Abstract
Recent advances in confocal microscopy, coupled with the development of numerous fluorescent reporters, provide us with a powerful tool to study the development of plants. Live confocal imaging has been used extensively to further our understanding of the mechanisms underlying the formation of roots, shoots and leaves. However, it has not been widely applied to flowers, partly because of specific challenges associated with the imaging of flower buds. Here, we describe how to prepare and grow shoot apices of Arabidopsis in vitro, to perform both single-point and time-lapse imaging of live, developing flower buds with either an upright or an inverted confocal microscope.
Keywords: Flower, flower development, flower meristem, plant development, confocal microscopy, live confocal imaging, sepals, floral organs
1. Introduction
Formation of the underground and aerial tissues of flowering plants is primarily post-embryonic. New tissues and organs are continuously produced by meristems, which are groups of undifferentiated cells with a subpopulation of stem cells, situated at the tip of the roots and the shoots. All of the structures that grow below ground in the plant derive from the root apical meristem (RAM), while all of the structures that grow above ground derive from the shoot apical meristem (SAM). During vegetative growth, the SAM generates leaves on it flanks, while during the reproductive phase, it produces flower meristems (FMs), which develop into flowers. The FM, like the SAM, consists of a group of undifferentiated cells, with stem cells at its center. However, unlike the SAM, which produces lateral organs one at a time, the FM generates 16 floral organs with four different identities (four sepals, four petals, six stamens and two fused carpels), in a partially synchronous manner, before stem cells cease to be maintained (Smyth et al., 1990). Thus, the FM is a much more crowded space than the SAM, in which multiple developmental programs take place simultaneously, only separated by narrow boundaries.
Many key developmental regulators are expressed in only a subset of cells at a particular time in development. For decades, the spatial and temporal expression patterns of developmental genes were determined by techniques such as in situ hybridization and analysis of transgenic plants that contain fusions with promoters driving the expression of non-fluorescent reporters. While these techniques have allowed for considerable progress in our understanding of the mechanisms controlling the development of plants, they generally lack cellular resolution, and do not allow for the easy characterization of the expression of several genes in the same biological samples. These techniques can also only be applied to dead and fixed samples, which limit our capacity to fully grasp the dynamic processes of development. The recent advances in laser scanning confocal microscopy of live samples, combined with the development of multiple fluorescent reporters, provide us with a formidable tool to overcome these restrictions, as they give us the possibility to monitor the expression of multiple genes, in live tissues, with a fine cellular resolution. Live confocal imaging has been extensively used to further our understanding of the mechanisms underlying root and shoot growth, but with the exception of a few studies (e.g. Chandler et al., 2011; Roeder et al., 2010; Urbanus et al., 2009), it has not been widely applied to the study of flower development.
While the RAM is easily accessible, confocal imaging of the SAM or the FMs during reproductive growth requires the prior removal of siliques and older flowers to uncover the SAM and the youngest flower buds. Therefore, imaging the SAM and the FMs involves a similar procedure. However, imaging developing flower buds presents specific challenges, notably the presence of the sepals, which quickly grow to cover the FM and filter out the fluorescence from underlying tissues (Fig. 1A). Here, we explain how to prepare and grow shoot apices in vitro for both one-time and time-lapse confocal imaging of live flower buds, using either an upright or an inverted microscope.
