Abstract
Over the past several decades, the biology of the developing lens has been investigated using molecular genetics-based approaches in various vertebrate model systems. These efforts, involving target gene knockouts or knockdowns, have led to major advances in our understanding of lens morphogenesis and the pathological basis of cataracts, as well as of other lens related eye defects. In particular, we now have a functional understanding of regulators such as Pax6, Six3, Sox2, Oct1 (Pou2f1), Meis1, Pnox1, Zeb2 (Sip1), Mab21l1, Foxe3, Tfap2a (Ap2-alpha), Pitx3, Sox11, Prox1, Sox1, c-Maf, Mafg, Mafk, Hsf4, Fgfrs, Bmp7, and Tdrd7 in this tissue. However, whether these individual regulators interact or their targets overlap, and the significance of such interactions during lens morphogenesis, is not well defined. The arrival of high-throughput approaches for gene expression profiling (microarrays, RNA-sequencing (RNA-seq), etc.), which can be coupled with chromatin immunoprecipitation (ChIP) or RNA immunoprecipitation (RIP) assays, along with improved computational resources and publically available datasets (e.g. those containing comprehensive protein-protein, protein-DNA information), presents new opportunities to advance our understanding of the lens tissue on a global systems level. Such systems-level knowledge will lead to the derivation of the underlying lens gene regulatory network (GRN), defined as a circuit map of the regulator-target interactions functional in lens development, which can be applied to expedite cataract gene discovery. In this review, we cover the various systems-level approaches such as microarrays, RNA-seq, and ChIP that are already being applied to lens studies and discuss strategies for assembling and interpreting these vast amounts of high-throughput information for effective dispersion to the scientific community. In particular, we discuss strategies for effective interpretation of this new information in the context of the rich knowledge obtained through the application of traditional single-gene focused experiments on the lens. Finally, we discuss our vision for integrating these diverse high-throughput datasets in a single web-based user-friendly tool iSyTE (integrated Systems Tool for Eye gene discovery) – a resource that is already proving effective in the identification and characterization of genes linked to lens development and cataract. We anticipate that application of a similar approach to other ocular tissues such as the retina and the cornea, and even other organ systems, will significantly impact disease gene discovery.
Keywords: Lens, Cataract, iSyTE, Gene Regulatory Networks, Bioinformatics, Transcription factors, RNA binding proteins
Systems biology is the study of how individual components collaborate with each other in regulatory networks to develop and maintain a cell or tissue (Chuang et al., 2010). These networks constitute a genetic circuitry to form and control cell/tissue behavior, and are represented in a graphical manner analogous to an electric circuit with switches that turn “on” or turn “off”, or turn “on at differential levels” (analogous to a light dimmer) regulators that control their targets to produce a specific output (Cvekl and Duncan, 2007; Lachke and Maas, 2010; Peter and Davidson, 2011). Systems biology requires the application of technologies that allow, on a high-throughput level, the measurement of changes in: genomic sequence (genomics, e.g. DNA sequencing, array-CGH, etc.), RNA (transcriptomics, e.g. RNA-seq, microarrays, etc.), proteins (proteomics, e.g. two-dimensional difference gel electrophoresis (2DIGE) coupled with mass spectrometry (MS); Yeast or mammalian two hybrid assays, etc.) and carbohydrates (metabolomics, e.g. nuclear magnetic resonance (NMR)). These approaches can be used to measure the “molecular state” of a tissue or “molecular interactions” within a tissue. For example, RNA-seq or microarrays can be applied to measure the total number and type of transcripts (messenger RNAs (mRNAs) and non-coding RNAs (ncRNAs)) expressed in a wild-type tissue at a specific developmental stage, defined as its “transcriptome”. They can also be applied to compare a mutant tissue with a normal control. While the former reveals the molecular state of a tissue by cataloging its transcriptome in normal development, the latter provides first-order insights into the potential interactions of “regulator” molecules (e.g. mutant gene/protein) with “target” molecules (e.g. transcripts altered in mutant tissue), albeit these perturbations may not all result from direct physical interactions between the regulators and its targets.
In addition to the well-established regulatory molecules such as proteins that control gene expression (e.g. transcription factors (TFs)), recent findings have identified ncRNAs as factors that control a cell’s proteome (Morris and Mattick, 2014; Pauli et al., 2011). For example, small ncRNAs (e.g. microRNAs (miRNAs)) facilitate mRNA decay or silencing (translational inhibition) (Brosnan and Voinnet, 2009), whereas long ncRNAs (lncRNAs) modulate chromatin, among other regulatory functions (Hu et al., 2012; Kugel and Goodrich, 2012). Further insights into the regulator/target relationship are obtained through ChIP-seq (Chromatin immunoprecipitation followed by DNA-sequencing) if the regulator is a DNA-binding protein, or RIP-seq/CLIP-seq (RNA immunoprecipitation or Cross link immunoprecipitation followed by RNA-sequencing) if the regulator is an RNA-binding protein (RBP). These approaches can identify direct targets of a regulator in vivo.
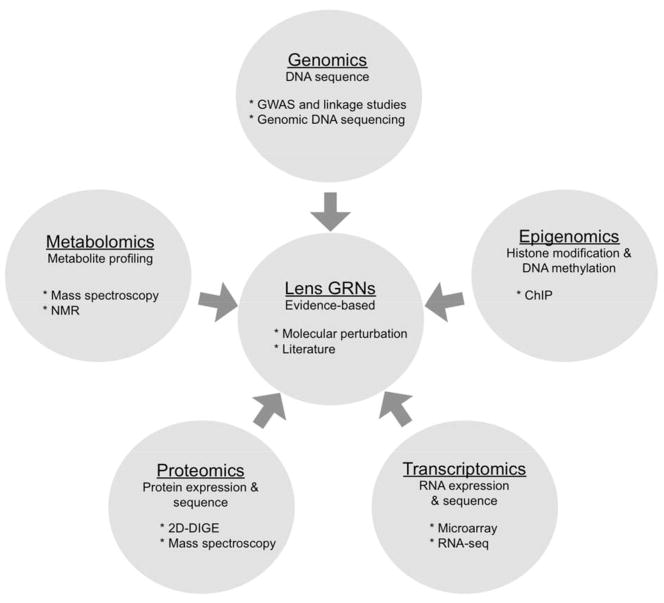
Thus far, the various systems-level approaches including genomics, transcriptomics, proteomics and metabolomics have all been applied to characterize lens biology and cataract (Fig. 1), several of which are discussed in detail in later sections, and for which the following studies are representative (Chauhan et al., 2002b; Hammond et al., 2001; Hawse et al., 2003; Lachke et al., 2012b; Lampi et al., 2002; Sousounis et al., 2013; Tsentalovich et al., 2015; Ueda et al., 2002; Yanshole et al., 2014). These approaches have generated immense quantities of data. For example, a single gene expression microarray chip (e.g. Affymetrix GeneChip® Mouse Genome 430 2.0 Array) has thousands of probe-datasets informing on the expression of 39,000 transcripts that are representative of ~22,000 genes (130 megabytes data). Further, a single RNA-seq experiment using next-generation sequencing technology generates >400 million sequence-reads that are each 100 nucleotides long (~10 gigabytes data) (Scholz et al., 2012). However, analysis of these large datasets for expediting discovery of new genes linked to lens development and cataracts has been limited (Lachke et al., 2012b; Sousounis and Tsonis, 2012).
Figure 1. Systems-level approaches to study lens biology.
Information from genomics, epigenomics, transcriptomics, proteomics, metabolomics approaches are already being applied to study lens biology. Integration of these various high-throughput data will require the development of a web-based community resource. Such a resource will enable the derivation, visualization, and analysis of the spatio-temporal gene regulatory networks associated with lens development and homeostasis.
In addition to systems-level approaches, lens research has involved the functional characterization of individual regulatory molecules. However, the application of this rich molecular knowledge to broaden our understanding of the relationship between newly-identified regulators with established regulators on a global network-level, in a spatio-temporal context, has not been attempted. For example, presently there are no tools to address how the newly identified cataract-linked TF Mafg (Agrawal et al., 2015) and the RBP Tdrd7 (Lachke et al., 2011) fit within the context of existing lens regulatory knowledge. We propose that integration of lens gene-expression knowledge with functional data from molecular perturbation analyses can lead to an analytical resource-tool. Such a resource-tool will allow a comprehensive analysis of any regulatory data in the broader context of lens biology and will predict new regulators and targets (e.g. genes and their product RNA or proteins defined as “nodes”) and their relationships (termed as “edges” between nodes) in the lens.
We articulate a strategy for building this resource-tool, by reviewing: (1) the current functional understanding of key lens regulators, (2) genome-level expression profiling of the lens developmental transcriptome in normal and pathological states, and (3) ChIP and RIP approaches that provide insights on lens GRNs. We then highlight approaches to integrate this rich information for effective interpretation of new lens data from perturbation experiments. In particular, we discuss how the existing wealth of functional data from molecular genetics-based approaches can be converted into a systems-level representation, which will allow derivation of a “core” lens developmental GRN (gene regulatory network). We anticipate that a detailed spatio-temporal core lens GRN will serve as a base for overlaying other high-throughput data, leading to the predictions of new nodes and edges, which will impact both the basic understanding of lens development as well as cataract gene discovery and pathways for therapeutic targets. We also anticipate that a similar derivation of core GRNs for other eye components will expedite ocular gene discovery and provide insights into the molecular cross-talk between these developing tissues.
1. Molecular biology of lens development: a brief overview
During development, precise control of gene expression is achieved through distinct levels of regulation. These include modulation of chromatin structure and epigenetic memory, transcriptional regulation, and post-transcriptional control of RNA stability and translation. Thus far, molecular analysis of gene expression control in vertebrate lens development has identified several chromatin regulators and transcription factors (TFs) with early function during lens induction (Cvekl and Ashery-Padan, 2014; Lachke and Maas, 2010; Ogino et al., 2012).
Early in lens development, the lysine acetyltransferases CBP and p300 that function to modify core histones proteins are necessary in establishing cell identify (Wolf et al., 2013b). These epigenetic modifications facilitate expression of key TFs Six3, Pax6 and Sox2 that function to initiate lens placode formation (Cvekl and Ashery-Padan, 2014; Wolf et al., 2013b). The transcriptional co-activator Pygo2 also functions to positively control Pax6 expression in a Wnt-independent pathway during lens induction (Song et al., 2007). Further, signaling from the optic vesicle, reflecting the activities of Raldh2, Hes1, Lhx2, Mab21l2, Rx, Fgf, Bmp4, and Bmp7, positively regulate Pax6 expression during lens induction (Cvekl and Ashery-Padan, 2014; Lachke and Maas, 2010). Further, the precise control of ERK signaling by the GTPase-activating protein Nf1 is also important for lens induction (Carbe and Zhang, 2011). Pax6 is also regulated by TALE-family TFs Meis1 and Pknox1 (Prep1) (Rowan et al., 2010; Zhang et al., 2002) and by Six3, the Sox2-Oct1 complex, or by its own activity, in positive feedback loops (Donner et al., 2007a; Liu et al., 2006). In turn, Pax6 regulates the TFs Sox11 and AP-2α that are involved in lens placode invagination and lens vesicle separation, respectively (Pontoriero et al., 2008; Wurm et al., 2008), while another TF Pitx3 is required for maintenance of the anterior epithelium of the lens (AEL) (Ho et al., 2009). Pax6 also positively regulates the TF Mab21l1 that in turn up-regulates the AEL-restricted TF Foxe3, which is also up-regulated by a zinc-finger TF Zeb2 (Sip1) (Manthey et al., 2014a; Yoshimoto et al., 2005) and the TF Msx2 (Zhao et al., 2012). In later stages, the TFs Foxe3 and Prox1, in AEL and transition zone/fiber cells, respectively, function to control of epithelial cell-cycle exit and fiber cell differentiation (Audette et al., 2016; Blixt et al., 2000; Brownell et al., 2000; Landgren et al., 2008; Medina-Martinez et al., 2005; Wigle et al., 1999; Yamada et al., 2003).
