Abstract
Global climate change has a significant effect on extreme environments and a profound influence on species survival. However, little is known of the genome-wide pattern of livestock adaptations to extreme environments over a short time frame following domestication. Sheep (Ovis aries) have become well adapted to a diverse range of agroecological zones, including certain extreme environments (e.g., plateaus and deserts), during their post-domestication (approximately 8–9 kya) migration and differentiation. Here, we generated whole-genome sequences from 77 native sheep, with an average effective sequencing depth of ∼5× for 75 samples and ∼42× for 2 samples. Comparative genomic analyses among sheep in contrasting environments, that is, plateau (>4,000 m above sea level) versus lowland (<100 m), high-altitude region (>1500 m) versus low-altitude region (<1300 m), desert (<10 mm average annual precipitation) versus highly humid region (>600 mm), and arid zone (<400 mm) versus humid zone (>400 mm), detected a novel set of candidate genes as well as pathways and GO categories that are putatively associated with hypoxia responses at high altitudes and water reabsorption in arid environments. In addition, candidate genes and GO terms functionally related to energy metabolism and body size variations were identified. This study offers novel insights into rapid genomic adaptations to extreme environments in sheep and other animals, and provides a valuable resource for future research on livestock breeding in response to climate change.
Keywords: extreme environment, rapid adaptation, Ovis aries, whole-genome sequencing, climate change.
Introduction
Global climate change has a considerable impact on organisms, including livestock (Brown and Funk 2008; Hoffmann 2010). In particular, the influence of climate change on living organisms is exacerbated at the interface of extreme environments, such as on plateaus and in desert regions (Easterling et al. 2000). Within this context, it is important to understand the genetic basis of well-adapted local livestock breeds in extreme environments to develop appropriate breeding programs under scenarios of future climate change (Howden et al. 2007; Lobell et al. 2008). After domestication in the Fertile Crescent ∼8,000–9,000 years ago (8–9 kya; Legge 1996), sheep (Ovis aries) spread and became adapted to a wide range of agroecological conditions, especially those distributed on plateaus or in desert regions, which are sensitive to climate change (Easterling et al. 2000; Lv et al. 2014). Thus, these animals provide an excellent model to gain novel insights into genetic mechanisms underlying the rapid adaptations of livestock to extreme environments within a short period of time.
In recent years, to characterize adaptive genetic variations, whole-genome sequencing studies have been performed on a wide range of organisms that live in harsh or extreme environments (Simonson et al. 2010; Mackelprang et al. 2011; Scheinfeldt et al. 2012; Cho et al. 2013; Ge et al. 2013; Ma et al. 2013; Qu et al. 2013; Liu et al. 2014). Studies conducted on livestock are limited, although they include work on adaptations to high altitudes in yak (Bos grunniens; Qiu et al. 2012), Tibetan mastiff (Canis lupus familiaris; Gou et al. 2014) and Tibetan chicken (Wang et al. 2015), hot and arid environments in goat and sheep (Kim et al. 2016), severe desert conditions in domestic Bactrian camel (Jirimutu et al. 2012; Wu et al. 2014) and subarctic cold environments in Yakutian horse (Librado et al. 2015). However, to our knowledge, no study has characterized the rapid genetic adaptations of livestock to various extreme environments based on whole-genome sequences.
Out of the dispersal center in the Mongolian region, Chinese native sheep breeds only have diverged for several thousands of years (e.g., Lv et al. 2015). Previous investigations showed significant differences in hematologic parameters (e.g., erythrocyte count, hematocrit, and hemoglobin; Jiang et al. 1991; Liu and Chen 2006) and body size (Du 2011) between Tibetan sheep on the Qinghai–Tibetan Plateau and those at relatively low altitudes as well as robust morphology (i.e., good body constitution; Du 2011) in sheep originating from the Taklimakan Desert region.
In this study, we sequenced the whole genomes of 77 sheep (O. aries) including those from habitats in extreme (or harsh) environments: Tibetan areas on the Qinghai–Tibetan Plateau (defined as ‘plateau’, altitude >4,000 m above sea level), high-altitude region (altitude >1,500 m), Taklimakan Desert region (defined as ‘desert’, average annual precipitation <10 mm), and arid zone (<400 mm, representing arid and semi-arid regions; Piao et al. 2010; fig. 1A and B, supplementary tables S1 and S2, Supplementary Material online). The set of samples represented 21 native breeds of different genetic and geographic origins in China (supplementary table S1, Supplementary Material online). Through comparisons of the genomes of sheep from extreme environments with those from contrasting environments, we aimed to identify the candidate genes, functional Gene Ontology (GO) categories and signaling pathways responsible for the rapid adaptations (i.e., over thousands of years) of sheep to plateau and desert environments. In addition, to elucidate the evolutionary history of Chinese native sheep, a comprehensive analysis of the genomic diversity, population structure and demographic history of these animals was performed based on genomic data.
Fig. 1.
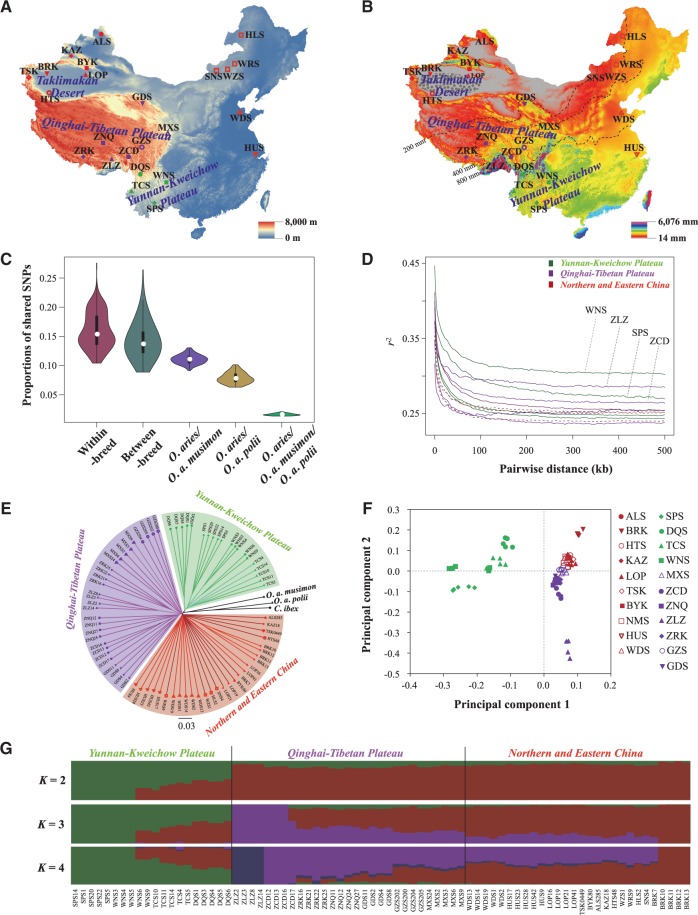
Geographic distribution and population genetics analyses of 21 native sheep breeds. (A) Sampling sites in this study. A total of 77 Chinese sheep representing 21 native breeds were included. The elevation (m) of the study area is also visualized. (B) Geographic variation of the annual mean precipitation (mm). Precipitation data during the 21 years from 1981 to 2001 were retrieved from the Abdus Salam International Center for Theoretical Physics, Italy (ICTP, https://www.ictp.it, last accessed January 5, 2015). The 200, 400, and 800 mm average annual precipitation lines are also visualized. (C) Diagram of the average shared SNPs between O. aries individuals as well as between O. aries individuals and the individuals of the three wild species (O. a. musimon, O. a. polii, and C. ibex). (D) Decay of linkage disequilibrium (LD) in the Chinese native sheep breeds, with one line per breed. (E) NJ tree constructed using p-distances between individuals. The three wild species are used as outgroups. (F) Principal components 1 and 2 for the 77 native sheep. (G) Population genetic structure of the 77 native sheep inferred from the program FRAPPE v1.1. The length of each colored segment represents the proportion of the individual genome inferred from ancestral populations (K = 2–4). See supplementary table S1, Supplementary Material online for the abbreviations of the breeds and individuals.
Results and Discussion
Genomic Variants
We generated the genomic sequences of 77 native sheep and, for genomic comparison and as phylogenetic outgroups, three wild species of the subfamily Caprinae, viz O. a. musimon, Ovis ammon polii and Capra ibex (one animal from each species). This yielded 1,150 Gb of aligned high-quality genomic data at a total of 438× effective sequence depth for the subsequent analyses.
An average of 94.66% (92.14–95.30%) of the SNPs identified in the 77 native sheep were validated in the sheep dbSNP database (supplementary table S3, Supplementary Material online), indicating the high reliability of the called SNP variations in this study (e.g., Park et al. 2015). For the 3,720,550 SNPs not confirmed by dbSNP database, 1,582,868 SNPs were further validated by their presence in 2–77 individuals of the samples. Of the common SNPs for our sequencing data and the Ovine 50K Beadchip array in the 33 native sheep (7,200–15,016 SNPs in each individual), an average 95.51% (89.46–96.21%) of the SNP genotypes were consistent (supplementary table S4 and fig. S1, Supplementary Material online), thereby demonstrating the high quality of our SNP calling. The 4.49% discrepancy was resulted from genotyping errors and missing data in the whole genome sequencing process (supplementary table S5, Supplementary Material online). Moreover, a large number of shared SNPs (15.58–17.48 million) between different sheep groups from different regions (i.e., Qinghai–Tibetan Plateau, Yunnan–Kweichow Plateau, Northern and Eastern China; supplementary table S6, Supplementary Material online) also support for the validity of the SNP calling.
We obtained 21.26 million high-quality SNPs for 77 native sheep, 2.77 million for O. a. musimon, 4.71 million for O. a. polii, and 10.10 million for C. ibex (supplementary table S7, Supplementary Material online). There were 15.81 million high-quality SNPs unique to the native sheep (supplementary table S7, Supplementary Material online). Based on one individual per species, the overall numbers of SNPs per bp in the exonic, intronic, intergenic regions and chromosomes of domestic sheep (averaged over 77 individuals) were higher than those of O. a. musimon, but lower than those of O. a. polii and C. ibex (supplementary table S8, Supplementary Material online). Of the three identified genetic groups of native sheep, Northern and Eastern Chinese breeds possessed the largest amounts of the total (19,227,904) and unique (784,796) SNPs (supplementary table S6 and fig. S2A, Supplementary Material online), indicating their overall relatively high genomic diversity and early population history (Lv et al. 2015). Most of the high-quality SNPs in the individual genome of sheep were located in intergenic regions (2.678 million, 71.83%), with only 0.76% (0.028 million) located in exonic regions (supplementary table S8, Supplementary Material online). A total of 11,167 nonsynonymous SNPs and 16,879 synonymous SNPs were located within exons, which resulted in a nonsynonymous/synonymous ratio of 0.662. This ratio was within the range of ratios (0.40–1.42) for humans and other animals (e.g., mouflon, argali, ibex, and goat; supplementary table S9, Supplementary Material online).
