Abstract
Intermediate filament proteins are structural components of the cellular cytoskeleton with cell-type specific expression and function. Glial fibrillary acidic protein (GFAP) is a type III intermediate filament protein and is up-regulated in glia of the nervous system in response to injury and during neurodegenerative diseases. In the retina, GFAP levels are dramatically increased in Müller glia and are thought to play a role in the extensive structural changes resulting in Müller cell hypertrophy and glial scar formation. In spite of similar changes to the morphology of Xenopus Müller cells following injury, we found that Xenopus lack a gfap gene. Other type III intermediate filament proteins were induced following rod photoreceptor ablation and retinal ganglion cell axotomy. The recently available X. tropicalis and X. laevis genomes indicate a small deletion most likely resulted in the loss of the gfap gene during evolution. Lastly, a survey of representative species from all three extant amphibian orders including the Anura (frogs, toads), Caudata (salamanders, newts), and Gymnophiona (caecilians) suggests that deletion of the gfap locus occurred after the common ancestor of the Anura and Caudata. Our results demonstrate that extensive changes in Müller cell morphology following retinal injury do not require GFAP in Xenopus, and other type III intermediate filament proteins may be involved in the gliotic response.
Introduction
Intermediate filaments (IFs) are constituents of the cell cytoskeleton. IFs are composed of one or more intermediate filament proteins (IFPs), a large family consisting of over 70 genes in humans (Lowery et al., 2015; Szeverenyi et al., 2008). Vertebrate IFPs are currently categorized into six different classes based on their sequence homology and structural motifs (Gray et al., 2015; Szeverenyi et al., 2008). Class I and II IFPs are the acidic and basic keratins and include over 50 genes. Class III proteins are most commonly expressed in the nervous system or muscle and include Glial Fibrillary Acidic Protein (GFAP), Vimentin (VIM), Peripherin (PRPH), and Desmin (DES). Class IV IFPs are expressed mostly in neurons and include the neurofilaments, α-Internexin (INA), and Nestin (NES). Class V and VI are the nuclear lamins that form intranuclear filaments and two beaded filament structural proteins in the lens, respectively. As part of the cytoskeleton, IFPs provide mechanical support for the cell. Recent accumulating evidence suggests that IFPs also function in other ways: organelle stabilization, cell migration, mechanotransduction, and signal transduction (Chernoivanenko et al., 2015; Goldman et al., 1996; Guo et al., 2014; Helfand et al., 2003; Ivaska et al., 2007; Matveeva et al., 2015).
The Class III IFPs, GFAP and VIM, are key players in reactive gliosis, a response to injury and disease thought to be an attempt to protect neural tissues from further damage. In the retina, Müller glia become reactive following injury and in the course of retinal degenerations. Müller cell gliosis is a complex response that involves changes in cell physiology, gene expression, and morphology. Although restricted gliosis is proposed to benefit the damaged retina, extensive, proliferative gliosis has the opposite effect, contributing to further retinal degeneration (Bringmann et al., 2006; Bringmann et al., 2009; Bringmann and Wiedemann, 2012). An early and prominent molecular change in response to retinal injury or disease is the upregulation of GFAP and VIM (Bignami and Dahl, 1979; Davidson et al., 1990; Erickson et al., 1987; Grosche et al., 1997; Lewis et al., 1989; Okada et al., 1990). Evidence suggests that excess expression of these IFPs results in glial scar formation, which drives abnormal retinal remodeling and inhibits regeneration of the damaged tissue. Consistent with this idea, Gfap−/−/Vim−/− double knockout mice exhibit reduced reactive gliosis in the spinal cord and improved axonal regeneration in the hypothalamus following injury (Menet et al., 2003; Wilhelmsson et al., 2004). Gfap−/−/Vim−/− mice also displayed reduced Müller cell gliosis following retinal detachment or photoreceptor degeneration, and cells transplanted into the Gfap−/−/Vim−/− retina migrate, integrate and form neurites more efficiently than in wild-type controls (Kinouchi et al., 2003; Nakazawa et al., 2007; Verardo et al., 2008). Together, these results underscore a functional importance for IFs during reactive gliosis.
Class III Vim, Prph, Des, and Class IV Ina and Nes have been identified in X. laevis. As in other vertebrates, X. laevis Vim is expressed in cells of mesenchymal origin including the gut connective tissue (Herrmann et al., 1989a; Sharpe, 1988). In the nervous system, Vim is first detected at the time of neural tube closure, and its expression increases in the nervous system during development (Dent et al., 1989). Des expression is restricted to the muscles, and Ina to neurons (Herrmann et al., 1989b; Zhao and Szaro, 1997a). The expression patterns of X. laevis and mammalian prph however, are distinct. While both Xenopus and mouse prph are expressed in axon-elaborating postmitotic neurons, Xenopus prhp is also expressed in radial glia and in regions of the nervous system actively proliferating (Gervasi et al., 2000). The transcriptional expression pattern of Xenopus gfap has not been reported. Multiple reports have described a GFAP or GFAP-like protein expression patterns in Xenopus using immunohistochemistry. However, cross-reactivity of mammalian GFAP antibodies with Xenopus IFPs, and large differences in the molecular mass of putative Xenopus and known mammalian GFAP proteins have been noted and bring into question the identity of the detected proteins (Godsave et al., 1986; Messenger and Warner, 1989; Szaro and Gainer, 1988).
We previously reported that X. laevis Müller glia undergo hypertrophy, and form a glial scar in response to rod photoreceptor ablation (Choi et al., 2011). Although GFAP-like immunoreactivity is observed in the normal and injured retina, we present evidence demonstrating commonly used GFAP antibodies recognize multiple X. laevis IFPs, including Vim, Prph, Des, and Ina. The recently available X. tropicalis and X. laevis genomes indicate the gfap locus was most likely lost due to a small deletion. A survey of the three extant amphibian orders suggests gfap was most likely lost after the last common ancestor shared by the Caudata and Anura. Using two injury paradigms (rod photoreceptor ablation and retinal ganglion cell (RGC) axotomy), we show that X. laevis Müller glia upregulate vim and prph suggesting, in the absence of Gfap, Vim and/or Prph, may participate in the Müller glia response in X. laevis.
Methods
Animals
Xenopus embryos were obtained by in vitro fertilization following standard protocols and developmental stages were determined according to (Nieuwkoop and Faber, 1994). XOPNTR (Xl.Tg(rho:Eco.nfsB)zuber) transgenic tadpoles were obtained by fertilization of eggs from F2 females with wild-type sperm. Generation of XOPNTR transgenic line was previously described (Choi et al., 2011). The Committee for the Humane Use of Animals at SUNY Upstate Medical University approved all procedures.
Metronidazole-induced rod ablation, genotyping and axotomy
Metronidazole (Mtz) treatments and genotyping were done as previously described (Choi et al., 2011). Axotomy was performed on one eye of anesthetized stage 50 embryos using forceps and a 26-gauge needle and collected at 1 and 3 days post-axotomy (Viczian and Zuber, 2014).
Degenerate PCR for identification of GFAP and Vimentin orthologs
Genomic DNA was isolated using DNAeasy Blood and Tissue kit (Qiagen, Valencia, CA). GFAP and Vimentin primers were designed to conserved sequence regions specific to GFAP or Vimentin (Table S8). Cycling conditions for the first round of PCR were an initial denaturation step at 95°C for 3 min, 40 cycles of denaturing at 95°C for 30 sec, annealing at 42°C–56°C (see Table S9 and S10) for 30 sec, extension at 72°C for 30 sec, and a final extension step at 72°C for 5 min. Second round nested PCR amplification was performed using the first PCR reaction diluted 1:10 as template with cycling conditions identical to the first round PCR. Annealing temperatures used are listed in Tables S9 and S10. All PCR products of the expected amplicon size were gel extracted, TA cloned, and verified by sequencing.
Phylogenetic and molecular evolution analysis
When referring to the gene/mRNA or protein of a specific species the symbol convention for that species is used. When referring to multiple species with distinct symbol conventions, we have used gfap and GFAP for gene/mRNA and proteins, respectively. Nucleotide sequences of all clones obtained were aligned using TranslatorX (Abascal et al., 2005) using MAFFT version 7.245 (Katoh et al., 2002; Katoh and Toh, 2008) to compute the protein alignments. Obvious alignment ambiguities and frame shifts were checked and corrected by hand. Deduced amino acid alignments of clone nucleotide sequences were profile-aligned to the master amino acid alignment containing IFP sequences, as well as transcriptomic sequence data of caecilian species from an unpublished, ongoing project by DSM. The best-fit model of amino acid substitution was then determined with ProtTest version 3.4 (Abascal et al., 2005; Darriba et al., 2011) using the Bayesian information criterion (BIC). JTT (Jones et al., 1992) + Γ (Yang, 1994) + I (Reeves, 1992) were selected.
The resulting alignment was analyzed using maximum likelihood (ML; Felsenstein, 1981) and Bayesian inference (BI; Huelsenbeck et al., 2001), which are currently the standard methods for molecular phylogenetic inference (reviewed in San Mauro and Agorreta, 2010). Analyses were run on the CIPRES Science Gateway (Miller et al., 2010). ML analyses were performed with RAxML version 8.2.4 (Stamatakis, 2006) using the rapid hill-climbing algorithm (Stamatakis et al., 2007). BI analyses were performed with MrBayes version 3.2.6 (Ronquist et al., 2012) conducting two independent MCMC runs (with four chains each) for 10 million generations, sampling every 1000 generations, and discarding the first 25% of samples as burn-in. Adequate convergence of the BI runs was judged by plots of scores versus generation time, low standard deviation of split frequencies, as well as convergence diagnostics (Estimated Sample Size [ESS], Potential Scale Reduction Factor [PSRF]), as implemented in MrBayes. Support for internal branches was evaluated by non-parametric bootstrapping with 1,000 replicates (RAxML) and posterior probabilities (MrBayes).
Plasmid construction
Mouse Gfap variant 2 (IMAGE; NM_010277; MmGFAP.pGEM-T) (advertised as variant 1) was obtained from Sino Biologicals, Inc (Beijing, China). X. laevis des.S (IMAGE: 5514607; NM_001093564; XlaDesmin.pCMVSport6), peripherin (IMAGE: 5543232; NM_001087060; Xla.Pprh.pCMVSport6), and vim.L (IMAGE: 4888155; NM_001087439; Xla.Vim-a.pCMVSport6) were obtained from Source Bioscience (Nottingham, UK). X. laevis ina.L (NM_001087060; Xla.Ina.pGEM4) was kindly provided by B. Szaro (Zhao and Szaro, 1997a). The coding sequence for each intermediate filament protein was PCR amplified using the primers listed in Table S8, TA cloned, and sequence verified. Coding sequences were then excised and ligated in frame into pCS2+MT. All plasmids and sequences are available upon request. Xla.Nestin.pBSKS- was kindly provided by R. Harland (Hemmati-Brivanlou et al., 1992).
RNA synthesis and microinjection
Capped RNAs were synthesized in vitro from NotI linearized plasmids using the SP6 mMessage Machine Kit (Ambion/ThermoFisher Scientific, Waltham, MA). cRNAs were injected (500 pg) into both blastomeres at the 2-cell stage or into one dorsal blastomere at the 4-cell stage and cultured to stage 15 or 35/36.
Probes and in situ hybridization
Digoxigenin (DIG) labeled antisense riboprobes were in vitro transcribed from XlaDesmin.pGEMTEZ (NcoI, SP6), XlaPrph.pGEMTEZ (NcoI, SP6), XlaVim-a.pGEMTEZ (NcoI, SP6), XlaIna.pGEMTEZ (NcoI, SP6), and Xla.Nestin.pBSKS- (NotI, T7) (Hemmati-Brivanlou et al., 1992) using RNA Polymerase Plus (Ambion/ThermoFisher Scientific, Waltham, MA) according to manufacturer’s instructions. In situ hybridization on tissue sections was performed as previously described (Hemmati-Brivanlou et al., 1992; Matsuda and Kondoh, 2014; Viczian et al., 2006).
Immunohistochemistry
Tadpoles were euthanized as previously described (Choi et al., 2011). Dissected eyes and tailbud stage embryos were fixed in Dent’s overnight at −20°C, rehydrated in PBS, embedded in 15% cold water fish gelatin/15% sucrose, and cryostat sectioned (16 μm). Immunohistochemistry was done as previously described (Martinez-De Luna et al., 2013; Viczian et al., 2003). A complete list of antibodies and dilutions at which they were used are in Supplementary Tables S11 and S12.
Western blotting
Injected stage 15 embryos were lysed and protein extracted using intermediate filament enriching buffer (Szaro and Gainer, 1988) and protease inhibitors were replaced with cOmplete protease inhibitor cocktail (Roche, Basel, Switzerland) and PhosphoSTOP phosphatase inhibitor cocktail (Roche, Basel, Switzerland). Samples were centrifuged at 16,000 × g for 90 minutes and supernatant and pellet fractions were collected. Amount of total protein was determined using Pierce 660nm protein assay kit according to manufacturer’s instructions (ThermoFisher Scientific, Waltham, MA). For chemiluminescence detection, 10 μg of supernatant or 30 μg of pellet fractions were separated by SDS-PAGE, transferred, and stained per antibody specifications as previously described (Wong et al., 2015). Images were captured by Gel Doc and quantified using Image Lab 5.0 (BioRad). For fluorescence-based detection, 30–90 μg of pellet fractions were used, blots were blocked using Odyssey Blocking Buffer and stained per manufacturer’s instructions (Li-Cor, Lincoln, NE). Images were captured using ODYSSEY CLx and signal quantitation was done with Image Studio 4.0 (Li-Cor, Lincoln, NE). Complete list of primary and secondary antibodies used are listed in Tables S11 and S12.
