Abstract
The development of resistance to cisplatin (cDDP) is commonly accompanied by reduced drug uptake or increased efflux. Previous studies in yeast and murine embryonic fibroblasts have reported that the copper (Cu) transporters and chaperones participate in the uptake, efflux, and intracellular distribution of cDDP. However, there is conflicting data from studies in human cells. We used CRISPR-Cas9 genome editing to individually knock out the human copper transporters CTR1 and CTR2 and the copper chaperones ATOX1 and CCS. Isogenic knockout cell lines were generated in both human HEK-293T and ovarian carcinoma OVCAR8 cells. All knockout cell lines had slowed growth compared to parental cells, small changes in basal Cu levels, and varying sensitivities to Cu depending on the gene targeted. However, all of the knockouts demonstrated only modest 2 to 5-fold changes in cDDP sensitivity that did not differ from the range of sensitivities of 10 wild type clones grown from the same parental cell population. We conclude that, under basal conditions, loss of CTR1, CTR2, ATOX1, or CCS does not produce a change in cisplatin sensitivity that exceeds the variance found within the parental population, suggesting that they are not essential to the mechanism by which cDDP enters these cell lines and is transported to the nucleus.
Keywords: ATOX1, cisplatin, CCS, copper, CRISPR, CTR1, CTR2
Introduction
Cisplatin (cDDP) is an important chemotherapeutic agent that is an essential member of treatment programs for many types of cancer. Although initially effective, many tumors gradually become resistant during therapy with cDDP, and resistance is often accompanied by reduced drug uptake. cDDP is highly polar and its ability to diffuse across plasma membranes is thought to be limited, suggesting the presence of a transport system. Previous studies have suggested a role for copper (Cu) transporters in the uptake and efflux of cDDP 1.
Copper is essential to the growth of eukaryotic cells but can also be toxic due to its ability to undergo redox reactions that generate oxygen free radicals. A complex system of transporters and chaperones has evolved to manage Cu homeostasis. The Cu proteome is well conserved from yeast to humans with many of the proteins sharing conserved motifs that bind Cu. Cu is imported into the cell predominantly through the high-affinity transporter CTR1, which is a surface membrane-bound receptor that forms a homotrimer 2, 3. Structural models predict that the extracellular domain of CTR1 forms a pore that directly imports Cu 4. Increasing the extracellular concentration of Cu causes CTR1 internalization, which limits additional Cu import and potential toxicity 5. A second Cu transporter, CTR2, has also been identified, although its role in Cu homeostasis is less clear. CTR2 has been proposed to form a heterotrimer with CTR1 on the cell surface to support pore formation 6, and is additionally localized to endosomes and lysosomes 7. Yeast CTR2 has been shown to regulate internal pools of Cu under low Cu conditions 8, 9, suggesting that the human homolog may have a similar Cu regulatory function. Recent studies found that CTR2 plays a role in mast cell maturation and hemostasis 10, and that CTR1 protects CTR2 protein from degradation 11.
Once inside the cell, the concentration and location of Cu is tightly controlled as it is shuttled throughout subcellular compartments via interactions with Cu chaperones. The human copper chaperone for superoxide dismutase 1 (CCS) directly donates Cu to the enzyme SOD1 to support antioxidant activity 12, 13. Antioxidant 1 (ATOX1) donates Cu to the efflux Golgi-associated transporters ATP7A and ATP7B, which incorporate Cu into proteins, such as ceruloplasmin and lysyl oxidase, or into vesicles for secretion or export of excess Cu when levels are elevated 14, 15. Recent reports have also shown that Cu-laden ATOX1 migrates to the nucleus and may have transcription-factor like activity that supports cell growth 16. Whether ATOX1 and CCS directly acquire Cu from CTR1 or CTR2 or another intermediate Cu chaperone is unclear, although the two chaperones have been shown to directly interact and transfer Cu 17. Data recently published by Wang et al. show that ATOX1 or CCS knockdown decreases cell proliferation and tumor growth by modulating ATP levels and lipogenesis, and that an ATOX1/CCS targeting small molecule has similar effects 18.
Data from several labs has suggested that proteins within the Cu homeostasis pathway mediate uptake and efflux of cDDP. CTR1 gene deletion in yeast and mouse embryonic fibroblasts (MEFs) decreased cDDP uptake and increased resistance, suggesting it may be the primary transporter for drug influx 19, 20. CTR1 mRNA and protein expression levels correlate with cDDP response and post-treatment survival 21-23. In contrast to CTR1, the role of CTR2 in platinum drug cellular pharmacology is less well characterized. Expression levels correlate with treatment success in patients 24, 25 and cDDP sensitivity 26, and CTR2 knockdown or knockout cells have increased levels of cDDP uptake 6, 27. cDDP also binds the Cu chaperone COX17 in solution, and COX17 mediates mitochondrial delivery which, in turn, affects cDDP sensitivity through non-DNA related apoptosis pathways 28. cDDP has also been shown to bind ATOX1 in solution 29, 30 and in cells 31. ATOX1 knockout mutations in both Drosophila and MEFs engendered resistance to cDDP 32, 33. Consequentially, ATOX1 was hypothesized to bind and transport cDDP to the nucleus. ATOX1 may also play a role in cDDP detoxification through transfer to ATP7A or ATP7B. ATP7A and ATP7B protein levels were reported to be upregulated in cDDP resistant ovarian 34, 35 and oral squamous cell carcinoma cell lines 36. ATP7B overexpression in epidermoid carcinoma KB-3-1 cells also engendered cDDP resistance 37. Although no data has been reported for the direct interaction of CCS with cDDP, ATOX1, CCS and the transporters ATP7A and ATP7B share a conserved CXXC Cu chelating motif that coordinated cDDP in the crystal structure of ATOX1 29 and the soluble first domain of ATP7A 38. Despite all of these studies, there remains controversy as to the ability of the Cu homeostasis system to participate in the transport of cDDP or modulate sensitivity to this important drug 39, 40.
We used CRISPR-Cas9 genome editing to individually knockout CTR1, CTR2, ATOX1, and CCS in neoplastic human HEK-293T and ovarian cancer OVCAR8 cells. We report here that knockout of CTR1, CTR2, ATOX1, and CCS produced only a modest change in sensitivity to cDDP. These isogenic knockout cell lines targeting multiple steps within the same pathway provide compelling evidence that these Cu transporters and chaperones are not the primary uptake and transport mechanism for cDDP in these cell lines.
Methods
Cell culture and reagents
Human Embryonic Kidney 293T (HEK-293T) cells were obtained from ATCC (Manassas, VA). The wild type 293T and all sublines were cultured in DMEM medium (HyClone, Logan, UT) supplemented with 10% fetal bovine serum (Lonza, Basel, Switzerland), 1 mM sodium pyruvate (Lonza, Basel, Switzerland), 2 mM glutamine (Thermo Scientific, Waltham, MA) and 1X penicillin/streptomycin (Corning Life Sciences, Corning, NY). Ovarian Carcinoma 8 (OVCAR8) cells were received from Dr. Tom Hamilton, Fox Chase Cancer Center (Philadelphia, PA). All OVCAR8 cell lines were cultured in RPMI 1640 medium (HyClone) supplemented with 10% FBS and 1X penicillin/streptomycin. Both cell lines were grown at 37°C with 5% CO2 and tested negative for mycoplasma contaminated with a MycoSensor PCR kit (Agilent, Santa Clara, CA).
CRISPR-Cas9 genome engineering and knockout screening
Generation of the CTR1 and CTR2 knockout clones has been previously described 11. For ATOX1 and CCS knockout, single guide RNA (sgRNA) design and delivery was performed as previously described 41. The online CRISPR design tool (http://crispr.mit.edu/) was used to design guide RNAs targeting ATOX1 exon 2 on the coding strand and CCS exon 1 on the coding strand. Complementary guide oligonucleotides (ATOX1 5’-TCTCGGGTCCTCAATAAGCTTGG-3; CCS 5’- CACCGCTTCGGATTCGGGGAACCA-3’) were custom synthesized by Integrated DNA Technologies (Coralville) and ligated into pSpCas9(BB)-2A-GFP (pX458) (Addgene, Cambridge, MA). CRISPR plasmids were transiently transfected into HEK-293T or OVCAR8 cells with Lipofectamine 2000 (Life Technologies, Inc., Grand Island, NY). The transfected population was FACS sorted for GFP expression 48 h after transfection. Single cells were sorted into 96 well plates and grown into populations over a period of 3 weeks. Populations were expanded, and DNA was harvested from confluent 12-well plates with a QIAmp DNA mini kit (Qiagen, Valencia, CA). Genomic DNA was amplified with ATOX1 primers (F 5’-TCCTGCCCAGTC TCTCTGTCTT-3’; R:5’-CCATGGCTCCAGAGCTACTCCT-3’) or CCS primers (F 5’- GGGTTACTAAGGCAACCAGGA-3’; R 5’- GAGGCTTCTGGACTGTCTGC -3’) spanning the target cut site. Amplicons were digested with BamHI and HindIII (New England Biolabs, Ipswich, MA), ligated into pUC19 (Invitrogen, Grand Island, NY), and transformed into DH5alpha bacteria (Invitrogen) using standard molecular biology techniques. A minimum of 10 bacterial clones were sequenced per each knockout cell line.