There were 15.98% (10.40–27.56%) of SNPs shared between individuals within breeds, 14.20% (8.86–26.41%) between breeds, 11.08% (9.13–13.05%) for O. aries–O. a. musimon, 7.87% (6.28–10.13%) for O. aries–O. a. polii, and 1.60% (1.32–2.07%) for O. aries–O. a. musimon–O. a. polii (fig. 1C). For the three wild species, the shared proportions of SNPs were 6.93%, 2.65%, 4.28%, and 0.64% for O. a. musimon–O. a. polii, O. a. musimon–C. ibex, O. a. polii–C. ibex and O. a. musimon–O. a. polii-C. ibex, respectively. These statistics showed the proportions of SNPs with a given taxonomic range (i.e., Ovine species, O. aries and breeds) and thus of different origins (i.e., from predating the divergence of ovine species to predomestic, and then to postdating breed divergence). Moreover, O. aries–O. aries showed the largest average number of shared SNPs per pair of individuals, followed by O. aries–O. a. musimon, O. aries–O. a. polii, and O. aries–C. ibex (supplementary table S7, Supplementary Material online). The average numbers of private SNPs per pair of individuals ranged from 2.11 million for O. aries–O. a. musimon to 9.77 million for O. a. musimon–C. ibex (supplementary table S10, Supplementary Material online).
In addition, we identified an average of 264,871 indels (<100 bp) per individual for the native sheep, 192,315 for O. a. musimon, 354,619 for O. a. polii and 755,867 for C. ibex (supplementary table S11, Supplementary Material online). For the two high-coverage samples (ZNQ24 and LOP41), we detected 177,371 copy number variations (CNVs, 200 bp–5 Mb; Freeman et al. 2006) (supplementary table S12, Supplementary Material online) and 156,134 structural variations (SVs, 100 bp–chromosome level; Alkan et al. 2011) which included insertions, deletions, inversions, intra-chromosomal translocations and inter-chromosomal translocations (supplementary table S13, Supplementary Material online). Most of the indels, CNVs and SVs were located in intergenic regions.
Patterns of Genomic Variation and Linkage Disequilibrium
The genome-wide average θπ value for the native sheep was 2.28 × 10−3 (1.9–2.5 × 10−3), which was within the ranges of the parameters for other animals and humans (supplementary table S14, Supplementary Material online). The genomic variation parameters [average θπ value, average number of regions of homozygosity (ROHs) and mean ROH size] for the three sheep groups showed congruent patterns. In other words, the Northern and Eastern Chinese breeds showed the greatest genomic diversity (θπ) and exhibited the fewest ROH number (879) and the smallest average ROH size (171.34 kb), whereas the lowest genomic variations were observed in the breeds from the Yunnan–Kweichow Plateau (supplementary tables S15 and S16 and fig. S2B–G, Supplementary Material online). Inbred individuals were not observed in the 77 native sheep samples according to the IBS score (IBS <0.9). The breeds from the Qinghai–Tibetan Plateau (e.g., ZLZ and ZCD) and the Yunnan–Kweichow Plateau (e.g., WNS and SPS) showed an overall slow decay rate and a high level of LD, whereas the breeds from Northern and Eastern China exhibited a rapid decay rate and a low level of LD (fig. 1D). The differences in the genome-wide LD patterns between breeds likely reflected larger effective population sizes (Ne) for the Northern and Eastern Chinese breeds compared with the other breeds. Overall, the patterns of genomic diversity and LD strongly suggested a northern origin for Chinese sheep, from which Qinghai–Tibetan and Yunnan–Kweichow breeds were subsequently derived. This result is consistent with the migration routes of eastern Eurasian sheep, being from northern China to southern China, which were deduced in our recent mitogenomic study (Lv et al. 2015).
Population Genetic Structure
The neighbor-joining (NJ) tree revealed strong clustering of the native sheep into three genetic groups that presented a low level of genetic differentiation (mean FST = 0.014–0.040; supplementary table S17, Supplementary Material online): Qinghai–Tibetan breeds (n = 29, ZCD, ZNQ, ZRK, ZLZ, GDS, GZS, and MXS), Yunnan–Kweichow breeds (n = 20, TCS, WNS, SPS, and DQS) and Northern and Eastern Chinese breeds (n = 28, BRK, LOP, NMS, HUS, WDS, ALS, BYK, KAZ, HTS, and TSK; fig. 1E, supplementary fig. S3, Supplementary Material online). A principal component analysis (PCA) and a Bayesian model-based clustering analysis provided additional corroborating evidence for these groupings (fig. 1F and G, supplementary figs. S4 and S5, Supplementary Material online). In the clustering analysis, when K = 2, the native sheep were genetically divided into the Yunnan–Kweichow breeds and the remaining breeds; when K = 4, the Northern and Eastern Chinese breeds, the Qinghai–Tibetan breeds and population ZLZ of the Tibetan sheep formed three additional genetic clusters (fig. 1G). We observed a clear signature of genetic admixture between the sheep breeds from the Qinghai–Tibetan Plateau and Northern and Eastern China as well as genetic introgression from Northern and Eastern Chinese breeds into the Yunnan–Kweichow breeds (fig. 1G, supplementary fig. S5, Supplementary Material online). Compared with the traditional view that Chinese native sheep were derived from three old breeds (i.e., Kazakh, Mongolian and Tibetan sheep; Du 2011), our results provided evidence that breeds from the Qinghai–Tibetan Plateau, Yunnan–Kweichow Plateau, and Northern and Eastern China are genetically distinct (fig. 1G). This finding is consistent with a recent study on 10 representative Chinese sheep breeds genotyped using the 50K SNP Beadchip (Wei et al. 2015).
Demographic History and Population Admixture
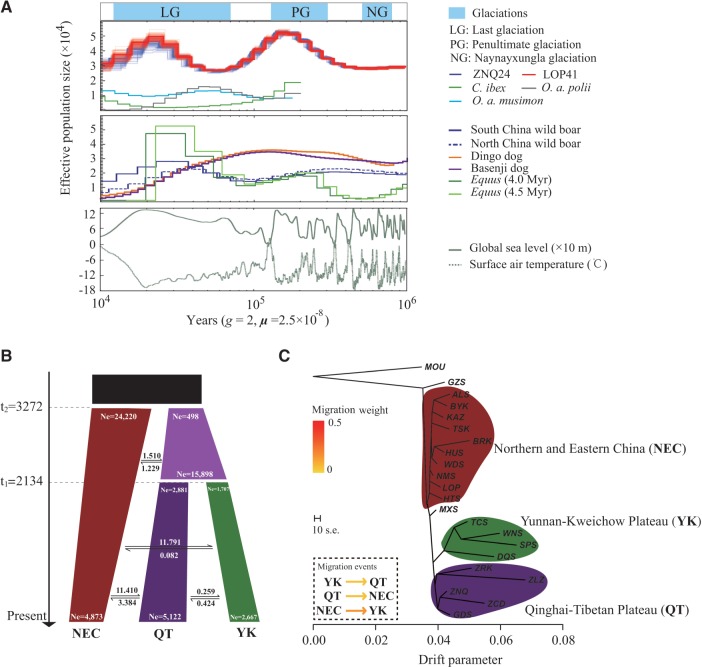
The two high-coverage samples (ZNQ24 and LOP41) exhibited concordant demographic trajectories, with two apparent expansions and two severe bottlenecks that mirrored the glacial cycles and sea level fluctuations that affected the areas (fig. 2A). The first bottleneck occurred ∼0.6 Mya (million years ago; fig. 2A) and is consistent with the Naynayxungla glaciation (0.78–0.50 Mya), which was the most extensive glaciation during the Quaternary Period (Zheng et al. 2002; Lehmkuhl and Owen 2005; Ehlers and Gibbard 2007). At that time, the sea level frequently fluctuated and was maintained at an overall low level (fig. 2A). After the retreat of the Naynayxungla glaciation, the ancestral sheep population expanded and reached a pinnacle (∼0.18 Mya; fig. 2A) during the Penultimate glaciation (0.30–0.13 Mya; Zheng et al. 2002; Lehmkuhl and Owen 2005; Ehlers and Gibbard 2007). The cold-climate interval and rising sea level at this stage could have contributed to a population expansion because an increase in grassland was likely under such environmental conditions (Lorenzen et al. 2011). The second bottleneck (∼70 kya; fig. 2A) was detected towards the end of the interglacial period (0.13–0.07 Mya; Zheng et al. 2002; Lehmkuhl and Owen 2005; Ehlers and Gibbard 2007), which presented environmental conditions similar to that of the present (Orlando et al. 2013). The ancestral sheep population size then peaked again at ∼15 kya during the Last Glacial Maximum (LGM) because the glaciations were less extensive and the grasslands had expanded (Zheng et al. 2002; Lehmkuhl and Owen 2005; Ehlers and Gibbard 2007; fig. 2A). Subsequently, the population size gradually decreased to a small number (fig. 2A), which was most likely the result of climate warming and associated grassland contractions after the LGM (Lorenzen et al. 2011). Despite relatively low estimates of Ne, the pairwise sequentially Markovian coalescent (PSMC) results for the low-coverage samples (supplementary fig. S6, Supplementary Material online) showed demographic trends that were consistent with those inferred from the two high-coverage samples (fig. 2A). In general, our findings are consistent with previous results, indicating historical population expansion events for Chinese sheep as inferred from mtDNA sequence variability (Wang et al. 2007; Zhao et al. 2013). Compared with other animals (e.g., horse, Orlando et al. 2013; wild boar, Li et al. 2013; dog, Freedman et al. 2014; mouflon, argali, and ibex in this study), the demographic trend of sheep is most similar to horse (e.g., close timings of peaks and bottlenecks; fig. 2A) and mouflon (supplementary fig. S6, Supplementary Material online). These similar demographic trends might have been attributed to the similar habitat requirements for these herbivores. The differences in demographic shapes such as the extent of the changes in effective population sizes and timing of expansion/contraction were observed between sheep and the two wild Caprinae species (i.e., argali and ibex; supplementary fig. S6, Supplementary Material online). This may result from smaller Ne and higher susceptibility to environmental changes (e.g., habitat fragmentation) of the wild herbivores compared with sheep and various environmental fluctuations in geographic regions where they inhabited.
