SJ, Cynamon MH, Kilburn JO, Klemens SP, Glickman SE, Grega KC, Hendges SK, Toops DS, Ford CW, Zurenko GE. Journal of medicinal chemistry. 1996;39:680–685. doi: 10.1021/jm950956y. [DOI] [PubMed] [Google Scholar]
- 92.Jo YW, Im WB, Rhee JK, Shim MJ, Kim WB, Choi EC. Bioorganic & medicinal chemistry. 2004;12:5909–5915. doi: 10.1016/j.bmc.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 93.Gravestock MB, Acton DG, Betts MJ, Dennis M, Hatter G, McGregor A, Swain ML, Wilson RG, Woods L, Wookey A. Bioorg. Med. Chem. Lett. 2003;13:4179–4186. doi: 10.1016/j.bmcl.2003.07.033. [DOI] [PubMed] [Google Scholar]
- 94.Phillips OA, Udo EE, Ali AA, Al-Hassawi N. Bioorganic & medicinal chemistry. 2003;11:35–41. doi: 10.1016/s0968-0896(02)00423-6. [DOI] [PubMed] [Google Scholar]
- 95.Swaney SM, Aoki H, Ganoza MC, Shinabarger DL. Antimicrobial agents and chemotherapy. 1998;42:3251–3255. doi: 10.1128/aac.42.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kisgen JJ, Mansour H, Unger NR, Childs LM. Am. J. Health Syst. Pharm. 2014;71:621–633. doi: 10.2146/ajhp130482. [DOI] [PubMed] [Google Scholar]
- 97.Locke JB, Zurenko GE, Shaw KJ, Bartizal K. Clin. Infect. Dis. 2014;58(Suppl 1):S35–42. doi: 10.1093/cid/cit616. [DOI] [PubMed] [Google Scholar]
- 98.Zurenko GE, Gibson JK, Shinabarger DL, Aristoff PA, Ford CW, Tarpley WG. Curr. Opin. Pharmacol. 2001;1:470–476. doi: 10.1016/s1471-4892(01)00082-0. [DOI] [PubMed] [Google Scholar]
- 99.Schumacher A, Trittler R, Bohnert JA, Kummerer K, Pages JM, Kern WV. The Journal of antimicrobial chemotherapy. 2007;59:1261–1264. doi: 10.1093/jac/dkl380. [DOI] [PubMed] [Google Scholar]
- 100.Roberts MC. Curr. Drug Targets Infect. Disord. 2004;4:207–215. doi: 10.2174/1568005043340678. [DOI] [PubMed] [Google Scholar]
- 101.Roberts MC. FEMS Microbiol. Lett. 2008;282:147–159. doi: 10.1111/j.1574-6968.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- 102.Zhanel GG, Love R, Adam H, Golden A, Zelenitsky S, Schweizer F, Gorityala B, Lagace-Wiens PR, Rubinstein E, Walkty A, Gin AS, Gilmour M, Hoban DJ, Lynch JP, 3rd, Karlowsky JA. Drugs. 2015;75:253–270. doi: 10.1007/s40265-015-0352-7. [DOI] [PubMed] [Google Scholar]
- 103.Stryjewski ME, Corey GR. Clin. Infect. Dis. 2014;58(Suppl 1):S10–19. doi: 10.1093/cid/cit613. [DOI] [PubMed] [Google Scholar]
- 104.Locke JB, Zuill DE, Scharn CR, Deane J, Sahm DF, Denys GA, Goering RV, Shaw KJ. Antimicrobial agents and chemotherapy. 2014;58:6592–6598. doi: 10.1128/AAC.03493-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morales G, Picazo JJ, Baos E, Candel FJ, Arribi A, Pelaez B, Andrade R, de la Torre MA, Fereres J, Sanchez-Garcia M. Clin. Infect. Dis. 2010;50:821–825. doi: 10.1086/650574. [DOI] [PubMed] [Google Scholar]
- 106.Chahine EB, Sucher AJ, Knutsen SD. Consult. Pharm. 2015;30:386–394. doi: 10.4140/TCP.n.2015.386. [DOI] [PubMed] [Google Scholar]
- 107.Durkin MJ, Corey GR. Ther. Clin. Risk Manag. 2015;11:857–862. doi: 10.2147/TCRM.S64553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hall RG, 2nd, Michaels HN. Infect Drug Resist. 2015;8:75–82. doi: 10.2147/IDR.S56691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morata L, Mensa J, Soriano A. Curr. Opin. Pharmacol. 2015;24:45–51. doi: 10.1016/j.coph.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 110.Kanafani ZA, Corey GR. Expert Opin Investig Drugs. 2012;21:515–522. doi: 10.1517/13543784.2012.660250. [DOI] [PubMed] [Google Scholar]