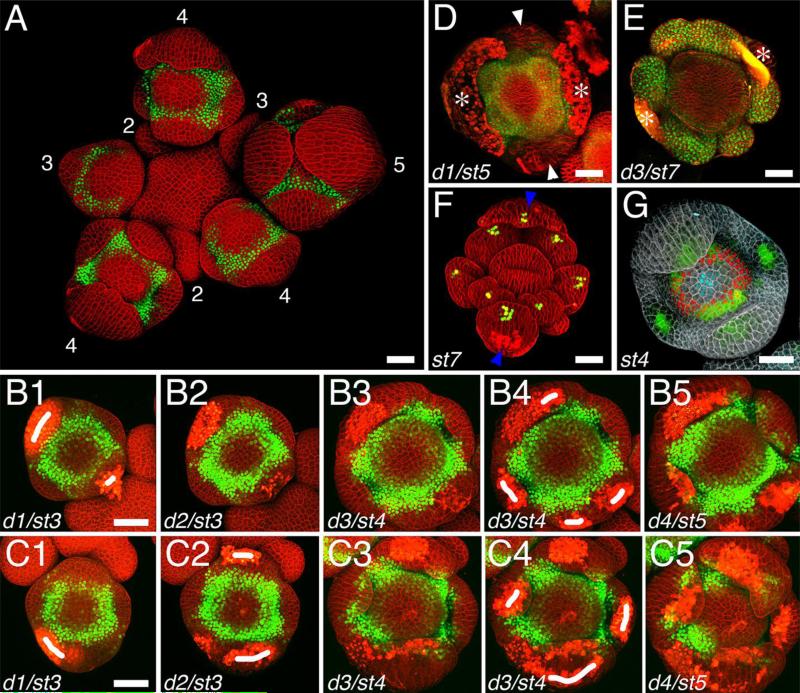
Fig. 1.
Maximum intensity projections of confocal z-stacks of live flower buds. (A-E) flowers expressing a Venus reporter for the APETALA3 gene (green); cell walls were stained with propidium iodide (red). (A) Inflorescence; numbers indicate floral stages; sepals in stage 4 and 5 flowers filter out the fluorescence of the Venus reporter, which normally forms a ring. Note that some flower buds appear tilted compared to the inflorescence. (B1-B5) 4-day time-lapse of an individual flower bud from stage 3 to stage 5; laser ablations (marked as white traits) performed on day 1 and day 3 were sufficient to prevent the sepals from covering the center of the flower bud at stage 5. (C1-C5) 4-day time-lapse of an individual flower bud from stage 3 to stage 5; laser ablations performed on day 1, 2 and 3 were insufficient to prevent the sepals from covering the center of the flower bud. (D-E) Individual flower bud after manual removal of the abaxial and adaxial sepals (D), and of all sepals (E); white arrowheads indicate remaining sepals; white asterisks indicate scars resulting from the removal of the sepals. (F) stage 7 ap1-1 flower expressing a DR5-3xVenusN7 reporter (green; (Vernoux et al., 2011); plasma membranes were stained with FM4-64 (red); blue arrowheads indicate the leaf-like structures that replace sepals and do not cover the flower bud. (G) Stage 4 flower buds expressing fluorescent a GFP reporter for DORNROSCHEN-LIKE (green; (Chandler et al., 2011), a Venus reporter for SUPERMAN (red) and a dsRed reporter for CLAVATA3 (cyan; (Zhou et al., 2015); cells walls were stained with propidium iodide (grey). d: day; st: stage. Bars = 25 μm.
2. Material
Tweezers (e.g. Dumont #5). Before use, sharpen the tweezers using a sharpening stone (e.g. Arkansas Sharpening Stone, Translucent, Grobet USA) and a drop of oil. Making the tweezers blade-like rather than pointed allows for better leverage on the flower buds to be removed.
Pin vise with straight stainless steel needles for dissecting sepals.
P10 and P1000 pipettes with appropriate tips.
Tissue paper (e.g. Kimwipes, Kimtech).
MS plates (1 × Murashige and Skoog basal salt mixture without vitamins, 0.8% agar, pH 5.8 with potassium hydroxide solution) for seed germination.
Dissecting dishes. Round dishes approximately 6 cm wide and 2 cm deep (e.g. plastic box, round, RD2, Electron Microscopy Science), filled with approximately 0.5 cm of 1% agarose.
Imaging dishes. For imaging with an upright confocal microscope, use a plastic box with a transparent lid (e.g. rectangular hinged boxes, 2-7/8” long, 2” wide, 1-1/4” deep, Durphy Packaging Co.), filled with 0.5 cm of imaging medium. For imaging with an inverted confocal microscope, use a small Petri dish (e.g. easy grip Petri dish, polystyrene, 3.5 cm wide, 1 cm deep, Falcon), filled exactly to the brim with imaging medium.