Several TFs that function in fiber cell differentiation and homeostasis are identified. Prox1, Sox1, and the large Maf protein, Maf (c-Maf) positively regulate crystallin-encoding genes in fiber cells (Audette et al., 2016; Donner et al., 2007b; Kawauchi et al., 1999; Kim et al., 1999; Nishiguchi et al., 1998; Ring et al., 2000; Wigle et al., 1999). Interestingly, Mafg and Mafk are linked to early-onset age-related cataract and control non-crystallin cataract genes (Agrawal et al., 2015). Not surprisingly, several TF mutations (e.g. FOXE3, HSF4, MAF, PAX6, PITX3) are directly linked to cataract in humans (Bu et al., 2002; Glaser et al., 1994; Jamieson et al., 2002; Semina et al., 2001, 1998).
In addition to TFs, several signaling factors (e.g. Fgf, Bmp, etc.) function in lens development and fiber differentiation (Belecky-Adams et al., 2002; Faber et al., 2001; Furuta and Hogan, 1998; Garcia et al., 2011, 2011, 2005; Huang et al., 2015, 2003; Kuracha et al., 2011; Li et al., 2014; Madakashira et al., 2012; Rajagopal et al., 2008; Simirskii et al., 2007; Sugiyama et al., 2013; Upadhya et al., 2013; Wawersik et al., 1999; Zhang et al., 2016; Zhao et al., 2008). These topics are covered elsewhere (Cvekl and Ashery-Padan, 2014).
Investigation of ncRNAs in lens biology has been initiated (Frederikse et al., 2006; Li and Piatigorsky, 2009; Shaham et al., 2013; Wolf et al., 2013a; Xu, 2009). miRNAs are a subset of ncRNAs involved in post-transcriptional regulation by direct binding of target mRNAs to initiate their decay or inhibit translation (Brosnan and Voinnet, 2009; Morris et al., 2010). Lens- and cornea-specific Dicer1 null mice exhibit severe lens and corneal defects, indicating the requirement of miRNAs in their development (Li and Piatigorsky, 2009; Wolf et al., 2013a).
miRNA function in FGF2-induced fiber cell differentiation has been investigated in a rat lens epithelial explant culture model. This has identified 131 miRNAs, some of which (miR-143, miR-155, miR-301a) down-regulate c-Maf in reporter assays (Wolf et al., 2013a). Another miRNA, miR-204, has been investigated in Medaka fish and mouse lens development. miR-204 knockdown in Medaka causes Meis2 mis-regulation and lens defects (Conte et al., 2010). In mouse lens vesicle, Pax6 positively regulates miR-204 that in turn controls expression of several targets including negative regulation of Sox11 (Shaham et al., 2013). miRNAs expression is also described in rat cataractous lenses (Kubo et al., 2013). Besides miRNAs, the newborn mouse lens expresses >250 lncRNAs, a third of which are differentially regulated in AEL and fibers (Hoang et al., 2014). Differential expression of lncRNAs has also been investigated in human age-related cataract lenses (Shen et al., 2016), and recently ncRNAs have been profiled in mouse lens development (Khan et al., 2016). Future investigations should further define ncRNA function in lens development and homeostasis.
Finally, two conserved RBPs Tdrd7 and Caprin2 are linked to the lens-associated defects congenital cataracts and Peters anomaly, respectively. While Tdrd7 mediates post-transcriptional regulation in fiber cells (Lachke et al., 2011), Caprin2 functions to control the lens-cornea separation and the central fiber cell “nucleus” zone area (Dash et al., 2015). RBPs in lens development are discussed in a separate review article (Dash et al. 2016). For space limitations, we will refrain from discussing proteomics and metabolomics approaches, which have provided important information, especially regarding the post-translational modifications and physiological changes associated with cataract (Lampi et al., 2002; Tsentalovich et al., 2015; Ueda et al., 2002; Yanshole et al., 2014). Together, these studies have identified the various signaling factors, TFs, RBPs, and miRNAs that are important in lens development and homeostasis.
2. Genome-level expression profiling of lens development
Over the past 15 years, genome-wide expression profiling of the lens has been performed in several vertebrate species, initially by serial analysis of gene expression (SAGE) and microarrays, and recently by RNA-seq. These efforts have generated large expression datasets for normal and defective lens development (Agrawal et al., 2015; Anand et al., 2015; Audette et al., 2016; Chauhan et al., 2002a, 2002b; Chauss et al., 2014; De Maria and Bassnett, 2015; Greiling et al., 2009; Hawse et al., 2005, 2004, 2003; Hoang et al., 2014; Ivanov et al., 2005; Khan et al., 2016, 2015; Kumar et al., 2015; Lachke et al., 2012b, 2011; Manthey et al., 2014a; Sousounis et al., 2015, 2013; Sousounis and Tsonis, 2012; Wistow et al., 2002; Wolf et al., 2013b; Wride et al., 2003; Xiao et al., 2006). Although highly informative, early microarray studies used platforms representing only a subset of genes (n=9,700) in the mouse genome (Chauhan et al., 2002a, 2002b). Subsequent development of commercial microarray platforms by Affymetrix and Illumina offered a comprehensive representation of the mouse genome (n=~22,000) (Lachke et al., 2012b, 2011). However, even these platforms, which are inherently limited by probe-sequences for known transcripts, are not representative of all the possible transcripts encoded by the genome.
Because it is not limited to detecting only known transcripts, RNA-seq allows identification of new transcripts, while providing a deep coverage of both protein-coding and non-coding transcripts, as well as alternate splice forms. Further, RNA-seq can be applied to profile smaller tissue samples from early lens developmental stages or pools of isolated AEL or fiber cells. Indeed, recent studies describe RNA-seq profiling of isolated epithelium and fiber cells from newborn mouse lenses (Hoang et al., 2014; Sun et al., 2015a, 2015b). Moreover, these high-throughput approaches combined with laser capture micro-dissection can profile at the cell region-specific level (i.e. outer cortical fiber cells, inner cortical fiber cells, etc.) (De Maria and Bassnett, 2015; Ivanov et al., 2005; Wolf et al., 2013b; Xie et al., 2013). In addition to normal lens development, high-throughput expression profiling has been applied for characterizing lens defects from mouse knockouts for genes encoding chromatin regulators (CBP, p300), TFs (Tfap2a (Ap2-alpha), Hsf4, Mafg, Mafk, Pax6, Prox1, Zeb2 (Sip1)), RBPs (Tdrd7), miRNA processors (Dicer1), and glycoproteins (Sparc), some of which are discussed below.
Initial efforts applied custom cDNA microarrays to compare Pax6+/+ and Pax6+/− mouse lenses, identifying >500 differentially expressed genes (DEGs), including Pitx3, Cryab and Cryba1/3 among the down-regulated genes (Chauhan et al., 2002b). Affymetrix microarray-based analysis of P1 stage Pax6+/− mouse lenses has identified >550 DEGs, including Tgfb2, suggesting that a subset of Pax6-controlled targets are common between the lens and forebrain (Wolf et al., 2009). Microarrays on Tfap2a (Ap2-alpha) conditional-knockout (cKO) mouse lenses revealed >400 DEGs including down-regulation of AEL-expressed genes (Pontoriero et al., 2008). Conversely, in mice overexpressing Foxe3 in fiber cells, microarray profiling identified down-regulation of fiber differentiation genes (Landgren et al., 2008).
Microarrays on Mafg−/−:Mafk+/− compound mouse lenses identified mis-regulation of several non-crystallin-encoding cataract-associated genes from the oxidative stress and sterol synthesis pathways (Agrawal et al., 2015). This is interesting because lanosterol synthase gene mutations cause inherited cataracts in humans and sterol-based compounds can reverse lens protein-aggregation in animal models (Makley et al., 2015; Zhao et al., 2015). Thus, Mafg−/−:Mafk+/− mice represent a new model for age-related cataract, and may offer insights into regulation of sterol pathway genes in the lens.
Further, Dicer1-cKO mouse lens microarray profiles reveal down-regulation of several crystallins and other lens factors, while key lens TFs (Pax6, Prox1, Pitx3, c-Maf) are unaltered (Li and Piatigorsky, 2009). Microarray analysis of mouse mutant lenses of a post-transcriptional regulator gene Tdrd7 identified several DEGs including the heat shock protein Hspb1 (Hsp27) (Lachke et al., 2011), while microarrays on mouse lenses with a deletion in the matricellular glycoprotein gene Sparc showed altered expression of genes encoding matrix and adhesion proteins, cytoskeletal proteins and signaling molecules (Greiling et al., 2009).
Microarray-based comparative analysis of human aged-cataract and normal lenses has identified >900 down-regulated genes, among which are cataract-linked genes such as TDRD7 (Hawse et al., 2003). Clustering analysis of down-regulated candidates in cataract lenses reveal enrichment of genes for protein synthesis, oxidative stress, heat shock/chaperone activity and lens structural components. In addition to RNA-seq on newborn mouse lens isolated epithelium and fiber cells (Hoang et al., 2014; Sun et al., 2015a, 2015b), microarrays have been performed on these cells types from P13 mouse lenses (Nakahara et al., 2007). Finally, miRNA and ncRNA profiling has been performed on normal lens (Karali et al., 2010; Khan et al., 2016, 2015; Kubo et al., 2013; Wu et al., 2012) or lens cell lines (Tian et al., 2010). Thus, even a brief overview highlights the wealth of genome-level expression data that is available on lenses from different stages in normal and defective developmental states.
3. Protein-nucleic acid interaction-profiling for lens GRNs
The Cvekl laboratory has spear-headed the efforts on ChIP analysis on the lens. ChIP combined with downstream analyses such as ChIP-Chip (“Chip”, microarrays), ChIP-seq (seq, high-throughput DNA-sequencing) or ChIP-PCR identifies the genomic regions occupied in vivo by a DNA-binding protein, thus informing on its direct targets.
A systematic approach of genome-wide measurement of ChIP-based protein-DNA interactions compared with RNA expression profiling in appropriate mutant lenses has revealed critical information on the Pax6-controlled transcriptional regulatory network in the lens. For example, comparative analysis of Pax6-target genomic regions identified in the lens by ChIP-Chip assays with microarray data on Pax6+/− lenses prioritized 76 Pax6-direct targets that are validated by reporter assays (Wolf et al., 2009; Xie et al., 2013). The application of ChIP-seq and formaldehyde-assisted identification of regulatory elements followed by DNA-seq (FAIRE-seq) to analyze newborn mouse lens chromatin have extended these findings (Sun et al., 2015a, 2015b). This approach identified three Pax6 cis-binding motifs that reflect binding through its paired domain, homeodomain, or a combined use of both domains in the lens in vivo (Sun et al., 2015b).
