Abstract
Violacein is a natural purple pigment of Chromobacterium violaceum with potential medical applications as antimicrobial, antiviral, and anticancer drugs. The initial step of violacein biosynthesis is the oxidative conversion of l-tryptophan into the corresponding α-imine catalyzed by the flavoenzyme l-tryptophan oxidase (VioA). A substrate-related (3-(1H-indol-3-yl)-2-methylpropanoic acid) and a product-related (2-(1H-indol-3-ylmethyl)prop-2-enoic acid) competitive VioA inhibitor was synthesized for subsequent kinetic and x-ray crystallographic investigations. Structures of the binary VioA·FADH2 and of the ternary VioA·FADH2·2-(1H-indol-3-ylmethyl)prop-2-enoic acid complex were resolved. VioA forms a “loosely associated” homodimer as indicated by small-angle x-ray scattering experiments. VioA belongs to the glutathione reductase family 2 of FAD-dependent oxidoreductases according to the structurally conserved cofactor binding domain. The substrate-binding domain of VioA is mainly responsible for the specific recognition of l-tryptophan. Other canonical amino acids were efficiently discriminated with a minor conversion of l-phenylalanine. Furthermore, 7-aza-tryptophan, 1-methyl-tryptophan, 5-methyl-tryptophan, and 5-fluoro-tryptophan were efficient substrates of VioA. The ternary product-related VioA structure indicated involvement of protein domain movement during enzyme catalysis. Extensive structure-based mutagenesis in combination with enzyme kinetics (using l-tryptophan and substrate analogs) identified Arg64, Lys269, and Tyr309 as key catalytic residues of VioA. An increased enzyme activity of protein variant H163A in the presence of l-phenylalanine indicated a functional role of His163 in substrate binding. The combined structural and mutational analyses lead to the detailed understanding of VioA substrate recognition. Related strategies for the in vivo synthesis of novel violacein derivatives are discussed.
Keywords: crystal structure, enzyme mechanism, flavoprotein, site-directed mutagenesis, substrate specificity, L-tryptophan oxidase, enzyme-inhibitor complex, redox chemistry, violacein
Introduction
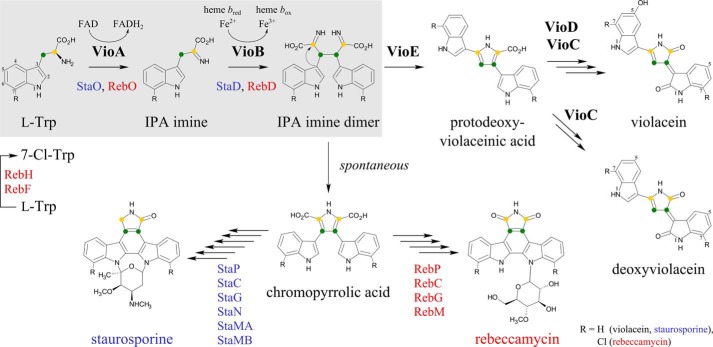
The amino acid tryptophan (Trp) is a versatile metabolite that is shunted into several microbial secondary pathways. The oxidative dimerization of two Trp scaffolds is an essential step for the synthesis of the natural bisindole compounds violacein, rebeccamycin, and staurosporine. The water-insoluble purple pigment violacein is mainly produced by the bacteria Chromobacterium violaceum and Janthinobacterium lividum in tropical habitats where it might be responsible for the shielding against UV radiation (1–3). Various biological activities of violacein, including antibacterial, antiviral, as well as cytotoxic effects against several tumor cell lines, have been demonstrated (4). The related compounds rebeccamycin and staurosporine show strong antitumorigenic activity because of DNA topoisomerase or protein kinase inhibition (5, 6).
Synthesis of violacein, rebeccamycin, or staurosporine is based on a closely related initial pathway in which orthologous enzymes catalyze the oxidation of a Trp moiety into the related indole-3-pyruvic acid (IPA)2 imine by the enzymes VioA, RebO, or StaO (Fig. 1) (7–9). Subsequently, oxidative coupling of two imines by VioB, RebD, or StaD results in the formation of a short-lived compound that was proposed to be an IPA imine dimer (7, 10). For the synthesis of rebeccamycin and staurosporine, this reactive intermediate is spontaneously converted into chromopyrrolic acid (11–13). By contrast, violacein biosynthesis requires a key intramolecular rearrangement. The postulated IPA imine dimer is the substrate of VioE, which is catalyzing a [1,2]-shift of the indole ring to produce protodeoxyviolaceinic acid (7, 14). Fig. 1 gives a schematic overview about the related pathways as follows: common enzymatic reactions and the involved cofactors are highlighted (gray shading), subsequent steps for the synthesis of violacein (VioE, VioD, VioC, and one autocatalytic step) (3, 7), rebeccamycin (RebP, RebC, RebG, and RebM) (15, 16), and staurosporine (StaP, StaC, StaG, StaN, StaMA, and StaMB) (16) are indicated.
FIGURE 1.
Biosynthetic pathway of violacein (black), staurosporine (blue), and rebeccamycin (red). Orthologous enzymes VioA, StaO, or RebO and VioB, StaD, or RebD are involved in the initial pathway for the synthesis of violacein, staurosporine, and rebeccamycin, respectively (highlighted in gray). A Trp moiety (l-Trp) is converted into indole-3-pyruvic acid imine (IPA imine) and subsequently dimerized into a short-lived compound proposed as IPA imine dimer. Synthesis of rebeccamycin initially requires the chlorination of l-Trp to 7-Cl-Trp by virtue of RebH and RebF. Violacein biosynthesis then includes a VioE-dependent [1,2]-shift of the indole ring (curved arrow) resulting in protodeoxyviolaceinic acid. Subsequent VioD and VioC catalysis followed by a final autocatalytic oxidation step leads to the product violacein. A deoxyviolacein derivative is synthesized without involvement of VioD. For staurosporine and rebeccamycin biosynthesis, the IPA imine dimer is spontaneously converted into chromopyrrolic acid. Subsequent enzymes StaP, StaC, StaG, StaN, StaMA, StaMB or RebP, RebC, RebG, RebM then facilitate the synthesis of staurosporine or rebeccamycin.
Synthesis of violacein starts with the FAD-dependent oxidation of l-Trp to IPA imine catalyzed by VioA. The VioA protein from C. violaceum shares a substantial degree of sequence conservation with RebO or StaO proteins (ranging from 18 to 22% identity; Clustal Omega (17)). Furthermore, sequence identity values of 14–22% were observed for the comparison of VioA with l-amino acid oxidases (LAAOs) (3, 18). LAAO-catalyzed two-electron oxidations are well studied from prokaryotic and eukaryotic enzyme sources (19, 20). However, the synthesized imines are subsequently deaminated by virtue of an attacking water molecule into the respective α-keto acids (21, 22).
By contrast, violacein biosynthesis relies on the reactive IPA imine as a direct substrate of VioB. Furthermore, the postulated IPA imine dimer reaction product is also labile, which might reflect the need for an activated substrate for the unusual [1,2]-shift of the indole ring during VioE catalysis. However, present date in vitro investigations revealed that the direct interaction of VioA and VioB (or of VioB and VioE) is not an absolute prerequisite for protodeoxyviolaceinic acid synthesis (7).
In a recent publication, 50% FAD occupancy was determined for recombinantly purified VioA protein. Kinetic characterization of this protein was performed in a tandem peroxidase assay with an optimal pH of 9.25. Formation of the unstable IPA imine goes along with a reduced flavin on VioA, which is subsequently reoxidized by molecular oxygen leading to stoichiometric peroxide formation. The detection of hydrogen peroxide revealed kcat and Km(Trp) values of 203 min−1 and 31 μm for VioA (7).
The heme-containing oxidase VioB catalyzes the coupling of two IPA imine molecules to form the postulated intermediate IPA imine dimer (7). For this molecule, only indirect experimental evidence is available due to the short-lived nature of this compound (often referred to as compound X in the literature) (7, 18). Catalase activity has been demonstrated for VioB, and it was proposed that the heme iron center is responsible for VioB catalysis (7).
In the absence of VioE, the off-pathway metabolite chromopyrrolic acid is observed. This non-enzymatic chromopyrrolic acid formation is an essential step during staurosporine or rebeccamycin biosynthesis (Fig. 1) (3, 11, 13). In the presence of VioE, the reactive IPA imine dimer instead is converted to protodeoxyviolaceinic acid. Structural and biochemical investigations for VioE revealed that the sophisticated [1,2]-indole rearrangement is catalyzed in the absence of any cofactor or metal ion (67).
Violacein biosynthesis is completed by sequential action of two flavin-dependent oxygenases, VioD and VioC. VioD hydroxylates one indole ring at the 5-position to yield protoviolaceinic acid. Subsequently, VioC hydroxylates the second indole ring at the 2-position and spontaneous oxidative decarboxylation then leads to the formation of violacein (Fig. 1) (7).
Under physiological conditions, the violacein biosynthetic pathway also results in the formation of deoxyviolacein (23). Deoxyviolacein, lacking the hydroxyl group at position 5, is synthesized without involvement of VioD, which might indicate a certain degree of flexibility for VioC substrate recognition (Fig. 1).
In this study, recombinantly produced VioA from C. violaceum is analyzed in a combined biochemical and x-ray crystallographic approach. Structure-based site-directed mutagenesis along with kinetic experiments in the presence of artificial substrates or active site inhibitors reveal the molecular mechanism of VioA.
Results
Production and Purification of VioA
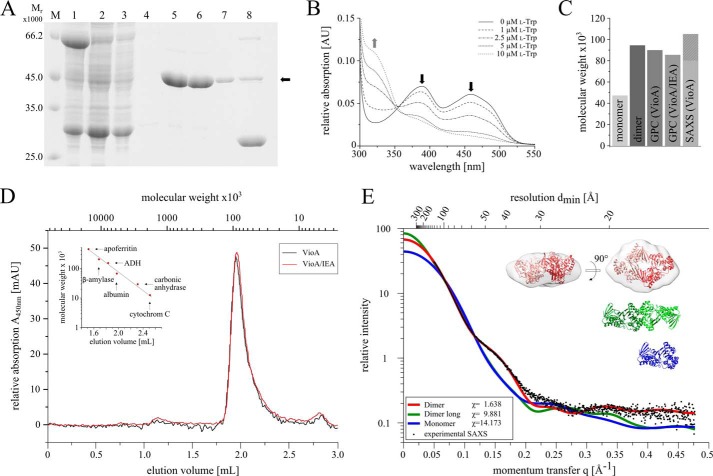
The l-Trp oxidase VioA from C. violaceum was efficiently overproduced in Escherichia coli as a soluble GST-VioA fusion protein (Fig. 2A, lane 1). The tagged protein was immobilized on a glutathione-agarose column. Subsequently, the target VioA protein was liberated by on-column cleavage with PreScissionTM protease (Fig. 2A, lanes 2–7). Overall, 40 mg of the homogeneous target protein was obtained from 1 liter of cell culture. SDS-PAGE analyses indicated a relative molecular weight of 45,000 (Fig. 2A, lanes 5–7, calculated molecular mass, 47,159 Da) and the integrity of purified VioA was confirmed by N-terminal sequencing.
FIGURE 2.
Purification and characterization of VioA analyzed by SDS-PAGE, UV-visible spectroscopy, analytical gel permeation chromatography and SAXS. A, SDS-PAGE analysis of GST-tagged VioA affinity purification using Protino® glutathione-agarose 4B. Lane M, molecular mass marker; relative molecular masses (×1000) are indicated. Lane 1, supernatant after ultracentrifugation; lane 2, flow-through of the GST affinity chromatography; lanes 3 and 4, washing steps; lanes 5–7, elution fractions of untagged VioA with a calculated molecular mass of 47,159 Da (black arrow) after on-column cleavage with PreScission protease; lane 8, GST tag elution with buffer containing reduced glutathione. B, UV-visible absorption spectra of the prosthetic FAD/FADH2 group of VioA. Purified VioA (15 μm) was incubated with different concentrations of l-Trp (0–10 μm) under anaerobic conditions. Increasing l-Trp concentrations resulted in a sequential decrease of the characteristic absorption maxima of FAD at 387 and 457 nm (black arrows) and an increased absorption at 328 nm (gray arrow) due to the formation of FADH2. C, bar chart comparing the calculated molecular weight of a VioA monomer or dimer with the experimentally derived values obtained from analytical gel permeation chromatography (GPC) experiments (also in the presence of the inhibitor IEA) and small angle x-ray scattering experiments (SAXS). D, analytical gel permeation chromatography of purified VioA (75 μm) using a SuperdexTM 200 Increase 5/150 GL column (GE Healthcare) on an Äkta purifier system. Protein standards were apoferritin (Mr = 443,000), β-amylase (Mr = 200,000), alcohol dehydrogenase (Mr = 150,000), albumin (Mr = 66,000), carbonic anhydrase (Mr = 29,000), and cytochrome c (Mr = 12,000). Column calibration (inset) and protein sample analysis were performed at a flow rate of 0.45 ml min−1. Elution volumes were recorded at 280 nm (for protein standards) or at 450 nm (for the VioA·FAD complex). Samples of VioA were measured in the absence (black line) or presence (red line) of 500 μm IEA. E, SAXS analysis of the ternary VioA structure. Experimental SAXS data obtained from a soluble VioA sample (black dots) were related to the simulated SAXS curves of a potential monomer (blue), a more globular dimer (red), or an elongated dimer (green). Theoretical SAXS curves were calculated on the basis of the binary VioA crystal structure. The χ values reflect the fit quality with respect to the experimental data (maximum fit quality observed for the globular dimer). Superposition of the three-dimensional low resolution electron density (gray, calculated from the SAXS scattering curve) with the globular dimer (red, calculated from the binary VioA x-ray structure) indicates a high degree of structural complementarity.
