Abstract
Up to 30% of patients with obsessive-compulsive disorder (OCD) exhibit an inadequate response to serotonin reuptake inhibitors (SRIs). To date, genetic predictors of OCD treatment response have not been systematically investigated using genome-wide association study (GWAS). To identify specific genetic variations potentially influencing SRI response, we conducted a GWAS study in 804 OCD patients with information on SRI response. SRI response was classified as “response” (n = 514) or “non-response” (n = 290), based on self-report. We used the more powerful Quasi-Likelihood Score Test (the MQLS test) to conduct a genome-wide association test correcting for relatedness, and then used an adjusted logistic model to evaluate the effect size of the variants in probands. The top SNP was rs17162912 (P = 1.76×10−8) which is near the DISP1 gene on 1q41-q42, a microdeletion region implicated in neurological development. The other six SNPs showing suggestive evidence of association (P <10−5) were rs9303380, rs12437601, rs16988159, rs7676822, rs1911877, and rs723815. Among them, two SNPs in strong linkage disequilibrium, rs7676822 and rs1911877, located near the PCDH10 gene, gave p-values of 2.86×10−6 and 8.41×10−6, respectively. The other 35 variations with signals of potential significance (P <10−4) involve multiple genes expressed in the brain, including GRIN2B, PCDH10, and GPC6. Our enrichment analysis indicated suggestive roles of genes in the glutamatergic neurotransmission system (FDR = 0.0097) and the serotonergic system (FDR = 0.0213). While the results presented may provide new insights into genetic mechanisms underlying treatment response in OCD, studies with larger sample sizes and detailed information on drug dosage and treatment duration are needed.
Keywords: Obsessive-Compulsive Disorder, Serotonin Reuptake Inhibitors, Genome Wide Association Study, Pharmacogenetics
INTRODUCTION
Approximately 1-3% of the US population suffers from obsessive-compulsive disorder (OCD), a neuropsychiatric disorder characterized by recurrent obsessions and/or compulsions that cause marked distress and impairment.1 OCD often aggregates in families, and results from segregation analysis and twin studies support significant genetic influence.2 A genome-wide linkage study identified several OCD susceptibility loci (i.e., 3q, 7p, 1q, 15q and 6q).3 Variants in several genes have been associated with OCD, including SLC1A14, SLC6A45, 6, and GRIN2B7-9. Prior to the advent of the GWAS platform, association studies targeted a set of candidate genes that were inconsistently reported to be associated with OCD.10 More recently, two genome-wide association studies have identified PTPRD, DLGAP1, CDH10, and GRIK2 as potential OCD susceptible loci.11, 12
Individuals affected with OCD are typically treated with a combination of exposure response prevention (ERP) and medications; serotonin reuptake inhibitors (SRIs) are the first-line pharmacotherapy option for the treatment of OCD. However, up to 30% of patients treated with these medications show poor or no response to standard treatment; and some patients cannot tolerate adverse effects of medications.13 The literature on genetic predictors of SRI treatment response in OCD is sparse.14-16 Therefore, elucidation of genetic variants influencing treatment response is needed.
SRIs inhibit the reuptake of the neurotransmitter serotonin by presynaptic cells, thereby increasing extracellular levels of serotonin in the synaptic cleft and allowing serotonin to more easily bind to the postsynaptic receptor.6, 17 More than 60 proteins are known to play a role in the serotonin signaling pathway. Among these, the serotonin transporter gene SLC6A4 may impact SRI response.6 In addition, genetic variants in several other genes (i.e., CYP2D6, SLC1A1, SLC6A4, HTR1B receptor, 5-HT2A receptor, and BDNF) have been reported to influence SRI response in OCD.16 However, many of these studies were hampered by small sample sizes and a limited number of known genetic variations in candidate genes. Additionally, analytical approaches vary widely among different studies, which may have led to inconsistent results. In 2012, Tansey et al. reported results from the first genome wide association study (GWAS) of SRI response in major depression.18 However, to our knowledge, no GWAS study of medication response in OCD has been reported. Therefore, an important unexplored research question is whether genetic variations influence SRI treatment response in OCD.
To address this question, we performed a whole genome association analysis on response to SRIs in 804 OCD cases, using a novel, more powerful Quasi-Likelihood Score Test to correct for the relatedness. Here we report our findings of genome-wide association analysis of therapeutic response of OCD as well as the results of enrichment analysis of nervous system pathways.
MATERIALS AND METHODS
Subject recruitment and data collection
The sample for the current analysis was recruited as part of the OCD Collaborative Genetics Association Study (OCGAS). Detailed methods for OCD diagnosis and sample description have been previously described.19, 20 In brief, the evaluation of OCD and drug response was conducted by PhD-level clinical psychologists using a semi-structured diagnostic instrument (SCID), and included the Yale -Brown Obsessive Compulsive Scale (YBOCS) OCD symptom checklist and YBOCS OCD severity scale.21 Final DSM-IV (The Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition) OCD diagnosis was assigned by consensus of clinicians at each study site and reviewed at Johns Hopkins University.
