Abstract
Nanoscale distance measurements by pulse dipolar Electron paramagnetic resonance (EPR) spectroscopy allow new insights into the structure and dynamics of complex biopolymers. EPR detection requires site directed spin labeling (SDSL) of biomolecule(s), which remained challenging for long RNAs up-to-date. Here, we demonstrate that novel complementary-addressed SDSL approach allows efficient spin labeling and following structural EPR studies of long RNAs. We succeeded to spin-label Hepatitis C Virus RNA internal ribosome entry site consisting of ≈330 nucleotides and having a complicated spatial structure. Application of pulsed double electron–electron resonance provided spin–spin distance distribution, which agrees well with the results of molecular dynamics (MD) calculations. Thus, novel SDSL approach in conjunction with EPR and MD allows structural studies of long natural RNAs with nanometer resolution and can be applied to systems of biological and biomedical significance.
INTRODUCTION
Electron paramagnetic resonance (EPR) spectroscopy is actively used in studies of structure, dynamics and conformational changes of nucleic acids and their complexes (1–28). In this regard, RNAs evoke enormous interest of researchers because these biopolymers are extremely structurally dynamic macromolecules able to generate a wide set of conformations (29) and to form a variety of complexes with proteins (30). Site directed spin labeling (SDSL) of nucleic acids is an integral part of structural studies exploiting EPR spectroscopy, because the specific attachment of a spin label exactly to the target site allows obtaining unambiguous structural information. To date, a large number of different methods for spin labeling of RNA has been described. Three main approaches underlie all the methods available for SDSL, namely spin labeling during oligonucleotide synthesis, post-synthetic labeling and non-covalent labeling (10,31). Moreover, for SDSL of long RNA, spin labeling during oligonucleotide synthesis can be combined with enzymatic ligation utilizing DNA splints to bring together 3′- and 5′-termini of RNA fragments and merge them (21,22). This method suggested more than two decades ago (32) and widely used for RNA labeling critically requires homogeneous 3′-ends of RNAs, which are obtained, as a rule, by T7 transcription. The main disadvantage of the method is generally low yield of ligation, especially for large RNAs, which is likely caused by low extent of formation of correct duplexes between the RNA fragments and the DNA splint (33). Thus, elaboration of approaches for effective introduction of spin labels in desired locations of long natural structured RNAs remains an attractive task.
Recently, a versatile approach to SDSL of RNA has been proposed and implemented in model 10-mer RNAs (34). The key step of this approach is site-directed alkylation of RNA using a specially designed 4-[N-(2-chloroethyl)-N-methylamino]benzylphosphoramide derivative of oligodeoxyribonucleotide, which must be complementary to the only sequence adjacent to the target site. The following steps include the release of aliphatic amino group by hydrolysis of phosphoramide bond in covalent adduct formed and, finally, the selective coupling of spin label to this amino group via acylation with the respective derivative of N-hydroxysuccinimide ester. Doubly spin-labeled model RNA duplexes prepared using the above-mentioned complementary-addressed SDSL approach were investigated using Q-band (34 GHz) double electron–electron resonance (PELDOR (35), also termed DEER), and the obtained distance distributions corresponded well to the expected values. Despite the success of that work and anticipations that new approach can as well be used for RNAs of any size, its applicability to long RNAs was not yet experimentally evidenced.
In this work, we demonstrate for the first time SDSL of long RNA with intricate folded tertiary structure using this novel complementary-addressed approach. For this purpose, we selected the Hepatitis C Virus (HCV) genomic RNA internal ribosome entry site (IRES), whose structure has been well studied previously (36). This RNA consists of up to 350 nucleotides and has a complicated spatial structure. It is thus much longer and more complex than any RNA that was spin labeled prior to this work. Exploiting the proposed SDSL approach, we obtained HCV RNA IRES derivative containing pair of spin labels at definite locations in its domain II. As will be shown below, distance distribution obtained using Q-band DEER clearly demonstrates the applicability of novel SDSL approach to long RNAs, thus opening new possibilities for application of EPR spectroscopy in structural studies of RNAs existing in living organisms.
MATERIALS AND METHODS
Spin label
The spin label 3-Carboxy-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-1-oxyl succinimidyl ester (NHS-M) was prepared according to the procedure described in Ref. (37) and kindly provided by Dr Igor A. Kirilyuk.
Oligonucleotides and their alkylating derivatives
Oligodeoxyribonucleotides 5′-pTAGACGCTTTCTGCGTGAAGA-3′ (DNA1), 5′-pTCTGCGTGAAGACAGTAG-3′ (DNA2) and 5′-CACTCAATACTAACGCCATG-3′ (helper) were synthesized by the amidophosphite method and purified by high performance liquid chromatography (HPLC) in the Laboratory of medicinal chemistry at ICBFM SB RAS. 4-[N-(2-Chloroethyl)-N-methylamino]benzylphosphoramide derivatives of oligodeoxyribonucleotides (DNA-NHCH2RCl) were synthesized and purified as described in Ref. (38).
Preparation of HCV RNA IRES
The fragment corresponding to HCV RNA nucleotides 40–372 (HCV RNA IRES) was obtained by in vitro transcription of DNA generated by polymerase chain reaction using plasmid pXL40-372.NS (39) containing the sequence corresponding to the 5′-UTR of the HCV genome RNA as template. RNA transcript was synthesized by incubation of 2 μg of DNA template in 200 μl of reaction mixture containing 200 mM HEPES-KOH (pH 7.5), 30 mM MgCl2, 300 mM trimethylamine N-oxide, 2 mM spermidine, 40 mM 1,4-dithiothreitol, 6 mM nucleoside triphosphates, 500 U of T7 RNA polymerase and 0.2 U of inorganic pyrophosphatase (Sigma) at 37°C for 3 h. The DNA template was then degraded by incubation of the reaction mixture with 1 U of DNase (Ambion) at 37°C for 30 min and RNA was purified by gel-filtration on a Sephadex G-75 column.
Site-specific modification of HCV RNA IRES and identification of the alkylated nucleotides
A solution of HCV RNA IRES (5 nmol) in 400 μl of 20 mM Tris–HCl buffer, pH 7.5 containing 200 mM KCl, 20 mM MgCl2 and 0.5 mM ethylenediaminetetraacetic acid (EDTA) (buffer A) was incubated with alkylating derivative of oligodeoxyribonucleotide (25 nmol) in the presence of helper DNA oligomer (25 nmol) for 18 h at 25°C. After the incubation, the reaction mixture was supplemented by 2.5 volumes of ice-cold ethanol and centrifuged at 14 000 g for 15 min at 4°C. The pellet was dissolved in formamide containing 0.1% bromophenol and 0.1% xylenecyanol and subjected to 8% polyacrylamide gel electrophoresis (PAGE) in the presence of 10% formamide. The RNA bands in the gel were visualized by UV-shadowing and the band corresponding to covalent adduct formed as result of the alkylation (migrated slower than unmodified RNA) was excised. The covalent adduct was eluted from the gel by incubation with 400 μl of 300 mM NaOAc buffer, pH 4.5 containing 0.5% sodium dodecyl sulphate (SDS) and 1 mM EDTA at 25°C for 18 h; the eluate was then separated from the gel, and the covalent adduct was ethanol precipitated as described above. The obtained sample was dissolved in 100 μl of bidistilled water and optical density of the solution at A260 nm was measured. The amount of covalent adduct recovered from the gel was from 1 up to 1.3 nmol. The extent of the alkylation, whose typical values will be discussed below in Table 1, was determined in a separate experiment in 500-fold reduced scale. To do this, the gel after separation of covalent adduct from unmodified RNA was stained by Toluidine O and scanned on a Molecular Imager Pro FX (BioRad, USA) with subsequent quantification of stained RNA bands using the Software QuantityOne.