Imaging medium. For one-time imaging, use 1% agarose. For time-course experiments, use apex growth medium (Fernandez et al., 2010): 0.5 × Murashige and Skoog basal salt mixture without vitamins, 1% sucrose, 0.8% agarose, pH 5.8 with potassium hydroxide solution, with vitamins (0.01% myo-inositol, 0.0001% nicotinic acid, 0.0001% pyridoxine hydrochloride, 0.001% thiamine hydrochloride, 0.0002% glycine) and cytokinins (500 nM N6-benzyladenine).
Propidium iodide (1 mg/mL stock) or FM4-64 (80 μg/mL solution) for staining of the cell walls or plasma membranes, respectively.
Stereomicroscope (e.g. Zeiss Discovery V8) with sufficient working space and magnification (a maximum magnification of 80-90x works well) for dissecting the shoot apices and sepals.
Confocal microscope. An upright confocal microscope is more convenient for imaging live flower buds, but an inverted microscope can also be used.
40x dipping lens, with long working distance (e.g. W Plan Apochromat 40X/1.0 DIC, Zeiss, 2.5 mm working distance). Once dipped in water, such a lens allows for the imaging of the sample without a coverslip.
Laser ablation system (e.g. MicroPoint, Andor Technology) for sepal ablation.
Plants for imaging (any accession works).
3. Methods
3.1. Plant growth
It is easier to dissect the inflorescence of plants with a large SAM. The following protocol works well to grow vigorous plants, which have a larger SAM:
Germinate seeds on horizontal MS plates with appropriate selection under long day (16h light/day) at 20°C.
Transplant seedlings to soil two weeks after sowing, so that plants are well space out in the pots.
Grow plants under short day (8 hours light/day), 16-20°C conditions for three weeks.
Transfer plants to either long day (16h light/day) or continuous day, 16-20°C conditions. Growing plants for more than 3 weeks in short day conditions results in the formation of an inflorescence that is less vigorous.
3.2. Dissection of the shoot apex
Shoot apices are easiest to dissect when the inflorescence is approximately to 2 to 8 cm long. As the inflorescence elongates, the stem gets narrower and the inflorescence becomes more difficult to dissect.
Water the plants the evening before dissection to ensure that they are turgid. We observed that watering the plants prior to dissection is also important to get strong fluorescent signals when imaging the apices.
Remove siliques and older flowers by pushing the base of the peduncle with tweezers until it breaks. Cutting the peduncle instead of breaking it usually leaves part of the peduncle attached to the stem and should be avoided. Remove as many flowers as possible without magnification (Fig. 2, A and B).
Immediately transfer the tip of the inflorescence to a dissecting dish. Use the tweezers to make a small vertical hole in the agarose at the center of the dissecting dish. Cut the inflorescence approximately 1 cm below the SAM, and stick the cut end into the agarose in the dissecting dish, so that only the SAM and young flower buds remain above the surface of the agarose.
Fill the dissecting dish with sterile, de-ionized water. Dissecting the shoot apex in water prevents the sample from dehydrating. However, as long as one proceeds quickly and puts a drop of sterile water at the base of the sample, it is possible to dissect without submerging the inflorescence.
Under the stereomicroscope, remove the air trapped around the inflorescence (Fig. 2, C and D) by creating water movement with a P1000 pipette.
Under the stereomicroscope, remove the flower buds you do not wish to image by pushing the base of the peduncle or the top of the bud with tweezers (Fig. 2E). If you only wish to image flower buds stage 5 and younger (stages as described in Smyth et al., 1990), it is easier to remove the water before removing the last few buds (stages 6-8).
If needed, remove sepals of flowers stage 5 and older under the stereomicroscope. See Sepal ablation section.