Western blotting
Cells were washed with ice cold PBS, then lysed in ice-cold RIPA buffer (50 mM Tris-HCl pH 8.0; 150 mM NaCl; 1 mM EDTA, 1% Triton X-100; 0.1% SDS; 0.5% sodium deoxycholate) supplemented with 1X Halt protease inhibitor cocktail (Thermo Fisher Scientific). Protein contents of the lysates were determined with the DC-Protein assay kit (Bio-Rad; Hercules, CA). Fifty micrograms of total protein was boiled with Laemmli dye for 5-10 minutes, then loaded on Tris-Glycine or Tris-Tricine gels and electrotransferred to low fluorescence PVDF membranes (EMD Millipore, Billerica, MA). Blots were stained with 1:1000 anti-CTR1 (gift from Dr. Marcus Kuo42), 1:1000 mouse anti-CTR2 (National Cancer Institute43), 1:1000 rabbit anti-ATOX1 (Abcam, Cabridge, MA) or 1:1000 rabbit anti-CCS (Santa Cruz Biotechnology, Dallas, TX) and 1:1000 mouse β-actin (Cell Signaling Technologies, Danvers, MA) primary antibodies overnight at 4°C. The blots were counter stained with 1:10,000 goat-anti rabbit 680LT and 1:5,000 goat-anti-mouse-800CW secondary antibodies (Li-Cor Biosciences, Lincoln, NE) and imaged with a Li-Cor Odyssey Imager (Li-Cor Biosciences). Images were quantified with Image J software (NIH, http://imagej.nih.gov/ij/).
cDDP cytotoxicity assay
Cells were plated in sextuplicate in 96-well plates at a density of 3,000 cells/well. The cells were incubated for 16-24 hours and then treated with increasing doses of cisplatin (0-100 μM; Teva Parenteral Medicines, Inc., Irvine, CA) for 1 hour. After 1 hour, the cDDP media was aspirated and replaced with fresh media. The plates were incubated for an additional 96 hours, and the cell viability was determined with CCK-8 assay kit (Clonetech, Inc., Mountain View, CA). Five microliters of CCK-8 reagent was added per well, the plates were incubated at 37°C for 4 hours, and absorbance was read at 450 nm with an Molecular Devices microplate reader (Sunnyvale, CA).
Copper sensitivity assay
Cells were plated in quadruplicate in 96-well plates at a density of 3,000 cells/well. The cells were incubated for 16-24 hours and then treated with increasing doses of CuSO4 continuously for 96 hours. Cell viability was determined with CCK-8 assay kit (Clonetech, Inc., Mountain View, CA) as described above. The data were fit with a linear trend line to calculate and compare Cu sensitivity.
Cell growth curve assay
Cells were plated in quadruplicate in 96-well plates at a density of 2,000 cells/well. The cells were incubated for 1-7 days, and the cell viability was quantified every 24 hours with CCK-8 reagent as above. Cell growth was log10 transformed and graphed over time. The linear portion of the data was fit with a linear regression to calculate and compare cell growth rates.
Basal copper levels (ICP-MS)
HEK-293T cells were plated at a cell density of 0.5×10*6 (wildtype) − 0.75*10^6 cells (KOs) in two 6-well plates per cell line and incubated for 48-72 hours until 80-90% confluent. One 6 well plate was used to measure protein concentrations and the second was for Cu. The protein control plate was washed once with ice-cold PBS then lysed with 100 uL of ice-cold RIPA-Buffer supplemented with 1X Halt protease Inhibitor cocktail. Protein samples were stored at −80°C and the protein concentration was quantified with a BD BioRad protein quantification kit. The protein concentration was averaged for the 6 repeat wells. For the Cu samples, the cells were washed with ice-cold Cu-free PBS three times. Cell pellets were dissolved in 215 uL of 70% nitric acid at room temperature overnight, then diluted the next morning in 6 mL of ICP-MS buffer (0.1% Triton X, 1% Nitric acid, 1 ppb Indium, diluted in Cu-free water). 64Cu levels were measured with an iCAP inductively coupled plasma mass spectrometer at the Scripps Institute of Oceanography (Thermo Scientific, Waltham, MA). Cu concentrations were normalized to the average protein concentration for that cell line.
Nude mice xenografts
All animal work was approved by the UCSD Institutional Animal Care and Use Committee (IACUC). Female BALB/c nu/nu mice 6-8 weeks old were injected subcutaneously with 1×106 cells (150 uL) diluted 1:1 in PBS:Matrigel (BD Biosciences, Franklin Lakes, NJ). Mice were injected bilaterally at the left and right shoulder and flank regions (4 inoculation sites per mouse, 2-4 mice per group, n=8 to 16 injections per cell line). Once palpable, tumor volume was recorded twice per week, log10 transformed and graphed over time. Linear regression was used to calculate and compare the tumor growth rates.
Statistical analyses
Data presented represent the mean ± SEM of at least three independent experiments performed in at least triplicates. Student’s t-test p-values of the individual KOs compared to WT were calculated with GraphPad Prism (GraphPad, La Jolla, CA), and P-values < 0.05 were considered significant.
Results
Knockout cell line generation with CRISPR-Cas9 genome editing
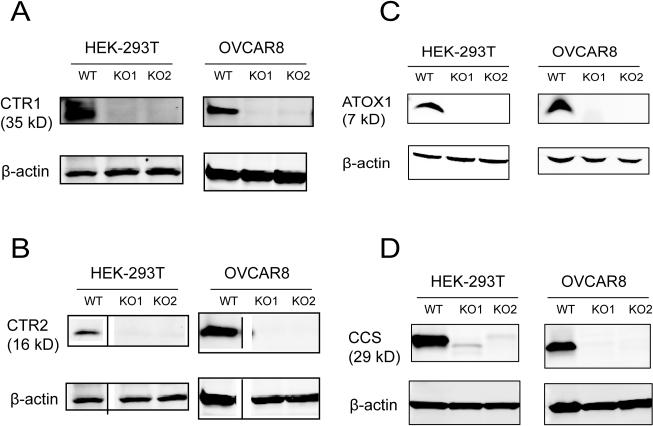
The bacterial CRISPR-Cas9 system was used to generate human isogenic knockout cell lines that completely lack detectable levels of the target protein. The CTR1, CTR2, ATOX1, and CCS genes were independently edited using the CRISPR-Cas9 technique and the tools initially described by the Zhang laboratory 41. Generation of the CTR1 and CTR2 knockouts was previously described 11. Guide RNAs were designed with the online CRISPR design tool (http://crispr.mit.edu/) and cloned into the pSpCas9(BB)-2A-GFP (pX458) vector. Exon 2 was targeted in CTR1, exon 3 in CTR2, exon 2 in ATOX1, and exon 1 in CCS. We edited each gene in two different human cell lines, the neoplasticHEK-293T cell line and the OVCAR8 human ovarian carcinoma line. Parental cell populations were transiently transfected with the appropriate vector, and GFP-positive single cells were sorted by FACS into individual wells 48 h after transfection. For each gene targeted genes, approximately 50 single cell clones were expanded into populations over the course of 3 weeks. PCR primers flanking the targeted editing site were designed to generate an amplicon, and sequencing of the expanded populations indicated numerous clones (>40%) had mutations in the individual target genes. Candidate mutant clones were further screened by Western blotting with whole cell lysates to assess target protein expression. Multiple clones that lacked protein expression were identified for each gene indicating that CRISPR-Cas9 targeting and knockout was successful. Two clones per gene (KO1 and KO2) for each cell type were further characterized using standard molecular cloning techniques to sequence the individual alleles. Sequencing indicated that the KO clones contained small insertions or deletions near the targeted CRISPR cut site on both alleles (Table 1). All of the DNA in/dels in the 2 clones selected for each gene were predicted to cause a frameshift mutation and introduce an early downstream stop codon on both alleles resulting in a severely truncated, out of frame, or unstable protein (Table 2). A total of 16 clonal cell lines were generated for comparison: two knockout clones per gene (CTR1, CTR2, ATOX1, and CCS) in each of two cell lines (HEK-293T and OVCAR8). Figure 1 shows Western blot analyses of each KO cell line confirming the absence of target protein expression.