Fig. 2.
Demographic history of the sheep population. (A) PSMC analysis results for the representative individuals sequenced at a high read coverage (∼42×) exhibit inferred variations in Ne over the last 106 years. The Ne of mouflon, argali, ibex, dog, horse, and wild boar are also rescaled. The glaciation periods, atmospheric surface air temperature (°C) and global relative sea level data over the last 106 years are included. (B) ∂a∂i analysis showing the demographic history of Chinese native sheep from ∼4,000 years ago to the present. Two population divergences occurred at 3,272 (t1) and 2,134 (t2) years ago. The average number of migrants per year between groups is shown between the black arrows. (C) Phylogenetic network of the inferred relationships among the 21 native breeds with three inter-group migration edges. The colored regions in the phylogenetic tree represent three inferred genetic groups. Arrows indicate migration events, and a spectrum of heat colors indicate the migration weights of the migration events. The scale bar shows 10× the average standard error of the entries in the sample covariance matrix. NEC, Northern and Eastern Chinese breeds; QT, Qinghai–Tibetan breeds; YK, Yunnan–Kweichow breeds. For the abbreviations of the breeds, see supplementary table S1, Supplementary Material online.
Among the five diffusion approximation for demographic inference (∂a∂i) models tested, model 3 achieved a maximum log-likelihood value of −16643 and was selected as the optimal model (supplementary table S18, Supplementary Material online). This model showed that Northern and Eastern Chinese breeds diverged from the remaining breeds ∼3,200 years ago, with an estimated Ne of 24,220 for Northern and Eastern Chinese breeds and 498 for the other breeds (fig. 2B, supplementary table S19, Supplementary Material online). Approximately 2,100 years ago, breeds in areas other than Northern and Eastern China further diverged into the Qinghai–Tibetan breeds and Yunnan–Kweichow breeds, both of which have gradually increased and exhibited estimated Ne values from 2,881 to 5,122 and 1,707 to 2,667, respectively (fig. 2B, supplementary table S19, Supplementary Material online). The current Ne value for Northern and Eastern Chinese breeds is 4,873, which has obviously decreased since their initial appearance (24,220; fig. 2B, supplementary table S19, Supplementary Material online). An overall low to moderate level of gene flow (0.082–11.791 migrants per year) was detected between the sheep groups (fig. 2B, supplementary table S19, Supplementary Material online).
The maximum-likelihood (ML) tree without migration events inferred from the TreeMix analysis divided the 77 native sheep into three clusters (fig. 2C), which are similar to the population structuring patterns identified from the phylogenetic tree, PCA and genetic structure analysis (fig. 1E–G). When potential migration edges (i.e., migration events between the branches) were added to the ML tree, strong migration events were detected among the three clusters. Up to 98.77% of the variance between breeds was explained by a model with five migration events (supplementary fig. S7, Supplementary Material online). In the model, we observed three migration edges among clusters from Northern and Eastern China to the Yunnan–Kweichow Plateau, from the Yunnan–Kweichow Plateau to the Qinghai–Tibetan Plateau and from the Qinghai–Tibetan Plateau to Northern and Eastern China (fig. 2C). This result is consistent with the inter-group gene flow found in the ∂a∂i analysis and the genetic admixture between sheep groups observed in the genetic structure analysis.
Genome-Wide Selective Sweep Test
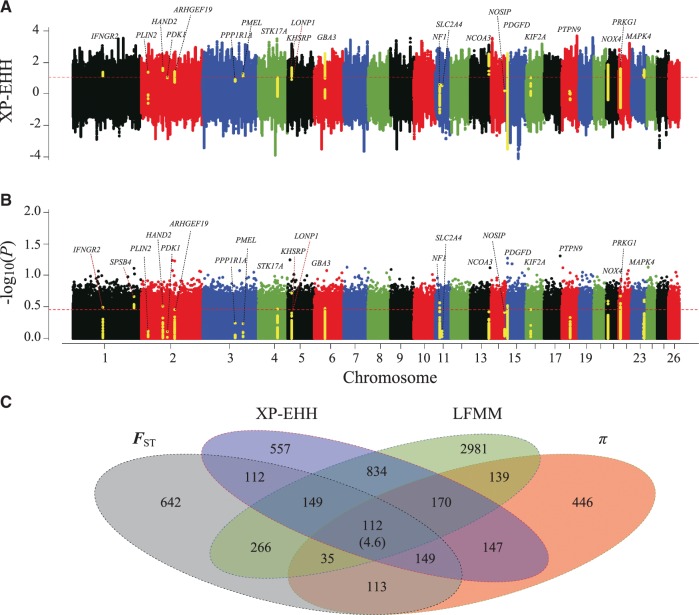
Using the top 5% of FST values and θπ ratio cutoffs (Z(FST) > 1.834, 1.859, 1.731, and 1.823 and log2(θπ ratio) > 0.352, 0.277, 0.247 and 0.198 for the Control group (HUS and WDS from Eastern China; see supplementary table S1, Supplementary Material online)/Tibetan group, Control group/Taklimakan Desert group, Low-altitude group/High-altitude group and Humid group/Arid group, respectively; figs. 3A and 4A), we identified 731 candidate genes associated with plateau adaptations (>4,000 m; supplementary table S20, Supplementary Material online), 452 candidate genes associated with desert adaptations (average annual precipitation <10 mm; supplementary table S21, Supplementary Material online), 465 candidate genes involved in high-altitude adaptations (>1,500 m; supplementary table S22, Supplementary Material online) and 603 candidate genes involved in arid adaptations (average annual precipitation <400 mm; supplementary table S23, Supplementary Material online) for the native sheep. In addition, 261 (top 5% outliers, XP-EHH value >1.076) and 145 genes (top 5% outliers, XP-EHH value >0.897) were positively selected in XP-EHH analysis, and 147 (top 1% outliers, −log10(P) >0.433) and 95 genes (top 1% outliers, −log10(P) >0.456) were positively selected in LFMM analysis regarding the plateau and desert environments, respectively. Overall, 112 and 75 genes were positively selected across all the four methods under the plateau and desert environments, respectively (figs. 5 and 6, supplementary tables S20 and S21, Supplementary Material online).
Fig. 3.
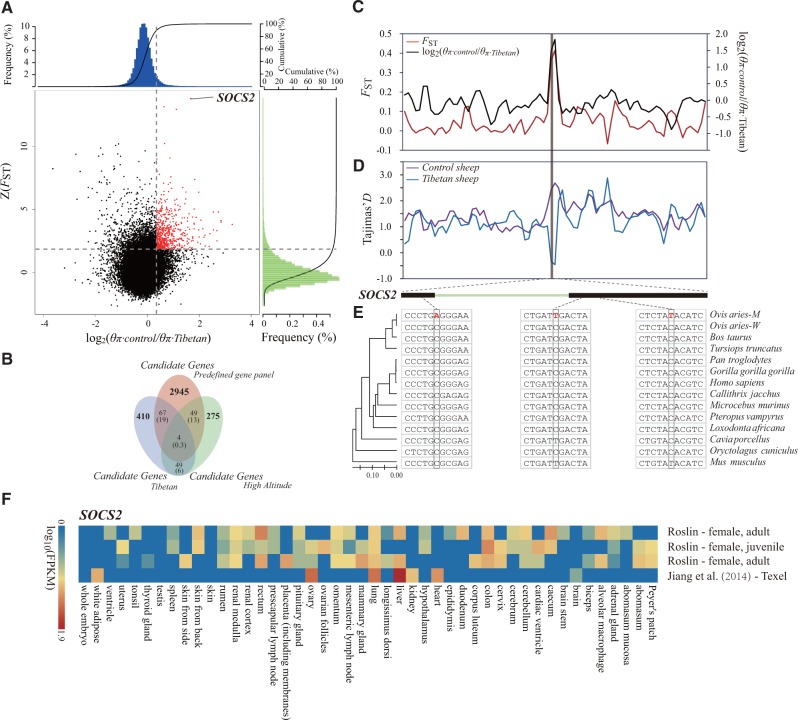
Genomic regions with strong selective signals in Tibetan sheep. (A) Distribution of log2(θπ ratios) and Z(FST) values calculated in 100-kb sliding windows with 50-kb increments between Tibetan group (including breeds ZNQ, ZCD, and ZRK from the plateau environment) and control group (including breeds HUS and WDS from East China). The data points in red (corresponding to the top 5% of the empirical log2(θπ ratios) ratio distribution with values >0.35 and the top 5% of the empirical Z(FST) distribution with values >1.83) are genomic regions under selection in Tibetan sheep. (B) Comparison between the overlap of candidate genes and the overlap expected by chance. Numbers in the intersection regions are the observed overlapping genes among the candidate genes in Tibetan sheep, sheep breeds from the high-altitude regions and the predefined gene panel (i.e., the previously published candidate genes in other mammalian species under the high-altitude environment, including human, dog, wolf, yak, pig, and Tibetan antelope). Numbers in parentheses show the number of genes expected by chance. The total numbers of genes for the sheep and the gene panel involved in the test are 18,013 and 65,029, respectively. (C) log2(θπ ratios) and FST values around the SOCS2 locus. The black and red lines represent the log2(θπ ratios) and FST values, respectively. (D) Tajima’s D values around the SOCS2 locus. The blue and purple lines represent the Tibetan sheep and control sheep, respectively. (E) Evolutionary analysis of the SOCS2 gene. The inter-species NJ tree is derived from the 12 vertebrate orthologous sequences, and the mutations are marked in red. O. aries M., namely O. arise mutant, represents Tibetan sheep in which the mutations were observed. O. aries W., namely O. aries wild, refers to other sheep breeds in which the nucleotides were conserved. (F) Gene expression of SOCS2 in different sheep tissues is based on four different experiments deposited in the EBI Gene Expression Atlas database. The FPKM (fragments per kilobase of transcript per million mapped reads) value is used to measure the expression level.
Fig. 4.