Image acquisition and processing
Images were collected using the 20X HC PLAN APO (0.7NA) objective lens on a conventional Leica DM6000 B upright microscope (Leica Microsystems, Bannockburn, IL) fitted with a Retiga SRV camera (Q-Imaging, Surrey, BC, Canada). Tissue sections were scanned using a motorized XY stage then stitched together using Volocity Software version 6.3 (Perkin Elmer, Waltham, MA).
Results
GFAP-like immunoreactivity in Müller Glia following retinal injuries
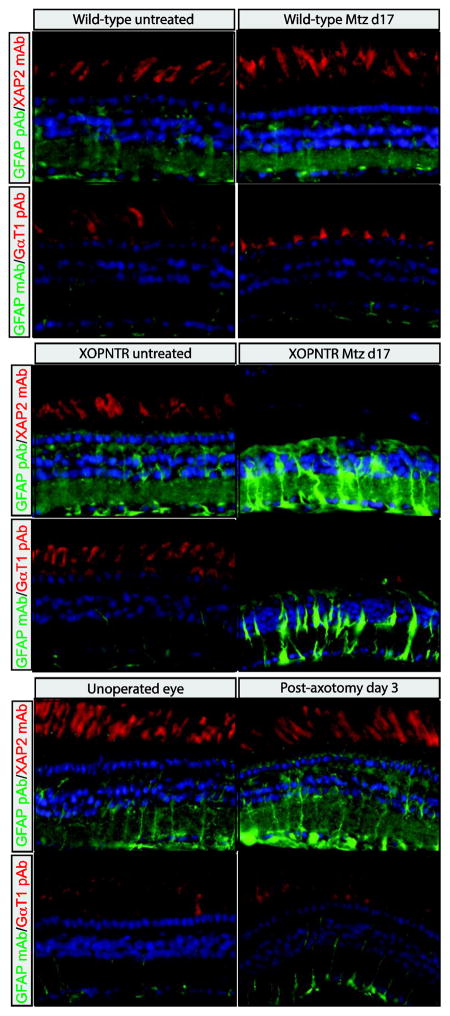
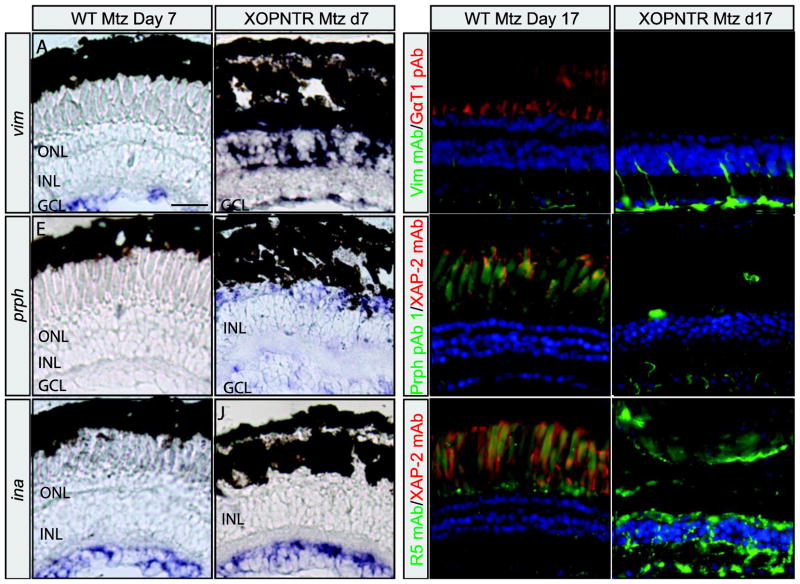
GFAP expression and reactive gliosis is induced in mammalian Müller glia following injury and during retinal degeneration. Following rod photoreceptor ablation in X. laevis, the R5 antibody labels filamentous components of Müller cells in a pattern reminiscent of GFAP (Choi et al., 2011; Sakaguchi et al., 1989). Unfortunately, the epitope for R5 is unknown (Dräger et al., 1984). In an attempt to determine if X. laevis Müller cells upregulate Gfap expression in response to rod ablation, we treated stage 50 XOPNTR tadpoles with Metronidazole (Mtz) for 17 days (when gliosis is severe Choi et al., 2011), and stained retinal sections using two commercially available anti-GFAP antibodies. The GFAP pAb (Dako, Product Number Z0334) and GFAP mAb (Sigma-Aldrich, clone G-A-5, Product Number G3893) both faintly stained the retinas of wild-type (untreated as well as Mtz-treated) and untreated XOPNTR tadpoles (Fig. 1A–E, G). Stained structures were filamentous and spanned the retina, consistent with the morphology of Müller glia. The GFAP pAb and mAb staining patterns were not identical however. In addition to the Müller glia, the GFAP pAb labelled both the inner and outer plexiform layers, while the GFAP mAb was much more restricted to the Müller glia and their processes (Compare Fig. 1A, E, I to C, G and K). Following rod ablation, there was a dramatic increase in the GFAP pAb and mAb immunoreactivity, which extended well into the outer nuclear and ganglion cell layers (Fig. 1E, G versus F, H). To determine if an upregulation of GFAP-like immunoreactivity was also observed following retinal axotomy, we unilaterally severed the axons of retinal ganglion cells (RGCs) in age matched wild-type tadpoles. Although much less pronounced than the response to rod ablation, GFAP-like immunoreactivity was clearly induced when the unoperated and operated sides of the same tadpoles were compared (Fig. 1I, K versus J, L). The increase in immunoreactivity was particularly obvious in the ganglion cell layer, where Müller glia endfeet are located (asterisks in Fig. 1I, K versus J, L). As with untreated and unoperated retinas, the staining pattern of the GFAP pAb and mAb in injured retinas were not identical (Fig. 1J versus L). Although the GFAP mAb staining pattern appeared restricted to the Müller glia and their endfeet in the RGC layer (Fig. 1L), an increase in GFAP pAb immunoreactivity was also evident in retinal ganglion cells (Fig. 1J). Together, these results indicate that retinal injury via rod photoreceptor ablation or RGC axotomy results in an increase in GFAP pAb and mAb immunoreactivity within Müller cells, but also indicates the GFAP pAb and mAb staining patterns are distinct, suggesting that they may not recognize the same antigen(s).
Figure 1. GFAP-like immunolabeling in the normal and injured X. laevis retina.
Retinal sections of wild-type (A–D, I–L) and XOPNTR transgenic (E–H) tadpoles were co-stained with GFAP pAb and XAP2 mAb (A, B, E, F, I, J) or GFAP mAb and GαT1 pAb (C, D, G, H, K, L). Green fluorescent secondary antibodies were used to visualize GFAP antibody staining, while red fluorescent secondary antibodies detect XAP2 (unknown epitope) and GαT1 (Transducin) antibodies, which stain rod photoreceptors. Wild-type controls and XOPNTR transgenic tadpoles were treated for 17 days with either DMSO-alone (A, C, E, G) or Mtz (B, D, F, H) to ablate rod photoreceptors. Retinal sections of wild-type tadpoles were stained three days following retinal axotomy. The left, unoperated eyes (I and K) are compared to the right, operated eyes (J and L) of the same animals. Hoescht (blue) stains nuclei of the ONL, INL and GCL, which are the outer nuclear, inner nuclear, and ganglion cell layers (asterisks in I–L), respectively. Scale bar = 25 μm.
No Xenopus gfap transcript or protein sequences annotated in public databases
We attempted to identify gfap transcripts that would allow us to confirm the upregulation of Gfap following injury (Fig. 1). However, blastn, blastp and tblastn searches of the NCBI (www.ncbi.nlm.nih.gov) and Ensembl databases (www.ensembl.org/) using the reference nucleotide and protein sequences (including isoforms) of the human, mouse, chicken and zebrafish GFAP failed to identify any X. laevis Gfap ortholog. The most similar reference transcripts and proteins coded for the Xenopus laevis Type III Intermediate Filament Proteins (IFPs) vimentin (Vim), desmin (Des), peripherin (Prph) and Type IV internexin neuronal intermediate filament protein, alpha (Ina; Table S1). A search of all available X. laevis ESTs (at NCBI and Xenbase http://xenbase.org/) also failed to identify any gfap transcripts (not shown). Similarly, we were unable to identify any nucleotide or protein sequence corresponding to a Xenopus tropicalis Gfap ortholog (not shown). Although X. laevis and X. tropicalis Unigene records for vim, des, prph and ina were identified, none exist for gfap (Table S1).
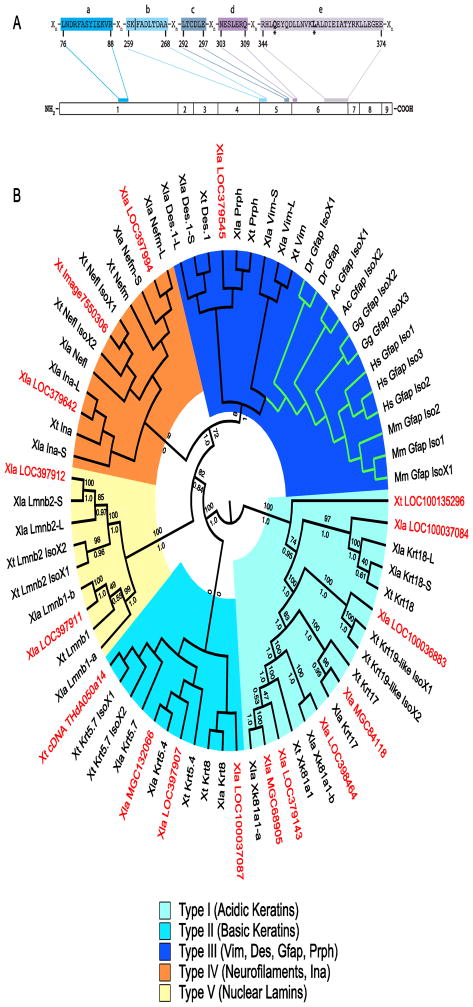
To identify regions conserved among GFAP orthologs, we aligned the human, mouse, chicken, green anole (Anolis carolinensis), X. tropicalis, X. laevis, and zebrafish, IFPs GFAP, VIM, DES, PRPH and INA, as well as their isoforms. The GFAP consensus contains five regions, ranging in size from 6 to 31 sequential residues that are invariant among GFAP orthologs, but vary at one or more positions in the IFPs VIM, DES, PRPH and/or INA (Figure 2A and S1). No X. laevis nor X. tropicalis protein or mRNA sequence was identified that included all of these GFAP consensus regions including the glutamine (Q) and leucine (L) residues that are invariant and unique to GFAP orthologs (Figure 2A, S1 and not shown).
Figure 2. Sequences most similar to GFAP in Xenopus.
A GFAP consensus used to distinguish candidate GFAP sequences from other intermediate filament proteins. Conserved regions (labelled a–e) are present in all GFAP orthologs identified and separated by regions of varying length (Xn). The gfap exon in which each region is coded is shown. The numbering system below the consensus is for human GFAP Isoform 1 (NP_002046). All residues shown are invariant in the GFAP orthologs aligned (Figure S1). The residues (*) at positions 347 (Q) and 357 (L) are unique to GFAP. Consensus region b is coded for in two exons (4 and 5). Vertical line (|) demarcates sequence coding for exons 4 and 5. B Cladogram of GFAP orthologs, Xenopus IFPs and untitled Xenopus proteins with similarity to GFAP. Midpoint rooted Polar Tree based on MAFFT alignment of GFAP orthologs, selected known X. laevis and X. tropicalis IFPs and the 17 untitled X. laevis and X. tropicalis sequences showing greatest similarity to GFAP (Katoh and Standley, 2013). Untitled X. laevis (Xla) and X. tropicalis (Xt) sequences are shown in red. Green branches show location of the GFAP clade. Groupings of IFP types are shown. Maximum likelihood bootstrap (above) and Bayesian inference posterior probabilities (below) for each branch are included (Felsenstein, 1981; Huelsenbeck et al., 2001). Accession numbers for each sequence in the tree can be found in Supplementary Table S2.
We identified multiple unnamed X. laevis and X. tropicalis protein (and translated DNA) records with similarity to the GFAP consensus sequence (Table S1 and not shown). To determine if one or more of these unnamed sequences might be Gfap, we aligned the 17 most highly similar to GFAP, with the GFAP orthologs from human, mouse, chicken, green anole, and zebrafish. None of the unnamed sequences contained the complete GFAP consensus sequence (Figure S2). We also built phylogenetic trees to compare the relative similarity of the unnamed records to GFAP orthologs and other Xenopus IFPs. None of the unnamed records grouped in the GFAP clade, rather, all showed greater sequence similarity to other Xenopus IFPs including Type I and II keratins, Type IV neurofilaments, and Type V lamins in addition to the IFPs described above (Fig. 2B). Clade assignments were recovered (often with high support) irrespective of the alignment algorithm (Clustal Omega, MAFFT or MUSCLE), method of inference (Bayesian inference or Maximum likelihood) or if GBlocks masking was used prior to alignment (Fig. 2B and Supplementary Figs. S3 – S14).
The GFAP gene in human consists of 9 coding exons, and 23 GFAP splice variants have been identified. Alternative splice variants might explain why no single Xenopus record contained all 5 GFAP consensus regions. Therefore, we identified the exons containing each of the five consensus GFAP regions (Fig. 2A). GFAP conserved region b spans exons 4 and 5, therefore we used the coding regions of exons 1, 5 and 6 (from human, mouse, chicken, green anole, and zebrafish) independently as probes to search for Xenopus gfap. No reference nucleotide nor protein was identified that matched the consensus GFAP sequences from exons 1, 5 or 6 (Table S3). All similar sequences were identified as another intermediate filament protein or one of the unnamed sequences sharing greater sequence similarity to other Xenopus IFPs described above (Fig. 2B, S1, S2, Table S1). Finally, none of the GFAP consensus sequences of Figure 2A were detected in any Xenopus (laevis or tropicalis) expressed sequence tag (EST) present in NCBI (not shown).