While several approaches identify open chromatin structure, application of FAIRE-seq identifies nucleosome-free regions that potentially support TF-binding, and therefore can identify regulatory cis-regions (Giresi et al., 2007). Thus, FAIRE-seq-mapped regions, combined with post-translational histone modification and RNA Polymerase II occupancy data, provide insights into the extent of transcription of specific loci and the TFs involved in their regulation in vivo. Indeed, FAIRE-seq demonstrated that while Cryaa is the most highly-expressed gene in AEL, Hsf4 is among the highly expressed TFs in lens fibers (Sun et al., 2015a).
These experiments have been challenging because of the large volume of lens tissue required. The recent molecular characterization of mouse lens epithelial cell lines (Terrell et al., 2015) and the establishment of protocols for iPSc-based derivation of lens cells (Anchan et al., 2014; Yang et al., 2010) may facilitate their use in ChIP assays, addressing this problem. Modified assays such as “recovery via protection” (RP)-ChIP-seq and favored amplification RP-ChIP-seq, have recently been developed to capture low-abundance chromatin from a single lens, or from a few hundred cells (~500) (Zheng et al., 2015). Because these assays work on single lens tissues, their potential application to embryonic lens tissue now offers new opportunities to investigate epigenetic regulation in early lens development.
Analogous to identifying lens cis-regulatory targets of DNA-binding proteins, the recent identification of RBPs Tdrd7 and Caprin2 in lens development (Dash et al., 2015; Lachke et al., 2011) has initiated the application of assays that identify protein-RNA targets. RIP and CLIP assays combined with microarrays or RNA-seq allow genome-wide analysis of RBP-RNA interactions (Jain et al., 2011; König et al., 2011; Li et al., 2015; Milek et al., 2012; Modic et al., 2013; Re et al., 2014). RIP analysis on the lens epithelial cell line 21EM15 identified Hspb1 (Hsp27) among Tdrd7-RNA targets (Lachke et al., 2011). As new lens RBPs are identified, performing these assays on genome-level on lens tissue will be necessary. Together, these approaches will identify the direct transcriptional and post-transcriptional regulator-target interactions in lens development.
4. Future of lens research: toward lens systems biology
Mammalian lens morphogenesis involves complex molecular and embryonic events, including coordinate interactions with the developing retina and cornea (Cvekl and Ashery-Padan, 2014; Donner et al., 2006). Defects in these processes cause structural birth defects – including congenital cataract and Peters anomaly, among others – that can be inherited as a primary defect or as part of syndromes. Identification of cataract-linked genes has been challenging. For example, the majority of ~30 genes associated with isolated (non-syndromic) human congenital cataracts were identified over a 20-year period (Shiels et al., 2010; Shiels and Hejtmancik, 2013). Besides studies on animal models such as chicken, Xenopus and zebrafish, eye gene discovery has advanced by characterization of mouse ocular mutants in forward-genetics screens or targeted deletions and mapping of human inherited cataracts. However, these approaches face a common challenge - that of prioritization of candidate genes, namely in the mapped genomic regions, which persists even in this age of exome-sequencing, because it is not straightforward to recognize the causative mutation among the many variants identified in the proband. Furthermore, identification of several important genes in eye development has benefitted from their initial characterization in Drosophila, which is not always feasible (Charlton-Perkins et al., 2011; Gehring, 2014; Wawersik and Maas, 2000). Thus, new approaches to expedite eye gene discovery are necessary. Toward this objective, we had hypothesized in the past that comprehensive gene expression analysis of eye development will yield key information for identifying ocular disease-associated genes (Lachke and Maas, 2010). In the following sections, we review the progress so far toward this goal and propose future directions to achieve a comprehensive systems-level understanding of the lens.
4.1 iSyTE: integrated Systems Tool for Eye gene discovery
A few years ago, we and others recognized that genome-wide expression profiling holds the potential to impact gene discovery in lens development and disease, and therefore generated microarray data on mouse and human lenses in both normal and cataract conditions (Chauhan et al., 2002a, 2002b; Hawse et al., 2005, 2004, 2003; Ivanov et al., 2005; Lachke et al., 2012b, 2011; Lachke and Maas, 2010; Xiao et al., 2006). Besides these efforts, high-throughput expression profiling was performed on lenses in other animal models (Chauss et al., 2014; Sousounis et al., 2015, 2013). However, the strength of high-throughput expression profiling – providing genome-wide data – perhaps poses its greatest challenge, because these approaches identify thousands of transcripts that are scored as “expressed”. Thus, it is challenging to identify the select genes that are important for morphogenesis and homeostasis of a specific tissue, and are associated with developmental defects. Further, gene expression analyses are comparative and are effective when an appropriate reference is available (Control vs. Treatment). However, in normal developing tissue, comparisons are challenging as there is no obvious “control” dataset.
To address these challenges through generation of developmental cell/tissue-enriched expression profiles, a strategy termed “whole-embryo body (WB) in silico subtraction” was applied to develop a resource-tool called iSyTE (http://bioinformatics.udel.edu/Research/iSyTE) (Lachke et al., 2012b). The underlying principle is tissue-enrichment (and not just tissue-expression) may reflect importance. The enrichment strategy is based on the identification of transcripts with significantly high molar concentrations in a cell/tissue of interest, as compared to WB. This strategy is advantageous because it identifies transcripts that may not necessarily have high absolute expression, but yet have high enrichment in a tissue. This is because transcripts with low absolute expression in a tissue can still have high molar concentration in that tissue compared to WB and can therefore be scored as “tissue-enriched”. Thus, Pax6 mRNA, which is expressed at significantly lower absolute levels than housekeeping genes, but nevertheless is highly lens-enriched (high molar concentration in lens compared to WB), and therefore effectively identified as a high-priority candidate, while housekeeping genes are not (as they are constitutively expressed and not lens-enriched).
Because iSyTE in silico subtraction makes no a priori assumptions on candidate genes and is based solely on their lens-enrichment scores, it can provide unprecedented insights into lens biology. This is appreciated from the following example. iSyTE identified two new lens RBPs Tdrd7 and Caprin2, thereby driving post-transcriptional regulation studies in lens development (Dash et al., 2015; Lachke et al., 2011; Lachke and Maas, 2011). These findings are significant also because although the human genome encodes ~1500 RBPs, few are directly linked to developmental disorders (Neelamraju et al., 2015).
Within the past four years iSyTE has facilitated the identification and characterization of several new genes encoding TFs, RBPs, cell adhesion proteins, and selenoproteins associated with cataract or lens defects (Tdrd7, Pvrl3, Sep15, MafG, MafK) (Agrawal et al., 2015; Dash et al., 2015; Kasaikina et al., 2011; Lachke et al., 2012a, 2012b, 2011) and has contributed to the understanding of many other important regulatory pathways in the lens (e.g. Sip1 (Zeb2), CBP, p300, Prox1, etc.) (Audette et al., 2016; Manthey et al., 2014a; Wolf et al., 2013b). Further, it has impacted the characterization of lens cell lines (Terrell et al., 2015) and establishment of protocols for processing lens microarray and RNA-seq data (Anand et al., 2015; Manthey et al., 2014b).
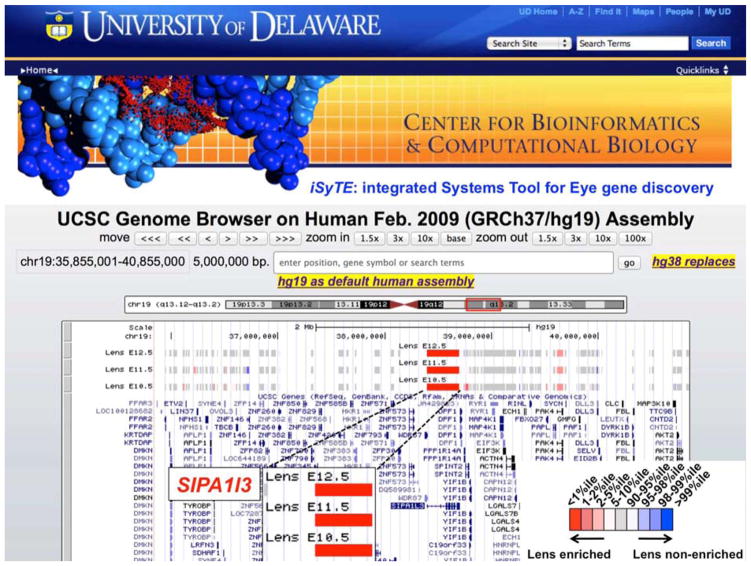
In addition to directly predicting cataract-linked genes, iSyTE has provided fundamental biological insights into challenging clinical eye-related cases wherein the causative mutation was elusive. Even with deep-sequencing, identifying the causative mutation is tricky, especially if the mutation is non-exonic. Thus, such analysis benefits from additional biological insights through iSyTE. Indeed, several studies have successfully applied iSyTE in their investigations of eye and lens defects in human patients, namely for: 1) prioritization of novel candidate genes for pediatric cataract (Aldahmesh et al., 2012); 2) prioritization of promising candidates for anophthalmia and microphthalmia (Schilter et al., 2013); 3) linking ADAMTS18 in ocular syndrome in microcornea and myopia (Aldahmesh et al., 2013), 4) linking STX3 to congenital cataract (Chograni et al., 2014), and 5) identifying ASPH mutations that are associated with Traboulsi syndrome with ocular lens dislocation (Patel et al., 2014). Further, in 2012 iSyTE correctly predicted SIPA1l3 as a cataract-linked gene (Fig. 2) (Lachke et al., 2012b), which was recently confirmed by two groups who independently described SIPA1l3 mutations or deficiency to cause cataracts in human, zebrafish and mouse (Evers et al., 2015; Greenlees et al., 2015). Thus, the first version of iSyTE has strongly impacted lens and cataract research.
Figure 2. iSyTE predicts SIPA1l3 as a cataract gene.
iSyTE predicts SIPA1l3 as high-priority candidate gene in the lens among a 5Mb interval on human chromosome 19. Also see: (Lachke et al. 2012b). The color gradation in the inset offers an estimate of enriched expression of the genes in mouse lens tissue at embryonic day (E)10.5, E11.5 and E12.5. SIPA1l3 is among the top 1% of lens-enriched genes. This prediction was independently proven by two groups that recently reported SIPA1l3 mutations in human congenital cataract (Evers et al. 2015, Greenlees et al. 2015).
4.2 Integration of new high-throughput lens expression data
Although effective in lens gene discovery, iSyTE’s first version is based on only three mouse embryonic-stage lens microarray datasets. However, there is a wealth of lens whole-genome expression data from wild-type and gene-deletion mouse mutants over a range of stages that can be integrated into iSyTE to increase its efficacy. Presently, mouse lens microarray data is available on various wild-type embryonic and postnatal stages (E9.5, E10.5, E11.5, E12.5, E15.5, E16.5, E17.5, E19.5, P0, P2, P4, P8, P12, P20, P28, P30, P42, P52, P56, P60) on Affymetrix 430 2.0 and Illumina mouse WG6 chips. Further, mouse lens RNA-seq data is available on stages E13.5, E15, E15.5, E18, P0, P3, P6 and P9 (Audette et al., 2016; Hoang et al., 2014; Khan et al., 2016, 2015; Manthey et al., 2014a; Sun et al., 2015a, 2015b). Moreover, RNA-seq data is available from newborn mouse isolated lens epithelium and fiber cells (Hoang et al., 2014; Sun et al., 2015a, 2015b). RNA-seq data has also been generated from distinct cellular regions of the E13 chicken lens, such as the central epithelium, equatorial epithelium, cortical fiber cells, and central fiber cells (Chauss et al., 2014). Furthermore, microarray or RNA-seq data on the following mouse mutants with lens defects are available for distinct stages (in parenthesis): CBP, p300 (E9.5, E10.5), Pax6 (E9.5, E10.25), Prox1 (E13.5), Zeb2/Sip1 (E15.5), Brg1 (E15.5), Notch2 (E19.5), Hsf4 (P0), E2f123 (E17.5, P0), Foxe3 (P2; overexpression mutant), Sparc (P28; lens epithelium), Tdrd7 (P4, P30) and Mafg/k (P60).