Table 1. Modification of HCV RNA IRES by alkylating derivatives of complementary oligodeoxyribonucleotides.
| DNA derivative | Relative modification extent | Cross-linking site in HCV IRES | |
|---|---|---|---|
| with DNA helper | |||
| DNA1-NHCH2RCl | 0.1 | 0.5 | C83 (N3) |
| DNA2-NHCH2RCl | 0.2 | 0.6 | A73 (N1) |
Sequence of DNA derivative and complementary region in HCV IRES are pTAGACGCTTTCTGCGTGAAGA (61–81) for DNA1-NHCH2RCl and pTCTGCGTGAAGACAGTAG (55-72) for DNA2-NHCH2RCl.
Then the covalent adduct was additionally purified by gel-filtration on Sephadex G-75 in bidistilled water, and the ethanol precipitation in the presence of 300 mM NaOAc, pH 4.5 was carried out again. The obtained sample was dissolved in 50 μl of 300 mM NaOAc buffer, pH 4.1 and hydrolysis of phosphoramide bond in the covalent adduct was performed by incubation of the solution at 50°C for 6 h. The alkylated RNA together with the released oligonucleotide was then ethanol precipitated, following the addition of NP-40 detergent up to 0.2% to prevent sorption of the alkylated RNA on tube walls. The pellet was dissolved in 6 M Urea and the modified RNA was separated from the oligonucleotide by RNA Clean Up Kit -5 (Zymo Research) with the subsequent ethanol precipitation as described above. The final sample was dissolved in water and stored at −20°C. Final yield of the alkylated RNA was about 60–70% of the starting amount of the covalent adduct.
To introduce a second amino linker at another nucleotide position, the singly-alkylated HCV RNA IRES (0.9 nmol) was incubated with another alkylating DNA derivative (4.5 nmol) in 70–100 μl of buffer A and exposed to all procedures described in this section previously in the same conditions. The yield of the RNA sample containing amino linkers at two defined locations was similar to that of the singly-alkylated RNA.
Nucleotides of the HCV RNA IRES cross-linked to the oligodeoxyribonucleotides derivatives were determined by application of primer extension as described in Ref. (40) using AMV reverse transcriptase (Invitrogen). As a primer, oligodeoxyribonucleotide complementary to HCV RNA IRES sequence 103–120 was used. As the reaction was completed, the samples were treated and analyzed as described in Ref. (41).
Attachment of spin labels to alkylated HCV RNA IRES
A total of 0.2 nmol of doubly-alkylated HCV RNA IRES was dissolved in 22 μl of 50 mM Hepes-KOH (pH 8.5) and mixed with an equal volume of 50 mM NHS-M in dimethyl sulfoxide; the reaction mixture was then incubated at ambient temperature for 2 h. The spin-labeled RNA was precipitated by adding 2.5-3 volumes of ice-cold ethanol in the presence of 300 mM NaOAc pH 4.5 and centrifuged as described in the previous section; the supernatant was thoroughly removed and the pellet was rinsed with 200 μl of ice-cold 70% ethanol, dried and dissolved in 50 μl of H2O. Ethanol precipitation was repeated once again; the final pellet was dissolved in deuterium oxide (D2O) up to a concentration of 4 × 10−5 M and then reactivated by heating to 90°C with subsequent cooling to room temperature. An aliquot of the sample was analyzed by 8% denaturing PAGE. For EPR experiments, the RNA sample was transferred into buffer of 20 mM Tris–HCl pH 7.5 containing 100 mM KCl and 2.5 mM MgCl2 prepared in D2O. Glycerol-d8 was added to the solution up to 40–50% concentration for EPR analysis. The concentration of HCV RNA IRES molecules in the sample used for the analysis was approximately 25 μM, which was estimated by the measurement of the optical density of the solution at λ = 260 nm (OD260) before Glycerol-d8 addition, accepting that 1 OD260 Unit corresponds to 300 pmol of the RNA and considering the subsequent dilution with the buffer and glycerol.
Functional assay of doubly spin-labeled HCV RNA IRES
dslRNA was labeled with 7.5 μCi [5′-32P]pCp using T4 RNA ligase according to (42). HCV RNA IRES, which preliminary passed the same processing steps as dslRNA but without reagents used for dslRNA preparation, was labeled in the similar way and was then applied as control RNA. 48S pre-initiation complex with 28 pmol of either dslRNA or control RNA was obtained in 100 μl of rabbit reticulocyte lysate (RRL) in the presence of 2 mM GMPPNP according to (43) and isolated by ultracentrifugation in 15–30% sucrose density gradient (rotor SW40, 75 000 g, 4°C, 18 h). After centrifugation, gradient fractions were collected and the optical density at λ = 260 nm and the radioactivity of each fraction were measured. The fractions containing radioactivity were combined and ethanol-precipitated. The pellet was resuspended in 0.1% SDS containing 1 mM EDTA, incubated at 37°C for 10 min and phenol deproteinated. Resulting total RNA was dissolved in 10 μl of H2O, and ester bond in aminoacyl-tRNA was hydrolyzed by incubation in a buffer containing 150 mM NaOAc, pH 5.5 and 10 mM CuSO4 at 30°C for 25 min. Then, RNA was 3′-end-post-labeled with 7.5 μCi of [5′-32P] pCp using T4 RNA ligase according to (42) and resolved by 10% denaturing PAGE as described above. The gel was dried and autoradiographed on a phosphoroimager screen.
EPR experiments
Samples were placed in glass capillary tubes (OD 1.5 mm, ID 0.9 mm, with the sample volume being ca. 10 μl). Continuous wave (CW) EPR experiments were carried out at X-band (9 GHz) at 300 K using a commercial Bruker EMX spectrometer. The experimental spectra were simulated using EasySpin (44,45).
For DEER measurements samples were shock-frozen in liquid nitrogen and investigated at T = 50 K. The data were collected at the Q-band (34 GHz) using a Bruker Elexsys E580 pulse/cw EPR spectrometer equipped with 150 W TWT amplifier, EN5107D2 resonator and Oxford Instruments temperature control system. A standard four-pulse DEER sequence (46) was used with pulse lengths of 12 ns (both π and π/2) for probe (νprobe) and 24 ns for pump (νpump) frequency. The measurements were done at field position of ∼2.2 mT higher than the maximum of the spectrum, thus using Δν = (νpump–νprobe) = 65 MHz led to the pump pulse applied at the spectral maximum. All obtained DEER traces were background corrected by exponential function (3D homogeneous background) and analyzed with Tikhonov regularization using DeerAnalysis program (47).