The individuals in the current study had participated in one of two multisite, collaborative family/genetic studies of OCD, which have been described in detail elsewhere. In brief, the OCD Collaborative Genetics Study (OCGS) (2001-2006), targeted recruitment on families with OCD-affected sibling pairs, and extended these when possible through affected first- and second-degree relatives.19 The OCD Collaborative Genetic Association Study (OCGAS) (2007-2012) targeted recruitment on trios (i.e., an affected proband and both parents), but also included pedigrees with a proband and unaffected sibling, as well as families with multiple-affected members.12 Participants were recruited into the studies from outpatient and inpatient clinics, referrals from clinicians in the community, web sites, media advertisements, self-help groups, and annual conventions of the International Obsessive Compulsive Foundation.
As part of the treatment history section of the clinical interview, examiners asked participants about their duration of medication use, maximum dosage, and response to each of several SRI medications (if received), including clomipramine, citalopram, escitalopram, fluoxetine, fluvoxamine, paroxetine, sertraline, venlafaxine, and duloxetine. Examiners also asked about response to other medications and behavioral therapy.
Treatment response was initially assessed using a five-point scale of “no response”, “could not tolerate”, “minimal”, “moderate improvement”, and “total remission”. For the current analyses, we dichotomized treatment response into “response” (“moderate improvement” or “total remission”) and “non-response” (“no response” or “minimal response”) categories; those reporting “couldn't tolerate” and those with missing data on drug response were excluded from the analyses. For those patients who were treated with multiple SRI medications, response was based on the medication to which the best response was reported.
Blood samples were collected from affected probands, their parents, and their affected relatives. DNA samples were extracted using the Qiagen DNA extraction Kit and stored at −80°C for further genotyping. The current analyses included 804 OCD cases who were participants of the OCGAS GWAS. The study was approved by the institutional review board at each participating institution.
Genotyping and quality control
Genotyping was conducted with the Illumina HumanOmniExpress-12v1 (San Diego, CA, USA), in which genotyping was attempted for 730,525 SNPs. Quality control was performed in PLINK to remove poorly genotyped SNPs and individuals. Details of the quality control on the original data have been previously described.12 To detail, technically failed SNPs were removed. Individuals with sex discrepancy were either removed or updated. The relationship information was corrected and individuals with excess of genotyping errors were removed. Outliers were removed based on a multidimensional scaling (MDS) analysis. We included all OCD patients from the initial quality controlled dataset for subsequent study. Further quality re-assurance and filtering were conducted, including SNPs with Hardy-Weinberg equilibrium (HWE) test p-value <10−6 were excluded; SNPs with genotyped rate <98% or with minor allele frequency (MAF) <0.05 were excluded; SNPs violating Mendelian errors were treated as missing. After genotyping quality control, 53 individuals who could not tolerate the medications were removed, along with 741 individuals that did not have any information on drug effect. The affected individuals with informative drug response data were subjected to an association test. Finally, a total of 804 individuals (including 514 responders and 290 non-responders) were subjected to statistical analysis.
Statistical methods and integrated analysis using bioinformatics resource
We used the more powerful Quasi-Likelihood Score test, termed MQLS test22, to conduct association tests correcting for the relatedness coefficients (based on identity-by-descent, IBD). A sex- and age-adjusted logistic model was used for the evaluation of effect size in probands using the PLINK software.23 Since the association test could underestimate true signals of association with SRI response due to limited statistical power, all variations with MQLS test p-values <10−4 with relatively large effect-size (odds ratios ≥1.50 for the risk allele) were reported. All statistical procedures were conducted using in-house R scripts on a GentOS based Cluster computer.
SNP annotation was conducted using a web-based software SNP-NEXUS24 (http://www.snp-nexus.org) based on dbSNP135/hg19. Cross references to other GWAS association studies were explored using the NHGRI GWAS Catalogue.25 Neurobiological evidence was examined in peer-reviewed publications in the PubMed database. LD plots were completed using the LocusZoom software based on 1000 genome CEU population data (hg19/1000 Genomes Mar 2012 EUR).26
Imputation around one SNP of interest was conducted using the Impute2 software (URL: https://mathgen.stats.ox.ac.uk/impute/impute_v2.html) using 1000 Genomes Phase 1 as reference panel (Jun 2011 version). After the imputation, data quality control (QC) was performed to exclude imputed SNPs with genotyping rate <0.95, or significantly deviate from HWE test (p-value <10−6), or minor allele frequency (MAF) <0.01. Genotypes detected with Mendelian error were set to missing. After QC, MQLS test was performed using the same options described above.
Power calculation was conducted using the GPC software27 (URL: http://pngu.mgh.harvard.edu/~purcell/gpc/cc2.html) to indicate the power of the association test given our sample size. We performed a pathway analysis using all SNPs passing QC checkup. Functional enrichment analysis was conducted in ten nervous system pathways defined by the KEGG (Kyoto Encyclopedia of Genes and Genomes) databases using DAVID Bioinformatics Resource v6.7 (URL: http://david.abcc.ncifcrf.gov/).28 A total of 8,182 genes in both our dataset and pathway databases were used as the reference background list. Gene-level p-values were calculated by summarizing the SNP-level statistics using MAGENTA software (version 2.4)29, corrected for total number of SNPs in the gene, gene size, as well as the LD patterns in the genes (URL: http://www.broadinstitute.org/mpg/magenta/). A gene was classified as “significant” if its p-value is less than 0.001. Each pathway is then tested for whether it contains more “significant” genes than expected by chance using a modified Fisher exact test.