In conclusion, we were unable to identify a transcript or protein sequence coding for a Xenopus gfap in the publicly accessible sequence databases, suggesting Xenopus gfap is either not transcribed or not present in the X. laevis and X. tropicalis genomes.
Syntenic analyses indicate a chromosomal rearrangement resulted in the loss of Xenopus gfap gene
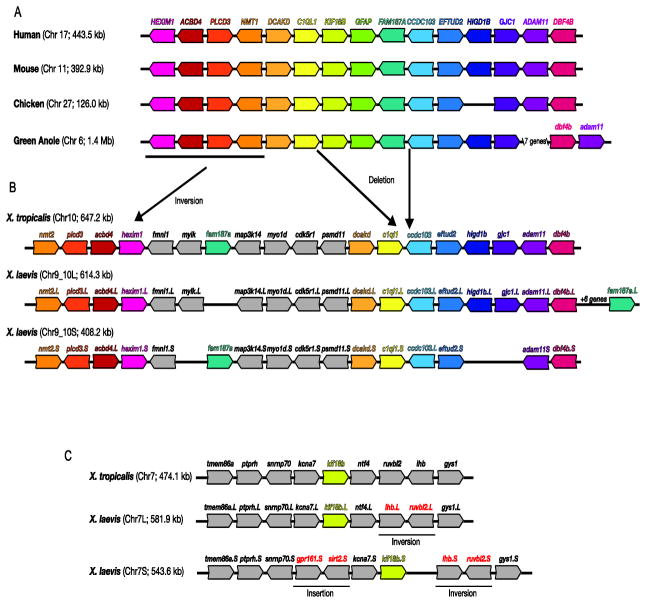
To determine if the gfap gene is present in the Xenopus genome, we first identified and compared the regions surrounding the gfap gene in the human, mouse, chicken and green anole genomes to that of X. tropicalis (Fig. 3A, B). Both the gene order and orientation were well conserved in the genomic region surrounding the gfap genes from humans to green anole. Exceptions were the lack of HIGD1B in chicken and a rearrangement of the region including the adam11 and dbf4b genes, which appears in a reverse orientation in green anole (Fig. 3A). In the corresponding region of the X. tropicalis genome however, the gfap locus is notably absent (Fig. 3B). A region flanked by the c1ql1 and ccdc103 genes, which includes kif18b, gfap and fam187a, is not syntenic with that of the other species. In amniotes fam187a is located immediately 3′ to gfap. In contrast, X. tropicalis fam187a (Chr10) is inverted in orientation and located 5′ to its position in other species (Fig. 3B). kif18b is located 5′ to GFAP in amniotes, while X. tropicalis kif18b is located on a different chromosome altogether (Chr7; Fig. 3C). Despite the relative close proximity of gfap to kif18b and fam187a in other species (between 1.0 and 84.1 kb), we did not detect GFAP consensus sequences on either the kif18b or fam187a containing chromosomes 7 and 10, respectively.
Figure 3. Syntenic analyses of gfap genomic regions.
The genomic regions surrounding gfap genes in select vertebrates are illustrated. For clarity, homologous genes have been similarly colored. Non-coding RNAs and pseudogenes were not included. Although human FAM187a is not annotated on the most recent reference assembly (GRCh38), it was included on previous assemblies and its location is included here. Chromosome number as well as the size of the regions depicted in the schematic are shown in parenthesis. The reverse complement of sequences were used so the gfap gene appears 5′ (left) to 3′ (right) in all species. The location of hypothesized deletions, duplications and inversions are also illustrated. The sequence source and locations used to build these syntenic schematics can be found in Table S4.
Similar disruptions encompassing syntenic regions in X. laevis were also observed (Fig. 3B, C). A genome allopolyploidization event between two ancestral 18-chromosome species resulted in nearly double the number of X. laevis chromosomes (N=36), relative to X. tropicalis (N=20) (Bisbee et al., 1977; Chain and Evans, 2006; Evans et al., 2004; Hellsten et al., 2007). Consequently, two X. laevis regions syntenic to each X. tropicalis region were identified. Minor genomic rearrangements including small deletions, inversions and insertions distinguish between the two Xenopus species. Nevertheless, X. laevis chromosomes including the c1ql1/ccdc103 (Chr9_10L and S; Fig. 3B) and kif18b (Chr7L and S; Fig. 3C) genes, were generally syntenic with the corresponding X. tropicalis regions, and also lacked consensus sequences for GFAP. These results, and the absence of any transcript or protein sequences homologous to GFAP, suggest both these Xenopus species, lack a gfap gene, and the chromosomal rearrangement resulting in gfap deletion occurred before these species diverged from their common ancestor 64 to 45 million years ago (Evans et al., 2004; Wiens, 2007).
Commonly used GFAP antibodies are nonspecific and detect multiple X. laevis Intermediate Filament Proteins
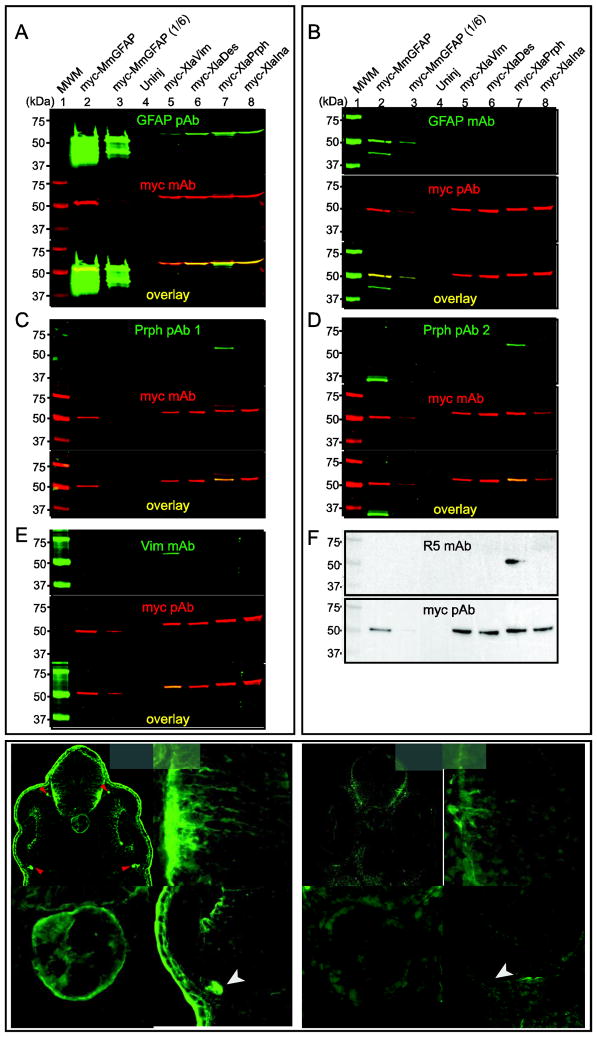
GFAP-like immunoreactivity is detected following retinal injury (Fig. 1), however, syntenic analyses indicates the X. laevis genome lacks a gfap gene (Fig. 3). We used Western blots to determine if the GFAP antibodies only detect GFAP (Fig. 4A and B). If nonspecific, we reasoned GFAP antibodies would most likely detect closely related, Type III IFPs (Fig. 2). Therefore, we expressed myc-tagged versions of mouse GFAP (as a positive control), X. laevis IFPs Vim, Des, Prph, as well as the Type IV IFP Ina in Xenopus embryos to test the specificity of the GFAP pAb and mAb (Fig. 4). Myc antibodies detected all of the myc-IFP fusion proteins in extracts from injected embryos, confirming they were successfully expressed (Fig. 4A and B, middle panels, lanes 2, 3, and 5–8). The GFAP pAb and mAb both detected products consistent with the size of myc-MmGFAP, that was co-labeled with the myc antibody (Fig. 4A and B, lanes 2 and 3). The GFAP pAb did not detect a product in uninjected embryos (Fig. 4A, lane 4), but did detect proteins in embryos expressing myc-XlaVim, myc-XlaDes, myc-XlaPrph and myc-XlaIna, which ran at the same position as the myc-tagged IFPs (Fig. 4A, lanes 5–8). The GFAP mAb by contrast, only detected the overexpressed myc-MmGFAP, and did not cross-react with any of the myc-XlaIFPs tested (Fig. 4B).
Figure 4. Specificity of intermediate filament protein antibodies.
Western blots were used to determine the specificity of GFAP pAb (A), GFAP mAb (B), Prph pAb 1 (C), Prph pAb 2 (D), Vim mAb (E), and R5 mAb (F). Extracts were prepared from embryos injected with mRNA coding for the indicated myc-tagged IFP. Blots were probed with the indicated combination of primary antibodies. Green fluorescent secondary antibodies were used to detect the IFP and myc primary antibodies, respectively (A–E). Red fluorescent signals were pseudocolored to make visualization easier. Merged images indicate myc-IFP proteins detected by both antibodies (A–E, yellow). Enhanced chemoluminescence was used to test the specificity of the R5 mAb and myc pAb (F). One-sixth the volume of extract from myc-MmGFAP expressing embryos in lane 2 was used in lane 3. Immunohistochemistry was use to compare the staining pattern of the GFAP pAb (G–G‴) and GFAP mAb (H–H‴) in sections of stage 35/36 X. laevis embryos. G′/H′, G″/H″ and G‴/H‴ show magnified views of the brain, notochord and retina, respectively. Arrow and arrow heads, indicate the location of skin epidermis and ocular motorneuron projections dorsal and ventral to the optic cup, respectively. Scale bar, 50 μm.
The ability of other IFP antibodies to detect GFAP, Vim, Des, Prph and Ina was also determined (Fig. 4C–F). Polyclonal antibodies generated in two rabbits using the same X. laevis Prph C-terminal peptide both detected myc-XlaPrph, but no other IFP tested (Fig. 4C and D; Gervasi et al., 2000). Similarly, a mouse monoclonal antibody (14h7) generated using extracts of the X. laevis A6 kidney epithelial cell line as an immunogen and later shown to detect vimentin, detected myc-XlaVim, but none of other IFPs tested (Fig. 4E; Dent et al., 1989; Klymkowsky et al., 1987). The monoclonal antibody R5 was generated using isolated ganglion cell layers from adult mice as the immunogen (Dräger et al., 1984). The R5 antigen was not identified, but labels filamentous components of predominantly non-neural cell types in mouse in a pattern similar, but not identical, to the distribution of the IFP vimentin. In mouse retina R5 strongly labels the regularly spaced Müller glia, astroglia of the optic fiber layer, and axonless horizontal cells, while only Müller glia appear stained in the X. laevis retina (Dräger et al., 1984; Sakaguchi et al., 1989). By Western blot R5 mAb detected myc-XlaPrph but not myc-XlaVim or any of the other IFPs (Fig. 4F).
The different retinal staining patterns of the GFAP pAb and mAb suggest they detect different antigens (Fig. 1). To test this hypothesis, X. laevis embryos were cultured to tailbud stages (stg. 35/36) and immunolabeled with either GFAP pAb or mAb (Fig. 4G–H‴). Both GFAP antibodies showed bilaterally symmetric staining patterns (Fig. 4G and H). However the GFAP pAb and mAb did not stain identical tissues. The GFAP pAb had a filamentous labeling pattern in the brain and retina, labelled the notochord, the cortex of cells in both layers of the skin epidermis, as well as the processes of ocular motor neurons immediately dorsal and ventral to the optic cup (Fig. 4G–G‴). Although the GFAP mAb also had a filamentous labeling pattern in the brain (most intensely staining ventrolateral regions; Fig. 4H′), staining of the retina, notochord, skin and extra-ocular structures was greatly reduced or absent, relative to the GFAP pAb (Fig. 4H–H‴).
Together, these results indicate the GFAP mAb, two Prph pAbs and the Vim mAb recognize their respective mouse and X. laevis IFP targets, but not the other IFPs when tested by Western blot. Furthermore, we conclude that XlaPrph is an R5 mAb antigen. Importantly, the GFAP pAb is not specific for GFAP, but also detects the four additional Xenopus IFPs tested here by Western blot (summarized in Table S5). The staining patterns of the GFAP pAb and mAb are distinct, also suggesting they do not exclusively recognize the same antigen. In combination with our inability to detect a Xenopus gfap gene, transcript or protein, these observations suggest the GFAP-like immunoreactivity detected in the Xenopus tadpole retina following injury may be Vim, Des, Prph, Ina or another as yet unidentified antigen.
Expression patterns of the IFPs vim, des, prph, ina and nes in pre-metamorphic X. laevis
Since X. laevis lack gfap, and the GFAP pAb antibody recognized multiple IFPs, we wondered which IFPs are expressed in the retina and induced following retinal injury. We first used in situ hybridization to determine the normal expression patterns of the IFPs in central retinal sections of stage 50 tadpole retina, and adjacent spinal cord and brain regions. The type IV intermediate filament protein nestin (nes) was previously shown to be upregulated in mouse, rat and chicken retina following mechanical or chemical injury (Fischer and Omar, 2005; Kohno et al., 2006; Luna et al., 2010; Xue et al., 2006; Xue et al., 2010). Therefore, in addition to the X. laevis type III intermediate filament proteins vim, des, prph, and type IV ina we also determined the expression pattern of nes.
In the uninjured retina, vim expression was detected in the ciliary marginal zone (CMZ), cells dispersed within the inner nuclear and ganglion cell layers, the optic nerve head as well as the optic nerve (Fig. 5A). Elsewhere in the eye, vim was detected in the lens epithelium and the epithelia lining the outside of the peripheral retina (Fig. 5A). Expression was also detected in cells of the spinal cord and brain ventricular zones (Fig. 5B and C). In contrast to vim (and all other IFPs tested), des was not detected in retinal, spinal or brain tissues (Fig. 5D–F). However, consistent with previous findings, des expression was observed in muscle tissues, including the muscles surrounding the eye and the somites surrounding the spinal cord (Fig. 5D and not shown; Herrmann et al., 1989b).