Molecular dynamics simulations
For MD calculations, the structure of the HCV IRES domain II fragment C69–G98 (PDB ID: 1P5N (48)) has been chosen. To stabilize base-pairing of the 5′- and 3′-terminal nucleotides in the structure, two additional terminal GC-pairs were added. The simulations were performed using the Amber 12 software package (49). The study was carried out using the ff12SB force field in explicit solvent (TIP3P water, cuboid periodic box, edge 12 Å). The SHAKE algorithm for hydrogen involving bonds was applied, allowing a 2 fs time step to be set. Long range electrostatics was calculated using particle mesh ewald, with a 1 Å grid. Molecular dynamics (MD) trajectories were generated in the isothermal–isobaric (NPT) ensemble. Pressure of the system was maintained at 1 bar and an Anderson (strong collision) temperature regulation scheme was used. Non-bonded cutoff of 10 Å was applied. A harmonic potential for heavy atoms (1 kcal/mol/Å2) was applied in MD simulations for the nucleotides G67-G71, G75-A85, C88-U91 and U94-C100 to prevent changes in the unmodified part of RNA. MD productive trajectory simulation contained 10 series of 100 ns trajectories with different initial atoms’ speed. The analysis of the MD trajectories was performed using the cpptraj program. Structures and library files of spin-labeled nucleotides were prepared using AmberTools 12. Structure of nucleotides was optimized and atom charges were calculated using the Hartree−Fock method and the 6–31G* basis set in Gaussian’09. The particular atoms charges for MD libraries were calculated using the RESP method in antechamber. More details of the simulation procedures are given in Supplementary Data.
RESULTS AND DISCUSSION
Site-directed introduction of amino linkers into HCV RNA IRES
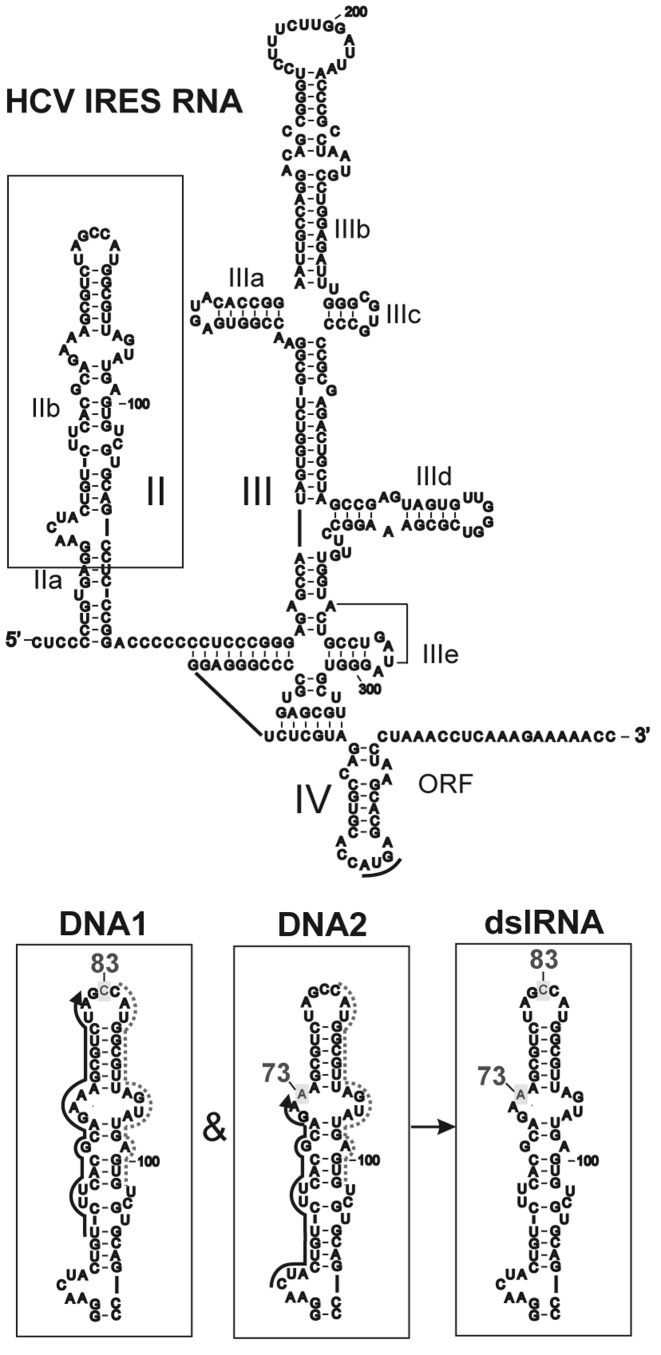
The RNA strand of 341 nt length containing sequence corresponding to the HCV RNA IRES was synthesized in vitro by T7 transcription. The secondary structure of the HCV IRES comprises three independently folded domains (II, III and IV) (50) (Figure 1), and the domain II was chosen as a region for SDSL. Two sites in this region, namely the apex loop and a strand of the internal loop of subdomain IIb, were targeted for site specific alkylation of the HCV RNA IRES with 4-[N-(2-chloroethyl)-N-methylamino]benzyl-5′-phosphoramide derivatives of oligodeoxyribonucleotides (DNA) (Figure 2). The corresponding DNA derivatives were complementary to the HCV RNA IRES sequences 61–81 (DNA1-NHCH2RCl) or 55–72 (DNA2-NHCH2RCl) (Figure 1). Alkylation of the HCV RNA IRES with DNA derivatives was carried out at 25°C long enough for practically complete conversion of 2-chloroethylamino group into active intermediate—ethylenimmonium cation (51). This resulted in formation of covalent adducts of the RNA with the respective DNA derivatives, which could be easily separated from unmodified RNA and unattached DNA derivatives by denaturing PAGE (Supplementary Figure S1a). The extents of the HCV RNA IRES alkylation by derivatives of DNA1 and DNA2 were approximately 0.1 and 0.2 of oligomer residue per mol of RNA, respectively. These values were determined using Toluidine Blue O staining technique and defined as the ratio of intensity of covalent adduct band to total intensity of the RNA-containing bands (Supplementary Figure S1a).
Figure 1.
(Top) The secondary structure of the HCV IRES showing its domains and subdomains. (bottom) The part of RNA framed in rectangle above; black solid lines indicate regions complementary to alkylating derivatives of oligodeoxyribonucleotides (DNA1 and DNA2), gray dotted lines show the strand complementary to helper DNA-oligomer. Nucleotides of HCV IRES modified by the respective alkylating DNA derivatives are highlighted and marked by serial numbers. The result of the sequential alkylation by the corresponding derivatives is shown in the panel dslRNA.
Figure 2.