- 111.Shen K, Yoshikawa K, Tanaka T, et al. presented in part at the 43rd Society of Critical Care Medicine Critical Care Congress; San Francisco, CA. 2014. [Google Scholar]
- 112.Tanaka T, Hayashi Y, Okumura K, et al. presented in part at the 24th European Congress of Clinical Microbiology and Infectious Diseases; Barcelona, Spain. 2014. [Google Scholar]
- 113.Locke JB, Finn J, Hilgers M, Morales G, Rahawi S, Picazo CKG,JJ, Im W, Shaw KJ, Stein JL. Antimicrobial agents and chemotherapy. 2010;54:5337–5343. doi: 10.1128/AAC.00663-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Locke J, Finn J, Hilgers M, et al. presented in part at the 50th Interscience Conference on Antimicrobial Agents and Chemotherapy; Boston, MA. 2010. [Google Scholar]
- 115.Michalska K, Karpiuk I, Krol M, Tyski S. Bioorganic & medicinal chemistry. 2013;21:577–591. doi: 10.1016/j.bmc.2012.11.036. [DOI] [PubMed] [Google Scholar]
- 116.Shaw KJ, Poppe S, Schaadt R, Brown-Driver V, Finn J, Pillar CM, Shinabarger D, Zurenko G. Antimicrobial agents and chemotherapy. 2008;52:4442–4447. doi: 10.1128/AAC.00859-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yum JH, Choi SH, Yong D, Chong Y, Im WB, Rhee DK, Lee K. Antimicrobial agents and chemotherapy. 2010;54:5381–5386. doi: 10.1128/AAC.00728-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Prokocimer P, Bien P, Deanda C, Pillar CM, Bartizal K. Antimicrobial agents and chemotherapy. 2012;56:4608–4613. doi: 10.1128/AAC.00458-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brown SD, Traczewski MM. Antimicrobial agents and chemotherapy. 2010;54:2063–2069. doi: 10.1128/AAC.01569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schaadt R, Sweeney D, Shinabarger D, Zurenko G. Antimicrobial agents and chemotherapy. 2009;53:3236–3239. doi: 10.1128/AAC.00228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen H, Yang Q, Zhang R, He W, Ma X, Zhang J, Xia F, Zhao F, Cao J, Liu Y, Wu W, Hu D, Wang Q, Zhao C, Zhang F, Wang X, Wang Z, Li H, Wang H. Int. J. Antimicrob. Agents. 2014;44:276–277. doi: 10.1016/j.ijantimicag.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 122.Jones RN, Moet GJ, Sader HS, Mendes RE, Castanheira M. The Journal of antimicrobial chemotherapy. 2009;63:716–720. doi: 10.1093/jac/dkp021. [DOI] [PubMed] [Google Scholar]
- 123.Shaw KJ, Barbachyn MR. Ann. N. Y. Acad. Sci. 2011;1241:48–70. doi: 10.1111/j.1749-6632.2011.06330.x. [DOI] [PubMed] [Google Scholar]
- 124.Ippolito J, Kanyo B, Wimberly B, et al. presented in part at the 45th Interscience Conference on Antimicrobial Agents and Chemotherapy; Washington, DC. 2005. [Google Scholar]
- 125.Wang D, Sherer E, Duffy E. presented in part at the 45th Interscience Conference on Antimicrobial Agents and Chemotherapy; Washington, DC. 2005. [Google Scholar]
- 126.Zhou J, Bhattacharjee A, Chen S, Chen Y, Duffy E, Farmer J, Goldberg J, Hanselmann R, Ippolito JA, Lou R, Orbin A, Oyelere A, Salvino J, Springer D, Tran J, Wang D, Wu Y, Johnson G. Bioorg. Med. Chem. Lett. 2008;18:6179–6183. doi: 10.1016/j.bmcl.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 127.Zhou J, Bhattacharjee A, Chen S, Chen Y, Duffy E, Farmer J, Goldberg J, Hanselmann R, Ippolito JA, Lou R, Orbin A, Oyelere A, Salvino J, Springer D, Tran J, Wang D, Wu Y, Johnson G. Bioorg. Med. Chem. Lett. 2008;18:6175–6178. doi: 10.1016/j.bmcl.