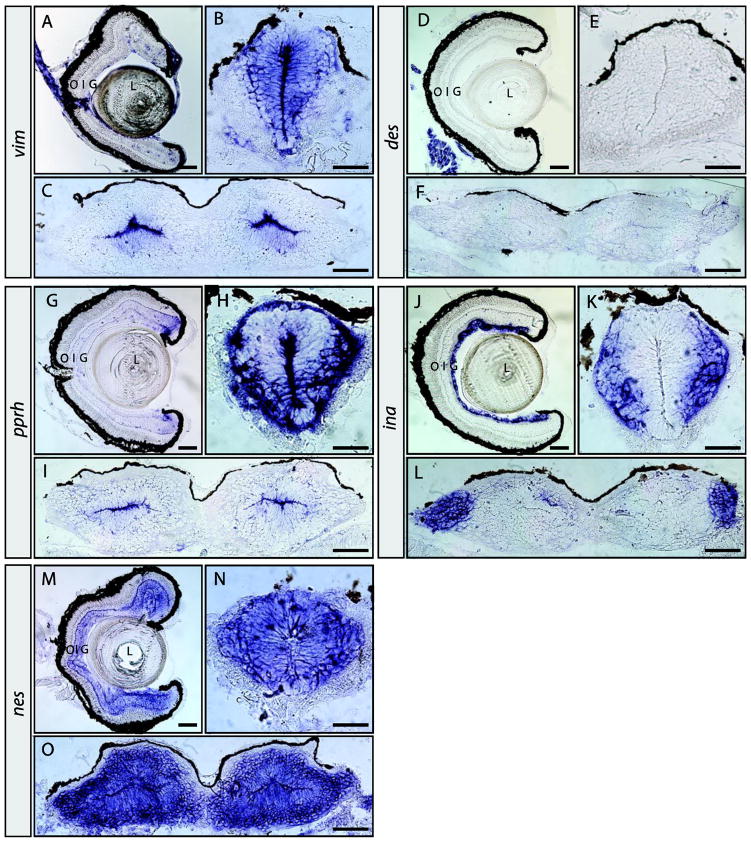
Figure 5. Expression patterns of vim, des, prph, ina, and nes in the pre-metamorphic tadpole nervous system.
In situ hybridization on retinal, brain and spinal cord sections for vim (A–C), des (D–F), prph (G–I), ina (J–L), and nes (M–O). Sections were obtained from pre-metamorphic stage 50 tadpoles. Location of the lens (L), outer (O), inner (I) and ganglion (G) cell layers are indicated. Scale bars, 100 μm in A, D, G, J and M; all others are 50 μm.
In the pre-metamorphic tadpole retina prph expression is detected at the ciliary marginal zone (CMZ), but not in the stem cells located at the very periphery (Fig. 5G). prph expression is most abundantly detected in newly born ganglion and other retinal cells adjacent to the CMZ, consistent with previous findings (Gervasi et al., 2000). Transcripts were also detected at the inner and outer plexiform layers of the peripheral retina (Fig. 5G). In the spinal cord, prph is expressed in cells of the ventricular zone lining the central canal, but its expression is strongest in a patch of ventral neurons and the marginal zone (Fig. 5H). Expression was also observed in the dorsal root ganglia (not shown, and Gervasi et al., 2000). In the brain, prph transcripts appeared restricted to cells of the ventricular zone (Fig. 5I). Northern blot analyses of X. laevis tissues indicated ina was expressed in the nervous system, and in situ hybridization confirmed that ina is expressed in the brain, retina, and spinal cord (Zhao and Szaro, 1997b). Retinal expression of ina at stage 50 is restricted to the ganglion cell layer (Fig. 5J and Matsuda and Kondoh, 2014). Expression was also observed in a few cells dispersed in the inner nuclear layer, possibly Müller gila, amacrine or displaced ganglion cells (Fig. 5J). In the spinal cord ina was expressed in differentiated cells outside the ventricular zone (Fig. 5K). Similar to prph, ina expression was also observed in the DRG (not shown, and Matsuda and Kondoh, 2014). ina expression was also detected in the lateral brain (Fig. 5L). Nestin had the most extensive expression pattern of the IFPs investigated. In the stage 50 retina nes was detected in the CMZ, ganglion cell, inner and outer nuclear layers, as well as both plexiform layers (Fig. 5M). In both the spinal cord and brain, nes expression was detected in the ventricular and marginal zones (Fig. 5N, O).
In summary, the expression patterns of the IFPs determined here are distinct and only partially overlap in the stage 50 tadpole retina, brain and spinal cord. In the retina, expression of the IFPs vim, prph, ina and nes, but not des, are detected.
vim, prph and ina expression are altered in response to retinal injuries in X. laevis
To determine which, if any, IFPs were upregulated follow retinal injury, we first examined the expression of vim, prph, ina and nes by in situ hybridization on retinal sections of XOPNTR tadpoles treated with Mtz for seven and seventeen days to ablate rod photoreceptors (Fig. 6). These time points were selected for in situ hybridization and immunohistochemistry since preliminary results indicated peak transcript expression at day 7, while Müller cell hypertrophy peaks at day 17 (Choi et al., 2011). Rod photoreceptors and vim expression in wild-type animals were unaffected by Mtz-treatment (Fig. 6A and not shown; n=4). Mtz-treated XOPNTR tadpoles lacked rod outer segments and an increase in vim expression was observed in both the inner and outer nuclear layers, where expression of vim in wild-type animals was below detectable levels (Fig. 6A vs B, white asterisk, n=4). The morphology of the INL cells expressing vim was consistent with that of Müller glia (Fig. 6B). To distinguish between the RPE and the signal produced by vim expression, we treated retinal sections with hydrogen peroxide. Bleaching of the RPE pigment revealed that vim expression was dramatically increased in the subretinal space of XOPNTR animals lacking rod outer segments (Supplementary Fig. S15, white asterisk). Although much less dramatic in comparison to the INL and ONL, there was also a detectable increase in vim in the ganglion cell layer (Fig. 6A, B and S15A, B). At day 17, vim expression was no longer detected in the INL or subretinal space of Mtz-treated XOPNTR tadpoles (not shown, n=4).
Figure 6. Retinal expression of intermediate filament proteins after rod photoreceptor ablation.
vim (A and B), prph (E and F), and ina (I and J) in situ hybridization in retinal sections from wild-type (A, E, and I) and XOPNTR (B, F, and J) tadpoles treated with Mtz for 7 days. Dashed white lines indicate the boundary between the RPE (dark pigment) and neural retina. White asterisks in B and F indicates expression of vim (B) and prph (F) adjacent to the RPE in the subretinal space. Immunolabeling of retinal sections with anti-Vim mAb (C and D), Prph pAb 1 (G and H), and R5 mAb (K and L) in Mtz-treated wild-type (C, G, and K) or XOPNTR (D, H, and L) retinas. Retinal sections were co-stained with DAPI to visualize nuclei. Scale bars, 50 μm.
prph expression in the Mtz-treated wild-type tadpole retina was only observed at the CMZ and expression was not detected in the central retina (Fig. 6E and not shown; n=2). Rod ablation in XOPNTR tadpoles increased prph levels in the GCL, INL, as well as subretinal space of the central retina (Fig. 6F). Compared to the expression of vim, prph subretinal expression was reduced, yet detectable by in situ hybridization (Fig. 6F, white asterisk). Similar to vim, prph expression was not detected in the retina of rod ablated XOPNTR tadpoles by day 17 (not shown, n=2).
In contrast to the increase observed with vim and prph transcripts, the expression patterns of ina and nes were unchanged in response to rod ablation at day 7 and 17 (Fig. 6I, J and not shown). These results indicate that vim and prph transcript levels are increased, but neither ina nor nes are altered following rod loss in the X. laevis retina.
We next asked if an increase in vimentin- and peripherin-immunoreactivity could be detected following rod loss. Control and rod ablated retinas were stained by immunohistochemistry using Vim mAb, Prph pAb 1 and 2, as well as the R5 mAb. In control, Mtz-treated wild-type tadpoles, Vim mAb immunoreactivity was detected in Müller glia from the end-feet at the inner limiting membrane, through the ganglion cell layer, but only extended into the first half of INL (Fig. 6C). Vim-labeled processes were not observed at the outer limiting membrane in wild-type tadpoles (Fig. 6C). In rod ablated XOPNTR retinas however, Müller glia processes were detected in the inner as well as outer limiting membranes, with staining intense enough to trace individual Müller cells and their processes through the entire thickness of the retina (Fig. 6D).
Prph pAb 1 immunolabeling was weak, but detectable in control retinas (Fig. 6G). In the absence of injury, only short, faintly labeled processes were observed in the GCL (Fig. 6G). In rod ablated retinas however, Müller glia processes were easily detected using Prph pAb 1, with processes extended from the GCL to the OLM (Fig. 6H). In spite of detecting myc-XlaPrph by Western blot (Figs. 4D) we were unable to detect Müller glia in either the uninjured or rod ablated retina using Prph pAb 2 (not shown).
R5 mAb stained Müller cell processes in wild-type control retinas, but only faint, thin Müller cell processes were detected, extending from the GCL into the INL (Fig. 6K). Consistent with our previous results, rod ablation resulted in a dramatic increase in Müller cell immunoreactivity, with staining observed from the Müller cell endfeet to the OLM (Fig. 6L and Choi et al., 2011).
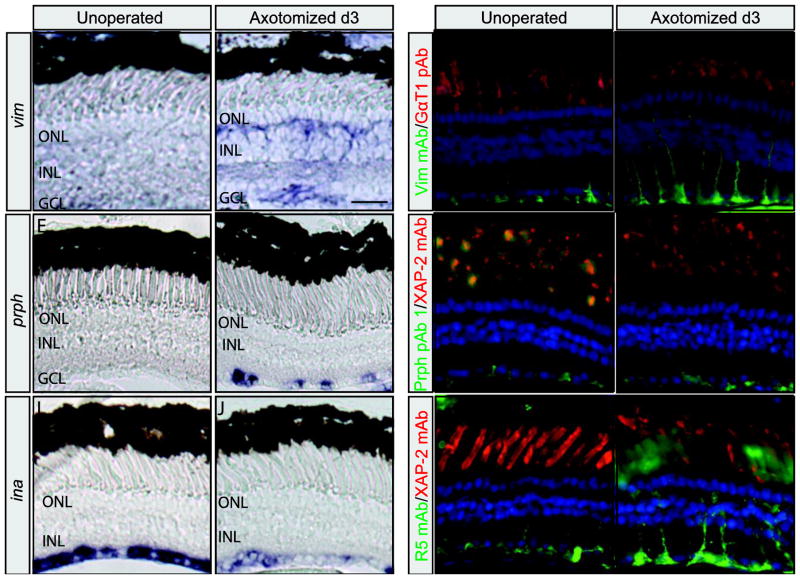
To determine if changes in IFP gene expression also occurred when neurons on the opposite side of the retina were damaged, we performed RGC axotomy on stage matched (stg. 50) pre-metamorphic tadpoles (Fig. 7). Changes in vim, prph, ina, and nes expression were identified by comparing the in situ expression patterns of retinal sections from unoperated tadpoles, to the unoperated (control; left) and operated (optic nerve severed; right) eyes of operated tadpoles 3 days post surgery. vim, prph, ina, and nes expression patterns on the unoperated left eyes of operated animals (Fig. 7A, E, I, and not shown), were indistinguishable from the expression in unoperated animals (Fig. 5A, G, J and M). However, we observed an increase in vim and prph expression when the operated and unoperated eyes of the same animals were compared (Fig. 7A vs B, n=5 and E vs F, n=4). vim expression was detected in both the GCL and INL (Fig. 7B). Expression was present in filamentous processes extending from the ILM to the base of the ONL, where vim transcripts appeared to accumulate (Fig. 7B). The vim pattern of expression was consistent with the morphology of Müller cells, with strong expression also detected throughout the INL (Fig. 7B). The increase in prph expression was detected in distinct spots along the GCL and the optic nerve (Fig. 7F and not shown). These findings are consistent with the upregulation of prph observed in the GCL of post-metamorphic frogs following optic nerve crush (Gervasi et al., 2003). Interestingly, the location of both vim and prph expression was injury dependent. Rod ablation resulted in increased expression in the outer nuclear layer (Fig. 6B, E), while RGC axotomy intensified vim and prph expression in GCL (Fig. 7B, F).
Figure 7. Retinal expression of intermediate filament proteins after retinal ganglion cell axotomy.
vim (A and B), prph (E and F), and ina (I and J) in situ hybridization on retinal sections from control unoperated (A, E, and I) and operated (retinal axotomy) (B, F, and J) eyes. Vim mAb (C and D), Prph pAb 1 (G and H), and R5 mAb (K and L) immunolabeling of retinal sections from control (C, G, and K) and operated (D, H, and L) eyes. Rod photoreceptors were labelled with either GαT1(Transducin) pAb (C and D) or XAP-2 mAb (G, H, K, and L). Sections were counterstained with DAPI to visualize nuclei. Scale bars, 50 μm.
ina was strongly expressed throughout the GCL of the wild-type and unoperated eye (Figs. 5J and 7I; n=6). Contrary to vim and prph, ina expression was reduced in the GCL of the operated versus control eye 3 days post-axotomy (Fig. 7I vs J; n=6). Downregulation of ina was also reported following optic nerve crush in post-metamorphic frogs (Gervasi et al., 2003). In contrast to other IFPs, we detected no change in the expression of nes following RGC axotomy (not shown).