Table 1.
DNA allele sequences of the ATOX1 and CCS knockout cell lines generated with CRISPR-Cas9 targeting technology.
| Cell Line | Allele 1 nucleotide sequence | Allele 2 nucleotide sequence |
|---|---|---|
|
ATOX1
(Exon 2 Targeted CRISPR site) |
||
| ATOX1 WT | GCTGAAGCTGTCTCTCGGGTCCTCAATAAGCTTGGAGGTGAGTGAGTGG | |
| HEK-293T ATOX1 KO1 |
GCTGAAGCTGTCTCTCGGGTCCTCAATAAAGCTTGGAGGTGAGTGAGTGG (+1 nt) |
|
| HEK-293T ATOX1 KO2 |
GCTGAAGCTGTCTCTCGGGTC- - - - - - - -GCTTGGAGGTGAGTGAGTGG (Δ8 nt) |
|
| OVCAR8 ATOX1 KO1 |
G- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -AGTGAGTGG (Δ39 nt) |
GCTGAAG- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -G (Δ41 nt) |
| OVCAR8 ATOX1 KO2 |
GCTGAAGCTGTCTCTCGGGTCCTCAA TAAAGCTT GGAGGTGAGTGAGTGG (+1 nt) |
GCTGAAGCTGTCTCTCGGGTCCTCA ATA- GCTT GGAGGTGAGTGAGTGG (Δ1 nt) |
|
CCS
(Exon 1 Targeted CRISPR site) |
||
| CCS WT | GGGTCCAGAATGGCTTCGGATTCGGGGAACCAGGGG | |
| HEK-293T CCS KO1 |
GGGTCCAGAATGGCTTCGGA TTCGGGG - ACCAGGGG (Δ 1 nt) |
GGG - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -GG (Δ 31 nt) |
| HEK-293T CCS KO2 |
GGGTCCAGAATGGCTTCGGA TTCGGGG - ACCAGGGG (Δ 1 nt) |
GGGTCCAGAATGGCTTCGGA TTCGGG - - ACCAGGGG (Δ 2 nt) |
| OVCAR8 CCS KO1 |
GGGTCCAGAATGGCTTCGGA TTCGGGG - ACCAGGGG (Δ 1 nt) |
GGGTCCAGAATGGCTTCGGA TTCGGGGAA - CAGGGG (Δ 1 nt) |
| OVCAR8 CCS KO2 |
GGGTCCAGAATGGCTTCGGA TTCGGGG - ACCAGGGG (Δ 1 nt) |
GGGTCCAGAATGG- - - - - - - - - - - - - - - - - - - - - -G (Δ 22 nt) |
Table 2.
Predicted protein amino acid sequences of the ATOX1 and CCS knockout cell lines.
| Cell Line | Allele 1 amino acid sequence | Allele 2 amino acid sequence |
|---|---|---|
| ATOX1 | ||
| ATOX1 WT | MPKHEFSVDMTCGGCAEAVSRVLNKLGGVKYDIDLPNKKVCIESEHSMDTLLATLKKTG KTVSYLGLE |
|
| HEK-293T ATOX1 KO1 |
MPKHEFSVDMTCGGCAEAVSRVLNKAWRS* | |
| HEK-293T ATOX1 KO2 |
MPKHEFSVDMTCGGCAEAVSRVAWRS* | |
| OVCAR8 ATOX1 KO1 |
n/a§ | n/a§ |
| OVCAR8 ATOX1 KO2 |
MPKHEFSVDMTCGGCAEAVSRVLNK AWRS* |
MPKHEFSVDMTCGGCAEAVSRVLNSLEELSM TLTCPTRRSALNLSTAWTLCLQP¶ |
| CCS | ||
| CCS WT | MASDSGNQGTLCTLEFAVQMTCQSCVDAVRKSLQGVAGVQDVEVHLEDQMVLVHTTL PSQEVQALLEGTGRQAVLKGMGSGQLQNLGAAVAILGGPGTVQGVVRFLQLTPERCLIE GTIDGLEPGLHGLHVHQYGDLTNNCNSCGNHFNPDGASHGGPQDSDRHRGDLGNVRAD ADGRAIFRMEDEQLKVWDVIGRSLIIDEGEDDLGRGGHPLSKITGNSGERLACGIIARSAG LFQNPKQICSCDGLTIWEERGRPIAGKGRKESAQPPAHL |
|
| HEK-293T CCS KO1 |
MASDSGTRGPSARWSSRCR* | n/a# |
| HEK-293T CCS KO2 |
MASDSGTRGPSARWSSRCR* | MASDSGPGDPLHVGVRGADDLSELCGRGAQI PARGGRCPGCGGALGGPDGLGTHHSTQPGGA GSPGRHGAAGGTQGHGQRPVAESGGSSGHPG GAWHRAGGGALPTADP§ |
| OVCAR8 CCS KO1 |
MASDSGTRGPSARWSSRCR* | MASDSGNRGPSARWSSRCR* |
| OVCAR8 CCS KO2 |
MASDSGTRGPSARWSSRCR* | MGPSARWSSRCR* |
Early stop codon causes translation termination
Sequence shows less than 50% homology to WT protein
CRISPR induced lesion removes translation start codon
CRISPR induced lesion removes exon/intron boundary; is predicted to impair mRNA splicing.
Figure 1.
Western blot analysis of clones in which CTR1, CTR2, ATOX1, and CCS were knocked out. Basal protein expression levels in the wild type (WT) HEK293T and OVCAR8 and the knockout (KO) clones. A, CTR1; B, CTR2; C, ATOX1; and, D, CCS.
Cisplatin sensitivity
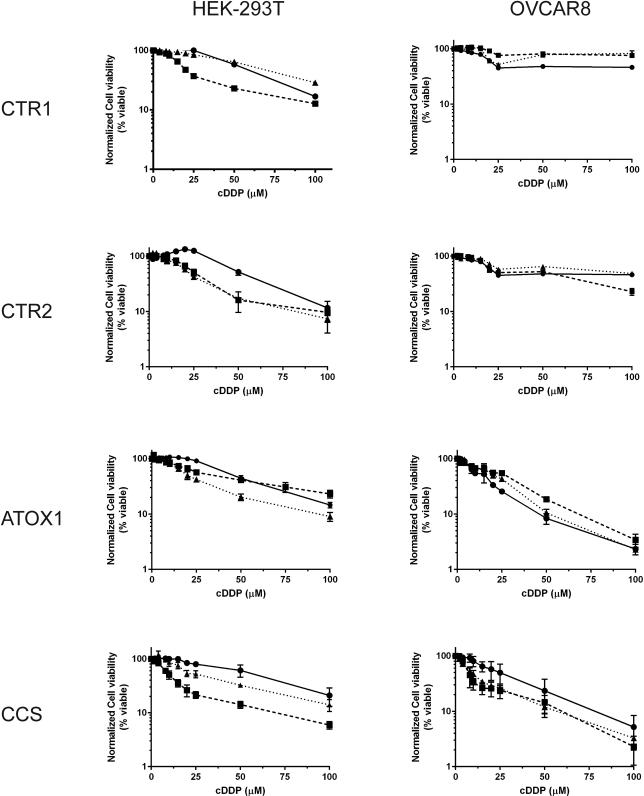
Concentration survival curves were generated with wild-type (WT) HEK-293T and OVCAR8 cells and compared to each knockout clone using a cell viability assay after 1 hour cDDP exposure followed by 96 hour culture in drug-free medium. Figure 2 shows the dose-response curves for all the cell lines. Interestingly, different clones from the same parental cell line in which the same gene was knocked out showed substantial differences in cDDP sensitivity. With 1 hour of cDDP exposure the 293T CRT1 KOs had a variable phenotype; whereas KO1 had a 0.4-fold decrease in IC50, KO2 had a 1.2-fold increase. All of the CTR2, ATOX1, and CCS KOs were more sensitive to cDDP compared to the WT HEK-293Ts. The CTR2 KOs had a 0.5- and 0.4-fold decrease in IC50, while the ATOX1 KOs a 0.7- and 0.4-fold decrease; the CCS KOs had a 0.2 and 0.5-fold decrease in IC50 value (Figure 2 and Table 3). Collectively, the data suggest that knockout of CTR1, CTR2, ATOX1, or CCS modestly increases cDDP sensitivity in the HEK-293T cell line.