UV-visible absorption spectroscopy of a purified VioA sample revealed characteristic absorption maxima at 387 and 457 nm (Fig. 2B, solid line), as is characteristic for FAD-containing flavoproteins (24). Spectroscopically, an overall FAD content of 55% was determined using the molar extinction coefficient of the oxidized flavin, in agreement with the outcome of a recent publication (7). In addition, the FAD content of purified VioA was also estimated on the basis of single-turnover VioA activity experiments. Strict anoxic conditions were required to prevent the reoxidation of the resulting FADH2 cofactor due to electron transfer onto molecular oxygen. Purified samples of VioA at a concentration of 15 μm were incubated in the presence of varying l-Trp concentrations ranging from 0 to 10 μm. The recorded absorption spectra revealed a clear decrease of the characteristic FAD signal (Fig. 2B, compare solid line versus dotted lines). When the remaining FAD concentrations were plotted against the applied l-Trp concentration, an FAD content of ∼50% was estimated for the recombinantly purified VioA.
Tight binding of the FAD cofactor was demonstrated in an aerobic dialysis experiment. Under the employed turnover conditions, the FAD cofactor is constantly regenerated due to the presence of O2. A VioA sample was incubated in the presence of a large volume of buffer containing 40 mm l-Trp. The resulting VioA dialysate after a subsequent buffer exchange step (to remove VioA educt and product) did not reveal a decreased FAD content (data not shown).
Inhibition of VioA and Synthesis of a Substrate- and Product-related Inhibitor
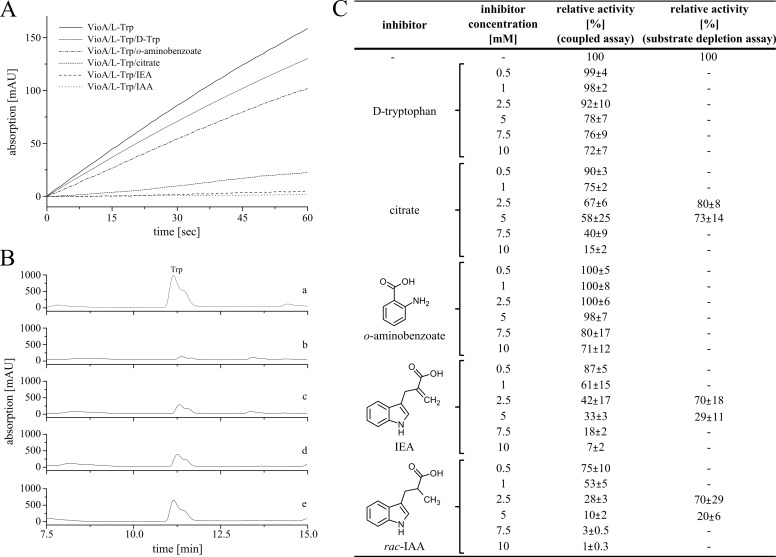
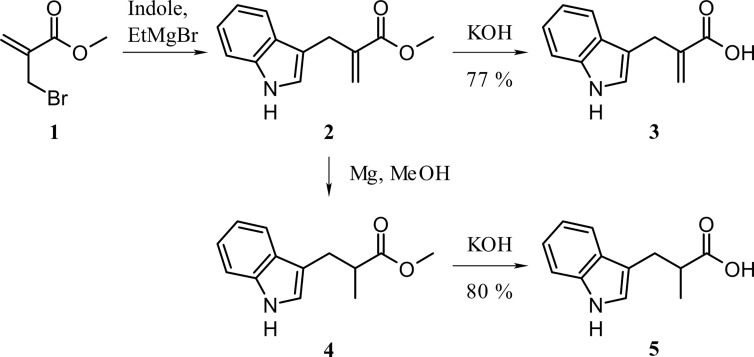
The identification of efficient active site inhibitors is a well established strategy for the detailed understanding of enzyme mechanisms. Inhibitor concentrations of up to 10 mm were analyzed in the coupled VioA activity assay (9) or alternatively in VioA substrate depletion kinetics (compare Fig. 3, A–C). Experiments in the presence of d-Trp revealed a moderate VioA inhibition (72% relative activity, 10 mm d-Trp). In the present literature, citrate and o-aminobenzoate have been described as efficient inhibitors of LAAOs (21, 25, 26). In agreement with these distantly related investigations, a relative activity of 15% in the presence of 10 mm citrate was observed. By contrast, in the presence of 10 mm o-aminobenzoate only a moderate inactivation indicated by a residual activity of 71% was determined (Fig. 3C). Because citrate does not closely resemble the l-Trp molecule, a substrate-related and a product-related l-Trp derivative was newly synthesized for subsequent inhibition experiments. In compound 3-(1H-indol-3-yl)-2-methylpropanoic acid (IAA), the amino group of l-Trp is replaced by a methyl group, whereas the non-chiral compound 2-(1H-indol-3-ylmethyl)prop-2-enoic acid (IEA) is carrying a methylene group, instead. Accordingly, the IEA molecule is sharing a closely related spatial orientation (due to the sp2-hybridized carbon) as the IPA imine reaction product of VioA (Fig. 3C). IEA and racemic IAA were synthesized from indole and methyl 2-(bromomethyl)acrylate (1) following a procedure from Holzapfel et al. (27). Methyl 2-(bromomethyl)acrylate was obtained in two steps from methyl acrylate and paraformaldehyde, followed by bromination with PBr3 (28, 29). Reduction of the methylene group was performed using magnesium in MeOH, and saponification of the corresponding esters 2 and 4 led to the desired products in good yields (Fig. 4) (30, 31). Kinetic experiments revealed a residual specific VioA activity of 61 and 53% in the presence of 1 mm IEA and IAA. At inhibitor concentrations of 10 mm, residual activities of 7 and 1% were determined. Results for the efficient inhibitors citrate, IEA, and IAA were independently confirmed in substrate depletion activity assays (Fig. 3C). The results of these inhibition experiments were further substantiated in a direct biomolecular interaction study.
FIGURE 3.
Identification of VioA inhibitors. d-Trp, citrate, o-aminobenzoate, IEA, and rac-IAA were analyzed as potential inhibitors in the coupled VioA horseradish peroxidase assay containing 0.2 μm VioA in the presence of 1 mm l-Trp (A). Alternatively, potential inhibitors were analyzed in an HPLC-based substrate depletion assay using 1 μm VioA and 500 μm l-Trp (B). A, representative inactivation kinetics monitoring the formation of the quinonimine dye at a wavelength of 505 nm. A VioA activity assay (solid line) and competition experiments in the presence of 10 mm d-Trp, o-aminobenzoate, citrate, IEA, and IAA, respectively, are displayed (gray line and consecutive dotted lines). B, representative substrate depletion assays analyzed on a Jasco HPLC system using μBondapakTM C18 3.9 × 150 mm column. Reactions were incubated for 3 min at 30 °C. A control experiment in the absence of VioA (a), a standard VioA activity experiment (b), and inhibition experiments in the presence of 5 mm citrate (c), 5 mm IEA (d), or 5 mm rac-IAA (e) are displayed. C, relative activities of VioA inhibition experiments (coupled assay and depletions assay) using different inhibitor concentrations. The activity of the standard VioA assay in the presence of 1 mm l-Trp was set as 100%. Dashes indicate experiments not performed. Results are presented as means ± S.D. of three independent biological samples, measured as triplicates.
FIGURE 4.
Synthesis of potential VioA inhibitors IEA and rac-IAA.
Microscale Thermophoresis
Thermophoresis is a phenomenon that describes the movement of molecules along a temperature gradient (also termed Ludwig-Soret effect) (32). In this study, we made use of microscale thermophoresis to analyze the direct binding of citrate, IEA, and IAA to VioA (labeled with the fluorescence dye NT-647-NHS). Fluorescence was measured at a spot in the capillary that was heated by an IR laser beam. Inhibitor binding was observed as a change in the thermophoretic properties of the fluorescently labeled protein due to complex formation with VioA at various ligand concentrations. The following dissociation constants were determined in agreement with the results of the above mentioned inhibition study: IAA 32.2 ± 10.3 μm ≤ IEA 42.6 ± 13.6 μm ≤ citrate 45.3 ± 7.8 μm. According to these results, the newly synthesized compounds IEA and IAA were considered as promising candidates for the co-crystallization of VioA.
Determination of the Binary and Ternary VioA Structure
The sitting-drop vapor diffusion technique was used to crystallize VioA, and the phase problem was resolved using single-wavelength anomalous diffraction data of a samarium nitrate-derivatized crystal. Electron density in the active center of the native VioA structure indicated the presence of the flavin cofactor (occupancy 100%). Crystals of VioA were also grown in the presence of IEA. The molecular replacement technique was used to solve the structure of the ternary VioA complex in the presence of the flavin and IEA. Table 1 summarizes the data collection and refinement statistics. The finalized models with a resolution of 1.8 and 2.4 Å (native structure and ternary complex) were deposited in the Protein Data Bank with accession numbers 5G3T and 5G3U.
TABLE 1.
Data collection and refinement statistics
The values in parentheses account for the shells of highest resolution. AU, asymmetric unit.
| Dataset | Samarium nitrate derivative | Native VioA | VioA·IEA |
|---|---|---|---|
| Synchrotron source, beamline | SLS, X06DA | Desy, Petra III P1 1 | BESSY, BL 14.1 |
| Wavelength (Å) | 1.770 | 1.033 | 0.918 |
| Temperature (K) | 100 | 100 | 100 |
| dmax − dmin (Å) | 19.92–2.08 (2.15–2.08) | 19.98–1.80 (1.86–1.80) | 45.98–2.38 (2.42–2.38) |
| Space group | P21 | P21 | P212121 |
| Unit cell parameters | |||
| a, b, c (Å) | 67.88, 87.07, 78.02 | 67.09, 89.167, 144.43 | 69.27, 81.46, 167.12 |
| α, β, γ (°) | 90.00, 112.95, 90.00 | 90.00, 92.66, 90.00 | 90.00, 90.00, 90.00 |
| Unique reflections | 49,742 (4,528) | 157,109 (15,444) | 38,865 (3,822) |
| Completeness | 0.98 (0.91) | 0.98 (0.97) | 1.00 (1.00) |
| Multiplicity | 24.4 (19.6) | 6.9 (6.9) | 6.6 (6.9) |
| Mean I/σ(I) | 26.3 6.6) | 10.9 (2.0) | 11.0 (2.0) |
| Rmerge | 0.1097 (0.5100) | 0.1034 (0.9474) | 0.1616 (0.7014) |
| CC1/2 | 0.999 (0.961) | 0.998 (0.817) | 0.998 (0.932) |
| Rwork | 0.1721 (0.2010) | 0.1567 (0.2271) | 0.1769 (0.2243) |
| Rfree | 0.1903 (0.2389) | 0.1926 (0.2694) | 0.2133 (0.2549) |
| No. of protein molecules per AU | 2 | 4 | 2 |
| No. of amino acid residues ordered/total | 819/850 | 1,652/1,700 | 824/850 |
| No. of non-hydrogen atoms per AU | |||
| Total | 6,908 | 14,478 | 6,837 |
| Protein | 6,357 | 12,858 | 6,398 |
| Ligands | 168 | 259 | 142 |
| Average B (Å2) | 28.8 | 26.5 | 31.2 |
| Root mean square deviation from ideal | |||
| Bonds (Å) | 0.002 | 0.008 | 0.004 |
| Angles (°) | 0.59 | 0.94 | 0.94 |
| Ramachandran plot | |||
| Favored (%) | 97.3 | 98.1 | 97.4 |
| Outliers (%) | 2.0 | 0.0 | 0.0 |
| PDB code | 5G3S | 5G3T | 5G3U |
Identification of the Physiological VioA Dimer
Analytical size exclusion chromatography revealed a dimeric structure of VioA as indicated by a relative molecular mass of 94.000 ± 7.000 (Fig. 2D). Identical chromatographic results were also obtained in the presence of 500 μm IEA (Fig. 2D). Examination of potential VioA contacts in the ternary crystal structure (and in the binary structure) of VioA using the PISA server (Protein Interfaces, Surfaces and Assemblies) (33) revealed a globular dimer or alternatively an elongated dimer burying a surface area of 721.7 or 667.3 Å2 (compare Fig. 2E, inset: top, globular dimer; middle, elongated dimer; bottom, monomer). Identical dimers were also observed for VioA·FADH2.