RESULTS
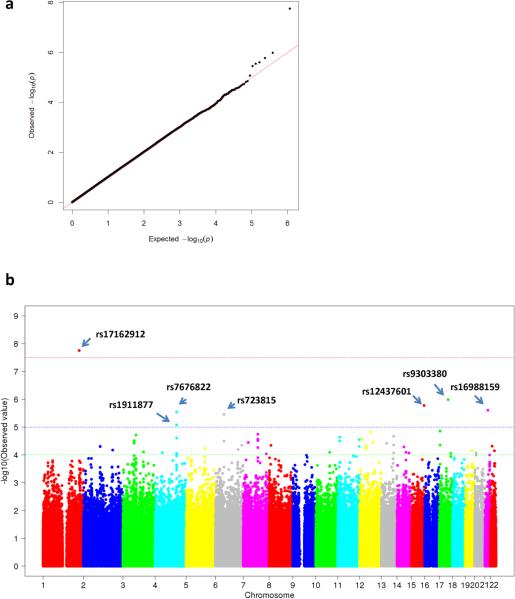
The demographic and clinical characteristics of the samples are summarized in Table 1. After data quality control, 597,847 SNPs (81.8% of the total SNPs attempted in the array) were successfully genotyped. A total of 804 individuals with informative drug effect data (514 responders and 290 non-responders) had a set of high quality genotyped data with a call rate of 99.9%. Figure 1a shows a Q-Q plot. Of the 42 SNPs identified with a p-value <10−4, one SNP met the genome-wide significance level (P = 1.76 ×10−8) for SRI treatment response; six SNPs showed suggestive evidence of association at the level of P <10−5; and 35 SNPs showed signals of association at the level of P<10−4 (Figure 1b and Table 2).
Table 1.
Characteristics of OCD participants
| Group | Subgroup | Count (N = 804) | Frequency |
|---|---|---|---|
| Sex | |||
| Male | 312 | 0.39 | |
| Female | 492 | 0.61 | |
| Agea | |||
| 7-9 | 19 | 0.02 | |
| 10-19 | 189 | 0.23 | |
| 20-29 | 170 | 0.21 | |
| 30-39 | 173 | 0.22 | |
| 40-49 | 159 | 0.20 | |
| 50-78 | 94 | 0.12 | |
| Age at onset of OC symptoms | |||
| 5-9 | 518 | 0.64 | |
| 10-19 | 237 | 0.30 | |
| 20-44 | 118 | 0.06 | |
| SRI responseb | |||
| “No response” | 290 | 0.36 | |
| “Response” | 514 | 0.64 | |
Age unknown for 5 participants.
“Couldn't tolerate” and “Unknown” were excluded from data analysis.
Figure 1.
Genome-wide association study of genetic variations and treatment response. (a) Q-Q plot for the association test of genetic variations. (b) Manhattan plot for the association test of genetic variations and SRI response. MQLS test was performed to test the association of variants associated with drug response. A red line indicates genome-wide significance (5×10−8); a blue line indicates the level of suggestive evidence for association (1×10−5).
Table 2.
Top loci associated with treatment response in OCD patients
| SNP | Chr. Position | A1/A2a | Resp.b | Non-Resp.b | Pc | OR(95%CI)d | Region | Nearest Gene (distance/bp) | |
|---|---|---|---|---|---|---|---|---|---|
| rs17162912 | 1 | 222974926 | C/T | 0.06 | 0.15 | 1.76×10−8 | 0.39(0.26-0.58) | intergenic | DISP1(13505) |
| rs9303380 | 17 | 54117492 | A/G | 0.03 | 0.07 | 1.03×10−6 | 0.37(0.21-0.64) | intergenic | ANKFN1(113344) |
| rs12437601 | 15 | 98687330 | C/T | 0.09 | 0.03 | 1.66×10−6 | 4.07(2.16-7.66) | intergenic | ARRDC4(170262) |
| rs16988159 | 21 | 32727653 | C/T | 0.3 | 0.42 | 2.48×10−6 | 0.57(0.45-0.73) | intronic | TIAM1 |
| rs7676822 | 4 | 132252355 | G/T | 0.28 | 0.39 | 2.86×10−6 | 0.65(0.51-0.83) | intergenic | PCDH10(1818115) |
| rs723815 | 6 | 52519203 | A/C | 0.2 | 0.11 | 3.50×10−6 | 2.06(1.46-2.9) | intergenic | LOC730101(9996) |
| rs1911877 | 4 | 132298239 | C/T | 0.3 | 0.4 | 8.41×10−6 | 0.66(0.52-0.84) | intergenic | PCDH10(1772231) |
| rs8081611 | 17 | 4813365 | C/T | 0.12 | 0.05 | 1.40×10−5 | 2.59(1.6-4.19) | intergenic | CHRNE(6996) |
| rs7972963 | 12 | 66646199 | T/G | 0.08 | 0.14 | 1.50×10−5 | 0.54(0.37-0.78) | UTR3 | IRAK3 |
| rs17253738 | 13 | 94874089 | A/G | 0.14 | 0.21 | 2.13×10−5 | 0.59(0.43-0.82) | intronic | GPC6 |
| rs2706652 | 11 | 12289058 | A/G | 0.42 | 0.33 | 2.30×10−5 | 1.5(1.18-1.92) | intergenic | MICAL2(3727) |