Figure 2.
cDDP concentration-survival curves for the wild type HEK293T and OVCAR8 cell lines and each of the knockout clones. Cells were seeded in 96 well plates and treated with 0-100 μM cDDP for 1 h and then incubated in drug-free medium for an additional 4 days. The data represent the mean ± SEM of three independent experiments plated in sextuplicate for each drug concentration. For all cell lines circles (λ, solid line) represent WT, squares represent KO1 (ν, dashed line), and triangles (π, dotted line) represent KO2.
Table 3.
Cisplatin IC50 values.
| HEK-293T | OVCAR8 | ||||||
|---|---|---|---|---|---|---|---|
| IC50 (μM) mean ± SEM* |
Fold change |
P-value | IC50 (μM) mean ± SEM* |
Fold change |
P-value | ||
| WT | 53.1 ± 3.3 | - | - | 25.6 ± 4.5 | - | - | |
| CTR1/KO1 | 19.3 ± 1.0 | 0.4 | 0.412 | 112.3 ± 25.9 | 4.4 | 0.341 | |
| CTR1/KO2 | 65.3 ± 4.5 | 1.2 | 0.408 | 23.6 ± 2.6 | 0.9 | 0.165 | |
| CTR2/KO1 | 26.1 ± 0.3 | 0.5 | 0.043 | 31.5 ± 7.9 | 1.2 | 0.915 | |
| CTR2/KO2 | 21.9 ± 0.1 | 0.4 | <0.01 | 34.4 ± 11.8 | 1.3 | 0.398 | |
| ATOX1/KO1 | 36.8 ± 8.4 | 0.7 | 0.440 | 26.9 ± 1.8 | 1.1 | 0.025 | |
| ATOX1/KO2 | 20.7 ± 1.9 | 0.4 | <0.01 | 17.2 ± 4.0 | 0.7 | 0.287 | |
| CCS/KO1 | 10.5 ± 1.8 | 0.2 | 0.047 | 5.6 ± 0.6 | 0.2 | <0.01 | |
| CCS/KO2 | 26.9 ± 5.5 | 0.5 | 0.214 | 8.2 ± 0.7 | 0.3 | 0.04 | |
The data represent the mean ± SEM of at least three independent experiments plated in sextuplicate
Clonal variability was also observed among the OVCAR8 cell lines. The OVCAR8 CTR1 KOs had a 4.4- and 0.9-fold decrease in IC50, while the CTR2 KOs had a 1.2- and 1.3-fold decrease in IC50. The ATOX1 OVCAR8 KOs had a 1.1-fold increase and 0.7-fold decrease in IC50, while the CCS KOs had a 0.2 and 0.3-fold decrease (Figure 2 and Table 3). Collectively, these results suggest that the effect of CTR1, CTR2, or ATOX1 knockout was variable among individual clones and the two cell lines, while knockout of CCS modestly increased cDDP sensitivity in both cell types.
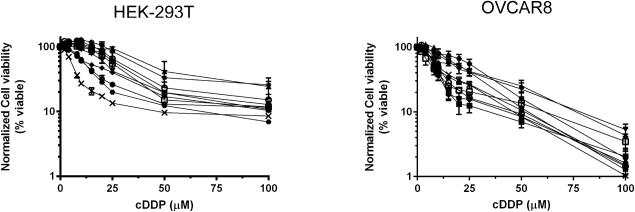
Notable variability in cDDP sensitivity was observed among the individual KO clones for each gene, suggesting that clonal variability and genetic drift within the population may have a large effect. To assess the background variability of the WT population, we isolated 10 sub-clones from our transfected HEK-293T and OVCAR8 populations that were negative for CRISPR editing (WT gene sequence) and determined their cDDP sensitivity. The IC50 values in the HEK-293T WT sub-clones ranged from 6.1-50.2 μM, and 9.1-22.7 μM in the WT OVCAR8 sub-clones (Figure 3). These changes in sensitivity are similar to the magnitude changes in our knockout clones, indicating that the variability observed in the CTR1, CTR2, ATOX1, and CCS KO clones was less than the clonal variability of the starting population in these cell lines.
Figure 3.
cDDP concentration-survival curves for ten wild type HEK293T or OVCAR8 subclones. Ten subclones were sorted from the parental population. Cells were seeded in 96 well plates and treated with 0-100 μM cDDP for 1 h and then incubated in drug-free medium for an additional 4 days. The data represent the mean ± SEM of two independent experiments plated in sextuplicate for each drug concentration.
Cellular growth rate
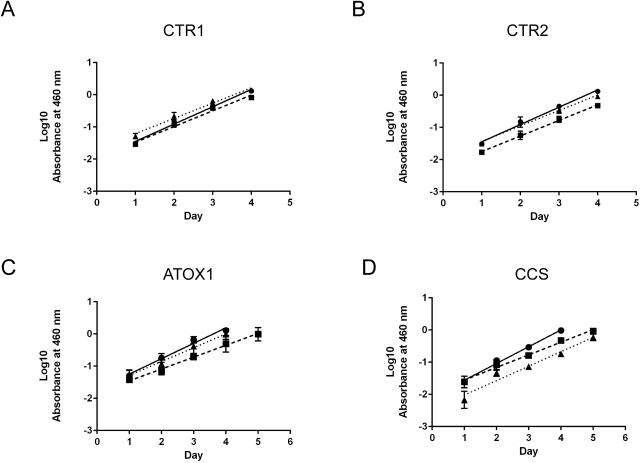
The growth rates of the KO clones were compared to the WT cells to assess the impact of protein deletion on in vitro growth. Previous studies in endothelial cells indicated that silencing of CTR1 decreased proliferation by 2-fold 44. ATOX1 and CCS silencing in H1299 lung cancer cells was reported to impair cellular growth by decreasing ATP production, decreasing cytochrome c oxidase activity, and reducing lipid biosynthesis 18. Cell density was determined as a function of time over a period of 7 days in culture and compared among the WT and KO cells. Compared to the WT HEK-293T cells, both CTR1 KOs and CTR2 KOs had a slightly decreased rate of growth (p<0.05; Figure 4A and 4B). Both ATOX KO1 and KO2 had an increased lag time, where the cells took longer to establish growth after passaging; however, once established, these cells had a similar cell growth rate compared to the WT cells (p>0.05; Figure 4C). Similarly, both CCS KOs had a longer lag time; however, KO1 had a similar rate of growth compared to WT cells, while KO2 had a 1.2-fold decrease in rate (p=0.01) (Figure 4D). Collectively, the cellular growth assays showed that all the KO cells are viable in culture and most have similar growth rates to the parental cells once they reach a threshold cell density.
Figure 4.
Growth rates of the parental HEK-293T and knockout cells in culture. A) CTR1, B) CTR2, C) ATOX1, and D) CCS. Cells were seeded in 96 well plates and cell viability was quantified every 24 hours. The data were log10 transformed and the linear data for cell growth were fit with a linear trend line shown. All data represent the mean ± SEM of three independent experiments plated in quadruplicate. For all cell lines circles (λ, solid line) represent WT, squares represent KO1 (ν, dashed line), and triangles (π, dotted line) represent KO2.