Subsequently, small angle x-ray scattering (SAXS) experiments were performed to characterize the dimer of VioA in solution. This technique makes use of a dilute protein solution and allows for the reconstruction of a low resolution electron density map. Almost identical scattering curves for VioA and for VioA in the presence of 3.75 mm IEA were obtained. In Fig. 2E, the comparison of the experimental VioA scattering curve (black dots) with the theoretical scattering curves for the globular dimer (red), the elongated dimer (green), and for the VioA monomer (blue) is shown. A significantly higher quality of fit was obtained for the globular dimer (χ = 1.638) when compared with the elongated dimer (χ = 9.881) or the monomer (χ = 14.173). From SAXS measurements, an apparent molecular mass of 80,000–105,000 was determined, in agreement with the related gel filtration experiments (Fig. 2C). The ab initio model derived from the SAXS experiments described the shape of the globular dimer well (Fig. 2E, inset). These findings allowed us to clearly assign the globular dimer with a rather small interface as the physiological unit of VioA.
Overall Structure of VioA
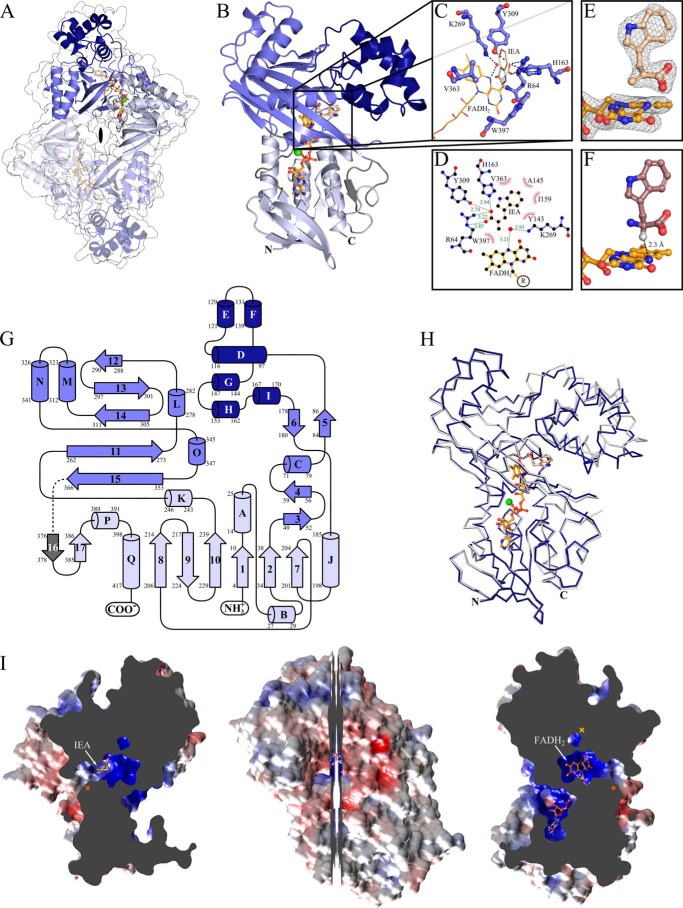
Each VioA monomer is composed of 17 α-helices and 17 β-strands and folds into three distinct domains. The FAD-binding domain is composed of residues 4–38, 185–256, and 380–418 and forms a classical nucleotide-binding fold (with the P-loop sequence motif 11GXGXXG(X)21D38 (34)) of the glutathione reductase family 2. The substrate-binding domain includes residues 49–86, 177–180, and 262–366, and the helical domain contains residues 87–175 (Fig. 5, A, B, and G).
FIGURE 5.
Crystal structure of VioA from C. violaceum. A, schematic representation of the physiological dimer of the VioA·FADH2·IEA complex viewed along the 2-fold symmetry dimer axis (black lens). Individual VioA domains are colored light blue (FAD-binding domain), blue (substrate-binding domain), and dark blue (helical domain); the respective domains of the second protomer are rendered transparent. The co-crystallized FADH2 (orange) and the inhibitor IEA (wheat) are shown as ball-and-stick model and the Mg2+ ion is displayed as a green sphere. The transparent van der Waals surface is depicted in white. B, schematic representation of an isolated VioA protomer from A with indicated N and C termini. C, close-up of the active site of VioA. The IEA inhibitor (wheat), FADH2 (orange), and key VioA residues (blue ball-and-sticks) and the respective hydrogen bonding and salt bridge interactions are shown (dashed lines); water molecule is displayed as red sphere. D, LIGPLOT (66) representation of the active site showing non-polar and polar contacts (with indicated distances in green (Å)). E, conformation of the co-crystallized IEA and the flavo moiety of FADH2. Portion of the 2Fo − Fc electron density map is shown as gray mesh at a contour level of 1.5 σ. F, the substrate l-Trp (violet) was modeled into the VioA active site. The distance between the hydrogen atom located at the sp3-hybridized Cα atom (light gray) and atom N5 of FAD (2.3 Å) is indicated (dashed line). G, topology diagram of VioA. Helices are depicted as cylinders and β-strands as arrows (color code as in A). Undefined regions are colored in gray and unresolved residues 369–371 are indicated by the dotted line. H, superposition of the binary (light gray) and ternary (dark blue) VioA structure (same orientation as in B). The ribbon representation is showing significant movement of the helical domain toward the FAD-binding domain. I (center), a longitudinal section of the VioA monomer represented as van der Waals surface colored according to its surface electrostatics (−10 (red) to +10 (blue) KbT/ec). The left half is highlighting the substrate-binding pocket with the bound IEA molecule and the proposed substrate entry path (indicated by an orange asterisk). The right half additionally shows the FAD/FADH2-binding pocket and the proposed channel for the second substrate O2 (indicated by a yellow ×) required for the oxidative half-reaction of VioA. The orientation of this right half is roughly as in B and H).
The VioA dimer is associated via a discontinuous protein interface that involves amino acid residues of the FAD-binding and of the substrate-binding domain. The dimer assembly is based on two symmetry-related interfaces mainly mediated by residues Arg319, Ala323, Glu324, with His4, Arg35, Tyr206, Asp226, respectively. Among these, conserved residues Arg319, Tyr206, and Asp226 are engaged in a polar network, including a bidentate salt bridge interaction of Arg319 and Asp226. This “loosely associated” dimer forms a central cavity (along the 2-fold symmetry of the dimer) with a volume of ∼13 × 10 × 8 Å3 (Fig. 5A). From this cavity, the adenosyl moiety of the tightly bound FAD cofactor is accessible.
A Dali search of the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (35) indicated the strongest structural similarity to the LAAO from Calloselasma rhodostoma (Protein Data Bank code 1F8S). Both proteins share a sequence identity of only 14%. However, structural superposition revealed a root mean square deviation of solely 2.7 Å (calculated via Dali server; 483 residues included). Most relevant structural differences are localized in the helical domain of VioA and include the deletion of two strands and one helical segment (following strand 5 of VioA) as well as the insertion of the short helix I (residues 167–170). Beside this, the substrate-binding domain of VioA is linked to the FAD-binding domain via loop region 367–375 (containing unresolved residues 369–371) and strand 16 (residues 376–378) instead of a helical segment found in LAAO. In Fig. 5G, the topology diagram (with labeled residues of each secondary structural element) is highlighting the FAD-binding domain (light blue), substrate-binding domain (blue), and the helical domain (dark blue).
FAD Binding
In the binary and ternary VioA structure, the prosthetic group FAD is bound and deeply buried in an identical extended conformation as observed for several LAAO enzymes (Fig. 5, A, B, H, and I) (21). The FAD molecule forms an array of polar contacts, including main chain atoms of amino acids Ser15, Met39, Arg46, Gly63, Arg64, Leu208, and Met398, side chain atoms of residues Ser15, Asp38, Arg46, and Asp389, and also a series of hydrogen bonding interactions via spatially conserved water molecules (data not shown). A partially related polar network is observed in the structure of LAAO from C. rhodostoma. However, these interactions rely on the conservation of only three residues (Asp38, Arg46, and Arg64 VioA numbering) but otherwise make use of a closely related array of polar contacts involving several structurally conserved water molecules that are positioned via equivalent main chain atoms of non-conserved amino acids. As described for most flavoproteins, an Mg2+ ion is involved in ionic bonds to the diphosphate group of FADH2. The isoalloxazine ring of FAD points toward the substrate-binding domain. The dimethylbenzene ring is positioned in a partially hydrophobic pocket surrounded by highly conserved VioA residues Trp359, Val363, and Gly61. In both VioA complex structures, atom N5 of the isoalloxazine ring is in direct contact with a structurally conserved water molecule that is bound to Lys269 (Fig. 5, C and F). For the structure of LAAO from C. rhodostoma, the equivalent water molecule was discussed as a potential nucleophile for subsequent imine deamination (21). However, synthesis of violacein is strictly based on the formation of the IPA imine as the final product of VioA catalysis. Accordingly, a nucleophile function of the water molecule must be excluded for VioA. Instead, this water might be essential for the appropriate positioning of the isoalloxazine ring of FAD.
Active Site of VioA
The structure of VioA in the presence of the flavin cofactor and IEA reveals a detailed picture of VioA catalysis. In the final electron density map, the isoalloxazine ring adopts a bent conformation (Fig. 5E) as observed for many reduced flavoproteins, including the complex structure of LAAO from C. rhodostoma (36). For crystallization purposes, the VioA·cofactor complex was always purified in the presence of the reducing agent DTT, which results in a reduced VioA·FADH2 sample. Crystal growth under oxic conditions then resulted in faintly yellow crystals indicative of the minor oxidation of the FAD complex. However, subsequent reduction of the prosthetic FAD group due to radiation damage during synchrotron data collection has been well described in the literature (37). Accordingly, the observed crystal structure was referred to as VioA·FADH2·IEA. For this structure, no differences of the overall flavin geometry were observed when compared with the binary VioA·FADH2 complex.
In the ternary structure, atom N5 of the prosthetic group resides out of the plane of the isoalloxazine ring and points toward the IEA ethene group. This inhibitor was synthesized as a potential transition state analog that closely resembles the IPA imine reaction product. Accordingly, the VioA·FADH2·IEA structure might represent a snapshot in the course of VioA catalysis. The electron density of the IEA ligand clearly revealed the sp2-hybridized ethene group atoms of IEA. By contrast, the natural l-Trp substrate contains an sp3-Cα atom (compare Fig. 5E and the structure with a modeled l-Trp ligand depicted in Fig. 5F).
The substrate-binding site of VioA is located in a deeply buried cavity (Fig. 5, A, B, H, and I). IEA is accessible via a substrate entry channel that is delineated by residues Met155, Asp158, Ile159, Phe365, Cys395, and Trp397. The isoalloxazine ring of the cofactor is located at the re face of the inhibitor, and the imidazole moiety of IEA is extending away from the cofactor with the N1 atom pointing toward the entry path. The inhibitor's carboxylate forms a salt bridge with the guanidinium group of Arg64 and is further engaged in hydrogen bonds with the hydroxyl of Tyr309 and with the imidazole ring of His163 (Fig. 5, C and D). A closely related carboxylate binding mode is also observed in the structure of LAAO (33). The C2 atom of IEA (representing the site of oxidative attack in the VioA substrate) is positioned at a distance of ∼3.4 Å from the N5 atom of the isoalloxazine ring. Such distance often is observed for flavoenzymes performing a direct hydride transfer mechanism. The methylene group of IEA (as part of the imine mimic) points toward the planar indole ring of Trp397. The indole moiety of IEA resides in a hydrophobic pocket that is mainly formed by side chains of residues Tyr143, Ala145, Ile159, and Val363.