| rs7972211 | 12 | 14269986 | G/A | 0.16 | 0.23 | 2.71×10−5 | 0.65(0.49-0.87) | intergenic | GRIN2B(136964) |
| rs318982 | 11 | 131415267 | T/C | 0.21 | 0.29 | 2.82×10−5 | 0.65(0.5-0.86) | intronic | NTM |
| rs6918918 | 6 | 52515078 | T/C | 0.22 | 0.13 | 3.17×10−5 | 1.94(1.4-2.68) | intergenic | LOC730101(14121) |
| rs11022029 | 11 | 11806317 | C/T | 0.13 | 0.2 | 3.18×10−5 | 0.65(0.48-0.87) | intergenic | USP47(56653) |
| rs881499 | 7 | 30976064 | C/T | 0.25 | 0.36 | 3.58×10−5 | 0.55(0.42-0.72) | intergenic | AQP1(10933) |
| rs905690 | 3 | 68725295 | T/C | 0.35 | 0.26 | 3.79×10−5 | 1.56(1.2-2.02) | intergenic | FAM19A4(55620) |
| rs12561532 | 13 | 52108978 | G/A | 0.06 | 0.12 | 3.81×10−5 | 0.48(0.32-0.72) | intergenic | MIR4703(17747) |
| rs9516369 | 13 | 94868584 | G/A | 0.14 | 0.21 | 4.38×10−5 | 0.61(0.44-0.84) | intronic | GPC6 |
| rs7214776 | 17 | 4811615 | C/T | 0.12 | 0.06 | 4.39×10−5 | 2.4(1.51-3.83) | intergenic | CHRNE(5246) |
| rs9365319 | 6 | 162114707 | T/C | 0.13 | 0.21 | 4.49×10−5 | 0.57(0.42-0.77) | intronic | PARK2 |
| rs7004833 | 8 | 11840011 | G/A | 0.05 | 0.1 | 4.53×10−5 | 0.47(0.3-0.75) | intronic | DEFB135 |
| rs4768165 | 12 | 40025034 | A/G | 0.25 | 0.34 | 4.79×10−5 | 0.66(0.51-0.85) | intronic | C12orf40 |
| rs6005451 | 22 | 27852183 | C/T | 0.09 | 0.16 | 4.85×10−5 | 0.53(0.37-0.75) | intergenic | MN1(292082) |
| rs10894396 | 11 | 131326035 | A/G | 0.41 | 0.29 | 4.91×10−5 | 1.72(1.34-2.22) | intronic | NTM |
| rs2293223 | 2 | 103035468 | T/C | 0.15 | 0.24 | 4.92×10−5 | 0.6(0.44-0.8) | intronic | IL18RAP |
| rs1403552 | 2 | 103088777 | A/G | 0.15 | 0.24 | 5.00×10−5 | 0.59(0.44-0.79) | upstream | SLC9A4 |
| rs11158347 | 14 | 61930678 | A/G | 0.33 | 0.21 | 5.18×10−5 | 1.83(1.39-2.41) | intronic | PRKCH |
| rs7706447 | 5 | 116513164 | C/A | 0.04 | 0.1 | 5.83×10−5 | 0.36(0.23-0.59) | intergenic | LOC728342(238044) |
| rs11611119 | 12 | 40166257 | C/T | 0.35 | 0.26 | 5.83×10−5 | 1.59(1.22-2.07) | intronic | SLC2A13 |
| rs4596498 | 6 | 139540103 | A/G | 0.24 | 0.16 | 6.36×10−5 | 1.73(1.26-2.36) | intergenic | TXLNB(21096) |
| rs7565966 | 2 | 179742232 | C/T | 0.45 | 0.33 | 6.69×10−5 | 1.63(1.28-2.08) | intronic | CCDC141 |
| rs12974044 | 19 | 42368629 | G/A | 0.37 | 0.27 | 7.07×10−5 | 1.56(1.2-2.02) | intronic | RPS19 |
| rs139531 | 22 | 41676176 | G/A | 0.3 | 0.2 | 7.07×10−5 | 1.69(1.28-2.24) | intronic | RANGAP1 |
| rs1471659 | 3 | 126812577 | G/A | 0.11 | 0.17 | 7.74×10−5 | 0.61(0.43-0.85) | intergenic | PLXNA1(56342) |
| rs4933958 | 10 | 85821027 | C/T | 0.29 | 0.21 | 8.04×10−5 | 1.56(1.18-2.05) | intergenic | GHITM(78158) |
| rs10013818 | 4 | 44293409 | T/C | 0.26 | 0.18 | 8.34×10−5 | 1.6(1.19-2.15) | intronic | KCTD8 |
| rs3891616 | 13 | 94866849 | C/A | 0.14 | 0.2 | 8.39×10−5 | 0.63(0.46-0.87) | intronic | GPC6 |
| rs722665 | 20 | 8508604 | C/T | 0.4 | 0.29 | 8.47×10−5 | 1.61(1.25-2.08) | intronic | PLCB1 |
| rs2295394 | 14 | 93412743 | T/C | 0.04 | 0.08 | 8.55×10−5 | 0.48(0.29-0.79) | NA | NA |
| rs351098 | 4 | 132409029 | T/C | 0.22 | 0.3 | 8.77×10−5 | 0.67(0.51-0.86) | intergenic | PCDH10(1661441) |
| rs12532545 | 7 | 141875267 | A/C | 0.17 | 0.25 | 9.21×10−5 | 0.63(0.48-0.84) | intronic | LOC100124692 |
Abbreviations: Chr, chromosome number; A1/A2 OR, odds ration; CI confidence interval; MQLS, a more powerful quasi-likelihood score test.