All the above data can be processed by in silico subtraction using WB reference datasets on appropriate platforms for identifying lens-enriched genes in embryonic, early postnatal and adult stages. This will generate a comprehensive lens-expression database whose application to analyze genome-wide expression perturbation data from regulator mutants will prioritize targets and provide insights into the stage-specific dynamics of such interactions (Fig. 3). It will also identify targets shared between a new regulator and known regulators. New sequence alignment tools to reduce false positives among lens RNA-seq-identified DEGs will be useful in this analysis (Kumar et al., 2015). Finally, lens-enriched expression data can be analyzed in the context of ChIP data for Pax6 (Sun et al., 2015b; Xie et al., 2013) or other lens chromatin data (Sun et al., 2015a; Wolf et al., 2013b) for prioritizing lens-linked candidates.
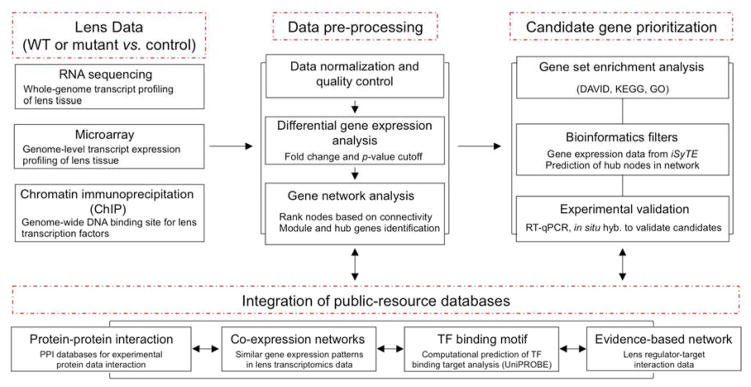
Figure 3. An integrated approach flow chart to analyze lens transcriptomics data.
High-throughput lens expression and functional data analysis involves distinct steps. The lens transcriptomics data (RNA-seq and microarray) and ChIP data requires data preprocessing steps (normalization and quality control), identification of differentially expressed genes (DEGs) and network analysis. Candidate gene prioritization of DEGs includes functional pathway analysis (functional annotation clustering by DAVID tool and KEGG pathway analysis) and relevance to the lens (iSyTE-analysis). Development of new hypotheses requires integration of knowledge within various publically available informative resources such as protein-protein interactions databases, derivation of co-expression networks (high-throughput expression data of normal lens development), computational prediction of TF-binding motifs (UniPROBE data) and published lens-literature (experimentally validated molecular evidence-based data). This integrative analysis approach of high-throughput gene expression data from wild-type and mutant lens tissue from targeted gene deletion experiments will facilitate prioritization of candidate genes and pathways in the lens.
4.3 Derivation of “core” lens GRNs
In addition to expression data, our knowledge of lens development still largely derives from conventional – but effective – application of molecular genetics, published as peer-reviewed research, the advantage of which is the data are experimentally validated. To extract biologically-relevant information from microarrays or RNA-seq, it is necessary to assemble the wealth of experimentally-validated knowledge as a systems-friendly platform. Single gene perturbation-derived regulatory data-points, currently present in the published literature with limited connectivity, hold the potential for such a representation (Djordjevic et al., 2014; O’Connell et al., 2012). The potential connectivity in isolated lens molecular data, if analyzed, assembled, and processed by an algorithm effectively, can lead to the development of evidence-based “core” developmental GRNs (Djordjevic et al., 2014; Lachke and Maas, 2010; O’Connell et al., 2012). Similar approaches have generated evidence-based GRNs in tooth and heart development (Djordjevic et al., 2014; O’Connell et al., 2012). Thus, such evidence-based core lens developmental GRNs can be assembled by essentially converting molecular biology data into systems-level representation.
In such GRNs, the nodes (factors such as protein, RNA) and their edges (nature of the connection/relationship with other nodes, e.g. DNA-binding protein directly binding to target genomic region) are purely based on experimental evidences from lens, and therefore provide a strong foundation to further expand the network using other informational datasets. Thus, application of protein-protein, protein-DNA and protein-RNA interaction data (e.g. Human protein interactome, BIND, REACTOME, etc.) (Rolland et al., 2014) can expand the core lens GRN. Such new expanded relationships can be analyzed with iSyTE lens expression, which will serve to filter the predicted edges that are irrelevant to lens tissue (e.g. a new protein-protein edge that was predicted based on their interactions in a different tissue, but the newly predicted target protein is found to be absent in the lens), and retain the predicted edges that are potentially significant (e.g. a new protein-protein edge in which the newly predicted target protein exhibits lens expression or enriched-expression). Thus, this integrative approach can be applied to “grow” the core lens GRNs.
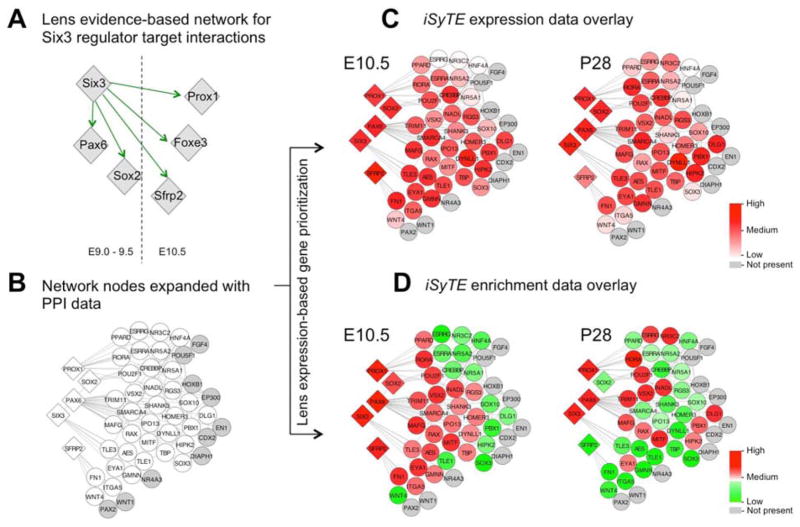
This approach is illustrated by the expansion of a lens sub-network for Six3. Experimental evidence indicates that Six3 deficiency causes reduced expression of Pax6 and Sox2 in the surface ectoderm between E9.0–9.5, and in the reduced expression of Foxe3, Sfrp2 and Prox1 in the lens at E10.5 (Fig. 4A). This basic Six3 lens sub-network can be “grown” by incorporating protein-protein interaction data (Fig. 4B), which predicts new candidates linked to the targets that are affected in the Six3 mutant lens. Overlaying iSyTE lens-expression further strengthens these predictions by retaining candidates expressed in the lens (Fig. 4C). Because lens-enrichment is indicative of function in the lens, overlaying iSyTE lens-enrichment serves to further refine candidates as “high-priority” in the lens (Fig. 4D). Because Six3 is AEL-enriched in later stages, isolated lens epithelium data can be applied (Fig. 4C, D). This prioritizes several nodes (e.g. Pou2f1 (Oct1), Mafg, Eya1) that offer new insights into the pathobiology of the Six3-mutant lens defect (Fig. 4). This approach demonstrates that integration of core lens GRNs and iSyTE lens expression can predict new nodes for future experimental validation. Other datasets such as DNA-binding motif data from UniPROBE (Hume et al., 2015) can be applied for further predictions. The Six3 case-study suggests that a comprehensive core lens GRN will present greater opportunities for new predictions in the context of established knowledge in lens biology.
Figure 4.
Integrated network analysis to identify lens-specific Six3 interacting candidates. (A) Six3 lens perturbation data from published literature (evidence-based). (B) Expansion of Six3 evidence-based network using human protein-protein interaction (PPI) data for SIX3 to identify its immediate interacting partners (nodes). (C) Overlay of iSyTE expression data from E10.5 mouse lens tissue and P28 mouse lens isolated epithelium tissue on SIX3 PPI nodes, allowing the identification of high-priority lens relevant candidate nodes. (D) Overlay of iSyTE enriched expression data from E10.5 mouse lens tissue and P28 mouse lens isolated epithelium tissue on SIX3 PPI nodes, further refining the identification of high-priority candidates.
4.4 Deciphering cross-talks between lens GRNs and other eye tissue networks
Coordinate interactions between distinct ocular tissues influence their morphogenesis. For example, Bmp4, Raldh2, Hes1, Lhx2, Mab21l2 and Rx activities in the optic vesicle are necessary for lens development (Cvekl and Ashery-Padan, 2014; Lachke and Maas, 2010), while the lens is necessary for proper retinal placement (Ashery-Padan et al., 2000). At an earlier stage in chicken, the pre-lens ectoderm is required for optic cup morphogenesis (Hyer et al., 2003). Chicken studies also suggest that lens ectoderm-derived Bmp activity specifies retinal cells (Pandit et al., 2015). Further, Dr. David Beebe’s findings indicate that the lens is necessary for organizing the anterior segment through control of corneal endothelium (Beebe and Coats, 2000). These data were reinforced by an in situ screen that additionally showed lens-based induction of iris and ciliary body-expressed factors (Thut et al., 2001). Application of lens GRN approaches can derive other eye tissue-GRNs, contributing to the “oculome” (Lachke and Maas, 2010), whose spatiotemporal analysis will unravel the molecular interplay that coordinates ocular morphogenesis. Dr. David Beebe’s group has already contributed toward this goal by generating isolated optic cup and lens placode microarrays on wild-type and various gene-targeted mice (Dr. Beebe, personal communication).
4.5 An integrative approach to investigate lens molecular defects
RNA-seq and microarrays are increasingly applied to identify DEGs underlying lens defects. However, it is important to identify functional relationships between the identified DEGs and prioritize those candidates that are relevant to the observed phenotype. This can be addressed by the following analytical workflow: data normalization and quality-control to identify DEGs, followed by functional pathway annotation, and filtering with tissue-specific molecular profiling data (e.g. iSyTE). Such an integrative workflow can connect DEGs into interacting nodes, the edges between which can represent features such as protein-protein interaction, co-expression, and presence of cis-regulatory motifs, as explained below (Fig. 3). Application of commercial pathway analysis software or public tools such as DAVID (Database for Annotation, Visualization and Integrated Discovery) (Huang et al., 2009) can identify functionally clustered candidates among DEGs. Thereafter, functional gene ontology enrichment can be enhanced by expanding the relationships of DEGs by overlaying publically available protein-protein (Rolland et al., 2014) and protein-DNA (Hume et al., 2015), protein-RNA interaction data (Hashemikhabir et al., 2015), along with iSyTE lens expression and evidence-based data. This approach recently applied to microarray-identified DEGs in Mafg−/−:MafKk+/− mouse lenses led to the derivation of a small Maf regulatory sub-network that primarily regulates non-crystallin genes linked to cataract (Agrawal et al., 2015; Anand et al., 2015). Thus, integrative analysis of mutant lens-derived genome-wide expression data can provide significant insights into the molecular pathology of cataracts.
