Key Points
Chromosomal abnormalities predict outcome after relapse in BCP-ALL, and high-risk cytogenetics takes precedence over clinical risk factors.
Patients with mutations or deletions targeting TP53, NR3C1, BTG1, and NRAS were associated with clinical high risk and an inferior outcome.
Abstract
Somatic genetic abnormalities are initiators and drivers of disease and have proven clinical utility at initial diagnosis. However, the genetic landscape and its clinical utility at relapse are less well understood and have not been studied comprehensively. We analyzed cytogenetic data from 427 children with relapsed B-cell precursor ALL treated on the international trial, ALLR3. Also we screened 238 patients with a marrow relapse for selected copy number alterations (CNAs) and mutations. Cytogenetic risk groups were predictive of outcome postrelapse and survival rates at 5 years for patients with good, intermediate-, and high-risk cytogenetics were 68%, 47%, and 26%, respectively (P < .001). TP53 alterations and NR3C1/BTG1 deletions were associated with a higher risk of progression: hazard ratio 2.36 (95% confidence interval, 1.51-3.70, P < .001) and 2.15 (1.32-3.48, P = .002). NRAS mutations were associated with an increased risk of progression among standard-risk patients with high hyperdiploidy: 3.17 (1.15-8.71, P = .026). Patients classified clinically as standard and high risk had distinct genetic profiles. The outcome of clinical standard-risk patients with high-risk cytogenetics was equivalent to clinical high-risk patients. Screening patients at relapse for key genetic abnormalities will enable the integration of genetic and clinical risk factors to improve patient stratification and outcome. This study is registered at www.clinicaltrials.org as #ISCRTN45724312.
Medscape Continuing Medical Education online
This activity has been planned and implemented through the joint providership of Medscape, LLC and the American Society of Hematology.
Medscape, LLC is accredited by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC), to provide continuing education for the healthcare team.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.00 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at http://www.medscape.org/journal/blood; and (4) view/print certificate. For CME questions, see page 1021.
Disclosures
Laurie Barclay, freelance writer and reviewer, Medscape, LLC, owns stock, stock options, or bonds from Pfizer. Associate Editor Jacob M. Rowe and the authors declare no competing financial interests.
Learning objectives
Describe the spectrum of chromosomal abnormalities, copy number alterations (CNAs), and sequence mutations observed in patients with relapsed B-cell precursor acute lymphoblastic leukemia (BCP-ALL).
Determine the prognostic relevance after first relapse in BCP-ALL of individual genetic abnormalities and predefined cytogenetic and CNA risk groups.
Evaluate possible improvements to the current clinical risk algorithm to stratify patients with relapsed BCP-ALL by integrating genetic biomarkers.
Release date: August 18, 2016; Expiration date: August 18, 2017
Introduction
Acute lymphoblastic leukemia (ALL) is a heterogeneous disease at the genetic level, and the spectrum of somatic aberrations range from chromosomal abnormalities to submicroscopic copy number alterations (CNAs) and sequence mutations.1 In pediatric ALL, chromosomal abnormalities and CNA define subgroups associated with risk of relapse and death.2,3 High-risk chromosomal abnormalities are used to risk-stratify treatment, ensuring patients receive the most appropriate intensity of chemotherapy.4 Although genetic abnormalities are prognostic biomarkers of outcome at initial diagnosis, their clinical applicability after relapse has rarely been studied systemically within the context of a large clinical trial.
Currently, 3 parameters define risk groups at relapse: (1) duration of first remission (CR1); (2) site of relapse; and (3) immunophenotype.5 Shorter CR1, T-ALL, and marrow involvement are associated with poor outcome postrelapse.5 In contrast, patients who have late or isolated extra-medullary relapses have better outcomes.5 Studies investigating the genetic profile of relapse samples have identified important similarities and differences with the matched diagnostic sample. Primary genetic abnormalities that define distinct subtypes, including ETV6-RUNX1, KMT2A (MLL) translocations, TCF3-PBX1, and high hyperdiploidy (HeH), are stable between diagnosis and relapse.6,7 In contrast, cooperating or secondary aberrations are lost, enriched, or gained as the leukemia evolves under the selection pressure of frontline chemotherapy and the surviving subclones expand, resulting in relapse.8-11 Several abnormalities are more prevalent at relapse (eg, TP5312 and NR3C18,13 alterations). Recent studies suggest that some genetic abnormalities (eg, IKZF1, TP53, KRAS abnormalities) are prognostic biomarkers that can refine current risk stratification algorithms.10,12,14 These biomarkers need independent validation to ensure they are disease-specific, rather than trial-dependent.
The aims of this study were to determine (1) the spectrum and frequency of chromosomal abnormalities, CNA, and sequence mutations among patients with relapsed B-cell precursor ALL (BCP-ALL); (2) the prognostic relevance of individual genetic abnormalities and predefined cytogenetic and CNA risk groups for predicting outcome after first relapse; and (3) whether the current clinical risk algorithm used to stratify relapsed ALL patients can be improved through the integration of genetic biomarkers.
Methods
Patients
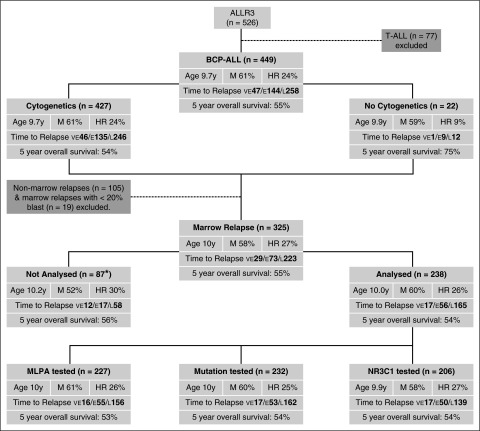
Between January 2003 and October 2013, 449 children aged 1 to 18 years from the United Kingdom, Ireland, The Netherlands, Australia, and New Zealand diagnosed with relapsed BCP-ALL were enrolled on the ALLR3 trial (Figure 1).15 The study was approved by the relevant institutional ethics committee(s) and written informed consent was obtained for each patient from parents, legal guardians, or the patients themselves.15
Figure 1.
CONSORT diagram depicting the patient cohorts used in this study with their key demographic and clinical features. “Age” refers to the mean age at relapse in years. *Material not available. E, early; F, female; HR, clinical high risk; L, late; M, male; VE, very early.
Procedures
Detailed treatment protocols and the main trial results have been published.15-17 An outline of the risk stratification algorithm and treatment is provided in supplemental Figure 1 and supplemental Table 1, available on the Blood Web site. An initial randomization during induction between idarubicin and mitoxantrone was stopped in favor of mitoxantrone.15 Patients were classified as standard, intermediate, or high risk (supplemental Figure 1). Patients with late (>6 months after stopping frontline therapy) isolated extra-medullary (EM) relapses were classified as standard risk (SR). Intermediate risk (IR) comprised BCP-ALL patients with late relapses involving the bone marrow (BM) or early (<6 months from stopping frontline therapy) isolated EM and combined relapses, as well as T-ALL patients with early isolated EM relapses. All remaining patients were classified as high risk (HR) and included: (1) patients with a very early relapse (<18 months from initial diagnosis); (2) T-ALL relapses involving the marrow; and (3) BCP-ALL patients with an early isolated BM relapse. HR patients were eligible for an allogenic stem cell transplant (SCT), and a small cohort (n = 44) received clorafabine. Patients were evaluated for minimal residual disease (MRD) by real-time quantitative polymerase chain reaction (qPCR) analysis of immunoglobulin and T-cell receptor gene rearrangements.18 IR patients with a late marrow relapse or an early combined relapse who had MRD levels ≥1 × 10−4 at the end of induction were eligible for an allogenic SCT. IR patients with MRD levels <1 × 10−4 at end of induction and SR patients with isolated extramedullary relapses received chemotherapy. Patients with extramedullary disease who were not transplanted also received radiotherapy. Only 26 BCP-ALL patients were classified as SR, and none were assessed for CNA or mutations because, by definition, there was no marrow involvement. In keeping with the current international relapsed ALL trial, we combined SR and IR, and throughout this manuscript they are referred to as “SR.”
Routine cytogenetic and fluorescence in situ hybridization (FISH) testing at diagnosis and/or relapse was used to classify patients into 3 previously defined mutually exclusive cytogenetic risk groups: (1) good risk—ETV6-RUNX1 and HeH (CYTO-GR); (2) intermediate risk—TCF3-PBX1, IGH translocations, B-other (none of these established abnormalities) (CYTO-IR); and (3) high risk—BCR-ABL1, KMT2A translocations, near haploidy, low hypodiploidy iAMP21, TCF3-HLF (CYTO-HR)3 (supplemental Figure 2). None of the 245 patients tested at both diagnosis and the time of marrow relapse was discordant for these classifying chromosomal abnormalities.