A1/A2, in which “A1” is minor allele, “A2” is major allele.
Resp., minor allele frequence (MAF) for the patients response to SSRIs; Non-Resp., MAF for the patients non-response to SSRIs.
MQLS_Robust p-value, cut-off p-value threshold was set 1×10−4 for the risk allele.
Logistic regression model was performed on probands, adjusted by sex, age. Cut-off threshold was set at OR ≥1.5 for the risk allele.
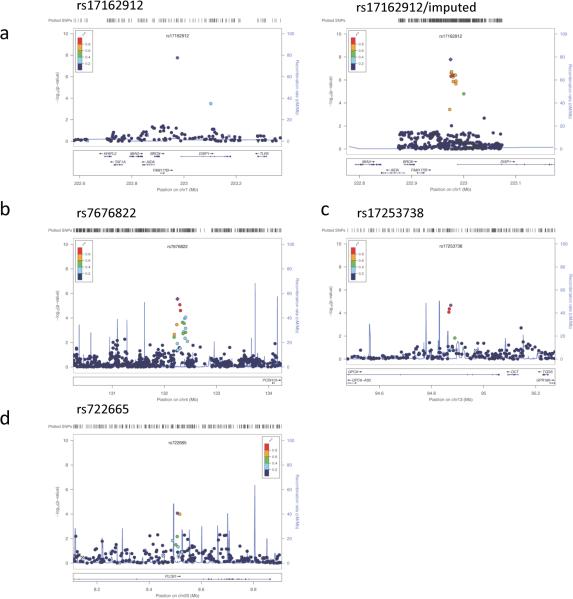
The top-ranked SNP, rs17162912, is located in proximity (within a distance of ~13kb) to the Dispatched 1 gene (DISP1) (P = 1.76×10−8; OR = 0.39 [95%CI 0.26-0.58]) (Table 2 and Figure 2a left panel). Since there were no nearby markers with complete LD with rs17162912, we imputed genotypes in the left and right regions flanking rs17162912 (up to 250 kb) and carried out the association test. The results indicated that SNPs with strong LD with rs17162912 also presented suggestive association signals (Figure 2a right panel). We explored the integrated ENCODE regulation databases and found that rs17162912 is close to a peak (approximately 13kb) of the H3K27AC protein binding score, suggesting that this region encompasses the promoter of DISP1. DISP1 encodes a twelve trans-membrane domain protein that is required for long-range sonic hedgehog (Shh) secretion and transporting, which is important for the establishment of cell-cell contact and crucial for spinal cord development.30
Figure 2.
Regional association plot with LD illustrated for significant SNPs. (a) SNAP plot of rs17162912 for the association test (left) and for the association test after the imputed SNPs were included (right). (b) SNAP plot of rs7676822, rs17253738 (c) and rs722665 (d).
Among the suggestive signals, rs7676822 and rs1911877 located near the PCDH10 gene (distance = 1,818kb and 1,772kb respectively) showed p-values of 2.86 ×10−6 (OR = 0.65 [95%CI 0.51-0.83]), and 8.41×10−6 (OR = 0.66 [95%CI 0.52–0.84]), respectively (Figure 2b). Due to the LD relationship between rs7676822 and rs1911877, these two SNPs should be counted as one hit. It is worth mentioning that PCDH10 belongs to a protocadherin gene family consisting of the largest subgroup of the cadherin superfamily and mediates cell-cell adhesion and intracellular signaling. Most PCDHs (Protocadherins) are predominantly expressed in the central nervous system and have been suggested play pivotal roles in the formation and maintenance of synaptic functions.31
In order to comprehensively evaluate the role of some known pathways in the nervous system, we performed an enrichment analysis to test whether there are any genes significantly enriched in neuron signaling pathways. The results indicated that the glutamatergic neurotransmission pathway and the serotonergic neurotransmission pathway displayed more than two-fold enrichment. The glutamatergic signaling pathway had the highest enrichment score (Enrichment score = 3.38) and the best false discovery rate (FDR) (FDR = 0.0097), and the serotonergic neurotransmission pathway gave the second best enrichment score (Enrichment score = 2.39, FDR = 0.0213) (Table 3 and Supplementary Figure S1).
Table 3.