Transfer the dissected apex to an imaging dish. Insert the basal end of the inflorescence into the imaging medium so that it is perpendicular to the surface of the medium, with only the apex and the young flower buds protruding from the agarose (Fig. 2F). If the inflorescence protrudes too far above the agarose, it will cause difficulties when staining the sample, and imaging it with an inverted confocal microscope. It is worth noting that flower buds are not always oriented exactly the same way as the stem. Depending on which flower bud is to be imaged, it might be necessary to stick the shoot apex in the growth medium in a tilted fashion (i.e. not perpendicular to the agar surface).
Fill the imaging dish with sterile, de-ionized water to a depth of 5 mm (Fig.2F). Immersing the samples for a few minutes after dissection prevents dehydration of the shoot apices and ensures that they recover properly.
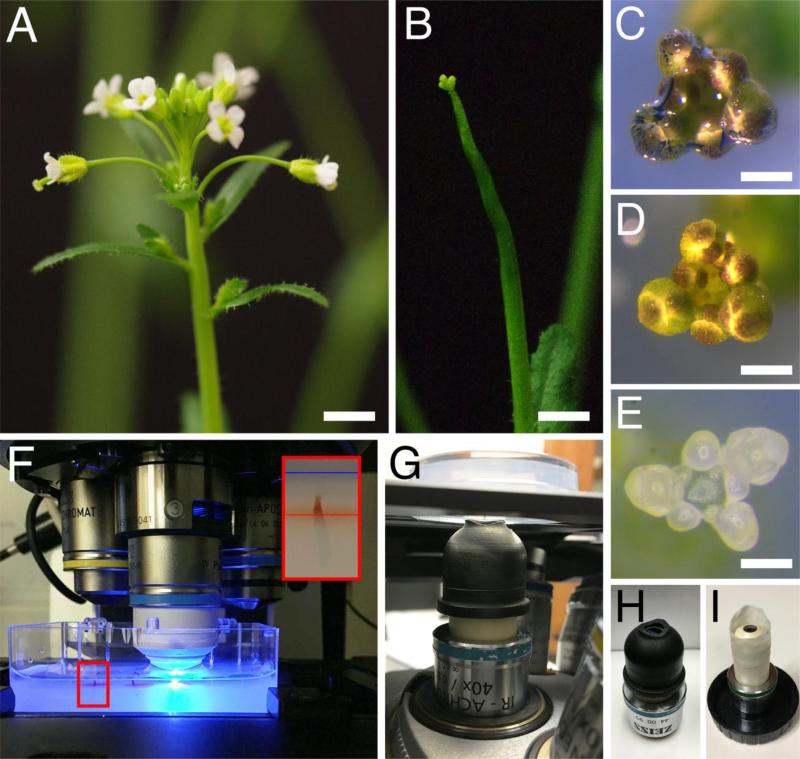
Fig 2.
Preparation of the shoot apices for imaging. (A-E) Dissection of a shoot apex for imaging. Inflorescence before (A) and after (B) removal of siliques and older flowers. (C-D) Shoot apex immersed in water in the imaging dish, with an air bubble trapped at the tip (C) and after removal of the air bubble (D). (E) Shoot apex in the imaging dish, after dissection of flower buds older than stage 5. (F-G) View of shoot apices in the imaging dish, on the stage of an upright (F) and inverted (G) confocal microscopes. In (F), a 40x dipping lens is positioned above one of the apices, with the tip of the lens immersed in water. In (G), the shoot apex is positioned upside down above the 40x dipping lens, with a water column connecting the imaging medium to the tip of the lens. The smaller panel in (F) shows a higher magnification view of the area in the red rectangle, with a shoot apex inserted in imaging medium; red and blue lines indicate the surface of the medium and the water, respectively. (H-I) Examples of custom-made devices that allow adding more water at the tip of the dipping lens: a rubber sleeve (H), and a makeshift sleeve made from the finger of a powderless latex glove (G). Bars = 0.5 cm in (A) and (B), 0.1 cm in (C) and (D), and 100 μm in (E).
It is also possible to image flower buds from shoot apices still attached to the whole plant, although it is much harder to do so with an inverted microscope. For a description of how to proceed, see Heisler and Ohno (2014).