We also used indirect immunohistochemistry to detect changes in Vim mAb, Prph pAb 1, and R5 mAb immunoreactivity following retinal axotomy. In the unoperated contralateral eye, Vim immunoreactivity was observed mostly at the Müller glia endfeet and in thin Müller cell processes extending into the INL, but not beyond (Fig. 7C; 100%, n=5). In the operated eye, Vim immunoreactivity was increased in the Müller glia (Fig. 7D; 60%, n=5). Strong Vim staining was observed at the glial endfeet, which extended through most of the GCL. Müller cell processes in the injured retina appeared enlarged and in most cases, spanned the entire width of the retina (Fig. 7D; 60%, n=5). Similar to Vim, Prph pAb 1 immunoreactivity was also increased after the optic nerve was severed, but to a lesser extent and not in every retina. In the unoperated contralateral eye, Prph immunoreactivity was mostly detected at the Müller glia endfeet (Fig. 7G; 100%, n=5). In contrast, at 3 days post-axotomy, Prph pAb1 immunoreactivity was observed at the endfeet, but also in Müller glia processes extending into the INL (Fig. 7H; 40%, n=5). R5 immunoreactivity was also detected mostly at the endfeet in the unoperated eye, although faintly labeled processes were also observed as far as the outer plexiform layer (Fig. 7K; 50%, n=6). Following axotomy, R5 strongly labeled the Müller cell endfeet and processes into the ONL (Fig. 7L).
A summary of the IFP in situ and immunohistochemistry expression results are shown in Table S6. The extent of Vim and Prph immunoreactivity in response to axotomy varied somewhat from animal to animal, possibly due to undetectable differences in surgery success. Nevertheless these results demonstrate that transcripts for vim and prph were both higher following RGC axotomy as well as rod photoreceptor ablation. Interestingly, although ina was unchanged after photoreceptor ablation, ina expression was reduced following RGC axotomy. The increases in Vim and Prph immunoreactivity, however, paralleled the changes in vim and prph transcript levels, supporting the conclusion that both were higher in response to retinal damage. The IFP response also appeared injury dependent. For example, peripherin levels appeared to increase in Müller cells following rod ablation, but in Müller and RGCs following axotomy. The differences observed in the expression patterns of vim and ina also suggest the response was injury-dependent since transcript as well as immunoreactivity locations were dependent on the injury paradigm used and appeared more localized to the injured retinal layer.
gfap detected in representative species from Caudata and Gymnophiona, but not Anura, amphibian Orders
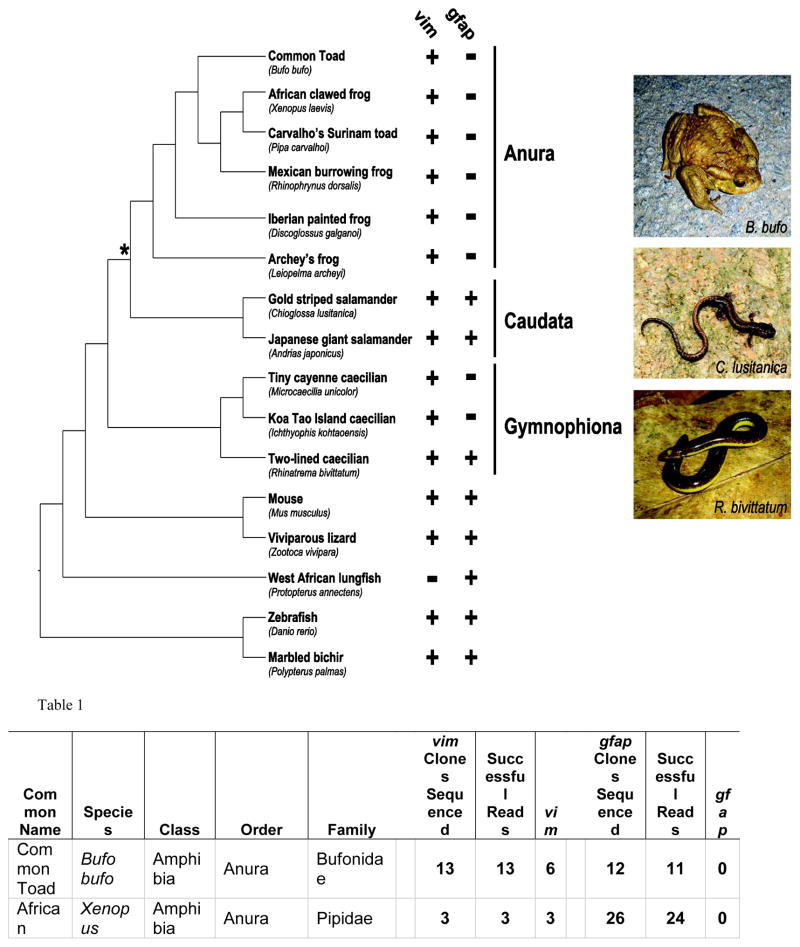
In the absence of gfap, gliosis and/or glial scar formation might be reduced or delayed, providing a permissive environment for retinal regeneration in Xenopus. In addition to X. laevis, other amphibians can also regenerate retina (reviewed in Araki, 2007; Barbosa-Sabanero et al., 2012; Chiba, 2014; Mitashov, 1997). To determine if gfap loss was restricted to X. laevis and X. tropicalis, we designed degenerate PCR primers and attempted to amplify gfap and vim from multiple amphibian species. Template gDNA was isolated from representatives of the three living Orders of amphibians including six Anura (frogs and toads), two Caudata (salamanders and newts) and three limbless Gymnophiona (caecilians) species (Fig. 8). Despite multiple attempts, and the fact that vim was amplified from the same gDNA of all six anuran species, we were unable to amplify gfap (Fig. 8 and Table 1). Degenerate gfap primers were designed using human, mouse, chicken, anole and zebrafish sequences, and were successfully used to amplify gfap from mouse and zebrafish gDNA (Fig. 8 and Table 1). In addition, gfap was also successfully amplified from other non-anuran species, including Viviparous lizard, West African lungfish and Marbled bichir, suggesting the failure to amplify gfap from anuran species did not represent a false negative. In contrast to the lack of gfap in anurans, we were able to amplify gfap from both salamanders and one of the three caecilian species (Fig. 8 and Table 1).
Figure 8. Schematic representation of the evolutionary relatedness of species in the survey for gfap and vim genes.
Orders for amphibian species are indicated. Asterisk illustrates last common ancestor shared by Anura and Caudata. + and - indicate presence or absence of indicated gene in the PCR surveys, respectively. Photo credits: B. bufo, R. bivittatum (DSM), C. lusitanica (Benny Trapp: https://commons.wikimedia.org/wiki/User:Benny_Trapp)
Table 1. Results of PCR survey to identify gfap and vim sequences in the genomes of representative amphibian and other vertebrate species.
See Supplementary Fig. S16 for detailed phylogenetic analysis used to confirm the vim (column 8) and gfap (column 11) sequences identified in PCR survey.
| Common Name | Species | Class | Order | Family | vim Clones Sequenced | Successful Reads | vim | gfap Clones Sequenced | Successful Reads | gfap |
|---|---|---|---|---|---|---|---|---|---|---|
| Common Toad | Bufo bufo | Amphibia | Anura | Bufonidae | 13 | 13 | 6 | 12 | 11 | 0 |
| African clawed frog | Xenopus laevis | Amphibia | Anura | Pipidae | 3 | 3 | 3 | 26 | 24 | 0 |
| Carvalho’s Surinam toad | Pipa carvalhoi | Amphibia | Anura | Pipidae | 7 | 7 | 6 | 13 | 12 | 0 |
| Mexican burrowing frog | Rhinophrynus dorsalis | Amphibia | Anura | Rhinophrynidae | 7 | 7 | 7 | 12 | 12 | 0 |
| Iberian painted frog | Discoglossus galganoi | Amphibia | Anura | Discoglossidae | 8 | 7 | 7 | 11 | 11 | 0 |
| Archey’s frog | Leiopelma archeyi | Amphibia | Anura | Leiopelmatidae | 24 | 13 | 9 | 12 | 12 | 0 |
| Gold striped salamander | Chioglossa lusitanica | Amphibia | Caudata | Salamandridae | 12 | 10 | 7 | 25 | 12 | 11 |
| Japanese giant salamander | Andrias japonicus | Amphibia | Caudata | Cryptobranchidae | 7 | 7 | 7 | 7 | 7 | 7 |
| Tiny cayenne caecilian | Microcaecilia unicolor | Amphibia | Gymnophiona | Siphonopidae | 7 | 6 | 6 | 23 | 22 | 0 |
| Koa Tao Island caecilian | Ichthyophis kohtaoensis | Amphibia | Gymnophiona | Ichthyophiidae | 7 | 7 | 7 | 8 | 7 | 0 |
| Two-lined caecilian | Rhinatrema bivittatum | Amphibia | Gymnophiona | Rhinatre matidae | 11 | 9 | 6 | 8 | 8 | 8 |
| Mouse | Mus musculus | Mammalia | Rodentia | Muridae | 3 | 3 | 3 | 6 | 6 | 6 |
| Viviparous lizard | Zootoca vivipara | Sauropsida | Squamata | Lacertidae | 12 | 9 | 7 | 7 | 6 | 6 |
| West African lungfish | Protopterus annectens | Sarcopterygii | Lepidosireniformes | Protopteidae | 13 | 13 | 0 | 9 | 7 | 7 |
| Zebra fish | Daniorerio | Actinopterygii | Cypriniformes | Cyprinidae | 3 | 3 | 2 | 3 | 3 | 3 |
| Marbled bichir | Polypterus palmas | Actinopterygii | Polypteriformes | Polypteridae | 7 | 6 | 6 | 11 | 10 | 7 |
We used phylogenetic analyses to test if the PCR products amplified were most similar to gfap and vimentin or to other IFP orthologs (Supplementary Fig. S16). Although support was low in the more internal parts of the tree, both GFAP and VIM clusters are well differentiated and strongly (nearly maximally) supported. Support values of some nodes within these two clusters are low, but this lack of support is unsurprising given that amino acid sequences of the clones are short compared to the other sequences of the full matrix, and overlap is relatively small (for example, only 37 amino acid positions shared between GFAP and VIM clones). In any case, the results clearly and confidently support the hypothesis that gfap is present in the genome of both caecilians and salamanders, having been amplified from one caecilian (R. bivittatum) and two salamander (A. japonicus and C. lusitanica) representatives. Amino acid sequences of the clones obtained were all normal and there is no reason to suspect that they are not functional. No frameshifts or irregular stop codons were identified, and a similar amino acid composition for GFAP orthologs from other vertebrates was observed. All these phylogenetic results, plus the absence of gfap positive clones in the anuran representatives, suggest that the deletion of gfap most likely occurred in the ancestor of all Anura after its divergence from the Caudata ancestor around 290 million years ago (San Mauro, 2010).
Discussion
We report that despite an increase in GFAP-like immunoreactivity following retinal injury, Xenopus lacks a gfap gene. The epitope(s) detected by the GFAP antibodies remain unknown. However, the GFAP pAb cross-reacts with multiple related IFPs, including Vim and Prph, which are both transcriptionally elevated in Müller glia following retinal injury. The antigen recognized by the GFAP mAb remains unknown. Interestingly, vim and prph transcription as well as Vim and Prph immunoreactivity were induced in an injury-dependent manner. We provide evidence that a chromosomal rearrangement, most likely an unbalanced translocation, resulted in deletion of the gfap gene during amphibian evolution. Deletion of the gfap locus does not appear to be restricted to Xenopus, but most likely occurred early in the anuran lineage, since gfap was not detected in basal anuran species, but was amplified from species of the two other amphibian Orders.
GFAP and cross-species antibody specificity
The GFAP pAb was generated using GFAP isolated from cow spinal cord (Dako, Carpinteria, CA). The epitope has not been reported, making determination of the specificity of the antibody problematic, particularly with respect to its cross-species use (Bordeaux et al., 2010; Saper, 2009). Perhaps unsurprisingly, we found by Western blot the GFAP pAb was non-specific and recognized X. laevis Vim, Prph, Des, and Ina, as well as mouse GFAP (Fig. 4). The GFAP mAb was generated using pig GFAP, also isolated from spinal cord (Debus et al., 1983). The GFAP mAb epitope was mapped to the rod domain in the C-terminus of the protein (Sigma-Aldrich Technical Service, personal communication). The rod domain is highly conserved in size, sequence and secondary structure (Chernyatina et al., 2015; Kornreich et al., 2015; Strelkov et al., 2002). Within the rod domain is a highly conserved sequence known as the intermediate filament consensus sequence present in all IFPs (Hatzfeld and Weber, 1992). The mapped epitope for the GFAP mAb includes the IF consensus sequence, suggesting this antibody might recognize other IFPs. Yet despite having the IF consensus, the GFAP mAb failed to cross-react with any of the Xenopus IFPs tested and only detecting mouse GFAP by Western blot (Fig. 4).
Since both the pAb and mAb had been previously used for IHC on X. laevis brain, spinal cord and retinal tissues, we directly compared their expression patterns by immunohistochemistry. Their distinct expression patterns suggest they are unlikely to be recognizing a single common antigen. The GFAP mAb epitope shares between 63% to 66% sequence identity with the corresponding regions of X. laevis Vim, Prph, Des and Ina (not shown). However, additional work will be necessary to identify the antigen detected by the GFAP mAb.
The specificity problems we encountered using the GFAP pAb and mAb reemphasized to us an important issue for work in Xenopus and other non-mammalian species. Most antibodies used in Xenopus are generated against mammalian proteins and therefore specificity can be an issue. For example, disparities in specificity have been previously observed for antibodies raised against mammalian IFPs used to immunolabel X. laevis CNS structures (Szaro and Gainer, 1988). Our current results extend previous findings that raise caution to the use of mammalian antibodies in X. laevis without thorough characterization. One example is the antibody R5, which was generated in mice using homogenized mouse ganglion cell layers as the immunogen (Dräger et al., 1984). By Western blot R5 detected X. laevis Prph (Fig. 4). Furthermore, prph transcripts and R5 immunoreactivity were both upregulated in Müller cells following retina injury (Fig. 6). However, R5 was only tested against five IFPs by Western blot. We can conclude that X. laevis Prph is an R5 immunogen. We cannot however, discount the possibility that R5 may also detect other, unidentified proteins in reactive Müller cells.