Enrichment analysis results in ten neurologically-relevant pathways
| Pathways examined | Genes Enriched | Enrichment Score | P-value | FDR |
|---|---|---|---|---|
| Glutamatergic signaling | 14 | 3.38 | 0.0009 | 0.0097 |
| Serotonergic signaling | 11 | 2.39 | 0.0047 | 0.0213 |
| Long-term potentiation | 6 | 1.55 | 0.0058 | 0.0213 |
| Neurotrophin signaling pathway | 8 | 1.54 | 0.0120 | 0.0330 |
| Long-term depression | 4 | 1.04 | 0.0280 | 0.0512 |
| GABAergic signaling | 7 | 1.12 | 0.0340 | 0.0512 |
| Dopaminergic synapse | 7 | 1.02 | 0.0346 | 0.0511 |
| Retrograde endocannabinoid signaling | 6 | 0.88 | 0.0372 | 0.0512 |
| Cholinergic signaling | 4 | 0.67 | 0.5720 | 0.6292 |
| Synaptic vesicle cycle | 3 | 0.34 | 0.9280 | 0.9281 |
In the glutamatergic signaling pathway, there was a SNP rs7972211 near GRIN2B (N-methyl-D-aspartate receptor subunit 2B), a pivotal component of the glutamatergic neurotransmission system, showing a signal of association with SRI response, with P = 2.71×10−5 (OR = 0.65 [95%CI 0.49-0.87]) (Table 2). In addition to GRIN2B, GPC6 (Glypican 6), another gene of the glutamatergic neurotransmission system, has three SNPs (rs17253738, rs9516369, rs3891616) exhibiting association signals, with P = 2.13×10−5 (OR = 0.59 [95%CI 0.43-0.82]); P = 4.38×10−5 (OR = 0.61 [95%CI 0.44-0.84]), and P = 8.39×10−5 (OR = 0.63 [95%CI 0.46-0.87]), respectively (Table 2 and Figure 2c). Due to the tight LD among these three SNPs, they serve as one hit. GPC6 promotes the glutamate receptor clustering and receptivity and induces the formation of postsynaptic signaling in the central nervous system (CNS) synapses. Depletion of GPC6 significantly reduces its function to induce postsynaptic activity.32 It was also interesting to observe that DLGAP1 and DLGAP2 support the enrichment (Supplementary Figure S1a). Of note, DLGAP1 has been recently suggested as an OCD susceptibility gene.11
In the serotonergic neurotransmission system, two within-LD (R2 = 0.6) variants (rs722665 and rs2423366) in the PLCB1 gene showed association at P = 8.47×10−5 (OR = 1.61 [95%CI 1.25-2.08]) and P = 0.0001, respectively. In addition, the protein kinase PKC harbors a SNP, rs11158347, showing association with P = 5.18×10−5 (OR = 1.83 [95%CI 1.39-2.41]) (Table 2 and Figure 2d). Furthermore, several well-established genes including HTR2A and SLC6A4 appeared to support the enrichment (Supplementary Figure S1b).
DISCUSSION
In this study, we tested the association between genetic variations and treatment response in OCD. To date, this is the largest study on treatment response of OCD. While replication is warranted, this study represents an important step towards the comprehension of how genetic variants may contribute to the drug response in OCD treatment. The GWAS top SNP hit identified in this study is rs17162912 located near DISP1. In addition, our enrichment analysis indicated the roles of genes in the glutamatergic neurotransmission system (FDR = 0.0097) and the serotonergic system (FDR = 0.0213).
DISP1 is located in the 1q41-q42 locus which harbors a microdeletion related to a syndrome characterized by significant mental retardation, behavior problems, seizures, and characteristic dysmorphic features.33 While rs17162912 is not within the gene regulators, it was found in close proximity of a promoter of the DISP1 gene in ENCODE databases.
In addition to DISP1, another gene involved in cell-cell contact is PCDH10, an autism-spectrum disorders (ASD) related gene34, provided suggestive level of association with SRI response. GWAS studies have shown that several PCDH genes are associated with neuropsychiatric disorders, including autism, bipolar disease and schizophrenia.35 In our published OCD GWAS study12, cadherin 10, type 2 (CDH10) was also reported as the second strongest association signal for OCD susceptibility. Collectively, these findings suggest that the cell-cell contact molecules might be involved in SRI response in OCD patients. However, due to the lack of adequate biological evidence in OCD to support this data-driven notion, future investigations are warranted.
Among the genes in the glutamatergic neurotransmission system, GRIN2B, an the NMDA (N-methyl-D-aspartate) glutamate receptor, emerged as one of the genes relevant to OCD and SRI response with some nominally significant SNPs. At least three previous genetic studies reported a significant association between a variant in GRIN2B and OCD.7-9 Volumetric magnetic resonance imaging suggested that genetic variations in GRIN2B are associated with regional volumetric brain abnormalities in OCD.36 Preliminary results also suggest that GRIN2B variations interact with variations in SLC1A137, the susceptibility gene consistently replicated in OCD. However, our GWAs analysis did not provide strong evidence for any single variant association in the glutamatergic and serotonergic neurotransmission systems contributing to SRI response.
On the other hand, our enrichment analysis indicated that multiple genes in the glutamatergic and serotonergic neurotransmission system might jointly contribute to the outcome of SRI treatment in OCD (Table 3 and Supplementary Figure S1). More genes nominated occurred in the glutamatergic pathways than the ones in the serotonergic pathways. These genes are indicated in Supplementary Figures S1. However, we recognize that our study is under powered to identify all neuropathogenic SNPs for enrichment.
Despite obtaining one genome-wide significant hit and two suggestive pathway enrichment scores, several potential limitations of this study should be acknowledged. First, drug response was based on retrospective self-report. Second, given the rarity of large OCD samples with drug response information, the analysis was based on the limited sample size available. Third, there was lack of detailed information on the dosage and duration of SRI medications, as well as receipt of behavioral therapy. Future studies, which measure treatment received in greater detail, and which evaluate response using reliable measures of symptom reduction within the first few months of treatment initiation, are needed to support a firm relationship between genetic variants and pathways and the SRI treatment effect in OCD.