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 128.Lawrence L, Danese P, DeVito J, Franceschi F, Sutcliffe J. Antimicrobial agents and chemotherapy. 2008;52:1653–1662. doi: 10.1128/AAC.01383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Skripkin E, McConnell TS, DeVito J, Lawrence L, Ippolito JA, Duffy EM, Sutcliffe J, Franceschi F. Antimicrobial agents and chemotherapy. 2008;52:3550–3557. doi: 10.1128/AAC.01193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gordeev MF, Yuan ZY. Journal of medicinal chemistry. 2014;57:4487–4497. doi: 10.1021/jm401931e. [DOI] [PubMed] [Google Scholar]
- 131.Li CR, Zhai QQ, Wang XK, Hu XX, Li GQ, Zhang WX, Pang J, Lu X, Yuan H, Gordeev MF, Chen LT, Yang XY, You XF. Antimicrobial agents and chemotherapy. 2014;58:2418–2421. doi: 10.1128/AAC.01526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kali A, Charles MV, Srirangaraj S. Australas Med J. 2015;8:253–262. doi: 10.4066/AMJ.2015.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gordeev MF, Hackbarth C, Barbachyn MR, Banitt LS, Gage JR, Luehr GW, Gomez M, Trias J, Morin SE, Zurenko GE, Parker CN, Evans JM, White RJ, Patel DV. Bioorg. Med. Chem. Lett. 2003;13:4213–4216. doi: 10.1016/j.bmcl.2003.07.021. [DOI] [PubMed] [Google Scholar]
- 134.Hubschwerlen C, Specklin JL, Baeschlin DK, Borer Y, Haefeli S, Sigwalt C, Schroeder S, Locher HH. Bioorg. Med. Chem. Lett. 2003;13:4229–4233. doi: 10.1016/j.bmcl.2003.07.028. [DOI] [PubMed] [Google Scholar]
- 135.Hubschwerlen C, Specklin JL, Sigwalt C, Schroeder S, Locher HH. Bioorganic & medicinal chemistry. 2003;11:2313–2319. doi: 10.1016/s0968-0896(03)00083-x. [DOI] [PubMed] [Google Scholar]
- 136.Locher HH, Caspers P, Bruyere T, Schroeder S, Pfaff P, Knezevic A, Keck W, Ritz D. Antimicrobial agents and chemotherapy. 2014;58:901–908. doi: 10.1128/AAC.01831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rashid MU, Dalhoff A, Weintraub A, Nord CE. Anaerobe. 2014;28:216–219. doi: 10.1016/j.anaerobe.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 138.Rashid MU, Lozano HM, Weintraub A, Nord CE. Anaerobe. 2013;20:32–35. doi: 10.1016/j.anaerobe.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 139.Locher HH, Seiler P, Chen X, Schroeder S, Pfaff P, Enderlin M, Klenk A, Fournier E, Hubschwerlen C, Ritz D, Kelly CP, Keck W. Antimicrobial agents and chemotherapy. 2014;58:892–900. doi: 10.1128/AAC.01830-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Baldoni D, Gutierrez M, Timmer W, Dingemanse J. The Journal of antimicrobial chemotherapy. 2014;69:706–714. doi: 10.1093/jac/dkt401. [DOI] [PubMed] [Google Scholar]
- 141.Miller AA, Bundy GL, Mott JE, Skepner JE, Boyle TP, Harris DW, Hromockyj AE, Marotti KR, Zurenko GE, Munzner JB, Sweeney MT, Bammert GF, Hamel JC, Ford CW, Zhong WZ, Graber DR, Martin GE, Han F, Dolak LA, Seest EP, Ruble JC, Kamilar GM, Palmer JR, Banitt LS, Hurd AR, Barbachyn MR. Antimicrobial agents and chemotherapy. 2008;52:2806–2812. doi: 10.1128/AAC.00247-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Basarab GS, Kern GH, McNulty J, Mueller JP, Lawrence K, Vishwanathan K, Alm RA, Barvian K, Doig P, Galullo V, Gardner H, Gowravaram M, Huband M, Kimzey A, Morningstar M, Kutschke A, Lahiri SD, Perros M, Singh R, Schuck VJ, Tommasi R, Walkup G, Newman JV. Sci. Rep. 2015;5:11827. doi: 10.1038/srep11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Unemo M, Ringlander J, Wiggins C, Fredlund H, Jacobsson S, Cole M. Antimicrobial agents and chemotherapy. 