Possible Channel for the Oxidative Half-reaction of VioA
VioA catalysis is based on the FAD-dependent formation of the IPA imine (reductive half-reaction) in a process that is coupled to the O2-dependent formation of H2O2 (oxidative half-reaction). Because of the tightly bound prosthetic flavin group, the second substrate O2 must gain access to the active site of VioA. In the ternary VioA structure, the catalytic cavity is completely locked due to the binding of IEA. Accordingly, the deeply buried flavin appears to be inaccessible to O2 via the substrate entrance channel. Inspection of the VioA protein surface (for both crystal structures) indicated a second water-filled channel (constriction radius ∼2.0 Å) (Fig. 5I, right) giving direct access to the structurally conserved water molecule (3.1 Å) involved in a bridging interaction between atom N5 of the isoalloxazine and Lys269. This channel is mainly delineated by conserved residues Lys269 and Tyr309 and by the partially conserved residues Glu59, Tyr87, and Phe307. The observed channel might suggest a direct path for O2 to the catalytic site. Indeed, the position of the conserved water molecule would be also appropriate for the binding of O2 and the subsequent formation of H2O2. Analogously, a related LAAO structural analysis revealed a (spatially unrelated) substrate channel for the oxidative half-reaction (36).
Domain Movement of VioA
Structural comparison of the overall VioA·FADH2·IEA and the VioA·FADH2 complex reveals a significant domain movement (compare gray and blue Cα representation in Fig. 5H). The structural alterations observed in the presence of IEA can be roughly described as a movement of the helical domain toward the FAD-binding domain, resulting in a partial closure of the substrate entry path as indicated by a 3.5-Å closer positioning of the Cα atoms of Met155 and Phe365. These overall movements result in slight alterations of the side chain conformation of active site residues Val363 and Trp397. Most significant changes are observed for residues His163 and Arg64, which are located in a 0.4-Å closer position in the VioA·FADH2·IEA structure. The observed conformational alterations might be relevant for catalysis and/or for the appropriate timing of the IPA imine product release. The presented structures of VioA have been further explored in biochemical experiments to provide a solid basis for the understanding of the catalytic mechanism and the substrate recognition of VioA.
Substrate Specificity of VioA
A series of recently described activity assays (9) with the 20 canonical l-amino acids revealed solely the conversion of l-Trp. This is in agreement with our experiments in the presence of l-tyrosine, l-valine, l-proline, l-histidine, l-leucine, and l-alanine (detection limit <9.9 pmol min−1 mg−1, data not shown). Such efficient discrimination might be essential under physiological conditions, because VioA catalysis results in highly toxic peroxide formation.
Our experiments using l-phenylalanine revealed a relative activity of 1.8% when compared with the natural substrate. To some extent, the benzyl group of l-phenylalanine might adopt a related spatial conformation in the active site of VioA under the employed in vitro assay conditions. Efficient discrimination of d-Trp was observed in agreement with a stereospecific hydride abstraction mechanism.
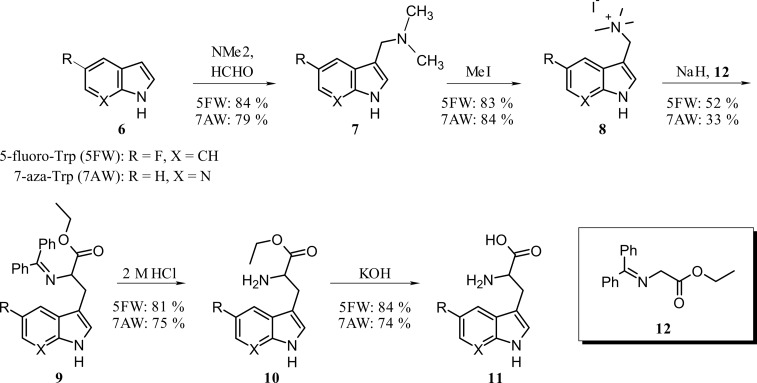
To further elucidate the substrate specificity of VioA, a series of four artificial substrates (modified at C1, C5, and C7; compare Fig. 7A) were analyzed in the coupled enzyme assay. For this purpose, 5-fluoro-Trp and 7-aza-Trp were synthesized starting from commercially available 5-fluoroindole and 7-azaindole, respectively. First, a Mannich reaction to the corresponding gramines was performed, followed by addition of methyl iodide (38). The resulting tetramethylammonium iodide salts were obtained with very good yields. Introduction of the amino acid side chain was realized using N,N-diphenylethylglycinate (39). Deprotection of the amino group and saponification led to the desired product in good yields (Fig. 6).
FIGURE 7.
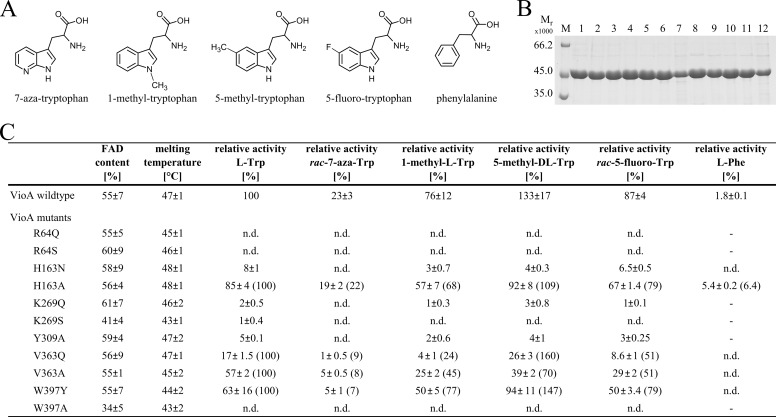
Characterization of VioA mutant proteins. A, Trp analogs as artificial substrates for VioA. B, SDS-PAGE analysis of purified recombinant VioA wild type and mutant proteins. Lane M, molecular mass marker, relative molecular masses (×1000) are indicated. Lane 1, VioA wild type, and lanes 2–12 showing VioA mutants R64Q, R64S, H163N, H163A, K269Q, K269S, Y309A, V363Q, V363A, W397Y, and W397A. C, relative activities of VioA mutant proteins in the presence of Trp analogs shown in A. The activity of the wild type protein in the presence of l-Trp was set as 100%. The FAD content and the melting temperature (obtained from thermal shift experiments) for all proteins are indicated. For mutants H163A, V363Q, V363A, and W397Y, the activity in the presence of l-Trp was also set as 100%, and the activities from substrate analog kinetics were related to that (percent values in parentheses). Activities below the detection limit of the employed assay are indicated as n.d. (not detectable). Dashes indicate experiments not performed. Results are presented as means ± S.D. of three independent biological samples, measured as triplicates.
FIGURE 6.
Synthesis of tryptophan analogs 5-fluoro-Trp and 7-aza-Trp.
For compound 7-aza-Trp, a relative VioA activity of 23% was determined when compared with the natural substrate (Fig. 7, A and C). Interference of the 7-aza modification of the indolyl ring with the side chain of Ile159 might be responsible for the decreased activity. A significantly higher VioA activity of 76% was observed for 1-methyl-Trp in agreement with the ternary VioA structure (Fig. 7, A and C). This modification does not result in any steric clashes because a substituent at N1 extends into the active site entry channel of VioA (Fig. 5I, left). The most efficient VioA substrates were compounds 5-methyl-Trp and 5-fluoro-Trp showing activities of 133 and 87% (Fig. 7, A and C). Obviously, a methyl or a fluoro substituent (providing inverse inductive effects) is well tolerated in the active site pocket. A certain degree of flexibility for the side chain of residue Asp311 might be responsible for the broadened substrate specificity of VioA. These results are further supported by a recent investigation showing a relative activity of 86% in the presence of the artificial substrate 5-hydroxy-Trp (7). Basically, substrate discrimination only occurs in the presence of natural amino acids, whereas all chemically synthesized l-Trp derivatives were efficiently converted by VioA.
Mutagenesis of Catalytic Residues of VioA
In a structure-based mutagenesis approach, the impact of conservative (or alternatively of a more drastic) mutations was analyzed with respect to amino acids Arg64, His163, Lys269, Tyr309, Val363, and Trp397. The integrity of all site-directed mutant proteins was confirmed with thermal shift experiments, and the amount of co-purified FAD cofactor was determined (Fig. 7, B and C).
The bidentate salt bridge of the substrate's carboxylate with the guanidinium group of Arg64 (Fig. 5, C and D) is of prime importance because mutant proteins R64Q and R64S did not reveal any detectable VioA activity (Fig. 7C). The spatial positioning of this carboxylate by additional hydrogen bonding interactions to Tyr309 and His163 is further supported by residual activities of 5, 8, and 85% for variant proteins Y309A, H163N, and H163A (Fig. 7C). The moderately reduced activity of mutant H163A clearly indicates that VioA catalysis does not rely on an imidazole-mediated general acid/base mechanism.
For mutant W397A, no enzymatic activity was determined. The ternary VioA structure indicates the shielding of the substrate's amino group via the planar indole ring system of Trp397 (Fig. 5, C and D). Obviously, this Trp plays a substantial role for the accurate positioning of the substrate as a prerequisite for subsequent enzyme catalysis. This hypothesis is in agreement with the observed residual activity of 63% for mutant W397Y (Fig. 7C). To some extent, the planar tyrosyl side chain might be able to cope with the spatial orientation of the indole side chain of Trp397.
In both VioA structures, Lys269 is in contact with N5 of the isoalloxazine ring of the cofactor via a bridging water molecule (Fig. 5, C and D). This indirect hydrogen bonding interaction of Lys269 is of central importance for the catalytic mechanism of VioA because mutant proteins K269Q and K269S revealed residual activities of solely 2 and 1% (Fig. 7C). This residue might be essential for the accurate positioning of the isoalloxazine ring. However, a potential role during the O2-dependent reoxidation of FADH2 must also be considered (40).
Val363 is part of the defined binding pocket that is relevant for the specific recognition of the hydrophobic indole ring of the substrate (Fig. 5, C and D). This is supported by our mutational analysis, because variants V363Q and V363A yielded relative activities of 17 and 57%. Replacing Val363 by a larger and more polar glutamine residue might result in a steric restriction of the hydrophobic active site. By contrast, the related alanine mutation might result in a widening of this pocket, reflected by an increased activity of 57% (Fig. 7C). Nevertheless, integrity of the overall hydrophobic active site pocket of VioA might be relevant to obtain full enzymatic activity.
Mutant Activities in the Presence of Non-natural Substrates
As summarized in the table depicted in Fig. 7C, the activity of mutants R64Q, R64S, H163N, K269Q, K269S, Y309A, and W397A significantly decreased using artificial substrates 7-aza-Trp, 1-methyl-Trp, 5-methyl-Trp, and 5-fluoro-Trp so that values partially below the detection limit were determined. When the activity of the “more active” mutants H163A, V363Q, V363A, and W397Y in the presence of l-Trp was set as 100% (values in parentheses, respectively) the following order of substrate activities was obtained: 5-methyl-Trp (70–160%) > 5-fluoro-Trp (51–79%) > 1-methyl-l-Trp (24–77%) > 7-aza-Trp (7–22%). Basically, these mutant activities mirror the outcome of the wild type enzyme assays in the presence of the different artificial substrates. Nevertheless, the individually determined values do not only reflect the simple addition of a mutant and of an artificial substrate effect.
Site-directed mutagenesis of key residues often goes along with an altered specificity of the respective enzymes (41). Accordingly, active mutant proteins V363Q, V363A, H163A, and W397Y were further analyzed with respect to substrate specificity.
Widening the Substrate Specificity of VioA
VioA proteins H163N, V363Q, V363A, and W397Y were analyzed in the presence of l-tyrosine, l-valine, l-proline, l-histidine, l-leucine, and l-alanine and l-phenylalanine, respectively. None of these combinations resulted in any detectable enzymatic activity, indicative of a preserved VioA substrate specificity. By contrast, for variant H163A, discrimination of l-tyrosine, l-valine, l-proline, l-histidine, l-leucine, and l-alanine was shown, whereas a relative enzymatic activity of 5.4% was determined in the presence of l-phenylalanine, corresponding to a 3-fold increase with respect to the wild type enzyme (Fig. 7C). This indicates that His163 plays an essential role for the appropriate positioning of the (artificial) substrate in the active site of VioA. A slightly altered network for the recognition of the substrate's carboxylate group might cause an improved binding of l-phenylalanine in the active site of VioA.
Discussion
VioA and orthologous enzymes StaO and RebO catalyze the FAD-dependent oxidation of a Trp scaffold within the biosynthesis of important natural bisindole compounds (Fig. 1). The catalytic cycle for the VioA-dependent IPA imine formation involves the reoxidation of the prosthetic group by O2 with formation of H2O2. The resulting imine reaction product is susceptible for subsequent deamination as it is observed for the related catalysis of LAAO enzymes (21, 42). By contrast, violacein biosynthesis requires efficient transfer of the labile intermediate from the active site of VioA to the subsequent enzyme of the pathway.
This investigation reveals a detailed picture for the understanding of VioA substrate specificity. Efficient discrimination of the canonical amino acids might be essential under in vivo conditions because the H2O2 “side product” and also unwanted imine products might be deleterious for the bacterial cell. However, in the presence of unnatural artificial VioA substrates, substantial flexibility due to the conversion of 7-aza-Trp, 1-methyl-Trp, 5-methyl-Trp, and 5-fluoro-Trp was indicated. Obviously, evolution of the VioA active site did not rely on clearly defined determinants for the recognition of the indole ring system.