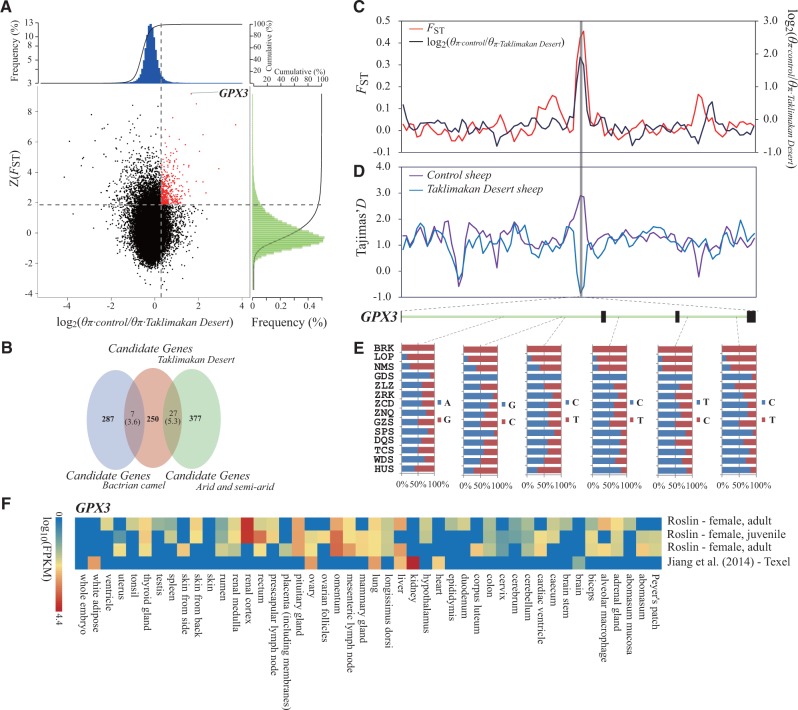
Genomic regions with strong selective signals in sheep breeds from the Taklimakan Desert region. (A) Distribution of log2(θπ ratios) and Z(FST) values calculated in 100-kb sliding windows with 50-kb increments between Taklimakan Desert group (including breeds LOP and BRK from the desert environment) and control group (including breeds HUS and WDS from East China). The data points in red (corresponding to the top 5% of empirical log2(θπ ratios) ratio distribution with values >0.28 and the top 5% of empirical Z(FST) distribution with values >1.86) are genomic regions under selection in the sheep breeds from the Taklimakan Desert region. (B) Comparison between the overlap of candidate genes and the overlap expected by chance. Numbers in the intersection regions are the observed overlapping genes among the candidate genes in sheep breeds from the Taklimakan Desert region, sheep breeds from the arid regions and the predefined gene panel (i.e., the previously published candidate genes in other mammalian species under the arid environment, including Bactrian camel). Numbers in parentheses show the number of genes expected by chance. The total numbers of genes for sheep and Bactrian camel involved in the test are 18,013 and 20,251, respectively. (C) log2(θπ ratios) and FST values around the GPX3 locus. The black and red lines represent log2(θπ ratios) and FST values, respectively. (D) Tajima’s D values around the GPX3 locus. The blue and purple lines represent the Taklimakan Desert sheep and control sheep, respectively. (E) Allele frequencies of six SNPs within the GPX3 gene across Chinese native sheep breeds. The desert breeds include BRK and LOP, whereas the non-desert breeds comprise all other breeds. (F) Gene expression of GPX3 in different sheep tissues is based on four different experiments deposited in the EBI Gene Expression Atlas database. The FPKM (fragments per kilobase of transcript per million mapped reads) value is used to measure the expression level. For the abbreviations of the breeds, see supplementary table S1, Supplementary Material online.
Fig. 5.
Analysis of the signatures of positive selection in the genome of Tibetan sheep. Genomic landscape of the (A) XP-EHH values and (B) P-values in the LFMM analysis in the genome of Tibetan sheep. The genes visualized in (A) and (B) are the candidate genes from the signaling pathways of Tibetan sheep in fig. 7A. (C) Number of candidate genes identified in Tibetan sheep by the four methods listed in each of the Venn diagram components. Numbers in parentheses show the number of genes expected by chance.
Fig. 6.
Analysis of the signatures of positive selection in the genome of sheep breeds from the Taklimakan Desert region. Genomic landscape of the (A) XP-EHH values and (B) P-values in the LFMM analysis in the genome of Taklimakan Desert sheep. The genes visualized in (A) and (B) are the candidate genes from the signaling pathways of Taklimakan Desert sheep in fig. 7B. (C) Number of candidate genes identified in Taklimakan Desert sheep by the four methods listed in each of the Venn diagram components. Numbers in parentheses show the number of genes expected by chance.
The phylogenetic tree reconstructed based on the SNPs of the candidate genes of Tibetan sheep showed that most of the 77 native sheep (71/77, 92.21%) were correctly assigned into separated clades between the high-altitude (i.e., the Qinghai–Tibetan Plateau and Yunnan–Kweichow Plateau) and low-altitude regions (supplementary fig. S8A, Supplementary Material online). Similarly, the phylogenetic tree that included the SNPs of the candidate genes of breeds from the Taklimakan Desert region largely divided the 77 sheep (74/77, 96.10%) into two clades between arid (i.e., average annual precipitation <200 mm) and non-arid regions (supplementary fig. S8B, Supplementary Material online). This topological pattern was different from that inferred from the genome-wide SNPs (fig. 1E), which supported the credibility of the candidate genes under selection. Regarding the large-effect SNP analyses of the two categories of candidate genes, we observed significantly (P < 0.05) higher frequency differences between the contrasting groups than between the other pairwise groups (supplementary fig. S9, Supplementary Material online). Moreover, the Z(FST) values (supplementary fig. S10, Supplementary Material online) and log2(θπ ratio) values (supplementary fig. S11, Supplementary Material online) were much higher for the selected genomic regions of the sheep from the plateau region, desert region, high-altitude area and arid zone than the values at the whole-genome level. Compared with the overlap expected from chance, there was a significant excess of overlapping selective signals shared between the candidate genes identified here and the predefined gene panel (i.e., the previously published candidate genes in other mammalian species under similar extreme environments; figs. 3B and 4B). Similarly, the number of overlapping candidate genes among the FST, θπ ratio, XP-EHH, and LFMM analyses significantly exceeded those expected by chance (figs. 5C and 6C). These results provided further support that the candidate genes were reliable.
Adaptive Mechanisms in Plateau Environments
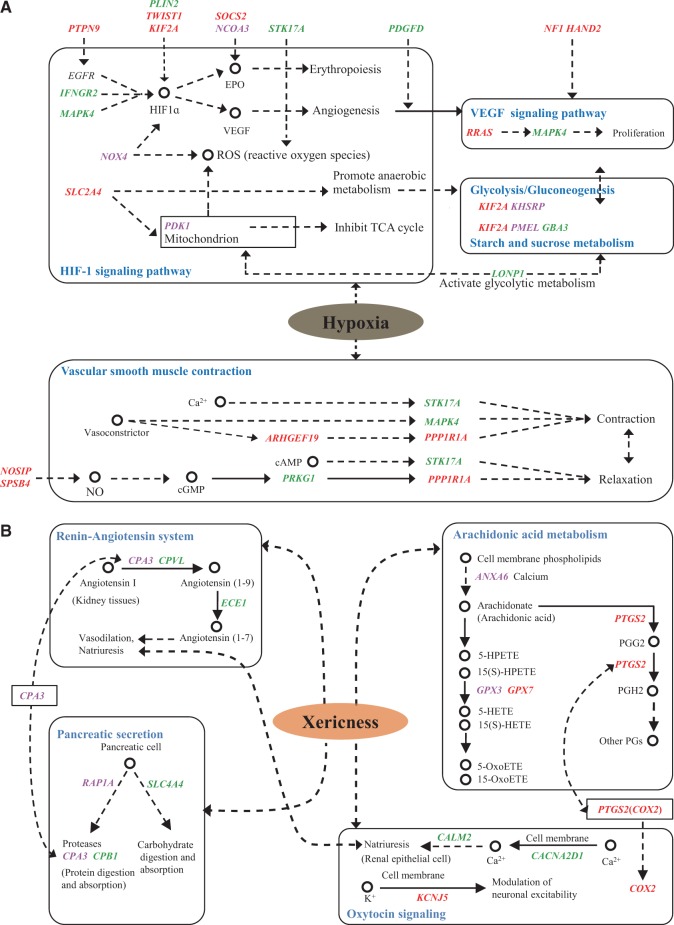
Among the candidate genes for the plateau adaptations, five (IFNGR2, MAPK4, NOX4, SLC2A4, and PDK1) were located in the classical HIF-1 (hypoxia-induced factors) pathway, which plays a central role in regulating cellular responses to hypoxia (Frede and Fandrey 2013); four were found in the corresponding downstream vascular endothelial growth factor (VEGF; RRAS and MAPK4) and glycolysis/gluconeogenesis (KIF2A and KHSRP) pathways; and five (ARHGEF19, STK17A, MAPK4, PPP1R1A and PRKG1) were found in the vascular smooth muscle contraction (VSMC) pathway, which adjusts the diameter of blood vessels and the delivery of blood oxygen (fig. 7A). We also found nine positively selected genes (PLIN2, TWIST1, KIF2A, PTPN9, SOCS2, NCOA3, STK17A, PDGFD, and LONP1) regulating specific genes in the HIF-1 pathway, two genes (NF1 and HAND2) affecting the VEGF signaling pathway, one gene (LONP1) influencing the glycolysis/gluconeogenesis pathway and two genes (NOSIP and SPSB4) regulating the nitric oxide (NO) compound in the VSMC pathway (fig. 7A). The network of these relevant pathways indicated that hypoxia-induced factors, angiogenesis, vasodilatation and glycolysis metabolism are the most important factors that allow sheep to manage extreme hypoxic environmental pressure. Furthermore, the functions of the proposed pathway genes are related to the regulation of the cardiovascular system and energy metabolism under hypoxic conditions in human and murine cells (e.g., Mora et al. 2003; Tang et al. 2003; Kim et al. 2006; Jian et al. 2010; Sawada et al. 2012; Pinti et al. 2015). For a detailed description of the relevant functions of the pathway candidate genes, please see supplementary information, Supplementary Material online.
Fig. 7.
Schematic mechanisms of signaling pathways for genetic adaptations to extreme environments in sheep. (A) Functionally important pathways and associated candidate genes for the genetic adaptations of Tibetan sheep to the plateau environment. (B) Functionally important pathways and associated candidate genes for the genetic adaptations of sheep breeds from the Taklimakan Desert region to the desert environment. The names of the KEGG pathways are shown in blue. The candidate genes positively selected in the two methods of FST and θπ ratio tests are shown in red, in the three methods of FST, θπ ratio, and XP-EHH tests or FST, θπ ratio, and LFMM tests are shown in green, and in all of the four selection tests are shown in purple. Dotted arrows indicate an indirect effect.