Basal copper levels and copper sensitivity
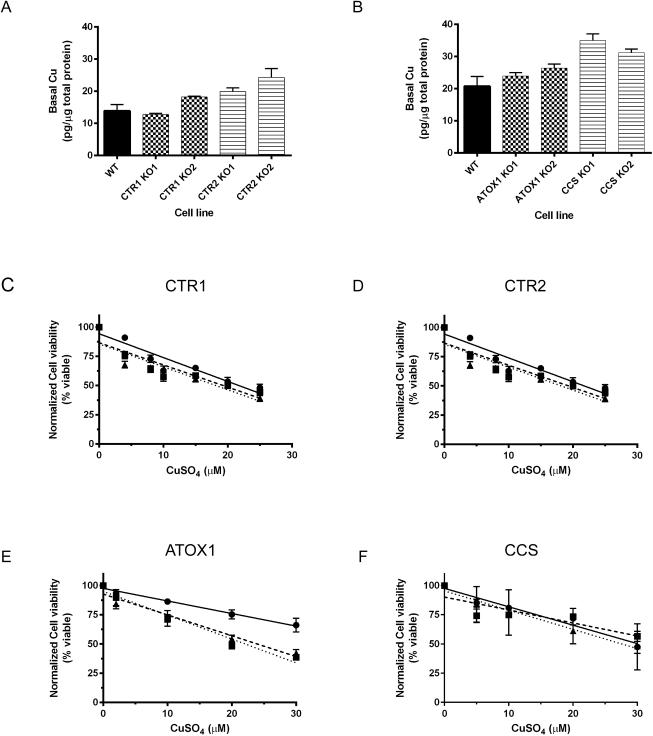
We initially hypothesized that the observed growth defects in the KO lines may be caused by defects in Cu homeostasis, such as decreased uptake or failure to shuttle intracellular Cu to essential enzymes and cellular compartments. To test this, the total cellular basal Cu content of HEK-293T cells was determined by inductively coupled plasmon mass spectrometry (ICP-MS). Compared to the WT cells, the CTR1 knockouts had no significant change in basal Cu, while the CTR2 KOs had a 1.4 and 1.8-fold increase in Cu (p=0.06 and 0.04, respectively) (Figure 5A). ATOX1 protein KO resulted in small 1.2 and 1.3-fold increases in basal Cu level (Figure 5B; p=0.20 and 0.06, respectively), which agrees with previous data reported in ATOX1−/− mouse embryonic fibroblasts 45. Strikingly, the CCS KO cells had significantly higher levels of basal Cu compared to the WT HEK-293T cells; Cu levels were elevated 1.7-fold in CCS KO1 and 1.5-fold in CCS KO2 (Figure 5B; both p<0.01), in contrast to previous reports with in which CCS was knocked down in MEFs 46. Thus, the growth defects observed in all the KOs were not mediated by defects in total basal cellular Cu.
Figure 5.
Copper homeostasis in HEK-293T parental and knockout cell lines. A, B) Basal whole cell Cu concentrations in the wild-type (WT) and all knockout (KO) cell lines. P-value WT vs.: CTR1 KO1 = 0.62; CTR1 KO2 = 0.09; CTR2 KO1 = 0.06; CTR2 KO2 = 0.04. P-value WT vs.: ATOX KO1 = 0.02; ATOX1 KO2 = 0.06; CCS KO1 <0.01; CCS KO2 <0.01. Cells were plated in in 6-well plates, and each well was harvested for basal Cu quantification with ICP-MS. Copper concentrations were normalized to the average total protein concentration from a second 6-well plate. The data represent the mean ± SEM of three independent experiments plated in sextuplicate. C-F) Cu sensitivity stress test in the C) 293T CTR1 knockout cell lines, D) 293T CTR2 KO cell lines, E) 293T ATOX1 KO cell lines, and F) 293T CCS KO cell lines. For all cell lines circles (λ, solid line) represent WT, squares represent KO1 (ν, dashed line), and triangles (π, dotted line) represent KO2. Cells were plated in 96 well plates in media supplemented with 0-30 μM CuSO4 and incubation in culture for 4 days. The data represent the mean ± SEM for three independent experiments plated in triplicate.
Previous reports with ATOX−/− MEFs showed that the knockout cells were more sensitive to elevated Cu levels 32 due to impaired ATP7B-mediated Cu efflux 14. To test the effect of elevated Cu, we repeated the cellular growth experiments using medium supplemented with Cu. Cells were grown for 4 days in the presence of medium supplemented with 0-30 μM CuS04 following which cellular viability was quantified. Compared to the WT HEK-293T cells, both ATOX1 KOs had a significantly decreased cell viability that correlated with increased concentrations of Cu (p < 0.01 and = 0.02; Figure 5E). In contrast, the CTR1, CTR2, and CCS KOs had no significant change in viability compared to the WT cells (Figure 5C, D, F; all p > 0.05). These data are consistent with previous findings that ATOX1 deficient cells are sensitive to Cu, presumably due to export defects via ATOX1 interactions with ATP7B 14. Moreover, these data suggest that the slower growth observed in all the KO cells was not due to a cellular Cu deficiency, as even low concentrations of CuSO4 failed to stimulate growth.
Tumor growth in vivo
Studies have consistently shown that tumors have elevated Cu levels, and that Cu is closely associated with key steps in angiogenesis and tumor growth 47. Cu chelators are effective in slowing tumor formation in mice, and the treatment of breast cancer patients with tetrathiomolybdate was reported to produce a benefit in survival in a small study 48. Recent data published by Wang et al. showed that targeting ATOX1 and CCS with a small molecule significantly decreased both lung H1299 and leukemia K562 tumor xenograft growth 18. We wondered if any of the individual copper transporters or chaperones is a potential drug target and tested the in vivo tumorigenicity of our KO cell lines in nude mice.
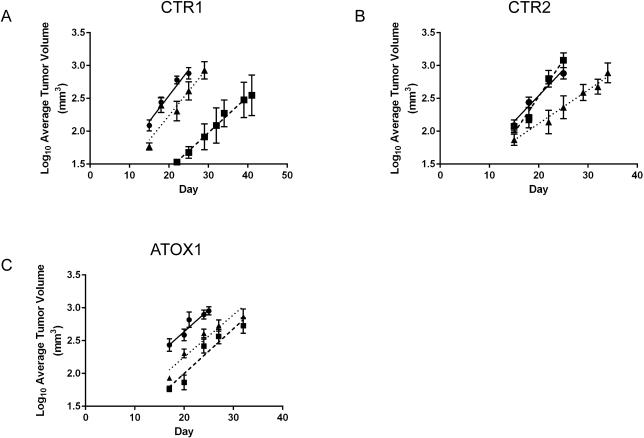
Xenografts were established by injecting female nu/nu mice subcutaneously with either the HEK-293T WT, CTR1, CTR2, or ATOX1 KO cell lines (both knockouts per each gene). Tumor growth was measured over a period of 30-45 days, and the logarithmic tumor growth rate was compared among the WT and KO cell lines to assess the impact of loss of gene function on tumor establishment and growth. The CTR1 KOs had a variable phenotype. CTR1 KO1 had an increased lag time and took much longer to establish than KO2 or the WT cells and had a significantly decreased growth rate (p = 0.04), while KO2 had a slightly increased lag time compared to WT, yet a similar growth rate (p = 0.34) (Figure 6A). The CTR2 KOs also had a variable phenotype; CTR2 KO1 had a similar lag time and a significantly increased growth rate compared to the WT cells (p < 0.01), and CTR2 KO2 had a similar lag time and growth rate (p = 0.91) (Figure 6B). Interestingly, the in vivo data for the HEK-293T ATOX1 KOs mirrored the cell culture growth results. Compared to the HEK-293T WT cells, both ATOX1 KO cell lines had an increased lag time to tumor establishment. However, once the KO tumors were established, their growth rate was not different from the WT tumors (Figure 6C; p = 0.98 and 0.51). Thus, targeting ATOX1, CTR1, or CTR2 alone did not appear to be a viable antineoplastic therapeutic strategy, as these KO tumors continued to exhibit robust, albeit delayed growth in vivo.
Figure 6.
HEK-293T wild type and CTR1, CTR2, and ATOX1 knockout tumor growth in nude mice. Six-week old female nu/nu mice were injected with 1 ×106 cells. Tumor growth was measured over a period of 30-50 days and log10 transformed. A) CTR1 KOs, B) CTR2 KOs, and C) ATOX1 KOs. For all cell lines circles (λ, solid line) represent WT, squares represent KO1 (ν, dashed line), and triangles (π, dotted line) represent KO2. The linear growth rates of the transformed data were calculated and statistically compared among the 293T WT and KO cell lines. N=5-8 tumors CTR1, 8 tumors CTR2, and 15-16 tumors for ATOX1.