Response of X. laevis Müller glia cells to retinal injuries
Müller glia become reactive in virtually every known disease and injury of the retina (Bringmann et al., 2006). The adverse response of Müller glia to retinal injury includes upregulation of IFPs and formation of a gliotic scar, which are not only implicated in progressive retinal degeneration, but also in the reduced capacity of new retinal neurons to integrate and differentiate in the damaged retina. Mice deficient in GFAP and vimentin exhibit decreased reactive gliosis and improved neural regeneration following injury, indicating IFPs are not only a marker, but might have a causal link to reactive gliosis (Kinouchi et al., 2003; Menet et al., 2001; Menet et al., 2003; Nakazawa et al., 2007; Verardo et al., 2008; Wilhelmsson et al., 2004). GFAP expression is induced after retinal injury in fish and mammals and thought to be important for cytoskeletal changes that drive alterations in cell morphology (Bignami and Dahl, 1979; Davidson et al., 1990; Erickson et al., 1987; Grosche et al., 1997; Lewis et al., 1989; Lu et al., 2011; Lundkvist et al., 2004; MacDonald et al., 2015; Okada et al., 1990; Raymond et al., 2006; Sethi et al., 2005). X. laevis Müller glia also respond to retinal injury by changing their morphology, swelling (hypertrophy), and forming a gliotic scar (Choi et al., 2011). The question is, what mediates these changes in Xenopus Müller cells that lack a GFAP? Other IFPs are the logical possibility. One possibility is that Müller cell gliosis in Xenopus may be mediated by different IFPs. Based on our current results, two logical candidates for mediating the observed morphological changes following injury are Vim and Prph (Fig. 6 and 7).
In mammals and fish vimentin immunoreactivity is induced in Müller glia after injury (Cerdà et al., 1998; Lewis and Fisher, 2003; Sethi et al., 2005). Prior to injury, vimentin immunoreactivity in the retina nears undetectable levels, with staining only present in endfeet of mammalian Müller glia (Cerdà et al., 1998; Lewis and Fisher, 2003). Following either rod ablation or RGC axotomy vimentin transcripts and immunoreactivity were significantly upregulated in X. laevis Müller glia suggesting, like in mammals, vimentin might be involved in the morphological changes observed in Müller glia after injury (Fig. 6 and 7).
Prph is a second IFP with the potential to mediate the gliotic response in X. laevis. Prph is an interesting candidate because in mammals and zebrafish, it is only expressed in neurons (Fuchs and Weber, 1994; McLenachan et al., 2008), while Xenopus Prph is expressed in neurons, proliferating cells as well as glia (Gervasi et al., 2000). In mammals, peripherin is abundant in PNS neurons and is also expressed in RGCs (McLenachan et al., 2008; Wang et al., 2007). The peripherin response to retinal injury has not been determined in mammals. In the goldfish retina, Prph is expressed in ganglion cells and after optic nerve crush, its expression is induced in RGCs (Fuchs and Weber, 1994; Glasgow et al., 1992). Interestingly, and distinct from the response in mammals and fish, in X. laevis, increased prph expression was dependent on the method used to injure the retina. After rod photoreceptor ablation, prph expression appeared to be enriched in Müller cells (Fig. 6). In contrast, ganglion cell axotomy caused elevated prph expression in both Müller and ganglion cells (Fig. 7). The increased expression following both injury paradigms is consistent with peripherin also potentially participating in the gliotic response of Xenopus Müller glia. Because Prph shows a more restricted expression pattern in other species, it is interesting to consider the possibility that the broader expression domain of Prph, may have been an evolutionary adaptation in Anurans to compensate for the loss of gfap. Together, Vim and Prph may function to replace the role GFAP plays in mammalian glia to support the changes in morphology associated with injury induced gliosis. Further work is necessary to define the role of these IFPs in the response of X. laevis Müller glia to retinal injury.
Possible consequences of gfap gene loss in anuran species
The PCR-based survey of the three extant amphibian orders, suggests the gfap gene was deleted in an anuran ancestor (Fig. 8). The event resulting in gfap loss, likely took place early in the Anura lineage, since two of the species surveyed (D. galganoi and L. archeyi) are from Families that include the most primitive frogs (San Mauro, 2014).
GFAP is clearly not required for frog survival. Similarly, Gfap−/− mice are born and survive, exhibiting only subtle defects (Liedtke et al., 1996; McCall et al., 1996; Pekny et al., 1995). Interestingly, mice deficient for both GFAP and VIM also survive but have attenuated reactive gliosis in the brain, spinal cord and retina following injury or disease (Giménez Y Ribotta et al., 2000; Menet et al., 2003; Nakazawa et al., 2007; Wang et al., 1997; Wilhelmsson et al., 2004). Xenopus have a remarkable regenerative capacity, with the ability to generate retina following the ablation of single retinal cell types, partial, and even complete retinectomy (Araki, 2007; Choi et al., 2011; Filoni, 2009; Martinez-De Luna et al., 2011; Martinez-De Luna and Zuber, 2014; Vergara and Del Rio-Tsonis, 2009). Might a reduction in the gliotic response following retinal injury, due to the loss of gfap, result in the ability of X. laevis to regenerate retina?
One argument against this hypothesis would be that fish do not lack gfap, yet can regenerate retina. Gfap is induced in the damaged zebrafish retina, however, glial scars do not form. Instead of inhibiting regeneration, Müller cells are the source of new retinal neurons in fish. In response to retinal injury, fish Müller glia re-enter the cell cycle, then differentiate to generate new retinal cells that ultimately regenerate the retina (reviewed in Lenkowski and Raymond, 2014). A more compelling argument against the hypothesis that deletion of gfap allows retinal regeneration is that some Caudata, a sister Order to the Anura that includes salamanders, also regenerate the retina, yet they also have a gfap gene (Fig. 8 and Table 1). Caudata, like fish, may have evolved an independent mechanism to control the gliotic response in Müller cells, thereby permitting regeneration. It should be noted, that it is not known if gold-stripped or Japanese giant salamanders can regenerate retina. Therefore it will be important to determine if the gfap gene is present and functional in Caudata that can regenerate retina. Although loss of gfap may be a contributing factor, is unlikely to be the sole explanation for retinal regeneration in Anura.
In spite of no gfap and the remarkable regenerative capacity of the X. laevis retina, upregulation of IFPs, gliotic scar formation, and progressive retinal degeneration still follow rod photoreceptor ablation in Xenopus (Fig. 6, 7 and Choi et al., 2011). Additional work is needed to determine how these events are linked. In addition to providing mechanical support to cells, IFPs also function in cell migration, mechanotransduction, and signal transduction (Chernoivanenko et al., 2015; Goldman et al., 1996; Guo et al., 2014; Helfand et al., 2003; Ivaska et al., 2007; Matveeva et al., 2015). To better understand the role of IFPs in these processes, it will be important to know if reducing or eliminating other IFPs, such as Vim and Prph, alters the gliotic response, progressive retinal degeneration and promotes more robust retinal regeneration.
Supplementary Material
A consensus sequence (GFAP Consensus, Fig. 2A) was generated by aligning intermediate filament proteins GFAP, vimentin, desmin, peripherin and internexin neuronal intermediate filament protein, alpha. IFPs (and known isoforms) from human (Hs), mouse (Mm), chicken (Gg), green anole (Ac), X. laevis (Xla), X. tropicalis (Xt) and zebrafish (Dr) were aligned using the Clustal W algorithm (Thompson et al., 1994). For simplicity, only regions corresponding to residues 76 – 374 of human GFAP Isoform 1 (NP_002046) are shown. Residues identical to the GFAP Consensus are shaded black. Dashes represent gaps that have been inserted into sequences to increase their similarity in the alignment. Accession numbers for each sequence in the alignment can be found in Supplementary Table S2.
Clustal W alignment of GFAP orthologs from human, mouse, chicken, green anole, and zebrafish and the 17 most similar, yet untitled X. laevis and X. tropicalis sequences. For simplicity, only regions corresponding to residues 76 – 374 of human GFAP Isoform 1 (NP_002046) are shown. Residues identical to the GFAP consensus are shaded black. Dashes represent gaps that have been inserted into sequences to increase their similarity in the alignment. Accession numbers for each sequence in the alignment can be found in Supplementary Table S2.
Midpoint rooted Polar Trees based on Clustal Omega (Figs. S3, S4, S9 and S10), MAFTT (S5, S6, S11 and S12) or MUSCLE (S7, S8, S13 and S14) alignment of GFAP orthologs (Edgar, 2004a; Edgar, 2004b; Katoh and Standley, 2013; Sievers et al., 2011). GBlocks was used prior to alignments (Figs. S3, S5, S7, S9, S11, S13) (Castresana, 2000; Talavera and Castresana, 2007). Substitution rates are indicated by branch length variation. Bayesian inference posterior probabilities (Figs. S3 – S8) and Maximum likelihood (S9 – S14) bootstrap support values are shown for each branch (Felsenstein, 1981; Huelsenbeck et al., 2001). Accession numbers for each protein sequence in the trees can be found in Supplementary Table S2.
Wild-type (A) and XOPNTR (B) stage 50 tadpoles were treated for seven days with Mtz. Retinal sections were stained by in situ hybridization for vim expression and then bleached with hydrogen peroxide to clear RPE pigment. White dashed lines in both panels mark the boundary between RPE and photoreceptors. White asterisk in B labels vim expression in the subretinal space in response to rod ablation. Scale bar, 50 μm.
Maximum likelihood phylogeny of IFP sequences from GenBank (black), PCR survey clones (green) and unpublished transcriptomic data (red). Branch lengths represent substitutions per site. Bootstrap proportions (above the branch) and posterior probabilities (below the branch) are indicated only for those branches receiving significant support (i.e. bootstrap support > 70%, posterior probability > 0.95). Naming nomenclature in tree: Species name clone# Gene Symbol.
List of the six most highly similar reference proteins in Xenopus laevis (Xla) to the orthologs of human (Homo sapiens: Hs), mouse (Mus musculus: Mm), chicken (Gallus gallus: Gg) and zebrafish (Danio rerio: Dr) GFAP proteins and their known isoforms. Evalue was determined using blastp of each species isoform against NCBI X. laevis reference sequence. Highlighted in pink are X. laevis NCBI records for which the ortholog has not yet been assigned. X. laevis and X. tropicalis IFPs and their corresponding Unigene numbers are shown.
Table S2. Accession numbers for proteins of Figures 2 and Supplementary Figures S1–S14.
List of the six most highly similar reference proteins in Xenopus laevis (Xla) and Xenopus tropicalis (Xt) to the orthologs of human (Homo sapiens: Hs), mouse (Mus musculus: Mm), chicken (Gallus gallus: Gg), anole (Anolis carolinensis: Ac) and zebrafish (Danio rerio: Dr) GFAP proteins coded on exons 1, 5 and 6. E-value was determined using blastp of each species against NCBI X. laevis and X. tropicalis reference sequence. NCBI Gene ID numbers are shown.
NCBI and Xenbase accession numbers, position and assembly version used to generate syntenic schematic of Figure 3.
Table S5. Summary of Western blot results presented in Figures 4.
Table S6. Summary of the response of IFP expression to rod photoreceptor ablation and RGC axotomy presented in Figures 5, 6 and 7.
Table S7. Origin of biological samples used for PCR survey.
Table S8. Primer sequences.
Table S9. Annealing temperatures for GFAP amplification.
Table S10. Annealing temperatures for Vimentin amplification.
Table S11. Primary antibodies.
Table S12. Secondary antibodies.
Highlights.
GFAP-like immunoreactivity is upregulated in X. laevis Müller glia following retinal injury.
X. laevis and X. tropicalis lack the gene for gfap.
Antibodies commonly used to detect GFAP are non-specific.
Injury-dependent induction of vimentin and peripherin are observed in the X. leavis retina.
Anura species (frogs, toads) lack, while Caudata (salamanders, newts) and Gymnophiona (caecilians) amphibians have a gfap gene.