The copy number and mutational status of key genes were tested using DNA extracted from marrow relapse samples, which comprised ≥20% blasts derived from representative patient cohorts (Figure 1). The copy number status of IKZF1, CDKN2A/B, PAX5, EBF1, ETV6, BTG1, RB1, and PAR1 were determined using the SALSA multiplex ligation-dependent probe amplification (MLPA) kit P335 (MRC Holland, The Netherlands).19 The copy number status of these 8 loci was used to classify patients into predefined CNA profiles2 (supplemental Figure 2). NR3C1 copy number status was determined using TaqMan Copy Number Assays using primer and probe sets directed to NR3C1 (intron 4) and 2 references genes (ribonuclease P RNA component H1 [14q11.2] and telomerase reverse transcription [5q15.33]). NR3C1 deletions were confirmed by FISH using probes generated from the BAC clones (RP11-138C1 [5p13.1] and RP11-278J6 [5q31.3]), where suitable material was available. Key exons of TP53, NRAS, KRAS, PTPN11, FLT3, and CBL genes were assessed for mutations by denaturing high-performance liquid chromatography and Sanger or next-generation sequencing (supplemental Tables 2 and 3).10,12 Germline material was available for 10 patients harboring PTPN11 (n = 7), KRAS (n = 2), and TP53 (n = 3) mutations; all mutations were confirmed to be somatic.
Statistical analysis
Survival analysis considered 2 end points—progression-free (PFS) and overall survival (OS).15 PFS was calculated as the time from first relapse, using the date of registration onto ALLR3, until reinduction failure, second relapse, second malignancy, or death, censoring at last contact. OS was defined as time from first relapse to death, censoring at last contact. The median follow-up time for patients without an event was 4.3 years. Survival rates were calculated using Kaplan-Meier methods. The risk associated with the presence of each genetic abnormality was determined using a binary variable in univariate or multivariate Cox regression models. The prognostic impact of cytogenetic risk group was also assessed using a Cox regression model using 3-way categorical variable comparing CYTO-GR and CYTO-HR with CYTO-IR. Other comparisons were performed using the χ2 or Fisher exact test as appropriate. All analyses were performed using Intercooled Stata 13.0 (Stata Corporation, USA).
Results
Spectrum and frequency of chromosomal abnormalities, gene mutations, and CNA
A total of 427 of 449 (95%) patients with relapsed BCP-ALL had successful cytogenetic/FISH analysis at initial diagnosis or relapse. The spectrum of chromosomal abnormalities observed was similar to that seen at initial diagnosis, but the proportions were different (supplemental Figure 3). Patients were classified into predefined cytogenetic risk groups (supplemental Figure 2), which have been validated in consecutive frontline treatments protocols.2,3 The distribution of relapsed patients across the 3 risk groups was: CYTO-GR (48%), CYTO-IR (38%), and CYTO-HR (13%). The frequency of ETV6-RUNX1 and HeH was lower among these relapsed patients compared with newly diagnosed patients treated on ALL973: 83 of 427 (19%) vs 368 of 1451 (25%), P = .012; and 124 of 427 (29%) vs 562 of 1486 (38%), P = .001. In contrast, after the exclusion of BCR-ABL1 patients, there was a higher proportion of CYTO-HR patients among ALLR3 patients compared with the frontline trial ALL97: 55 of 425 (13%) vs 124 of 1505 (8%), P = .004.
We screened 190 to 222 patients for CNAs and sequence mutations affecting 15 genes (Figure 1; supplemental Table 4). The frequency of secondary abnormalities ranged from 1% to 41%, with the most prevalent aberrations targeting CDKN2A/B (41%), IKZF1 (23%), and PAX5 (20%). Among 173 patients successfully tested for all 15 aberrations, 80% harbored ≥1 abnormality: 1 (29%), 2 (24%), 3 (16%), and 4+ (10%) abnormalities. The secondary abnormalities were not distributed evenly across cytogenetic risk groups or individual abnormalities (Table 1). Overall, the frequencies of individual CNA and mutations and their correlation with chromosomal abnormalities strongly reflected the genomic landscape observed at diagnosis.20-22 IKZF1 and CDKN2A/B deletions were associated with B-other ALL and were less prevalent among CYTO-GR patients. ETV6 deletions were associated with ETV6-RUNX1 and iAMP21. TP53 mutation/loss was more frequent among CYTO-HR (35% vs 8%, P < .001) and clinical HR patients (25% vs 7%, P < .001). NRAS mutations were associated with HeH (14/62 [23%] vs 12/149 [8%], P = .003). Specific secondary abnormalities rarely coexisted more often than chance would predict (supplemental Figure 4). Deletions/mutations of CDKN2A/B-PAX5, CDKN2A/B–KRAS, and PAX5–IKZF1 were more common than expected, and the majority of patients with each pair had B-other ALL (64%, 56%, and 85%, respectively), which is in line with the distribution of individual CNA and mutations by chromosomal abnormality (Table 1). Association between clinical parameters and CNA/mutations were rare (supplemental Table 4) but IKZF1-deleted patients were older (median, 12.2 vs 8.7 years, P = .004) and NRAS/KRAS-mutated patients were younger (median, 7.4 vs 10.5 years, P = .002) compared with other patients.
Table 1.
Frequency of copy number alterations and mutations at the time of marrow relapse in childhood acute lymphoblastic leukemia (ALL) stratified by chromosomal abnormality and cytogenetic risk group
| Copy number alteration/mutation | Total number of cases | Overall frequency, %† | Frequency of copy number alterations and mutations by chromosomal abnormality and cytogenetic risk group | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytogenetic risk group | Chromosomal abnormality | |||||||||||||||
| Good | Intermediate | High | ETV6-RUNX1 | High Hyperdiploid | B‐other | t(1;19) | IGH translocation | KMT2A translocation | iAMP21 | Haploid | Low hypodiploid‡ | t(17;19) | t(9;22) | |||
| Total number of cases tested§ | 238 | 100 | 107 | 80 | 27 | 45 | 66 | 74 | 4 | 7 | 12 | 10 | 4 | 3 | 3 | 2 |
| CDKN2A/B deletion | 90 | 41 | 36%* | 54%* | 30%* | 34% | 37% | 59%** | 25% | 14% | 10%* | 20% | — | 100% | 33% | 100% |
| IKZF1 deletion | 52 | 23 | 11%** | 40%** | 26% | 2%** | 17% | 42%** | 25% | 29% | 20% | 40% | — | 50% | 0% | 0% |
| PAX5 alteration | 44 | 20 | 14% | 28% | 19% | 18% | 11%* | 32%** | 0% | 0% | 20% | 20% | — | 50% | 0% | 0% |
| ETV6 deletion | 36 | 16 | 18% | 11% | 26% | 34%** | 6%* | 10% | 50% | 0% | 0% | 60%** | — | 0% | 0% | 50% |
| TP53 alteration¶ | 29 | 10 | 7% | 7% | 28%** | 5% | 8% | 7% | 17% | 13% | 12% | 27% | 100%** | 40% | 0% | 0% |
| KRAS mutation | 26 | 12 | 11% | 14% | 10% | 0%** | 18% | 14% | 0% | 17% | 18% | 0% | 0% | 0% | 33% | 0% |
| NRAS mutation | 26 | 12 | 15% | 9% | 13% | 3%* | 23%** | 7% | 25% | 17% | 9% | 25% | 25% | 0% | 0% | 0% |
| P2RY8‐CRLF2 | 21 | 10 | 8% | 14% | 7% | 14% | 3%* | 12% | 25% | 29% | 10% | 10% | — | 0% | 0% | 0% |
| NR3C1 deletion | 18 | 9 | 9% | 6% | 22% | 14% | 6% | 7% | 0% | 0% | 18% | 11% | — | 50% | 67%** | 0% |
| PTPN11 mutation | 17 | 8 | 11% | 5% | 0% | 0% | 18%** | 6% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% |
| BTG1 deletion | 11 | 5 | 4% | 6% | 7% | 9% | 0%* | 6% | 0% | 14% | 0% | 0% | — | 50% | 33% | 0% |
| RB1 deletion | 11 | 5 | 3% | 10% | 0% | 2% | 3% | 12%** | 0% | 0% | 0% | 0% | — | 0% | 0% | 0% |
| EBF1 deletion | 10 | 5 | 4% | 6% | 0% | 7% | 2% | 6% | 0% | 14% | 0% | 0% | — | 0% | 0% | 0% |
| FLT3 mutation | 9 | 4 | 4% | 5% | 3% | 0% | 6% | 6% | 0% | 0% | 0% | 13% | 0% | 0% | 0% | 0% |
| CBL1 mutation | 2 | 1 | 0% | 3% | 0% | 0% | 0% | 3%* | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% |
P values from χ2 tests: *P < .05; **P < .01.
Not all patients were successfully tested for all abnormalities.
Among a total of 5 low hypodiploid patients, 3 were tested for CNA and/or mutations, but all 5 were assessed for TP53 alterations (see ¶ footnote).
Only patients with a marrow relapse and with >20% blasts in the marrow at relapse were eligible for MLPA and mutation testing.
Among BCP‐ALL with a marrow relapse, 243 patients had successful cytogenetics, whereas 210 patients were successfully screened for mutations. The frequency of sequence mutations and deletions is <10%, so we used a denominator of comprising cases successfully tested for at least one of the abnormalities (n = 290).