Discussion
The Cu homeostasis pathway has been proposed to function as a transport system for cDDP. The effect of CTR1 overexpression and/or knockdown on cDDP uptake and sensitivity has been conflicting. Yeast CTR1 null mutants and mouse CTR1−/− embryonic fibroblasts (MEFs) had increased cDDP resistance and decreased cDDP uptake 19, and re-expression of CTR1 in these MEFs restored cDDP sensitivity 49. Similarly, overexpression of human CTR1 in SCLC and SR2 cells modestly increased cDDP uptake 50. However, additional groups reported no correlation between cDDP sensitivity and uptake in cells overexpressing human CTR139, 51. Our CTR1 KOs did not have a consistent phenotype due to clonal variation. The relatively modest changes in cDDP sensitivity observed here (3 to 4-fold) agree with the previous literature and suggest that CTR1 may not be the primary uptake transport mechanism in HEK-293T and OVCAR8 cell lines.
Whereas CTR1 deletion has been linked with resistance, CTR2 knockdown in MEFs has been reported to increase cDDP uptake and increase cellular sensitivity to cDDP 26. CTR2−/− xenograft tumors grew much slower and acquired significantly more cDDP 52. Similarly, CTR2 knockdown in human 2008 cancer cells rendered the cells 2 to 3-fold hypersensitive to cDDP 43. The CTR2 KOs here had modest, similar-fold changes in cDDP IC50. The HEK-293T KOs appeared more sensitive while the OVCAR8 KO cells were resistant, possibly due to differences in CTR2 function in varying cell lines.
Several groups have documented direct binding of cDDP to ATOX1 in solution 30, 53 and in cells 31. Drosophila ATOX1 knockout cells were found to be resistant to 1 mM cDDP 33, while ATOX1−/− MEFs had a 1.5-fold increase in IC50 32. Because ATOX1 has been shown to directly transfer cDDP to ATP7A and ATP7B in solution 14, 15, and to modulate Cu detoxification, it may similarly modulate cDDP efflux. Studies regarding direct cDDP transfer between ATOX1 and the metallo-binding domains of ATP7A or ATP7B in solution are conflicting depending on the reducing agent present 54, 55. Recent studies suggests that Cu binding influences cDDP binding to ATOX1 56 and may influence transfer of platinum drugs from ATOX1 57. While ATP7A and ATP7B protein levels were noted to be elevated in cDDP resistant paired cell lines, no changes in ATOX1 expression were seen 34. Our ATOX KO cells varied 2-fold in cDDP sensitivity compared to the parental cells. Collectively, these data suggest that ATOX1 is not a major mediator of cDDP toxicity and is not critical to the nuclear import of cDDP. Similar fold changes in cDDP sensitivity were also seen with 96 hour continuous drug exposure assays (data not shown), suggesting that ATOX1 does not mediate cDDP detoxification even though these ATOX1 deficient cells were significantly more sensitive to Cu compared to the other knockouts. However, because ATOX1 and CCS can cross-transfer Cu and are both present in the nucleus they may have redundant activities. Knockout of both genes may produce clearer results; however, deletion of both genes simultaneously may be lethal.
To our knowledge, no reports have described interactions between cDDP and CCS. Knockout of CCS in both cell lines in this study resulted in a 2 to 5-fold increase in cDDP sensitivity. Previous studies have shown that glutathione (GSH) can compensate for CCS deletion to support Cu-mediated SOD1 function 58. It’s possible that changes in GSH or the cellular oxidation state can compensate for the loss of CCS. Additional studies are needed to determine whether the changes in cDDP sensitivity seen here are specific or mediated by changes in SOD1 activity and ROS balance.
This study exposed a major issue with the use of individual KO clones for studies of cellular pharmacology. The changes in cDDP sensitivity observed in the KO cells were less than those observed between independently isolated wild type clones derived from the parental HEK-293T and OVCAR8 populations. However, similar magnitude changes in cDDP sensitivity have been reported in cells deficient in DNA damage repair pathways 59-61, as well as cDDP resistant cells where upregulated ATPase genes important for maintaining tumor cell pH, which influences cDDP aquation and activity, were knocked down 62. Constant genetic drift of cancer cells, especially cells in tissue culture, pose a challenge to the use of isogenic knockout clones to assess the effect of loss of function of a given gene. The clonal variability observed in this study mandates the use of numerous knockout clones in future studies.
It is important to note that variation in drug sensitivity among the knockouts may be mediated by off-target CRISPR-Cas9 editing. Cas9 mutants with increased fidelity have now been engineered that have off-target effects below the limit of detection 63, 64. Improvements in Cas9 design may allow for bulk sorting of a transfected population to more rapidly study the effects of gene editing and protein deletion, thus bypassing the arduous task of characterizing individual clones.
Conclusion
The mechanisms of cDDP uptake and transport remain largely unknown, despite widespread clinical use as an anti-neoplastic agent for the past 50 years. Initial studies described correlations between cDDP response and patient survival with copper transport protein expression, suggesting that the two metals may utilize similar pathways to enter and traffic through cells. However, protein knockdown studies in cells from several species have been conflicting. The generation of isogenic knockout cell lines in 4 Cu transport/chaperones genes represents a clean system to address this question.
Knockout of any of the 4 genes reported here resulted in variable changes in sensitivity to cDDP. The fold changes range from 2 to 5-fold. However, our data indicate that knockout of these Cu transporters and chaperones failed to produce changes in cDDP sensitivity that were greater than the variance in cDDP sensitivity observed among non-edited clones isolated from the same parental population. The mechanisms of cellular entry and nuclear translocation of cDDP remain unknown. Genome-wide CRISPR-Cas9 screens directed at identifying genes that modulate cDDP sensitivity are currently underway.
Significance to Metallomics.
Prior studies have suggested a role for Cu homeostasis proteins in the cellular pharmacology of the platinum-containing drugs. However, using the more definitive approach of CRISPR-Cas9-mediated gene knockout this study shows that neither the loss of Cu transport proteins (CTR1 or CTR2), nor the knockout of two Cu chaperones (ATOX1 and CCS), modulated sensitivity to the chemotherapeutic agent cisplatin in two different types of neoplastic cell lines.
Acknowledgements
This work was supported by grants R01-CA152185 and T32CA121938 from the National Institutes of Health. We would like to thank Professor James Day the Scripps Institute of Oceanography for his help with ICP-MS.