2015;59:5220–5225. doi: 10.1128/AAC.00786-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Huband MD, Bradford PA, Otterson LG, Basarab GS, Kutschke AC, Giacobbe RA, Patey SA, Alm RA, Johnstone MR, Potter ME, Miller PF, Mueller JP. Antimicrobial agents and chemotherapy. 2015;59:467–474. doi: 10.1128/AAC.04124-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Biedenbach DJ, Bouchillon SK, Hackel M, Miller LA, Scangarella-Oman NE, Jakielaszek C, Sahm DF. Antimicrobial agents and chemotherapy. 2016;60:1918–1923. doi: 10.1128/AAC.02820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Singh SB. Bioorg. Med. Chem. Lett. 2014;24:3683–3689. doi: 10.1016/j.bmcl.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 147.Bax BD, Chan PF, Eggleston DS, Fosberry A, Gentry DR, Gorrec F, Giordano I, Hann MM, Hennessy A, Hibbs M, Huang J, Jones E, Jones J, Brown KK, Lewis CJ, May EW, Saunders MR, Singh O, Spitzfaden CE, Shen C, Shillings A, Theobald AJ, Wohlkonig A, Pearson ND, Gwynn MN. Nature. 2010;466:935–940. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 148.Widdowson K, Hennessy A. Future Med. Chem. 2010;2:1619–1622. doi: 10.4155/fmc.10.250. [DOI] [PubMed] [Google Scholar]
- 149.Lemaire S, Van Bambeke F, Tulkens PM. Int. J. Antimicrob. Agents. 2011;38:52–59. doi: 10.1016/j.ijantimicag.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 150.Wohlert S-E, Jaetsch T, Gallenkamp B, Knops HJ, Lui N, Preiss M, Haebich D, Labischinski H. presented in part at the 48th Interscience Converence on Antimicrobial Agents and Chemotherapy (ICAAC); Washington, D.C.. October 25-28, 2008. [Google Scholar]
- 151.Adam HJ, Laing NM, King CR, Lulashnyk B, Hoban DJ, Zhanel GG. Antimicrobial agents and chemotherapy. 2009;53:4915–4920. doi: 10.1128/AAC.00078-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Qin X, Huang H. Drug Des. Devel. Ther. 2014;8:765–774. doi: 10.2147/DDDT.S63581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li ZX, Liu YN, Wang R, Li AM. Clin. Ter. 2015;166:e374–380. doi: 10.7417/T.2015.1903. [DOI] [PubMed] [Google Scholar]
- 154.Chen YH, Liu CY, Lu JJ, King CH, Hsueh PR. The Journal of antimicrobial chemotherapy. 2009;64:1226–1229. doi: 10.1093/jac/dkp370. [DOI] [PubMed] [Google Scholar]
- 155.Nilius AM, Shen LL, Hensey-Rudloff D, Almer LS, Beyer JM, Balli DJ, Cai Y, Flamm RK. Antimicrobial agents and chemotherapy. 2003;47:3260–3269. doi: 10.1128/AAC.47.10.3260-3269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Van Bambeke F. Future Microbiol. 2015;10:1111–1123. doi: 10.2217/fmb.15.39. [DOI] [PubMed] [Google Scholar]
- 157.Lemaire S, Tulkens PM, Van Bambeke F. Antimicrobial agents and chemotherapy. 2011;55:649–658. doi: 10.1128/AAC.01201-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.So W, Crandon JL, Nicolau DP. J. Urol. 2015;194:563–570. doi: 10.1016/j.juro.2015.01.094. [DOI] [PubMed] [Google Scholar]
- 159.Duffy EM, DeVito JA, Remy J, Burak ES. presented in part at the 50th Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC); Boston, MA. September 12-15, 2010. [Google Scholar]
- 160.Brodersen DE, Clemons WM, Jr., Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 161.Sum PE, Petersen P. Bioorg. Med. Chem. Lett. 1999;9:1459–1462. doi: 10.1016/s0960-894x(99)00216-4. [DOI] [PubMed] [Google Scholar]