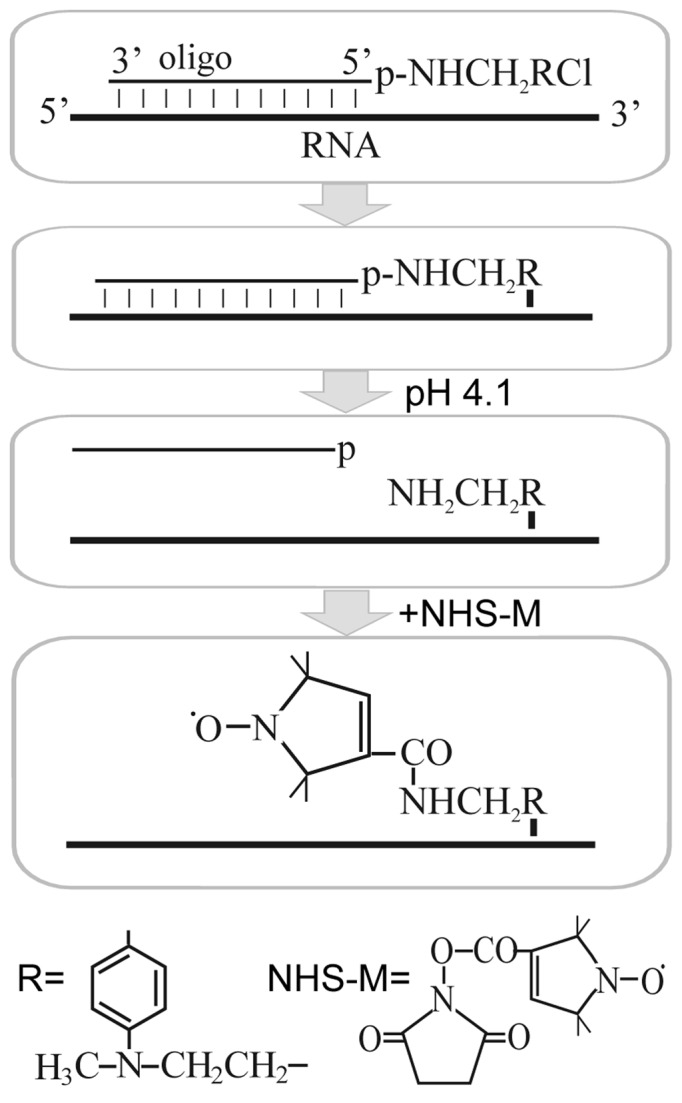
Scheme of site-specific introduction of spin labels into definite RNA sites based on the complementary-addressed alkylation of the RNA with [4-(N-2-chloroethyl-N-methylamino)benzyl]-phosphoramides of oligodeoxyribonucleo-tides, hydrolysis of phosphoramide bond in the covalent adduct formed and selective acylation of the released aliphatic amino group by N-hydroxysuccinimidyl derivative of the spin label.
In both cases of DNA1-NHCH2RCl and DNA2-NHCH2RCl, we attempted to increase the extent of alkylation of HCV RNA IRES (RNA) utilizing helper DNA-oligomer. In this approach, helper facilitates unfolding of RNA structure in the target sequence region and makes it accessible for binding with the oligonucleotide moiety of the DNA derivative (see Ref. (52) and Refs therein). With the use of helper complementary to sequence 84–102 (Figure 1), the extents of the alkylation of RNA by DNA1-NHCH2RCl and DNA2-NHCH2RCl were increased to approximately ∼0.5 and 0.6 mol of oligomer residue per mol of RNA, respectively (see also Supplementary Figure S1a).
The 4-[N-(2-chloroethyl)-N-methylamino]benzylphosphoramidе moiety attached to the 5′-terminal nucleotide of DNA might cross-link, as a rule, either to the last 3′-terminal RNA base in DNA•RNA heteroduplex or to the unpaired RNA base adjacent to it (52–54). In the former case, modification can occur only at atoms N7 of G or N3 of A that are not involved in the Watson–Crick base-pairing. In the latter case, any base in this position (except for uracil, which cannot be alkylated by chloroethylarylamines under the reaction conditions (55)) could be cross-linked with the DNA derivatives, leading to modification of N3 of C or N1 of A in addition to the above mentioned targets (see Ref. (56) and Refs therein). To determine the nucleotides of RNA cross-linked to the alkylating DNA derivatives, the reverse transcription was applied that allows detection of modified nucleotide by stop or pause of primer extension (40). During extension of 32P-labeled primer on the modified RNA, enzyme makes a stop at 3′-nucleotides proximal to the modified sites, if alkylation occurred at the nitrogen atoms involved in Watson–Crick base-pairing. This leads to accumulation of the DNA product of the respective length and enhancement of the radioactive signal at this nucleotide in radioautograph of the PAGE separation of the reaction products. If, however, alkylation occurred at the atoms untouched by Watson–Crick interaction (N7G or N3A), the enzyme makes a pause right at the modified nucleotide due to steric hindrances, giving a strong signal at this nucleotide and sometimes weaker signals at nucleotides 3′ and 5′ of it. Considering this, we could conclude that the main cross-linking target for DNA2-NHCH2RCl was N1 atom in A73 adjoining 3′ the nucleotide involved in the binding to DNA2-NHCH2RCl (Supplementary Figure S2). The main cross-linking site for DNA1-NHCH2RCl, N3 atom in C83, was determined using the described method (51). Results on site-directed alkylation of the HCV RNA IRES by two DNA derivatives are summarized in Table 1.
Note that our choice of nucleotide positions for SDSL was based on the preliminary analysis of nuclear magnetic resonance (NMR)-derived structure of HCV IRES domain II (PDB ID: 1P5N (48)): the pair C83 and A73 was selected to provide spin–spin distances appropriate for following DEER studies (<80 Å). The doubly-modified RNA derivative was prepared by sequential alkylation of RNA by DNA1-NHCH2RCl and DNA2-NHCH2RCl (at nucleotide positions C83 and A73, respectively; Figure 1). The isolation of alkylated products at each step allowed obtaining pure doubly-alkylated RNA derivative, free of any admixtures of singly- or non-alkylated precursors (Supplementary Figure S1b). The selective attachment of nitroxide spin labels to amino linker-containing RNA was carried out by treatment with the NHS-M (see Figure 2). Finally, the target doubly spin-labeled (dsl) derivative of the HCV RNA IRES (dslRNA) was purified by ethanol precipitation and its integrity was confirmed by denaturing PAGE. Functional activity of dslRNA was validated by examining its capability of the 48S pre-initiation complex formation in a mammalian cell-free protein synthesis system. It should be noted that upon binding of HCV RNA IRES to the 40S ribosomal subunit, its domain II is involved in the interaction with ribosomal protein uS7, providing the placement of the RNA start AUG codon in the ribosomal P site (see (57,58) for its subsequent recognition by initiator Met-tRNAiMet, which results in the formation of the 48S pre-initiation complex. Therefore, the modification of domain II in the HCV RNA IRES might prevent 48S complex assembly on this RNA. The results presented in Supplementary Figure S3 show that functional activity of dslRNA remains similar to that of the unmodified RNA.