In addition, six candidate genes (FSTL1, PVR, EXT2, ALT4, SOX6, and HAND2) ranking within the top 20 Z(FST) values and two additional candidate genes (PDGFD and BMPR2; supplementary table S20, Supplementary Material online) were functionally involved in bone, muscle, craniofacial, limb, skin, and embryonic development in vertebrates based on the NCBI annotations and Ensembl databases as well as previous functional studies in humans, mice and zebrafish (e.g., Norton et al. 2005; Kayserili et al. 2009; Geng et al. 2011; Long et al. 2015; supplementary information, Supplementary Material online). These findings indicated that these genes may have played a dominant role in regulating body morphology in Tibetan sheep, suggesting that selection on morphology is an important adaptive mechanism under the plateau environment, which is consistent with the small body size observed in Tibetan sheep (Du 2011; supplementary fig. S12, Supplementary Material online).
Notably, we observed markedly higher Z(FST) and log2(θπ ratio) values (fig. 3C) but lower Tajima’s D values (fig. 3D) for the target gene SOCS2 compared with those in the adjacent genomic regions, indicating that a strong selective sweep occurred in this gene. SOCS2 encodes a member of the suppressor of cytokine signaling (SOCS) family. The SOCS family proteins form part of a classical negative feedback system that regulates cytokine signal transduction. An important paralog of the SOCS2 gene, SOCS6, closely interacts with the EPO gene, which regulates erythrocyte differentiation and plays an important role in the classic HIF-1 pathway (Stopka et al. 1998). Mutations in SOCS2 can cause primary polycythemia, essential thrombocythemia, and cardiovascular diseases (Rico-Bautista et al. 2006). Moreover, SOCS2 is one of the main regulators of GH/IGF-1 signaling and has an important role in the regulation of growth and metabolism (Turnley 2005). The inter-species NJ tree of SOCS2 sequences revealed that three evolutionarily conserved nucleotides have mutated in Tibetan sheep but remained invariant in other sheep breeds and species (fig. 3E). The expression levels of SOCS2 in sheep were significantly higher in the liver, a primary organ for energy metabolism, than in other organs (fig. 3F). In ruminants, the liver is mainly involved in gluconeogenesis metabolism, which uses propionate (a volatile fatty acid) as the source of carbon (Jiang et al. 2014). This may reflect the importance of glycolysis and volatile fatty acids in the adaptation of sheep to plateau environments. Overall, our results support that SOCS2 and its specific mutations are important for the responses of Tibetan sheep to extreme high-altitude stress through their regulation of the EPO gene in the HIF-1 pathway and contribution towards more efficient energy metabolism.
We also found 22 significantly over-represented GO categories (binomial distribution test, P < 0.05) for Tibetan sheep (supplementary table S24, Supplementary Material online). The GO clusters were primarily associated with metabolic processes (triacylglycerol, fatty acid, purine nucleobase, and amino acid), oxidative phosphorylation, respiratory electron transport chain and response to stimulus. These functional clusters are biologically relevant to the plateau adaptations because they are involved in energy metabolism, oxidation reaction, and stress–response, which are important regulating factors for animals to respond to extreme hypoxic environments.
Our integrated analyses provide the first evidence of hypoxia responses, energy metabolism, and body size changes for sheep living at high plateaus. We detected several positively selected genes within or regulating the HIF-1 pathway. These genes may account for the significant differences in hematologic parameters between Tibetan sheep and low-altitude breeds (Jiang et al. 1991; Liu and Chen 2006). In addition, we found that the VEGF pathway downstream of HIF-1 and the VSMC pathway also significantly contributed to the plateau adaptations. The multiple pathways involved suggest a complex genetic mechanism underlying hypoxia responses in Tibetan sheep. In addition, we found that glycolysis is an important mechanism of energy metabolism for sheep under extreme hypoxic conditions, which is consistent with previous findings in the Tibetan antelope (Ge et al. 2013). Also, lipid metabolism is a common mechanism for energy metabolism as observed in the over-represented GO terms from our analyses of the high-altitude samples as well as from the analyses of other species at high altitudes (Qiu et al. 2012; Cho et al. 2013; Qu et al. 2013). Coupled with evidence from Tibetan antelope (Ge et al. 2013), yak (Qiu et al. 2012; Zhang et al. 2016), and climate-mediated selection in sheep (Lv et al. 2014), our results suggest that energy metabolism is a possible adaptive mechanism for ruminants that inhabit the plateau environments. Consistent with the development-related genes identified here, the small body size of Tibetan sheep (Du 2011; supplementary fig. S12, Supplementary Material online) is not unexpected and indicates the importance of body size variations in the plateau adaptation. This result also provides additional evidence for the primary role of energy metabolism in the adaptation of sheep to the plateau environments because small body sizes require less energy consumption and represent an advantageous phenotype at high elevations.
Adaptive Mechanisms in Desert Environments
Among the candidate genes for desert environment adaptations, four (ANXA6, GPX3, GPX7, and PTGS2) were located in the arachidonic acid metabolism pathway, three (CPA3, CPVL, and ECE1) were involved in the renin–angiotensin system pathway and four (CALM2, CACNA2D1, KCNJ5, and COX2) were located in the oxytocin signaling pathway (fig. 7B), and they were all functionally related to regulating water retention and reabsorption in renal cells and blood vessels in the kidney. In addition, we detected four genes (RAP1A, SLC4A4, CPA3, and CPB1) that belonged to the pancreatic secretion pathway and presented protein and carbohydrate digestion and absorption functions (fig. 7B). Because of the extreme xericity, the saline–alkaline soil and the poor food resources in the focus environment, it is reasonable that these pathways are functionally important for the adaptations of sheep to desert environment stress. Moreover, the functions of the pathway candidate genes are also explicitly related to desert environment adaptations, such as renal vasodilation, ion transmembrane transport, water–salt metabolism and bicarbonate absorption (e.g., Maquire et al. 1997; Breyer and Breyer 2000; Gatalica et al. 2008; supplementary information, Supplementary Material online).
The target gene GPX3 has the largest Z(FST) value in the Taklimakan Desert group. This gene exhibited much higher Z(FST) and log2(θπ ratio) values (fig. 4C) but lower Tajima’s D values (fig. 4D) than adjacent genomic regions, indicating a strong selective sweep. GPX3 belongs to the glutathione peroxidase family, which is responsible for the detoxification of hydrogen peroxide (Esworthy et al. 1991). Remarkably, GPX3 is located in the arachidonic acid metabolism pathway, one of the enriched pathways in their functional overrepresentation analysis (KEGG accession code 00590, P < 0.05), and plays an important role in converting arachidonic acid into 19(S)-HETE. Because 19(S)-HETE efficiently dilates renal preglomerular vessels and stimulates water reabsorption (Carroll et al. 1996; Jirimutu et al. 2012), it is reasonable to assume that GPX3 is important for the survival of sheep in desert environments. This finding is consistent with the results of a previous genomic study on Bactrian camel (Jirimutu et al. 2012), in which the arachidonic acid metabolism pathway was reported to be an important molecular mechanism for desert adaptations, indicating the occurrence of convergent evolution between sheep and Bactrian camel inhabiting similar desert environments. Within the GPX3 gene, specific allele of six SNPs in breeds from the Taklimakan Desert region (i.e., BRK and LOP) had significantly higher frequencies than those observed in the other breeds (fig. 4E), thereby suggesting the importance of specific advantageous alleles for sheep living in desert environments. In addition, the expression levels of GPX3 in the sheep and human tissues were significantly higher in the kidney and renal cortex (fig. 4F, supplementary fig. S13, Supplementary Material online), the primary organs for water retention and reabsorption, than in other organs, implying that GPX3 may play a central role in desert environment adaptations.
We also identified 56 significantly over-represented GO categories (binomial distribution test, P < 0.05) for sheep breeds from the desert environments of the Taklimakan Desert region (supplementary table S25, Supplementary Material online). The GO clusters were primarily enriched in the categories of organ (e.g., muscle organ) and system (e.g., nervous system) development, metabolic process, and response to stimulus, indicating the importance of body size, energy optimization, and stress–response for sheep survival in desert environments. In fact, the functions of these GO terms are quite consistent with the small body size and high saline-alkali tolerance (Du 2011) observed in sheep breeds from the Taklimakan Desert region (e.g., BRK and LOP; supplementary fig. S12, Supplementary Material online).
Adaptations to High-Altitude and Arid Environments
The functional enrichment analysis identified 24 and 67 significantly over-represented GO categories (binomial distribution test, P < 0.05) for high-altitude (supplementary table S26, Supplementary Material online) and arid (supplementary table S27, Supplementary Material online) environment adaptations, respectively. The GO clusters were primarily associated with metabolism, catalytic activity and response to stimulus in both environments (supplementary table S28, Supplementary Material online). Although the specific metabolic mechanisms were not similar, for example, triacylglycerol/fatty acid/purine metabolism for the high-altitude adaptations but RNA-related processes for the arid adaptations (supplementary table S28, Supplementary Material online), the contribution of metabolism for sheep survival in the high-altitude and arid environments was supported by at least two lines of evidence. First, environmental factors (e.g., climate) can have a direct effect on herbivores (e.g., sheep), such as their effect on thermoregulation (Parker and Robbins 1985), whereas plant quality and biomass are expected to have indirect effects on herbivores (Mysterud et al. 2001, 2008). Indeed, a recent genomic study demonstrated the considerable impact of climate on the energy metabolic adaptations across sheep breeds (Lv et al. 2014). In humans and Drosophila melanogaster, several studies reported a correlation between energy metabolism and climate and latitude (Oakeshott et al. 1988; Sezgin et al. 2004; Hancock et al. 2011). Second, in the high-altitude adaptations, the candidate genes SLC10A5 and GPCPD1 play potential roles in the metabolism of lipids and the lipoprotein pathway based on the Reactome database. Also, ENPP2 has a relevant function in the starch and sucrose metabolism pathway, and NAT8 is located in the glyceride metabolism pathway according to the KEGG database. Moreover, ME1 is functionally involved in the PPAR pathway (KEGG), which activates a variety of downstream lipid metabolism and energy metabolism pathways. Similarly, in the arid adaptations, the candidate gene AKR1A1 is involved in glyceride metabolism and glycolysis pathways (KEGG).