- 162.Bergeron J, Ammirati M, Danley D, James L, Norcia M, Retsema J, Strick CA, Su WG, Sutcliffe J, Wondrack L. Antimicrobial. 1996;40:2226–2228. doi: 10.1128/aac.40.9.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Petersen PJ, Jacobus NV, Weiss WJ, Sum PE, Testa RT. Antimicrob. Agents Chemother. 1999;43:738–744. doi: 10.1128/aac.43.4.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Garrison MW, Neumiller JJ, Setter SM. Clin. Ther. 2005;27:12–22. doi: 10.1016/j.clinthera.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 165.Barden TC, Buckwalter BL, Testa RT, Petersen PJ, Lee VJ. Journal of medicinal chemistry. 1994;37:3205–3211. doi: 10.1021/jm00046a003. [DOI] [PubMed] [Google Scholar]
- 166.Eliopoulos GM, Wennersten CB, Cole G, Moellering RC. Antimicrobial agents and chemotherapy. 1994;38:534–541. doi: 10.1128/aac.38.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jenner L, Starosta AL, Terry DS, Mikolajka A, Filonava L, Yusupov M, Blanchard SC, Wilson DN, Yusupova G. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3812–3816. doi: 10.1073/pnas.1216691110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Macone AB, Caruso BK, Leahy RG, Donatelli J, Weir S, Draper MP, Tanaka SK, Levy SB. Antimicrobial agents and chemotherapy. 2014;58:1127–1135. doi: 10.1128/AAC.01242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Honeyman L, Ismail M, Nelson ML, Bhatia B, Bowser TE, Chen J, Mechiche R, Ohemeng K, Verma AK, Cannon EP, Macone A, Tanaka SK, Levy S. Antimicrobial agents and chemotherapy. 2015;59:7044–7053. doi: 10.1128/AAC.01536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xiao XY, Hunt DK, Zhou J, Clark RB, Dunwoody N, Fyfe C, Grossman TH, O'Brien WJ, Plamondon L, Ronn M, Sun C, Zhang WY, Sutcliffe JA. Journal of medicinal chemistry. 2012;55:597–605. doi: 10.1021/jm201465w. [DOI] [PubMed] [Google Scholar]
- 171.Cammarata A, Yau SJ. Journal of medicinal chemistry. 1970;13:93–97. doi: 10.1021/jm00295a024. [DOI] [PubMed] [Google Scholar]
- 172.Sutcliffe JA, O'Brien W, Fyfe C, Grossman TH. Antimicrobial agents and chemotherapy. 2013;57:5548–5558. doi: 10.1128/AAC.01288-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Grossman TH, Starosta AL, Fyfe C, O'Brien W, Rothstein DM, Mikolajka A, Wilson DN, Sutcliffe JA. Antimicrobial agents and chemotherapy. 2012;56:2559–2564. doi: 10.1128/AAC.06187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhanel GG, Cheung D, Adam H, Zelenitsky S, Golden A, Schweizer F, Gorityala B, Lagace-Wiens PR, Walkty A, Gin AS, Hoban DJ, Karlowsky JA. Drugs. 2016;76:567–588. doi: 10.1007/s40265-016-0545-8. [DOI] [PubMed] [Google Scholar]
- 175.Sivapalasingam S, Steigbigel NH. In: Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. 8th edn. Bennett JE, Dolin R, Blaser MJ, editors. Vol. 1. Saunders - Elsevier, Inc.; Philadelphia, PA: 2014. pp. 358–376. ch. 29. [Google Scholar]
- 176.Bryskier A. Clin. Microbiol. Infect. 2000;6:661–669. doi: 10.1046/j.1469-0691.2000.00185.x. [DOI] [PubMed] [Google Scholar]
- 177.Okamoto H, Miyazaki S, Tateda K, Ishii Y, Yamaguchi K. The Journal of antimicrobial chemotherapy. 2000;46:797–802. doi: 10.1093/jac/46.5.797. [DOI] [PubMed] [Google Scholar]
- 178.Chollet R, Chevalier J, Bryskier A, Pages JM. Antimicrobial agents and chemotherapy. 2004;48:3621–3624. doi: 10.1128/AAC.48.9.3621-3624.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Xiong L, Shah S, Mauvais P, Mankin AS. Mol. Microbiol. 1999;31:633–639. doi: 10.1046/j.1365-2958.1999.01203.x. [DOI] [PubMed] [Google Scholar]