Continuous wave EPR study of the spin-labeled domain II of HCV RNA IRES
To confirm the attachment of spin labels to RNA, we compared the room-temperature EPR spectrum with the spectra of (i) free nitroxide NHS-M in water/glycerol, and (ii) free NHS-M in water/glycerol in the presence of RNA dissolved in the same concentration as dslRNA (Figure 3). The EPR spectra of free NHS-M in the presence/absence of RNA are identical and can be simulated in the model of isotropic motion with rotational correlation time τcorr = 0.18 ns; therefore, it is evident that non-covalent interactions between spin label and RNA are absent for the concentrations used. Contrary, the spectrum of dslRNA is drastically different from the corresponding spectrum of free label in the presence of RNA. The rotational correlation time becomes much longer, τcorr = 1.02 ns and the spectral shape can only be simulated assuming anisotropic motion of label in the orienting potential (59) (details in Supplementary Data). This indicates partial ordering of spin label within its local environment occurring due to the covalent attachment to RNA. Thus, both observations, namely spectral change compared to free nitroxide and the spectral shape itself, unambiguously confirm that spin labels have been successfully attached to amino linker-containing RNA. To confirm that this attachment is site-specific, we perform DEER distance measurements as described below.
Figure 3.
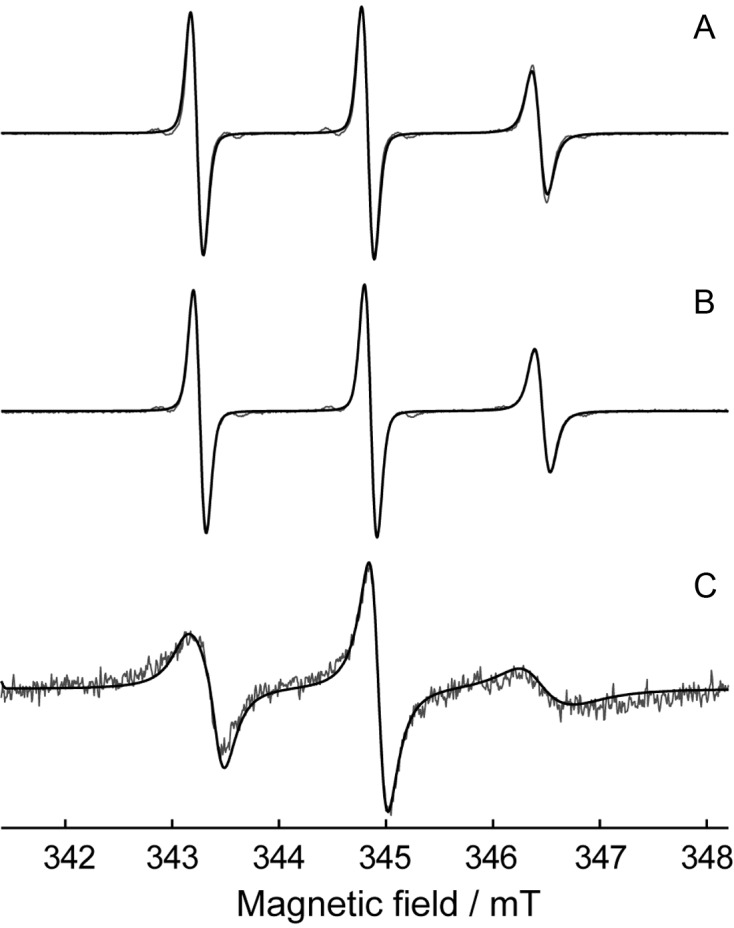
Room-temperature continuous wave X-band EPR spectra of liquid-phase samples: (A) free label NHS-M in water/glycerol; (B) free label NHS-M in the presence of RNA in water/glycerol; (C) dslRNA in water/glycerol. Gray lines show experiment, black lines show the simulations with the following parameters used: g = [2.0091 2.0061 2.0022], A = [14 14 107.1] and [14 14 107.5] MHz in (A and B) and (C), respectively; τcorr = 0.18 ns and 1.02 ns in (A and B) and (C), respectively. Parameters of the orienting potential in (C) λ2,0 and λ2,2 are 0.53 and 0.55, respectively. For more details see Supplementary Data.
Measurement of interspin distances in the spin-labeled domain II of HCV RNA IRES
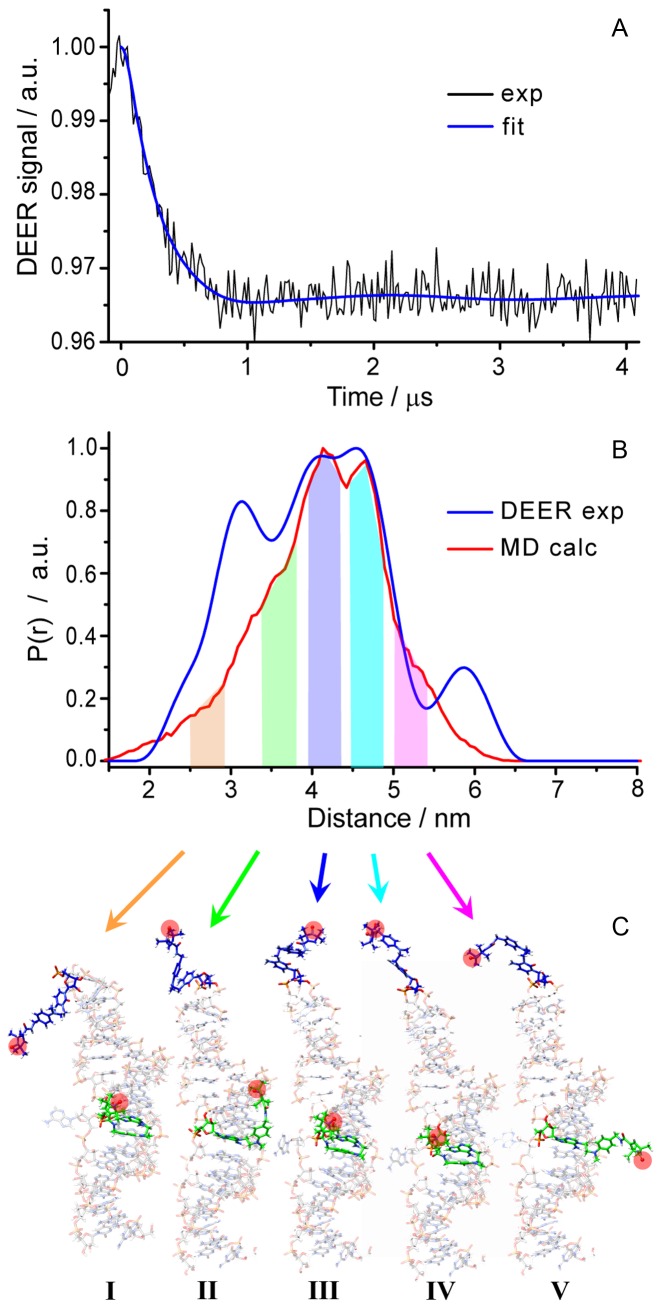
Figure 4A shows the background-corrected Q-band DEER time trace obtained for dslRNA, which clearly demonstrates the occurrence of dipolar oscillation. The absolute modulation depth is ∼3.5%. Typical modulation depth achieved with our experimental setup on standard nitroxide biradicals at similar conditions is ∼20–25%. As was mentioned above, the doubly-alkylated RNA derivative can be isolated in its pure form; therefore relatively small modulation depth should be ascribed to the low efficiency of spin label attachment to the alkylated RNA derivative and/or to the reversibility of this reaction. Based on the DEER modulation depth and observed spin concentration in CW EPR, we have estimated the factual spin labeling efficiency as 20% and absolute concentration of doubly spin-labeled RNA as 2.4 μM (see Supplementary Data). This means that only 10% of the total number of HCV RNA IRES molecules in the sample were doubly spin-labeled, being still sufficient for the EPR measurements.