On the other hand, several strengths of the current study should be noted. First, the rigorous semi-structured clinical examination and diagnostic best-estimation procedures support phenotypic reliability. Secondly, given the clinical, and assumed genetic heterogeneity of OCD, the OCGAS sample attempted to increased homogeneity by targeting recruitment on OCD-affected individuals with early age at onset. The fact that up to 30% OCD patients show minimal clinical improvement may reflect the biological heterogeneity of OCD phenotypes. Thus, consideration of the subgroups of OCD patients defined by drug response might provide a relatively more homogeneous population for clarification of the pathogenesis.38 Finally, it is worth noting that study participants came from two studies, one of which was a family-based linkage study while the other was a trios-based association study. Although relatedness might confound association tests and odds ratio estimation, the MQLS test developed by Thornton and McPeek offers a better way to conduct a robust association test that corrects for the relatedness coefficients within pedigrees, using a kinship matrix (identity-by-descent, IBD) calculated from genotype data.22
Further research is warranted to replicate the current findings on genetic variations related to SRI response in OCD-affected individuals. We anticipate that next-generation sequencing (NGS) methods, which facilitate the analysis of multiple genes including the effects of both common and rare variants39, will provide further understanding of the mechanisms of OCD treatment response, and lead to more effective treatments for OCD.
Supplementary Material
ACKNOWLEDGEMENTS
This project is a multiple sites collaborative project of OCD Collaborative Genetics Association Study (OCGAS), which is a collaboration among investigators at seven sites in the United States (namely Brown University, Columbia University, University of Southern California, Johns Hopkins University, Massachusetts General Hospital, University of California at Los Angeles, and the National Institute of Mental Health) funded by NIMH Grant Numbers: MH071507, MH079489, MH079487, MH079488, and MH079494. Qin and Shugart are both supported by IRP (Project number MH002930-04).
The views expressed in this presentation do not necessarily represent the views of the NIMH, NIH, HHS, or the United States Government.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest related to this work.
Supplementary information is available at Molecular Psychiatry's website.
REFERENCES
- 1.American Psychiatric Association, editor. Diagnostic and statistical manual of mental disorders (DSM-IV) Psychiatric Press; Washington: 1994. [Google Scholar]
- 2.Nestadt G, Grados M, Samuels JF. Genetics of obsessive-compulsive disorder. Psychiatr Clin North Am. 2010;33(1):141–158. doi: 10.1016/j.psc.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shugart YY, Samuels J, Willour VL, Grados MA, Greenberg BD, Knowles JA, et al. Genomewide linkage scan for obsessive-compulsive disorder: evidence for susceptibility loci on chromosomes 3q, 7p, 1q, 15q, and 6q. Mol Psychiatry. 2006;11(8):763–770. doi: 10.1038/sj.mp.4001847. [DOI] [PubMed] [Google Scholar]
- 4.Shugart YY, Wang Y, Samuels JF, Grados MA, Greenberg BD, Knowles JA, et al. A family-based association study of the glutamate transporter gene SLC1A1 in obsessive-compulsive disorder in 378 families. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(6):886–892. doi: 10.1002/ajmg.b.30914. [DOI] [PubMed] [Google Scholar]
- 5.Voyiaziakis E, Evgrafov O, Li D, Yoon HJ, Tabares P, Samuels J, et al. Association of SLC6A4 variants with obsessive-compulsive disorder in a large multicenter US family study. Mol Psychiatry. 2012;16(1):108–120. doi: 10.1038/mp.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy DL, Lesch KP. Targeting the murine serotonin transporter: insights into human neurobiology. Nat Rev Neurosci. 2008;9(2):85–96. doi: 10.1038/nrn2284. [DOI] [PubMed] [Google Scholar]
- 7.Arnold PD, Rosenberg DR, Mundo E, Tharmalingam S, Kennedy JL, Richter MA. Association of a glutamate (NMDA) subunit receptor gene (GRIN2B) with obsessive-compulsive disorder: a preliminary study. Psychopharmacology (Berl) 2004;174(4):530–538. doi: 10.1007/s00213-004-1847-1. [DOI] [PubMed] [Google Scholar]
- 8.Stewart SE FJ, Moorjani J, Jenike E, Beattie K, Illmann C, Delorme R, Leboyer M, Sedovic M, Smoller J, Jenike M, Pauls D. Family-Based Association between Obsessive-Compulsive Disorder and Glutamate Receptor Candidate Genes. World Congress of Psychiatric Genetics; New York: 2007. [Google Scholar]
- 9.Alonso P, Gratacos M, Segalas C, Escaramis G, Real E, Bayes M, et al. Association between the NMDA glutamate receptor GRIN2B gene and obsessive-compulsive disorder. J Psychiatry Neurosci. 2012;37(4):273–281. doi: 10.1503/jpn.110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor S. Molecular genetics of obsessive-compulsive disorder: a comprehensive meta-analysis of genetic association studies. Mol Psychiatry. 2013;18(7):799–805. doi: 10.1038/mp.2012.76. [DOI] [PubMed] [Google Scholar]