In addition, based on the annotations in the NCBI and Ensembl databases, certain positively selected genes that are functionally relevant to a specific environment (i.e., STK17A, EIF2AK3, TMTC2, PTPN9, and MREG in high-altitude environments; RAB11FIP2, CPVL, and MFSD6 in arid environments) could also facilitate high-altitude and arid adaptive processes (supplementary information, Supplementary Material online).
Methodological Considerations
The strategy of combining alternative statistical approaches to detect selective signatures can provide sound results through decreasing the false-positive rates (Oleksyk et al. 2010; Frichot et al. 2013). In this study, we combine the FST, θπ ratio, XP-EHH, and LFMM methods for robust selection results. The number of overlapping candidate genes among different methods is significantly higher than those expected by chance (figs. 5C and 6C). Although demographic factors, crossbreeding, and artificial selection may potentially obscure genomic patterns of selection, we believe that they would be minimal in our study for the following reasons. First, we evaluated the genetic attributes of the candidate genes relative to the genomic background. It is clear that the candidate genes are much more strongly differentiated (supplementary fig. S10, Supplementary Material online) than those at the genome-wide scale. Also, the topology of the phylogenetic trees based on the candidate genes (supplementary fig. S8, Supplementary Material online) is distinct from that inferred from the whole genome data (fig. 1E) and the scenario of best-supported demographic model (fig. 2B). These two lines of evidence indicate that the selective signatures identified here are likely to reflect selection rather than demographic changes which should have impacted across the whole genome. Second, as the three wild species were well located in separate clades from the native sheep in the phylogenetic tree (fig. 1E) and no migration was detected between them (fig. 2C), outcrossing seems impossible to distort the genomic selective sweep results. Similarly, crossbreeding is not likely to impact the selective signals because only very low migration rates were observed between different groups (fig. 2B and C, supplementary fig. S7, Supplementary Material online). Third, artificial selection would impose a minimal effect, if any, on the selective signatures since the samples used in this study are all from old and autochthonous breeds, which have inhabited local environments for several thousands of years. Furthermore, only a small number of candidate genes under the extreme environments were overlapped with the 416 published candidate genes under artificial selection in sheep (supplementary table S29, Supplementary Material online), and none of the overlapped candidate genes was located in the proposed signaling pathways. Also, the selective regions identified under each environment were only overlapped with two out of the 31 artificial-related sheep genomic regions reported in Kijas et al. (2012). Taken together, these results support the hypothesis that human-mediated artificial selection has had only a slight effect, if any, on the candidate genes reported here.
Conclusions
In conclusion, comparisons of the whole genomes of native sheep from extreme environments and contrasting reference conditions revealed a variety of novel genes, important pathways and GO categories associated with local adaptations of sheep in plateau and desert environments. Specifically, the candidate genes, pathways and GO terms were functionally related to hypoxia responses in the plateau environment, water reabsorption in the desert environment, and energy metabolism and body size in both environments. These results advance our understanding of the genetic mechanisms underlying the rapid adaptations of sheep and other livestock species, particularly small ruminants to survive in similar extreme environments. The population genomic analyses of 77 Chinese native sheep and three wild species provided new insights into sheep domestication, evolution, and demographic history. In particular, we found strong genomic evidence for the partitioning of Chinese native sheep into three genetic groups (Qinghai–Tibetan, Yunnan–Kweichow, and Northern and Eastern Chinese breeds) as well as for their divergence and gene flow. We also detected climate-driven population size fluctuations of ancestral sheep population over the past million years. The genomic data generated in this study will serve as a valuable resource for genomics-assisted breeding to develop new tolerant sheep breeds in the face of global climate change.
Materials and Methods
Detailed descriptions of samples and methods are provided in supplementary information, Supplementary Material online.
Sample Preparation and Sequencing
We sampled 77 Chinese native sheep (O. aries) that include 21 representative breeds adapted to various local environments, such as extreme environments from Tibetan area on the Qinghai–Tibetan Plateau (defined as ‘plateau’, altitude >4,000 m), high-altitude region (altitude >1,500 m), Taklimakan Desert region (defined as ‘desert’, average annual precipitation <10 mm) and arid zone (average annual precipitation <400 mm, representing arid and semi-arid regions; Piao et al. 2010), as well as contrasting environments in Eastern China (altitude <100 m and average annual precipitation >600 mm), low-altitude region (altitude <1,300 m) and humid zone (average annual precipitation >400 mm, representing humid and semi-humid regions; Piao et al. 2010; fig. 1A and B, supplementary tables S1 and S2, Supplementary Material online). The environments encompassed here represent the typical environments that sheep inhabit. In addition, we also collected three wild species of the subfamily Caprinae, viz O. aries musimon from the Island of Sappi, Finland (descendant from the population in the Mediterranean Island of Sardinia), O. a. polii and C. ibex from Kashgar, Xinjiang, China (one animal from each species; supplementary table S1, Supplementary Material online). We sequenced genomes of the 80 samples using next-generation sequencing technology on an Illumina HiSeq 2000 platform (Illumina, San Diego, CA, USA). High-quality reads were aligned against the reference sheep genome assembly Oar_v3.1.75 using Burrows-Wheeler Aligner (BWA; Li and Durbin 2009) software. The program SAMtools v0.1.19 (Li et al. 2009) was used to identify SNPs and short insertions and deletions (indels, length <100 bp). The high-quality SNPs obtained here were subsequently regarded as the ‘called high-quality SNPs’ and used in the SNP summary, regions of homozygosity, linkage disequilibrium, selective sweep, GO and target gene analyses. The overall analysis pipeline is detailed in supplementary figure S14, Supplementary Material online.
Validation of the Called SNPs
To examine the accuracy of our SNPs, we first compared the SNPs identified here with those from Build 143 of the sheep dbSNP database in the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/SNP, last accessed November 12, 2015). We then compared the genotypes of the called SNPs with those on the Ovine 50K BeadChip array (Illumina, San Diego, CA) for 33 native sheep with available chip data. The chip-based SNPs were filtered using the same criteria as those adopted in this study, and genomic locations of the SNPs were adjusted according to the sheep reference genome assembly Oar_v3.1.75.
Analyses of the ROHs, Inbreeding, and LD
We measured the regions of homozygosity (ROHs) for each breed/group using the ‘runs of homozygosity’ function in the program PLINK v1.07 (Purcell et al. 2007). We estimated the value of identity-by-state (IBS) for all samples using the program PLINK v1.07 (Purcell et al. 2007). Based on the non-inbred individuals (IBS < 0.9), we calculated the LD pattern for different breeds via the squared correlation coefficient (r2) between pairwise SNPs using the software Haploview v4.2 (Barrett et al. 2005).
Population Genetics Analysis
We conducted genetic diversity, population structure, and demographic history analyses using the genotype likelihoods (GL) and associated SNP calls calculated from the program ANGSD v.0.902 (Analysis of Next Generation Sequencing Data; Korneliussen et al. 2014). This strategy accounts for genotype uncertainty by using GL and an ML estimate of the site frequency spectrum (SFS; Nielsen et al. 2012) as a prior information and thus can facilitate unbiased interpretations of low-coverage data. The pairwise nucleotide diversity (θπ) for each sheep breed/group was estimated using a sliding-window approach (100-kb windows with 50-kb increments). An individual-based NJ tree was constructed for the 77 native sheep based on the p-distance, with either one outgroup (i.e., O. a. musimon) or three outgroups (i.e., O. a. musimon, O. a. polii, and C. ibex), using the software TreeBeST v1.9.2 (http://treesoft.sourceforge.net/treebest.shtml, last accessed August 6, 2013). A PCA analysis was performed on an individual-scale for the 77 native sheep using the smartpca program in the package EIGENSOFT v5.0 (Patterson et al. 2006). The population genetic structure of the native sheep was examined via an expectation maximization algorithm implemented in the program FRAPPE v1.170 (Tang et al. 2005). The number of assumed genetic clusters K ranged from 2 to 9, with 10,000 iterations for each run. Population genetic differentiation between the three identified groups of sheep breeds (Qinghai–Tibetan, Yunnan–Kweichow, and Northern and Eastern Chinese breeds; see Results) was measured by pairwise FST values (Weir and Cockerham 1984).
Demographic History and Population Admixture Analyses
We used the PSMC (Li and Durbin 2011) method to estimate changes in the effective population size (Ne) of sheep over the last one million years. The PSMC analysis was implemented in the two samples sequenced at a high read depth (ZNQ24 and LOP41; each ∼42×). The parameters were set as follows: -N30 -t15 -r5 -p ‘4 + 25*2 + 4 + 6’. An average mutation rate (μ) of 2.5 × 10−8 per base per generation and a generation time (g) of 2 years (de Magalhães and Costa 2009) were used for the analysis.
We also inferred the recent demographic history (e.g., < 4,000 years) of the three identified sheep groups (Qinghai–Tibetan, Yunnan–Kweichow, and Northern and Eastern Chinese breeds; see Results) using the diffusion approximation for demographic inference (∂a∂i) approach (Gutenkunst et al. 2009). To ensure selective neutrality, we only considered the SNPs within the intergenic regions of autosomal chromosomes. Five divergence models were constructed and tested, and the model with the highest log-likelihood value was considered to be optimal. The SFS of all 80 samples and the 77 native sheep were estimated in ANGSD (Nielsen et al. 2012; Korneliussen et al. 2014). A population-level admixture analysis was conducted in the TreeMix v.1.12 program (Pickrell and Pritchard 2012). The program inferred the ML tree for 21 native sheep breeds (77 individuals) and an outgroup (O. a. musimon), then the residuals matrix was used to identify pairs of populations that showed poor fits in the ML tree. These populations were regarded as candidates around which we added potential migration edges, and new arrangements of the ML tree accounting for migration events were generated (Pickrell and Pritchard 2012). From one to 16 migration events were gradually added to the ML tree until 98% of the variance between the breeds could be explained. The command was ‘-i input -bootstrap -k 10000 -m migration events -o output’.