Our structural investigation showed a well defined network for the specific interaction of the polar “head group” of the VioA substrate (Fig. 5, C and D). Such type of a fixed l-Trp only shows limited conformational flexibility at the Cβ atom. The hydrophobic indole moiety is not specifically recognized in the hydrophobic part of the active site pocket. Obviously, conformational restraints at Cβ in combination with the rigid and planar indoyl substituent play a key role for VioA substrate recognition.
With respect to the overall biosynthetic pathway, compounds 7-aza-Trp, 1-methyl-Trp, 5-methyl-Trp, and 5-fluoro-Trp are interesting candidates for the in vivo synthesis of new violacein derivatives. As outlined in Fig. 1, variant Trp molecules with a modified indole ring at position 1, 5, or 7 are theoretically processed into the respective violacein (or deoxyviolacein) derivatives (compare numbering indicated for violacein and deoxyviolacein). Initial in vivo pathway reconstitution experiments in our laboratory already indicated the conversion of synthetic artificial Trp derivatives to new enzymatic products and intermediates. Structural elucidation of the novel compounds produced and investigations toward their possible biosynthetic pathway are ongoing. On the basis of these results, synthetic biology approaches for the engineering of variant violacein, rebeccamycin, or staurosporine pathways might be a promising strategy for the synthesis of novel bisindole therapeutics. Successful engineering of staurosporine and rebeccamycin variant molecules has been demonstrated recently (43, 44).
The outcome of the combined structural and biochemical investigation allowed us to propose a detailed molecular mechanism of VioA catalysis. Enzyme kinetics of mutant proteins functionally identified Arg64, His163, Lys269, Tyr309, and Trp397 as essential residues of VioA catalysis. Inspection of the spatial orientation of these amino acids in both structures of VioA did not reveal an appropriate catalytic base for the specific deprotonation of the Cα-H-bond of the l-Trp substrate. This is inconsistent with an acid-base mechanism via a substrate carbanion. However, the Cα of l-Trp (as indicated by the bound IEA molecule) is well oriented with respect to a direct hydride transfer. In Fig. 5F, the Cα-H-bond (of the modeled substrate complex) is pointing toward the N5 atom of the cofactor exactly as observed for many related flavoenzymes performing a hydride transfer mechanism (21, 45, 46). Accordingly, the following Reaction sequence for VioA catalysis was postulated. The zwitterionic l-Trp substrate approaches the catalytic site via the substrate entry channel (compare orange asterisk in Fig. 5I, left and right). Subsequently, deprotonation of the α-amino group is required for the formation of the Michaelis complex. This proton abstraction might be supported by the optimal pH of 9.25 (7) and also by the active site architecture where the polar α-amino group points perpendicularly to the hydrophobic indole ring system of Trp397. Then, the exact orientation of Cα in the Michaelis complex facilitates the oxidative attack. This transition state might be further activated by distortion of the substrate. By analogy to the VioA·FADH2·IEA complex, the tetrahedral Cα of the substrate might adopt a more planar geometry with partial sp2 character. Such a distorted geometry would not only reflect the sterical characteristics of the IPA imine reaction product but would also explain the observed competitive inhibition in the presence of the transition state analog IEA. Formation of this activated ternary complex is accompanied by protein domain movements as indicated by the structural transition from the VioA·FADH2 to the VioA·FADH2·IEA complex. These conformational rearrangements result in a partial closure of the active site cavity and also include the repositioning of residues His163 and Arg64. These alterations might be a prerequisite for the direct transfer of the Cα hydride onto N5 of the isoalloxazine ring. This process is guided by the movement of the lone pair of electrons from the α-amino nitrogen to the Cα, leading to the formation of the protonated imine reaction product. To some extent, the ternary VioA complex also resembles a well protected enzyme·product complex in which the IPA imine is shielded from the deamination via attacking water molecules. Subsequently, channeling of the IPA imine only in the presence of the subsequent enzyme of the pathway would be an elegant solution to the metabolic problem. However, characterization of such a type of transient interaction might require the trapping of the overall VioA protein dynamics. With respect to this hypothesis, the conformational rearrangements of VioA might also be relevant for the appropriate timing of the release of the IPA imine reaction product.
The second half-reaction of VioA catalysis is responsible for the reoxidation of the prosthetic group by O2. For this purpose, a smaller entry channel gives direct access to the bridging water molecule that links Lys269 to the N5 atom of FADH2 (compare Fig. 5, C and D, and yellow × in Fig. 5I, right). The position of this water molecule (found in both structures) might also represent the site for H2O2 generation as a result of flavin reoxidation. Mutagenesis of Lys269 might indicate almost complete VioA inactivation due to a significant influence on the reductive and also on the oxidative half-reaction of VioA. A related binding position for the oxidative half-reaction of LAAO from C. rhodostoma has been also described in the literature (36). Additional experimental work is required to further elucidate the oxidative half-reaction and the precise sequential order of VioA catalysis.
Experimental Procedures
Plasmid Construction and Site-directed Mutagenesis
The vioA gene from C. violaceum ATCC 12472 was amplified from genomic DNA (German Collection of Microorganisms and Cell Cultures (DSMZ)) using primers CAC GGA TCC ATG AAG CAT TCT TCC GAT ATC and GTA CTC GAG TCA CGC GGC GAT GCG CTG C (restriction sites underlined) and cloned into the BamHI and XhoI sites of pGEX-6P-1 (GE Healthcare), yielding the plasmid pGEX-vioA. Mutations in the vioA gene were introduced using the QuikChangeTM site-directed mutagenesis kit (Stratagene) using the following oligonucleotides (exchanged nucleotides in bold): R64Q, GAG CTG GGC GCG GGG CAG TAC TCC CCG CAG C and GCT GCG GGG AGT ACT GCC CCG CGC CCA GCT C; R64S, GAG CTG GGC GCG GGG TCT TAC TCC CCG CAG C and GCT GCG GGG AGT AAG ACC CCG CGC CCA GCT C; H163N, CGA CAT CGT CGG CAA GAA CCC GGA AAT CCA GAG CG and CGC TCT GGA TTT CCG GGT TCT TGC CGA CGA TGT CG; H163A, CGA CAT CGT CGG CAA GGC GCC GGA AAT CCA GAG CG and CGC TCT GGA TTT CCG GCG CCT TGC CGA CGA TGT CG; K269Q, GGC TCG CTG CCG CTG TTC CAG GGT TTC CTC ACC TAC GG and CCG TAG CTG AGG AAA CCC TGG AAC AGC GGC AGC GAG CC; K269S, GGC TCG CTG CCG CTG TTC TCT GGT TTC CTC ACC TAC GG and CCG TAG GTG AGG AAA CCA GAG AAC AGC GGC AGC GAG CC; Y309F, GGG CGA CAA GTA CCT GTT CTT CTT TAC CGA CAG CGA GAT GGC C and GGC CAT CTC GCT GTC GGT AAA GAA GAA CAG GTA CTT GTC GCC C; Y309A, GGG CGA CAA GTA CCT GTT CTT CGC GAC CGA CAG CGA GAT GGC C and GGC CAT CTC GCT GTC GGT CGC GAA GAA CAG GTA CTT GTC GCC C; V363Q, CAA GTA TTG GGC GCA TGG CCA GGA GTT CTG CCG CGA CAG CG and CGC TGT CGC GGC AGA ACT CCT GGC CAT GCG CCC AAT ACT TG; V363A, CAA GTA TTG GGC GCA TGG CGC GGA GTT CTG CCG CGA CAG CG and CGC TGT CGC GGC AGA ACT CCG CGC CAT GCG CCC AAT ACT TG; W397Y, CAC CGA GCA CTG CGG CTA TAT GGA GGG CGG CCT GC and GCA GGC CGC CCT CCA TAT AGC CGC AGT GCT CGG TG; and W397A, CAC CGA GCA CTG CGG CGC GAT GGA GGG CGG CCT GC and GCA GGC CGC CCT CCA TCG CGC CGC AGT GCT CGG TG.
Heterologous Production and Purification of VioA
Recombinant VioA was produced in E. coli BL21 (λDE3) cells containing plasmid pGEX-vioA. An overnight culture was used to inoculate 500 ml of LB medium containing 100 μg ml−1 ampicillin. Protein production was induced at an A578 of ∼0.5 by addition of 50 μm isopropyl 1-thio-β-d-galactopyranoside. After 21 h at 25 °C and 180 rpm, cells were harvested by centrifugation (15 min, 3000 × g, 4 °C) and washed with 25 ml of buffer 1 (50 mm Tris-HCl, pH 9.0, and 100 mm NaCl), and the sedimented cells were stored at −20 °C. The pellet was thawed on ice, resuspended in 7.5 ml of buffer 1, and BenzonaseTM (Merck Millipore, Darmstadt, Germany) was added according to the manufacturer's instructions. Cells were disrupted by a single passage through a French press cell at 19,200 p.s.i. After ultracentrifugation for 1 h at 110,000 × g at 4 °C, the cell-free extract was incubated for 2 h with 2 ml of Protino® glutathione-agarose 4B (Macherey & Nagel). After washing two times with 10 ml of buffer 1, the VioA-loaded column was incubated with 200 units of PreScissionTM protease (GE Healthcare) in 2 ml of buffer 1 for 16 h at 4 °C under slight agitation to liberate the target protein from the GST tag. VioA was eluted in 6 ml of buffer 1, and protein containing fractions were identified by SDS-PAGE. The target protein was stored in buffer 1 containing 5% (v/v) glycerol at 4 °C or at −20 °C. For subsequent crystallization experiments, VioA purification was performed in the presence of 2 mm DTT.
Determination of Protein Concentration
The concentration of purified VioA was determined at 280 nm using an extinction coefficient of ϵ280 = 71,695 m−1 cm−1.
N-terminal Amino Acid Sequence Determination
Automated Edman degradation was used to confirm the identity of purified VioA (47–49).
Thermal Shift Experiments
The temperature of the unfolding transition midpoint was determined in thermal shift experiments as described elsewhere (50).
FAD Cofactor Quantification
The FAD content of purified VioA samples was quantified by UV-visible absorption spectroscopy using an extinction coefficient of ϵ450 = 11,300 m−1 cm−1.
Alternatively, single turnover experiments were performed under anoxic conditions (oxygen partial pressure below 1 ppm, oxygen detector, Coy Laboratories, Grass Lake, MI) to estimate the amount of co-purified FAD. All buffers were prepared by repeated cycles of evaporation and nitrogen flushing. Purified samples of VioA at a concentration of 15 μm were supplemented with increasing concentrations of l-Trp (0–10 μm) in a total volume of 600 μl and incubated for 2 min at 22 °C. The absorption spectrum of all samples was measured using sealed cuvettes (550–250 nm). Employed concentrations of l-Trp where then plotted versus the photometrically determined FAD concentration to estimate the amount of copurified FAD in the original VioA sample.
Synthesis of VioA Inhibitors and Trp-derivatives, General Remarks
Commercially available starting material and solvents were purchased from Sigma, Fisher/Acros, Alfa Aesar, or Carl Roth and were used without further purification. Technical solvents were distilled before use. All reactions involving water-sensitive chemicals were carried out in heat gun-dried glass equipment with magnetic stirring under an argon atmosphere. TLC was performed on Polygram® SIL G/UV254 plates (Macherey-Nagel) with detection by UV (254 nm) or by immersion in aqueous solution containing sulfuric acid, ammonium molybdate tetrahydrate, and Cer(IV)sulfate tetrahydrate, followed by heating. Flash chromatography was performed on Silica Gel M60 (0.04–0.063 mm, 230–400 mesh ASTM) (Macherey & Nagel) under nitrogen pressure. NMR spectra were recorded on a Bruker DRX-400 spectrometer (1H 400 MHz, 13C 101 MHz). Chemical shifts are reported in parts per million relative to tetramethylsilane in case of CDCl3 and MeOD, and relative to trimethylsilylpropanoic acid in case of D2O, as an internal standard (δ = 0). Coupling constants J are given in Hertz.