3.3. Sepal ablation
As they grow and cover the FM, sepals attenuate the light emission from underlying tissue (Fig. 1A). Thus, to properly image the FM after early stage 4, it is necessary to either prevent the sepals from growing over the FM by laser ablation, or to manually remove them afterwards. It is extremely difficult to manually remove the sepals of flower buds younger than stage 5 without inadvertently damaging the rest of the flower. Thus, the most reliable way to remove the sepals is by laser ablation.
Laser ablation of the tip of the sepals at stage 3-4
Ablation of cells at the tip of the emerging sepal primordia prevents them from subsequently covering the FM, and allows for unhindered imaging of the flower bud. This can be achieved with a laser ablation system, or alternatively, with a 2-photon microscope. Two examples of laser ablation of sepals, one successful (Fig. 1, B1-B5) and one unsuccessful (Fig. 1, C1-C5), are shown in Figure 1.
Place the imaging dish under the microscope and proceed to focus on the flower bud whose sepals are to be ablated, following the same steps as to image the sample (see Imaging section).
Using the laser ablation software, designate the cells to be ablated. Typically, ablating cells alongside a line following the crest of the developing sepal ensures that it does not grow to cover the FM.
Adjust the laser power and dwelling time to such level that it destroys cells without causing too much damage to your sample.
Proceed to ablation.
Ablation of the tip of the abaxial and adaxial sepal primordia must be performed at stage 3, before they start covering the FM (Fig. 1, B1). Ablation of the tip of the lateral sepal primordia, which form later, can be done as late as mid stage 4 (Fig. 1, B4). If the initial ablation proves insufficient, an additional ablation can be performed the following days (Fig. 1, B4).
Manual dissection of the sepals with a pin vise and a micro metal pin after stage 5
Sepals can also be manually removed under a stereomicroscope using a pin vise with a microscopy metal pin. In the following description, “tangentially” and “radially” refer to the positioning of the pin related to the whole inflorescence, not to the flower bud. An example of flower buds growing after manual dissection of the sepals is shown in Fig. 1, D and E.
Place the dissecting dish with the shoot apex under the stereomicroscope, and set the magnification to maximum.
Holding the pin vise, position the pin tangentially, on top of the abaxial sepal.
Push the abaxial sepal towards the periphery of the inflorescence. The sepal should break cleanly at its base.
Proceed similarly with the adaxial sepal, but push the sepal towards the shoot apical meristem.
Proceed similarly with the lateral sepals, this time positioning the pin radially, and pushing the sepal towards the neighboring flower.
Manual dissection of the sepals can be performed either with the sample covered in water or without water. If not using water, it is recommended to fill the imaging dish with water for 2 minutes after removing each sepal, before proceeding to the next one, to prevent dehydration and keep the sample turgid. It is extremely difficult to manually remove sepals before late stage 5, because they are extremely small and elastic. After stage 5, the sepals become more brittle and thus are easier to remove.
If studying the central part of the flower, it is possible to use the apetala1-1 (ap1-1) mutant background instead of removing the sepals. ap1-1 mutant flowers lack sepals (which are replaced by leaf-like organs that do not cover the bud) and petals, but the inner whorls develop normally (Fig. 1F; Irish and Sussex, 1990).
3.4. Staining
If not using transgenic lines with a fluorescent protein addressed to the plasma membrane (Cutler et al., 2000; Reddy et al., 2004), it is necessary to stain either the plasma membranes or the cell walls with a fluorescent dye to have a good cellular resolution when imaging shoot apices and flower buds. Plasma membranes can be stained using the water-soluble, lipophilic dye FM4-64. Cell walls can be stained with propidium iodide.
Drain water from the imaging dish with the dissected shoot apex. Dab the periphery of the dish with tissue paper to ensure the surface of the imaging medium is dry. If it is still wet, the dye will be sucked away from the shoot apex, resulting in weak, heterogeneous staining.