Clinical relevance of chromosomal abnormalities, gene mutations, and CNA
The clinical and outcome features of ETV6-RUNX1 and HeH patients were similar with OS rates 67% (49-80) and 68% (54-79), respectively (supplemental Table 3). These chromosomal abnormalities define the GR-CYTO group and, collectively, they had a superior outcome postrelapse compared with other patients (Table 2; Figure 2A-B). Although all CYTO-HR patients had a poor outcome, there were demographic and clinical differences by individual abnormality (Table 2). Patients with KMT2A translocations were <10 years old at relapse and 87% had a very early or early relapse. In contrast, iAMP21 patients were older, and 11 of 19 (58%) had relapsed late. Hence, the majority of KMT2A patients (74%) were classified as HR, whereas the majority of iAMP21 patients were classified as SR (84%). Although both sets of patients had a high second relapse rate (44% and 31%, respectively), iAMP21 patients had a lower OS rate (16% vs 42%), despite similar proportions having a transplant—12/23 KMT2A patients and 12/19 iAMP21 patients. There was evidence of outcome heterogeneity within the CYTO-IR group. Among 9 patients with TCF3-PBX1, 8 relapsed within 2.5 years of initial diagnosis. All 7 TCF3-PBX1 patients who had a marrow relapse died, whereas 2 patients with an isolated central nervous system (CNS) relapse remain alive 3+ years post-relapse. TCF3-PBX1 patients had an increased risk of second event and death compared with other patients: hazard ratios—PFS 2.85 (95% confidence interval [CI], 1.34-6.06), P = .007 and OS 3.30 (1.55-7.05), P = .002. Ten patients harbored an IGH translocation and were older than other patients (median, 16 vs 9 years, P < .001). Even though the majority (6/10) had late relapses, postrelapse outcomes were poor with an increased risk of second event and death: hazard ratio PFS 2.05 (0.96-4.35), P = .06 and OS 2.48 (1.16-5.29), P = .019. Interestingly, only 2 of 10 IGH translocation patients suffered a second relapse, so this poor outcome was driven by treatment-related mortality.
Table 2.
Demographic and clinical features of patients with relapsed B‐cell precursor ALL harboring specific chromosomal abnormalities
| Total | Clinical risk group | Cytogenetic risk group | Individual chromosomal abnormalities | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Standard | High | Good | Intermediate | High | ETV6-RUNX1 | High hyperdiploid | t(1;19)(q21;p13) | IGH translocation† | B‐other | KMT2A | iAMP21 | Haploidy | Low hypodploidy | t(17;19)(q23p13) | t(9;22)(q34;q11) | ||
| Total, n (%) | 449 | 343 (76) | 106 (24) | 207 (48) | 163 (38) | 57 (13) | 83 (19) | 124 (29) | 9 (2) | 10 (3) | 144 (34) | 23 (5) | 19 (4) | 5 (1) | 5 (1) | 3 (1) | 2 (1) |
| Sex (male), % | 61 | 61 | 62 | 62 | 64 | 49 | 71 | 56 | 33 | 40 | 68 | 48 | 63 | 40 | 20 | 33 | 50 |
| Age at relapse, n (%) | |||||||||||||||||
| 1‐9 y | 259 (58) | 194 (57) | 65 (61)*** | 143 (69) | 74 (45) | 30 (53)*** | 58 (70)* | 85 (69)*** | 4 (44) | 1 (10)*** | 69 (48)** | 23 (100)*** | 0 (0)*** | 3 (60) | 2 (40) | 1 (33)* | 1 (50) |
| 10‐14 y | 123 (27) | 107 (31) | 16 (15) | 52 (25) | 51 (31) | 14 (25) | 20 (24) | 32 (26) | 4 (44) | 3 (30) | 44 (31) | 0 (0) | 11 (58) | 1 (20) | 1 (20) | 0 (0) | 1 (50) |
| 15‐18 y | 67 (15) | 42 (12) | 25 (24) | 12 (6) | 38 (23) | 13 (23) | 5 (6) | 7 (6) | 1 (11) | 6 (60) | 31 (22) | 0 (0) | 8 (42) | 1 (20) | 2 (40) | 2 (67) | 0 (0) |
| Time to first relapse, n (%)‡ | |||||||||||||||||
| Very early | 47 (10) | 0 (0) | 47 (44) | 2 (1) | 27 (17) | 17 (30)*** | 2 (2)*** | 0 (0)*** | 5 (56)*** | 1 (10) | 21 (15)* | 12 (52)*** | 0 (0) | 2 (40) | 2 (40) | 1 (33) | 0 (0) |
| Early | 144 (32) | 86 (25) | 58 (55) | 55 (27) | 57 (35) | 23 (40) | 18 (22) | 37 (30) | 3 (33) | 3 (30) | 51 (35) | 8 (35) | 8 (42) | 2 (40) | 2 (40) | 2 (67) | 1 (50) |
| Late | 258 (57) | 257 (75) | 1 (1) 7 | 150 (72) | 79 (48) | 17 (30) | 63(76) | 87(70) | 1(11) | 6(60) | 72 (50) | 3(13) | 11(58) | 1 (20) | 1 (20) | 0 (0) | 1 (50) |
| Site of first relapse, n (%)§ | |||||||||||||||||
| Isolated BM | 273 (61) | 188 (55) | 85 (80) | 126 (61) | 96 (59) | 42 (74) | 51 (61) | 75 (60) | 5 (56) | 9 (90) | 82 (57) | 16 (70) | 12 (63) | 5 (100) | 5 (100) | 3 (100) | 1 (50) |
| Isolated EM | 105 (23) | 93 (27) | 12 (11) | 48 (23) | 37 (23) | 9 (16) | 17(20) | 31(25) | 2(22) | 1(10) | 34 (24) | 5(22) | 4(21) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Combined | 71 (16) | 62 (18) | 9 (8) | 33 (16) | 30 (18) | 6 (11) | 15(18) | 18(15) | 2(22) | 0 (0) | 28 (19) | 2(9) | 3(16) | 0 (0) | 0 (0) | 0 (0) | 1 (50) |
| Clinical risk group, n (%)¶ | |||||||||||||||||
| Standard | 343 (76) | — | — | 186 (90) | 112 (69) | 25 (44)*** | 76 (92)*** | 110 (89)*** | 2 (22)*** | 6 (60) | 104 (72) | 6 (26)*** | 16 (84) | 4 (80)** | 4 (80)** | 0 (0) | 1 (50) |
| High | 106 (24) | — | — | 21 (10) | 51 (31) | 32 (56) | 7 (8) | 14 (11) | 7 (78) | 4 (40) | 40 (28) | 17 (74) | 3 (16) | 1 (20) | 1 (20) | 3 (100)** | 1 (50) |
| Minimal residual disease, n (%)‖ | |||||||||||||||||
| Positive | 144 (59) | 107 (57) | 37 (65) | 65 (57) | 55 (61) | 18 (62) | 26 (54) | 39 (58) | 3 (75) | 3 (100) | 49 (59) | 4 (44) | 8 (73) | 3 (100) | 3 (75) | 0 (0) | 0 (0) |
| Negative | 100 (41) | 80 (43) | 20 (35) | 50 (43) | 35 (39) | 11 (38) | 22 (46) | 28 (42) | 1 (25) | 0 (0) | 34 (41) | 5 (56) | 3 (27) | 0 (0) | 1 (25) | 1 (100) | 1 (100) |
| Unknown/not done | 205 | 156 | 49 | 92 | 73 | 28 | 35 | 57 | 5 | 7 | 61 | 14 | 8 | 2 | 1 | 2 | 0 |
| Outcome of induction therapy# | |||||||||||||||||
| Induction failure | 25 (6) | 6 (2) | 19 (18)*** | 3 (1) | 13 (8) | 9 (16)*** | 1(1)* | 2(2)* | 3(33)* | 1(10)* | 9 (6) | 3 (13) | 1(5) | 2 (40)** | 1 (20) | 2 (67)*** | 0 (0) |
| Induction death | 19 (4) | 11 (3) | 8 (8) | 4 (2) | 8 (5) | 6 (11) | 1 (1) | 3 (2) | 0 (0) | 2 (20) | 6 (4) | 2 (9) | 2 (11) | 0 (0) | 1 (20) | 0 (0) | 1 (50) |
| Second CR | 405 (90) | 326 (95) | 79 (75) | 200 (97) | 142 (87) | 42 (74) | 81 (98) | 119 (96) | 6 (67) | 7 (70) | 129 (90) | 18 (78) | 16 (84) | 3 (60) | 3 (60) | 1 (33) | 1 (50) |
| Outcome of patients achieving a second remission | |||||||||||||||||
| Relapsed | 108 (27) | 72 (22) | 36 (46)*** | 50 (25) | 38 (27) | 18 (43)*** | 21 (26) | 29 (24) | 3 (50) | 2 (29) | 33 (26) | 8 (44) | 5 (31)** | 3 (100)* | 2 (67) | 0 (0)* | 0 (0) |
| Died in CR | 54 (13) | 38 (12) | 16 (20) | 17 (9) | 24 (17) | 9 (21) | 6 (7) | 11 (9) | 1 (17) | 2 (29) | 21 (16) | 2 (11) | 6 (38) | 0 (0) | 0 (0) | 1 (100) | 0 (0) |
| Continuing second CR | 243 (60) | 216 (66) | 27 (34) | 133 (67) | 80 (56) | 15 (36) | 54 (67) | 79 (66) | 2 (33) | 3 (43) | 75 (58) | 8 (44) | 5 (31) | 0 (0) | 1 (33) | 0 (0) | 1 (100) |
| Survival at 5 y, % (95% CI) | |||||||||||||||||
| Progression-free | 48% (43‐53) | 56% (50‐62) | 24% (16‐33) | 58% (50‐65) | 43% (35‐52) | 20% (10‐33) | 57% (44‐68) | 58% (48‐67) | 22% (3‐51) | 17% (9‐50) | 47% (37‐55) | 31% (13‐52) | 16% (3‐37) | 0% | 20% (8‐58) | 0% | 50% (1‐91) |
| Overall | 55% (50‐60) | 65% (59‐70) | 26% (17‐35) | 68% (60‐75) | 47% (38‐56) | 26% (14‐40) | 67% (54‐77) | 68% (58‐77) | 22% (3‐51) | 16% (9‐49) | 52% (42‐60) | 42% (22‐61) | 16% (3‐39) | 0% | 40% (5‐75) | 0% | 50% (1‐91) |
| Hazard ratio (95% CI), P | |||||||||||||||||
| Progression-free | — | 1 | 3.45 (2.60‐4.60), <.001 | 0.58 (0.43‐0.80), .001 | 1 | 2.30 (1.59‐3.35), <.001 | 0.62 (0.42‐0.92), .017 | 0.57 (0.41‐0.80), .001 | 2.85 (1.34‐6.06), .007 | 2.05 (0.96‐4.35), .06 | 1.06 (0.79‐1.41),.7 | 2.15 (1.27‐3.81), .004 | 2.54 (1.47‐4.38), .001 | 6.67 (2.70‐16.47), <.001 | 3.11 (1.15‐8.37),.03 | 17.06 (5.28‐55.13), <.001 | 1.23 (0.17‐8.76),.8 |
| Overall | — | 1 | 4.15 (3.07‐5.61), <.001 | 0.48 (0.34‐0.67), <.001 | 1 | 2.17 (1.47‐3.20), <.001 | 0.51 (0.33‐0.81), .004 | 0.50 (0.34‐0.73), <.001 | 3.30 (1.55‐7.05), .002 | 2.48 (1.16‐5.29), .019 | 1.15 (0.84‐1.56),.4 | 2.20 (1.27‐3.81), .005 | 2.52 (1.43‐4.45), .001 | 6.85 (2.77‐16.91), <.001 | 2.71 (0.86‐8.50),.08 | 17.69 (5.49‐57.00), <.001 | 1.53 (0.21‐10.90),.7 |
P values from χ2 or log-rank tests as appropriate are represented as: *P < .05; **P < .01; ***P < .001.