Synthesis of Inhibitors IEA and IAA
IEA (2-(1H-Indol-3-ylmethyl)prop-2-enoic Acid) (3)
To a stirred solution of ester 2 (750 mg, 3.48 mmol) in 10 ml of MeOH was added slowly a solution of KOH (590 mg, 10.52 mmol) in 3 ml of water and 5 ml of MeOH at 0 °C. The mixture was stirred for 30 min with cooling. After stirring for 24 h at room temperature, the solvent was evaporated under reduced pressure. The aqueous phase was acidified with HCl to pH 4 and extracted with ethyl acetate. After drying with MgSO4 and filtration, the solvent was evaporated under reduced pressure. Purification on a silica gel column (pentane/ethyl acetate 2:1, Rf = 0.23) gave the desired product as a brownish solid (541 mg, 2.69 mmol, 77%). 1H NMR (ppm) (400 MHz, CDCl3): 11.00 (br.s., 1H), 7.97 (s, 1H), 7.55–7.51 (m, 1H), 7.35 (ddd, 3J = 8.1, 4J = 1.0, 5J = 0.8, 1H), 7.19 (ddd, 3J = 8.3, 3J = 6.9, 4J = 1.4, 1H), 7.11 (ddd, 3J = 7.8, 3J = 7.1, 4J = 1.0, 1H), 7.02 (d, 3J = 2.3, 1H), 6.36–6.34 (m, 1H), 5.63–5.60 (m, 1H), 3.79–3.76 (m, 2H). 13C NMR (ppm) (100 MHz, CDCl3): 172.7 (Cq), 138.9 (Cq), 136.4 (Cq), 128.0 (=CH2), 127.2 (Cq), 122.9 (CHarom), 122.1 (CHarom), 119.4 (CHarom), 119.0 (CHarom), 112.7 (Cq), 111.1 (CHarom), 27.2 (-CH2-).
Methyl 3-(1H-Indol-3-yl)-2-methylpropanoate (4)
Following the general method described by Youn et al. (30), magnesium turnings (180 mg, 7.41 mmol) were added to a solution of 2 (750 mg, 3.48 mmol) in 20 ml of absolute MeOH and stirred for 30 min with gas development under an argon atmosphere. The temperature was then kept at 10 °C for 2 h. This mixture was poured into a cooled HCl solution (3 m) and stirred until the turbidity disappeared. Adjusting the pH to 8.5–9 by addition of 3 m ammonium hydroxide solution was followed by extraction with diethyl ether. The organic phase was dried with MgSO4, and the solvent evaporated under reduced pressure. The procedure yielded a yellow liquid that was used in the next step without further purification.
IAA (rac-3-(1H-Indol-3-yl)-2-methylpropanoic Acid) (5)
To a stirred solution of KOH (424 mg, 7.55 mmol) in 2 ml of MeOH and 1.2 ml water, a solution of 4 (546 mg, 2.52 mmol) in 4 ml of MeOH was added at 0 °C. The reaction was performed as described for 3. Purification on a silica gel column (pentane/ethyl acetate 4:1 to 1:1) gave the pure product (409 mg, 2.01 mmol, 80%) as a beige-colored solid. 1H NMR (ppm) (400 MHz, MeOD): 7.55–7.52 (m, 1H), 7.33 (ddd, 3J = 8.0, 4J = 1.0, 5J = 0.8, 1H), 7.08 (ddd, 3J = 8.1, 3J = 6.9, 4J = 1.2, 1H), 7.00 (ddd, 3J = 7.9, 3J = 7.0, 4J = 1.2, 1H), 7.03 (s, 1H), 3.19–3.10 (m, 1H), 2.87–2.77 (m, 2H), 1.17 (d, 3J = 6.6, 3H). 13C NMR (ppm) (100 MHz, MeOD): 180.7 (Cq), 137.8 (Cq), 128.6 (Cq), 123.7 (-CH2-), 122.1 (CHarom), 119.4 (CHarom), 119.2 (CHarom), 113.5 (Cq), 112.1 (CHarom), 41.8 (-CH-), 30.3 (-CH2-), 17.4 (-CH3).
Synthesis of Trp-derivatives 7-Aza-Trp and 5-Fluoro-Trp
7-Azagramine (7A) was synthesized according to the procedure of Pierce et al. (38). Purification by flash column chromatography (CHCl3/MeOH/NH4OH 90:10:1 → 40:10:1) yielded the product as a yellow solid (2.34 g, 13.34 mmol, 79%). Analytical data were in agreement with the literature (38).
5-Fluorogramine (7F)
A solution of 5-fluoroindole (1.0 g, 7.40 mmol) in 1 ml of AcOH was added to a solution of dimethylamine (567 mg, 1.42 ml, 12.58 mmol, 40% aqueous solution) and formaldehyde (333 mg, 0.9 ml, 11.09 mmol, 37% aqueous solution) in 5 ml of acetic acid at 0 °C under an argon atmosphere. The mixture was stirred without cooling for 3 h, poured onto 20 ml of ice-water, and adjusted to pH 11 with 2 m NaOH. The resulting suspension was extracted with diethyl ether and dried with MgSO4, and the solvent was evaporated under reduced pressure. Column chromatography on silica (CHCl3/MeOH/NH4OH 90:10:1 → 40:10:1) gave the product (1.33 g, 6.92 mmol, 93%) as a beige-colored solid. 1H NMR (ppm) (400 MHz, CDCl3): 8.47 (s, 1H), 7.36–7.31 (m, 1H), 7.25–7.20 (m, 1H), 7.12 (d, 3J = 2.6, 1H), 6.95–6.88 (m, 1H), 3.58 (s, 2H), 2.28 (s, 6H). 13C NMR (ppm) (100 MHz, CDCl3): 157.5 (d, 1J = 234.4, F-Cq), 132.4 (Cq), 128.0 (d, 3J = 9.9, Cq), 125.1 (CHarom), 113.2 (d, 4J = 4.9, Cq), 111.3 (d, 3J = 9.7, CHarom), 110.0 (d, 2J = 26.4, CHarom), 103.87 (d, 2J = 23.5, CHarom), 54.1 (-CH2-), 44.9 (2x-CH3).
Tetramethylammonium Iodide Salts (8A, 8F)
Based on the method of Wartmann and Lindel (39), 1.5 eq of methyl iodide was added at 0 °C to an 0.15 m solution of the corresponding gramine in THF and stirred for 5 min at this temperature. The reaction mixture was stirred for 1 h at room temperature and then diluted to a tripled volume with pentane. The resulting suspension was filtered, and the residue washed with pentane. The products were used without further purification.
Addition of the Amino Acid Side Chain, rac-Ethyl 2-((diphenylmethylene)amino)-3-(5-fluoro-1H-indol-3-yl)propanoate(9F)
Based on the procedure of Wartmann and Lindel (39), ethyl N-(diphenylmethylene)glycinate (902 mg, 3.38 mmol) was added at 0 °C under argon atmosphere to a suspension of NaH (135 mg, 3.38 mmol, 60% suspension in mineral oil) in 21 ml of THF, and the resulting mixture was stirred for 30 min under cooling. A suspension of 7F (1.13 g, 3.38 mmol) in 10 ml of THF was added and stirred for an additional 3 h at 0 °C. The reaction was quenched with saturated aqueous NH4Cl solution and extracted with ethyl acetate. After drying with MgSO4 and filtration, the solvent was evaporated under reduced pressure. Flash column chromatography (pentane/ethyl acetate 5:1 → 3:1, Rf = 0.4) gave the diphenyl protected ester 9F (722 mg, 1.74 mmol, 52%) as a clear yellow resin-like liquid. 1H NMR (ppm) (400 MHz, CDCl3): 8.19, 8.02 (s), 7.83–7.79 (m), 7.63–7.56 (m), 7.52–7.45 (m), 7.39–7.33 (m), 7.33–7.22 (m), 7.21–7.14 (m), 7.11–7.08 (d), 6.99 (d), 6.96–6.81 (m), 6.66–6.59 (m), 4.35 (dd), 4.23–4.09 (m), 3.38 (ddd), 3.24–2.99 (m), 1.25 (t), 1.23 (t). 13C-NMR (ppm) (100 MHz, CDCl3): 172.1 (Cq), 170.6 (Cq), 157.6 (d, 1J = 234.40, F-Cq), 139.4 (Cq), 137.6 (Cq), 135.8 (Cq), 132.4 (CHarom), 130.0 (CHarom), 128.8 (CHarom), 128.3 (CHarom), 128.1 (d, 3J = 11.70, Cq), 127.9 (CHarom), 127.5 (CHarom), 124.9 (CHarom), 112.2 (d, 4J = 4.71, Cq), 111.4 (d, 3J = 9.65, CHarom), 110.1 (d, 2J = 26.37, CHarom), 103.8 (d, 2J = 23.46, CHarom), 65.1 (-CH-), 60.9 (-CH2-), 29.2 (-CH2-), 14.2 (-CH3).
rac-Ethyl 2-((Diphenylmethylene)amino)-3-(1H-pyrrolo[2,3-b]pyridin-3-yl)propanoate (9A)
As described for 9F, 7-azagramine salt (8A) (850 mg, 2.67 mmol) was used to yield the desired product (350 mg, 0.88 mmol, 33%) after column chromatography (pentane/ethyl acetate 1:1 → 1:2, Rf = 0.4) as a clear brown viscous liquid. 1H NMR (ppm) (400 MHz, CDCl3): 9.57, 9.24 (s), 8.29, 8.22 (dd),7.95 (dd), 7.83–7.89 (m), 7.62–7.55 (m), 7.52–7.45 (m), 7.40–7.34 (m), 7.33–7.23 (m), 7.22–7.12 (m), 7.10–7.05 (m), 6.88 (dd), 6.60 (dd), 4.37–4.32 (m), 4.24–4.00 (m), 3.80 (dd), 3.45–3.04 (m), 1.28–1.19 (m). 13C NMR (ppm) (100 MHz, CDCl3): 172.0 (Cq), 170.6 (Cq), 142.8 (CHarom), 139.3 (Cq), 137.6 (Cq), 135.8 (Cq), 132.4 (CHarom), 130.3 (CHarom), 130.1 (CHarom), 128.8 (CHarom), 128.3 (CHarom), 128.1 (CHarom), 127.5 (CHarom), 127.4 (CHarom), 123.3 (CHarom), 120.2 (Cq), 115.7 (CHarom), 110.8 (Cq), 66.1 (-CH-), 61.0 (-CH2-), 29.3 (-CH2-), 14.2 (-CH3).
Deprotection of the Amino Group
As described by Wartmann et al. (39), the intermediate esters 9A and 9F were dissolved in THF (0.1 m), treated with 2 m HCl (3.5 eq), and stirred for 1 h at room temperature. The reaction mixture was diluted with ethyl acetate and neutralized with saturated aqueous NaHCO3. The aqueous phase was extracted three times with ethyl acetate, and the combined organic phases were dried with MgSO4. The solvent was evaporated under reduced pressure.
Ethyl 2-amino-3-(5-fluoro-1H-indol-3-yl)propanoate (10F)
Following the above described procedure, protected 5-fluoroamino ester 9F (520 mg, 1.26 mmol) gave the desired product after flash column chromatography (DCM/MeOH 7:1) as a yellow viscous oil (260 mg, 1.04 mmol, 83%). 1H NMR (ppm) (400 MHz, CDCl3): 8.19 (s, NH, 1H), 7.27–7.23 (m, 2H), 7.11 (d, 4J = 2.4, 1H), 6.93 (dt, 3J = 9.0, 4J = 2.4), 4.16 (dq, 3J = 7.2, 2J = 2.5, 2H), 3.78 (dd, 3J = 7.5, 3J = 5.0, 1H), 3.21 (dd, 2J = 14.45, 3J = 4.98, 1H), 3.03 (dd, 2J = 14.49, 3J = 7.47, 1H), 1.25 (t, 3J = 7.15). 13C NMR (ppm) (100 MHz, CDCl3): 175.2 (Cq), 157.9 (d, 1J = 234.5, F-Cq), 132.8 (Cq), 127.9 (d, 3J = 9.7, Cq), 124.7 (CHarom), 111.8 (d, 3J = 9.6, CHarom), 111.5 (d, 4J = 4.7, Cq), 110.6 (d, 2J = 26.4, CHarom), 103.8 (d, 2J = 23.6, CHarom), 61.0 (-CH2-), 54.9 (-CH-), 30.6 (-CH2-), 14.2 (-CH3).
Ethyl 2-amino-3-(1H-pyrrolo[2,3-b]pyridin-3-yl)propanoate (10A3)
Following the above described procedure, protected 7-aza-amino ester 9A (340 mg, 0.86 mmol) gave the desired product after flash column chromatography (CHCl3/MeOH/NH4OH 90:10:1 → 40:10:1) as a brownish clear solid (150 mg, 0.64 mmol, 75%). 1H NMR (ppm) (400 MHz, CDCl3): 10.20 (s, NH, 1H), 8.28 (dd, 3J = 4.8, 4J = 1.5, 1H), 7.95 (dd, 3J = 7.9, 4J = 1.6, 1H), 7.22 (s, 1H), 7.08 (dd, 3J = 7.9, 3J = 4.8, 1H), 4.15 (dq, 3J = 7.2, 2J = 2.1, 2H), 3.81 (dd, 3J = 7.2, 3J = 5.3, 1H), 3.24 (dd, 2J = 14.6, 3J = 5.2, 1H), 3.09 (dd, 2J = 14.5, 3J = 7.5, 1H), 1.22 (t, 3J = 7.2, 3H). 13C NMR (ppm) (100 MHz, CDCl3): 175.1 (Cq), 148.7 (Cq), 142.8 (CHarom), 127.4 (CHarom), 123.6 (CHarom), 120.2 (Cq), 115.5 (CHarom), 109.7 (Cq), 61.0 (-CH2-), 55.1 (-CH-), 30.7 (-CH2-), 14.2 (-CH3).