Acknowledgments
We would like to thank the following for providing plasmids and antibodies: Ben Szaro (Xla.Ina.pGEM4, Prph pAb 1 and 2) and Richard Harland (Xla.Nestin.pBSKS-). We also thank Karisa Rawlins for technical assistance and those who kindly provided the DNA and tissue samples for the phylogenetic and evolutionary analysis (see Table S7 for complete list). Research reported in this work was supported in part by the National Eye Institute of the National Institutes of Health under award numbers R01EY017964 and R01EY015748 (MEZ), Hendricks Bridge Grant Award (MEZ), Ministry of Economy and Competitiveness of Spain under awards RYC-2011-09321 CGL2012-40082 (DSM), a Research to Prevent Blindness unrestricted grant to the Upstate Medical University Department of Ophthalmology, and the Lions Club of Central New York.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Araki M. Regeneration of the amphibian retina: Role of tissue interaction and related signaling molecules on RPE transdifferentiation. Dev Growth Differ. 2007;49:109–120. doi: 10.1111/j.1440-169X.2007.00911.x. [DOI] [PubMed] [Google Scholar]
- Barbosa-Sabanero K, Hoffmann A, Judge C, Lightcap N, Tsonis PA, Del Rio-Tsonis K. Lens and retina regeneration: new perspectives from model organisms. Biochem J. 2012;447:321–334. doi: 10.1042/BJ20120813. [DOI] [PubMed] [Google Scholar]
- Bignami A, Dahl D. The radial glia of Müller in the rat retina and their response to injury. An immunofluorescence study with antibodies to the glial fibrillary acidic (GFA) protein. Exp Eye Res. 1979;28:63–69. doi: 10.1016/0014-4835(79)90106-4. [DOI] [PubMed] [Google Scholar]
- Bisbee CA, Baker MA, Wilson AC, Haji-Azimi I, Fischberg M. Albumin phylogeny for clawed frogs (Xenopus) Science. 1977;195:785–787. doi: 10.1126/science.65013. [DOI] [PubMed] [Google Scholar]
- Bordeaux J, Welsh A, Agarwal S, Killiam E, Baquero M, Hanna J, Anagnostou V, Rimm D. Antibody validation. Biotechniques. 2010;48:197–209. doi: 10.2144/000113382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann A, Iandiev I, Pannicke T, Wurm A, Hollborn M, Wiedemann P, Osborne NN, Reichenbach A. Cellular signaling and factors involved in Müller cell gliosis: neuroprotective and detrimental effects. Prog Retin Eye Res. 2009;28:423–451. doi: 10.1016/j.preteyeres.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Bringmann A, Pannicke T, Grosche J, Francke M, Wiedemann P, Skatchkov SN, Osborne NN, Reichenbach A. Müller cells in the healthy and diseased retina. Prog Retin Eye Res. 2006;25:397–424. doi: 10.1016/j.preteyeres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Bringmann A, Wiedemann P. Müller glial cells in retinal disease. Ophthalmologica. 2012;227:1–19. doi: 10.1159/000328979. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cerdà J, Conrad M, Markl J, Brand M, Herrmann H. Zebrafish vimentin: molecular characterization, assembly properties and developmental expression. Eur J Cell Biol. 1998;77:175–187. doi: 10.1016/S0171-9335(98)80105-2. [DOI] [PubMed] [Google Scholar]
- Chain FJ, Evans BJ. Multiple mechanisms promote the retained expression of gene duplicates in the tetraploid frog Xenopus laevis. PLoS Genet. 2006;2:e56. doi: 10.1371/journal.pgen.0020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoivanenko IS, Matveeva EA, Gelfand VI, Goldman RD, Minin AA. Mitochondrial membrane potential is regulated by vimentin intermediate filaments. FASEB J. 2015;29:820–827. doi: 10.1096/fj.14-259903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyatina AA, Guzenko D, Strelkov SV. Intermediate filament structure: the bottom-up approach. Curr Opin Cell Biol. 2015;32:65–72. doi: 10.1016/j.ceb.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Chiba C. The retinal pigment epithelium: An important player of retinal disorders and regeneration. Exp Eye Res. 2014;123:107–114. doi: 10.1016/j.exer.2013.07.009. [DOI] [PubMed] [Google Scholar]
- Choi RY, Engbretson GA, Solessio EC, Jones GA, Coughlin A, Aleksic I, Zuber ME. Cone degeneration following rod ablation in a reversible model of retinal degeneration. Invest Ophthalmol Vis Sci. 2011;52:364–373. doi: 10.1167/iovs.10-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson M, Nasisse M, Kornegay J. Intermediate filament complement of the normal and gliotic canine retina. J Comp Pathol. 1990;103:125–134. doi: 10.1016/s0021-9975(08)80169-7. [DOI] [PubMed] [Google Scholar]
- Debus E, Weber K, Osborn M. Monoclonal antibodies specific for glial fibrillary acidic (GFA) protein and for each of the neurofilament triplet polypeptides. Differentiation. 1983;25:193–203. doi: 10.1111/j.1432-0436.1984.tb01355.x. [DOI] [PubMed] [Google Scholar]
- Dent JA, Polson AG, Klymkowsky MW. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development. 1989;105:61–74. doi: 10.1242/dev.105.1.61. [DOI] [PubMed] [Google Scholar]
- Dräger UC, Edwards DL, Barnstable CJ. Antibodies against filamentous components in discrete cell types of the mouse retina. J Neurosci. 1984;4:2025–2042. doi: 10.1523/JNEUROSCI.04-08-02025.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004a;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004b;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson PA, Fisher SK, Guérin CJ, Anderson DH, Kaska DD. Glial fibrillary acidic protein increases in Müller cells after retinal detachment. Exp Eye Res. 1987;44:37–48. doi: 10.1016/s0014-4835(87)80023-4. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. A mitochondrial DNA phylogeny of African clawed frogs: phylogeography and implications for polyploid evolution. Mol Phylogenet Evol. 2004;33:197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Filoni S. Retina and lens regeneration in anuran amphibians. Semin Cell Dev Biol. 2009;20:528–534. doi: 10.1016/j.semcdb.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Omar G. Transitin, a nestin-related intermediate filament, is expressed by neural progenitors and can be induced in Müller glia in the chicken retina. J Comp Neurol. 2005;484:1–14. doi: 10.1002/cne.20406. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- Gervasi C, Stewart CB, Szaro BG. Xenopus laevis peripherin (XIF3) is expressed in radial glia and proliferating neural epithelial cells as well as in neurons. J Comp Neurol. 2000;423:512–531. [PubMed] [Google Scholar]
- Gervasi C, Thyagarajan A, Szaro BG. Increased expression of multiple neurofilament mRNAs during regeneration of vertebrate central nervous system axons. J Comp Neurol. 2003;461:262–275. doi: 10.1002/cne.10695. [DOI] [PubMed] [Google Scholar]
- Giménez Y Ribotta M, Langa F, Menet V, Privat A. Comparative anatomy of the cerebellar cortex in mice lacking vimentin, GFAP, and both vimentin and GFAP. Glia. 2000;31:69–83. doi: 10.1002/(sici)1098-1136(200007)31:1<69::aid-glia70>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Glasgow E, Druger RK, Levine EM, Fuchs C, Schechter N. Plasticin, a novel type III neurofilament protein from goldfish retina: increased expression during optic nerve regeneration. Neuron. 1992;9:373–381. doi: 10.1016/0896-6273(92)90175-d. [DOI] [PubMed] [Google Scholar]
- Godsave SF, Anderton BH, Wylie CC. The appearance and distribution of intermediate filament proteins during differentiation of the central nervous system, skin and notochord of Xenopus laevis. J Embryol Exp Morphol. 1986;97:201–223. [PubMed] [Google Scholar]
- Goldman RD, Khuon S, Chou YH, Opal P, Steinert PM. The function of intermediate filaments in cell shape and cytoskeletal integrity. J Cell Biol. 1996;134:971–983. doi: 10.1083/jcb.134.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–85. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosche J, Grimm D, Clemens N, Reichenbach A. Retinal light damage vs. normal aging of rats: altered morphology, intermediate filament expression, and nuclear organization of Müller (glial) cells. J Hirnforsch. 1997;38:459–470. [PubMed] [Google Scholar]
- Guo M, Ehrlicher AJ, Jensen MH, Renz M, Moore JR, Goldman RD, Lippincott-Schwartz J, Mackintosh FC, Weitz DA. Probing the stochastic, motor-driven properties of the cytoplasm using force spectrum microscopy. Cell. 2014;158:822–832. doi: 10.1016/j.cell.2014.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld M, Weber K. A synthetic peptide representing the consensus sequence motif at the carboxy-terminal end of the rod domain inhibits intermediate filament assembly and disassembles preformed filaments. J Cell Biol. 1992;116:157–166. doi: 10.1083/jcb.116.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand BT, Mendez MG, Pugh J, Delsert C, Goldman RD. A role for intermediate filaments in determining and maintaining the shape of nerve cells. Mol Biol Cell. 2003;14:5069–5081. doi: 10.1091/mbc.E03-06-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Khokha MK, Grammer TC, Harland RM, Richardson P, Rokhsar DS. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biol. 2007;5:31. doi: 10.1186/1741-7007-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Mann RW, Harland RM. A protein expressed in the growth cones of embryonic vertebrate neurons defines a new class of intermediate filament protein. Neuron. 1992;9:417–428. doi: 10.1016/0896-6273(92)90180-l. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Fouquet B, Franke WW. Expression of intermediate filament proteins during development of Xenopus laevis. I. cDNA clones encoding different forms of vimentin. Development. 1989a;105:279–298. doi: 10.1242/dev.105.2.279. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Fouquet B, Franke WW. Expression of intermediate filament proteins during development of Xenopus laevis. II. Identification and molecular characterization of desmin. Development. 1989b;105:299–307. doi: 10.1242/dev.105.2.299. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kinouchi R, Takeda M, Yang L, Wilhelmsson U, Lundkvist A, Pekny M, Chen DF. Robust neural integration from retinal transplants in mice deficient in GFAP and vimentin. Nat Neurosci. 2003;6:863–868. doi: 10.1038/nn1088. [DOI] [PubMed] [Google Scholar]
- Klymkowsky MW, Maynell LA, Polson AG. Polar asymmetry in the organization of the cortical cytokeratin system of Xenopus laevis oocytes and embryos. Development. 1987;100:543–557. doi: 10.1242/dev.100.3.543. [DOI] [PubMed] [Google Scholar]
- Kohno H, Sakai T, Kitahara K. Induction of nestin, Ki-67, and cyclin D1 expression in Müller cells after laser injury in adult rat retina. Graefes Arch Clin Exp Ophthalmol. 2006;244:90–95. doi: 10.1007/s00417-005-0030-7. [DOI] [PubMed] [Google Scholar]
- Kornreich M, Avinery R, Malka-Gibor E, Laser-Azogui A, Beck R. Order and disorder in intermediate filament proteins. FEBS Lett. 2015;589:2464–2476. doi: 10.1016/j.febslet.2015.07.024. [DOI] [PubMed] [Google Scholar]
- Lenkowski JR, Raymond PA. Müller glia: Stem cells for generation and regeneration of retinal neurons in teleost fish. Prog Retin Eye Res. 2014;40:94–123. doi: 10.1016/j.preteyeres.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GP, Erickson PA, Guérin CJ, Anderson DH, Fisher SK. Changes in the expression of specific Müller cell proteins during long-term retinal detachment. Exp Eye Res. 1989;49:93–111. doi: 10.1016/0014-4835(89)90079-1. [DOI] [PubMed] [Google Scholar]
- Lewis GP, Fisher SK. Up-regulation of glial fibrillary acidic protein in response to retinal injury: its potential role in glial remodeling and a comparison to vimentin expression. Int Rev Cytol. 2003;230:263–290. doi: 10.1016/s0074-7696(03)30005-1. [DOI] [PubMed] [Google Scholar]
- Liedtke W, Edelmann W, Bieri PL, Chiu FC, Cowan NJ, Kucherlapati R, Raine CS. GFAP is necessary for the integrity of CNS white matter architecture and long-term maintenance of myelination. Neuron. 1996;17:607–615. doi: 10.1016/s0896-6273(00)80194-4. [DOI] [PubMed] [Google Scholar]
- Lowery J, Kuczmarski ER, Herrmann H, Goldman RD. Intermediate Filaments Play a Pivotal Role in Regulating Cell Architecture and Function. J Biol Chem. 2015;290:17145–17153. doi: 10.1074/jbc.R115.640359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YB, Iandiev I, Hollborn M, Korber N, Ulbricht E, Hirrlinger PG, Pannicke T, Wei EQ, Bringmann A, Wolburg H, Wilhelmsson U, Pekny M, Wiedemann P, Reichenbach A, Kas JA. Reactive glial cells: increased stiffness correlates with increased intermediate filament expression. FASEB J. 2011;25:624–631. doi: 10.1096/fj.10-163790. [DOI] [PubMed] [Google Scholar]
- Luna G, Lewis GP, Banna CD, Skalli O, Fisher SK. Expression profiles of nestin and synemin in reactive astrocytes and Muller cells following retinal injury: a comparison with glial fibrillar acidic protein and vimentin. Mol Vis. 2010;16:2511–2523. [PMC free article] [PubMed] [Google Scholar]
- Lundkvist A, Reichenbach A, Betsholtz C, Carmeliet P, Wolburg H, Pekny M. Under stress, the absence of intermediate filaments from Müller cells in the retina has structural and functional consequences. J Cell Sci. 2004;117:3481–3488. doi: 10.1242/jcs.01221. [DOI] [PubMed] [Google Scholar]
- MacDonald RB, Randlett O, Oswald J, Yoshimatsu T, Franze K, Harris WA. Müller glia provide essential tensile strength to the developing retina. J Cell Biol. 2015;210:1075–1083. doi: 10.1083/jcb.201503115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-De Luna RI, Kelly LE, El-Hodiri HM. The Retinal Homeobox (Rx) gene is necessary for retinal regeneration. Dev Biol. 2011;353:10–18. doi: 10.1016/j.ydbio.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-De Luna RI, Ku RY, Lyou Y, Zuber ME. Maturin is a novel protein required for differentiation during primary neurogenesis. Dev Biol. 2013;384:26–40. doi: 10.1016/j.ydbio.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-De Luna RI, Zuber ME. Putting regeneration into regenerative medicine. J Ophthalmic Vis Res. 2014;9:126–133. [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Kondoh H. Dkk1-dependent inhibition of Wnt signaling activates Hesx1 expression through its 5′ enhancer and directs forebrain precursor development. Genes Cells. 2014 doi: 10.1111/gtc.12136. [DOI] [PubMed] [Google Scholar]