5. Lens systems biology as a paradigm for eye disease gene discovery
Systems biology of the lens has initiated derivation of its developmental GRNs (Cvekl and Ashery-Padan, 2014; Lachke and Maas, 2010) and the web-based resource-tool iSyTE (Lachke et al., 2012b). Presently, iSyTE focuses on only one eye component, the lens, and thus is limited to a single eye disease, cataract. The iSyTE approach can be extended to other ocular tissues by: (1) integrating all available gene expression datasets on retinal, corneal and other eye tissue, (2) assembling a database for comprehensive evidence-based GRNs for these tissues, and (3) developing an eye-specific user-friendly web interface. Presently, there is a wealth of microarray and RNA-Seq data on several eye tissues. Similar to lens iSyTE, WB-subtraction can be performed for existing retina and cornea expression datasets. In addition to their application to advance ocular development, these eye tissue-specific knowledge-maps will be effective for the interpretation of patient whole genome/exome sequencing data for ocular disorders such as retinitis pigmentosa and cone-rod dystrophy, among others.
Other systems approaches have also impacted ocular gene discovery. In particular, ChIP-seq on the photoreceptor TF CRX has led to a novel strategy to rank photoreceptor-disease gene candidates based on CRX cis-binding sites abundance. This approach termed cis-regulatory mapping, combined with patient exome-seq data, identified a new retinitis pigmentosa-linked gene (Ozgül et al., 2011). ChIP-seq data on Pax6 and other lens TFs, and its application for cis-regulatory mapping (similar to CRX), combined with iSyTE enrichment-scores, will further expedite gene-discovery in the lens.
6. Conclusion
In the past, the lens has served as a paradigm for tissue induction, cellular differentiation, transcriptional gene expression control, and structural protein biochemistry. Systems biology of lens development can similarly serve as a model for defining comprehensive GRNs in ocular morphogenesis. In 2010, we proposed that a systems-level understanding of the lens can impact cataract gene discovery, and if this approach is extended to the eye, can lead to the “developmental oculome” (Lachke and Maas, 2010). In the past 5 years, applications of systems-level approaches to investigate lens and eye biology have significantly increased, and have led to the development of a web resource-tool iSyTE (Lachke et al., 2012b). Application of iSyTE, in turn, has identified several new cataract-linked genes and contributed to the characterization of lens defects in mouse mutants. Further, iSyTE-mediated gene discoveries have established the lens as a model for RBP-mediated post-transcriptional control in eye development.
The advent of CRISPR/Cas9 present new opportunities for gene deletion in animal models (Peng et al., 2015; Shalem et al., 2015; Singh et al., 2015), which, combined with iSyTE-based information, can accelerate eye disease discovery. The resulting new functional data, in combination with known evidence-based lens/eye GRNs, will allow an effective analysis of molecular data on lens/eye morphogenesis at the systems-level. Further, these resources, in combination with patient-exome-seq, will allow physician-scientists to identify new genes that impact lens/eye diseases. Thus, these approaches will provide a comprehensive picture of the genetic circuitry involved in the spatiotemporal control of the lens/eye proteome, which is central to understanding of lens/eye biology and pathology. This is best appreciated from the following example. Recent studies reported sterol compounds to reverse lens protein aggregation and reduce cataracts in animal models (Makley et al., 2015; Zhao et al., 2015). The focus on sterols was initiated by lanosterol synthase gene mutations that cause inherited cataracts in human and rat (Mori et al., 2006; Zhao et al., 2015). Thus, it is important to investigate the molecular basis of these cataracts, regulation of the lens sterol pathway, and the mechanism of sterol-based intervention (Hejtmancik, 2015; Quinlan, 2015). Toward this goal and supporting these findings, an integrative approach-based molecular characterization of Mafg−/−:Mafk+/− mice independently identified mis-regulation of sterol pathway genes in mutant lenses that develop early-onset age-related cataracts (Agrawal et al., 2015; Anand et al., 2015). Because their lens-crystallin levels are largely normal, these animals represent a new model for investigating the pathological mechanism of non-crystallin proteins, namely, sterol pathway mis-regulation, in early-onset age-related cataracts. Thus, the integrative approaches outlined here, while expanding the basic biology of the lens, can provide novel insights into regulation of pathways that are potential targets of new therapeutic strategies for cataract.
iSyTE (integrated Systems Tool for Eye gene discovery) is a user friendly web tool
Application of iSyTE has expedited lens developmental and cataract gene discovery
Microarray and RNA-seq gene expression data for several lens stages are now available
Integration of expression, molecular interaction, ChIP data will allow GRN assembly
iSyTE in silico subtraction approach can be extended to retina and cornea
Acknowledgments
Grant support: Supported by National Institutes of Health (NIH) Grant R01 EY021505 to Drs. David C. Beebe and Salil A. Lachke. SAL is a Pew Scholar in Biomedical Sciences.
The findings from the Lachke and Beebe laboratories mentioned in this article was supported by the National Eye Institute of the National Institutes of Health under Award Number R01EY021505 on which Drs. David C. Beebe and Salil A. Lachke were PIs during the funding period of 2011–2015. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Salil A. Lachke is a Pew Scholar in Biomedical Sciences. This review is respectfully dedicated to the memory of Dr. David C. Beebe, whose enthusiasm, scientific achievements, mentorship, and encouragement of young investigators will always serve as an inspiration to advance eye research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agrawal SA, Anand D, Siddam AD, Kakrana A, Dash S, Scheiblin DA, Dang CA, Terrell AM, Waters SM, Singh A, Motohashi H, Yamamoto M, Lachke SA. Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum Genet. 2015;134:717–735. doi: 10.1007/s00439-015-1554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldahmesh MA, Alshammari MJ, Khan AO, Mohamed JY, Alhabib FA, Alkuraya FS. The syndrome of microcornea, myopic chorioretinal atrophy, and telecanthus (MMCAT) is caused by mutations in ADAMTS18. Hum Mutat. 2013;34:1195–1199. doi: 10.1002/humu.22374. [DOI] [PubMed] [Google Scholar]
- Aldahmesh MA, Khan AO, Mohamed JY, Hijazi H, Al-Owain M, Alswaid A, Alkuraya FS. Genomic analysis of pediatric cataract in Saudi Arabia reveals novel candidate disease genes. Genet Med. 2012;14:955–962. doi: 10.1038/gim.2012.86. [DOI] [PubMed] [Google Scholar]
- Anand D, Agrawal S, Siddam A, Motohashi H, Yamamoto M, Lachke SA. An integrative approach to analyze microarray datasets for prioritization of genes relevant to lens biology and disease. Genom Data. 2015;5:223–227. doi: 10.1016/j.gdata.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anchan RM, Lachke SA, Gerami-Naini B, Lindsey J, Ng N, Naber C, Nickerson M, Cavallesco R, Rowan S, Eaton JL, Xi Q, Maas RL. Pax6- and Six3-mediated induction of lens cell fate in mouse and human ES cells. PLoS ONE. 2014;9:e115106. doi: 10.1371/journal.pone.0115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audette DS, Anand D, So T, Rubenstein TB, Lachke SA, Lovicu FJ, Duncan MK. Prox1 and fibroblast growth factor receptors form a novel regulatory loop controlling lens fiber differentiation and gene expression. Development. 2016;143:318–328. doi: 10.1242/dev.127860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe DC, Coats JM. The lens organizes the anterior segment: specification of neural crest cell differentiation in the avian eye. Dev Biol. 2000;220:424–431. doi: 10.1006/dbio.2000.9638. [DOI] [PubMed] [Google Scholar]
- Belecky-Adams TL, Adler R, Beebe DC. Bone morphogenetic protein signaling and the initiation of lens fiber cell differentiation. Development. 2002;129:3795–3802. doi: 10.1242/dev.129.16.3795. [DOI] [PubMed] [Google Scholar]
- Blixt A, Mahlapuu M, Aitola M, Pelto-Huikko M, Enerbäck S, Carlsson P. A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev. 2000;14:245–254. [PMC free article] [PubMed] [Google Scholar]
- Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Brownell I, Dirksen M, Jamrich M. Forkhead Foxe3 maps to the dysgenetic lens locus and is critical in lens development and differentiation. Genesis. 2000;27:81–93. doi: 10.1002/1526-968x(200006)27:2<81::aid-gene50>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Bu L, Jin Y, Shi Y, Chu R, Ban A, Eiberg H, Andres L, Jiang H, Zheng G, Qian M, Cui B, Xia Y, Liu J, Hu L, Zhao G, Hayden MR, Kong X. Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet. 2002;31:276–278. doi: 10.1038/ng921. [DOI] [PubMed] [Google Scholar]
- Carbe C, Zhang X. Lens induction requires attenuation of ERK signaling by Nf1. Hum Mol Genet. 2011;20:1315–1323. doi: 10.1093/hmg/ddr014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins M, Brown NL, Cook TA. The lens in focus: a comparison of lens development in Drosophila and vertebrates. Mol Genet Genomics. 2011;286:189–213. doi: 10.1007/s00438-011-0643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan BK, Reed NA, Yang Y, Cermák L, Reneker L, Duncan MK, Cvekl A. A comparative cDNA microarray analysis reveals a spectrum of genes regulated by Pax6 in mouse lens. Genes Cells. 2002a;7:1267–1283. doi: 10.1046/j.1365-2443.2002.00602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan BK, Reed NA, Zhang W, Duncan MK, Kilimann MW, Cvekl A. Identification of genes downstream of Pax6 in the mouse lens using cDNA microarrays. J Biol Chem. 2002b;277:11539–11548. doi: 10.1074/jbc.M110531200. [DOI] [PubMed] [Google Scholar]
- Chauss D, Basu S, Rajakaruna S, Ma Z, Gau V, Anastas S, Brennan LA, Hejtmancik JF, Menko AS, Kantorow M. Differentiation state-specific mitochondrial dynamic regulatory networks are revealed by global transcriptional analysis of the developing chicken lens. G3 (Bethesda) 2014;4:1515–1527. doi: 10.1534/g3.114.012120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chograni M, Alkuraya FS, Ourteni I, Maazoul F, Lariani I, Chaabouni HB. Autosomal recessive congenital cataract, intellectual disability phenotype linked to STX3 in a consanguineous Tunisian family. Clin Genet. 2014 doi: 10.1111/cge.12489. [DOI] [PubMed] [Google Scholar]