Genome-Wide Selective Sweep Test
We calculated the genome-wide distribution of FST values (Weir and Cockerham 1984) and θπ ratios (i.e., θπ-Control/θπ-Tibetan, θπ-Control/θπ-Taklimakan Desert, θπ-Low-altitude/θπ-High-altitude, and θπ-Humid/θπ-Arid) for four extreme-control group pairs, which included the Tibetan group versus the Control group (Hu sheep and Wadi sheep, five samples from each breed from Eastern China), the Taklimakan Desert group versus the Control group, the High-altitude group versus the Low-altitude group and the Arid group versus the Humid group (supplementary table S2, Supplementary Material online), using a sliding-window approach (100-kb windows with 50-kb increments). The FST values were Z-transformed, and the θπ ratios were log2-transformed. We considered the windows with the top 5% values for the Z(FST) and log2(θπ ratio) simultaneously as the candidate outliers under strong selective sweeps. All of the outlier windows were assigned to corresponding SNPs and genes. Furthermore, we estimated the cross-population extended haplotype homozygosity (XP-EHH; Sabeti et al. 2007) statistic for the Tibetan group and the Taklimakan Desert group, using the Control group as a reference. The genetic map was assumed to be 1 cM/Mb for the sheep genome (Kijas et al. 2012). Also, we used the program LFMM (Frichot et al. 2013) to calculate the correlations between the genetic variants of native sheep and climate variables (i.e., altitude and precipitation). The z scores for all variants were calculated using a burn-in of 100, 1,000 sweeps, and K = 3 latent factors on the basis of the results from the population structure analyses. The threshold for identifying candidate genes in the XP-EHH and LFMM analyses was set to the top 5% and top 1% percentile outliers, respectively.
Candidate Gene Analysis
Using the 77 native sheep, we constructed NJ trees and performed large-effect SNP analyses (i.e., nonsynonymous SNPs in candidate genes) for the candidate genes of Tibetan sheep and Taklimakan Desert region breeds. We also compared the Z(FST) and log2(θπ ratio) values of the selective genomic regions with those at the whole-genome scale for the sheep from Tibet, the Taklimakan Desert region, high-altitude areas and arid zones. The overlap between the candidate genes identified in the extreme environments and the predefined gene panel (i.e., the previously published candidate genes in other mammalian species under similar extreme environments) was compared with the overlap expected by chance.
We performed GO and functional pathway analyses on the candidate genes (i.e., all of the annotated genes in the outlier windows with the top 5% Z(FST) and log2(θπ ratio) values) using the PANTHER Classification System version 10 (Mi et al. 2013, 2016). We also positioned the candidate genes on known KEGG pathways (http://www.kegg.jp, last accessed October 16, 2015) and Reactome pathways (http://www.reactome.org, last accessed October 16, 2015). The functions of the candidate genes were consulted based on the annotations in the NCBI (http://www.ncbi.nlm.nih.gov, last accessed October 2, 2015) and Ensembl (http://www.ensembl.org, last accessed October 2, 2015) databases. The expression levels of the candidate genes from the proposed network of pathways (see Results) in different sheep organs were examined based on four functional genomics experiments in the EBI Gene Expression Atlas database (Petryszak et al. 2014).
For the target genes, that is, SOCS2 in Tibetan sheep and GPX3 in Taklimakan Desert region breeds, we compared the Z(FST) and log2(θπ ratio) values of the two target genes with their adjacent genomic regions and examined the expression levels of the target genes in specific sheep and human tissues using the EBI Gene Expression Atlas database (Petryszak et al. 2014). As Tajima’s D (Tajima 1989) value is an informative indicator of selective signature, we used Tajima’s D to test the target genes. Furthermore, we constructed an inter-species NJ tree for the SOCS2 gene based on the orthologous sequences of 12 other vertebrates, and summarized the allele frequencies of the SNPs in the GPX3 gene across the native sheep breeds.
Data Access
The sequencing data reported in this article are available upon request for research purpose.
Supplementary Material
Supplementary information, figures S1–S16, and tables S1–S30 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This study was financially supported by the External Cooperation Program of Chinese Academy of Sciences (No. 152111KYSB20150010), the International S&T Cooperation Program of China (i.e., ISTCP, No. 2014DFA30907), the Breakthrough Project of Strategic Priority Program of the Chinese Academy of Sciences (No. XDB13000000), the National High Technology Research and Development Program of China (i.e., 863 Program, No. 2013AA102506), the National Transgenic Breeding Project of China (No. 2014ZX0800952B), the Taishan Scholars Program of Shandong Province (No. ts201511085) and the grants from the National Natural Science Foundation of China (Nos. 31272413 and U1303284).
References
- Alkan C, Coe BP, Eichler EE. 2011. Genome structural variation discovery and genotyping. Nat Rev Genet. 12:363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. [DOI] [PubMed] [Google Scholar]
- Breyer MD, Breyer RM. 2000. Prostaglandin receptors: their role in regulating renal function. Curr Opin Nephrol Hypertens. 9:23–29. [DOI] [PubMed] [Google Scholar]
- Brown ME, Funk CC. 2008. Food security under climate change. Science 319:580–581. [DOI] [PubMed] [Google Scholar]
- Carroll MA, Balazy M, Margiotta P, Huang DD, Falck JR, McGiff JC. 1996. Cytochrome P-450-dependent HETEs: profile of biological activity and stimulation by vasoactive peptides. Am J Physiol. 271:R863–R869. [DOI] [PubMed] [Google Scholar]
- Cho YS, Hu L, Hou H, Lee H, Xu J, Kwon S, Oh S, Kim H-M, Jho S, Kim S, et al. 2013. The tiger genome and comparative analysis with lion and snow leopard genomes. Nat Commun. 4:2433.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhães JP, Costa J. 2009. A database of vertebrate longevity records and their relation to other life-history traits. J Evol Biol. 22:1770–1774. [DOI] [PubMed] [Google Scholar]
- Du L-X. 2011. Animal genetic resources in China. Beijing: China Agriculture Press. [Google Scholar]
- Easterling DR, Meehl GA, Parmesan C, Changnon SA, Karl TR, Mearns LO. 2000. Climate extremes: observations, modeling, and impacts. Science 289:2068–2074. [DOI] [PubMed] [Google Scholar]
- Ehlers J, Gibbard PL. 2007. The extent and chronology of Cenozoic global glaciation. Quatern Int. 164–165:6–20. [Google Scholar]
- Esworthy RS, Chu FF, Paxton RJ, Akman S, Doroshow JH. 1991. Characterization and partial amino acid sequence of human plasma glutathione peroxidase. Arch Biochem Biophys. 286:330–336. [DOI] [PubMed] [Google Scholar]
- Frede S, Fandrey J. 2013. Cellular and molecular defenses against hypoxia In: Swenson ER, Bärtsch P, editors. High altitude: human adaptation to hypoxia. New York: Springer Press; p. 26. [Google Scholar]
- Freedman AH, Gronau I, Schweizer RM, Ortega-Del Vecchyo D, Han E, Silva PM, Galaverni M, Fan Z, Marx P, Lorente-Galdos B, et al. 2014. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet 10:e1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME. 2006. Copy number variation: new insights in genome diversity. Genome Res. 16:949–961. [DOI] [PubMed] [Google Scholar]
- Frichot E, Schoville SD, Bouchard G, François O. 2013. Testing for associations between loci and environmental gradients using latent factor mixed models. Mol Biol Evol. 30:1687–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatalica Z, Lilleberg SL, Koul MS, Vanecek T, Hes O, Wang B, Michal M. 2008. COX-2 gene polymorphisms and protein expression in renomedullary interstitial cell tumors. Hum Pathol. 39:1495–1504. [DOI] [PubMed] [Google Scholar]
- Ge R-L, Cai Q, Shen Y-Y, San A, Ma L, Zhang Y, Yi X, Chen Y, Yang L, Huang Y, et al. 2013. Draft genome sequence of the Tibetan antelope. Nat Commun. 4:1858.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Dong Y, Yu M, Zhang L, Yan X, Sun J, Qiao L, Geng H, Nakajima M, Furuichi T, et al. 2011. Follistatin-like 1 (Fstl1) is a bone morphogenetic protein (BMP) 4 signaling antagonist in controlling mouse lung development. Proc Natl Acad Sci USA. 108:7058–7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, Wang Z, Li N, Qiu F, Xu Z, Yan D, Yang S, Jia J, Kong X, Wei Z, et al. 2014. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 24:1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5:e1000695.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Clark VJ, Qian Y, Di Rienzo A. 2011. Population genetic analysis of the uncoupling proteins supports a role for UCP3 in human cold resistance. Mol Biol Evol. 28:610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann I. 2010. Climate change in context: implications for livestock production and diversity In: Odongo NE, Garcia M, Viljoen GJ, editors. Sustainable improvement of animal production and health. Vienna: IAEA-FAO; p. 33–44. [Google Scholar]
- Howden SM, Soussana JF, Tubiello FN, Chhetri N, Dunlop M, Meinke H. 2007. Adapting agriculture to climate change. Proc Natl Acad Sci USA. 104:19691–19696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian B, Wang D, Chen D, Voss J, Chaudry I, Raju R. 2010. Hypoxia-induced alteration of mitochondrial genes in cardiomyocytes—role of Bnip3 and Pdk1. Shock 34:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J-C, He M-L, Karma R, Penpa T. 1991. A comparative study on several hematologic values of Tibet Plateau sheep at different altitudes. Chinese J Anim Sci. 27:11–14. [Google Scholar]
- Jiang Y, Xie M, Chen W, Talbot R, Maddox JF, Faraut T, Wu C, Muzny DM, Li Y, Zhang W, et al. 2014. The sheep genome illuminates biology of the rumen and lipid metabolism. Science 344:1168–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirimutu WZ, Ding G, Chen G, Sun Y, Sun Z, Zhang H, Wang L, Hasi S, Zhang Y, Li J, et al. 2012. Genome sequences of wild and domestic Bactrian camels. Nat Commun. 3:1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayserili H, Uz E, Niessen C, Vargel I, Alanay Y, Tuncbilek G, Yigit G, Uyguner O, Candan S, Okur H, et al. 2009. ALX4 dysfunction disrupts craniofacial and epidermal development. Hum Mol Genet. 18:4357–4366. [DOI] [PubMed] [Google Scholar]