- Matveeva EA, Venkova LS, Chernoivanenko IS, Minin AA. Vimentin is involved in regulation of mitochondrial motility and membrane potential by Rac1. Biol Open. 2015;4:1290–1297. doi: 10.1242/bio.011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall MA, Gregg RG, Behringer RR, Brenner M, Delaney CL, Galbreath EJ, Zhang CL, Pearce RA, Chiu SY, Messing A. Targeted deletion in astrocyte intermediate filament (Gfap) alters neuronal physiology. Proc Natl Acad Sci U S A. 1996;93:6361–6366. doi: 10.1073/pnas.93.13.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLenachan S, Goldshmit Y, Fowler KJ, Voullaire L, Holloway TP, Turnley AM, Ioannou PA, Sarsero JP. Transgenic mice expressing the Peripherin-EGFP genomic reporter display intrinsic peripheral nervous system fluorescence. Transgenic Res. 2008;17:1103–1116. doi: 10.1007/s11248-008-9210-7. [DOI] [PubMed] [Google Scholar]
- Menet V, Gimenez y Ribotta M, Chauvet N, Drian MJ, Lannoy J, Colucci-Guyon E, Privat A. Inactivation of the glial fibrillary acidic protein gene, but not that of vimentin, improves neuronal survival and neurite growth by modifying adhesion molecule expression. J Neurosci. 2001;21:6147–6158. doi: 10.1523/JNEUROSCI.21-16-06147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet V, Prieto M, Privat A, Giménez y Ribotta M. Axonal plasticity and functional recovery after spinal cord injury in mice deficient in both glial fibrillary acidic protein and vimentin genes. Proc Natl Acad Sci U S A. 2003;100:8999–9004. doi: 10.1073/pnas.1533187100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenger NJ, Warner AE. The appearance of neural and glial cell markers during early development of the nervous system in the amphibian embryo. Development. 1989;107:43–54. doi: 10.1242/dev.107.1.43. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees in Proceedings of the Gateway Computing Environments Workshop (GCE). 2011 TeraGrid Conference: Extreme Digital Discovery; 2010. pp. 1–8. [Google Scholar]
- Mitashov VI. Retinal regeneration in amphibians. Int J Dev Biol. 1997;41:893–905. [PubMed] [Google Scholar]
- Nakazawa T, Takeda M, Lewis GP, Cho KS, Jiao J, Wilhelmsson U, Fisher SK, Pekny M, Chen DF, Miller JW. Attenuated glial reactions and photoreceptor degeneration after retinal detachment in mice deficient in glial fibrillary acidic protein and vimentin. Invest Ophthalmol Vis Sci. 2007;48:2760–2768. doi: 10.1167/iovs.06-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus Laevis (Daudin): A Systematical & Chronological Survey of the Development from the Fertilized Egg till the End of the Fertilized Egg Till the End of Metamorp) Garland Science; New York: 1994. [Google Scholar]
- Okada M, Matsumura M, Ogino N, Honda Y. Müller cells in detached human retina express glial fibrillary acidic protein and vimentin. Graefes Arch Clin Exp Ophthalmol. 1990;228:467–474. doi: 10.1007/BF00927264. [DOI] [PubMed] [Google Scholar]
- Pekny M, Leveen P, Pekna M, Eliasson C, Berthold CH, Westermark B, Betsholtz C. Mice lacking glial fibrillary acidic protein display astrocytes devoid of intermediate filaments but develop and reproduce normally. EMBO J. 1995;14:1590–1598. doi: 10.1002/j.1460-2075.1995.tb07147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves JH. Heterogeneity in the substitution process of amino acid sites of proteins coded for by mitochondrial DNA. J Mol Evol. 1992;35:17–31. doi: 10.1007/BF00160257. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi DS, Moeller JF, Coffman CR, Gallenson N, Harris WA. Growth cone interactions with a glial cell line from embryonic Xenopus retina. Dev Biol. 1989;134:158–174. doi: 10.1016/0012-1606(89)90086-9. [DOI] [PubMed] [Google Scholar]
- San Mauro D. A multilocus timescale for the origin of extant amphibians. Mol Phylogenet Evol. 2010;56:554–561. doi: 10.1016/j.ympev.2010.04.019. [DOI] [PubMed] [Google Scholar]
- San Mauro D. Amphibians: land conquest by vertebrates. In: Vargas P, Zardoya R, editors. The tree of life: evolution and classification of living organisms. Sinauer Associates, Inc; Sunderland, MA: 2014. pp. 494–501. [Google Scholar]
- San Mauro D, Agorreta A. Molecular systematics: A synthesis of the common methods and the state of knowledge. Cell Mol Biol Lett. 2010;15:311–341. doi: 10.2478/s11658-010-0010-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper CB. A guide to the perplexed on the specificity of antibodies. J Histochem Cytochem. 2009;57:1–5. doi: 10.1369/jhc.2008.952770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi CS, Lewis GP, Fisher SK, Leitner WP, Mann DL, Luthert PJ, Charteris DG. Glial remodeling and neural plasticity in human retinal detachment with proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2005;46:329–342. doi: 10.1167/iovs.03-0518. [DOI] [PubMed] [Google Scholar]
- Sharpe CR. Developmental expression of a neurofilament-M and two vimentin-like genes in Xenopus laevis. Development. 1988;103:269–277. doi: 10.1242/dev.103.2.269. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Blagojevic FN, Antonopoulos DC. Exploring new search algorithms and hardware for phylogenetics: RAxML meets the IBM Cell. The Journal of VLSI Signal Processing. 2007;48:271–286. [Google Scholar]
- Strelkov SV, Herrmann H, Geisler N, Wedig T, Zimbelmann R, Aebi U, Burkhard P. Conserved segments 1A and 2B of the intermediate filament dimer: their atomic structures and role in filament assembly. EMBO J. 2002;21:1255–1266. doi: 10.1093/emboj/21.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaro BG, Gainer H. Immunocytochemical identification of non-neuronal intermediate filament proteins in the developing Xenopus laevis nervous system. Brain Res. 1988;471:207–224. doi: 10.1016/0165-3806(88)90100-9. [DOI] [PubMed] [Google Scholar]
- Szeverenyi I, Cassidy AJ, Chung CW, Lee BT, Common JE, Ogg SC, Chen H, Sim SY, Goh WL, Ng KW, Simpson JA, Chee LL, Eng GH, Li B, Lunny DP, Chuon D, Venkatesh A, Khoo KH, McLean WH, Lim YP, Lane EB. The Human Intermediate Filament Database: comprehensive information on a gene family involved in many human diseases. Hum Mutat. 2008;29:351–360. doi: 10.1002/humu.20652. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verardo MR, Lewis GP, Takeda M, Linberg KA, Byun J, Luna G, Wilhelmsson U, Pekny M, Chen DF, Fisher SK. Abnormal reactivity of muller cells after retinal detachment in mice deficient in GFAP and vimentin. Invest Ophthalmol Vis Sci. 2008;49:3659–3665. doi: 10.1167/iovs.07-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara MN, Del Rio-Tsonis K. Retinal regeneration in the Xenopus laevis tadpole: a new model system. Mol Vis. 2009;15:1000–1013. [PMC free article] [PubMed] [Google Scholar]
- Viczian AS, Bang AG, Harris WA, Zuber ME. Expression of Xenopus laevis Lhx2 during eye development and evidence for divergent expression among vertebrates. Dev Dyn. 2006;235:1133–1141. doi: 10.1002/dvdy.20708. [DOI] [PubMed] [Google Scholar]
- Viczian AS, Vignali R, Zuber ME, Barsacchi G, Harris WA. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development. 2003;130:1281–1294. doi: 10.1242/dev.00343. [DOI] [PubMed] [Google Scholar]
- Viczian AS, Zuber ME. A simple behavioral assay for testing visual function in Xenopus laevis. J Vis Exp. 2014 doi: 10.3791/51726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Kunzevitzky NJ, Dugas JC, Cameron M, Barres BA, Goldberg JL. Disease gene candidates revealed by expression profiling of retinal ganglion cell development. J Neurosci. 2007;27:8593–8603. doi: 10.1523/JNEUROSCI.4488-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Messing A, David S. Axonal and nonneuronal cell responses to spinal cord injury in mice lacking glial fibrillary acidic protein. Exp Neurol. 1997;148:568–576. doi: 10.1006/exnr.1997.6702. [DOI] [PubMed] [Google Scholar]
- Wiens JJ. Global patterns of diversification and species richness in amphibians. Am Nat. 2007;170(Suppl 2):S86–106. doi: 10.1086/519396. [DOI] [PubMed] [Google Scholar]
- Wilhelmsson U, Li L, Pekna M, Berthold CH, Blom S, Eliasson C, Renner O, Bushong E, Ellisman M, Morgan TE, Pekny M. Absence of glial fibrillary acidic protein and vimentin prevents hypertrophy of astrocytic processes and improves post-traumatic regeneration. J Neurosci. 2004;24:5016–5021. doi: 10.1523/JNEUROSCI.0820-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KA, Trembley M, Abd Wahab S, Viczian AS. Efficient retina formation requires suppression of both Activin and BMP signaling pathways in pluripotent cells. Biol Open. 2015;4:573–583. doi: 10.1242/bio.20149977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Ding P, Xiao L, Hu M, Hu Z. Nestin, a new marker, expressed in Müller cells following retinal injury. Can J Neurol Sci. 2010;37:643–649. doi: 10.1017/s0317167100010830. [DOI] [PubMed] [Google Scholar]
- Xue LP, Lu J, Cao Q, Kaur C, Ling EA. Nestin expression in Müller glial cells in postnatal rat retina and its upregulation following optic nerve transection. Neuroscience. 2006;143:117–127. doi: 10.1016/j.neuroscience.2006.07.044. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Szaro BG. Xefiltin, a new low molecular weight neuronal intermediate filament protein of Xenopus laevis, shares sequence features with goldfish gefiltin and mammalian alpha-internexin and differs in expression from XNIF and NF-L. J Comp Neurol. 1997a;377:351–364. doi: 10.1002/(sici)1096-9861(19970120)377:3<351::aid-cne4>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Szaro BG. Xefiltin, a Xenopus laevis neuronal intermediate filament protein, is expressed in actively growing optic axons during development and regeneration. J Neurobiol. 1997b;33:811–824. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A consensus sequence (GFAP Consensus, Fig. 2A) was generated by aligning intermediate filament proteins GFAP, vimentin, desmin, peripherin and internexin neuronal intermediate filament protein, alpha. IFPs (and known isoforms) from human (Hs), mouse (Mm), chicken (Gg), green anole (Ac), X. laevis (Xla), X. tropicalis (Xt) and zebrafish (Dr) were aligned using the Clustal W algorithm (Thompson et al., 1994). For simplicity, only regions corresponding to residues 76 – 374 of human GFAP Isoform 1 (NP_002046) are shown. Residues identical to the GFAP Consensus are shaded black. Dashes represent gaps that have been inserted into sequences to increase their similarity in the alignment. Accession numbers for each sequence in the alignment can be found in Supplementary Table S2.
Clustal W alignment of GFAP orthologs from human, mouse, chicken, green anole, and zebrafish and the 17 most similar, yet untitled X. laevis and X. tropicalis sequences. For simplicity, only regions corresponding to residues 76 – 374 of human GFAP Isoform 1 (NP_002046) are shown. Residues identical to the GFAP consensus are shaded black. Dashes represent gaps that have been inserted into sequences to increase their similarity in the alignment. Accession numbers for each sequence in the alignment can be found in Supplementary Table S2.
Midpoint rooted Polar Trees based on Clustal Omega (Figs. S3, S4, S9 and S10), MAFTT (S5, S6, S11 and S12) or MUSCLE (S7, S8, S13 and S14) alignment of GFAP orthologs (Edgar, 2004a; Edgar, 2004b; Katoh and Standley, 2013; Sievers et al., 2011). GBlocks was used prior to alignments (Figs. S3, S5, S7, S9, S11, S13) (Castresana, 2000; Talavera and Castresana, 2007). Substitution rates are indicated by branch length variation. Bayesian inference posterior probabilities (Figs. S3 – S8) and Maximum likelihood (S9 – S14) bootstrap support values are shown for each branch (Felsenstein, 1981; Huelsenbeck et al., 2001). Accession numbers for each protein sequence in the trees can be found in Supplementary Table S2.
Wild-type (A) and XOPNTR (B) stage 50 tadpoles were treated for seven days with Mtz. Retinal sections were stained by in situ hybridization for vim expression and then bleached with hydrogen peroxide to clear RPE pigment. White dashed lines in both panels mark the boundary between RPE and photoreceptors. White asterisk in B labels vim expression in the subretinal space in response to rod ablation. Scale bar, 50 μm.
Maximum likelihood phylogeny of IFP sequences from GenBank (black), PCR survey clones (green) and unpublished transcriptomic data (red). Branch lengths represent substitutions per site. Bootstrap proportions (above the branch) and posterior probabilities (below the branch) are indicated only for those branches receiving significant support (i.e. bootstrap support > 70%, posterior probability > 0.95). Naming nomenclature in tree: Species name clone# Gene Symbol.
List of the six most highly similar reference proteins in Xenopus laevis (Xla) to the orthologs of human (Homo sapiens: Hs), mouse (Mus musculus: Mm), chicken (Gallus gallus: Gg) and zebrafish (Danio rerio: Dr) GFAP proteins and their known isoforms. Evalue was determined using blastp of each species isoform against NCBI X. laevis reference sequence. Highlighted in pink are X. laevis NCBI records for which the ortholog has not yet been assigned. X. laevis and X. tropicalis IFPs and their corresponding Unigene numbers are shown.
Table S2. Accession numbers for proteins of Figures 2 and Supplementary Figures S1–S14.
List of the six most highly similar reference proteins in Xenopus laevis (Xla) and Xenopus tropicalis (Xt) to the orthologs of human (Homo sapiens: Hs), mouse (Mus musculus: Mm), chicken (Gallus gallus: Gg), anole (Anolis carolinensis: Ac) and zebrafish (Danio rerio: Dr) GFAP proteins coded on exons 1, 5 and 6. E-value was determined using blastp of each species against NCBI X. laevis and X. tropicalis reference sequence. NCBI Gene ID numbers are shown.
NCBI and Xenbase accession numbers, position and assembly version used to generate syntenic schematic of Figure 3.
Table S5. Summary of Western blot results presented in Figures 4.
Table S6. Summary of the response of IFP expression to rod photoreceptor ablation and RGC axotomy presented in Figures 5, 6 and 7.
Table S7. Origin of biological samples used for PCR survey.
Table S8. Primer sequences.
Table S9. Annealing temperatures for GFAP amplification.
Table S10. Annealing temperatures for Vimentin amplification.
Table S11. Primary antibodies.
Table S12. Secondary antibodies.