Figure 4.
Distance measurements on dslRNA. (A) Background-corrected Q-band DEER/PELDOR time trace (exp) and DeerAnalysis fitting (fit); (B) obtained distance distribution using Tikhonov regularization parameter 1000 (DEER exp) and calculated MD distribution (MD calc). (C) Typical conformations of spin labels corresponding to the selected ranges of distances (highlighted by colored bars in (B) and pointed out by corresponding arrows). Red circles indicate the NO group of the label, for clarity. Spin-labeled C83 (top) is shown in blue, spin-labeled A73 (middle) is shown in green.
The obtained distribution (Figure 4B) is quite broad corresponding to a mean distance <rDEER> = 4.16 nm and a standard deviation parameter σ = 1.10 nm. According to NMR data, the distance between respective N atoms of label attachment sites is 3.26 nm (48). Taking into account the length of the linker, the obtained mean spin–spin distance is quite reasonable. However, to gain further insights into spin label conformations and more solid proof on selectivity of SDSL, we performed MD studies.
Theoretical (MD) calculations readily provide a distance distribution for dslRNA, which is reasonably close to the experimental one and reproduces all major peaks (Figure 4B). Figure 4C shows the conformations of spin label corresponding to different ranges of spin–spin distances, as indicated in Figure 4B by colored bars. The spin label of C83 located at the top of the hairpin loop is relatively mobile and the linker is largely extended in most of the conformations. In contrast, the spin label of A73 can bind to the minor groove (structures I–IV), where linker is partly (II) or strongly (I, III, IV) compacted, or it can be disposed outside the helix with the stretched linker (structure V). The shortest distances occur when the spin label of C83 is directed along the RNA hairpin toward the A73, whereas the spin label of A73 is localized in the minor groove. Contrary, the longest distances are found when the spin label of A73 is directed outside of the groove, spin label of C83 is turned outward of the hairpin and the linkers are maximally extended (structure V).
The side peaks of experimental DEER distance distribution (≈3.1 and 5.9 nm) are somewhat underestimated in MD calculations (Figure 4B), however, in our opinion, general agreement is quite fair for the given accuracy of experimental data. Thus, the observed agreement of experimental and theoretical distributions clearly confirms that the attachment of spin labels via complementary-addressed SDSL approach indeed occurred at the target sites, namely C83 and A73 positions of the domain II of HCV RNA IRES.
The distance distribution obtained for dslRNA here is rather broad. Based on the analysis of MD data we conclude that the overall spin–spin distance distribution width (σMD ≈ 0.8 nm) is contributed by ≈0.3 nm from intrinsic mobility of RNA and by ≈0.5 nm from mobility of two labels (Supplementary Data). The value of σ ≈ 0.5 nm for the label-induced broadening is in perfect agreement with previous value σ = 0.54 nm obtained for more rigid 10-mer RNA duplex labeled at its termini using similar method (34). Although the other approaches for spin-labeling of nucleic acids suffer less from intrinsic label mobility and allow obtaining much narrower distance distributions (60,61), they are not as versatile and are not applicable for long natural RNAs. In addition, proper selection of the labeling sites in the future might allow obtaining narrower distributions (σ < 0.5 nm) using complementary-addressed SDSL (Supplementary Data).
CONCLUSIONS AND OUTLOOK
In this work we have for the first time demonstrated the applicability of novel complementary-addressed SDSL approach to long structured RNAs. For the validation purposes, HCV RNA IRES with known structure and expected distances has been spin-labeled in two definite positions and studied using EPR/DEER spectroscopy. The central step of the above SDSL procedure is the targeted introduction of aliphatic amino group into RNA. A similar method was also applied previously to long RNAs with complex tertiary structures, although in radioactive picoscale variant where only small amounts of the sample were required (38,52–54,62). The version of the method suitable for SDSL was first proposed by us recently and tested on short oligoribonucleotides (34). The present work finally provides clear evidence for the crucial advantage of new SDSL approach, i.e. its applicability to long native RNAs. The distance distributions in doubly spin-labeled HCV RNA IRES have been measured using Q-band DEER, and the values obtained correspond well to those predicted by MD simulations.
Compared to the method based on enzymatic RNA ligation utilizing DNA splints, complementary-addressed SDSL approach is less sensitive to the RNA purity and does not require homogeneity of its ends; at the same time, it is also laborious and results in low yield of the final product. While ligation has clearly better performance for short RNAs, this will necessarily drop for the long RNA strands, especially when label attachment needs to be performed in the middle part of the RNA with two ligation steps required. It is difficult to predict whether the ligation or complementary-addressed SDSL method would perform better for a particular RNA; however it is important to note that novel SDSL method is conceptually very different from the ligation approach. From this point of view, the SDSL strategy described by us can be considered as valuable alternative to the above mentioned ligation approach.
The applicability of complementary-addressed SDSL approach to long natural RNAs opens up a variety of opportunities for structural DEER studies of these RNAs alone and their complexes with proteins functioning in living organisms. For instance, conformational changes of viral IRES elements upon their binding to the small subunit of the mammalian ribosome are of great interest, since they might allow novel insights into initial steps of translation initiation of the respective viral RNAs. We therefore believe that the new complementary-addressed SDSL approach in conjunction with pulse dipolar EPR spectroscopy has a great potential for future studies of complex biological systems involving long RNAs.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Science Foundation [14-14-00922]. Funding for open access charge: Russian Science Foundation [14-14-00922].
Conflict of interest statement. None declared.