Follow‐up was short for patients with IGH translocations. The PFS rate was quoted at 3.8 years and the OS rate at 4.8 years.
Time to first relapse was defined as very early (<18 months after initial diagnosis), early (<6 months after the end of treatment), or late (>6 months after the end of treatment).
Site of relapse was defined as isolated marrow, isolated extramedullary (EM), or other, which included all combined relapses.
Patients with very early relapses or early isolated marrow relapses were classified as clinically high risk, whereas all other patients were classified as clinical intermediate/standard risk.
Minimal residual disease (MRD) was evaluated by real‐time qPCR analysis of immunoglobulin and T‐cell receptor gene rearrangements on marrow samples obtained at relapse and day 35 of induction, and classified as positive (>1 × 10–4) or negative (<1 × 10–4).
Induction failure was defined as ≥5% blasts, persistence of cerebrospinal fluid (CSF) blasts, or nonregression of testicular enlargement by day 35. Patients were defined as having achieved a second complete remission (CR) if they had <5% blasts in the marrow or no blasts in the CSF at the end of phase 1.
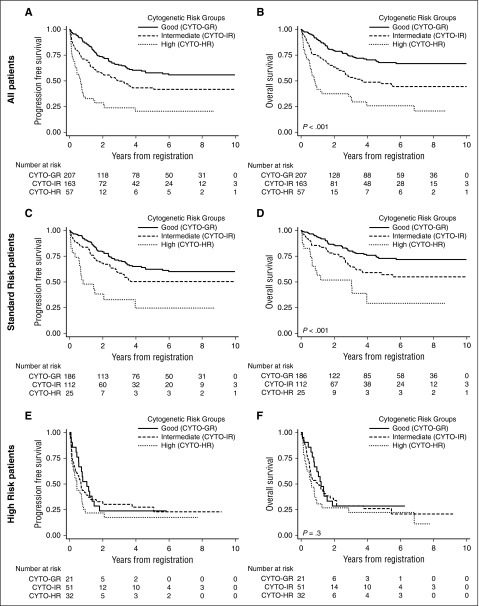
Figure 2.
Progression-free and overall survival of relapsed B-cell precursor ALL patients stratified by cytogenetic risk and clinical risk group. Kaplan-Meier survival graphs depicting the PFS and OS of relapsed childhood. Patients with ALL treated on ALLR3 and stratified by cytogenetic risk group.
Univariate analysis revealed that only TP53 alterations, NR3C1 and BTG1 deletions, were significantly associated with outcome (supplemental Table 4). Patients with a TP53 alteration were more likely to have had a very early relapse and hence be classified and treated as HR. Although their initial response was not poorer than other cases, they did have a higher relapse rate (52%) and consequently a higher hazard ratio for PFS and OS: 2.36 (1.51-3.70), P < .001; and 2.56 (1.61-4.06), P < .001. NR3C1 and BTG1 deletions were linked with induction failure/death, second relapse, and OS. Because both genes are implicated in resistance to glucocorticoids and the deletions are mutually exclusive, we also considered the effect of the deletions together: hazard ratios for PFS and OS were 2.15 (1.32-3.48), P = .002, and 1.91 (1.13-3.22), P = .015. Patients with a NRAS mutation appeared to have lower PFS and OS rates as well as higher rates of induction failure and second relapse, but this did not reach statistical significance.
We have previously shown that the prognostic impact of CNA at initial diagnosis was strongest when combined into a profile rather than individually (supplemental Figure 2).2 Applying the same criteria to CNA data generated at relapse identified that 58% patients harbored an IR/PR CNA profile, which was higher than in the original diagnostic ALL97 cohort (39%).2 Patients with an IR/PR CNA profile were older but did not have an inferior outcome (supplemental Table 4).
Refining the risk classification by integrating genetic and clinical risk factors
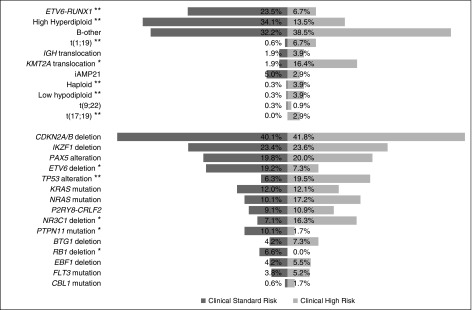
The current international trial for children with relapsed ALL—IntReALL2010—comprises 2 separate protocols, one for clinically defined SR patients and one for HR patients. Therefore, we reexamined the prognostic effect of genetic factors within clinical SR and HR groups to identify: (1) SR patients who have a poor outcome and may benefit from HR therapy, (2) HR patients who have a dismal outcome and maybe benefit from experimental therapy, and (3) HR patients with a good outcome who may be spared HR therapy. The cytogenetic, CNA, and mutational profile of SR and HR patients was distinctive (Figure 3). SR patients were enriched for ETV6-RUNX1, HeH, ETV6 and RB1 deletions, and PTPN11 mutations. In contrast, TCF3-PBX1, KMT2A translocations, haploidy, low hypodiploidy, TCF3-HLF, TP53 alterations, and NR3C1 deletions were more frequent among HR patients. CDKN2A/B, IKZF1, PAX5, and KRAS alterations were equally prevalent among SR and HR patients.
Figure 3.
Cytogenetic, copy number, and mutational profile of relapsed acute lymphoblastic leukemia patients stratified by clinical risk group. Frequency of individual chromosomal abnormalities, copy number alterations and sequence mutations among clinical standard and high-risk B-cell precursor ALL patients treated in ALLR3. *P < .05; **P < .01.
Among clinical SR patients, those with CYTO-GR and CYTO-HR had superior or inferior risks of progression/death compared with CYTO-IR patients (Table 3). The most prevalent secondary abnormalities among SR patients were deletions or alterations of CDKN2A/B, IKZF1, and PAX5, but were not associated with response or outcome. SR patients with a TP53 alteration had an inferior outcome; in particular, an increased risk of death: OS hazard ratio 2.56 (95% CI, 1.61-4.06), P < .001. SR patients with an NR3C1 deletion had a borderline adverse outcome, and combining NR3C1 and BTG1 deletions did reveal an increased risk of death: hazard ratio 1.91 (95% CI, 1.13-3.22), P = .015. Correlations between the primary chromosomal subgroup and the prevalence of specific secondary abnormalities raise the prospect of pleiotropic effects (Table 1). B-other ALL is enriched for CDKN2A/B, IKZF1, and PAX5 alterations, but their presence was not associated with outcome among SR patients. Similarly, the good outcome of ETV6-RUNX1 and HeH was not adversely affected by ETV6 deletions and PTPN11 mutations (data not shown). However, the presence of NRAS mutations was associated with an increased risk of progression/death among SR HeH patients: hazard ratio 3.17 (95% CI, 1.15-8.71), P = .026, and 3.41 (95% CI, 1.11-10.43), P = .032. The 5-year PFS and OS rates for the 10 SR patients with HeH and an NRAS mutation were 32% (6-63) and 26% (1-65). Among SR patients, the prognostic effect of MRD was borderline (supplemental Table 5) and did not correlate with the presence of specific genetic abnormalities (Table 3). A multivariate Cox regression model with variables for both MRD and high-risk genetics suggests both factors are associated with a higher risk of second relapse or death; however, neither reached statistical significance. (supplemental Table 5).