Under the stereomicroscope, use a P10 pipette to apply 20-30 μL of either a 80 μg/mL FM4-64 solution, or a 1 mg/mL propidium iodide solution to the shoot apex. It is important that the whole shoot sample is covered with dye for the staining to be homogeneous.
Stain for 20 minutes if using FM4-64, 2 minutes if using propidium iodide.
Rinse twice with sterile, de-ionized water.
3.5. Setting the sample on the microscope
With an upright confocal
Fill the imaging dish so that the samples are immersed in approximately 5 mm sterile, de-ionized water.
Place the imaging dish onto the stage (Fig. 2F).
Lower the dipping lens until it touches the surface of the water. If there is air trapped under the lens, remove it by creating water movement with a P1000 pipette.
Under epifluorescence illumination, position the sample within the field of the objective with the XY controls.
Looking through the eyepieces, focus on the shoot apex using the Z control.
Switch to confocal mode. Using the confocal software, zoom onto the flower bud you wish to image.
Proceed to imaging your sample (see Imaging parameters section).
For time-lapse experiments, return the imaging dish with the samples to either long day (16h light/day) or continuous day, 16-20°C conditions during the intervals between the imaging times.
With an inverted confocal
Using an inverted confocal microscope to image flower buds is both more complicated and time-consuming than using an upright microscope, as it requires forming a fragile water column between the sample and the lens, and this setup needs to be re-established for each sample. It is nonetheless possible to use an inverted microscope by proceeding as follows:
Put a drop of sterile, de-ionized water on the tip of the lens with a P1000 pipette.
Hold the imaging dish upside down, and add a drop of sterile, de-ionized water to the base of the shoot apex using a P1000. A small drop of water should cover the dissected inflorescence including the inflorescence apex.
Place the imaging dish upside down on the stage (Fig. 2G). Make sure that the stage is high enough, in order not to crush the sample against the lens in the process.
Lower the stage until the tip of the shoot apex makes contact with the water on the lens. A water column should form between the lens and the growth medium (Fig. 2G). If it does not, carefully add water to the sample using a P1000 pipette.
Under epifluorescence illumination, position the sample within the field of the objective with the XY controls.
Looking through the eyepieces, focus on the shoot apex using the Z control.
Switch to confocal mode. Using the confocal software, zoom onto the flower bud you wish to image.
Proceed to imaging your sample (see Imaging parameters section).
There are several points to take into consideration when imaging shoot apices with an inverted microscope:
It is important that the imaging dish is filled exactly to the brim with imaging medium. If overfilled, the medium, rather than the dish, will be resting on the stage. Because the medium is soft, this will cause the sample to drift during the imaging process. Conversely, if the imaging dish is not fully filled with medium, water tends to be sucked away from the sample, which makes it harder to establish and maintain the water column.
Make sure that only the tip of the inflorescence sticks out of the imaging medium. If the length of the sample sticking out is too long, the water column is harder to establish, and more likely to break during the imaging process.
Check the water column regularly while imaging. Depending on the microscope, generating a full confocal stack of a flower bud with multiple channels can take more than 30 minutes, and the water column slowly dries up in the process. If the water column becomes too thin, pause the stack and carefully add water to the sample with a P1000 pipette, then resume acquisition.
Some inverted confocal microscopes do not have much working space underneath the stage, which complicates establishing and maintaining the water column. When using such a microscope, adding a custom-made device around the tip of the lens so that it can hold more water can be very useful (Fig. 2, H and I). For instance, a sleeve can be made out of the finger of a non-powdered latex glove (Fig. 2I).
For time lapse-experiments, it is necessary to store the imaging dish with the sample in a plastic box with a transparent lid (e.g. rectangular hinged boxes, 2-7/8” long, 2” wide, 1-1/4” deep, Durphy Packaging Co.) in either long day (16h light/day) or continuous day, 16-20°C conditions to prevent dehydration during the intervals between imaging times.