REFERENCES
- 1.Schiemann O., Weber A., Edwards T.E., Prisner T.F., Sigurdsson S.T. Nanometer distance measurements on RNA using PELDOR. J. Am. Chem. Soc. 2003;125:3434–3435. doi: 10.1021/ja0274610. [DOI] [PubMed] [Google Scholar]
- 2.Schiemann O., Piton N., Mu Y.G., Stock G., Engels J.W., Prisner T.F. A PELDOR-based nanometer distance ruler for oligonucleotides. J. Am. Chem. Soc. 2004;126:5722–5729. doi: 10.1021/ja0393877. [DOI] [PubMed] [Google Scholar]
- 3.Borbat P.P., Davis J.H., Butcher S.E., Freed J.H. Measurement of large distances in biomolecules using double-quantum filtered refocused electron spin-echoes. J. Am. Chem. Soc. 2004;126:7746–7747. doi: 10.1021/ja049372o. [DOI] [PubMed] [Google Scholar]
- 4.Cai Q., Kusnetzow A.K., Hubbell W.L., Haworth I.S., Gacho G.P.C., Van Eps N., Hideg K., Chambers E.J., Qin P.Z. Site-directed spin labeling measurements of nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nucleic Acids Res. 2006;34:4722–4730. doi: 10.1093/nar/gkl546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant G.P.G., Qin P.Z. A facile method for attaching nitroxide spin labels at the 5 ' terminus of nucleic acids. Nucleic Acids Res. 2007;35:e77. doi: 10.1093/nar/gkm240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piton N., Mu Y.G., Stock G., Prisner T.F., Schiemann O., Engels J.W. Base-specific spin-labeling of RNA for structure determination. Nucleic Acids Res. 2007;35:3128–3143. doi: 10.1093/nar/gkm169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schiemann O., Prisner T.F. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Quart. Rev. Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 8.Schiemann O., Piton N., Plackmeyer J., Bode B.E., Prisner T.F., Engels J.W. Spin labeling of oligonucleotides with the nitroxide TPA and use of PELDOR, a pulse EPR method, to measure intramolecular distances. Nat. Protoc. 2007;2:904–923. doi: 10.1038/nprot.2007.97. [DOI] [PubMed] [Google Scholar]
- 9.Qin P.Z., Haworth I.S., Cai Q., Kusnetzow A.K., Grant G.P.G., Price E.A., Sowa G.Z., Popova A., Herreros B., He H. Measuring nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nat. Protoc. 2007;2:2354–2365. doi: 10.1038/nprot.2007.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sowa G.Z., Qin P.Z. Site-directed spin labeling studies on nucleic acid structure and dynamics. Prog. Nucleic Acid Res. Mol. Biol. 2008;82:147–197. doi: 10.1016/S0079-6603(08)00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X., Cekan P., Sigurdsson S.T., Qin P.Z. Studying RNA using site-directed spin-labeling and continuous-wave electron paramagnetic resonance spectroscopy. Methods Enzymol. 2009;469:303–328. doi: 10.1016/S0076-6879(09)69015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunsicker-Wang L., Vogt M., DeRose V.J. EPR methods to study specific metal-ion binding sites in RNA. Methods Enzymol. 2009;468:335–367. doi: 10.1016/S0076-6879(09)68016-2. [DOI] [PubMed] [Google Scholar]
- 13.Schiemann O., Cekan P., Margraf D., Prisner T.F., Sigurdsson S.Th. Relative orientation of rigid nitroxides by PELDOR: beyond distance measurements in nucleic acids. Angew. Chem. Int. Ed. 2009;48:3292–3295. doi: 10.1002/anie.200805152. [DOI] [PubMed] [Google Scholar]
- 14.Grohmann D., Klose D., Klare J.P., Kay C.W.M., Steinhoff H.J., Werner F. RNA-binding to archaeal RNA polymerase subunits F/E: a DEER and FRET study. J. Am. Chem. Soc. 2010;132:5954–5955. doi: 10.1021/ja101663d. [DOI] [PubMed] [Google Scholar]
- 15.Kim N.K., Bowman M.K., DeRose V.J. Precise mapping of RNA tertiary structure via nanometer distance measurements with double electron-electron resonance spectroscopy. J. Am. Chem. Soc. 2010;132:8882–8884. doi: 10.1021/ja101317g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sicoli G., Wachowius F., Bennati M., Hobartner C. Probing secondary structures of spin-labeled RNA by pulsed EPR spectroscopy. Angew. Chem. Int. Ed. 2010;49:6443–6447. doi: 10.1002/anie.201000713. [DOI] [PubMed] [Google Scholar]
- 17.Krstic I., Frolow O., Sezer D., Endeward B., Weigand J. E., Suess B., Engels J.W., Prisner T.F. PELDOR spectroscopy reveals preorganization of the neomycin-responsive riboswitch tertiary structure. J. Am. Chem. Soc. 2010;132:1454–1455. doi: 10.1021/ja9077914. [DOI] [PubMed] [Google Scholar]
- 18.Krstic I., Hansel R., Romainczyk O., Engels J.W., Dotsch V., Prisner T.F. Long-range distance measurements on nucleic acids in cells by pulsed EPR spectroscopy. Angew. Chem. Int. Ed. 2011;50:5070–5074. doi: 10.1002/anie.201100886. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X., Tung C.-S., Sowa G.Z., Hatmal M.M., Haworth I.S., Qin P.Z. Global structure of a three-way junction in a Phi29 packaging RNA dimer determined using site-directed spin labeling. J. Am. Chem. Soc. 2012;134:2644–2652. doi: 10.1021/ja2093647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen P., Qin P.Z. RNA dynamics: perspectives from spin labels. WIREs RNA. 2012;3:62–72. doi: 10.1002/wrna.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duss O., Michel E., Yulikov M., Schubert M., Jeschke G., Allain F.H.T. Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature. 2014;509:588–592. doi: 10.1038/nature13271. [DOI] [PubMed] [Google Scholar]
- 22.Duss O., Yulikov M., Jeschke G., Allain F.H.T. EPR-aided approach for solution structure determination of large RNAs or protein-RNA complexes. Nat. Commun. 2014;5:3669. doi: 10.1038/ncomms4669. [DOI] [PubMed] [Google Scholar]
- 23.Esquiaqui J.M., Sherman E.M., Ye J.-D., Fanucci G.E. Site-directed spin-labeling strategies and electron paramagnetic resonance spectroscopy for large riboswitches. Methods Enzymol. 2014;549:287–311. doi: 10.1016/B978-0-12-801122-5.00013-1. [DOI] [PubMed] [Google Scholar]
- 24.Esquiaqui J.M., Sherman E.M., Ionescu S.A., Ye J.-D., Fanucci G.E. Characterizing the dynamics of the leader-linker interaction in the glycine riboswitch with site-directed spin labeling. Biochemistry. 2014;53:3526–3528. doi: 10.1021/bi500404b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shevelev G.Yu, Krumkacheva O.A., Kuzhelev A.A., Lomzov A.A., Rogozhnikova O.Yu., Trukhin D.V., Troitskaya T.I., Tormyshev V.M., Fedin M.V., Pyshnyi D.V., et al. Physiological-temperature distance measurement in nucleic acid using triarylmethyl-based spin labels and pulsed dipolar EPR spectroscopy. J. Am. Chem. Soc. 2014;136:9874–9877. doi: 10.1021/ja505122n. [DOI] [PubMed] [Google Scholar]