Table 3.
Key risk factors in patients with standard-risk relapsed B-cell precursor ALL
| Total | Cytogenetic goodrisk | Cytogenetic intermediate risk | Cytogenetic high risk | ETV6-RUNX1 | High hyperdiploidy | B-other | iAMP21 | CDKN2A/B deletion | IKZF1 deletion | PAX5 alteration | ETV6 deletion | KRAS mutation | NRAS mutation | PTPN11 mutation | P2RY8-CRLF2 | TP53 alteration | RB1 deletion | NR3C1 deletion | NR3C1/BTG1 deletion | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total, n (%)† | 343 | 186 | 112 | 25 | 76 | 110 | 104 | 16 | 67 | 39 | 33 | 32 | 19 | 16 | 16 | 15 | 13 | 11 | 10 | 17 |
| Sex ratio (% males) | 61% | 60% | 66% | 48% | 71%* | 53%* | 68% | 56% | 61% | 46% | 58% | 53% | 42% | 63% | 31%* | 60% | 38% | 73% | 50% | 59% |
| Age at relapse, n (%) | ||||||||||||||||||||
| 1-9 y | 194 (57) | 127 (68) | 46 (41) | 9 (36) | 52 (68)* | 75 (68)** | 46 (44)** | 0 (0) | 31 (46) | 11 (28) | 18 (55) | 15 (47) | 12 (63) | 12 (75) | 5 (31) | 9 (60) | 5 (38) | 4 (36) | 6 (60) | 7 (41) |
| 10-14 y | 107 (31) | 49 (26) | 42 (38) | 10 (40) | 20 (26) | 29 (26) | 38 (37) | 10 (63) | 26 (39) | 18 (46) | 11 (33) | 12 (38) | 4 (21) | 3 (19) | 8 (50) | 3 (20) | 4 (31) | 4 (36) | 2 (20) | 5 (29) |
| 15-18 y | 42 (12) | 10 (5) | 24 (21) | 6 (24) | 4 (5) | 6 (5) | 20 (19) | 6 (38)*** | 10 (15) | 10 (26) | 4 (12) | 5 (16) | 3 (16) | 1 (6) | 3 (19) | 3 (20) | 4 (31) | 3 (27) | 2 (20) | 5 (29) |
| Time to first relapse, n (%)‡ | ||||||||||||||||||||
| Very early | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Early | 86 (25) | 36 (19) | 34 (30) | 8 (32) | 13 (17) | 23 (21) | 33 (32) | 5 (31) | 10 (15) | 2 (5) | 6 (18) | 4 (13) | 1 (5) | 0 (0) | 1 (6) | 3 (20) | 2 (15) | 0 (0) | 1 (10) | 1 (6) |
| Late | 257 (75) | 150 (81) | 78 (70) | 17 (68) | 63 (82) | 87 (79) | 71 (68)* | 11 (69) | 57 (85)** | 37 (95) | 27 (82) | 28 (88) | 18 (95) | 16 (100) | 15 (94) | 12 (80)* | 11 (85) | 11 (100) | 9 (90) | 16 (94) |
| Site of first relapse, n (%)§ | ||||||||||||||||||||
| Isolated BM | 188 (55) | 107 (58) | 59 (53) | 14 (56) | 45 (59) | 62 (56) | 53 (51) | 9 (56) | 44 (66) | 35 (90) | 22 (67) | 21 (66) | 15 (79) | 12 (75) | 14 (88) | 9 (60) | 9 (69) | 9 (82) | 6 (60) | 12 (71) |
| Isolated EM | 93 (27) | 47 (25) | 30 (27) | 6 (24) | 16 (21) | 31 (28) | 29 (28) | 4 (25) | — | — | — | — | — | — | — | — | — | — | — | — |
| Combined | 62 (18) | 32 (17) | 23 (21) | 5 (20) | 15 (20) | 17 (15) | 22 (21) | 3 (19) | 23 (34)* | 4 (10)* | 11 (33) | 11 (34) | 4 (21) | 4 (25) | 2 (13) | 6 (40) | 4 (31) | 2 (18) | 4 (40) | 5 (29) |
| Minimal residual disease, n (%)¶ | ||||||||||||||||||||
| Positive | 107 (57) | 56 (55) | 35 (56) | 10 (77) | 22 (50) | 34 (59) | 33 (56) | 5 (63) | 29 (62) | 22 (69) | 19 (67) | 12 (50) | 7 (46) | 7 (70) | 9 (69) | 4 (44) | 6 (60) | 7 (78) | 7 (88) | 9 (69) |
| Negative | 80 (43) | 46 (45) | 27 (44) | 3 (23) | 22 (50) | 24 (41) | 26 (44) | 3 (38) | 18 (38) | 10 (31) | 9 (32) | 12 (50) | 6 (46) | 3 (30) | 4 (31) | 5 (55) | 4 (40) | 2 (22) | 1 (13) | 4 (31) |
| Unknown/not done | 156 | 84 | 50 | 12 | 32 | 52 | 45 | 8 | 20 | 7 | 5 | 8 | 6 | 6 | 3 | 6 | 3 | 2 | 2 | |
| Outcome of induction therapy‖ | ||||||||||||||||||||
| Induction failure | 6 (2) | 2 (1) | 3 (3) | 1 (4) | 1 (1) | 1 (1) | 3 (3) | 1 (6) | 2 (3) | 2 (5) | 3 (9) | 2 (6) | 0 (0) | 1 (6) | 0 (0) | 0 (0) | 1 (8) | 0 (0) | 0 (0) | 0 (0) |
| Induction death | 11 (3) | 2 (1) | 5 (4) | 3 (12) | 0 (0) | 2 (2) | 3 (3) | 2 (13) | 4 (6) | 4 (10) | 1 (3) | 1 (3) | 3 (16) | 1 (6) | 0 (0) | 2 (13) | 0 (0) | 0 (0) | 0 (0) | 1 (6) |
| Second CR | 326 (95) | 182 (98) | 104 (93) | 21 (84)* | 75 (99) | 107 (97) | 98 (94) | 13 (81)* | 61 (91) | 33 (85) | 29 (88) | 29 (91) | 16 (84) | 14 (88) | 16 (100) | 13 (87) | 12 (92) | 11 (100) | 10 (100) | 16 (94) |
| Outcome of patients achieving a second CR | ||||||||||||||||||||
| Relapsed | 72 (22) | 41 (23) | 22 (21) | 7 (33) | 18 (24) | 23 (22) | 20 (16) | 3 (23) | 12 (20) | 8 (24) | 8 (28) | 6 (21) | 2 (13) | 6 (43) | 2 (13) | 2 (15) | 4 (33) | 4 (36) | 5 (50) | 7 (44) |
| Died in CR | 38 (12) | 13 (7) | 17 (16) | 5 (24) | 5 (7) | 8 (7) | 16 (16) | 5 (38) | 7 (11) | 5 (15) | 1 (3) | 4 (14) | 2 (13) | 2 (14) | 2 (13) | 2 (15) | 2 (17) | 0 (0) | 1 (10) | 1 (6) |
| Continuing second CR | 216 (66) | 128 (70) | 65 (63) | 9 (43)* | 52 (69) | 76 (71) | 62 (63) | 5 (38)** | 42 (69) | 20 (61) | 20 (69) | 19 (66) | 12 (75) | 6 (43) | 12 (75) | 9 (69) | 6 (50) | 7 (64) | 4 (40) | 8 (50) |
| Survival at 5 y, % (95% CI) | ||||||||||||||||||||
| Progression-free | 56% (50-62) | 62% (54-70) | 50% (39-60) | 25% (8-46) | 60% (45-61) | 64% (52-73) | 53% (42-63) | 19% (4-44) | 60% (46-71) | 50% (33-64) | 56% (36-72) | 56% (36-72) | 61% (34-79) | 32% (10-56) | 74% (45-89) | 56% (25-78) | 43% (16-68) | 55% (18-80) | 35% (8-64) | 37% (12-62) |
| Overall | 65% (59-70) | 73% (65-80) | 57% (46-67) | 29% (9-52) | 71% (57-82) | 74% (63-82) | 61% (49-71) | 20% (3-46) | 65% (51-76) | 57% (39-71) | 68% (48-81) | 66% (45-80) | 67% (40-84) | 43% (15-69) | 73% (43-89) | 50% (16-77) | 43% (16-68) | 91% (51-99) | 53% (17-80) | 48% (19-72) |
| Hazard ratio (95% CI) from Cox model, P | ||||||||||||||||||||
| Progression-free | 0.66 (0.45-0.97), .034 | 1 | 2.49 (1.41-4.39), .002 | 0.77 (0.50-1.22), .3 | 0.64 (0.43-0.95), .026 | 1.15 (0.80-1.68), .4 | 3.62 (1.95-6.74), <.001 | 0.96 (0.58-1.59), .9 | 1.38 (0.81-2.36), .2 | 1.07 (0.58-1.96), .8 | 1.25 (0.68-2.31), .5 | 1.05 (0.48-2.30), .9 | 1.90 (0.96-3.74), .07 | 0.55 (0.20-1.51), .2 | 1.51 (0.65-3.50), .3 | 1.54 (0.71-3.35), .3 | 0.95 (0.35-2.62), .9 | 1.69 (0.72-3.95), .2 | 1.72 (0.85-3.47), .1 | |
| Overall | 0.54 (0.35-0.83), .005 | 1 | 2.30 (1.25-4.23), .008 | 0.64 (0.37-1.10), .1 | 0.57 (0.36-0.91), .018 | 1.29 (0.85-1.96), .2 | 3.75 (1.94-7.24), <.001 | 1.09 (0.72-1.64), .7 | 1.28 (0.82-2.02), .3 | 0.83 (0.49-1.41), .5 | 0.91 (0.52-1.61), .7 | 1.32 (0.72-2.41), .4 | 1.43 (0.82-2.48), .2 | 0.54 (0.22-1.33), .2 | 1.75 (0.93-3.30), .08 | 2.56 (1.61-4.06), <.001 | 0.17 (0.02-1.24), .08 | 1.71 (0.91-3.22), .098 | 1.91 (1.13-3.22), .015 | |
Only abnormalities present in 10 or more patients were considered
P values from χ2 or log-rank tests as appropriate: *P < .05; **P < .01; ***P < .001.