References
- 1.Abada P, Howell SB. Regulation of cisplatin cytotoxicity by Cu influx transporters. Met Based Drugs. 2010;2010:317581. doi: 10.1155/2010/317581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Feo CJ, Aller SG, Unger VM. A structural perspective on copper uptake in eukaryotes. Biometals. 2007;20:705–716. doi: 10.1007/s10534-006-9054-7. [DOI] [PubMed] [Google Scholar]
- 3.De Feo CJ, Aller SG, Siluvai GS, Blackburn NJ, Unger VM. Three-dimensional structure of the human copper transporter hCTR1. Proc Natl Acad Sci USA. 2009;106:4237–4242. doi: 10.1073/pnas.0810286106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsigelny IF, Sharikov Y, Greenberg JP, Miller MA, Kouznetsova VL, Larson CA, Howell SB. An All-Atom Model of the Structure of Human Copper Transporter 1. Cell Biochem Biophys. 2012;63:223–234. doi: 10.1007/s12013-012-9358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molloy SA, Kaplan JH. Copper-dependent recycling of hCTR1, the human high affinity copper transporter. J Biol Chem. 2009;284:29704–29713. doi: 10.1074/jbc.M109.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohrvik H, Nose Y, Wood LK, Kim BE, Gleber SC, Ralle M, Thiele DJ. Ctr2 regulates biogenesis of a cleaved form of mammalian Ctr1 metal transporter lacking the copper- and cisplatin-binding ecto-domain. Proc Natl Acad Sci USA. 2013;110:E4279–4288. doi: 10.1073/pnas.1311749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van den Berghe PV, Folmer DE, Malingre HE, van Beurden E, Klomp AE, van de Sluis B, Merkx M, Berger R, Klomp LW. Human copper transporter 2 is localized in late endosomes and lysosomes and facilitates cellular copper uptake. Biochem J. 2007;407:49–59. doi: 10.1042/BJ20070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rees EM, Thiele DJ. Identification of a vacuole-associated metalloreductase and its role in CTR2-mediated intracellular copper mobilization. J Biol Chem. 2007;282:21629–21638. doi: 10.1074/jbc.M703397200. [DOI] [PubMed] [Google Scholar]
- 9.Portnoy ME, Schmidt PJ, Rogers RS, Culotta VC. Metal transporters that contribute copper to metallochaperones in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;265:873–882. doi: 10.1007/s004380100482. [DOI] [PubMed] [Google Scholar]
- 10.Ohrvik H, Logeman B, Noguchi G, Eriksson I, Kjellen L, Thiele DJ, Pejler G. Ctr2 Regulates Mast Cell Maturation by Affecting the Storage and Expression of Tryptase and Proteoglycans. J Immunol. 2015;195:3654–3664. doi: 10.4049/jimmunol.1500283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai CY, Liebig JK, Tsigelny IF, Howell SB. The copper transporter 1 (CTR1) is required to maintain the stability of copper transporter 2 (CTR2) Metallomics. 2015;7:1477–1487. doi: 10.1039/c5mt00131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Culotta VC, Klomp LW, Strain J, Casareno RL, Krems B, Gitlin JD. The copper chaperone for superoxide dismutase. J Biol Chem. 1997;272:23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 13.Wong PC, Waggoner D, Subramaniam JR, Tessarollo L, Bartnikas TB, Culotta VC, Price DL, Rothstein J, Gitlin JD. Copper chaperone for superoxide dismutase is essential to activate mammalian Cu/Zn superoxide dismutase. Proc Natl Acad Sci USA. 2000;97:2886–2891. doi: 10.1073/pnas.040461197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamza I, Prohaska J, Gitlin JD. Essential role for Atox1 in the copper-mediated intracellular trafficking of the Menkes ATPase. Proc Natl Acad Sci USA. 2003;100:1215–1220. doi: 10.1073/pnas.0336230100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamza I, Schaefer M, Klomp L, Gitlin J. Interaction of the copper chaperone HAH1 with the Wilson disease protein is essential for copper homeostasis. Proc Natl Acad Sci USA. 1999;96:13363–13368. doi: 10.1073/pnas.96.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh S, Kim HW, Nakagawa O, Ozumi K, Lessner SM, Aoki H, Akram K, McKinney RD, Ushio-Fukai M, Fukai T. Novel role of antioxidant-1 (atox1) as a copper dependent transcription factor involved in cell proliferation. J Biol Chem. 2008;283:9157–9167. doi: 10.1074/jbc.M709463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petzoldt S, Kahra D, Kovermann M, Dingeldein AP, Niemiec MS, Aden J, Wittung-Stafshede P. Human cytoplasmic copper chaperones Atox1 and CCS exchange copper ions in vitro. Biometals. 2015;28:577–585. doi: 10.1007/s10534-015-9832-1. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Luo C, Shan C, You Q, Lu J, Elf S, Zhou Y, Wen Y, Vinkenborg JL, Fan J, Kang H, Lin R, Han D, Xie Y, Karpus J, Chen S, Ouyang S, Luan C, Zhang N, Ding H, Merkx M, Liu H, Chen J, Jiang H, He C. Inhibition of human copper trafficking by a small molecule significantly attenuates cancer cell proliferation. Nature Chem. 2015;7:968–979. doi: 10.1038/nchem.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishida S, Lee J, Thiele DJ, Herskowitz I. Uptake of the anticancer drug cisplatin mediated by the copper transporter Ctr1 in yeast and mammals. Proc Natl Acad Sci USA. 2002;99:14298–14302. doi: 10.1073/pnas.162491399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin X, Okuda T, Holzer A, Howell SB. The copper transporter CTR1 regulates cisplatin uptake in saccharomyces cerevisiae. Mol Pharmacol. 2002;62:1154–1159. doi: 10.1124/mol.62.5.1154. [DOI] [PubMed] [Google Scholar]
- 21.Ishida S, McCormick F, Smith-McCune K, Hanahan D. Enhancing Tumor-Specific Uptake of the Anticancer Drug Cisplatin with a Copper Chelator. Cancer Cell. 2010;17:574–583. doi: 10.1016/j.ccr.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang ZD, Long Y, Tsai WB, Fu S, Kurzrock R, Gagea-Iurascu M, Zhang F, Chen HH, Hennessy BT, Mills GB, Savaraj N, Kuo MT. Mechanistic basis for overcoming platinum resistance using copper chelating agents. Mol Cancer Ther. 2012;11:2483–2494. doi: 10.1158/1535-7163.MCT-12-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen HH, Yan JJ, Chen WC, Kuo MT, Lai YH, Lai WW, Liu HS, Su WC. Predictive and prognostic value of human copper transporter 1 (hCtr1) in patients with stage III non-small-cell lung cancer receiving first-line platinum-based doublet chemotherapy. Lung Cancer. 2012;75:228–234. doi: 10.1016/j.lungcan.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YY, Choi CH, Do IG, Song SY, Lee W, Park HS, Song TJ, Kim MK, Kim TJ, Lee JW, Bae DS, Kim BG. Prognostic value of the copper transporters, CTR1 and CTR2, in patients with ovarian carcinoma receiving platinum-based chemotherapy. Gynecol Oncol. 2011;122:361–365. doi: 10.1016/j.ygyno.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida H, Teramae M, Yamauchi M, Fukuda T, Yasui T, Sumi T, Honda K, Ishiko O. Association of copper transporter expression with platinum resistance in epithelial ovarian cancer. Anticancer Res. 2013;33:1409–1414. [PubMed] [Google Scholar]
- 26.Blair BG, Larson CA, Safaei R, Howell SB. Copper transporter 2 regulates the cellular accumulation and cytotoxicity of cisplatin and carboplatin. Clin Cancer Res. 2009;15:4312–4321. doi: 10.1158/1078-0432.CCR-09-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blair B, Larson C, Adams P, Abada P, Safaei R, Howell S. Regulation of copper transporter 2 expression by copper and cisplatin in human ovarian carcinoma cells. Mol Pharmacol. 2010;77:912–921. doi: 10.1124/mol.109.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao L, Cheng Q, Wang Z, Xi Z, Xu D, Liu Y. Cisplatin binds to human copper chaperone Cox17: the mechanistic implication of drug delivery to mitochondria. Chem Commun (Camb) 2014;50:2667–2669. doi: 10.1039/c3cc48847k. [DOI] [PubMed] [Google Scholar]
- 29.Boal AK, Rosenzweig AC. Crystal structures of cisplatin bound to a human copper chaperone. J Am Chem Soc. 2009;131:14196–14197. doi: 10.1021/ja906363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calandrini V, Nguyen TH, Arnesano F, Galliani A, Ippoliti E, Carloni P, Natile G. Structural biology of cisplatin complexes with cellular targets: the adduct with human copper chaperone atox1 in aqueous solution. Chemistry. 2014;20:11719–11725. doi: 10.1002/chem.201402834. [DOI] [PubMed] [Google Scholar]
- 31.Palm-Espling ME, Lundin C, Bjorn E, Naredi P, Wittung-Stafshede P. Interaction between the Anticancer Drug Cisplatin and the Copper Chaperone Atox1 in Human Melanoma Cells. Protein Pept Lett. 2014;21:63–68. doi: 10.2174/09298665113209990036. [DOI] [PubMed] [Google Scholar]