- Kijas JW, Lenstra JA, Hayes B, Boitard S, Neto LRP, San Cristobal M, Servin B, McCulloch R, Whan V, Gietzen K, et al. 2012. Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 10:e1001258.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E-S, Elbeltagy AR, Aboul-Naga AM, Rischkowsky B, Sayre B, Mwacharo JM, Rothschild MR. 2016. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity 116:255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV. 2006. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3:177–185. [DOI] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legge T. 1996. The beginnings of caprine domestication In: Harris DR, editor. The origins and spread of agriculture and pastoralism in Eurasia. New York: Smithsonian Institution Press; p. 238–262. [Google Scholar]
- Lehmkuhl F, Owen LA. 2005. Late Quaternary glaciation of Tibet and the bordering mountains: a review. Boreas 34:87–100. [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature 475:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Tian S, Jin L, Zhou G, Li Y, Zhang Y, Wang T, Yeung CK, Chen L, Ma J, et al. 2013. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet. 45:1431–1438. [DOI] [PubMed] [Google Scholar]
- Librado P, Der Sarkissian C, Ermini L, Schubert M, Jónsson H, Albrechtsen A, Fumagalli M, Yang MA, Gamba C, Sequin-Orlando A, et al. 2015. Tracking the origins of Yakutian horses and the genetic basis for their fast adaptation to subarctic environments. Proc Natl Acad Sci USA. 112:E6889–E6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F-Y, Chen Q-H. 2006. Effect of acute hypoxia on the parameters of blood gas in Tibetan Plateau sheep. Chinese J Zool. 41:48–52. [Google Scholar]
- Liu S, Lorenzen ED, Fumagalli M, Li B, Harris K, Xiong Z, Zhou L, Korneliussen TS, Somel M, Babbitt C, et al. 2014. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157:785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell DB, Burke MB, Tebaldi C, Mastrandrea MD, Falcon WP, Naylor RL. 2008. Prioritizing climate change adaptation needs for food security in 2030. Science 319:607–610. [DOI] [PubMed] [Google Scholar]
- Long L, Ormiston ML, Yang X, Southwood M, Gräf S, Machado RD, Mueller M, Kinzel B, Yung LM, Wilkinson JM, et al. 2015. Selective enhancement of endothelial BMPR-II with BMP9 reverses pulmonary arterial hypertension. Nat Med. 21:777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen ED, Nogués-Bravo D, Orlando L, Weinstock J, Binladen J, Marske KA, Ugan A, Borregaard MK, Gilbert MT, Nielsen R, et al. 2011. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479:359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv F-H, Agha S, Kantanen J, Colli L, Stucki S, Kijas JW, Joost S, Li M-H, Ajmone Marsan P. 2014. Adaptations to climate-mediated selective pressures in sheep. Mol Biol Evol. 31:3324–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv F-H, Peng W-F, Yang J, Zhao Y-X, Li W-R, Liu M-J, Ma Y-H, Zhao Q-J, Yang G-L, Wang F, et al. 2015. Mitogenomic meta-analysis identifies two phases of migration in the history of eastern Eurasian sheep. Mol Biol Evol. 32:2515–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Wang J, Zhou G, Yue Z, Hu Q, Chen Y, Liu B, Qiu Q, Wang Z, Zhang J, et al. 2013. Genomic insights into salt adaptation in a desert poplar. Nat Commun. 4:2797.. [DOI] [PubMed] [Google Scholar]
- Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ, Rubin EM, Jansson JK. 2011. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480:368–371. [DOI] [PubMed] [Google Scholar]
- Maquire JJ, Johnson CM, Mockridge JW, Davenport AP. 1997. Endothelin converting enzyme (ECE) activity in human vascular smooth muscle. Br J Pharmacol. 122:1647–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. 2013. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 8:1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. 2016. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44:D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A, Davies AM, Bertrand L, Sharif I, Budas GR, Jovanović S, Mouton V, Kahn CR, Lucocg JM, Gray GA, et al. 2003. Deficiency of PDK1 in cardiac muscle results in heart failure and increased sensitivity to hypoxia. Embo J. 22:4666–4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysterud A, Stenseth NC, Yoccoz NG, Langvatn R, Steinheim G. 2001. Nonlinear effects of large-scale climatic variability on wild and domestic herbivores. Nature 410:1096–1099. [DOI] [PubMed] [Google Scholar]
- Mysterud A, Yoccoz NG, Langvatn R, Pettorelli N, Stenseth NC. 2008. Hierarchical path analysis of deer responses to direct and indirect effects of climate in northern forest. Philos Trans R Soc Lond B Biol Sci. 363:2359–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Korneliussen T, Albrechtsen A, Li Y, Wang J. 2012. SNP calling, genotype calling, and sample allele frequency estimation from new-generation sequencing data. PLoS One 7:e37558.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton WHJ, Ledin J, Grandel H, Neumann CJ. 2005. HSPG synthesis by zebrafish Ext2 and Extl3 is required for Fgf10 signalling during limb development. Development 132:4963–4973. [DOI] [PubMed] [Google Scholar]
- Oakeshott JG, Wilson SR, Knibb WR. 1988. Selection affecting enzyme polymorphisms in enclosed Drosophila populations maintained in a natural environment. Proc Natl Acad Sci USA. 85:293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksyk TK, Smith MW, O’Brien SJ. 2010. Genome-wide scans for footprints of natural selection. Philos Trans R Soc Lond B Biol Sci. 365:185–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, et al. 2013. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499:74–78. [DOI] [PubMed] [Google Scholar]
- Park SD, Magee DA, McGettigan PA, Teasdale MD, Edwards CJ, Lohan AJ, Murphy A, Braud M, Donoghue MT, Liu Y, et al. 2015. Genome sequencing of the extinct Eurasian wild aurochs, Bos primigenius, illuminates the phylogeography and evolution of cattle. Genome Biol 16:234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker KL, Robbins CT. 1985. Thermoregulation in ungulates In: Hudson RJ, White RG, editors. Bioenergetics of wild herbivores. Boca Raton, FL: CRC Press, Inc; p. 161–182. [Google Scholar]
- Patterson N, Price AL, Reich D. 2006. Population structure and eigenanalysis. PLoS Genet. 2:e190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak R, Burdett T, Fiorelli B, Fonseca NA, Gonzalez-Porta M, Hastings E, Huber W, Jupp S, Keays M, Kryvych N, et al. 2014. Expression Atlas update—a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 42:D926–D932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao S, Ciais P, Huang Y, Shen Z, Peng S, Li J, Zhou L, Liu H, Ma Y, Ding Y, et al. 2010. The impacts of climate change on water resources and agriculture in China. Nature 467:43–51. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8:e1002967.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinti M, Gibellini L, Liu Y, Xu S, Lu B, Cossarizza A. 2015. Mitochondrial Lon protease at the crossroads of oxidative stress, aging and cancer. Cell Mol Life Sci. 72:4807–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, Cao C, Hu Q, Kim J, Larkin DM, et al. 2012. The yak genome and adaptation to life at high altitude. Nat Genet. 44:946–949. [DOI] [PubMed] [Google Scholar]
- Qu Y, Zhao H, Han N, Zhou G, Song G, Gao B, Tian S, Zhang J, Zhang R, Meng X, et al. 2013. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat Commun. 4:2071.. [DOI] [PubMed] [Google Scholar]
- Rico-Bautista E, Flores-Morales A, Fernández-Pérez L. 2006. Suppressor of cytokine signaling (SOCS) 2, a protein with multiple functions. Cytokine Growth Factor Rev. 17:431–439. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada J, Urakami T, Li F, Urakami A, Zhu W, Fukuda M, Li DY, Ruoslahti E, Komatsu M. 2012. Small GTPase R-Ras regulates integrity and functionality of tumor blood vessels. Cancer Cell 22:235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt LB, Soi S, Thompson S, Ranciaro A, Woldemeskel D, Beggs W, Lambert C, Jarvis JP, Abate D, Belay G, et al. 2012. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 13:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin E, Duvernell DD, Matzkin LM, Duan Y, Zhu CT, Verrelli BC, Eanes WF. 2004. Single locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics 168:923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, et al. 2010. Genetic evidence for high-altitude adaptation in Tibet. Science 329:72–75. [DOI] [PubMed] [Google Scholar]
- Stopka T, Zivny JH, Stopkova P, Prchal JF, Prchal JT. 1998. Human hematopoietic progenitors express erythropoietin. Blood 91:3766–3772. [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Peng J, Wang P, Risch NJ. 2005. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 28:289–301. [DOI] [PubMed] [Google Scholar]
- Tang M, Wang GR, Lu P, Karas RH, Aronovitz M, Heximer SP, Kaltenbronn KM, Blumer KJ, Siderovski DP, Zhu Y, et al. 2003. Regulator of G-protein signaling-2 mediates vascular smooth muscle relaxation and blood pressure. Nat Med. 9:1506–1512. [DOI] [PubMed] [Google Scholar]
- Turnley AM. 2005. Role of SOCS2 in growth hormone actions. Trends Endocrinol Metab. 16:53–58. [DOI] [PubMed] [Google Scholar]
- Wang M-S, Li Y, Peng M-S, Zhong L, Wang Z-J, Li Q-Y, Tu X-L, Dong Y, Zhu C-L, Wang L, et al. 2015. Genomic analyses reveal potential independent adaptation to high altitude in Tibetan chickens. Mol Biol Evol. 32:1880–1889. [DOI] [PubMed] [Google Scholar]
- Wang X, Ma YH, Chen H, Guan WJ. 2007. Genetic and phylogenetic studies of Chinese native sheep breeds (Ovis aries) based on mtDNA D-loop sequences. Small Ruminant Res. 72:232–236. [Google Scholar]
- Wei C, Wang H, Liu G, Wu M, Cao J, Liu Z, Liu R, Zhao F, Zhang L, Lu J, et al. 2015. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genomics 16:194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. [DOI] [PubMed] [Google Scholar]
- Wu H, Guang X, Al-Fageeh MB, Cao J, Pan S, Zhou H, Zhang L, Abutarboush MH, Xing Y, Xie Z, et al. 2014. Camelid genomes reveal evolution and adaptation to desert environments. Nat Commun. 5:5188.. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Xu D, Wang L, Hao J, Wang J, Zhou X, Wang W, Qiu Q, Huang X, Zhou J, et al. 2016. Convergent evolution of rumen microbiomes in high-altitude mammals. Curr Biol. 26(14):1873–1879. [DOI] [PubMed] [Google Scholar]
- Zhao E, Yu Q, Zhang N, Kong D, Zhao Y. 2013. Mitochondrial DNA diversity and the origin of Chinese indigenous sheep. Trop Anim Health Pro. 45:1715–1722. [DOI] [PubMed] [Google Scholar]
- Zheng B, Xu Q, Shen Y. 2002. The relationship between climate change and Quaternary glacial cycles on the Qinghai-Tibetan Plateau: review and speculation. Quatern Int. 97–98:93–101. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.