3.6. Imaging parameters
Given the tridimensional nature of flower buds, studying the dynamics of flower development requires the acquisition of confocal z-stacks at different floral stages. While several stages of flower development can be observed at any single time at the periphery of the SAM, and provide an overview of the dynamics of flower development, it is sometimes required to perform time-lapse experiments. Flower buds used for these experiments are potentially subjected to long and repeated exposure to laser illumination, and confocal parameters should be adjusted in order not to photobleach or kill the sample.
Confocal microscopes potentially allow for the imaging of multiple fluorophores in the same sample (for a description of fluorophores that can be used in plants, see Reddy and Roy-Chowdhury, 2009). It is technically possible, for instance, to image GFP, YFP, dsRed and propidium iodide in a single confocal stack (Fig. 1G). Imaging parameters must be adjusted and optimized each time plants with a different combination of reporters is used, through a trial and error process that involves a trade-off between the resolution and imaging time, and between the intensity and the proper separation of the signal from the different fluorophores. Below are some aspects to take into consideration when adjusting these parameters.
If using a regular detector, a choice of lasers and filters must be made according to the combination of fluorophores used. Some fluorophores emit fluorescence with sufficiently distinct wavelengths (e.g CFP and GFP, or GFP and propidium iodide), and fluorescence from such fluorophores can be collected simultaneously. However, other fluorophores emit fluorescence with partially overlapping wavelength (e.g. GFP and YFP, or dsRed and propidium iodide), and in such cases, fluorescence must be collected separately, which lengthens the duration of acquisition. Alternatively, some confocal microscopes come with a spectral detector that can be used to collect simultaneously, and separate, the fluorescence from multiple fluorophores.
Laser power should be minimized so as not to bleach the sample. To increase the intensity of the signal, it is possible to adjust the gain and the pinhole to some extent (an excessive gain or pinhole results in lower quality images due to background noise and acquisition of signal from non-focal planes, respectively). The quality and duration of acquisition of a z-stack also depends on the xy resolution and the step-size. A higher xy resolution and lower step-size results in a higher quality stacks with better cellular resolution, but also in an increased imaging time and exposure of the sample to laser illumination. We typically use a pinhole of 1 to 1.5 airy units, an xy resolution of 1024 × 1024 pixels and a step size of 0.5 to 1.5 μm.
While cells divide relatively fast in emerging floral organs, we did not observe cells dividing more than once every in 24 hours in the central part of the flower buds. While samples can be imaged every 6 hours, cell lineages in the FM can thus easily be tracked when imaging flower buds at 24-hour intervals.
3.7. Post-processing
The analysis of slice views, tridimensional views and maximum intensity projections of confocal z-stacks of live flower buds gives us access to qualitative information about flower development. However, confocal z-stacks also potentially provide us with a wealth of quantitative data. Confocal z-stacks can be used to quantify cell sizes, cell division rates and gene expression levels in individual cells, as well as the number of cell expressing different genes, and the volume of different domains within the flower. While the software associated with a confocal microscope usually offers tools to extract some of this quantitative information from confocal images, other software, both free-access (e.g. FiJi, MorphoGraphX, MARS-ALT [Barbier de Reuille et al., 2015; Barbier de Reuille et al., 2014; Fernandez et al., 2010]) and commercial (e.g. Imaris, Bitplane) software have been specifically designed for that purpose. Such software can also be used to perform cell segmentation and automatically track cell lineages from time-lapse experiments.
3.8. Considerations on the effect of the protocol on flower development
Growing detached and dissected inflorescence apices in vitro can potentially affect flower development. The apex growth medium used for time-lapse experiments was designed empirically by Fernandez et al. (2010) to ensure that both the plastochron and flower development were normal. Cytokinin was added to the medium to compensate for the lack of cytokinin produced in the lower parts of the plant, and different concentrations of cytokinin, either alone or in combination with auxin, were tested for the ability to allow normal flower development over several days. The growth medium described here allows for continued growth of both wild-type and mutant shoot apices for five days, and the proper development of flowers up to at least stage 10. Longer time-lapse experiments are possible but often result in bacterial and/or fungal contaminations. However, shoot apices can keep growing in vitro for several weeks, but most flowers senesce, and only very rarely produce seeds. Similarly, sepal removal does not appear to affect flower development or gene expression patterns. While this protocol works well, it may be advisable to perform live confocal on whole plants grown in vitro as described by Heisler and Ohno (2014) when investigating cytokinin-related processes, or phenotypes that are known to be affected by cytokinin levels.