- 180.Blasi F, Farrell DJ, Dubreuil L. Diagn. Microbiol. Infect. Dis. 2009;63:302–308. doi: 10.1016/j.diagmicrobio.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 181.Felmingham D, Farrell DJ. J. Infect. 2006;52:178–180. doi: 10.1016/j.jinf.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 182.Hammerschlag MR, Roblin PM, Bebear CM. The Journal of antimicrobial chemotherapy. 2001;48(Suppl T1):25–31. doi: 10.1093/jac/48.suppl_2.25. [DOI] [PubMed] [Google Scholar]
- 183.Georgopapadakou NH. Expert Opin Investig Drugs. 2014;23:1313–1319. doi: 10.1517/13543784.2014.954036. [DOI] [PubMed] [Google Scholar]
- 184.Barie PS. Surg. Infect. (Larchmt.) 2006;7:247–249. doi: 10.1089/sur.2006.7.247. [DOI] [PubMed] [Google Scholar]
- 185.Mathews AW. Wall St J (East Ed) 2006:A1, A12. [PubMed] [Google Scholar]
- 186.Mansour H, Chahine EB, Karaoui LR, El-Lababidi RM. Ann. Pharmacother. 2013;47:368–379. doi: 10.1345/aph.1R435. [DOI] [PubMed] [Google Scholar]
- 187.Kouvela EC, Kalpaxis DL, Wilson DN, Dinos GP. Antimicrobial agents and chemotherapy. 2009;53:1411–1419. doi: 10.1128/AAC.01425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nilius AM, Ma Z. Curr. Opin. Pharmacol. 2002;2:493–500. doi: 10.1016/s1471-4892(02)00198-4. [DOI] [PubMed] [Google Scholar]
- 189.Barrera CM, Mykietiuk A, Metev H, Nitu MF, Karimjee N, Doreski PA, Mitha I, Tanaseanu CM, Molina JM, Antonovsky Y, Van Rensburg DJ, Rowe BH, Flores-Figueroa J, Rewerska B, Clark K, Keedy K, Sheets A, Scott D, Horwith G, Das AF, Jamieson B, Fernandes P, Oldach D, Team S-OP. Lancet Infect. Dis. 2016;16:421–430. doi: 10.1016/S1473-3099(16)00017-7. [DOI] [PubMed] [Google Scholar]
- 190.Van Bambeke F, Tulkens PM. Expert Rev. Anti Infect. Ther. 2016;14:311–324. doi: 10.1586/14787210.2016.1138857. [DOI] [PubMed] [Google Scholar]
- 191.McGhee P, Clark C, Kosowska-Shick KM, Nagai K, Dewasse B, Beachel L, Appelbaum PC. Antimicrobial agents and chemotherapy. 2010;54:230–238. doi: 10.1128/AAC.01123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Llano-Sotelo B, Dunkle J, Klepacki D, Zhang W, Fernandes P, Cate JH, Mankin AS. Antimicrobial agents and chemotherapy. 2010;54:4961–4970. doi: 10.1128/AAC.00860-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhanel GG, Lawson CD, Zelenitsky S, Findlay B, Schweizer F, Adam H, Walkty A, Rubinstein E, Gin AS, Hoban DJ, Lynch JP, Karlowsky JA. Expert Rev. Anti Infect. Ther. 2012;10:459–473. doi: 10.1586/eri.12.25. [DOI] [PubMed] [Google Scholar]
- 194.Aggen JB, Armstrong ES, Goldblum AA, Dozzo P, Linsell MS, Gliedt MJ, Hildebrandt DJ, Feeney LA, Kubo A, Matias RD, Lopez S, Gomez M, Wlasichuk KB, Diokno R, Miller GH, Moser HE. Antimicrobial agents and chemotherapy. 2010;54:4636–4642. doi: 10.1128/AAC.00572-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Schneider P, Hawser S, Islam K. Bioorg. Med. Chem. Lett. 2003;13:4217–4221. doi: 10.1016/j.bmcl.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 196.Dale GE, Broger C, D'Arcy A, Hartman PG, DeHoogt R, Jolidon S, Kompis I, Labhardt AM, Langen H, Locher H, Page MG, Stuber D, Then RL, Wipf B, Oefner C. J. Mol. Biol. 1997;266:23–30. doi: 10.1006/jmbi.1996.0770. [DOI] [PubMed] [Google Scholar]