Not all patients were successfully tested for all abnormalities.
Time to first relapse was defined as very early (<18 months after initial diagnosis), early (<6 months after the end of treatment), or late (>6 months after the end of treatment).
Site of relapse was defined as isolated marrow, isolated CNS, or other, which included all combined relapses as well as other isolated extramedullary relapses (eg, testes).
Minimal residual disease (MRD) was evaluated by real-time qPCR analysis of immunoglobulin and T-cell receptor gene rearrangements on marrow samples obtained at relapse and day 35 of induction, and classified as positive (>1 × 10−4) or negative (<1 × 10−4).
Induction failure was defined as ≥5% blasts, persistence of CSF blasts, or nonregression of testicular enlargement by day 35. Patients were defined as having achieved a second CR if they had <5% blasts in the marrow or no blasts in the CSF at the end of phase 1.
The outcome of HR patients was universally poor and none of the genetic alterations was associated with a superior outcome (supplemental Table 6). However, none of the HR patients with a TP53 alteration survived 3 years, and the hazard ratio for risk of death was 2.56 (95% CI, 1.61-4.06), P < .001. Although the number of patients is small, the poor outcome appeared to be largely driven by a higher rate of second relapse. Nine of these 16 (56%) were CYTO-HR patients, 5 were CYTO-IR, and 2 CYTO-GR. HR patients with a NR3C1 or BTG1 deletion had an inferior outcome with an increased risk of progression: hazard ratio 3.40 (95% CI, 1.70-6.80), P = .001. Interestingly, 8 of 12 HR patients with NR3C1/BTG1 deletions failed or died during induction.
Discussion
This cohort represents the largest and most comprehensive genetic study of relapsed childhood BCP-ALL. Our previous studies have reported convincing evidence for the prognostic relevance of genetic aberrations at initial presentation.2,3 Here we provide compelling evidence that genetic biomarkers are important at predicting outcome in the relapse setting. The cytogenetic classification system, defined in our previous study, identified 3 risk groups with differential outcome; especially among SR patients. Observations from this study mirror those from initial disease presentation3 and argue for the prospective genetic stratification of patients. CYTO-HR patients had a uniformly poor outcome, with high rates of induction failure/death and second relapse and may benefit from alternative therapeutic strategies. The 5-year PFS and OS rates for SR patients with CYTO-HR were 25% (95% CI, 8%-46%) and 29% (95% CI, 9%-52%) (Figure 2), which are almost identical to HR patients overall (Table 2) and not different from CYTO-HR patients classified as HR (Table 4; Figure 2). Although there were differences between CYTO-HR patients classified as SR and HR—most notably the distribution of iAMP21 and KMT2A translocations—this did not translate into a survival difference. Nearly two thirds of SR patients with CYTO-HR had iAMP21 and their OS at 5 years was just 20% (Table 3). Despite the size of this study, we were limited in our ability to factor in the effect of MRD and treatment. Our analysis suggests that both postinduction MRD and genetics will be important risk factors in determining outcome in SR patients. This is the first study of relapsed ALL that has been able to examine the prognostic effect of CYTO-HR abnormalities by clinical risk group. Our data indicate that all CYTO-HR patients should be treated on future HR protocols regardless of clinical risk stratification.
Table 4.
Clinical, genetic, and outcome features of 57 patients with high-risk cytogenetics stratified by clinical risk
| Clinical risk group† | |||
|---|---|---|---|
| Total | Standard/Intermediate | High | |
| Total, n (%) | 57 | 25 | 32 |
| Sex, male (%) | 49% | 48% | 50% |
| Age at relapse, n (%) | |||
| 1‐9 y | 30 (53) | 9 (36) | 21 (66)* |
| 10‐14 y | 14 (25) | 10 (40) | 4 (13) |
| 15‐18 y | 13 (23) | 6 (24) | 7 (23) |
| Genetic subgroup, n (%) | |||
| Haploidy | 5 (9) | 1 (4) | 4 (13)** |
| Low hypodiploidy | 5 (9) | 1 (4) | 4 (13) |
| KMT2A translocation | 23 (40) | 6 (24) | 17 (53) |
| iAMP21 | 19 (33) | 16 (24) | 3 (9) |
| t(17;19) | 3 (5) | 0 (0) | 3 (9) |
| t(9;22) | 2 (4) | 1 (4) | 1 (3) |
| Time to first relapse, n (%)‡ | |||
| Very early | 17 (30) | 0 (0) | 17 (53)*** |
| Early | 23 (40) | 8 (32) | 15 (47) |
| Late | 17 (30) | 17 (68) | 0 (0) |
| Site of first relapse, n (%)§ | |||
| Isolated BM | 42 (74) | 14 (56) | 28 (88)* |
| Isolated CNS | 9 (16) | 6 (24) | 3 (9) |
| Combined | 6 (11) | 5 (20) | 1 (3) |
| MRD, n (%)¶ | |||
| Positive | 18 (62) | 10 (77) | 8 (50) |
| Negative | 11 (38) | 3 (23) | 8 (50) |
| Unknown/not done | 28 | 12 | 16 |
| Stem cell transplant, n (%)‖ | |||
| Yes | 28 (67) | 15 (71) | 13 (62) |
| No | 14 (33) | 6 (29) | 8 (38) |
| Induction drug randomization/allocation, n (%)# | |||
| Mitoxantrone | 30 (63) | 18 (72) | 12 (52) |
| Idarubicin | 18 (38) | 7 (28) | 11 (48) |
| Outcome of induction therapy†† | |||
| Induction failure | 9 (16) | 1 (4) | 8 (25) |
| Induction death | 6 (11) | 3 (12) | 3 (9) |
| Second CR | 42 (74) | 21 (84) | 21 (66) |
| Outcome of patients achieving a second remission | |||
| Relapsed | 18 (43) | 7 (33) | 11 (52) |
| Died in CR | 9 (21) | 5 (24) | 4 (19) |
| Continuing second CR | 15 (36) | 9 (42) | 6 (28) |
| Survival at 5 y, % (95% CI) | |||
| Progression-free | 20% (6‐33) | 25% (8‐46) | 18% (6‐33) |
| Overall | 26% (14‐40) | 29% (9‐52) | 22% (9‐39) |
| Hazard ratio (95% confidence interval), P | |||
| Progression-free | — | 1 | 1.57 (0.84‐2.95), .2 |
| Overall | — | 1 | 1.77 (0.91‐3.42), .09 |
P values from χ2 or log rank tests as appropriate: *P < .05; **P < .01; ***P < .001.
†Patients with very early relapses or early isolated marrow relapses were classified as clinically high risk with all other patients classified as clinical intermediate/standard risk.
Time to first relapse was defined as very early (<18 months after initial diagnosis), early (<6 months after the end of treatment) or late (>6 months after the end of treatment).
Site of relapse was defined as isolated marrow, isolated CNS, or other, which included all combined relapses as well as other isolated extramedullary relapses (eg, testes).
Minimal residual disease (MRD) was evaluated by real‐time qPCR analysis of immunoglobulin and T-cell receptor gene rearrangements on marrow samples obtained at relapse and day 35 of induction, and classified as positive (>0.01%) or negative (<0.01%).
Restricted to the 42 patients who achieved a second CR.