- 11.Stewart SE, Yu D, Scharf JM, Neale BM, Fagerness JA, Mathews CA, et al. Genome-wide association study of obsessive-compulsive disorder. Mol Psychiatry. 2013;18(7):788–798. doi: 10.1038/mp.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattheisen M, Samuels JF, Wang Y, Greenberg BD, Fyer AJ. McCracken JTet al. Genome-wide association study in obsessive-compulsive disorder: results from the OCGAS. Mol Psychiatry. 2014;20:337–344. doi: 10.1038/mp.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson JM. SSRI Antidepressant Medications: Adverse Effects and Tolerability. Prim Care Companion J Clin Psychiatry. 2001;3(1):22–27. doi: 10.4088/pcc.v03n0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Bella D, Erzegovesi S, Cavallini MC, Bellodi L. Obsessive-Compulsive Disorder, 5-HTTLPR polymorphism and treatment response. Pharmacogenomics J. 2002;2(3):176–181. doi: 10.1038/sj.tpj.6500090. [DOI] [PubMed] [Google Scholar]
- 15.Brandl EJ, Tiwari AK, Zhou X, Deluce J, Kennedy JL, Muller DJ, et al. Influence of CYP2D6 and CYP2C19 gene variants on antidepressant response in obsessive-compulsive disorder. Pharmacogenomics J. 2014;14(2):176–181. doi: 10.1038/tpj.2013.12. [DOI] [PubMed] [Google Scholar]
- 16.Brandl EJ, Muller DJ, Richter MA. Pharmacogenetics of obsessive-compulsive disorders. Pharmacogenomics. 2012;13(1):71–81. doi: 10.2217/pgs.11.133. [DOI] [PubMed] [Google Scholar]
- 17.Sangkuhl K, Klein TE, Altman RB. Selective serotonin reuptake inhibitors pathway. Pharmacogenet Genomics. 2009;19(11):907–909. doi: 10.1097/FPC.0b013e32833132cb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tansey KE, Guipponi M, Perroud N, Bondolfi G, Domenici E, Evans D, et al. Genetic predictors of response to serotonergic and noradrenergic antidepressants in major depressive disorder: a genome-wide analysis of individual-level data and a meta-analysis. PLoS Med. 2012;9(10):e1001326. doi: 10.1371/journal.pmed.1001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samuels JF, Riddle MA, Greenberg BD, Fyer AJ, McCracken JT, Rauch SL, et al. The OCD collaborative genetics study: methods and sample description. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(3):201–207. doi: 10.1002/ajmg.b.30224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nestadt G, Wang Y, Grados MA, Riddle MA, Greenberg BD, Knowles JA, et al. Homeobox genes in obsessive-compulsive disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;159B(1):53–60. doi: 10.1002/ajmg.b.32001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman WK, Price LH, Rasmussen SA, Mazure C, Fleischmann RL, Hill CL, et al. The Yale-Brown Obsessive Compulsive Scale. I. Development, use, and reliability. Arch Gen Psychiatry. 1989;46(11):1006–1011. doi: 10.1001/archpsyc.1989.01810110048007. [DOI] [PubMed] [Google Scholar]
- 22.Thornton T, McPeek MS. Case-control association testing with related individuals: a more powerful quasi-likelihood score test. Am J Hum Genet. 2007;81(2):321–337. doi: 10.1086/519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chelala C, Khan A, Lemoine NR. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics. 2009;25(5):655–661. doi: 10.1093/bioinformatics/btn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hindorff LA MJ, Morales J, Junkins HA, Hall PN, Klemm AK, Manolio TA. [Jan 23, 2013];A Catalog of Published Genome-Wide Association Studies. URL: www.genome.gov/gwastudies.
- 26.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 28.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 29.Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6(8) doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Etheridge LA, Crawford TQ, Zhang S, Roelink H. Evidence for a role of vertebrate Disp1 in long-range Shh signaling. Development. 2010;137(1):133–140. doi: 10.1242/dev.043547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SY, Chung HS, Sun W, Kim H. Spatiotemporal expression pattern of non-clustered protocadherin family members in the developing rat brain. Neuroscience. 2007;147(4):996–1021. doi: 10.1016/j.neuroscience.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 32.Allen NJ, Bennett ML, Foo LC, Wang GX, Chakraborty C, Smith SJ, et al. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486(7403):410–414. doi: 10.1038/nature11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jun KR, Hur YJ, Lee JN, Kim HR, Shin JH, Oh SH, et al. Clinical characterization of DISP1 haploinsufficiency: A case report. Eur J Med Genet. 2013 doi: 10.1016/j.ejmg.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321(5886):218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redies C, Hertel N, Hubner CA. Cadherins and neuropsychiatric disorders. Brain Res. 2012;1470:130–144. doi: 10.1016/j.brainres.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 36.Arnold PD, Macmaster FP, Hanna GL, Richter MA, Sicard T, Burroughs E, et al. Glutamate system genes associated with ventral prefrontal and thalamic volume in pediatric obsessive-compulsive disorder. Brain Imaging Behav. 2009;3(1):64–76. doi: 10.1007/s11682-008-9050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai J, Zhang W, Yi Z, Lu W, Wu Z, Chen J, et al. Influence of polymorphisms in genes SLC1A1, GRIN2B, and GRIK2 on clozapine-induced obsessive-compulsive symptoms. Psychopharmacology (Berl) 2013;230(1):49–55. doi: 10.1007/s00213-013-3137-2. [DOI] [PubMed] [Google Scholar]
- 38.Davis KL, Charney D, Coyle JT, Nemeroff C. Neuropsychopharmacology: The Fifth Generation of Progress. Lippincott Williams & Wilkins; Philadelphia: 2002. [Google Scholar]
- 39.Korf BR, Rehm HL. New approaches to molecular diagnosis. JAMA. 2013;309(14):1511–1521. doi: 10.1001/jama.2013.3239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.