- 197.Oefner C, Bandera M, Haldimann A, Laue H, Schulz H, Mukhija S, Parisi S, Weiss L, Lociuro S, Dale GE. The Journal of antimicrobial chemotherapy. 2009;63:687–698. doi: 10.1093/jac/dkp024. [DOI] [PubMed] [Google Scholar]
- 198.Sincak CA, Schmidt JM. Ann. Pharmacother. 2009;43:1107–1114. doi: 10.1345/aph.1L167. [DOI] [PubMed] [Google Scholar]
- 199.Laue H, Valensise T, Seguin A, Lociuro S, Islam K, Hawser S. Antimicrobial agents and chemotherapy. 2009;53:4542–4544. doi: 10.1128/AAC.00766-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Laue H, Weiss L, Bernardi A, Hawser S, Lociuro S, Islam K. The Journal of antimicrobial chemotherapy. 2007;60:1391–1394. doi: 10.1093/jac/dkm409. [DOI] [PubMed] [Google Scholar]
- 201.White SW, Zheng J, Zhang YM, Rock Annu. Rev. Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 202.Zhang YM, Lu YJ, Rock CO. Lipids. 2004;39:1055–1060. doi: 10.1007/s11745-004-1330-3. [DOI] [PubMed] [Google Scholar]
- 203.Marrakchi H, Zhang YM, Rock CO. Biochem. Soc. Trans. 2002;30:1050–1055. doi: 10.1042/bst0301050. [DOI] [PubMed] [Google Scholar]
- 204.Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO. J. Biol. Chem. 1999;274:11110–11114. doi: 10.1074/jbc.274.16.11110. [DOI] [PubMed] [Google Scholar]
- 205.Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. Nature. 2009;458:83–86. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 206.Balemans W, Lounis N, Gilissen R, Guillemont J, Simmen K, Andries K, Koul A. Nature. 2010;463:E3. doi: 10.1038/nature08667. discussion E4. [DOI] [PubMed] [Google Scholar]
- 207.Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. Nature. 2010;463:E4–E5. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 208.Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15378–15383. doi: 10.1073/pnas.1109208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Parsons JB, Rock CO. Curr. Opin. Microbiol. 2011;14:544–549. doi: 10.1016/j.mib.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mehboob S, Song J, Hevener KE, Su PC, Boci T, Brubaker L, Truong L, Mistry T, Deng J, Cook JL, Santarsiero BD, Ghosh AK, Johnson ME. Bioorg. Med. Chem. Lett. 2015;25:1292–1296. doi: 10.1016/j.bmcl.2015.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zheng CJ, Sohn MJ, Lee S, Kim WG. PLoS One. 2013;8:e78922. doi: 10.1371/journal.pone.0078922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hevener KE, Mehboob S, Su PC, Truong K, Boci T, Deng J, Ghassemi M, Cook JL, Johnson ME. J. Med. Chem. 2012;55:268–279. doi: 10.1021/jm201168g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mehboob S, Hevener KE, Truong K, Boci T, Santarsiero BD, Johnson ME. J. Med. Chem. 2012;55:5933–5941. doi: 10.1021/jm300489v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Schiebel J, Chang A, Lu H, Baxter MV, Tonge PJ, Kisker C. Structure. 2012;20:802–813. doi: 10.1016/j.str.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Gerusz V, Denis A, Faivre F, Bonvin Y, Oxoby M, Briet S, LeFralliec G, Oliveira C, Desroy N, Raymond C, Peltier L, Moreau F, Escaich S, Vongsouthi V, Floquet S, Drocourt E, Walton A, Prouvensier L, Saccomani M, Durant L, Genevard JM, Sam-Sambo V, Soulama-Mouze C. Journal of medicinal chemistry. 2012;55:9914–9928. doi: 10.1021/jm301113w. [DOI] [PubMed] [Google Scholar]