- Chuang H-Y, Hofree M, Ideker T. A decade of systems biology. Annu Rev Cell Dev Biol. 2010;26:721–744. doi: 10.1146/annurev-cellbio-100109-104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte I, Carrella S, Avellino R, Karali M, Marco-Ferreres R, Bovolenta P, Banfi S. miR-204 is required for lens and retinal development via Meis2 targeting. Proc Natl Acad Sci USA. 2010;107:15491–15496. doi: 10.1073/pnas.0914785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Ashery-Padan R. The cellular and molecular mechanisms of vertebrate lens development. Development. 2014;141:4432–4447. doi: 10.1242/dev.107953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Duncan MK. Genetic and epigenetic mechanisms of gene regulation during lens development. Prog Retin Eye Res. 2007;26:555–597. doi: 10.1016/j.preteyeres.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S, Dang CA, Beebe DC, Lachke SA. Deficiency of the RNA binding protein caprin2 causes lens defects and features of peters anomaly. Dev Dyn. 2015;244:1313–1327. doi: 10.1002/dvdy.24303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maria A, Bassnett S. Birc7: A Late Fiber Gene of the Crystalline Lens. Invest Ophthalmol Vis Sci. 2015;56:4823–4834. doi: 10.1167/iovs.15-16968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic D, Yang A, Zadoorian A, Rungrugeecharoen K, Ho JWK. How difficult is inference of mammalian causal gene regulatory networks? PLoS ONE. 2014;9:e111661. doi: 10.1371/journal.pone.0111661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AL, Episkopou V, Maas RL. Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev Biol. 2007a;303:784–799. doi: 10.1016/j.ydbio.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AL, Ko F, Episkopou V, Maas RL. Pax6 is misexpressed in Sox1 null lens fiber cells. Gene Expr Patterns. 2007b;7:606–613. doi: 10.1016/j.modgep.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AL, Lachke SA, Maas RL. Lens induction in vertebrates: variations on a conserved theme of signaling events. Semin Cell Dev Biol. 2006;17:676–685. doi: 10.1016/j.semcdb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Evers C, Paramasivam N, Hinderhofer K, Fischer C, Granzow M, Schmidt-Bacher A, Eils R, Steinbeisser H, Schlesner M, Moog U. SIPA1L3 identified by linkage analysis and whole-exome sequencing as a novel gene for autosomal recessive congenital cataract. Eur J Hum Genet. 2015 doi: 10.1038/ejhg.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber SC, Dimanlig P, Makarenkova HP, Shirke S, Ko K, Lang RA. Fgf receptor signaling plays a role in lens induction. Development. 2001;128:4425–4438. doi: 10.1242/dev.128.22.4425. [DOI] [PubMed] [Google Scholar]
- Frederikse PH, Donnelly R, Partyka LM. miRNA and Dicer in the mammalian lens: expression of brain-specific miRNAs in the lens. Histochem Cell Biol. 2006;126:1–8. doi: 10.1007/s00418-005-0139-0. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Hogan BL. BMP4 is essential for lens induction in the mouse embryo. Genes Dev. 1998;12:3764–3775. doi: 10.1101/gad.12.23.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia CM, Huang J, Madakashira BP, Liu Y, Rajagopal R, Dattilo L, Robinson ML, Beebe DC. The function of FGF signaling in the lens placode. Dev Biol. 2011;351:176–185. doi: 10.1016/j.ydbio.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia CM, Yu K, Zhao H, Ashery-Padan R, Ornitz DM, Robinson ML, Beebe DC. Signaling through FGF receptor-2 is required for lens cell survival and for withdrawal from the cell cycle during lens fiber cell differentiation. Dev Dyn. 2005;233:516–527. doi: 10.1002/dvdy.20356. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. The evolution of vision. Wiley Interdiscip Rev Dev Biol. 2014;3:1–40. doi: 10.1002/wdev.96. [DOI] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- Greenlees R, Mihelec M, Yousoof S, Speidel D, Wu SK, Rinkwitz S, Prokudin I, Perveen R, Cheng A, Ma A, Nash B, Gillespie R, Loebel DAF, Clayton-Smith J, Lloyd IC, Grigg JR, Tam PPL, Yap AS, Becker TS, Black GCM, Semina E, Jamieson RV. Mutations in SIPA1L3 cause eye defects through disruption of cell polarity and cytoskeleton organization. Hum Mol Genet. 2015;24:5789–5804. doi: 10.1093/hmg/ddv298. [DOI] [PubMed] [Google Scholar]
- Greiling TMS, Stone B, Clark JI. Absence of SPARC leads to impaired lens circulation. Exp Eye Res. 2009;89:416–425. doi: 10.1016/j.exer.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond CJ, Duncan DD, Snieder H, de Lange M, West SK, Spector TD, Gilbert CE. The heritability of age-related cortical cataract: the twin eye study. Invest Ophthalmol Vis Sci. 2001;42:601–605. [PubMed] [Google Scholar]
- Hashemikhabir S, Neelamraju Y, Janga SC. Database of RNA binding protein expression and disease dynamics (READ DB) Database (Oxford) 2015;2015:bav072. doi: 10.1093/database/bav072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawse JR, DeAmicis-Tress C, Cowell TL, Kantorow M. Identification of global gene expression differences between human lens epithelial and cortical fiber cells reveals specific genes and their associated pathways important for specialized lens cell functions. Mol Vis. 2005;11:274–283. [PMC free article] [PubMed] [Google Scholar]
- Hawse JR, Hejtmancik JF, Horwitz J, Kantorow M. Identification and functional clustering of global gene expression differences between age-related cataract and clear human lenses and aged human lenses. Exp Eye Res. 2004;79:935–940. doi: 10.1016/j.exer.2004.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawse JR, Hejtmancik JF, Huang Q, Sheets NL, Hosack DA, Lempicki RA, Horwitz J, Kantorow M. Identification and functional clustering of global gene expression differences between human age-related cataract and clear lenses. Mol Vis. 2003;9:515–537. [PMC free article] [PubMed] [Google Scholar]
- Hejtmancik JF. Ophthalmology: Cataracts dissolved. Nature. 2015;523:540–541. doi: 10.1038/nature14629. [DOI] [PubMed] [Google Scholar]
- Hoang TV, Kumar PKR, Sutharzan S, Tsonis PA, Liang C, Robinson ML. Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing. Mol Vis. 2014;20:1491–1517. [PMC free article] [PubMed] [Google Scholar]
- Ho H-Y, Chang K-H, Nichols J, Li M. Homeodomain protein Pitx3 maintains the mitotic activity of lens epithelial cells. Mech Dev. 2009;126:18–29. doi: 10.1016/j.mod.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics. 2009;Chapter 13(Unit 13.11) doi: 10.1002/0471250953.bi1311s27. [DOI] [PubMed] [Google Scholar]
- Huang J, Liu Y, Filas B, Gunhaga L, Beebe DC. Negative and positive auto-regulation of BMP expression in early eye development. Dev Biol. 2015;407:256–264. doi: 10.1016/j.ydbio.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J-X, Feldmeier M, Shui Y-B, Beebe DC. Evaluation of fibroblast growth factor signaling during lens fiber cell differentiation. Invest Ophthalmol Vis Sci. 2003;44:680–690. doi: 10.1167/iovs.01-1177. [DOI] [PubMed] [Google Scholar]
- Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2015;43:D117–122. doi: 10.1093/nar/gku1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyer J, Kuhlman J, Afif E, Mikawa T. Optic cup morphogenesis requires pre-lens ectoderm but not lens differentiation. Dev Biol. 2003;259:351–363. doi: 10.1016/s0012-1606(03)00205-7. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Dvoriantchikova G, Pestova A, Nathanson L, Shestopalov VI. Microarray analysis of fiber cell maturation in the lens. FEBS Lett. 2005;579:1213–1219. doi: 10.1016/j.febslet.2005.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Devine T, George AD, Chittur SV, Baroni TE, Penalva LO, Tenenbaum SA. RIP-Chip analysis: RNA-Binding Protein Immunoprecipitation-Microarray (Chip) Profiling. Methods Mol Biol. 2011;703:247–263. doi: 10.1007/978-1-59745-248-9_17. [DOI] [PubMed] [Google Scholar]
- Jamieson RV, Perveen R, Kerr B, Carette M, Yardley J, Heon E, Wirth MG, van Heyningen V, Donnai D, Munier F, Black GCM. Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet. 2002;11:33–42. doi: 10.1093/hmg/11.1.33. [DOI] [PubMed] [Google Scholar]
- Karali M, Peluso I, Gennarino VA, Bilio M, Verde R, Lago G, Dollé P, Banfi S. miRNeye: a microRNA expression atlas of the mouse eye. BMC Genomics. 2010;11:715. doi: 10.1186/1471-2164-11-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasaikina MV, Fomenko DE, Labunskyy VM, Lachke SA, Qiu W, Moncaster JA, Zhang J, Wojnarowicz MW, Jr, Natarajan SK, Malinouski M, Schweizer U, Tsuji PA, Carlson BA, Maas RL, Lou MF, Goldstein LE, Hatfield DL, Gladyshev VN. Roles of the 15-kDa selenoprotein (Sep15) in redox homeostasis and cataract development revealed by the analysis of Sep 15 knockout mice. J Biol Chem. 2011;286:33203–33212. doi: 10.1074/jbc.M111.259218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Takahashi S, Nakajima O, Ogino H, Morita M, Nishizawa M, Yasuda K, Yamamoto M. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J Biol Chem. 1999;274:19254–19260. doi: 10.1074/jbc.274.27.19254. [DOI] [PubMed] [Google Scholar]
- Khan SY, Hackett SF, Lee M-CW, Pourmand N, Talbot CC, Riazuddin SA. Transcriptome Profiling of Developing Murine Lens Through RNA Sequencing. Invest Ophthalmol Vis Sci. 2015;56:4919–4926. doi: 10.1167/iovs.14-16253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SY, Hackett SF, Riazuddin SA. Non-coding RNA profiling of the developing murine lens. Exp Eye Res. 2016;145:347–351. doi: 10.1016/j.exer.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Li T, Ho IC, Grusby MJ, Glimcher LH. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci USA. 1999;96:3781–3785. doi: 10.1073/pnas.96.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- Kubo E, Hasanova N, Sasaki H, Singh DP. Dynamic and differential regulation in the microRNA expression in the developing and mature cataractous rat lens. J Cell Mol Med. 2013;17:1146–1159. doi: 10.1111/jcmm.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel JF, Goodrich JA. Non-coding RNAs: key regulators of mammalian transcription. Trends Biochem Sci. 2012;37:144–151. doi: 10.1016/j.tibs.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PKR, Hoang TV, Robinson ML, Tsonis PA, Liang C. CADBURE: A generic tool to evaluate the performance of spliced aligners on RNA-Seq data. Sci Rep. 2015;5:13443. doi: 10.1038/srep13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuracha MR, Burgess D, Siefker E, Cooper JT, Licht JD, Robinson ML, Govindarajan V. Spry1 and Spry2 are necessary for lens vesicle separation and corneal differentiation. Invest Ophthalmol Vis Sci. 2011;52:6887–6897. doi: 10.1167/iovs.11-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Alkuraya FS, Kneeland SC, Ohn T, Aboukhalil A, Howell GR, Saadi I, Cavallesco R, Yue Y, Tsai ACH, Nair KS, Cosma MI, Smith RS, Hodges E, Alfadhli SM, Al-Hajeri A, Shamseldin HE, Behbehani A, Hannon GJ, Bulyk ML, Drack AV, Anderson PJ, John SWM, Maas RL. Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science. 2011;331:1571–1576. doi: 10.1126/science.1195970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Higgins AW, Inagaki M, Saadi I, Xi Q, Long M, Quade BJ, Talkowski ME, Gusella JF, Fujimoto A, Robinson ML, Yang Y, Duong QT, Shapira I, Motro B, Miyoshi J, Takai Y, Morton CC, Maas RL. The cell adhesion gene PVRL3 is associated with congenital ocular defects. Hum Genet. 2012a;131:235–250. doi: 10.1007/s00439-011-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Ho JWK, Kryukov GV, O’Connell DJ, Aboukhalil A, Bulyk ML, Park PJ, Maas RL. iSyTE: integrated Systems Tool for Eye gene discovery. Invest Ophthalmol Vis Sci. 2012b;53:1617–1627. doi: 10.1167/iovs.11-8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Maas RL. RNA Granules and Cataract. Expert Rev Ophthalmol. 2011;6:497–500. doi: 10.1586/eop.11.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Maas RL. Building the developmental oculome: systems biology in vertebrate eye development and disease. Wiley Interdiscip Rev Syst Biol Med. 2010;2:305–323. doi: 10.1002/wsbm.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampi KJ, Shih M, Ueda Y, Shearer TR, David LL. Lens proteomics: analysis of rat crystallin sequences and two-dimensional electrophoresis map. Invest Ophthalmol Vis Sci. 2002;43:216–224. [PubMed] [Google Scholar]