- 26.Shevelev G.Yu., Krumkacheva O.A., Lomzov A.A., Kuzhelev A.A., Trukhin D.V., Rogozhnikova O.Yu., Tormyshev V.M., Pyshnyi D.V., Fedin M.V., Bagryanskaya E.G. Triarylmethyl labels: toward improving the accuracy of EPR nanoscale distance measurements in DNAs. J. Phys. Chem. B. 2015;119:13641–13648. doi: 10.1021/acs.jpcb.5b03026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagryanskaya E.G., Krumkacheva O.A., Fedin M.V., Marque S.R.A. Development and Application of Spin Traps, Spin Probes, and Spin Labels. Methods Enzymol. 2015;563:365–396. doi: 10.1016/bs.mie.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Malygin A.A., Graifer D.M., Meschaninova M.I., Venyaminova A.G., Krumkacheva O.A., Fedin M.V., Karpova G.G., Bagryanskaya E.G. Doubly spin-labeled RNA as an EPR reporter for studying multicomponent supramolecular assemblies. Biophys. J. 2015;109:2637–2643. doi: 10.1016/j.bpj.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dethoff E.A., Chugh J., Mustoe A.M., Al-Hashimi H.M. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones S., Daley D.T., Luscombe N.M., Berman H.M., Thornton J.M. Protein-RNA interactions: a structural analysis. Nucleic Acids Res. 2001;29:943–954. doi: 10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shelke S.A., Sigurdsson S.T. Structural changes of an abasic site in duplex DNA affect noncovalent binding of the spin label c. Nucleic Acids Res. 2012;40:3732–3740. doi: 10.1093/nar/gkr1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore M.J., Sharp P.A. Site-specific modification of pre-messenger-RNA - the 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 33.Graifer D., Karpova G. General approach for introduction of various chemical labels in specific RNA locations based on insertion of amino linkers. Molecules. 2013;18:14455–14469. doi: 10.3390/molecules181214455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Babaylova E.S., Ivanov A.V., Malygin A.A., Vorobjeva M.A., Venyaminova A.G., Polienko Y.F., Kirilyuk I.A., Krumkacheva O.A., Fedin M.V., Karpova G.G., et al. A versatile approach for site-directed spin labeling and structural EPR studies of RNAs. Org. Biomol. Chem. 2014;12:3129–3136. doi: 10.1039/c3ob42154f. [DOI] [PubMed] [Google Scholar]
- 35.Milov A.D., Salikhov K.M., Shirov M.D. Application of ELDOR in electron-spin echo for paramagnetic center space distribution in solids. Fiz. Tverd. Tela. 1981;23:975–982. [Google Scholar]
- 36.Lukavsky P.J. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hankovszky H.O., Hideg K., Tigyi J. Nitroxides II. l-Oxyl-2, 2, 5, 5-tetramethyl pyrroline-3-carboxylic acid derivatives. Acta Chim. Acad. Sci. Hung. 1978;98:339–348. [Google Scholar]
- 38.Malygin A.A., Graifer D.M., Laletina E.S., Shatskii I.N., Karpova G.G. Approach to identifying the functionally important segments of RNA, based on complementation-addressed modification. Molek. Biol. 2003;37:1027–1034. [PubMed] [Google Scholar]
- 39.Reynolds J.E., Kaminski A., Kettinen H.J., Grace K., Clarke B.E., Carroll A.R., Rowlands D.J., Jackson R.J. Unique features of internal initiation of Hepatitis-C virus-RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wollenzien P. Isolation and identification of RNA cross-links. Methods Enzymol. 1988;164:319–329. doi: 10.1016/s0076-6879(88)64052-3. [DOI] [PubMed] [Google Scholar]
- 41.Malygin A.A., Kossinova O.A., Shatsky I.N., Karpova G.G. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res. 2013;41:8706–8714. doi: 10.1093/nar/gkt632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kossinova O., Malygin A., Krol A., Karpova G. A novel insight into the mechanism of mammalian selenoprotein synthesis. RNA. 2013;19:1147–1158. doi: 10.1261/rna.036871.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharifulin D., Babaylova E., Kossinova O., Bartuli Y., Graifer D., Karpova G. Ribosomal protein S5e is implicated in translation initiation through its interaction with the N-terminal domain of initiation factor eIF2α. Chembiochem. 2013;14:2136–2143. doi: 10.1002/cbic.201300318. [DOI] [PubMed] [Google Scholar]
- 44.Stoll S., Schweiger A. Easyspin: simulating CW ESR spectra. Biol. Magn. Reson. 2007;27:299–321. [Google Scholar]
- 45.Stoll S., Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 46.Pannier M., Veit S., Godt A., Jeschke G., Spiess H.W. Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 47.Jeschke G., Chechik V., Ionita G., Godt A., Zimmermann H., Banham J., Timmel C.R., Hilger D., Jung H. DeerAnalysis2006—a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006;30:473–498. [Google Scholar]
- 48.Lukavsky P.J., Kim I., Otto G.A., Puglisi J.D. Structure of HCVIRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 49.Case D.A., Darden T.A., Cheatham T.E. III, Simmerling C.L., Wang J., Duke R.E., Luo R., Walker R.C., Zhang W., Merz K.M., et al. AMBER 12. San Francisco: University of California; 2012. [Google Scholar]
- 50.Honda M., Beard M.R., Ping L.H., Lemon S.M. A phylogenetically conserved stem-loop structure at the 5 ' border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grineva N.I., Lomakina T.S., Tigeeva N.T., Chimitova T.A. Kinetics of ionization of the C-Cl bond in 4-(N-2-chloroethyl-N-methylamino)benzyl-5′-phosphamides of nucleosides and oligonucleotides. Bioorg. Khim. 1977;3:210–214. [Google Scholar]
- 52.Bulygin K., Malygin A., Karpova G., Westermann P. Site-specific modification of 4.5S RNA apical domain by complementary oligodeoxynucleotides carrying an alkylating group. Eur. J. Biochem. 1998;251:175–180. doi: 10.1046/j.1432-1327.1998.2510175.x. [DOI] [PubMed] [Google Scholar]
- 53.Babaylova E., Graifer D., Malygin A., Stahl J., Shatsky I., Karpova G. Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res. 2009;37:1141–1151. doi: 10.1093/nar/gkn1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laletina E., Graifer D., Malygin A., Ivanov A., Shatsky I., Karpova G. Proteins surrounding hairpin IIIe of the hepatitis C virus internal ribosome entry site on the human 40S ribosomal subunit. Nucleic Acids Res. 2006;34:2027–2036. doi: 10.1093/nar/gkl155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ross W.C J. Biological alkylating agents. London: Butterworths and Co., Ltd; 1962. pp. 51–63. [Google Scholar]
- 56.Lundblad R.L., MacDonald F.M. Handbook of biochemistry and molecular biology. 4th edn. Boca Raton: CRC Press Taylor & Francis group; 2010. Chemical Modification of Nucleic Acids; pp. 359–372. [Google Scholar]
- 57.Berry K.E., Waghray S., Mortimer S.A., Bai Y., Doudna J.A. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456–1466. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Filbin M.E., Vollmar B.S., Shi D., Gonen T., Kieft J.S. HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat. Struct. Mol. Biol. 2013;20:150–158. doi: 10.1038/nsmb.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moro G., Freed J.H. Efficient computation of magnetic-resonance spectra and related correlation-functions from stochastic liouville equations. J. Phys. Chem. 1980;84:2837–2840. [Google Scholar]
- 60.Prisner T.F., Marko A., Sigurdsson S.Th. Conformational Dynamics of Nucleic Acid Molecules Studied by PELDOR Spectroscopy with Rigid Spin Labels. J. Magn. Reson. 2015;252:187–198. doi: 10.1016/j.jmr.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 61.Schiemann O., Cekan P., Margraf D., Prisner T.F., Sigurdsson S.Th. Relative orientation of rigid nitroxides by PELDOR: beyond distance measurements in nucleic acids. Angew. Chem., Int. Ed. 2009;48:3292–3295. doi: 10.1002/anie.200805152. [DOI] [PubMed] [Google Scholar]
- 62.Zenkova M., Ehresmann C., Caillet J., Springer M., Karpova G., Ehresmann B., Romby P. A novel-approach to introduce site-directed specific cross-links within RNA-protein complexes - application to the escherichia-coli threonyl-transfer-RNA synthetase translational operator complex. Eur. J. Biochem. 1995;231:726–735. doi: 10.1111/j.1432-1033.1995.0726d.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.