- 216.Liu N, Cummings JE, England K, Slayden RA, Tonge PJ. The Journal of antimicrobial chemotherapy. 2011;66:564–573. doi: 10.1093/jac/dkq509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sampson PB, Picard C, Handerson S, McGrath TE, Domagala M, Leeson A, Romanov V, Awrey DE, Thambipillai D, Bardouniotis E, Kaplan N, Berman JM, Pauls HW. Bioorg. Med. Chem. Lett. 2009;19:5355–5358. doi: 10.1016/j.bmcl.2009.07.129. [DOI] [PubMed] [Google Scholar]
- 218.Ramnauth J, Surman MD, Sampson PB, Forrest B, Wilson J, Freeman E, Manning DD, Martin F, Toro A, Domagala M, Awrey DE, Bardouniotis E, Kaplan N, Berman J, Pauls HW. Bioorg. Med. Chem. Lett. 2009;19:5359–5362. doi: 10.1016/j.bmcl.2009.07.094. [DOI] [PubMed] [Google Scholar]
- 219.Karlowsky JA, Kaplan N, Hafkin B, Hoban DJ, Zhanel GG. Antimicrobial agents and chemotherapy. 2009;53:3544–3548. doi: 10.1128/AAC.00400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zheng CJ, Sohn MJ, Kim WG. The Journal of antimicrobial chemotherapy. 2009;63:949–953. doi: 10.1093/jac/dkp058. [DOI] [PubMed] [Google Scholar]
- 221.Lu H, Tonge PJ. Acc. Chem. Res. 2008;41:11–20. doi: 10.1021/ar700156e. [DOI] [PubMed] [Google Scholar]
- 222.Dhillon GS, Kaur S, Pulicharla R, Brar SK, Cledon M, Verma M, Surampalli RY. Int J Environ Res Public Health. 2015;12:5657–5684. doi: 10.3390/ijerph120505657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.McMurry LM, Oethinger M, Levy SB. Nature. 1998;394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 224.Flamm RK, Rhomberg PR, Kaplan N, Jones RN, Farrell DJ. Antimicrobial agents and chemotherapy. 2015;59:2583–2587. doi: 10.1128/AAC.05119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hafkin B, Kaplan N, Murphy B. Antimicrobial agents and chemotherapy. 2015;60:1695–1701. doi: 10.1128/AAC.01741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wittke F, Nicolas-Metral V, Menetrey A, et al. presented in part at the 55th Interscience Conference on Antimicrobial Agents and Chemotherapy; San Diego, CA. 2015. [Google Scholar]
- 227.Kaplan N, Albert M, Awrey D, Bardouniotis E, Berman J, Clarke T, Dorsey M, Hafkin B, Ramnauth J, Romanov V, Schmid MB, Thalakada R, Yethon J, Pauls HW. Antimicrobial agents and chemotherapy. 2012;56:5865–5874. doi: 10.1128/AAC.01411-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Karlowsky JA, Laing NM, Baudry T, Kaplan N, Vaughan D, Hoban DJ, Zhanel GG. Antimicrobial agents and chemotherapy. 2007;51:1580–1581. doi: 10.1128/AAC.01254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bogdanovich T, Clark C, Kosowska-Shick K, Dewasse B, McGhee P, Appelbaum PC. Antimicrobial agents and chemotherapy. 2007;51:4191–4195. doi: 10.1128/AAC.00550-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Park HS, Yoon YM, Jung SJ, Kim CM, Kim JM, Kwak JH. The Journal of antimicrobial chemotherapy. 2007;60:568–574. doi: 10.1093/jac/dkm236. [DOI] [PubMed] [Google Scholar]
- 231.Yum JH, Kim CK, Yong D, Lee K, Chong Y, Kim CM, Kim JM, Ro S, Cho JM. Antimicrobial agents and chemotherapy. 2007;51:2591–2593. doi: 10.1128/AAC.01562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Schiebel J, Chang A, Shah S, Lu Y, Liu L, Pan P, Hirschbeck MW, Tareilus M, Eltschkner S, Yu W, Cummings JE, Knudson SE, Bommineni GR, Walker SG, Slayden RA, Sotriffer CA, Tonge PJ, Kisker C. J. Biol. Chem. 2014;289:15987–16005. doi: 10.1074/jbc.M113.532804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. [2/10/2016]; allphasepharma.com, http://allphasepharma.com/dir/2015/12/23/2205/qidp-antibiotics-2015-year-end-update/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.