Saponification to Amino Acids
To a solution of KOH (3 eq) in MeOH/H2O (5:3) was added a solution of the respective ester in MeOH (0.6 m), and the resulting mixture was stirred overnight. MeOH was evaporated under reduced pressure, and the aqueous residue was washed with diethyl ether. The aqueous phase was acidified with AcOH to pH 4–5. Reversed phase column chromatography on RP-18 (100 g, MeOH/H2O) gave the desired amino acid.
2-Amino-3-(5-fluoro-1H-indol-3-yl)propanoic Acid (11F)
Reaction performed with amino ester 10F (156 mg, 0.623 mmol) yielded the desired product after purification on RP-18 (MeOH/H2O 60:40 → 50:50) as a slight yellow solid (84%). 1H NMR (ppm) (400 MHz, D2O): 7.51 (dd, 3J = 8.8, 4J = 4.6, 1H), 7.44 (dd, 2J = 10.0, 4J = 2.2, 1H), 7.13–7.03 (m, 1H), 4.07 (dd, 3J = 7.6, 3J = 4.8, 1H), 3.46 (dd, 2J = 15.2, 3J = 4.84, 1H), 3.32 (dd, 2J = 15.4, 3J = 7.9, 1H). No 13C NMR spectra were recorded due to low solubility.
2-Amino-3-(1H-pyrrolo[2,3-b]pyridin-3-yl)propanoic Acid (11A)
Amino ester 10A (140 mg, 0.600 mmol) was obtained as a yellow solid after purification on RP-18 (MeOH/H2O 70:30 → 50:50) (90 mg, 0.436 mmol, 75%). 1H NMR (ppm) (400 MHz, D2O): 8.22 (dd, 3J = 4.8, 4J = 1.4, 1H), 8.12 (dd, 3J = 8.0, 4J = 1.5, 1H), 7.18 (dd, 3J = 7.9, 3J = 4.9, 1H), 4.02 (dd, 3J = 7.5, 3J = 5.0, 1H), 3.41 (dd, 2J = 15.4, 3J = 5.0, 1H), 3.31 (dd, 2J = 15.3, 3J = 7.5, 1H). No 13C NMR spectra were recorded due to low solubility.
VioA Activity Assays
VioA enzyme kinetics were determined in a coupled peroxidase assay mainly as described previously (7, 9). A reaction mixture of 600 μl containing 50 mm Tris-HCl, pH 9.0, 0.2 μm VioA, 1 mm 4-aminoantipyrine, 1 mm phenol, and 15 units/ml horseradish peroxidase was pre-incubated at 30 °C. Then, VioA catalysis was initiated by the addition of 1 mm l-Trp, and the formation of the red quinoneimine dye was monitored at a wavelength of 505 nm in the linear range of the activity assay (∼3 min). The specific activity of VioA was determined by calculation of the slope in the linear range. An extinction coefficient of ϵ505 = 6400 m−1 cm−1 (9) was used to determine the specific activity of VioA.
Alternatively, the activity of VioA was directly measured with an HPLC-based substrate depletion assay as described elsewhere (7). Samples of 100 μl were taken from a reaction mixture (1 ml) containing 50 mm Tris-HCl, pH 9.0, 1 μm VioA, and 500 μm l-Trp at various time points (every 20 s for 3 min). Samples were quenched by the addition of 20 μl of 50% (v/v) trichloroacetic acid and centrifuged for 10 min at 12,100 × g, and the supernatant was analyzed on a μBondapakTM C18 3.9 × 150-mm column (125 Å 10 μm, Waters/Millipore) using a linear gradient of 0–50% acetonitrile over 15 min in 0.1% TFA at a flow rate of 0.5 ml min−1. The employed Jasco HPLC system was equipped with a diode array detector (MD-1515), which was used to monitor the absorbance from 200 to 900 nm. The amount of l-Trp of each sample was quantified using Borwin® peak integration routine.
Both types of activity assays were also performed in the presence of potential VioA inhibitors at concentrations ranging from 0.5 to 10 mm. All VioA activity assays were completed by control experiments in the absence of l-Trp. Tandem peroxidase assays of selected experiments were also performed with varying concentrations of horseradish peroxidase (15–150 units/ml) to ensure that catalysis of VioA is rate-determining.
VioA Dialysis under Turnover Conditions
FAD binding was investigated in a dialysis experiment under aerobic conditions. A 10 μm VioA sample (100 μl) was dialyzed against 1 liter of buffer 1 containing 40 mm l-Trp for 16 h using a Slide-A-Lyzer MINI dialysis unit (10,000 molecular weight cutoff, Thermo Scientific). l-Trp and the VioA reaction product were removed by a buffer exchanged step (illustraTM NAPTM-5 column SephadexTM, GE Healthcare, using buffer 1). The residual FAD content of the VioA sample was determined by UV-visible absorption spectroscopy.
Analytic Gel Permeation Chromatography
The native molecular mass of VioA was determined on an Äkta purifier system equipped with a SuperdexTM 200 Increase 5/150 GL column (GE Healthcare). The system was equilibrated with buffer 1 or alternatively with buffer 1 containing 500 μm Trp derivative IEA at a flow rate of 0.45 ml min−1. The column was calibrated using protein standards (gel filtration markers kit for protein molecular mass 12,000–200,000 Da, Sigma) according to the manufacturer's instructions. Samples of VioA (20 μl) at a concentration of 215 μm were injected. Alternatively VioA was pre-incubated in the presence of 500 μm IEA for 3 h. Elution was monitored at 280 and 450 nm, respectively.
Small-angle X-ray Scattering
SAXS experiments were carried out on beamline BM29 (51) of the European Synchrotron Radiation Facility (Grenoble, France) to elucidate the oligomeric architecture of VioA. Samples of VioA (or of VioA in the presence of 1 mm IEA) at concentrations ranging from 10 to 265 μm in buffer 1 were loaded by an automated sample changer to a quartz capillary mounted in vacuum and exposed to an x-ray energy of 12.5 keV. Scattering images covering a momentum transfer range of 0.0025–5 nm−1 were recorded with a Dectris Pilatus 1M hybrid photon counting detector. A total of 10 images were recorded per sample with an exposure time of 1 s per frame. To reduce radiation damage, the protein solution was kept under steady flow while being irradiated. The scattering functions derived from the images were normalized for exposure time and sample transmission and checked for effects of radiation damage in PRIMUS (52). Data unaffected by radiation damage were averaged, corrected for buffer scattering, normalized, and merged from the different concentrations. The comparison of the determined scattering data with the theoretical scattering curves of a VioA monomer or two potential dimers (33) was performed with CRYSOL (53). The radius of gyration (Rg) and the pair distance distribution P(r) were derived with GNOM (54). Twenty models were computed from SAXS data with DAMMIF (55) and used for the construction of a representative ab initio model using DAMAVER and SUPCOMP (56, 57). The models had a mean normalized spatial discrepancy (N̄SD) of 1.244 and a standard deviation of NSD (σNSD) of 0.189. One of the 20 models showed an NSD of greater than 1.622 (= N̄SD + 2 × σNSD) and was thus omitted for the calculation of the final ab initio model.
Micro Scale Thermophoresis
VioA-ligand/inhibitor interaction was measured on a Monolith NT.115 instrument (NanoTemper Technologies, GmbH). VioA (20 μm) was labeled using the Monolith NTTM protein labeling kit (NT-647-Red-NHS amine reactive fluorescent dye, NanoTemper Technologies, GmbH). The labeled protein was supplemented with 0.05% (w/v) Tween 20 and different concentrations of IAA, IEA, and citrate (30 nm to 20 mm) were titrated, respectively. Thermophoresis was measured in hydrophilic capillaries (NanoTemper Technologies, GmbH) using a laser intensity of 20% for 35 s.
Protein Crystallization
Purified VioA was concentrated to 150–200 μm using a VivaspinTM 4 centrifugal concentrator (30,000 molecular weight cutoff PES, Sartorius). Crystals were prepared as sitting drops by mixing 1 μl of protein solution with 1 μl of reservoir solution. Native VioA crystals grew at 4 °C from 80 mm Tris-HCl, pH 8.5, 24% (w/v) PEG 4000, 20% (v/v) glycerol after 5 weeks. A heavy metal derivative was produced by soaking VioA crystals with 10 mm Sm(NO3)3 for 2 weeks. Co-crystals of VioA and IEA grew after 5 days at 17 °C in sitting drops in the presence of 100 mm HEPES, pH 7.0, 10% PEG 6000, 100 mm lithium chloride, and 3.75 mm IEA. Crystals were cryo-protected in the presence of 20% glycerol (v/v) and flash-cooled in liquid nitrogen.
Data Collection, Structure Determination, and Refinement
Diffraction experiments were performed on the beamline X06DA, operated by the Paul Scherrer Institute at the Swiss Light Source (Villigen, Switzerland), on beamline P11, operated by DESY at PETRA III synchrotron (Hamburg, Germany) (58), and on beamline 14.1, operated by the Helmholtz-Zentrum Berlin at the BESSY II electron storage ring (Berlin-Adlershof, Germany) (59). Data processing was carried out with XDS (60), and the upper resolution limits were assessed through careful observation of I/σI and CC1/2 (61). The phase problem was solved by a single-wavelength anomalous diffraction experiment at λ = 1.77 Å using the anomalous signal arising from the proximity of samarium's L-III absorption edge. ShelxC/D/E (62) was employed to locate heavy metal sites, to compute a first electron density map, and to build an initial structural model. The other crystal forms were phased by molecular replacement, with Phaser (63), using the initial model obtained from the Sm derivative. With subsequent refinement in Phenix.refine (64) and manual rebuilding in Coot (65), the models were finalized. Data collection and refinement statistics are shown in Table 1.
Author Contributions
J. M., D. J., S. S., W. B., J. J. F., R. R., and J. K. designed the study and wrote the paper. J. J. F., J. M., R. R., J. K., K. E. R., and N. P. D. conducted the experiments and analyzed the results. R. R. synthesized inhibitors and artificial substrates. J. K. conducted crystal x-ray measurements and solved the structure. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Jutta Pruss for experimental assistance during mutagenesis and Beate Jaschok-Kentner for N-terminal protein sequencing.
This work was supported by the Government of Niedersachsen, Graduate School MINAS Microbial Natural Products. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5G3S, 5G3T, and 5G3U) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- IPA
- indole-3-pyruvic acid
- IAA
- 3-(1H-indol-3-yl)-2-methylpropanoic acid
- IEA
- 2-(1H-indol-3-ylmethyl)prop-2-enoic acid
- LAAO
- l-amino acid oxidase
- SAXS
- small angle x-ray scattering
- NSD
- normalized spatial discrepancy.