- 32.Safaei R, Maktabi MH, Blair BG, Larson CA, Howell SB. Effects of the loss of Atox1 on the cellular pharmacology of cisplatin. J Inorg Biochem. 2009;103:333–341. doi: 10.1016/j.jinorgbio.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hua H, Gunther V, Georgiev O, Schaffner W. Distorted copper homeostasis with decreased sensitivity to cisplatin upon chaperone Atox1 deletion in Drosophila. Biometals. 2011;24:445–453. doi: 10.1007/s10534-011-9438-1. [DOI] [PubMed] [Google Scholar]
- 34.Katano K, Kondo A, Safaei R, Holzer A, Samimi G, Mishima M, Kuo YM, Rochdi M, Howell SB. Acquisition of resistance to cisplatin is accompanied by changes in the cellular pharmacology of copper. Cancer Res. 2002;62:6559–6565. [PubMed] [Google Scholar]
- 35.Mangala LS, Zuzel V, Schmandt R, Leshane ES, Halder JB, Armaiz-Pena GN, Spannuth WA, Tanaka T, Shahzad MM, Lin YG, Nick AM, Danes CG, Lee JW, Jennings NB, Vivas-Mejia PE, Wolf JK, Coleman RL, Siddik ZH, Lopez-Berestein G, Lutsenko S, Sood AK. Therapeutic Targeting of ATP7B in Ovarian Carcinoma. Clin Cancer Res. 2009;15:3770–3780. doi: 10.1158/1078-0432.CCR-08-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshizawa K, Nozaki S, Kitahara H, Ohara T, et al. Copper efflux transporter (ATP7B) contributes to acquisition of cisplatin-resistance in human oral squamous cell lines. Oncol Rep. 2007;18:987–991. [PubMed] [Google Scholar]
- 37.Komatsu M, Sumizawa T, Mutoh M, Chen Z-S, Terada K, Furukawa T, Yang X-L, Gao H, Miura N, Sugiyama T, Akiyama S. Copper-transporting P-type adenosine triphosphatase (ATP7B) is associated with cisplatin resistance. Cancer Res. 2000;60:1312–1316. [PubMed] [Google Scholar]
- 38.Calandrini V, Arnesano F, Galliani A, Nguyen TH, Ippoliti E, Carloni P, Natile G. Platination of the copper transporter ATP7A involved in anticancer drug resistance. Dalton Trans. 2014;43:12085–12094. doi: 10.1039/c4dt01339e. [DOI] [PubMed] [Google Scholar]
- 39.Ivy KD, Kaplan JH. A re-evaluation of the role of hCTR1, the human high-affinity copper transporter, in platinum-drug entry into human cells. Mol Pharmacol. 2013;83:1237–1246. doi: 10.1124/mol.113.085068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maryon EB, Molloy SA, Zimnicka AM, Kaplan JH. Copper entry into human cells: progress and unanswered questions. Biometals. 2007;20:355–364. doi: 10.1007/s10534-006-9066-3. [DOI] [PubMed] [Google Scholar]
- 41.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo MT. Overcoming cisplatin-resistance by copper-lowering agents. Mol Cancer Ther. 2012;11:1221–1225. doi: 10.1158/1535-7163.MCT-11-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang CP, Fofana M, Chan J, Chang CJ, Howell SB. Copper Transporter 2 Regulates Intracellular Copper and Sensitivity to Cisplatin. Metallomics. 2014;26:654–661. doi: 10.1039/c3mt00331k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narayanan G, R BS, Vuyyuru H, Muthuvel B, Konerirajapuram Natrajan S. CTR1 Silencing Inhibits Angiogenesis by Limiting Copper Entry into Endothelial Cells. PLoS One. 2013;8:e71982. doi: 10.1371/journal.pone.0071982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McRae R, Lai B, Fahrni CJ. Copper redistribution in Atox1-deficient mouse fibroblast cells. J Biol Inorg Chem. 2010;15:99–105. doi: 10.1007/s00775-009-0598-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyayama T, Ishizuka Y, Iijima T, Hiraoka D, Ogra Y. Roles of copper chaperone for superoxide dismutase 1 and metallothionein in copper homeostasis. Metallomics. 2011;3:693–701. doi: 10.1039/c1mt00016k. [DOI] [PubMed] [Google Scholar]
- 47.Chen GF, Sudhahar V, Youn SW, Das A, Cho J, Kamiya T, Urao N, McKinney RD, Surenkhuu B, Hamakubo T, Iwanari H, Li S, Christman JW, Shantikumar S, Angelini GD, Emanueli C, Ushio-Fukai M, Fukai T. Copper Transport Protein Antioxidant-1 Promotes Inflammatory Neovascularization via Chaperone and Transcription Factor Function. Sci Rep. 2015;5:14780. doi: 10.1038/srep14780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain S, Cohen J, Ward MM, Kornhauser N, Chuang E, Cigler T, Moore A, Donovan D, Lam C, Cobham MV, Schneider S, Hurtado Rua SM, Benkert S, Mathijsen Greenwood C, Zelkowitz R, Warren JD, Lane ME, Mittal V, Rafii S, Vahdat LT. Tetrathiomolybdate-associated copper depletion decreases circulating endothelial progenitor cells in women with breast cancer at high risk of relapse. Ann Oncol. 2013;24:1491–1498. doi: 10.1093/annonc/mds654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larson CA, Blair BG, Safaei R, Howell SB. The Role of the Mammalian Copper Transporter 1 in the Cellular Accumulation of Platinum-based Drugs. Mol Pharmacol. 2009;75:324–330. doi: 10.1124/mol.108.052381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song I, Savaraj N, Siddik Z, Liu P, Wei Y, Wu C, Kuo M. Role of copper transporter Ctr1 in the transport of platinum-based antitumor agents in cisplatin-sensitive and resistant cells. Mol Cancer Ther. 2004;3:1543–1549. [PubMed] [Google Scholar]
- 51.Beretta GL, Gatti L, Tinelli S, Corna E, Colangelo D, Zunino F, Perego P. Cellular pharmacology of cisplatin in relation to the expression of human copper transporter CTR1 in different pairs of cisplatin-sensitive and -resistant cells. Biochem Pharmacol. 2004;68:283–291. doi: 10.1016/j.bcp.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 52.Blair BG, Larson CA, Adams PL, Abada PB, Pesce CE, Safaei R, Howell S. Copper Transporter 2 regulates endocytosis and controls tumor growth and sensitivity to cisplatin in vivo. Mol Pharmacol. 2011;79:157–166. doi: 10.1124/mol.110.068411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palm ME, Weise CF, Lundin C, Wingsle G, Nygren Y, Bjorn E, Naredi P, Wolf-Watz M, Wittung-Stafshede P. Cisplatin binds human copper chaperone Atox1 and promotes unfolding in vitro. Proc Natl Acad Sci USA. 2011;108:6951–6956. doi: 10.1073/pnas.1012899108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galliani A, Losacco M, Lasorsa A, Natile G, Arnesano F. Cisplatin handover between copper transporters: the effect of reducing agents. J Biol Inorg Chem. 2014 doi: 10.1007/s00775-014-1138-1. DOI: 10.1007/s00775-014-1138-1. [DOI] [PubMed] [Google Scholar]
- 55.Dolgova NV, Nokhrin S, Yu CH, George GN, Dmitriev OY. Copper chaperone Atox1 interacts with the metal-binding domain of Wilson disease protein in cisplatin detoxification. Biochem J. 2013;454:147–156. doi: 10.1042/BJ20121656. [DOI] [PubMed] [Google Scholar]
- 56.Xi Z, Guo W, Tian C, Wang F, Liu Y. Copper binding promotes the interaction of cisplatin with human copper chaperone Atox1. Chem Commun (Camb) 2013;49:11197–11199. doi: 10.1039/c3cc45905e. [DOI] [PubMed] [Google Scholar]
- 57.Xi Z, Guo W, Tian C, Wang F, Liu Y. Copper binding modulates the platination of human copper chaperone Atox1 by antitumor trans-platinum complexes. Metallomics. 2014;6:491–497. doi: 10.1039/c3mt00338h. [DOI] [PubMed] [Google Scholar]
- 58.Carroll MC, Girouard JB, Ulloa JL, Subramaniam JR, Wong PC, Valentine JS, Culotta VC. Mechanisms for activating Cu- and Zn-containing superoxide dismutase in the absence of the CCS Cu chaperone. Proc Natl Acad Sci USA. 2004;101:5964–5969. doi: 10.1073/pnas.0308298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Djit FJ, Fichtinger-Schepman AMJ, Berends F, Reedijk J. Formation and repair of cisplatin-induced addcts to DNA in cultured normal and repair-deficient human fibroblasts. Cancer Res. 1988;48:6058–6062. [PubMed] [Google Scholar]
- 60.Plooy AC, van Dijk M, Berends F, Lohman PH. Formation and repair of DNA interstrand cross-links in relation to cytotoxicity and unscheduled DNA synthesis induced in control and mutant human cells treated with cis-diamminedichloroplatinum(II) Cancer Res. 1985;45:4178–4184. [PubMed] [Google Scholar]
- 61.Poll EHA, Abrahams PJ, Arwert F, Eriksson AW. Host-cell reactivation of cis-diamminedichloroplatinum (II)- treated SV40 DNA in normal human, Fanconi anaemia and xeroderma pigmentosum fibroblasts. Mutation Res. 1984;132:181–187. doi: 10.1016/0167-8817(84)90036-1. [DOI] [PubMed] [Google Scholar]
- 62.Kulshrestha A, Katara GK, Ginter J, Pamarthy S, Ibrahim SA, Jaiswal MK, Sandulescu C, Periakaruppan R, Dolan J, Gilman-Sachs A, Beaman KD. Selective inhibition of tumor cell associated Vacuolar-ATPase 'a2' isoform overcomes cisplatin resistance in ovarian cancer cells. Mol Oncol. 2016 doi: 10.1016/j.molonc.2016.01.003. DOI: 10.1016/j.molonc.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2015;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]