- Landgren H, Blixt A, Carlsson P. Persistent FoxE3 expression blocks cytoskeletal remodeling and organelle degradation during lens fiber differentiation. Invest Ophthalmol Vis Sci. 2008;49:4269–4277. doi: 10.1167/iovs.08-2243. [DOI] [PubMed] [Google Scholar]
- Li H, Tao C, Cai Z, Hertzler-Schaefer K, Collins TN, Wang F, Feng G-S, Gotoh N, Zhang X. Frs2α and Shp2 signal independently of Gab to mediate FGF signaling in lens development. J Cell Sci. 2014;127:571–582. doi: 10.1242/jcs.134478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Uemura Y, Kawahara Y. Cross-linking and immunoprecipitation of nuclear RNA-binding proteins. Methods Mol Biol. 2015;1262:247–263. doi: 10.1007/978-1-4939-2253-6_15. [DOI] [PubMed] [Google Scholar]
- Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–5395. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Piatigorsky J. Targeted deletion of Dicer disrupts lens morphogenesis, corneal epithelium stratification, and whole eye development. Dev Dyn. 2009;238:2388–2400. doi: 10.1002/dvdy.22056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madakashira BP, Kobrinski DA, Hancher AD, Arneman EC, Wagner BD, Wang F, Shin H, Lovicu FJ, Reneker LW, Robinson ML. Frs2α enhances fibroblast growth factor-mediated survival and differentiation in lens development. Development. 2012;139:4601–4612. doi: 10.1242/dev.081737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makley LN, McMenimen KA, DeVree BT, Goldman JW, McGlasson BN, Rajagopal P, Dunyak BM, McQuade TJ, Thompson AD, Sunahara R, Klevit RE, Andley UP, Gestwicki JE. Pharmacological chaperone for α-crystallin partially restores transparency in cataract models. Science. 2015;350:674–677. doi: 10.1126/science.aac9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey AL, Lachke SA, FitzGerald PG, Mason RW, Scheiblin DA, McDonald JH, Duncan MK. Loss of Sip1 leads to migration defects and retention of ectodermal markers during lens development. Mech Dev. 2014a;131:86–110. doi: 10.1016/j.mod.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey AL, Terrell AM, Lachke SA, Polson SW, Duncan MK. Development of novel filtering criteria to analyze RNA-sequencing data obtained from the murine ocular lens during embryogenesis. Genomics Data. 2014b;2:369–374. doi: 10.1016/j.gdata.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Martinez O, Brownell I, Amaya-Manzanares F, Hu Q, Behringer RR, Jamrich M. Severe defects in proliferation and differentiation of lens cells in Foxe3 null mice. Mol Cell Biol. 2005;25:8854–8863. doi: 10.1128/MCB.25.20.8854-8863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milek M, Wyler E, Landthaler M. Transcriptome-wide analysis of protein-RNA interactions using high-throughput sequencing. Semin Cell Dev Biol. 2012;23:206–212. doi: 10.1016/j.semcdb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Modic M, Ule J, Sibley CR. CLIPing the brain: studies of protein-RNA interactions important for neurodegenerative disorders. Mol Cell Neurosci. 2013;56:429–435. doi: 10.1016/j.mcn.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Li G, Abe I, Nakayama J, Guo Z, Sawashita J, Ugawa T, Nishizono S, Serikawa T, Higuchi K, Shumiya S. Lanosterol synthase mutations cause cholesterol deficiency-associated cataracts in the Shumiya cataract rat. J Clin Invest. 2006;116:395–404. doi: 10.1172/JCI20797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD. Systematic analysis of posttranscriptional gene expression. Wiley Interdiscip Rev Syst Biol Med. 2010;2:162–180. doi: 10.1002/wsbm.54. [DOI] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara M, Nagasaka A, Koike M, Uchida K, Kawane K, Uchiyama Y, Nagata S. Degradation of nuclear DNA by DNase II-like acid DNase in cortical fiber cells of mouse eye lens. FEBS J. 2007;274:3055–3064. doi: 10.1111/j.1742-4658.2007.05836.x. [DOI] [PubMed] [Google Scholar]
- Neelamraju Y, Hashemikhabir S, Janga SC. The human RBPome: from genes and proteins to human disease. J Proteomics. 2015;127:61–70. doi: 10.1016/j.jprot.2015.04.031. [DOI] [PubMed] [Google Scholar]
- Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V. Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev. 1998;12:776–781. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell DJ, Ho JWK, Mammoto T, Turbe-Doan A, O’Connell JT, Haseley PS, Koo S, Kamiya N, Ingber DE, Park PJ, Maas RL. A Wnt-bmp feedback circuit controls intertissue signaling dynamics in tooth organogenesis. Sci Signal. 2012;5:ra4. doi: 10.1126/scisignal.2002414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino H, Ochi H, Reza HM, Yasuda K. Transcription factors involved in lens development from the preplacodal ectoderm. Dev Biol. 2012;363:333–347. doi: 10.1016/j.ydbio.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Ozgül RK, Siemiatkowska AM, Yücel D, Myers CA, Collin RWJ, Zonneveld MN, Beryozkin A, Banin E, Hoyng CB, van den Born LI, Bose R, Shen W, Sharon D, Cremers FPM, Klevering BJ, den Hollander AI, Corbo JC. Exome sequencing and cis-regulatory mapping identify mutations in MAK, a gene encoding a regulator of ciliary length, as a cause of retinitis pigmentosa. Am J Hum Genet. 2011;89:253–264. doi: 10.1016/j.ajhg.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit T, Jidigam VK, Patthey C, Gunhaga L. Neural retina identity is specified by lens-derived BMP signals. Development. 2015;142:1850–1859. doi: 10.1242/dev.123653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N, Khan AO, Mansour A, Mohamed JY, Al-Assiri A, Haddad R, Jia X, Xiong Y, Mégarbané A, Traboulsi EI, Alkuraya FS. Mutations in ASPH cause facial dysmorphism, lens dislocation, anterior-segment abnormalities, and spontaneous filtering blebs, or Traboulsi syndrome. Am J Hum Genet. 2014;94:755–759. doi: 10.1016/j.ajhg.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Zhou Y, Zhu S, Wei W. High-throughput screens in mammalian cells using the CRISPR-Cas9 system. FEBS J. 2015;282:2089–2096. doi: 10.1111/febs.13251. [DOI] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144:970–985. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontoriero GF, Deschamps P, Ashery-Padan R, Wong R, Yang Y, Zavadil J, Cvekl A, Sullivan S, Williams T, West-Mays JA. Cell autonomous roles for AP-2alpha in lens vesicle separation and maintenance of the lens epithelial cell phenotype. Dev Dyn. 2008;237:602–617. doi: 10.1002/dvdy.21445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan RA. DRUG DISCOVERY. A new dawn for cataracts. Science. 2015;350:636–637. doi: 10.1126/science.aad6303. [DOI] [PubMed] [Google Scholar]
- Rajagopal R, Dattilo LK, Kaartinen V, Deng C-X, Umans L, Zwijsen A, Roberts AB, Bottinger EP, Beebe DC. Functions of the type 1 BMP receptor Acvr1 (Alk2) in lens development: cell proliferation, terminal differentiation, and survival. Invest Ophthalmol Vis Sci. 2008;49:4953–4960. doi: 10.1167/iovs.08-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Re A, Joshi T, Kulberkyte E, Morris Q, Workman CT. RNA-protein interactions: an overview. Methods Mol Biol. 2014;1097:491–521. doi: 10.1007/978-1-62703-709-9_23. [DOI] [PubMed] [Google Scholar]
- Ring BZ, Cordes SP, Overbeek PA, Barsh GS. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307–317. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, Kamburov A, Ghiassian SD, Yang X, Ghamsari L, Balcha D, Begg BE, Braun P, Brehme M, Broly MP, Carvunis AR, Convery-Zupan D, Corominas R, Coulombe-Huntington J, Dann E, Dreze M, Dricot A, Fan C, Franzosa E, Gebreab F, Gutierrez BJ, Hardy MF, Jin M, Kang S, Kiros R, Lin GN, Luck K, MacWilliams A, Menche J, Murray RR, Palagi A, Poulin MM, Rambout X, Rasla J, Reichert P, Romero V, Ruyssinck E, Sahalie JM, Scholz A, Shah AA, Sharma A, Shen Y, Spirohn K, Tam S, Tejeda AO, Trigg SA, Twizere JC, Vega K, Walsh J, Cusick ME, Xia Y, Barabási AL, Iakoucheva LM, Aloy P, De Las Rivas J, Tavernier J, Calderwood MA, Hill DE, Hao T, Roth FP, Vidal M. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Siggers T, Lachke SA, Yue Y, Bulyk ML, Maas RL. Precise temporal control of the eye regulatory gene Pax6 via enhancer-binding site affinity. Genes Dev. 2010;24:980–985. doi: 10.1101/gad.1890410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilter KF, Reis LM, Schneider A, Bardakjian TM, Abdul-Rahman O, Kozel BA, Zimmerman HH, Broeckel U, Semina EV. Whole-genome copy number variation analysis in anophthalmia and microphthalmia. Clin Genet. 2013;84:473–481. doi: 10.1111/cge.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz MB, Lo C-C, Chain PSG. Next generation sequencing and bioinformatic bottlenecks: the current state of metagenomic data analysis. Curr Opin Biotechnol. 2012;23:9–15. doi: 10.1016/j.copbio.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Semina EV, Brownell I, Mintz-Hittner HA, Murray JC, Jamrich M. Mutations in the human forkhead transcription factor FOXE3 associated with anterior segment ocular dysgenesis and cataracts. Hum Mol Genet. 2001;10:231–236. doi: 10.1093/hmg/10.3.231. [DOI] [PubMed] [Google Scholar]
- Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, Reiter RS, Funkhauser C, Daack-Hirsch S, Murray JC. A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet. 1998;19:167–170. doi: 10.1038/527. [DOI] [PubMed] [Google Scholar]