References
- 1. Antônio R. V., and Creczynski-Pasa T. B. (2004) Genetic analysis of violacein biosynthesis by Chromobacterium violaceum. Genet. Mol. Res. 3, 85–91 [PubMed] [Google Scholar]
- 2. Durán N., and Menck C. F. (2001) Chromobacterium violaceum: a review of pharmacological and industrial perspectives. Crit. Rev. Microbiol. 27, 201–222 [DOI] [PubMed] [Google Scholar]
- 3. August P. R., Grossman T. H., Minor C., Draper M. P., MacNeil I. A., Pemberton J. M., Call K. M., Holt D., and Osburne M. S. (2000) Sequence analysis and functional characterization of the violacein biosynthetic pathway from Chromobacterium violaceum. J. Mol. Microbiol. Biotechnol. 2, 513–519 [PubMed] [Google Scholar]
- 4. Durán N., Justo G. Z., Ferreira C. V., Melo P. S., Cordi L., and Martins D. (2007) Violacein: properties and biological activities. Biotechnol. Appl. Biochem. 48, 127–133 [DOI] [PubMed] [Google Scholar]
- 5. Tamaoki T., Nomoto H., Takahashi I., Kato Y., Morimoto M., and Tomita F. (1986) Staurosporine, a potent inhibitor of phospholipid/Ca2+-dependent protein kinase. Biochem. Biophys. Res. Commun. 135, 397–402 [DOI] [PubMed] [Google Scholar]
- 6. Anizon F., Belin L., Moreau P., Sancelme M., Voldoire A., Prudhomme M., Ollier M., Sevère D., Riou J.-F., Bailly C., Fabbro D., and Meyer T. (1997) Syntheses and biological activities (topoisomerase inhibition and antitumor and antimicrobial properties) of rebeccamycin analogues bearing modified sugar moieties and substituted on the imide nitrogen with a methyl group. J. Med. Chem. 40, 3456–3465 [DOI] [PubMed] [Google Scholar]
- 7. Balibar C. J., and Walsh C. T. (2006) In vitro biosynthesis of violacein from l-tryptophan by the enzymes VioA-E from Chromobacterium violaceum. Biochemistry 45, 15444–15457 [DOI] [PubMed] [Google Scholar]
- 8. Nishizawa T., Aldrich C. C., and Sherman D. H. (2005) Molecular analysis of the rebeccamycin l-amino acid oxidase from Lechevalieria aerocolonigenes ATCC 39243. J. Bacteriol. 187, 2084–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kameya M., Onaka H., and Asano Y. (2013) Selective tryptophan determination using tryptophan oxidases involved in bis-indole antibiotic biosynthesis. Anal. Biochem. 438, 124–132 [DOI] [PubMed] [Google Scholar]
- 10. Howard-Jones A. R., and Walsh C. T. (2005) Enzymatic generation of the chromopyrrolic acid scaffold of rebeccamycin by the tandem action of RebO and RebD. Biochemistry 44, 15652–15663 [DOI] [PubMed] [Google Scholar]
- 11. Asamizu S., Kato Y., Igarashi Y., Furumai T., and Onaka H. (2006) Direct formation of chromopyrrolic acid from indole-3-pyruvic acid by StaD, a novel hemoprotein in indolocarbazole biosynthesis. Tetrahedron Lett. 47, 473–475 [Google Scholar]
- 12. Asamizu S., Hirano S., Onaka H., Koshino H., Shiro Y., and Nagano S. (2012) Coupling reaction of indolepyruvic acid by StaD and its product: implications for biosynthesis of indolocarbazole and violacein. ChemBioChem 13, 2495–2500 [DOI] [PubMed] [Google Scholar]
- 13. Spolitak T., and Ballou D. P. (2015) Evidence for catalytic intermediates involved in generating the chromopyrrolic acid scaffold of rebeccamycin by RebO and RebD. Arch. Biochem. Biophys. 573, 111–119 [DOI] [PubMed] [Google Scholar]
- 14. Sánchez C., Braña A. F., Méndez C., and Salas J. A. (2006) Reevaluation of the violacein biosynthetic pathway and its relationship to indolocarbazole biosynthesis. ChemBioChem 7, 1231–1240 [DOI] [PubMed] [Google Scholar]
- 15. Sánchez C., Butovich I. A., Braña A. F., Rohr J., Méndez C., and Salas J. A. (2002) The biosynthetic gene cluster for the antitumor rebeccamycin: characterization and generation of indolocarbazole derivatives. Chem. Biol. 9, 519–531 [DOI] [PubMed] [Google Scholar]
- 16. Onaka H., Taniguchi S., Igarashi Y., and Furumai T. (2003) Characterization of the biosynthetic gene cluster of rebeccamycin from Lechevalieria aerocolonigenes ATCC 39243. Biosci. Biotechnol. Biochem. 67, 127–138 [DOI] [PubMed] [Google Scholar]
- 17. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shinoda K., Hasegawa T., Sato H., Shinozaki M., Kuramoto H., Takamiya Y., Sato T., Nikaidou N., Watanabe T., and Hoshino T. (2007) Biosynthesis of violacein: a genuine intermediate, protoviolaceinic acid, produced by VioABDE, and insight into VioC function. Chem. Commun. 40, 4140–4142 [DOI] [PubMed] [Google Scholar]
- 19. Blanchard M., Green D. E., Nocito V., and Ratner S. (1944) l-Amino acid oxidase of animal tissue. J. Biochem. 155, 421–440 [Google Scholar]
- 20. Geueke B., Hummel W. (2002) A new bacterial l-amino acid oxidase with a broad substrate specificity: purification and characterization. Enzyme Microb. Technol. 31, 77–87 [Google Scholar]
- 21. Pawelek P. D., Cheah J., Coulombe R., Macheroux P., Ghisla S., and Vrielink A. (2000) The structure of l-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. EMBO J. 19, 4204–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Umhau S., Pollegioni L., Molla G., Diederichs K., Welte W., Pilone M. S., and Ghisla S. (2000) The x-ray structure of d-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation. Proc. Natl. Acad. Sci. U.S.A. 97, 12463–12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matz C., Webb J. S., Schupp P. J., Phang S. Y., Penesyan A., Egan S., Steinberg P., and Kjelleberg S. (2008) Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS ONE 3, e2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Macheroux P. (1999) UV-visible spectroscopy as a tool to study flavoproteins. Methods Mol. Biol. 131, 1–7 [DOI] [PubMed] [Google Scholar]
- 25. Faust A., Niefind K., Hummel W., and Schomburg D. (2007) The structure of a bacterial l-amino acid oxidase from Rhodococcus opacus gives new evidence for the hydride mechanism for dehydrogenation. J. Mol. Biol. 367, 234–248 [DOI] [PubMed] [Google Scholar]
- 26. Ida K., Kurabayashi M., Suguro M., Hiruma Y., Hikima T., Yamomoto M., and Suzuki H. (2008) Structural basis of proteolytic activation of l-phenylalanine oxidase from Pseudomonas sp. P-501. J. Biol. Chem. 283, 16584–16590 [DOI] [PubMed] [Google Scholar]
- 27. Holzapfel C. W., Bischofberger K., and Olivier J. (1994) A simple cycloaddition approach to a racemate of the natural sweetener monatin. Synth. Commun. 24, 3197–3211 [Google Scholar]
- 28. Yu C., Liu B., and Hu L. (2001) Efficient Baylis-Hillman reaction using stoichiometric base catalyst and an aqueous medium. J. Org. Chem. 66, 5413–5418 [DOI] [PubMed] [Google Scholar]
- 29. Borrell J. I., Teixidó J., Martínez-Teipel B., Matallana J. L., Copete M. T., Llimargas A., and García E. (1998) Synthesis and biological activity of 4-amino-7-oxo-substituted analogues of 5-deaza-5,6,7,8-tetrahydrofolic acid and 5,10-dideaza-5, 6,7,8-tetrahydrofolic acid. J. Med. Chem. 41, 3539–3545 [DOI] [PubMed] [Google Scholar]
- 30. Kwon Youn I., Hwan Yon G., and Siek Pak C. (1986) Magnesium-methanol as a simple convenient reducing agent for α,β-unsaturated esters. Tetrahedron Lett. 27, 2409–2410 [Google Scholar]
- 31. Mori K., and Kisida H. (1986) Synthesis of both the enantiomers of the heterocyclic pheromones isolated from the male swift moth Hepialus hecta L. Tetrahedron 42, 5281–5290 [Google Scholar]
- 32. Jerabek-Willemsen M., Wienken C. J., Braun D., Baaske P., and Duhr S. (2011) Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 9, 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baskaran K., Duarte J. M., Biyani N., Bliven S., and Capitani G. (2014) A PDB-wide, evolution-based assessment of protein-protein interfaces. BMC Struct. Biol. 14, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dym O., and Eisenberg D. (2001) Sequence-structure analysis of FAD-containing proteins. Protein Sci. 10, 1712–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moustafa I. M., Foster S., Lyubimov A. Y., and Vrielink A. (2006) Crystal structure of LAAO from Calloselasma rhodostoma with an l-phenylalanine substrate: insights into structure and mechanism. J. Mol. Biol. 364, 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yano J., Kern J., Irrgang K. D., Latimer M. J., Bergmann U., Glatzel P., Pushkar Y., Biesiadka J., Loll B., Sauer K., Messinger J., Zouni A., and Yachandra V. K. (2005) X-ray damage to the Mn4Ca complex in single crystals of photosystem II: a case study for metalloprotein crystallography. Proc. Natl. Acad. Sci. U.S.A. 102, 12047–12052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pierce L. T., Cahill M. M., and McCarthy F. O. (2011) Synthesis of novel 3,4-diaryl-5-aminopyrazoles as potential kinase inhibitors. Tetrahedron 67, 4601–4611 [Google Scholar]
- 39. Wartmann T., and Lindel T. (2013) l-Phototryptophan. Eur. J. Org. Chem. 2013, 1649–1652 [Google Scholar]
- 40. Henderson Pozzi M., and Fitzpatrick P. F. (2010) A lysine conserved in the monoamine oxidase family is involved in oxidation of the reduced flavin in mouse polyamine oxidase. Arch. Biochem. Biophys. 498, 83–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen H. S., Wang Y. M., Huang W. T., Huang K. F., and Tsai I. H. (2012) Cloning, characterization and mutagenesis of Russell's viper venom l-amino acid oxidase: Insights into its catalytic mechanism. Biochimie 94, 335–344 [DOI] [PubMed] [Google Scholar]
- 42. Wellner D., and Meister A. (1961) Studies on the mechanism of action of l-amino acid oxidase. J. Biol. Chem. 236, 2357–2364 [PubMed] [Google Scholar]
- 43. Sanchez C., Salas A. P., Brana A. F., Palomino M., Pineda-Lucena A., Carbajo R. J., Mendez C., Moris F., and Salas J. A. (2009) Generation of potent and selective kinase inhibitors by combinatorial biosynthesis of glycosylated indolocarbazoles. Chem. Commun. 27, 4118–4120 [DOI] [PubMed] [Google Scholar]
- 44. Cuenca-López M. D., Serrano-Heras G., Montero J. C., Corrales-Sánchez V., Gomez-Juarez M., Gascón-Escribano M. J., Morales J. C., Voisin V., Núñez L. E., Morís F., Bader G. D., Pandiella A., and Ocaña A. (2015) Antitumor activity of the novel multi-kinase inhibitor EC-70124 in triple negative breast cancer. Oncotarget 6, 27923–27937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tilocca A., Gamba A., Vanoni M. A., and Fois E. (2002) First-principles molecular dynamics investigation of the d-amino acid oxidative half-reaction catalyzed by the flavoenzyme d-amino acid oxidase. Biochemistry 41, 14111–14121 [DOI] [PubMed] [Google Scholar]
- 46. Kachalova G., Decker K., Holt A., and Bartunik H. D. (2011) Crystallographic snapshots of the complete reaction cycle of nicotine degradation by an amine oxidase of the monoamine oxidase (MAO) family. Proc. Natl. Acad. Sci. U.S.A. 108, 4800–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Edman P. (1950) Method for determination of the amino acid sequence in peptides. Acta Chem. Scand. 4, 283–293 [Google Scholar]
- 48. Edman P., and Begg G. (1967) A protein sequenator. Eur. J. Biochem. 1, 80–91 [DOI] [PubMed] [Google Scholar]
- 49. Niall H. D. (1973) Automated Edman degradation: the protein sequenator. Methods Enzymol. 27, 942–1010 [DOI] [PubMed] [Google Scholar]
- 50. Pantoliano M. W., Petrella E. C., Kwasnoski J. D., Lobanov V. S., Myslik J., Graf E., Carver T., Asel E., Springer B. A., Lane P., and Salemme F. R. (2001) High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 6, 429–440 [DOI] [PubMed] [Google Scholar]
- 51. Pernot P., Round A., Barrett R., De Maria Antolinos A., Gobbo A., Gordon E., Huet J., Kieffer J., Lentini M., Mattenet M., Morawe C., Mueller-Dieckmann C., Ohlsson S., Schmid W., Surr J., et al. (2013) Upgraded ESRF BM29 beamline for SAXS on macromolecules in solution. J. Synchrotron. Radiat. 20, 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 53. Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL–a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 54. Svergun D. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 55. Franke D., and Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Volkov V. V., and Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kozin M. B., and Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 58. Burkhardt A., Pakendorf T., Reime B., Meyer J., Fischer P., Stübe N., Panneerselvam S., Lorbeer O., Stachnik K., Warmer M., Rödig P., Göries D., and Meents A. (2016) Status of the crystallography beamlines at PETRA III. Eur. Phys. J. Plus 131, 56 [Google Scholar]
- 59. Mueller U., Förster R., Hellmig M., Huschmann F. U., Kastner A., Malecki P., Pühringer S., Röwer M., Sparta K., Steffien M., Ühlein M., Wilk P., and Weiss M. S. (2015) The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. EPJ Plus 130, 1–10 [Google Scholar]
- 60. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 63. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Laskowski R. A., and Swindells M. B. (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 [DOI] [PubMed] [Google Scholar]
- 67. Hirano S., Asamizu S., Onaka H., Shiro Y., and Nagano S. (2008) Crystal structure of VioE, a key player in the construction of the molecular skeleton of violacein. J. Biol. Chem. 283, 6459–6466 [DOI] [PubMed] [Google Scholar]