Excluded the 9 high-risk patients who received clofarabine.
Induction failure was defined as ≥5% blasts, persistence of CSF blasts, or nonregression of testicular enlargement by day 35. Patients were defined as having achieved a second complete remission CR if they had <5% blasts in the marrow or no blasts in the CSF at the end of phase 1.
Even though ETV6-RUNX1 and HeH are established good prognostic markers, they still accounted for ∼50% of BCP-ALL patients treated in ALLR3. This high incidence reflects both their prevalence at initial diagnosis and the fact that recruitment started in 2003 and hence captured “good risk” patients suffering late relapses from earlier protocols. The outcome of SR patients with CYTO-GR was >70%. Although ETV6-RUNX1 and HeH were rare among HR patients, they did have a poor outcome. One possible explanation for the poor outcome of HR HeH patients is the presence of RAS pathway mutations. The frequency of RAS mutations among HeH patients at diagnosis is higher compared with other subgroups.22,23 This study and the BFM study10 confirm that this association prevails at relapse. SR patients with HeH and an NRAS mutation had a threefold increased risk of death. In the HR group, 6 of 10 HeH patients had a NRAS/KRAS mutation and 5 have died. A recent BFM ALL-REZ 2002 study has reported that RAS pathway mutations can influence outcome.10 Moreover, it has recently been suggested that HeH clones are susceptible to RAS mutations, and specific combinations (eg, KRAS and CREBBP) might cooperate to drive relapse.24
Within the CYTO-IR subgroup, TCF3-PBX1 and IGH translocations were associated with a poor outcome. The prognostic impact of TCF3-PBX1 at initial diagnosis appears to be protocol-dependent, with historic studies reporting inconsistent results and recent intensive protocols achieving better outcomes.1,3,25 This study indicates that if TCF3-PBX1 patients do relapse, they tend to relapse very early and have a poor response to treatment. In contrast to the St. Jude’s group,25 we did not observe an association between TCF3-PBX1 and CNS relapse. The association of IGH translocations with age and poor outcome is consistent with our previous observations.17,26
Secondary abnormalities can be acquired, lost, or enriched between initial diagnosis and marrow relapse.8,9,12,27 Therefore, it is not surprising that the CNA risk profile proposed at initial diagnosis was not predictive of outcome when applied at relapse.2 The incidence of IKZF1 deletions was similar among SR and HR patients, but was not prognostic in either group or overall. The ALL-REZ BFM 2002 trial reported a negative prognostic impact of IKZF1 deletions at relapse.14 Although the 2 studies used the same detection strategy, there were differences in the composition of the cohorts: (1) the BFM study included more patients with BCR-ABL1 or Down syndrome (13/204 [6%] vs 4/224 [2%], P = .02); (2) the proportion of very early relapses was lower in ALLR3 overall (10% vs 18%, P = .005) and among IKZF1-deleted patients (8% vs 26%, P = .02). Hence, IKZF1-deleted patients in the BFM study were more likely to be associated with HR clinical risk factors, most notably shorter CR1 duration. This observation further supports the idea that the prognostic effect of IKZF1 deletions is context-dependent.28-30
Our analysis revealed that TP53 alterations were significantly more prevalent in the HR group but associated with an inferior outcome in both the SR and HR groups. Indeed within the HR group, none of the patients with a TP53 alteration survived. TP53 mutations and 17p13 deletions occurred at an increased frequency in the CYTO-HR groups at initial diagnosis31,32 and have been linked with a poor outcome at relapse.12 The overall frequency of TP53 alterations was similar in the ALLR3 and BFM cohorts (12%), as was the frequency of TP53 loss (9%); despite the use of different detection strategies (MLPA vs cytogenetics). The frequency of TP53 mutations was lower in the ALLR3 study (3% vs 8%, P = .02), which led to a lower number of ALLR3 cases harboring a defect in both TP53 alleles (ie, a double-hit [9 vs 3 cases]). Recent studies have demonstrated a strong correlation between TP53 mutations and selected cytogenetic subgroups, namely low hypodiploidy and MYC translocations.31,32 In this study, only one TP53 mutated case was found along with low hypodiploidy, whereas the remainder had various chromosomal abnormalities—HeH, ETV6-RUNX1, TCF3-PBX1, KMT2A, and B-other. This observation mirrored the BFM study,12 suggesting the distribution of TP53 mutations at relapse is different from initial diagnosis.
Loss of NR3C1 and BTG1 has been implicated in resistance to glucocorticoids, which is a key component of ALL therapy.33-35 NR3C1/BTG1 deletions were present in 12% of cases but were more common among cytogenetic and clinical high-risk patients. The deletions were associated with an inferior outcome in both SR and HR groups, but the effect appeared strongest in the HR group, where it correlated with a high rate of induction failure and death. These observations are consistent with the results of a recent BFM study showing an association between NR3C1 deletion and poor outcome in relapsed ETV6-RUNX1 ALL.36 Further studies are required to establish whether this poor outcome is linked directly to glucocorticoid resistance or whether it is part of a broader drug resistance profile.
One third of relapsed patients harbored a mutation in the RAS signaling pathway, with KRAS and NRAS mutations being the most prevalent (12% cases each). The incidence and distribution of RAS pathway mutations (88% G12/13 mutations; supplemental Table 3) was consistent with that observed by the BFM group.10 The BFM study suggested that KRAS mutations were stronger predictors of outcome, whereas our study implicated NRAS mutations especially in SR patients with HeH. It is not yet known whether KRAS and NRAS mutations have different functional consequences, and taken together, these 2 studies suggest that NRAS/KRAS mutations confer significant chemoresistant properties to the relapse clone. The in vitro and in vivo evidence that NRAS- and KRAS-mutated clones were sensitive to the MEK inhibitors selumetinib and trametinib, provides a potential targeted therapy strategy for this subset of patients.10,11
The outcome of patients with relapsed ALL remains unsatisfactory. Current risk stratification in BCP-ALL is based primarily on CR1 duration. The integration of genetic data into the current risk classification indicates that SR patients with CYTO-HR have a very poor outcome and therefore require alternative treatment. Furthermore, our data suggest that many patients with CYTO-GR are still chemosensitive. A combination of clinical and genetic risk factors could therefore be used to define a subgroup of relapsed patients that may be spared SCT. We have confirmed the adverse effect of TP53 alteration reported by a BFM study12 and provide evidence that the poor outcome associated with NR3C1 deletions extends beyond ETV6-RUNX1.36 In conclusion, the screening of patients at relapse for key genetic abnormalities will enable the integration of genetic and clinical risk factors to improve patient stratification and outcome.
Acknowledgments
The authors thank member laboratories of the UK Cancer Cytogenetic Group, the ANZCHOG cytogenetic laboratories, and the Dutch Children’s Oncology Group for providing cytogenetic data and material. Primary childhood leukemia samples used in this study were provided by the Bloodwise (formerly Leukaemia and Lymphoma Research) Childhood Leukaemia Cell Bank (UK), the Tumour Bank at Children’s Cancer Institute (Australia), and DCOG (The Netherlands). The authors also acknowledge the contribution of past and present members of the Leukaemia Research Cytogenetics Group for helping to establishing this dataset. Finally, they thank all the clinicians who entered patients into the trial and the children and families who agreed to take part.
This study was supported by Bloodwise (formerly Leukaemia and Lymphoma Research), Cancer Research UK, Sporting Chance Cancer Foundation and National Health and Medical Research Council Australia, Kinderen Kankervrij (KiKa), and the India Alliance Margdarshi Fellowship. ALLR3 was sponsored by Central Manchester University Hospitals Foundation Trust, UK. This research has received funding from the European Union’s Seventh Framework Programme for research, technological development, and demonstration (grant 278514–IntReALL).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.V.M., J.A.E.I., V.S., R.S., R.P.K., and C.J.H. conceived of and designed the study; A. Erhorn, L.M., N.C.V., T.L., J.Y., C.S., R.D., E.M., A.D., E.S., and M.L.d.B. performed experiments and undertook data analysis and interpretation; A. Enshaei performed statistical analyses; A. Erhorn, C.S., A.V.M., and C.J.H. collected and classified cytogenetic and FISH data; C.A.P., S.B.L., P.M.H., T.R., and V.S. were responsible for trial coordination and provision of clinical and outcome data; A.V.M., J.A.E.I., and V.S. wrote the manuscript; and all authors critically reviewed and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anthony V. Moorman, Newcastle University, Newcastle NE1 4LP, United Kingdom; e-mail: anthony.moorman@newcastle.ac.uk.