- Shaham O, Gueta K, Mor E, Oren-Giladi P, Grinberg D, Xie Q, Cvekl A, Shomron N, Davis N, Keydar-Prizant M, Raviv S, Pasmanik-Chor M, Bell RE, Levy C, Avellino R, Banfi S, Conte I, Ashery-Padan R. Pax6 regulates gene expression in the vertebrate lens through miR-204. PLoS Genet. 2013;9:e1003357. doi: 10.1371/journal.pgen.1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Dong L-F, Zhou R-M, Yao J, Song Y-C, Yang H, Jiang Q, Yan B. Role of long non-coding RNA MIAT in proliferation, apoptosis and migration of lens epithelial cells: a clinical and in vitro study. J Cell Mol Med. 2016 doi: 10.1111/jcmm.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels A, Bennett TM, Hejtmancik JF. Cat-Map: putting cataract on the map. Mol Vis. 2010;16:2007–2015. [PMC free article] [PubMed] [Google Scholar]
- Shiels A, Hejtmancik JF. Genetics of human cataract. Clin Genet. 2013;84:120–127. doi: 10.1111/cge.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simirskii VN, Wang Y, Duncan MK. Conditional deletion of beta1-integrin from the developing lens leads to loss of the lens epithelial phenotype. Dev Biol. 2007;306:658–668. doi: 10.1016/j.ydbio.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Schimenti JC, Bolcun-Filas E. A mouse geneticist’s practical guide to CRISPR applications. Genetics. 2015;199:1–15. doi: 10.1534/genetics.114.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N, Schwab KR, Patterson LT, Yamaguchi T, Lin X, Potter SS, Lang RA. pygopus 2 has a crucial, Wnt pathway-independent function in lens induction. Development. 2007;134:1873–1885. doi: 10.1242/dev.001495. [DOI] [PubMed] [Google Scholar]
- Sousounis K, Looso M, Maki N, Ivester CJ, Braun T, Tsonis PA. Transcriptome analysis of newt lens regeneration reveals distinct gradients in gene expression patterns. PLoS ONE. 2013;8:e61445. doi: 10.1371/journal.pone.0061445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousounis K, Qi F, Yadav MC, Millán JL, Toyama F, Chiba C, Eguchi Y, Eguchi G, Tsonis PA. A robust transcriptional program in newts undergoing multiple events of lens regeneration throughout their lifespan. Elife. 2015;4 doi: 10.7554/eLife.09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousounis K, Tsonis PA. Patterns of gene expression in microarrays and expressed sequence tags from normal and cataractous lenses. Hum Genomics. 2012;6:14. doi: 10.1186/1479-7364-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama Y, Shelley EJ, Wen L, Stump RJW, Shimono A, Lovicu FJ, McAvoy JW. Sfrp1 and Sfrp2 are not involved in Wnt/β-catenin signal silencing during lens induction but are required for maintenance of Wnt/β-catenin signaling in lens epithelial cells. Dev Biol. 2013;384:181–193. doi: 10.1016/j.ydbio.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Rockowitz S, Chauss D, Wang P, Kantorow M, Zheng D, Cvekl A. Chromatin features, RNA polymerase II and the comparative expression of lens genes encoding crystallins, transcription factors, and autophagy mediators. Mol Vis. 2015a;21:955–973. [PMC free article] [PubMed] [Google Scholar]
- Sun J, Rockowitz S, Xie Q, Ashery-Padan R, Zheng D, Cvekl A. Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic Acids Res. 2015b;43:6827–6846. doi: 10.1093/nar/gkv589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrell AM, Anand D, Smith SF, Dang CA, Waters SM, Pathania M, Beebe DC, Lachke SA. Molecular characterization of mouse lens epithelial cell lines and their suitability to study RNA granules and cataract associated genes. Exp Eye Res. 2015;131:42–55. doi: 10.1016/j.exer.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thut CJ, Rountree RB, Hwa M, Kingsley DM. A Large-Scale in Situ Screen Provides Molecular Evidence for the Induction of Eye Anterior Segment Structures by the Developing Lens. Developmental Biology. 2001;231:63–76. doi: 10.1006/dbio.2000.0140. [DOI] [PubMed] [Google Scholar]
- Tian L, Huang K, DuHadaway JB, Prendergast GC, Stambolian D. Genomic profiling of miRNAs in two human lens cell lines. Curr Eye Res. 2010;35:812–818. doi: 10.3109/02713683.2010.489182. [DOI] [PubMed] [Google Scholar]
- Tsentalovich YP, Verkhovod TD, Yanshole VV, Kiryutin AS, Yanshole LV, Fursova AZ, Stepakov DA, Novoselov VP, Sagdeev RZ. Metabolomic composition of normal aged and cataractous human lenses. Exp Eye Res. 2015;134:15–23. doi: 10.1016/j.exer.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Duncan MK, David LL. Lens proteomics: the accumulation of crystallin modifications in the mouse lens with age. Invest Ophthalmol Vis Sci. 2002;43:205–215. [PubMed] [Google Scholar]
- Upadhya D, Ogata M, Reneker LW. MAPK1 is required for establishing the pattern of cell proliferation and for cell survival during lens development. Development. 2013;140:1573–1582. doi: 10.1242/dev.081042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawersik S, Maas RL. Vertebrate eye development as modeled in Drosophila. Hum Mol Genet. 2000;9:917–925. doi: 10.1093/hmg/9.6.917. [DOI] [PubMed] [Google Scholar]
- Wawersik S, Purcell P, Rauchman M, Dudley AT, Robertson EJ, Maas R. BMP7 acts in murine lens placode development. Dev Biol. 1999;207:176–188. doi: 10.1006/dbio.1998.9153. [DOI] [PubMed] [Google Scholar]
- Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- Wistow G, Bernstein SL, Wyatt MK, Behal A, Touchman JW, Bouffard G, Smith D, Peterson K. Expressed sequence tag analysis of adult human lens for the NEIBank Project: over 2000 non-redundant transcripts, novel genes and splice variants. Mol Vis. 2002;8:171–184. [PubMed] [Google Scholar]
- Wolf L, Gao CS, Gueta K, Xie Q, Chevallier T, Podduturi NR, Sun J, Conte I, Zelenka PS, Ashery-Padan R, Zavadil J, Cvekl A. Identification and characterization of FGF2-dependent mRNA: microRNA networks during lens fiber cell differentiation. G3 (Bethesda) 2013a;3:2239–2255. doi: 10.1534/g3.113.008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf L, Harrison W, Huang J, Xie Q, Xiao N, Sun J, Kong L, Lachke SA, Kuracha MR, Govindarajan V, Brindle PK, Ashery-Padan R, Beebe DC, Overbeek PA, Cvekl A. Histone posttranslational modifications and cell fate determination: lens induction requires the lysine acetyltransferases CBP and p300. Nucleic Acids Res. 2013b;41:10199–10214. doi: 10.1093/nar/gkt824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf LV, Yang Y, Wang J, Xie Q, Braunger B, Tamm ER, Zavadil J, Cvekl A. Identification of pax6-dependent gene regulatory networks in the mouse lens. PLoS ONE. 2009;4:e4159. doi: 10.1371/journal.pone.0004159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wride MA, Mansergh FC, Adams S, Everitt R, Minnema SE, Rancourt DE, Evans MJ. Expression profiling and gene discovery in the mouse lens. Mol Vis. 2003;9:360–396. [PubMed] [Google Scholar]
- Wu C, Lin H, Wang Q, Chen W, Luo H, Chen W, Zhang H. Discrepant expression of microRNAs in transparent and cataractous human lenses. Invest Ophthalmol Vis Sci. 2012;53:3906–3912. doi: 10.1167/iovs.11-9178. [DOI] [PubMed] [Google Scholar]
- Wurm A, Sock E, Fuchshofer R, Wegner M, Tamm ER. Anterior segment dysgenesis in the eyes of mice deficient for the high-mobility-group transcription factor Sox11. Exp Eye Res. 2008;86:895–907. doi: 10.1016/j.exer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Xiao W, Liu W, Li Z, Liang D, Li L, White LD, Fox DA, Overbeek PA, Chen Q. Gene expression profiling in embryonic mouse lenses. Mol Vis. 2006;12:1692–1698. [PubMed] [Google Scholar]
- Xie Q, Yang Y, Huang J, Ninkovic J, Walcher T, Wolf L, Vitenzon A, Zheng D, Götz M, Beebe DC, Zavadil J, Cvekl A. Pax6 interactions with chromatin and identification of its novel direct target genes in lens and forebrain. PLoS ONE. 2013;8:e54507. doi: 10.1371/journal.pone.0054507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. microRNA expression in the eyes and their significance in relation to functions. Prog Retin Eye Res. 2009;28:87–116. doi: 10.1016/j.preteyeres.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Yamada R, Mizutani-Koseki Y, Hasegawa T, Osumi N, Koseki H, Takahashi N. Cell-autonomous involvement of Mab21l1 is essential for lens placode development. Development. 2003;130:1759–1770. doi: 10.1242/dev.00399. [DOI] [PubMed] [Google Scholar]
- Yang C, Yang Y, Brennan L, Bouhassira EE, Kantorow M, Cvekl A. Efficient generation of lens progenitor cells and lentoid bodies from human embryonic stem cells in chemically defined conditions. FASEB J. 2010;24:3274–3283. doi: 10.1096/fj.10-157255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanshole VV, Snytnikova OA, Kiryutin AS, Yanshole LV, Sagdeev RZ, Tsentalovich YP. Metabolomics of the rat lens: a combined LC-MS and NMR study. Exp Eye Res. 2014;125:71–78. doi: 10.1016/j.exer.2014.05.016. [DOI] [PubMed] [Google Scholar]
- Yoshimoto A, Saigou Y, Higashi Y, Kondoh H. Regulation of ocular lens development by Smad-interacting protein 1 involving Foxe3 activation. Development. 2005;132:4437–4448. doi: 10.1242/dev.02022. [DOI] [PubMed] [Google Scholar]
- Zhang X, Friedman A, Heaney S, Purcell P, Maas RL. Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes Dev. 2002;16:2097–2107. doi: 10.1101/gad.1007602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fan J, Ho JWK, Hu T, Kneeland SC, Fan X, Xi Q, Sellarole MA, de Vries WN, Lu W, Lachke SA, Lang RA, John SWM, Maas RL. Crim1 regulates integrin signaling in murine lens development. Development. 2016;143:356–366. doi: 10.1242/dev.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Yang T, Madakashira BP, Thiels CA, Bechtle CA, Garcia CM, Zhang H, Yu K, Ornitz DM, Beebe DC, Robinson ML. Fibroblast growth factor receptor signaling is essential for lens fiber cell differentiation. Dev Biol. 2008;318:276–288. doi: 10.1016/j.ydbio.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kawai K, Wang H, Wu D, Wang M, Yue Z, Zhang J, Liu Y-H. Loss of Msx2 function down-regulates the FoxE3 expression and results in anterior segment dysgenesis resembling Peters anomaly. Am J Pathol. 2012;180:2230–2239. doi: 10.1016/j.ajpath.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Chen XJ, Zhu J, Xi YB, Yang X, Hu LD, Ouyang H, Patel SH, Jin X, Lin D, Wu F, Flagg K, Cai H, Li G, Cao G, Lin Y, Chen D, Wen C, Chung C, Wang Y, Qiu A, Yeh E, Wang W, Hu X, Grob S, Abagyan R, Su Z, Tjondro HC, Zhao XJ, Luo H, Hou R, Perry JJP, Gao W, Kozak I, Granet D, Li Y, Sun X, Wang J, Zhang L, Liu Y, Yan YB, Zhang K. Lanosterol reverses protein aggregation in cataracts. Nature. 2015;523:607–611. doi: 10.1038/nature14650. [DOI] [PubMed] [Google Scholar]
- Zheng X, Yue S, Chen H, Weber B, Jia J, Zheng Y. Low-Cell-Number Epigenome Profiling Aids the Study of Lens Aging and Hematopoiesis. Cell Rep. 2015;13:1505–1518. doi: 10.1016/j.celrep.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]