Highlights.
Protocol for dissecting and growing shoot apices for live confocal imaging of flower buds.
How to remove sepals to image underlying tissues.
How to use either an upright or an inverted confocal microscope to image live flower buds.
Tips to set the imaging parameters.
Acknowledgements
The authors wish to thank Ann Lavanway at Dartmouth and Andres Collazo at the Biological Imaging Facility at Caltech for their help in solving technical issues with the live confocal imaging. Funding in the Meyerowitz Laboratory was provided by the Howard Hughes Medical Institute, the US National Institutes of Health through grant R01 GM104244 and the Gordon and Betty Moore Foundation through Grant GBMF3406. Funding in the Jack lab was provided by the US National Science Foundation through grant IOS-0926347.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: NP did the imagery and wrote the manuscript. TPJ and EMM edited the manuscript.
References
- Barbier de Reuille P, Robinson S, Smith RS. Quantifying cell shape and gene expression in the shoot apical meristem using MorphoGraphX. Methods Mol. Biol. 2014;1080:121–134. doi: 10.1007/978-1-62703-643-6_10. [DOI] [PubMed] [Google Scholar]
- Barbier de Reuille P, Routier-Kierzkowska AL, Kierzkowski D, Bassel GW, Schupbach T, Tauriello G, Bajpai N, Strauss S, Weber A, Kiss A, Burian A, Hofhuis H, Sapala A, Lipowczan M, Heimlicher MB, Robinson S, Bayer EM, Basler K, Koumoutsakos P, Roeder AH, Aegerter-Wilmsen T, Nakayama N, Tsiantis M, Hay A, Kwiatkowska D, Xenarios I, Kuhlemeier C, Smith RS. MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife. 2015;4:05864. doi: 10.7554/eLife.05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW, Jacobs B, Cole M, Comelli P, Werr W. DORNROSCHEN-LIKE expression marks Arabidopsis floral organ founder cells and precedes auxin response maxima. Plant Mol. Biol. 2011;76:171–185. doi: 10.1007/s11103-011-9779-8. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proceedings of the National Academy of Sciences of the USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez R, Das P, Mirabet V, Moscardi E, Traas J, Verdeil JL, Malandain G, Godin C. Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nat Methods. doi: 10.1038/nmeth.1472. [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C. Live-imaging of the Arabidopsis inflorescence meristem. Methods Mol. Biol. 2014;1110:431–440. doi: 10.1007/978-1-4614-9408-9_25. [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell. 1990;2:741–753. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004;131:4225–4237. doi: 10.1242/dev.01261. [DOI] [PubMed] [Google Scholar]
- Reddy GV, Roy-Chowdhury A. Live-Imaging and Image Processing of Shoot Apical Meristems of Arabidopsis thaliana. Methods Mol. Biol. 2009;553:305–316. doi: 10.1007/978-1-60327-563-7_15. [DOI] [PubMed] [Google Scholar]
- Roeder AH, Chickarmane V, Cunha A, Obara B, Manjunath BS, Meyerowitz EM. Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS biology. 2010;8:e1000367. doi: 10.1371/journal.pbio.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanus SL, de Folter S, Shchennikova AV, Kaufmann K, Immink RG, Angenent GC. In planta localisation patterns of MADS domain proteins during floral development in Arabidopsis thaliana. BMC Plant Biol. 2009;9:5. doi: 10.1186/1471-2229-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, Guedon Y, Armitage L, Picard F, Guyomarc'h S, Cellier C, Parry G, Koumproglou R, Doonan JH, Estelle M, Godin C, Kepinski S, Bennett M, De Veylder L, Traas J. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature. 2015;517:377–380. doi: 10.1038/nature13853. [DOI] [PMC free article] [PubMed] [Google Scholar]