References
- 1.Moorman AV. The clinical relevance of chromosomal and genomic abnormalities in B-cell precursor acute lymphoblastic leukaemia. Blood Rev. 2012;26(3):123–135. doi: 10.1016/j.blre.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Moorman AV, Enshaei A, Schwab C, et al. A novel integrated cytogenetic and genomic classification refines risk stratification in pediatric acute lymphoblastic leukemia. Blood. 2014;124(9):1434–1444. doi: 10.1182/blood-2014-03-562918. [DOI] [PubMed] [Google Scholar]
- 3.Moorman AV, Ensor HM, Richards SM, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11(5):429–438. doi: 10.1016/S1470-2045(10)70066-8. [DOI] [PubMed] [Google Scholar]
- 4.Moorman AV, Robinson H, Schwab C, et al. Risk-directed treatment intensification significantly reduces the risk of relapse among children and adolescents with acute lymphoblastic leukemia and intrachromosomal amplification of chromosome 21: a comparison of the MRC ALL97/99 and UKALL2003 trials. J Clin Oncol. 2013;31(27):3389–3396. doi: 10.1200/JCO.2013.48.9377. [DOI] [PubMed] [Google Scholar]
- 5.Bhojwani D, Pui CH. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 2013;14(6):e205–e217. doi: 10.1016/S1470-2045(12)70580-6. [DOI] [PubMed] [Google Scholar]
- 6.Wehrli LA, Braun J, Buetti LN, et al. Non-classical karyotypic features in relapsed childhood B-cell precursor acute lymphoblastic leukemia. Cancer Genet Cytogenet. 2009;189(1):29–36. doi: 10.1016/j.cancergencyto.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Vora AJ, Potter AM, Anderson LM, Lilleyman JS. Frequency and importance of change in blast cell karyotype in relapsing childhood lymphoblastic leukemia. Pediatr Hematol Oncol. 1994;11(4):379–386. doi: 10.3109/08880019409140537. [DOI] [PubMed] [Google Scholar]
- 8.Mullighan CG, Phillips LA, Su X, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322(5906):1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang JJ, Bhojwani D, Yang W, et al. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood. 2008;112(10):4178–4183. doi: 10.1182/blood-2008-06-165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irving J, Matheson E, Minto L, et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood. 2014;124(23):3420–3430. doi: 10.1182/blood-2014-04-531871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ariës IM, van den Dungen RE, Koudijs MJ, et al. Towards personalized therapy in pediatric acute lymphoblastic leukemia: RAS mutations and prednisolone resistance. Haematologica. 2015;100(4):e132–e136. doi: 10.3324/haematol.2014.112995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hof J, Krentz S, van Schewick C, et al. Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. J Clin Oncol. 2011;29(23):3185–3193. doi: 10.1200/JCO.2011.34.8144. [DOI] [PubMed] [Google Scholar]
- 13.Hogan LE, Meyer JA, Yang J, et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood. 2011;118(19):5218–5226. doi: 10.1182/blood-2011-04-345595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krentz S, Hof J, Mendioroz A, et al. Prognostic value of genetic alterations in children with first bone marrow relapse of childhood B-cell precursor acute lymphoblastic leukemia. Leukemia. 2013;27(2):295–304. doi: 10.1038/leu.2012.155. [DOI] [PubMed] [Google Scholar]
- 15.Parker C, Waters R, Leighton C, et al. Effect of mitoxantrone on outcome of children with first relapse of acute lymphoblastic leukaemia (ALL R3): an open-label randomised trial. Lancet. 2010;376(9757):2009–2017. doi: 10.1016/S0140-6736(10)62002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan S, Wade R, Moorman AV, et al. Temporal changes in the incidence and pattern of central nervous system relapses in children with acute lymphoblastic leukaemia treated on four consecutive Medical Research Council trials, 1985-2001. Leukemia. 2010;24(2):450–459. doi: 10.1038/leu.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masurekar AN, Parker CA, Shanyinde M, et al. Outcome of central nervous system relapses in childhood acute lymphoblastic leukaemia--prospective open cohort analyses of the ALLR3 trial. PLoS One. 2014;9(10):e108107. doi: 10.1371/journal.pone.0108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Velden VH, Cazzaniga G, Schrauder A, et al. European Study Group on MRD detection in ALL (ESG-MRD-ALL) Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia. 2007;21(4):604–611. doi: 10.1038/sj.leu.2404586. [DOI] [PubMed] [Google Scholar]
- 19.Schwab CJ, Jones LR, Morrison H, et al. Evaluation of multiplex ligation-dependent probe amplification (MLPA) as a method for the detection of copy number abnormalities in B-cell precursor acute lymphoblastic leukaemia. Genes Chromosomes Cancer. 2010;49(12):1104–1113. doi: 10.1002/gcc.20818. [DOI] [PubMed] [Google Scholar]
- 20.Schwab CJ, Chilton L, Morrison H, et al. Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: association with cytogenetics and clinical features. Haematologica. 2013;98(7):1081–1088. doi: 10.3324/haematol.2013.085175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rand V, Parker H, Russell LJ, et al. Genomic characterization implicates iAMP21 as a likely primary genetic event in childhood B-cell precursor acute lymphoblastic leukemia. Blood. 2011;117(25):6848–6855. doi: 10.1182/blood-2011-01-329961. [DOI] [PubMed] [Google Scholar]
- 22.Case M, Matheson E, Minto L, et al. Mutation of genes affecting the RAS pathway is common in childhood acute lymphoblastic leukemia. Cancer Res. 2008;68(16):6803–6809. doi: 10.1158/0008-5472.CAN-08-0101. [DOI] [PubMed] [Google Scholar]
- 23.Wiemels JL, Zhang Y, Chang J, et al. RAS mutation is associated with hyperdiploidy and parental characteristics in pediatric acute lymphoblastic leukemia. Leukemia. 2005;19(3):415–419. doi: 10.1038/sj.leu.2403641. [DOI] [PubMed] [Google Scholar]
- 24.Malinowska-Ozdowy K, Frech C, Schönegger A, et al. KRAS and CREBBP mutations: a relapse-linked malicious liaison in childhood high hyperdiploid acute lymphoblastic leukemia. Leukemia. 2015;29(8):1656–1667. doi: 10.1038/leu.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeha S, Pei D, Raimondi SC, et al. Increased risk for CNS relapse in pre-B cell leukemia with the t(1;19)/TCF3-PBX1. Leukemia. 2009;23(8):1406–1409. doi: 10.1038/leu.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell LJ, Enshaei A, Jones L, et al. IGH@ translocations are prevalent in teenagers and young adults with acute lymphoblastic leukemia and are associated with a poor outcome. J Clin Oncol. 2014;32(14):1453–1462. doi: 10.1200/JCO.2013.51.3242. [DOI] [PubMed] [Google Scholar]
- 27.Irving JA, Minto L, Bailey S, Hall AG. Loss of heterozygosity and somatic mutations of the glucocorticoid receptor gene are rarely found at relapse in pediatric acute lymphoblastic leukemia but may occur in a subpopulation early in the disease course. Cancer Res. 2005;65(21):9712–9718. doi: 10.1158/0008-5472.CAN-05-1227. [DOI] [PubMed] [Google Scholar]
- 28.Zaliova M, Zimmermannova O, Dörge P, et al. ERG deletion is associated with CD2 and attenuates the negative impact of IKZF1 deletion in childhood acute lymphoblastic leukemia. Leukemia. 2014;28(1):182–185. doi: 10.1038/leu.2013.282. [DOI] [PubMed] [Google Scholar]
- 29.Palmi C, Valsecchi MG, Longinotti G, et al. What is the relevance of Ikaros gene deletions as a prognostic marker in pediatric Philadelphia-negative B-cell precursor acute lymphoblastic leukemia? Haematologica. 2013;98(8):1226–1231. doi: 10.3324/haematol.2012.075432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enshaei A, Schwab CJ, Konn ZJ, et al. Long-term follow-up of ETV6-RUNX1 ALL reveals that NCI risk, rather than secondary genetic abnormalities, is the key risk factor. Leukemia. 2013;27(11):2256–2259. doi: 10.1038/leu.2013.136. [DOI] [PubMed] [Google Scholar]
- 31.Holmfeldt L, Wei L, Diaz-Flores E, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45(3):242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waanders E, Scheijen B, van der Meer LT, et al. The origin and nature of tightly clustered BTG1 deletions in precursor B-cell acute lymphoblastic leukemia support a model of multiclonal evolution. PLoS Genet. 2012;8(2):e1002533. doi: 10.1371/journal.pgen.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Galen JC, Kuiper RP, van Emst L, et al. BTG1 regulates glucocorticoid receptor autoinduction in acute lymphoblastic leukemia. Blood. 2010;115(23):4810–4819. doi: 10.1182/blood-2009-05-223081. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt S, Irving JA, Minto L, et al. Glucocorticoid resistance in two key models of acute lymphoblastic leukemia occurs at the level of the glucocorticoid receptor. FASEB J. 2006;20(14):2600–2602. doi: 10.1096/fj.06-6214fje. [DOI] [PubMed] [Google Scholar]
- 35.Donner KM, Hiltunen TP, Jänne OA, Sane T, Kontula K. Generalized glucocorticoid resistance caused by a novel two-nucleotide deletion in the hormone-binding domain of the glucocorticoid receptor gene NR3C1. Eur J Endocrinol. 2012;168(1):K9–K18. doi: 10.1530/EJE-12-0532. [DOI] [PubMed] [Google Scholar]
- 36.Bokemeyer A, Eckert C, Meyr F, et al. Copy number genome alterations are associated with treatment response and outcome in relapsed childhood ETV6/RUNX1-positive acute lymphoblastic leukemia. Haematologica. 2014;99(4):706–714. doi: 10.3324/haematol.2012.072470. [DOI] [PMC free article] [PubMed] [Google Scholar]